SPORE-FORMING BACTERIAL PATHOGENS
To survive adverse conditions, some bacterial species are capable of developing into a cell type, the “spore,” which exhibits minimal metabolic activity and remains viable in the presence of multiple environmental challenges. For some pathogenic bacteria, this developmental state serves as a means of survival during transmission from one host to another. The spores are the highly infectious form of these bacteria. Upon entrance into a host, specific signals facilitate germination into metabolically active replicating organisms, resulting in disease pathogenesis. In this chapter, we will review spore structure and function in well-studied pathogens of two genera, Bacillus and Clostridium, focusing on Bacillus anthracis and Clostridium difficile, and explore current data regarding the lifestyles of these bacteria outside the host and transmission from one host to another.
Bacillus anthracis and Other Pathogenic Bacillus Species
The Bacillus species are Gram-positive, spore-forming, facultative aerobes that are commonly found in the soil, sometimes associated with plants and nematodes. The Bacillus cereus sensu lato clade of this well-studied genus contains pathogens and nonpathogens, with a complex taxonomy that in recent years has been continuously modified to reflect DNA sequence data. In addition to DNA sequence similarities and gene synteny, horizontal transfer of closely related plasmids is apparent among these ubiquitous soil bacteria (1). Member species of the Bacillus cereus sensu lato group include B. cereus sensu stricto, B. anthracis, Bacillus thuringiensis, Bacillus mycoides, Bacillus pseudomycoides, Bacillus weihenstephanensis, and Bacillus cytotoxicus (2–10). Of these, the most well-studied species are B. anthracis, B. cereus sensu stricto, and B. thuringiensis. B. anthracis is the causative agent of anthrax in mammals, an often-lethal disease. Human disease is generally acquired accidentally during outbreaks of anthrax in domestic livestock and wildlife, but has also been associated with bioterrorism (11). Certain strains of B. cereus can cause food poisoning in humans, while some strains of B. thuringiensis are lethal for invertebrates and are used as insecticides (12, 13). Other species of the B. cereus sensu lato group are only occasionally cited as disease causing in mammals but their potential virulence factors are not known.
B. anthracis and anthrax
B. anthracis infection can manifest as a cutaneous or systemic disease. Entry of spores via a preexisting lesion in the skin can result in cutaneous anthrax. This disease is distinguished by the presence of a characteristic, black eschar on the skin, and typically remains a localized infection. Systemic anthrax can take several forms. One of these, gastrointestinal anthrax, is the result of ingesting spores, which may occur in grass-fed mammals that consume the spores in the soil, or in humans through the consumption of contaminated food products. The other systemic form, inhalational anthrax, is the most deadly form, in part because of its ambiguous set of symptoms and rapid onset. Spores are inhaled and deposited into the lung tissue, where they proceed to germinate and spread through lymph nodes, rapidly causing systemic disease, massive tissue damage, shock and death (14). Recently, a fourth form, injectional anthrax, has been recognized in heroin addicts whose drug supply was likely contaminated by contact with infected animal material (15, 16).
Virulent B. anthracis carry two virulence plasmids. The anthrax toxin genes are located on virulence plasmid pXO1, and the capsule biosynthetic operon is located on pXO2 (17). The anthrax toxin is composed of three proteins: lethal factor, edema factor, and protective antigen. Lethal factor is a zinc-dependent metalloprotease that targets the mitogen-activated kinase kinases (MAPKKs, or Meks). Lethal factor cleaves the N-terminus of these proteins, disrupting the ERK1/2, JNK/SAPK, and p38 signaling pathways in host cells. These pathways are critical to proper cell cycle regulation, cellular proliferation, and defense against cellular stresses. Edema factor is a calmodulin-dependent adenylate cyclase, which, as the name implies, causes the intense edema observed with the disease. This enzyme influences many cellular signaling pathways in the host cell by increasing cyclic AMP (cAMP) levels. Protective antigen is required for the delivery of both lethal factor and edema factor to host cells. Protective antigen is cleaved into a fragment that can multimerize, forming a heptameric structure that binds to receptors on the host cell (TEM8 and CMG2), forms a pore within the membrane, and delivers the toxin to the host cell cytosol (18, 19). The capsule of B. anthracis is unique among capsule-forming bacteria in that it is composed of poly-D-glutamic acid instead of the more common polysaccharide or hyaluronic acid capsules seen in other pathogens. As is true for other bacterial capsules, the B. anthracis capsule has antiphagocytic and anti-opsinogenic roles in infection. Recent reports have expanded the role of capsule, indicating that it can also act as an adhesion, suppress dendritic cell function, and may serve as a TLR2 agonist (20–22).
B. thuringiensis, the insect pathogen
B. thuringiensis is used extensively as a biological control agent for insect pests. During sporulation, certain B. thuringiensis strains produce parasporal crystals comprised of Cry proteins (also known as delta-endotoxins). These toxins are lethal for many invertebrates, including members of the orders Lepidoptera, Diptera, Coleoptera, some nematodes, mites, and protozoa (12, 13). The Cry toxins vary in degree of toxicity, but generally disrupt host cell membranes, including those of epithelial cells in the insect gut (23). Two models have been proposed for Cry-mediated death. Formation of pores in cell membranes causes an osmotic imbalance and/or the opening of ion channels activates the process of cell death (24). B. thuringiensis infection in insects includes three successive steps, virulence, necrotrophism, and sporulation, which are controlled by quorum sensing systems and multiple regulators. In addition to the Cry toxins, multiple other factors, including degradative enzymes, play roles in these different stages of infection (25).
B. thuringiensis is generally considered non-pathogenic to humans and other animals. However, there are some reports of association of the bacterium with human diseases including food-poisoning-associated diarrheas, periodontitis, and bacteremia. There are also rare instances of B. thuringiensis-associated ocular, burn, and wound infections (26, 27). Multiple secreted proteins of B. thuringiensis, including phospholipases, hemolysins, and enterotoxins, have been shown to contribute to disease in animal models of infection (28).
B. cereus and food poisoning
Some B. cereus sensu stricto strains are opportunistic human pathogens, and infection and intoxication can result from consumption of contaminated foods. Cereulide toxin-producing strains are responsible for an enteric syndrome that is acquired by eating contaminated rice or pasta. Diarrheal disease, linked with contaminated dairy products and vegetables, is caused by B. cereus sensu stricto strains producing three different toxins: cytotoxin K and the heat-labile protein complexes hemolysin BL and non-hemolytic enterotoxin (29, 30).
Pathogenic Clostridium Species
Many members of the Clostridiales, like the Bacillales, are Gram-positive sporulating soil bacteria. Yet unlike the Bacillales species that form spores, the known spore-forming Clostridiales species are obligate anaerobes and are exquisitely sensitive to oxygen. The major pathogenic Clostridiales species are a genetically diverse group including Clostridium difficile, Clostridium perfringens, Clostridium botulinum, Clostridium tetani, and Clostridium sordellii. (We note that C. difficile is properly referred to as Peptoclostridium difficile (31). However, for the purpose of this review, we will use the former and still very widespread nomenclature). C. perfringens is the second-most common bacterial source of foodborne illness in the United States (32) and the most common Clostridium species associated with gas gangrene. During the last decade, C. difficile has emerged over the past ten years as a causative agent of antibiotic-associated diarrhea (33). Other pathogenic Clostridium species include: C. tetanii, the agent of tetanus; C. botulinum, which causes botulism; and C. sordellii, which has been associated with rare post-abortion infections (34).
Hospital-acquired C. difficile infections
C. difficile is the leading cause of nosocomial diarrhea worldwide and the bacterium can cause life-threatening infections in patients undergoing antibiotic treatment. Spores are transmitted via the fecal-oral route and are very easily spread by health care workers. C. difficile is a toxin-producing extracellular pathogen. Typically, the treatment of a patient with antibiotics for an unrelated infection negatively affects the normal flora within the gut. These commensals provided colonization resistance, or the ability to prevent invasion by pathogens simply by colonizing the niche first. In the absence of these protective commensals, C. difficile spores can germinate, replicate, and produce toxins. Unlike other infections, C. difficile infected patients have a high relapse rate, further complicating the treatment of the disease. This relapse rate may be due to the ability of C. difficile to sporulate and re-germinate within the host environment, thereby persisting within the host. The ability of C. difficile to complete its life cycle within the host is one significant difference between C. difficile and B. anthracis, for which sporulation within the host has not be observed (35, 36).
The primary virulence factors of C. difficile are two enterotoxins – Toxin A (TcdA) and Toxin B (TcdB). These toxins are required for the development of clinical symptoms. These toxins act by glycosylating members of the Rho family of host proteins, which causes remodeling of the host cell cytoskeleton and leads to massive damage to the epithelium and induction of a strong inflammatory response (37, 38). The results of these events are intestinal tissue injury and acute inflammation, leading to the diarrheal symptoms observed clinically. The ability to produce toxins A and/or B is essential for virulence; highly toxinogenic strains are capable of causing severe disease, while non-toxin producing strains do not cause symptomatic disease (38). While the relative contributions of toxins A and B to disease are still an important area of investigation (39, 40), recent data suggest that toxin B is the primary contributor to disease (41). C. difficile produces a third toxin called CDT (also known as binary toxin). This toxin is an ADP-ribosyltransferase, much like the diphtheria, cholera, and pertussis toxins. The role of this toxin in the causation of disease is unclear, as many pathogenic strains do not possess the gene to encode it. However, one hypothesis is that CDT augments the activity of TcdA and TcdB. Interestingly, one study has implicated CDT as a significant predictor of recurrent disease (42).
C. perfringens infections
C. perfringens can cause human clostridial myonecrosis and food poisoning as well as several enterotoxemic and enteritis diseases of animals. Clostridial myonecrosis, also known as gas gangrene, arises when spores from contaminated soil enter muscle tissue, often as a result of traumatic injury (43). Spontaneous myonecrosis can also occur, but is usually associated with C. septicum or other related species (44). Clostridial myonecrosis in humans and other mammals spreads rapidly via tissue necrosis, often leading to systemic toxemia and shock with high mortality rates. C. perfringens gastroenteritis can be acquired from contaminated meat, including poultry. Spores, and vegetative cells from food stored in conditions that permit germination and multiplication, survive passage through the stomach and reach the intestine, where they can multiply and form toxin (32). Pathogenesis of C. perfringens is mediated by multiple secreted toxins, many of which are plasmid-encoded (45). Strains are distinguished by the combinations of toxins that they produce. Toxigenic types A, B, C, D, and E, are based on distinct combinations of four different toxins (alpha-, beta-, epsilon- and iota-toxins). Some strains can produce additional toxins, including the beta 2 enterotoxin and C. perfringens enterotoxin (CPE) (32, 46, 47).
THE SPORE AS AN INFECTIOUS PARTICLE
Spores of Bacillus and Clostridium represent a distinctive cell type that is made when vegetative cells experience a specific alteration in the environment that is usually associated with starvation (Fig. 1). Spores are distinguished from the vast majority of other cell types by their minimal metabolic activity and high resistance to environmental stressors (48). As a result, spores can survive in the environment for extended periods of time (49). In most cases where disease is initiated by a spore, the spore must germinate before the virulence factors that cause disease (such as toxins) are expressed (for an important exception, see discussion of C. perfringens, below). This is because the spore itself appears to be generally non-toxic to the host; rather, toxicity arises from factors produced after germination, when vegetative cell growth resumes.
Fig. 1.
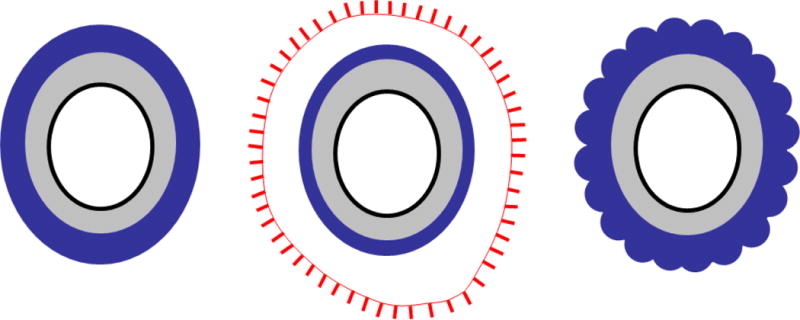
Schematic representation of spores of B. subtilis (left), B. anthracis (middle) and C. difficile (right) illustrating the major spore structures. The inner-most compartment (housing the spore DNA), the core, is white. The inner membrane (the location of the germinant receptors in B. subtilis and B. anthracis, but not C. difficile) is the black oval surrounding the core. The grey region is the spore peptidoglycan and, surrounding that, the dark blue is the coat. Although an outer membrane is present during spore formation, it is most likely not present in the mature spore (72) and, therefore, not shown here. The sublayers of the coat are not indicated for simplicity. B. anthracis (but not the other two species), (65) possesses an additional outer layer, the exosporium, indicated by a red line (the exosporium basal layer) and protrusions extending from the basal layer surface (the hair-like projections, or nap). The region between the coat and the exosporium is the interspace. The C. difficile coat has a scalloped appearance in cross-section, as indicated in the representation.
Spore Structure and Function
All spores produced by species in the Bacillales and Clostridiales have a common architectural plan; they are spherical or roughly ovoid cells composed of a series of concentric shells (Fig. 1).
