Abstract
Annotation of herpesvirus genomes has traditionally been undertaken through the detection of open reading frames and other genomic motifs, supplemented with sequencing of individual cDNAs. Second generation sequencing and high-density microarray studies have revealed vastly greater herpesvirus transcriptome complexity than is captured by existing annotation. The pervasive nature of overlapping transcription throughout herpesvirus genomes, however, poses substantial problems in resolving transcript structures using these methods alone. We present an approach that combines the unique attributes of Pacific Biosciences Iso-Seq long-read, Illumina short-read and deepCAGE (Cap Analysis of Gene Expression) sequencing to globally resolve polyadenylated isoform structures in replicating Epstein-Barr virus (EBV). Our method, Transcriptome Resolution through Integration of Multi-platform Data (TRIMD), identifies nearly 300 novel EBV transcripts, quadrupling the size of the annotated viral transcriptome. These findings illustrate an array of mechanisms through which EBV achieves functional diversity in its relatively small, compact genome including programmed alternative splicing (e.g. across the IR1 repeats), alternative promoter usage by LMP2 and other latency-associated transcripts, intergenic splicing at the BZLF2 locus, and antisense transcription and pervasive readthrough transcription throughout the genome.
INTRODUCTION
Epstein-Barr virus (EBV) is a human gammaherpesvirus that is endemic worldwide and is associated with a number of cancers including Hodgkin lymphoma, Burkitt and other non-Hodgkin lymphomas, nasopharyngeal carcinoma and gastric carcinoma (1,2). Initial infection leads to a complex progression through multiple viral gene expression programs after which time the virus typically transitions to a highly latent (type 0 latency), immunologically silent state in memory B cells where no viral protein coding genes are expressed. EBV's distinct latency gene expression programs serve various purposes during its infection cascade and are uniquely manifested in different diseases (for reviews see (3) and (4)). EBV-positive Burkitt lymphoma tumors show a restricted latency type I viral gene expression pattern in which the only viral protein expressed is EBNA1, which is essential for replication and segregation of the EBV genome during S-phase and mitosis. Type II latency, associated with Hodgkin lymphoma and nasopharyngeal carcinoma, is characterized by the expression of EBNA1 and the membrane proteins LMP1 and LMP2A/B. In type III latency, which is associated with post-transplant lymphoproliferative disease and in vitro EBV-immortalized lymphoblastoid cell lines (LCLs), the full spectrum of EBV latency proteins is expressed: LMP1, LMP2A, LMP2B, EBNA1 EBNA2, EBNA-LP, EBNA3A, EBNA3B and EBNA3C. Although the virus typically persists in the host in a predominantly latent form lytic reactivation is occasionally triggered, inducing widespread transcription of the viral genome and production of infectious virus.
When the EBV genome was first sequenced, gene annotation was based primarily on the detection of ORFs and salient genomic promoter and transcript termination features (5). Since then, full transcript structures have been determined on a primarily gene-by-gene basis through cloning and sequencing of individual cDNAs. Based on this cumulative work, there are ∼90 transcripts currently annotated in the EBV genome (see NCBI Reference Sequence NC_007605.1 and KC207813.1).
New technologies including tiling arrays and next generation RNA sequencing have revealed the presence of many more lytic transcripts than was previously known in EBV and related herpesviruses (6–9). Despite abundant evidence of antisense and intergenic transcription in herpesviruses however, extensive transcript overlap throughout the genome makes it difficult to demarcate transcript start, end and splicing features from tiling arrays or short-read sequencing. This problem has confounded attempts to definitively annotate transcript structures.
Here we report a new workflow that overcomes obstacles to transcript structure resolution in high gene-density genomes. Our approach, termed Transcriptome Resolution through Integration of Multi-platform Data (TRIMD), integrates unique complementary characteristics of three distinct sequencing technologies to discover, validate and annotate polyadenylated transcript structure features throughout a genome. The Pacific Biosciences Iso-Seq protocol for Single-Molecule Real-Time (SMRT) long-read sequencing of full-length RNA transcripts forms the basis of this method, with the integration of deepCAGE data to identify and validate transcript 5′ ends and Illumina short-read RNA-Seq data to identify and validate splice junctions and 3′ ends. To facilitate the general application of TRIMD, we have generated a flexible set of scripts that allow customized transcript resolution and annotation of other gene-dense as well as less complex genomes.
MATERIALS AND METHODS
Cell culture
Akata, Mutu I, JY and X50-7 cells were cultured in RPMI 1640 medium (Thermo Scientific, catalog no. SH30027) supplemented with 10% fetal bovine serum (FBS; Invitrogen-Gibco, catalog no. 16000) and 0.5% penicillin-streptomycin (pen/strep; Invitrogen-Gibco, catalog no. 15070) in a 37°C, 5% CO2 humidified incubator.
Lytic cycle induction
Near-saturation Akata and Mutu I cell cultures were diluted with equal volumes of fresh RPMI 1640 (with 10% FBS and 0.5% pen/strep) one day before induction. To induce reactivation, cells were pelleted and resuspended at a concentration of 106 cells/ml in fresh RPMI 1640 (with 10% FBS and 0.5% pen/strep) plus either 10μg/ml anti-human IgG (Sigma-Aldrich, catalog no. I2136) (for Akata cells) or 10 μg/ml anti-human IgM (Sigma-Aldrich, catalog no. I0759) (for Mutu cells). For Pacific Biosciences Iso-Seq, cells were harvested at 20 and 24 h post-induction, for Illumina RNA-Seq, cells were harvested at 0 min, 5 min, 30 min, 1 h, 2 h, 4 h, 8 h, 24 h and 48 h post-induction and for deepCAGE, cells were harvested at 24 h post-induction. For qRT-PCR, cells were harvested at 0 and 24 h (Akata) or 0 and 48 h (Mutu).
RNA extraction
Whole cell RNA was extracted with TRIzol reagent (Life Technologies, catalog no. 15596-018) according to the vendor's protocol. Nuclear and cytoplasmic RNA isolation for qRT-PCR was carried out using a cytoplasmic and nuclear RNA purification kit from Norgen Biotek (catalog no. 2100).
Sequencing
For PacBio SMRT Iso-Seq, polyadenylated RNA was selected using a Poly(A)Purist MAG kit (Life Technologies, catalog no. AM1922) and 7 and 3.3 µg of polyA RNA from 20 and 24 h induction time points were pooled. Library preparation and sequencing was performed according to the Pacific Biosciences Iso-Seq protocol by the Johns Hopkins Deep Sequencing and Microarray Core Facility, Baltimore, Maryland, USA. Eight SMRT cells were used: two with a 1–2 kb RNA fraction, two with a 2–3 kb RNA fraction and four with non-size-selected RNA.
For Illumina RNA-Seq, samples were treated with DNAse (Ambion AM1906) then poly(A)-selected and prepared using the TruSeq stranded protocol (Illumina, catalog no. RS-930-2001), then subjected to 2 × 101 base paired-end sequencing using an Illumina HiSeq 2000 instrument. Poly(A) selection, library preparation and sequencing were performed by the University of Wisconsin Biotechnology Center, Madison, Wisconsin, USA.
For deepCAGE, nAnT-iCAGE libraries (10) were prepared from RNA extracted from two parallel samples of induced (24 h) Akata cells. From each sample a portion of the RNA was treated with DNAse (Ambion AM1906) and a portion was left untreated, for four total samples. Samples were subjected to 50-base single-end sequencing using an Illumina HiSeq 2500 instrument. Library preparation and sequencing were performed by DNAform, Yokohama, Japan.
Sequence data have been deposited to NCBI GEO (GSE79337).
Data acquisition
Pacific Biosciences SMRT sequence data for type III latency LCLs was downloaded from NCBI SRA, accession number SRP036136 (11).
Previously published Illumina RNA-Seq data from induced and uninduced Akata cells was downloaded from NCBI GEO, series accession number GSE52490 (9). Previously published RNA-Seq data for JY cells was downloaded from NCBI SRA, accession number SRR364065 (12,13).
Sequence alignments
Pacific Biosciences SMRT consensus ‘full-length’ isoforms (CFLs) were aligned and mapped with GMAP (14) release 21 July 2014 to the human (hg19 assembly) and Akata EBV (NCBI accession number KC207813.1 (15)) genomes. For this analysis, the circular Akata EBV genome was ‘inverted’, or split between the BBRF3 and BGLF3 genes (between positions 107 954 and 107 955) rather than the terminal repeats to allow for the detection of LMP2 transcripts, which span the terminal repeats. Additionally, the CFLs were aligned to the un-split (standard) Akata genome to identify transcripts spanning the 107 954–107 955 junction. Full-length isoforms unpolished by Quiver were used in these analyses as we observed that Quiver polishing sometimes obscured introns and prevented discrimination of overlapping transcripts in the gene-dense EBV genome. Only reads mapping to a single location were retained (argument –n 1). Illumina RNA-Seq reads were aligned and mapped using STAR (16) to the human (hg19 assembly) and Akata EBV genomes (‘inverted’ and standard as described above) with default settings. To find Illumina reads mapping to splice junctions in the IR1 W repeat region (bases 75 265–98 628 on the inverted Akata genome), the STAR outFiltermultimapNmax argument was set to 100 to report alignments for reads that mapped up to 100 times. Transcript abundance estimates were generated from Illumina RNA-Seq reads using RSEM (17) with an annotation file including the human genome GRCh38 assembly and the updated Akata annotation. DeepCAGE reads were aligned with STAR using the parameters --outFiltermultimapNmax 100 and --outSAMprimaryFlag AllBestScore to identify start sites in repeat regions.
For Pacific Biosciences SMRT sequence data from type III latency LCLs, reads were first oriented using their poly(A) tails. Reads ending with AAAAAAA and reads beginning with TTTTTTT were extracted. Reads beginning with TTTTTTT and their quality scores were reversed to produce fastq files of ‘sense’ oriented RNA. These reads were then aligned with GMAP (14) release 21 July 2014 to the Akata EBV (NCBI accession number KC207813.1 (15)) genome, split as above between positions 107 954 and 107 955.
