Abstract
Mutations in RPGR (retinitis pigmentosa GTPase regulator) are the most common cause of X-linked RP, a severe blindness disorder. RPGR mutations result in clinically variable disease with early- to late-onset phenotypic presentation. Molecular mechanisms underlying such heterogeneity are unclear. Here we show that phenotypic expression of Rpgr-loss in mice is influenced genetically by the loss of Cep290, a human ciliopathy gene. We found that Rpgrko/Y mice with a heterozygous hypomorphic allele of Cep290 (Cep290rd16/+) but not of a heterozygous null allele of Cep290 (Cep290null/+) or of other ciliopathy genes, Rpgrip1, Nphp1, Nphp4 and Nphp5, exhibit relatively early onset (by 3 months of age) retinal degeneration and dysfunction when compared with the onset at ∼7 months of age in the Rpgrko/Y mice. We also observed disorganized photoreceptor outer-segment morphology and defective trafficking of opsins in the Rpgrko/Y::Cep290rd16/+ mice. Together with a physical interaction between RPGR and the C-terminal domain of CEP290, our data suggest that RPGR and CEP290 genetically interact and highlight the involvement of hypomorphic alleles of genes as potential modifiers of heterogeneous retinal ciliopathies.
Introduction
Retinitis Pigmentosa (RP) is a debilitating inherited blindness disorder, which results in a progressive and severe loss of rod and cone photoreceptors (1,2). X-linked forms of RP (XLRP) are the most severe forms of RP characterized by the loss of photoreceptor function starting as early as in the first decade of life and progressing into complete blindness by the third or fourth decade (3–5). Mutations in two genes, RPGR and RP2, account for a majority of XLRP cases (6–8). Of these, RPGR mutations are associated with >70% of XLRP and 15–20% of simplex RP cases, making it a common cause of RP worldwide (5,6,9,10).
Genotype–phenotype correlation studies revealed that RPGR mutations result in variable clinical manifestation with mild to severe rod followed by cone photoreceptor loss or a predominantly cone–rod degeneration (11–13). Moreover, even patients with same mutations exhibit heterogenic clinical phenotype, suggesting the influence of genetic modifiers (14). Variable severity associated with RPGR mutations is also recapitulated in animal models of Rpgr. The Rpgrko mouse, generated by deleting exons 4–6 (15), undergoes relatively delayed onset (around 7 months of age) retinal degeneration and mild defects in the trafficking of opsins to the light-sensing outer segment (OS; sensory cilium) compartment of photoreceptors. A naturally occurring Rpgr mutant mouse model rd9 (retinal degeneration 9) and a conditional allele of Rpgr exhibit a moderate progressing retinal degenerative disease (5,16). Two canine models of RPGR mutation, XLPRA1 and XLPRA2 have also been characterized. Whereas XLPRA1 exhibits a relatively mild phenotype, the XLPRA2 canine model undergoes a severe and fast progressing retinal degeneration (17).
Here, we sought to identify potential genetic modifiers of RPGR-associated retinal degeneration. Genetic modifiers should meet the criteria of being able to interact with the causative gene and of being themselves involved in retinal degeneration (18,19). RPGR is a ciliary protein and interacts with autosomal recessive ciliopathy-associated proteins, such as RPGR-interacting protein1 (RPGRIP1), CEP290 and Nephronophthisis (NPHP)-associated proteins NPHP1, NPHP4 and NPHP5 in mammalian retina (20–26). In this study, we assessed the effect of heterozygous alleles (not disease-causing by themselves) of these RPGR-interacting proteins on the progression of retinal degeneration in the Rpgrko mice.
Results
Generation and characterization of Rpgrko double-mutant mice
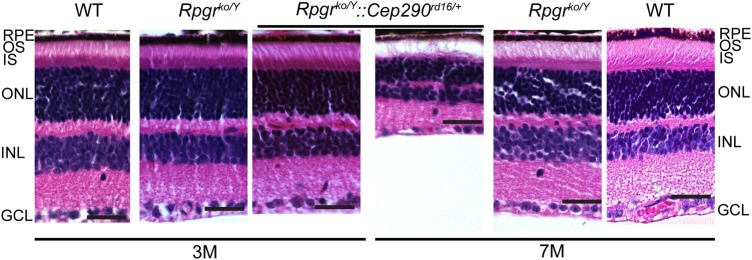
To assess the effect of allelic combinations of select RPGR-interacting proteins, we bred the Rpgrko mice with Rpgrip1−/− (25), Nphp1−/− (27), Nphp4−/− (Nphp4nmf192) (28), Nphp5−/− (C.R. and W.B., manuscript in preparation) and Cep290rd16 (20) mice and analyzed male mice of the following genotypes: Rpgrko/Y::Rpgrip1+/−, Rpgrko/Y::Nphp1+/−, Rpgrko/Y::Nphp4+/−, Rpgrko/Y::Nphp5+/− and Rpgrko/Y::Cep290+/rd16. All double-mutant mice were viable and fertile. As the tested genes are associated with autosomal recessive disorders, heterozygous alleles of Rpgrip1, Nphp1, Nphp4, Nphp5 and Cep290rd16 do not result in retinal degeneration or photoreceptor dysfunction. We next performed histological analysis of the double-mutant mouse retina and compared it with the age-matched Rpgrko/Y mouse retina. As shown in Figure 1, the Rpgrko/Y::Cep290rd16/+ mice exhibited a relatively faster degeneration of the photoreceptor layer (8–10 layers of photoreceptor nuclei by 3 months of age) when compared with age-matched Rpgrko mice, which had 10–12 nuclear layers. By 7 months of age, only 5–6 layers of nuclei in the outer nuclear layer were detected in the Rpgrko/Y::Cep290rd16/+ mice when compared with 8–9 layers in the Rpgrko/Y mice. No other double-mutant mice tested showed an effect on retinal morphology of the Rpgrko/Y mice (Supplementary Material, Fig. S1).
