Abstract
Modifications of the bacterial ribosome regulate the function of the ribosome and modulate its susceptibility to antibiotics. By modifying a highly conserved adenosine A2503 in 23S rRNA, methylating enzyme Cfr confers resistance to a range of ribosome-targeting antibiotics. The same adenosine is also methylated by RlmN, an enzyme widely distributed among bacteria. While RlmN modifies C2, Cfr modifies the C8 position of A2503. Shared nucleotide substrate and phylogenetic relationship between RlmN and Cfr prompted us to investigate evolutionary origin of antibiotic resistance in this enzyme family. Using directed evolution of RlmN under antibiotic selection, we obtained RlmN variants that mediate low-level resistance. Surprisingly, these variants confer resistance not through the Cfr-like C8 methylation, but via inhibition of the endogenous RlmN C2 methylation of A2503. Detection of RlmN inactivating mutations in clinical resistance isolates suggests that the mechanism used by the in vitro evolved variants is also relevant in a clinical setting. Additionally, as indicated by a phylogenetic analysis, it appears that Cfr did not diverge from the RlmN family but from another distinct family of predicted radical SAM methylating enzymes whose function remains unknown.
INTRODUCTION
As a critical component of the ribosome, ribosomal RNA (rRNA) plays a vital role in protein synthesis. In all living organisms rRNA carries wide range of physiological post-transcriptional modifications that facilitate both the assembly and the activity of the ribosome. Methylation of nucleotides, both at nucleobases and the 2′-hydroxyl of the ribose, is the most common rRNA modification in bacteria. Many of these modifications are located in the functional regions of the ribosome, such as the peptidyl transferase center (PTC) and decoding centers, as well as at the interface of ribosomal subunits. Although no single rRNA modification is critical for the survival of the cell, the presence of individual methylations confers advantages under certain growth or stress conditions (1). As the bacterial ribosome represents a major antibiotic target, methylation of rRNA has emerged as one of the most clinically relevant mechanisms of resistance to ribosome-targeting antibiotics (2,3). In most instances, methylation of rRNA alters the drug-binding site thus reducing the ability of drugs to inhibit the ribosome's translational activity. For example, mono- or dimethylation of the 23S rRNA nucleotide A2058 (E. coli numbering), located in the exit tunnel, by Erm methyltransferases interferes with the binding of macrolides and streptogramin B antibiotics (4,5). Less commonly, absence of methylation can also confer antibiotic resistance (2,3,6–8). For instance, the lack of methylation at A1518 and A1519 of 23S rRNA by KsgA confers resistance to kasugamycin (9).
Methylation of rRNA in bacteria is carried out by a number of methylating enzymes. Among them, enzymes that modify the PTC of the ribosome, the site of peptide bond formation, are particularly interesting, as these enzymes contribute to the control of protein biosynthesis and can also mediate resistance to antibiotics (8,10). RlmN and Cfr are two related bacterial enzymes that, by methylating 23S rRNA at an adenosine located in the PTC region (A2503), contribute to the post-transcriptional modifications of this important region of the ribosome. RlmN methylates A2503 at the C2 carbon (m2A), a modification that is nearly ubiquitous among bacterial species (11). While the role of this modification is yet to be fully understood, it has been suggested that it regulates interactions between the ribosome and the nascent peptide (12,13). Additionally, deletion of rlmN reduces the accuracy of protein synthesis, an effect attributed to the loss of methylation at A2503 (14). While the effects of the RlmN-mediated modification on bacterial phenotypes are yet to be fully characterized, lack of RlmN in S. aureus leads to an increase in linezolid resistance and provides a slight fitness advantage over the WT S. aureus cells (15).
While Cfr shares a nucleotide substrate with RlmN, it preferentially methylates the C8 position of A2503, leading to resistance to phenicols, lincosamides, oxazolidinones, pleuromutilins, streptogramin A, 16-membered macrolides, as well as aminocyclitol hygromycin A and nucleoside antibiotic A201A (7,16,17). Interestingly, Cfr can also methylate the C2 position of A2503 both in vivo (in ΔrlmN strain) and in vitro (with an in vitro transcribed rRNA fragment), thus forming a 2,8-dimethyl adenosine (m2m8A) (18,19). In addition to sharing a common nucleotide substrate and overlapping reaction specificity, RlmN and Cfr are both members of the radical SAM methylsynthase family sharing 34% sequence identity (E. coli RlmN versus Cfr isolated from S. aureus, hereafter S. aureus Cfr), and are considered to employ the same catalytic mechanism (18,20–25). These observations indicate common ancestry of the Cfr and RlmN families.
Unlike RlmNs, currently identified Cfrs are distributed among pathogenic species, with exception of three Cfr-like enzymes from the Bacillales order (26). Previous phylogenetic analysis of RlmN and Cfr-like enzymes indicated that Cfr-like enzymes form a well-defined clade within the RlmN family (27,28). Additionally, it was suggested that cfr may have evolved from an rlmN ancestor via gene duplication, but the lineage along which the duplication occurred is still unknown (27). In this study, we implemented directed evolution to examine if Cfr-like activity could be evolved from RlmN in response to antibiotic pressure. Upon selection we identified RlmN mutants conferring low-level resistance, but remarkably, rather than the expected enzymes with Cfr-like properties, these RlmN variants were catalytically inactive towards 23S rRNA. The mutations conferring resistance were specific, and did not include nonsense mutations. Interestingly, these evolved variants successfully compete with the endogenous E. coli RlmN for the A2503 of 23S rRNA in vivo, and as a result diminish the C2 methylation of 23S RNA. Our findings suggest that inhibition of C2 A2503 methylation is beneficial in the presence of certain antibiotics as previously suggested for S. aureus (15). Rapid rise and maintenance of these dominant negative variants through multiple rounds of evolution suggest that this resistance mechanism can easily appear among clinical pathogens. Our results expand the list of resistance mechanisms resulting from lack of natural rRNA modifications, and are compatible with the possibility that Cfr may not have diverged directly from an RlmN, but rather, from an RlmN related ancestor whose primary methylation target is not A2503.
MATERIALS AND METHODS
Phylogenetic analysis
Seventy species from the Firmicutes phylum, with the emphasis on the genus with higher coverage (i.e. Bacillus and Staphylococcus) and Clostridium, were selected from the Joint Genome Institute – Integrated Microbial Genomes/Expert Review (IMG-er/JGI) database. From this subgroup, RlmN and Cfr sequences were retrieved by BLAST using the RlmN sequence from B. subtilis (hereafter Bsub_RlmN) as query. The E-value was set to 1e-5 to allow for the selection of sequences that share low identity to the query. The sequences were aligned using MUSCLE (29). The phylogenetic reconstruction was done in PhyML (30), using the automatic model selection and selecting by the Akaike Information Criteria.
Plasmids and strains used for the transformation, expression, antibiotic susceptibility and methylation analysis
E. coli ER2267 was used in directed evolution experiments. E. coli BL21(DE3) was used for overexpression of the His6-tagged variants from either pET21a or pET15b vectors. E. coli BW25113 and BW25113/ΔrlmN strains were used in antibiotic susceptibility tests and in in vivo methylation assays. BW25113/ΔrlmN strain, where the rlmN was replaced with the kanamycin resistance cassette, was a generous gift from Dr Alexander Mankin, University of Illinois at Chicago, USA (31). The pZA vector was used in directed evolution experiments, antibiotic susceptibility and in vivo methylation analysis (32).
