TO THE EDITOR
Hermansky-Pudlak Syndrome (HPS) is a genetically heterogeneous, recessive disorder that results in defects of multiple cytoplasmic organelles, including melanosomes, platelet-dense granules and lysosomes (Tomita and Suzuki, 2004; Witkop et al., 1990). Salient clinical features of HPS include Oculocutaneous albinism (OCA), bleeding, lysosomal ceroid storage, nystagmus, strabismus, iris trans-illumination, foveal hypoplasia, bruising, and a significant reduction in visual acuity (Gahl et al., 1998; Witkop et al., 1990). In approximately half of HPS1 and HPS4 cases, pulmonary fibrosis also has been documented, with onset often in the early twenties to early forties (Gahl et al., 1998). To date, mutant alleles in nine genes have been linked to HPS (Wei and Li, 2013). The currently known HPS proteins have been categorized into three biogenesis of lysosome-related organelle (LRO) complexes, BLOC-1, -2 and -3 (Dell’Angelica, 2004; Huizing et al., 2008; Wei and Li, 2013; Wei, 2006). HPS2 also constitute a subunit of adaptor complex-3, which has critical functions in pigment-producing cells (Dell’Angelica et al., 1999). Together with other proteins, BLOC complexes participate in endolysosomal trafficking to facilitate LRO biogenesis (Wei et al., 2013).
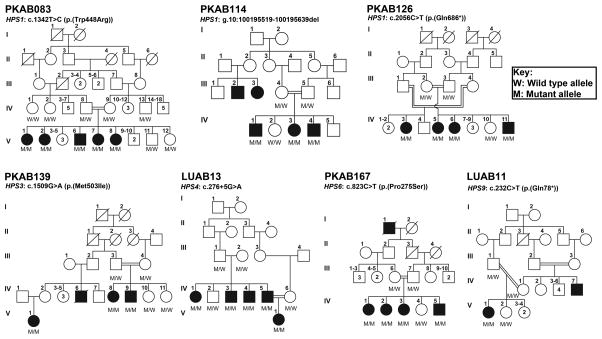
According to the HPSD, OMIM and HGMD databases, as of October 2015, only one mutation in HPS8 (BLOC1S3) was previously reported in the Pakistani population. Here, using whole exome sequencing (WES) we report seven pathogenic variants (Table S1) in five HPS genes segregating with the phenotype in seven large consanguineous Pakistani families (Figure 1; see also Supplementary Data online). In first family PKAB83, sequencing revealed a transition mutation (c.1342T>C, p.(Trp448Arg), Figure S1). Molecular modeling data revealed that the p.Trp448 residue is located in the β-strand of the secondary structure and replacing it with smaller, positively charged, less hydrophobic arginine residue will slightly destabilize the local conformation (Figure S2). We performed detailed hematological examinations (Table 1, Table S2) and chest computerized tomography (CT) scans of the affected individuals. All hematological parameters were within the normal range (Table S2). However, we observed reduced platelet aggregation among the affected individuals (Table 1). CT scans of affected individuals V:6 and V:7 revealed no evidence of diffuse interstitial lung disease, bronchiectasis, pulmonary hypertension, intra pulmonary/mediastinal mass or lymphadenopathy was found. As stated earlier, the lung involvement in HPS1 patients usually begins in adulthood and an absence of pulmonary fibrosis in these affected individuals might be due to their relative young age. Indeed, the CT images of the affected individual V:7 (30 yrs old) revealed small subsegmental fibrotic areas in the apico-posterior segment of the left upper lobe and in the apical segment of the right upper lobe. These clinical findings suggest milder form of HPS phenotype among the affected individual of family PKAB83.
Figure 1. HPS phenotypes in seven Pakistani families co-segregate with mutations in known HPS genes.
The filled and empty symbols represent the affected and unaffected individuals, respectively. A double line uniting two individuals represents a consanguineous marriage. Given also are the HPS genes mutations and two point LOD scores for each family, along with the genotypes for the mutated loci.
Table 1.
