Abstract
Linear dsDNA replicons with hairpin ends are found in the three domains of life, mainly associated with plasmids and viruses including the poxviruses, some phages and archaeal rudiviruses. However, their replication mechanism is not clearly understood. In this study, we find that the rudivirus SIRV2 undergoes multiple consecutive replication reinitiation events at the genomic termini. Using a strand-displacement replication strategy, the multiple reinitiation events from one parental template yield highly branched intermediates corresponding to about 30 genome units which generate exceptional ‘brush-like’ structures. Moreover, our data support the occurrence of an additional strand-coupled bidirectional replication from a circular dimeric intermediate. The multiple reinitiation process ensures rapid copying of the parental viral genome and will enable protein factors involved in viral genome replication to be specifically localised intracellularly, thereby helping the virus to avoid host defence mechanisms.
INTRODUCTION
In contrast to cellular organisms, viruses carry DNA or RNA genomes, which can be double-stranded or single-stranded with linear or circular structures. Consequently, their replication strategies show great diversity (1). Whereas the replication of circular DNA replicons follows two general replication models: θ replication and σ replication, named after the characteristic architecture of their replication intermediates (RIs) under electron microscope (2), the picture is more complex for viruses with linear DNA genomes such as phages λ, T4, φ20, PRD1 and some eukaryotic viruses including adenoviruses, herpesviruses and parvoviruses (3–6). These viruses have to solve the so-called ‘end-problem’ and the generation of concatemers is one of the strategies employed to solve it. While replication of some viruses (e.g. T7), generates concatemeric RIs with predominantly linear configuration, branched and complex concatemers have been observed for phage T4 and Herpes simplex virus-1 (HSV-1) (3,5). For T4, recombination-dependent replication is the mechanism of formation of networked concatemeric intermediates, while for HSV-1 neither the shape nor the mechanism of formation has been elucidated (3,5). For other viruses, such as poxviruses, the formation of multimeric RIs has been reported, but their structure is still unknown (7).
Linear dsDNA with covalently-closed ends (hairpin ends) has been found in all three domains of life, mainly associated with viruses. These include the genomes of eukaryotic viruses such as poxviruses, African Swine Fever virus and Chlorella viruses; the prophage genomes of bacteriophages N15, φKO2 and PY54; the plasmids and genomes of Borrelia, Agrobacterium tumefaciens and a group of mitochondria and plastids; and the genomes of Rudiviridae that infect hyperthermophilic archaea (8–10). Based on the telomere structure and results from biochemical studies of relevant proteins, different replication models have been proposed. However, in none of the cases is replication clearly understood.
Archaeal viruses exhibit a strikingly wide variety of morphologies, and the rod-shaped Rudiviridae family represents one of the most common morphotypes in hot terrestrial environments (11,12). This family contains four isolated members: SIRV1, SIRV2, ARV and SRV (11); and two additional candidate members (SMRV1 and ARV2) identified in hot spring metagenomes (13,14). Rudiviruses have linear dsDNA genomes of 25–35 kb with hairpin ends and inverted terminal repeats (ITRs) of up to 2 kb. Among rudiviruses, Sulfolobus islandicus rod-shaped virus 2 (SIRV2) is one of the most intensively studied, which has a genome of 35 498 bp with 1652 bp long ITRs (10).
Previous biochemical and structural studies by Oke et al. identified a candidate replication initiation protein (Rep), conserved in Rudiviridae members. Rep can introduce a nick at the SIRV genomic terminus, forming a covalent adduct with the newly created 5′end and releasing a 3′-OH terminus presumably used for priming DNA replication. Based on the in vitro data, it was proposed that the Rep protein initiates a strand-displacement replication (15).
In this work, we show that SIRV2 reinitiates strand-displacement replication consecutively from a single parental template, leading to the formation of large replication intermediates with unusual ‘brush-like’ structures. Moreover, a minor portion of the circular dimeric intermediate undergoes strand-coupled replication.
MATERIALS AND METHODS
Cell culture and virus propagation
Sulfolobus solfataricus 5E6 (16) or Sulfolobus islandicus LAL14/1 (17) were grown at 78°C, in SCVY and SCVYU media respectively (salt base medium described by Zillig et al. enriched with 0.2% sucrose, 0.2% Casamino Acids, 0.005% yeast extract, Wolin's vitamin mixture and 20 mg/l uracil (18)).
For SIRV2 propagation S. solfataricus 5E6 or S. islandicus LAL14 cultures were infected with SIRV2 at an optical density (A600) around 0.2. After 2–3 days incubation, the cells were removed by centrifugation at 4025 × g for 10 min. When indicated, supernatants obtained were used for further infection of cell cultures. Otherwise, virus present in the supernatant was precipitated with 10% polyethylene glycol 6000 and 1 M NaCl at 4°C overnight. Virus particles were recovered by centrifugation at 11 180 × g for 30 min and resuspended in 10 mM Tris–acetate pH 6. Cell debris was removed by centrifugation at 4025 × g for 5 min. When indicated, further purification of the virus particles by centrifugation in a CsCl density gradient was performed as described previously (19).
Preparation of plugs for neutral/neutral 2D-AGE, DNA spreading and PFGE
Sulfolobus solfataricus 5E6 or Sulfolobus islandicus LAL14 cultures were infected with SIRV2 with a multiplicity of infection (MOI) of approximately 10 or 30. Infected cultures at the indicated post-infection times were harvested by centrifugation at 7155 x g for 6 min at 4°C. Cells were washed twice with TEN solution (50 mM Tris, 50 mM EDTA, 100 mM NaCl; pH 8) and resuspended in TEN buffer to an optical density of 200. The cell suspension was briefly warmed to 37°C and mixed with an equal volume of pre-warmed 0.8% hi-strength low-melting point agarose (BioGene Ltd., Kimbolton, United Kingdom) and 80 μl of the mixture were poured into plug molds (Bio-Rad Laboratories, Hercules, CA, USA). Plugs were treated overnight at 37°C in NDS solution (0.5 M EDTA, 10 mM Tris, 0.55 M NaOH, 36.8 mM lauroyl sarcosine; pH 9) supplemented with 1 mg/ml Proteinase K (Sigma-Aldrich, St. Louis, MO, USA) and then transferred to fresh NDS solution (+1 mg/ml Proteinase K) for a second 37°C overnight incubation. The plugs were then washed for 5 times with TE buffer (100 mM Tris, 100 mM EDTA; pH 7.5) before being subjected to treatment with enzymes, 2D-AGE or PFGE; otherwise they were stored at 4°C in NDS solution.
Treatment with restriction enzymes
Before restriction digestion the plugs were washed 4–5 times each in 50 ml 1× restriction enzyme buffer for 1 h. Plugs were digested at 37°C overnight with 500 units of the corresponding enzyme in 500 μl of commercial restriction enzyme buffer (New England BioLabs, Ipswich, MA, USA; or Thermo Fisher Scientific, Waltham, MA, USA).
