Abstract
RNA is targeted in biological conflicts by enzymatic toxins or effectors. A vast diversity of systems which repair or ‘heal’ this damage has only recently become apparent. Here, we summarize the known effectors, their modes of action, and RNA targets before surveying the diverse systems which counter this damage from a comparative genomics viewpoint. RNA-repair systems show a modular organization with extensive shuffling and displacement of the constituent domains; however, a general ‘syntax’ is strongly maintained whereby systems typically contain: a RNA ligase (either ATP-grasp or RtcB superfamilies), nucleotidyltransferases, enzymes modifying RNA-termini for ligation (phosphatases and kinases) or protection (methylases), and scaffold or cofactor proteins. We highlight poorly-understood or previously-uncharacterized repair systems and components, e.g. potential scaffolding cofactors (Rot/TROVE and SPFH/Band-7 modules) with their respective cognate non-coding RNAs (YRNAs and a novel tRNA-like molecule) and a novel nucleotidyltransferase associating with diverse ligases. These systems have been extensively disseminated by lateral transfer between distant prokaryotic and microbial eukaryotic lineages consistent with intense inter-organismal conflict. Components have also often been ‘institutionalized’ for non-conflict roles, e.g. in RNA-splicing and in RNAi systems (e.g. in kinetoplastids) which combine a distinct family of RNA-acting prim-pol domains with DICER-like proteins.
INTRODUCTION
The established roles of RNA have expanded greatly in the last several decades beyond its initial characterization in the Central Dogma of molecular biology, wherein the messenger RNA (mRNA) acts as the informational template specifying the protein sequence (1). Other characterized classes of RNA include ribosomal RNAs (rRNAs), which form the framework around which ribosomes are assembled, while one of them (23S/28S rRNA) also directly catalyzes the peptide ligase reaction during protein synthesis (2). Transfer RNAs (tRNAs) act as amino-acylated intermediaries as well as the complementary ‘readers’ of the genetic code during initiation and extension of nascent polypeptide chains in translation of mRNAs (2). Beyond these three basic classes of RNA several other classes of RNA have since been characterized and range in their phyletic distributions from being universally-conserved across all branches of Life to being specific to particular lineages (3–6). Likewise they exhibit functional diversity which spans structural roles (e.g. long-non-coding RNAs in eukaryotes) (7), highly specific recognition of low-molecular weight substances (riboswitches) (8), catalysis (ribozymes) (9–12), and either direct or indirect transmission of information for various purposes (e.g. snRNA, various small RNAs of RNAi and CRISPR systems) (13–15). As a consequence a rich cellular ‘ecology’ of proteins has evolved around these RNAs, which has only recently come to light and is still being uncovered.
The strong constraints emerging from the indispensability of universally-present RNA classes to fundamental cellular pathways like translation has resulted in high conservation of their structure and sequence. As a result they appear to be less ‘evolvable’ than other cellular components, making them both susceptible to a range of insults from environmental factors and attractive targets for molecular weaponry deployed in biological conflicts (effectors) aiming to decisively disable a cell. Research in this direction over the past several decades has brought to light two key findings. First, RNA damage is prevalent in cells, with much of this damage likely inflicted by a range of RNA-targeting effectors from diverse sources (16–20). Second, diverse RNA repair systems exist in cellular and viral genomes (21–23), which are capable of specifically repairing or reversing the damages suffered by different RNAs.
The enormous variety and pervasiveness of systems involved in these opposing activities, namely, targeted RNA damage and RNA repair, is becoming increasingly clear with the continually-expanding availability of sequence data from diverse branches of Life. Thus, we felt it was appropriate to survey and classify the key players in this vast biological ‘battleground’ centered on RNA damage. To this end, we briefly summarize the agents, targets, and mechanisms of RNA toxicity followed by a detailed discussion of repair systems, with particular emphasis on the evolutionary relationships of their components, structural features, and mechanisms. Finally, we present potential directions for future research.
