Abstract
Sam68 is a known sequence-specific RNA binding protein that regulates alternative splicing events during the cell cycle and apoptosis. Sam68 has also been shown to influence transcription, but the molecular mechanism remains undefined. Herein we identify Sam68 as a transcriptional coactivator of the p53 tumor suppressor in response to DNA damage. Using CRISPR/Cas9 generated isogenic HCT116 Sam68−/− cell lines wild type or deficient for p53, we show that Sam68 is required for the efficient transactivation of p53 target genes. Consistently, Sam68 depletion caused defects in DNA damage-induced cell cycle arrest and apoptosis mediated by p53. Mechanistically, we demonstrate that Sam68 physically interacted with p53 in an RNA-dependent manner, and that this interaction was essential for the coactivator function of Sam68. Furthermore, we show that both Sam68 and p53 were recruited to promoters of p53-responsive genes, suggesting interdependence. Finally, Sam68 acted in concert with the p53 long noncoding RNA (lncRNA) target PR-lncRNA-1 for p53 recruitment, implicating a positive-feedback mechanism in which lncRNAs induced by the Sam68/p53 complex can enhance p53 transcriptional activity. These findings define a hitherto novel mechanism of action for Sam68 in governing p53 transcriptional activation, and represent the first report of Sam68 in the regulation of tumor suppressor activities.
INTRODUCTION
The Src associated substrate during mitosis of 68kDa (Sam68) is a KH-type RNA-binding protein (RBP) involved in signal transduction, pre-mRNA splicing, mRNA translation, cell cycle regulation and apoptosis (1,2). Predominantly nuclear, Sam68 has been shown to regulate the alternative splicing of CD44 (3), cyclin D1 (4), Bcl-x (5), neurexin-1 (6), and mTOR (7). Sam68 can also transiently localize to the cytoplasm during the initial phase of cell attachment, as well as in spermatocytes, where it regulates cell signalling and mRNA translation, respectively (8,9). Additionally, Sam68 has been shown to influence transcription. Sam68 in complex with the SWI/SNF family members is required for alternative splicing of variable exons, and can affect transcriptional rates (10). Sam68 has also been demonstrated to modulate transcription by associating with the coactivator CBP (11), the androgen receptor (12) and NF-κB (13). Nevertheless, the exact mechanism of action of Sam68 in transcription is not defined, nor is the role of its RNA binding activity.
Sam68 is over-expressed in several cancer types including breast and prostate cancers (12,14). Additionally, extensive post-translational modifications of Sam68, such as phosphorylation, can influence its RNA binding activity (3,5,15). Interestingly, Sam68 phosphorylation and/or its cytoplasmic localization has been associated with a significant risk factor for poor prognosis (16,17), suggesting that aberrant Sam68 regulation, including sequestration of Sam68 from its nuclear role or inactivation by phosphorylation contributes to exacerbate tumorigenesis.
Whole body Sam68 knockout mice are viable and do not develop spontaneous tumors (18), and its haploinsufficiency delays MMTV-PyMT mammary tumors (19); however, its pro-tumorigenic mode of action in this mouse model remains unknown. On the other hand, Sam68 has also been previously identified to have tumor suppressor-like activities using a screen in NIH3T3 cells (20), though the mechanism was not defined. Given the evidence of Sam68 in cancer, we questioned whether Sam68 could regulate transcription factors that are important in tumor development. One major protein of interest is the p53 tumor suppressor. In response to stress signals such as DNA damage, p53 is stabilized and activated to exert its function as a sequence-specific transcription factor, inducing genes involved in cell cycle arrest (P21), apoptosis (BAX, PUMA), as well as the expression of its negative regulator (MDM2) (21,22). In addition to protein-coding genes, a rising number of long non-coding RNAs (lncRNAs) have been recently identified as p53 targets, with roles in p53 regulation and effector functions (23). Mutations of p53 have been found in > 50% of human cancers (24), where specifically 90% of those mutations occur in the DNA binding domain (25), thus highlighting the importance of its transcriptional role. The p53-dependent transcriptional activity is controlled through complex regulatory mechanisms, including p53 post-translational modifications and recruitment of transcriptional cofactors, yet its regulation is not completely understood, especially pertaining to the roles of non-coding RNAs and RBPs.
Herein we generated isogenic p53+/+;Sam68−/− and p53−/−;Sam68−/− HCT116 colon carcinoma cell lines by CRISPR/Cas9. Using these cell lines, as well as RNAi, we report that Sam68 functions as a transcriptional coactivator of p53, as the absence of Sam68 attenuated the induction of p53 target genes and subsequent cellular functions. Sam68 physically interacted with p53 in an RNA-dependent manner upon DNA damage-induced activation, and was recruited along with p53 to target promoters with the lncRNA PR-lncRNA-1, thereby defining a new molecular role for Sam68.
MATERIALS AND METHODS
Cell culture, transfection, and drug treatments
HCT116 p53+/+ and p53−/− human colon carcinoma cell lines were obtained from B. Vogelstein (Johns Hopkins Oncology Center, Baltimore, MD) (26). Cells were grown in McCoy's 5A medium supplemented with 10% fetal bovine serum (FBS), sodium pyruvate, and penicillin-streptomycin at 37°C under 5% CO2. Transfections of plasmids and antisense oligonucleotides (ASOs) were done with Lipofectamine 2000 and RNAiMAX reagents (Invitrogen), respectively according to the manufacturer's instructions. At 24 h post-transfection, cells were mock treated or treated with doxorubicin or UV as indicated. For RNA interference, cells at 25% confluency were transfected with either siGFP as control or siSam68 (SMARTpool, Dharmacon) using RNAiMAX (Invitrogen) as per manufacturer's instructions. 48 h post-transfection, cells were mock-treated or treated with doxorubicin (1 μM) for 24 h to induce DNA damage before harvest.
CRISPR/Cas9 generation of Sam68−/− cells
Sequences of human codon optimized Cas9 and gRNAs targeting the SAM68 gene were obtained from Mali et al. (27). Cas9 and gRNA plasmids (IDT) were co-transfected with GFP into HCT116 p53+/+ and p53−/- cells using Lipofectamine 2000 (Invitrogen) as per manufacturer's instructions. Cells were harvested 48 h after transfection, and the top GFP-expressing cells were sorted into 96-well plates as single clones. To screen for clones with SAM68 gene disruption, total genomic DNA was extracted using AccuStart II Mouse Genotyping Kit (Quanta) following the manufacturer's protocol. Genomic PCR was performed with primers listed in Supplementary Table S1, which were designed according to the NCBI database sequence. PCR products were analyzed on 1% agarose gel stained with ethidium-bromide. Sequencing (McGill University and Génome Québec Innovation Centre) and immunoblots were performed to confirm SAM68 gene disruption and protein depletion, respectively.
RNA extraction and RT-qPCR
Total RNA was extracted using TRIzol (Invitrogen) according to the manufacturer's instructions from HCT116 cells that were mock- or doxorubicin-treated. Reverse transcription (RT) was performed with M-MLV reverse transcriptase (Promega) in a programmable thermal controller (MJ Research). Real-time PCR (qPCR), including primer design and efficiency tests, was carried out according to the Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) guidelines (28). Experiments were performed in a MicroAmp Fast Optical 96-well reaction plate (Life technologies) using SsoFast EvaGreen Supermix (Bio-Rad) following the manufacturer's instructions. Data were normalized to reference genes (GAPDH, ACTB, 18S) by the ΔΔCt method. Sequences of primers used are listed in Supplementary Table S1.
Protein extraction, immunoblots and immunoprecipitation
HCT116 cells were washed with ice-cold PBS and lysed on ice in lysis buffer (50 mM HEPES, 150 mM NaCl, 1% Triton X-100) supplemented with protease inhibitor cocktail (Roche). Cell debris was cleared by centrifugation. Whole cell extracts were resolved by 10% SDS-PAGE, transferred onto nitrocellulose membranes, and blotted with antibodies against Sam68 (07-415, Millipore), p53 (HAF1355, R&D), MDM2 (Ab-5, Millipore), p21 (C-19, Santa Cruz), Bax (P-19, Santa Cruz), Puma (4976, Cell Signaling), PARP (F-2, Santa Cruz) and β-actin (A3853, Sigma Aldrich). For immunoprecipitation, anti-Sam68 (07-415, Millipore), anti-p53 (1C12, Cell Signaling), anti-Myc (05-724, Sigma), anti-GFP (Anti-GFP, Roche), mouse or rabbit IgG (Santa Cruz) antibodies were incubated for 2 h with whole cell lysates, and 1 h with 50 μl of 50% slurry protein A/G beads (Sigma) at 4°C. Beads were washed three times with lysis buffer, and bound proteins were recovered by boiling in Laemmli sample buffer.
Luciferase assays
HCT116 cells were transiently transfected with firefly luciferase expression vectors driven by either the artificial p53 binding site repeat (PG13, Addgene) or p21 promoter (Addgene) that were co-transfected with renilla luciferase expression vectors (Promega) at a 10:1 (firefly: renilla) ratio. At 24 h post transfection, cells were mock treated or treated with doxorubicin for 24 h before harvest. Firefly luciferase and renilla luciferase activities were analyzed using the Dual-Luciferase Reporter Assay System kit (Promega) following the manufacturer's directions.
