Abstract
Understanding how mutations affect protein activity and organismal fitness is a major challenge. We used saturation mutagenesis combined with deep sequencing to determine mutational sensitivity scores for 1,664 single-site mutants of the 101 residue Escherichia coli cytotoxin, CcdB at seven different expression levels. Active-site residues could be distinguished from buried ones, based on their differential tolerance to aliphatic and charged amino acid substitutions. At nonactive-site positions, the average mutational tolerance correlated better with depth from the protein surface than with accessibility. Remarkably, similar results were observed for two other small proteins, PDZ domain (PSD95pdz3) and IgG-binding domain of protein G (GB1). Mutational sensitivity data obtained with CcdB were used to derive a procedure for predicting functional effects of mutations. Results compared favorably with those of two widely used computational predictors. In vitro characterization of 80 single, nonactive-site mutants of CcdB showed that activity in vivo correlates moderately with thermal stability and solubility. The inability to refold reversibly, as well as a decreased folding rate in vitro, is associated with decreased activity in vivo. Upon probing the effect of modulating expression of various proteases and chaperones on mutant phenotypes, most deleterious mutants showed an increased in vivo activity and solubility only upon over-expression of either Trigger factor or SecB ATP-independent chaperones. Collectively, these data suggest that folding kinetics rather than protein stability is the primary determinant of activity in vivo. This study enhances our understanding of how mutations affect phenotype, as well as the ability to predict fitness effects of point mutations.
Keywords: mutagenesis, deep sequencing, protein folding, fitness effect prediction
Introduction
The amino acid sequence of a protein determines its three dimensional structure, function and stability. Understanding and predicting the effects of mutations on protein structure, function and organismal fitness is a major challenge in biology. It has been suggested that most positions in a protein can tolerate mutations while retaining stability and function (DePristo et al. 2005; Bershtein et al. 2006). Other studies indicate that proteins acquire new functions at the cost of stability (Wang et al. 2002). Human single nucleotide polymorphisms analyses suggest that >80% of disease-causing mutations cause a loss of stability (Yue et al. 2005). The stability of the wildtype protein is believed to determine the nature and extent of mutations that can be tolerated. The distribution of fitness effects of mutations is thought to be primarily shaped by their effects on protein thermodynamic stability (Firnberg et al. 2014). It is widely believed that residues in the protein interior are important for protein shape and stability, and those on the surface for function/interaction (Ponder and Richards 1987; Bowie and Sauer 1989; Milla et al. 1994). However, there are few studies which exhaustively test this assertion. For example, in the case of Thioredoxin, the correlation between thermodynamic stability and biological activity was not evident for single mutants (Hellinga et al. 1992). Buried residues are less tolerant to mutations than nonactive-site surface exposed residues. Hence, the proportion of solvent exposed residues in a protein is an important determinant of its evolutionary rate (Lin et al. 2007). Further, it is difficult to determine whether deleterious mutations at nonactive-site residues act primarily through affecting thermodynamic stability or folding kinetics, because both factors can affect the amount of properly folded, functional protein in vivo. It has been suggested that mutations which are destabilizing beyond a certain threshold can render a protein dysfunctional and hence accumulation of such mutations can decrease organismal fitness (Yue et al. 2005; Bershtein et al. 2006; Randles et al. 2006).
In the past, the effects of mutations on protein stability were usually determined by creating a limited number of single-site mutants followed by protein expression, purification and characterization of the properties of each mutant, relative to the wildtype protein. Each of these steps is laborious and limits the number of mutants that can be studied. If a convenient phenotypic readout for protein function is available, this can be combined with deep sequencing to obtain relative activity estimates for large numbers of mutants (Tripathi and Varadarajan 2014). In cases where a phenotypic readout is unavailable, monitoring the levels of a reporter gene fused to the protein of interest can be used as a proxy for activity, although such fusions may also affect the stability and folding of the protein (Kim et al. 2013). The advent of next generation sequencing has provided a considerable amount of phenotypic data linked to mutations, but studies that aim at understanding the molecular basis of these phenotypes are limited. Many studies that employ site-saturation mutagenesis methodology have goals specific to a given protein such as to identify active-site residues (Melnikov et al. 2014; Romero et al. 2015), improve/alter protein properties (Wang et al. 2002; Deng et al. 2012; Whitehead et al. 2012; Starita et al. 2013), identify stabilizing mutations (Araya et al. 2012; Traxlmayr et al. 2012; Kim et al. 2013), determine affinity and specificity determinants of protein–protein interaction (DeBartolo et al. 2012; Dutta et al. 2013) or to study the fitness landscape (Hietpas et al. 2011, 2012; Melnikov et al. 2014; Thyagarajan and Bloom 2014; Sarkisyan et al. 2016). The readout in most cases is either qualitative (binding/no binding) or semi-quantitative, experiments are carried out at a single expression level, some cases sample a limited number of sites (Fowler et al. 2010; Hietpas et al. 2011; Deng et al. 2012; McLaughlin et al. 2012; Schlinkmann et al. 2012) and can involve metastable proteins with multiple functional conformations (Thyagarajan and Bloom 2014). Some of these studies sample multi-site as well as single mutations, complicating interpretation of the data (Hietpas et al. 2011; Deng et al. 2012) and in most cases, inferences from these analyses are not validated by detailed characterization of individual single mutants. Previously, attempts to obtain residue-specific contributions to activity with either a full length protein such as Ubiquitin (76 aa) (Roscoe et al. 2013) or with protein domains such as the hYAP65 WW domain (25-aa region) (Fowler et al. 2010) have been made, but in such cases it is difficult to separate the effect of single mutations on stability/folding from those that directly affect function, either because the system has multiple binding partners, such as in the case of Ubiquitin or due to a limited number of single mutants and presence of several double and triple mutants in the library (Fowler et al. 2010; Deng et al. 2012).
There have also been numerous prior attempts to understand and predict the functional consequences of mutations by using computational methods (Bloom et al. 2005; Parthiban et al. 2006; Moretti et al. 2013; Pires et al. 2014). While experimental approaches often measure changes in thermodynamic stability or activity of proteins upon mutation, computational methods typically predict stabilities, based on either sequence and/or structure. Some recent methods based on machine learning such as SNAP2 (Hecht et al. 2015) and SuSPect (Yates et al. 2014) take into account evolutionary information and other sequence and structure based features to predict functional consequences of mutations.
In the present study, we attempt to understand the contribution of every amino acid in a protein to its structure, stability and function, understand how mutations modulate protein activity in vivo, and use this information in predicting the functional effects of mutations computationally. We attempt to address the following issues: (1) Can we distinguish active-site residues from buried ones based solely on saturation mutagenesis phenotypes? (2) Are there consistent patterns in substitution preferences at buried sites? (3) What is the primary mechanism by which mutations at buried sites affect activity in vivo? (4) Can we predict functional effects of specific mutations at buried sites? We use the protein CcdB (Controller of Cell Death protein B) as an experimental test protein. CcdB is a homodimeric protein and each protomer contains 101 residues (Loris et al. 1999). CcdB is a part of the CcdAB toxin–antitoxin system present on the Escherichia coli F-plasmid and plays an important role in F-plasmid maintenance by killing plasmid free cells (Jaffe et al. 1985; Hayes 2003). Biophysical and thermodynamic studies of dimeric CcdB (Chakshusmathi 2002; Bajaj et al. 2004) indicate that the protein exists as a homodimer at neutral pH and undergoes a two-state unfolding process, with a free energy of unfolding of ∼21 kcal/mol at 298 K (Bajaj et al. 2004). CcdB has two primary ligands, its cognate antitoxin CcdA and cellular target, DNA Gyrase. The Kd of CcdB for CcdA37–72 is in the picomolar range, and is much smaller than for GyrA, which is ∼10 nM (De Jonge et al. 2009).
Phenotypes of 1,664 single-site mutants of CcdB were determined at seven different expression levels (designated as 2–8 in the order of increasing expression level) by using two different deep sequencing techniques, 454 (Adkar et al. 2012) and Illumina (this work). We describe a mutational sensitivity score derived from sequencing (MSseq) and use it to quantitatively rank order mutant effects on phenotype at both buried and exposed positions, and to distinguish buried from active-site residues based solely on mutational data. Two other systems for which experimentally derived mutational sensitivity scores were available, namely PDZ domain (PSD95pdz3) and IgG-binding domain of protein G (GB1) were used to compare the substitution preferences and determine if a coherent set of rules derived from a fraction of the CcdB mutational data can also be used for predicting the functional effects of other mutations in CcdB as well as the two additional test proteins.
To gain additional insights into the molecular determinants of phenotype for the nonactive-site mutants, ∼80 CcdB mutants with a range of in vivo activities were purified and characterized in vitro, to obtain insights into determinants of protein stability, solubility and activity. Effects of chaperone over-expression as well as chaperone and protease deletion on activities of individual mutants were also studied to rationalize the effect of mutations on protein folding and stability. The data suggest that mutational effects on folding rather than stability determine the in vivo phenotype of CcdB mutants.
In summary, this work has important implications for understanding the molecular basis of mutant phenotypes and for mutant phenotype prediction.
Results
Phenotypes Determined from 454 Sanger Sequencing Match Well with Phenotypes Determined by Illumina Sequencing
We have previously described a library consisting of approximately 1,000 single-site mutants of CcdB (Adkar et al. 2012) which was constructed by pooling single-site mutants and individually sequenced by 454 Sanger sequencing to obtain phenotypes (Bajaj et al. 2008). We have previously shown that phenotypes of individual mutants determined by growing them on plates at various repressor and inducer concentrations correlate well (r = 0.95) with those obtained from 454 deep sequencing (Adkar et al. 2012). In the present study, a fresh library for CcdB was prepared by individually randomizing each codon using an inverse PCR procedure (Jain and Varadarajan 2014). This library was transformed and screened at seven different expression levels, under identical conditions to those used for the earlier library. The relative population of each mutant as a fraction of repressor/inducer concentration was estimated using Illumina deep sequencing. In contrast to 454 sequencing where the read length was sufficient to cover the entire gene, each Illumina read provided only 50–70 bp of useful sequence. Hence it was necessary to create six PCR products to obtain complete sequence coverage for the whole gene. The key assumption here is that each mutant gene is mutant only at a single codon, thus we considered reads which contain exactly one mutant codon. We observed 78.5% of the reads to be wildtype, which is close to the expected 83.3% (5/6×100). Only 2.5% of the non wildtype reads (0.12% of total reads) had two mutations. Since the additional mutations will likely be randomly distributed and given that most single mutants show an active phenotype, the fraction of incorrectly assigned, inactive phenotypes is expected to be small. Since expression of active CcdB leads to cell death, the number of sequencing reads for a given mutant abruptly decreases at the expression level where the mutant shows an active phenotype. These expression levels are assigned numerical values from 2 to 8 (value of 9 is assigned to the mutants that show cell growth even at the highest expression level). The CcdB gene is amplified from colonies surviving at each expression level, and tagged with a Multiplex IDentifier sequence (MID) unique to each expression level. MSseq is the expression level at which the number of the sequencing reads for a particular mutant decreases by a factor of five or more compared to the previous expression level (Adkar et al. 2012; Sahoo et al. 2015). Based on this, phenotypes for a total of 1,664 single-site mutants in the two independent single-site libraries of CcdB were mapped collectively by the two deep sequencing methods, 454 and Illumina, respectively, which corresponds to 16.5 mutants per position (87.6% of all possible mutants). Of the 1,093 mutants analyzed by 454 sequencing and 1,342 by Illumina sequencing, 771 mutants were common, 625 mutants have the same MSseq value and the MSseq score differed by at most 1 for 59 mutants. In few cases, where the MSseq value differed between Illumina and 454, the lower value (higher activity) was taken. The high concordance between phenotypes derived from Illumina, 454 and plate based assays of individual mutants validates the deep sequencing based phenotypic identification.