BIOLOGICAL AGENTS OF RNA DAMAGE
Biological conflict systems and targeting of RNA
The moniker ‘conflict system’ applies to any molecular system which is deployed by genic entities across all organizational levels of Life in offensive and/or defensive strategies to establish and maintain their own fitness relative other directly competing entities (24–27). At the level of trophic interactions between organisms, protein effectors targeting RNA are commonly deployed in diverse predator-prey and host–parasite relationships. Yeasts such as Kluyveromyces and Millerozyma attack host plant cells via deployment of RNA-targeting toxins like zymocin, collectively referred to as ‘yeast killer toxins’ (Table 1) (28–31). Conversely, certain filamentous fungi defensively use α-sarcin-like and related RNA toxins (e.g. restrictocin and hirsutellin) against insects (32,33). Likewise, plant RNA toxins like ricin, abrin and saporin are produced in seeds to ward off potential animal predators (34). The closely-related Shiga toxins of certain bacteria are deployed against natural predators like Tetrahymena (35) and are also toxic to some animals (Table 1) (36). Inter-organismal conflict additionally arises during cooperative cell-colony formation: co-aggregating ‘cheater’ species/strains whose interests do not align completely with the cooperating species/strain are targeted by conflict effectors to eliminate or curb their growth (37). A large, recently-described collection of such systems which discriminate kin versus non-kin are the polymorphic toxin systems, which share the strong tendency for their toxin domains to recombine and undergo rapid sequence and structural divergence (Table 1), the majority of which are predicted to target RNA (38–41).
Table 1. Examples of conflict systems containing RNA-targeting effector domains.
| Conflict system class | RNA toxin effector examples | Targets | Phyletic distribution | Comments |
|---|---|---|---|---|
| Yeast killer toxin | Zymocin, PaT | Modified U34 (wobble uridine) base-containing tRNA. In Pichia acaciae, predominantly tRNAGln. | Yeast | Toxin domain fused to N-terminal secreted chitinase domain which might breach fungal cell wall |
| α-Sarcin-like | BECR fold domains (sarcin, restrictocin, hirsutellin, mitogillin, etc.) | Backbone cleavage in rRNA sarcin-ricin loop | Filamentous fungi | Several such RNases with 1–3 BECR domains are found across fungi |
| Ricin-like | Ricin, saporin, pokeweed antiviral protein, Shiga | N-glycoside hydrolysis of rRNA sarcin-ricin loop base | plants, ciliates, bacteria | Found in a class of ciliate toxin with architectural parallels to bacterial polymorphic toxins |
| Toxin-antitoxin (T-A) | BECR fold domains (Barnase, EndoU, Colicin-like, RelE-like) MazF/PemK/EndoA, PIN (VapC, etc.) domains, HEPN domains | mRNA (BECR, MazF/PemK/EndoA, RNase LS and RNase LsoA: HEPN), tRNAMet (VapC: PIN), tRNA (BECR), rRNA sarcin-ricin loop (PIN), rRNA S16 (Colicin E3, E4, E6: Colicin E3-like) | Bacteria, archaea | Widespread intra-genomic conflict systems |
| Polymorphic toxins | Various BECR fold domains, deaminase | tRNA, likely other targets | Bacteria, archaea | The toxin domains typical vary via replacement by alternative cassettes |
| Colicin-like | BECR fold domains, colicin E3-like | Probably tRNA (BECR), rRNA S16 (Colicin E3, E4, E6: Colicin E3-like) | Bacteria | Differ from above in being secreted by lysis of the producing cell and being encoded on plasmids |
| CRISPR/Cas | Cas2, HEPN, active RAMPs and Csx3. Cmr and Cas9 in some subtypes | mRNAs, Csx3 exonucleolytically targets terminal polyA tails | Bacteria, archaea | A wide range of actions which are both directed by complementary CRISPR spacer RNAs and independently of them |
| Restriction-modification systems | HEPN (PrrC-like RNases) | tRNALys | Bacteria | inactivation of R-M system by phage leads to activation of PrrC, which targets endogenous tRNA |
| Abortive infection (Abi) | HEPN (AbiA-CTD, AbiD, AbiF, AbiJ, AbiU2, AbiV) | Unknown | Bacteria | Part of an extensive antiphage defense which might also directly target phage components |
| Phage growth limitation (Pgl) | HEPN (RloC-like, pEK499_p136-like families), SNase | Unknown | Bacteria | Sporadic coupling to RNases could cleave phage RNAs in attacks complementary to Pgl DNA modification |
| Ter-dependent anti-phage system | HEPN (DUF4145-like) | Unknown | Bacteria | Sporadic coupling to HEPN domain could work with core system |
| Prokaryotic nucleotide or nucleotide-derived secondary messenger-based systems | HEPN | Unknown | Bacteria | Combines with CARF sensor and mCpol nucleotide secondary messenger synthetase |
| prokaryotic PIWI-based systems | PIWI/argonaute, HEPN | Unknown | Bacteria | In bacteria, pPIWI is adjacent to effector RNAse domains |
| Animal innate immunity | RNaseL – permuted version of HEPN domain | Viral RNA | Eukaryotes | Activated in vertebrates by oligoadenylate (OA) linear secondary messenger nucleotide |
Within a single cell, conflicts also arise between distinct genomes. Inter-genomic conflict can be seen in intracellular symbiotic relationships between bacteria and eukaryotes. It also includes the conflicts between the host genome and invading genomes, examples of which include viruses, plasmids and conjugative elements. Molecular weaponry deployed in these conflicts are among the best-studied examples of conflict systems, and several of these have been experimentally determined to target RNA. The Type IIIB and certain representatives of the TypeII (Cas9)-dependent CRISPR/Cas antiviral systems, subtypes within the large conglomeration of prokaryotic acquired immunity systems collectively termed CRISPR/Cas (15), target RNA via the crRNA-Cmr ribonucleoprotein effector complex (42,43), contrasting many other CRISPR/Cas subtypes which target DNA. Additionally, evidence continues to mount in support of secondary or complementary RNA-targeting effectors in some CRISPR/Cas system subtypes which provide a force-multiplier for the DNA-targeting elements by additionally targeting RNA (44,45) (Table 1).
