Abstract
Schistosomiasis affects millions of people across Africa. We detected eggs of Schistosoma mansoni in western lowland gorilla and central chimpanzee fecal samples in Loango National Park, Gabon. We analyzed nuclear and mitochondrial DNA, namely internal transcribed spacer and cytochrome c oxidase subunit 1 fragments, and the resulting maximum likelihood phylogenetic analyses and haplotype network of the ITS and COI, respectively, showed that the samples from gorillas and chimpanzees clustered clearly within the S. mansoni clade. This is the first confirmed record of S. mansoni from Gabon, which urges surveillance in the area and prompts questions regarding the extent of zoonotic transmission and the clinical impact.
Schistosomiasis affects almost 240 million people worldwide, with a majority of cases found in sub-Saharan Africa, and can have severe socioeconomic and public health impact. Two forms of the disease are recognized in humans—urogenital schistosomiasis caused by Schistosoma haematobium and intestinal schistosomiasis caused by Schistosoma mansoni, Schistosoma guineensis, Schistosoma mekongi, Schistosoma intercalatum, or Schistosoma japonicum.1 Most of the human schistosome species can produce patent infection in other mammals such as rodents, insectivores, ungulates, procyonids, carnivores, and nonhuman primates, hosts that can in turn serve as reservoirs of the infection.2
Despite the high prevalence of schistosomiasis in humans across sub-Saharan Africa and their proximity to populations of free-ranging great apes, there is a lack of information on schistosome infections in wild chimpanzees and gorillas. However, primates have been used extensively as experimental animals to study human schistosomiasis; thus, most of the current knowledge of this disease in nonhuman primates comes from experimental conditions.3,4 Recently, a semicaptive population of chimpanzees on Ngamba Island in the Lake Victoria (Uganda) was found to be infected by S. mansoni, probably as a result of contact with the S. mansoni–infected human population.5
Herein, we report finding S. mansoni in free-ranging western lowland gorillas (Gorilla gorilla gorilla) and central chimpanzees (Pan troglodytes troglodytes) in Loango National Park (LNP), Gabon.
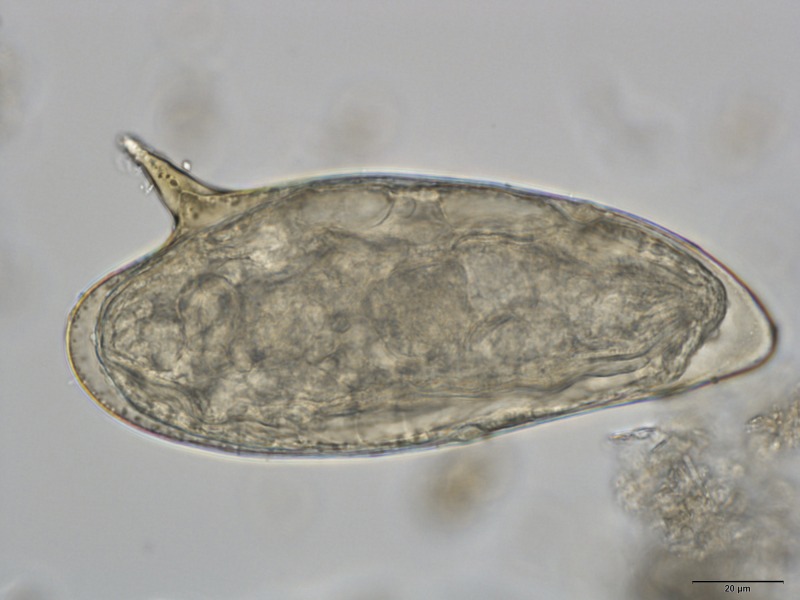
From May to July 2014, we opportunistically collected fecal samples of unhabituated lowland gorillas and central chimpanzees in a ±24,000-ha private ecotourism concession under development in LNP. We preserved subsamples in 10% formalin and 96% ethanol. We performed basic coproscopic examination using modified Sheather's flotation and merthiolate iodine formaldehyde concentration sedimentation.6,7 Aside from other parasites commonly found in gorillas and chimpanzees (e.g., strongylids, Strongyloides spp., or Entamoeba spp.), we detected trematode eggs with the morphology of S. mansoni (Figure 1 ) in five of eight gorilla and two of eight chimpanzee fecal samples.
Figure 1.
Egg of Schistosoma mansoni isolated from a chimpanzee fecal sample from Loango National Park, Gabon. Internal transcribed spacer sequence under accession no. KX011042 originates from the same sample (Scale bar = 20 μm).