Determination of the Active-Site Residues Solely from the Mutational Data
As a first step towards understanding and interpretation of the large amount of mutational data, we calculated residue-wise mutational tolerance, namely the fraction of active mutants for each residue at a given condition.
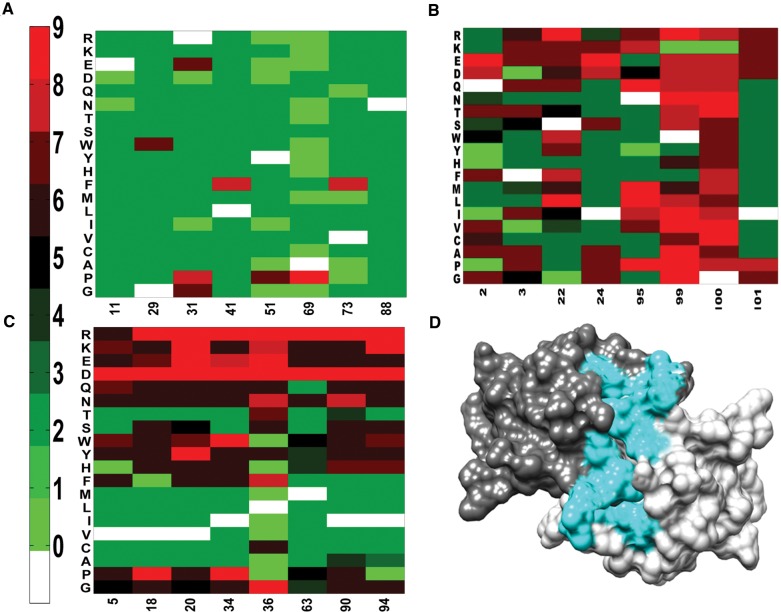
Residues with low mutational tolerance are mostly buried, whereas some are surface exposed. The latter are likely to be a part of the active-site (Wu et al. 2015). Active-site residues can be distinguished from buried ones, even in the absence of structural information, based on the pattern of mutational sensitivity. At buried positions, typically most aliphatic substitutions are tolerated, except when the wildtype residue is a small A or G residue, whereas polar and charged residues are poorly tolerated. In contrast, for active-site residues (which are typically exposed), mutations to aliphatic residues are often poorly tolerated, polar and charged residues are sometimes tolerated and the average mutational tolerance is typically lower than that of the buried residues. Based on these criteria, we can identify residues Q2, F3, Y6, S22, I24, N95, W99, G100 and I101 as putative active-site residues based solely on the mutational data (fig. 1). Upon examining the crystal structure of free CcdB (PDB ID 3VUB), all the active-site residues identified from the mutational phenotypes with the exception of Y6, are in close proximity to each other and line a surface groove, indicating that these eight residues are likely to be part of the active-site (fig. 1D). In the structure of CcdB bound to a fragment of GyrA (PDB ID 1X75), all eight residues are in proximity to GyrA, confirming that these are indeed part of the active-site. Y6 has an exposure of just 9%, and only the terminal OH group is exposed, suggesting that the low mutational tolerance at this position is likely to be primarily due to mutational effects on folding and stability, rather than due to direct effects on GyrA binding. In subsequent analyses, we focus primarily on effects of mutations at nonactive-site positions. Mutational effects on active-site residues involved in binding Gyrase will be discussed in more detail elsewhere.
Fig. 1.
Mutational effects on CcdB protein activity inferred from phenotypic screening and deep sequencing. (A), (B) and (C) show the MSseq values for representative exposed-site (accessibility >5%), all active-site and buried-site residues (accessibility ≤5%), respectively. On the vertical axis, residues are grouped into (G, P), aliphatic (A–M), aromatic (F–W), polar (S–Q) and charged (D–R) amino acids. Residue numbers and substitutions are indicated on the horizontal and vertical axes, respectively. Each heatmap is colored according to the MSseq value of the mutant. Green to red color gradation represents increasing MSseq values. Zero value (light green) indicates that the corresponding mutant was not observed in the library. WT residue at each position is indicated in white. Data for only representative residue positions are shown for clarity. (D) Active-site residues (highlighted in cyan) identified from the mutational phenotypes mapped onto the crystal structure of CcdB (PDB ID 3VUB).
Substitution Preferences at Buried Positions
There are 92 nonactive-site positions in CcdB, of which 21 positions are buried (accessibility ≤ 5%) and 71 are exposed (accessibility > 5%). Of the 21 buried residues, 18 are hydrophobic (table 1). Mutational tolerance increased with increasing expression level (supplementary fig. S2, Supplementary Material online) and was lower at buried positions compared with the exposed positions. At the lowest expression level (MID 2), the average mutational tolerance for the 14 buried residues that are not part of the dimer interface or active-site is 48.5% while for dimer-interface buried residues it is 47.5%, indicating that both classes of buried residues are equally sensitive to mutation (supplementary table S3, Supplementary Material online). Residue D19 is the only buried, potentially charged residue and yet surprisingly shows the highest mutational tolerance relative to other buried residues. Although the residue is largely buried, the side-chain points outwards towards solvent explaining its high tolerance to mutation. A subset of buried residues most sensitive to mutation was selected using the following criteria: tolerance at MID 2 < 40%, tolerance at MID 8 < 90% and phenotypic data for ≥ 15 mutants is available. Interestingly, this selected subset (V18, V20, I34, I90, and I94) clusters together in the interior of each monomer (supplementary fig. S1E, Supplementary Material online).
Table 1.
Mutational Tolerance at the Buried-Site Residues at Lowest and Highest Expression Levels.
| Amino acid | No. of mutants | Depth (Å) | ACCa (%) | Tol at MID2b (%) | Tol at MID8b (%) |
|---|---|---|---|---|---|
| V05 | 18 | 6.8 | 0 | 39 | 94 |
| F17 | 17 | 7.3 | 0.2 | 82 | 100 |
| V18 | 18 | 9.3 | 0 | 33 | 83 |
| D19 | 18 | 6.7 | 1.4 | 83 | 100 |
| V20c,d | 19 | 8.6 | 0 | 32 | 74 |
| Q21c,d | 19 | 6.5 | 1 | 63 | 100 |
| M32d | 17 | 7.8 | 0.3 | 76 | 100 |
| V33 | 19 | 6.5 | 1.4 | 68 | 95 |
| I34 | 19 | 7.9 | 0 | 37 | 79 |
| L36 | 12 | 7.2 | 0 | 0 | 67 |
| P52 | 17 | 5.4 | 3.5 | 41 | 100 |
| V54 | 15 | 5.6 | 0.4 | 73 | 100 |
| M63 | 19 | 8.1 | 0.1 | 47 | 89 |
| T65 | 9 | 7.9 | 0 | 44 | 100 |
| M68c,d | 12 | 6.6 | 0 | 33 | 100 |
| L83 | 19 | 5.8 | 1.5 | 53 | 100 |
| I90 | 19 | 7.4 | 0.1 | 26 | 89 |
| A93c,d | 14 | 6.0 | 0 | 36 | 100 |
| I94c,d | 18 | 7.9 | 0.6 | 33 | 83 |
| M97c,d | 16 | 7.5 | 0 | 56 | 94 |
| F98c,d | 19 | 7.7 | 0.7 | 37 | 79 |
aSide-chain accessibility.
bMutational tolerance at the lowest (MID 2) and highest (MID 8) expression levels.
cResidues within van der Waals distance of the active-site residues.
dResidues present at dimer interface.
On analyzing the mutational tolerance as a function of mutant amino acid at buried residues, we found that at the lowest expression level, D, R and P are the least tolerated mutations and tolerance decreases in the order aliphatic > aromatic, polar > charged. Interestingly, for charged and polar amino acids, smaller amino acids were consistently more poorly tolerated than larger ones (compare D, E; N, Q; S, T tolerances in supplementary table S2, Supplementary Material online). The opposite trend is observed for aromatic substitutions where tolerance decreases in order F > Y, H > W. D and R are the least tolerated substitutions (fig. 1C) though most other mutations are well tolerated at the highest expression level (supplementary table S2, Supplementary Material online). The poor tolerance for a buried Aspartate at all expression levels is likely due to the inability of the small charged side-chain to be solvated upon burial and reconfirms our earlier result (Bajaj et al. 2005), indicating that Aspartate mutant phenotypes are good indicators of residue burial.
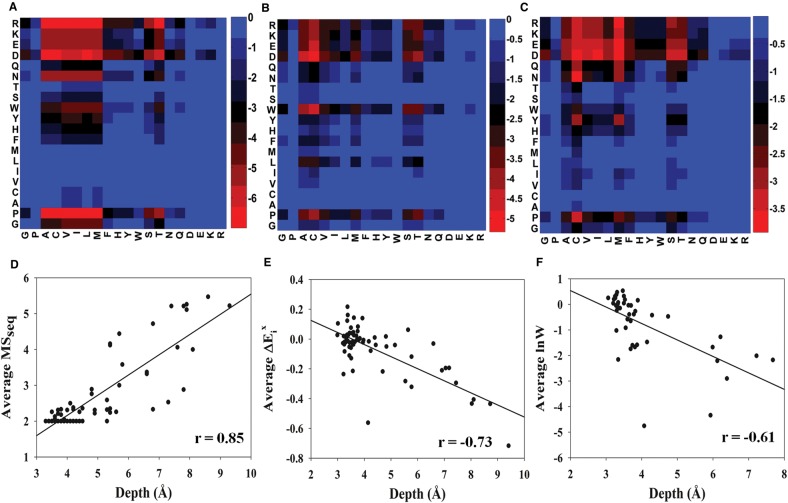
We further attempted to quantitate the relative preference for different substitutions for all buried positions, by incorporating phenotypic data at multiple expression levels. The distribution of MSseq values for introducing a specific residue “X” at every buried-site was obtained. Pair-wise comparisons of these distributions were made using a Wilcoxon signed-rank test. The heatmap (fig. 2A) indicates the log10 P value for the null hypothesis that the introduction of the row residues at a buried site does not reduce protein function significantly more than introduction of the corresponding column residue at the same site. It is important to note here that both the residues being compared are mutant residues. Unlike typical amino acid substitution matrices (Henikoff and Henikoff 1992) used for sequence alignment, our matrix is asymmetric. Aspartate and Arginine mutants possess significantly higher MSseq values than 18 and 16 other residues, respectively, indicating that they are the least tolerated mutations. Proline is the next most poorly tolerated mutation. P values for (D, E), (N, Q) and (S, T) (row, column) pairs are lower than for (E, D), (Q, N) and (T, S) indicating that on an average the order of tolerance is D < E, N < Q and S < T. Similarly, for aromatic residue tolerances, W < Y, H < F. In order to examine if these observations remain valid for systems other than CcdB, we examined previously published mutational sensitivity data for PSD95pdz3 (McLaughlin et al. 2012) and GB1 (Olson et al. 2014) (fig. 2B and C). The general trends were very similar and confirm our observation that for buried sites, smaller charged and polar residues are disfavored relative to larger ones, whereas the opposite is true for aromatic residues. Close examination of the log10 P values in figure 2A suggests that at buried sites, the substitution preference is approximately in the following order A,C,V,L,I,M > T>F > H,Y,S > Q,G,W > N>K,P,E > R>D. A similar (but not identical) trend is also visible in the PSD95pdz3 and GB1 data, though this is based on fewer buried positions and at a single expression level. Additional saturation mutagenesis studies on other systems using quantitative or semi-quantitative readouts would be useful in consolidating our observations.
