Abstract
PNLDC1 is a homologue of poly(A) specific ribonuclease (PARN), a known deadenylase with additional role in processing of non-coding RNAs. Both enzymes were reported recently to participate in piRNA biogenesis in silkworm and C. elegans, respectively. To get insights on the role of mammalian PNLDC1, we characterized the human and mouse enzymes. PNLDC1 shows limited conservation compared to PARN and represents an evolutionary related but distinct group of enzymes. It is expressed specifically in mouse embryonic stem cells, human and mouse testes and during early mouse embryo development, while it fades during differentiation. Its expression in differentiated cells, is suppressed through methylation of its promoter by the de novo methyltransferase DNMT3B. Both enzymes are localized mainly in the ER and exhibit in vitro specificity restricted solely to 3′ RNA or DNA polyadenylates. Knockdown of Pnldc1 in mESCs and subsequent NGS analysis showed that although the expression of the remaining deadenylases remains unaffected, it affects genes involved mainly in reprogramming, cell cycle and translational regulation. Mammalian PNLDC1 is a novel deadenylase expressed specifically in cell types which share regulatory mechanisms required for multipotency maintenance. Moreover, it could be involved both in posttranscriptional regulation through deadenylation and genome surveillance during early development.
INTRODUCTION
Deadenylases represent a diverse group of Mg2+-dependent 3′–5′ exonucleases, found in all eukaryotes (1,2). Their main role is the shortening of 3′ poly(A) tails, which is the first step during mRNA decay, thus regulating the translational rates (3,4). Their discovery was based on early observations showing that maternal mRNAs stability, controlled expression and clearance, was important during early development (5,6). Two deadenylase superfamilies (termed EEP and DEDD) exist, with several members each, and three major deadenylase activities, the CCR4–NOT (Carbon Catabolite Repressor 4 – Negative on TATA) complex, the poly (A) nuclease (PAN) complex and the poly(A) specific ribonuclease-PARN have been widely studied (6,7). In many cases, interactions of specific RNA-binding proteins with several cis-acting elements on mRNAs or miRNA-mediated targeting, trigger recruitment of deadenylases and ensure specificity (4,8–11). As a result, different combinations of interactions can either modulate deadenylase activity or can recruit deadenylase complexes for mRNA decay (4,11). The conservation of deadenylases varies among species and attempts to clarify the role of each member in several model organisms concluded that PAN and CNOT complexes are the major deadenylases responsible for mammalian cytoplasmic mRNA decay (12). On the other hand, PARN seems to have a more specific role in regulating discrete subsets of mRNAs and it was shown to have additional exonuclease activity beyond mRNA deadenylation (6). It is known that PARN catalyses the miRNA-mediated TP53 degradation and participates in the biogenesis of several non-coding RNAs, like snoRNAs, scaRNAs, miRNAs and the telomerase RNA moiety (TERC) (13–16). Most recently, PARN was reported as the enzyme which catalyses the 3′ pre-piRNA trimming in C. elegans (17). In the same report, the recombinant PARN-1 was characterized in vitro as a 3′-5′ exonuclease, as C. elegans has two genes for PARN (PARN-1 and -2) which, however, can both be severely knocked down without any obvious phenotype (18). The unexpected role of PARN beyond mRNA deadenylation, indicates that is evolved most likely to control specific mRNA functions required for specific events during early development, such as the meiotic process in oocyte maturation or plant embryogenesis (6).
Several homologous genes encoding putative deadenylases exist with elusive biological role (1,19). A new group of genes is represented by PNLDC1 [poly(A)-specific ribonuclease (PARN)-like domain containing 1 or PARN-like], a PARN homologue. PNLDC1 genes are detected in lower eukaryotes (i.e. Cnidaria) and co-exist with PARN in almost all amniotic vertebrates and in arthropods, including silkworm (B. mori). Interestingly, it is absent from model organisms like S. pombe (replaced possibly by Triman), D. melanogaster (replaced possibly by Nibbler) and C. elegans that expresses PARN-1 and PARN-2 (20,21). Interestingly, the activity responsible for 3′ pre-piRNA trimming in silkworm (B. mori), termed Trimmer, was attributed to PNLDC1 in a recent report (22). Trimmer depends on the presence of Papi for the piRNA maturation that takes place on the mitochondrial surface. In the same report, the authors conclude that PNLDC1 is Trimmer after knocking down each of the nucleases, but no further biochemical or imaging characterization was reported (22).
In the present study, we report the characterization of the human and mouse PNLDC1 in an effort to get insights on the properties and role of the enzyme in mammals. We show that PNLDC1 has 3′-5′ exonuclease in vitro activity and substrate specificity restricted only to DNA or RNA polyadenylates. Interestingly, attempts to detect PNLDC1 in various cells lines and tissues revealed specific expression in embryonic stem cells and testes. In addition, immunohistochemistry and immunofluorescence experiments and imaging analysis verified exclusive localization of PNLDC1 in the cytoplasm of stem and spermatogenic cells. The expression of PNLDC1 gradually diminishes during early mouse embryo development and is epigenetically suppressed during mESCs differentiation. Previous high-throughput analyses have suggested that PNLDC1 promoter region is target for methylation by the de novo methyltransferase DNMT3B (23,24). Treatment of HEK 293 cultures with 5-AZA-CdR, a specific methyltransferase inhibitor, restored PNLDC1 expression. Knockdown of Pnldc1 did not affect the expression of the major known deadenylases, including PARN, an observation that supports the previously reported functional individuality of deadenylases (12). Subsequent NGS and functional enrichment analysis indicated genes involved mainly in epigenetic reprogramming, chromatin assembly and regulation of cell cycle and translation. Based on the results presented and the notion that stem cells and germline must share common mechanisms to maintain multipotency, we propose that PNLDC1 could play role in both genome integrity maintenance and posttranscriptional regulation and surveillance, considering the recent discovery of polyadenylated piRNAs in mammalian early embryos (25–27).
MATERIALS AND METHODS
Cloning, expression and purification of recombinant proteins
Nucleotide sequences of both human isoforms and mouse PNLDC1 were obtained from NCBI nucleotide database (NM_001271862.1 NM_173516.2 and NM_001034866.1, respectively). All recombinant plasmids were constructed following standard cloning protocols. A PARN-His6 pET33a plasmid also used in previous studies was kindly provided by Dr A. Virtanen (Uppsala University) (28). Both full length human PNLDC1 isoforms and mouse Pnldc1 were cloned using as template cDNA from HEK 293 cells treated with 5-aza-2′-deoxycytidine (5-AZA-CdR, Sigma) and mESC cDNA, respectively. For the cloning procedure specific primers bearing NdeI and BamHI restriction sites were used (Supplementary Table S1). PCR products were cloned in all cases in pET28b plasmid for heterologous expression in E. coli BL21(DE3) strains (Novagen). For localization experiments cDNA of both full length isoforms were cloned in pcDNA™3.1 EGFP (Invitrogen) using specific primers (Supplementary Table S1). Polymerase chain reaction (PCR) products were cloned between NheI and KpnI restriction sites, for the human genes and between NheI and HindIII for the mouse gene. The strains with the recombinant plasmids were used to inoculate initially 5 ml of Luria–Bertani medium supplemented with 50 μg/ml kanamycin and grew overnight at 37°C. The cultures were diluted (1:100) in the same medium and grown until an OD600 of 0.4–0.6. The optimal expression of soluble His6-tagged HsPNLDC1 recombinant proteins was induced by the addition of 0.5 mM IPTG and growth at room temperature, overnight. Cells were harvested by centrifugation at 9000 xg, disrupted by sonication and the extract was further centrifuged at 100 000 xg. The soluble protein fraction was analysed on an SDS–PAGE. Both human and mouse His6-tagged PNLDC1 enzymes were purified with affinity chromatography using a 5 ml HisTrap HP column (GE Healthcare). To improve the purity of the recombinant proteins, 20 mM imidazole was added to the wash solution to eliminate non-specific binding of the bacterial proteins. Optimal soluble protein elution was obtained between 120–150 mM imidazole after applying a step gradient (50–250 mM imidazole). Further purification was achieved through size exclusion of contaminant proteins on a 16/40 Sephacryl S200 High Resolution gel filtration column attached to an ÄKTA P–500 FPLC. The elution was isocratic in the presence of 20 mM HEPES–KOH, pH 7.0, 100 mM NaCl, 1.5 mM MgCl2, 0.1 mM DTT, 0.1 mM EDTA, 10% (v/v) glycerol. The purity of the final products was estimated by SDS–PAGE. Immunoblot analysis was performed using current protocols and a commercially available antibody against PNLDC1 (Acris AP53365PU-N) following the manufacturer's instructions. Protein concentration was determined according to the Bradford method and activity of fractions was assessed as described (See Supplementary Materials and Methods).
