Abstract
The breast cancer gene, BRCA2, is essential for viability, yet patients with Fanconi anemia-D1 subtype are born alive with biallelic mutations in this gene. The hypomorphic nature of the mutations is believed to support viability, but this is not always apparent. One such mutation is IVS7+2T>G, which causes premature protein truncation due to skipping of exon 7. We previously identified a transcript lacking exons 4–7, which restores the open-reading frame, encodes a DNA repair proficient protein and is expressed in IVS7+2T>G carriers. However, because the exons 4–7 encoded region contains several residues required for normal cell-cycle regulation and cytokinesis, this transcript's ability to support viability can be argued. To address this, we generated a Brca2 knock-in mouse model lacking exons 4–7 and demonstrated that these exons are dispensable for viability as well as tumor-free survival. This study provides the first in vivo evidence of the functional significance of a minor transcript of BRCA2 that can play a major role in the survival of humans who are homozygous for a clearly pathogenic mutation. Our results highlight the importance of assessing protein function restoration by premature truncating codon bypass by alternative splicing when evaluating the functional significance of variants such as nonsense and frame-shift mutations that are assumed to be clearly pathogenic. Our findings will impact not only the assessment of variants that map to this region, but also influence counseling paradigms and treatment options for such mutation carriers.
Introduction
BRCA2, a tumor suppressor located on chromosome 13 encodes a 340 kDa protein consisting of 3418 amino acids (1). Mutation of BRCA2 is predominantly associated with an increased risk of familial breast cancer. Inheritance of a deleterious BRCA2 mutation increases the lifetime risk of developing breast and ovarian cancer to 88 and 27%, respectively (2) while also increasing the risk of developing primary contralateral breast cancer (3,4). Mutation carriers also have an increased lifetime risk of male breast cancer (6.8%), pancreatic (4–8%) and prostate cancer (10.9–20.2%) (5,6). Functionally, BRCA2 plays an important role in homologous recombination to repair DNA double-strand breaks (DSB) through its interaction with DNA recombinase RAD51 (7,8). BRCA2 is also required for the protection of stalled replication forks (9,10). Defects in these functions result in genomic instability that contributes to tumor development.
BRCA2 is also the gene (FANCD1) that underlies the FA-D1 subtype of Fanconi anemia (FA), which is a rare autosomal recessive disorder characterized by bone marrow failure and developmental abnormalities. A total of 19 FA complementation groups have been identified to date (11–13). Although FA-D1 accounts for only 2% of FA cases, patients exhibit more severe clinical symptoms compared with other FA complementation groups (14). BRCA2 mutations are usually found in a heterozygous state in breast, ovarian, pancreatic or other cancers, where the loss of wild-type allele facilitates neoplastic transformation. However, individuals with FA-D1 carry biallelic BRCA2 mutations, which can be either homozygous or compound heterozygous. Although such individuals succumb to hematopoietic disorders and cancers within the first decade of life (15–17), their ability to survive has been puzzling because studies in mice have demonstrated that BRCA2 is essential for normal embryonic development and viability (7,18). It is believed that at least one of the compound heterozygote mutant alleles of BRCA2 is hypomorphic, i.e. retains partial function, as mice carrying such alleles are viable and in some cases fertile (19,20).
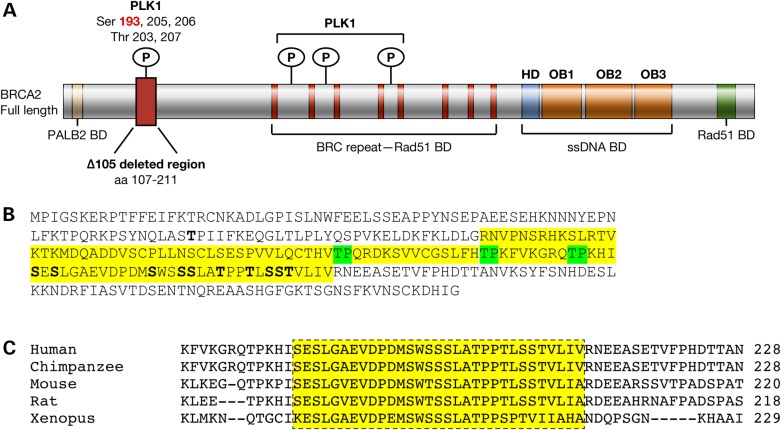
IVS7+2T>G is a splice-site mutation in intron 7 of BRCA2 that causes skipping of exon 7 resulting in a frame-shift, which generates a premature truncating stop codon in exon 9 (21). At least two children homozygous for IVS7+2T>G are reported to have been born alive (22). We previously identified a novel splice variant lacking exons 4–7 (BRCA2Δex4–7) that was expressed at relatively high levels in cells with the IVS7+2T>G mutation (23). This deletion was predicted to bypass the premature stop codon and result in a stable protein with an internal deletion of 105 amino acids (residues 107–211, BRCA2Δ105). These residues contain multiple evolutionarily conserved sites that are phosphorylated selectively during mitosis by Polo-like kinase1 (PLK1) (Fig. 1A–C) (24); the region also has several putative CDK1 phosphorylation sites. Knockdown of BRCA2 has been shown to abrogate G2 checkpoint causing cells to enter mitosis in the presence of DNA damage (25). This premature entry was blocked by inhibition of PLK1 activity (25). Also, phosphorylation of Serine193, which is lost in BRCA2Δ105, has been shown to be required for the localization of BRCA2 to the midbody and normal cytokinesis (26).
Figure 1.
Human BRCA2 protein showing the distribution of PLK1 phosphorylation sites and their evolutionary conservation. (A) Schematic representation of BRCA2 protein showing its functional domains and binding partners. The 105-amino acid region containing multiple PLK1 (Ser 193, a widely studied PLK1 phosphorylation site is highlighted in red) and CDK1 phosphorylation sites and region deleted in BRCA2Δ105 is marked by a red box. BD, binding domain; HD, Helicase Domain; OB1, OB2 and OB3, oligonucleotide-binding folds 1, 2 and 3. (B) A sequence of 284 N-terminal amino acids of BRCA2; residues highlighted in yellow are in BRCA2Δ105. CDK1 phosphorylation sites highlighted in green and PLK1 phosphorylation sites in bold. Residue T77, in bold, is phosphorylated by CDK1. (C) Evolutionary conservation of PLK1 phosphorylation sites.
Using a mouse embryonic stem (mES) cell-based functional assay, we demonstrated the hypomorphic nature of IVS7+2T>G based on the ability of BRCA2Δ105 to rescue the lethality of Brca2 null mES cells, albeit with reduced efficiency compared with WT BRCA2 (23). We found BRCA2Δ105 to be proficient in RAD51 recruitment as well as repair of DSB by homologous recombination. Furthermore, we found the BRCA2Δex4–7 transcript to be expressed in the fibroblasts of a patient who was compound heterozygous for IVS7+2T>G, but its expression was down-regulated in leukemic cells (23). Whether the loss of the BRCA2Δex4–7 transcript is the cause or effect of disease progression, is not known.
Based on our results demonstrating that the protein encoded by the BRCA2Δex4–7 transcript is proficient in DNA repair functions of BRCA2, we hypothesized that this transcript support the viability of humans who are homozygous for IVS7+2T>G mutation. To test this, we have now generated a knock-in mouse model with a targeted deletion of Brca2 exons 4–7. Our results show that at the level of expression achieved in these mice, the transcript lacking exons 4–7 can indeed support viability. Interestingly, on a Trp53 mutant background, these mice exhibited an increase in the incidence of lymphomas and an increase in γH2AX foci in hematopoietic progenitor cells. We used mES cells to mechanistically examine how the loss of the 105 amino acids can contribute to the phenotypes observed in mutant mice. The functional relevance of an alternatively spliced transcript described here will impact the risk assessment of truncating mutations identified not only in BRCA2, but other genes as well. Our findings also highlight the need to exercise caution when patients carrying potentially pathogenic splicing mutations are offered targeted therapy.