Figure 1.
Paraffin-embedded sections of mouse retina of indicated genotypes were stained with hematoxylin/eosin (H&E). Central retina from all sections was imaged. Scale bars: 10 μm. Age of the mice used in the experiment is indicated in months (M). A progressively faster thinning of the outer nuclear layer (ONL) was observed in the Rpgrko/Y::Cep290rd16/+ mice when compared with the Rpgrko/Y mice. RPE, retinal pigmented epithelium; OS, outer segment; IS, inner segment; GCL, ganglion cell layer.
Photoreceptor dysfunction in Rpgrko/Y::Cep290rd16/+
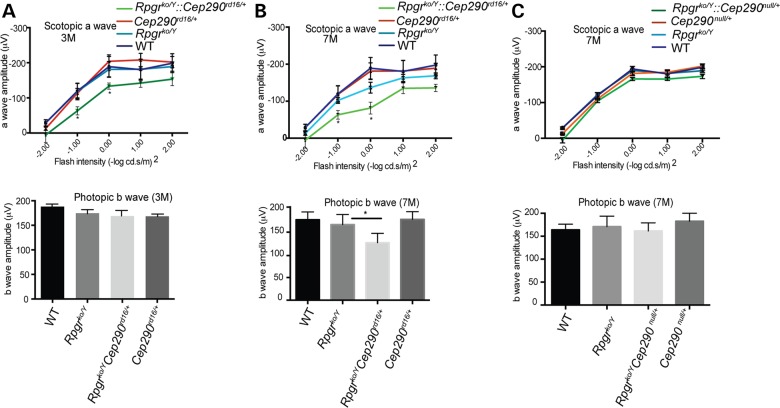
To assess photoreceptor dysfunction in the Rpgrko/Y::Cep290rd16/+ mice, we performed electroretinography (ERG) analysis. While the Rpgrko/Y mice did not exhibit a detectable defect in the scotopic a-wave (rod-mediated) amplitude when compared with the wild-type mice at 3 months of age, the Rpgrko/Y::Cep290rd16/+ mice revealed significant reduction (∼40% reduction; *P<0.0001) in the amplitude (Fig. 2A; Supplementary Material, Fig. S2A). However, no effect on the photopic (cone-mediated) b-wave amplitude was detected at this age. The Rpgrko/Y::Cep290rd16/+ mice exhibited a progressive and relatively severe decline in both scotopic and photopic ERG amplitudes of the Rpgrko/Y::Cep290rd16/+ mice at 7 months of age when compared with Rpgrko/Y mice (Fig. 2B; Supplementary Material, Fig. S2A). We also performed ERG analysis of Rpgrko/+::Cep290rd16/+ carrier female mice. As shown in Supplementary Material, Figure S2B, we did not detect a change in ERG a- and b-waveforms between wild-type, Rpgrko/+, Rpgrko/+::Cep290rd16/+ female mice. Rpgrko/Y mice carrying heterozygous mutant alleles of other Nphp or of Rpgrip1 genes did not show an effect on Rpgrko/Y ERGs, up to 7 months of age.
Figure 2.
Scotopic (dark-adapted a-wave; upper panel) and photopic (light-adapted b-wave; lower panel) ERG recordings were performed in WT, Rpgrko/Y, Cep290rd16/+ and Rpgrko/Y::Cep290rd16/+ at 3 months (A) and 7 months (B) of age. (C) ERG of the Rpgrko/Y::Cep290null/+ mice is shown at the latest time point (7 months). Five mice were used for each genotype. *P < 0.0001. M, age in months.
The Cep290rd16 mice carry in-frame deletion in the Cep290 gene, which results in the deletion of amino acids 1599–1897 of the myosin-tail homology domain (also called Deleted in rd16 domain; DRD) of the mouse CEP290 protein (20,29). This mutation is hypomorphic as the deleted variant of the CEP290 protein (ΔCEP290) lacking amino acids 1599–1897 is still expressed in these mice (20,29). The Cep290rd16/rd16 mice undergo relatively severe photoreceptor dysfunction and degeneration, starting as early as postnatal Day P18.
The faster progression of retinal degeneration in the Rpgrko/Y::Cep290rd16/+ mice could be attributed either to a deleterious effect of the presence of the ΔCEP290 protein in the absence of RPGR or to insufficient amount of the full-length CEP290 protein. To differentiate between these possibilities, we used previously reported Cep29null/null mice, which is a model of Joubert Syndrome and do not show a detectable expression of the CEP290 protein (30), to generate Rpgrko/Y::Cep290null/+ mice. ERG analysis of the 7-month-old double-mutant mice did not reveal any effect on the progression of photoreceptor dysfunction in the Rpgrko/Y mice (Fig. 2C). These results suggest that the presence of one copy of ΔCEP290 but not insufficiency of wild-type CEP290 alters the severity of RPGR-associated photoreceptor dysfunction, and that genetic interaction between RPGR and CEP290 may depend upon the nature of the allelic variants.