In a similar vein, RNA-targeting effectors are also observed as ‘backup’ components in some restriction-modification (R-M) systems and Phage growth limitation (Pgl) systems (46). In R-M systems the characterized versions of these effectors (e.g. PrrC-type proteins; Table 1) are known to be activated in the event of failure of the primary line of attack, namely the restriction enzyme (47–49). As opposed to the R-M systems, which modify the host genome to discriminate against invasive DNA, the Pgl systems methylate the invading phage genome to protect cells containing the Pgl system from future infection by the same phage (46,50,51). The RNA toxins genomically linked to Pgl systems are likely to provide a second prong in conflict by limiting phage growth via RNA degradation (Table 1) (44). A comparable mode of action might relate to the predicted RNA toxins found associated with the recently-described Ter-dependent stress/phage response systems (Table 1) (52). A distinct two-pronged mechanism utilizing RNA-targeting effectors has been proposed for the anti-phage abortive infection (Abi) systems (Table 1). Here, one of the domains (or components) directly targets particular components of the phage while the RNA toxin might help limit infection via host-cell death in the event of failure to contain the infection (44,53,54). Such coupling of effectors predicted to target RNA along with those targeting DNA or other macromolecules is also observed in some less-understood, recently uncovered intra-organismal prokaryotic conflict systems unified by the presence of PIWI superfamily modules. In these systems, PIWI modules, which are homologs of Argonaute (AGO) proteins involved in eukaryotic small RNA-dependent interference and related processes, are coupled via genome co-localization with an additional enzymatic effector. In a subset of these systems, both the PIWI and sometimes the additional effector domains are predicted to target RNA (Table 1) (14).
A recent study uncovered a range of previously-undescribed conflict systems centered on the production of a nucleotide- or nucleotide-derived second messenger in response to a conflict-initiated stimulus which is leveraged to activate downstream effectors (55). A subset of these systems, which might also feature the mCpol (minimal CRISPR polymerase) domain to likely generate an inducing cyclic nucleotide messenger, potentially target RNA (Table 1) (55). Parallel systems are also observed in counter-viral innate immune responses of eukaryotes. The vertebrate immune response initiated by detection of viral double-stranded RNA (dsRNA) by the oligoadenylate synthase (OAS) enzyme, generates OAS linear nucleotide secondary messengers, which in turn activates RNases that target invasive dsRNA (56,57) (Table 1). Additionally in animals, certain broad-specificity RNases are also believed to be part of the general immune response (58,59). In contrast, an important aspect of the immune response of some plants is the direct targeting of viral RNA by toxins related to the Shiga and ricin-like toxin. Experimentally characterized examples include the pokeweed antiviral protein, which protects the pokeweed plant (Phytolacca americana) from RNA virus attack (60,61).