Cell growth and flow cytometry
To determine cell growth rate, 0.3 × 106 cells were seeded per 6-well plate. At times indicated, cells were collected and counted using a Coulter Cell Counter (Beckman). For flow cytometry, cells were pulsed for 1 h with 10 μg/ml of bromodeoxyuridine (BrdU, BD), washed twice with PBS, and were allowed to fix overnight in 75% ethanol at −20°C. For BrdU staining, fixed cells were washed once with washing solution (0.5% BSA in PBS), denatured in 2 M HCl for 20 min at room temperature and neutralized with 0.1 M Na2B4O7. Cells were pelleted by centrifugation, resuspended in dilution buffer (0.5% BSA, 0.5% Tween-20 in PBS), and incubated with FITC-conjugated anti-BrdU (BD) antibody for 20 min at room temperature. Cells were then stained with propidium iodide (PI) containing RNase for 30 min at room temperature before analysis by FACS Calibur flow cytometry. Quantifications were performed using the FlowJo software.
Chromatin immunoprecipitation (ChIP)
ChIP was performed essentially as described previously (29). Briefly, HCT116 cells were grown to 50–60% confluency and were either untreated or treated with doxorubicin. After trypsinization, cells were cross-linked in 1% paraformaldehyde solution for 10 min at room temperature. Cross-linking was stopped by addition of glycine to 125 mM final concentration. Cells were washed twice with ice-cold PBS and chromatin was isolated with nuclei lysis buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 1% SDS) on ice. Samples were sonicated to yield fragments between 200–1500 base pairs. For immunoprecipitation, chromatin was precleared using 50% slurry of protein A/G beads (Sigma) before addition of antibodies (1:200) for overnight incubation at 4°C. Preblocked protein A/G beads were added to the reaction to couple the immunocomplexes. Beads were washed twice with dialysis buffer (2 mM EDTA, 50 mM Tris-HCl pH 8.0), four times with ChIP wash buffer (100 mM Tris-HCl pH 8.0, 500 mM LiCl, 1% NP40, 1% deoxycholic acid sodium salt) and immunocomplexes were eluted twice with elution buffers (50 mM sodium bicarbonate, 1% SDS). Cross-linking was reversed by adjusting to 200 mM NaCl and boiling for 15 min. RNase A was added and incubated for 30 min at 37°C. Finally, DNA was purified using QIAquick PCR Purification Kit (Qiagen) following the manufacturer's instructions. qPCR primers used for P21 and PUMA promoters were described previously (30,31), and are listed in Supplementary Table S1. Antibodies used: mouse/ rabbit IgG (Santa Cruz), anti-p53 (1C12, Cell Signaling), anti-Sam68 (07-415, Millipore) and anti-Myc antibodies (05-724, Sigma).
RNA immunoprecipitation (RIP)
To evaluate protein-RNA interactions, HCT116 cells were incubated with 100 μM 4-thiouridine (Sigma) 14 h prior to cross-linking. The cells were washed once with PBS, placed on ice, and irradiated uncovered with 0.15 J/cm2 of 365-nm UV light. Cells were harvested in lysis buffer (50 mM HEPES pH 7.5, 150 mM KCl, 2 mM EDTA, 1 mM NaF, 0.5% NP-40, 0.5 mM DTT, 0.125% SDS) supplemented with protease inhibitor cocktail (Roche) and 0.5 U/μl of RNasin (Promega). Cell lysates were immunoprecipitated using 2 μg of either anti-Sam68 antibodies (07-415, Millipore) or IgG (Santa Cruz), and complexes captured using protein A/G beads (Sigma). The immunoprecipitates were treated with 1 mg/ml of proteinase K in proteinase K buffer (100 mM Tris pH 7.5, 50 mM NaCl, 10 mM EDTA) for 30 min at 55°C. The bound RNA was isolated using TRIzol reagent (Invitrogen) as per the manufacturer's protocol. RT-qPCR was carried out as described above.
RESULTS
Generation of Sam68−/− HCT116 cell lines
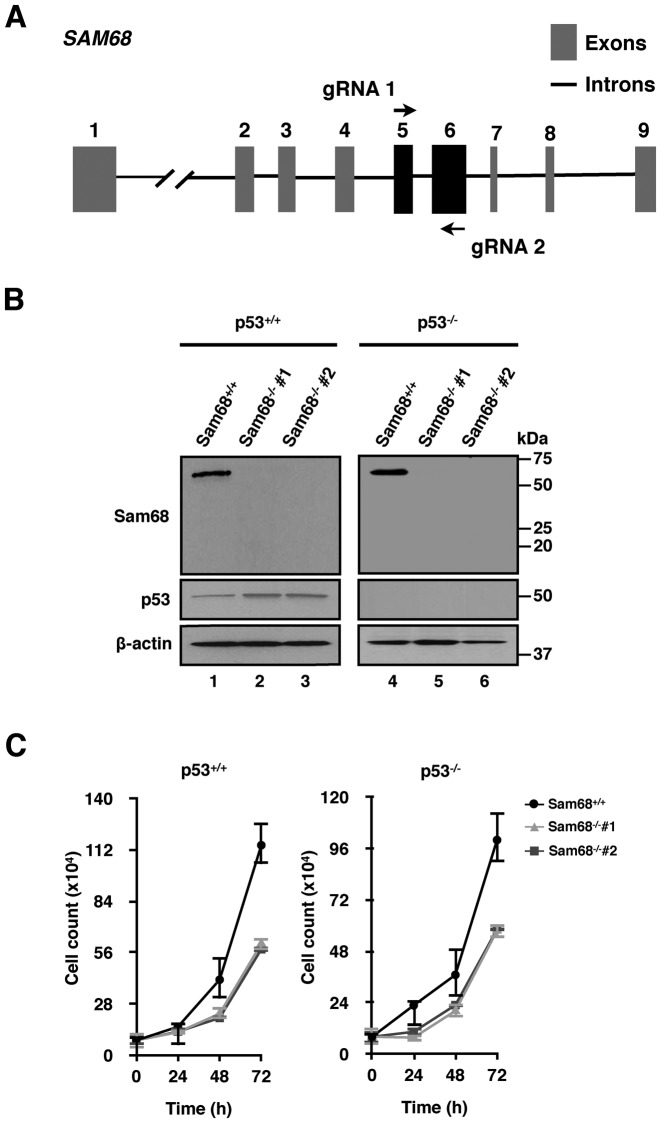
To investigate the role of Sam68 in the p53 tumor suppressor pathway, we first established isogenic systems by generating null alleles of the SAM68 gene in both p53+/+ and p53−/− HCT116 cell lines using the CRISPR/Cas9 technology (26,27). Guide RNAs (gRNA 1 and 2) were designed to target SAM68 exons 5 and 6 (Figure 1A), excising part of the KH domain of Sam68 (32). Two representative Sam68−/− clones (#1 and #2) derived from each p53+/+ and p53−/− HCT116 cell lines were chosen. Null alleles with deletions ranging from 515 to 899 base pairs were generated (Supplementary Table S2). The absence of Sam68, as well as p53 protein expression was confirmed by immunoblotting (Figure 1B). Interestingly, we observed a slight increase of basal p53 protein levels in p53+/+;Sam68−/− HCT116 clones #1 and #2 (Figure 1B, lanes 1–3), but the p53 mRNA levels were unaltered (not shown), suggesting that Sam68 may regulate p53 protein expression in the absence of cellular stress. The Sam68−/− cells were viable and had decreased growth rates regardless of their p53 status (Figure 1C), implying that Sam68 has a role in cell growth that is independent of p53.
Figure 1.
CRISPR/Cas9 deletion of Sam68 in isogenic HCT116 cells. (A) Schematic of CRISPR target sites on the SAM68 gene. Guide RNAs (gRNA 1 and gRNA 2) flanking the target region are indicated by arrows. (B) Immunoblot verification of HCT116 Sam68+/+ and Sam68−/− clones (#1 and #2) derived from p53+/+ and p53−/− cell lines with antibodies specific for Sam68 and p53. β-actin was used as loading control. (C) Growth rate of HCT116 Sam68+/+ and Sam68−/− cells over a 72 h period. Data are represented as mean ± S.D. from 3 independent experiments.