The inner membrane and its contents
The inner-most structure is always a membrane-bound compartment harboring the spore’s DNA, which is tightly associated with small peptides called small acid-soluble proteins, or SASPs (50). This compartment also contains cations and pyridine-2,6-dicarboxylic acid (or dipicolinic acid, DPA) in a 1:1 chelate with divalent calcium ions (Ca2+DPA), which serve to maintain the spore’s dormant state and resistance to certain DNA-damaging agents. The membrane of this compartment retards the diffusion of water (51, 52).
An additional important function of the inner membrane, at least in many species, is to house the germinant receptors, which initiate a signal transduction pathway ultimately leading to the resumption of metabolism (51, 53). The receptors bind to specific small-molecule “germinants,” which can be a subset of sugars, amino acids and ribonucleosides, as well as other small molecules, depending on the spore species (53). Not all germinant receptors are located in the inner membrane; the C. difficile germinant receptor CspC is likely associated with either the coat and/or cortex (54). CspC is distinct from the “canonical” germinant receptors, best studied in B. subtilis and present in a variety of Bacillaceae and C. perfringens (55). In particular, unlike the typical germinant receptors, CspC does not contain membrane-spanning sequences. As will be discussed later, this difference between CspC and the other known germinant receptors suggests that germinant receptors in other spore-forming bacteria may vary from the examples studied so far. As a result, searches for novel germinant receptors should not be based solely on bioinformatics-driven searches.
All known germinant receptors are located in deeper layers of the spore, suggesting that germinants must reach the receptors via diffusion. Therefore, it can be inferred that all the layers that encase the inner membrane (discussed below) are porous to molecules at least as large as the corresponding germinants. The porosity of the coat to small molecules has been demonstrated experimentally for a few Bacillus species and can be inferred for many others where data are available; these species possess sequences for canonical germinant receptors. The porosity of the cortex is a reasonable inference based on the known structure and composition of peptidoglycan layers.
The Cortex and germ cell wall
Surrounding the forespore membrane is another spore layer that is common to all species examined: a shell of peptidoglycan containing two sublayers, the cortex, and the germ cell wall. The germ cell wall is destined to become the nascent cell wall of the vegetative cell when outgrowth begins. The cortex encases the germ cell wall. In transmission electron micrographs, the cortex appears distinct from vegetative cell peptidoglycan and, at least in B. anthracis and B. subtilis, the cortex differs biochemically as well. A major role of the cortex is to retain the relatively dry state of the core. The cortex does this by constricting the core’s volume. In the absence of this girdle-like effect of the cortex (if, for example, the integrity of the cortex is compromised by first extracting the coat and then treating with lysozyme (56)), the core will immediately begin to expand and rehydrate.
The Spore coat
The next layer of the spore, the coat, is a protein shell that is universally present, but varies significantly in composition and morphology among species (57–63). In many species, including B. subtilis and C. difficile, but not B. anthracis and its close relatives, the coat is the external layer of the spore (Fig. 1) (64, 65). The coat is biochemically complex, comprising approximately 70 protein species organized into several layers (60, 66–69). The spore coat of B. anthracis is relatively thin; in contrast, it is much thicker in C. difficile (65). The coat has a larger surface area than the cortex on which it sits. As a result, while much of the inner surface of the coat associates with the outer cortex surface, the mismatch in surface areas results in folds or buckles in the coat (70–72). In fact, the coat is relatively flexible and can fold and unfold in response to changes in the spore’s degree of hydration. Such changes are evident in spores where cortex volume changes in response to alterations in ambient humidity (72–75).
As already mentioned, the coat acts as a sieve, restricting passage of large molecules. An important consequence of this constraint is that structures within the spore are protected from degradative enzymes such as mammalian lysozyme, which readily lyses the cortex, and other destructive enzymes common in the soil environment (57, 59). The coat also has roles in resistance to several types of small reactive chemicals (48). The mechanism(s) responsible for this type of resistance is (are) not yet known. The coat also provides resistance to digestion by eukaryotic microbes and nematodes (76, 77). Possibly, resistance to these organisms is due to exclusion of microbial degradative enzymes by sieving. It is reasonable to speculate that the evolution of resistance to degradative enzymes in the environment gave rise to the ability to exclude host-generated degradative enzymes during infection. In this view, the diversity in coat structures among pathogenic spores evolved prior to the appearance of pathogenic life styles in these lineages. As might be expected from its biochemical complexity, the coat probably has other functions, although these are still poorly understood (66). However, in B. subtilis, where some information on these functions is available, it has been shown that the coat plays complex roles in germination. Because germination is central to pathogenesis in many if not all cases where the spore is the infectious particle (see below), a deeper understanding of the role of the coat in germination in pathogenic species would likely provide mechanistic insights into how these species cause disease and new therapeutics to target them.
Studies of spore structure and function have been limited to a small number of species and the diversity in coat structure across the much larger number of known species remains poorly explored. Given that some species (such as the non-pathogenic species B. clausii) possess long spike-like appendages, and others (such as Clostridium taeniosporum) harbor intriguing propeller-like ribbons extending from one end, no doubt more exotic structural variations remain to be discovered (63, 78, 79). It is unlikely that we will be able to understand fully the roles of the coat in natural environments until we have better understanding of how coat architecture and composition varies among spore formers. One future challenge involves elucidating the environmental selective pressures that drive the diversity in coat morphology across species (80).
The Exosporium
The outermost structure of many spores (and some, but not all, pathogens) is a structurally distinct layer called the exosporium. The exosporium is defined by its location and morphology: it is separated from the coat by a “gap” (referred to as the interspace) and, importantly, the interspace thickness varies around the circumference of the spore (58, 60, 64). As a result, the contours of the exosporium do not follow the folds of the coat. While it is reasonable to infer that some material must connect the exosporium to the coat (and, specifically, guide exosporium deposition during spore assembly (60)), the identity of this material and that of the interspace contents remains unknown. Regardless of which specific molecules compose the interspace, their organization almost certainly must be different than that of molecules in the coat, given that variation in interspace width is so prevalent among exosporium-bearing spores (63).
Like the coat, the exosporium has been studied in only a few species, primarily B. anthracis and B. cereus (60, 81–96). Structural studies provide strong support for the view that, like the coat, the exosporium serves as a sieve that excludes large molecules (84). There is also evidence that the B. cereus exosporium provides resistance to specific chemicals including ethanol, toluene, chloroform, phenol and, in the context of a macrophage, nitric oxide (86, 97). Spores from which the exosporium has been removed, due to mutations in genes encoding exosporium proteins or other proteins involved in assembly, are able to germinate, but at levels that are altered relative to wild type spores (83, 98, 99).
Characterizing the roles for the coat and/or the exosporium in pathogenesis is an important ongoing area of work. Clearly, the presence or absence of an exosporium is not an indication of a pathogenic lifestyle, given that many exosporium-bearing species are not virulent, and some spores without an exosporium cause disease. It is also notable that B. anthracis, a pathogen with an exosporium, can cause disease in several animal models when the exosporium has been removed mechanically or by mutation (60, 81, 100). Importantly, this ability to cause disease when the exosporium is absent does not necessarily imply that the B. anthracis exosporium has no role in natural infection. In fact, the B. anthracis exosporium surface protein BclA has been shown to bind to macrophage receptors and to mediate interactions with epithelial cells and extracellular matrix proteins (100–103).
An additional reasonable inference from the lack of an essential role for the exosporium in disease is the view that the coat provides significant protection against host defenses. This view is supported by the well characterized roles of the B. subtilis coat in resistance to diverse chemical and environmental stresses, and the presence of spores of B. subtilis, C. difficile and other species without exosporia in the mammalian gastrointestinal tract (59, 65, 104). Other roles for the B. anthracis exosporium have been identified which, while not at this point directly connected with pathogenesis, could nonetheless plausibly facilitate infection. For example, the exosporium modulates germination, as already mentioned. It seems plausible that the exosporium could participate in a variety of other functions, including acting as a sieve to prevent entry of large molecules (and especially degradative enzymes) to the spore interior, and providing appropriate surface chemistry for optimal interactions with surfaces in the environment (105). Of course, in the cases of the spores harboring exosporia produced by the many non-pathogenic species, these other roles will also be important, albeit to a non-pathogenic lifestyle.
The Decision to Sporulate
For maximal efficiency of infection, B. anthracis and C. difficile, and most likely other spore-forming bacterial pathogens, usually enter the body as a spore, rather than a vegetative cell. The spore itself, however, is not the direct cause of disease; rather, symptoms initiate only after the spores germinate and produce vegetative cells that, in turn, produce the toxins and other virulence factors that are direct causes of pathology (bearing in mind the important exception of C. perfringens, already mentioned). Nonetheless, spore formation in the non-host environment, in the case of Bacillus, and sporulation in the context of the host, in the case of Clostridium, leads to further dissemination of disease.
Complex regulation of Bacillus sporulation
The precise identities of the factors triggering sporulation are not known; even in the best studied model, B. subtilis, these factors remain obscure. Nonetheless, it is known that sporulation is usually stimulated by depletion of nutrients and other molecules associated with growth. This environmental change is detected by a set of sensor kinases, which in turn, activate a phosphorylation cascade. This cascade appears to be a major part of a regulatory circuit responsible for pushing the cell towards one or another behavior, depending on the severity of the nutrient depletion and other environmental and cell-internal factors (106–108). Sporulation is one such choice. The specific circuitry that couples activation of the sensor kinases with the decision to sporulate is likely to vary significantly among species, as each species will have evolved to sporulate in a manner that is adaptive in its specific niche. For example, B. anthracis does not sporulate during infection; spores appear only after an animal that has died of anthrax is scavenged and, as a result, the vegetative cells that have accumulated in the blood are exposed to the environment (109). By remaining vegetative, the bacterial load and toxin production in the host is maximized. By restricting the initiation of sporulation to a point when the bacterial titer is highest, the number of spores that can ultimately disseminate the disease is also maximized. There is strong evidence that this is achieved by the specific configuration of the phosphorylation cascade in B. anthracis, has likely evolved to limit sporulation initiation during infection (110–113).
In C. difficile, the situation is different; spores are produced in the host during active infection, when the bacterium has colonized the gastrointestinal tract (36). It is not yet known how C. difficile initiates sporulation in the host. However, the genetic control of sporulation in this organism is being elucidated. An important lesson from these early studies is that the mechanism differs in important ways from what is known in B. subtilis (114, 115). As for the case of B. anthracis, it is likely that in C. difficile the control of sporulation initiation has adapted to support pathogenesis.
Spore development and toxin synthesis
Two very distinct examples of how sporulation can participate in the initiation of disease are provided by the gastrointestinal disease caused by the C. perfringens enterotoxin (CPE) and the insecticidal effect of B. thuringiensis (and several other members of the Bacillaceae, including Lysinibacillus sphaericus). In both organisms, a toxin produced concomitantly with the spore induces disease. In the case of C. perfringens, vegetative cells enter the body orally, often due to consumption of spoiled food that is stored anaerobically (such as when canned). The cells then sporulate in the gastrointestinal tract, resulting in the synthesis of toxin, an event that is governed by the sporulation developmental program. The result is the appearance of toxin in the mother cell compartment, and the liberation of toxin into the gastrointestinal tract upon mother cell envelope lysis. The diarrhea that results from the toxin facilitates spore dissemination and the next round of infection.
B. thuringiensis produces an insecticidal crystal toxin during sporulation and under the control of the sporulation transcription factors that direct mother cell gene expression (σE and/or σK) (116). The crystal is typically in close association with the spore, and can be underneath or outside of the exosporium (117–119). Unlike what usually results from the disease caused by the C. perfringens enterotoxin, B. thuringiensis kills its insect host and then replicates in the cadaver. A subset of these replicating cells goes on to sporulate (25, 120).
In both C. perfringens and B. thuringiensis, the association of a toxin with the spore facilitates disease and dissemination of spores after infection, although the resulting pathology and the mechanisms of dissemination differ. It is intriguing to consider that such similar mechanisms of toxin production and delivery arose in such phylogenetically different species.
Germination Dynamics and Signals
Germination is a multi-step process in which spores transition from dormancy to metabolically active, dividing cells. This process can be triggered in one of two ways: i) nutrient-triggered germination, or 2) non-nutrient-triggered germination. In nutrient-triggered germination, the response is initiated by the sensing of environmental signals/nutrients; this pathway is more likely followed by spores that germinate in the host. Non-nutrient triggered germination occurs in response to damage or degradation of the spore coat/cortex/inner membrane (121). This method is considered an attempt at vegetative growth when the spore is damaged. The process of germination is characterized by distinct and temporal structural and biochemical changes in spores. Because individual cells proceed asynchronously through these changes, and with varying rates and timing, germination in a spore population is understood to be heterogeneous. The stages of germination differ between B. anthracis and C. difficile. The process is best understood in B. subtilis, and thought to be similar in B. anthracis, given the conservation in germination proteins between the two species. In cases where pathogenic Bacillus species have been studied, results are comparable to those in B. subtilis.