The final level of genetic conflict is observed within a single genome, consisting of selfish elements seeking to maximize their fitness at the expense of other elements in the genome. These include several types of transposable elements and toxin-antitoxin (T-A) systems which are characteristically highly mobile and rapidly evolving. T-A systems, in particular, are rife with diverse RNA-targeting effectors whose action is unleashed in the absence of the less-stable antitoxin component (Table 1) (62). This mechanism enforces the maintenance of T-A systems in genomes (addiction) by inducing cell death. Moreover, the CRISPR/Cas systems also likely harbor a comparable ‘addiction’ component which probably enforces the maintenance of CRISPR/Cas systems in cellular genomes. This component, the Cas2 protein, one of the few core components shared across all CRISPR/Cas systems, targets RNA (63–65).
Catalytic mechanisms and diversity of RNA-targeting effector domains
The predominant type of catalytic domain found in RNA-targeting effectors is the RNase domain. There are two broad mechanisms for RNase activity: (i) the metal-independent RNases use a transesterification reaction yielding a 5′-hydroxyl at the 5′ end of the 3′ fragment and a 2′,3′-cyclic phosphate at the 3′ end of the 5′ fragment (Figure 1A). (ii) Metal-dependent RNases typically leave a 5′-phosphate at the 5′ end of the 3′ fragment and a 3′-hydroxyl at the 3′ end of 5′ fragment (Figure 1A). Beyond these common configurations certain RNases have been shown to generate fragments with a 3′-phosphate and a 5′-hydroxyl (Figure 1A). While most RNases in conflict systems catalyze an endonucleolytic cleavage, some might additionally catalyze further exonucleolytic cleavage (e.g. RloC).
Figure 1.
Known and predicted biochemical mechanisms of biological conflict-related RNA damage and repair. The methyl group added during Hen1-mediated methylation is in red in (B). Slash in (D) separates the two RNA 3′ end phosphate group configurations the RtcB ligase accepts as substrates. Green labels provide reaction step explanation/categorization. Blue dots: predicted enzymes or reaction steps predicted for the first time.
The two most widespread examples of the former are the BECR and HEPN fold RNases. The BECR fold is a recently described α/β fold that unifies diverse RNases previously thought to belong to disparate domains, including the sarcin and related toxins, several T-A system toxins like RelE, EndoU, Barnase and certain colicins (Table 1) (39). Scores of distinct, poorly-characterized BECR fold RNases families are also present in polymorphic toxin systems (39). The all α-helical HEPN RNase domain, often typified by a characteristic constellation of conserved active site residues, is likewise seen across diverse conflict systems (Table 1). In some instances, HEPN is deployed as the primary anti-RNA effector, e.g. in RNase L from animal innate immunity systems, several T-A systems and possibly in certain prokaryotic PIWI (pPIWI) systems (14). However, in other systems like CRISPR/Cas, R-M, and Abi, it is coupled with other effectors (44,66). While several other metal-independent RNase domains are seen across conflict systems, the more common ones among these include some RAMPs of the CRISPR/Cas systems, which are derived versions of the RRM fold, and the predominantly β-sheet colicin E3-like nucleases found in T-A systems (Table 1) (67).
Metal-dependent nucleases, less frequently observed in conflict systems, include the Rossmannoid fold PIN RNase superfamily (Table 1) (68–70). The PIWI domain of the RNaseH fold is another metal-dependent RNase or DNase, targeting RNA in some prokaryotic conflict systems and eukaryotic RNAi systems (Table 1) (14,71–73). The CRISPR/Cas systems contain the RRM fold nucleases Cas2 with a metal-chelating aspartate and Csx3 with manganese-dependent exonucleolytic deadenylation activity (74). Finally, it is possible that double-stranded RNA is subject to impairing backbone ‘nicks’ instead of outright cleavage (75–77). While effector domains capable of inflicting RNA backbone nicks have not yet been experimentally characterized the prediction of potential nicking endoRNases in certain polymorphic toxin and other conflict systems suggests this could be an unexplored mechanism that impairs RNA function (39).
Alternatives to RNase activity have also been identified across disparate conflict systems. The best-studied of such domains are the ricin-like toxins or the ribosome-inactivating proteins (Table 1) which catalyze N-glycoside hydrolase activity, targeting and removing a specific base from the RNA backbone (78). In addition to plant and bacterial toxins, ricin-like domains also occur in certain predicted ciliate toxins with architectures similar to prokaryotic polymorphic toxins (L Aravind, unpublished observations). Recent analyses have further identified divergent families of the deaminase superfamily, which counts as members the animal AID, ADAR and APOBEC enzymes, in a subset of polymorphic toxin systems (79). Deaminase domains in these systems are predicted to inactivate RNAs through an editing/mutation strategy targeting key RNA bases (Table 1). Finally, while as yet experimentally uncharacterized, it has been speculated that ADP ribosylation of bases in RNA catalyzed by nucleic-acid-ADP ribosylating domains comparable to the DNA-modifying enzymes CARP-1 and Pierisin could act in a distinct RNA-targeting strategy (80,81).