Ultrastructural analysis of Rpgrko/Y::Cep290rd16/+ photoreceptors
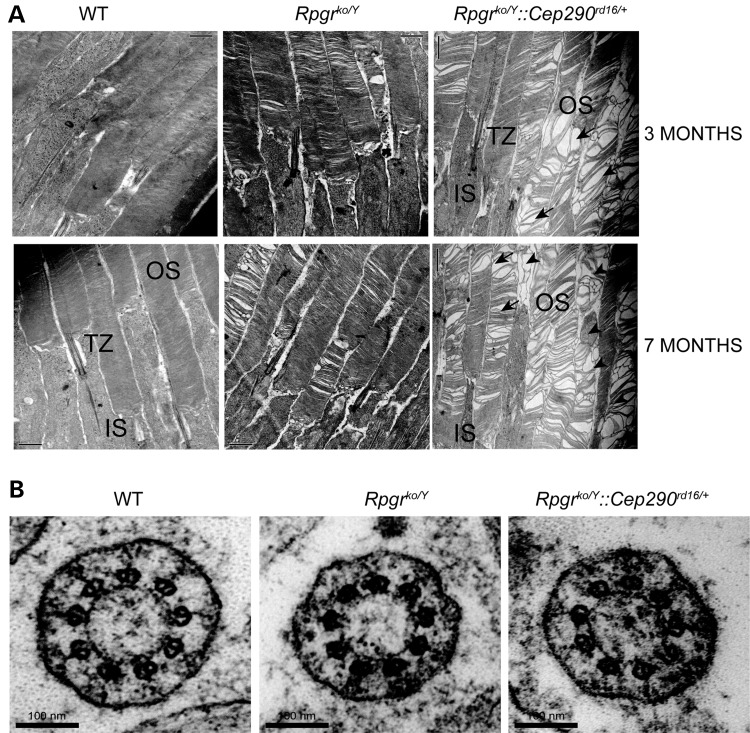
We then examined photoreceptor morphology of the Rpgrko/Y::Cep290rd16/+ mice. The Rpgrko mice exhibit OS degeneration starting around 7 months of age. Using transmission electron microscopy (TEM) at 3 months of age, we found that the photoreceptor OS discs of the Rpgrko/Y::Cep290rd16/+ mice exhibited abnormal ultrastructure. We found large empty spaces in the OS along with small areas of disorganization, as indicated by the appearance of whorls or spherical membranes in place of discs (indicated by arrowheads), in the Rpgrko/Y::Cep290rd16/+ mice. However, as reported earlier (15), only subtle alterations in the OS were observed in the 7-month-old Rpgrko/Y mice (Fig. 3A). We also examined the ultrastructure of the TZ of the mutant photoreceptors. As shown in Figure 3B, no morphological alterations in the 9 + 0 arrangement of microtubule doublets was observed between the Rpgrko/Y and Rpgrko/Y::Cep290rd16/+ mice.
Figure 3.
(A) TEM analysis of photoreceptors of WT, Rpgrko/Y and Rpgrko/Y::Cep290rd16/+ mice was performed at 3 months (upper panel) and 7 months (lower panel) of age. Scale bar: 1 μm. OS, outer segment; IS, inner segment; TZ, transition zone. Arrows point to the large empty spaces and arrowheads denote disorganized spaces and appearance of spherical membranous structures in the double-mutant photoreceptor OS. (B) Cross-section of the TZ of photoreceptors of the mutant mice revealed no detectable differences in the morphology of the TZ microtubule architecture among the indicated genotypes.
Opsin mislocalization in Rpgrko/Y::Cep290rd16/+ retina
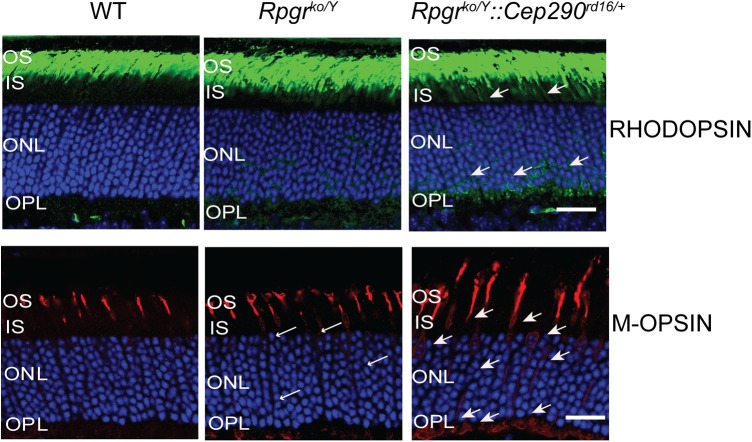
Both RPGR and CEP290 are ciliary proteins implicated in regulating the trafficking of proteins into the OS (20,21,31–34). We, therefore, investigated the trafficking of opsins to the OS in the double-mutant retina, when compared with the Rpgrko mice. Immunostaining of retina sections with anti-rhodopsin or anti-M-cone opsin antibody showed that both rod and cone opsins redistributed to the inner segment (IS), outer nuclear layer (ONL) and the outer plexiform layer (OPL) of the Rpgrko/Y::Cep290rd16/+ mouse retina at 1 month of age at which time the Rpgrko/Y::Cep290rd16/+ mice do not undergo degeneration and the Rpgrko mice do not exhibit appreciable rhodopsin or only show mild cone opsin trafficking defects (Fig. 4). Interestingly, abundant mislocalization of M-opsin was detected in the IS of the Rpgrko/Y::Cep290rd16/+ mice. However, we did not detect a change in the trafficking of another OS protein PERIPHERIN/RDS or rod Phosphodiesterase 6α (PDE6α) (Supplementary Material, Fig. S3).
Figure 4.
Immunofluorescence analysis of retinal cryosections from 1-month-old mice of indicated genotypes was performed using anti-rhodopsin (green) or anti-M-opsin (red) antibodies. Nuclei were stained with Hoechst (blue). Arrows indicate mislocalized opsins in the outer nuclear layer (ONL) and outer plexiform layer (OPL). Scale bars: 40 μm. OS, outer segment; IS, inner segment. Abundant mislocalized M-opsin is also detected in the IS of the Rpgrko/Y::Cep290rd16/+ mice.