Treatment with S1-nuclease
Plugs were washed for 3 times in S1-buffer (40 mM sodium acetate pH 4.5, 0.3M NaCl and 2 mM ZnSO4), cut into halves and incubated for 2 h at 37°C, 500 rpm agitation, with the indicated concentration of S1-nuclease (Thermo Fisher Scientific, Waltham, MA, USA). Finally, plugs were washed with TE buffer before being subjected to analysis by 2D-AGE or PFGE.
Neutral/neutral 2D agarose gel electrophoresis
Agarose plugs containing S. solfataricus 5E6 cells infected with SIRV2 (MOI 30) were subjected to digestion by the corresponding restriction enzymes. The digested agarose plugs were run on the first dimension in 0.4% agarose gel with no ethidium bromide at 1–1.3 V/cm for 40 h in 1× TBE (89 mM Tris–borate, 2 mM EDTA) at 4°C; and on the second dimension at 5.0 V/cm for 7 h in 1% agarose gel containing 0.3 μg/ml ethidium bromide. The same concentration of ethidium bromide was added to the 1× TBE running buffer for the second dimension electrophoresis. DNA was transferred to nylon membrane (Amersham Hybond-XL; GE Healthcare) by capillary transfer and cross-linked by UV irradiation.
Probe DNAs were PCR amplified using the primers listed in Supplementary Figure S1 and were α-32P- radiolabeled by random priming using NEBlot kit (New England BioLabs, Ipswich, MA, USA). Filters were probed and washed to a final stringency of 0.5× SSC (1× SSC: 0.15 M NaCl, 0.015 M sodium citrate; pH 6.35) in 0.1% sodium dodecyl sulfate at 65°C.
DNA spreading
Sulfolobus solfataricus 5E6 cultures were grown to an optical density (A600) of 0.2 and then infected with SIRV2 with a MOI of 3 and incubated until 3 hpi. Samples were taken at 1 or 3 hpi and cells were washed twice with SCVY medium and diluted. After dilution, 2 μl with 5–50 cells were spotted onto Superfrost®Plus slides (Thermo Fisher Scientific, Waltham, MA, USA) and let sit for 5 min. Then, 7 μl of spreading buffer (200 mM Tris–HCl pH 7.4, 50 mM EDTA, 2.5% sodium dodecyl sulfate) were added, the drop was mixed and spread into a circle of 0.5–1 cm with the tip of the pipette. The drop was left to air-dry and then fixed with methanol: acetic acid (3:1) for 2 min. The slide was dried again and subjected to fluorescence in situ hybridization.
Fluorescence in situ hybridization (FISH)
The probe used for FISH was PCR amplified from DNA extracted from infected cells using the primers P2F and P2R described above. The probe was labeled with digoxigenin using the DIG High Prime DNA labeling and Detection Starter Kit II (Roche, Basel, Switzerland) according to the instructions of the manufacturer.
Slides were washed once with TE buffer and washed twice with 70% ethanol, 5 min per wash. Afterward, the slides were dehydrated with 4 min washes with 70%, 90% and 100% ethanol, and air-dried. 150 μl of Hybridization solution (2× SSC, 50% formamide, 1% Tween-20, 10 mg/ml dextran sulfate) with 300 ng of DIG-P2 probe were added to the slide and a coverslip was placed on top. The DNA on the slide and the probe were denatured at 95°C for 90 s and then incubated overnight at 37°C in a hybridization chamber. After hybridization, the following washes were performed at 45°C, 4 min per wash: (i) 2X SSC: 50% formamide; (ii) 2× SSC; 3. 0.1× SSC. Afterward, the slides were washed twice with PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4) and blocked for 30 min with PBS:0.1% Tween-20:1% bovine serum albumin (BSA). Primary antibodies were diluted in PBST: 1% BSA. For visualization of the total DNA an anti-ssDNA monoclonal antibody (Merck Millipore, Billerica, MA, USA) was used at a dilution of 1:50. We found that, in our hands, this antibody could also recognize dsDNA, therefore the single- or double-stranded nature of the fibers visualized could not be determined. For detecting the DIG-P2 probe the anti-digoxigenin antibody coupled to fluorescein (Roche, Basel, Switzerland) was used at a dilution of 1:30. Slides were incubated with primary antibodies for 1 h and then washed for three times with PBS, 2 min/wash. Slides were incubated for 1 h with the secondary antibody Alexa Fluor® 350 anti-mouse (Invitrogen, Carlsbad, CA, USA) diluted to 1:100 in PBST: 1%BSA. After incubation, the slides were washed for three times with PBS and a coverslip was placed over the slide. Samples were analyzed by epifluorescence microscopy with a Zeiss Axio Imager.Z1 ApoTome coupled to a CCD Hamamatsu ORCA-ER camera (C4742-80). Image processing was done using Volocity® 6.3 software (Perkin Elmer, Waltham, MA, USA) and Adobe Photoshop CS6.
Pulse-field gel electrophoresis (PFGE) and Southern blotting
One third to one half slices from the agarose plugs were embedded in 1% Pulse Field Certified Agarose (Bio-Rad Laboratories Inc., Hercules, CA, USA) gels and subjected to electrophoresis in 0.5× TBE buffer using a CHEF-DR® III Variable Angle System (Bio-Rad Laboratories Inc., Hercules, CA, USA) using one of two different running conditions as indicated. For resolution of viral replication intermediates with low molecular weight the MidRange I PFG Marker (New England BioLabs, Ipswich, MA, USA) was used and samples were electrophoresed for 19–20 h at 6 V/cm, switch time 1–25 s, 14°C. For resolution of high molecular weight replication intermediates the Yeast Chromosome PFG Marker (New England BioLabs, Ipswich, MA, USA) was used and the samples were electrophoresed at 6 V/cm, 15°C, for 11 h with a switch time of 70 s and for 9 h with a switch time of 120 s. After electrophoresis, gels were stained with ethidium bromide and the DNA in the gel was depurinated by a 10 min treatment with 0.25 M HCl, denatured by a 15 min treatment with 0.5 M NaOH, 1.5 M NaCl and equilibrated in 1 M Tris–HCl pH 7.5, 1.5 M NaCl for 30 min. Then, DNA in gels were transferred to a IMMOBILON®-NY+ nylon membrane (Merck Millipore, Billerica, MA, USA) by capillary transfer and cross-linked with a UV Stratalinker™ 1800 (Stratagene, San Diego, CA, USA).
The probes P4 and P5 for Southern blot analysis (Supplementary Figure S1) were PCR amplified and labeled with digoxigenin using the DIG High Prime DNA labeling and Detection Starter Kit II (Roche, Basel, Switzerland) according to the instructions of the manufacturer. Membranes were subjected to Southern blotting using the DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Basel, Switzerland) and the DIG Wash and Block Buffer Set (Roche, Basel, Switzerland) according to the instructions of the manufacturer.