Determination of full-length coverage percentages of expressed cellular genes
All human RNA transcripts with RefSeq Reviewed or Validated status were evaluated. A transcript was considered to have full-length Iso-Seq coverage if a CFL could be identified with 5′ and 3′ ends mapping within 50 and 20 bp respectively of the annotated transcript's 5′ and 3′ ends. A transcript was considered to have incomplete coverage if a CFL's 3′ end aligned within 20 bp of the annotated transcript's 3′ end, its 5′ end did not map within 50 bases of the annotated transcript's 5′ end, and at least five Illumina RNA-Seq reads mapped within the first 100 bp of the first exon (this reduces false calls of incomplete coverage for non-expressed transcripts that share a 3′ end with expressed transcripts).
Identification of transcription start sites
Iso-Seq CFL 5′ end clusters were generated based on CFL 5′ ends mapping within 8 bp of each other. Only CFL ends that were not softclipped were considered (i.e. CFLs whose 5′ ends did not contain mismatches to the genomic sequence). The consensus transcription start site for each cluster was determined by calculating weighted (based on the number of SMRT circular consensus sequence reads starting at each coordinate) averages of the start coordinates of CFLs within the cluster.
Start site clusters in mapped deepCAGE data were extracted using Paraclu (18) with the parameters (i) a minimum of 15 tags/cluster, (ii) (maximum density/baseline density) ≥ 2 and (iii) 1–20 base cluster length. Consensus transcription start sites were determined by calculating weighted (based on the number of CAGE tags starting at each coordinate) averages of the start coordinates of each read in a cluster. Only consensus start sites appearing within two bases of each other in at least three of the four CAGE samples were used to validate Iso-Seq consensus transcription start sites.
Iso-Seq consensus transcription start sites that were within three bases of CAGE consensus transcription start sites were considered validated.
Identification of splice junctions
Splice junctions were identified using GMAP for Iso-Seq CFLs and STAR for Illumina reads. Splice junctions detected by both Iso-Seq and Illumina short-read sequencing were considered validated. Because 101-bp reads cannot be assigned definitively to specific splice junctions within the W repeat region, a repeat splice junction was considered to have been detected by Illumina RNA-Seq if any of the set of possible alignments was reported by STAR. Illumina RNA-Seq read depth for repeat splice junctions was normalized by dividing by the number of equivalent genomic alignments possible.
Identification of polyadenylation sites
Non-softclipped Iso-Seq CFL 3′ ends aligning within 8 bp of each other on the genome were considered a single candidate polyadenylation site. The CFL consensus polyadenylation sites were determined by calculating weighted (based on the number of SMRT circular consensus sequence reads ending at each coordinate) averages of the end coordinates.
Illumina reads containing putative poly(A) tails were extracted from SAM alignment files using the following criteria: reads identified by FLAG code as being first-of-pair (for paired-end sequencing) that end with a run of at least five As, at least two of which are softclipped (plus strand) or that start with a run of at least five Ts, at least two of which are softclipped (minus strand). The alignment position of the softclipped-adjacent base was taken to represent a candidate polyadenylation site, with sites situated within 8 bp of each other considered single candidate polyadenylation sites. The Illumina consensus polyadenylation site was determined using a read-end-depth weighted average as for the Iso-Seq CFL 3′ ends (above). Candidate polyadenylation sites were considered validated if they were supported by at least five SMRT reads and the presence of an Illumina candidate polyadenylation site on the same strand within four bases upstream or ten bases downstream.
Isoform validation
Each Iso-Seq CFL was examined to determine whether its 5′ end, 3′ end and splice junctions (if any) met the validation criteria described above. When reporting isoforms, 5′ and 3′ ends that formed part of validated consensus transcription start sites and polyadenylation sites were adjusted to match the validated consensus sites, if necessary. Finally, CFLs that had matching validated transcription start sites, polyadenylation sites and splice junctions (if any) were collapsed into ‘validated transcripts’. Validated transcripts were compared to annotated genes in the Akata genome (NCBI accession number KC207813.1) to determine whether they had been previously annotated.
Naming of new transcripts
The naming scheme for novel transcripts is based on the existing naming scheme for EBV genes. The first two letters correspond to the genomic BamHI restriction fragment in which transcription initiates: e.g. BC is BamHI fragment C and Ba is BamHI fragment a. The next letter is R for Rightward transcripts (i.e. annotated on the plus strand) or L for Leftward transcripts (i.e. annotated on the minus strand). The final letter in our naming scheme is T for Transcript: the original naming scheme is based on protein-coding genes and uses F for reading Frame. We begin numbering our validated transcripts where the GenBank annotation numbering ends for each fragment. For example, BBRF3 is a previously annotated gene and we present here novel transcripts that we designate BBRT4, BBRT5, etc. This will allow easy gene name conversion for any transcripts that are later determined to be coding by simply changing the name to the ‘F’ version (e.g. BBRT4 to BBRF4).
Calculation of coding potential
Sequences of validated isoforms were analyzed for coding potential and the presence of open reading frames with the Coding Potential Assessment Tool (19).
qRT-PCR
cDNA was synthesized from 400 ng of RNA using an iScript cDNA synthesis kit (Bio-Rad catalog no. 170–8891) according to the vendor's protocol. Quantitative polymerase chain reaction (PCR) was performed using iQ SYBR green Supermix (Bio-Rad, catalog no. 170–8882) on a Bio-Rad CFX96 instrument. About 1 μl of cDNA product was denatured for 3 min at 95°C and amplified for 40 cycles of 15 s denaturation at 95°C and 1 min annealing/extension at 58°C. Total RNA transcript abundance was quantified using the comparative CT method (2−ΔΔCT) normalized to ACTB. Nuclear to cytoplasmic ratios were calculated as 2−(NuclearCt-CytoplasmicCt). The following primers were used:
ACTB: CACTCTTCCAGCCTTCCTTC and GTACAGGTCTTTGCGGATGT
Zta: CACGACGTACAAGGAAACCA and GAAGCCACCTCACGGTAGTG
W1–W1: TCGGGCCAGAGCCTAGGG and TGGTCCAGGGACTTCACTTC
W1–BHRF1: AGGGGAGACCGAAGTGAAGT and CCCTTGTTGAATAGGCCATC
W1–W2: AGGGGAGACCGAAGTGAAGT and CCTTCTACGGACTCGTCTGG
LMP2A exon 1 to exon 2: CCTACTCTCCACGGGATGAC and CGGTGTCAGCAGTTTCCTTT
Junction A: GCAGGTCAGACTTGGTGCTT and GAGTTGTTTCCGCCATCGT
Junction C: GCCCGAGGAGCTGTAGACC and GAGTTGTTTCCGCCATCGT
Junction D: CGATAGAGGGCCAGGTAGTG and GAGTTGTTTCCGCCATCGT
Junction E: GCAAAGGCAGGTCTTTCTCA and GAGTTGTTTCCGCCATCGT
Strand-specific qRT-PCR
The method of Feng et al. (20) was used for strand-specific quantitative reverse-transcription PCR. cDNA was synthesized from 400 ng RNA at 65°C for 50 min using gene-specific sequence modifying primers (or non-sequence modifying reverse primers for Zta and Kcnq1ot1) and ThermoScript reverse transcriptase (Life Technologies, catalog no. 12236-022) according to the manufacturer's protocol. Quantitative PCR was performed using iQ SYBR green Supermix (Bio-Rad, catalog no. 170–8882) on a Bio-Rad CFX96 instrument. About 1 μl of cDNA product was denatured for 3 min at 95°C and amplified for 40 cycles of 15 s denaturation at 95°C and 1 min annealing/extension at 58°C. Melting-curve analysis was performed from 58 to 95°C with a ramp of 0.5°C/5 s to confirm strand specificity. Total RNA transcript abundance was quantified using the comparative CT method (2−ΔΔCT) normalized to ACTB. Nuclear to cytoplasmic ratios were calculated as 2−(NuclearCt-CytoplasmicCt). The following primers were used:
ACTB: Sequence-modifying RT primer: GTACAGGTCTTTGCGGATGTttAtaTaACACTTCATG and qPCR primers CACTCTTCCAGCCTTCCTTC and GTACAGGTCTTTGCGGATGT
Zta: CACGACGTACAAGGAAACCA and GAAGCCACCTCACGGTAGTG
BCLT2/3: Sequence-modifying RT primer GTTCAGTGCGTCGAGTGCTcgCggcGGAACAG and qPCR primers CGCCAACAAGGTTCAATTTT and GTTCAGTGCGTCGAGTGCT
Kcnq1ot1: TACCGGATCCAGGTTTGCAGTACA and GCTGATAAAGGCACCGGAAGGAAA
FISH and immunofluorescence
Fluorescence in situ hybridization (FISH) was performed with custom Stellaris RNA FISH probes (Biosearch Technologies) using CAL Fluor Red 610. Immunolabeling was performed simultaneously using a modified version of the Stellaris protocol with mouse anti-EBV EA-D-p52/50 antibody (EMD Millipore catalog no. MAB8186) and Alexa Fluor 488 goat anti-mouse secondary antibody (Life Technologies catalog no. A11001). 10 × 106 24 h-induced or uninduced Akata cells were washed in phosphate buffered saline (PBS), fixed for 10 min in freshly made fixation buffer (3.7% formaldehyde in 1× PBS), washed in PBS and permeabilized for ∼24 h in 70% ethanol. Cells were washed in wash buffer (10% formamide in 2× saline sodium citrate (SSC) buffer), hybridized overnight at 37°C using freshly made hybridization buffer (100 mg/ml dextran sulfate and 10% formamide in 2× SSC) with 50 nM FISH probe or no probe, then incubated with the primary antibody (diluted 1:200) for 3 h at room temperature, washed in wash buffer and incubated for 30 min with the secondary antibody (diluted 1:500) and 5 ng/ml DAPI at 37°C in the dark. Cells were then washed in wash buffer a final time, mounted on slides with Prolong Diamond mounting medium (Life Technologies catalog no. P36961) and cured in the dark for 2 days. Imaging was performed using a Zeiss Axioplan 2 upright microscope and Z-stacks were deconvolved using Slidebook software (version 6) (Intelligent Imaging Innovations).
