Abstract
Exposure of newborns to the maternal vaginal microbiota is interrupted with cesarean birthing. Babies delivered by Cesarean section (C-section) acquire a microbiota that differs from that of vaginally delivered infants, and C-section delivery has been associated with increased risk for immune and metabolic disorders. Here we conducted a pilot study in which infants delivered by C-section are exposed to maternal vaginal fluids at birth. Similar to vaginally delivered babies, the gut, oral, and skin microbiome of these newborns during the first 30 days of life was enriched in vaginal bacteria underrepresented in unexposed C-section infants, albeit similarity to vaginally-delivered infants was higher in oral and skin than in anal samples. Although the long-term health consequences of restoring the microbiota of C-section born infants remain unclear, our results demonstrate that vaginal microbes can be partially restored at birth in C-section delivered babies.
Mode of delivery is a major determinant of microbiota composition in newborns. Vaginally delivered infants harbor bacterial communities resembling those of the maternal vagina, while C-section infants are enriched in skin microbiota1. The microbiome that colonizes the body of newborns can play a determinant role in educating the immune system2. Early interaction with commensal microbes is essential for healthy immune development and metabolic programming, and aberrant microbial colonization in newborns has been associated with long-term effects on host metabolism3 or impaired immune development2. Epidemiological studies, although not showing causality, have reported associations between C-section delivery and increased risk of obesity, asthma, allergies, and immune deficiencies4–7. Rates of cesarean delivery are increasing worldwide and in some countries exceed 50% of total births8–10, substantially over the estimated 15% of births requiring C-section delivery to protect the health of the mother or baby11.
In this study, C-section delivered infants were exposed to their maternal vaginal fluids at birth, and the composition of their microbiota was determined longitudinally to assess whether they developed more similarly to vaginally born babies than to unexposed C-section infants. We collected samples from 18 infants and their mothers, including 7 born vaginally and 11 delivered by scheduled C-section, of which 4 were exposed to the maternal vaginal fluids at birth (Supplementary Table 1). Briefly, the microbial restoration procedure consists of incubating a sterile gauze in the vagina of mothers that were negative for Group B streptococcus (GBS), had no signs of vaginosis, and had a vaginal pH below 4.5, during the hour preceding the C-section. Within the first two minutes of birth, babies were exposed to their maternal vaginal contents by being swabbed with the gauze, starting with the mouth, then the face, and finally the rest of the body (Fig. 1a). A total of 1,519 samples were obtained from anal, oral and skin sites of infants and mothers at six time points during the first month of life (1, 3, 7, 14, 21 and 30 days after birth; Supplementary Table 2), Microbiome composition was characterized by sequencing the V4 region of 16S rRNA gene as previously described12, and 1,016 samples were utilized for analysis after quality filtering (Supplementary Methods).
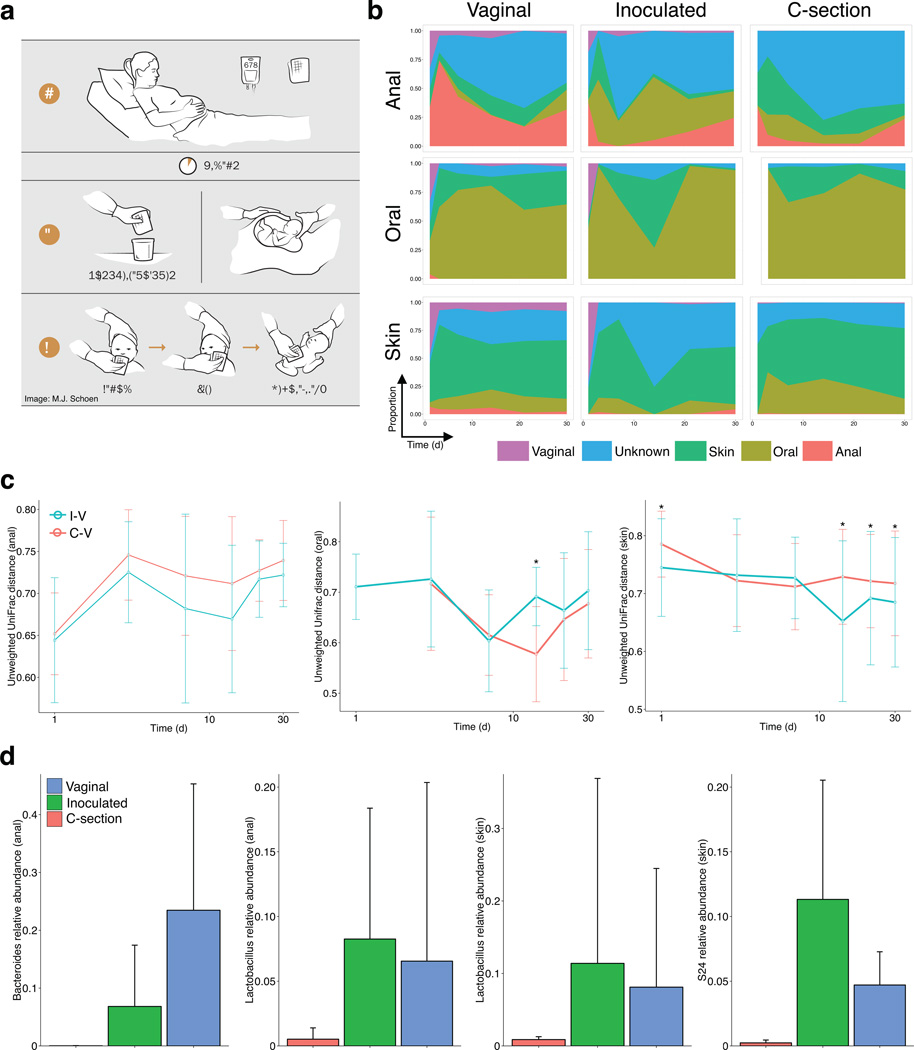
Figure 1. Restoring the maternal microbiota in infants born by C-section.