Clinical findings of Hermansky-Pudlak syndrome in the Pakistani families
| Family | ID | Age (yrs) | Gene | Allele | Status | Hair color | Skin tone | Iris color | Visual acuity | Refraction error | Fundus | FVH | PP | NYS | GI problems# | BT (2-7 min) | CT (6-7 min) | PT (10-14 sec) | APTT (≤32sec) | P.A.T. (80-100%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Right | Left | ||||||||||||||||||||
| PKAB083 | IV:13 | 45 | HPS1 | c.1342T>C | Norm. | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | No | 2m | 4m 40s | NA | NA | NA |
| V:1 | 8 | Aff. | Golden | White | Hazel green | 6/60 | 6/60 | NA | Albinotic | Yes | Yes | Yes | Yes | 2m | 3m | NA | NA | 52% | |||
| V:2 | 4 | Aff. | Golden | White | Hazel green | 6/60 | 6/60 | NA | Albinotic | Yes | Yes | Yes | Yes | 3m 15s | 3m | NA | NA | 15% | |||
| V:6 | 32 | Aff. | Golden | White | Hazel green | 6/60 | 6/60 | NA | Albinotic | Yes | Yes | Yes | Yes | 2m 15s | 6m 20s | 14s (12s) | 32s | 33% | |||
| V:7 | 30 | Aff. | Golden | White | Hazel green | 6/60 | 6/60 | NA | Albinotic | Yes | Yes | Yes | Yes | 3m 20s | 5m 15s | 12s (12s) | 30s | 46% | |||
| V:8 | 28 | Aff. | Golden | White | Hazel green | 6/60 | 6/60 | NA | Albinotic | Yes | Yes | Yes | Yes | 3m 15s | 5m 50s | NA | NA | 29% | |||
| PKAB114 | III:5 | 63 | HPS1 | genomic deletion | Norm. | NA | NA | NA | NA | NA | NA | NA | NA | NA | No | No | 3m 30s | 3m 15s | NA | 43s | 85% |
| III:4 | 58 | Norm. | NA | NA | NA | NA | NA | NA | NA | NA | NA | No | No | NA | NA | NA | NA | 93% | |||
| IV:3 | 25 | Aff. | Brown | White | Grey | NA | NA | NA | NA | NA | NA | No | Yes | 3m 40s | 3m 30s | NA | 39s | 10% | |||
| IV:4 | 23 | Aff. | Brown | White | Green | NA | NA | NA | NA | NA | NA | No | Yes | 6m 20s | 7m 30s | NA | 49s | 32% | |||
| IV:1 | 10 | Aff. | Brown | White | Green | NA | NA | NA | NA | NA | NA | No | Yes | NA | NA | NA | NA | 22% | |||
| PKAB126 | III:3 | 58 | HPS1 | c.2056C>T | Norm. | Black | Wheat | Brown | 6/6 | 6/6 | Presbyopic | Normal | No | No | No | No | 3m 30s | 4m | NA | 41s | 81% |
| III:2 | 16 | Norm. | Black | Wheat | Black | 6/6 | 6/6 | Emmetropic | Normal | No | No | No | No | NA | NA | NA | NA | 88% | |||
| IV:11 | 10 | Aff. | Yellow brown | Pink white | Hazel green | 6/18 | 6/18 | +3.25/−3.50*22 +2.50/−3.75*176 |
Albinotic | Yes | Yes | Yes | No | 5m 30s | 4m 30s | NA | 50s | 53% | |||
| IV:6 | 15 | Aff. | Yellow brown | Pink white | Hazel green | 6/18 | 6/18 | +5.25/−4.75*16 +5.25/−2.50*4 |
Albinotic | Yes | Yes | Yes | No | 5m | 4m 30s | NA | 44s | 35% | |||
| IV:3 | 12 | Aff. | Yellow brown | Pink white | Hazel green | CF | CF | +7.25/−5.25*24 +6.0/−1.25*180 |
Albinotic | Yes | Yes | Yes | No | NA | NA | NA | NA | 35% | |||
| PKAB139 | IV:9 | 16 | HPS3 | c.1509G>A | Aff. | Dark brown | White | Brown | 6/24 | 6/24 | −3.25/−2.25*168 -4.75/−3.25*173 |
Normal | No | Yes | Yes | No | 3m (≤5m) | 8m (≤11m) | NA | NA | NA |
