Abstract
Angiogenesis is recognized as an important hallmark of cancer. Although telomerase is thought to be involved in tumor angiogenesis, the evidence and underlying mechanism remain elusive. Here, we demonstrate that human telomerase reverse transcriptase (hTERT) activates vascular epithelial growth factor (VEGF) gene expression through interactions with the VEGF promoter and the transcription factor Sp1. hTERT binds to Sp1 in vitro and in vivo and stimulates angiogenesis in a manner dependent on Sp1. Deletion of the mTert gene in the first generation of Tert null mice compromised tumor growth, with reduced VEGF expression. In addition, we show that hTERT expression levels are positively correlated with those of VEGF in human gastric tumor samples. Together, our results demonstrate that hTERT facilitates tumor angiogenesis by up-regulating VEGF expression through direct interactions with the VEGF gene and the Sp1 transcription factor. These results provide novel insights into hTERT function in tumor progression in addition to its role in telomere maintenance.
INTRODUCTION
Human telomerase is a ribonucleoprotein enzyme complex that is minimally composed of an RNA template (hTR or hTERC) and the telomerase reverse transcriptase (hTERT) for telomeric DNA synthesis (1). Most human somatic cells have undetectable telomerase activity due to transcriptional repression or alterations in hTERT splicing during fetal development, but telomerase is either up-regulated or reactivated during tumorigenesis in over 90% of human primary tumors (2–4), as well in as murine tumors, even though mouse telomeres are significantly longer compared to humans (5,6). Thus, it remains unclear why telomerase would be up-regulated in mouse tumors if telomeres are not rate limiting. Telomerase plays a pivotal role in the pathology of cancer by maintaining the ends of chromosomes and providing for the unlimited cell proliferation potential required of almost all advanced cancers (7–10). However, emerging evidence indicates that the telomerase reverse transcriptase protein may have non-telomeric functions in tumor devolvement, although the underlying mechanisms remain elusive (10–13). Many published studies can be interpreted differently. For example, telomerase may exert telomere-independent functions in a number of fundamental cellular processes that promote tumorigenesis, such as the regulation of cell growth and proliferation (14,15), EMT (16), stem cells (17,18) and oncogenesis (19–21). However, the direct connection and the underlying mechanism(s) involved in the telomere-independent functions of telomerase in cancer progression are not clear at present. The inhibition of telomerase in vascular endothelial cells has been reported to disrupt tumor angiogenesis in glioblastoma xenografts (22). In addition, suppression of hTERT correlates with decreases in angiogenesis and tumor growth (23). These observations suggest that telomerase may be involved in angiogenesis in brain gliomas, but again, the mechanisms involved are unknown.
Vascular endothelial growth factor (VEGF) is the most potent endothelial mitogen and a critical component of tumor growth, angiogenesis and metastasis (24–28). Previously, we found that hTERT induces the expression of VEGF (29). However, the underlying regulatory mechanism and the biological significance of VEGF up-regulation are not known. In the present study, we demonstrate that hTERT plays a telomere-independent role in angiogenesis by up-regulating VEGF expression through its interaction with the transcription factor Sp1. Our results provide new possibilities for cancer therapeutic strategies targeted to both telomerase and VEGF, which would be effective against both neoplastic cell proliferation through telomere attrition and tumor angiogenesis by inhibiting VEGF.
MATERIALS AND METHODS
Cell culture, transfection and antibodies
HeLa and 293T cell lines were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) at 37°C and 5% CO2. Human umbilical vein endothelial cells (HUVECs) were cultured in endothelial cell medium (Invitrogen, Carlsbad, CA, USA) with 5% FBS at 37°C and 5% CO2. Transfections were performed with Lipofectamine 2000. β-actin antibody was purchased from Cell Signaling (Beverly, MA, USA); hTERT, VEGF, CD31, Ki67 and Sp1 antibodies were purchased from Abcam (Cambridge, UK); IgG antibody and protein A/G beads were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA); and His and Flag antibodies and anti-Flag resin were purchased from Sigma (St. Louis, MO, USA).
Plasmid constructs and siRNAs
Flag-tagged hTERT, Flag-tagged Sp1 expression plasmids and the Flag-tagged deletion mutants were obtained by polymerase chain reaction (PCR) amplification and were subsequently cloned into the p3xFlag-CMV 10 vector. The GST-Sp1 expression plasmid was constructed by cloning the full-length Sp1 cDNA into the pGEX-6p-1 vector. The luciferase reporter pSp1-luc was constructed by cloning the sequence 5′-ATTCGATCGGGGCGGGGCGAGATTAGATTCGATCGGGGCGGGGCGAG-3′ into the pGL6-TA vector. pVEGF1-5 was constructed by cloning the different sequences of the human VEGF promoter into the pGL2-Basic vector (29). pVEGF4-Sp1 mut was constructed by cloning the VEGF promoter containing three mutant Sp1 binding sites into the pGL2-Basic vector. hTERT siRNA was purchased from Thermo Scientific (L-003547-00-0020, ON-TARGET plus SMART pool, Human TERT; Waltham, MA, USA). Control siRNA (siNC) and Sp1 siRNAs were purchased from GenePharma (Shanghai, China). The sequences of the three Sp1 siRNA (mix) are as follows: 5′-CCAGCAACAUGGGAAUUAUTT-3′, 5′-GUGCAAACCAACAGAUUAUTT-3′ and 5′-CCUGGAGUGAUGCCUAAUATT-3′.
Quantitative real-time PCR (qRT-PCR)
Trizol reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA, followed by cDNA preparation with M-MLV reverse transcriptase (Promega, Madison, WI, USA) according to the manufacturer's protocol. Real-time PCR (RT-PCR) reactions were performed in triplicate with SYBR Green Supermix (Bio-Rad, Hercules, CA, USA). GAPDH was measured as an internal control. Three independent experiments were performed. The sequences of the primers used for RT-PCR are described in the Supplementary Information, Table S1.
Chromatin immunoprecipitation (ChIP)
Chromatin immunoprecipitation (ChIP) assays were performed with a Chromatin Immunoprecipitation Kit (Millipore, Billerica, MA, USA). The Flag antibody and Sp1 antibody were used to precipitate DNA fragments. IgG was used as the negative control. The protein–DNA complexes were collected with protein G. The primers used to amplify the VEGF promoter were 5′-GAGCTTCCCCTTCATTGCGG-3′ and 5′-CGGCTGCCCCAAGCCTC-3′, and the primers for the GAPDH promoter were 5′-TACTAGCGGTTTTACGGGCG-3′ and 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′. The enrichment of the VEGF promoter was determined by PCR. The GAPDH promoter was used as the negative control.