Results
Brca2Δex4–7 knock-in mice are viable and fertile
We assessed the ability of the BRCA2Δex4–7 transcript to support viability of humans by generating a knock-in mouse model. We replaced exons 4–7 with a loxP-Neo-loxP cassette in mES cells (Supplementary Material, Fig. S1A and B). We used targeted ES cells to generate Brca2Δex4–7-Neo heterozygous mice. We, then, excised the Neo cassette by crossing these mice with β-actin Cre transgenic mice and obtained Brca2Δex4–7/+ mice (Supplementary Material, Fig. S1C). Expression of Brca2 transcript with proper splicing from exon 3 to exon 8 was confirmed by sequencing (Supplementary Material, Fig. S1D and E). Brca2Δex4–7/+ mice were intercrossed to obtain Brca2Δex4–7/Δex4–7 mice. These mice were obtained at expected Mendelian ratio, which supports our hypothesis that the Brca2Δex4–7 transcript, at least when expressed at the levels of endogenous Brca2, can support viability in mice (Supplementary Material, Table S1). Both males and females were fertile and phenotypically indistinguishable from WT and Brca2Δex4–7/+ littermates. To examine the effect of expressing Brca2Δex4–7 allele in a hemizygous state, we also crossed Brca2Δex4–7/+ mice with Brca2Ko/+ mice that carry Brca2 null allele (referred to as Ko) to obtain Brca2Δex4–7/Ko mice (7). These mice were obtained at the expected Mendelian ratio (Supplementary Material, Fig. S2 and Table S2), and were also viable, fertile with no apparent developmental defect.
We examined the mouse embryonic fibroblasts (MEFs) derived from Wt and Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/ko embryos and found normal RAD51 foci formation (Supplementary Material, Fig. S3A and B), which was consistent with previous observations in mES cells (23).
Loss of exons 4–7 does not affect bone marrow hematopoietic homeostasis
To examine if loss of exons 4–7 has any deleterious effect on the hematopoietic system, we examined the peripheral blood as well as the bone marrow cells. Complete peripheral blood analysis of 8-week-old mice revealed that the red blood cell, white blood cell, monocyte and neutrophil counts in Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice were within the normal range and similar to the control groups (Supplementary Material, Fig. S4A). To examine the effect of exons 4–7 loss on bone marrow homeostasis, we isolated mononuclear cells from femur marrow of 8-week-old mice (Wt, Brca2+/Ko, Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko) and analyzed them by flow cytometry. We did not observe any difference in the bone marrow cellularity, lineage negative or progenitor cells across various genotypes. In addition, the percentage of immature, pre-B and B-cell populations, myeloid cells and erythrocytes at various stages of maturation were similar, suggesting that differentiation of Hematopoietic stem cells (HSCs) into the various lineages was not affected by the loss of the 105-amino acid region (Supplementary Material, Fig. S4B–D).
We also examined the effect of irradiation (IR) on the bone marrow stem and progenitor cells of mutant and control mice. While there was no difference in the stem cell compartment in terms of DNA damage and proliferation across various genotypes (Supplementary Material, Fig. S5A, left), a significant increase in DNA damage was seen in more committed progenitors (Lin−Sca-1−Kit+) of Brca2Δex4–7/Ko mice compared with WT (P = 0.0133) and Brca2Δex4–7/Δex4–7 (P = 0.0301) mice, (Supplementary Material, Fig. S5A, top left). An increase in Ki67 staining intensity indicating increased proliferation was also observed in Brca2Δex4–7/Ko compared with WT mice (P = 0.0423) (Supplementary Material, Fig. S5A, lower left). However, the increase in DNA damage and proliferation in Brca2Δex4–7/Ko progenitor cells did not reveal any significant difference in the growth kinetics of mononuclear cells isolated from femurs of unirradiated (Supplementary Material, Fig. S5B, top) compared with irradiated mice of various genotypes (Supplementary Material, Fig. S5B, lower).
To examine if the increase in IR-induced DNA damage observed in vitro in more committed progenitors (Lin−Sca-1−Kit+) cells affected the survival of mice, we exposed mice to 8 Gy whole body IR. About 50% of Wt mice die at this dose. We found no difference in the survival of Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice compared with Wt littermates (Supplementary Material, Fig. S5C). Similarly, we observed no defect in the ability of stem cells to repopulate the bone marrow in response to the depletion of proliferating hematopoietic cells by treating the mice with 5-fluorouracil (5-FU) (Supplementary Material, Fig. S5D).
Loss of exons 4–7 does not affect tumor susceptibility of Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice
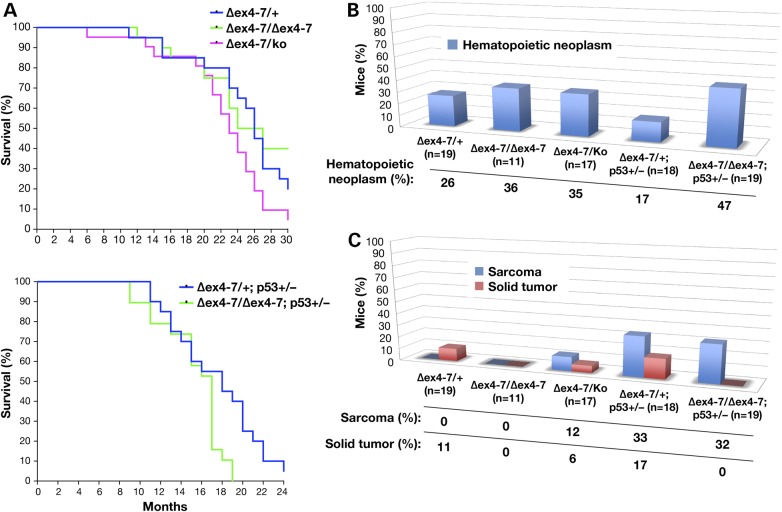
To examine the effect of the deletion of exons 4–7 on tumorigenesis, we aged a cohort of Brca2Δex4/+, Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice, 20 mice of each genotype for 30 months. These mice exhibited a median survival of 23–26 months. We observed no significant difference in the tumor-free survival of mice of the three genotypes (Fig. 2A, upper). Overall, the tumor spectrum of these mice was also similar (Fig. 2B and C). We concluded that the loss of the region encoding the 105 amino acids is not essential for the tumor suppressor function of BRCA2. It is known that the effect of BRCA2 loss on tumorigenesis is more evident on a Trp53 heterozygous background, which provides a cellular environment that promotes cell proliferation (27). Therefore, we crossed mice lacking exons 4–7 to Trp53Ko/+ mice to obtain Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice. Brca2Δex4–7/Ko mice were not included in this study because we have not observed any significant difference in Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice. Twenty mice of each genotype were aged for 24 months. Mice on a Trp53Ko/+ background exhibited shorter survival rate, as the median survival time was 17–18 months, which is 6–8 months less than the Trp53+/+ cohort (Fig. 2A, lower). However, the survival of Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice was not significantly different. Interestingly, when we examined the cause of death, we found that 47% of Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice succumbed to hematopoietic neoplasm, solely lymphomas, while 11 and 6% of Brca2Δex4–7/+;Trp53Ko/+ mice developed lymphoma and histiocytic sarcoma, respectively (Fig. 2B). Sarcoma was the second leading cause of death, however, no difference in incidence was seen between genotypes. Interestingly, while 17% of Brca2Δex4–7/+;Trp53Ko/+ mice developed solid tumors none was seen in Brca2Δex4–7/Δex4–7;Trp53Ko/+ or Brca2Δex4–7/Δex4–7 mice. A widespread increase in hematopoietic neoplasia in multiple organs was also observed in Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice compared with other genotypes (Supplementary Material, Fig. S6).
Figure 2.
Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice exhibit increased occurrence of lymphomas, solid tumors and other hematopoietic and proliferative abnormalities. (A) The Kaplan–Meier plot showing tumor-free survival of Brca2Δex4–7/+, Brca2Δex4–7/Δex4–7 and Brca2Δex4–7/Ko mice (10 males and 10 females/genotype) on a Trp53+/+ (upper panel; Over all: χ2 = 4.226, P = 0.1209) and Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ (lower panel; over all: χ2 = 1.730, P = 0.4210). (B) Incidence of hematopoietic neoplasm (Brca2Δex4–7/+ versus Brca2Δex4–7/Ko, P = 0.7206; Brca2Δex4–7/+;Trp53Ko/+ versus Brca2Δex4–7/Δex4–7;Trp53Ko/+, P = 0.0789), (C) Sarcoma and solid tumor (Brca2Δex4–7/+;Trp53Ko/+ versus Brca2Δex4–7/Δex4–7;Trp53Ko/+, P = 0.1050 for solid tumor). N = confirmed pathology per genotype.
Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice exhibit signs of bone marrow attrition with age
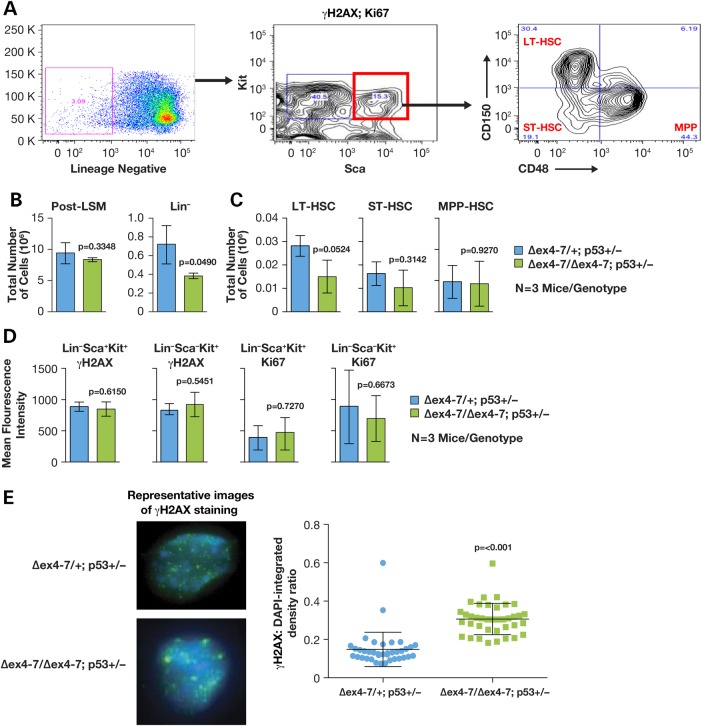
We examined bone marrow of older mice to determine whether the increased incidence of lymphoma in Brca2Δex4–7/Δex4–7;Trp53Ko/+ could be attributed to an increase in bone marrow attrition. We assessed the cellularity, DNA damage (γH2AX staining and foci formation) and proliferation (Ki67 staining) in the HSC compartment of the bone marrow of three 16–18-month-old Brca2Δex4–7/+;Trp53Ko/+ and three Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice aged 14–21 months (Fig. 3A). We found that while the bone marrow cellularity was similar between the two groups, Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice had a 1.9-fold reduction in the number of lineage-negative cells (Fig. 3B), which translated to a significant reduction in the long-term stem cells, and a mild reduction in the short-term stem cells as well as multipotent progenitor cells (Fig. 3C). Although flow cytometric analysis was unable to discern differences in levels of DNA damage between the two groups (Fig. 3D), immunofluorescence analysis of γH2AX foci formation in multipotent progenitors, however, showed a 2-fold increase in γH2AX:DNA ratio demonstrating an increase in DNA damage in Brca2Δex4–7/Δex4–7;Trp53Ko/+ aged mice (Fig. 3E).
Figure 3.
Brca2Δex4–7/Δex4–7;Trp53Ko/+ mice undergo HSC depletion. (A) Representative FACS analysis of bone marrow mononuclear cells stained with antibodies against lineage markers [to identify lineage (Lin) negative cells, c-Kit and Sca (to identify stem and progenitor cells), γH2AX and Ki67. CD150 and CD48 were used to analyze and sort Lin− Sca+ Kit+ cells into long-term (LT), short-term (ST) and multipotent progenitors (MPP). (B) Comparison of total number of mononuclear cells harvested from bone marrow following gradient separation (post-LSM) and percentage of Lin− cells in Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ (n = 3 mice/genotype). (C) Comparison of LT, ST and MPP cell population, extrapolated as part of total mononuclear cells harvested from whole bone marrow, between Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ (n = 3 mice/genotype). (D) Comparison of the presence of DNA damage (γH2AX) and proliferation (Ki67) staining in stem (Lin− Sca+ Kit+) and progenitor (Lin− Sca− Kit+) cells of Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ (n = 3 mice/genotype). (E) Representative images of Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ FACS sorted MPP cells stained with γH2AX-Alexa488 and DAPI (left panel). Right panel shows ratio of Alexa488:DAPI as a measure of levels of DNA damage present in Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ MPP cells (n = 2 mice/genotype).
ES cells expressing BRCA2Δ105 reveal aberrant cell-cycle regulation and genomic instability after IR
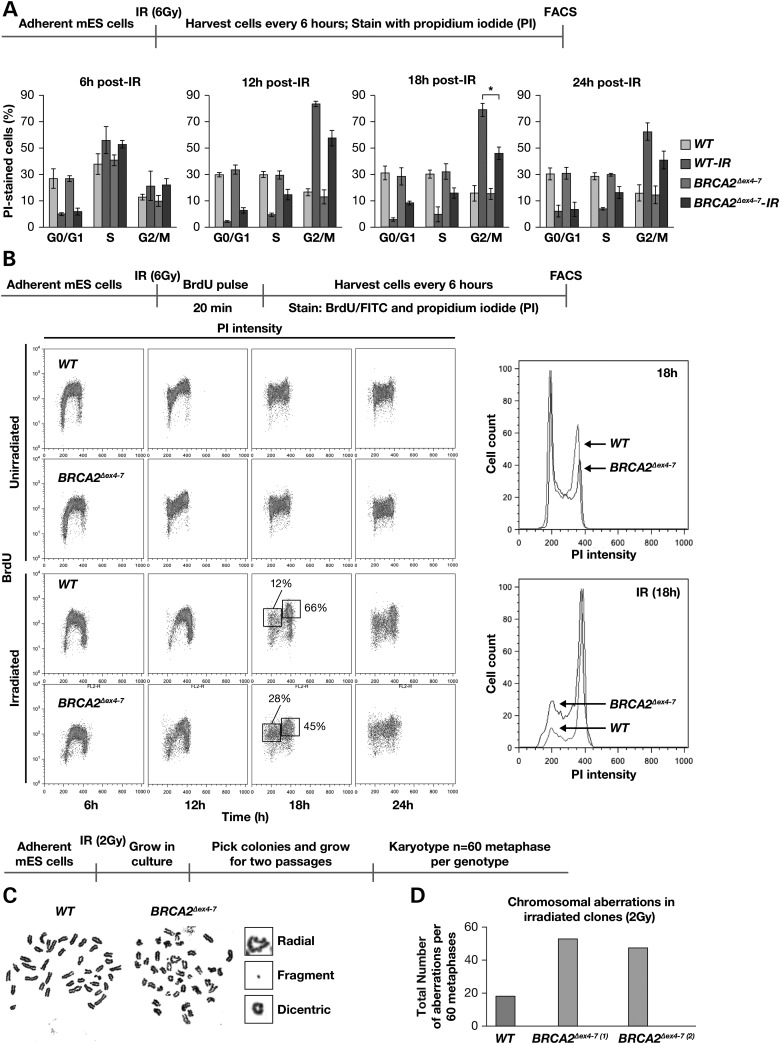
Given the normal RAD51-mediated DNA repair in cells expressing BRCA2 lacking 105 amino acids, an increase in γH2AX foci in multipotent progenitors was puzzling. We used mES cells that lack endogenous mouse Brca2, but express human WT BRCA2 or BRCA2Δex4–7 transgenes cloned in a bacterial artificial chromosome (BAC) to determine the cause of increase in γH2AX foci formation. Two previously generated Brca2Ko/Ko;BRCA2Δex4–7 BAC Tg mES cell clones (here on referred to as BRCA2Δex4–7) and Brca2Ko/Ko;BRCA2WT BAC Tg control cells (here on referred to as WT) were irradiated (6 Gy), harvested at 6 h intervals and assessed for cell-cycle progression by flow cytometry after PI staining (23). The percentage of cells in various phases of cell cycle at all time points were similar in unirradiated asynchronous cultures of WT and BRCA2Δex4–7 mutant cells. However, while the majority of WT cells accumulate at G2/M phase following IR, a significantly reduced the number of BRCA2Δex4–7 cells were in G2/M phase (84 versus 61% at 12 h and 80 versus 51% at 18 h post-IR P ≤ 0.0001) (Fig. 4A). This suggests that BRCA2Δex4–7 cells are less prone to arrest cell-cycle progression in response to IR induced DNA damage.