Quantitative PCR
S. solfataricus 5E6 cultures were grown to an optical density of 0.2 and infected with SIRV2 with a MOI ≥ 5. Samples were taken at the indicated post-infection times and washed three times to remove excessive virus in the supernatant. Purified SIRV2 particles were used as a reference. All samples were first treated with 1 unit DNase I (Thermo Fisher Scientific, Waltham, MA, USA) at 37°C for 1 h to remove any contaminating DNA from the solutions, and DNase I was then inactivated by incubation at 75°C for 15 min. Then DNA in the samples was purified using the DNeasy Blood & Tissue Kit (QIAGEN N.V., Hilden, Germant). For amplification of the terminal ends of SIRV2 the following pair of primers was used gp01/gp54F: 5′-TGT TCA AGT CGG TCA AAC AAG G-3′and gp01/gp54R: 5′-TCC TAA CTT TTC TTA CAC TGA CTC C-3′. For amplification of the internal region of SIRV2 genome the primers used were gp26F 5′-GGC AAA AGG TCA CAC ATC AAG AAG-3′ and gp26R 5′-GTC TGG AAA TTT TGT TGG GCA AC-3′. Quantitative PCR reactions were performed in 10 μl mixtures containing 5 μl iQ SYBR Green Supermix (Bio-Rad Laboratories, Hercules, CA, USA), 1 μM of primers and 4 μl (4pg) of DNA. Samples were amplified in a CFX96 Real-Time Detection System (Bio-Rad Laboratories, Hercules, CA, USA) using the following cycling parameters: 95°C for 3 min, 35 cycles of 95°C for 10 s, 55°C for 10 s for the gp01/gp54 primers or 57°C for the gp26 primers, and 72°C for 15 s. Each reaction was run in triplicate and the average values were used for quantification. Data was analyzed with the Bio-Rad CFX manager software. The gp01/gp26 ratio was estimated by relative quantification using the Pfaffl method (20). Viral DNA extracted from CsCl-purified SIRV2 particles was used as the reference for normalization and its gp01/gp26 ratio was assumed to be 2:1. Supplementary Figure S2 shows the amplification efficiencies of the target and reference sequences.
RESULTS
Multimeric and highly branched ‘brush-like’ structures of SIRV2 replication intermediates
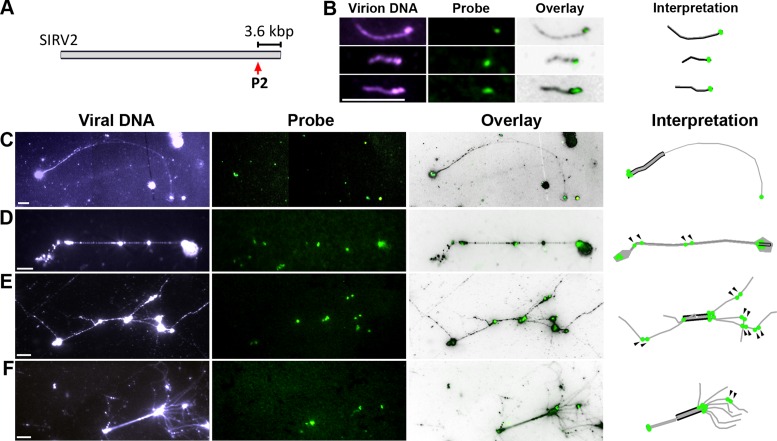
To investigate the DNA replication mechanism of SIRV2, the geometry of individual RIs was analyzed by DNA spreading (21) and fluorescence microscopy. S. solfataricus 5E6 cells, infected with SIRV2 at 1 and 3 hours post-infection (hpi), were lysed on SuperFrost®Plus slides and the DNA was spread on the spot. Fluorescence in-situ hybridization was then employed to verify the viral nature of the DNA using a digoxigenin-labeled probe generated from a sequence located ∼3.6 kb from the right terminus, outside of the ITR (Figure 1A). Moreover, the probe also provides information on the number and orientation of the viral genome units within each RI. The evidence for a single probe-binding site in each viral monomer is shown in Figure 1B, using DNA extracted from CsCl-purified SIRV2 virions. Representative fibers derived from the RIs from SIRV2-infected cells are shown in Figure 1C–F. Linear fibers comprising more than one viral genome unit were frequently observed (Figure 1C and D) and, moreover, the fibers showed complex branching (Figure 1E and F). The presence of multiple probe-binding sites confirmed that multimeric genomes were formed producing a ‘brush-like’ structure (Figure 1E and F). The topologies of the fibres are shown in the right-most panel (Figure 1B–F) and additional representative images of multimeric and branched RIs are also included (Supplementary Figure S3). In summary, these results show that SIRV2 forms diverse RIs including multimeric and branched ‘brush-like’ structures.
Figure 1.
Multimeric replication intermediates from SIRV2-infected cells. (A). P2-probe binding site on the SIRV2 genome. The size between P2-binding site and the right terminus is shown. (B). Presence of a single probe binding site in the monomeric SIRV2 genome. DNA extracted from SIRV2 virions that were purified by cesium chloride gradient ultracentrifugation was used for DNA spreading and FISH. (C–F). Representative RIs from SIRV2-infected cells. Infected cells were directly lysed on the slide before DNA spreading and FISH. Panels from left to right: entire fiber DNA shown in blue visualized by an anti-DNA antibody (viral DNA); FISH signal in green revealed by site-specific probe DIG-P2 (probe); entire fiber and probe signals are superimposed (overlay); and interpretation of the topology of each RI (interpretation). In the Interpretation panel, single black lines indicate a monomeric dsDNA genome (1n) and double black lines represent the genome dimer (2n) whereas daughter DNA is depicted in grey. The P2-probe binding site is shown by green dots; arrowheads indicate two adjacent probe signals in an internal segment. Bars, 6 μm.
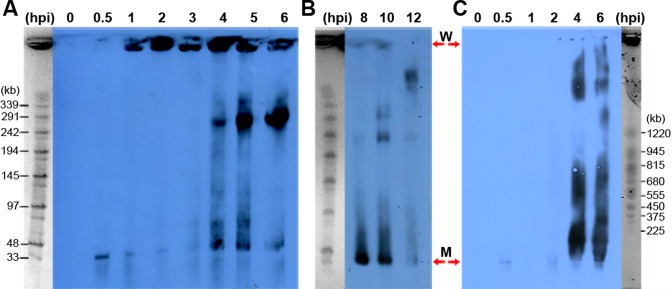
Subsequently, we performed pulse-field gel electrophoresis (PFGE) analysis to determine the sizes of the viral RIs and to follow the replication dynamics during the virus life cycle. S. islandicus LAL14/1 cells infected with SIRV2 at 0.5 to 12 hpi were embedded in agarose plugs, lysed by treatment with detergent and Proteinase K and subjected to PFGE. The viral RIs were then identified by Southern hybridization using the digoxigenin-labelled probe P4 (Supplementary Figure S1). A band of approximately 35 kb corresponding to the size of the monomeric genome is present in the infected samples at all time-points from 0.5 hpi and it increased in yield from 4 hpi (Figure 2A and B). Remarkably, a strong signal is also seen at the loading wells from 1 hpi. Treatment of viral RIs embedded in agarose plugs with restriction enzymes did not significantly diminish the amount of DNA retained in the loading wells, but digestion with the ssDNA-specific endonuclease S1 nuclease led to loss of DNA from the loading wells and a strong increase in the yields of smaller DNA (<145 kb) (Supplementary Figure S4A-B). We concluded that the viral RIs carry extensive regions of ssDNA. To determine the size of the viral DNA retained in the loading wells the PFGE running conditions were modified (Figure 2C) to resolve the DNA in the gels. Most of the DNA was then resolved as intermediates longer than 1.2 Mb at about 4–6 hpi (Figure 2C) with smaller amounts of these intermediates at 1–2 hpi (Supplementary Figure S5). In summary, the viral RIs are multimeric extending to sizes greater than 1.2 Mb.