Germinants
Germination is activated when the spore senses the appropriate external signals or molecules, termed germinants, to initiate the germination pathway. Germinants interact with their corresponding germinant receptors to initiate the germination cascade. The germinants for B. anthracis and C. difficile are distinct, and each pathogen germinates with different dynamics. Studies of B. anthracis germination in vitro have revealed major germinants for initiation of the process and functions of key spore proteins associated with development of the vegetative cell. The major germinants for B. anthracis are alanine or purine ribonucleotides. One or more additional amino acids are required to act as co-germinants. In vitro, B. anthracis requires a minimum of two germinants, but the number of germinants affects the required concentration of each germinant. In other words, the more germinants are present, the less of each is necessary to trigger germination initiation. In B. anthracis, six distinct germination pathways have been described (122–126). These pathways require different combinations of amino acids, sugars, and ions to initiate germination. Interestingly, D-amino acids are unable to trigger germination, while L-amino acids can (127).
Germinant receptors
The germinant receptors (GRs) are encoded by ger genes in Bacillus, located on the inner membrane, and composed of three or four subunits (GerA, GerB, GerC, and GerD). In general, GRs are thought to be present at very low protein levels (few copies per cell) (51, 128). GRs are clustered in one spot on the inner membrane. The cluster of GRs is termed the germinosome. GerD is important to the formation of the germinosome (cluster). Spores lacking GerD have GRs dispersed throughout the inner membrane. (129). Because GRs are located on the inner membrane, germinants must travel through the spore’s outer layers to initiate germination. However, spores germinate within minutes after contact with germinants, so it is reasonable to hypothesize that germinant penetration of the outer layers is somehow selective or specific. The gerP operon (where “P” denotes permeation) may be involved in this process. First discovered in B. cereus (130), gerPABCDEF is regulated by σK, suggesting that it is expressed in the mother cell during sporulation (130, 131)). This operon in B. anthracis is 99% similar by sequence similarity and 97% protein identity. (132). Disruption of GerP operon leads to a germination defect, but the functions of the genes in the operon are unknown. They are not predicted to resemble any known enzymes. Mutants on gerP make a normal, stable spore, suggesting that this operon does not affect the structure of the spore. GerP might function to enable the Ca2+ dipicolinic acid (CaPDA) release (133).
To date, there have been no in vitro studies showing the direct binding of a germinant receptor to a germinant. Thus, there is no definitive proof of the function of GRs. However, there is other strong evidence to support their function as receptors for germinants. Genetic studies involving point mutants of GRs result in altered response to germinants, most frequently demonstrated by changes in the concentration of germinate needed to trigger germination. These studies strongly suggest that GRs and germinants do interact (134, 135).
gerX, a tricistronic operon found on the pXO1 plasmid, is expressed exclusively in the developing spore. Deletion of the gerX locus altered germination in vivo (136, 137). However, no in vitro phenotype was uncovered. Many other ger genes have been discovered through whole genome sequencing and annotation. These GRs are chromosomally encoded; gerX remains the only identified germinant receptor locus that is found on a virulence plasmid in B. anthracis.
Although the primary means of germination seems to be through response to germinants via GRs, germination can also occur in response to peptidoglycan fragments released by nearby bacteria (138). This response seems to be completely independent of the nutrient pathway, as no GRs are used to initiate germination. Instead of GRs, PrkC, a membrane-associated Ser/Thr kinase binds directly to peptidoglycan fragments shed by neighboring vegetative cells. This phenomenon was shown in B. subtilis and B. anthracis. Although the role of PrkC in this germination pathway was well demonstrated, the signal transduction thereafter is not yet understood. It is reasonable to hypothesize that spores might use the presence of actively growing bacteria in the environment as a signal of favorable growth conditions; however, it is unclear whether this pathway has a role in anthrax pathogenesis.
Computationally, orthologs of the GerA family of GRs found in B. anthracis cannot be predicted in the C. difficile genome. Furthermore, even within Clostridium species, the conserved pathways are differentially regulated. Of the five pathogenic species of Clostridium, C. perfringens is the best studied with respect to germination. However, some recent research has shed some light on the unique properties of germination in C. difficile. In contrast to B. subtilis and C. perfringens, which sense nucleosides, sugars, amino acids, and ions, C. difficile, germinates in response to bile salts and derivatives, including cholate taurocholate, glycocholate, and deoxycholate (139, 140). In addition to bile salts, the amino acids L-glycine or L-histidine serve as co-germinants (139, 141, 142). Germination in C. difficile is inhibited by chenodeoxycholic acid derivatives (139, 143), including muricholic acids (54).
Commitment to germination
Commitment to germination can be defined as the point at which the dissociation of a nutrient germinant by a strong competitive inhibitor or acidification to a pH of ~ 4.5 no longer blocks completion of germination. It is hypothesized that germinant receptor:nutrient binding results in commitment (126). Commitment is characterized by major changes in the permeability and structure of the inner membrane. As a result, the spore releases monovalent cations, including Na+, K+, and H+. Once this reaction has occurred, the spore is committed to germinate even if the germinant is displaced from the receptor (126, 144–146). Recently, Wang et al. measured germination in individual spores and found that a spore commits to germination at or very near the time at which the spore’s inner membrane permeability changes (147).
The period between the moment germinants are mixed with spores and the initiation of germination is termed the lag period. The lag period ranges in length from a few minutes to more than 24 hours. Spores that have extremely long lag periods are termed superdormant (SD). Superdormant spores represent only a small fraction of the total spore population. While superdormancy is not well understood, it appears to be due to very low levels of GRs present in the inner membrane (148–150). The variation may be epistatic in nature. Regardless of the length of the lag period, very little is known about what happens during this time in either species (151, 152)
Within the Bacillales spore core, water makes up only 25–50% of the total weight. Meanwhile, Ca2+DPA makes up 10% of the dry weight of the spore. These biochemical properties change drastically in Stage I. Stage I is characterized by a series biophysical events that involve rapid release of Ca2+DPA and the rapid influx of water molecules into the spore. These events are triggered by the interaction of the GRs to germinants. Stage II encompasses the events from Ca2+DPA release to expansion of the core, until the cell contains similar amounts of water equivalent to growing cells (80%). During Stage II, spore cortex-lytic enzymes, or SCLEs, degrade the peptidoglycan (PG) cortex. The increase in water enables the metabolism to resume.
The dynamics of germination in C. difficile are distinct from those of the Bacillales. Francis et al. (2015) compared the biochemical dynamics of germination in C. difficile to those in B. subtilis. They found that, in B. subtilis, Ca2+DPA release occurs first, followed by release of cortex fragments. These findings were similar to those reported previously. However, in C. difficile, the opposite is true; cortex fragments can be detected in the medium before Ca2+DPA. Additionally, deletion of hydrolase genes from B. subtilis prevented cortex fragment release, but not Ca 2+DPA release (123).
Outgrowth
Outgrowth is the conversion of a germinated spore to a growing cell in several distinct steps. During outgrowth, the initial production of the components needed to generate a vegetative cell take place, leading eventually to a binary fission event and therefore resumed vegetative growth. Outgrowth initiates with the removal of the spore cortex. The SCLEs lie dormant within the spore and upon activation allow full core rehydration and cell outgrowth. SCLEs recognize the muramic-δ-lactam (MAL) molecule, which is found exclusively in cortex PG. This is one reason why SCLEs can degrade the cortex PG, but not the spore’s cell wall PG, which is chemically identical to cell wall in vegetative cells (152).
In B. subtilis, two SCLEs, SleB and CwlJ, serve partially redundant roles in cortex hydrolysis and spore germination (153). SleB in a lytic transglycosylase that recognizes and hydrolyzes the modified bond between N-acetyl muramic acid and N-acetyl glucosamine (154). While the activity of SleB seems to be activated by the Ca2+DPA signal, the mechanism of activation is not yet known. CwlJ has not be shown to have specific enzymatic activity, but it bears one catalytic domain with homology to SleB (53), is located in the spore coat, and is necessary for spore germination by exogenous Ca2+DPA treatment (155).
In B. subtilis and B. anthracis, the sleB gene is located within a bicistronic operon with ypeB. YpeB is essential for the proper assembly of SleB during sporulation (154, 156). One hypothesis is that YpeB serves as an inhibitory molecule to SleB during sporulation, and proteolysis of YpeB during germination allows SleB proteolytic activity to resume (157). Similarly, in both organisms, cwlJ is located in an operon with gerQ, which is required for CwlJ activity (158). The B. anthracis genome, in contrast with the B. subtilis and B. cereus, contains a second copy of gerQ (gerQ-like) and cwlJ (cwlJ2). SleB and CwlJ are required for full virulence in a murine model for inhalation anthrax, but deletion of cwlJ2 alone does not significantly affect virulence (159). Deletion of cwlJ2 along with sleB and cwlJ reduces virulence synergistically.
As mentioned above, the proteins involved in germination of Clostridium strains are distinctly different from those found in Bacillus, so much so that computational analysis could not predict the genes in Clostridium species based upon those known in Bacillus (37). In C. perfringens, the Csp family of subtilin-like serine proteases – CspA, CspB, and CspC – act to degrade the core. One protein of the Csp family cleaves SleC, the spore-cortex lytic enzyme (SleB in B. anthracis), which in turn degrades the core. In C. difficile, CspC and CspA are predicted to be catalytically dead because two of the three residues of the catalytic triad are mutated in each (160). Instead, CspC acts as a germinant receptor for bile salt (54). This interaction between germinant and germinant receptor is thought to important for virulence, as a cspC mutant is attenuated in a Syrian hamster model of C. difficile infection (59). Furthermore, mutation of CspC (G457R) altered the specificity of this receptor for bile salt (54). Lastly, cspC-null C. difficile spores were able to germinate in the presence of chenodeoxycholate, which normally inhibits taurocholate-mediated germination (a competitive inhibitor).
CspB is the only Csp protease with an intact catalytic triad. One hypothesis is that, in response to bile salts, CspC activates CspB, which processes SleB to activate it for cortex degradation. Unlike the SleC-CwlJ system in Bacillus, CspC and SleB are not activated by Ca2+DPA. In fact, the signals that activate the system in C. difficile are unknown (37).
Germination in the Host
For Bacillus and Clostridium species, it is likely that successful pathogenesis depends on restricting germination to a specific point during infection; otherwise, the spore could become susceptible to host defenses before reaching a location in the host where survival in the vegetative form is feasible. Therefore, in considering the evolution of pathogenesis in spore-forming species, it is very important to analyze adaptations of the germination system to the host.
Although B. anthracis can exist in outside of the mammalian host as spores or vegetative cells, spores are more highly infectious than cells by most routes of infection. Vegetative cells have been only shown to cause anthrax disease when inoculated directly into the bloodstream (161, 162). In contrast, spores were pathogenic by every infection route tested. Entry of spores by the inhalation route is most deadly, and therefore most-studied. A landmark study by Ross (163) demonstrated that spores are phagocytosed quickly by phagocytes, which detach from the lung and migrate to the lymph nodes. This work was done in an inhalation anthrax model using guinea pigs as the host. Ross was able to document a large number of spore-containing phagocytes and some germinated bacterial cells within the host cells en route to the lymph node. B. anthracis cells can also be phagocytosed by dendritic cells, which are also able to capture spores and deliver them to lymph nodes (164). However, differences have been observed between strains. While the Sterne strain tends to germinate quickly (165)), the Ames strain has been shown to remain in the lung in an ungerminated state for 96 hours (166). These differences are not fully understood, but suggest that the timing of germination is not dependent solely upon the presence of germinants. Phagocytes can bind and phagocytose B. anthracis efficiently, but neither event is necessary for germination – spores can germinate in cell-free, macrophage conditioned media (125), suggesting that host cells add small-molecule germinants to the medium.
Despite these reports that capture snapshots in the establishment phase of infection, no conclusive study shows whether spores germinate before or after phagocytosis. Both scenarios have been demonstrated in vitro. However, there is evidence for the presence of at least one bottleneck during infection. From an inoculum containing equal numbers of three B. anthracis strains, each expressing a different fluorescent protein, only one fluorescent protein was detected in distal organs following aerosol infection (167). Another study proposed two bottlenecks dependent on the infection route. Infections of the nasal mucosa-associated lymphoid tissue resulted in a bottleneck at the cervical lymph node, whereas lung-based infections revealed a bottleneck in a focal region of growth within the lung (168).
For C. difficile, germination in the host depends heavily on the gut microbial metabolism (169, 170). The gut microbiota is responsible for the conversion of bile acids into secondary bile acids, which inhibit germination of C. difficile. Antibiotic treatment alters the structure of the gut microbiome and thereby alters the ability of these bacteria to synthesize secondary bile acids. Without such inhibitory molecules present, C. difficile can germinate, grow, produce toxin and cause C. difficile infection, known as CDI. Use of fecal transplants to regenerate the natural flora of the gut has proved successful for treatment of resistant and recurrent CDI (171).