Figure 4.
Defects in maintaining G2/M arrest and genomic stability in mES cells expressing BRCA2Δex4–7. (A) FACS analysis of PI stained mES cells expressing WT and BRCA2Δex4–7. (B) Cell-cycle profile of BrdU and PI-stained unirradiated and 6 Gy irradiated mES cells expressing WT and BRCA2Δex4–7 (left panel). DNA content analysis of the G2/M fraction of BrdU and PI-stained cells. (C) Representative metaphase spread of irradiated WT and BRCA2Δex4–7 mES cells. (D) Quantification of chromosomal aberrations observed in mES cells expressing WT and BRCA2Δex4–7 (WT versus BRCA2Δex4–7, P = <0.001); N = 60 metaphase per genotype. Scheme of experimental design is shown at the top of each panel.
Next, we examined whether the reduction in the number of mutant cells at G2/M was because the cells progressed through the cell cycle in the presence of DNA damage. We pulse labeled the cells with BrdU for 20 min post-IR (6 Gy), harvested every 6 h and analyzed the cell cycle profile after BrdU and PI staining (Fig. 4B). While unirradiated asynchronous WT and BRCA2Δex4–7 cells exhibited similar cell-cycle profile, upon IR 28% of BrdU labeled BRCA2Δex4–7 cells were in G0/G1 phase in contrast to only 12% of WT cells, indicating that BRCA2Δex4–7 cells continued to cycle even in the presence of damaged DNA.
To determine whether the precocious exit from G2/M phase affected genomic stability, both WT and BRCA2Δex4–7 mouse ES cells were given a low dose of IR (2 Gy) because 6 Gy affected viability of cells. Surviving colonies were grown for two passages and global chromosomal analysis was performed. BRCA2Δex4–7 cells exhibited a marked increase in chromosomal aberrations compared with WT cells (Fig. 4C and D, Supplementary Material, Fig. S7). Taken together, these results suggest that the failure of BRCA2Δex4–7 ES cells to effectively arrest in G2/M phase in response to DNA damage results in chromosomal abnormalities leading to genomic instability.
BRCA2Δex4–7 mES cells have aberrant activation of PLK1 after IR
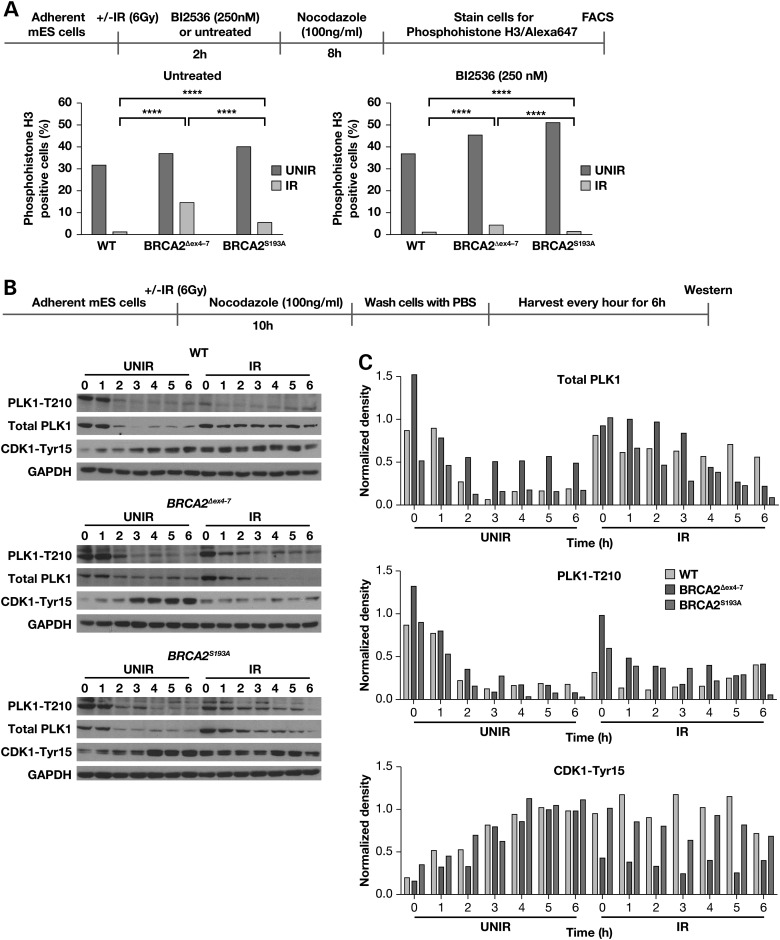
PLK1 is activated via phosphorylation at residue Thr210 during G2/M progression. Phosphorylated PLK1 activates CDC25 phosphatases that remove the inhibitory phosphorylation on the CDK1/Cyclin B complex that drives cells through mitosis (28). To determine whether PLK1 activity contributes to the premature exit from IR induced cell cycle arrest in BRCA2Δex4–7 cells, both WT and BRCA2Δex4–7 cells were irradiated (6 Gy) and treated with 250 nm PLK1 inhibitor (BI2536) for 2 h, and mitotic cells were trapped in 100 ng/ml nocodazole for 8 h (25). Cells were stained for the mitotic marker phospho-histone H3 (Ser10) and analyzed using flow cytometry to detect mitotic entry (29). Flow cytometric analysis of asynchronous unirradiated cultures showed 31–36% of cells of both genotypes were phospho-histone H3 positive. However, after IR only 0.7% of WT cells were positive for phospho-histone H3, whereas 14% of BRCA2Δex4–7 stained positive (Fig. 5A, left panel). While PLK1 inhibition resulted in a mild increase in phospho-histone H3-positive cells (36–45%) in unirradiated cultures across cell lines, it caused a significant reduction in irradiated cultures. BRCA2Δex4–7 cells showed a 3.5-fold decrease in phospho-histone H3 positive cells (4%) while, similar to irradiated culture without PLK1 inhibition, almost no phospho-histone three positive cells (0.5%) were seen in WT cells (Fig. 5A, right panel). These results suggest a defect in PLK1-mediated regulation in BRCA2Δex4–7 cells.
Figure 5.
Aberrant activation of PLK1 following IR in BRCA2Δex4–7 expressing mES cells. (A) FACS analysis of phospho-histone H3 (Ser10) stained mES cells expressing WT, BRCA2Δex4–7 and BRCA2S193A with or without PLK1 inhibitor treatment, following IR. (B) Western blot analysis of total cell lysate from mES cells expressing WT, BRCA2Δex4–7 and BRCA2S193A showing total PLK1, activated PLK1-Thr210 and CDK1-Tyr15 levels at different time following release from nocodazole. (C) Densitometric analysis of protein bands shown in (B), normalized against glyceraldehyde-3-phosphate dehydrogenase. The scheme of experimental design is shown at the top of each panel.