To test whether disruption of RPGR and CEP290 proteins results in alteration of localization of other ciliary TZ proteins, we tested the localization of CENTRIN-2 and NPHP1 in the Rpgrko/Y::Cep290rd16/+ and compared it with the Rpgrko/Y mice. As shown in Supplementary Material, Figure S4, there was no detectable difference in the localization of CENTRIN-2 or NPHP1 in the double-mutant photoreceptors when compared with Rpgrko/Y or WT mice, as indicated by co-staining with acetylated α-tubulin. As predicted, the Rpgrko/Y::Cep290null/+ mice also did not show an appreciable change in the localization of these proteins. We then tested whether absence of RPGR alters the previously reported association of CEP290 and BBS6 (35) in the Rpgrko/Y::Cep290rd16/+ mice. As shown in Supplementary Material, Figure S5, IP using anti-BBS6 antibody precipitated CEP290 from retina extracts of Rpgrko/Y::Cep290rd16/+ mice. Our results suggest that RPGR–CEP290 complex is involved in the trafficking of selected proteins, such as opsins, highlighting the involvement of distinct regulators of ciliary trafficking in photoreceptors (36,37).
Physical interaction between RPGR and CEP290
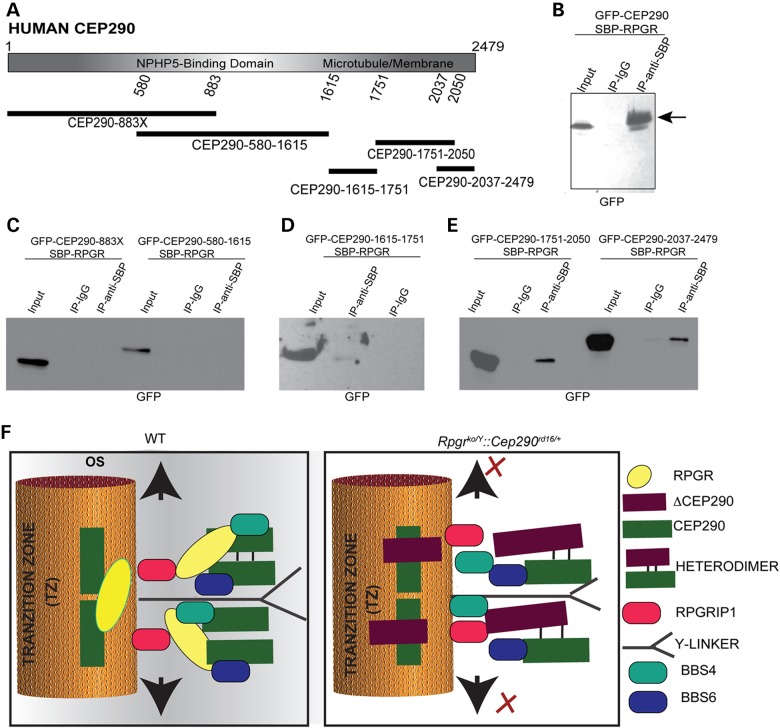
We had previously shown that RPGR and CEP290 are part of multi-protein complexes in mouse photoreceptors (20,23). Given a potential genetic modification of RPGR-associated disease by CEP290, we investigated the interaction of human RPGR with human CEP290. To this end, we generated mammalian expression constructs encoding recombinant full-length SBP (Streptavidin-Binding Protein)-tagged human RPGR and GFP-tagged full length and deleted variants of human CEP290 (Fig. 5A). We then tested the interaction of recombinant RPGR and CEP290 proteins in a co-transfection followed by co-immunoprecipitation (IP) assay. As shown in Figure 5B, IP of protein extracts from cells expressing SBP-RPGR and full-length GFP-CEP290 using anti-SBP antibody followed by immunoblotting revealed anti-GFP immunoreactive bands. IP using control immunoglobulin did not reveal any signal. We then sought to determine the region of CEP290 that interacts with RPGR. Co-IP assay revealed GFP-immunoreactive bands only in the samples from cells expressing CEP290-1751-2050 amino acids and CEP290-2037-2479 amino acids but not in the samples expressing GFP-CEP290-883X, GFP-CEP290-580-1615 or GFP-CEP290-1615-1751 (Fig. 5C–E), indicating that the C-terminal domain of CEP290 is involved in its binding to RPGR. It should be noted that a lack of detectable interaction with GFP-CEP290-883X and GFP-CEP290-580-1615 may be because of relatively lower levels of the encoded proteins, likely due to their labile nature. Potential effect of the involvement of additional regions of CEP290 in its interaction with RPGR is discussed in the next section.
Figure 5.
(A) Schematic representation of the human CEP290 protein depicting the NPHP5-Binding domain: amino acids 696–896 and Microtubule/Membrane binding domain: amino acids 1695–1966. The different domains tested for interaction with RPGR are shown under the diagram. (B–E) Co-transfection followed by co-IP was performed to examine the interaction of RPGR with full-length GFP-CEP290 (B) or its domains (C, D and E) in human embryonic kidney (HEK) 293 cells. One hundred micrograms of protein extracts was used for IP. The Input lane represents 5% of the extract used for IP. Immunoblots of the precipitated proteins were probed with anti-GFP antibody. IP with IgG was used as negative control. (F) A potential model is depicted that represents the interaction of RPGR and CEP290 in the transition zone (TZ) of mouse photoreceptors in the WT mice (Left panel). RPGR is linked to the TZ by RPGRIP1. Some RPGR–CEP290 complexes may also be indirectly associated with the TZ. Overall, the RPGR–CEP290 complexes retain the morphology of the TZ and the Y-linkers, thus facilitating trafficking of cargo (represented by black arrows). However, in the Rpgrko/Y::Cep290rd16/+ mice, the CEP290–ΔCEP290 heterodimer may lead to severe alterations in the TZ integrity and the morphology of the Y-linkers, thus perturbing cargo trafficking (represented by red X). OS, outer segment.