Sam68-deficiency compromises DNA damage induction of p53 targets
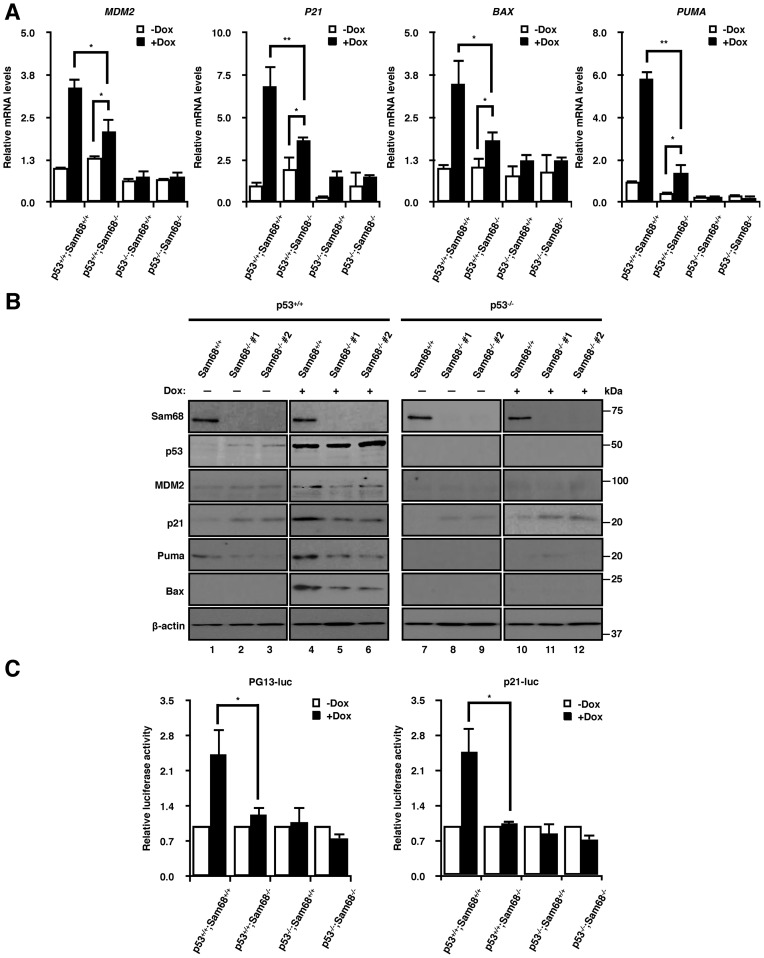
To determine whether Sam68 expression modulates p53 transactivation function, we monitored the expression of known p53 target genes by RT-qPCR. As expected, treatment with doxorubicin for 24 h led to an increase in MDM2, P21, BAX and PUMA mRNAs in p53+/+;Sam68+/+ cells (Figure 2A). This increase was severely attenuated in p53+/+;Sam68−/− cells (Figure 2A). These findings were also confirmed in HCT116 and MCF-7 cells depleted of Sam68 by RNAi (Supplementary Figure S1), suggesting that Sam68 is required for p53 transcriptional activity in response to DNA damage in several cell types, as verified by both RNA interference and CRISPR/Cas9. Consistent with the requirement of p53 for activation, MDM2, P21, BAX and PUMA remained at low levels in p53−/− cells upon DNA damage treatment (Figure 2A). Despite these lower baseline levels there were increases observed for P21 and BAX in response to DNA damage independent of the status of Sam68 (Figure 2A). We further confirmed these observations at the protein level by immunoblotting. While MDM2, p21, Bax, and Puma were strongly induced in p53+/+;Sam68+/+ cells in response to DNA damage, their up-regulation was compromised in p53+/+;Sam68−/− cells (Figure 2B), as quantified by 3 independent immunoblotting experiments (Supplementary Figure S2); however, Sam68 depletion did not affect doxorubicin-induced p53 stabilization (Figure 2B, lanes 4–6). It is also important to note that although p53+/+;Sam68−/− cells displayed slightly higher basal p53 levels (Figure 2B, lanes 1–3), the cells were defective in p53 transactivation in response to the DNA damaging agent doxorubicin (Figure 2A and B). Together, these results show that p53 transcriptional regulation of MDM2, P21, BAX and PUMA requires Sam68.
Figure 2.
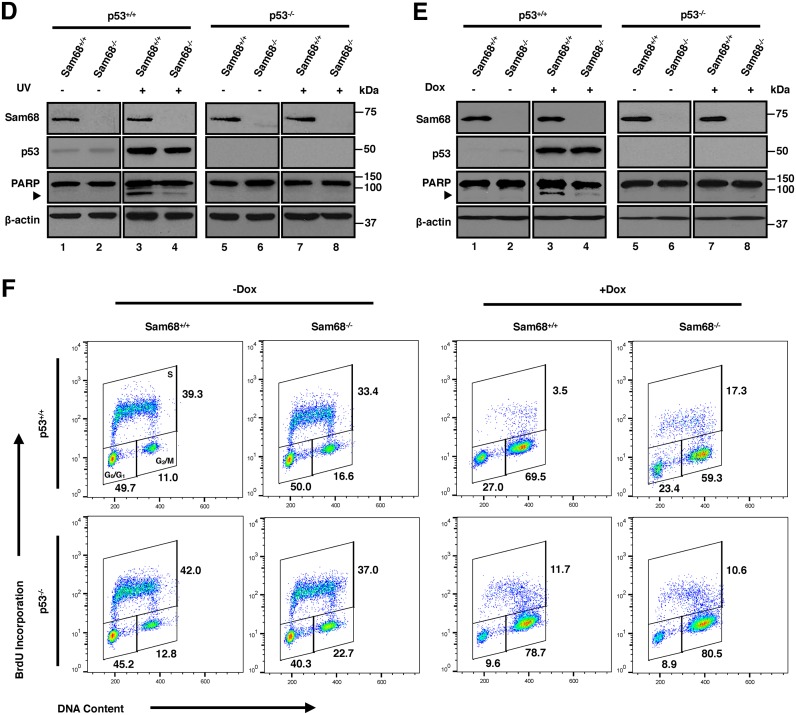
Sam68 is required for p53 transcriptional activity and cellular functions in response to DNA damage. (A) RT-qPCR of p53 target genes in HCT116 cells untreated (-Dox) or treated with 1 μM doxorubicin (+Dox) for 24 h. RT-qPCR was performed with primers specific for MDM2, P21, BAX, and PUMA, where levels were normalized to GAPDH expression. Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. (B) Immunoblot of HCT116 cells that were either untreated (-) or treated with 1 μM doxorubicin (+) for 24 h before protein extraction. Analysis was performed using Sam68, p53, MDM2, p21, Bax and Puma-specific antibodies. β-actin was used as loading control. (C) Luciferase assay of cells transiently transfected with a PG13 (PG13-luc) or a p21 promoter controlled (p21-luc) firefly luciferase reporter. At 24 h post-transfection, cells were mock treated (-Dox) or treated with 1 μM doxorubicin (+Dox) for 24 h before determining luciferase activity. Firefly luciferase activities were normalized to renilla luciferase levels. Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. (D) Immunoblot of HCT116 cells untreated (-) or treated with either 60 J/m2 UV (+) or (E) 1μM doxorubicin (+). Cells were harvested at 24 h after initial treatment. Analysis was performed using Sam68, p53, and PARP-specific antibodies, where arrow indicates cleaved PARP. β-actin was used as loading control. (F) Flow cytometry analysis of cell cycle. HCT116 cells were either untreated (-Dox) or treated with 0.2 μM doxorubicin (+Dox) for 24 h. BrdU pulsed cells were fixed in ethanol, then stained with anti-BrdU antibodies and PI before analysis. Statistical significance was calculated with Student's t-test (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005).
Interestingly, we observed slightly elevated p21 protein levels in Sam68−/− cells compared with Sam68+/+ cells in the absence of p53 activation (Figure 2B, lanes 1–3, 7–12). To address this point, we monitored the half-life of P21 mRNA in the presence of actinomycin D. The loss of Sam68 expression increased P21 mRNA stability in a p53-independent manner, while it had no significant effect on MDM2 transcript half-life (Supplementary Figure S3). The up-regulation of p21 in the absence of Sam68 has been observed previously (14); however, we now show that this is a result of Sam68 regulating P21 mRNA stability.
We next examined whether Sam68 could influence the ability of p53 to activate a luciferase reporter under the control of an artificial p53-responsive promoter containing thirteen p53 consensus sites (PG13-luc) or the P21 promoter (p21-luc). Transient transfection of PG13-luc or p21-luc into p53+/+;Sam68+/+ cells led to significant increases in luciferase expression following DNA damage, but almost no induction was observed for p53+/+;Sam68−/− cells (Figure 2C). Moreover, there were no noticeable differences in luciferase expression between p53−/−;Sam68+/+ and p53−/−;Sam68−/− cells (Figure 2C). Taken together, these data suggest that Sam68 is required to regulate p53-dependent transcription of target genes and is therefore a positive transcriptional regulator of p53.
Sam68-deficiency reduces p53-mediated cellular functions
The major outcomes of p53 activation are apoptosis and cell cycle arrest (21). To determine whether Sam68 contributes to p53-mediated apoptosis, isogenic HCT116 cells were treated with UV or doxorubicin to induce DNA damage. Flow cytometry analysis showed that although Sam68 had little effect on cell death in untreated HCT116 cells (1.49% versus 2.07% for p53+/+ cells; 2.75% versus 3.67% for p53−/− cells) (Supplementary Figure S4), the sub-G1 population was dramatically reduced in p53+/+;Sam68−/− cells (15.4%) compared to p53+/+;Sam68+/+ cells (27.8%) following UV treatment (Supplementary Figure S4). Concomitantly, immunoblotting analysis revealed that while UV and doxorubicin potently induced caspase-mediated cleavage of poly(ADP-ribose) polymerase (PARP) in p53+/+;Sam68+/+ cells, an early marker of apoptosis (33), it was reduced in p53+/+;Sam68−/− cells (Figure 2D and E). As expected for a p53-specific response, no significant difference in sub-G1 population (9.43% versus 9.25%) and PARP cleavage was observed between p53−/−;Sam68+/+ cells and p53−/−;Sam68−/− cells (Figure 2D, E and Supplementary Figure S4). These results suggest that Sam68 participates in the DNA damage-induced p53-mediated cell apoptosis.