Figure 2.
Large viral replication intermediates detected by PFGE during the SIRV2 life cycle. The viral RI sizes are determined for the early/mid phases (A) and late phase (B) of the life cycle. A size marker is placed at one side of each blot. (C). SIRV2 replication intermediates reach the size of >1.2 Mb. MidRange I PFG Marker was used in (A) and (B), while Yeast Chromosome PFG Marker was used in (C). W, wells; M, viral genome monomer.
A marked increase of viral RIs with sizes between 2 and 10 genome units was observed near the middle of the infection cycle (4 hpi) (Figure 2B) which we infer results from processing of the large RIs. Later, between 10 and 12 hpi, the yield of viral RIs decreases (Figure 2B), coincident with the onset of cell lysis and release of viral progeny (22).
Strand-displacement replication intermediates derive from both genomic termini of SIRV2
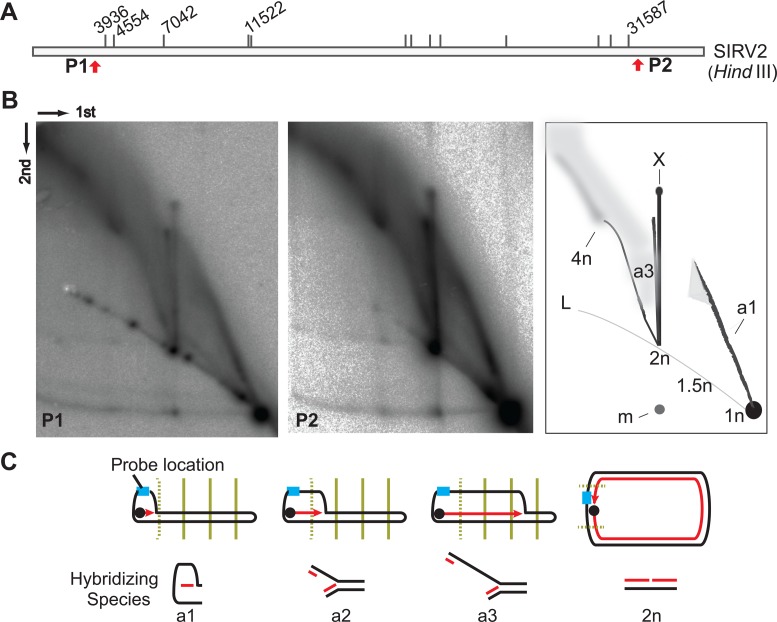
Although it has been proposed by Oke et al. (15) that rudiviruses initiate replication from genomic termini at the site of a nick generated by the Rep protein, the strand-displacement replication remains to be studied experimentally. Therefore, to determine the mechanism underlying generation of the ‘brush-like’ RIs, neutral-neutral 2D agarose gel electrophoresis (2D-AGE) was performed. Agarose plugs containing infected cells were digested with HindIII, generating fragments of about 4 kb from both genomic termini (Figure 3A). After 2D gel electrophoresis and Southern blotting, membranes were probed for the left terminal genomic region. A complex population of species was detected (Figure 3B) and some species had an apparent size in the first 2D gel dimension of between 1 and 2 times the initial fragment length (n). Moreover, consistent with the strand-displacement model (15), no standard replication fork arcs were visualized. Instead, there was a well-defined structural species termed ‘a1’ ascending from the 1n spot to a point that, in the first dimension, is about 1.5 times the size (1.5n) of the unit-length fragment (Figure 3B, right panel). Moreover, at this point the signal is diffuse, suggesting that RIs of similar size exhibit different structures.
Figure 3.
Strand-displacement replication of SIRV2. (A) HindIII digestion map of SIRV2. The HindIII recognition sites are indicated with short and vertical lines with the relevant sites labeled in nucleotides above the bar. Red arrows indicate the locations of the two terminal probes P1 and P2. (B) RIs revealed at both genomic termini. Left panel, 2D-AGE pattern of the left terminal fragment revealed with probe P1; middle panel, 2D-AGE pattern of the right terminal fragment (P2); right panel, diagrammatic illustrations of the gels: L, linear arc; 1n, unit-length linear fragment; 1.5n, 2n, 4n, fragments of 1.5, 2 and 4 times the unit-length, respectively; arc a1, bubble-like form intermediates ranging in size from 1n to around 1.5n; X, X-shaped molecules; m, 1n fragments derived from melted 2n species. Dimensions of the 2D-AGE (first and second) are indicated. (C) Illustration of early intermediates from strand-displacement replication. SIRV2 genome is depicted as black rods. Light blue rectangle indicates the probe position. The four HindIII recognition sites at the left side of the genome are indicated by vertical lines (dotted for the first site). The dark blue dot illustrates the Rep protein while the red arrowed line indicates the newly synthesized DNA strand. Arc a1 is the semi-bubble-like form as depicted in B, arcs a2 and a3 are described in the text.
Rep protein-mediated initiation at the termini of SIRV2 genome, followed by strand-displacement replication, will generate a partial ssDNA bubble-like species with only one strand replicated (Figure 3C). Restriction digestion of intermediates, the replication of which has not reached the first restriction site will produce partial ssDNA bubbles with sizes of up to 1.5n (species a1). After replication proceeds through the restriction site, the new dsDNA can be digested but not the displaced ssDNA. Moreover, since the genomic ends form hairpins, the ssDNA will tether the replicated terminal fragment (which will hybridize to the radiolabeled probe) to the restriction fragments carrying the site of synthesis. This will generate branched species where one arm is short and double stranded, the other significantly longer with ssDNA proximal to the junction, and the junction distal region will be dsDNA. Thus, species a2 (Figure 3C), corresponds to strand displacement synthesis between restriction sites at 3.9 and 4.5 kb (Figure 3A). This species spans only 600 bp and is a transient and low abundance species, and thus difficult to detect. As DNA synthesis proceeds further along the template, the ssDNA region will become longer, as exemplified by species a3, and account for the diffuse hybridization observed for the larger structured species.