Discussion
In this study, we investigated the involvement of genetic modifiers in modulating the severity of XLRP due to RPGR. We found that the severity of RPGR-associated disease and effect on opsin trafficking are exacerbated by the presence of a single copy of the hypomorphic Cep290rd16 allele. Loss of a single copy of none of the other tested NPHP proteins or RPGRIP1 affected the severity of RPGR disease in mice. Our data, therefore, suggest that distinct combinations of multiprotein complexes are involved in optimal ciliary trafficking of proteins in photoreceptors. Earlier studies have revealed that genetic interactions between distinct ciliopathy protein complexes can alter ciliary trafficking and function (27,38). As CEP290 and RPGR are part of several protein complexes, including BBS and MKS proteins (20–22,35,39–43) and both RPGR and CEP290 function at the ciliary TZ likely by regulating the entry or exit of cargo (32,34,44), our studies suggest that functional interactions across distinct ciliopathy protein complexes may collaborate to maintain photoreceptor ciliary architecture and function.
It is possible that the differences in the genetic backgrounds and associated unknown modifiers mice may play a role in the interpretation of our data. However, we did not observe any effect on RPGR disease in the Rpgrko/Y::Cep290null/+ or Rpgrko/Y::Rpgrip1ko/+ (mixed C57BL6/J and 129Sv/Ev) mice, suggesting that the mixed background also may not affect the retinal phenotype. All other mouse mutants including the Rpgrko mice are in C57BL6/J background. However, a subtle involvement of other genetic modifiers or additional allelic combinations of RPGR-interacting proteins may alter the severity (exacerbate or protect) of RPGR-associated disease. Additional studies are needed to test these possibilities.
An interesting observation in our study was that insufficiency of CEP290 (Cep290null/+) did not affect the progression of RPGR-associated dysfunction. Thus, predicted hypomorphic alleles of CEP290 in patients are potential candidates for genetic interaction with RPGR. Previous reports also showed the involvement of hypomorphic in genes (including RPGRIP1L and AHI1) as modifiers of retinal degeneration in ciliopathies (18,19,27). The additional Nphp and Rpgrip1 alleles tested in this study are also null alleles. Thus, it is possible that hypomorphic variants in these proteins can affect the severity of RPGR-associated disease.
We found that RPGR preferentially binds to the C-terminus of CEP290 (preferentially between amino acids 2037 and 2050), which does not coincide with its microtubule/membrane-binding domain (amino acids 1695–1966) (45) or NPHP5-binding domain (amino acids 696–896) (46) (Fig. 5A). We, however, cannot exclude the possibility that other regions of CEP290 provide a stabilizing or inhibitory domain for interaction with RPGR. Support of this hypothesis comes from two previous observations: (i) absence of the DRD in the Cep290rd16/rd16 mice results in increased association with RPGR and sequestration of RPGR in the IS of photoreceptors (20); and (ii) the N- and C-terminal domains of CEP290 can form homo- or heterodimers, assisting in regulating the function of its binding proteins (46).
How might ΔCEP290 alter photoreceptor function in the Rpgrko mice? We and others have shown that CEP290 interacts with other ciliary proteins, including NPHP5, BBS4, BBS6, MKS proteins and RKIP (29,35,39–42). The ΔCEP290 variant although no longer interacts with BBS6 or RKIP, can still associate with BBS4, and likely with NPHP5. Thus, the ΔCEP290 protein may exacerbate the dysfunction of the ciliary gate in the Rpgrko mice by perturbing the organization or composition of the multiprotein complexes (47,48). A model representing this hypothesis is depicted in Figure 5F. As RPGR interacts with NPHP proteins (22,23,26), such effects are probably tolerated in the presence of RPGR. Further studies are needed to test this hypothesis.
Overall, our studies suggest that RPGR–CEP290 complexes may be distinct in composition and/or function when compared with complexes of RPGR with NPHP-associated proteins and with RPGRIP1. Hence, investigations of the discrete complexes of ciliopathy proteins in a tissue-specific manner may hold key to understand the pleiotropic nature of such devastating diseases.
Materials and Methods
Mice and ERG
All animal procedures were performed according to the approved guidelines of Institutional Animal Care and Use Committee. Mice were housed on standard diet and in a 12 h light to 12 h dark cycle. The female Rpgrko mice (on C57BL6/J background) were bred to Rpgrip1−/− (mixed C57BL6/J and 129SvEv background), Nphp1−/−, Nphp4−/− (Nphp4nmf192) mice (procured from Jackson Labs and on C57BL6/J background) and Nphp5−/− (C57BL6/J background; truncation after exon 4 of the Nphp5 gene resulting in a null allele; C.R. and W.B., manuscript in preparation) to produce double-mutant mice. All mice were genotyped for the required combination of genes and also excluded for rd1 and rd8 alleles. Primers and protocols for genotyping of Rpgrko, Rpgrip1−/−, Nphp1−/−, Nphp4−/− and Cep290rd16 mice have been described previously (15,20,25,27,28). The Nphp5−/− mice were genotyped using Nphp5 F: (5′<CCTTTAGGGTGATAGTAGCCAATTCC 3′>), Nphp5 R:wt (5′<AGGAACTAAGCTGTGAAATGGACC 3′>) and Nphp5 mut (5′<CAACGGGTTCTTCTGTTAGTCC 3′>) primers. The WT allele produced a PCR product of 452 bp, whereas the mutant allele produced a 294 bp pCR product.
Scotopic and photopic ERG was performed as previously reported (49). For each experiment, five mice of each genotype were used.
Antibodies and plasmids
Antibody against rhodopsin was procured from EMD Millipore (Billerica, MA). The anti-M-opsin antibody was a gift of Dr Cheryl M. Craft (University of Southern California) (50). Ant-GFP and anti-SBP antibodies were obtained from Abcam (Cambridge, MA) and Agilent technologies (La Jolla, CA); anti-CENTRIN-2 was procured from Sigma and anti-BBS6 was obtained from Abnova. Anti-NPHP1 was kindly provided by Dr Gregory Pazour (UMASS Medical School). Constructs encoding GFP-tagged CEP290 fragments and SBP-tagged RPGR were generated by cloning human CEP290 or RPGR in pEGFPN1 (Clontech, Mountain View, CA) and pNTAP (Agilent technologies). Fragments encoding variants of RPGR and CEP290 were obtained by site-directed mutagenesis.