Transcript knockdown with GapmeR antisense oligonucleotides
GapmeRs targeted to the BZLT12-22 transcripts (sequence: TTTGGCCAGTCTTAAT) were designed and ordered from Exiqon. For knockdown, Akata cells were pelleted and resuspended in RPMI 1640 medium supplemented with 10% FBS (no antibiotic) and maintained in antibiotic-free medium for at least 2 days. For transfection, 3 × 106 cells per treatment were pelleted and resuspended in 100 μl Nucleofector Solution R (Lonza catalog no. VVCA-1001) with 600 pmol targeted GapmeR or negative control GapmeR A (Exiqon, catalog no. 300613-04). Cells were electroporated using program G-16 and transferred to a 6-well plate containing warm RPMI 1640 + 10% FBS. On the following day an equal volume of RPMI 1640 + 10% FBS, with 10 μg/ml anti-IgG or no anti-IgG was added to each well. Twenty-four hours later the cells were harvested and RNA extracted and subjected to poly(A)-selection and single-end Illumina 101 base sequencing as above.
RESULTS
Long-read sequencing of the EBV lytic transcriptome
To interrogate the EBV lytic transcriptome, we used a B-cell receptor mediated reactivation model in the Burkitt lymphoma cell line Akata (21). B-cell receptor crosslinking was induced by exposing Akata cells to an anti-IgG antibody and 20–24 h later, whole-cell RNA was prepared and subjected to poly(A) selection. Sequencing library preparation and Pacific Biosciences Single-Molecule Real-Time (SMRT) long-read sequencing were performed using the Iso-Seq method (22). Initial data analysis was performed using RS_IsoSeq on SMRTPortal v. 1 (22). From eight sequencing cells, 144 409 SMRT consensus ‘full-length’ isoforms (CFLs) were generated, supported by 376 548 circular consensus sequence SMRT reads that were determined to be full-length based on the presence of 5′ and 3′ cDNA primer sequences. The CFLs were aligned to the human (hg19) and EBV-Akata (NCBI accession number KC207813.1, (15)) genomes using GMAP (14). Approximately 6% of the CFLs mapped to the EBV genome (Figure 1A), in line with the proportion of Illumina EBV reads mapped previously in anti-IgG treated Akata cells (9,12). Mapped CFLs ranged in length from 300 to 16 430 bases, with a mean of 1335 bases. Size fractionation of the cDNA before sequencing helped reduce the bias toward sequencing shorter transcripts (Supplementary Figure S1). Length distributions for human and EBV CFLs were found to be similar (Figure 1B).
Figure 1.
Long-read sequencing data and validation strategy. (A) Percentages of consensus full-length isoforms mapped to cellular or EBV genomes. (B) Length distribution of consensus full-length isoforms mapped to cellular or EBV genomes. Blue boxes represent second and third quartiles, horizontal black lines indicate mean. (C) Distribution of proportion of annotated transcripts, by length, that are represented by full-length sequenced isoforms. (D) Strategy for data integration to validate full-length sequenced transcripts. (E) Example validated cellular transcripts.
To gauge the extent to which full-length transcripts were captured, we assessed the proportion of expressed (as determined by Illumina short read sequencing: see ‘Materials and Methods’ section) annotated cellular transcripts of different sizes that were represented by CFLs. The majority of shorter annotated transcripts (up to about 1000 bases) were found to be captured by CFLs while the capture of full-length transcripts dropped off with increasing transcript length (Figure 1C).
Limitations in attributing bona fide transcript structures from Iso-Seq data
Examination of CFLs that mapped to well-characterized cellular genes revealed those that agree with RefSeq annotation as well as those that appear to reflect novel variants. Nevertheless, we noted apparent anomalies that confounded the direct attribution of CFLs to true transcript structures. Most notable was the prevalence of shorter CFLs with 5′ ends that map progressively further downstream of annotated start sites, suggestive of 5′ truncations (Supplementary Figure S2). For the bulk of shorter CFLs, the 5′ ends were not supported by deepCAGE (Cap Analysis of Gene Expression) peaks (see below) affirming the contention that these CFLs do not represent full-length transcripts. These truncations likely result from strand invasion during cDNA synthesis (23), an issue that becomes more pervasive in longer transcripts due to inefficiencies in reverse transcriptase processivity. We also noted occasional CFLs with apparent splicing at non-canonical splice junctions (Supplementary Figure S2). These candidate splice junctions were usually represented by only one SMRT read and generally could not be validated by Illumina short-read sequencing data. It therefore appears that deletions can occasionally be introduced during library preparation or sequencing.
Transcriptome resolution through integration of multi-platform data (TRIMD)
While certain assumptions could potentially be made to impute ambiguously presenting CFL structural features (e.g. assuming the 5′-most Iso-Seq finding as the true 5′ end), such approaches are problematic for high gene-density genomes containing widespread functional overlapping genes. To overcome these obstacles we developed TRIMD, a validation strategy that utilizes biologically complementary datasets to interrogate each of the key transcript structure features (5′ and 3′ ends and splice junctions) of CFLs, thereby facilitating more accurate global transcript annotation (Figure 1D).
To assess whether the 5′ end of each CFL reflects a true transcription initiation site, we used deepCAGE (Cap Analysis of Gene Expression (24,25)) sequencing which incorporates the CAP Trapper method (26) to acquire RNA fragments containing the 5′ cap. We reasoned that its use of a distinct mechanism to capture 5′ ends (versus Iso-Seq's utilization of SMART (Switching Mechanism At 5′ end of RNA Template) cDNA synthesis (27)) would make deepCAGE well suited for testing the veracity of CFL 5′ ends. DeepCAGE tags were clustered using Paraclu (18) and putative transcription start sites were calculated as the average chromosomal start coordinate weighted by CAGE tag depth. Iso-Seq 5′ end clusters were identified as described in the ‘Materials and Methods’ section and the start coordinate for each cluster was calculated as the weighted average of SMRT read depth. Candidate Iso-Seq transcription start sites were considered validated if they were within 3 bases of a deepCAGE-identified transcription start site.
Illumina short-read (101 base) sequencing data was used to validate CFL splice junctions and 3′ ends. Iso-Seq splice junctions were considered validated if exact splice junction donor and acceptor coordinates were identified in Illumina RNA-Seq data. To verify CFL 3′ ends, Illumina short reads containing poly(A) tails were extracted, 3′ end clusters were identified, and weighted chromosomal transcript end coordinate averages were compared to weighted chromosomal end coordinate averages of Iso-Seq 3′ end clusters (see ‘Materials and Methods’ section).
Using these strategies, the 5′ end, intron junctions and 3′ end of each CFL were interrogated and CFLs in which all structural features were validated were considered to represent bona fide polyadenylated transcripts. Applying this method to the cellular transcriptome, we found that it appropriately validated annotated cellular genes while also revealing novel splice variants, alternative transcription start sites and alternative polyadenylation sites (Figure 1E and Supplementary Figure S3). TRIMD scripts are available at https://github.com/flemingtonlab/public and can easily be used to assess transcript structures for other viral and even cellular genomes.
Transcription start site identification in the EBV lytic transcriptome
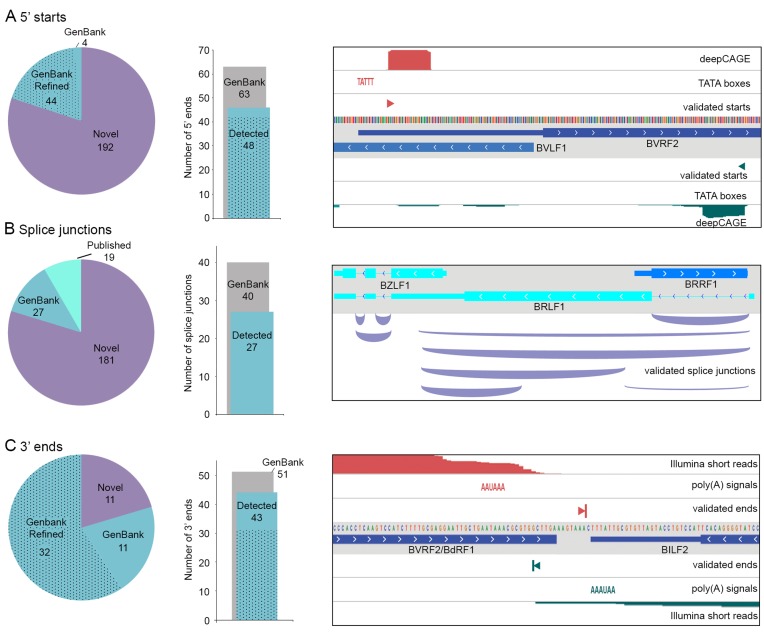
Using TRIMD, we identified 240 transcript initiation sites in EBV during reactivation, substantially more than the 67 5′ start sites for polyadenylated transcripts annotated in the GenBank Akata genome (Figure 2A). (See Supplementary File 1 for list of start sites; file is in BED format for visualization in a spreadsheet program or graphically on a genome browser in conjunction with EBV genome fasta and annotation files found at https://github.com/flemingtonlab/public). One hundred ninety-two of these were found to be novel while 48 GenBank-annotated 5′ ends were validated. The majority of the GenBank-annotated 5′ ends were originally annotated based on the coordinates of TATA box findings in the genome sequence (15,28). Using TRIMD we were able to provide true 5′ start coordinates based on empirical evidence (e.g. see Figure 2A, right panel, BVRF2). For many other EBV genes, the 5′ UTRs were not originally annotated in GenBank due to the absence of canonical TATA boxes upstream of their open reading frames. Again, TRIMD determined the transcription start sites for many of these genes (e.g. see Figure 2A, right panel, BVLF1). Most of the GenBank-annotated 5′ start sites not validated in our study fall into one of two classes: start sites of longer transcripts that aren't reliably covered by CFLs and start sites of latency-associated transcripts (Supplementary Figure S4). While we and others have observed lytic expression of latency-associated transcripts (9,12,29,30), the failure to detect some annotated latent transcription start sites is due to the use of alternative unannotated lytic promoters during reactivation (see below, Figure 6).
Figure 2.