| LUAB13 | III:1 | 55 | HPS4 | c.276+5G>A | Norm. | NA | NA | NA | NA | NA | NA | NA | NA | No | No | No | 1m 30s (≤3m) | 5m (≤11m) | 15s | 32s | NA |
| IV:4 | 34 | Aff. | Brown | Pink white | Brown | NA | NA | NA | NA | NA | Yes | Yes | No | 2m (≤3m) | 6m (≤11m) | 14s | 36s | NA | |||
| IV:5 | 38 | Aff. | Brown | Pink white | Brown | NA | NA | NA | NA | NA | Yes | Yes | No | 1m 30s (≤3m) | 4m 30s (≤11m) | 12s | 32s | NA | |||
| PKAB167 | IV:4 | 18 | HPS6 | c.823C>T | Norm. | Black | Wheat | Brown | 6/60 | 6/24 | +10.25/−4.75*177 +10.0/−3.75*6 |
Normal | No | No | No | No | NA | NA | NA | NA | NA |
| IV:3 | 10 | Aff. | Red | Pink white | Dark brown | 6/60 | 6/36 | +2.0/−1.25*161 +2.0/−1.0*149 |
Albinotic | Yes | Yes | Yes | Yes | 2m 45s (≤5m) | 4m 50s (≤11m) | NA | NA | NA | |||
| IV:2 | 14 | Aff. | Red | Pink white | Dark brown | 6/18 | 6/18 | +0.25/−0.50*63 +0.25/−0.50*65 |
Albinotic | Yes | Yes | No | Yes | NA | NA | NA | NA | NA | |||
| LUAB11 | VI:4* | 04 | HPS9 | c.232C>T | Aff. | Golden white | Pink white | Light brown | NA | NA | NA | NA | NA | Yes | Yes | Yes | >3m (≤3m) | >11m (≤11m) | >1m | >2m | NA |
Normal ranges for tests are given in parenthesis. Norm: Normal; Aff: Affected; NA: Not Available; CF: Count Fingers. FVH.: Foveal hypoplasia; PP: photophobia; NYS: Nystagmus; BT: Bleeding time; CT: Clotting time; PT: Prothrombin time; APTT: Activated Partial Thromboplastin time; P.A.T.: Platelet aggregation test; Y: Yes; N: No.
Gastrointestinal problems including nausea, vomiting, abdominal pain, diarrhea etc.
Had platelet count was 74×103/uL (normal range 150 – 400 103/uL) at the time of clinical investigation.
In family PKAB126, a new nonsense mutation (p.(Gln686*)) in the HPS1 gene was segregating (Figure 1 and S1) with the HPS phenotype (Table 1 and Table S2). The affected individual had reduced vision and platelet aggregation abilities besides OCA (Table 1). On the other hand, the affected individuals of family PKAB114 were homozygous for a 121 bp genomic deletion (Table S3) that results in the partial loss of intron and a splice acceptor site for second coding exon of HPS1 (Figure 1 and S3). Human Splicing Finder program predicted cryptic splicing and premature truncation of the HPS1 protein (p.(Pro41Aspfs*12)) due to this deletion mutation.
In contrast, the affected individuals of family PKAB139 were homozygous for a transition mutation (c.1509G>A) in the HPS3 (Figure 1, Table 1), which is predicted to replace an evolutionarily conserved methionine (p.(Met503Ile); Figure S4). Molecular modeling of central region of HPS3 revealed that p.Met503 residue is located in an μ-helix and replacing it with isoleucine that does not prefer μ-helices as secondary structure might lead to loss of interactions (Figure S2). Although limited clinical information was available for family PKAB139, the sixteen-year-old affected individual had no history of gastrointestinal problems, bleeding diathesis and his bleeding time and clotting time were normal (Table 1).