Cell culture, transfection and immunoprecipitation assays
HEK293 cells were maintained at 37°C with 5% CO2 in DMEM (Invitrogen) supplemented with 10% fetal bovine serum and penicillin/streptomycin. Transient transfection was carried out using Lipofectamine 2000 (Invitrogen) in serum-free medium, as described (21). Transfected cells were lysed and proteins harvested in phosphate buffered saline (pH 7.4) containing protease inhibitors. Samples were subjected to co-IP as described (21) or according to manufacturer's instructions (Agilent Technologies).
Immunofluorescence and TEM analyses
Mouse eyes were enucleated and fixed with 4% paraformaldehyde in PBS followed by cryosectioning and staining, as previously described (51). For TEM analysis, enucleated eyes were fixed in 2% glutaraldehyde, 2% paraformaldehyde in 0.1 M sodium cacodylate buffer (pH 7.2). The samples were processed as described previously (51).
Supplementary Material
Funding
This research is supported by grants from the National Institutes of Health (NIH) [EY022372 (to H.K.), EY08123, EY019298 (to W.B.) and EY014800-039003 (NEI core grant)]; Foundation Fighting Blindness (to H.K.), University of Massachusetts Center for Clinical and Translational Sciences (UMCCTS) (H.K.) and Massachusetts Lions Eye Research Funds (H.K.). W.B. is the recipient of an RPB Nelson Trust Award and an award from the Retina Research Foundation (Alice McPherson, MD), Houston.
Supplementary Material
Acknowledgements
We thank Drs Branch Craige and George Witman for helpful discussions about this work; Vishesh Khanna for critical reading of the manuscript and Manisha Anand for help with immunofluorescence and preparation of the figure depicting the model of RPGR-CEP290 association.
Conflict of Interest statement. None declared.
References
- 1.Hartong D.T., Berson E.L., Dryja T.P. (2006) Retinitis pigmentosa. Lancet, 368, 1795–1809. [DOI] [PubMed] [Google Scholar]
- 2.Heckenlively J.R., Yoser S.L., Friedman L.H., Oversier J.J. (1988) Clinical findings and common symptoms in retinitis pigmentosa. Am. J. Ophthalmol., 105, 504–511. [DOI] [PubMed] [Google Scholar]
- 3.Bird A.C. (1975) X-linked retinitis pigmentosa. Br. J. Ophthalmol., 59, 177–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fishman G.A., Farber M.D., Derlacki D.J. (1988) X-linked retinitis pigmentosa. Profile of clinical findings. Arch. Ophthalmol., 106, 369–375. [DOI] [PubMed] [Google Scholar]
- 5.Huang W.C., Wright A.F., Roman A.J., Cideciyan A.V., Manson F.D., Gewaily D.Y., Schwartz S.B., Sadigh S., Limberis M.P., Bell P. et al. (2012) RPGR-associated retinal degeneration in human X-linked RP and a murine model. Invest. Ophthalmol. Vis. Sci., 53, 5594–5608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Churchill J.D., Bowne S.J., Sullivan L.S., Lewis R.A., Wheaton D.K., Birch D.G., Branham K.E., Heckenlively J.R., Daiger S.P. (2013) Mutations in the X-linked retinitis pigmentosa genes RPGR and RP2 found in 8.5% of families with a provisional diagnosis of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci., 54, 1411–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meindl A., Dry K., Herrmann K., Manson F., Ciccodicola A., Edgar A., Carvalho M.R., Achatz H., Hellebrand H., Lennon A. et al. (1996) A gene (RPGR) with homology to the RCC1 guanine nucleotide exchange factor is mutated in X-linked retinitis pigmentosa (RP3). Nat. Genet., 13, 35–42. [DOI] [PubMed] [Google Scholar]
- 8.Roepman R., van Duijnhoven G., Rosenberg T., Pinckers A.J., Bleeker-Wagemakers L.M., Bergen A.A., Post J., Beck A., Reinhardt R., Ropers H.H. et al. (1996) Positional cloning of the gene for X-linked retinitis pigmentosa 3: homology with the guanine-nucleotide-exchange factor RCC1. Hum. Mol. Genet., 5, 1035–1041. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan L.S., Bowne S.J., Reeves M.J., Blain D., Goetz K., Ndifor V., Vitez S., Wang X., Tumminia S.J., Daiger S.P. (2013) Prevalence of mutations in eyeGENE probands with a diagnosis of autosomal dominant retinitis pigmentosa. Invest. Ophthalmol. Vis. Sci., 54, 6255–6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Branham K., Othman M., Brumm M., Karoukis A.J., Atmaca-Sonmez P., Yashar B.M., Schwartz S.B., Stover N.B., Trzupek K., Wheaton D. et al. (2012) Mutations in RPGR and RP2 account for 15% of males with simplex retinal degenerative disease. Invest. Ophthalmol. Vis. Sci., 53, 8232–8237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharon D., Bruns G.A., McGee T.L., Sandberg M.A., Berson E.L., Dryja T.P. (2000) X-linked retinitis pigmentosa: mutation spectrum of the RPGR and RP2 genes and correlation with visual function. Invest. Ophthalmol. Vis. Sci., 41, 2712–2721. [PubMed] [Google Scholar]
- 12.Sharon D., Sandberg M.A., Rabe V.W., Stillberger M., Dryja T.P., Berson E.L. (2003) RP2 and RPGR mutations and clinical correlations in patients with X-linked retinitis pigmentosa. Am. J. Hum. Genet., 73, 1131–1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu D.M., Khanna H., Atmaca-Sonmez P., Sieving P.A., Branham K., Othman M., Swaroop A., Daiger S.P., Heckenlively J.R. (2010) Long-term follow-up of a family with dominant X-linked retinitis pigmentosa. Eye (Lond), 24, 764–774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Walia S., Fishman G.A., Swaroop A., Branham K.E., Lindeman M., Othman M., Weleber R.G. (2008) Discordant phenotypes in fraternal twins having an identical mutation in exon ORF15 of the RPGR gene. Arch. Ophthalmol., 126, 379–384. [DOI] [PubMed] [Google Scholar]