Immunoprecipitation (IP) and Western blotting
Transfected cells were lysed in lysis buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 10% glycerol, 1 mM EDTA, 1% NP-40 and a cocktail of proteinase inhibitors). For immunoprecipitation (IP), the cell lysates were cleared using centrifugation and incubated with proteinA/G beads (Santa Cruz, CA, USA) or M2 anti-Flag resin (Sigma, St. Louis, MO, USA) for 2–3 h. The beads were boiled after extensive washing, the proteins were resolved using SDS-PAGE gel electrophoreses and the proteins were transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) followed by Western blotting. The proteins were detected using the VersaDoc Imaging System (Bio-Rad), and quantification was performed using Quantity One 1-D Analysis Software.
In vitro pull-down assay
GST-Sp1 fusion proteins were expressed in Escherichia coli BL21 and were purified with glutathione–Sepharose. His-hTERT fusion proteins were expressed in 293T cells and were purified with Ni-NTA agarose. In vitro pull-down assays were performed by incubating equal amounts of GST or GST-Sp1 fusion proteins immobilized onto glutathione–Sepharose beads with His-hTERT. The mixture was placed on a rocking platform for 2 h in binding buffer (20 mM Tris-HCl, pH 7.5, 1.5 mM MgCl2, 100 mM NaCl, 0.05% NP-40) and then washed three times. Bound proteins were detected by immunoblotting with anti-His antibodies.
Electrophoretic mobility shift assay (EMSA)
Nuclear extracts from HeLa cells were prepared with a Nuclear Extract Kit (Active Motif, Carlsbad, CA, USA) as previously described (30). The sequences of double-stranded oligonucleotides used as probes labeled with biotin in the electrophoretic mobility shift assay (EMSA) were as follows: synthetic consensus Sp1 probe, 5′-ATTCGATCGGGGCGGGGCGAGC-3′ and VEGF probe (–89 to –50 bp of the human VEGF promoter), 5′-CCCGGGGCGGGCCGGGGGCGGGGTCCCGGCGGGGCGGAG-3′. The sequence of cold unlabeled double-stranded DNA used as a competitor was 5′-ATTCGATCGGGGCGGGGCGAGC-3′. For competition experiments, the nuclear extract was incubated with a 100 times higher concentration of unlabeled DNA probe compared with biotin-labeled DNA probe in binding buffer for 15 min and was then incubated with the biotin-labeled DNA probe for 20 min at room temperature. For supershift experiments, the nuclear extract was incubated with 1 μg of antibody against Sp1 or hTERT in binding buffer for 15 min and then incubated with the biotin-labeled DNA probe for 20 min at room temperature. Samples were subjected to native 4% polyacrylamide gel electrophoresis (PAGE) and then transferred to nylon membranes. The membranes were incubated with HRP-streptavidin and then visualized with a VersaDoc Imaging System (Bio-Rad).
Endothelial cell capillary-like tube formation assay
The endothelial cell capillary-like tube formation assay was performed as previously described (31). Briefly, matrigel (BD Biosciences, Franklin Lakes, NJ, USA) was pipetted into pre-chilled 96-well plates and polymerized at 37°C. The HUVECs were collected and placed onto the Matrigel layer. After 6–8 h, the endothelial cells were photographed using an inverted microscope. The tube network was quantified using ImageJ software.
Lewis lung carcinoma tumor formation
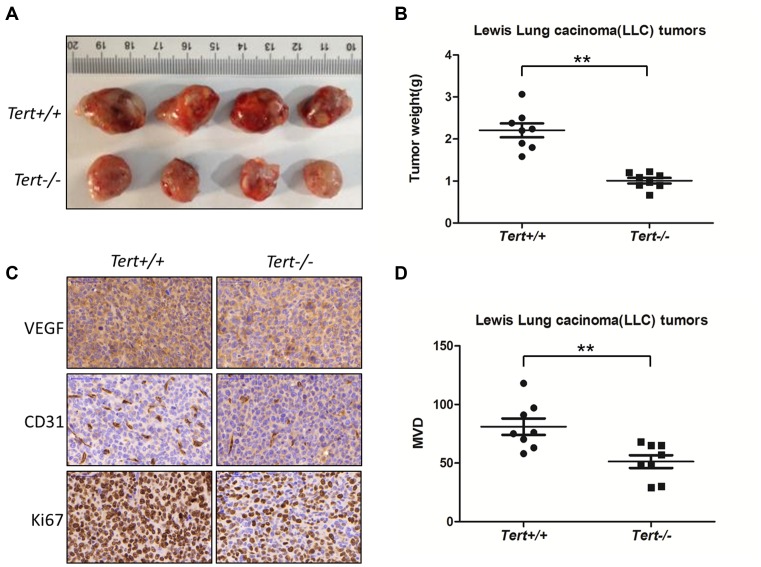
Eight-week-old wild-type and Tert knockout mice (C57BL/6) received subcutaneous implantations of 1 × 106 Lewis lung carcinoma cells in the flank as described (32). Twenty one days after inoculation, all mice were euthanized. Eight wild-type and eight Tert KO mice were examined. The tumor masses were excised, weighed, photographed and subjected to immunohistochemistry using VEGF, CD31 and Ki67 antibodies (Abcam, USA).
Immunohistochemistry
Immunohistochemical staining was performed by Core Facilities, Zhejiang University School of Medicine. Tumor sections were immunostained with specific anti-VEGF, anti-Ki67 and anti-CD31 antibodies. The images were captured using a Pannoramic MIDI scanner.
Quantification of microvessel density
Tumor sections were immunostained with a specific anti-CD31 antibody for blood vessels. Tumor microvessel density (MVD) was determined using Pannoramic Viewer software as described (33). Briefly, the area of most intense neovascularization was selected by Pannoramic Viewer software at low magnification (10–100x). Individual microvessels were counted at 200x magnification (0.152 mm2/field). For each section, five areas were selected from the vascularity of tumor areas. Any brown-stained, nucleus-containing endothelial cell that was clearly separated from adjacent larger vessels, tumor cells and other connective tissue elements was considered a single CD31-positive tumor-associated microvessel. Sixteen micrographs were obtained from multiple tumor sections. Each dot represents the average of several vessels found in a micrograph. Eight wild-type and eight Tert KO mice were examined.
Animal studies
Tert+/− mice on the C57BL/6/129/SvJ genetic background were kindly provided by Dr Yie Liu (NIA, Baltimore, MD, USA). Heterozygous (Tert+/−) pairs were intercrossed to generate first-generation Tert+/+ (WT) and Tert−/− (KO) strains. The Animal Care and Ethics Committee at Hangzhou Normal University approved all animal experiments in our study. All mice were maintained in a pathogen-free environment and fed a standard diet.