Validation of transcript features. (A) Validation of 5′ starts. Pie chart indicates annotation status of validated 5′ starts. ‘Refined’ includes start sites annotated at TATA boxes that are more accurately identified in this study. Bar chart indicates the number of GenBank-annotated 5′ starts validated in this study (stippled = refined). Genome browser panel shows example validated 5′ starts. (B) Validation of splice junctions. Pie chart indicates annotation status of validated splice junctions. Bar chart indicates the number of GenBank-annotated splice junctions validated in this study. Genome browser panel shows example validated splice junctions. (C) Validation of 3′ ends. Pie chart indicates annotation status of validated 3′ ends. ‘Refined’ includes end sites annotated at canonical polyadenylation signals that are more accurately identified in this study. Bar chart indicates the number of GenBank-annotated 3′ ends validated in this study (stippled = refined). Genome browser panel shows example validated 3′ ends.
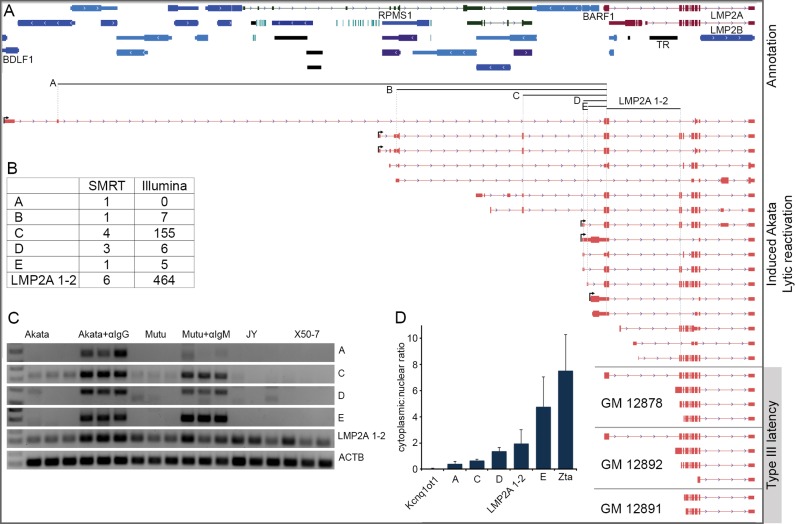
Figure 6.
Complex lytic promoter usage for LMP2 transcripts. (A) Genome browser visualization of CFLs mapping to the LMP2 exons in induced Akata cells and LCLs. Arrows positioned at the beginning of reads signify those with validated 5′ ends. (B) Splice junction read depth for SMRT circular consensus and Illumina short-read sequencing. Labels A through E refer to junctions indicated below GenBank-annotated gene track in (A). (C) PCR using junction-spanning primers in Akata, Mutu, JY and X50-7 cells. Akata + αIgG and Mutu + αIgM refer to Akata and Mutu cells induced for 24 and 48 h, respectively. (D) qRT-PCR of nuclear and cytoplasmic fractions of induced Akata cells (24 h).
Notably, though our study substantially increases the number of known transcription start sites in lytic EBV, our findings of additional deepCAGE clusters that were not supported by Iso-Seq 5′ ends (Supplementary Figure S5), due in part to low Iso-Seq 5′ start site coverage for longer transcripts, suggests that these data still under-represent the total number of EBV lytic promoters.
Splice junction identification in the lytic EBV transcriptome
Through TRIMD, a total of 227 splice junctions associated with the polyadenylated lytic EBV transcriptome were identified (Figure 2B and Supplementary Files 2 and 5). All 7 GenBank-annotated lytic gene splice junctions were identified as well as 20 GenBank-annotated splice junctions associated with latency transcripts. Of the validated splice junctions that are not in the GenBank annotation, 19 have been previously reported using Illumina RNA-Seq or other methods (9,12,31–34) leaving a total of 181 novel splice junctions. Importantly, many of the novel splice junctions identified here are detected with sequencing depths that are comparable to already known splice junctions (e.g. see Figure 2B, right panel).
Polyadenylation site identification in the lytic EBV transcriptome
A total of 54 lytic viral transcript 3′ ends were identified through TRIMD (Figure 2C and Supplementary File 3), the majority of which (43) are already represented in GenBank annotation. Like transcript 5′ ends, the GenBank annotation of many EBV transcript 3′ ends is based on genomic features rather than empirical evidence, with most transcripts being annotated as ending at the nearest downstream polyadenylation signal (AAUAAA/AUUAAA) (15,28). Using TRIMD we were able to refine the annotation for many of these informatically-annotated 3′ ends (Figure 2C).
EBV lytic transcript annotation
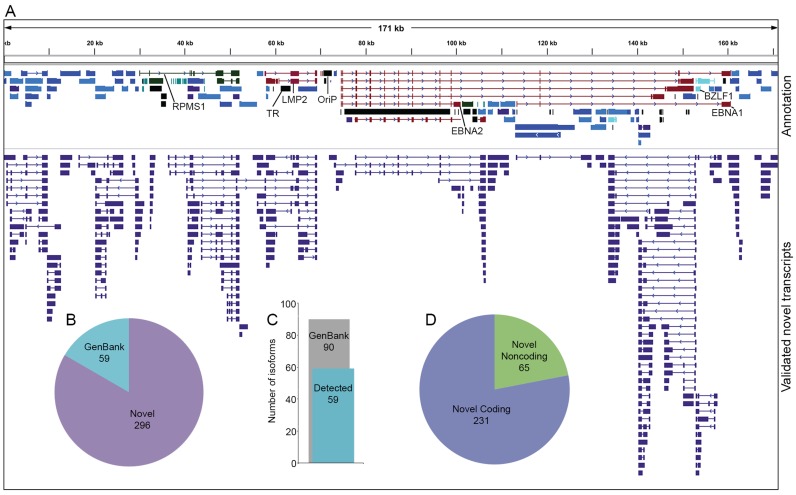
While validation of CFL 5′ ends, 3′ ends and splice junctions (Figure 2) each required support from another platform, the validation of whole transcript structures is more stringent, requiring that each structural feature of a CFL be validated. Nevertheless, we were able to identify 296 novel polyadenylated EBV transcript structures in addition to 59 GenBank-annotated transcript structures (Figure 3 and Supplementary Files 4 and 5). A total of 31 (34%) GenBank annotated transcripts were not represented in our analysis although as we observed in the validation of transcript features, most of these are either very long and/or are latency-associated.
Figure 3.
Novel validated transcripts. (A) Top track contains EBV-Akata GenBank annotation that has been refined and updated in this study. Bottom track contains novel EBV transcripts validated in this study. (B) Annotation status of validated transcripts. (C) Number of GenBank-annotated transcripts validated in this study. (D) Coding potential of novel EBV transcripts as determined by CPAT (19).
Novel EBV transcripts identified by TRIMD are distributed throughout the genome and include a wide range of lengths, structures and expression levels. Using the Coding Potential Assessment Tool (CPAT (19)), 22% of novel transcripts are predicted to be non-coding RNAs, a relatively under-investigated class of viral transcripts. Some newly identified transcripts are wholly novel, with unannotated transcription start sites and polyadenylation sites and being transcribed from genomic regions previously believed to be intergenic (see below, Figure 4). Others are splice variants of previously reported transcripts, skipping exons to produce altered UTRs or ORFs (see below, Figures 5 and 6). Many novel transcripts share polyadenylation sites and/or splice junctions with annotated transcripts but use different transcription start sites (see below, Figure 6). Finally, readthrough transcription resulting from failure to terminate at polyadenylation signals occurs throughout the genome, in many cases leading to intergenic splicing and the generation of chimeric transcripts (see below, Figure 7).
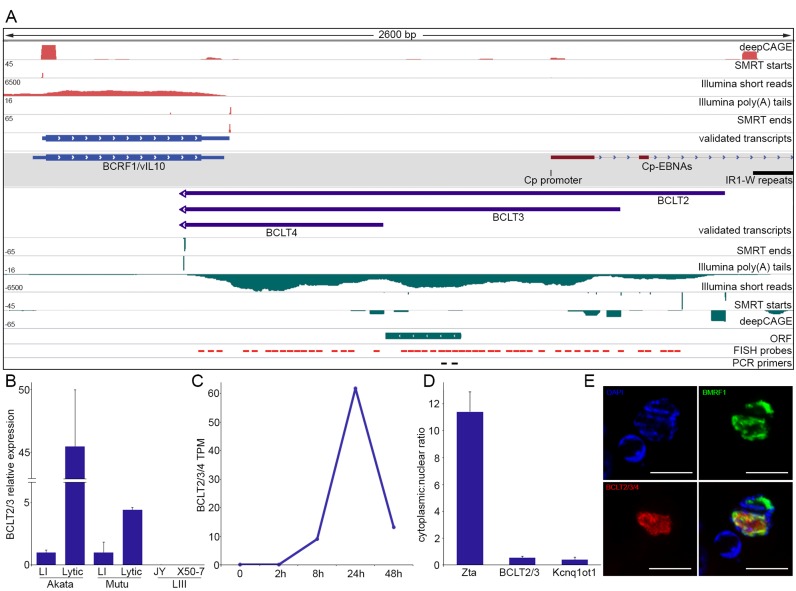
Figure 4.
Novel intergenic transcripts. (A) Genome browser visualization of BCLT2-4 transcripts and supporting evidence. Gray shaded track displays GenBank-annotated features. (B) Strand-specific qRT-PCR of BCLT2/3 in Akata, Mutu, JY and X50-7 cells. LI = type I latency, LIII = type III latency. Error bars are standard deviation. (C) Normalized Illumina RNA-Seq read counts of BCLT2/3/4 at multiple time points after induction. TPM = transcripts per million. (D) Strand-specific qRT-PCR of nuclear and cytoplasmic fractions of induced Akata cells (24 h). Error bars are standard deviation. (E) FISH and immunofluorescence of BCLT2/3/4 and EBV nuclear protein BMRF1.
Figure 5.