PNLDC1 activity assay and kinetic characterization
The enzymatic activity of human and mouse PNLDC1 was measured using the following synthetic oligoRNA substrates: N9A15, A12, U12, C12, G12 and dA12 (VBC Biotech, Figure 1D and E). The optimal activity conditions were found to be pH 7.0, 100 mM NaCl, 1.5 mM MgCl2 and 37°C. All subsequent measurements were performed in the standard reaction buffer (20 mM HEPES–KOH, pH 7.0, 1.5 mM MgCl2, 100 mM NaCl, 0.1 mM EDTA, 0.1 mM DTT, 0.2 U RNasin and 10% v/v glycerol) and [γ–32P] ATP (3000 Ci/mmol; IZOTOP Hungary) labeled substrate supplemented with unlabeled substrate to a final concentrations varying from 0.01–1 μM (total reaction volume 20 μl). The recombinant PNLDC1 concentration in the reaction mixture was 5–50 nM and with the exception of time plots, the reactions were taking place for 45 min. Activity modulation experiments were performed in the presence of 0.1–10 μM m7G(5′)ppp(5′)G cap analogue (AM8048, Ambion). All reactions were stopped with 3 μl of 20 mM EDTA followed by phenol extraction and ethanol precipitation. Products were suspended in loading buffer (90% formamide, 50 mM EDTA pH 8.0, 0.05 bromophenol blue, 0.05 xylene cyanol) and separated on a 25% polyacrylamide (19:1 acrylamide/bisacrylamide) gel in the presence of 4 M urea. The gels were visualized on a Fujifilm FLA–3000 phosphoimager and analysed with the AIDA Image Analyzer software (Raytest). The activity of PNLDC1 was determined measuring the optical density as the percentage of the remaining radioactive band corresponding to each substrate before and after the addition of the enzyme. For the determination of kinetic values, measurements from at least three independent experiments were used (Supplementary Table S2).
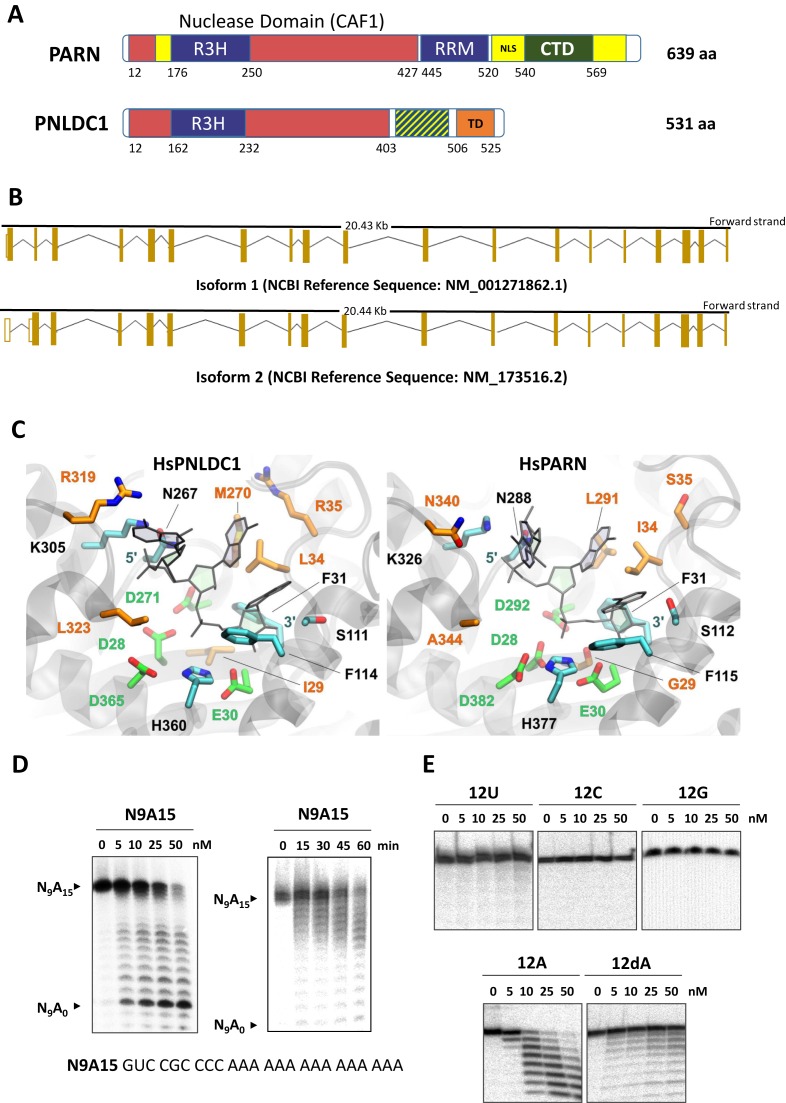
Figure 1.
(A) Comparison of PNLDC1 and PARN domain architecture. PARN contains a characteristic nuclear localisation signal (NLS, residues 520–540), a carboxy terminal domain (CTD, 540–639) which are not conserved in PNLDC1 and additional regions (represented in yellow) which are absent in PNLDC1. The RRM domain of PARN (445–520) is partially conserved in PNLDC1. Residues 506–525 in HsPNLDC1-1 represent a putative transmembrane domain (TD) (B) Human PNLDC1-1 and PNLDC1-2 exons arrangement. (C) Close-up view of HsPNLDC1 active site model in comparison with the poly(A)-bound X-ray structure of HsPARN nuclease domain (PDB ID: 2A1R). The catalytic DDED-motif residues are illustrated with green C atoms, the conserved and non-conserved RNA-interacting residues are shown with cyan and orange C atoms, respectively. All other atoms are blue for N, red for O and yellow for S. The bound trinucleotide is shown with gray sticks. (D) Autoradiography of in vitro PNLDC1 reaction products analysed as described in the materials and methods section. The reactions took place in the presence of either 5′ end [32P]-labeled N9A15 synthetic RNA substrate or in the presence of 5′ end [32P]-labeled poly(U), poly(C), poly(G), poly(A) and poly(dA) synthetic substrates.
Confocal microscopy analysis
To study the localization of both human PNLDC1 isoforms and the mouse homologue, we co-transfected HEK 293 cells with plasmids encoding C–terminal EGFP-tagged PNLDC1 together with the ER targeting mTurquoise2–ER vector (kindly provided by Dr I. Mylonis, University of Thessaly). HEK 293 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), at 37°C under 5% CO2. Cells were seeded at 3 × 105 cells·ml−1 in a 6–well or a 35 mm plate, for 24 h and transiently transfected with the recombinant plasmid DNA using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. For the co-localization experiments, the transfected cells were incubated at 37°C for 48 h, washed twice with phosphate buffered saline (PBS) (pH 7.5), fixed with 4% paraformaldehyde, stained with DRAQ5 (Abcam) and mounted with Mowiol 488 mounting media for microscopy (Sigma). For co–staining of mitochondria the same procedure was performed on cells treated with 20 nM MitoTracker Red CMXRos (Thermo Scientifc). Fluorescent image acquisition was conducted at the Functional Imaging Unit of the University of Patras. A Leica TCS SP5 microscope equipped with a 63 × 1.4 NA oil-immersion lens was used. For the co–localization study we used Coloc2 plugin of ImageJ.