To explore the possible role of Sam68 in regulating cell-cycle checkpoint responses, we treated isogenic HCT116 cells with a low dose of doxorubicin for 24 h, then analyzed cells dually labeled with PI and anti-BrdU by flow cytometry. Sam68-depletion in unstressed p53+/+ cells led to a slight increase in G2/M phase from 11.0% to 16.6% (Figure 2F), while in p53−/− cells this increase was more substantial from 12.8% to 22.7% (Figure 2F). Moreover, a minor decrease in S phase was observed in the absence of Sam68 (39.3% versus 33.4% for p53+/+ cells; 42.0% versus 37.0% for p53−/− cells) (Figure 2F). These findings implicate a p53-independent function of Sam68 in cell cycle progression, especially pertaining to the G2/M phase (Figure 2F). Upon doxorubicin-induced p53 activation, however, while p53+/+;Sam68+/+ cells displayed normal G2/M checkpoint arrest (Figure 2F, 69.5%), this response was reduced in p53+/+;Sam68−/− cells (Figure 2F, 59.3%). Concomitantly, p53+/+;Sam68−/− cells also displayed ∼5-fold increase in S phase cells (Figure 2F, 3.5% versus 17.3%). These data reveal that Sam68 is required for the efficient induction of G2/M cell cycle arrest in response to DNA damage. This attenuated effect in Sam68 depleted cells in the presence of doxorubicin was not observed in p53−/− cells (Figure 2F, 78.7% versus 80.5%). Taken together, our data suggest that Sam68 selectively enhances cell-cycle-checkpoint responses that are mediated by p53.
The N-terminus and KH domain of Sam68 are necessary to interact with p53
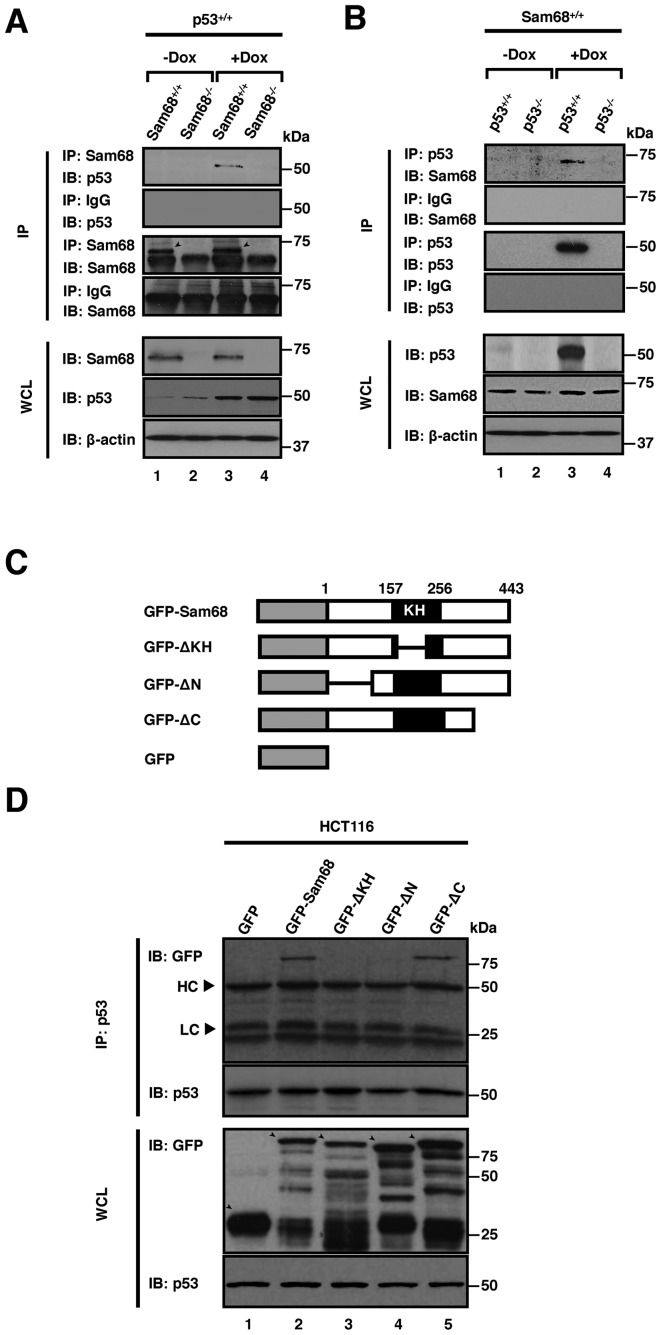
To test whether Sam68 interacts with p53, we first investigated if they co-immunoprecipitated. HCT116 cells of the indicated genotypes were either untreated or treated with doxorubicin, followed by cell lysis and immunoprecipitation using anti-Sam68 antibodies. The bound proteins were resolved by SDS-PAGE and immunoblotted with anti-p53 antibodies pre-conjugated to horseradish peroxidase, as p53 migrates close to the size of the immunoglobulin heavy chains. Indeed, we detected p53 co-immunoprecipitation with endogenous Sam68 when cells were treated with doxorubicin (Figure 3A, lane 3). This interaction was not observed using p53+/+;Sam68−/− cells (Figure 3A, lane 4) nor in undamaged cells that served as negative controls (Figure 3A, lanes 1 and 2). The converse was also observed as anti-p53 immunoprecipitates contained Sam68 in p53+/+;Sam68+/+ cells treated with doxorubicin (Figure 3B, lane 3). This interaction was not detected using p53−/−;Sam68+/+ cells (Figure 3B, lane 4), nor in undamaged cells that served as a negative controls (Figure 3B, lanes 1 and 2).
Figure 3.
The Sam68 N-terminus and KH domain are required for p53 interaction. (A and B) Immunoprecipitation of endogenous Sam68 and p53 with antibodies specific for Sam68 or p53 in untreated (−Dox) or doxorubicin treated (+Dox) HCT116 cells for the indicated genotypes. IgG is a non-specific antibody used as control. (C) Schematic of GFP-tagged Sam68-coding constructs. (D) p53 immunoprecipitation in doxorubicin-treated HCT116 cells that were transiently transfected with either control plasmid (GFP), GFP-Sam68 or the mutants (GFP-ΔKH, GFP-ΔN, GFP-ΔC). IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate; HC, heavy chain; LC, light chain.
To map the region of Sam68 required for interaction with p53, we transiently transfected wild type HCT116 cells with expression vectors encoding either the green fluorescent protein (GFP), GFP-tagged full-length Sam68 (GFP-Sam68), or truncated Sam68 lacking the KH (GFP-ΔKH; RNA binding defective), the N-terminal (GFP-ΔN; RNA binding proficient) or the C-terminal domain (GFP-ΔC) (Figure 3C). Cells were treated with doxorubicin, then lysed and immunoprecipitated with anti-p53 antibodies, and the bound proteins were visualized by immunoblotting with anti-GFP antibodies. GFP-Sam68 and GFP-ΔC co-immunoprecipitated with p53, while GFP, GFP-ΔN and GFP-ΔKH did not (Figure 3D). These findings suggest that Sam68 interacts with p53 via protein–protein and RNA–protein interactions, as the integrity of both the N-terminus and its KH domain was required to maintain association with p53.
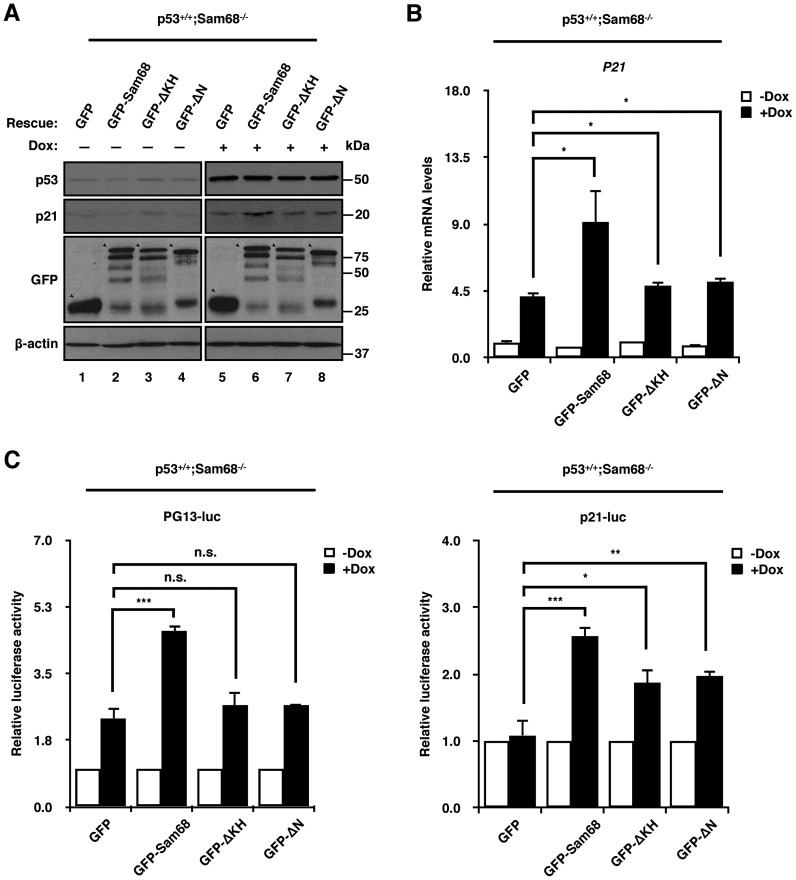
Ectopic expression of Sam68 rescues p53+/+;Sam68−/− cells from defective p53 gene transactivation
To further determine the functional significance of the Sam68/p53 interaction, we performed rescue experiments in p53+/+;Sam68−/− HCT116 cells by transfecting expression vectors encoding GFP, GFP-Sam68 or the mutants GFP-ΔKH and GFP-ΔN, both of which were defective in p53 interaction (Figure 3D). Transfection of GFP-Sam68, but not GFP-ΔKH or GFP-ΔN, fully rescued p21 protein levels following doxorubicin treatment in p53+/+;Sam68−/− cells (Figure 4A, compare lane 6 with lanes 5, 7 and 8). GFP protein expression was verified by immunoblotting, as well as the induction of p53 with doxorubicin (Figure 4A). RT-qPCR analysis confirmed that P21 mRNA levels were restored with GFP-Sam68, but not GFP-ΔKH or GFP-ΔN, in p53+/+;Sam68−/− cells treated with doxorubicin (Figure 4B). GFP-Sam68 increased doxorubicin-induced luciferase activities of both PG13-luc and p21-luc reporters in p53+/+;Sam68−/− cells, while GFP-ΔKH and GFP-ΔN partially increased the p21-luc reporter above GFP (Figure 4C). Together, these data show that Sam68 requires the integrity of its N-terminus and KH domain to regulate p53 transcriptional activity.