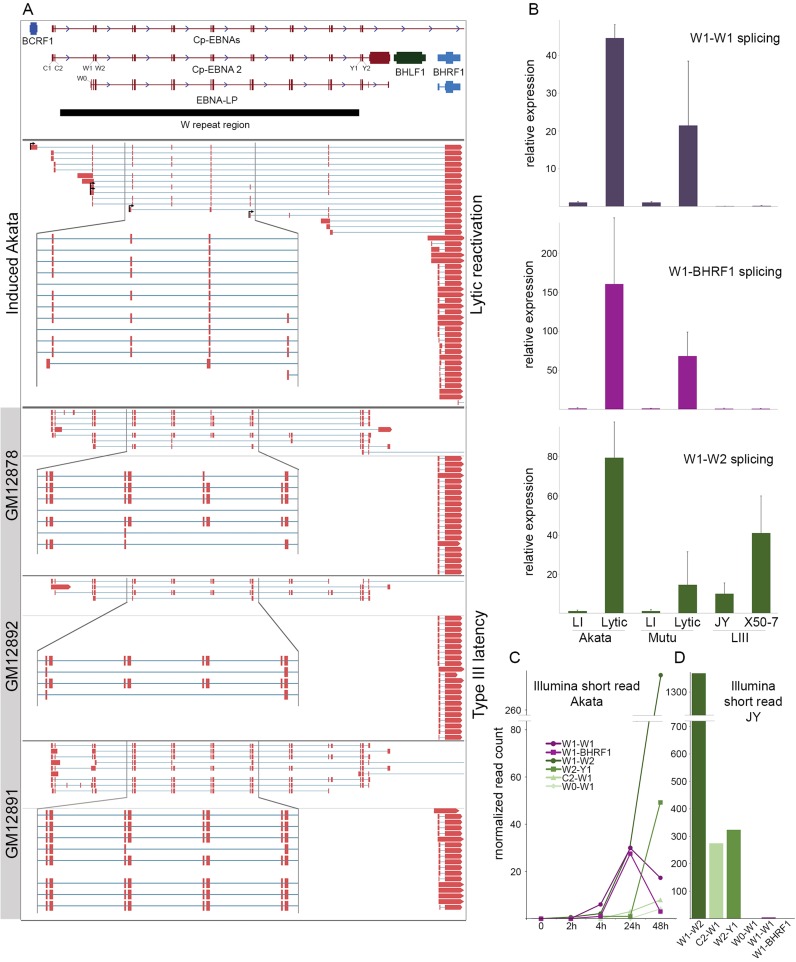
Programmed exon skipping in the W repeat region. (A) Genome browser visualization of CFLs mapping to the W repeat region and/or BHRF1 gene in induced Akata cells and lymphoblastoid cell lines (LCLs). (B) qRT-PCR using primers spanning the indicated splice junctions in Akata, Mutu, JY and X50-7 cells. LI = type I latency, LIII = type III latency, Lytic refers to 24 or 48 h induction in Akata and Mutu cells. (C) Time course analysis of splice junction reads in polyA+ RNA from Akata cells. (D) Splice junction reads detected in polyA+ RNA from the type III latency cell line, JY.
Figure 7.
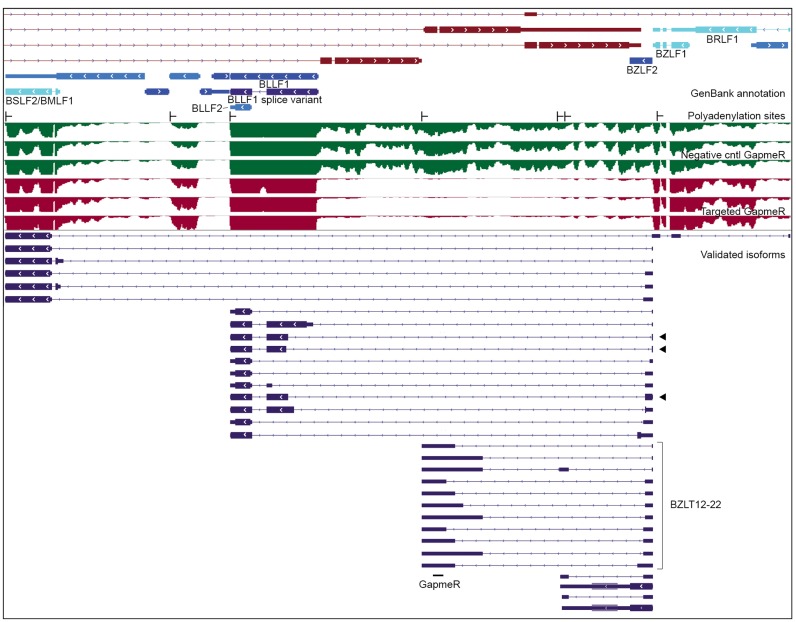
Readthrough transcription and intergenic splicing at the BZLF2 locus. From top: GenBank gene annotation, TRIMD-validated polyadenylation sites, Illumina short-read coverage of induced Akata cells with negative control GapmeR (green tracks) and induced Akata cells with GapmeR targeting BZLT12-22 (red tracks), novel validated isoforms (blue transcript features). Black arrows indicate transcripts whose largest ORF is an in-frame fusion.
Novel intergenic transcripts
Despite previous findings of pervasive transcription of the EBV genome outside of GenBank-annotated genes (9,12,30,35,36), the structures of transcripts derived from these regions could not be directly ascertained. Using TRIMD we provide structures for transcripts spanning unannotated loci. Figure 4A illustrates a group of newly identified overlapping transcripts, BCLT2-4 (BamHI C fragment Leftward Transcripts (BCLT)–2–4) that arise from transcription start sites near, but antisense to, the latency-associated Cp promoter, are transcribed leftward through an intergenic region, and terminate using a shared polyadenylation signal antisense to the lytic viral IL-10 homolog BCRF1. DeepCAGE and Iso-Seq support for the three novel transcription start sites is comparable to that for the refined BCRF1 transcription start site (Figure 4A). Likewise, the novel polyadenylation site is supported by a depth of Illumina poly(A) tail reads and Iso-Seq reads that is comparable to the depth observed for the BCRF1 gene (Figure 4A).
To further investigate BCLT2/3 transcripts, we used strand-specific qRT-PCR (20) using primers that amplify a region common to the two isoforms. Higher expression of BCLT2/3 was observed in the Akata and Mutu cell lines after B-cell receptor (BCR) crosslinking while little or no BCLT2/3 expression was observed in the type III latency LCLs JY and X50-7 (Figure 4B). Pacific Biosciences long-read cDNA sequencing of the type III latency cell lines GM12878, GM12891 and GM12892 failed to detect evidence of BCLT expression, further supporting the lytic restriction of BCLT2-4 expression ((11) and data not shown). Using Illumina time course RNA-Seq data, the combined normalized read values for BCLT2, 3 and 4 were found to be highest at 24 h after B-cell receptor crosslinking, coincident with the expression of viral Late protein-coding genes (Figure 4C).
To gain further insights into the functional nature of BCLT transcripts we examined their coding potential and subcellular location. Despite the presence of a short open reading frame in BCLT2 and BCLT3, all three transcripts are predicted to be non-coding by CPAT (Figure 4A and data not shown). Strand-specific qRT-PCR of nuclear and cytoplasmic RNA fractions showed enrichment of BCLTs in the nuclear fraction (Figure 4D), further supporting a non-coding function. FISH confirmed their nuclear localization and showed partial overlap with the viral nuclear protein BMRF1 (Figure 4E). As BMRF1 is a component of the viral DNA replication machinery that also associates with newly-synthesized viral genomes (37,38), this partial colocalization raises the possibility that some of the BCLT transcripts are in the proximity of replicating and/or newly synthesized genomes and potentially play a role in viral genome processes such as replication, gene expression or viral genome processing.
Programmed exon skipping in W-repeat/BHRF1 transcripts
The EBNA transcripts expressed in type III latency contain a common 5′ structure that is composed of exon pairs from each repeat of the W repeat region (Figure 5A). This iterative joining of identical repeated 66-base W1 exons and 132-base W2 exons is followed by splicing to the non-repeat Y1 and Y2 exons, and finally to the unique exons of the EBNA genes (39,40) (Figure 5A). Analysis of Iso-Seq CFLs from reactivated EBV revealed transcripts containing W repeat exons but with the consistent omission of the W2 exon (W1–W1 splicing), with subsequent splicing directly to the viral Bcl2 homolog BHRF1 (Figure 5A). BHRF1 transcripts containing W1 and W2 exons have been detected previously in latently infected cells (41–43) so the omission of the W2 exon observed here may be unique to the setting of reactivation. Analysis of Pacific Biosciences long-read cDNA sequencing from the latent GM12878, GM12892 and GM12891 cell lines shows the expected type III latency W1–W2–Y1–Y2 splicing pattern with no evidence of W1–W1 splicing (Figure 5A, lower panels). Notably, although BHRF1-containing reads were detected in the LCLs none of these reads contained W-derived exons, instead likely initiating from one or more proximal promoters ((11) and Figure 5A).
We further investigated the programming of W1–W1 splicing by qRT-PCR. W1–W1 splicing was found to increase substantially upon reactivation in both type I latency cell lines Akata and Mutu (Figure 5B). In contrast, little or no evidence of W1–W1 splicing was observed in the type III latency JY and X50-7 cell lines (Figure 5B). A similar lytic restriction was observed using PCR primers spanning the W1–BHRF1 splice junction (Figure 5B). qRT-PCR with primers spanning the annotated W1–W2 junction revealed, as expected, abundant W1–W2 splicing in type III latency JY and X50-7 cell lines, and low levels of W1–W2 splicing in untreated type I latency Akata and Mutu cells. Strikingly, however, although we didn't detect W1–W2 splicing in any Iso-Seq CFLs from induced Akata cells, qRT-PCR revealed high levels of W1–W2 splicing in reactivated Akata and Mutu cells (Figure 5B), a result that was substantiated by Illumina RNA-Seq data (Figure 5C). While this is an apparent discrepancy, we postulate that while W1–W1 splicing may be restricted to BHRF1-containing transcripts, splicing to W2 may be specific to the EBNA transcripts. Since all EBNA transcripts except EBNA-LP are substantially longer (ranging from 3361 to 8265 bases) than BHRF1 transcripts (1447–2062 bases), the lack of Iso-Seq CFLs with W1–W2 splicing in induced Akata cells may be related to the less efficient capture of the long EBNA transcripts by Iso-Seq.