Primary tissue samples
Primary gastric tumor tissues were obtained from Sir Run Run Shaw Hospital after patients signed informed consent forms, and the studies were approved by the Medical Ethics Committee of the Medical School, Zhejiang University, China.
Statistical analysis
All data for statistical analysis are presented as the mean ± S.D. Student's t-test was used to determine statistical significance. Correlation analyses between VEGF, CD31 and hTERT expression in gastric cancer samples were performed using linear regression. Statistical analyses were performed with Prism 5.0 software.
RESULTS
hTERT increases the expression of VEGF in HeLa cells via Sp1 activity
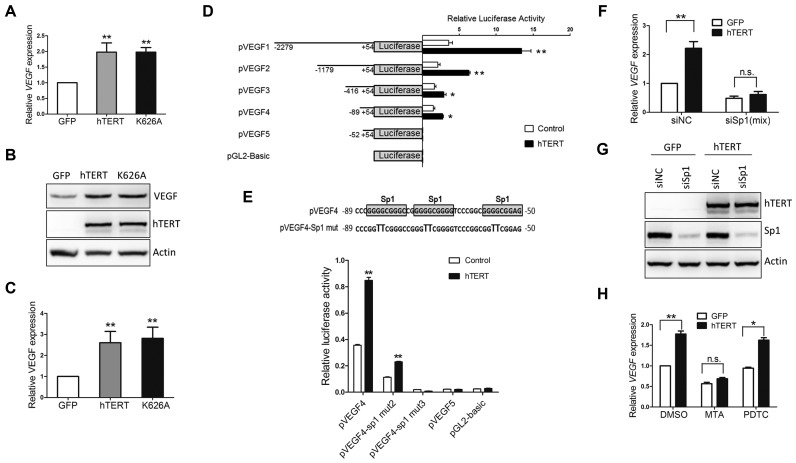
We previously reported that hTERT induces VEGF expression in human fibroblasts, WI-38 cells and HeLa cells (29). Specifically, we showed that both transient transfection of an hTERT expression construct or retroviral transduction of hTERT results in the up-regulation of VEGF expression independent of hTERT telomerase activity and that knocking down hTERT, but not hTR, reduces VEGF expression (29). To further explore the regulation of VEGF expression by hTERT, we manipulated the expression of hTERT in HeLa cells. Consistent with our previous observations, ectopically expressed hTERT and a catalytically inactive hTERT K626A mutant (30) deficient in telomerase activity induced the up-regulation of VEGF mRNA (Figure 1A) and VEGF protein expression (Figure 1B and C) independent of telomerase activity. To investigate whether hTERT regulates VEGF expression at the transcriptional level, a set of VEGF promoter luciferase constructs was co-transfected into HeLa cells with an hTERT expression vector (pcDNA-hTERT) or control vector (pcDNA3). Promoter luciferase assays showed that hTERT increased VEGF promoter activity (Figure 1D), suggesting that hTERT transcriptionally up-regulated VEGF expression. Interestingly, reporter luciferase assays showed that the sequences responding to hTERT-induced promoter activity are within −89 to −54 nt of the VEGF promoter. This suggested, but did not prove, that the sequences from −89 to −54 nt in the VEGF promoter may be responsible for hTERT-induced VEGF promoter activity (Figure 1D). Sequence analysis revealed three Sp1 transcription factor-binding sites in this region (Figure 1E). Mutations in either two or all three of the Sp1 binding sites deprived hTERT-induced VEGF promoter activity, due to loss of the basal activity of the VEGF promoter (Figure 1E). This indicates that the Sp1 is essential for the VEGF promoter activity. Furthermore, in HeLa cells that stably expressed either GFP (HeLa-GFP) or hTERT (HeLa-hTERT), depletion of Sp1 by a mix of three Sp1 siRNAs (siSp1), but not control siRNA (siNC), abrogated the hTERT-induced VEGF mRNA expression (Figure 1F and G). In addition, Mithramycin A (MTA), a chemical that binds GC-rich promoters and has shown some specificity for Sp1 binding sites, blocked hTERT-induced VEGF mRNA expression. But no such inhibition was observed with an NF-κB inhibitor Pyrrolidine dithiocarbamate (PDTC) (Figure 1H). Together, these results can be interpreted to suggest that hTERT increases the expression of VEGF through Sp1 activity.
Figure 1.
hTERT up-regulates VEGF expression in HeLa cells through Sp1 activity. (A) HeLa cells were transduced with lentiviruses expressing hTERT, K626A and GFP as a control. At 48 h after infection, VEGF mRNA levels were quantified by qPCR. (B) The protein levels of VEGF, hTERT and Actin were determined by immunoblotting. (C) Quantitative expression of VEGF from three independent experiments was analyzed using Quantity One 1-D Analysis Software and normalized to Actin levels. (D and E) HeLa cells were transiently transfected with pcDNA3-hTERT (hTERT) and pcDNA3 as a control, along with luciferase reporter plasmids containing different regions of the VEGF promoter or mutations in two or all three Sp1 binding sites. Luciferase activity was determined at 48 h after transfection. (F and G) Knockdown of Sp1 decreased the endogenous levels of VEGF mRNA. HeLa-GFP and HeLa-hTERT stable cells were transfected with control siRNA (siNC) or siRNA (mix) against Sp1. The mRNA levels of VEGF were analyzed by qPCR, and the levels of hTERT and Sp1 protein expression were determined by Western blot analysis using hTERT and Sp1 antibodies. (H) HeLa-GFP and HeLa-hTERT stable cells were treated with MTA (100 nM), PDTC (100 μM) or DMSO for an additional 24 h. VEGF mRNA levels were quantified by qPCR. The data are presented as the mean ± S.D. (Student's t-test). *P < 0.05; **P < 0.01 (n = 3); n.s., no significance.