TRANSMISSION AND LIFESTYLES OUTSIDE OF THE HUMAN HOST
For Bacillus and Clostridium species, the infectious form of the bacterium is the dormant spore. Thus, pathogenesis depends upon survival of spores in the non-host environment prior to entry into susceptible hosts. Yet a fundamental difference between pathogenic Clostridium and Bacillus species is the state of the bacterium upon exit from a mammalian host. C. difficile and C. perfringens sporulate within the mammalian gut during infection and bacteria are released from the colonic tract to the non-host environment as a dormant spore. As an obligate anaerobe, vegetative cells of Clostridium cannot survive for more than a few hours outside of the anaerobic host environment (172, 173). Thus, fecal-oral transmission of Clostridium species from person to person requires persistence of the bacterium in the dormant spore form in the non-host environment. By contrast, B. anthracis is not released from mammalian hosts until death of the infected host, at which time vegetative cells of the bacterium are found in blood and other body tissues. Reasons for lack of sporulation in within dead or dying hosts are not clear. It has been suggested that once nutrients are depleted the oxygen tension is too low for sporulation (174). Sporulation in vivo may also be repressed by the virulence gene regulator AtxA (175). Blood from dead B. anthracis-infected animals does not clot, and can drain from orifices into the non-host environment. In the wild, vegetative cells from infected tissues can also be released through damage imposed by scavenging carnivores. Sporulation of B. anthracis is initiated when vegetative cells are exposed to the air (176).
Transition from Host to Non-host Environments
The developmental state of spore-forming bacteria is dependent upon the ability to sense niche-specific signals. Transition from the host to non-host environment presents a major change in nutrient availability, temperature, and moisture. In the metabolically inactive spore state, the pathogens can withstand these challenges and others, including significant changes in pressure, pH, and ultraviolet light (177, 178). In the case of C. difficile, spores are intrinsically resistant to antibiotics and the host immune system (179). Upon shedding from the host, spores persist because they are resistant to inactivation by non-bleach disinfectants that are used commonly in hospital settings (180). Resistance of B. anthracis spores to the challenges of the non-host environment is also considered essential for persistence, yet some studies suggest that the B. anthracis sporulation - germination cycle outside of the host plays a role transmission.
Pathogenic Bacillus Lifestyles Outside of the Host
B. cereus group species exist in soil and water, but can also be isolated from a diverse array of mammals, birds, reptiles, and invertebrates. The bacteria can be found in the intestinal tracts of poultry and turtles, and on udders of cows, (9, 181–185). Multiple strains of B. cereus group species have been reported to exist as commensal inhabitants of invertebrate intestines, including the gut of termites, millipedes, sow bugs, and cockroaches (186, 187). The species may also associate with amoeba (188). It is not clear to what degree the pathogenic Bacillus species persist upon release from these organisms, as opposed to potential commensal lifestyles within complex ecosystems of these varied hosts.
The epidemiology of human anthrax has livestock, wildlife, soil, and water components. Blood from B. anthracis-infected mammals can contain as much as 109 vegetative cells per milliliter (189). Processed animal products, such as leather and wool, can carry large numbers of spores, and human cases of anthrax generally result from direct contact with contaminated animal products. A few anthrax cases have been reported to result from insect bites, which are presumed to be due to feeding of insects on infected animals or carcasses (190, 191). In the soil, spores can remain viable for decades, especially when deposited 15 centimeters below the upper soil levels (176). Herbivores are most likely to be exposed to spores during grazing, and infection may occur via inhalation or ingestion.
B. cereus strains have been reported in multiple foods and are most often associated with rice and dairy products (184, 192). It is likely that the spores survive pasteurization and heating. They are also somewhat resistant to gamma-ray irradiation, which is used to reduce food pathogens. Spore hydrophobicity is thought to contribute to adherence to surfaces. In addition, biofilms of B. cereus are a recurrent problem in milk tanks at dairies (193–195).
B. anthracis germination in the non-host environment
The lifestyle of B. anthracis in the non-host environment is complex and somewhat controversial. Anthrax ecology is influenced by climate, but it is difficult to draw firm conclusions from various reports because of large variations in the timing and severity of outbreaks even within single ecosystems (109, 196, 197). Some studies suggest that spore germination in the soil can be triggered by alkaline pH, high organic content, moisture, and temperatures in excess of 15 °C (177). Germination may also occur in response to components of exudates from grass plants. B. anthracis, B. cereus, and B. thuringiensis have been reported to germinate in the plant rhizosphere (198, 199). Cycles of germination, proliferation, and sporulation would ultimately result in amplification of the bacterium outside of its host. Yet it has been suggested that replication of B. anthracis vegetative cells in non-host environments reduces virulence, possibly via loss of the virulence plasmid pXO2 that encodes the B. anthracis capsule. This plasmid, unlike the toxin-encoding plasmid pXO1, is unstable during growth in the absence of selection for the antiphagocytic property of capsule (200). Moreover, it is likely, but has not been clearly demonstrated, that vegetative cells fall prey to other microbial species and protozoans in the soil (177).
B. anthracis and other B. cereus group species have been reported to germinate and replicate in response to factors excreted by the amoeba (188). The bacteria can colonize the surface of amoeba as micro-colonies, resisting phagocytosis. At high amoeba densities, the bacteria form long filaments that cannot be ingested due to size exclusion. It has been proposed that these different cell morphologies may be of significance not only outside of the host, but also during infection in the context of immune evasion (188).
Interestingly, genomic analyses indicate that B. cereus group species cannot efficiently utilize complex plant carbohydrates (201). Thus, in contrast to the common soil-borne non-pathogenic species B. subtilis, the B. cereus species appear to be adapted for use of animal-derived material. And, while the concept of B. anthracis germination in the soil is controversial, this difference in catabolism suggests that even in the event of germination in the non-host environment, these species may proliferate to a lesser extent that the non-pathogenic Bacillus species.
Clostridium Species in the Non-host Environment
A number of reports suggest that Clostridium species are ubiquitous in multiple non-host environments, including soil and water close to urban areas (202). C. perfringens has been reported in wastewater treatment systems and can be isolated from animal intestinal tracks (5, 203). C. botulinum spores are found in animal carcasses and even algal mats, where accumulation of botulinum toxin is central to human disease occurrence (204). Multiple strains of C. difficile, including those associated with disease and those less commonly found in patients, have been isolated from water and sediments. The genetic diversity of C. difficile isolates found in close proximity and the complex network of their prophages have been suggested to contribute to the emergence of new strains in clinics (205).
The relative abundance of C. difficile spores in clinical settings is in part due to infected patients entering a “super-shedder” state in which high quantities of spores are released in feces. The spores can persist in these settings for extended periods (206). Therefore, measures to control and prevent C. difficile infection include enhanced compliance with cleaning and disinfection procedures. Decontamination protocols for patient rooms often include use of dilute hypochlorite solutions and other sporicidal products. Recently, ultraviolet (UV) light and hydrogen peroxide vapor systems have been introduced as “hands off” methods of disinfection (33). Nevertheless, there are few reports assessing the impact of these methods on C. difficile transmission.
IMPLICATIONS AND QUESTIONS
Sporulation as an Enabler of Pathogen Transmission
The ability of spores to resist diverse environmental stresses and to remain viable for extended periods in the absence of nutrients make spores highly suited to the dissemination of disease. It is important to emphasize, however, that only a small number of spore-forming species are known to cause disease in humans or other animals. Most likely, the ability to form a spore was a relatively early event in the evolution of the firmicutes (the phylum containing the Bacillales and Clostridiales) and, only after their appearance did pathogenic lifestyles emerge for a subset of the spore-forming species. In those cases where survival in a host was beneficial, the resistance characteristics of spores may have provided the opportunity for rapid evolution of pathogenesis by, for example, acquisition of toxin genes. It is plausible that for a spore-forming species to initially acquire the ability to cause disease, the spore itself does not necessarily have to evolve, given its impressive resistance properties in most if not all species. Nonetheless, specific adaptations of spore structures that facilitate pathogenesis are possible, and their study remains an important open area of investigation. Regardless of whether the spore itself has adapted to facilitate disease, specific spore resistance properties and, therefore, specific spore structures must be considered in understanding the mechanistic basis of dissemination and infection by spores.
Spore – Vegetative Cell Interactions
Spore-forming bacteria are found in almost every environment where they have been sought. Given that the resistance properties of the spore are likely to have had a significant impact on survival in diverse niches, it is tempting to propose that spore formation is specifically an adaptation to the stresses of these environments. However, an important possible prediction of this view, that only spores, and not vegetative cells, would be found in the soil, is not borne out. The view that spores and vegetative cells coexist in the environment is suggested by analyses of biofilms in the pathogen C. difficile and the model spore-forming (and non-pathogenic) species B. subtilis (207, 208). In both species, spores are formed from vegetative cells as the biofilm matures, and both cell types are present in the biofilm for a significant period during biofilm development. Therefore it is plausible that during an infection, both spores and vegetative cells are present in the host and that, in fact, the presence of both cell types contributes to survival in the host even when only the vegetative cell produces essential virulence factors such as toxins.
Gaps in Knowledge of the Ecology of Spore Formers
For pathogenic spore formers, increased understanding the mechanisms for sporulation, germination, and interaction with non-host environments will facilitate development of strategies for more effective disease prevention and treatment. In particular, our poor understanding of the signals and pressures of the non-host environment is directly related to the difficulty of establishing laboratory assays that authentically mimic natural environments. Further, the bio-complexity of non-host environment is likely to affect spore/cell viability and the developmental cycle. The nature of the mixed microbial communities and the threat of predation is another largely unknown area related to the lifestyles of spore formers outside of their host organisms. Development of a comprehensive understanding of the natural ecology of spore formers will enhance our understanding of these diverse developmental pathogens.