Library construction and selection
Random mutagenesis was performed by polymerase chain reaction (PCR) using an error-prone polymerase (GeneMorph Mutazyme, Stratagene). Wild-type (WT) Bsub_RlmN, or the pool of Bsub_RlmN genes from the previous round, was used as a template with primers that flank the enzyme's open reading frame. The protocol was optimized to 1–3 mutations per gene. The mutated Bsub_RlmN genes were recloned into the pZA vector and transformed into E. coli ER2267. Selection for the evolved variants was performed on the LB agar plates containing different concentrations of tiamulin (Wako Chemicals USA). Additionally, each plate contained ampicillin (100 μg/ml) and anhydrotetracycline (AHT; Sigma-Aldrich; 20 ng/ml). For each round of selection, the total transformation (1 ml) was divided into three aliquots and plated onto LB agar plates containing varying concentrations of tiamulin. The tiamulin concentration was increased in 25 μg/ml increments. For example, in the first round of evolution the transformation was plated on the 75, 100 and 125 μg/ml tiamulin plates, in the last round we selected on 150, 175 and 200 μg/ml tiamulin plates. Two microliters were plated on tiamulin deficient plates in order to determine transformation efficiency. Cells were grown at 37°C for up to 48 h. Between the first and second rounds of evolution (mutation and selection) we included an additional cycle of selection with no mutagenesis (transformation, selection and plasmid extraction). At the end of each round, 6–10 randomly chosen clones, from each tiamulin concentration, were isolated and sequenced.
Antibiotic susceptibility tests
Drug susceptibility was tested on the LB agar plates. Each plate contained 100 μg/ml of ampicillin for the selection of the plasmid and 20 ng/ml of AHT for the expression of the enzymes. Plates also contained 50 μg/ml of kanamycin when antibiotic susceptibility was tested in the E. coli ΔrlmN strain. In a standard experiment, 2 ml of LB media containing appropriate antibiotics were inoculated from a fresh colony. Cultures were grown at 37°C for ∼3 h, at which point OD600 was measured and the cultures were diluted to 108, 106 and 104 cells. A 3 μl aliquot of each dilution was spotted on the agar plates. Plates were incubated at 37°C for 24–48 h. MICs of the antibiotics were determined by the broth microdilution assay following ref (33). LB medium was inoculated with single colonies harboring plasmids with WT Bsub_RlmN or Bsub_RlmN variant genes and incubated at 37°C for 4–5 h. The cultures were diluted to OD600 = 0.001 and 50 μl of diluted culture was mixed with 50 μl of antibiotic solution prepared in LB medium. Expression of enzymes was induced by addition of AHT to the antibiotics solution (final concentration 20 ng/ml). The tested concentration ranges were: for tiamulin, 250 to 1800 μg/ml (in 50 μg/ml steps from 250 to 600 μg/ml followed by 200 μg/ml steps to 1800 μg/ml); for clindamycin (TCI America), 50 to 1200 μg/ml (in 50 μg/ml steps from 50 to 400 μg/ml followed by 100 μg/ml steps to 800 μg/ml and 200 μg/ml steps to 1200 μg/ml); for chloramphenicol (Acros), 0.5 to 32 μg/ml (in 2-fold concentration steps); for virginiamycin M1 (Cayman Chemical), 100 to 1200 μg/ml (in 50 to 100 μg/ml concentration steps to 600 μg/ml followed by 200 μg/ml steps to 1200 μg/ml); for trimethoprim (Sigma-Aldrich), 0.125 to 2 μg/ml (in 2-fold concentration steps). The MIC values were determined after an overnight incubation (16–18 h) at 37°C by measuring the optical density at 600 nm with a microtiter plate reader (SpectraMax M5, Molecular Devices). The MIC was defined as the drug concentration with no visible growth. Each value is a replicate of at least three independent experiments.
Expression and purification of Bsub_RlmN evolved variants
WT Bsub_RlmN and evolved variants were expressed, purified and reconstituted for their iron-sulfur clusters using modified versions of previously published protocols (18,34,35). Briefly, enzymes were overexpressed and purified by Talon chromatography (Clontech). After chemical reconstitution of the iron-sulfur cluster, proteins were further purified by FPLC either on a Superdex 200 10/30 column or on a HiLoad 26/60 Superdex 75 Prep grade column (GE Healthcare Life Sciences) using 10 mM HEPES (pH 7.5) buffer containing 500 mM NaCl, 10% glycerol and 5 mM DTT. The fractions containing protein were combined and concentrated before being stored at −80°C.
Preparation of truncated rRNA substrates and tRNAs for the in vitro methylation assay
E. coli 23S rRNA fragments 2447–2625 and 2018–2625 used in in vitro methylation assay were generated by in vitro transcription following previously published methods (18,35). pGOV4 vectors containing genes for tRNAAspGUC, tRNAGlnUUG, tRNAGluUUC, tRNAHisGUG and tRNAGlyCCC were purchased from Gene Oracle. T7 promotor sequence was inserted in front of these gene, while BamHI sequence was inserted at the end of the gene. Prior to the in vitro transcription reaction, vector was linearized with BamHI (New England Biolabs) at 37°C for 14 h. Linearized vector was purified using Qiagen PCR cleanup kit prior to in vitro transcription reaction following the same procedure as for the rRNA fragments.
In vitro methylation assay
Methylation activity of the evolved variants was assessed by monitoring the radioactivity incorporation into RNA. Reactions were performed in 100 μl volumes under the following conditions: 100 mM HEPES pH 8.0, 100 mM KCl, 10 mM MgCl2, 2 mM DTT, 20 μM Flavodoxin, 2 μM Flavodoxin reductase, 4 μM RNA and 0.14 μCi [14C-methyl]-SAM (58 mCi/mmoL) and 1.3–14 μM enzyme. Reactions were initiated by addition of NADPH (final concentration 1 mM), and were allowed to proceed at 37°C for 1–1.5 h. The RNA was recovered from the reaction mixture using the RNA Clean & Concentrator kit (Zymo Research) and added to the vials containing Ultima Gold scintillation fluid. The amount of radioactivity incorporated in the product was measured using Beckman–Coulter LS6500 multipurpose scintillation counter (Fullerton, CA, USA). Each value represents the average of at least triplicate (for reactions with rRNA) or duplicate (for reactions with tRNA) measurements, with one standard deviation (SD) indicated.
HPLC separation and identification of methylated adenosines
The methylated rRNA from WT Bsub_RlmN assay mixtures was purified using the RNA Clean & Concentrator kit (Zymo Research). Subsequently, the purified rRNA was enzymatically digested to mononucleosides using nuclease P1 (Sigma-Aldrich), snake venom phosphodiesterase (Sigma-Aldrich) and antarctic phosphatase (New England Biolabs). The digested samples were separated on HPLC using Luna analytical C18 column (10 μm, 4.6 mm × 250 mm) (Phenomenex, Torrance, CA, USA) and previously published protocol (18). The mononucleosides and the synthetic methyladenosines were detected by their UV absorption at 256 nm, while the 14C-labled mehyladenosines were detected by a Packard radiomatic 515TR flow scintillation analyzer (Perkin–Elmer).
Preparation of rRNA fragments and MALDI analysis
For isolation of total RNA, E. coli BW25113 or BW25113/ΔrlmN strains harboring appropriate pZA plasmid were grown in LB medium at 37°C in the presence of inducer, AHT (final concentration 20 ng/ml) to an OD600 of 0.6–0.8. The total RNA was purified using RNeasy Midi Kit (Qiagen) following manufacturer's recommendations. The 41-mer fragment encompassing A2503 (C2480–C2520), was isolated using an established complementary oligodeoxynucleotide procedure (35,36). A total of 25–30 pmol of isolated RNA fragment was mixed with 0.5 volumes of 0.5 M 3-hydroxypicolinic acid (3-HPA; Sigma-Aldrich) and 500 U of RNase T1 (US Biological) and left to digest at 37°C for 3 h. To linearize cyclic phosphates that result from the RNase T1 digestion, 0.25 volumes of 0.5 M HCl were added and the mixture was left at room temperature for 30 min. The samples were lyophilized, and then re-dissolved in water. One microliter of the sample was spotted onto the target plate and mixed with 1 μl of 0.5 M 3-HPA. Spectra were recorded in a reflector and positive ion mode either on a Voyager Elite STR MALDI-TOF mass spectrometer (Applied Biosystems/Life Technologies) or AXIMA Performance MALDI TOF/TOF Mass Spectrometer (Shimadzu).