(a) Infants born by C-section were swabbed with a gauze that was incubated in the maternal vagina 30–60 min prior to the C-section. All mothers delivering by C-section received antibiotics (ABX) as part of standard of care. The gauze was extracted prior to the procedure, kept in a sterile container, and used to swab the newborn within the first one to three minutes after birth, starting with the mouth, face, and rest of the body. (b) Proportion of each sample estimated to originate from different maternal sources (using bacterial sourcetracking) of anal (top row), oral (middle), and skin (bottom) samples in infants delivered either vaginally (left column, n = 7 subjects sampled at six time points), by C-section (unexposed) (right, n = 8 × 6), or by C-section and exposed to vaginal fluids (middle, n = 4 × 6). (c) Bacterial community distances in anal (left), oral (middle), and skin (right) samples between vaginally delivered and C-section-delivered exposed (I-V) or not exposed (C-V) to the vaginal gauze, during the first month of life (Unweighted UniFrac distances). Bars indicate standard deviation from the mean. Distances between vaginal and exposed C-section infants were significantly smaller than from unexposed C-section infants (ANOVA and Tukey’s honest significant difference test. * P < 0.01) (d) Representative bacterial taxa enriched in infants with perinatal exposure to vaginal fluids during the first month of life. Bars indicate standard deviation from the mean.
Bacterial sourcetracking13 of the infant microbiome revealed that the four C-section infants exposed to vaginal fluids resembled vaginally delivered infants, particularly so during the first week of life (Fig. 1b). The distance between samples from exposed and vaginal newborns was lower in anal and skin samples (Fig. 1c). At day 1, and regardless of body site, the microbiome of babies delivered vaginally or by C-section but exposed to vaginal fluids was more similar to the maternal vaginal microbiome than that of C-section infants (Supplementary Fig. 1). A progression towards a body-specific configuration was observed in all body sites, either gradually (anus) or quickly (oral, skin), but both vaginally delivered and exposed newborns exhibit a vaginal-like signature that is absent in C-section babies (Supplementary Fig. 2). Although variations in microbiome composition per subject exist within each of the groups (Supplementary Figs. 3 to 7), we confirmed the differences between unexposed and exposed C-section infants by building a Random Forest classifier for each body site14. Samples from unexposed C-section and vaginally delivered infants could be classified with high accuracy, confirming that delivery mode shapes microbial communities of the infants (Supplementary Table 1). In relation to C-section infants unexposed to vaginal fluids, anal, oral, and skin samples from exposed C-section infants were classified less frequently as samples from C-section and more often as samples from vaginally delivered infants (Supplementary Table 3), with oral and skin samples more accurately classified than anal samples. A Random Forest classifier built from predicted metagenome content was equally able to distinguish vaginal and C-section infants, with exposed infants being classified as vaginal in oral and skin samples, and as C-section in anal samples but again less frequently than unexposed C-section infants (Supplementary Table 4).