An essentially identical intermediate species pattern was detected with a probe specific for the other genome end (Figure 3B, middle panel), indicating that strand displacement synthesis can initiate at both termini although it is unknown whether this can occur on a single genome. Nevertheless, given the genomic hairpin ends, ssDNA displaced in the initiation reaction will be replicated to generate circular dimeric genomes and, therefore, we observe hybridization to a species at the 2n position on the linear arc. In addition, a strong vertical signal emanates from the 2n position (Figure 3B), termed the ‘X-spike’ corresponding to an X-shaped molecule. Given that the 2n species is a perfect palindrome, centred on the terminal hairpin, it is likely that the X-shaped species arise spontaneously and migrate during electrophoresis. This would be compatible with species ‘m’ corresponding to generation of a 1n sized monomer molecule subsequent to the first dimension electrophoresis.
A second species emerges from the 2n spot, in addition to the X-spike, yielding an arc of increasing size and extends to a smear of large sizes (Figure 3C). Apart from the a3-like species from round one of strand displacement replication, intermediates from the rolling-circle mode of continuous strand-displacement synthesis that follow formation of the circular dimer chromosome, may also contribute to the diffuse hybridization products. The displaced strand will hybridize to the probes employed and be resistant to restriction enzyme digestion. Thus, regardless of the position of the replication complex on the DNA, the restriction fragment that it occupies, at any given time point, will be tagged with a single-stranded copy of the displaced strand. Therefore, the latter will hybridize to the probe and yield the high molecular weight signal (Figure 3C).
Strand-coupled replication of the duplex genome
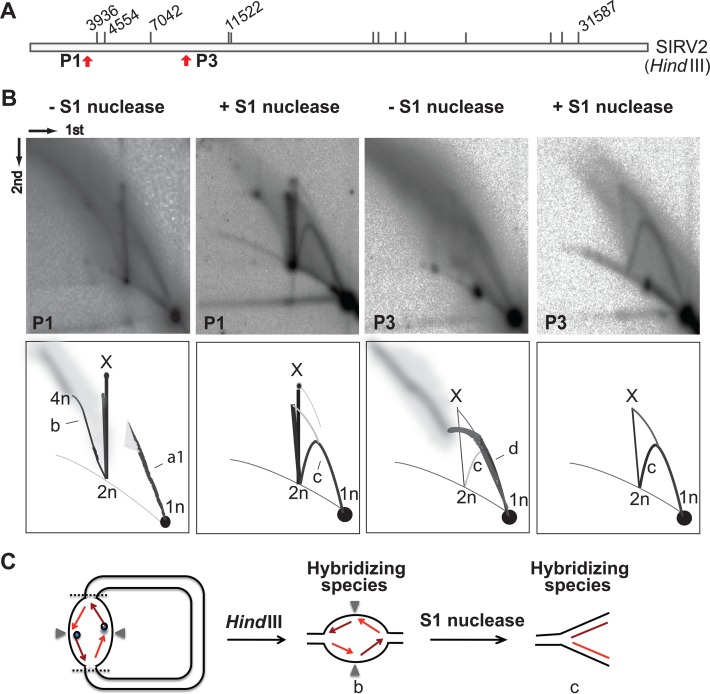
The previous result led to the prediction that the yield of the high molecular weight species will be diminished by treatment with a ssDNA-specific nuclease. Therefore, to test for the presence of ssDNA in the RIs (Figure 3), HindIII-digested agarose plugs were treated with S1 nuclease, followed by 2D analysis of the left terminal fragment (Figure 4B). Species a1, and those larger than 2n, were stronglly reduced by S1 nuclease treatment, consistent with the presence of ssDNA but unexpectedly a standard Y arc was formed indicative of conventional strand-coupled replication (Figure 4B, species c).
Figure 4.
Strand-coupled replication of SIRV2 genome. (A) HindIII digestion map of SIRV2 as in Figure 3A. The locations of the left terminal probes P1 and the internal probe P3 are indicated with red arrows. (B) 2D patterns of S1 nuclease treated plugs. Plugs were treated with or without S1 nuclease after HindIII digestion and before 2D gel electrophoresis. Dimensions of the 2D-AGE (first and second) are indicated. (C) Illustration of strand-coupled replication intermediates for the left terminal fragment. The dark blue dot indicates the Rep protein while the dark red arrowed line indicates the leading strand and light red arrowed lines depict the lagging strand. The gray triangles indicate the position of the genomic termini. Species 1n, 2n, 4n and X are as depicted in Figure 3; arc b, bubble form intermediates ranging in size from 2n to 4n; arc c, Y-shaped molecules; arc d, diffused molecules containing long ssDNA.
To determine the origin of the standard Y arc, 2D gel blots were hybridized with a probe (P3) generated from an internal HindIII fragment (Figure 4A). Before S1 nuclease treatment, high molecular weight RIs were observed located far from the linear arc and spanning between 1n and 2n (species d) and extending beyond 2n. In contrast to the 2D pattern generated from the terminal fragments, a very weak standard Y arc (species c) was produced from the internal fragment that was obscured by the slightly diffuse signal of species d. After S1 nuclease treatment, species d disappeared, indicating that it contained ssDNA. In contrast, the standard Y arc was unchanged and was therefore dsDNA.
The presence of both standard Y arc and nuclease S1 sensitive arcs strongly suggests that two different replication mechanisms, strand displacement (Figure 3C) and strand-coupled (Figure 4C), operate during SIRV2 genome replication. We infer that a sub-population of genomes initiate bidirectional strand-coupled synthesis at the end of the dimer chromosome, generating a bubble-shaped species b emanating from 2n position and ending at the 4n position. The presence of the Rep protein, covalently attached to the 5′-end of the leading strand, will prevent its immediate ligation to the lagging strand. The resultant discontinuity would then produce sensitivity to nuclease S1 and lead to formation of the Y-shaped structure upon S1 digestion (Figure 4C).
Specific loading of the lagging strand synthesis complex on the dimer chromosome is likely to be facilitated by special structural features including Holliday junctions, or other secondary structures resulting from recombination in the perfectly palindromic termini. Moreover, simultaneous initiation of the two Rep-mediated leading strand synthesis reactions on the dimer (2n) produces a different terminal conformation from that generated from a single genome undergoing strand-displacement replication, and may play a role in strand-coupled replication initiation.