RESULTS
To date, all known Cfr enzymes capable of C8 methylation at A2503 and associated with antibiotic resistance belong to the Firmicutes phylum. As it has been previously speculated that Cfr evolved from RlmN (27,28), we hypothesized that Cfr-like functionality could have evolved from an RlmN belonging to this bacterial phylum. To test this hypothesis, we examined the RlmN of the model organism for gram-positive Firmicutes species, namely B. subtilis (Bsub_RlmN).
Bsub_RlmN shares slightly higher sequence identity to known Cfrs than E. coli RlmN (Ecoli_RlmN; 39% and 34%, respectively). A previous study showed that in vivo Bsub_RlmN methylates C2 position of A2503 in 23S rRNA from E. coli (28), which we confirmed both in vivo and in vitro (described below). Additionally, as observed with other RlmNs, no C8 methylation of A2503 was detected with Bsub_RlmN, even at high enzyme to substrate ratio (e.g. 7:1). Promiscuous activities are considered the raw material for evolutionary innovations (37). Nonetheless, we endeavored to examine whether, under antibiotic selection pressure, Cfr-like functionality could evolve.
Evolved Bsub_RlmN variants mediate increased antibiotic resistance
Using error-prone PCR, we introduced random 1–3 mutations per bsub_rlmN gene cloned into a medium copy number plasmid, pZA (32). A library comprising of ∼107 WT E. coli transformants was subjected to selection under increasing amounts of tiamulin, a PTC antibiotic to which Cfr confers resistance (Supplementary Figure S1). Subsequently, the bsub_rlmN genes were recovered from all surviving colonies and were subjected to amplification with further error-prone mutagenesis and selection. The selection pressure was gradually strengthened by increasing the concentration of the antibiotic from 75 to 125 μg/ml and up to 200 μg/ml. In total, three rounds of random mutagenesis and selection were performed to gradually increase the ability of E. coli to survive in the presence of tiamulin.
As early as the first round of evolution we noticed different colony sizes, which were not observed in a control plate containing solely E. coli transformants with WT Bsub_RlmN, indicating that certain RlmN mutations were advantageous to cell survival in the presence of tiamulin. Accordingly, the surviving clones did not carry the wild-type RlmN gene and were enriched in mutations in certain positions (e.g. S168 and G201). By the third round, the sequences of randomly chosen variants indicated convergence, with a few specific mutations dominating the surviving pool (Supplementary Table S1). Most of the prevailing amino acid substitutions were located far from the active site (>12 Å; Figure 1A). One of the few exceptions was the A348T mutation (Figure 1A), located in the highly conserved C-terminal region of RlmN and a part of the active site upon substrate binding (38). More than 50% of the tiamulin selected clones contained S168C and G201D, two mutations that usually appeared together (Supplementary Tables S1 and S2). Q89R was also one of the more prevalent mutations after the first round of evolution, usually occurring together with G201D (Supplementary Table S1). This mutation was not observed in the second round, yet it reemerged in the third round. Importantly, nonsense mutations (frame shifts and stop codons) were not observed in the selected variants.
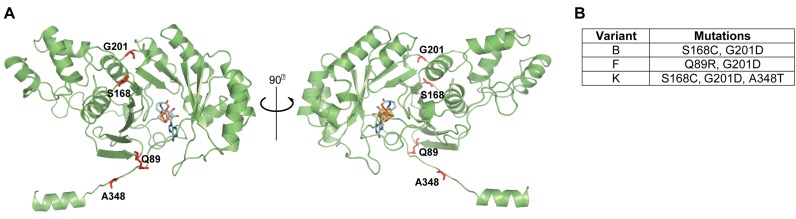
Figure 1.
Evolved variants of Bsub_RlmN contain specific mutations distal from the active site. (A) A homology model of the structure of Bsub_RlmN was generated by I-TASSER. Shown in sticks are the iron-sulfur center (gold) and the SAM cofactor (light blue) borrowed from the aligned template structure (PDB 3RFA). The most dominant mutations present in the evolved variants are shown as red sticks. Among the mutated residues present in the Bsub_RlmN variants studied herein, only A348, part of the C-terminal region that undergoes conformational change upon substrate binding, is a part of the active site, while other residues are located >12 Å from SAM's methyl group (38). (B) Mutations present in the 3 evolved Bsub_RlmN variants analyzed in this study.
We consequently focused on the in vivo and in vitro functional characterization of the three evolved variants that represented the most prevalent mutational combinations present in the tiamulin resistant variants (dubbed Bsub_RlmN variants B, F and K; Figure 1B). An antibiotic susceptibility test clearly indicated that E. coli cells expressing the Bsub_RlmN B, F and K variants grew better in the presence of tiamulin than cells carrying the WT Bsub_RlmN or an empty plasmid (Figure 2A). The selective survival of cells expressing evolved variants was observed at tiamulin concentrations up to 200 μg/ml (Figure 2A). Tiamulin resistance was also dose-dependent with respect to the enzymes’ expression levels, as determined by varying concentration of the pZA plasmid inducer, AHT (Supplementary Figure S2). Even though E. coli cells expressing the evolved Bsub_RlmN variants show growth advantage in comparison to cells carrying the WT Bsub_RlmN or an empty plasmid, their minimum inhibitory concentration (MIC) values are lower in comparison to cells expressing S. aureus Cfr (18,19) (Supplementary Figure S3 and Table S3).
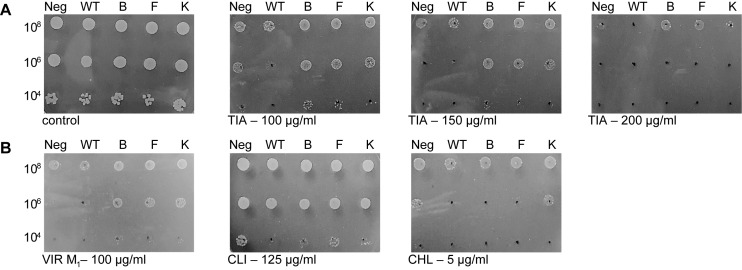
Figure 2.
Antibiotic susceptibility of E. coli transformed with plasmid-encoded Bsub_RlmN variants. (A) Dose-dependent antibiotic susceptibility test towards tiamulin. (B) Antibiotic susceptibility test toward several PTC-targeting antibiotics. All plates contained ampicillin (the plasmid resistance marker) and AHT to induce the expression of the Bsub_RlmN variants. Cells were plated at three densities, and plates were recorded after 24 h. Abbreviations: Neg = empty pZA; WT = pZA_WT_Bsub_RlmN; B = pZA_BsubB; F = pZA_BsubF; K = pZA_BsubK; TIA = tiamulin; VIR M1 = virginiamycin M1; CLI = clindamycin; CHL = chloramphenicol.
We subsequently examined if these variants confer survival advantage in the presence of other PTC antibiotics known to be affected by Cfr-mediated rRNA methylation. Three additional clinically important PTC antibiotics were tested: a lincosamide antibiotic, clindamycin; a phenicol, chloramphenicol; and a streptogramin A, virginiamycin M1 (Supplementary Figure S1). While preferential growth of strains expressing Bsub_RlmN variants was observed in the presence of virginiamycin M1 (VIR M1; Figure 2B and Supplementary Table S3), no growth advantage was observed in the presence of clindamycin and chloramphenicol (Figure 2B). Additionally, MIC values obtained by broth microdilution method suggest the same trend (Supplementary Table S3). MIC values of tiamulin for cells expressing evolved variants are ∼1.5-fold higher than those expressing WT Bsub_RlmN. For virginiamycin M1, this effect is more subtle (400 μg/ml for variants and 300 μg/ml for WT Bsub_RlmN) yet reproducible. No difference in MIC values was observed for clindamycin and chloramphenicol, which is in accord with the results obtained from the antibiotic susceptibility test on agar plates (Figure 2B).