hTERT binds to the VEGF promoter and activates Sp1-mediated transcription of the VEGF gene
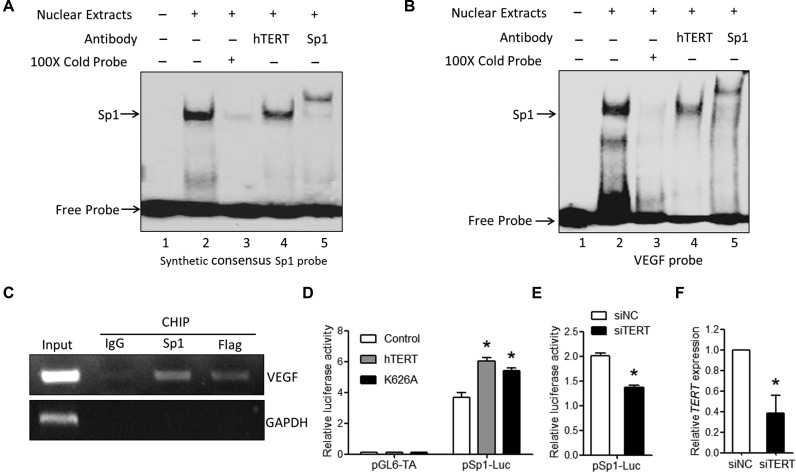
To understand whether hTERT is directly involved in VEGF transcriptional regulation, we preformed EMSA and supershift assays with HeLa nuclear extracts using synthetic consensus Sp1 or the VEGF probe containing three Sp1 binding sites (−89 to −50 nt of the human VEGF promoter). As shown in Figure 2A and B, the Sp1 binding shift was readily detectable after incubation of the biotin-labeled synthetic consensus Sp1 or the VEGF probe with the HeLa nuclear extracts (Figure 2A, line 2; Figure 2B, line 2); the addition of excess unlabeled probe resulted in the ablation of this binding (Figure 2A, line 3; Figure 2B, line 3). In addition, incubation with the Sp1 antibody resulted in a supershift of the Sp1 binding, whereas incubation with the hTERT antibody resulted in decreased Sp1 binding. This EMSA profile was very similar to those observed in studies investigating the interaction between hTERT and the NF-κB p65 transcription factor on the IL-6 promoter, for which the addition of hTERT antibodies significantly disrupted the NF-κB complexes (34). In addition, ChIP analysis further confirmed that hTERT can be recruited to the VEGF promoter (Figure 2C). Consistently, we showed that transient transfection with hTERT or mutant hTERT K626A expression constructs activated the Sp1 luciferase reporter (Figure 2D), whereas hTERT knockdown with siRNAs resulted in a reduction in Sp1 luciferase reporter activity (Figure 2E and F). Taken together, these results support the interpretation that hTERT may interact with Sp1 to enhance Sp1 transcriptional activity and thereby activate VEGF transcription independent of TERT enzymatic and telomere-based activity.
Figure 2.
hTERT binds to the VEGF promoter. (A and B) HeLa cell nuclear extracts were prepared 48 h after cells were transfected with hTERT expression vectors. EMSA supershift analyses using the (A) synthetic consensus Sp1 probe or the VEGF probe containing (B) three Sp1 binding sites were performed via the addition of specific hTERT or Sp1 antibodies. (C) HeLa cells were transfected with vectors expressing Flag-hTERT, and ChIP assays were performed using the Flag antibody and Sp1 antibody. IgG was used as the negative control. The signal enrichment with each antibody was measured by PCR using primers specific for VEGF or GAPDH as the negative control. The data shown are representative of two independent experiments. (D) HeLa cells were transiently transfected with the indicated vectors, along with the luciferase reporter pSp1-luc or pLG6-TA as a control. (E) siTERT or control siRNA was transfected into HeLa cells, along with pSp1-luc. Luciferase activity was determined 48 h after transfection. (F) The knockdown efficiency of hTERT expression by hTERT siRNAs was evaluated by qPCR analysis.
hTERT interacts with Sp1 both in vitro and in vivo
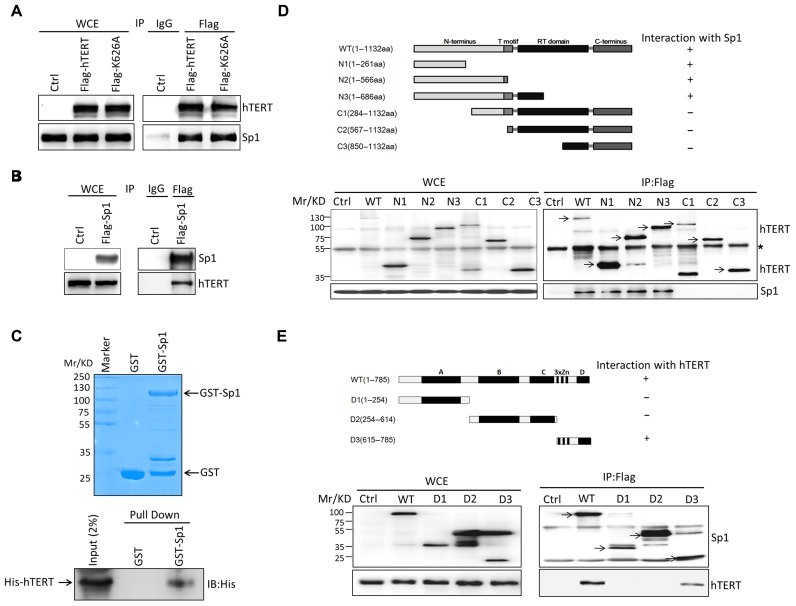
To test whether hTERT can interact with Sp1 using IP, we co-transfected the Flag-tagged hTERT or Flag-tagged hTERT K626A expression construct with the untagged Sp1 construct (pcDNA-Sp1), or co-transfected the Flag-tagged Sp1 expression construct with the untagged hTERT construct (pcDNA-hTERT), respectively. IP experiments were performed using the Flag antibody, and IgG as control. The cell lysates and immunoprecipitates were then analyzed with antibodies against hTERT or Sp1. This reciprocal IP showed an interaction of hTERT with Sp1 (Figure 3A and B). Furthermore, we have performed IP experiments with Anti-Flag M2 affinity gel. 293T cells were transiently transfected with plasmids expressing Flag-hTERT or Flag-K626A. The presence of endogenous Sp1 in immunoprecipitates with M2 anti-Flag resin was analyzed by immunoblotting using the Sp1 antibody. Both hTERT and hTERT K626A were found to co-immunoprecipitate with endogenous Sp1 (Supplementary Data S1A). In addition, Flag-tagged Sp1 was found to co-immunoprecipitate with endogenous hTERT in 293T cells (Supplemental Data S1B). This reciprocal IP indicates a physical interaction between hTERT and Sp1.