References
- 1.Cardazzo B, Negrisolo E, Carraro L, Alberghini L, Patarnello T, Giaccone V. Multiple-locus sequence typing and analysis of toxin genes in Bacillus cereus food-borne isolates. Applied and environmental microbiology. 2008;74:850–860. doi: 10.1128/AEM.01495-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ash C, Farrow JA, Dorsch M, Stackebrandt E, Collins MD. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. International journal of systematic bacteriology. 1991;41:343–346. doi: 10.1099/00207713-41-3-343. [DOI] [PubMed] [Google Scholar]
- 3.Helgason E, Caugant DA, Olsen I, Kolsto AB. Genetic structure of population of Bacillus cereus and B. thuringiensis isolates associated with periodontitis and other human infections. Journal of clinical microbiology. 2000;38:1615–1622. doi: 10.1128/jcm.38.4.1615-1622.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helgason E, Tourasse NJ, Meisal R, Caugant DA, Kolsto AB. Multilocus sequence typing scheme for bacteria of the Bacillus cereus group. Applied and environmental microbiology. 2004;70:191–201. doi: 10.1128/AEM.70.1.191-201.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drean P, McAuley CM, Moore SC, Fegan N, Fox EM. Characterization of the spore-forming Bacillus cereus sensu lato group and Clostridium perfringens bacteria isolated from the Australian dairy farm environment. BMC microbiology. 2015;15:38. doi: 10.1186/s12866-015-0377-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guinebretiere MH, Auger S, Galleron N, Contzen M, De Sarrau B, De Buyser ML, Lamberet G, Fagerlund A, Granum PE, Lereclus D, De Vos P, Nguyen-The C, Sorokin A. Bacillus cytotoxicus sp. nov. is a novel thermotolerant species of the Bacillus cereus Group occasionally associated with food poisoning. International journal of systematic and evolutionary microbiology. 2013;63:31–40. doi: 10.1099/ijs.0.030627-0. [DOI] [PubMed] [Google Scholar]
- 7.Guinebretiere MH, Velge P, Couvert O, Carlin F, Debuyser ML, Nguyen-The C. Ability of Bacillus cereus group strains to cause food poisoning varies according to phylogenetic affiliation (groups I to VII) rather than species affiliation. Journal of clinical microbiology. 2010;48:3388–3391. doi: 10.1128/JCM.00921-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Soufiane B, Cote JC. Discrimination among Bacillus thuringiensis H serotypes, serovars and strains based on 16S rRNA, gyrB and aroE gene sequence analyses. Antonie van Leeuwenhoek. 2009;95:33–45. doi: 10.1007/s10482-008-9285-4. [DOI] [PubMed] [Google Scholar]
- 9.Jensen GB, Hansen BM, Eilenberg J, Mahillon J. The hidden lifestyles of Bacillus cereus and relatives. Environmental microbiology. 2003;5:631–640. doi: 10.1046/j.1462-2920.2003.00461.x. [DOI] [PubMed] [Google Scholar]
- 10.Tourasse NJ, Helgason E, Okstad OA, Hegna IK, Kolsto AB. The Bacillus cereus group: novel aspects of population structure and genome dynamics. Journal of applied microbiology. 2006;101:579–593. doi: 10.1111/j.1365-2672.2006.03087.x. [DOI] [PubMed] [Google Scholar]
- 11.Bengis RG, Frean J. Anthrax as an example of the One Health concept. Revue scientifique et technique. 2014;33:593–604. doi: 10.20506/rst.33.2.2309. [DOI] [PubMed] [Google Scholar]
- 12.Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler DR, Dean DH. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiology and molecular biology reviews: MMBR. 1998;62:775–806. doi: 10.1128/mmbr.62.3.775-806.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Frankenhuyzen K, Liu Y, Tonon A. Interactions between Bacillus thuringiensis subsp. kurstaki HD-1 and midgut bacteria in larvae of gypsy moth and spruce budworm. Journal of invertebrate pathology. 2010;103:124–131. doi: 10.1016/j.jip.2009.12.008. [DOI] [PubMed] [Google Scholar]
- 14.Moayeri M, Leppla SH, Vrentas C, Pomerantsev AP, Liu S. Anthrax Pathogenesis. Annu Rev Microbiol. 2015;69:185–208. doi: 10.1146/annurev-micro-091014-104523. [DOI] [PubMed] [Google Scholar]
- 15.Grunow R, Klee SR, Beyer W, George M, Grunow D, Barduhn A, Klar S, Jacob D, Elschner M, Sandven P, Kjerulf A, Jensen JS, Cai W, Zimmermann R, Schaade L. Anthrax among heroin users in Europe possibly caused by same Bacillus anthracis strain since 2000. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2013;18 [PubMed] [Google Scholar]
- 16.Ringertz SH, Hoiby EA, Jensenius M, Maehlen J, Caugant DA, Myklebust A, Fossum K. Injectional anthrax in a heroin skin-popper. Lancet. 2000;356:1574–1575. doi: 10.1016/s0140-6736(00)03133-0. [DOI] [PubMed] [Google Scholar]
- 17.Keim P, Gruendike JM, Klevytska AM, Schupp JM, Challacombe J, Okinaka R. The genome and variation of Bacillus anthracis. Molecular aspects of medicine. 2009;30:397–405. doi: 10.1016/j.mam.2009.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu S, Moayeri M, Leppla SH. Anthrax lethal and edema toxins in anthrax pathogenesis. Trends in microbiology. 2014;22:317–325. doi: 10.1016/j.tim.2014.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe DE, Glomski IJ. Cellular and physiological effects of anthrax exotoxin and its relevance to disease. Frontiers in cellular and infection microbiology. 2012;2:76. doi: 10.3389/fcimb.2012.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jelacic TM, Chabot DJ, Bozue JA, Tobery SA, West MW, Moody K, Yang D, Oppenheim JJ, Friedlander AM. Exposure to Bacillus anthracis capsule results in suppression of human monocyte-derived dendritic cells. Infection and immunity. 2014;82:3405–3416. doi: 10.1128/IAI.01857-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jeon JH, Lee HR, Cho MH, Park OK, Park J, Rhie GE. The Poly-gamma-d-Glutamic Acid Capsule Surrogate of the Bacillus anthracis Capsule Is a Novel Toll-Like Receptor 2 Agonist. Infection and immunity. 2015;83:3847–3856. doi: 10.1128/IAI.00888-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Piris-Gimenez A, Corre JP, Jouvion G, Candela T, Khun H, Goossens PL. Encapsulated Bacillus anthracis interacts closely with liver endothelium. The Journal of infectious diseases. 2009;200:1381–1389. doi: 10.1086/644506. [DOI] [PubMed] [Google Scholar]
- 23.Soberon M, Gill SS, Bravo A. Signaling versus punching hole: How do Bacillus thuringiensis toxins kill insect midgut cells? Cell Mol Life Sci. 2009;66:1337–1349. doi: 10.1007/s00018-008-8330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melo AL, Soccol VT, Soccol CR. Bacillus thuringiensis: mechanism of action, resistance, and new applications: a review. Critical reviews in biotechnology. 2015:1–10. doi: 10.3109/07388551.2014.960793. [DOI] [PubMed] [Google Scholar]
- 25.Verplaetse E, Slamti L, Gohar M, Lereclus D. Cell Differentiation in a Bacillus thuringiensis Population during Planktonic Growth, Biofilm Formation, and Host Infection. MBio. 2015;6:e00138–00115. doi: 10.1128/mBio.00138-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hernandez E, Ramisse F, Ducoureau JP, Cruel T, Cavallo JD. Bacillus thuringiensis subsp. konkukian (serotype H34) superinfection: case report and experimental evidence of pathogenicity in immunosuppressed mice. Journal of clinical microbiology. 1998;36:2138–2139. doi: 10.1128/jcm.36.7.2138-2139.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samples JR, Buettner H. Corneal ulcer caused by a biologic insecticide (Bacillus thuringiensis) American journal of ophthalmology. 1983;95:258–260. doi: 10.1016/0002-9394(83)90028-4. [DOI] [PubMed] [Google Scholar]
- 28.Celandroni F, Salvetti S, Senesi S, Ghelardi E. Bacillus thuringiensis membrane-damaging toxins acting on mammalian cells. FEMS microbiology letters. 2014;361:95–103. doi: 10.1111/1574-6968.12615. [DOI] [PubMed] [Google Scholar]
- 29.Arslan S, Eyi A, Kucuksari R. Toxigenic genes, spoilage potential, and antimicrobial resistance of Bacillus cereus group strains from ice cream. Anaerobe. 2014;25:42–46. doi: 10.1016/j.anaerobe.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 30.Ehling-Schulz M, Fricker M, Scherer S. Bacillus cereus, the causative agent of an emetic type of food-borne illness. Molecular nutrition & food research. 2004;48:479–487. doi: 10.1002/mnfr.200400055. [DOI] [PubMed] [Google Scholar]
- 31.Yutin N, Galperin MY. A genomic update on clostridial phylogeny: Gram-negative spore formers and other misplaced clostridia. Environmental microbiology. 2013;15:2631–2641. doi: 10.1111/1462-2920.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grass JE, Gould LH, Mahon BE. Epidemiology of foodborne disease outbreaks caused by Clostridium perfringens, United States, 1998-2010. Foodborne pathogens and disease. 2013;10:131–136. doi: 10.1089/fpd.2012.1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barbut F. How to eradicate Clostridium difficile from the environment. The Journal of hospital infection. 2015;89:287–295. doi: 10.1016/j.jhin.2014.12.007. [DOI] [PubMed] [Google Scholar]
- 34.Aronoff DM. Clostridium novyi, sordellii, and tetani: mechanisms of disease. Anaerobe. 2013;24:98–101. doi: 10.1016/j.anaerobe.2013.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Kelly CP, LaMont JT. Clostridium difficile–more difficult than ever. The New England journal of medicine. 2008;359:1932–1940. doi: 10.1056/NEJMra0707500. [DOI] [PubMed] [Google Scholar]
- 36.Rupnik M, Wilcox MH, Gerding DN. Clostridium difficile infection: new developments in epidemiology and pathogenesis. Nat Rev Microbiol. 2009;7:526–536. doi: 10.1038/nrmicro2164. [DOI] [PubMed] [Google Scholar]
- 37.Paredes-Sabja D, Shen A, Sorg JA. Clostridium difficile spore biology: sporulation, germination, and spore structural proteins. Trends in microbiology. 2014;22:406–416. doi: 10.1016/j.tim.2014.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shields K, Araujo-Castillo RV, Theethira TG, Alonso CD, Kelly CP. Recurrent Clostridium difficile infection: From colonization to cure. Anaerobe. 2015;34:59–73. doi: 10.1016/j.anaerobe.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kuehne SA, Cartman ST, Heap JT, Kelly ML, Cockayne A, Minton NP. The role of toxin A and toxin B in Clostridium difficile infection. Nature. 2010;467:711–713. doi: 10.1038/nature09397. [DOI] [PubMed] [Google Scholar]
- 40.Kuehne SA, Collery MM, Kelly ML, Cartman ST, Cockayne A, Minton NP. Importance of toxin A, toxin B, and CDT in virulence of an epidemic Clostridium difficile strain. J Infect Dis. 2014;209:83–86. doi: 10.1093/infdis/jit426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carter GP, Chakravorty A, Pham Nguyen TA, Mileto S, Schreiber F, Li L, Howarth P, Clare S, Cunningham B, Sambol SP, Cheknis A, Figueroa I, Johnson S, Gerding D, Rood JI, Dougan G, Lawley TD, Lyras D. Defining the Roles of TcdA and TcdB in Localized Gastrointestinal Disease, Systemic Organ Damage, and the Host Response during Clostridium difficile Infections. MBio. 2015;6:e00551. doi: 10.1128/mBio.00551-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stewart DB, Berg A, Hegarty J. Predicting recurrence of C. difficile colitis using bacterial virulence factors: binary toxin is the key. Journal of gastrointestinal surgery: official journal of the Society for Surgery of the Alimentary Tract. 2013;17:118–124. doi: 10.1007/s11605-012-2056-6. discussion p 124-115. [DOI] [PubMed] [Google Scholar]
- 43.Stevens DL, Aldape MJ, Bryant AE. Life-threatening clostridial infections. Anaerobe. 2012;18:254–259. doi: 10.1016/j.anaerobe.2011.11.001. [DOI] [PubMed] [Google Scholar]
- 44.Bryant AE, Stevens DL. Clostridial myonecrosis: new insights in pathogenesis and management. Current infectious disease reports. 2010;12:383–391. doi: 10.1007/s11908-010-0127-y. [DOI] [PubMed] [Google Scholar]
- 45.Uzal FA, Freedman JC, Shrestha A, Theoret JR, Garcia J, Awad MM, Adams V, Moore RJ, Rood JI, McClane BA. Towards an understanding of the role of Clostridium perfringens toxins in human and animal disease. Future microbiology. 2014;9:361–377. doi: 10.2217/fmb.13.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fisher DJ, Fernandez-Miyakawa ME, Sayeed S, Poon R, Adams V, Rood JI, Uzal FA, McClane BA. Dissecting the contributions of Clostridium perfringens type C toxins to lethality in the mouse intravenous injection model. Infection and immunity. 2006;74:5200–5210. doi: 10.1128/IAI.00534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li J, Adams V, Bannam TL, Miyamoto K, Garcia JP, Uzal FA, Rood JI, McClane BA. Toxin plasmids of Clostridium perfringens. Microbiology and molecular biology reviews: MMBR. 2013;77:208–233. doi: 10.1128/MMBR.00062-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Setlow P. Spores of Bacillus subtilis: their resistance to and killing by radiation, heat and chemicals. J Appl Microbiol. 2006;101:514–525. doi: 10.1111/j.1365-2672.2005.02736.x. [DOI] [PubMed] [Google Scholar]
- 49.Nicholson WL, Munakata N, Horneck G, Melosh HJ, Setlow P. Resistance of Bacillus endospores to extreme terrestrial and extraterrestrial environments. Microbiol Mol Biol Rev. 2000;64:548–572. doi: 10.1128/mmbr.64.3.548-572.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Driks A. Proteins of the spore core and coat. In: Sonenshein AL, Hoch JA, Losick R, editors. Bacillus subtilis and its closest relatives. American Society for Microbiology; Washington, D.C.: 2002. pp. 527–536. [Google Scholar]