RNA targets of effectors
In both eukaryotes and prokaryotes, deployment of effectors against RNA follows a strategic dichotomy: either they directly attack RNA emanating from the competing genomic element or invasive entity in response to recognition of an ongoing or imminent attack, or they target endogenous, i.e. ‘self’ RNA. The latter triggers either cell suicide to limit the replication/spread of the invasive agent or cell dormancy until the threat is eliminated or overcome by other defenses (82). Examples of the former strategy include animal innate immunity, defensive and predatory deployment of fungal toxins (e.g. yeast killer and α-sarcin like toxins), certain CRISPR/Cas systems, and several polymorphic toxin systems. In the two-pronged systems described above including Abi, Ter, R-M and CRISPR/Cas systems (Table 1), the latter strategy is often coupled to an additional conflict system directly attacking the opposing entity in the conflict (52,83). Such self tRNA-targeting, which limits tRNA availability to the virus, is catalyzed by HEPN-containing PrrC-like proteins when the primary effector activity of its coupled R-M system is compromised (47,84,85). Similarly, Cas2, which along with the Cas2-inhibiting Cas1 protein comprise the only domains found across all CRISPR/Cas systems, is predicted to function as both as the CRISPR/Cas addiction module via self RNA targeting and as means of inducing suicide or dormancy in the event of CRISPR/Cas system failure (82).
There is strong convergence across disparate conflict systems featuring anti-RNA effector domains with diverse mechanisms in terms of their RNA targets, namely attack on RNA classes associated with translation—mRNA, tRNA and rRNA. Several distinct effector domains have converged on targeting of the 23S sarcin-ricin loop (SRL), a universally conserved and exposed structural feature in the completely-assembled ribosome which is essential for elongation factor interaction during translation (86). The titular sarcin-like and ricin-like toxin domains both target the SRL and inactivate ribosomes by preventing elongation factor binding, albeit through distinct mechanisms with sarcin catalyzing loop cleavage (87) and ricin catalyzing removal of a conserved adenine base (88,89). Additionally, certain T-A system PIN domains also transect the sarcin-ricin loop in a metal-dependent manner (Table 1) (90).
Paralleling rRNA targeting at the sarcin-ricin loop, effectors targeting tRNA typically attack at or near its best-conserved structural feature: the anticodon loop. These effectors typically target specific tRNAs or tRNA families; for example, the HEPN-containing RloC and PrrC domains both target tRNALys. While PrrC merely cleaves the anticodon loop, RloC catalyzes a further exonucleolytic cut to free the wobble nucleotide from the cloven end. Other RNases are more promiscuous in their tRNA targeting; for example, yeast killer toxins have been reported to target multiple tRNA families with wobble uridine-modified bases near the anticodon loop (30) while the T-A system BECR fold-containing colicin E5 and D toxins and VapC4 toxins of the PIN domain have been linked to a range of tRNA substrates (Table 1) (16,69,91).
Similar to both tRNA and rRNA, mRNAs are targeted by disparate effector domains. In contrast to the targeting of specific structural features in rRNA and tRNA, several recognition strategies have been experimentally elucidated for mRNA. For example, the MazF/PemK/EndoA-like RNases target specific sequence features (92), while the HEPN domain RNase LS and related toxin effectors indiscriminately target bulk mRNA (93,94). A distinct mRNA targeting strategy seen in the RelE-like BECR fold RNases involves recognition of specific structural features to occupy the ribosome A-site and cleave ribosomal-bound mRNA with apparently little sequence specificity (95). Expression of the AbrB/MazE-like SymE toxin protein also results in widespread mRNA degradation, although the exact mechanism of its action awaits further clarification (96). Transcribed RNA also forms RNA-DNA hybrid duplexes which can act as targets for pPIWI conflict system nucleases. In some of these systems, RNA fragments derived from invasive plasmids are utilized by pPIWI nucleases to selectively cleave exogenous DNA sources via a RNA–DNA hybrid (71), while the pPIWI-RE family is likely to directly target the naturally-forming RNA–DNA duplexes, termed R-loops (97), of invasive elements (98).