Immunohistochemical analysis and immunofluorescence microscopy
Slides of paraffin-embedded tissue from human testes with intratubular germ cell neoplasia, a premalignant neoplastic condition of testes, were retrieved from the collection of the Department of Anatomy, Histology and Embryology of the University Hospital of Patras. Experiments using human tissue samples were designed and performed according to the institutional bioethical guidelines and have been approved by the bioethical committee of the University of Patras University Hospital, following the directions of the Declaration of Helsinki. Standard immunohistochemistry was performed following deparaffinization in xylene and rehydration in graded ethanol solutions. The slides were boiled in 10 mM sodium citrate pH 6.0 to retrieve the antigens, treated with 1% H2O2 for 15 min to quench the activity of endogenous peroxidase and incubated further for 1 h in protein blocking solution composed of 2% milk powder in PBS. Sections were incubated overnight with 1:50 dilution of anti-PNLDC1 (AP53365PU-N Acris, Germany). Specific binding was detected with the Envision detection kit (Dako, Germany) and the colour reaction visualized with diaminobenzidine. The slides were counter stained with haematoxylin, dehydrated in graded ethanol solutions and mounted. For immunefluorescence detection, a testes tissue sample from a 12-week-old wild-type mouse was fixed in 4% paraformaldehyde in PBS after overnight incubation and processed for paraffin embedding. After dewaxing, paraffin sections (5 μm thickness) were blocked with 5% normal horse serum in 0.1% BSA/PBS (blocking solution) for 60 min before overnight incubation at 4°C using Goat anti-Pnldc1 as primary antibody (SC-243835, Santa Cruz Biotechnology). The protein was visualized by using an anti-Goat secondary antibody conjugated to Alexafluor-594 (Invitrogen), diluted 1:500 in 0.1% BSA/PBS for 60 min at room temperature. Subsequently, sections were rinsed with the blocking solution and stained with DRAQ5 (5 μM) in PBS for 10 min to label nuclei. Fluorescent image acquisition was conducted as described above.
Induction of PNLDC1 and silencing of PARN and PNLDC1 expression
For induction of PNLDC1 expression, HEK 293 cells were treated with 10 μM 5-AZA-CdR (5′-aza-2′-deoxycytinine, Sigma). The treatment took place for 3 days to allow the inhibitor to be incorporated into DNA and the medium was changed every day to maintain the inhibitor's stability during treatment. Total RNA was extracted before and after the treatment to examine the expression levels of PNLDC1 using RT–qPCR (See Supplementary Materials and Methods). For PARN and Pnldc1 silencing, HEK 293 cells grown into 6-well plates were transfected with pLKO.1 (MISSION® shRNA, Sigma) plasmids expressing shRNAs targeting PARN (NM_002582, kindly provided by Dr N. A. Balatsos, University of Thessaly), or scrambled shRNAs (SHC016, Sigma) as negative control, using Lipofectamine 2000 (Invitrogen) according to manufacturer's instructions. Puromycin (3 μg/ml) was added for selection 12 h after transfection. Mouse E14 ESC cells were cultured in GMEM BHK-12 (Gibco) supplemented with 10% ES Cell Qualified FBS, 2 mM GlutaMAX, MEM nonessential amino acids, βME (Gibco), tylosin, 1% Pen/Strep (Gibco) and 100 U/ml Leukemia Inhibitory Factor (LIF; ESGRO Chemicon) on 0.2% gelatin-coated dishes (Gibco). Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Lipofectamine® RNAiMAX Transfection Reagent (Invitrogen) was used to transfect the cells with the siRNAs according to the manufacturer's instructions. Cells were collected 24, 48 and 72 h after transfection. Reduction of either gene or protein expression was confirmed by RT-qPCR or by immunofluorescence analysis, respectively.
Differentiation of mouse embryonic stem cells
Mouse E14 ESC cells were cultured as described above. Cells were maintained at 37°C in a humidified atmosphere of 5% CO2. Differentiation experiments were performed either by withdrawing LIF (for spontaneous differentiation) or by culturing the cells in N2B27 medium [DMEM: F12 1:1 25 μg/ml insulin, 100 μg/ml apotransferrin, 6 ng/ml progesterone, 16 μg/ml putrescine, 30 nM sodium selenite and 50 μg/ml BSA fraction V, combined 1:1 with Neurobasal medium supplemented with B27 (Gibco)] under the conditions described above, for neuronal differentiation. The expression levels of Pnldc1 were monitored by RT-qPCR.
Analysis of gene expression by RNA-Seq after PNLDC1 silencing in mESCs
Mouse E14 ESC cells transfected with esiRNAs targeting Pnldc1 (EMU079411, Sigma) or Luc (Luciferase; EHURLUC, Sigma) were used for total RNA extraction, as described above. The majority of contaminant rRNA was removed from 3 μg total RNA preparation using the RiboMinus kit (Life Technologies). Quality and quantity of the remaining RNA was confirmed using the Agilent 2100 bioanalyzer (Agilent). The rRNA-depleted samples were subsequently used for RNA-seq analysis on an Ion Torrent PGM platform (Department of Biochemistry and Molecular Biology, National and Kapodistrian University of Athens) with the use of total RNAseq kit v2 according to the manufacturer's instructions (Life Technologies). Templated ion sphere particles were generated using the Ion OneTouch 200 template kit v2. Sequencing was performed on an IonTorrent 318 chip using an Ion PGM 200 sequencing kit (Read length 100 bp). The data were exported in Fastq files, uploaded to the galaxy server and processed further (https://galaxyproject.org/). The output files containing the quality data of the sequences were processed by Fastq groomer (Sanger format) and by Fasta converter (Fasta format). The sequences were aligned on the whole mouse genome sequence [Mus musculus: mm10 GRCm38.p4)] using TopHat software (29). Changes in gene expression profiles were determined using Cuffdiff software (30). Analysis resulted in 244 significantly differential expressed genes (P-value < 0.005) which were subjected for further functional enrichment analysis using the PANTHER classification platform of the Gene Ontology Consortium tool (http://geneontology.org/) (31).
RESULTS
PNLDC1 is a phylogenetically distinct exonuclease with specificity to polyadenylates
The protein sequence alignment of PNLDC1 with PARN homologues from many representative organisms showed that share sequence similarities mainly in the nuclease (CAF1) domain that contains the characteristic DEDD motif of the active site as expected (Figure 1A and Supplementary Figure S1), but the overall identity is low. A survey for the the detection of either PNLDC1 or PARN showed that either one is present in very low metazoa (i.e. PARN in Placozoa and PNLDC1 in Cnidaria) and both of them are present in arthropods (including B. mori) and mainly in amniotic vertebrates. A phylogenetic tree produced based on an extensive sequence alignment and using the Trichoplax adhaerens PARN as root, shows that both enzymes have a common ancestor, but the bootstrap values indicate an early separation (Supplementary Figure S2 red dots). PNLDC1 enzymes from higher eukaryotes form an evolutionary distinct group from that of PARN (Supplementary Figure S2 red and blue box, respectively). A separate group is also formed by the PNLDC1 from arthropods (Supplementary Figure S2 green box). Interestingly, C. elegans PARN-1 and PARN-2 cluster with the clade that gave rise to PNLDC1 in higher eukaryotes, but form a separate group.
Further bioinformatics analysis predicted that HsPNLDC1 produces two isoforms by alternative splicing of the full length transcript that contains 19 exons (Figure 1B). The peptide residues 1–25 (MDVGADEFEESLPLLQELVQEADFV) of isoform 1 are replaced by the peptide residues 1–14 (MFCTRGLLFFAFLA) in isoform 2 (according to the NCBI annotation). The sequences of isoform 1 (termed PNLDC1-1, 531 aa, average MW 61.3 KDa) and isoform 2 (termed PNLDC1-2, 520 aa, average MW 60.1 KDa,) exhibit 21.4% and 20.6% identity with human PARN, respectively. Mouse PNLDC1 has one predicted isoform (531 aa, average MW 61.3 KDa) with 22.2% sequence identity to mouse PARN (Supplementary Figure S1). As inferred from the X-ray crystal structures of HsPARN and MmPARN (6), the corresponding nuclease domain of HsPNLDC1 residues 1–142 and 243–414 displays sequence identity of 31% (98/314 residues) and only two 4-residue gaps (<4% total gaps) with respect to HsPARN (Supplementary Figure S3). Therefore, a homology model of HsPNLDC1 nuclease domain homodimer was generated, refined and examined with molecular dynamics simulations (see Supplementary Data and Figures S4–S7 for details). Comparative interface analysis of the crystallographic HsPARN dimer and the model of HsPNLDC1 nuclease domain exhibited similar characteristic properties (Supplementary Table S3), suggesting that HsPNLDC1 can probably form homodimers. Examination of the RNA-binding sites of HsPNLDC1 and HsPARN revealed similarities in the spatial arrangement of the catalytic DEDDh-motif and other RNA-interacting residues (Figure 1C). However, a number of non-conserved residues that potentially interact with the substrate might be responsible for a different selectivity profile of HsPNLDC1 with respect to HsPARN (Figure 1C and Supplementary Figure S8). It is also worth mentioning that the N-terminal residues 1–18 of HsPNLDC1 (isoform 2) form a putative signal peptide, indicating possible localization of HsPNLDC1 to the ER.