Figure 3.
hTERT interacts with Sp1. (A) 293T cells were co-transfected with the Flag-tagged hTERT or Flag-tagged hTERT K626A expression construct and the untagged Sp1 construct (pcDNA-Sp1). Transfected cells were lysed and immunoprecipitated with anti-IgG or anti-FLAG antibody and protein A/G beads, then the immunoprecipitates and the whole cell extracts (WCE) were analyzed by immunoblotting using anti-hTERT or anti-Sp1 antibody. (B) 293T cells were co-transfected with the Flag-tagged Sp1 expression construct and the untagged hTERT construct (pcDNA-hTERT). Transfected cells were lysed for immunoprecipitation and immunoblotting assay as described in (A). (C) Purified GST and GST-Sp1 were detected by Coomassie blue staining (upper panel). His-hTERT was incubated with GST or GST-Sp1. Bound proteins were detected by immunoblotting with anti-His antibodies (lower panel). (D) Sp1 interacts with the N-terminus of hTERT. Flag-tagged full-length hTERT or 6 truncation mutant constructs were transiently transfected into 293T cells. Flag-tagged full-length and mutant hTERT were immunoprecipitated with anti-Flag resin and then immunoblotted with Flag and Sp1 antibodies. (E) hTERT interacts with the C-terminus of Sp1. Flag-tagged full-length Sp1 or 3 truncation mutant constructs were transiently transfected into 293T cells. Flag-tagged full-length and mutant Sp1 were immunoprecipitated with anti-Flag resin and then immunoblotted with Flag and hTERT antibodies. *, non-specific band.
To further confirm the interaction between hTERT and Sp1, in vitro pull-down assays were performed by incubating equal amounts of E. coli-purified GST or GST-Sp1 fusion proteins immobilized onto glutathione-sepharose beads with His-hTERT that was partially purified from 293T cells (Figure 3C). Bound proteins were detected by immunoblotting with anti-His antibodies. As shown in Figure 3C (lower panel), GST-Sp1, but not GST, interacts with His-hTERT, suggesting that hTERT can directly interact with Sp1.
To identify the domains within hTERT and Sp1 required for their interaction, we mapped the regions by IP with different truncated forms of both proteins and found that the N-terminus of hTERT (amino acids 1–261) interacted with the C-terminus (amino acids 615–785) of Sp1, which contains three zinc finger motifs (Figure 3D and E). Interestingly, using the available crystal structure of the TEN domain of TERT from T. thermophila (PDB ID 2B2A) (35), we were able to perform molecular docking with the modeled Sp1-dsDNA structure to predict the interaction between TEN and Sp1 (Arg615-Phe785), which includes the Sp1 DNA-binding sequence (Supplementary Data S2A and B). The docking model shows that one of the zinc finger motifs of Sp1 and the end of the dsDNA fit cleanly into the cleft formed between the positively charged C-terminus (Val172-Asn191) and the flanking fragment (Met78-Tyr121) of the TEN domain, which is composed of one α helix and two short anti-parallel β strands (Supplementary Figure S2B and C). Overall, this model supports our experimental evidence that hTERT interacts with Sp1 on the VEGF promoter to activate VEGF expression. To functionally test whether the interaction of hTERT with Sp1 is required for the hTERT-induced up-regulation of VEGF, full-length hTERT or four truncated mutant constructs were transiently transfected into HeLa cells. VEGF mRNA levels were quantified by qPCR, and VEGF protein levels in the cell-free supernatants were evaluated by ELISA. The results showed that full-length hTERT, as well as two truncated mutant constructs containing the N-terminus of hTERT (N1 and N2), but not the C-terminus of hTERT that does not interact with Sp1 (C1 and C2), can up-regulate VEGF mRNA levels (Supplementary Figure S3A) and VEGF protein levels in the culture medium (Supplementary Figure S3B). These results indicate that hTERT transactivates expression through its interaction with Sp1.
hTERT promotes vascular tube formation of HUVECs
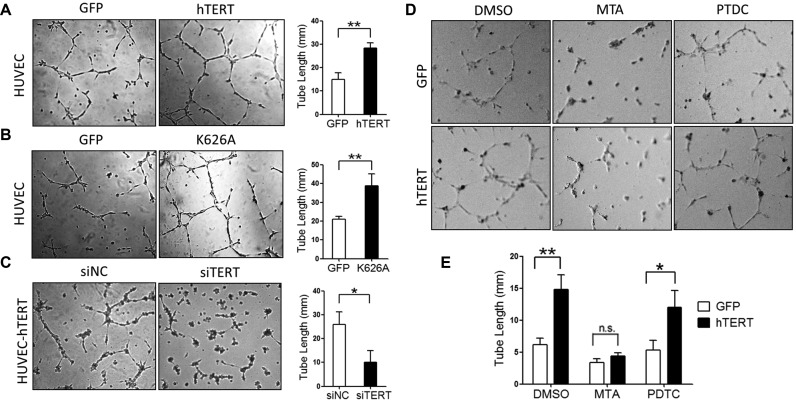
Angiogenesis, the growth of new blood vessels from pre-existing vessels, is a vital process in development, wound healing, inflammation and tumorigenesis (24,26,28,36). VEGF is the most potent endothelial mitogen and a critical component of tumor growth, angiogenesis and metastasis (24–28). To evaluate the potential role of TERT in angiogenesis, we investigated the effect of TERT on vascular endothelial tube formation in vitro. HUVECs were transduced with the indicated lentivirus (GFP, hTERT or hTERT K626A), which resulted in modest levels of hTERT expression and increased levels of VEGF mRNA (Supplementary Data S4A and B). These cells were then plated on Matrigel®, and capillary/tube-like structure formation was examined. We found that both hTERT and the catalytically inactive mutant hTERT K626A significantly promoted vascular tube formation compared with control cells (Figure 4A and B). Depletion of hTERT by hTERT siRNAs reduced VEGF mRNA expression in HUVEC-hTERT cells (Supplementary Data S4C and D) and significantly impaired vascular tube formation compared with control cells (Figure 4C). Furthermore, treatment with MTA, which blocks Sp1 activity, but not the NF-κB inhibitor PDTC inhibited vascular tube formation in hTERT-expressing HUVECs, suggesting that hTERT increased vascular tube formation in HUVECs through Sp1 activity but not through NF-κB signaling (Figure 4D and E). In addition, the effects of hTERT on 23 known angiogenesis-related genes were examined by qPCR analysis and revealed that only VEGF expression was significantly up-regulated by hTERT (Supplementary Data S5), suggesting that the up-regulation of VEGF expression critically contributes to the effect of hTERT on vascular tube formation. Endothelial cell migration is a key step in angiogenesis (26). We then determined whether hTERT affects endothelial cell migration. Our data showed that the ectopic expression of hTERT or catalytically inactive mutant hTERT K626A significantly increased HUVEC migration in a wound-healing assay (Supplementary Data S6A and B). Similar results were obtained from a transwell assay (data not shown). Taken together, these data suggest that hTERT up-regulates VEGF expression and promotes vascular tube formation and migration by HUVECs through Sp1 activity, which may contribute to angiogenesis in vivo.