Figure 4.
The KH domain and N-terminus of Sam68 are required for its role as a p53 transcriptional cofactor. (A–C) RT-qPCR, immunoblot, and luciferase assay analyses of HCT116 p53+/+;Sam68−/− cells transiently transfected with control plasmid (GFP), GFP-Sam68, GFP-ΔKH or GFP-ΔN. Cells were mock treated (−) or treated (+) with 1 μM doxorubicin 24 h post-transfection before mRNA extraction. P21 expression levels were normalized to GAPDH. Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. Immunoblot was performed with p21, p53 and GFP-specific antibodies, and β-actin was used as loading control. Luciferase levels were normalized to renilla activity levels. Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. Statistical significance was calculated with Student's t-test (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005).
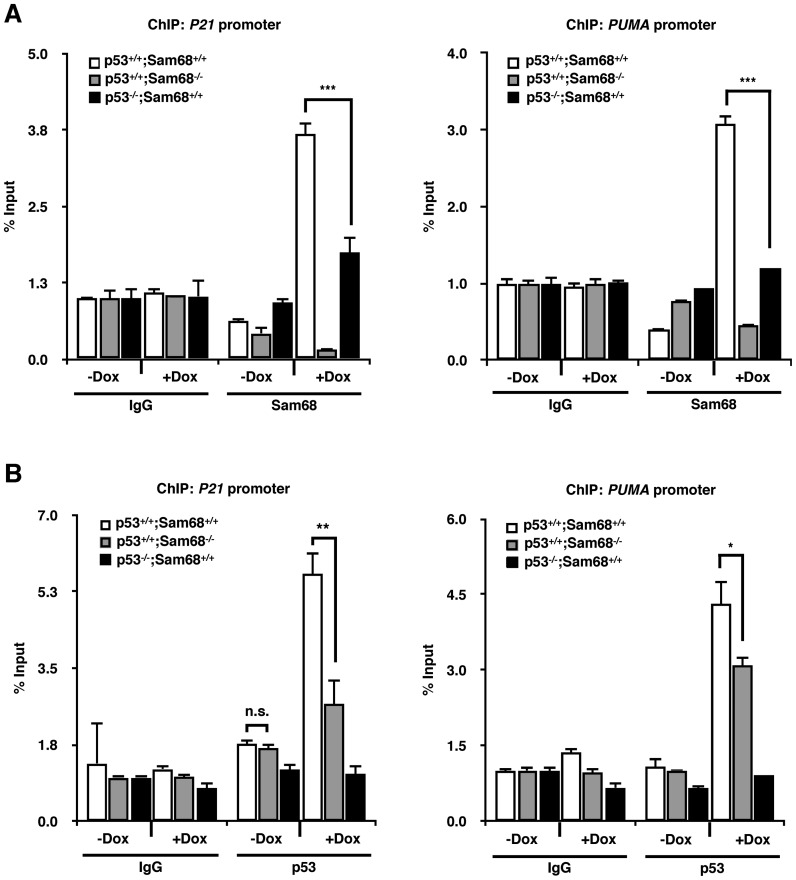
Sam68 and p53 recruitment to the P21 and PUMA promoters are interdependent
The association of Sam68 with p53 implied that Sam68 might be recruited to p53-dependent promoters along with p53. To investigate this possibility, we performed chromatin immunoprecipitation (ChIP) experiments with anti-Sam68 and -p53 antibodies in untreated and doxorubicin treated isogenic HCT116 cells. We used primers that encompassed the p53 consensus binding sites within the promoter regions of P21 and PUMA. We found that Sam68 was recruited to the P21 and PUMA promoters with doxorubicin treatment in p53+/+;Sam68+/+ cells, but this recruitment was impaired in p53−/−;Sam68+/+ cells (Figure 5A), suggesting that the recruitment of Sam68 is p53-dependent. It cannot be excluded that the absence of Sam68 recruitment observed in p53−/− cells is due to the low transcriptional activity of p53 target promoters under these conditions. Moreover, Sam68 was not recruited in the absence of DNA damage in p53+/+;Sam68+/+ cells nor in p53+/+;Sam68−/− cells (Figure 5A). Together, these data suggest that Sam68 is recruited by p53 at target promoters.
Figure 5.
Sam68 and p53 are recruited to promoters in an interdependent manner. (A and B) ChIP-qPCR analysis of HCT116 cells. Cell lysates from untreated and doxorubicin-treated cells were immunoprecipitated with either non-specific IgG, p53- or Sam68-specific antibodies. Precipitated DNA was quantified by qPCR with primers specific for p53-response elements of P21 and PUMA promoters. Data, replicated in 3 independent experiments, are represented as mean ± S.D. of technical triplicates. Statistical significance was calculated with Student's t-test (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005).
In parallel, p53 enrichment was observed at its consensus binding sites on the P21 and PUMA promoters following doxorubicin treatment in p53+/+;Sam68+/+ cells (Figure 5B). Interestingly, the recruitment of p53 to the P21 and PUMA promoters was severely compromised in DNA damage-induced p53+/+;Sam68−/− cells (Figure 5B). Altogether, these results suggest that the presence of Sam68 is required for optimal p53 recruitment to its target promoters in a mutually dependent manner.
Sam68 cooperates with PR-lncRNA-1 for efficient p53 recruitment to the P21 promoter
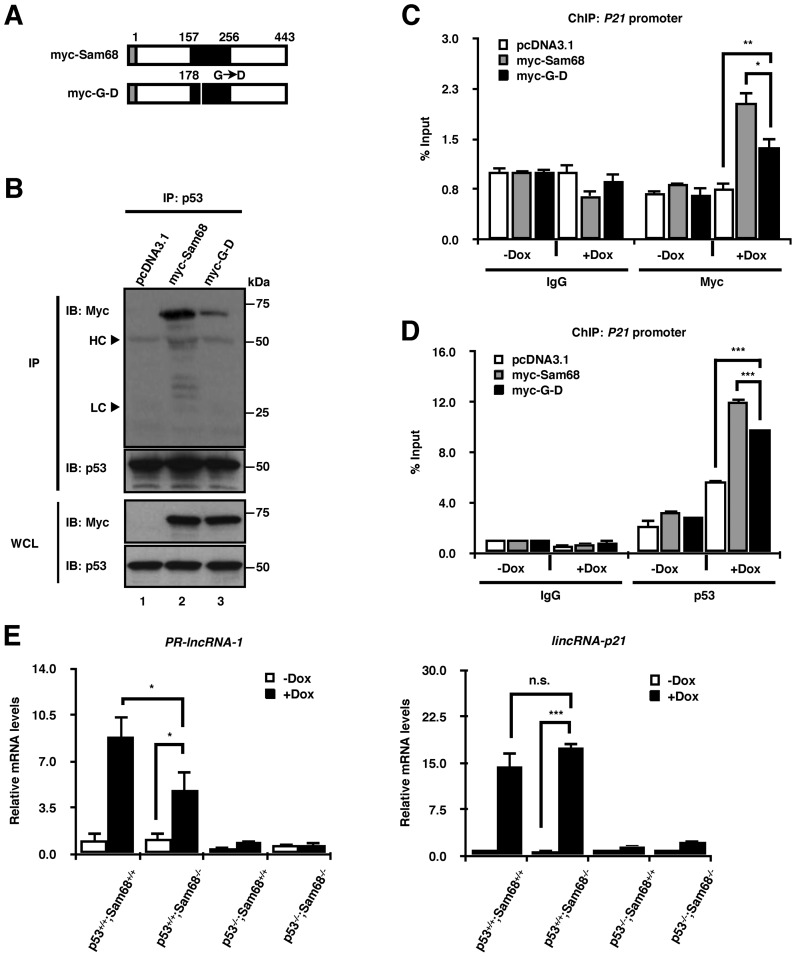
To delve deeper into the mechanism, and given that the KH domain of Sam68 is crucial for its coactivator function, we first investigated whether RNA is required for Sam68/p53 interaction. To address this, we transfected p53+/+;Sam68−/− cells with either the control vector (pcDNA3.1), plasmids encoding a myc-tagged full-length Sam68 (myc-Sam68), or an RNA-binding-deficient form of Sam68 bearing a point mutation in the KH domain (myc-G-D) (Figure 6A), and then performed immunoprecipitation using anti-p53 antibodies. We found that while p53 readily bound to full-length Sam68 (Figure 6B, lane 2), this association was severely dampened with the RNA-binding deficient mutant (Figure 6B, lane 3), suggesting that RNA is required for the Sam68/p53 interaction. Moreover, this interaction was also reduced by RNase treatment (Supplementary Figure S5), suggesting that RNA contributes to their association.
Figure 6.