Fig. 2.
Relative tolerance for substitutions at buried positions. (A) Mutational sensitivity data at all buried positions, obtained at different expression levels for CcdB was used to obtain the distribution of MSseq values for a given mutant residue. The distributions for row and column residues were compared using a Wilcoxon signed-rank test and the corresponding P values were calculated. A log10 of the P values is indicated. Gradation from red to blue indicates increasing values log10 P, i.e., decreasing destabilizing effect of the row residue w.r.t. column residue. A lower P value implies that introduction of the row residue at a buried site is typically more destabilizing than introduction of the corresponding column residue. (B and C) Similar plot, but using values derived from saturation mutagenesis of the PDZ domain (PSD95pdz3) and lnW values from saturation mutagenesis of IgG Binding domain of protein G (GB1), respectively. (D–F) Correlation of the average MSseq values, values and lnW values with side-chain depth for all nonactive-site residues of CcdB, PSD95pdz3 and GB1, respectively. Accessibility and depth values were calculated based on the crystal structure of WT homodimeric CcdB (PDB ID 3VUB), PSD95pdz3 (PDB ID 1BE9) and GB1 (PDB ID 1PGA). A residue was defined as buried if the side-chain accessibility is ≤5%.
Substitution preferences at active-site residues should be different than those at buried sites, because protein:protein interfaces are more polar than protein interiors (Janin et al. 1988; Tsai et al. 1997) and are also likely to display a greater context dependence. Extensive analysis of a large amount of mutational data would be required to decipher these substitution preferences. In the case of CcdB, data for only 142 active-site mutants is available. Hence, we did not attempt to predict mutational sensitivities at active-site residues.
Mutational Tolerance as a Function of Depth
Mutational tolerances at the lowest (MID 2) and highest (MID 8) expression levels for all nonactive-site residues are listed (supplementary table S2, Supplementary Material online, and fig. 1). At the lowest expression level, mutational tolerance increased with increasing accessibility while at the highest expression level it is less sensitive to accessibility and most mutants show an active phenotype. Most substitutions are tolerated at exposed, nonactive-site residues both at low and high expression levels (fig. 1A and supplementary fig. S1A, Supplementary Material online). However, a few mutants with accessibility > 40% were found to show an inactive phenotype. These exposed inactive, nonactive-site substitutions are typically either aromatic residues or proline (supplementary table S4, Supplementary Material online). These exposed, aromatic substitutions probably affect the folding of CcdB protein as they show high propensity to aggregation, although Tm’s are somewhat comparable to the wildtype (see mutants G29W, L41F and V73F in supplementary table S5, Supplementary Material online).
Cation–π interactions are thought to contribute to protein stability (Gallivan and Dougherty 1999) though an earlier study (Prajapati et al. 2006) shows these contribute little to the stability of Maltose Binding Protein. We find that all the 19 and 11 mutations at the 13th and 14th positions respectively, involed in cation–π interaction, including the charge reversal mutant R13D were well tolerated even at the lowest expression levels (supplementary table S6, Supplementary Material online). Salt-bridges are another possible stabilizing noncovalent electrostatic interaction in proteins. In case of CcdB, five salt-bridges are present between the following pairs of residues: D19-R31, D23-R31, E59-R40, E79-K4 and D89-R86. All amino acids participating in salt-bridges are solvent exposed except for D19, in which only the terminal oxygens are exposed. Mutations at all these positions are well tolerated even at the lowest expression level (supplementary table S6, Supplementary Material online), suggesting that none of the salt-bridges in CcdB contributes significantly to the stability or activity of the protein.
We also examined the correlation of average MSseq values with residue depth for all nonactive-site positions in CcdB (PDB ID 3VUB) (fig. 2D). Similar calculations were performed for PSD95pdz3 and GB1 using the phenotypic data obtained from (McLaughlin et al. 2012) (PDB ID 1BE9) and (Olson et al. 2014) (PDB ID 1PGA), respectively. In these studies, the ability of these proteins to bind their cognate ligands is quantitatively linked to a phenotypic readout. In all the three cases, average phenotypic effect was observed to increase with residue depth (Correlation coefficients of 0.85, −0.73 and −0.61 for CcdB, PSD95pdz3 and GB1, respectively) (fig. 2D–F). Buried positions with small (A or G) wildtype residues were not included in the correlations. These positions are unusually sensitive to mutation because all substitutions result in large steric overlap. These data suggest that a large fraction of the average sensitivity to mutation at nonactive-site residues is governed by a single parameter, the residue depth. This is a remarkably simple metric that provides an alternative to the sector based models used to analyze mutational data for PSD95pdz3 as well as other proteins (McLaughlin et al. 2012).
One alternative approach to estimating burial preferences of amino acids is measuring free energies of transfer of amino acid side-chain analogs from water to cyclohexane (Wolfenden et al. 2015). Another approach is to measure accessible surface areas of the side-chains averaged over a large database of protein structures and either infer free energies of transfer from aqueous solution into the protein interior as described previously (Rose et al. 1985), or construct environment dependent substitution matrices from such data (Overington et al. 1992). Relative ΔΔG’s of burial from the first two approaches are shown in supplementary figure S1C and D, Supplementary Material online. Both of these show some qualitative similarities with the mutational data in figure 2A–C, but there are several notable differences. For example, the relative ΔΔG’s of burial inferred from the free energy of transfer approaches show that the introduction of W at buried positions is clearly favored over Y and H, unlike the situation for the experimental mutational data. In addition, the transfer data predict that mutation to G and P will be largely tolerated, whereas the experimental mutational data suggest that substitutions to G or P are rarely tolerated. It is also observed in the mutational data-sets that at buried sites, smaller charged and polar residues are disfavored relative to larger ones, whereas the opposite trend is observed for aromatic residues. In case of ΔΔG transfer data, the trend is preserved for polar and charged residues but clearly not for aromatic residues.
Prediction of Mutational Sensitivity Score (MSpred) Using Penalties Derived from the CcdB Data
We further determined whether the above observations regarding substitution preferences could be employed for prediction of functional consequences of individual mutations. To this end, we developed a predictive model using a coherent set of rules derived from a randomly chosen subset of the CcdB mutational data containing 60% of the mutants and tested its applicability in predicting the mutational sensitivities of the remaining 40% mutants as well as two other proteins, PSD95pdz3 and GB1. The predicted score is denoted as MSpred.
For CcdB mutational data, a mutational sensitivity score (MSseq) of 2 is indicative of wild-type like behavior in the mutant and higher values of MSseq indicate higher mutational sensitivity. Therefore, a base MSpred value of 2 was assigned to all the mutants in the test set, and penalties were subsequently added according to the nature of the substitution, taking into account the wildtype residue identity. As exposed nonactive-site positions tolerated almost all substitutions, penalties were calculated only for buried positions. We also observed that buried side-chains that point outwards with respect to the protein core are less sensitive to mutations compared with the ones that point inside. These residues were identified by their side-chain depth values (see “Materials and Methods” section) and were not considered for penalty calculation.
Substitutions were divided into categories based on the nature of the wildtype and mutant residue. Each wildtype and mutant residue was assigned to one of six categories, namely, aliphatic, aromatic, polar, charged, G and P, resulting in a total of 34 (36−2 [G→G and P→P]) types of substitutions. The CcdB data was randomly divided into training (60% data) and test sets (40% data). The category penalty for each type of substitution was calculated using only the training data set by averaging the MSseq values observed for each category of substitution and subtracting the base MSpred value of 2 from the average MSseq. Additional “residue-specific penalties” were also derived to account for the residue-size-wise substitution preferences; e.g., smaller polar residues being more destabilizing than larger ones (Materials and Methods, supplementary table S7, Supplementary Material online). Penalties for proline substitutions (both buried and exposed) were derived using the flowchart described previously (Bajaj et al. 2007). Next, MSpred values were calculated for all buried positions based on these penalties (MSpred = 2 + category penalty + residue-specific penalty) and all exposed, nonactive-site positions were assigned an MSpred of 2. Active-site residues were not considered in the analysis. The predicted mutational sensitivity scores (MSpred) for the test data set showed a high Pearson’s correlation (r = 0.69) with the experimental MSseq values and a SD of 1.26 (table 2). We also derived the Matthews correlation coefficient in order to evaluate the performance of MSpred in classifying mutants as neutral and nonneutral (see “Materials and Methods” section). It was observed to be 0.65 (table 2).
Table 2.
Mutant Phenotype Prediction by MSpred, SNAP2 and SuSPect.
| Protein | Prediction method | Pearson’s correlation coefficienta | Matthews correlation coefficientb | Sensitivityc(%) | Specificityd(%) | Accuracye(%) |
|---|---|---|---|---|---|---|
| CcdB | MSpredf | 0.69 | 0.65 | 69 | 95 | 90 |
| SNAP2g | 0.27 | 0.19 | 100 | 11 | 37 | |
| SuSPecth | 0.29 | 0.14 | 100 | 8 | 30 | |
| PSD95pdz3 | MSpredf | 0.57 | 0.53 | 61 | 93 | 88 |
| SNAP2g | 0.24 | 0.15 | 100 | 7 | 34 | |
| SuSPecth | 0.6 | 0.61 | 87 | 87 | 87 | |
| GB1 | MSpredf | 0.65 | 0.49 | 44 | 96 | 79 |
| SNAP2g | 0.27 | 0.11 | 100 | 3 | 42 | |
| SuSPecth | 0.08 | −0.03 | 73 | 24 | 38 |
aModulus of the correlation coefficient.
bMathews correlation coefficient = , where TP, TN, FP, FN are True Positives, True Negatives, False Positives and False Negatives, respectively.
cSensitivity = .
dSpecificity = .
eAccuracy = .
fMutant was classified as nonneutral if MSpred > 2 and neutral if the score = 2. Mutants were classified into true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN).
gMutant was classified as nonneutral if SNAP2 score > 50 and neutral if the score < −50. −50 < Score < 50: low reliability predictions and were omitted. Mutants were classified into TP, TN, FP and FN.
hMutant was classified as nonneutral if SuSPect score > 75 and neutral if the score < 25. 25 < Score < 75: low reliability predictions and were omitted. Mutants were classified into TP, TN, FP and FN.