Evaluation of in vitro activity of Bsub_RlmN evolved variants
We next evaluated the in vitro activity of the evolved Bsub_RlmN variants. Our hypothesis was that, similar to Cfr, these enzymes would methylate A2503 at the C8 position, given their ability to confer an increase in antibiotic resistance. All three selected variants (Figure 1B) were successfully expressed and purified, and their catalytic iron–sulfur clusters were readily reconstituted. Using an established in vitro assay (35), we investigated the activities of the evolved enzyme variants by monitoring the accumulation of radioactivity from S-adenosyl- l-[methyl-14C]methionine ([14C-methyl]-SAM) in a fragment of 23S rRNA encompassing A2503 (Figure 3). While, as expected, WT Bsub_RlmN successfully methylated C2 position of A2503 in 23S rRNA in vitro (Figure 3 and Supplementary Figure S4), none of the evolved variants could methylate the rRNA fragments (Figure 3). We then examined if these mutations might have changed the substrate specificity of the Bsub_RlmN evolved variants, based on the report that E. coli RlmN can also methylate a subset of tRNAs (14). An in vitro activity assay showed that neither the WT Bsub_RlmN (Supplementary Figure S5A) nor any of the evolved variants can methylate the tested tRNAs, which is in accord with no identified tRNA A37 methylation in B. subtilis tRNAs (39) (Supplementary Figure S5B).
Figure 3.
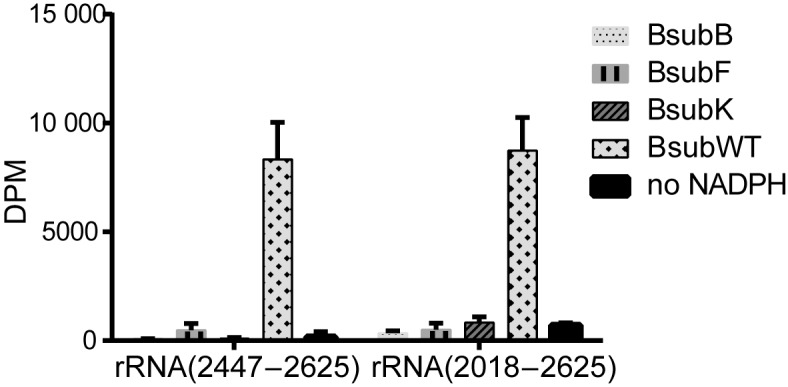
Methylation of 23S rRNA by WT Bsub_RlmN and its evolved variants. In vitro measurement of the methylation activity of Bsub_RlmN variants toward fragments of E. coli 23S rRNA. The assay monitored incorporation of the radioactivity from [14C-methyl]-SAM (24.9 μM) into the rRNA fragment (4 μM), after 1 h incubation with 14 μM enzyme at 37°C. Error bars (n ≥ 3), S.D.
These findings suggest that the evolved variants not only lacked Cfr-like reactivity, but have also lost the RlmN-like activity, implying that inactivation of RlmN is beneficial under tiamulin selection pressure. Interestingly, the inactivation of Bsub_RlmN was not achieved through random insertions of stop codons or frame shifts or via other severely deleterious mutations that typically cause misfolding or aggregation. Such mutations are frequent amongst the repertoire of random mutations. Rather full length, soluble and folded, albeit inactive variants of Bsub_RlmN were selected for.
Lack of methylation at A2503 decreases antibiotic susceptibility
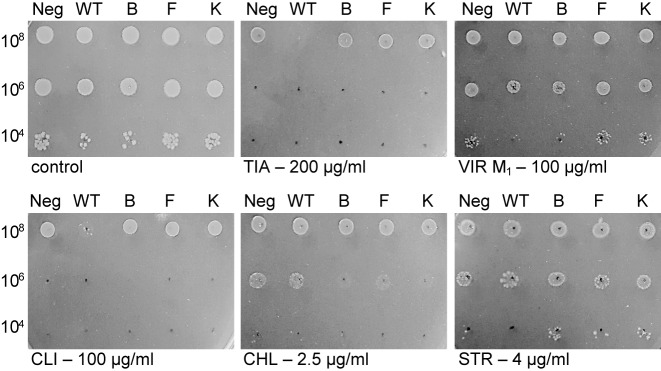
Several previous studies attempted to examine if the lack of RlmN is beneficial under antibiotic pressure (11,15). Deletion of rlmN in E. coli leads to a slight increase in susceptibility toward several PTC antibiotics (11). In contrast, inactivation of rlmN in S. aureus leads to an increase in linezolid resistance. Furthermore, in co-growth experiments under linezolid pressure, S. aureus lacking functional RlmN outcompetes the strain with an active RlmN (15). To evaluate the link between RlmN and low-level resistance to PTC antibiotics in E. coli, and to determine the effect of the evolved variants on antibiotic susceptibility, we examined antibiotic sensitivity of the rlmN knockout strain (E. coli BW25113/ΔrlmN) expressing an empty plasmid, WT Bsub_RlmN or the evolved variants (Figure 4, Supplementary Figure S6 and Supplementary Table S3). Our results show that RlmN knockout cells that carry an empty pZA plasmid, and thus are not methylated at A2503, survive equally well as knockout cells expressing evolved variants in the presence of tiamulin, clindamycin and virginiamycin M1, three antibiotics that target PTC. The only exception are cells expressing the BsubB variant in the presence of virginiamycin M1 where it is not clear if expression of this particular variant is advantageous. In contrast, cells expressing the WT Bsub_RlmN and thus have their rRNA methylated at the C2 position of A2503, are more susceptible to tiamulin and slightly more to clindamycin, while this susceptibility is not observed for chloramphenicol, another PTC-targeting antibiotic (Figure 4). To further explore these effects we measured the MIC values by the broth microdilution method. MICs of tiamulin and virginiamycin M1 against RlmN knockout cells that carry an empty plasmid, or are expressing evolved variants, are higher than MIC values for these antibiotics when WT Bsub_RlmN is expressed in the same strain (Supplementary Table S3). However, the MIC values of clindamycin and chloramphenicol for E. coli BW25113/ΔrlmN cells expressing WT Bsub_RlmN are not significantly different from MIC values obtained for these cells when expressing the evolved variants or carrying an empty plasmid. No difference in the cell growth was observed for streptomycin, an aminoglycoside that targets 30S subunit (Figure 4). Similarly, no difference in the MIC values of trimethoprim, an antibiotic that interferes with folate metabolism, was observed (Supplementary Table S3).
Figure 4.
Antibiotic susceptibility of E. coli ΔrlmN strain expressing plasmid-encoded wild-type and evolved Bsub_RlmN variants. All plates contained ampicillin (the plasmid selection marker), kanamycin for selection of the ΔrlmN strain and AHT to induce the expression of the RlmN variants. Cells were plated at three densities, and plates were recorded after 24 h. Abbreviations: Neg = empty pZA; WT = pZA_WT_Bsub_RlmN; B = pZA_BsubB; F = pZA_BsubF; K = pZA_BsubK; TIA = tiamulin; VIR M1 = virginiamycin M1; CLI = clindamycin; CHL = chloramphenicol; STR = streptomycin.
Overall, the comparison between the cell survival of E. coli ΔrlmN expressing the WT Bsub_RlmN and those containing the empty plasmid indicates that the lack of C2 methylation at A2503 is beneficial in the presence of a subset of PTC antibiotics. Furthermore, the similarity in antibiotics tolerance between ΔrlmN cells carrying the empty plasmid and WT cells expressing the evolved variants suggest that although the evolved variants do not modify A2503 of 23S rRNA as described above, they may act as dominant negative proteins, potentially via binding of the rRNA substrate, thus preventing methylation of A2503 by the endogenous RlmN.