To further examine whether the failure of BRCA2Δex4–7 cells to arrest in G2/M in response to IR is indeed due to a defect in PLK1-mediated regulation, we generated mES cells expressing BRCA2 with a serine to alanine change at position 193 (BRCA2S193A). S193 has been shown to be a PLK1 phosphorylation site (24). Similar to the WT and BRCA2Δex4–7, 39% of asynchronous unirradiated cells expressing BRCA2S193A were phospho-histone H3 positive. After IR 5% BRCA2S193A cells stained positive and again PLK1 inhibitor treatment reduced the percentage of phospho-histone H3-positive cells to 0.9% (Fig. 5A). Interestingly, compared with BRCA2Δex4–7 (4%) almost all BRCA2S193A (0.9%) cells were suppressed following IR and PLK1 inhibition. These results suggest that a defect in PLK1-mediated regulation of BRCA2 may contribute to the defect in cell-cycle regulation observed in cells expressing BRCA2Δex4–7.
To determine how the expression and activation of PLK1 and its downstream target, CDK1, is differentially regulated following DNA damage, WT, BRCA2Δex4–7 and BRCA2S193A cells were irradiated (6 Gy) and mitotic cells were trapped in 100 ng/ml nocodazole for 10 h. Nocodazole was then removed and cells were cultured in regular media, and harvested every hour for 6 h and analyzed for activated PLK1 (PLK1-Thr210) and CDK1 (CDK1-Tyr15) levels by western blot analysis (Fig. 5B and C). In unirradiated culture, WT, BRCA2Δex4–7 and BRCA2S193A cells underwent cell-cycle progression, upon release from nocodazole, as seen by decreasing levels of total and activated PLK1, and increasing levels of CDK1-Tyr15. Irradiated WT cells, however, remained arrested with 3-fold lower levels of activated PLK1-T210, up to 2 h post release and consistent high levels of inhibitory CDK1-Tyr15 (Fig. 5B). In addition, total PLK1 levels were similar to levels in unirradiated cells at 0 h post-release, and were maintained up to 6 h. This suggests that in the WT cells, PLK1 protein is present, but in an inactive form following DNA damage (Fig. 5B and C). In contrast, irradiated BRCA2Δex4–7 cells have high levels of PLK-Thr210 upon nocodazole release, which decreased with time, and a combined decrease in total PLK1 and CDK1-Tyr15 indicate mitotic progression (Fig. 5B and C). Irradiated BRCA2S193A cells, however, exhibited reduced total PLK1 and PLK1-Thr210 levels, but maintained higher levels of CDK1-Tyr15 protein compared with BRCA2Δex4–7, suggesting that less number of cells progressed through mitosis after IR. This observation was corroborated with the lower number of phospho-histone H3-positive cells found in irradiated BRCA2S193A (Fig. 5A).
BRCA2 has also been reported to play a role in cytokinesis. However, the precise role of BRCA2 is unclear as some studies have ruled out such a function (30) while others show defects in assembly of midbody related structures and in the time taken for abscission (26,31,32). In particular, it has been reported that disruption of S193 PLK1 phosphorylation site, prevents localization of BRCA2 to the midbody, which is predicted to affect the midbody formation and cytokinesis (26). Therefore, we examined the effect of loss of exons 4–7 on cytokinesis in MEFs derived from Wt and BRCA2Δex4–7/Δex4–7mice. We found no difference in the time taken from anaphase to abscission between Wt and Brca2Δex4–7/Δex4–7 MEFs (Supplementary Material, Fig. S8A and B); the formation of midbody was also not affected (Supplementary Material, Fig. S8C).
Taken together, our results show that BRCA2Δ105 is proficient in RAD51 recruitment and cytokinesis, and supports viability of whole organism. The loss of the 105 amino acids lead to cell-cycle perturbations in response to DNA damage, which may play a chronic rather than an acute role in bone marrow attrition under physiological condition.
Discussion
BRCA2 has been extensively sequenced in patients with a family history of breast and/or ovarian cancer as a means of identifying deleterious mutations that may account for the disease. Inheritance of a deleterious or cancer-causing mutation avails the mutation carrier to not only conventional surgery and chemotherapy, but also to adjuvant or neoadjuvant therapy options, such as Poly (ADP-ribose) Polymerase (PARP) inhibitors (33). Mutations that affect normal splicing and result in premature protein truncation due to skipping of exon(s) are considered deleterious mutations (34,35). While this is generally the case, occasionally multiple splice variants are generated including some that restore open-reading frame and result in a partially functional protein (36). Classification of such variants is challenging (37). IVS7+2T>G is one such splice site mutation, which is predicted to be a null allele because it results in premature protein truncation due to skipping of exon 7. Yet, some homozygous individuals are born alive. It has been demonstrated that at least one aberrant transcript lacking exons 4–7 is expressed in IVS7+2T>G carriers and the protein encoded by this transcript is functional (23).
Our observation of reduced viability seen in cells expressing BRCA2Δex4–7 (23) raised a serious concern whether this mutant was indeed capable of supporting the viability of individuals who are IVS7+2T>G homozygous. To address this, we generated a knock-in mouse model with a deletion of exons 4–7. Surprisingly, not only are the BRCA2Δex4–7/Δex4–7 as well as BRCA2Δex4–7/Ko mice viable and born at expected Mendelian ratios, they do not exhibit any developmental abnormalities. Both males and females are fertile and exhibit normal fecundity. These findings show that exons 4–7 of BRCA2 are dispensable, and the preserved functions of BRCA2Δex4–7 are sufficient for viability, fertility and normal development of mice.
IVS7+2T>G mutation has been found in patients with FA, a disease characterized by bone marrow failure resulting in bone marrow exhaustion leading to leukemia, and an increased susceptibility to multi-site tumor development. To date the best mouse model for the FA-D1 subtype is the one with a targeted deletion of exon 27 of Brca2 (Brca2Δ27) (38). Homozygous mutant mice are viable and fertile, but have significantly increased overall tumor incidence and decreased survival. These mice exhibit a marked proliferation defect in the hematopoietic progenitors and in the self-renewing HSCs (39). In contrast, in Brca2Δex4–7/Δex4–7 and Brca2Δex4-/Ko mice, homeostasis of peripheral blood components, bone marrow cellularity, and homeostasis of hematopoietic stem cell and committed progenitors were similar to Wt mice when unchallenged. In irradiated mice, we observed an increase in DNA damage and proliferation in the progenitor cells of Brca2Δex4–7/Ko mice, but again, this increase did not contribute to a difference in in vitro growth of mononuclear cells from irradiated animals, or survival of animals following whole-body IR, or after serial 5-FU treatment.
We observed no significant difference in the survival, tumor susceptibility or tumor spectrum of Brca2Δex4–7/Δex4–7 and Brca2Δex4-/Ko compared with Brca2Δex4–7/+ mice, which suggests that the loss of exons 4–7 has no effect on tumor suppressor function(s) of BRCA2. Even on a Trp53Ko/+ background, no difference in tumor-free survival was observed between Brca2Δex4–7/+ and Brca2Δex4–7/Δex4–7 mice. Although the precise reason for the increased incidence of lymphomas and hematopoietic neoplasia in Brca2Δex4–7/Δex4–7;Trp53Ko/+ is not understood, decrease in lineage-negative cells and increased DNA damage in multipotent progenitors in these mice suggests that, under physiological conditions, activation of the bone marrow to maintain hematopoietic homeostasis may lead to bone marrow attrition. Studies have shown that treatments that mimic physiological stress can force stem cells to exit quiescence, a process which elicits DNA damage, HSC attrition and bone marrow collapse (40,41).