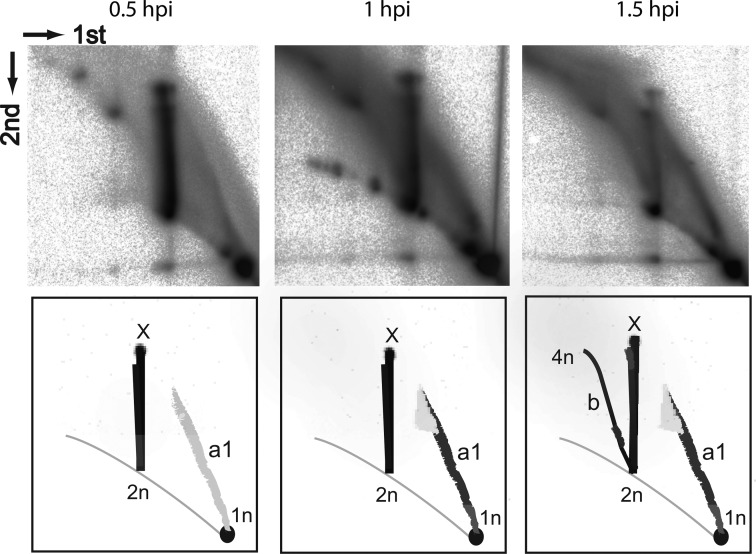
A time course experiment of 2D gel electrophoresis on samples taken up to 1.5 hpi (Figure 5) revealed very weak signals from intermediate species a1 at 0.5 hpi, and a signal from strand displacement replication at 1 hpi. At 1.5 hpi, we observed an additional signal of intermediates arising from strand-coupled replication. This indicates, in agreement with the above model, that most initial replication of SIRV2 is mediated by the strand-displacement with strand-coupled synthesis first occurring after the initial rounds of replication.
Figure 5.
SIRV2 replication dynamics. The left terminal probe P1 was used for all 3 time points, 30, 60 and 90 min post-infection. Species 1n, 2n, 4n and X are as depicted in Figure 4; arcs are as depicted in Figures 3 and 4. Dimensions of the 2D-AGE (first and second) are indicated. Arc a1, intermediates from strand-displacement replication ranging in size between 1n to about 1.5n; arc b, bubble form intermediates from strand-coupled replication ranging in size from 2n to 4n.
Multiple reinitiations from single parental template take place during SIRV2 DNA replication
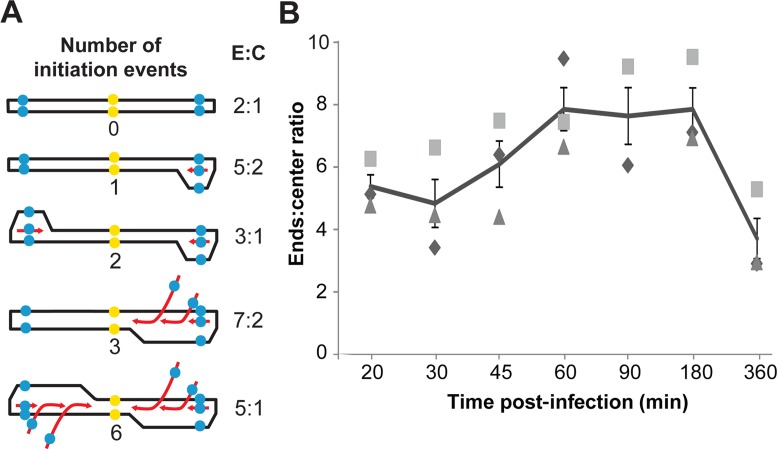
The architecture of SIRV2 RIs (Figure 1E and F), and the complexity of the high-molecular weight species, could be explained by the occurrence of multiple replication reinitiation events. To test this hypothesis, we used quantitative PCR to determine the ratio between the putative origins of replication (at the genomic ends within ITRs) and the central part of the genome in cells infected with SIRV2 with a MOI ≥ 5. In non-replicating viral genomes this ratio is 2:1. In molecules where a single initiation event takes place the ends:centre ratio will be lower than 3:1 if replication could start from both genomic ends (Figure 6A). To detect both ends simultaneously, an ITR sequence was selected as target and a gene gp26 (NP_666560) was selected as a target for the central region. The threshold cycle (Ct) values of samples, with three biological replicates, are shown in Supplementary Table S1. The results are normalized with respect to DNA extracted from purified SIRV2 virion particles for which the ends:center ratio is assumed to be 2:1. At 20 min p.i., the ends:center ratio was ∼5 and it increased steadily reaching a maximum of ∼8 at 1–1.5 hpi, and decreased thereafter (Figure 6B). These results indicate that the yield of replication origins in infected cells is 4-fold higher than for non-replicating genomes, and about 3-fold higher than expected for genomes with a single replication initiation event per end. This result supports the hypothesis that multiple reinitiation events occur during SIRV2 genome replication.
Figure 6.
Multiple replication reinitiation events during SIRV2 DNA replication. (A) Model of multiple replication reinitiation. The parental DNA is depicted in black and the daughter DNA is presented in red; the blue dots indicate the qPCR target in ITRs (ends) while the yellow dots indicate the center. (B) The ends:center ratio in SIRV2-infected cultures at different times post-infection. Results from three independent biological replicates are shown. The line indicates the average ratio at each time point and the error bars indicate the standard error. E:C, ends:center ratio.
Model for SIRV2 genome replication
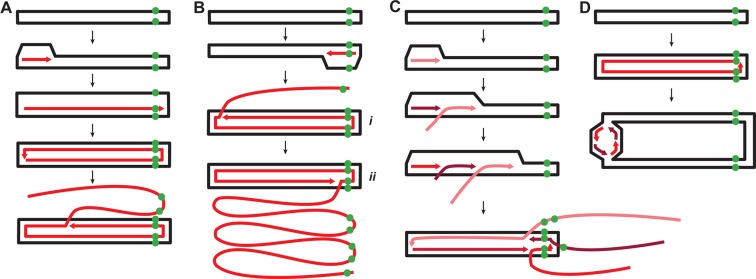
A predominant role of strand-displacement replication and multiple reinitiation events during SIRV2 genomic replication is directly supported by the topology of the replication intermediates visualized by DNA spreading and fluorescence microscopy. A fiber containing two closely adjacent probe signals in an internal segment (indicated by dark arrowheads in Figure 1D–F) is inferred to be ssDNA produced by strand-displacement replication that has traversed the right genomic terminus, whereas strand-displacement replication initiating at the right terminus generated ssDNA with a single probe signal at the tip (Figures 1C and 7B species i and ii). Within monomeric dsDNA, a single probe signal is expected as the two strands are very closely localized even after heat denaturation (Figure 1B). For dimeric dsDNA in a circular form (Figure 1D) a single spot signal is a more likely product than two adjacent ones because plectonemic supercoiling of covalently closed circular DNA will limit its spreading on the slide surface. Thus, the fibre present in Figure 1C is interpreted as a replication intermediate that initiated at the right terminus and reached the left terminus after 2n formation (Supplementary Figure S6A and species i in Figure 7B). In contrast, the fiber in Figure 1D is inferred to be a replication intermediate that also initiated at the right terminus but then passed the right terminus at least twice (Supplementary Figure S6B and species ii in Figure 7B) with the compacted dimeric dsDNA possibly located at the right side. The DNA species present in Figure 1E comprises five ssDNA branches with replication initiated at the left terminus and all products have traversed the right terminus at least once (Supplementary Figure S6C). The replication intermediate in Figure 1F comprises numerous ssDNA branches initiated at the left terminus; all except one are approaching the right terminus of the dimeric dsDNA (Supplementary Figure S6D). Notably, the stretch level of the linear dsDNA on the glass slide appears more or less constant (Figure 1B) while that of the ssDNA fibers is highly variable. Thus, the lengths of the ssDNA fibers could not be used to judge the size of the RIs.