Bsub_RlmN evolved variants act as dominant negative enzymes
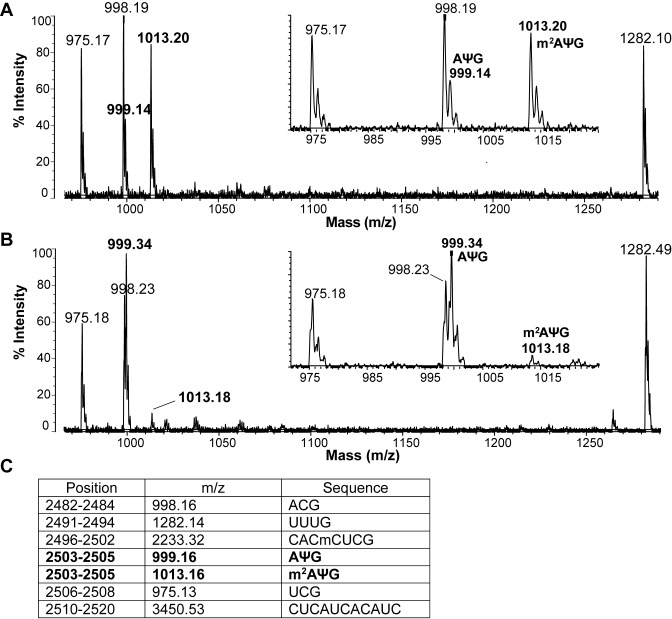
To elucidate if the evolved Bsub_RlmN variants interfere with the E. coli RlmN-catalyzed methylation at C2 position of A2503, we analyzed the extent of methylation of A2503 by matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) mass spectrometry. Specifically, using an oligonucleotide protection assay, a fragment of 23S rRNA (2480–2520) that includes A2503 was isolated, digested to ≥ 3-nt long oligonucleotide fragments and analyzed by MALDI-TOF mass spectrometry (Figure 5). Utilizing this approach, we examined RNA fragments isolated from the WT E. coli strain (that contains an endogenous copy of RlmN; Figure 5A), and from WT E. coli that also expresses BsubB variant (Figure 5B). We focused on the BsubB variant since this variant best represented the most prevalent mutations found in the tiamulin resistant clones. Analysis of samples obtained from the strain expressing only the endogenous copy of RlmN indicated that the peak at m/z 1013.20, corresponding to m2A2503, is more prominent than the peak at m/z 999.14 that corresponds to unmethylated A2503. This finding indicates that A2503 is predominately methylated by the endogenous RlmN in WT E. coli cells. However, the ratio between the peak at m/z 999.34 and the peak at m/z 1013.18 changes from approximately 1:4 in WT E. coli cells to 9:1 in cells where the evolved BsubB variant is expressed (Figure 5B). These findings support the hypothesis that the evolved variant BsubB acts as a dominant negative protein, preventing methylation by the endogenous RlmN.
Figure 5.
Reduction of in vivo methylation at A2503 upon expression of BsubB supports the dominant negative function of the evolved variants. MALDI-TOF mass spectrum of the C2480–C2520 fragment of E. coli 23S rRNA isolated from the WT E. coli BW25113 strain carrying an (A) empty plasmid, or expressing (B) evolved BsubB variant. RNA was digested by RNase T1 and analyzed by MALDI-TOF. Comparison between the insets shows a significant reduction in the amount of methylated 2503-m2AΨG-2505 fragment at m/z 1013.16 when the BsubB variant is overexpressed. (C) A list of the expected RNase T1 digestion fragments based on the rRNA sequence with presently known nucleotide modifications. Cm is methylated cytosine, m2A is 2-methyladenosine, and Ψ indicates pseudouridine.
To further corroborate that the evolved Bsub_RlmN variants do not methylate A2503, we examined the methylation status of A2503 in 23S rRNA isolated from ΔrlmN cells expressing either WT Bsub_RlmN, BsubB evolved variant or S. aureus Cfr. While mono-methylation of A2503 (peak at m/z 1013.32) was observed in cells expressing WT Bsub_RlmN, clearly indicating that 23S rRNA of E. coli is a substrate for this enzyme, no methylation was observed in cells expressing the evolved BsubB variant (Supplementary Figure S7). These findings confirm that the evolved variants lack methylation activity toward the 23S rRNA. Both mono- and dimethylation of A2503 (peaks at m/z 1013.34 and 1027.37, respectively) were observed in cells expressing S. aureus Cfr, recapitulating previously published in vitro data (18) (Supplementary Figure S7D).
Phylogenetic analysis of the Firmicutes RlmN and Cfr families
Our findings indicate that directed evolution of Bsub_RlmN under antibiotic pressure did not readily yield enzymes with Cfr-like activities. Instead, upon selection for higher resistance to PTC antibiotics, rather than evolving toward C8 methylation of A2503, Bsub_RlmN evolved to confer resistance by obstructing C2 methylation of A2503. In addition, neither we nor others could detect promiscuous levels of C8 methylation of A2503 in RlmNs, neither in the broadly studied Ecoli_RlmN, nor in the Bsub_RlmN examined here (18,28). We thus reexamined the RlmN-Cfr phylogeny in order to reevaluate the hypothesis that Cfr may have evolved from RlmN under selection for survival in the presence of high levels of PTC binding antibiotics.
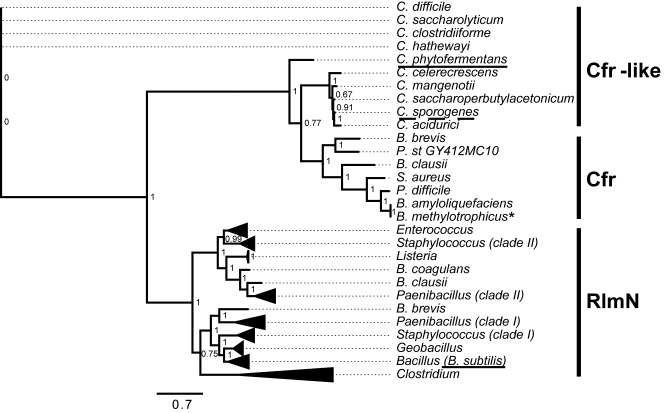
Previous phylogenetic analyses indicated that known RlmNs and Cfrs comprise two distinct clades (27,28). Additionally, a separate clade containing Cfr-like sequences that are particularly abundant in the Clostridia class was detected (28). To further examine the relationships between these three families, we performed a phylogenetic analysis of the Cfr-like and RlmN-like orthologous from selected Firmicutes species, with an emphasis on the Clostridia class. Sequences were retrieved from the Integrated Microbial Genomes – Joint Genome Institute (IMG/JGI) database using BLAST, and aligned prior to the generation of the phylogenetic tree by maximum likelihood method (PhylML) (Figure 6). Overall, our tree corroborated previously reported trees where the Cfr sequences from clinical and veterinary samples appear as a separate subgroup that branches out of the RlmN family (27,28). However, the closest clade to known Cfr enzymes are not RlmNs but rather a group of clostridial genes currently annotated as Cfr-like proteins (Figure 6, Supplementary Figure S8).
Figure 6.
A schematic phylogenetic tree of RlmN and Cfr sequences from selected Firmicutes species. Known Cfrs, known and putative RlmNs and Cfr-like sequences are marked. Underlined with a continuous black line are the enzymes characterized in this study: RlmN from B. subtilis (Bsub_RlmN) that is part of the Bacillus clade, and a Cfr-like from C. phytofermentas (ClCp) in the divergent clostridial clade. A discontinued underline indicates the previously characterized Cfr-like enzyme from C. sporogenes (27). An asterisk specifies a functionally uncharacterized enzyme from B. methylotrophicus that clades with known Cfr enzymes. A Paenibacillus species that possesses two putative RlmN paralogues is shown, and the paralogues that appear to belong to two separate Cfr clades are marked as I and II. Two putative paralogous RlmNs from Staphylococcous species that belong to these clades are also marked. The non-collapsed version of the phylogenetic tree with the full names and IMG/JGI database identifiers is shown in Supplementary Figure S8.