- 51.Setlow P. Spore germination. Curr Opin Microbiol. 2003;6:550–556. doi: 10.1016/j.mib.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 52.Knudsen SM, Cermak N, Feijo Delgado F, Setlow B, Setlow P, Manalis SR. Water and Small Molecule Permeation of Dormant Bacillus subtilis Spores. J Bacteriol. doi: 10.1128/JB.00435-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moir A. How do spores germinate? J Appl Microbiol. 2006;101:526–530. doi: 10.1111/j.1365-2672.2006.02885.x. [DOI] [PubMed] [Google Scholar]
- 54.Francis MB, Allen CA, Shrestha R, Sorg JA. Bile acid recognition by the Clostridium difficile germinant receptor, CspC, is important for establishing infection. PLoS Pathog. 2013;9:e1003356. doi: 10.1371/journal.ppat.1003356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ross C, Abel-Santos E. The Ger receptor family from sporulating bacteria. Curr Issues Mol Biol. 2010;12:147–158. [PMC free article] [PubMed] [Google Scholar]
- 56.Cutting SM, Vander Horn PB. Molecular Biological Methods for Bacillus. John Wiley & Sons Ltd; Chichester, United Kingdom: 1990. [Google Scholar]
- 57.Aronson AI, Fitz-James P. Structure and morphogenesis of the bacterial spore coat. Bacteriol Rev. 1976;40:360–402. doi: 10.1128/br.40.2.360-402.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holt SC, Leadbetter ER. Comparative ultrastructure of selected aerobic spore-forming bacteria: a freeze-etching study. Bacteriol Rev. 1969;33:346–378. doi: 10.1128/br.33.2.346-378.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Driks A. The Bacillus subtilis spore coat. Microbiol Mol Biol Rev. 1999;63:1–20. doi: 10.1128/mmbr.63.1.1-20.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Giorno R, Bozue J, Cote C, Wenzel T, Moody K-S, Ryan M, Wang R, Zielke R, Maddock JM, Friedlander A, Welkos S, Driks A. Morphogenesis of the Bacillus anthracis spore. J Bacteriol. 2007;189:691–705. doi: 10.1128/JB.00921-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Henriques AO, Moran CP., Jr Structure and assembly of the bacterial endospore coat. Methods. 2000;20:95–110. doi: 10.1006/meth.1999.0909. [DOI] [PubMed] [Google Scholar]
- 62.Henriques AO, Costa TV, Martins LO, Zilhao R. The functional architecture and assembly of the coat. In: Ricca RE, Henriques AO, Cutting SM, editors. Bacterial spore formers: probiotics and emerging applications. Horizon Biosciences; Norfolk: 2004. pp. 65–86. [Google Scholar]
- 63.Traag BA, Driks A, Stragier P, Bitter W, Broussard G, Hatfull G, Chu F, Adams KN, Ramakrishnan L, Losick R. Do Mycobacteria Produce Endospores? Proc Natl Acad Sci USA. 2010;107:878–881. doi: 10.1073/pnas.0911299107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Driks A. Maximum shields: the armor plating of the bacterial spore. Trends Microbiol. 2002;10:251–254. doi: 10.1016/s0966-842x(02)02373-9. [DOI] [PubMed] [Google Scholar]
- 65.Semenyuk EG, Laning ML, Foley J, Johnston PF, Knight KL, Gerding D, Driks A. Spore formation and toxin production in Clostridium difficile biofilms. PLoS ONE. 2014;9:e87757. doi: 10.1371/journal.pone.0087757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.McKenney PT, Driks A, Eichenberger P. The Bacillus subtilis endospore: assembly and functions of the multilayered coat. Nat Rev Microbiol. 2013;11:33–44. doi: 10.1038/nrmicro2921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim H, Hahn M, Grabowski P, McPherson DC, Wang R, Ferguson C, Eichenberger P, Driks A. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006;59:487–502. doi: 10.1111/j.1365-2958.2005.04968.x. [DOI] [PubMed] [Google Scholar]
- 68.McKenney PT, Driks A, Eskandarian HA, Grabowski P, Guberman J, Wang KH, Gitai Z, Eichenberger P. A distance-weighted interaction map reveals a previously uncharacterized layer of the B. subtilis spore coat. Current Biology. 2010;29:934–938. doi: 10.1016/j.cub.2010.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Henriques AO, Moran CP. Structure, assembly and function of the spore surface layers. Ann Rev Microbiol. 2007;61:555–588. doi: 10.1146/annurev.micro.61.080706.093224. [DOI] [PubMed] [Google Scholar]
- 70.Driks A. The dynamic spore. Proc Natl Acad Sci USA. 2003;100:3007–3009. doi: 10.1073/pnas.0730807100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chada VG, Sanstad EA, Wang R, Driks A. Morphogenesis of Bacillus spore surfaces. J Bacteriol. 2003;185:6255–6261. doi: 10.1128/JB.185.21.6255-6261.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sahin O, Yong E-H, McPherson DC, Giorno R, Buhr TL, Mahadevan L, Driks A. Adaptive flexibility of the Bacillus spore coat regulates mechanical events during dormancy, germination and outgrowth. 2010 In preparation. [Google Scholar]
- 73.Westphal AJ, Price PB, Leighton TJ, Wheeler KE. Kinetics of size changes of individual Bacillus thuringiensis spores in response to changes in relative humidity. Proc Natl Acad Sci USA. 2003;100:3461–3466. doi: 10.1073/pnas.232710999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen X, Goodnight D, Gao Z, Cavusoglu AH, Sabharwal N, DeLay M, Driks A, Sahin O. Scaling up nanoscale water-driven energy conversion into evaporation-driven engines and generators. Nat Commun. 2015:7346. doi: 10.1038/ncomms8346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen X, Mahadevan L, Driks A, Sahin O. Bacillus spores as building blocks for stimuli-responsive materials and nanogenerators. Nat Nanotechnol. 2014;9:137–141. doi: 10.1038/nnano.2013.290. [DOI] [PubMed] [Google Scholar]
- 76.Laaberki MH, Dworkin J. Role of spore coat proteins in the resistance of Bacillus subtilis spores to Caenorhabditis elegans predation. J Bacteriol. 2008;190:6197–6203. doi: 10.1128/JB.00623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Klobutcher LA, Ragkousi K, Setlow P. The Bacillus subtilis spore coat provides “eat resistance” during phagocytic predation by the protozoan Tetrahymena thermophila. Proc Natl Acad Sci U S A. 2006;103:165–170. doi: 10.1073/pnas.0507121102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Walker JR, Gnanam AJ, Blinkova AL, Hermandson MJ, Karymov MA, Lyubchenko YL, Graves PR, Haystead TA, Linse KD. Clostridium taeniosporum spore ribbon-like appendage structure, composition and genes. Mol Microbiol. 2007;63:629–643. doi: 10.1111/j.1365-2958.2006.05494.x. [DOI] [PubMed] [Google Scholar]
- 79.Driks A. Surface appendages of bacterial spores. Mol Microbiol. 2007;63:623–625. doi: 10.1111/j.1365-2958.2006.05564.x. [DOI] [PubMed] [Google Scholar]
- 80.Qin H, Driks A. Contrasting evolutionary patterns of spore coat proteins in two Bacillus species groups are linked to a difference in cellular structure. BMC Evol Biol. 2013;13:261. doi: 10.1186/1471-2148-13-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Giorno R, Bozue J, Mallozzi M, Moody K-S, Slack A, Wang R, Friedlander A, Welkos S, Driks A. Characterization of a Bacillus anthracis spore protein with roles in exosporium morphology. Microbiology. 2009;155:1133–1145. doi: 10.1099/mic.0.023333-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Todd SJ, Moir AJ, Johnson MJ, Moir A. Genes of Bacillus cereus and Bacillus anthracis encoding proteins of the exosporium. J Bacteriol. 2003;185:3373–3378. doi: 10.1128/JB.185.11.3373-3378.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Severson KM, Mallozzi M, Bozue J, Welkos SL, Cote CK, Knight KL, Driks A. Roles of the Bacillus anthracis spore protein ExsK in exosporium maturation and germination. J Bacteriol. 2009;191:7587–7596. doi: 10.1128/JB.01110-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ball DA, Taylor R, Todd SJ, Redmond C, Couture-Tosi E, Sylvestre P, Moir A, Bullough PA. Structure of the exosporium and sublayers of spores of the Bacillus cereus family revealed by electron crystallography. Mol Microbiol. 2008;68:947–958. doi: 10.1111/j.1365-2958.2008.06206.x. [DOI] [PubMed] [Google Scholar]
- 85.Kailas L, Terry C, Abbott N, Taylor R, Mullin N, Tzokov SB, Todd SJ, Wallace BA, Hobbs JK, Moir A, Bullough PA. Surface architecture of endospores of the Bacillus cereus/anthracis/thuringiensis family at the subnanometer scale. Proc Natl Acad Sci U S A. 2011;108:16014–16019. doi: 10.1073/pnas.1109419108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Johnson MJ, Todd SJ, Ball DA, Shepherd AM, Sylvestre P, Moir A. ExsY and CotY are required for the correct assembly of the exosporium and spore coat of Bacillus cereus. J Bacteriol. 2006;188:7905–7913. doi: 10.1128/JB.00997-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Redmond C, Baille L, Hibbs S, Moir A, Charlton S. Characterization of the exosporium of Bacillus anthracis. Abstract, 4th International Confernece on Anthrax; Annapolis, MD. 2001. [Google Scholar]
- 88.Redmond C, Baillie LW, Hibbs S, Moir AJ, Moir A. Identification of proteins in the exosporium of Bacillus anthracis. Microbiology. 2004;150:355–563. doi: 10.1099/mic.0.26681-0. [DOI] [PubMed] [Google Scholar]
- 89.Charlton S, Moir AJ, Baillie L, Moir A. Characterization of the exosporium of Bacillus cereus. J Appl Microbiol. 1999;87:241–245. doi: 10.1046/j.1365-2672.1999.00878.x. [DOI] [PubMed] [Google Scholar]
- 90.Rodenburg CM, McPherson SA, Turnbough CL, Jr, Dokland T. Cryo-EM analysis of the organization of BclA and BxpB in the Bacillus anthracis exosporium. J Struct Biol. 2014;186:181–187. doi: 10.1016/j.jsb.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan L, Turnbough CL., Jr Sequence motifs and proteolytic cleavage of the collagen-like glycoprotein BclA required for its attachment to the exosporium of Bacillus anthracis. J Bacteriol. 2010;192:1259–1268. doi: 10.1128/JB.01003-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steichen C, Chen P, Kearney JF, Turnbough CL., Jr Identification of the immunodominant protein and other proteins of the Bacillus anthracis exosporium. J Bacteriol. 2003;185:1903–1910. doi: 10.1128/JB.185.6.1903-1910.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Steichen CT, Kearney JF, Turnbough CL., Jr Characterization of the exosporium basal layer protein BxpB of Bacillus anthracis. J Bacteriol. 2005;187:5868–5876. doi: 10.1128/JB.187.17.5868-5876.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Steichen CT, Kearney JF, Turnbough CL., Jr Non-uniform assembly of the Bacillus anthracis exosporium and a bottle cap model for spore germination and outgrowth. Mol Microbiol. 2007;64:359–367. doi: 10.1111/j.1365-2958.2007.05658.x. [DOI] [PubMed] [Google Scholar]
- 95.Boydston JA, Chen P, Steichen CT, Turnbough CL., Jr Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J Bacteriol. 2005;187:5310–5317. doi: 10.1128/JB.187.15.5310-5317.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boydston JA, Yue L, Kearney JF, Turnbough CL., Jr The ExsY protein is required for complete formation of the exosporium of Bacillus anthracis. J Bacteriol. 2006;188:7440–7448. doi: 10.1128/JB.00639-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Weaver J, Kang TJ, Raines KW, Cao GL, Hibbs S, Tsai P, Baillie L, Rosen GM, Cross AS. Protective role of Bacillus anthracis exosporium in macrophage-mediated killing by nitric oxide. Infect Immun. 2007;75:3894–3901. doi: 10.1128/IAI.00283-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liang L, He X, Liu G, Tan H. The role of a purine-specific nucleoside hydrolase in spore germination of Bacillus thuringiensis. Microbiology. 2008;154:1333–1340. doi: 10.1099/mic.0.2007/014399-0. [DOI] [PubMed] [Google Scholar]
- 99.Chesnokova ON, McPherson SA, Steichen CT, Turnbough CL., Jr The spore-specific alanine racemase of Bacillus anthracis and its role in suppressing germination during spore development. J Bacteriol. 2009;191:1303–1310. doi: 10.1128/JB.01098-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Brahmbhatt TN, Janes BK, Stibitz ES, Darnell SC, Sanz P, Rasmussen SB, O’Brien AD. Bacillus anthracis exosporium protein BclA affects spore germination, interaction with extracellular matrix proteins, and hydrophobicity. Infect Immun. 2007;75:5233–5239. doi: 10.1128/IAI.00660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Bozue J, Moody KL, Cote CK, Stiles BG, Friedlander AM, Welkos SL, Hale ML. Bacillus anthracis spores of the bclA mutant exhibit increased adherence to epithelial cells, fibroblasts, and endothelial cells but not to macrophages. Infect Immun. 2007;75:4498–4505. doi: 10.1128/IAI.00434-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Oliva C, Turnbough CL, Jr, Kearney JF. CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proc Natl Acad Sci U S A. 2009;106:13957–13962. doi: 10.1073/pnas.0902392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Oliva CR, Swiecki MK, Griguer CE, Lisanby MW, Bullard DC, Turnbough CL, Jr, Kearney JF. The integrin Mac-1 (CR3) mediates internalization and directs Bacillus anthracis spores into professional phagocytes. Proc Natl Acad Sci U S A. 2008;105:1261–1266. doi: 10.1073/pnas.0709321105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hong HA, Khaneja R, Tam NM, Cazzato A, Tan S, Urdaci M, Brisson A, Gasbarrini A, Barnes I, Cutting SM. Bacillus subtilis isolated from the human gastrointestinal tract. Res Microbiol. 2009;160:134–143. doi: 10.1016/j.resmic.2008.11.002. [DOI] [PubMed] [Google Scholar]