We also performed a detailed functional characterization of mES cells expressing the BRCA2Δex4–7 variant and demonstrated that the deletion of the 105 amino acid region containing multiple PLK1 and CDK1 phosphorylation sites renders cells defective in their ability to arrest in cell cycle in response to DNA damage. To date, PLK1 and BRCA2 regulation, and the possible role of BRCA2 in cell-cycle regulation has only been shown in vitro using BRCA2 peptides (24,42) and by siRNA-mediated knockdown of BRCA2 (25). Using BRCA2 peptides, it was shown that mutating one, or a combination of, PLK1 sites within the 105-amino acid region results in the loss of PLK1 phosphorylation (24). In this study, we confirm that this change results in loss of the ability of BRCA2Δex4–7 cells to arrest cell-cycle progression in the presence of DNA damage. The higher proportion of cells entering mitosis in BRCA2Δex4–7 compared with BRCA2S193A, and the ability of PLK1 inhibition to suppress the progression in irradiated BRCA2S193A almost to the level seen in the WT cells suggest that while Ser193 is an important regulatory residue, other sites may play a non-redundant role in PLK1/BRCA2 interaction. The loss of these additional sites exacerbates the premature activation of PLK1, as seen in the increase in cells entering mitosis in the BRCA2Δex4–7 mutant in the presence of DNA damage. Also, suppression of premature activation of PLK1, in the presence of DNA damage by PLK1 inhibitors, suggests a possible feedback regulation of the two proteins to prevent premature entry of cells with DNA damage from entering mitosis. It is also possible that the 105-amino acid region may play a role in cell-cycle regulation via PLK1 independent pathways, which further highlights the functional importance of this region.
Considering the significant defects observed in mES cells in cell-cycle regulation and genomic stability in response to IR, it is surprising that we did not observe any increase in tumorigenesis in Brca2Δex4–7/Δex4–7 or Brca2Δex4–7/Ko mice compared with Brca2Δex4–7/+ mice. It is also surprising that not only in unchallenged mice, but also in response to acute stress such as IR or 5-FU treatment there was no effect on their overall survival. We conclude that the loss of exons 4–7 may have subtle effects in vivo, e.g. on the maintenance of bone marrow homeostasis and hematopoiesis in response to chronic physiological stress in older mice. If this is true, then why do individuals who are homozygous or compound heterozygous for IVS7+2T>G mutation have severe developmental abnormalities and succumb to malignancies within few years after birth? The difference may be attributed to the nature of the two mutations. In the case of mice expressing the Brca2Δex4–7 allele, the transcript lacking exons 4–7 is the predominant form. However, in the case of humans carrying IVS7+2T>G, the predominant transcript is the one lacking exon 7. For normal BRCA2 function, it is critical that skipping of exons 4–7 occurs at levels that can generate sufficient BRCA2Δ105 for normal BRCA2 function in cells. However, if the levels are below a certain threshold, it will result in a ‘functionally BRCA2-null’ state. Because splicing is a complex and tightly regulated process, any physiological, genetic or epigenetic perturbation may affect the levels of Δexons 4–7 transcripts with severe consequences (43).
Our findings described here will have a significant impact on the classification of BRCA2 variants, especially those that result in a premature stop codon in exons 4–7. The Breast Cancer Information Core database includes over a hundred variants in this region, including 54 potential splice site variants and 15 non-sense or frame-shift mutations. We have previously shown that W194X (c.589G>A), a nonsense mutation, expresses Δexons 4–7 transcript in mES cells as well as human lymphoblasts (23). The expression of alternatively spliced transcripts must be examined to determine their potential to generate a functional protein before classifying any non-sense or frame-shift mutation as clearly deleterious or pathogenic. In addition, it is also possible that the presence of such splice variants may contribute to resistance to targeted therapy, e.g. PARP inhibitors that induce synthetic lethality in cells that are defective in homologous recombination. Our findings suggest that caution must be exercised when counseling individuals who inherit such mutations, as well as when considering treatment options.
Materials and Methods
mES cell culture and differentiation
Mouse ES cells were cultured on feeder cells in supplemented M15 media, as described previously (44). ES cells were differentiated into embryoid bodies in supplemented Dulbecco's modified Eagle's medium (1×) media as described previously (45). Five to seven days old embryoid bodies were used to generate fibroblasts.
mES cells expressing BRCA2 variants
The S193A variant (c.577T>G) was recombineered into the human BRCA2 cloned in BAC CTD-2342K5, as described previously (46). Recombinant BAC was electroporated into PL2F7 mES cells, and the resulting recombinant mES clones were characterized as described by Kuznetsov et al. in 2008. Oligonucleotides used for generating the mutation are listed in Supplementary Material, Table S3.
Bone marrow stem and progenitor cell culture
For bone marrow stem and progenitor cells culture, marrows from 8 to 12-week-old mice were harvested from femurs. Mononuclear cell population was isolated by gradient separation using lymphocyte separation media, and cultured in Iscove's Modified Dulbecco's medium media supplemented with 10% heat-inactivated fetal bovine serum, glutamine penicillin streptomycin, 100 ng/ml mouse Stem Cell Factor, 100 ng/ml human Thrombopoeitin, 100 ng/ml hFlt3L, 50 ng/ml mIL-6 and 30 ng/ml mIL-3.
Functional studies in mice
Mice on 129SvJae X C57BL/6J mixed genetic background, aged between 8 and 12 weeks and littermate controls were used in all studies. To assess the effect of IR on bone marrow compartment, mice were given a single-IR dose of 6 Gy and femur marrow was harvested 4-day post-IR for in vitro culture, as described above. For survival, mice were either given a single dose of 8 Gy or given serial intraperitoneal injection of 150 mg/kg of clinical grade 5-FU (NDC 633323-117-10, Fresenius Kabi, USA, LLC) on Days 0, 7 and 14. Mice were followed for survival, and sacrificed when moribund.
For aging studies, Brca2Δex4–7/Δex4–7 mice were crossed with Trp53Ko/+ mice to generate Brca2Δex4–7/+;Trp53Ko/+ and Brca2Δex4–7/Δex4–7;Trp53Ko/+ cohort. Mice were monitored for survival until they became moribund or developed visible tumors after which they were sacrificed for pathological evaluation. All experimental protocols were approved by the NCI-Frederick Animal Care and Use Committee.
Flow cytometry
For cell-cycle analysis, asynchronous mES cells were given a single dose of 6 Gy IR and mitotic cells were trapped with 100 ng/ml nocodazole for 10 h or treated with 250 nm BI2536, a PLK inhibitor, for 2 h followed by nocodazole treatment for 8 h. Cells were then fixed with BD CytoFix/CytoPerm solution (Becton Dickinson) and stained with propidium iodide (PI) or Alexa647-phosphohistone H3 (Ser10) (Cell Signaling, #3458) for analysis. BI2536 predominantly inhibits the kinase activity of PLK1, but also affects PLK2 and PLK3 activity, while its effect on PLK4 has not been determined (47).
To detect DNA synthesis in the presence of DNA damage, mES cells were irradiated (6 Gy) and given a BrdU (31 µg/ml) pulse for 20 min before harvesting at 6, 12, 18 and 24 h post-IR for analysis. BrdU incorporation was detected using fluorescein isothiocyanate-conjugated anti-BrdU antibody (eBioscience, 11-6071-71) as described in (48).
For assessment of the bone marrow compartment, femur marrow was harvested from aged mice, unirradiated mice or from irradiated mice 4 days post 6 Gy IR. Mononuclear cells were isolated and stained for flow cytometric analysis with a combination antibodies described in the Supplementary Material, Methods section.