Previous examination of one of these genes, a Cfr-like gene from C. sporogenes, indicated that this enzyme is unable to methylate rRNA (28). We have examined another gene from this clade, a Cfr-like gene from C. phytofermentas – ClCp, and also could not observe methylation of A2503 in 23S rRNA either in vivo or in vitro (Supplementary Figure S9). Furthermore, our attempts to evolve the Cfr-like functionality in ClCp, namely C8 methylation of A2503, were also unsuccessful. Several libraries derived from WT ClCp were constructed and selected as described for Bsub_RlmN, yet colonies expressing ClCp variants that confer higher resistance to tiamulin compared to the cells expressing WT ClCp were not observed. In addition, we did not observe enrichment of specific mutations in sequenced variants or the dominant negative variants, as it was the case with Bsub_RlmN. We cannot completely exclude the possibility that ClCp has RlmN/Cfr-like activity, as this protein may be able to methylate the Clostridium 23S rRNA and not the E. coli 23S rRNA. However, this seems unlikely given the high sequence identity between the 23S rRNA of E. coli and C. phytofermentas (80%). Additionally, for one of the known Cfrs from clinical isolates of pathogen Peptoclostridium difficile, C8 methylation of A2503 was successfully recapitulated in E. coli (40). It therefore appears that Cfr-like genes comprise a distinct family of enzymes that noticeably differ in function from both RlmN and Cfr and whose methylation target(s), if any, remain unknown.
DISCUSSION
In the present study, we investigated how a specific post-transcriptional modification in 23S rRNA affects susceptibility to PTC-targeting antibiotics. Specifically, we focused on the methylation of A2503 in 23S rRNA, a nucleotide located in the PTC region of the ribosome. This base is subject to methylation by two evolutionary related enzymes, RlmN that catalyzes methylation at the C2 position of A2503, and Cfr that is responsible for the C8 methylation of A2503. While it has been shown that the methylation at C8 position leads to multi-antibiotic resistance, the effect of C2 methylation by RlmN on antibiotic resistance remains unclear. Further, the evolutionary origins of Cfr remain unknown. RlmN is widely spread, whereas Cfr is sporadically observed in high resistance strains and only in specific bacterial clades. The most plausible hypothesis is thus that Cfr evolved from an RlmN (27,28), but this hypothesis has not been validated. The results of our laboratory evolution experiment indicate that antibiotic pressure leads to evolution of catalytically inactive variants of RlmN. Rather than methylating the C8 position of A2503, these variants act as dominant negative proteins to prevent C2 methylation of A2503 by the endogenous E. coli RlmN, providing an immediate selective advantage under tiamulin pressure. These observations highlight the critical role of methylation of A2503 in regulating the response to antibiotics as it appears that both lack of the methylation at C2 position, and C8 methylation by acquisition of Cfr, lead to the antibiotic resistance (7,16).
The evolved variants likely exert their dominant negative function by competing with the Ecoli_RlmN for 23S rRNA. Since RlmN only modifies the free 23S rRNA, and not the large ribosomal subunit nor fully assembled ribosome (18), methylation must occur during the narrow time frame when 23S rRNA is available prior to its incorporation into the large ribosomal subunit. The evolved variants likely interfere with this process by blocking the access of Ecoli_RlmN to 23S rRNA. The effectiveness of each of the variants in preventing the methylation likely depends both on their affinity for 23S rRNA and their stability and thereby on their cellular concentrations. Once incorporated into the large subunit, lack of methylation cannot be corrected by endogenous RlmN as A2503 in 23S rRNA is no longer accessible for methylation. The requirement for the competition with Ecoli_RlmN during the selection may also rationalize the observation that our evolution experiments, performed in the WT E. coli, yielded stable, full length, yet catalytically inactive proteins. While nonsense and frameshift mutations would also yield catalytically inactive proteins, they would be unlikely to compete with the WT RlmN for binding to the substrate.
Many rRNA methylating enzymes function as antibiotic resistance determinants by modifying various nucleotides (Supplementary Table S4). Only in a few instances, however, was the absence of rRNA methylation found to increase antibiotic resistance (41–44). Consistent with the previous study on S. aureus RlmN (15), our findings indicate that the lack of RlmN-mediated modification of A2503 in 23S rRNA similarly yields decreased susceptibility to tiamulin and virginiamycin M1. How lack of methylation at A2503 leads to an antibiotic resistance phenotype is not completely clear. From the available bacterial ribosome crystal structures, it is evident that A2503 belongs to a region that defines the binding pocket for the PTC-targeting antibiotics. It is therefore possible that the lack of methylation by RlmN changes the local structure of PTC, subsequently affecting binding of some PTC-targeting antibiotics but not others. Furthermore, our observations that substrates of Ecoli_RlmN, 23S rRNA and several tRNAs are not modified by the evolved variants strongly supports the notion that the evolved proteins are not catalytically active. We cannot, however, exclude the possibility that the evolved variants might be catalytically active toward currently unknown targets, although it is unclear how modification of other possible RNAs might contribute to the resistance toward PTC-targeting antibiotics.
From the perspective of protein evolution, our findings do not support the prevailing hypothesis that Cfr-like function diverged from RlmN, and that this divergence proceeded via a gradual change in the methylation specificity, namely, from exclusive methylation of C2 of A2503 in RlmN, to C8-A2503 (with residual C2-A2503 methylation as observed in Cfrs). Rather, our observations indicate that despite their sequence homology, and the overlap in substrate specificity, RlmNs and Cfrs are only distantly related. Similar conclusions were also reached in a recent study by Ntokou et al. who explored the functional linkage between Cfr and RlmN by swapping different amino acid segments in their respective active sites (45). A distant evolutionary connection between RlmNs and Cfrs is also suggested from the phylogenetic analysis since the closest family to Cfr does not exhibit either RlmN or Cfr activity (Supplementary Figure S9) (28). The functional divergence of the radical SAM methylsynthase family is also suggested from the phylogenetic analysis where we detected the presence of several species with paraloguous RlmNs. Specifically, most Paenibacillus species possess two or more putative RlmNs, and a maximum of 4 paralogous RlmN/Cfr-like enzymes were detected in P. durum (data not shown). Multiple paralogs present an added difficulty in selection of a starting point for directed evolution experiments.
Our findings also hold important clinical implications. Loss-of-function mutations are far more frequent than gain-of-function. The fact that loss-of-function mutations arose rapidly in our directed evolution experiment and were maintained through multiple rounds of evolution suggests that similar mutations may be easily acquired in a clinic setting. Such mutations may become more common with the increased use of antibiotics. Interestingly, an amino acid insertion into a highly conserved C-terminal region of RlmN was observed in methicillin resistant Staphylococcus aureus (MRSA) isolates from a patient with a persistent and recurrent infection that was resistant to oxazolidinone linezolid, a PTC-targeting antibiotic (46). A causative relationship between this genetic alteration of rlmN and antibiotic susceptibility was confirmed by introduction of the insertion mutant into the parent strain which recapitulated linezolid resistance. This insertion was subsequently shown to inactivate RlmN, linking the loss of C2 methylation of A2503 to the clinical linezolid low-level resistance (15). Mutations in rlmN have also been reported in clinical isolates of linezolid-resistant S. capitis, although their individual contributions to antibiotic susceptibility are difficult to discern given the presence of other alterations associated with linezolid resistance (47). These observations indicate the importance of monitoring RlmN mutation status along with other commonly monitored resistance mechanisms such as acquisition of cfr, mutations in domain V of 23S rRNA and mutations in ribosomal proteins L3 and L4, in clinical isolates of linezolid-resistant Staphylococci (7,48–53). Together, our laboratory evolution experiments and the clinical observations illuminate a possibility that mutations that lead to a loss of RlmN-mediated methylation of A2503 could readily elicit resistance to antibiotics that target the PTC of the ribosome. From the perspective of clinical use of PTC antibiotics, this is particularly worrisome since the lack of methylation by RlmN has a minimal effect on the cell fitness (15), and inactivating mutations, according to our findings, easily emerge after exposure to antibiotics.