Recombinant human and mouse enzymes were isolated after heterologous expression and optimization of the expression conditions, from E. coli soluble fractions (Supplementary Figure S9A). Initial purification schemes included affinity and gel filtration chromatography. The elution profiles after gel filtration chromatography were consistent with a dimeric form of PNLDC1 (Supplementary Figure S9B). Unfortunately, from the 3 commercially available antibodies that were tested using immunoblotting, only one could detect the overexpressed His6-tagged recombinant enzymes but not the native forms of the enzyme (Supplementary Figure S9A) and a different one was suitable for the detection of both native human and mouse enzymes in immunohistochemical and immunofluorescence experiments (Figure 3D and E). Dynamic light scattering analysis yielded a HsPNLDC1 hydrodynamic diameter of about 10.1 nm and a calculated molecular weight of about 150 kDa (Supplementary Figure S9E), verifying that the dimeric form of the protein is the prevalent species in solution (monomer MW 62 kDa). Preliminary assays and time plots in the presence of either human or mouse PNLDC1 showed similar activity profiles, using as substrate a synthetic 24-mer RNA molecule bearing 3 codons and a 15-mer poly(A) tail. The optimal reaction conditions were pH 7.0, 100 mM NaCl, 1.5 mM MgCl2 and 37°C and, as expected, PNLDC1 activity was observed only in the presence of Mg2+ ions (Supplementary Figure S9C). HsPNLDC1-1 had better purification yields after two purification steps and was used for further characterization together with the mouse enzyme. Both enzymes exhibited specific deadenylation activity in vitro, with very high specificity for recognition of poly(A) or poly(dA) (Figure 1E). Interestingly, PNLDC1 enzymes cannot hydrolyse substrates that are not polyadenylated, a striking difference compared to PARN enzymes, which can additionally degrade quite efficiently poly(U) and to some extent other polymer substrates (32). To test whether PNLDC1 could use RNA substrates with more complex secondary structures, we used as substrate a pre-tRNASer (Supplementary Figure S9D). Under optimum conditions tested, PNLDC1 didn't show any activity on tRNA neither could trim its 3′ trailer sequence (Supplementary Figure S9D). Therefore, we concluded that the in vitro PNLDC1 substrate recognition and activity is restricted solely to single-stranded RNA or DNA polyadenylates. Next, we determined the kinetic values for both enzymes that were found similar, both in affinity and turnover rates (Supplementary Table S2). Mammalian PNLDC1 degrades poly(A) in a processive mode, as can be judged by the time course progress of the reaction (Figure 1D). Finally, when we used a 5′cap analogue in a broad range of concentrations we couldn't observe any modulation of PNLDC1 activity. The 5′ cap structure has been reported to stimulate PARN activity even when supplied in trans as an allosteric regulator in low concentrations and as an inhibitor in higher concentrations (33). Both human and mouse PNLDC1 lack at least one of the conserved residues of PARN (W468 and W475 in mouse and human PARN, respectively) responsible for stabilization of m7G interaction with the RRM domain, which is only partially represented in PNLDC1 sequence. This observation supports further the absence of cap-binding ability of mammalian PNLDC1 (Supplementary Figures S1 and S3).
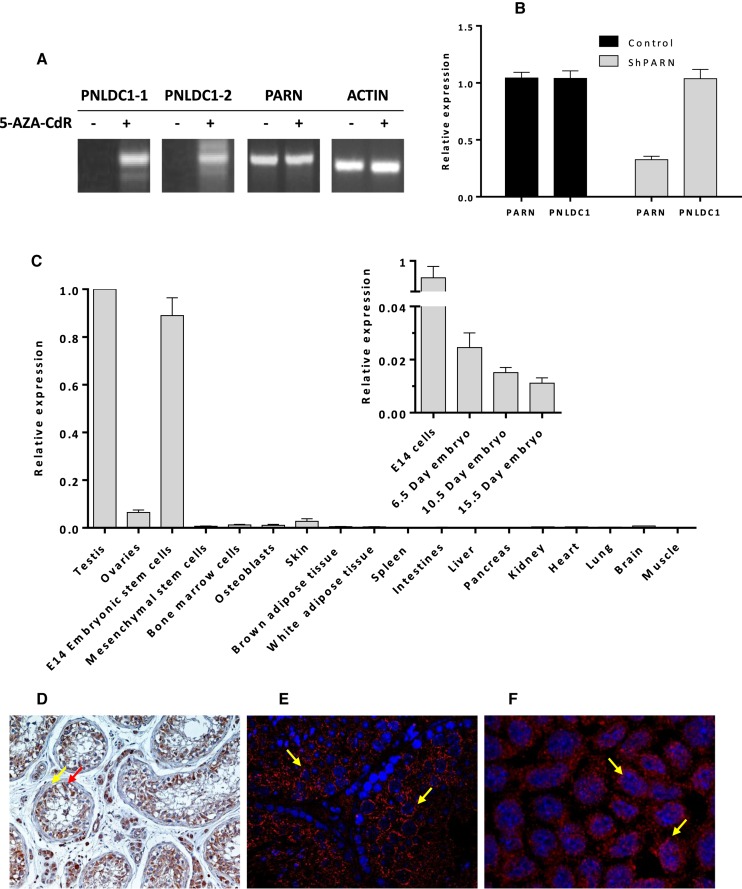
Figure 3.
(A) HEK 293 cells were treated with 100 μM of 5-AZA-CdR for 72 h to achieve maximum inhibitory effect of the promoter methylation. Total RNA was extracted and RT-qPCR was performed to detect expression of PNLDC1 and PARN. The expression of ACTIN was used as a positive control. The products were analysed on a 1.5% agarose gel. Both PNLDC1 isoforms were detected only after demethylation. (B) RT-qPCR results after PARN silencing in HEK 293 cells expressing PNLDC1 after treatment with 5-AZA-CdR for 72 h. PARN knockdown does not affect the expression levels of PNLDC1. (C) Relative expression levels of Pnldc1 among different mouse tissues and during embryogenesis (inset) compared to undifferentiated stem cells. (D) Immunohistochemistry image showing PNLDC1 localization in human testes samples with intratubular germ cell neoplasia. PNLDC1 is detected in the cytoplasm of almost all the neoplastic spermatogenic cells (red arrow). PNLDC1 does not show any immunostaining in the fibromyocytes (yellow arrow) that surrounds the spermatic testicular tubules (original magnification 20X). (E) Immunofluorescence of mouse testes sample. The expression of Pnldc1 is evident in the cells of all the spermatogenic series with higher expression in meiotic spermatocytes (yellow arrows). (F) Immunofluorescence analysis of mouse stem cells. Pnldc1 is shown in red and is localized in the cytoplasm (yellow arrows).
PNLDC1 is localized predominantly in the ER
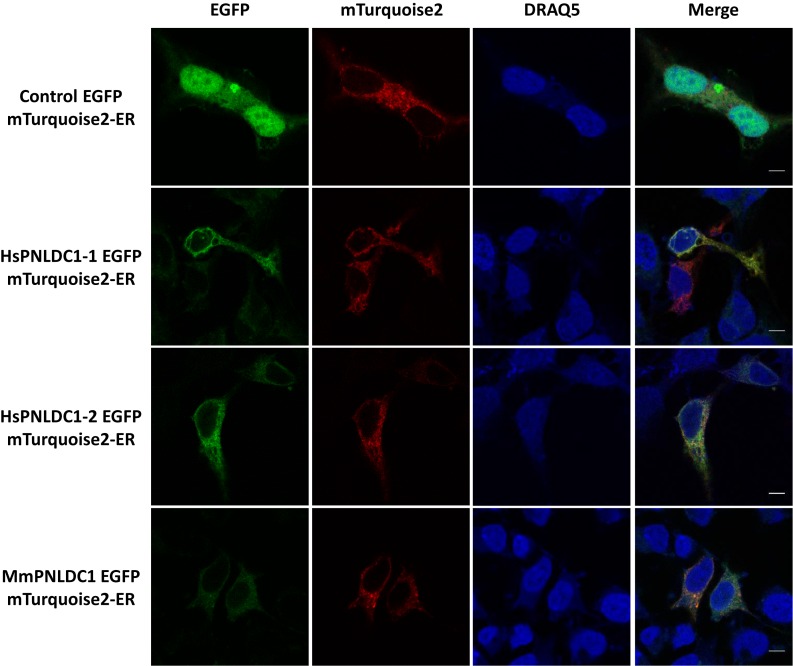
In a next step, we transfected HEK 293 cells with either human (both isoforms) or mouse PNLDC1-EGFP recombinant proteins to monitor their subcellular localization. The EGFP tagged the respective C-termini to avoid background signals from random sorting. Both human isoforms, as well as the mouse enzyme are localized exclusively outside the nucleus (Figure 2). Initial observations and subsequent quantification and calculation of the Mander's co-localization coefficient value indicated significant ER distribution and subsequent co-localization experiments were performed with the mTurquoise2 protein tagged with an ER sorting peptide signal. The confocal microscopy analysis in transfected HEK 293 cells showed that PNLDC1 is localized mainly in the ER (∼80%), with a certain degree of diffusion in the cytoplasm (∼20%). In addition, PNLDC1 does not shown any specific co-localization on the mitochondrial surface when transfected in HEK 293 cells (Supplementary Figure S10). However, the possibility that a portion of the protein could also be associated to the mitochondrial surface through interactions with partner proteins (as has been suggested for BmPNLDC1) cannot be excluded. The co-localization results clearly show that PNLDC1 is a deadenylase that is excluded from the nucleus and most likely, its function occurs mainly in the ER. This result distinguishes PNLDC1 from PARN that has been reported to shuffle between the nucleus and the cytoplasm, using similar methodology (12,34). During the analysis, both human recombinant isoforms were localized within the same context and intensity, an observation that requires further investigation in order to clarify possible intracellular specific roles. As for the endogenous enzymes, they most likely exhibit similar localization pattern, since PNLDC1 is absent from many cell lines and the antibody specificity that was used for immunohistochemical and fluorescence analysis cannot discriminate between the two isoforms. Therefore, which human isoform is dominating (either in a cell-specific or a tissue-specific manner), remains an open question.