We tested the performance of MSpred on two other proteins. The MSpred values for PSD95pdz3 and GB1 agreed well with the experimental mutational sensitivity data with Pearson’s correlation coefficients of −0.57 and −0.65 and Matthews correlation coefficients of 0.53 and 0.49, respectively (table 2).
We also carried out mutational sensitivity predictions for CcdB, PSD95pdz3 and GB1 using two frequently used methods, SNAP2 (Hecht et al. 2015) and SuSPect (Yates et al. 2014). Both SNAP2 and SuSPect show poorer correlation with the experimental mutational sensitivity data than MSpred (except SuSPect predictions for PSD95pdz3, table 2). Both the methods show a very high sensitivity but a very low specificity value compared with MSpred. Thus, MSpred which is derived based on very simple rules compares favorably with the popular machine learning based methods, SNAP2 and SuSPect. This approach should work to rank order mutational effects at buried sites for other globular proteins. While a three dimensional structure is not essential, it is important to have residue burial information, because predictions have been optimized for buried residues. A saturation mutagenesis data set is also not required. However, it is important to have experimental data on the functional effects of multiple point mutants, to decide on the cutoff value of MSpred that would result in an observable phenotype. This value would likely depend on factors such as intrinsic protein stability, expression level and gene essentiality, that would vary from one protein to another (Miosge et al. 2015).
In Vitro Determined Apparent Tm’s Correlate Better with in Vivo Solubility than with Relative Activity Derived from Deep Sequencing
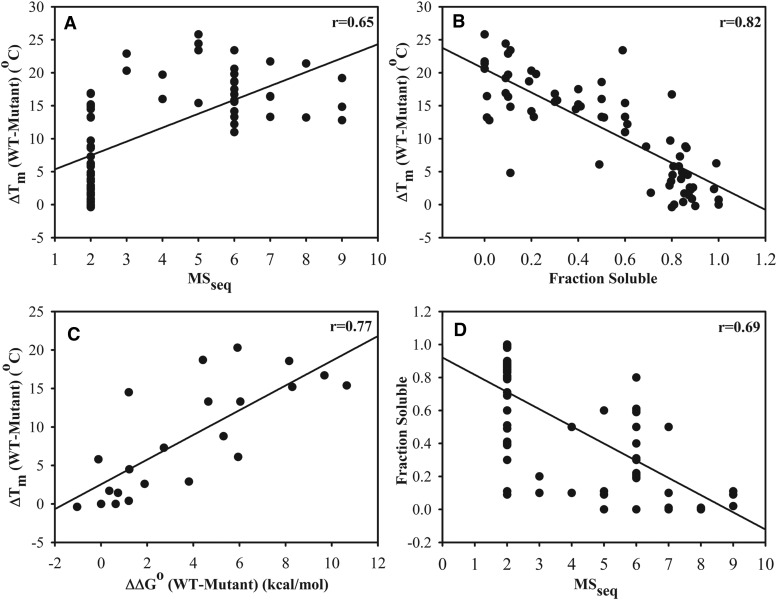
To experimentally probe the molecular basis for mutant phenotypes at nonactive-site positions, around 80 single-site mutants of CcdB were selected from the saturation mutagenesis library (Bajaj et al. 2008) based on MSseq and accessibility class (Adkar et al. 2012) (supplementary table S5, Supplementary Material online). All the mutants were purified by affinity purification against immobilized ligands, GyrA or CcdA. Each purified protein was subjected to thermal denaturation, monitored using Sypro orange dye (Niesen et al. 2007) and the apparent Tm was calculated for each mutant (supplementary fig. S3A and table S5, Supplementary Material online). During purification of various CcdB mutants, it is possible that the protein may be inactivated by aggregation or misfolding. Hence the ability of purified protein to bind CcdA was examined by monitoring the thermal denaturation of each mutant in the absence and presence of a CcdA peptide that contains CcdB binding residues (residues 46–72). If the mutant binds the CcdA peptide, this should result in an increase in its apparent Tm (supplementary fig. S3B, Supplementary Material online) (Fukada et al. 1983; Brandts and Lin 1990; Gonzalez et al. 1999). There were nine mutants, e.g., V05S, V05L, Y06G and F17D that did not show an increase in apparent Tm in the presence of CcdA peptide (supplementary table S5, Supplementary Material online), suggesting that these are misfolded or aggregated, hence these mutants were removed from the analysis. Most of these mutants are largely found in inclusion bodies and have Tm’s between 40 °C and 50 °C, in contrast to WT CcdB which has a Tm of 68.4 °C. Further studies were restricted to the remaining 71 mutants that showed an enhancement in thermal stability in the presence of CcdA peptide (supplementary fig. S3B, Supplementary Material online). Mutants showed a range of apparent Tm’s (supplementary table S5, Supplementary Material online). When in vitro determined thermal stability was compared with in vivo phenotypes (MSseq) determined by deep sequencing, a moderate correlation (r = 0.65) was obtained (fig. 3A). However, there were many mutants that showed similar activity, but differed substantially in their stability, such as L16S, V18T, D19N, V54E (supplementary table S5, Supplementary Material online). Conversely, there were also mutants (e.g., V33D, M32N) that showed similar thermal stability to wildtype, but had substantially lower activity in vivo. This shows that the in vivo activity of a protein depends on many factors inside a cell which assist in proper folding and maintaining an active conformation. Since the apparent Tm determined by the thermal shift assay may not reflect the true thermodynamic stability of the protein, a subset of 21 mutants was also subjected to GdnHCl chemical denaturation. These mutants were chosen to span a range of Tm and MSseq values. These measurements were done to see if the two measures of stability, i.e., thermal and chemical denaturation, correlate with one another. It was found that both measures of stability were highly correlated (fig. 3C and supplementary table S8, Supplementary Material online).
Fig. 3.
Correlation between apparent in vitro Tm, in vivo solubility and activity (MSseq value) for CcdB mutants. Correlations of ΔTm [Tm (WT)−Tm (Mutant)] for 67 single-site mutants with (A) in vivo activity and (B) in vivo fraction of soluble protein, respectively. (C) Correlation of relative thermal stability (ΔTm) of mutants with ΔΔGo of unfolding estimated by GdnHCl denaturation. (D) Correlation of fraction of protein in the soluble fraction with in vivo activity of mutants.
Various mutations have different effects on protein stability and activity. Properly folded proteins are found in the soluble fraction of the cell lysate, whereas misfolded proteins often form insoluble aggregates called inclusion bodies. Hence, misfolding reduces the amount of active, soluble and functional protein, though studies have shown that some amount of protein in the soluble fraction can also be misfolded (Liu et al. 2014). To study the relation between in vivo solubility of CcdB mutants with in vitro determined thermal stability, E. coli strain CSH501 (which has a mutation in the gyrA gene, and is hence resistant towards CcdB action) was transformed individually with the mutants and the amount of protein in both the soluble fraction and in inclusion bodies was estimated. Surprisingly, for a few mutants, although very little protein was found in the soluble fraction, these showed an active phenotype with an MSseq of 2 (fig. 3D). Hence, for these mutants the small amount of protein present in the soluble fraction is properly folded and sufficient to cause cell death in a CcdB sensitive strain. In some cases, different mutants have similar fractions of soluble protein in vivo, but have different in vivo activity and in vitro thermal stability (supplementary table S5, Supplementary Material online). The overall thermal stabilities of mutants correlated well with the in vivo amount of soluble protein (fig. 3B). This indicates that protein stability is an important determinant of proper folding in vivo. The moderate correlation of stability or solubility with in vivo activity likely arises because only a small amount of properly folded soluble protein is sufficient to result in an active phenotype.
One reason for the lack of a better correlation between solubility and in vivo activity is that for each mutant, various conformational forms of the protein can partition differently in the soluble and insoluble fractions of the cell lysate. The soluble fraction can comprise both of folded protein which is active, and soluble aggregates/partially misfolded protein which are inactive (Liu et al. 2014). Moreover, this partitioning can be influenced by perturbations in the cytosolic proteostasis network. To study the relation between in vivo activity and solubility, the ability of four selected CcdB mutants (V33K and Y06G as examples of active but insoluble mutants and R31G and V80N as examples of soluble but inactive mutants) in the soluble fraction of the cell lysate to bind Gyrase was monitored by surface plasmon resonance (supplementary fig. S4A and B, Supplementary Material online). Mutants with only a small amount of protein in the soluble fraction but displaying an active phenotype in vivo (V33K, Y06G), showed binding to Gyrase comparable to the wild-type in this surface plasmon resonance assay, showing that the protein is well folded. Whereas in cases where a mutant is mostly in the soluble fraction but shows an inactive phenotype in vivo (R31G, V80N), the in vitro binding with Gyrase was also negligible compared with the wild-type (supplementary fig. S4C, Supplementary Material online).
Refolding and Unfolding Kinetics
Refolding and unfolding kinetics for 10 mutants that have similar thermal stability but different in vivo solubility and activity were monitored by time-course fluorescence spectroscopy at 25 °C. Refolding and unfolding were carried out at pH 7.4 at final GdnHCl concentrations of 0.6 and 3.2 M, respectively. Of the 10 selected mutants, four (V05S, I56G, V18R and V18H) could not be studied for their refolding profiles due to high precipitation immediately following purification. Further, for these mutants the proportion in the soluble fraction in vivo was low, ranging from 0.1 to 0.3 (supplementary table S5, Supplementary Material online). Of these V05S and I56G are active (MSseq = 2), whereas V18R and V18H show an inactive phenotype (MSseq of 9 and 6, respectively). Most mutants (except V80N) showed slower refolding kinetics than the wild-type, indicating that these mutants are folding defective (table 3). Refolding for the wildtype occurs with a significant burst phase (k > 0.5 s − 1) and a slow phase. Mutants typically show a much smaller burst phase, an intermediate phase and a slow phase of much higher amplitude than the wildtype. Most mutants show unfolding kinetics similar to the wildtype, except V54E, which shows a much higher unfolding rate. The ability of the refolded mutants to bind to the cognate ligand, GyrA or the CcdA peptide (residues 46–72) was also monitored. Binding of refolded mutants to immobilized GyrA, on Amine Reactive Second Generation (AR2G) biosensors was monitored using Bio-layer interferometry (Sultana and Lee 2015), and the binding to CcdA peptide was monitored using Thermal Shift Assay (Niesen et al. 2007). Active mutants (L16S, V18T) retained their binding to both GyrA and CcdA upon refolding, even though their refolding kinetics was slow (table 3). Surprisingly, V54E which is also an active mutant failed to bind GyrA and CcdA upon refolding even though the native protein showed binding (supplementary fig. S6, Supplementary Material online). On the other hand, the inactive mutants R31G and M63N did not bind to GyrA and CcdA after refolding (table 3), showing that their refolded state is nonnative. Interestingly, the native V80N mutant did not show any binding to GyrA, but the refolded protein binds weakly to both ligands. Two of these mutants V80N and V54E also show formation of higher order oligomers (supplementary fig. S5, Supplementary Material online). Overall, the data indicate that slower refolding in vitro is qualitatively correlated with targeting to inclusion bodies in vivo. Further, mutants with low activity in vivo, often refold to an inactive state in vitro. Finally, some mutants which show high aggregation propensity in vitro, show an active phenotype in vivo, presumably because of the presence of chaperones which help in folding to the native state.