Figure 4.
hTERT positively regulates vascular tube formation by HUVECs and is dependent on Sp1 activity. (A and B) HUVECs were transduced with the indicated lentivirus. Tubular structures were photographed, and tube length was calculated using ImageJ software. (C) HUVEC-hTERT stable cells were transfected with hTERT siRNAs (siTERT) or control siRNAs (siNC), and tubular structures were photographed and quantitatively analyzed as in A. (D and E) HUVECs transduced with the indicated lentiviruses were treated with MTA (100 nM) or PDTC (100 μM). Tubular structures were photographed (D) and analyzed as in A (E). The data are presented as the mean ± S.D. (Student's t-test). *P < 0.05; **P < 0.01. (n = 3). n.s., no significance.
TERT deficiency compromises tumorigenesis and tumor vascular development
To test whether TERT deficiency may affect ectopic tumor growth in tumor xenograft assays, we utilized the Lewis lung carcinoma (LLC) tumor development model (32) in first-generation (G1) Tert wild-type and Tert knockout mice, which have no detectable telomere dysfunction phenotype (37), to avoid effects associated with telomere shortening.
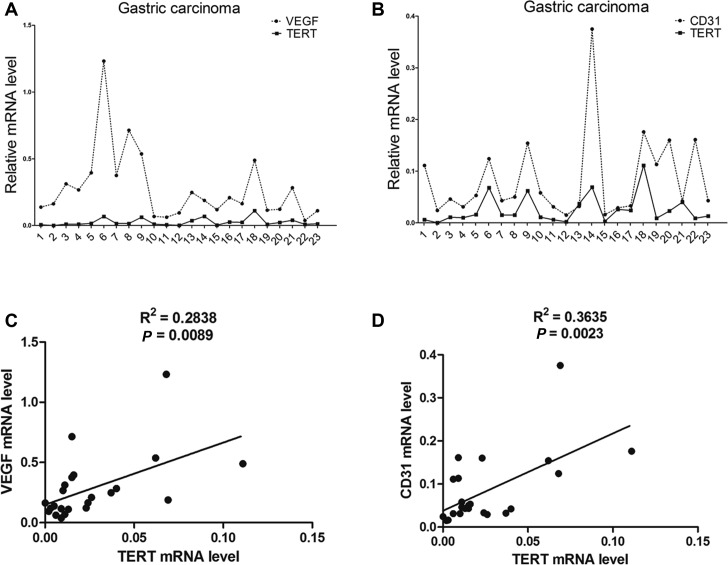
The 8-week-old Tert wild-type G1 and Tert knockout G1 mice (Supplementary Data S7A and B) received subcutaneous implantations of Lewis lung carcinoma cells in the flank. Tumor formation was observed and tumor weight was measured in these groups. As predicted, tumor size was considerably reduced in Tert knockout mice compared with Tert wild-type mice (Figure 5A). The average tumor weight of LLC tumors in Tert knockout mice was significantly lower than that in Tert wild-type mice (Figure 5B). Thus, TERT deficiency compromises tumor progression through telomere-independent mechanisms. In addition, immunohistochemical analysis of the expression of Ki67, a marker for tumor cell proliferation, revealed that the expression of Ki67 in LLC tumors from Tert wild-type mice was higher than that from Tert knockout mice. Significantly, immunohistochemical analysis indicated that the levels of VEGF and CD31 expression were significantly reduced in LLC tumors derived from Tert knockout G1 mice compared with those from the wild-type mice (Figure 5C and D). These results revealed that TERT deficiency compromises tumorigenesis and tumor vascular development. To test our findings in a clinical context, we analyzed the expression profiles of hTERT, VEGF and CD31 in 23 gastric tumor samples by qPCR. Consistent with previous studies in brain tumors, our results showed that hTERT expression levels were closely correlated with those of VEGF and CD31 in human gastric tumor samples (Figure 6). These observations suggest that the up-regulation of hTERT in tumor cells critically contributes to tumorigenesis not only through telomere maintenance, which feeds the unlimited proliferation potential of neoplastic cells, but also via enhanced tumor angiogenesis through the regulation of VEGF expression.
Figure 5.
TERT deficiency inhibits tumor growth and vascular development. (A) Tert+/+ and Tert−/− mice were implanted with 1 × 106 Lewis lung carcinoma cells at the flank, and the tumors were excised at day 21. (B) The scatter plot summarizes the weight of the tumors. (n = 8, **P < 0.01). (C) LLC tumor sections were stained for VEGF, CD31 and Ki67. Scale bars, 50 μm. (D) Quantification of blood vessels in LLC tumors. The scatter plot summarizes the microvessel density (MVD), which was determined by immunohistochemical staining for CD31. (n = 8, **P < 0.01). The data are presented as the mean ± S.D. (Student's t-test).
Figure 6.
hTERT expression correlates with VEGF and CD31 in gastric tumors. (A and B) qRT-PCR analysis of (A) VEGF, (B) CD31 and hTERT expression in 23 human gastric mucosa tissues. (C) Regression analysis of the correlation between VEGF and hTERT expression. (D) Regression analysis of the correlation between CD31 and hTERT expression. Each point represents one cancer sample.
DISCUSSION
Our studies support telomere-independent mechanisms of telomerase in cancer. We demonstrated that hTERT activates VEGF expression through its interaction with the transcription factor Sp1. Sp1 plays an essential role in the regulation of VEGF through the Sp1-binding sites near the transcriptional start site of the VEGF gene (38). Using the crystal structure of the TEN domain of TERT from T. thermophila (35), we were able to obtain a simulated complex with a modeled Sp1-dsDNA structure to predict the interaction between TEN and Sp1 (Arg615-Phe785), which includes the Sp1 DNA-binding sequence. The structural analysis supports our biological experimental results that hTERT interacts with Sp1 on the VEGF promoter to activate VEGF expression.
VEGF is the most potent endothelial mitogen and a critical component of angiogenesis. Functionally, we showed that hTERT promotes angiogenesis independently of its telomerase activity in the vascular tube formation assay. In addition, we found that hTERT promotes endothelial cell migration in a wound-healing assay. Consistently, we found that Tert knockout G1 mice, which have no telomere dysfunction phenotype, exhibited compromised LLC tumor growth in a tumor xenograft assay. This result suggests that the angiogenesis effects of TERT observed in vitro are not due to overexpression artifacts. Additionally, we found that the endogenous levels of hTERT expression were associated with those of VEGF and CD31 in human gastric tumors. Together, these results support a role for TERT in angiogenesis and tumor progression.
We showed that hTERT promotes VEGF expression in both cancer and normal cell lines. However, most human somatic cells have undetectable telomerase activity and telomerase is either up-regulated or reactivated during tumorigenesis in over 90% of human primary tumors. In addition, angiogenesis is an important factor in the progression of cancer. Therefore, inhibition of hTERT expression or activity would target both telomerase and VEGF, which would be effective against both neoplastic cell proliferation through telomere attrition and tumor angiogenesis by inhibiting VEGF.