In a family of Sindh origin, LUAB13, WES revealed a putative +5 splice site mutation (c.276+5G>A) in intron 4 of HPS4 segregating with the phenotype (Figure 1). HPS4 together with HPS1 constitute BLOC-3, a Rab guanine nucleotide exchange factor (GEF) that is essential for the biogenesis of LRO (Gerondopoulos et al., 2012). In silico prediction suggests that the skipping of exon 4 due to c.276+5G>A change results in a reading frameshift and premature truncation. However, as the mutation is not present at the canonical splice donor site, an in vivo c.276+5G>A change may result in a leaky splicing error. Hematological analyses of the three affected individuals ranging from 34 to 55 yrs old revealed milder phenotype with no platelet abnormalities and history of bleeding diathesis (Table 1).
Multiple isoforms are transcribed at the HPS4 locus (Anderson et al., 2003). The full-length HPS4 isoform encodes a 708 amino acid polypeptide (Figure S5). The Genome Browser annotation of the human HPS4 locus (NCBI build GRCh37) also contains a smaller cDNA clone (DB104830), isolated from the human thymus and transcribed by the first two coding exons of HPS4 and is predicted to encode a polypeptide of 89 amino acids (Figure S5). The open reading of the shorter isoform is predicted to encounter a stop codon at the beginning of intron 4 (Figure S5) and encode only two additional evolutionarily conserved amino acids beyond exon 4 (Figure S4). The mutation segregating in LUAB13 is predicted to replace the serine present at the end of this shorter transcript with an asparagine (Figure S4). However, the differential activities of the various isoforms of HPS4 are currently not well understood.
To date, nine mutations have been reported in the HPS6 gene that are responsible for the HPS phenotype in humans (Huizing et al., 2009). In family PKAB167, our sequencing revealed a novel homozygous missense mutation (p.(Pro275Ser)) in the HPS6 (Figure 1 and S4). The p.(Pro275Ser) change was predicted disease causing by at least two prediction programs with relatively moderate CADD scores (Table S1). Our molecular modeling data suggest that the p.Pro275 residue is part of interpro domain termed as Bloc-2 complex, Hps6 subunit (IPR017218). Prolines are known to be very rigid and therefore induce a special backbone conformation and replacing it with a smaller, less hydrophobic residue serine is predicted to disturb this special conformation (Figure S2). Clinical evaluation of the available affected individuals revealed no history of bleeding disorders and her bleeding and clotting times were within the normal range (Table 1).
Finally, our genetic analysis of the LUAB11 family revealed a nonsense mutation (p.(Gln78*); Figure 1 and S4) in the PLDN (HPS9). The same mutation was previously identified in two individuals, including a 9-month-old Indian male and a 17-year-old Italian female (Badolato et al., 2012; Cullinane et al., 2011). Both affected individuals had OCA and nystagmus but no hemorrhagic problems (Badolato et al., 2012; Cullinane et al., 2011). However, the Italian female had a history of recurrent cutaneous infections (Badolato et al., 2012). In family LUAB11, the 4-year-old female had OCA, photophobia, nystagmus, prolonged bleeding and clotting times (Table 1), which indicate platelet dysfunction.
In summary, our data suggest a genotype-phenotype correlation in which presumably the missense alleles identified are hypomorphic and associated with a milder phenotype, whereas truncating alleles result in classical severe HPS. The results provided here will improve our knowledge of the molecular epidemiology of HPS, diagnoses and genetic counseling. We expect that further analyses of the missense alleles identified here in the HPS genes are expected to increase our understanding of mutation-structure and structure-function relationships, and therefore the pathogenesis of HPS.
Supplementary Material
Acknowledgments
We thank the participants for their cooperation. The authors acknowledge support of the National Human Genome Research Institute (NHGRI) and the National Heart, Lung, and Blood Institute (NHLBI). Funding was provided by HG006504 (Yale Center for Mendelian Disorders), HG006542 (Baylor-Hopkins Center for Mendelian Genomics) and HG006493 (University of Washington Center for Mendelian Genomics). This study was supported by National Institute of Arthritis and Musculoskeletal and Skin Diseases (NIAMS/NIH) research grant R01AR065483 to ZMA.