Supplementary Material
Acknowledgments
The authors thank Lindsey Pack and other members of the Fujimori lab for helpful discussions and comments on the manuscript. The authors thank Dr Alexander Mankin, University of Illinois at Chicago, Dr Christopher T. Walsh, Harvard Medical School and Dr Dragana Despotović, Weizmann Institute, for their comments on the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EMBO Fellowship [to V.S.]; CONACYT grant [#203740 to L.N.G.]; Martin Kurshner Fellowship, Weizmann Institute of Science [to L.N.G.]; National Institutes of Health [NIAID R01AI095393 to D.G.F.]; Adelis Foundation [to D.S.T.]; UCSF Research Resource Program Shared Equipment Award funded by the Chancellor [AXIMA Performance MALDI TOF/TOF Mass Spectrometer]. Funding for open access charge: National Institutes of Health [NIAID R01AI095393].
Conflict of interest statement. None declared.
REFERENCES
- 1.Sergeeva O.V., Bogdanov A.A., Sergiev P.V. What do we know about ribosomal RNA methylation in Escherichia coli? Biochimie. 2015;117:110–118. doi: 10.1016/j.biochi.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 2.Vester B., Long K.S. In: DNA and RNA Modification Enzymes: Structure, Mechanism, Function and Evolution. Grosjean H, editor. Austin: Landes Bioscience; 2009. pp. 537–549. [Google Scholar]
- 3.Wilson D.N. Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 2014;12:35–48. doi: 10.1038/nrmicro3155. [DOI] [PubMed] [Google Scholar]
- 4.Skinner R., Cundliffe E., Schmidt F.J. Site of action of a ribosomal RNA methylase responsible for resistance to erythromycin and other antibiotics. J. Biol. Chem. 1983;258:12702–12706. [PubMed] [Google Scholar]
- 5.Leclercq R., Courvalin P. Bacterial resistance to macrolide, lincosamide, and streptogramin antibiotics by target modification. Antimicrob. Agents Chemother. 1991;35:1267–1272. doi: 10.1128/aac.35.7.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poehlsgaard J., Douthwaite S. The bacterial ribosome as a target for antibiotics. Nat. Rev. Microbiol. 2005;3:870–881. doi: 10.1038/nrmicro1265. [DOI] [PubMed] [Google Scholar]
- 7.Long K.S., Poehlsgaard J., Kehrenberg C., Schwarz S., Vester B. The Cfr rRNA methyltransferase confers resistance to Phenicols, Lincosamides, Oxazolidinones, Pleuromutilins, and Streptogramin A antibiotics. Antimicrob. Agents Chemother. 2006;50:2500–2505. doi: 10.1128/AAC.00131-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCusker K.P., Fujimori D.G. The chemistry of peptidyltransferase center-targeted antibiotics: enzymatic resistance and approaches to countering resistance. ACS Chem. Biol. 2012;7:64–72. doi: 10.1021/cb200418f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poldermans B., Goosen N., Van Knippenberg P.H. Studies on the function of two adjacent N6,N6-dimethyladenosines near the 3′ end of 16 S ribosomal RNA of Escherichia coli. I. The effect of kasugamycin on initiation of protein synthesis. J. Biol. Chem. 1979;254:9085–9089. [PubMed] [Google Scholar]
- 10.Wilson D.N. The A-Z of bacterial translation inhibitors. Crit. Rev. Biochem. Mol. Biol. 2009;44:393–433. doi: 10.3109/10409230903307311. [DOI] [PubMed] [Google Scholar]
- 11.Toh S.M., Xiong L., Bae T., Mankin A.S. The methyltransferase YfgB/RlmN is responsible for modification of adenosine 2503 in 23S rRNA. RNA. 2008;14:98–106. doi: 10.1261/rna.814408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vazquez-Laslop N., Ramu H., Klepacki D., Kannan K., Mankin A.S. The key function of a conserved and modified rRNA residue in the ribosomal response to the nascent peptide. EMBO J. 2010;29:3108–3117. doi: 10.1038/emboj.2010.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ramu H., Vazquez-Laslop N., Klepacki D., Dai Q., Piccirilli J., Micura R., Mankin A.S. Nascent peptide in the ribosome exit tunnel affects functional properties of the A-site of the peptidyl transferase center. Mol. Cell. 2011;41:321–330. doi: 10.1016/j.molcel.2010.12.031. [DOI] [PubMed] [Google Scholar]
- 14.Benitez-Paez A., Villarroya M., Armengod M.E. The Escherichia coli RlmN methyltransferase is a dual-specificity enzyme that modifies both rRNA and tRNA and controls translational accuracy. RNA. 2012;18:1783–1795. doi: 10.1261/rna.033266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LaMarre J.M., Howden B.P., Mankin A.S. Inactivation of the indigenous methyltransferase RlmN in Staphylococcus aureus increases linezolid resistance. Antimicrob. Agents Chemother. 2011;55:2989–2991. doi: 10.1128/AAC.00183-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smith L.K., Mankin A.S. Transcriptional and translational control of the mlr operon, which confers resistance to seven classes of protein synthesis inhibitors. Antimicrob. Agents Chemother. 2008;52:1703–1712. doi: 10.1128/AAC.01583-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Polikanov Y.S., Starosta A.L., Juette M.F., Altman R.B., Terry D.S., Lu W., Burnett B.J., Dinos G., Reynolds K.A., Blanchard S.C., et al. Distinct tRNA Accommodation Intermediates Observed on the Ribosome with the Antibiotics Hygromycin A and A201A. Mol. Cell. 2015;58:832–844. doi: 10.1016/j.molcel.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yan F., LaMarre J.M., Rohrich R., Wiesner J., Jomaa H., Mankin A.S., Fujimori D.G. RlmN and Cfr are radical SAM enzymes involved in methylation of ribosomal RNA. J. Am. Chem. Soc. 2010;132:3953–3964. doi: 10.1021/ja910850y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giessing A.M., Jensen S.S., Rasmussen A., Hansen L.H., Gondela A., Long K., Vester B., Kirpekar F. Identification of 8-methyladenosine as the modification catalyzed by the radical SAM methyltransferase Cfr that confers antibiotic resistance in bacteria. RNA. 2009;15:327–336. doi: 10.1261/rna.1371409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boal A.K., Grove T.L., McLaughlin M.I., Yennawar N.H., Booker S.J., Rosenzweig A.C. Structural basis for methyl transfer by a radical SAM enzyme. Science. 2011;332:1089–1092. doi: 10.1126/science.1205358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grove T.L., Benner J.S., Radle M.I., Ahlum J.H., Landgraf B.J., Krebs C., Booker S.J. A radically different mechanism for S-adenosylmethionine-dependent methyltransferases. Science. 2011;332:604–607. doi: 10.1126/science.1200877. [DOI] [PubMed] [Google Scholar]