Figure 2.
Confocal fluorescence microscopy images showing the subcellular localization of EGFP (control), HsPNLDC1-1-EGFP, HsPNLDC1-2-EGFP and MmPNLDC1-EGFP in comparison with the localization of the ER targeted protein mTurquoise-ER in transfected HEK 293 cells. The nuclei were stained with DRAQ5 (blue colour). In order to assist visual interpretation mTurquoise-ER was artificially stained with red colour. The merge channels (with 5 μm scale bar) reveal co-localization of the ER targeted protein and PNLDC1.
Differentiated cells express PNLDC1 only after demethylation
Efforts to detect the expression of PNLDC1 both at the protein or at the mRNA level in several cell lines (HEK 293, Jurkat, HeLa, A549, H23, Met5a) were unsuccessful. Previous studies regarding (i) high throughput screening of promoter methylation of genes in and around the lung cancer susceptibility locus 6q23-25 in human, and (ii) analysis of the upregulated genes after the loss of the essential de novo methyltransferase Dnmt3b in mice, suggested a possible correlation between promoter methylation by DNMT3B and suppression of PNLDC1 expression (23,24). This result prompted us to look for possible epigenetic control of PNLDC1 expression that could explain its apparent absence in differentiated cells. A closer examination of the promoter region of PNLDC1 revealed a 34 bp CpG island that is potential target of DNMT3B and thus, demethylation could induce the expression of PNLDC1. Therefore, we treated HEK 293 cells with 5-AZA-CdR, a compound that inhibits DNA methylation and has been studied as potent inhibitor of DNMT3B (35,36). Upon demethylation, PNLDC1 expression was induced and this observation clearly suggests an epigenetic regulation mechanism that has not been reported before for a deadenylase (Figure 3A). Our observation was verified with the use of isoform-specific primers to distinguish the expression of both isoforms after demethylation. Interestingly, during the induction of PNLDC1 expression, the expression of PARN remained unaffected, an observation that indicates lack of expression synchronization or feedback correlation between the two deadenylases. To verify further our observations, we knocked down PARN using specific shRNAs in HEK 293 cells treated with 5-AZA-CdR but again, we didn't notice any change in PNLDC1 expression levels (Figure 3B).
PNLDC1 is present in stem and germ cells and fades during embryogenesis and early differentiation
The observation that PNLDC1 expression is epigenetically supressed in differentiated cells led us to look for possible tissue-specific expression. After screening of several mouse tissues using RT-qPCR, Pnldc1 was found highly expressed in mouse embryonic stem cells (E14) and testes, whereas expression in skin, late embryos and ovaries was almost undetectable (albeit slightly higher in ovaries than other organs) (Figure 3C). This result indicates that Pnldc1 is present in early mouse development and in a next step, we examined for a detectable similar expression pattern during early embryo development. After we examined mouse embryos at different developmental time points (6.5, 10.5 and 15.5 days), we observed that Pnldc1 although present very early shows a gradual and significant reduction of relative expression, compared to the expression in mESCs (Figure 3C inset). This behaviour could be more likely attributed to epigenetic reprogramming that takes place during early development in association with various RNA binding proteins and might be related to the regulation of transition from pluripotency to multipotency (37). In addition, the presence of PNLDC1 at the protein level was verified by immunofluorescence in mESCs and mouse testes and by immunohistochemistry analysis in human testes. Histological analysis of sections obtained from human testes with intratubular germ cell neoplasia unveiled that HsPNLDC1 was strongly expressed in the cytoplasm of the spermatogenic tumor cells (Figure 3D). Intratubular germ cells neoplasia is characterized by the presence of neoplastic cells which show some degree of immaturity that is characteristic of undifferentiated cell state. This type of neoplasia is a pre-malignant condition that under specific circumstances can progress to fully developed cancer. Therefore, HsPNLDC1 upregulation is expected due to undifferentiated-pluripotent state of the neoplastic cells in the examined tissue sections. Notably, mature fibromyocytes that surround the seminiferous tubules were devoid of PNLDC1 immunoreactivity. Immunofluorescence analysis in mouse testes revealed that PNLDC1 is highly expressed in meiotic spermatocytes showing a perinuclear granular pattern (Figure 3E). The presence of PNLDC1 in mESCs was also verified by immunofluorescence and found again in the cytoplasm (Figure 3F).
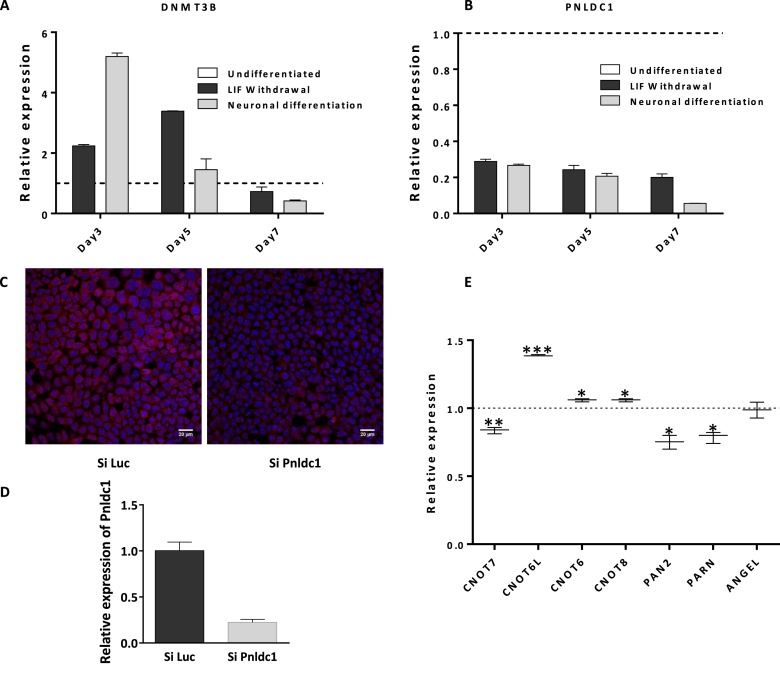
To obtain further information, we examined the Pnldc1 expression pattern during mESCs differentiation after, either withdrawal of LIF or under conditions which induce differentiation to neuronal cells. Initiation and progression of mESCs differentiation in both conditions was examined by monitoring with RT-qPCR the levels of Klf4, Nanog, Sox2, Oct4, Myc and Lin28 as pluripotency markers (found down-regulated) and Bmp2 and Shh as differentiation markers (found up-regulated, Supplementary Figure S11). The expression of Pnldc1 was monitored under the same conditions, in correlation with the expression levels of Dnmt3b in order to clarify the correlation between both genes. At day 3 Pnldc1 expression was reduced, while Dnmt3b was upregulated until day 5 (Figure 4A and B). The expression patterns observed for both genes during mESCs differentiation, is in agreement with previous studies and our previous findings which suggest that methylation of Pnldc1 promoter by Dnmt3b is possibly the mechanism that regulates transcriptional suppression of the corresponding gene.