Table 3.
Kinetic Parameters for In Vitro Refolding and Unfolding of Selected, Moderately Stable CcdB Mutantsa, b.
| Mutant | Fraction soluble | MSseq | ΔTm (Wt-mutant) (°C) |
Refolding |
Unfolding |
CcdA binding to refolded protein (TSA) | Gyrase binding to refolded protein (BLI) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
Fast phase |
Slow phase |
||||||||||||
| a0 | a1 | k1 (s−1) | a2 | k2 (s−1) | A0 | A1 | K1 (s−1) | ||||||
| L16S | 0.4 | 2 | 16.7 | 0.04 | 0.72 | 0.07 | 0.24 | 0.02 | 0.83 | 0.17 | 0.06 | ++ ++ | +++ |
| V18T | 0.7 | 2 | 9 | 0.04 | 0.7 | 0.1 | 0.26 | 0.02 | 0.8 | 0.2 | 0.16 | ++ ++ | ++ ++ |
| R31G | 0.6 | 6 | 11 | 0.05 | 0.8 | 0.2 | 0.15 | 0.02 | 0.85 | 0.15 | 0.02 | –c | –c |
| V54E | 0.4 | 2 | 14.5 | 0.14 | 0.17 | 0.28 | 0.68 | 0.04 | 1 | – | – | –c | –c |
| M63N | 0.2 | 6 | 15.2 | 0.15 | – | – | 0.85 | 0.08 | 0.84 | 0.16 | 0.07 | –c | –c |
| V80N | 0.8 | 6 | 17.5 | 0.8 | – | > 0.5 | 0.2 | 0.04 | 0.76 | 0.24 | 0.07 | + | + |
| WT | 1 | 2 | – | 0.84 | – | > 0.5 | 0.16 | 0.046 | 0.62 | 0.38 | 0.04 | ++ ++ | ++ ++ |
aThe mutants chosen for refolding studies have similar stability and different solubility and activity (MSseq). Four other selected mutants, could not be used for refolding studies due to very low solubility and high protein precipitation under the given reaction conditions. These had MSseq values of 2, 2, 9 and 6, respectively.
bThe traces were fit to a 5-parameter equation for exponential decay for refolding (f = y0 + a×e(−bx)+c×e(−dx)), yielding fast (k1) and slow phase rate constants (k2), with associated amplitudes a1 and a2, respectively, and to a 3-parameter exponential rise for unfolding (f = y0 + a×e(b×x)) yielding the rate constant k1 with associated amplitude change, A1. a0 and A0 are the amplitudes for the burst phase for refolding and unfolding, respectively. Errors for all the observed parameters were ≤ 10% of the measured experimental value.
cNo observable binding.
Over-Expression of Chaperones Rescues Folding Defects of Mutants
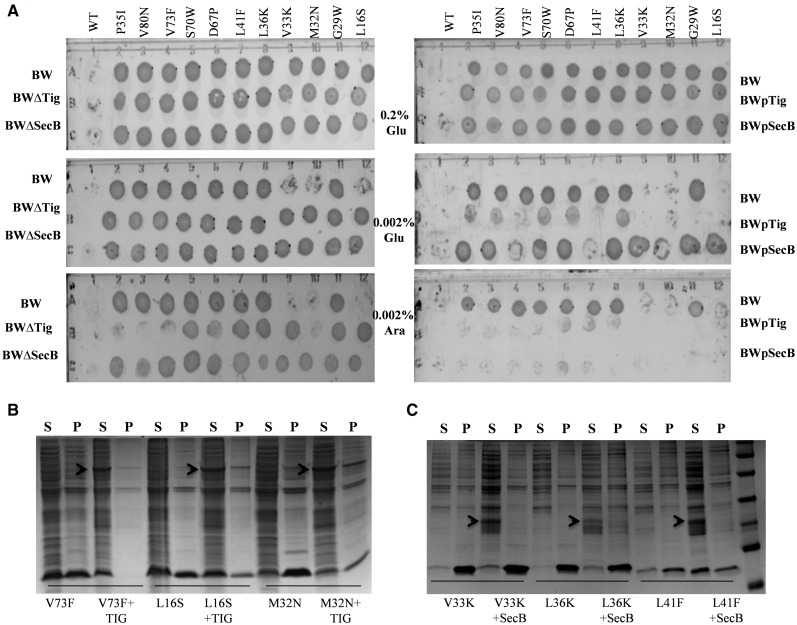
Various factors within the cell influence the proper folding of proteins to the native state. Folding assistance by various chaperones and other quality control mechanisms can buffer mutational effects on protein stability and function (Bershtein et al. 2013). To study this, the in vivo activity of CcdB mutants was assayed in various chaperone and protease deleted strains, as well as chaperone over-expressing strains (see “Materials and Methods” section). Eleven CcdB mutants with a range of solubility and activity were chosen, to study if the over-expression or deletion of chaperones and proteases affects both the in vivo solubility and activity of the mutants. Of these mutants L16S, V33K, L36K and V80N had low Tm’s (<55 °C), but they differ in their in vivo activity, whereas mutants G29W, D67P and V73F show a higher Tm (>56 °C) but are inactive. In vivo activity of these mutants was monitored both in chaperone and protease deletion strains to delineate effects on protein folding or stability. Mutants were transformed in different strains, and cells were plated in the presence of different repressor (glucose) and inducer (arabinose) concentrations to modulate CcdB expression. Over-expression of ATP-dependent chaperones (DnaJ, DnaK, GroEL, and ClpB) did not lead to a change in the in vivo activity of CcdB mutants. A few mutants showed a decrease in the activity in protease deletion strains BWΔlon, BWΔclpP, BWΔhchA (supplementary table S9, Supplementary Material online), but a consistent effect on the activity was not observed, probably due to direct involvement of proteases in the process of CcdB mediated cell death (Van Melderen et al. 1996). Many of these proteases have also been shown to have chaperone-like activity (Gottesman et al. 1997) which can further complicate interpretation of the observed phenotypes. Over-expression of two ATP-independent chaperones namely, Trigger Factor and SecB showed substantial and consistent effect on mutant activity, probably due to their ability to cooperate in the folding of newly synthesized cytosolic proteins (Ullers et al. 2004; Maier et al. 2005). Most mutants show an increase in activity upon over-expression of these two chaperones, whereas they become less active in BWΔtig and BWΔsecB strains, relative to the parent BW25113 strain (fig. 4A and B and table 4). An increase in the in vivo solubility of the mutants was also observed upon chaperone over-expression, the effect being larger for Trigger Factor over-expression (fig. 4B and C and table 4). These effects suggest that for many of these mutants, inactivity primarily results from folding defects which can be rescued by over-expression of chaperones. Interestingly, this is also the case for mutants which show similar stability to wildtype but lower solubility (V73F, D67P, and G29W). This further indicates that defects in folding, rather than stability are the primary causes for inactivity. Previous studies have shown that GroEL/ES chaperonins when over-expressed can not only buffer destabilizing and adaptive mutations, shown in E. coli enzymes during in vitro mutational drift experiments, but can have significant effects on the E. coli proteome evolution through their modulation of protein folding (Tokuriki and Tawfik 2009; Williams and Fares 2010). The observation that folding defects in CcdB mutants are rescued solely by the SecB and Trigger Factor chaperones implies that these defects occur at an early stage of folding, and once the misfolding occurs it cannot be rescued by the ATP-dependent chaperones, such as GroEL and DnaK, as described above. This could also be because for the ATP-dependent chaperones, multiple chaperones may need to be over-expressed as they may have to cooperate to disaggregate misfolded mutants (Mogk et al. 2015).
Fig. 4.
In vivo activity and solubility of CcdB mutants, in presence and absence of ATP-independent chaperones. (A) The activity of the selected mutants was monitored in chaperone deleted (BWΔtig and BWΔsecB) as well as in chaperone over-expression strains (BWpTig and BWpSecB) under seven different repressing or activating conditions for the expression of mutants and the condition where growth ceased was reported as the active condition. (B and C) The fraction of protein for cells grown at 37 °C and induced for CcdB with 0.2% arabinose, in both supernatant (soluble) and pellet (insoluble), with or without over-expression of chaperones, Trigger Factor and SecB, respectively, determined following SDS–PAGE and Coomasie staining using Quantity One software (Bio-Rad). S and P are supernatant and pellet, respectively. Data for representative mutants is shown. The relative estimates of protein present in the soluble fraction and inclusion bodies for all mutants are shown in table 4. The arrow indicates the band for the induced chaperone.
Table 4.
In Vivo Activity and Solubility of CcdB Mutants in Presence and Absence of ATP-Independent Chaperones.
| Mutant |
Strain |
Fraction soluble |
Fractional increase in solubilitya |
|||||
|---|---|---|---|---|---|---|---|---|
| BW25113 | BWΔtig | BWΔsecB | BWpTig | BWpSecB | Tig | SecB | ||
| WT | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 |
| L16S | 4 | 7 | 7 | 2 | 3 | 0.4 | 1.5 | 1.7 |
| G29W | 8 | 8 | 8 | 2 | 4 | 0.6 | 1.2 | 1.1 |
| M32N | 4 | 6 | 6 | 2 | 3 | 0.1 | 3 | 2 |
| V33K | 4 | 7 | 6 | 2 | 3 | 0.1 | 2 | 2 |
| P35I | 8 | 7 | 8 | 5 | 5 | 0.6 | 1.3 | 1.1 |
| L36K | 8 | 8 | 8 | 3 | 5 | 0.05 | 4 | 2 |
| L41F | 7 | 8 | 8 | 3 | 3 | 0.4 | 1.8 | 2.5 |
| D67P | 6 | 8 | 8 | 2 | 4 | 0.2 | 0.8 | 0.5 |
| S70W | 6 | 8 | 8 | 2 | 4 | 0.5 | 1 | 0.4 |
| V73F | 7 | 7 | 8 | 2 | 3 | 0.5 | 2 | 1.3 |
| V80N | 6 | 6 | 7 | 3 | 4 | 0.6 | 1.3 | 1.2 |
aRatio of the soluble fraction of the protein in the presence of over-expressed chaperone (Trigger Factor and SecB, respectively) to the soluble fraction of the protein under normal conditions.