- 22.Yan F., Fujimori D.G. RNA methylation by radical SAM enzymes RlmN and Cfr proceeds via methylene transfer and hydride shift. Proc. Natl. Acad. Sci. U.S.A. 2011;108:3930–3934. doi: 10.1073/pnas.1017781108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McCusker K.P., Medzihradszky K.F., Shiver A.L., Nichols R.J., Yan F., Maltby D.A., Gross C.A., Fujimori D.G. Covalent intermediate in the catalytic mechanism of the radical S-adenosyl-L-methionine methyl synthase RlmN trapped by mutagenesis. J. Am. Chem. Soc. 2012;134:18074–18081. doi: 10.1021/ja307855d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grove T.L., Livada J., Schwalm E.L., Green M.T., Booker S.J., Silakov A. A substrate radical intermediate in catalysis by the antibiotic resistance protein Cfr. Nat. Chem. Biol. 2013;9:422–427. doi: 10.1038/nchembio.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Silakov A., Grove T.L., Radle M.I., Bauerle M.R., Green M.T., Rosenzweig A.C., Boal A.K., Booker S.J. Characterization of a cross-linked protein-nucleic acid substrate radical in the reaction catalyzed by RlmN. J. Am. Chem. Soc. 2014;136:8221–8228. doi: 10.1021/ja410560p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hansen L.H., Planellas M.H., Long K.S., Vester B. The order Bacillales hosts functional homologs of the worrisome cfr antibiotic resistance gene. Antimicrob. Agents Chemother. 2012;56:3563–3567. doi: 10.1128/AAC.00673-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaminska K.H., Purta E., Hansen L.H., Bujnicki J.M., Vester B., Long K.S. Insights into the structure, function and evolution of the radical-SAM 23S rRNA methyltransferase Cfr that confers antibiotic resistance in bacteria. Nucleic Acids Res. 2010;38:1652–1663. doi: 10.1093/nar/gkp1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Atkinson G.C., Hansen L.H., Tenson T., Rasmussen A., Kirpekar F., Vester B. Distinction between the Cfr methyltransferase conferring antibiotic resistance and the housekeeping RlmN methyltransferase. Antimicrob. Agents Chemother. 2013;57:4019–4026. doi: 10.1128/AAC.00448-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Edgar R.C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 31.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wellner A., Raitses Gurevich M., Tawfik D.S. Mechanism of protein sequence divergence and incompatibility. PLoS Genet. 2013;9:e1003665. doi: 10.1371/journal.pgen.1003665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 34.Lanz N.D., Grove T.L., Gogonea C.B., Lee K.H., Krebs C., Booker S.J. RlmN and AtsB as models for the overproduction and characterization of radical SAM proteins. Methods Enzymol. 2012;516:125–152. doi: 10.1016/B978-0-12-394291-3.00030-7. [DOI] [PubMed] [Google Scholar]
- 35.Stojkovic V., Fujimori D.G. Radical SAM-Mediated Methylation of Ribosomal RNA. Methods Enzymol. 2015;560:355–376. doi: 10.1016/bs.mie.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andersen T.E., Porse B.T., Kirpekar F. A novel partial modification at C2501 in Escherichia coli 23S ribosomal RNA. RNA. 2004;10:907–913. doi: 10.1261/rna.5259404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Khersonsky O., Tawfik D.S. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 2010;79:471–505. doi: 10.1146/annurev-biochem-030409-143718. [DOI] [PubMed] [Google Scholar]
- 38.Schwalm E.L., Grove T.L., Booker S.J., Boal A.K. Crystallographic capture of a radical S-adenosylmethionine enzyme in the act of modifying tRNA. Science. 2016;352:309–312. doi: 10.1126/science.aad5367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Machnicka M.A., Milanowska K., Osman Oglu O., Purta E., Kurkowska M., Olchowik A.J.W., Kalinowski S., Dunin-Horkawicz S., Rother K.M., Helm M., et al. MODOMICS: a database of RNA modification pathways: 2012 update. Nucleic Acids Res. 2013;41:D262–D267. doi: 10.1093/nar/gks1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hansen L.H., Vester B. A cfr-like gene from Clostridium difficile confers multiple antibiotic resistance by the same mechanism as the cfr gene. Antimicrob. Agents Chemother. 2015;59:5841–5843. doi: 10.1128/AAC.01274-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Connolly K., Rife J.P., Culver G. Mechanistic insight into the ribosome biogenesis functions of the ancient protein KsgA. Mol. Microbiol. 2008;70:1062–1075. doi: 10.1111/j.1365-2958.2008.06485.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Okamoto S., Tamaru A., Nakajima C., Nishimura K., Tanaka Y., Tokuyama S., Suzuki Y., Ochi K. Loss of a conserved 7-methylguanosine modification in 16S rRNA confers low-level streptomycin resistance in bacteria. Mol. Microbiol. 2007;63:1096–1106. doi: 10.1111/j.1365-2958.2006.05585.x. [DOI] [PubMed] [Google Scholar]
- 43.Lazaro E., Rodriguez-Fonseca C., Porse B., Urena D., Garrett R.A., Ballesta J.P. A sparsomycin-resistant mutant of Halobacterium salinarium lacks a modification at nucleotide U2603 in the peptidyl transferase centre of 23 S rRNA. J. Mol. Biol. 1996;261:231–238. doi: 10.1006/jmbi.1996.0455. [DOI] [PubMed] [Google Scholar]
- 44.Gustafsson C., Persson B.C. Identification of the rrmA gene encoding the 23S rRNA m1G745 methyltransferase in Escherichia coli and characterization of an m1G745-deficient mutant. J. Bacteriol. 1998;180:359–365. doi: 10.1128/jb.180.2.359-365.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ntokou E., Hansen L.H., Kongsted J., Vester B. Biochemical and computational analysis of the substrate specificities of Cfr and RlmN methyltransferases. PLoS One. 2015;10:e0145655. doi: 10.1371/journal.pone.0145655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gao W., Chua K., Davies J.K., Newton H.J., Seemann T., Harrison P.F., Holmes N.E., Rhee H.W., Hong J.I., Hartland E.L., et al. Two novel point mutations in clinical Staphylococcus aureus reduce linezolid susceptibility and switch on the stringent response to promote persistent infection. PLoS Pathog. 2010;6:e1000944. doi: 10.1371/journal.ppat.1000944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takaya A., Kimura A., Sato Y., Ishiwada N., Watanabe M., Matsui M., Shibayama K., Yamamoto T. Molecular characterization of linezolid-resistant CoNS isolates in Japan. J. Antimicrob. Chemother. 2015;70:658–663. doi: 10.1093/jac/dku443. [DOI] [PubMed] [Google Scholar]
- 48.Farrell D.J., Morrissey I., Bakker S., Buckridge S., Felmingham D. In vitro activities of telithromycin, linezolid, and quinupristin-dalfopristin against Streptococcus pneumoniae with macrolide resistance due to ribosomal mutations. Antimicrob. Agents Chemother. 2004;48:3169–3171. doi: 10.1128/AAC.48.8.3169-3171.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Endimiani A., Blackford M., Dasenbrook E.C., Reed M.D., Bajaksouszian S., Hujer A.M., Rudin S.D., Hujer K.M., Perreten V., Rice L.B., et al. Emergence of linezolid-resistant Staphylococcus aureus after prolonged treatment of cystic fibrosis patients in Cleveland, Ohio. Antimicrob. Agents Chemother. 2011;55:1684–1692. doi: 10.1128/AAC.01308-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Besier S., Ludwig A., Zander J., Brade V., Wichelhaus T.A. Linezolid resistance in Staphylococcus aureus: gene dosage effect, stability, fitness costs, and cross-resistances. Antimicrob. Agents Chemother. 2008;52:1570–1572. doi: 10.1128/AAC.01098-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gales A.C., Sader H.S., Andrade S.S., Lutz L., Machado A., Barth A.L. Emergence of linezolid-resistant Staphylococcus aureus during treatment of pulmonary infection in a patient with cystic fibrosis. Int. J. Antimicrob. Agents. 2006;27:300–302. doi: 10.1016/j.ijantimicag.2005.11.008. [DOI] [PubMed] [Google Scholar]
- 52.Gu B., Kelesidis T., Tsiodras S., Hindler J., Humphries R.M. The emerging problem of linezolid-resistant Staphylococcus. J. Antimicrob. Chemother. 2013;68:4–11. doi: 10.1093/jac/dks354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mendes R.E., Deshpande L.M., Farrell D.J., Spanu T., Fadda G., Jones R.N. Assessment of linezolid resistance mechanisms among Staphylococcus epidermidis causing bacteraemia in Rome, Italy. J. Antimicrob. Chemother. 2010;65:2329–2335. doi: 10.1093/jac/dkq331. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.