Figure 4.
(A) Expression levels of Pnldc1 and (B) Dnmt3b, during mESCs differentiation. The expression of Pnldc1 diminishes during the first three days of differentiation. A simultaneous increase of Dnmt3b level is observed during the same period, followed by an equilibrated expression profile until day 7. (C) Effective knockdown of Pnldc1 in mESCs using esiRNA was confirmed by immunofluorescence and (D) RT-qPCR (E) Expression pattern of Cnot7, Cnot6l, Cnot6, Cnot8, Pan2, Parn and Angel analysed by RT-qPCR after Pnldc1 knockdown. (*P value < 0.05, **P value < 0.01; ***P value < 0.001).
The specific expression of Pnldc1 and its possible significance for early development, was examined using siRNAs to knockdown its expression in mESCs. The silencing in mESCs was very effective as can be judged by immunofluorescence and RT-qPCR analysis (almost 77%, Figure 4C and D). After Pnldc1 silencing, we monitored the expression of all the major dedadenylases via RT-qPCR. In addition, cDNA libraries prepared from total RNA from the same experiment (before and after knockdown) were used for NGS analysis. We observed that the expression levels of the majority of deadenylases remained essentially unaffected. The only noticeable change concerned the expression of Cnot6l which exhibited moderate upregulation (∼1.5-fold) (Figure 4E). As was initially observed for PARN, the expression profile of all major deadenylases is independent from the expression of Pnldc1, an observation that supports our results for specific spatial and temporal expression of PNLDC1. Finally, after the NGS analysis the functional enrichment of genes with statistical significant change of their expression showed that Pnldc1 silencing affects mainly the expression of genes that participate in the epigenetic reprogramming, chromatin assembly and organization and regulation of cell cycle and translation (Figure 5, Supplementary Figure S12). In addition, to the genes that encode important proteins including several histones, Trim proteins, ribosomal proteins, translation initiation factors and Poly(A) binding protein (PABP), few important non-coding RNAs were also found upregulated, such as the RNA subunits of telomerase (TERC), RNAse P and RNAse MRP and the long non-coding RNA DANCR (Supplementary Table S7). Taken together, our results point towards a central role of PNLDC1 during early development which is discussed below.
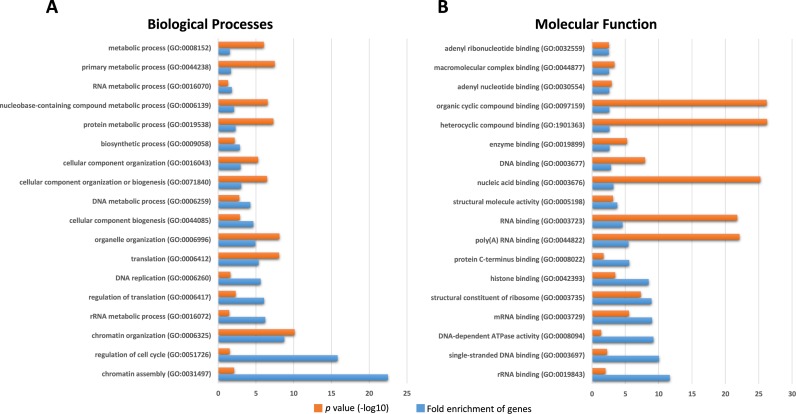
Figure 5.
Classes of biological processes and molecular functions related to the genes with significant differential expression in mESCs after Pnldc1 knockdown. Gene ontology enrichment analysis was performed using the PANTHER classification system as described and P-values were subjected to Bonferroni correction for multiple testing.
DISCUSSION
Although the role of major deadenylases in mRNA decay is established, their expanded repertoire in the biogenesis of non-coding RNAs indicates equally essential roles for many cellular processes. There is plenty of evidence suggesting that among deadenylases, PARN is mainly responsible for the decay of specific mRNA subsets as well as the biogenesis of several non-coding RNAs (6). This is elaborated by the existence of several regulatory elements and protein interactions that regulate PARN's activity and fine-tune its specificity (38). It is also supportive of the general notion that many more deadenylases must exist which, either as individual enzymes or as part of larger complexes may participate simultaneously in different networks or cellular events, exhibiting temporal and spatial expression across tissues and cell types. Hence, most likely depending on the organism, deadenylases could acquire roles in a genome-dependent, tissue-dependent or cell-type dependent context (4).
PNLDC1 represents a new entry in the DEDD exonuclease superfamily. Given the simultaneous presence of PNLDC1 together with PARN in all higher eukaryotes, we characterized the human and mouse enzymes in an effort to get insights on its possible role in mammals. PNLDC1 is evolutionarily distant from PARN and a PARN gene duplication event has been proposed to explain its existence (19). Mammalian PNLDC1 is a Mg2+-dependent exonuclease that degrades substrates in a processive mode. The reaction kinetic parameters between the human and mouse enzymes are comparable and similar to other deadenylases. Although PNLDC1 shows low similarity of the corresponding characteristic PARN-type RRM motif, and despite the absence of many residues that contact the 5′ cap of mRNAs in PARN, it exhibits narrow specificity which is restricted only to RNA or DNA polyadenylates, under the in vitro conditions tested (Supplementary Figure S1, Figure 1D and E). The molecular dynamics simulation of the putative active site of PNLDC1 also supports our observations at the structural level. Whether this is the case also in vivo, remains a question for further studies. Therefore, it seems that at least in vitro, PNLDC1 is a more poly(A) specific deadenylase than PARN.
Another interesting feature of PNLDC1 comes from the observation that it is exclusively localized in the cytoplasm. PARN on the other hand, shuffles between nucleus (found in the cytoplasm, the nucleolus and Cajal bodies) and cytoplasm (as part of RNA granule containing exosome). Recently, both PARN and PNLDC1 were reported to provide the elusive 3′ pre-piRNA trimming activities in C. elegans and B. mori, respectively (17,22). Based on previous annotated enrichment of PNLDC1 in mouse testes in relevant databases, both reports implied that MmPNLDC1 could be the elusive trimmer in the mammalian piRNA pathway (39,40). Our data confirm the expression of Pnldc1 in testes and more specifically in meiotic spermatocytes where the pachytene piRNAs are in their peak during spermatogenesis. It has been reported that BmPNLDC1 in association with PAPI is anchored on the mitochondrial surface, where piRNAs trimming takes place. In the case of the mammalian PNLDC1 a putative transmembrane domain (residues 506–525 in HsPNLDC1) could facilitate such an anchoring with the help of partner proteins. In the same report, co-expression of MmPNLDC1 with the MmTdrkh (a Papi homologue) can reconstitute pre-piRNA trimming activity in 293T cell extracts (22). However, previous studies reporting immunoprecipitation of TDRKH, MIWI and MILI, from mouse testes and subsequent proteomics analysis failed to identify PNLDC1 as a binding partner (41,42). Therefore, based on previous reports and our results, a possible role of PNLDC1 in mammalian piRNA biogenesis, although strongly suggested, awaits further verification. Finally, the localization of the protein may also be facilitated either by other interactions or by substrate abundance as has been described for PARN that can accumulate in cellular compartments with high substrate concentration by self-association (43).
The most interesting finding of this study is that Pnldc1 is expressed only early in development and upon differentiation, the expression gradually diminishes as a result of epigenetic events. In HEK 293 cells, PNLDC1 was detected only upon treatment with 5-AZA-CdR (5-Aza-2′-deoxycytidine, decitabine) and correlated with the expression of DNMT3B methyltransferase which is essential for de novo methylation during mammalian development and responsible for gene silencing in human cancer cells (44,45). Our results verified previous and recent reports demonstrating that the promoter of Pnldc1 was among the targets of DNMT3B (23,24,46). This is the first report of a deadenylase expression regulated through epigenetic silencing. It is important to note that additional experimentation following mESCs differentiation, under two different conditions (LIF withdrawal, or neuronal differentiation), showed the same correlation. The gradual silencing of Pnldc1 expression coincides with an increase in Dnmt3b expression and underlines the possible involvement of this synchronization in the transition from pluripotency to multipotency.