We next investigated W-splicing using Illumina RNA-Seq data from a time course of reactivation in Akata cells (Figure 5C) and from latent JY cells (Figure 5D). This analysis confirmed the expected splicing pattern in JY cells: abundant splicing from W1–W2, C1–W1 and W2–Y1 but little or no splicing from W1–W1 or W1–BHRF1 (Figure 5D). Illumina RNA-Seq data from RNA isolated from Akata cells at several time points after BCR crosslinking shows that the novel W1–W1 and W1–BHRF1 splicing events both increase after BCR crosslinking, reaching a peak at 24 h after which time their levels decrease. The latency splice junctions W1–W2, W2–Y1, C2–W1 and W0–W1 however increase throughout the time course, even at the 48 h time point when overall EBV transcription has decreased from its peak at 24 h (Figure 5C and data not shown). This is indicative of a likely regulatory distinction between the W1–W1–BHRF1 transcripts and the W1–W2 containing EBNA transcripts during reactivation.
Together these results indicate that there is a unique and specific alternative splicing program across the W repeats during reactivation. W1–W1 splicing appears to be specifically programmed rather than stochastic because (i) it is tissue/reactivation specific and (ii) while W1–W2 splicing is observed during reactivation, lytic CFLs that contain W1–W1 junctions uniformly contain only W1–W1 splicing rather than a combination of W1–W1 and W1–W2 splicing events. Interestingly, while two different open reading frames extend across the repeated W1 exons, we did not detect any AUG start codons corresponding to either of these reading frames. This splicing configuration may instead confer alternative 5′ UTR functionality rather than serving in a protein-coding capacity.
Complex lytic promoter usage for LMP2 transcripts
Microarray studies previously showed that the latency-associated LMP2 gene is induced during reactivation in type I latency cells (29,30). More recent RNA-Seq work from our lab revealed extensive alternative splicing of LMP2 transcripts during reactivation that is not observed in type III latency LMP2 transcripts (12), possibly representing a means of increasing the functional diversity of this locus during reactivation. Analysis of lytic LMP2-overlapping RNAs using TRIMD validated extensive alternative splicing and revealed the use of a complex set of novel lytic promoters (Figure 6). Further, while LMP2-overlapping CFL structures from type III latency LCLs are consistent with initiation at the latency LMP2A promoter, none of the CFLs or deepCAGE peaks from reactivated Akata cells support initiation from either of the latency LMP2 promoters (Figure 6A and data not shown (deepCAGE)). Although the 5′ ends of many of the lytic LMP2-overlapping CFLs were not supported by deepCAGE starts (likely representing non-full length cDNAs), four novel upstream promoters were detected (TRIMD-validated start sites shown with arrows in Figure 6A): one that corresponds to the sharing of the BARF1 promoter, one that is antisense to the BALF1 gene, one located in an intron of the latency RPMS1 non-coding RNA locus and one that initiates antisense to the BDLF1 gene, 46 kb upstream from the latency LMP2A start site.
Initiation of transcripts from the upstream lytic promoters adds complexity to the LMP2 lytic transcriptome with LMP2-overlapping CFLs containing numerous different exon combinations and a remarkably complex collection of open reading frames, including full-length LMP2A and LMP2B ORFs, truncated or expanded LMP2 ORFs that preserve the reading frame and even, through intergenic splicing, the ORF of the upstream immune-modulating gene BARF1 (Figure 6A). Most of the upstream splice junctions that are specific to the novel lytic promoter initiated transcripts were supported by Illumina RNA-Seq data from reactivated Akata cells (Figure 6B and data not shown). PCR using junction-spanning primer pairs demonstrated elevated levels of transcripts containing these junctions in reactivated Akata and Mutu cells, but not in JY or X50-7 cells (Figure 6C). qRT-PCR of nuclear and cytoplasmic fractions of reactivated Akata cells showed that the BDLF1- and the RPMS1-initiated transcripts (junctions A and B) have a more nuclear distribution than transcripts containing junction E, for which there is a CFL that contains the complete LMP2A ORF (Figure 6D). This supports the idea that some lytic LMP2-overlapping isoforms play non-coding nuclear functions while others are transported to the cytoplasm for translation. Together, these findings illustrate the complex nature of the lytic LMP2 transcriptome arising from alternative promoter usage, alternative splicing and differential cellular localization, producing a diverse group of LMP2-overlapping transcripts likely with both coding and non-coding functions.
Intergenic splicing of BZLF2 transcripts arising from readthrough transcription
While we observed poly(A) signal readthrough transcription throughout the genome (including at the LMP2 locus), transcripts originating from the BZLF2 promoter (BZLF2p) were remarkable with respect to the diversity of transcripts initiating from a single promoter including coding, non-coding and chimeric transcripts. The BZLF2 gene lacks a canonical TATA box and polyadenylation signal and was therefore previously annotated based only on its open reading frame. Using TRIMD we identified a transcription start site 12 bases upstream of the translation initiation codon (Figure 7). Two transcripts initiating at this promoter contain the entire BZLF2 open reading frame (encoding the membrane glycoprotein gp42) and terminate about 2 kilobases downstream from the translation stop codon at a pair of novel polyadenylation sites located 23 bases apart. These transcripts likely represent the 2.5 kb transcript noted in GenBank record V01555. Notably, both transcripts are potentially bicistronic, containing a second, larger downstream open reading frame that we previously reported based on RNA-Seq data (9). This ORF is conserved in the B95-8 strain of EBV and so we have named it, and a transcript containing it, Be2LF1 (Supplementary File 4).
We identified six leftward oriented polyadenylation sites within BZLF2p-initiated transcripts (Figure 7), with the generation of some these transcripts requiring the readthrough of as many as five polyadenylation signals. The differential use of polyadenylation signals combined with extensive alternative splicing results in over 30 different validated transcript structures. Five novel splice donors within the BZLF2 open reading frame join to both novel splice acceptors located in regions of the genome without previous gene annotation and to known splice acceptors in the BLLF1 and BSLF2/BMLF1 genes. Complete or partial BLLF1 and BSLF2/BMLF1 reading frames are retained in some of these intergenic transcripts. For three of the BZLF2-BLLF1 chimeric transcripts the reading frames are maintained across the splice junction suggesting that BZLF2-BLLF1 fusion proteins may be produced (these transcripts are indicated by arrows in Figure 7).
The structures identified by TRIMD at the BZLF2 locus help illustrate the diversity of transcripts that arise from readthrough transcription and alternative splicing. While the entire locus of BZLF2p-derived transcripts spans more than 19 kb, reverse transcriptase processivity limitations may have precluded the detection of additional longer BZLF2 isoforms. For example, RNA-Seq analysis of induced Akata cells treated with a GapmeR targeting novel exons of the BZLT12-22 transcripts (see Figure 7) did not result in the selective reduction of polyadenylated transcript coverage across BZLT12-22 exons, but rather a uniform knockdown of a large 12 kb region surrounding these exons. This suggests the presence of an abundant 12 kb unspliced transcript that was not detected by TRIMD analysis, originating from BZLF2p and terminating at the BLLF1 polyadenylation site (Figure 7).
DISCUSSION
Integrating data from multiple different platforms has allowed us to identify and validate nearly 300 novel polyadenylated viral transcripts expressed during EBV reactivation (Figure 3 and Supplementary Files 4 and 5). In addition to identifying novel transcripts, we have refined the annotation for nearly two thirds of annotated EBV genes, providing empirical evidence to precisely determine 5′ and 3′ transcript ends that had previously been annotated based on genomic sequence motifs (Figure 3 and Supplementary File 6). Further, we have fully resolved transcripts that had previously only been observed as splice junctions or cDNA fragments (9,12,31,33). TRIMD analysis of the cellular transcriptome also reveals an abundance of previously unannotated isoforms (unpublished), including previously unannotated splice variants and alterations of transcript ends. Since cellular isoform expression is known to be regulated across different tissues and conditions (44–46) TRIMD-based transcript structure resolution may have an important application in globally resolving tissue, developmental and disease specific cellular transcript structures.
Although we have more than quadrupled the number of known polyadenylated EBV transcripts, our analysis of cellular transcript capture rates (Figure 1C) suggests that our annotation of the lytic transcriptome is incomplete, particularly for longer transcripts (Figure 1C). Substantiating this contention, our deepCAGE analysis identified a number of start sites that are not supported by Iso-Seq CFLs (Supplementary Figure S5). Assuming that the percentage of full-length transcripts captured by Iso-Seq is similar between cellular and viral transcripts, we can use the capture rates shown in Figure 1C to predict the total number of polyadenylated viral transcripts expressed during reactivation. Based on this analysis, we estimate that there may be more than 900 polyadenylated EBV transcripts expressed during reactivation (Supplementary Figure S6). Even this number may under-represent the full complexity of the EBV lytic transcriptome because it only accounts for polyadenylated transcripts. This may be significant because we have previously reported deeper and more extensive coverage of the EBV genome during reactivation with Illumina RNA-Seq data from ribodepleted RNA compared to poly(A)-selected RNA (9).
A hallmark of viruses is their compact nature, with an uncanny density of functions encoded in relatively small amounts of genomic sequence. The work presented here helps highlight some of the many strategies utilized by EBV to achieve diverse functions with limited genetic material.
Coincident with the finding of 181 novel lytic splice junctions, we observed extensive alternative splicing (e.g. see Figures 5–7). We know that at least some of this alternative splicing is specified through cell and/or viral programing. For example, splicing of the LMP2 coding exons is consistently sequential during type III latency whereas during reactivation, these same LMP2 exons display an array of different splicing combinations (Figure 6 and (12)). Similarly, the W1–W1–BHRF1 splicing configuration is specific to the reactivation setting.
The extent to which the lytic restriction of alternative splicing is controlled by unique trans factors versus cis-acting regulatory elements is unclear. The use of distinct promoters to drive LMP2 expression during reactivation gives rise to unique 5′ sequences that may govern alternative splicing in cis. In contrast, our Iso-Seq data show that some of the W1–W1–BHRF1 transcripts likely originate from the latency Cp promoter, the same promoter that is utilized during type III latency where no W1–W1 or W1–BHRF1 splicing could be observed. This suggests that this unique splicing configuration may be primarily specified by tissue specific cellular and/or viral trans factors (although it does not rule out the possibility that unique cis elements may be formed by an initial W1–W1 splicing event, for example, that then dictates subsequent W1–W1 splicing and/or W1–BHRF1 splicing).