We isolated individual eggs from the sediment using a simple pipette created from a glass capillary tube and rubber tubing, subsequently washed them in a drop of distilled water, and mechanically ruptured them under the control of microscope. We extracted genomic DNA using the Genomic DNA Mini Kit (Tissue) (Geneaid Biotech Ltd., Taipei, Taiwan). We performed polymerase chain reaction (PCR) with 5 μL of genomic extract using PPP Master Mix (Top-Bio, s.r.o., Praha, Czech Republic) following previously published PCR protocols.8,9 We amplified and cycle sequenced (Macrogen Europe, Amsterdam, The Netherlands) a ∼955–base pair (bp) fragment of the ribosomal DNA ITS1–5.8S–ITS2 gene region (internal transcribed spacer [ITS]) and a ∼1,148-bp fragment of cytochrome c oxidase subunit 1 (COI) with the primers BD1 and BD2 and Cox1_Schist_5′ and Cox1_Schist_3′, respectively.8,10 We used DNA isolated from the cercariae of Trichobilharzia regenti (originally isolated from birds from the southern part of the Czech Republic, provided by the team of Petr Horak, Department of Parasitology, Faculty of Science, Charles University in Prague) as a positive control.
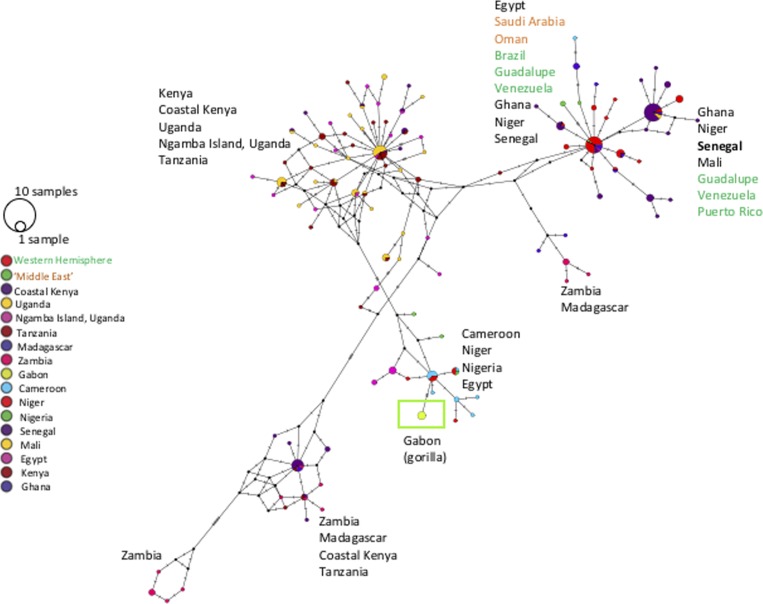
We obtained five identical ITS sequences (accession no. KX011041) and five identical COI sequences (accession no. KX011043) originating from gorillas and one ITS sequence (accession no. KX011042) originating from chimpanzee. Maximum likelihood phylogenetic analysis of the ITS region, using Schistosoma rodhaini as the outgroup, showed that the samples from both gorillas and chimpanzees clustered within the S. mansoni clade. We reconstructed a haplotype network with gorilla schistosome COI sequences obtained as part of this study and those downloaded from GenBank to infer schistosome haplotype and geographic relationships (Figure 2 ).
Figure 2.
Minimum spanning network inferred from Schistosoma mansoni cytochrome c oxidase subunit 1 (COI) sequences created in POPART.11 Eleven input sequences were defined by unique haplotype, identical haplotypes collapsed into a single haplotype. We started with 253 isolates of COI (311 base pairs [bp]) S. mansoni (from humans, Biomphalaria, chimps) that was transformed into a haplotype input file and had 111 unique haplotypes defined by 81 bp of unique nucleotide sites. In the key, the circle size is shown for one sample and 10 samples. Also, colored circles defined by ‘traitlabels’ in POPART represent country of origin(s) of the sequence except where some were collapsed (Western Hemisphere = Brazil, Guadalupe, Venezuela, Puerto Rico; Middle East = Saudi Arabia, Oman). For ease of visualization, country of origin is labeled next to the major clusters of those haplotypes (text color matches key). The samples of S. mansoni from gorillas in Gabon are highlighted by a green circle and box outline. The S. mansoni samples from previous reports of chimps on Ngamba Island, Uganda, did not form a unique cluster, nor did it cluster with the samples from Gabon.