A small number of other studies have reported that telomerase has a regulatory function in the expression of genes involved in diverse cellular processes (11,12). We and others have recently reported that hTERT regulates MMP-family genes, as well as IL-6 and TNF-α, by directly targeting NF-κB-dependent transcription (30,34), and we showed here that hTERT transactivated VEGF expression. Together, these findings lead us to propose that alterations in the expression profile of cancer-related genes, such as VEGF, MMPs and select NF-κB-target genes, associated with telomerase activation may also influence the tumor microenvironment to favor the invasive growth and metastasis of cancer cells.
The transcription factor Sp1 is ubiquitously expressed and binds to GC-boxes of target gene promoters. Sp1 is regulated by post-translational modifications as well as by protein–protein interactions. Our results showed that hTERT increases VEGF expression through interacting with Sp1. Interestingly, among 23 known angiogenesis-related genes including several Sp1-regulated genes (39), such as c-Myc, c-Jun, CyclinD1, PDGF, PDGFR, EGFR, only VEGF was affected by hTERT. Similarly, it has been reported that hTERT selectively regulates a subset of NF-κB target genes through its interaction with the NF-κB p65 (34). It is therefore possible that hTERT recruits different proteins (Sp1, p65, Brg1 or β-catenin) (34,40) and forms different protein complexes to regulate specific gene expression dependent on the target gene promoters and the cellular context. Future extended investigation is required to uncover the detailed mechanisms by which hTERT participates in gene expression regulation.
In summary, the present study revealed that hTERT plays a telomere maintenance-independent role in angiogenesis and tumor progression by up-regulating VEGF expression through interactions with the Sp1 transcription factor. Our results provide mechanistic evidence for a physiological, telomere-independent role of hTERT in tumor angiogenesis and progression and provide important insights into specific strategies for the therapeutic manipulation of telomerase in human cancer.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Ye Liu from the National Institute on Aging, Baltimore, Maryland, USA, for the Tert knockout mice and to Dr Katsuya Tsuchihara from the National Cancer Center Hospital East, Chiba, Japan for the VEGF promoter luciferase reporter constructs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Basic Research Program of China [2012CB911203]; National Natural Science Foundation of China [31371398, 31571409, 31401163, 31071200]. Funding for open access charge: National Basic Research Program of China [2012CB911203].
Conflict of interest statement. None declared.
REFERENCES
- 1.Blackburn E.H. Telomere states and cell fates. Nature. 2000;408:53–56. doi: 10.1038/35040500. [DOI] [PubMed] [Google Scholar]
- 2.Shay J.W., Bacchetti S. A survey of telomerase activity in human cancer. Eur. J. Cancer. 1997;33:787–791. doi: 10.1016/S0959-8049(97)00062-2. [DOI] [PubMed] [Google Scholar]
- 3.Wong M.S., Wright W.E., Shay J.W. Alternative splicing regulation of telomerase: a new paradigm? Trends Genet. 2014;30:430–438. doi: 10.1016/j.tig.2014.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cong Y.S., Wright W.E., Shay J.W. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bednarek A., Budunova I., Slaga T.J., Aldaz C.M. Increased telomerase activity in mouse skin premalignant progression. Cancer Res. 1995;55:4566–4569. [PubMed] [Google Scholar]
- 6.Broccoli D., Godley L.A., Donehower L.A., Varmus H.E., de Lange T. Telomerase activation in mouse mammary tumors: lack of detectable telomere shortening and evidence for regulation of telomerase RNA with cell proliferation. Mol. Cell. Biol. 1996;16:3765–3772. doi: 10.1128/mcb.16.7.3765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shay J.W., Wright W.E. Hallmarks of telomeres in ageing research. J. Pathol. 2007;211:114–123. doi: 10.1002/path.2090. [DOI] [PubMed] [Google Scholar]
- 8.Xu L., Li S., Stohr B.A. The role of telomere biology in cancer. Annu. Rev. Pathol. 2013;8:49–78. doi: 10.1146/annurev-pathol-020712-164030. [DOI] [PubMed] [Google Scholar]
- 9.Blasco M.A., Hahn W.C. Evolving views of telomerase and cancer. Trends Cell Biol. 2003;13:289–294. doi: 10.1016/s0962-8924(03)00085-0. [DOI] [PubMed] [Google Scholar]
- 10.Martinez P., Blasco M.A. Telomeric and extra-telomeric roles for telomerase and the telomere-binding proteins. Nat. Rev. Cancer. 2011;11:161–176. doi: 10.1038/nrc3025. [DOI] [PubMed] [Google Scholar]
- 11.Cong Y., Shay J.W. Actions of human telomerase beyond telomeres. Cell Res. 2008;18:725–732. doi: 10.1038/cr.2008.74. [DOI] [PubMed] [Google Scholar]
- 12.Ding D., Zhou J., Wang M., Cong Y.S. Implications of telomere-independent activities of telomerase reverse transcriptase in human cancer. FEBS J. 2013;280:3205–3211. doi: 10.1111/febs.12258. [DOI] [PubMed] [Google Scholar]
- 13.Low K.C., Tergaonkar V. Telomerase: central regulator of all of the hallmarks of cancer. Trends Biochem. Sci. 2013;38:426–434. doi: 10.1016/j.tibs.2013.07.001. [DOI] [PubMed] [Google Scholar]
- 14.Smith L.L., Coller H.A., Roberts J.M. Telomerase modulates expression of growth-controlling genes and enhances cell proliferation. Nat. Cell Biol. 2003;5:474–479. doi: 10.1038/ncb985. [DOI] [PubMed] [Google Scholar]
- 15.Choi J., Southworth L.K., Sarin K.Y., Venteicher A.S., Ma W., Chang W., Cheung P., Jun S., Artandi M.K., Shah N., et al. TERT promotes epithelial proliferation through transcriptional control of a Myc- and Wnt-related developmental program. PLoS Genet. 2008;4:e10. doi: 10.1371/journal.pgen.0040010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Li Q., Li K., Chen L., Li W., Hou M., Liu T., Yang J., Lindvall C., Bjorkholm M., et al. Telomerase reverse transcriptase promotes epithelial-mesenchymal transition and stem cell-like traits in cancer cells. Oncogene. 2013;32:4203–13. doi: 10.1038/onc.2012.441. [DOI] [PubMed] [Google Scholar]