Discussion
Saturation mutagenesis is a useful tool to study the contribution of each amino acid in a protein to its structure, stability and function and in understanding the relation between genotype and phenotype. In the present study, a saturation mutagenesis library of single-site mutants of CcdB was used to understand the molecular basis of mutant phenotypes and to derive a simple procedure to predict such phenotypes. While there have been other saturation mutagenesis studies published in the recent past (Abriata et al. 2015; Kowalsky et al. 2015; Romero et al. 2015; Starita et al. 2015), the present study examines multiple expression levels, effects of multiple chaperones and proteases and employs extensive in vitro characterization to understand how mutations affect phenotype. The tolerance of each residue to various substitutions at multiple expression levels was calculated and mapped on the crystal structure of CcdB (Loris et al. 1999). Mutational tolerance depended on both protein expression level and structural context, as noted by us earlier (Bajaj et al. 2005). Virtually all mutants which showed an inactive phenotype at low expression levels show an active phenotype when over-expressed. This is in contrast with other studies that showed growth defects in the presence of misfolded proteins in a dosage dependent manner (Geiler-Samerotte et al. 2011; Bershtein et al. 2012). In these studies, when destabilized mutants of YFP or DHFR were expressed at high levels, increased aggregation and growth defects were observed. In the case of the CcdB system, increasing expression results in an increased total amount of active protein inside a cell that is available for binding and inhibiting the function of DNA-Gyrase (Bajaj et al. 2008). A similar observation was made in another study which showed increased activity of Hsp90 mutants upon over-expression (Jiang et al. 2013). In the case of TEM-1β lactamase protein it has been found that deleterious effects of mutations primarily arise from a decrease in specific protein activity and not cellular protein levels (Firnberg et al. 2014), contrary to the results of the present study.
For CcdB, at exposed, nonactive-site residues, virtually all mutations are tolerated. At a few highly exposed positions (≥ 40% accessibility) aromatic residues and proline are not tolerated (supplementary table S4, Supplementary Material online), presumably because of aggregation or misfolding. Previous experimental studies have shown that the removal of one methylene group from the protein interior destabilizes a protein by ∼5 kJ/mol and suggested that loss of packing interactions is the major contributor to the increase in stability (Main et al. 1998; Chakravarty et al. 2002; Loladze et al. 2002) though the relative contributions of packing and the hydrophobic effect to protein stabilization remain a matter of debate.
Residue substitution penalties derived from analysis of the CcdB mutant data (supplementary table S7, Supplementary Material online) indicate that substitutions of the aliphatic to aliphatic category are well tolerated. In contrast, aliphatic to aromatic changes are poorly tolerated even when the volume change is equivalent to a single methylene group such as going from I, L or M to F (Richards 1977). This is likely due to the difference in shape between aliphatic and aromatic side-chains, and suggests that while small increases in volume can be tolerated, changes in shape of the side-chains require more reorganization of the neighboring residues that in turn incur a higher energetic penalty.
While there have been many studies that address the stability effects associated with large to small substitutions (Main et al. 1998; Loladze et al. 2002), there are relatively few studies which have quantitated effects of small to large substitutions, particularly substitutions to aromatic residues (Liu et al. 2000; Tanaka et al. 2010). In fact, some studies have shown that very significant increases in residue size of up to three methylene groups can be well tolerated (Hellinga et al. 1992; Wynn et al. 1996), that energetic effects are highly context dependent (Main et al. 1998; Liu et al. 2000) and that such substitutions can even be stabilizing (Lim et al. 1994; Liu et al. 2000). In the current Protherm database (Kumar et al. 2006) (http://www.abren.net/protherm/, last accessed 31 August 2016) 4,805 single buried site mutants from 180 proteins were available. About 1,667 mutants belonged to the aliphatic to aliphatic category, nearly half of them being mutations to alanine. Only 154 aliphatic to aromatic substitutions were available. About 50 aliphatic to aliphatic and 8 aliphatic to aromatic substitutions had similar volume increases with average ΔΔGH2O values of −0.43 and −2.75 kcal/mol, respectively. Thus, consistent with our mutational data, aromatic substitutions are more destabilizing than aliphatic ones involving similar volume changes.
Burial of polar groups in the nonpolar interior of a protein are highly destabilizing, and the degree of destabilization depends on the relative polarity of the group (Main et al. 1998). Interestingly, in the saturation mutagenesis data for charged and polar amino acids at buried positions, smaller amino acids were consistently more poorly tolerated than larger ones, whereas the opposite trend is observed for aromatic substitutions. Surprisingly, mutations at residues involved in cation-π and salt-bridge interactions were well tolerated, indicating that these interactions do not contribute significantly to the stability and function of CcdB.
By combining phenotypic data at multiple expression levels, at all buried positions, it was possible to approximately rank order mutational effects of substitutions at buried positions. The results obtained for CcdB were remarkably similar with those of other proteins, PSD95pdz3 and GB1 for which saturation mutagenesis data were also available (McLaughlin et al. 2012; Olson et al. 2014) and differed from trends observed in free energy of transfer data (compare fig. 2A–C with supplementary fig. S1C and D, Supplementary Material online). Prediction of mutational sensitivity score (MSpred) for other proteins (PSD95pdz3 and GB1) using penalties derived from the CcdB data, taking into account the wildtype residue identity (table 2) gave encouraging results and shows the potential for the use of sequencing based phenotypic data obtained from saturation mutagenesis in understanding and predicting the functional effects of mutations. The present approach compared favorably with known computational predictors (SNAP2 and SuSPect) showing more consistent results and higher specificity (table 2). These and data from other saturation mutagenesis studies can be used to improve predictions of effects of nonsynonymous single nucleotide polymorphisms on protein activity (Guerois et al. 2002; Randles et al. 2006; Yue and Moult 2006; Bromberg et al. 2008; Radivojac et al. 2013) as well as for protein threading applications to guide structure prediction (Shen and Sali 2006; Yang et al. 2015).
To obtain further insights into determinants of phenotypes, a set of ∼ 80 mutants were expressed and purified. They showed a range of stabilities. Thermal stabilities measured by thermal shift assay (Niesen et al. 2007) and equilibrium chemical denaturation were well correlated. Mutations affect both the thermodynamic stability and aggregation propensity of proteins by enhancing misfolding. Both these factors lead to a decrease in the amount of properly folded, active protein. Thermal stabilities of CcdB mutants correlated better with the amount of soluble protein present in a cell (r = 0.82) than with in vivo phenotype (r = 0.65). In some cases, despite being highly soluble, mutants show low activity in vivo, suggesting that a significant fraction of soluble mutant protein is misfolded, and that fraction differs between mutants. In other cases, mutants show high or moderate in vivo activity but differ in in vivo solubility. Both these observations could be rationalized by monitoring in vitro binding of CcdB mutants in the soluble fraction of the cell lysate with Gyrase, using surface plasmon resonance. Mutants with high solubility but low activity also show low binding to Gyrase, whereas partially soluble mutants with high in vivo activity bind well to Gyrase in this assay (supplementary fig. S4, Supplementary Material online). This shows that even a small amount of well folded protein results in sufficient activity to cause cell death even at the lowest level of expression, despite low solubility and stability. Refolding and unfolding kinetics for a subset of mutants, suggest that slow refolding rates measured in vitro correlate with the tendency to form inclusion bodies in vivo. Additionally, several inactive mutants fail to refold to a functional state in vitro as well. In contrast to the refolding rates, most mutants studied had similar unfolding rates to wild type.
The ability of a mutant to fold to the native state is affected by many parameters that include the crowded environment of the cell, folding assistance by various chaperones that buffer mutational effects on protein stability, and quality control mechanisms which are involved in degradation and removal of misfolded proteins from a cell. These factors are likely responsible for the less than perfect correlation between in vitro stability and in vivo activity. To study these effects, the cellular proteostasis machinery was perturbed by either over-expression or depletion of various chaperones and proteases. Interestingly, the most significant changes in the in vivo activity of many mutants were observed upon perturbing the levels of two ATP-independent chaperones, SecB and Trigger Factor, both of which act on their targets while the nascent polypeptide chain is being synthesized at the ribosome. This suggests that many of the CcdB mutants are targeted to inclusion bodies due to defects early in the folding pathway. Over expression of these chaperones lead to an increase in the amount of folded protein in the cell as well as increased in vivo activity and solubility for several formerly inactive mutants, whereas chaperone deletion lead to a corresponding decrease in the activity. These chaperones have previously been shown to increase soluble protein expression by rescuing folding defects (Nishihara et al. 2000). Since these chaperones are ATP-independent, the data clearly show that rescuing folding defects, without additional energy input or protein stabilization, results in increased activity in vivo.
In conclusion, comprehensive analyses of a CcdB saturation mutagenesis library reveal the contribution of each residue to protein activity and function. Protein activity was found to depend monotonically on expression level and was related to stability and solubility in a complex fashion, but correlated well with the ability of mutant protein in the soluble fraction of the cell lysate to bind DNA Gyrase. The moderate correlation of stability with activity, the high in vivo activity of several destabilized mutants, and the ability of the ATP-independent chaperones SecB and Trigger Factor to enhance mutant activity, all suggest that mutational effects on folding, rather than on solubility or stability are the primary determinant of CcdB activity and fitness in vivo. Despite this apparent mechanistic complication, the data demonstrate consistent preferences in accommodating specific residues at buried positions. Besides enhancing our understanding of how mutations affect phenotype, these data can be used to enhance predictions of fitness effects of Single Nucleotide Polymorphisms and to guide protein design and structure prediction efforts.
Materials and Methods
Information about all the strains used in this study is available in supplementary table S1, Supplementary Material online.
Mutant Library Preparation
Previously, a total of 1,430 single-site mutants of CcdB (∼75% of possible mutants) were generated by using a mega-primer based method (Bajaj et al. 2005, 2008). In the present study, an inverse-PCR based approach was used and mutagenesis was carried out by using adjacent nonoverlapping forward and reverse primers. The forward primer contained the mutant codon NNK in the middle of the primer (N is A/C/G/T, and K is G/T in equimolar ratio). The individual products were pooled, gel purified, phosphorylated, subjected to intramolecular ligation and transformed to generate the mutant library (Jain and Varadarajan 2014).
In Vivo Activity of Individual Single-Site Mutants
Escherichia coli strain, TOP10pJAT was individually transformed with mutant CcdB plasmids and activity was assayed by plating the transformation mix on LB-amp plates in the presence of the following concentrations of glucose (repressor) or arabinose (inducer); 2 × 10 − 1% glucose, 4 × 10 − 2% glucose, 7 × 10 − 3% glucose, 0% glucose/arabinose, 2 × 10 − 5% arabinose, 7 × 10 − 5% arabinose and 2 × 10 − 2% arabinose at 37 °C. Since active CcdB protein kills the cells, colonies were obtained only for mutants that showed an inactive phenotype. Plate data was analyzed and compared with relative activity estimates obtained by deep sequencing.
Determination of Active Fraction of the Protein in the Cell Lysate Using Surface Plasmon Resonance
Cultures of E. coli strain CSH501 transformed with the mutant of interest were grown in LB media, induced with 0.2% (w/v) arabinose at an OD600 of 0.6 and grown for 3 h at 37 °C. Cells were centrifuged (1,800×g, 10 min, RT). The pellet was resuspended in PBS buffer pH 7.4 and sonicated, followed by centrifugation at 11,000×g, 10 min, 4°C. Various dilutions of supernatant were passed over GyrA14 fragment immobilized on the surface of a CM5 chip and binding was monitored as change in resonance units per unit time. Analysis was carried out on a Biacore 3000 instrument (Biacore, GE Healthcare).