Zoonotic transmission of pathogens between populations of free-ranging primates and humans is a critical environmental health issue. Primates, and especially apes, are reservoirs for zoonotic pathogens ranging from those with relatively minor (e.g., nematodes) to severe (e.g., Ebola) health consequences. The reverse is also true where human pathogens (e.g., respiratory viruses) have had devastating effects on ape populations and are considered among the primary threats to these endangered species.12 Our morphological and molecular analyses confirm that gorillas and chimpanzees in LNP are hosts of the schistosome S. mansoni. Our results represent the first record of schistosomes in gorillas and the first record of S. mansoni occurring conclusively in free-ranging chimpanzees.
Schistosoma mansoni schistosomiasis is a zoonosis, and in tropical Africa, S. mansoni was reported to infect rodents, insectivores, artiodactylids (waterbuck), and nonhuman primates including baboons (Papio spp.), vervet monkey (Cercopithecus aethiops), and Sykes monkey (Cercopithecus mitis).2 The local epidemiology of schistosomiasis in Ngamba Island Chimpanzee Sanctuary in Uganda was classified as “a shifting balance between anthropozoonotic and zoonotic cycles.”5
Although S. mansoni is broadly distributed across equatorial Africa,2 to our knowledge, there are no reliable data on its occurrence in Gabon or its neighboring countries (e.g., Equatorial Guinea or the Republic of the Congo). The only reported cases of schistosomiasis in Gabon were caused by S. haematobium and S. guineensis (formerly S. intercalatum) and are only known from human population.13
Our study revealed the existence of a new focus of S. mansoni in tropical Africa. The haplotype analysis showed that analyzed gorilla schistosomes carry a unique haplotype which differs from other schistosomes sequenced to date (Figure 2). Tentatively, this haplotype appears within a haplogroup containing other haplotypes found in the region (e.g., Cameroon and Nigeria). However, a conclusive statement that this haplotype represents an ape-specific lineage is premature due to a lack of available sequence data from the region and the confounding issue of human-mediated global dispersal of schistosome lineages. Even a recent introduction of S. mansoni to the Loango area cannot yet be ruled out—particularly in a world where “recent” changes in human ecology (e.g., forest encroachment and increased regional and international connectivity by air) can have significant impacts on the epidemiology and dispersal of infectious diseases.14
Clinical studies confirmed that the course and morbidity of S. mansoni infection in chimpanzees strongly resembles that reported for humans, including severe liver fibrosis.3–5 In case of LNP gorillas and chimpanzees, the data on clinical significance of the schistosomiasis are indeed missing and possible impact of this disease on already endangered apes urgently requires further investigation. A detailed epidemiological study assessing the occurrence of S. mansoni in humans and great apes, as well as in its snail host, in and around LNP is critical for uncovering the ecology of this disease and its significance for local human population and LNP visitors.
ACKNOWLEDGMENTS
We are thankful for research authorization to the Centre National de la Recherche Scientifique et Technologique (permit number N°AR0010/14/MESRS/CENAREST/CG/CST/CSAR) and the Agence National des Parcs Nationaux (permit number N°AE140009/PR/ANPN/SE/CS/AEPN). We thank SFM Safari Gabon for hosting this research and seeing the value of health monitoring as part of the development of ape tourism programs. We thank our field assistants Pierre Bukosso and Kharl Remanda.
Footnotes
Financial support: This publication is an outcome of the HPI-lab (Laboratory for Infectious Diseases Common to Human and Non-Human Primates) cofinanced by European Social Fund and state budget of the Czech Republic (project OPVK CZ.1.07/2.3.00/20.0300) The study was also cofinanced by institutional support of Institute of Vertebrate Biology Academy of Sciences of the Czech Republic (RVO: 68081766).
Authors' addresses: Barbora Červená and David Modrý, Department of Pathology and Parasitology, Faculty of Veterinary Medicine, University of Veterinary and Pharmaceutical Sciences Brno, Brno, Czech Republic, E-mails: bara.cervena@gmail.com and modryd@vfu.cz. Sara Vanessa Brant, Museum of Southwestern Biology, Department of Biology, University of New Mexico, Albuquerque, NM, E-mail: sbrant@unm.edu. Emilie Fairet and Matthew H. Shirley, Sustainable Forestry Management–Safari Gabon, Libreville, Gabon, E-mails: efairet@sfmafrica.com and mshirley@sfmafrica.com. Klára Judita Petrželková, Institute of Vertebrate Biology, Academy of Sciences of the Czech Republic, Brno, Czech Republic, E-mail: petrzelkova@ivb.cz.
References
- 1.World Health Organization Schistosomiasis. 2016. http://www.who.int/schistosomiasis/en/ Available at. Accessed February 24, 2016.