While the functions of these newly discovered alternatively spliced lytic transcripts are still unknown, it is likely that alternative splicing specifies distinct functions. For example, while at least some of the LMP2 transcripts bearing sequentially spliced coding exons localize to the endoplasmic reticulum where they are translated, many of the alternative LMP2 isoforms are predicted to be non-coding and some of these are localized to the nucleus (Figure 6D). Some alternative splicing occurs within coding sequences, giving rise to transcripts encoding proteins with different repertoires of functional domains. Further, intergenic splicing, for example between BZLF2 and BLLF1, may encode fusion proteins with select functional domains of each respective gene. This may represent a conservation strategy involving the multi-purposing of functional domains to accomplish distinct tasks in the viral infection cycle.
The expression of several latency genes during reactivation has been shown previously to occur through distinct lytic promoters (e.g. EBNA1 (47,48), EBNA3A (9) and LMP1 (49)) and we now show that similarly, LMP2 is expressed through multiple lytic promoters. The use of distinct promoters may be a means of facilitating cleaner transcriptional regulation through the segregation of latency- and lytic-specific promoter elements. What is likely more relevant is that this configuration imparts distinct 5′ sequences to the lytic isoforms that facilitate unique post-transcriptional roles in localization, partner interactions and/or function. As mentioned above, this seems to be the case with LMP2 transcripts initiating from different lytic promoters, some of which appear to play non-coding roles in the nucleus while others are exported to the cytoplasm for translation. The use of multiple promoters to drive expression of common downstream sequences is not unique to the LMP2 locus or latency genes in general but is common among purely lytic genes as well. For example, transcripts containing BZLF1 sequences are derived from previously known and novel upstream promoter elements (Supplementary Figure S7). This leads to the generation of likely non-coding transcripts (BRLT5 and BRLT6) and truncated BZLF1 reading frames that are in frame with novel 5′ coding sequences (BRLT2 and BRLT3). In other cases, upstream promoter usage leads to the addition of extra 5′ UTR sequences (e.g. BZLT3-6) that may alter the translation efficiency of the BZLF1 reading frame. We also note downstream promoter usage (e.g. BZLT8) generating transcripts that are likely non-coding (Supplementary Figure S7). Alternative promoter usage is observed across the genome and likely contributes to the generation of substantial diversity in transcript function.
The readthrough of polyadenylation signals generates a substantial amount of transcript diversity during EBV reactivation (e.g. see Figures 6 and 7) and is pervasive across the EBV genome. Interestingly, a recent report showed that HSV-1 infection results in massive transcriptional readthrough in the cellular genome (50). A comparable finding was recently made in neural cells in response to osmotic stress (51), raising the possibility that HSV-1 taps into existing cellular metabolic pathways (it is notable that osmotic stress-induced readthrough was dependent on intracellular Ca2+ release, a process critical to both HSV-1 infection (52) and EBV reactivation (53)). We too observed increases in readthrough transcription of cellular genes in reactivating Akata cells (data not shown) suggesting that B-cell receptor signaling and/or viral factors similarly induce polyadenylation site readthrough. Although the functional significance of readthrough transcription of the cellular genome is unclear at this time, a global induction of polyadenylation site readthrough mechanisms may be essential for the generation of transcript diversity during the EBV replication cascade.
Beyond the functional relevance of the growing number of EBV transcripts being identified, pervasive overlapping lytic transcription has practical implications for experimental design. Investigations into viral gene function through genome modification need to take into account the full repertoire of transcripts potentially impacted by the introduced genetic alterations. At a minimum, great care should be taken to introduce alterations that are expected to have minimal impact on overlapping transcripts and/or that experimental results are interpreted in light of potential impact on overlapping transcripts. Further, even antisense directed knockdown strategies can have unintended influences on overlapping transcripts. While attempting to knock down BZLT12-22 transcripts using a GapmeR targeting common exon sequences, we unwittingly knocked down what appears to be a previously unknown 12 kb unspliced transcript whose expression is substantially greater than the BZLT12-22 transcripts combined (Figure 7). It is clear that investigations into EBV gene function require continued efforts to fully resolve the viral transcriptome as well as careful experimental design and interpretation of results.
The complex nature of the EBV lytic transcriptome also has implications for lytic transcript quantification. Accurately quantifying the abundance of coexpressed cellular gene isoforms using short-read sequencing data has been a challenge, although significant progress has been made (17,54). Can we expect to achieve reasonable accuracy in isoform discrimination/quantification from short-read data alone with such extensive levels of transcript overlap in the EBV genome? At a minimum, it will be important to fully resolve the EBV transcriptome to better facilitate transcript quantification. Simulations can then be performed to benchmark the accuracy of viral transcript quantification using the most complete transcript structure information. Nevertheless, we may find that lytic transcript quantification can only be achieved through the implementation of novel strategies using long-read data or a combination of platforms similar to the approaches used here for transcript resolution.
Supplementary Material
Acknowledgments
The authors thank the University of Wisconsin Biotechnology Center DNA Sequencing Facility for Illumina sequencing, the Johns Hopkins Deep Sequencing and Microarray Core Facility for Iso-Seq SMRT sequencing and DNAform for deepCAGE.
ACCESSION NUMBER
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [F31CA180449 to T.O., R01AI101046, R01AI106676 to E.K.F., P20GM103518 to Prescott Deininger]. Funding for open access charge: NIH [R01AI101046, R01AI106676].
Conflict of interest statement. None declared.
REFERENCES
- 1.Pattle S.B., Farrell P.J. The role of Epstein-Barr virus in cancer. Expert Opin. Biol. Ther. 2006;6:1193–1205. doi: 10.1517/14712598.6.11.1193. [DOI] [PubMed] [Google Scholar]
- 2.Henle W., Henle G. Epstein-Barr virus and human malignancies. Cancer. 1974;34(Suppl. S4):1368–1374. doi: 10.1002/1097-0142(197410)34:8+<1368::aid-cncr2820340806>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 3.Kang M.S., Kieff E. Epstein-Barr virus latent genes. Exp. Mol. Med. 2015;47:e131. doi: 10.1038/emm.2014.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Longnecker R., Kieff E., Cohen J.I. Epstein-Barr Virus. In: Knipe DM, Howley PM, editors. Fields Virology. 6th edn. Philadelphia: Wolters Kluwer Health/Lippincott WIlliams & Wilkins; 2013. pp. 1898–1959. [Google Scholar]
- 5.Baer R., Bankier A.T., Biggin M.D., Deininger P.L., Farrell P.J., Gibson T.J., Hatfull G., Hudson G.S., Satchwell S.C., Seguin C., et al. DNA sequence and expression of the B95-8 Epstein-Barr virus genome. Nature. 1984;310:207–211. doi: 10.1038/310207a0. [DOI] [PubMed] [Google Scholar]
- 6.Johnson L.S., Willert E.K., Virgin H.W. Redefining the genetics of murine gammaherpesvirus 68 via transcriptome-based annotation. Cell Host Microbe. 2010;7:516–526. doi: 10.1016/j.chom.2010.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gatherer D., Seirafian S., Cunningham C., Holton M., Dargan D.J., Baluchova K., Hector R.D., Galbraith J., Herzyk P., Wilkinson G.W., et al. High-resolution human cytomegalovirus transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2011;108:19755–19760. doi: 10.1073/pnas.1115861108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arias C., Weisburd B., Stern-Ginossar N., Mercier A., Madrid A.S., Bellare P., Holdorf M., Weissman J.S., Ganem D. KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 2014;10:e1003847. doi: 10.1371/journal.ppat.1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Grady T., Cao S., Strong M.J., Concha M., Wang X., Splinter Bondurant S., Adams M., Baddoo M., Srivastav S.K., Lin Z., et al. Global bidirectional transcription of the Epstein-Barr virus genome during reactivation. J. Virol. 2014;88:1604–1616. doi: 10.1128/JVI.02989-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murata M., Nishiyori-Sueki H., Kojima-Ishiyama M., Carninci P., Hayashizaki Y., Itoh M. Detecting expressed genes using CAGE. Methods Mol. Biol. 2014;1164:67–85. doi: 10.1007/978-1-4939-0805-9_7. [DOI] [PubMed] [Google Scholar]
- 11.Tilgner H., Grubert F., Sharon D., Snyder M.P. Defining a personal, allele-specific, and single-molecule long-read transcriptome. Proc. Natl. Acad. Sci. U.S.A. 2014;111:9869–9874. doi: 10.1073/pnas.1400447111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Concha M., Wang X., Cao S., Baddoo M., Fewell C., Lin Z., Hulme W., Hedges D., McBride J., Flemington E.K. Identification of new viral genes and transcript isoforms during Epstein-Barr virus reactivation using RNA-Seq. J. Virol. 2012;86:1458–1467. doi: 10.1128/JVI.06537-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin Z., Puetter A., Coco J., Xu G., Strong M.J., Wang X., Fewell C., Baddoo M., Taylor C., Flemington E.K. Detection of murine leukemia virus in the Epstein-Barr virus-positive human B-cell line JY, using a computational RNA-Seq-based exogenous agent detection pipeline, PARSES. J. Virol. 2012;86:2970–2977. doi: 10.1128/JVI.06717-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu T.D., Watanabe C.K. GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics. 2005;21:1859–1875. doi: 10.1093/bioinformatics/bti310. [DOI] [PubMed] [Google Scholar]