One common theme uniting several of these enzymatic effectors is that, regardless of the mechanism or RNA target, RNA damage ultimately appears to block translation through disabling or ‘jamming’ the ribosome. In addition to direct targeting of rRNA (90,99), mRNA, and presumably tRNA, attacks that take place at the ribosome effectively jam the ribosome and render it incapable of further translocation (95). Such attacks have the dual advantage of not only rapidly blocking a vital cellular process but also slowing any repair responses that are dependent on fresh translation.
MAJOR CLASSES OF PROTEIN DOMAINS INVOLVED IN RNA REPAIR
Given the diversity observed in RNA toxin effectors, their targets, and systems which deploy them, it is not surprising that a parallel world of diverse RNA repair domains and systems have evolved. Systems directly repairing RNase-induced damage are the best studied to date. Any systems which might potentially reverse other types of damage caused by RNA toxins, such as base-removal and deamination (Table 1), await discovery and characterization. Despite the first identification of a counter-RNase repair system over four decades ago and several advances in understanding the mechanisms of repair across distinct repair systems, much of their biology remains comparatively poorly-understood; particularly in terms of their genomic contexts, their specific in vivo RNA substrates, and their dynamics in course of biological conflicts.
Nevertheless, a few widespread features have been identified across RNA repair systems: (i) ligase modules which join RNA fragments created by endoRNase action; (ii) template-independent or dependent nucleotidyltransferases (NTases) with RNA polymerase activity which can elongate RNA termini; (3) one or more other domains, which can be functionally categorized as ‘cleaning’ enzymes that process RNA termini typically in preparation for ligation or elongation by NTases, or modify them for protection; (iv) cofactors which act as scaffolds or binding proteins or help enhance catalysis of the above components. In the following sections, we discuss in detail these modules and their mechanisms of action: both those which have been previously experimentally-characterized and those which are genomically-linked yet await experimental characterization in the context of RNA repair, as genome contextual associations in prokaryotes are excellent indicators of functional association (100,101).
RNA ligase catalytic modules
RNA ligases of the ATP-grasp fold
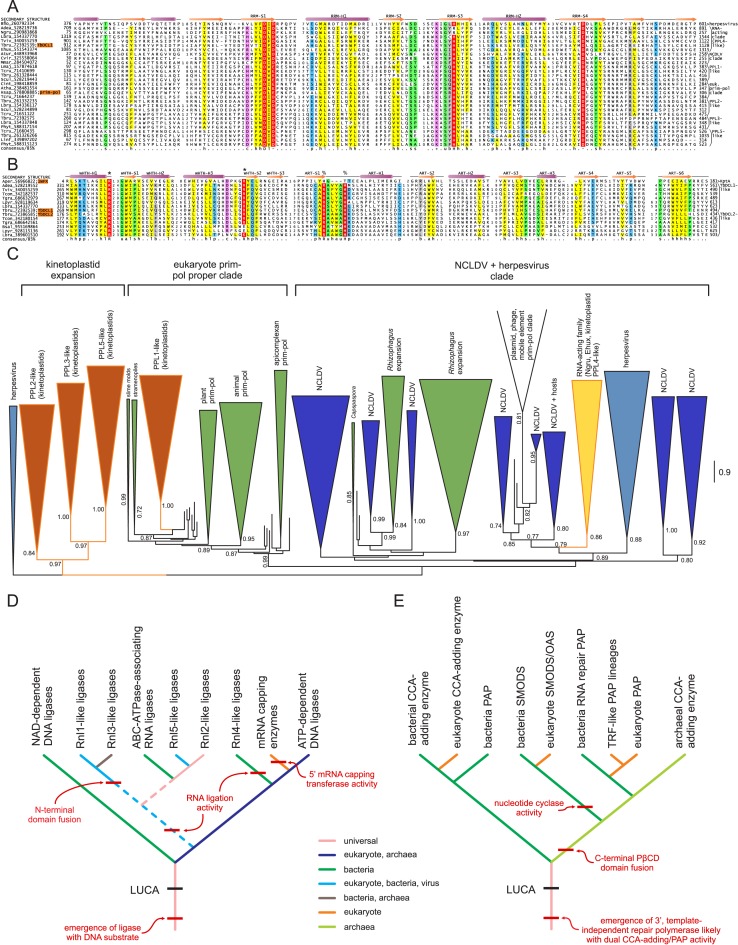