- 105.Chen G, Driks A, Tawfiq K, Mallozzi M, Patil S. Bacillus anthracis and Bacillus subtilis Spore Surface Properties as analyzed by transport analysis. Colloids and Surfaces B: Biointerfaces. 2010;76:512–518. doi: 10.1016/j.colsurfb.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 106.Grossman AD. Genetic networks controlling the initiation of sporulation and the development of genetic competence in Bacillus subtilis. Annu Rev Genet. 1995;29:477–508. doi: 10.1146/annurev.ge.29.120195.002401. [DOI] [PubMed] [Google Scholar]
- 107.Hoch JA. Regulation of the phosphorelay and the initiation of sporulation in Bacillus subtilis. Annu Rev Microbiol. 1993;47:441–465. doi: 10.1146/annurev.mi.47.100193.002301. [DOI] [PubMed] [Google Scholar]
- 108.van Gestel J, Vlamakis H, Kolter R. Division of Labor in Biofilms: the Ecology of Cell Differentiation. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MB-0002-2014. MB-0002-2014. [DOI] [PubMed] [Google Scholar]
- 109.Hugh-Jones M, Blackburn J. The ecology of Bacillus anthracis. Mol Aspects Med. 2009;30:356–367. doi: 10.1016/j.mam.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 110.Perego M, Hoch JA. Commingling regulatory systems following acquisition of virulence plasmids by Bacillus anthracis. Trends Microbiol. 2008;16:215–221. doi: 10.1016/j.tim.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 111.Brunsing RL, La Clair C, Tang S, Chiang C, Hancock LE, Perego M, Hoch JA. Characterization of sporulation histidine kinases of Bacillus anthracis. J Bacteriol. 2005;187:6972–6981. doi: 10.1128/JB.187.20.6972-6981.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bongiorni C, Stoessel R, Shoemaker D, Perego M. Rap phosphatase of virulence plasmid pXO1 inhibits Bacillus anthracis sporulation. J Bacteriol. 2006;188:487–498. doi: 10.1128/JB.188.2.487-498.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.White AK, Hoch JA, Grynberg M, Godzik A, Perego M. Sensor domains encoded in Bacillus anthracis virulence plasmids prevent sporulation by hijacking a sporulation sensor histidine kinase. J Bacteriol. 2006;188:6354–6360. doi: 10.1128/JB.00656-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Edwards AN, McBride SM. Initiation of sporulation in Clostridium difficile: a twist on the classic model. FEMS Microbiol Lett. 2014;358:110–118. doi: 10.1111/1574-6968.12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Edwards AN, Nawrocki KL, McBride SM. Conserved oligopeptide permeases modulate sporulation initiation in Clostridium difficile. Infect Immun. 2014;82:4276–4291. doi: 10.1128/IAI.02323-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Aronson A. Sporulation and delta-endotoxin synthesis by Bacillus thuringiensis. Cell Mol Life Sci. 2002;59:417–425. doi: 10.1007/s00018-002-8434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Debro L, Fitz-James PC, Aronson A. Two different parasporal inclusions are produced by Bacillus thuringiensis subsp. finitimus. J Bacteriol. 1986;165:258–268. doi: 10.1128/jb.165.1.258-268.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Ammons DR, Reyna A, Granados JC, Ventura-Suarez A, Rojas-Avelizapa LI, Short JD, Rampersad JN. A novel Bacillus thuringiensis Cry-like protein from a rare filamentous strain promotes crystal localization within the exosporium. Appl Environ Microbiol. 2013;79:5774–5776. doi: 10.1128/AEM.01206-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chai PF, Rathinam X, Solayappan M, Ahmad Ghazali AH, Subramaniam S. Microscopic analysis of a native Bacillus thuringiensis strain from Malaysia that produces exosporium-enclosed parasporal inclusion. Microscopy (Oxf) 2014;63:371–375. doi: 10.1093/jmicro/dfu022. [DOI] [PubMed] [Google Scholar]
- 120.Dubois T, Faegri K, Perchat S, Lemy C, Buisson C, Nielsen-LeRoux C, Gohar M, Jacques P, Ramarao N, Kolsto AB, Lereclus D. Necrotrophism is a quorum-sensing-regulated lifestyle in Bacillus thuringiensis. PLoS Pathog. 2012;8:e1002629. doi: 10.1371/journal.ppat.1002629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Fisher N, Carr KA, Giebel JD, Hanna PC. Anthrax Spore Germination. John Wiley & Sons; Hoboken, New Jersey: 2011. [Google Scholar]
- 122.Fisher N, Hanna P. Characterization of Bacillus anthracis germinant receptors in vitro. J Bacteriol. 2005;187:8055–8062. doi: 10.1128/JB.187.23.8055-8062.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Francis MB, Allen CA, Sorg JA. Spore Cortex Hydrolysis Precedes Dipicolinic Acid Release during Clostridium difficile Spore Germination. J Bacteriol. 2015;197:2276–2283. doi: 10.1128/JB.02575-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ireland JA, Hanna PC. Amino acid- and purine ribonucleoside-induced germination of Bacillus anthracis DeltaSterne endospores: gerS mediates responses to aromatic ring structures. J Bacteriol. 2002;184:1296–1303. doi: 10.1128/JB.184.5.1296-1303.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Weiner MA, Hanna PC. Macrophage-mediated germination of Bacillus anthracis endospores requires the gerH operon. Infection and immunity. 2003;71:3954–3959. doi: 10.1128/IAI.71.7.3954-3959.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yi X, Setlow P. Studies of the commitment step in the germination of spores of bacillus species. J Bacteriol. 2010;192:3424–3433. doi: 10.1128/JB.00326-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Atluri S, Ragkousi K, Cortezzo DE, Setlow P. Cooperativity between different nutrient receptors in germination of spores of Bacillus subtilis and reduction of this cooperativity by alterations in the GerB receptor. J Bacteriol. 2006;188:28–36. doi: 10.1128/JB.188.1.28-36.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Bergman NH, Anderson EC, Swenson EE, Niemeyer MM, Miyoshi AD, Hanna PC. Transcriptional profiling of the Bacillus anthracis life cycle in vitro and an implied model for regulation of spore formation. J Bacteriol. 2006;188:6092–6100. doi: 10.1128/JB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Griffiths KK, Zhang J, Cowan AE, Yu J, Setlow P. Germination proteins in the inner membrane of dormant Bacillus subtilis spores colocalize in a discrete cluster. Molecular microbiology. 2011;81:1061–1077. doi: 10.1111/j.1365-2958.2011.07753.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Behravan J, Chirakkal H, Masson A, Moir A. Mutations in the gerP locus of Bacillus subtilis and Bacillus cereus affect access of germinants to their targets in spores. J Bacteriol. 2000;182:1987–1994. doi: 10.1128/jb.182.7.1987-1994.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–529. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 132.Read TD, Peterson SN, Tourasse N, Baillie LW, Paulsen IT, Nelson KE, Tettelin H, Fouts DE, Eisen JA, Gill SR, Holtzapple EK, Okstad OA, Helgason E, Rilstone J, Wu M, Kolonay JF, Beanan MJ, Dodson RJ, Brinkac LM, Gwinn M, DeBoy RT, Madpu R, Daugherty SC, Durkin AS, Haft DH, Nelson WC, Peterson JD, Pop M, Khouri HM, Radune D, Benton JL, Mahamoud Y, Jiang L, Hance IR, Weidman JF, Berry KJ, Plaut RD, Wolf AM, Watkins KL, Nierman WC, Hazen A, Cline R, Redmond C, Thwaite JE, White O, Salzberg SL, Thomason B, Friedlander AM, Koehler TM, Hanna PC, et al. The genome sequence of Bacillus anthracis Ames and comparison to closely related bacteria. Nature. 2003;423:81–86. doi: 10.1038/nature01586. [DOI] [PubMed] [Google Scholar]
- 133.Paidhungat M, Ragkousi K, Setlow P. Genetic requirements for induction of germination of spores of Bacillus subtilis by Ca(2+)-dipicolinate. J Bacteriol. 2001;183:4886–4893. doi: 10.1128/JB.183.16.4886-4893.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Christie G, Gotzke H, Lowe CR. Identification of a receptor subunit and putative ligand-binding residues involved in the Bacillus megaterium QM B1551 spore germination response to glucose. J Bacteriol. 2010;192:4317–4326. doi: 10.1128/JB.00335-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Mongkolthanaruk W, Cooper GR, Mawer JS, Allan RN, Moir A. Effect of amino acid substitutions in the GerAA protein on the function of the alanine-responsive germinant receptor of Bacillus subtilis spores. J Bacteriol. 2011;193:2268–2275. doi: 10.1128/JB.01398-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Guidi-Rontani C, Pereira Y, Ruffie S, Sirard JC, Weber-Levy M, Mock M. Identification and characterization of a germination operon on the virulence plasmid pXO1 of Bacillus anthracis. Molecular microbiology. 1999;33:407–414. doi: 10.1046/j.1365-2958.1999.01485.x. [DOI] [PubMed] [Google Scholar]
- 137.McKevitt MT, Bryant KM, Shakir SM, Larabee JL, Blanke SR, Lovchik J, Lyons CR, Ballard JD. Effects of endogenous D-alanine synthesis and autoinhibition of Bacillus anthracis germination on in vitro and in vivo infections. Infection and immunity. 2007;75:5726–5734. doi: 10.1128/IAI.00727-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Sorg JA, Sonenshein AL. Bile salts and glycine as cogerminants for Clostridium difficile spores. J Bacteriol. 2008;190:2505–2512. doi: 10.1128/JB.01765-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ridlon JM, Kang DJ, Hylemon PB. Bile salt biotransformations by human intestinal bacteria. J Lipid Res. 2006;47:241–259. doi: 10.1194/jlr.R500013-JLR200. [DOI] [PubMed] [Google Scholar]
- 141.Howerton A, Ramirez N, Abel-Santos E. Mapping interactions between germinants and Clostridium difficile spores. J Bacteriol. 2011;193:274–282. doi: 10.1128/JB.00980-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wheeldon LJ, Worthington T, Lambert PA. Histidine acts as a co-germinant with glycine and taurocholate for Clostridium difficile spores. Journal of applied microbiology. 2011;110:987–994. doi: 10.1111/j.1365-2672.2011.04953.x. [DOI] [PubMed] [Google Scholar]
- 143.Sorg JA, Sonenshein AL. Chenodeoxycholate is an inhibitor of Clostridium difficile spore germination. J Bacteriol. 2009;191:1115–1117. doi: 10.1128/JB.01260-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Foster SJ, Johnstone K. The use of inhibitors to identify early events during Bacillus megaterium KM spore germination. Biochem J. 1986;237:865–870. doi: 10.1042/bj2370865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Stewart GS, Johnstone K, Hagelberg E, Ellar DJ. Commitment of bacterial spores to germinate. A measure of the trigger reaction. Biochem J. 1981;198:101–106. doi: 10.1042/bj1980101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Venkatasubramanian P, Johnstone K. Biochemical analysis of the Bacillus subtilis 1604 spore germination response. J Gen Microbiol. 1989;135:2723–2733. doi: 10.1099/00221287-135-10-2723. [DOI] [PubMed] [Google Scholar]
- 147.Wang S, Shen A, Setlow P, Li YQ. Characterization of the Dynamic Germination of Individual Clostridium difficile Spores Using Raman Spectroscopy and Differential Interference Contrast Microscopy. J Bacteriol. 2015;197:2361–2373. doi: 10.1128/JB.00200-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ghosh S, Setlow P. Isolation and characterization of superdormant spores of Bacillus species. J Bacteriol. 2009;191:1787–1797. doi: 10.1128/JB.01668-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Yi X, Liu J, Faeder JR, Setlow P. Synergism between different germinant receptors in the germination of Bacillus subtilis spores. J Bacteriol. 2011;193:4664–4671. doi: 10.1128/JB.05343-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Chen Y, Ray WK, Helm RF, Melville SB, Popham DL. Levels of germination proteins in Bacillus subtilis dormant, superdormant, and germinating spores. PLoS ONE. 2014;9:e95781. doi: 10.1371/journal.pone.0095781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Setlow P. When the sleepers wake: the germination of spores of Bacillus species. Journal of applied microbiology. 2013;115:1251–1268. doi: 10.1111/jam.12343. [DOI] [PubMed] [Google Scholar]
- 152.Setlow P. Germination of spores of Bacillus species: what we know and do not know. J Bacteriol. 2014;196:1297–1305. doi: 10.1128/JB.01455-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Heffron JD, Orsburn B, Popham DL. Roles of germination-specific lytic enzymes CwlJ and SleB in Bacillus anthracis. J Bacteriol. 2009;191:2237–2247. doi: 10.1128/JB.01598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Boland FM, Atrih A, Chirakkal H, Foster SJ, Moir A. Complete spore-cortex hydrolysis during germination of Bacillus subtilis 168 requires SleB and YpeB. Microbiology. 2000;146(Pt 1):57–64. doi: 10.1099/00221287-146-1-57. [DOI] [PubMed] [Google Scholar]
- 155.Bagyan I, Setlow P. Localization of the cortex lytic enzyme CwlJ in spores of Bacillus subtilis. J Bacteriol. 2002;184:1219–1224. doi: 10.1128/jb.184.4.1219-1224.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Chirakkal H, O’Rourke M, Atrih A, Foster SJ, Moir A. Analysis of spore cortex lytic enzymes and related proteins in Bacillus subtilis endospore germination. Microbiology. 2002;148:2383–2392. doi: 10.1099/00221287-148-8-2383. [DOI] [PubMed] [Google Scholar]