References
- Anderson PD, Huizing M, Claassen DA, White J, Gahl WA. Hermansky-Pudlak syndrome type 4 (HPS-4): clinical and molecular characteristics. Hum Genet. 2003;113:10–7. doi: 10.1007/s00439-003-0933-5. [DOI] [PubMed] [Google Scholar]
- Badolato R, Prandini A, Caracciolo S, Colombo F, Tabellini G, Giacomelli M, Cantarini ME, Pession A, Bell CJ, Dinwiddie DL, et al. Exome sequencing reveals a pallidin mutation in a Hermansky-Pudlak-like primary immunodeficiency syndrome. Blood. 2012;119:3185–7. doi: 10.1182/blood-2012-01-404350. [DOI] [PubMed] [Google Scholar]
- Cullinane AR, Curry JA, Carmona-Rivera C, Summers CG, Ciccone C, Cardillo ND, Dorward H, Hess RA, White JG, Adams D, et al. A BLOC-1 mutation screen reveals that PLDN is mutated in Hermansky-Pudlak Syndrome type 9. Am J Hum Genet. 2011;88:778–87. doi: 10.1016/j.ajhg.2011.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Dell’angelica EC. The building BLOC(k)s of lysosomes and related organelles. Curr Opin Cell Biol. 2004;16:458–64. doi: 10.1016/j.ceb.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Dell’angelica EC, Shotelersuk V, Aguilar RC, Gahl WA, Bonifacino JS. Altered trafficking of lysosomal proteins in Hermansky-Pudlak syndrome due to mutations in the beta 3A subunit of the AP-3 adaptor. Mol Cell. 1999;3:11–21. doi: 10.1016/s1097-2765(00)80170-7. [DOI] [PubMed] [Google Scholar]
- Gahl WA, Brantly M, Kaiser-Kupfer MI, Iwata F, Hazelwood S, Shotelersuk V, Duffy LF, Kuehl EM, Troendle J, Bernardini I. Genetic defects and clinical characteristics of patients with a form of oculocutaneous albinism (Hermansky-Pudlak syndrome) N Engl J Med. 1998;338:1258–64. doi: 10.1056/NEJM199804303381803. [DOI] [PubMed] [Google Scholar]
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–9. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Helip-Wooley A, Westbroek W, Gunay-Aygun M, Gahl WA. Disorders of lysosome-related organelle biogenesis: clinical and molecular genetics. Annu Rev Genomics Hum Genet. 2008;9:359–86. doi: 10.1146/annurev.genom.9.081307.164303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huizing M, Pederson B, Hess RA, Griffin A, Helip-Wooley A, Westbroek W, Dorward H, O’brien KJ, Golas G, Tsilou E, et al. Clinical and cellular characterisation of Hermansky-Pudlak syndrome type 6. J Med Genet. 2009;46:803–10. doi: 10.1136/jmg.2008.065961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomita Y, Suzuki T. Genetics of pigmentary disorders. Am J Med Genet C Semin Med Genet. 2004;131C:75–81. doi: 10.1002/ajmg.c.30036. [DOI] [PubMed] [Google Scholar]
- Wei AH, He X, Li W. Hypopigmentation in Hermansky-Pudlak syndrome. J Dermatol. 2013;40:325–9. doi: 10.1111/1346-8138.12025. [DOI] [PubMed] [Google Scholar]
- Wei AH, Li W. Hermansky-Pudlak syndrome: pigmentary and non-pigmentary defects and their pathogenesis. Pigment Cell Melanoma Res. 2013;26:176–92. doi: 10.1111/pcmr.12051. [DOI] [PubMed] [Google Scholar]
- Wei ML. Hermansky-Pudlak syndrome: a disease of protein trafficking and organelle function. Pigment Cell Res. 2006;19:19–42. doi: 10.1111/j.1600-0749.2005.00289.x. [DOI] [PubMed] [Google Scholar]
- Witkop CJ, Almadovar C, Pineiro B, Nunez Babcock M. Hermansky-Pudlak syndrome (HPS). An epidemiologic study. Ophthalmic Paediatr Genet. 1990;11:245–50. doi: 10.3109/13816819009020986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.