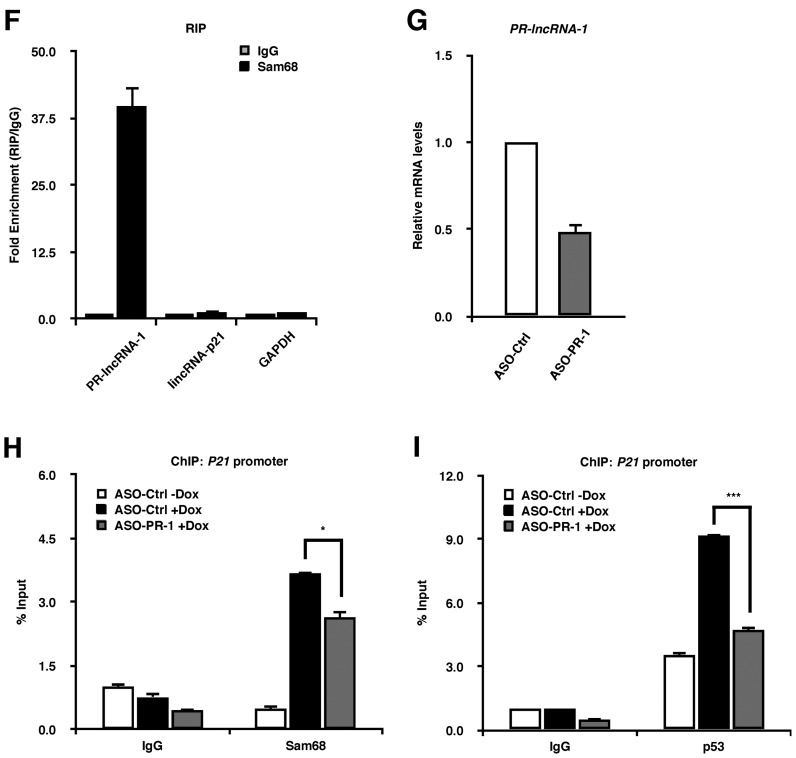
Sam68 associates with p53 in an RNA-dependent manner and coordinates PR-lncRNA-1 for efficient p53 promoter recruitment. (A) Schematic of myc-tagged Sam68-coding constructs. (B) p53 immunoprecipitation in doxorubicin-treated HCT116 cells, which were transiently transfected with either empty vector (pcDNA3.1), myc-Sam68 or myc-G-D mutant. IP, immunoprecipitation; IB, immunoblot; WCL, whole cell lysate; HC, heavy chain; LC, light chain. (C and D) ChIP-qPCR analysis of untreated and doxorubicin-treated HCT116 cells transfected with the indicated constructs. Lysates from cells were immunoprecipitated with either nonspecific IgG, myc- or p53-specific antibodies. Precipitated DNA was quantified by qPCR with primers specific for p53-response elements of the P21 promoter. Data, replicated in 3 independent experiments, are represented as mean ± S.D. of technical triplicates. (E) RT-qPCR analysis of p53-regulated lncRNAs in HCT116 cells untreated (−Dox) or treated with 1 μM doxorubicin (+Dox) for 24 h. RT-qPCR was performed with primers specific for PR-lncRNA-1 and lincRNA-p21, where levels were normalized to GAPDH expression. Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. (F) RIP assay in doxorubicin-induced HCT116 cells using anti-Sam68 antibodies, followed by RT-qPCR with primers for the indicated lncRNAs. IgG and GAPDH RNA both serve as negative controls. Data are represented as mean ± S.D. from 3 independent experiments. (G) RT-qPCR assessment of PR-lncRNA-1 depletion in HCT116 cells transfected with non-targeting ASOs (ASO-Ctrl) or PR-lncRNA-1-targeting ASOs (ASO-PR-1). Data are represented as mean ± S.D. from 3 independent experiments done in biological triplicates. (H and I) ChIP-qPCR analysis of Sam68 and p53 enrichment on the P21 promoter in HCT116 cells transfected with ASO-Ctrl or ASO-PR-1. Cells were untreated or treated with doxorubicin 36 h following ASO transfection. Precipitated DNA was quantified by qPCR with primers specific for p53-response elements of the P21 promoter. Data, replicated in 3 independent experiments, are represented as mean ± S.D. of technical triplicates. Statistical significance was calculated with Student's t-test (*P ≤ 0.05, **P ≤ 0.005, ***P ≤ 0.0005).
Given that Sam68 and p53 recruitment to target promoters is interdependent, we next investigated whether the Sam68 RNA binding activity would be required for its recruitment. Consistent with our co-immunoprecipitation data, ChIP analysis of the P21 promoter showed that both Sam68 and p53 recruitment was compromised in response to doxorubicin treatment when transfected with the RNA-binding deficient mutant, while their recruitment was more robust in the presence of full-length Sam68 (Figure 6C and D). Taken together, our data suggest that the RNA-binding integrity of Sam68 is required for enhanced association with p53, and that this interaction is important for their efficient target promoter recruitment.
RBPs play key roles in directing lncRNA functions, mainly through RBP-lncRNA interactions (34,35). Many lncRNAs have been shown to influence p53 function (23). We focused on PR-lncRNA-1 and lincRNA-p21, as their depletion results in defective p53 gene transactivation (31,35), consistent with what we observe in Sam68-deficient cells. Interestingly, RT-qPCR analysis showed that while the presence of Sam68 was required for maximum induction of PR-lncRNA-1 expression with doxorubicin, it did not significantly influence the upregulation of lincRNA-p21 (Figure 6E). We then performed RNA immunoprecipitation (RIP) following UV cross-linking with an anti-Sam68 antibody in wild type HCT116 cells subjected to DNA damage. We observed an enrichment of PR-lncRNA-1, but not lincRNA-p21 nor an unrelated RNA (GAPDH) with the Sam68 antibody compared to IgG control (Figure 6F). To investigate the functional significance of this association, we depleted PR-lncRNA-1 expression using antisense oligonucleotides (ASOs), as previously described (31) (Figure 6G). Our ChIP data revealed that depletion of PR-lncRNA-1 significantly decreased doxorubicin-induced Sam68 recruitment to the P21 promoter (Figure 6H). Similarly, p53 recruitment to the P21 promoter was also impaired in doxorubicin treated cells depleted of PR-lncRNA-1 (Figure 6I), consistent with the interdependence between Sam68 and p53, as well as findings from previous publication (31). Altogether, our results suggest that Sam68 acts in concert with PR-lncRNA-1 for efficient p53 promoter recruitment and transcription.
DISCUSSION
In this study, we identify Sam68 as a transcriptional coactivator of p53. We show using CRISPR/Cas9-generated HCT116 cell lines, as well as RNAi, that p53 requires Sam68 to stimulate expression of direct downstream genes involved in negative feedback (MDM2), cell cycle arrest (P21) and apoptosis (BAX, PUMA) in response to DNA damaging agents, as well as to enhance p53-mediated cellular responses. This was further supported by the inability of p53 to transactivate luciferase reporters containing p53-binding sites (PG13, P21 promoter) in Sam68−/− cells. Sam68 physically interacted with p53 in an RNA-dependent manner, and was recruited at p53-responsive promoters along with p53. Specifically, we identify PR-lncRNA-1 as an important Sam68-associated RNA critical for Sam68/p53 promoter recruitment. Hence, our findings define a new role for Sam68 in p53 transactivation.
But how exactly does Sam68 enhance p53-dependent transcription? Early studies have shown that p53 binding to its cognate sequences is not a stable event, therefore p53 can dissociate from DNA in the absence of other factors (36,37). Our data suggest that following DNA damage-induced interaction of p53 and Sam68, which is mediated through both the N-terminus and KH domain of Sam68, we observed increased loading onto target promoters, notably the P21 and PUMA promoters. We speculate that it is most likely due to increased stability/assembly of p53 promoter complexes facilitated by Sam68. This model is supported by previous studies of other p53 transcriptional cofactors as well (38,39). Sam68 might also enhance p53 transcriptional activity by functioning as an adaptor molecule to recruit additional coactivators such as CBP or PRMT1 (11,40), similar to its transcriptional regulation of the MLL complex (41), as well as localization of other p53 coactivators like ASPP1 (42,43). It is interesting to note that Sam68 depletion had a lesser effect on p53 recruitment for the PUMA promoter compared to the P21 promoter in doxorubicin treated cells, most probably attributed to their differences in transcriptional requirements (21,44). Although we have not investigated the possibility of Sam68 in regulating p53 activation per se, our results did show that Sam68 depletion did not alter DNA damage-induced p53 stabilization. In addition to the transcriptional role of Sam68, which absolutely requires functional p53, we observed that under normal cell culture conditions, Sam68 depletion slightly increased p53 protein levels. This implicates Sam68 in maintaining p53 levels in the absence of DNA damage, most likely by taking part in the negative feedback loop with MDM2 (45). Moreover, in a p53-independent manner, Sam68 depletion in undamaged HCT116 cells specifically led to increased p21 mRNA and protein expression, consistent with previous observations in breast cancer cells (14). We also show that Sam68 regulates p21 mRNA stability in a p53-independent manner. Since Sam68 is known to bind a bipartite UU/AAA RNA element (46), this notion was further supported by the presence of a consensus Sam68 binding site (nucleotides 727–736; UUAAUUUAAA) in its 3′-untranslated region (3′ UTR). Indeed, the stability of p21 mRNA can be modulated by binding to its 3′ UTR, as shown previously for RBPs such as PCBP4 (47). Moreover, Sam68 may also have ‘successor’ activity to p53 for checkpoint control similar to hnRNPA0 (48). Together, these data elucidate distinct Sam68 functions that are dictated by the p53 status. Our findings suggest that Sam68, like hnRNPA0, can participate in alternative pathways under p53-deficient contexts, though more in-depth investigations are required to decipher the exact mechanisms of action.