- 15.Hong D.H., Pawlyk B.S., Shang J., Sandberg M.A., Berson E.L., Li T. (2000) A retinitis pigmentosa GTPase regulator (RPGR)-deficient mouse model for X-linked retinitis pigmentosa (RP3). Proc. Natl Acad. Sci. USA, 97, 3649–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson D.A., Khan N.W., Othman M.I., Chang B., Jia L., Grahek G., Wu Z., Hiriyanna S., Nellissery J., Li T. et al. (2012) Rd9 is a naturally occurring mouse model of a common form of retinitis pigmentosa caused by mutations in RPGR-ORF15. PLoS One, 7, e35865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Acland G.M., Wu W.X., Johnson J.L., Pearce-Kelling S., Tulloch B., Vervoort R., Wright A.F., Aguirre G.D. (2002) Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum. Mol. Genet., 11, 993–1003. [DOI] [PubMed] [Google Scholar]
- 18.Fahim A.T., Bowne S.J., Sullivan L.S., Webb K.D., Williams J.T., Wheaton D.K., Birch D.G., Daiger S.P. (2011) Allelic heterogeneity and genetic modifier loci contribute to clinical variation in males with X-linked retinitis pigmentosa due to RPGR mutations. PLoS One, 6, e23021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Khanna H., Davis E.E., Murga-Zamalloa C.A., Estrada-Cuzcano A., Lopez I., den Hollander A.I., Zonneveld M.N., Othman M.I., Waseem N., Chakarova C.F. et al. (2009) A common allele in RPGRIP1L is a modifier of retinal degeneration in ciliopathies. Nat. Genet., 41, 739–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang B., Khanna H., Hawes N., Jimeno D., He S., Lillo C., Parapuram S.K., Cheng H., Scott A., Hurd R.E. et al. (2006) In-frame deletion in a novel centrosomal/ciliary protein CEP290/NPHP6 perturbs its interaction with RPGR and results in early-onset retinal degeneration in the rd16 mouse. Hum. Mol. Genet., 15, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna H., Hurd T.W., Lillo C., Shu X., Parapuram S.K., He S., Akimoto M., Wright A.F., Margolis B., Williams D.S. et al. (2005) RPGR-ORF15, which is mutated in retinitis pigmentosa, associates with SMC1, SMC3, and microtubule transport proteins. J. Biol. Chem., 280, 33580–33587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murga-Zamalloa C., Swaroop A., Khanna H. (2010) Multiprotein complexes of retinitis pigmentosa GTPase regulator (RPGR), a ciliary protein mutated in X-Linked retinitis pigmentosa (XLRP). Adv. Exp. Med. Biol., 664, 105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Murga-Zamalloa C.A., Desai N.J., Hildebrandt F., Khanna H. (2010) Interaction of ciliary disease protein retinitis pigmentosa GTPase regulator with nephronophthisis-associated proteins in mammalian retinas. Mol. Vis., 16, 1373–1381. [PMC free article] [PubMed] [Google Scholar]
- 24.Roepman R., Bernoud-Hubac N., Schick D.E., Maugeri A., Berger W., Ropers H.H., Cremers F.P., Ferreira P.A. (2000) The retinitis pigmentosa GTPase regulator (RPGR) interacts with novel transport-like proteins in the outer segments of rod photoreceptors. Hum. Mol. Genet., 9, 2095–2105. [DOI] [PubMed] [Google Scholar]
- 25.Zhao Y., Hong D.H., Pawlyk B., Yue G., Adamian M., Grynberg M., Godzik A., Li T. (2003) The retinitis pigmentosa GTPase regulator (RPGR)-interacting protein: subserving RPGR function and participating in disk morphogenesis. Proc. Natl Acad. Sci. USA, 100, 3965–3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Otto E.A., Loeys B., Khanna H., Hellemans J., Sudbrak R., Fan S., Muerb U., O'Toole J.F., Helou J., Attanasio M. et al. (2005) Nephrocystin-5, a ciliary IQ domain protein, is mutated in Senior–Loken syndrome and interacts with RPGR and calmodulin. Nat. Genet., 37, 282–288. [DOI] [PubMed] [Google Scholar]
- 27.Louie C.M., Caridi G., Lopes V.S., Brancati F., Kispert A., Lancaster M.A., Schlossman A.M., Otto E.A., Leitges M., Grone H.J. et al. (2010) AHI1 is required for photoreceptor outer segment development and is a modifier for retinal degeneration in nephronophthisis. Nat. Genet., 42, 175–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Won J., Marin de Evsikova C., Smith R.S., Hicks W.L., Edwards M.M., Longo-Guess C., Li T., Naggert J.K., Nishina P.M. (2011) NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet., 20, 482–496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murga-Zamalloa C.A., Ghosh A.K., Patil S.B., Reed N.A., Chan L.S., Davuluri S., Peranen J., Hurd T.W., Rachel R.A., Khanna H. (2011) Accumulation of the Raf-1 kinase inhibitory protein (Rkip) is associated with Cep290-mediated photoreceptor degeneration in ciliopathies. J. Biol. Chem., 286, 28276–28286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lancaster M.A., Gopal D.J., Kim J., Saleem S.N., Silhavy J.L., Louie C.M., Thacker B.E., Williams Y., Zaki M.S., Gleeson J.G. (2011) Defective Wnt-dependent cerebellar midline fusion in a mouse model of Joubert syndrome. Nat. Med., 17, 726–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghosh A.K., Murga-Zamalloa C.A., Chan L., Hitchcock P.F., Swaroop A., Khanna H. (2010) Human retinopathy-associated