ATP-dependent RNA ligases are members of the expansive ATP-grasp protein fold (102–104), emerging through ancestral fusion of protein-kinase C-terminal-like and RAGNYA domain folds (Figure 2A) (105). Nucleic acid ligases are a distinct superfamily within this fold, uniting several families catalyzing related reactions including cellular and viral mRNA capping, DNA ligation, and RNA ligation, utilizing either ATP or NAD+ co-substrates for their nucleotidyltransferase activity (Figure 1B) (106,107). All characterized RNA ligases of this superfamily use ATP as the co-substrate. Ligation proceeds through an absolutely-conserved lysine residue from the RAGNYA domain, which attacks the α-phosphate of the ATP, releasing pyrophosphate and forming a covalent lysyl-N-AMP intermediate. The AMP is then transferred to the 5′ phosphate of the 3′ RNA fragment, followed by attack of a free 3′ hydroxyl group from the 3′ end of the 5′ RNA fragment on the RNA-adenylate thereby re-joining the RNA (108). Therefore, the ATP-grasp RNA ligase requires a phosphate group at the 5′ end of the 3′ fragment and a 3′ OH group at the 3′ end of the 5′ fragment as prerequisites for ligation (Figure 1B). In this respect they closely resemble the reaction mechanism of the ATP-dependent DNA-ligases and in both sequence and structure searches appear to be closer to them than the NAD+-utilizing DNA ligases. The RNA ligases might have been derived on more than one occasion from the ATP-dependent DNA ligases: of the previously described RNA-ligases, the Rnl1-like and Rnl3-like clades are united by a common four-stranded N-terminal domain, which appears to be critical for RNA-binding. These are probably further united into a higher order clade with Rnl2-like ligases which include the Rnl2-proper, Rnl5-like and a novel ligase clade described below. In contrast, Rnl4-like ligases show closer sequence similarity to the ATP-dependent DNA ligases and appear to have been more recently derived from those.
Figure 2.
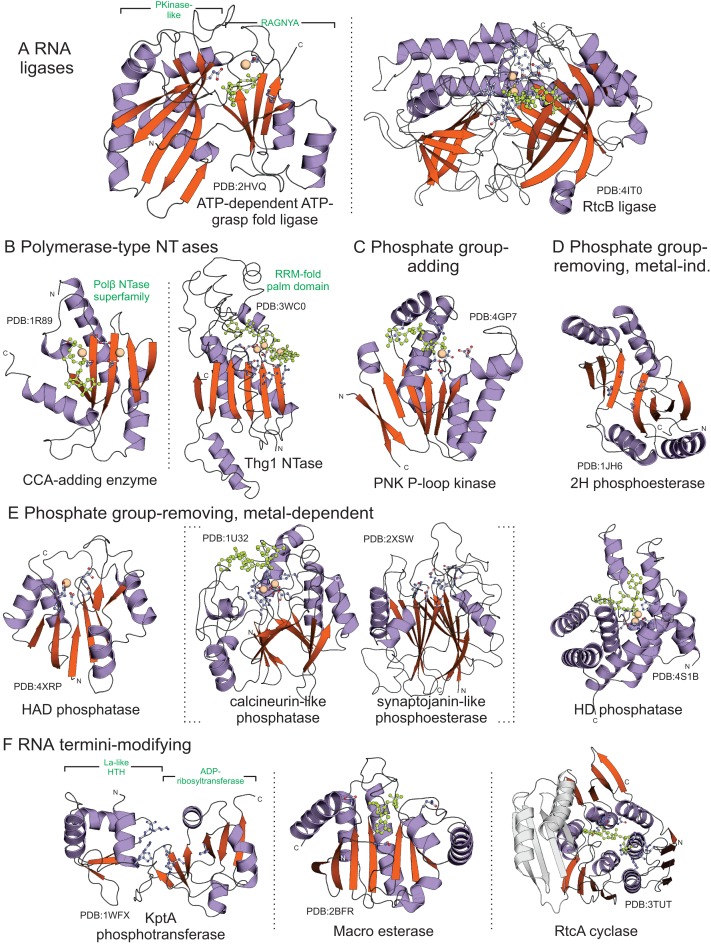
Structures of catalytic domains repairing RNA damaged in biological conflicts. Structures are grouped as appearing in the text and are labeled by protein name and Protein Databank ID; green labels provide additional domain information. Dotted lines separate domains performing similar functions but with different protein folds; dotted bracket grouping calcineurin-like and synaptojanin-like domains denotes higher-order relationship. Ligands: light green and ball-and-stick. Conserved residues involved in catalysis or substrate recognition: ball-and-stick (carbon, light blue; nitrogen, dark blue; oxygen, red; cysteine, orange), metal ions: spheres.