The fact that PNLDC1 was found specifically expressed in mESCs and human and mouse testes, was also an interesting observation. As described previously, deadenylases regulate cell's fate in early development where the dynamics of maintenance and clearance of maternal mRNAs, can either maintain multipotency and self-renewal or induce differentiation. Something similar has been shown for the CCR4-NOT complex that is involved in maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. In addition, it exhibits a distinct expression pattern during neural development and contributes to generation of induced pluripotent stem cells in mice (47–49). Moreover, during late spermatogenesis, pachytene piRNAs derived from non-transposon intergenic regions in pachytene spermatocytes (where PNLDC1 is highly expressed) mediate a synchronized massive degradation of mRNAs through a piRNA-induced silencing complex (pi-RISC) and recruitment of CAF1 deadenylase, in a process similar to that in Drosophila (50). For this purpose, pachytene piRNA-guided MIWI-CAF1 complexes (and possibly additional protein partners) are selectively assembled in elongating spermatids to mediate mRNA deadenylation and decay via a mechanism that resembles the action of miRNAs in somatic cells (50). The recent identification of 3′ polyadenylated piRNAs in mammalian early embryos (where PNLDC1 is also present) and the very rapid developmental adaptation due to changes in poly(A)-tail length could also involve PNLDC1 in mechanisms similar to that of pachytene piRNA induced silencing (26,51).
The effective knockdown of Pnldc1 in mESCs did not affect the expression of the remaining deadenylases, with the exception of a moderate upregulation observed for Cnot6l. The fact that Pnldc1 is specifically expressed in pluripotent cell types (or cells that retain their pluripotency to some point) points towards a specific role in common mechanisms shared by both stem and germ cells. In the same line, our immunohistochemical analysis revealed that PNLDC1 is detected in the cytoplasm of human neoplastic spermatogenic cells (deriving from surgically removed testes of patients with intraepithelial germ cell neoplasia) which are less differentiated. The possible involvement of PNLDC1 in early development through regulation of transcripts that are important for re-programming and translational regulation is also evident from the functional enrichment analysis that followed NGS experiments. The genes that are affected participate predominantly in chromatin assembly and regulation of translation and cell cycle. In accordance to the previous, the enriched molecular functions included genes with role in DNA-dependent ATPase activity and histone, rRNA, single strand DNA, mRNA and poly(A) binding. It is also noteworthy that some snoRNAs known to guide 2′-O-methyl modifications in rRNA, the RNA component of telomerase, the RNA subunit of RNase P and MRP and the long non-coding DANCR RNA are also found significantly upregulated. Although these observations require further investigation, they consist a strong indication of the central role of PNLDC1, beyond mRNA decay at least in some (if not all) of the above pathways. It is interesting to note that previous reports showed that DANCR RNA increases stemness in association with CTNNB1 (β-catenin) which in turn, through the β-catenin-dependent WNT signalling pathway regulates several diverse cell behaviours, including cell fate, proliferation and differentiation (52,53). Finally, the modulation of PNLDC1 activity by RNA structural elements and/or binding proteins, as has been suggested for PARN, cannot be excluded. Overall, it seems that PNLDC1 has distinct and independent intracellular activity and moreover, it affects the expression of pathways that are mainly involved in multipotency maintenance, genome surveillance and re-programming in early development.
ACCESSION NUMBERS
Raw sequencing and summarised data are available in the GEO repository with accession number GSE83405.
Supplementary Material
Acknowledgments
The authors would like to thank the Advanced Light Microscopy facility of the Medical School, University of Patras (Prof. Z. Lygerou and N. Giakoumakis), Prof. A. Scorilas and Dr C. Kontos for the use of the NGS facility (Department of Biochemistry and Molecular Biology, National and Kapodistrian University of Athens), Dr P. Giastas (Pasteur Institute, Athens) for help with the DLS analysis, Dr G. Amoutzias for advices on bioinformatics, Dr M. Apostolidi (University of Patras) for protocols, Prof G. Spyroulias (Department of Pharmacy, University of Patras) for discussions and advises on molecular dynamics, Prof D. Drainas for providing materials and encouragement and Dr K. Nika for critical reading of the manuscript. A-N.S is a recipient of ‘Bodossakis Foundation’ postgraduate fellowship, which is gratefully acknowledged.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Implemented under the ‘ARISTEIA I’ (EXCELLENCE I) Action of the ‘OPERATIONAL PROGRAMME EDUCATION AND LIFELONG LEARNING’ and is co-funded by the European Social Fund (ESF) and National Resources [MIS 1225, No D608 to C.S., in part].
Conflict of interest statement. None declared.
REFERENCES
- 1.Goldstrohm A.C., Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat. Rev. Mol. Cell Biol. 2008;9:337–344. doi: 10.1038/nrm2370. [DOI] [PubMed] [Google Scholar]
- 2.Parker R., Song H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 2004;11:121–127. doi: 10.1038/nsmb724. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell P., Tollervey D. mRNA turnover. Curr. Opin. Chem. Biol. 2001;13:320–325. doi: 10.1016/s0955-0674(00)00214-3. [DOI] [PubMed] [Google Scholar]
- 4.Yan Y.B. Deadenylation: enzymes, regulation, and functional implications. Wiley Interdiscip. Rev. RNA. 2014;5:421–443. doi: 10.1002/wrna.1221. [DOI] [PubMed] [Google Scholar]
- 5.Harnisch C., Moritz B., Rammelt C., Temme C., Wahle E. Activity and Function of Deadenylases. Enzymes. 2012;31:181–211. doi: 10.1016/B978-0-12-404740-2.00009-4. [DOI] [PubMed] [Google Scholar]
- 6.Virtanen A., Henriksson N., Nilsson P., Nissbeck M. Poly(A)-specific ribonuclease (PARN): An allosterically regulated, processive and mRNA cap-interacting deadenylase. Crit. Rev. Biochem. Mol. Biol. 2013;48:192–209. doi: 10.3109/10409238.2013.771132. [DOI] [PubMed] [Google Scholar]
- 7.Zuo Y., Deutscher M.P. Exoribonuclease superfamilies: Structural analysis and phylogenetic distribution. Nucleic Acids Res. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weill L., Belloc E., Bava F.-A., Mendez R. Translational control by changes in poly(A) tail length: recycling mRNAs. Nat. Struct. Mol. Biol. 2012;19:577–585. doi: 10.1038/nsmb.2311. [DOI] [PubMed] [Google Scholar]
- 9.Doma M.K., Parker R. RNA quality control in eukaryotes. Cell. 2007;131:660–668. doi: 10.1016/j.cell.2007.10.041. [DOI] [PubMed] [Google Scholar]
- 10.Houseley J., Tollervey D. The many pathways of RNA degradation. Cell. 2009;136:763–776. doi: 10.1016/j.cell.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 11.Jonas S., Izaurralde E. Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 2015;16:421–433. doi: 10.1038/nrg3965. [DOI] [PubMed] [Google Scholar]
- 12.Yamashita A., Chang T.C., Yamashita Y., Zhu W., Zhong Z., Chen C.Y., Shyu A.B. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struct. Mol. Biol. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 13.Zhang X., Devany E., Murphy M.R., Glazman G., Persaud M., Kleiman F.E. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic Acids Res. 2015;43:10925–10938. doi: 10.1093/nar/gkv959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berndt H., Harnisch C., Rammelt C., Stohr N., Zirkel A., Dohm J.C., Himmelbauer H., Tavanez J.P., Huttelmaier S., Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. RNA. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. PARN mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nguyen D., Grenier St-Sauveur V., Bergeron D., Dupuis-Sandoval F., Scott M.S., Bachand F. A polyadenylation-dependent 3′ end maturation pathway is required for the synthesis of the human telomerase RNA. Cell Rep. 2015;13:2244–2257. doi: 10.1016/j.celrep.2015.11.003. [DOI] [PubMed] [Google Scholar]