Protein expression
For western blot analysis, proteins were extracted in lysis buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES] [pH 7.5], 100 mM NaCl, 1 mM EDTA, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM NaF, 1 mM Dithiothreitol[DTT], 0.1% Triton X-100, 1 mM PMSF, protease inhibitor cocktail [Roche]), separated on 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and detected using the ECL Plus western blotting detection system (Amersham). Levels of total PLK1 [PLK1 (F-8), Santa Cruz, sc-17783], activated PLK1 (pT210; Becton Dickinson, 558400), CDC2/CDK1-Tyr15 (Cell signaling, #4539) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Cell Signaling, #2118) were assessed. Relative expression levels were measured by normalizing the levels of the respective proteins to GAPDH using GeneTools from Syngene, Frederick.
Immunofluorescence
To stain bone marrow cells, sorted cells grown in culture for 36 h were dropped onto polylysine-coated slides and left to adhere for 1 h, fixed and stained with antibodies against γH2AX. Staining and quantification of γH2AX staining density is described in Supplementary Material.
Statistical methods
Animal survival data were analyzed using Prism 6, and P-values were estimated using the Gehan–Breslow–Wilcoxon test. Statistical significance for Rad51 and γH2AX foci in MEFs, and FACS data were evaluated using two-way analysis of variance and T-test, respectively. Statistical significance for incidence of hematopoietic neoplasm and solid tumor was evaluated using two-tailed Fischer's exact test.
Supplementary Material
Funding
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E and the Intramural Research Program, Center for Cancer Research, U.S. National Cancer Institute. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Supplementary Material
Acknowledgements
We thank Drs Jairaj Acharya, Xia Ding and Suzanne Hartford for critical review of the manuscript. We also thank Stacey Stauffer and Susan Reid for technical help; Allen Kane (Leidos Biomedical Research, Inc., Scientific Publications, Graphics and Media Department) for illustrations. We thank Dr Satyendra Singh for technical advice.
Conflict of Interest statement. None declared.
References
- 1.Wooster R., Neuhausen S.L., Mangion J., Quirk Y., Ford D., Collins N., Nguyen K., Seal S., Tran T., Averill D. et al. (1994) Localization of a breast cancer susceptibility gene, BRCA2, to chromosome 13q12-13. Science, 265, 2088–2090. [DOI] [PubMed] [Google Scholar]
- 2.Ford D., Easton D.F., Stratton M., Narod S., Goldgar D., Devilee P., Bishop D.T., Weber B., Lenoir G., Chang-Claude J. et al. (1998) Genetic heterogeneity and penetrance analysis of the BRCA1 and BRCA2 genes in breast cancer families. The Breast Cancer Linkage Consortium. Am. J. Hum. Genet., 62, 676–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Malone K.E., Begg C.B., Haile R.W., Borg A., Concannon P., Tellhed L., Xue S., Teraoka S., Bernstein L., Capanu M. et al. (2010) Population-based study of the risk of second primary contralateral breast cancer associated with carrying a mutation in BRCA1 or BRCA2. J. Clin. Oncol., 28, 2404–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Molina-Montes E., Perez-Nevot B., Pollan M., Sanchez-Cantalejo E., Espin J., Sanchez M.J. (2014) Cumulative risk of second primary contralateral breast cancer in BRCA1/BRCA2 mutation carriers with a first breast cancer: a systematic review and meta-analysis. Breast, 23, 721–742. [DOI] [PubMed] [Google Scholar]
- 5.Tai Y.C., Domchek S., Parmigiani G., Chen S. (2007) Breast cancer risk among male BRCA1 and BRCA2 mutation carriers. J. Natl Cancer Inst., 99, 1811–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Habbe N., Langer P., Sina-Frey M., Bartsch D.K. (2006) Familial pancreatic cancer syndromes. Endocrinol. Metab. Clin. North Am., 35, 417–430, xi. [DOI] [PubMed] [Google Scholar]
- 7.Sharan S.K., Morimatsu M., Albrecht U., Lim D.S., Regel E., Dinh C., Sands A., Eichele G., Hasty P., Bradley A. (1997) Embryonic lethality and radiation hypersensitivity mediated by Rad51 in mice lacking Brca2. Nature, 386, 804–810. [DOI] [PubMed] [Google Scholar]
- 8.Patel K.J., Yu V.P., Lee H., Corcoran A., Thistlethwaite F.C., Evans M.J., Colledge W.H., Friedman L.S., Ponder B.A., Venkitaraman A.R. (1998) Involvement of Brca2 in DNA repair. Mol. Cell, 1, 347–357. [DOI] [PubMed] [Google Scholar]
- 9.Schlacher K., Christ N., Siaud N., Egashira A., Wu H., Jasin M. (2011) Double-strand break repair-independent role for BRCA2 in blocking stalled replication fork degradation by MRE11. Cell, 145, 529–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schlacher K., Wu H., Jasin M. (2012) A distinct replication fork protection pathway connects Fanconi anemia tumor suppressors to RAD51-BRCA1/2. Cancer Cell, 22, 106–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walden H., Deans A.J. (2014) The Fanconi anemia DNA repair pathway: structural and functional insights into a complex disorder. Annu. Rev. Biophys., 43, 257–278. [DOI] [PubMed] [Google Scholar]
- 12.Wang A.T., Kim T., Wagner J.E., Conti B.A., Lach F.P., Huang A.L., Molina H., Sanborn E.M., Zierhut H., Cornes B.K. et al. (2015) A dominant mutation in human RAD51 reveals its function in DNA interstrand crosslink repair independent of homologous recombination. Mol. Cell, 59, 478–490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sawyer S.L., Tian L., Kahkonen M., Schwartzentruber J., Kircher M., University of Washington Centre for Mendelian, G., Consortium, F.C., Majewski J., Dyment D.A., Innes A.M. et al. (2015) Biallelic mutations in BRCA1 cause a new Fanconi anemia subtype. Cancer Discov., 5, 135–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Myers K., Davies S.M., Harris R.E., Spunt S.L., Smolarek T., Zimmerman S., McMasters R., Wagner L., Mueller R., Auerbach A.D. et al. (2012) The clinical phenotype of children with Fanconi anemia caused by biallelic FANCD1/BRCA2 mutations. Pediatr. Blood Cancer, 58, 462–465. [DOI] [PubMed] [Google Scholar]
- 15.Howlett N.G., Taniguchi T., Olson S., Cox B., Waisfisz Q., De Die-Smulders C., Persky N., Grompe M., Joenje H., Pals G. et al. (2002) Biallelic inactivation of BRCA2 in Fanconi anemia. Science, 297, 606–609. [DOI] [PubMed] [Google Scholar]
- 16.Reid S., Renwick A., Seal S., Baskcomb L., Barfoot R., Jayatilake H., Pritchard-Jones K., Stratton M.R., Ridolfi-Luthy A., Rahman N. et al. (2005) Biallelic BRCA2 mutations are associated with multiple malignancies in childhood including familial Wilms tumour. J. Med. Genet., 42, 147–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Alter B.P., Rosenberg P.S., Brody L.C. (2007) Clinical and molecular features associated with biallelic mutations in FANCD1/BRCA2. J. Med. Genet., 44, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hakem R., de la Pompa J.L., Mak T.W. (1998) Developmental studies of Brca1 and Brca2 knock-out mice. J. Mammary Gland Biol. Neoplasia, 3, 431–445. [DOI] [PubMed] [Google Scholar]