Microbial colonization of body sites in the newborn occurs quickly and changes proceed during the first month in all groups. In anal samples from exposed infants and vaginally delivered infants, there is an early enrichment of Lactobacillus followed by a bloom of Bacteroides from week 2 that is not observed in newborns not exposed to vaginal fluids (Fig. 1d and Supplementary Fig. 2). The anal microbiota of newborns in our study, as previously reported, remained distinct from that of adults even at one month of life15,16. The infant skin and oral microbiota, however, acquired a more adult-like configuration after the first week of life in all three groups. Unexposed C-section infants, however, lacked vaginal bacteria that were restored with the gauze or present in vaginally delivered infants, particularly anal and skin Lactobacillus early in life, anal Bacteroides, and members of the Bacteroidales family S24–7 in the skin (Fig. 1d and Supplementary Fig. 2). It has been recently shown that maturation of the infant gut microbiome occurs with the cessation of breast-feeding, with no differences appreciated between exclusive or formula-supplemented breastfeeding until the fourth month of life15. Since all infants in our study received breast milk either exclusively or supplemented with formula during the first month of life (Supplementary Table 1), the microbiome composition profiles observed in each group do not appear to be due to feeding mode.
Neonatal bacterial diversity was highest at birth in anal and oral sites, and had declined by the third day (Supplementary Figs. 8a, 8b). Although this phenomenon is not yet well understood, we have previously observed it in the gut microbiome of mice17. While the post-natal decrease in digestive diversity might reflect the selective effect of milk on the gut and oral microbiome, the initially higher diversity could be explained by in utero colonization of the neonate18,19. The skin of newborns, on the other hand, was lowest at birth and gradually increased during the first month of life (Supplementary Fig. 8c).
Since C-section delivered infants were exposed to vaginal fluids through the use of sterile gauzes, we determined how similar the microbiome of the gauzes was to that of samples obtained from maternal body sites at day 1. We confirmed that the microbiota of the gauzes incubated in the maternal vagina were closest to the vaginal samples (Fig. 2a), with both enriched in Lactobacillus iners (Supplementary Fig. 9 and Supplementary Methods). Distance of each gauze sample to their own maternal vaginal sample was smaller than to those of other mothers, although these differences were not significant (Supplementary Fig. 10). Power analysis estimated that it would require 35 samples per group to detect the observed effect size (Cohen’s d = 0.68) with a power of 80% and a significance level of 0.05. UniFrac distances from the gauzes to vaginal samples were significantly smaller than from the gauzes to other body sites (ANOVA, P < 0.01, Fig. 2b) and bacterial sourcetracking further confirmed that the microbiota of the gauzes is mostly of vaginal origin (Fig. 2c).
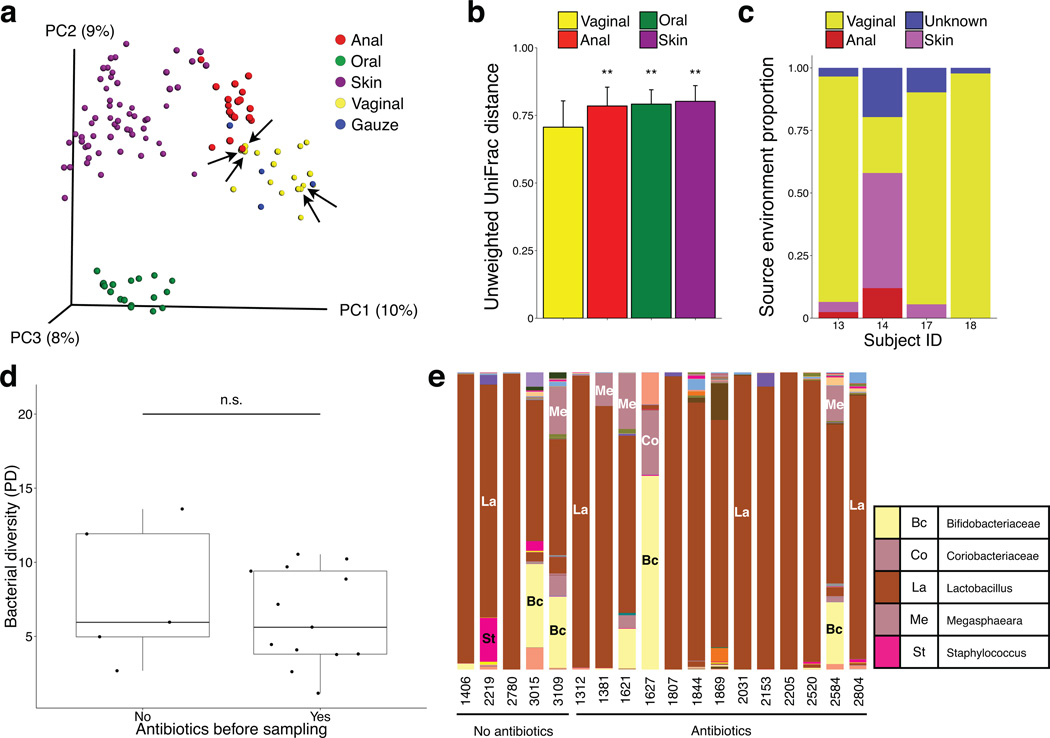
Figure 2. Transmission of maternal vaginal microbes to the gauze.