ciliary protein retinitis pigmentosa GTPase regulator mediates cilia-dependent vertebrate development. Hum. Mol. Genet., 19, 90–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khanna H. (2015) Photoreceptor sensory cilium: traversing the ciliary gate. Cells, 4, 674–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murga-Zamalloa C.A., Atkins S.J., Peranen J., Swaroop A., Khanna H. (2010) Interaction of retinitis pigmentosa GTPase regulator (RPGR) with RAB8A GTPase: implications for cilia dysfunction and photoreceptor degeneration. Hum. Mol. Genet., 19, 3591–3598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao K.N., Li L., Anand M., Khanna H. (2015) Ablation of retinal ciliopathy protein RPGR results in altered photoreceptor ciliary composition. Sci. Rep., 5, 11137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rachel R.A., May-Simera H.L., Veleri S., Gotoh N., Choi B.Y., Murga-Zamalloa C., McIntyre J.C., Marek J., Lopez I., Hackett A.N. et al. (2012) Combining Cep290 and Mkks ciliopathy alleles in mice rescues sensory defects and restores ciliogenesis. J. Clin. Invest., 122, 1233–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cheng T., Peachey N.S., Li S., Goto Y., Cao Y., Naash M.I. (1997) The effect of peripherin/rds haploinsufficiency on rod and cone photoreceptors. J. Neurosci., 17, 8118–8128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tian G., Ropelewski P., Nemet I., Lee R., Lodowski K.H., Imanishi Y. (2014) An unconventional secretory pathway mediates the cilia targeting of peripherin/rds. J. Neurosci., 34, 992–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yee L.E., Garcia-Gonzalo F.R., Bowie R.V., Li C., Kennedy J.K., Ashrafi K., Blacque O.E., Leroux M.R., Reiter J.F. (2015) Conserved genetic interactions between ciliopathy complexes cooperatively support ciliogenesis and ciliary signaling. PLoS Genet., 11, e1005627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anand M., Khanna H. (2012) Ciliary transition zone (TZ) proteins RPGR and CEP290: role in photoreceptor cilia and degenerative diseases. Expert Opin. Ther. Targets, 16, 541–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barbelanne M., Hossain D., Chan D.P., Peranen J., Tsang W.Y. (2015) Nephrocystin proteins NPHP5 and Cep290 regulate BBSome integrity, ciliary trafficking and cargo delivery. Hum. Mol. Genet., 24, 2185–2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barbelanne M., Song J., Ahmadzai M., Tsang W.Y. (2013) Pathogenic NPHP5 mutations impair protein interaction with Cep290, a prerequisite for ciliogenesis. Hum. Mol. Genet., 22, 2482–2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., Seo S., Bhattarai S., Bugge K., Searby C.C., Zhang Q., Drack A.V., Stone E.M., Sheffield V.C. (2014) BBS mutations modify phenotypic expression of CEP290-related ciliopathies. Hum. Mol. Genet., 23, 40–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gorden N.T., Arts H.H., Parisi M.A., Coene K.L., Letteboer S.J., van Beersum S.E., Mans D.A., Hikida A., Eckert M., Knutzen D. et al. (2008) CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am. J. Hum. Genet., 83, 559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Craige B., Tsao C.C., Diener D.R., Hou Y., Lechtreck K.F., Rosenbaum J.L., Witman G.B. (2010) CEP290 tethers flagellar transition zone microtubules to the membrane and regulates flagellar protein content. J. Cell. Biol., 190, 927–940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Drivas T.G., Holzbaur E.L., Bennett J. (2013) Disruption of CEP290 microtubule/membrane-binding domains causes retinal degeneration. J. Clin. Invest., 123, 4525–4539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schafer T., Putz M., Lienkamp S., Ganner A., Bergbreiter A., Ramachandran H., Gieloff V., Gerner M., Mattonet C., Czarnecki P.G. et al. (2008) Genetic and physical interaction between the NPHP5 and NPHP6 gene products. Hum. Mol. Genet., 17, 3655–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nachury M.V., Loktev A.V., Zhang Q., Westlake C.J., Peranen J., Merdes A., Slusarski D.C., Scheller R.H., Bazan J.F., Sheffield V.C. et al. (2007) A core complex of BBS proteins cooperates with the GTPase Rab8 to promote ciliary membrane biogenesis. Cell, 129, 1201–1213. [DOI] [PubMed] [Google Scholar]
- 48.Sang L., Miller J.J., Corbit K.C., Giles R.H., Brauer M.J., Otto E.A., Baye L.M., Wen X., Scales S.J., Kwong M. et al. (2011) Mapping the NPHP-JBTS-MKS protein network reveals ciliopathy disease genes and pathways. Cell, 145, 513–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li L., Anand M., Rao K.N., Khanna H. (2015) Cilia in photoreceptors. Methods Cell. Biol., 127, 75–92. [DOI] [PubMed] [Google Scholar]
- 50.Zhu X., Brown B., Li A., Mears A.J., Swaroop A., Craft C.M. (2003) GRK1-dependent phosphorylation of S and M opsins and their binding to cone arrestin during cone phototransduction in the mouse retina. J. Neurosci., 23, 6152–6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li L., Rao K.N., Zheng-Le Y., Hurd T.W., Lillo C., Khanna H. (2015) Loss of retinitis pigmentosa 2 (RP2) protein affects cone photoreceptor sensory cilium elongation in mice. Cytoskeleton (Hoboken), 72, 447–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.