In Vivo Activity of CcdB Mutants in Presence and Absence of Chaperones and Proteases
Escherichia coli BW25113 strain was transformed with plasmids expressing the following chaperones; Trigger factor and SecB (both ATP-independent), ClpB, DnaK, DnaJ GroEL, (all ATP dependent chaperones). The resulting strains were referred to as BWpTig, BWpSecB, BWpClpB, BWpDnaK, BWpDnaJ, and BWpGroEl. In addition, BW25113 strains deleted for the following proteases Lon, ClpP, HslU, HslV and HchA were also used and referred to as BWΔlon, BWΔclpP, BWΔhslU, BWΔhslV and BWΔhchA, respectively. Competent cells of each of these E. coli strains were prepared (Chung et al. 1989) and individually transformed with selected mutant CcdB plasmids and grown in deep well plates. Transformation with pUC19 was used as a transformation efficiency control. Activity of the mutants was assayed by spotting the transformation mix on LB-Amp plates in the presence of the following concentrations of glucose (repressor) or arabinose (inducer): 2 × 10 − 1% glucose, 2 × 10 − 2% glucose, 2 × 10 − 3% glucose, 0% glucose/arabinose, 2 × 10 − 3% arabinose, 2 × 10 − 2% arabinose and 2 × 10 − 1% arabinose at 30 °C, as many of these strains are temperature sensitive. In case of chaperone over-expression strains, medium used for recovery following transformation was LB + Chl (35 µg/ml) as the chaperone expressing plasmids are ChlR. After 60 min of incubation at 30°C in the above medium, cultures were spotted on LB + Amp plates containing 0.5 mM IPTG to induce chaperone expression, and various concentrations of glucose and arabinose as described above to modulate CcdB expression. Since active CcdB protein kills the cells, colonies are obtained only for mutants that show an inactive phenotype under the conditions examined. Plates were imaged, data was analyzed and the condition where each of the mutants became active in presence or absence of the chaperone was tabulated.
Supplementary Material
Supplementary figures S1–S6, tables S1–S9, CcdB MSseq data (S1_Appendix.xlsx) and Materials and Methods are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Dr Abhijit Sarkar (Center for DNA Fingerprinting and Diagnosis, India) for providing chaperone and protease deletion strains. This work was supported by the Department of Biotechnology (grant number NO.BT/COE/34/SP15219/2015, DT.20/11/2015) and the Department of Science and Technology, Government of India (grant number FNo.SB/SO/BB-0099/2013 DTD 24.6.14). The authors declare that no competing interests exist.
References
- Abriata LA, Palzkill T, Dal Peraro M. 2015. How structural and physicochemical determinants shape sequence constraints in a functional enzyme. PLoS One 10:e0118684.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adkar BV, Tripathi A, Sahoo A, Bajaj K, Goswami D, Chakrabarti P, Swarnkar MK, Gokhale RS, Varadarajan R. 2012. Protein model discrimination using mutational sensitivity derived from deep sequencing. Structure 20:371–381. [DOI] [PubMed] [Google Scholar]
- Araya CL, Fowler DM, Chen W, Muniez I, Kelly JW, Fields S. 2012. A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function. Proc Natl Acad Sci U S A. 109:16858–16863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj K, Chakrabarti P, Varadarajan R. 2005. Mutagenesis-based definitions and probes of residue burial in proteins. Proc Natl Acad Sci U S A. 102:16221–16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj K, Chakshusmathi G, Bachhawat-Sikder K, Surolia A, Varadarajan R. 2004. Thermodynamic characterization of monomeric and dimeric forms of CcdB (controller of cell division or death B protein). Biochem J. 380:409–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajaj K, Dewan PC, Chakrabarti P, Goswami D, Barua B, Baliga C, Varadarajan R. 2008. Structural correlates of the temperature sensitive phenotype derived from saturation mutagenesis studies of CcdB. Biochemistry 47:12964–12973. [DOI] [PubMed] [Google Scholar]
- Bajaj K, Madhusudhan MS, Adkar BV, Chakrabarti P, Ramakrishnan C, Sali A, Varadarajan R. 2007. Stereochemical criteria for prediction of the effects of proline mutations on protein stability. PLoS Comput Biol. 3:e241.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Serohijos AW, Zhou J, Shakhnovich EI. 2013. Protein quality control acts on folding intermediates to shape the effects of mutations on organismal fitness. Mol Cell. 49:133–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Mu W, Shakhnovich EI. 2012. Soluble oligomerization provides a beneficial fitness effect on destabilizing mutations. Proc Natl Acad Sci U S A. 109:4857–4862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bershtein S, Segal M, Bekerman R, Tokuriki N, Tawfik DS. 2006. Robustness-epistasis link shapes the fitness landscape of a randomly drifting protein. Nature 444:929–932. [DOI] [PubMed] [Google Scholar]
- Bloom JD, Silberg JJ, Wilke CO, Drummond DA, Adami C, Arnold FH. 2005. Thermodynamic prediction of protein neutrality. Proc Natl Acad Sci U S A. 102:606–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowie JU, Sauer RT. 1989. Identifying determinants of folding and activity for a protein of unknown structure. Proc Natl Acad Sci U S A. 86:2152–2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandts JF, Lin LN. 1990. Study of strong to ultratight protein interactions using differential scanning calorimetry. Biochemistry 29:6927–6940. [DOI] [PubMed] [Google Scholar]
- Bromberg Y, Yachdav G, Rost B. 2008. SNAP predicts effect of mutations on protein function. Bioinformatics 24:2397–2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarty S, Bhinge A, Varadarajan R. 2002. A procedure for detection and quantitation of cavity volumes proteins. Application to measure the strength of the hydrophobic driving force in protein folding. J Biol Chem. 277:31345–31353. [DOI] [PubMed] [Google Scholar]
- Chakshusmathi G. 2002. Temperature sensitive mutants of CcdB:in vivo and in vitro studies. PhD thesis. Indian Institute of Science, Bangalore, India.
- Chung CT, Niemela SL, Miller RH. 1989. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 86:2172–2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonge N, Garcia-Pino A, Buts L, Haesaerts S, Charlier D, Zangger K, Wyns L, De Greve H, Loris R. 2009. Rejuvenation of CcdB-poisoned gyrase by an intrinsically disordered protein domain. Mol Cell. 35:154–163. [DOI] [PubMed] [Google Scholar]
- DeBartolo J, Dutta S, Reich L, Keating AE. 2012. Predictive Bcl-2 family binding models rooted in experiment or structure. J Mol Biol. 422:124–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Z, Huang W, Bakkalbasi E, Brown NG, Adamski CJ, Rice K, Muzny D, Gibbs RA, Palzkill T. 2012. Deep sequencing of systematic combinatorial libraries reveals beta-lactamase sequence constraints at high resolution. J Mol Biol. 424:150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL. 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nat Rev Genet. 6:678–687. [DOI] [PubMed] [Google Scholar]
- Dutta S, Chen TS, Keating AE. 2013. Peptide ligands for pro-survival protein Bfl-1 from computationally guided library screening. ACS Chem Biol. 8:778–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firnberg E, Labonte JW, Gray JJ, Ostermeier M. 2014. A comprehensive, high-resolution map of a gene’s fitness landscape. Mol Biol Evol. 31:1581–1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. 2010. High-resolution mapping of protein sequence-function relationships. Nat Methods. 7:741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukada H, Sturtevant JM, Quiocho FA. 1983. Thermodynamics of the binding of L-arabinose and of D-galactose to the L-arabinose-binding protein of Escherichia coli. J Biol Chem. 258:13193–13198. [PubMed] [Google Scholar]
- Gallivan JP, Dougherty DA. 1999. Cation-pi interactions in structural biology. Proc Natl Acad Sci U S A. 96:9459–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiler-Samerotte KA, Dion MF, Budnik BA, Wang SM, Hartl DL, Drummond DA. 2011. Misfolded proteins impose a dosage-dependent fitness cost and trigger a cytosolic unfolded protein response in yeast. Proc Natl Acad Sci U S A. 108:680–685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez M, Argarana CE, Fidelio GD. 1999. Extremely high thermal stability of streptavidin and avidin upon biotin binding. Biomol Eng. 16:67–72. [DOI] [PubMed] [Google Scholar]
- Gottesman S, Wickner S, Maurizi MR. 1997. Protein quality control: triage by chaperones and proteases. Genes Dev. 11:815–823. [DOI] [PubMed] [Google Scholar]
- Guerois R, Nielsen JE, Serrano L. 2002. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol. 320:369–387. [DOI] [PubMed] [Google Scholar]
- Hayes F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496–1499. [DOI] [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. 2015. Better prediction of functional effects for sequence variants. BMC Genomics 16(Suppl 8):S1.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellinga HW, Wynn R, Richards FM. 1992. The hydrophobic core of Escherichia coli thioredoxin shows a high tolerance to nonconservative single amino acid substitutions. Biochemistry 31:11203–11209. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG. 1992. Amino acid substitution matrices from protein blocks. Proc Natl Acad Sci U S A. 89:10915–10919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas R, Roscoe B, Jiang L, Bolon DN. 2012. Fitness analyses of all possible point mutations for regions of genes in yeast. Nat Protoc. 7:1382–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hietpas RT, Jensen JD, Bolon DN. 2011. Experimental illumination of a fitness landscape. Proc Natl Acad Sci U S A. 108:7896–7901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe A, Ogura T, Hiraga S. 1985. Effects of the ccd function of the F plasmid on bacterial growth. J Bacteriol. 163:841–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain PC, Varadarajan R. 2014. A rapid, efficient, and economical inverse polymerase chain reaction-based method for generating a site saturation mutant library. Anal Biochem. 449:90–98. [DOI] [PubMed] [Google Scholar]
- Janin J, Miller S, Chothia C. 1988. Surface, subunit interfaces and interior of oligomeric proteins. J Mol Biol. 204:155–164. [DOI] [PubMed] [Google Scholar]
- Jiang L, Mishra P, Hietpas RT, Zeldovich KB, Bolon DN. 2013. Latent effects of Hsp90 mutants revealed at reduced expression levels. PLoS Genet. 9:e1003600.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim I, Miller CR, Young DL, Fields S. 2013. High-throughput analysis of in vivo protein stability. Mol Cell Proteomics 12:3370–3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalsky CA, Klesmith JR, Stapleton JA, Kelly V, Reichkitzer N, Whitehead TA. 2015. High-resolution sequence-function mapping of full-length proteins. PLoS One 10:e0118193.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar MD, Bava KA, Gromiha MM, Prabakaran P, Kitajima K, Uedaira H, Sarai A. 2006. ProTherm and ProNIT: thermodynamic databases for proteins and protein-nucleic acid interactions. Nucleic Acids Res. 34:D204–D206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim WA, Hodel A, Sauer RT, Richards FM. 1994. The crystal structure of a mutant protein with altered but improved hydrophobic core packing. Proc Natl Acad Sci U S A. 91:423–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YS, Hsu WL, Hwang JK, Li WH. 2007. Proportion of solvent-exposed amino acids in a protein and rate of protein evolution. Mol Biol Evol. 24:1005–1011. [DOI] [PubMed] [Google Scholar]