- 15.Lin Z., Wang X., Strong M.J., Concha M., Baddoo M., Xu G., Baribault C., Fewell C., Hulme W., Hedges D., et al. Whole-genome sequencing of the Akata and Mutu Epstein-Barr virus strains. J. Virol. 2013;87:1172–1182. doi: 10.1128/JVI.02517-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323–339. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frith M.C., Valen E., Krogh A., Hayashizaki Y., Carninci P., Sandelin A. A code for transcription initiation in mammalian genomes. Genome Res. 2008;18:1–12. doi: 10.1101/gr.6831208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang L., Park H.J., Dasari S., Wang S., Kocher J.P., Li W. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 2013;41:e74. doi: 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Feng L., Lintula S., Ho T.H., Anastasina M., Paju A., Haglund C., Stenman U.H., Hotakainen K., Orpana A., Kainov D., et al. Technique for strand-specific gene-expression analysis and monitoring of primer-independent cDNA synthesis in reverse transcription. Biotechniques. 2012;52:263–270. doi: 10.2144/0000113842. [DOI] [PubMed] [Google Scholar]
- 21.Takada K., Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J. Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pacific Biosciences of California, I. 2015 http://www.pacb.com/ [Google Scholar]
- 23.Tang D.T., Plessy C., Salimullah M., Suzuki A.M., Calligaris R., Gustincich S., Carninci P. Suppression of artifacts and barcode bias in high-throughput transcriptome analyses utilizing template switching. Nucleic Acids Res. 2013;41:e44. doi: 10.1093/nar/gks1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurosawa J., Nishiyori H., Hayashizaki Y. Deep cap analysis of gene expression. Methods Mol. Biol. 2011;687:147–163. doi: 10.1007/978-1-60761-944-4_10. [DOI] [PubMed] [Google Scholar]
- 25.Shiraki T., Kondo S., Katayama S., Waki K., Kasukawa T., Kawaji H., Kodzius R., Watahiki A., Nakamura M., Arakawa T., et al. Cap analysis gene expression for high-throughput analysis of transcriptional starting point and identification of promoter usage. Proc. Natl. Acad. Sci. U.S.A. 2003;100:15776–15781. doi: 10.1073/pnas.2136655100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Carninci P., Kvam C., Kitamura A., Ohsumi T., Okazaki Y., Itoh M., Kamiya M., Shibata K., Sasaki N., Izawa M., et al. High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics. 1996;37:327–336. doi: 10.1006/geno.1996.0567. [DOI] [PubMed] [Google Scholar]
- 27.Chenchik A., Zhu Y.Y., Diatchenko L., Li R., Hill J., Siebert P.D. Generation and use of high-quality cDNA from small amounts of total RNA by SMART PCR. In: Siebert PD, Larrick JW, editors. Gene cloning and analysis by RT-PCR. Natick: Biotechniques Books; 1998. pp. 305–319. [Google Scholar]
- 28.Farrell P.J. Epstein-Barr virus. The B95-8 strain map. Methods Mol. Biol. 2001;174:3–12. doi: 10.1385/1-59259-227-9:3. [DOI] [PubMed] [Google Scholar]
- 29.Lu C.C., Jeng Y.Y., Tsai C.H., Liu M.Y., Yeh S.W., Hsu T.Y., Chen M.R. Genome-wide transcription program and expression of the Rta responsive gene of Epstein-Barr virus. Virology. 2006;345:358–372. doi: 10.1016/j.virol.2005.09.064. [DOI] [PubMed] [Google Scholar]
- 30.Yuan J., Cahir-McFarland E., Zhao B., Kieff E. Virus and cell RNAs expressed during Epstein-Barr virus replication. J. Virol. 2006;80:2548–2565. doi: 10.1128/JVI.80.5.2548-2565.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao S., Strong M.J., Wang X., Moss W.N., Concha M., Lin Z., O'Grady T., Baddoo M., Fewell C., Renne R., et al. High-throughput RNA sequencing-based virome analysis of 50 lymphoma cell lines from the Cancer Cell Line Encyclopedia project. J. Virol. 2015;89:713–729. doi: 10.1128/JVI.02570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edwards R.H., Marquitz A.R., Raab-Traub N. Epstein-Barr virus BART microRNAs are produced from a large intron prior to splicing. J. Virol. 2008;82:9094–9106. doi: 10.1128/JVI.00785-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sadler R.H., Raab-Traub N. Structural analyses of the Epstein-Barr virus BamHI A transcripts. J. Virol. 1995;69:1132–1141. doi: 10.1128/jvi.69.2.1132-1141.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Smith P.R., de Jesus O., Turner D., Hollyoake M., Karstegl C.E., Griffin B.E., Karran L., Wang Y., Hayward S.D., Farrell P.J. Structure and coding content of CST (BART) family RNAs of Epstein-Barr virus. J. Virol. 2000;74:3082–3092. doi: 10.1128/jvi.74.7.3082-3092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dresang L.R., Teuton J.R., Feng H., Jacobs J.M., Camp D.G. 2nd, Purvine S.O., Gritsenko M.A., Li Z., Smith R.D., Sugden B., et al. Coupled transcriptome and proteome analysis of human lymphotropic tumor viruses: insights on the detection and discovery of viral genes. BMC Genomics. 2011;12:625–640. doi: 10.1186/1471-2164-12-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cao S., Moss W., O'Grady T., Concha M., Strong M.J., Wang X., Yu Y., Baddoo M., Zhang K., Fewell C., et al. New noncoding lytic transcripts derived from the Epstein-Barr virus latency origin of replication, oriP, are hyperedited, bind the paraspeckle protein, NONO/p54nrb, and support viral lytic transcription. J. Virol. 2015;89:7120–7132. doi: 10.1128/JVI.00608-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Daikoku T., Kudoh A., Fujita M., Sugaya Y., Isomura H., Shirata N., Tsurumi T. Architecture of replication compartments formed during Epstein-Barr virus lytic replication. J. Virol. 2005;79:3409–3418. doi: 10.1128/JVI.79.6.3409-3418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sugimoto A., Sato Y., Kanda T., Murata T., Narita Y., Kawashima D., Kimura H., Tsurumi T. Different distributions of Epstein-Barr virus early and late gene transcripts within viral replication compartments. J. Virol. 2013;87:6693–6699. doi: 10.1128/JVI.00219-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bodescot M., Perricaudet M., Farrell P.J. A promoter for the highly spliced EBNA family of RNAs of Epstein-Barr virus. J. Virol. 1987;61:3424–3430. doi: 10.1128/jvi.61.11.3424-3430.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sample J., Hummel M., Braun D., Birkenbach M., Kieff E. Nucleotide sequences of mRNAs encoding Epstein-Barr virus nuclear proteins: a probable transcriptional initiation site. Proc. Natl. Acad. Sci. U.S.A. 1986;83:5096–5100. doi: 10.1073/pnas.83.14.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Austin P.J., Flemington E., Yandava C.N., Strominger J.L., Speck S.H. Complex transcription of the Epstein-Barr virus BamHI fragment H rightward open reading frame 1 (BHRF1) in latently and lytically infected B lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 1988;85:3678–3682. doi: 10.1073/pnas.85.11.3678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bodescot M., Perricaudet M. Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 1986;14:7103–7114. doi: 10.1093/nar/14.17.7103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pearson G.R., Luka J., Petti L., Sample J., Birkenbach M., Braun D., Kieff E. Identification of an Epstein-Barr virus early gene encoding a second component of the restricted early antigen complex. Virology. 1987;160:151–161. doi: 10.1016/0042-6822(87)90055-9. [DOI] [PubMed] [Google Scholar]
- 44.FANTOM Consortium and the RIKEN PMI and CLST (DGT) Forrest A.R., Kawaji H., Rehli M., Baillie J.K., de Hoon M.J., Haberle V., Lassmann T., et al. A promoter-level mammalian expression atlas. Nature. 2014;507:462–470. doi: 10.1038/nature13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mallinjoud P., Villemin J.P., Mortada H., Polay Espinoza M., Desmet F.O., Samaan S., Chautard E., Tranchevent L.C., Auboeuf D. Endothelial, epithelial, and fibroblast cells exhibit specific splicing programs independently of their tissue of origin. Genome Res. 2014;24:511–521. doi: 10.1101/gr.162933.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayr C., Bartel D.P. Widespread shortening of 3′ UTRs by alternative cleavage and polyadenylation activates oncogenes in cancer cells. Cell. 2009;138:673–684. doi: 10.1016/j.cell.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schaefer B.C., Strominger J.L., Speck S.H. The Epstein-Barr virus BamHI F promoter is an early lytic promoter: lack of correlation with EBNA 1 gene transcription in group 1 Burkitt's lymphoma cell lines. J. Virol. 1995;69:5039–5047. doi: 10.1128/jvi.69.8.5039-5047.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nonkwelo C., Skinner J., Bell A., Rickinson A., Sample J. Transcription start sites downstream of the Epstein-Barr virus (EBV) Fp promoter in early-passage Burkitt lymphoma cells define a fourth promoter for expression of the EBV EBNA-1 protein. J. Virol. 1996;70:623–627. doi: 10.1128/jvi.70.1.623-627.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hudson G.S., Farrell P.J., Barrell B.G. Two related but differentially expressed potential membrane proteins encoded by the EcoRI Dhet region of Epstein-Barr virus B95-8. J. Virol. 1985;53:528–535. doi: 10.1128/jvi.53.2.528-535.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rutkowski A.J., Erhard F., L'Hernault A., Bonfert T., Schilhabel M., Crump C., Rosenstiel P., Efstathiou S., Zimmer R., Friedel C.C., et al. Widespread disruption of host transcription termination in HSV-1 infection. Nat. Commun. 2015;6:7126–7141. doi: 10.1038/ncomms8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vilborg A., Passarelli M.C., Yario T.A., Tycowski K.T., Steitz J.A. Widespread inducible transcription downstream of human genes. Mol. Cell. 2015;59:449–461. doi: 10.1016/j.molcel.2015.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheshenko N., Del Rosario B., Woda C., Marcellino D., Satlin L.M., Herold B.C. Herpes simplex virus triggers activation of calcium-signaling pathways. J. Cell Biol. 2003;163:283–293. doi: 10.1083/jcb.200301084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Faggioni A., Zompetta C., Grimaldi S., Barile G., Frati L., Lazdins J. Calcium modulation activates Epstein-Barr virus genome in latently infected cells. Science. 1986;232:1554–1556. doi: 10.1126/science.3012779. [DOI] [PubMed] [Google Scholar]
- 54.Patro R., Mount S.M., Kingsford C. Sailfish enables alignment-free isoform quantification from RNA-seq reads using lightweight algorithms. Nat. Biotechnol. 2014;32:462–464. doi: 10.1038/nbt.2862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.