To further understand how Sam68 functions as a p53 transcriptional modulator in response to DNA damage, and more specifically to prove the validity of our CRISPR system, we performed rescue experiments in CRISPR-generated Sam68−/− cells using GFP-tagged constructs that encode either full-length, ΔKH- or ΔN-truncated forms of Sam68. Our data demonstrated that while full-length Sam68 effectively rescued p53-dependent gene transactivation in doxorubicin treated cells, the ΔKH and ΔN mutants did not. Thus, the rescue experiments provided crucial evidence to not only confirm that the observed phenomenon was Sam68-specific, but also highlighted the importance of the KH domain and the N-terminus in Sam68/p53 complex formation, consistent with previous work demonstrating the requirement of RPS3 KH domain for p53 interaction (45). These results implicate that Sam68 associates with p53 through both protein-protein and RNA-dependent interactions, and suggest that this complex formation is essential for Sam68 to function as a transcriptional coactivator of p53.
To add another layer of complexity, recent genome-wide studies have implicated lncRNAs as p53 targets involved in p53 regulation or effector functions (23). In our study, we show that Sam68 increases its association with p53 through RNA-binding. Specifically, we identify Sam68 as an interacting partner of PR-lncRNA-1, a well-characterized p53 target required for p53 chromatin recruitment (31), and identified a consensus Sam68 binding site (nucleotides 1149–1159; UUAAUAUUUAA) in the available sequence. Interestingly, the presence of Sam68 selectively enhanced DNA damage-induced expression of PR-lncRNA-1, and the upregulation of PR-lncRNA-1 in turn promoted increased Sam68 and p53 loading onto the target promoter. Although Sam68 was required for the optimal expression of PR-lncRNA-1 (Figure 6E), depletion of PR-lncRNA-1 alone cannot fully recapitulate the defective p53 transactivation observed in Sam68-deficient cells. For example, MDM2 gene expression and p53 recruitment to the MDM2 promoter was not altered in cells depleted of PR-lncRNA-1 (31), but was significantly reduced in Sam68-deficient cells (Figure 2A and Supplementary Figure S6). Based on our data, we propose that PR-lncRNA-1 acts in concert with Sam68 to stimulate p53-mediated transcription, perhaps by changing local chromatin conformation. These findings suggest a positive-feedback mechanism in which lncRNAs induced by the Sam68/p53 complex enhances p53 transcriptional activity. The significance of Sam68-lncRNA interaction has also been observed previously for INXS during Sam68-mediated BCL-XS splicing (49).
Sam68−/− mice, like p21−/− and Bax−/− mice, are not tumor prone, suggesting that the tumor suppressor activity of p53 is not mediated through Sam68 alone (18,50–52), while also implicating that there may be some redundancies with other KH-type RBPs (53). Indeed, it has been recently shown that QUAKING, an RBP closely related to Sam68, can exhibit tumor suppressive roles (54,55). Consistent with this notion, overexpression of Sam68 in normal fibroblasts triggered cell cycle arrest and apoptosis that are characteristic of tumor suppressors (56). On the other hand, tyrosine phosphorylation has been demonstrated to drastically alter Sam68 function, where it switches Sam68 from pro-apoptotic to anti-apoptotic (5). This was further confirmed in an independent study where prostate cancer cells transfected with a Sam68 mutant lacking tyrosine phosphorylation sites were augmented in their sensitivity to apoptosis (57), suggesting that altered Sam68 function contributes to tumorigenesis. Nevertheless, we do not exclude the possibility that Sam68 could enhance the activity of other tumor suppressors. In conclusion, our findings reveal a new role for Sam68 as an essential component of the p53 response pathway, where its presence and interaction is crucial for p53 transactivation to regulate tumor suppression.
Supplementary Material
Acknowledgments
The authors thank Zhenbao Yu and Gillian Vogel for helpful discussions and for critically reading the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
CIHR/FRSQ training grant in cancer research of the McGill Integrated Cancer Research Training Program [FRN53888 to N.L.]; Canadian Institute of Health Research [MOP-123531 to S. R.]. Funding for open access charge: Canadian Institute of Health Research [MOP-123531].
Conflict of interest statement. None declared.
REFERENCES
- 1.Richard S. Reaching for the stars: Linking RNA binding proteins to diseases. Adv. Exp. Med. Biol. 2010;693:142–157. [PubMed] [Google Scholar]
- 2.Bielli P., Busa R., Paronetto M.P., Sette C. The RNA-binding protein Sam68 is a multifunctional player in human cancer. Endocr. Relat. Cancer. 2011;18:R91–R102. doi: 10.1530/ERC-11-0041. [DOI] [PubMed] [Google Scholar]
- 3.Matter N., Herrlich P., Konig H. Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature. 2002;420:691–695. doi: 10.1038/nature01153. [DOI] [PubMed] [Google Scholar]
- 4.Paronetto M.P., Cappellari M., Busa R., Pedrotti S., Vitali R., Comstock C., Hyslop T., Knudsen K.E., Sette C. Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res. 2010;70:229–239. doi: 10.1158/0008-5472.CAN-09-2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paronetto M.P., Achsel T., Massiello A., Chalfant C.E., Sette C. The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J. Cell Biol. 2007;176:929–939. doi: 10.1083/jcb.200701005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iijima T., Wu K., Witte H., Hanno-Iijima Y., Glatter T., Richard S., Scheiffele P. SAM68 regulates neuronal activity-dependent alternative splicing of neurexin-1. Cell. 2011;147:1601–1614. doi: 10.1016/j.cell.2011.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huot M.E., Vogel G., Zabarauskas A., Ngo C.T., Coulombe-Huntington J., Majewski J., Richard S. The Sam68 STAR RNA-binding protein regulates mTOR alternative splicing during adipogenesis. Mol. Cell. 2012;46:187–199. doi: 10.1016/j.molcel.2012.02.007. [DOI] [PubMed] [Google Scholar]
- 8.Huot M.E., Brown C.M., Lamarche-Vane N., Richard S. An adaptor role for cytoplasmic Sam68 in modulating Src activity during cell polarization. Mol. Cell. Biol. 2009;29:1933–1943. doi: 10.1128/MCB.01707-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paronetto M.P., Messina V., Bianchi E., Barchi M., Vogel G., Moretti C., Palombi F., Stefanini M., Geremia R., Richard S., et al. Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J. Cell Biol. 2009;185:235–249. doi: 10.1083/jcb.200811138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Batsche E., Yaniv M., Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
- 11.Hong W., Resnick R.J., Rakowski C., Shalloway D., Taylor S.J., Blobel G.A. Physical and functional interaction between the transcriptional cofactor CBP and the KH domain protein Sam68. Mol. Cancer Res. 2002;1:48–55. [PubMed] [Google Scholar]
- 12.Rajan P., Gaughan L., Dalgliesh C., El-Sherif A., Robson C.N., Leung H.Y., Elliott D.J. The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J. Pathol. 2008;215:67–77. doi: 10.1002/path.2324. [DOI] [PubMed] [Google Scholar]
- 13.Fu K., Sun X., Zheng W., Wier E.M., Hodgson A., Tran D.Q., Richard S., Wan F. Sam68 modulates the promoter specificity of NF-κB and mediates expression of CD25 in activated T cells. Nat. Commun. 2013;4:1909. doi: 10.1038/ncomms2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Song L., Wang L., Li Y., Xiong H., Wu J., Li J., Li M. Sam68 up-regulation correlates with, and its down-regulation inhibits, proliferation and tumourigenicity of breast cancer cells. J. Pathol. 2010;222:227–237. doi: 10.1002/path.2751. [DOI] [PubMed] [Google Scholar]
- 15.Chen T., Damaj B.B., Herrera C., Lasko P., Richard S. Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol. Cell. Biol. 1997;17:5707–5718. doi: 10.1128/mcb.17.10.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aubele M., Walch A.K., Ludyga N., Braselmann H., Atkinson M.L., Luber B., Auer G., Tapio S., Cooke T., Bartlett J.M. Prognostic value of protein tyrosine kinase 6 (PTK6) for long-term survival of breast cancer patients. Br. J. Cancer. 2008;99:1089–1095. doi: 10.1038/sj.bjc.6604660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z., Yu C.P., Zhong Y., Liu T.J., Huang Q.D., Zhao X.H., Huang H., Tu H., Jiang S., Zhang Y., et al. Sam68 expression and cytoplasmic localization is correlated with lymph node metastasis as well as prognosis in patients with early-stage cervical cancer. Ann. Oncol. 2012;23:638–646. doi: 10.1093/annonc/mdr290. [DOI] [PubMed] [Google Scholar]
- 18.Richard S., Torabi N., Franco G.V., Tremblay G.A., Chen T., Vogel G., Morel M., Cleroux P., Forget-Richard A., Komarova S., et al. Ablation of the Sam68 RNA binding protein protects mice from age-related bone loss. PLoS Genet. 2005;1:e74. doi: 10.1371/journal.pgen.0010074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Richard S., Vogel G., Huot M.E., Guo T., Muller W.J., Lukong K.E. Sam68 haploinsufficiency delays onset of mammary tumorigenesis and metastasis. Oncogene. 2008;27:548–556. doi: 10.1038/sj.onc.1210652. [DOI] [PubMed] [Google Scholar]