- 17.Sarin K.Y., Cheung P., Gilison D., Lee E., Tennen R.I., Wang E., Artandi M.K., Oro A.E., Artandi S.E. Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature. 2005;436:1048–1052. doi: 10.1038/nature03836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Flores I., Cayuela M.L., Blasco M.A. Effects of telomerase and telomere length on epidermal stem cell behavior. Science. 2005;309:1253–1256. doi: 10.1126/science.1115025. [DOI] [PubMed] [Google Scholar]
- 19.Hahn W.C., Counter C.M., Lundberg A.S., Beijersbergen R.L., Brooks M.W., Weinberg R.A. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- 20.Stewart S.A., Hahn W.C., O'Connor B.F., Banner E.N., Lundberg A.S., Modha P., Mizuno H., Brooks M.W., Fleming M., Zimonjic D.B., et al. Telomerase contributes to tumorigenesis by a telomere length-independent mechanism. Proc. Natl. Acad. Sci. U.S.A. 2002;99:12606–12611. doi: 10.1073/pnas.182407599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koh C.M., Khattar E., Leow S.C., Liu C.Y., Muller J., Ang W.X., Li Y., Franzoso G., Li S., Guccione E., et al. Telomerase regulates MYC-driven oncogenesis independent of its reverse transcriptase activity. J. Clin. Invest. 2015;125:2109–2122. doi: 10.1172/JCI79134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Falchetti M.L., Mongiardi M.P., Fiorenzo P., Petrucci G., Pierconti F., D'Agnano I., D'Alessandris G., Alessandri G., Gelati M., Ricci-Vitiani L., et al. Inhibition of telomerase in the endothelial cells disrupts tumor angiogenesis in glioblastoma xenografts. Int. J. Cancer. 2008;122:1236–1242. doi: 10.1002/ijc.23193. [DOI] [PubMed] [Google Scholar]
- 23.George J., Banik N.L., Ray S.K. Combination of hTERT knockdown and IFN-gamma treatment inhibited angiogenesis and tumor progression in glioblastoma. Clin. Cancer Res. 2009;15:7186–7195. doi: 10.1158/1078-0432.CCR-09-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carmeliet P., Jain R.K. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee S., Chen T.T., Barber C.L., Jordan M.C., Murdock J., Desai S., Ferrara N., Nagy A., Roos K.P., Iruela-Arispe M.L. Autocrine VEGF signaling is required for vascular homeostasis. Cell. 2007;130:691–703. doi: 10.1016/j.cell.2007.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olsson A.K., Dimberg A., Kreuger J., Claesson-Welsh L. VEGF receptor signalling - in control of vascular function. Nat. Rev. Mol. Cell Biol. 2006;7:359–371. doi: 10.1038/nrm1911. [DOI] [PubMed] [Google Scholar]
- 27.Plate K.H., Breier G., Weich H.A., Risau W. Vascular endothelial growth factor is a potential tumour angiogenesis factor in human gliomas in vivo. Nature. 1992;359:845–848. doi: 10.1038/359845a0. [DOI] [PubMed] [Google Scholar]
- 28.Bergers G., Benjamin L.E. Tumorigenesis and the angiogenic switch. Nat. Rev. Cancer. 2003;3:401–410. doi: 10.1038/nrc1093. [DOI] [PubMed] [Google Scholar]
- 29.Zhou L., Zheng D., Wang M., Cong Y.S. Telomerase reverse transcriptase activates the expression of vascular endothelial growth factor independent of telomerase activity. Biochem. Biophys. Res. Commun. 2009;386:739–743. doi: 10.1016/j.bbrc.2009.06.116. [DOI] [PubMed] [Google Scholar]
- 30.Ding D., Xi P., Zhou J., Wang M., Cong Y.S. Human telomerase reverse transcriptase regulates MMP expression independently of telomerase activity via NF-kappaB-dependent transcription. FASEB J. 2013;27:4375–4383. doi: 10.1096/fj.13-230904. [DOI] [PubMed] [Google Scholar]
- 31.Wang R., Wang Y., Liu N., Ren C., Jiang C., Zhang K., Yu S., Chen Y., Tang H., Deng Q., et al. FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res. 2013;23:803–819. doi: 10.1038/cr.2013.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawada J., Urakami T., Li F., Urakami A., Zhu W., Fukuda M., Li D.Y., Ruoslahti E., Komatsu M. Small GTPase R-Ras regulates integrity and functionality of tumor blood vessels. Cancer Cell. 2012;22:235–249. doi: 10.1016/j.ccr.2012.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen H., Li L., Wang S., Lei Y., Ge Q., Lv N., Zhou X., Chen C. Reduced miR-126 expression facilitates angiogenesis of gastric cancer through its regulation on VEGF-A. Oncotarget. 2014;5:11873–11885. doi: 10.18632/oncotarget.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh A., Saginc G., Leow S.C., Khattar E., Shin E.M., Yan T.D., Wong M., Zhang Z., Li G., Sung W.K., et al. Telomerase directly regulates NF-kappaB-dependent transcription. Nat. Cell Biol. 2012;14:1270–1281. doi: 10.1038/ncb2621. [DOI] [PubMed] [Google Scholar]
- 35.Jacobs S.A., Podell E.R., Cech T.R. Crystal structure of the essential N-terminal domain of telomerase reverse transcriptase. Nat. Struct. Mol. Biol. 2006;13:218–225. doi: 10.1038/nsmb1054. [DOI] [PubMed] [Google Scholar]
- 36.Folkman J. Angiogenesis: an organizing principle for drug discovery? Nat. Rev. Drug Discov. 2007;6:273–286. doi: 10.1038/nrd2115. [DOI] [PubMed] [Google Scholar]
- 37.Liu Y., Snow B.E., Hande M.P., Yeung D., Erdmann N.J., Wakeham A., Itie A., Siderovski D.P., Lansdorp P.M., Robinson M.O., et al. The telomerase reverse transcriptase is limiting and necessary for telomerase function in vivo. Curr. Biol. 2000;10:1459–1462. doi: 10.1016/s0960-9822(00)00805-8. [DOI] [PubMed] [Google Scholar]
- 38.Beishline K., Azizkhan-Clifford J. Sp1 and the 'hallmarks of cancer'. FEBS J. 2015;282:224–258. doi: 10.1111/febs.13148. [DOI] [PubMed] [Google Scholar]
- 39.Wierstra I. Sp1: emerging roles–beyond constitutive activation of TATA-less housekeeping genes. Biochem. Biophys. Res. Commun. 2008;372:1–13. doi: 10.1016/j.bbrc.2008.03.074. [DOI] [PubMed] [Google Scholar]
- 40.Park J.I., Venteicher A.S., Hong J.Y., Choi J., Jun S., Shkreli M., Chang W., Meng Z., Cheung P., Ji H., et al. Telomerase modulates Wnt signalling by association with target gene chromatin. Nature. 2009;460:66–72. doi: 10.1038/nature08137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.