(a) Principal coordinate analysis of unweighted UniFrac community distances for maternal anal, oral, skin, and vaginal microbiota (n = 95 samples) and gauze (n = 4) microbiota at day one. Vaginal gauze bacteria resemble vaginal communities. Arrows indicate vaginal samples from mothers exposed to antibiotics. (b) Bacterial community distances between gauzes and each maternal body site at day one. Bars indicate standard deviation from the mean. (c) Proportion of gauze samples estimated to originate from different maternal sources using bacterial sourcetracking. Each stacked bar represents a gauze sample from a different mother. Oral samples were not found to be a potential source for any gauze and are not indicated in the legend. (d) Bacterial diversity (Faith’s phylogenetic diversity) of maternal vaginal microbiota in mothers that received (n = 13) or did not receive (n = 5) antibiotics prior to vaginal sampling before delivery. (e) Relative abundance of bacterial genera in the vaginal microbiota in mothers that received (n = 13) or did not receive (n = 5) antibiotics prior to vaginal sampling before delivery.
Although our sample size is limited and sampling extends only through the first month after birth, our results suggest that, by exposing the infant to the maternal vaginal microbiota, the bacterial communities of newborns delivered by C-section can be partially restored to resemble that of vaginally delivered babies. The partial microbiota restoration provided by the gauze might be due to the compounded effects of the antibiotic treatments accompanying C-section and suboptimal bacterial transfer from the vagina to the gauze and then to the baby. However, there was no apparent clustering of the vaginal microbiota based on exposure to antibiotics (Fig. 2a, arrows indicate mothers unexposed to antibiotics). Bacterial diversity was not lower in the vaginal microbiota of mothers who received antibiotics (Fig. 2d), and no clear differences were observed in taxonomic composition either (Fig. 2e). The abundance of Lactobacillus was not depleted in the vaginal microbiota of antibiotic-exposed mothers compared to that of unexposed mothers (Student’s t-test, P = 0.618). Both exposed and unexposed C-section infants were comparable in terms of antibiotic exposure and feeding (Supplementary Table 1), suggesting the differences observed in microbiome composition between these two groups can be most parsimoniously explained by the exposure to the vaginal gauze. A study of larger sample size will be needed to clarify the effect of perinatal antibiotics on the vaginal microbiome and, subsequently, on the microbiome of the gauze incubated in the maternal vagina and the bacterial load received by the infant. Determining more effective approaches to transfer the maternal microbiota to newborns or, more importantly, establishing which keystone species newborn infants should acquire at birth, will be important to maximize the beneficial effects provided by vaginal delivery in C-section infants. The partial microbial restoration observed in our study could be due to the fact that infants are exposed a single time to topical application of vaginal fluids. Furthermore, particular body sites (mouth, skin) were more amenable to inoculation than others. A modified protocol that exposed newborns repeatedly to the gauze would more faithfully reproduce the extended exposure that vaginally delivered infants receive, and might potentially improve microbial restoration, although this hypothesis remains to be tested. Enteral administration of key bacterial species could further supplement the method here described, however extensive research would be required to ensure the safety and efficacy of such approach. We stress that our work represents a proof of principle on a small cohort and with limited follow up in time. Labor is a complex process that cannot be fully recaptured by our procedure, and that encompasses multiple factors beyond the mere transmission of microbes from mother to infant. Finally, extended longitudinal analysis of larger cohorts will be needed to determine if this procedure has any effects on diseases later in life.