- 2.Standley CJ, Dobson AP, Stothard JR. Out of animals and back again: Schistosomiais as a zoonosis in Africa. In: Rokni MB, editor. Schistosomiasis. Rijeka, Croatia: INTECH Open Access Publisher; 2012. pp. 209–230.http://www.intechopen.com/books/schistosomiasis/out-of-animals-and-back-again-schistosomiasis-as-a-zoonosis-in-africa Available at. Accessed August 19, 2015. [Google Scholar]
- 3.Sadun EH, Von Lichtenberg F, Hickman RL, Bruce JI, Smith JH, Schoenbechler MJ. Schistosomiasis mansoni in the chimpanzee: parasitologic, clinical, serologic, pathologic and radiologic observations. Am J Trop Med Hyg. 1966;15:496–506. doi: 10.4269/ajtmh.1966.15.496. [DOI] [PubMed] [Google Scholar]
- 4.von Lichtenberg F, Sadun EH. Experimental production of bilharzial pipe-stem fibrosis in the chimpanzee. Exp Parasitol. 1968;22:264–278. doi: 10.1016/0014-4894(68)90102-1. [DOI] [PubMed] [Google Scholar]
- 5.Standley CJ, Mugisha L, Adriko M, Arinaitwe M, Rukundo J, Ajarova L, Mopya S, Betson M, Kabatereine NB, Stothard JR. Intestinal schistosomiasis in chimpanzees on Ngamba Island, Uganda: observations on liver fibrosis, schistosome genetic diversity and praziquantel treatment. Parasitology. 2013;40:285–295. doi: 10.1017/S0031182012001576. [DOI] [PubMed] [Google Scholar]
- 6.Sheather AL. The detection of intestinal protozoa and mange parasites by a flotation technique. J Comp Pathol Ther. 1923;36:266–275. [Google Scholar]
- 7.Blagg W, Schloegel EL, Mansour NS, Khalaf GI. A new concentration technique for the demonstration of protozoa and helminth eggs in feces. Am J Trop Med Hyg. 1955;4:23–28. doi: 10.4269/ajtmh.1955.4.23. [DOI] [PubMed] [Google Scholar]
- 8.Lockyer AE, Olson PD, Østergaard P, Rollinson D, Johnston DA, Attwood SW, Southgate VR, Horak P, Snyder SD, Le TH, Agatsuma T, McManus DP, Carmichael AC, Naem S, Littlewood DT. The phylogeny of the Schistosomatidae based on three genes with emphasis on the interrelationships of Schistosoma Weinland, 1858. Parasitology. 2003;126:203–224. doi: 10.1017/s0031182002002792. [DOI] [PubMed] [Google Scholar]
- 9.Morgan JAT, DeJong RJ, Lwambo NJS, Mungai BN, Mkoji GM, Loker ES. First report of a natural hybrid between Schistosoma mansoni and S. rodhaini. J Parasitol. 2003;89:416–418. doi: 10.1645/0022-3395(2003)089[0416:FROANH]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 10.Morgan JAT, DeJong RJ, Kazibwe F, Mkoji GM, Loker ES. A newly-identified lineage of Schistosoma. Int J Parasitol. 2003;33:977–985. doi: 10.1016/s0020-7519(03)00132-2. [DOI] [PubMed] [Google Scholar]
- 11.Leigh JW, Bryant D. POPART: full feature software for haplotype network construction. Methods Ecol Evol. 2015;6:1110–1116. [Google Scholar]
- 12.Leendertz FH, Pauli G, Maetz-Rensing K, Boardman W, Nunn C, Ellerbrok H, Jensen SA, Junglen S, Christophe B. Pathogens as drivers of population declines: the importance of systematic monitoring in great apes and other threatened mammals. Biol Conserv. 2006;131:325–337. [Google Scholar]
- 13.Ngouema MR, Milama KM, Kombila M, Richard-Lenoble D, Tisseyre P, Ibikounlé M, Moné H, Mouahid G. Morphometric and molecular characterizations of schistosome populations in Estuaire province Gabon. J Helminthol. 2010;84:81–85. doi: 10.1017/S0022149X09990289. [DOI] [PubMed] [Google Scholar]
- 14.Pigott DM, Golding N, Mylne A, Huang Z, Henry AJ, Weiss DJ, Brady OJ, Kraemer MU, Smith DL, Moyes CL, Bhatt S, Gething PW, Horby PW, Bogoch II, Brownstein JS, Mekaru SR, Tatem AJ, Khan K, Hay SI. Mapping the zoonotic niche of Ebola virus disease in Africa. eLife. 2014;3:e04395. doi: 10.7554/eLife.04395. [DOI] [PMC free article] [PubMed] [Google Scholar]