- 20.Liu K., Li L., Nisson P.E., Gruber C., Jessee J., Cohen S.N. Neoplastic transformation and tumorigenesis associated with sam68 protein deficiency in cultured murine fibroblasts. J. Biol. Chem. 2000;275:40195–40201. doi: 10.1074/jbc.M006194200. [DOI] [PubMed] [Google Scholar]
- 21.Zilfou J.T., Lowe S.W. Tumor suppressive functions of p53. Cold Spring Harb. Perspect. Biol. 2009;1:a001883. doi: 10.1101/cshperspect.a001883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vousden K.H., Prives C. Blinded by the light: The growing complexity of p53. Cell. 2009;137:413–431. doi: 10.1016/j.cell.2009.04.037. [DOI] [PubMed] [Google Scholar]
- 23.Zhang A., Xu M., Mo Y.Y. Role of the lncRNA-p53 regulatory network in cancer. J. Mol. Cell Biol. 2014;6:181–191. doi: 10.1093/jmcb/mju013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hollstein M., Sidransky D., Vogelstein B., Harris C.C. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 25.Hainaut P., Hollstein M. p53 and human cancer: the first ten thousand mutations. Adv. Cancer Res. 2000;77:81–137. doi: 10.1016/s0065-230x(08)60785-x. [DOI] [PubMed] [Google Scholar]
- 26.Bunz F., Dutriaux A., Lengauer C., Waldman T., Zhou S., Brown J.P., Sedivy J.M., Kinzler K.W., Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 27.Mali P., Yang L., Esvelt K.M., Aach J., Guell M., DiCarlo J.E., Norville J.E., Church G.M. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., Mueller R., Nolan T., Pfaffl M.W., Shipley G.L., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 29.Espinosa J.M., Verdun R.E., Emerson B.M. p53 functions through stress- and promoter-specific recruitment of transcription initiation components before and after DNA damage. Mol. Cell. 2003;12:1015–1027. doi: 10.1016/s1097-2765(03)00359-9. [DOI] [PubMed] [Google Scholar]
- 30.Jackson J.G., Pereira-Smith O.M. p53 is preferentially recruited to the promoters of growth arrest genes p21 and GADD45 during replicative senescence of normal human fibroblasts. Cancer Res. 2006;66:8356–8360. doi: 10.1158/0008-5472.CAN-06-1752. [DOI] [PubMed] [Google Scholar]
- 31.Sanchez Y., Segura V., Marin-Bejar O., Athie A., Marchese F.P., Gonzalez J., Bujanda L., Guo S., Matheu A., Huarte M. Genome-wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat. Commun. 2014;5:5812. doi: 10.1038/ncomms6812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukong K.E., Richard S. Sam68, the KH domain-containing superSTAR. Biochim. Biophys. Acta. 2003;1653:73–86. doi: 10.1016/j.bbcan.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 33.Kaufmann S.H., Desnoyers S., Ottaviano Y., Davidson N.E., Poirier G.G. Specific proteolytic cleavage of poly(ADP-ribose) polymerase: an early marker of chemotherapy-induced apoptosis. Cancer Res. 1993;53:3976–3985. [PubMed] [Google Scholar]
- 34.Zhang A., Zhou N., Huang J., Liu Q., Fukuda K., Ma D., Lu Z., Bai C., Watabe K., Mo Y.Y. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dimitrova N., Zamudio J.R., Jong R.M., Soukup D., Resnick R., Sarma K., Ward A.J., Raj A., Lee J.T., Sharp P.A., et al. LincRNA-p21 activates p21 in cis to promote Polycomb target gene expression and to enforce the G1/S checkpoint. Mol. Cell. 2014;54:777–790. doi: 10.1016/j.molcel.2014.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prives C., Hall P.A. The p53 pathway. J. Pathol. 1999;187:112–126. doi: 10.1002/(SICI)1096-9896(199901)187:1<112::AID-PATH250>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 37.Banerjee S., Kumar B.R., Kundu T.K. General transcriptional coactivator PC4 activates p53 function. Mol. Cell. Biol. 2004;24:2052–2062. doi: 10.1128/MCB.24.5.2052-2062.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bates G.J., Nicol S.M., Wilson B.J., Jacobs A.M., Bourdon J.C., Wardrop J., Gregory D.J., Lane D.P., Perkins N.D., Fuller-Pace F.V. The DEAD box protein p68: a novel transcriptional coactivator of the p53 tumour suppressor. Nucleic Acids Res. 2005;3:543–553. doi: 10.1038/sj.emboj.7600550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moumen A., Masterson P., O'Connor M.J., Jackson S.P. hnRNP K: an HDM2 target and transcriptional coactivator of p53 in response to DNA damage. Cell. 2005;123:1065–1078. doi: 10.1016/j.cell.2005.09.032. [DOI] [PubMed] [Google Scholar]
- 40.An W., Kim J., Roeder R.G. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 41.Cheung N., Chan L.C., Thompson A., Cleary M.L., So C.W. Protein arginine-methyltransferase-dependent oncogenesis. Nat. Cell Biol. 2007;9:1208–1215. doi: 10.1038/ncb1642. [DOI] [PubMed] [Google Scholar]
- 42.Thornton J.K., Dalgleish C., Venables J.P., Sergeant K.A., Ehrmann I.E., Lu X., Saunders P.T.K., Elliott D.J. The tumour-suppressor protein ASPP1 is nuclear in human germ cells and can modulate ratios of CD44 exon V5 spliced isoforms in vivo. Oncogene. 2006;25:3104–3112. doi: 10.1038/sj.onc.1209341. [DOI] [PubMed] [Google Scholar]
- 43.Samuels-Lev Y., O'Connor D.J., B D., Trigiante G., Hsieh J.K., Zhong S., Campargue I., Naumovski L., Crook T., Lu X. ASPP proteins specifically stimulate the apoptotic function of p53. Mol. Cell. 2001;8:781–794. doi: 10.1016/s1097-2765(01)00367-7. [DOI] [PubMed] [Google Scholar]
- 44.Beckerman R., Prives C. Transcriptional regulation by p53. Cold Spring Harb. Perspect. Biol. 2010;2:a000935. doi: 10.1101/cshperspect.a000935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yadavilli S., Mayo L.D., Higgins M., Lain S., Hegde V., Deutsch W.A. Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair (Amst) 2009;8:1215–1224. doi: 10.1016/j.dnarep.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Galarneau A., Richard S. The STAR RNA binding proteins GLD-1, QKI, SAM68 and SLM-2 bind bipartite RNA motifs. BMC Mol. Biol. 2009;10:47. doi: 10.1186/1471-2199-10-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Scoumanne A., Cho S.J., Zhang J., Chen X. The cyclin-dependent kinase inhibitor p21 is regulated by RNA-binding protein PCBP4 via mRNA stability. Nucleic Acids Res. 2011;39:213–224. doi: 10.1093/nar/gkq778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cannell I.G., Merrick K.A., Morandell S., Zhu C.Q., Braun C.J., Grant R.A., Cameron E.R., Tsao M.S., Hemann M.T., Yaffe M.B. A pleiotropic RNA-binding protein controls distinct cell cycle checkpoints to drive resistance of p53-defective tumors to chemotherapy. Cancer Cell. 2015;28:623–637. doi: 10.1016/j.ccell.2015.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeOcesano-Pereira C., Amaral M.S., Parreira K.S., Ayupe A.C., Jacysyn J.F., Amarante-Mendes G.P., Reis E.M., Verjovski-Almeida S. Long non-coding RNA INXS is a critical mediator of BCL-XS induced apoptosis. Nucleic Acids Res. 2014;42:8343–8355. doi: 10.1093/nar/gku561. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 50.Macleod K.F., Jacks T. Insights into cancer from transgenic mouse models. J. Pathol. 1999;187:43–60. doi: 10.1002/(SICI)1096-9896(199901)187:1<43::AID-PATH246>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 51.Deng C., Zhang P., Harper J.W., Elledge S.J., Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 52.Knudson C.M., Tung K.S., Tourtellotte W.G., Brown G.A., Korsmeyer S.J. Bax-deficient mice with lymphoid hyperplasia and male germ cell death. Science. 1995;270:96–99. doi: 10.1126/science.270.5233.96. [DOI] [PubMed] [Google Scholar]
- 53.Di Fruscio M., Chen T., Richard S. Characterization of Sam68-like mammalian proteins SLM-1 and SLM-2: SLM-1 is a Src substrate during mitosis. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2710–2715. doi: 10.1073/pnas.96.6.2710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zong F.Y., Fu X., Wei W.J., Luo Y.G., Heiner M., Cao L.J., Fang Z., Fang R., Lu D., Ji H., et al. The RNA-binding protein QKI suppresses cancer-associated aberrant splicing. PLoS Genet. 2014;10:e1004289. doi: 10.1371/journal.pgen.1004289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bandopadhayay P., Ramkissoon L.A., Jain P., Bergthold G., Wala J., Zeid R., Schumacher S.E., Urbanski L., O'Rourke R., Gibson W.J., et al. MYB-QKI rearrangements in angiocentric glioma drive tumorigenicity through a tripartite mechanism. Nat. Genet. 2016;48:273–282. doi: 10.1038/ng.3500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Taylor S.J., Resnick R.J., Shalloway D. Sam68 exerts separable effects on cell cycle progression and apoptosis. BMC Cell Biol. 2004;5:5. doi: 10.1186/1471-2121-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Busa R., Paronetto M.P., Farini D., Pierantozzi E., Botti F., Angelini D.F., Attisani F., Vespasiani G., Sette C. The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene. 2007;26:4372–4382. doi: 10.1038/sj.onc.1210224. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.