Online methods
Study design
The inclusion criteria for mothers participating in this study were: healthy mothers as assessed by their doctors who were delivering vaginally or by scheduled C-section. Mothers scheduled to have a C-section were offered to participate in the study, and were divided into the two groups based on their willingness to have their newborns swabbed with the gauze. For the group of C-section delivered infants exposed to maternal vaginal fluids, mothers had to have negative results for the standard of care tests of STDs, including HIV, Chlamydia and Group B Streptococcus (GBS, standard test at 36 weeks by culturing), no signs of vaginosis or viral infections as determined by their obstetrician, and a vaginal pH<4.5 at 1–2 h preceding the procedure. Of the 14 mothers whose infants were not exposed to the gauze (7 vaginal and 7 C-section delivery), three were GBS positive, and out of which two were delivered by C-section and one was vaginal delivery (Supplementary Table 1). All mothers received standard of care treatment, including preventive perinatal antibiotics (Beta lactams: mostly Cephalosporins or Penicillin. See Supplementary Table 1 for details) for mothers who underwent Cesarean section or for vaginally delivering GBS positive mothers. The study was approved by the Institutional Review Boards from University of Puerto Rico Medical Science (A9710112) and from Rio Piedras (1011-107) campus. All C-sections in this study were due to previous C-sections, and the procedure was conducted at the University Hospital, University of Puerto Rico Medical Science campus. Written informed consent was obtained from all participants.
Microbial restoration procedure
Within the hour previous to the procedure, maternal vaginal pH was measured using a sterile swab and a paper pH strip (Fisher). Once the pH was confirmed to be <4.5, a 8×8 cm four layered gauze (Fisherbrand Cat # 22028558) was folded like a fan, and then in half, wet with sterile saline solution and inserted in the vagina for one hour. Right before the C-section surgery started the gauze was extracted and placed in a sterile collector and kept at room temperature. As soon as the baby was brought to the neonate lamp and within 1 minute after delivery, the infant was swabbed with the gauze, starting on the lips, followed by the face, thorax, arms, legs, genitals and anal region, and finally the back. The swabbing took approximately 15 seconds. The neonatologist then proceeded to perform the standard detailed examination of the newborn.
Sample collection and processing
Sampling with sterile swabs in different body sites took place within the first 5 minutes after birth in all babies (including the vaginal gauze exposed C-section group, who were sampled after the gauze swabbing procedure), then at day 3, and weekly for the first month. Sampled body sites included oral mucosa, forehead, right arm, right foot and anal region of the baby, and the same sites of the mother plus a vaginal swab (Supplementary Table 2). Gauze samples were obtained from one cm2 from the center of the gauze. All samples were transported to the laboratory with ice packs within two hours of collection, and stored at −80C until further processing. DNA was extracted from samples using the MoBio Powersoil Kit following manufacturers instructions modified as described in the Earth Microbiome Project protocol (http://www.earthmicrobiome.org/emp-standard-protocols/dna-extraction-protocol/).
Sequencing and data processing
Sequencing of the swabs and gauzes was performed at NYU Genome Technology Center using the Illumina MiSeq sequencing instrument, with v2 reagents and 2 × 250 cartridge. Raw reads were de-multiplexed and quality filtered using QIIME v1.8.0 with default parameters20. Quality-filtered reads were clustered into Operational Taxonomic Units (OTUs) using an open-reference algorithm algorithm21 and Greengenes v13_8 as a reference set22. Samples that had at least 1,000 sequences (n=1,016) were further analyzed, resulting in a total of 6,515,724 sequences (mean 6,413±4,593; median 5,360 sequences). Alpha diversity on rarefied tables was estimated using Faith’s phylogenetic diversity23, and beta diversity using unweighted UniFrac24.