Figure 7.
Model of the genome replication mechanism of SIRV2. (A) Strand-displacement replication initiated from the left end of the genome. (B) Rolling-circle mode of strand-displacement replication initiated from the right end. (C) Multiple replication reinitiation leading to the brush-like replication. (D) Strand-coupled replication. The parental DNA is shown in black and the newly synthesized DNA is depicted in shades of red. The green dots indicate the position of the probe DIG-P2 at the right end of the genome.
Based on the data obtained, we propose the following model for SIRV2 genome replication. Replication starts at the ends of the genome by a self-priming mechanism mediated by the covalently-bound Rep protein (15). It then proceeds asymmetrically via strand-displacement generating circular dimer intermediates. Then replication continues via two alternative mechanisms: by strand displacement or by strand-coupled replication. Strand-displacement replication predominates and, after formation of the circular dimer, multimers of ssDNA are produced by rolling-circle replication. Finally, multiple reinitiation events at the termini of the parental genome template, and strand-displacement replication, produce ‘brush-like’ structures.
DISCUSSION
Among the diverse replication strategies employed by DNA viruses and plasmids, only two have been well-defined, θ replication and σ replication. The formation of multimeric and branched replication intermediates has been reported for viruses infecting bacteria (e.g. phage T4 (3)) and eukaryotes (e.g. HSV-1 (23)) as well as plasmids (e.g. mitochondrial plasmid mp1 (24)). Moreover, recombination-dependent replication has been proposed for the formation of the complex networked branches in phage T4 (3). However, to date, no defined structures have been described. Our present work presents a novel replication model with a clear structure defined for highly branched replication intermediates. The model we propose could be commonly employed by a broad range of viruses with linear dsDNA genomes and is consistent with the multimeric and branched intermediates observed for both bacterial and eukaryotic viruses.
Poxviruses and rudiviruses have similar genomic structure and organization (10,25). Moreover, head-to-head and tail-to-tail terminally linked concatemers have been detected for both. Furthermore, a similar resolution mechanism by a virus-encoded Holliday junction resolvase has been proposed for both families (7,25). However, the identification of a primase and an endonuclease encoded in the poxvirus genome hint to a RNA primer-dependent replication initiation and the occurrence of lagging-strand synthesis (26). This is different to the Rep-mediated initiation mechanism proposed for rudiviruses, in which the initiatior protein Rep makes a nick close to the genomic terminus, thus providing a 3′-OH to prime DNA synthesis (15).
We demonstrate in this study that SIRV2 employs strand-displacement, rolling circle and strand-coupled replication mechanisms. This raises a question as to whether all three replication mechanisms are essential for SIRV2 replication and how these different replication mechanisms are coordinated. We hypothesize that the strand-coupled replication requires a different set of proteins for replication initiation, and future genetic analyses shall help to elucidate which host and/or viral proteins regulate the process.
Our experimental approach of DNA spreading on slides, combined with fluorescence microscopy, allowed the visualization of viral RIs obtained directly from lysed cells and minimized the potential for intermolecular annealing of ssDNA RIs. Moreover, it avoided the extensive preparative steps required for electron microscopy which might generate artifacts or produce RI damage. This approach has proven powerful in the analysis of the replication dynamics of eukaryotic cellular chromosomes but has rarely been employed for studying viral DNA replication (21). Thus, future application of the technique on viral RIs is likely to provide novel insights into the detailed replication mechanisms of viral DNA.
In general, the 2D-AGE signals from samples without S1 treatment are more diffuse (Figures 3 and 4). This could be partially explained by the multiple replication reinitiations. For example, the diffuse signal of species a1 around the 1.5n position (Figure 3B) is possibly due to the presence of molecules of similar sizes but containing different numbers of ssDNA branches, which migrate differently in the second dimension.
Due to the branched and multimeric nature of the intermediates and the widespread presence of ssDNA, processing such complex structures into mature genome units poses a challenge for both the replication and maturation/resolution machineries. We suggest that DNA recombination may play a key role in these processes. The rudivirus-encoded Holliday-junction resolvase (Hjr) has been proposed to play a role in the resolution of the SIRV2 replication intermediates, and in the assembly of progeny virions due to its interaction with the major coat protein (27). Moreover, a conserved rudiviral operon, encoding ssDNA binding (SIRV2 gp17), ssDNA annealing (SIRV2 gp18) and ssDNA nuclease (SIRV2 gp19) activities, has been suggested to play a role in DNA recombination and/or genome maturation (28). We note that the ssDNA binding activity of gp17 could help protect the ssDNA regions arising during viral replication. Further, sequence analysis of the SIRV genomes had previously suggested the occurrence of recombination (10). Further studies to address the role of recombination during SIRV2 replication will be needed.
SIRV2 replication intermediates reach a size of >1200 kb (Figure 2 and Supplementary Figure S5) corresponding to around 34 viral genome units. The sizes of these species might not be precisely estimated due to the potentially anomalous electrophoretic mobility of branched DNA, but the value correlates with the estimated burst size of around 30 (±10) SIRV2 virions per cell (22). This implies that daughter genome units synthesized from a parental template remain linked throughout the replication process and their conversion into monomeric viral genome units occurs concomitantly with packaging into progeny virions. Indeed, multiple viral genomes were often found to be joined together in SIRV2 RIs (Figure 1F and Supplementary Figure S3).
Interestingly, SIRV2 progeny form 1–3 bundles of virions intracellularly before cell lysis (22) which could imply that daughter genome linkage is a mechanism for concentrating replication intermediates and facilitating their maturation and packaging. Moreover, this would be similar to the formation of viral replication factories by some eukaryotic viruses (29). In addition, multiple replication reinitiation events from a single parental template facilitate rapid viral genome replication. In summary, this replication model provides a spatial and temporal control of viral genome replication that may facilitate viral escape from host defence mechanisms.
Supplementary Material
Acknowledgments
We thank Rachel Samson for technical advice on 2D analysis, Yang Guo for preparing part of the samples used in qPCR and Roger Garrett for critical revision of this manuscript. The funding institutions had no role in any decision taken about the design, development or publication of the present work.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Union FP7 collaborative project HotZyme [265933]; X.P. was funded by EMBO short term fellowship; L.M was supported by a doctoral scholarship from Consejo Nacional de Ciencia y Tecnología [312536]. Funding for open access charge: University internal fund.
Conflict of interest statement. None declared.