- Liu R, Baase WA, Matthews BW. 2000. The introduction of strain and its effects on the structure and stability of T4 lysozyme. J Mol Biol. 295:127–145. [DOI] [PubMed] [Google Scholar]
- Liu Y, Tan YL, Zhang X, Bhabha G, Ekiert DC, Genereux JC, Cho Y, Kipnis Y, Bjelic S, Baker D, et al. 2014. Small molecule probes to quantify the functional fraction of a specific protein in a cell with minimal folding equilibrium shifts. Proc Natl Acad Sci U S A. 111:4449–4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loladze VV, Ermolenko DN, Makhatadze GI. 2002. Thermodynamic consequences of burial of polar and non-polar amino acid residues in the protein interior. J Mol Biol. 320:343–357. [DOI] [PubMed] [Google Scholar]
- Loris R, Dao-Thi MH, Bahassi EM, Van Melderen L, Poortmans F, Liddington R, Couturier M, Wyns L. 1999. Crystal structure of CcdB, a topoisomerase poison from E. coli. J Mol Biol. 285:1667–1677. [DOI] [PubMed] [Google Scholar]
- Maier T, Ferbitz L, Deuerling E, Ban N. 2005. A cradle for new proteins: trigger factor at the ribosome. Curr Opin Struct Biol. 15:204–212. [DOI] [PubMed] [Google Scholar]
- Main ER, Fulton KF, Jackson SE. 1998. Context-dependent nature of destabilizing mutations on the stability of FKBP12. Biochemistry 37:6145–6153. [DOI] [PubMed] [Google Scholar]
- McLaughlin RN, Jr., Poelwijk FJ, Raman A, Gosal WS, Ranganathan R. 2012. The spatial architecture of protein function and adaptation. Nature 491:138–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnikov A, Rogov P, Wang L, Gnirke A, Mikkelsen TS. 2014. Comprehensive mutational scanning of a kinase in vivo reveals substrate-dependent fitness landscapes. Nucleic Acids Res. 42:e112.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milla ME, Brown BM, Sauer RT. 1994. Protein stability effects of a complete set of alanine substitutions in Arc repressor. Nat Struct Biol. 1:518–523. [DOI] [PubMed] [Google Scholar]
- Miosge LA, Field MA, Sontani Y, Cho V, Johnson S, Palkova A, Balakishnan B, Liang R, Zhang Y, Lyon S, et al. 2015. Comparison of predicted and actual consequences of missense mutations. Proc Natl Acad Sci U S A. 112:E5189–E5198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogk A, Kummer E, Bukau B. 2015. Cooperation of Hsp70 and Hsp100 chaperone machines in protein disaggregation. Front Mol Biosci. 2:22.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti R, Fleishman SJ, Agius R, Torchala M, Bates PA, Kastritis PL, Rodrigues JP, Trellet M, Bonvin AM, Cui M, et al. 2013. Community-wide evaluation of methods for predicting the effect of mutations on protein-protein interactions. Proteins 81: 1980–1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc. 2:2212–2221. [DOI] [PubMed] [Google Scholar]
- Nishihara K, Kanemori M, Yanagi H, Yura T. 2000. Overexpression of trigger factor prevents aggregation of recombinant proteins in Escherichia coli. Appl Environ Microbiol. 66:884–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson CA, Wu NC, Sun R. 2014. A comprehensive biophysical description of pairwise epistasis throughout an entire protein domain. Curr Biol. 24:2643–2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overington J, Donnelly D, Johnson MS, Sali A, Blundell TL. 1992. Environment-specific amino acid substitution tables: tertiary templates and prediction of protein folds. Protein Sci. 1:216–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parthiban V, Gromiha MM, Schomburg D. 2006. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 34:W239–W242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires DE, Ascher DB, Blundell TL. 2014. DUET: a server for predicting effects of mutations on protein stability using an integrated computational approach. Nucleic Acids Res. 42:W314–W319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponder JW, Richards FM. 1987. Tertiary templates for proteins. Use of packing criteria in the enumeration of allowed sequences for different structural classes. J Mol Biol. 193:775–791. [DOI] [PubMed] [Google Scholar]
- Prajapati RS, Sirajuddin M, Durani V, Sreeramulu S, Varadarajan R. 2006. Contribution of cation-pi interactions to protein stability. Biochemistry 45:15000–15010. [DOI] [PubMed] [Google Scholar]
- Radivojac P, Clark WT, Oron TR, Schnoes AM, Wittkop T, Sokolov A, Graim K, Funk C, Verspoor K, Ben-Hur A, et al. 2013. A large-scale evaluation of computational protein function prediction. Nat Methods 10:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randles LG, Lappalainen I, Fowler SB, Moore B, Hamill SJ, Clarke J. 2006. Using model proteins to quantify the effects of pathogenic mutations in Ig-like proteins. J Biol Chem. 281:24216–24226. [DOI] [PubMed] [Google Scholar]
- Richards FM. 1977. Areas, volumes, packing and protein structure. Annu Rev Biophys Bioeng. 6:151–176. [DOI] [PubMed] [Google Scholar]
- Romero PA, Tran TM, Abate AR. 2015. Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc Natl Acad Sci U S A. 112:7159–7164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roscoe BP, Thayer KM, Zeldovich KB, Fushman D, Bolon DN. 2013. Analyses of the effects of all ubiquitin point mutants on yeast growth rate. J Mol Biol. 425:1363–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose GD, Geselowitz AR, Lesser GJ, Lee RH, Zehfus MH. 1985. Hydrophobicity of amino acid residues in globular proteins. Science 229:834–838. [DOI] [PubMed] [Google Scholar]
- Sahoo A, Khare S, Devanarayanan S, Jain PC, Varadarajan R. 2015. Residue proximity information and protein model discrimination using saturation-suppressor mutagenesis. Elife 4:e09532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkisyan KS, Bolotin DA, Meer MV, Usmanova DR, Mishin AS, Sharonov GV, Ivankov DN, Bozhanova NG, Baranov MS, Soylemez O, et al. 2016. Local fitness landscape of the green fluorescent protein. Nature 533:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlinkmann KM, Honegger A, Tureci E, Robison KE, Lipovsek D, Pluckthun A. 2012. Critical features for biosynthesis, stability, and functionality of a G protein-coupled receptor uncovered by all-versus-all mutations. Proc Natl Acad Sci U S A. 109:9810–9815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen MY, Sali A. 2006. Statistical potential for assessment and prediction of protein structures. Protein Sci. 15:2507–2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Pruneda JN, Lo RS, Fowler DM, Kim HJ, Hiatt JB, Shendure J, Brzovic PS, Fields S, Klevit RE. 2013. Activity-enhancing mutations in an E3 ubiquitin ligase identified by high-throughput mutagenesis. Proc Natl Acad Sci U S A. 110:E1263–E1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita LM, Young DL, Islam M, Kitzman JO, Gullingsrud J, Hause RJ, Fowler DM, Parvin JD, Shendure J, Fields S. 2015. Massively Parallel Functional Analysis of BRCA1 RING Domain Variants. Genetics 200:413–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sultana A, Lee JE. 2015. Measuring protein-protein and protein-nucleic acid interactions by biolayer interferometry. Curr Protoc Protein Sci. 79:19 25 11–19 25 26. [DOI] [PubMed] [Google Scholar]
- Tanaka M, Chon H, Angkawidjaja C, Koga Y, Takano K, Kanaya S. 2010. Protein core adaptability: crystal structures of the cavity-filling variants of Escherichia coli RNase HI. Protein Pept Lett. 17:1163–1169. [DOI] [PubMed] [Google Scholar]
- Thyagarajan B, Bloom JD. 2014. The inherent mutational tolerance and antigenic evolvability of influenza hemagglutinin. Elife 3:e03300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuriki N, Tawfik DS. 2009. Chaperonin overexpression promotes genetic variation and enzyme evolution. Nature 459:668–673. [DOI] [PubMed] [Google Scholar]
- Traxlmayr MW, Hasenhindl C, Hackl M, Stadlmayr G, Rybka JD, Borth N, Grillari J, Ruker F, Obinger C. 2012. Construction of a stability landscape of the CH3 domain of human IgG1 by combining directed evolution with high throughput sequencing. J Mol Biol. 423:397–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi A, Varadarajan R. 2014. Residue specific contributions to stability and activity inferred from saturation mutagenesis and deep sequencing. Curr Opin Struct Biol. 24:63–71. [DOI] [PubMed] [Google Scholar]
- Tsai CJ, Lin SL, Wolfson HJ, Nussinov R. 1997. Studies of protein-protein interfaces: a statistical analysis of the hydrophobic effect. Protein Sci. 6:53–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullers RS, Luirink J, Harms N, Schwager F, Georgopoulos C, Genevaux P. 2004. SecB is a bona fide generalized chaperone in Escherichia coli. Proc Natl Acad Sci U S A. 101:7583–7588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Melderen L, Thi MH, Lecchi P, Gottesman S, Couturier M, Maurizi MR. 1996. ATP-dependent degradation of CcdA by Lon protease. Effects of secondary structure and heterologous subunit interactions. J Biol Chem. 271:27730–27738. [DOI] [PubMed] [Google Scholar]
- Wang X, Minasov G, Shoichet BK. 2002. Evolution of an antibiotic resistance enzyme constrained by stability and activity trade-offs. J Mol Biol. 320:85–95. [DOI] [PubMed] [Google Scholar]
- Whitehead TA, Chevalier A, Song Y, Dreyfus C, Fleishman SJ, De Mattos C, Myers CA, Kamisetty H, Blair P, Wilson IA, et al. 2012. Optimization of affinity, specificity and function of designed influenza inhibitors using deep sequencing. Nat Biotechnol. 30:543–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TA, Fares MA. 2010. The effect of chaperonin buffering on protein evolution. Genome Biol Evol. 2:609–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfenden R, Lewis CA, Jr., Yuan Y, Carter CW., Jr. 2015. Temperature dependence of amino acid hydrophobicities. Proc Natl Acad Sci U S A. 112:7484–7488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu NC, Olson CA, Du Y, Le S, Tran K, Remenyi R, Gong D, Al-Mawsawi LQ, Qi H, Wu TT, et al. 2015. Functional Constraint Profiling of a Viral Protein Reveals Discordance of Evolutionary Conservation and Functionality. PLoS Genet. 11:e1005310.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wynn R, Harkins PC, Richards FM, Fox RO. 1996. Mobile unnatural amino acid side chains in the core of staphylococcal nuclease. Protein Sci. 5:1026–1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. 2015. The I-TASSER Suite: protein structure and function prediction. Nat Methods 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates CM, Filippis I, Kelley LA, Sternberg MJ. 2014. SuSPect: enhanced prediction of single amino acid variant (SAV) phenotype using network features. J Mol Biol. 426:2692–2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue P, Li Z, Moult J. 2005. Loss of protein structure stability as a major causative factor in monogenic disease. J Mol Biol. 353:459–473. [DOI] [PubMed] [Google Scholar]
- Yue P, Moult J. 2006. Identification and analysis of deleterious human SNPs. J Mol Biol. 356:1263–1274. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.