Bacterial sourcetracking
To estimate the sources of the microbial communities observed in each of the three infant groups at different body sites and time points, we used SourceTracker (v1.0), a Bayesian approach for bacterial sourcetracking13. Samples from each body site in the infants were designated as sinks, and samples from all body sites of the corresponding mother were tagged as sources. Sourcetracking of bacterial communities in the gauzes were performed similarly, designating gauzes as sinks and paired maternal body sites as potential sources.
Supervised learning classification
A Random Forest classifier was built for each body site using 500 trees and a leave-one-out error model as previously described14. To account for sub-sampling variability, the input tables were rarefied 10 times and results averaged over the total. The confusion matrices represent the mean and standard deviation percentage of samples from the true class assigned to each of the possible classes (vaginal, exposed, C-section) for each body site.
Predicted metagenome
OTU tables were filtered to remove de novo OTUs (i.e. not found in Greengenes). Metagenomic content was estimated using PICRUSt25 first normalizing each OTU in the filtered tables by its corresponding copy number and then predicting abundance of functional traits from the normalized OTU counts.
Deblurring
In order to identify with higher resolution the Lactobacillus present in the samples we used deblurring, a novel denoising method for Illumina based amplicon sequencing (manuscript in preparation). Using the raw reads as input, deblurring processes each sample independently and tries to remove all sequences derived from sequencing read errors or PCR errors, based on an upper bound on error probabilities. Briefly, the deblurring algorithm steps and parameters used for the current study are:
Trim the all reads to constant length (150bp) and de-replicate, retaining the total number of reads per unique sequence.
Subsample to 4,000 reads (per sample).
Discard all singleton reads.
Remove sequences containing known sequencing artifacts (PhiS or adapter sequences)
Perform multiple sequence alignment using MAFFT v7.130b26.
- Perform the actual deblurring: Iterate on all sequences from highest frequence to lowest.
- For each sequence, reduce the frequency of neighboring sequences (based on hamming distance + indel) according to the distance dependent maximal error profile. The error profile used ranges from 6% maximal read error for hamming distance 1, 2% for distance 2, down to 0.1% for hamming distance 10.
- If the resulting frequency is lower than 0, remove the sequence from the list.
Remove chimeras from the resulting sequences using de-novo chimera detection with usearch 5.2.236 and the parameter –uchime_denovo
The performance of deblurring has been validated in simulations and mock mixtures, and was shown to retain the exact actual sequences in the sample while removing most of the sequencing/PCR error derived sequences, with the ability to detect sequences differing by as little as 1 nucleotide over the entire sequenced region. Additional details on deblurring and the source code can be found at https://github.com/biocore/deblur
Statistical analyses
All technicians processing the samples were blinded to the group allocations. Computational analyses were not blinded due to the use of supervised learning methods, which require knowledge of the groups. Since our study was designed as a proof-of-concept, sample size was not estimated a priori. Statistical tests were performed using QIIME 1.8.0 with default parameters and R 3.2.2.
Supplementary Material
Acknowledgments
This work was partially supported by the C&D Research Fund (M.G.D.B.), U.S. National Institutes of Health R01 DK090989 (M.G.D.B), the Crohn’s and Colitis Foundation of America grant #362048 (J.C.C.), and the Sinai Ulcerative Colitis: Clinical, Experimental & Systems Studies philanthropic grant (J.C.C.). Sequencing at the New York University Genome Technology Center was partially supported by the Cancer Center Support Grant, P30CA016087, at the Laura and Isaac Perlmutter Cancer Center. Computing was partially supported by the Department of Scientific Computing at the Icahn School of Medicine at Mount Sinai. We acknowledge the contribution of students who participated in obtaining the samples and the metadata: S.M. Rodriguez, J.F. Ruiz, N. Garcia, and J.L. Rivera Correa. We also thank M.J. Blaser for inspiring discussions and critical comments, and three anonymous reviewers for their suggestions to improve this manuscript
Footnotes
Accession codes
Sequence data have been deposited to EBI under study accession number ERP012216 and submission accession number ERA486171.
Author contributions
M.G.D.B. designed the study. M.G.D.B., K.M.D.J.L., J.I.R.V., and K.M. collected and processed specimens. M.G.D.B. sequenced and generated data. N.S., L.M.C., A.A., A.G., N.A.B., S.J.S., M.H., and J.C.C. performed experiments. M.G.D.B., N.S., L.M.C., A.A., A.G., N.A.B., S.J.S., M.H., R.K., and J.C.C. analyzed data. M.G.D.B. and J.C.C. drafted the manuscript. All authors reviewed the final manuscript.