REFERENCES
- 1.Koonin E.V., Dolja V.V. A virocentric perspective on the evolution of life. Curr. Opin. Virol. 2013;3:546–557. doi: 10.1016/j.coviro.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.del Solar G., Giraldo R., Ruiz-Echevarría M.J., Espinosa M., Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. MMBR. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weigel C., Seitz H. Bacteriophage replication modules. FEMS Microbiol. Rev. 2006;30:321–381. doi: 10.1111/j.1574-6976.2006.00015.x. [DOI] [PubMed] [Google Scholar]
- 4.Hoeben R.C., Uil T.G. Adenovirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5:a013003. doi: 10.1101/cshperspect.a013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weller S.K., Coen D.M. Herpes simplex viruses: mechanisms of DNA replication. Cold Spring Harb. Perspect. Biol. 2012;4:a013011. doi: 10.1101/cshperspect.a013011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Berns K.I. Parvovirus replication. Microbiol. Rev. 1990;54:316–329. doi: 10.1128/mr.54.3.316-329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Moss B. Poxvirus DNA replication. Cold Spring Harb. Perspect. Biol. 2013;5 doi: 10.1101/cshperspect.a010199. doi:10.1101/cshperspect.a010199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ravin N.V. Replication and maintenance of linear phage-plasmid N15. Microbiol. Spectr. 2015;3 doi: 10.1128/microbiolspec.PLAS-0032-2014. doi:10.1128/microbiolspec.PLAS-0032-2014. [DOI] [PubMed] [Google Scholar]
- 9.Hinnebusch J., Tilly K. Linear plasmids and chromosomes in bacteria. Mol. Microbiol. 1993;10:917–922. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 10.Peng X., Blum H., She Q., Mallok S., Brügger K., Garrett R.A., Zillig W., Prangishvili D. Sequences and replication of genomes of the archaeal rudiviruses SIRV1 and SIRV2: relationships to the archaeal lipothrixvirus SIFV and some eukaryal viruses. Virology. 2001;291:226–234. doi: 10.1006/viro.2001.1190. [DOI] [PubMed] [Google Scholar]
- 11.Dellas N., Snyder J.C., Bolduc B., Young M.J. Archaeal viruses: diversity, replication, and structure. Annu. Rev. Virol. 2014;1:399–426. doi: 10.1146/annurev-virology-031413-085357. [DOI] [PubMed] [Google Scholar]
- 12.Prangishvili D., Forterre P., Garrett R.A. Viruses of the Archaea: a unifying view. Nat. Rev. Microbiol. 2006;4:837–848. doi: 10.1038/nrmicro1527. [DOI] [PubMed] [Google Scholar]
- 13.Servín-Garcidueñas L.E., Peng X., Garrett R.A., Martínez-Romero E. Genome sequence of a novel archaeal rudivirus recovered from a mexican hot spring. Genome Announc. 2013;1:e0016413. doi: 10.1128/genomeA.00040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gudbergsdóttir S.R., Menzel P., Krogh A., Young M., Peng X. Novel viral genomes identified from six metagenomes reveal wide distribution of archaeal viruses and high viral diversity in terrestrial hot springs. Environ. Microbiol. 2016;18:863–874. doi: 10.1111/1462-2920.13079. [DOI] [PubMed] [Google Scholar]
- 15.Oke M., Kerou M., Liu H., Peng X., Garrett R.A., Prangishvili D., Naismith J.H., White M.F. A dimeric Rep protein initiates replication of a linear archaeal virus genome: implications for the Rep mechanism and viral replication. J. Virol. 2011;85:925–931. doi: 10.1128/JVI.01467-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okutan E., Deng L., Mirlashari S., Uldahl K., Halim M., Liu C., Garrett R.A., She Q., Peng X. Novel insights into gene regulation of the rudivirus SIRV2 infecting Sulfolobus cells. RNA Biol. 2013;10:875–885. doi: 10.4161/rna.24537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zillig W., Arnold H.P., Holz I., Prangishvili D., Schweier A., Stedman K., She Q., Phan H., Garrett R., Kristjansson J.K. Genetic elements in the extremely thermophilic archaeon Sulfolobus. Extrem. Life Extreme Cond. 1998;2:131–140. doi: 10.1007/s007920050052. [DOI] [PubMed] [Google Scholar]
- 18.Zillig W., Kletzin A., Schleper C., Holz I., Janekovic D., Hain J., Lanzendörfer M., Kristjansson J.K. Screening for sulfolobales, their plasmids and their viruses in icelandic solfataras. Syst. Appl. Microbiol. 1993;16:609–628. [Google Scholar]
- 19.Bettstetter M., Peng X., Garrett R.A., Prangishvili D. AFV1, a novel virus infecting hyperthermophilic archaea of the genus acidianus. Virology. 2003;315:68–79. doi: 10.1016/s0042-6822(03)00481-1. [DOI] [PubMed] [Google Scholar]
- 20.Pfaffl M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parra I., Windle B. High resolution visual mapping of stretched DNA by fluorescent hybridization. Nat. Genet. 1993;5:17–21. doi: 10.1038/ng0993-17. [DOI] [PubMed] [Google Scholar]
- 22.Bize A., Karlsson E.A., Ekefjärd K., Quax T.E.F., Pina M., Prevost M.-C., Forterre P., Tenaillon O., Bernander R., Prangishvili D. A unique virus release mechanism in the Archaea. Proc. Natl. Acad. Sci. U.S.A. 2009;106:11306–11311. doi: 10.1073/pnas.0901238106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Severini A., Scraba D.G., Tyrrell D.L. Branched structures in the intracellular DNA of herpes simplex virus type 1. J. Virol. 1996;70:3169–3175. doi: 10.1128/jvi.70.5.3169-3175.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Backert S. R-loop-dependent rolling-circle replication and a new model for DNA concatemer resolution by mitochondrial plasmid mp1. EMBO J. 2002;21:3128–3136. doi: 10.1093/emboj/cdf311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blum H., Zillig W., Mallok S., Domdey H., Prangishvili D. The genome of the archaeal virus SIRV1 has features in common with genomes of eukaryal viruses. Virology. 2001;281:6–9. doi: 10.1006/viro.2000.0776. [DOI] [PubMed] [Google Scholar]
- 26.De Silva F.S., Lewis W., Berglund P., Koonin E.V., Moss B. Poxvirus DNA primase. Proc. Natl. Acad. Sci. U.S.A. 2007;104:18724–18729. doi: 10.1073/pnas.0709276104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gardner A.F., Guan C., Jack W.E. Biochemical characterization of a structure-specific resolving enzyme from Sulfolobus islandicus rod-shaped virus 2. PLoS One. 2011;6:e23668. doi: 10.1371/journal.pone.0023668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Guo Y., Kragelund B.B., White M.F., Peng X. Functional characterization of a conserved archaeal viral Operon Revealing Single-Stranded DNA Binding, Annealing and Nuclease Activities. J. Mol. Biol. 2015;427:2179–2191. doi: 10.1016/j.jmb.2015.03.013. [DOI] [PubMed] [Google Scholar]
- 29.Netherton C., Moffat K., Brooks E., Wileman T. A guide to viral inclusions, membrane rearrangements, factories, and viroplasm produced during virus replication. Adv. Virus Res. 2007;70:101–182. doi: 10.1016/S0065-3527(07)70004-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.