- 157.Li Y, Butzin XY, Davis A, Setlow B, Korza G, Ustok FI, Christie G, Setlow P, Hao B. Activity and regulation of various forms of CwlJ, SleB, and YpeB proteins in degrading cortex peptidoglycan of spores of Bacillus species in vitro and during spore germination. J Bacteriol. 2013;195:2530–2540. doi: 10.1128/JB.00259-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ragkousi K, Eichenberger P, van Ooij C, Setlow P. Identification of a new gene essential for germination of Bacillus subtilis spores with Ca 2+-dipicolinate. J Bacteriol. 2003;185:2315–2329. doi: 10.1128/JB.185.7.2315-2329.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Giebel JD, Carr KA, Anderson EC, Hanna PC. The germination-specific lytic enzymes SleB, CwlJ1, and CwlJ2 each contribute to Bacillus anthracis spore germination and virulence. J Bacteriol. 2009;191:5569–5576. doi: 10.1128/JB.00408-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Adams CM, Eckenroth BE, Putnam EE, Doublie S, Shen A. Structural and functional analysis of the CspB protease required for Clostridium spore germination. PLoS Pathog. 2013;9:e1003165. doi: 10.1371/journal.ppat.1003165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Koch R. On the etiology of anthrax. Greenwood Press; New York: 1881. [Google Scholar]
- 162.Koch R. Mitt. Kaisserliche Gesundheitsamte. 1881;1:174–206. [Google Scholar]
- 163.Ross J. The pathogenesis of anthrax following the administration of spores by the respiratory route. J Pathol Bacteriol. 1957;73:485–494. [Google Scholar]
- 164.Cleret A, Quesnel-Hellmann A, Vallon-Eberhard A, Verrier B, Jung S, Vidal D, Mathieu J, Tournier JN. Lung dendritic cells rapidly mediate anthrax spore entry through the pulmonary route. J Immunol. 2007;178:7994–8001. doi: 10.4049/jimmunol.178.12.7994. [DOI] [PubMed] [Google Scholar]
- 165.Sanz P, Teel LD, Alem F, Carvalho HM, Darnell SC, O’Brien AD. Detection of Bacillus anthracis spore germination in vivo by bioluminescence imaging. Infection and immunity. 2008;76:1036–1047. doi: 10.1128/IAI.00985-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Cote CK, Van Rooijen N, Welkos SL. Roles of macrophages and neutrophils in the early host response to Bacillus anthracis spores in a mouse model of infection. Infection and immunity. 2006;74:469–480. doi: 10.1128/IAI.74.1.469-480.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Plaut RD, Kelly VK, Lee GM, Stibitz S, Merkel TJ. Dissemination bottleneck in a murine model of inhalational anthrax. Infection and immunity. 2012;80:3189–3193. doi: 10.1128/IAI.00515-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Lowe DE, Ernst SM, Zito C, Ya J, Glomski IJ. Bacillus anthracis has two independent bottlenecks that are dependent on the portal of entry in an intranasal model of inhalational infection. Infection and immunity. 2013;81:4408–4420. doi: 10.1128/IAI.00484-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Theriot CM, Koenigsknecht MJ, Carlson PE, Jr, Hatton GE, Nelson AM, Li B, Huffnagle GB, J ZL, Young VB. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat Commun. 2014;5:3114. doi: 10.1038/ncomms4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Theriot CM, Young VB. Interactions Between the Gastrointestinal Microbiome and Clostridium difficile. Annu Rev Microbiol. 2015;69:445–461. doi: 10.1146/annurev-micro-091014-104115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bowman KA, Broussard EK, Surawicz CM. Fecal microbiota transplantation: current clinical efficacy and future prospects. Clin Exp Gastroenterol. 2015;8:285–291. doi: 10.2147/CEG.S61305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Deakin LJ, Clare S, Fagan RP, Dawson LF, Pickard DJ, West MR, Wren BW, Fairweather NF, Dougan G, Lawley TD. The Clostridium difficile spo0A gene is a persistence and transmission factor. Infection and immunity. 2012;80:2704–2711. doi: 10.1128/IAI.00147-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Jump RL, Pultz MJ, Donskey CJ. Vegetative Clostridium difficile survives in room air on moist surfaces and in gastric contents with reduced acidity: a potential mechanism to explain the association between proton pump inhibitors and C. difficile-associated diarrhea? Antimicrobial agents and chemotherapy. 2007;51:2883–2887. doi: 10.1128/AAC.01443-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Roth NG, Livery DH, Hodge HM. Influence of oxygen uptake and age of culture on sporulation of Bacillus anthracis and Bacillus globigii. Journal of bacteriology. 1955;69:455–459. doi: 10.1128/jb.69.4.455-459.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mignot T, Mock M, Robichon D, Landier A, Lereclus D, Fouet A. The incompatibility between the PlcR- and AtxA-controlled regulons may have selected a nonsense mutation in Bacillus anthracis. Molecular microbiology. 2001;42:1189–1198. doi: 10.1046/j.1365-2958.2001.02692.x. [DOI] [PubMed] [Google Scholar]
- 176.Goel AK. Anthrax: A disease of biowarfare and public health importance. World journal of clinical cases. 2015;3:20–33. doi: 10.12998/wjcc.v3.i1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Dragon DC, Rennie RP. The ecology of anthrax spores: tough but not invincible. The Canadian veterinary journal La revue veterinaire canadienne. 1995;36:295–301. [PMC free article] [PubMed] [Google Scholar]
- 178.Dutz W, Kohout-Dutz E. Anthrax. International journal of dermatology. 1981;20:203–206. doi: 10.1111/j.1365-4362.1981.tb00461.x. [DOI] [PubMed] [Google Scholar]
- 179.Baines SD, O’Connor R, Saxton K, Freeman J, Wilcox MH. Activity of vancomycin against epidemic Clostridium difficile strains in a human gut model. The Journal of antimicrobial chemotherapy. 2009;63:520–525. doi: 10.1093/jac/dkn502. [DOI] [PubMed] [Google Scholar]
- 180.Ali S, Moore G, Wilson AP. Spread and persistence of Clostridium difficile spores during and after cleaning with sporicidal disinfectants. The Journal of hospital infection. 2011;79:97–98. doi: 10.1016/j.jhin.2011.06.010. [DOI] [PubMed] [Google Scholar]
- 181.Andersson A, Ronner U, Granum PE. What problems does the food industry have with the spore-forming pathogens Bacillus cereus and Clostridium perfringens? International journal of food microbiology. 1995;28:145–155. doi: 10.1016/0168-1605(95)00053-4. [DOI] [PubMed] [Google Scholar]
- 182.Barbosa TM, Serra CR, La Ragione RM, Woodward MJ, Henriques AO. Screening for bacillus isolates in the broiler gastrointestinal tract. Applied and environmental microbiology. 2005;71:968–978. doi: 10.1128/AEM.71.2.968-978.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jones TO, Turnbull PC. Bovine mastitis caused by Bacillus cereus. The Veterinary record. 1981;108:271–274. doi: 10.1136/vr.108.13.271. [DOI] [PubMed] [Google Scholar]
- 184.McAuley CM, McMillan K, Moore SC, Fegan N, Fox EM. Prevalence and characterization of foodborne pathogens from Australian dairy farm environments. Journal of dairy science. 2014;97:7402–7412. doi: 10.3168/jds.2014-8735. [DOI] [PubMed] [Google Scholar]
- 185.Nfor NN, Lapin CN, McLaughlin RW. Isolation of Bacillus cereus Group from the Fecal Material of Endangered Wood Turtles. Current microbiology. 2015;71:524–527. doi: 10.1007/s00284-015-0875-x. [DOI] [PubMed] [Google Scholar]
- 186.Feinberg L, Jorgensen J, Haselton A, Pitt A, Rudner R, Margulis L. Arthromitus (Bacillus cereus) symbionts in the cockroach Blaberus giganteus: dietary influences on bacterial development and population density. Symbiosis. 1999;27:109–123. [PubMed] [Google Scholar]
- 187.Margulis L, Jorgensen JZ, Dolan S, Kolchinsky R, Rainey FA, Lo SC. The Arthromitus stage of Bacillus cereus: intestinal symbionts of animals. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:1236–1241. doi: 10.1073/pnas.95.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Beeton ML, Atkinson DJ, Waterfield NR. An amoeba phagocytosis model reveals a novel developmental switch in the insect pathogen Bacillus thuringiensis. Journal of insect physiology. 2013;59:223–231. doi: 10.1016/j.jinsphys.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 189.Turnbull PC. Introduction: anthrax history, disease and ecology. Current topics in microbiology and immunology. 2002;271:1–19. doi: 10.1007/978-3-662-05767-4_1. [DOI] [PubMed] [Google Scholar]
- 190.Bradaric N, Punda-Polic V. Cutaneous anthrax due to penicillin-resistant Bacillus anthracis transmitted by an insect bite. Lancet. 1992;340:306–307. doi: 10.1016/0140-6736(92)92395-v. [DOI] [PubMed] [Google Scholar]
- 191.Turell MJ, Knudson GB. Mechanical transmission of Bacillus anthracis by stable flies (Stomoxys calcitrans) and mosquitoes (Aedes aegypti and Aedes taeniorhynchus) Infection and immunity. 1987;55:1859–1861. doi: 10.1128/iai.55.8.1859-1861.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Martinez-Blanch JF, Sanchez G, Garay E, Aznar R. Development of a real-time PCR assay for detection and quantification of enterotoxigenic members of Bacillus cereus group in food samples. International journal of food microbiology. 2009;135:15–21. doi: 10.1016/j.ijfoodmicro.2009.07.013. [DOI] [PubMed] [Google Scholar]
- 193.Christiansson A, Bertilsson J, Svensson B. Bacillus cereus spores in raw milk: factors affecting the contamination of milk during the grazing period. Journal of dairy science. 1999;82:305–314. doi: 10.3168/jds.S0022-0302(99)75237-9. [DOI] [PubMed] [Google Scholar]
- 194.Vissers MM, Te Giffel MC, Driehuis F, De Jong P, Lankveld JM. Minimizing the level of Bacillus cereus spores in farm tank milk. Journal of dairy science. 2007;90:3286–3293. doi: 10.3168/jds.2006-873. [DOI] [PubMed] [Google Scholar]
- 195.Zottola EA, Sasahara KC. Microbial biofilms in the food processing industry–should they be a concern? International journal of food microbiology. 1994;23:125–148. doi: 10.1016/0168-1605(94)90047-7. [DOI] [PubMed] [Google Scholar]
- 196.Blackburn JK, McNyset KM, Curtis A, Hugh-Jones ME. Modeling the geographic distribution of Bacillus anthracis, the causative agent of anthrax disease, for the contiguous United States using predictive ecological [corrected] niche modeling. The American journal of tropical medicine and hygiene. 2007;77:1103–1110. [PubMed] [Google Scholar]
- 197.Hampson K, Lembo T, Bessell P, Auty H, Packer C, Halliday J, Beesley CA, Fyumagwa R, Hoare R, Ernest E, Mentzel C, Metzger KL, Mlengeya T, Stamey K, Roberts K, Wilkins PP, Cleaveland S. Predictability of anthrax infection in the Serengeti, Tanzania. The Journal of applied ecology. 2011;48:1333–1344. doi: 10.1111/j.1365-2664.2011.02030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Halverson LJ, Clayton MK, Handelsman J. Variable stability of antibiotic-resistance markers in Bacillus cereus UW85 in the soybean rhizosphere in the field. Molecular ecology. 1993;2:65–78. doi: 10.1111/j.1365-294x.1993.tb00001.x. [DOI] [PubMed] [Google Scholar]
- 199.Saile E, Koehler TM. Bacillus anthracis multiplication, persistence, and genetic exchange in the rhizosphere of grass plants. Applied and environmental microbiology. 2006;72:3168–3174. doi: 10.1128/AEM.72.5.3168-3174.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Green BD, Battisti L, Koehler TM, Thorne CB, Ivins BE. Demonstration of a capsule plasmid in Bacillus anthracis. Infection and immunity. 1985;49:291–297. doi: 10.1128/iai.49.2.291-297.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ivanova N, Sorokin A, Anderson I, Galleron N, Candelon B, Kapatral V, Bhattacharyya A, Reznik G, Mikhailova N, Lapidus A, Chu L, Mazur M, Goltsman E, Larsen N, D’Souza M, Walunas T, Grechkin Y, Pusch G, Haselkorn R, Fonstein M, Ehrlich SD, Overbeek R, Kyrpides N. Genome sequence of Bacillus cereus and comparative analysis with Bacillus anthracis. Nature. 2003;423:87–91. doi: 10.1038/nature01582. [DOI] [PubMed] [Google Scholar]
- 202.Marcheggiani S, D’Ugo E, Puccinelli C, Giuseppetti R, D’Angelo AM, Gualerzi CO, Spurio R, Medlin LK, Guillebault D, Weigel W, Helmi K, Mancini L. Detection of emerging and re-emerging pathogens in surface waters close to an urban area. International journal of environmental research and public health. 2015;12:5505–5527. doi: 10.3390/ijerph120505505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Schneeberger CL, O’Driscoll M, Humphrey C, Henry K, Deal N, Seiber K, Hill VR, Zarate-Bermudez M. Fate and transport of enteric microbes from septic systems in a coastal watershed. Journal of environmental health. 2015;77:22–30. [PubMed] [Google Scholar]
- 204.Espelund M, Klaveness D. Botulism outbreaks in natural environments - an update. Frontiers in microbiology. 2014;5:287. doi: 10.3389/fmicb.2014.00287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hargreaves KR, Clokie MR. A Taxonomic Review of Clostridium difficile Phages and Proposal of a Novel Genus, “Phimmp04likevirus”. Viruses. 2015;7:2534–2541. doi: 10.3390/v7052534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Gerding DN, Muto CA, Owens RC., Jr Measures to control and prevent Clostridium difficile infection. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2008;46(Suppl 1):S43–49. doi: 10.1086/521861. [DOI] [PubMed] [Google Scholar]
- 207.Cairns LS, Hobley L, Stanley-Wall NR. Biofilm formation by Bacillus subtilis: new insights into regulatory strategies and assembly mechanisms. Molecular microbiology. 2014;93:587–598. doi: 10.1111/mmi.12697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Pantaleon V, Bouttier S, Soavelomandroso AP, Janoir C, Candela T. Biofilms of Clostridium species. Anaerobe. 2014;30:193–198. doi: 10.1016/j.anaerobe.2014.09.010. [DOI] [PubMed] [Google Scholar]