Competing financial interests
New York University has filed an U.S. patent application (number 62161549) on behalf of M.G.D.B., related to methods for restoring the microbiota of newborns.
References
- 1.Dominguez-Bello MG, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Olszak T, et al. Microbial exposure during early life has persistent effects on natural killer T cell function. Science. 2012;336:489–493. doi: 10.1126/science.1219328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox LM, et al. Altering the intestinal microbiota during a critical developmental window has lasting metabolic consequences. Cell. 2014;158:705–721. doi: 10.1016/j.cell.2014.05.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thavagnanam S, Fleming J, Bromley A, Shields MD, Cardwell CR. A meta-analysis of the association between Caesarean section and childhood asthma. Clinical and experimental allergy : journal of the British Society for Allergy and Clinical Immunology. 2008;38:629–633. doi: 10.1111/j.1365-2222.2007.02780.x. [DOI] [PubMed] [Google Scholar]
- 5.Pistiner M, Gold DR, Abdulkerim H, Hoffman E, Celedon JC. Birth by cesarean section, allergic rhinitis, and allergic sensitization among children with a parental history of atopy. J Allergy Clin Immunol. 2008;122:274–279. doi: 10.1016/j.jaci.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huh SY, et al. Delivery by caesarean section and risk of obesity in preschool age children: a prospective cohort study. Archives of disease in childhood. 2012;97:610–616. doi: 10.1136/archdischild-2011-301141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sevelsted A, Stokholm J, Bonnelykke K, Bisgaard H. Cesarean section and chronic immune disorders. Pediatrics. 2015;135:e92–e98. doi: 10.1542/peds.2014-0596. [DOI] [PubMed] [Google Scholar]
- 8.Gibbons L, et al. The Global Numbers and Costs of Additionally Needed and Unnecessary Caesarean Sections Performed per Year: Overuse as a Barrier to Universal Coverage. World Health Report. 2010;30 [Google Scholar]
- 9.Finger C. Caesarean section rates skyrocket in Brazil. Many women are opting for caesareans in the belief that it is a practical solution. Lancet. 2003;362:628. doi: 10.1016/s0140-6736(03)14204-3. [DOI] [PubMed] [Google Scholar]
- 10.Barber EL, et al. Indications contributing to the increasing cesarean delivery rate. Obstetrics and gynecology. 2011;118:29–38. doi: 10.1097/AOG.0b013e31821e5f65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Appropriate technology for birth. Lancet. 1985;2:436–437. [PubMed] [Google Scholar]
- 12.Clemente JC, et al. The microbiome of uncontacted Amerindians. Science Advances. 2015;1 doi: 10.1126/sciadv.1500183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knights D, et al. Bayesian community-wide culture-independent microbial source tracking. Nat Methods. 2011;8:761–763. doi: 10.1038/nmeth.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knights D, Parfrey LW, Zaneveld J, Lozupone C, Knight R. Human-associated microbial signatures: examining their predictive value. Cell Host Microbe. 2011;10:292–296. doi: 10.1016/j.chom.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Backhed F, et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe. 2015;17:690–703. doi: 10.1016/j.chom.2015.04.004. [DOI] [PubMed] [Google Scholar]
- 16.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pantoja-Feliciano IG, et al. Biphasic assembly of the murine intestinal microbiota during early development. Isme J. 2013;7:1112–1115. doi: 10.1038/ismej.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fardini Y, Chung P, Dumm R, Joshi N, Han YW. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infection and immunity. 2010;78:1789–1796. doi: 10.1128/IAI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aagaard K, et al. The placenta harbors a unique microbiome. Science translational medicine. 2014;6:237ra265. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caporaso JG, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rideout JR, et al. Subsampled open-reference clustering creates consistent, comprehensive OTU definitions and scales to billions of sequences. Peer J. 2014;2:e545. doi: 10.7717/peerj.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. Isme J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faith DP, Baker AM. Phylogenetic diversity (PD) and biodiversity conservation: some bioinformatics challenges. Evolutionary bioinformatics online. 2006;2:121–128. [PMC free article] [PubMed] [Google Scholar]
- 24.Lozupone C, Knight R. UniFrac: a new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–8235. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Langille MG, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular biology and evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.