- 19.Connor F., Bertwistle D., Mee P.J., Ross G.M., Swift S., Grigorieva E., Tybulewicz V.L.J., Ashworth A. (1997) Tumorigenesis and a DNA repair defect in mice with a truncating Brca2 mutation. Nat. Genet., 17, 423–430. [DOI] [PubMed] [Google Scholar]
- 20.Donoho G., Brenneman M.A., Cui T.X., Donoviel D., Vogel H., Goodwin E.H., Chen D.J., Hasty P. (2003) Deletion of Brca2 exon 27 causes hypersensitivity to DNA crosslinks, chromosomal instability, and reduced life span in mice. Genes Chromosomes Cancer, 36, 317–331. [DOI] [PubMed] [Google Scholar]
- 21.Pyne M.T., Brothman A.R., Ward B., Pruss D., Hendrickson B.C., Scholl T. (2000) The BRCA2 genetic variant IVS7+2T -> G is a mutation. J. Hum. Genet., 45, 351–357. [DOI] [PubMed] [Google Scholar]
- 22.Wagner J.E., Tolar J., Levran O., Scholl T., Deffenbaugh A., Satagopan J., Ben-Porat L., Mah K., Batish S.D., Kutler D.I. et al. (2004) Germline mutations in BRCA2: shared genetic susceptibility to breast cancer, early onset leukemia, and Fanconi anemia. Blood, 103, 3226–3229. [DOI] [PubMed] [Google Scholar]
- 23.Biswas K., Das R., Alter B.P., Kuznetsov S.G., Stauffer S., North S.L., Burkett S., Brody L.C., Meyer S., Byrd R.A. et al. (2011) A comprehensive functional characterization of BRCA2 variants associated with Fanconi anemia using mouse ES cell-based assay. Blood, 118, 2430–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin H.R., Ting N.S., Qin J., Lee W.H. (2003) M phase-specific phosphorylation of BRCA2 by Polo-like kinase 1 correlates with the dissociation of the BRCA2-P/CAF complex. J. Biol. Chem., 278, 35979–35987. [DOI] [PubMed] [Google Scholar]
- 25.Menzel T., Nahse-Kumpf V., Kousholt A.N., Klein D.K., Lund-Andersen C., Lees M., Johansen J.V., Syljuasen R.G., Sorensen C.S. (2011) A genetic screen identifies BRCA2 and PALB2 as key regulators of G2 checkpoint maintenance. EMBO Rep., 12, 705–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takaoka M., Saito H., Takenaka K., Miki Y., Nakanishi A. (2014) BRCA2 phosphorylated by PLK1 moves to the midbody to regulate cytokinesis mediated by nonmuscle myosin IIC. Cancer Res., 74, 1518–1528. [DOI] [PubMed] [Google Scholar]
- 27.Jonkers J., Meuwissen R., van der Gulden H., Peterse H., van der Valk M., Berns A. (2001) Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nat. Genet., 29, 418–425. [DOI] [PubMed] [Google Scholar]
- 28.van Vugt M.A., Medema R.H. (2005) Getting in and out of mitosis with Polo-like kinase-1. Oncogene, 24, 2844–2859. [DOI] [PubMed] [Google Scholar]
- 29.Hans F., Dimitrov S. (2001) Histone H3 phosphorylation and cell division. Oncogene, 20, 3021–3027. [DOI] [PubMed] [Google Scholar]
- 30.Lekomtsev S., Guizetti J., Pozniakovsky A., Gerlich D.W., Petronczki M. (2010) Evidence that the tumor-suppressor protein BRCA2 does not regulate cytokinesis in human cells. J. Cell Sci., 123, 1395–1400. [DOI] [PubMed] [Google Scholar]
- 31.Daniels M.J., Wang Y., Lee M., Venkitaraman A.R. (2004) Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science, 306, 876–879. [DOI] [PubMed] [Google Scholar]
- 32.Mondal G., Rowley M., Guidugli L., Wu J., Pankratz V.S., Couch F.J. (2012) BRCA2 localization to the midbody by filamin A regulates cep55 signaling and completion of cytokinesis. Dev. Cell, 23, 137–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Livraghi L., Garber J.E. (2015) PARP inhibitors in the management of breast cancer: current data and future prospects. BMC Med., 13, 188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomassen M., Pedersen I.S., Vogel I., Hansen T.V., Brasch-Andersen C., Brasen C.L., Cruger D., Sunde L., Nielsen F.C., Jensen U.B. et al. (2011) A BRCA2 mutation incorrectly mapped in the original BRCA2 reference sequence, is a common West Danish founder mutation disrupting mRNA splicing. Breast Cancer Res. Treat., 128, 179–185. [DOI] [PubMed] [Google Scholar]
- 35.Sanz D.J., Acedo A., Infante M., Duran M., Perez-Cabornero L., Esteban-Cardenosa E., Lastra E., Pagani F., Miner C., Velasco E.A. (2010) A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin. Cancer Res., 16, 1957–1967. [DOI] [PubMed] [Google Scholar]
- 36.Skandalis A., Uribe E. (2004) A survey of splice variants of the human hypoxanthine phosphoribosyl transferase and DNA polymerase beta genes: products of alternative or aberrant splicing? Nucleic Acids Res., 32, 6557–6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenthal E.T., Bowles K.R., Pruss D., van Kan A., Vail P.J., McElroy H., Wenstrup R.J. (2015) Exceptions to the rule: Case studies in the prediction of pathogenicity for genetic variants in hereditary cancer genes. Clin. Genet., 88, 533–541. [DOI] [PubMed] [Google Scholar]
- 38.McAllister K.A., Bennett L.M., Houle C.D., Ward T., Malphurs J., Collins N.K., Cachafeiro C., Haseman J., Goulding E.H., Bunch D. et al. (2002) Cancer susceptibility of mice with a homozygous deletion in the COOH-terminal domain of the Brca2 gene. Cancer Res., 62, 990–994. [PubMed] [Google Scholar]
- 39.Navarro S., Meza N.W., Quintana-Bustamante O., Casado J.A., Jacome A., McAllister K., Puerto S., Surralles J., Segovia J.C., Bueren J.A. (2006) Hematopoietic dysfunction in a mouse model for Fanconi anemia group D1. Mol. Ther., 14, 525–535. [DOI] [PubMed] [Google Scholar]
- 40.Rossi D.J., Bryder D., Seita J., Nussenzweig A., Hoeijmakers J., Weissman I.L. (2007) Deficiencies in DNA damage repair limit the function of haematopoietic stem cells with age. Nature, 447, 725–729. [DOI] [PubMed] [Google Scholar]
- 41.Walter D., Lier A., Geiselhart A., Thalheimer F.B., Huntscha S., Sobotta M.C., Moehrle B., Brocks D., Bayindir I., Kaschutnig P. et al. (2015) Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature, 520, 549–552. [DOI] [PubMed] [Google Scholar]
- 42.Lee M., Daniels M.J., Venkitaraman A.R. (2004) Phosphorylation of BRCA2 by the Polo-like kinase Plk1 is regulated by DNA damage and mitotic progression. Oncogene, 23, 865–872. [DOI] [PubMed] [Google Scholar]
- 43.Kornblihtt A.R., Schor I.E., Allo M., Dujardin G., Petrillo E., Munoz M.J. (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat. Rev. Mol. Cell Biol., 14, 153–165. [DOI] [PubMed] [Google Scholar]
- 44.Kuznetsov S.G., Liu P., Sharan S.K. (2008) Mouse embryonic stem cell-based functional assay to evaluate mutations in BRCA2. Nat. Med., 14, 875–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hopfl G., Gassmann M., Desbaillets I. (2004) Differentiating embryonic stem cells into embryoid bodies. Methods Mol. Biol., 254, 79–98. [DOI] [PubMed] [Google Scholar]
- 46.Sharan S.K., Thomason L.C., Kuznetsov S.G., Court D.L. (2009) Recombineering: a homologous recombination-based method of genetic engineering. Nat. Protoc., 4, 206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steegmaier M., Hoffmann M., Baum A., Lenart P., Petronczki M., Krssak M., Gurtler U., Garin-Chesa P., Lieb S., Quant J. et al. (2007) BI 2536, a potent and selective inhibitor of polo-like kinase 1, inhibits tumor growth in vivo. Curr. Biol., 17, 316–322. [DOI] [PubMed] [Google Scholar]
- 48.Cecchini M.J., Amiri M., Dick F.A. (2012) Analysis of cell cycle position in mammalian cells. J. Vis. Exp., 59, e3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.