- 17.Tang W., Tu S., Lee H.C., Weng Z., Mello C.C. The RNase PARN-1 trims piRNA 3′ ends to promote transcriptome surveillance in C. elegans. Cell. 2016;164:974–984. doi: 10.1016/j.cell.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Reverdatto S.V., Dutko J.A., Chekanova J.A., Hamilton D.A., Belostotsky D.A. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–1214. doi: 10.1261/rna.7540204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlopoulou A., Vlachakis D., Balatsos N.A.A., Kossida S. A comprehensive phylogenetic analysis of deadenylases. Evol. Bioinform. Online. 2013;9:491–497. doi: 10.4137/EBO.S12746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marasovic M., Zocco M., Halic M. Argonaute and Triman generate dicer-independent priRNAs and mature siRNAs to initiate heterochromatin formation. Mol. Cell. 2013;52:173–183. doi: 10.1016/j.molcel.2013.08.046. [DOI] [PubMed] [Google Scholar]
- 21.Feltzin V.L., Khaladkar M., Abe M., Parisi M., Hendriks G.J., Kim J., Bonini N.M. The exonuclease Nibbler regulates age-associated traits and modulates piRNA length in Drosophila. Aging cell. 2015;14:443–452. doi: 10.1111/acel.12323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Izumi N., Shoji K., Sakaguchi Y., Honda S., Kirino Y., Suzuki T., Katsuma S., Tomari Y. Identification and Functional Analysis of the Pre-piRNA 3′ Trimmer in Silkworms. Cell. 2016;164:962–973. doi: 10.1016/j.cell.2016.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tessema M., Willink R., Do K., Yu Y.Y., Yu W., Machida E.O., Brock M., Van Neste L., Stidley C.A., Baylin S.B., et al. Promoter methylation of genes in and around the candidate lung cancer susceptibility locus 6q23-25. Cancer Res. 2008;68:1707–1714. doi: 10.1158/0008-5472.CAN-07-6325. [DOI] [PubMed] [Google Scholar]
- 24.Hlady R.A., Novakova S., Opavska J., Klinkebiel D., Peters S.L., Bies J., Hannah J., Iqbal J., Anderson K.M., Siebler H.M., et al. Loss of Dnmt3b function upregulates the tumor modifier Ment and accelerates mouse lymphomagenesis. J. Clin. Invest. 2012;122:163–177. doi: 10.1172/JCI57292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Juliano C., Wang J., Lin H. Uniting germline and stem cells: the function of Piwi proteins and the piRNA pathway in diverse organisms. Annu. Rev. Genet. 2011;45:447–469. doi: 10.1146/annurev-genet-110410-132541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Neff A.T., Lee J.Y., Wilusz J., Tian B., Wilusz C.J. Global analysis reveals multiple pathways for unique regulation of mRNA decay in induced pluripotent stem cells. Genome Res. 2012;22:1457–1467. doi: 10.1101/gr.134312.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Roovers E.F., Rosenkranz D., Mahdipour M., Han C.T., He N., Chuva de Sousa Lopes S.M., van der Westerlaken L.A., Zischler H., Butter F., Roelen B.A., et al. Piwi proteins and piRNAs in mammalian oocytes and early embryos. Cell Rep. 2015;10:2069–2082. doi: 10.1016/j.celrep.2015.02.062. [DOI] [PubMed] [Google Scholar]
- 28.Balatsos N.A., Anastasakis D., Stathopoulos C. Inhibition of human poly(A)-specific ribonuclease (PARN) by purine nucleotides: kinetic analysis. J. Enzyme. Inhib. Med. Chem. 2009;24:516–523. doi: 10.1080/14756360802218763. [DOI] [PubMed] [Google Scholar]
- 29.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:1–13. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mi H., Muruganujan A., Casagrande J.T., Thomas P.D. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013;8:1551–1566. doi: 10.1038/nprot.2013.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henriksson N., Nilsson P., Wu M., Song H., Virtanen A. Recognition of adenosine residues by the active site of poly(A)-specific ribonuclease. J. Biol. Chem. 2010;285:163–170. doi: 10.1074/jbc.M109.043893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martinez J., Ren Y.G., Nilsson P., Ehrenberg M., Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. J. Biol. Chem. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- 34.Körner C.G., Wormington M., Muckenthaler M., Schneider S., Dehlin E., Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. EMBO J. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Christman J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 36.Cui M., Wen Z., Chen J., Yang Z., Zhang H. 5-Aza-2′-deoxycytidine is a potent inhibitor of DNA methyltransferase 3B and induces apoptosis in human endometrial cancer cell lines with the up-regulation of hMLH1. Med. Oncol. 2010;27:278–285. doi: 10.1007/s12032-009-9204-1. [DOI] [PubMed] [Google Scholar]
- 37.Ye J., Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15:271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Balatsos N.A., Maragozidis P., Anastasakis D., Stathopoulos C. Modulation of poly(A)-specific ribonuclease (PARN): Current knowledge and perspectives. Curr. Med. Chem. 2012;19:4838–4849. doi: 10.2174/092986712803341539. [DOI] [PubMed] [Google Scholar]
- 39.Weick E.M., Miska E.A. piRNAs: from biogenesis to function. Development. 2014;141:3458–3471. doi: 10.1242/dev.094037. [DOI] [PubMed] [Google Scholar]
- 40.Petryszak R., Keays M., Tang Y.A., Fonseca N.A., Barrera E., Burdett T., Füllgrabe A., Fuentes A.M., Jupp S., Koskinen S., et al. Expression Atlas update–an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res. 2016;44:D746–D752. doi: 10.1093/nar/gkv1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vagin V.V., Wohlschlegel J., Qu J., Jonsson Z., Huang X., Chuma S., Girard A., Sachidanandam R., Hannon G.J., Aravin A.A. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen C., Jin J., James D.A., Adams-Cioaba M.A., Park J.G., Guo Y., Tenaglia E., Xu C., Gish G., Min J., et al. Mouse Piwi interactome identifies binding mechanism of Tdrkh Tudor domain to arginine methylated Miwi. Proc. Natl. Acad. Sci. U.S.A. 2009;106:20336–20341. doi: 10.1073/pnas.0911640106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He G.J., Yan Y.B. Self-association of poly(A)-specific ribonuclease (PARN) triggered by the R3H domain. Biochim. Biophys. Acta. 2014;1844:2077–2085. doi: 10.1016/j.bbapap.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 44.Okano M., Bell D.W., Haber D.A., Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 45.Rhee I., Bachman K.E., Park B.H., Jair K.W., Yen R.W., Schuebel K.E., Cui H., Feinberg A.P., Lengauer C., Kinzler K.W., et al. DNMT1 and DNMT3b cooperate to silence genes in human cancer cells. Nature. 2002;416:552–556. doi: 10.1038/416552a. [DOI] [PubMed] [Google Scholar]
- 46.Ramos M-P., Wijetunga N.A., McLellan A.S., Suzuki M., Greally J.M. DNA methylation by 5-aza-2′-deoxycytidine is imprinted, targeted to euchromatin, and has limited transcriptional consequences. Epigenetics Chromatin. 2015;8:11–27. doi: 10.1186/s13072-015-0004-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rouget C., Papin C., Boureux A., Meunier A.C., Franco B., Robine N., Lai E.C., Pelisson A., Simonelig M. Maternal mRNA deadenylation and decay by the piRNA pathway in the early Drosophila embryo. Nature. 2010;467:1128–1132. doi: 10.1038/nature09465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen C., Ito K., Takahashi A., Wang G., Suzuki T., Nakazawa T., Yamamoto T., Yokoyama K. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem. Biophys. Res. Commun. 2011;411:360–364. doi: 10.1016/j.bbrc.2011.06.148. [DOI] [PubMed] [Google Scholar]
- 49.Zukeran A., Takahashi A., Takaoka S., Mohamed H.M., Suzuki T., Ikematsu S., Yamamoto T. The CCR4-NOT deadenylase activity contributes to generation of induced pluripotent stem cells. Biochem. Biophys. Res. Commun. 2016;474:233–239. doi: 10.1016/j.bbrc.2016.03.119. [DOI] [PubMed] [Google Scholar]
- 50.Gou L.T., Dai P., Yang J.H., Xue Y., Hu Y.P., Zhou Y., Kang J.Y., Wang X., Li H., Hua M.M., et al. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 2014;24:680–700. doi: 10.1038/cr.2014.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subtelny A.O., Eichhorn S.W., Chen G.R., Sive H., Bartel D.P. Poly(A)-tail profiling reveals an embryonic switch in translational control. Nature. 2014;508:66–71. doi: 10.1038/nature13007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yuan S.X., Wang J., Yang F., Tao Q.F., Zhang J., Wang L.L., Yang Y., Liu H., Wang Z.G., Xu Q.G., et al. Long noncoding RNA DANCR increases stemness features of hepatocellular carcinoma by derepression of CTNNB1. Hepatology. 2016;63:499–511. doi: 10.1002/hep.27893. [DOI] [PubMed] [Google Scholar]
- 53.Anastas J.N., Moon R.T. WNT signalling pathways as therapeutic targets in cancer. Nat. Rev. Cancer. 2013;13:11–26. doi: 10.1038/nrc3419. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.