ATP-grasp RNA ligase modules are sometimes further characterized by presence of distinctive C-terminal extensions (109), although certain clades of these ligases are predicted to lack any such extension. Comparative analyses revealed that the extensions, when present, are predominantly α-helical in nature, typically containing at least two α-helices although four or more can be present (Figure 3B–Q, Supplementary Material). Sequence analysis does not reveal any relationship between these extensions; however, given the rapid sequence divergence typically observed even among otherwise closely-related versions of these ligases, we cannot rule out rapid divergence from a common α-helical ancestral extension. The subset of C-terminal extensions studied to date in structural complexes with RNA substrates have roles in RNA recognition, including tRNA (110) and duplex RNA structures (111,112). The additional C-terminal extensions recognized in this work could similarly participate in RNA recognition (Figure 3B–Q, Supplementary Material).
Figure 3.
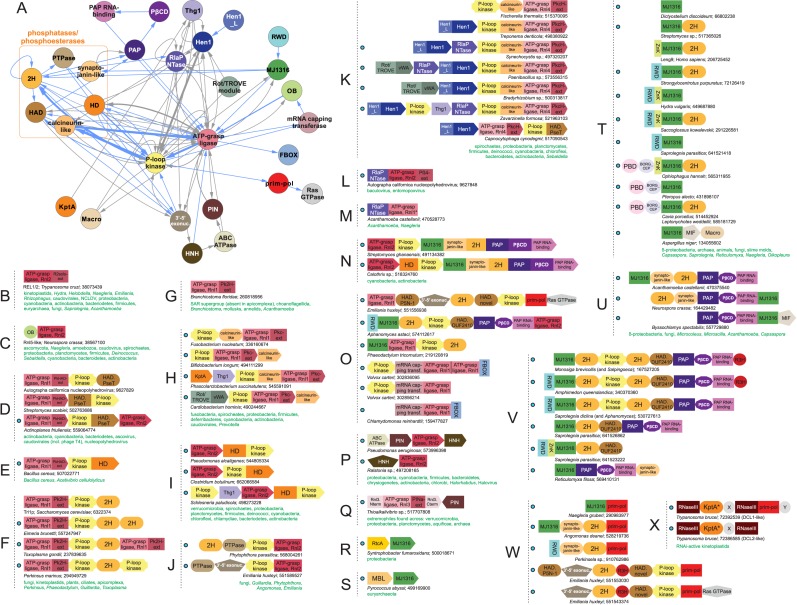
Contextual connections of ATP-grasp ligase-based and related RNA repair systems. (A) Contextual network constructed as described in Supplementary Material. Protein domain nodes are colored according to general functional category. Phosphoesterase/phosphotransferase domains are further demarcated by dotted orange box. (B–Q) Representative depictions of conserved domain architectures and gene neighborhoods. Domain architectures are depicted as adjoining shapes, not drawn to scale. Gene neighborhoods are depicted as directed boxes, genes within neighborhood encoding multiple domains contain individually-colored boxes for each domain. All contexts are labeled with organism name and NCBI gene identifier (gi) number. Green lettering: phyletic distributions for each group of systems. Blue dots: novel predicted RNA repair systems or systems containing a previously-unrecognized component. (R–X) Representatives of conserved domain architectures and gene neighborhoods containing the MJ1316 domain. All domain and organism expansions are provided in Supplementary Material.
RtcB-like ligases
RtcB-like RNA ligases, first described in a series of studies in 2011 (23,113–115), contrasted sharply with the known properties of the ATP-grasp ligases. The RtcB catalytic domain contains a distinctive α/β fold comprised of two core β-sheets with no other known members. The active site with a two metal center is nestled between the two sheets (Figure 2A) (116,117). An absolutely-conserved histidine residue in this active site initiates ligation by attacking the α-β linkage between the phosphates of a GTP molecule, forming a covalent histidinyl-N-GMP intermediate. RtcB then hydrolyzes cyclic 2′,3′-phosphate 3′ ends of the 5′ RNA fragment by activating water via one of the active site metals; the freed phosphate group then receives the GMP from the histidine. This phosphate group is then attacked by the free 5′ OH group of the 3′ RNA fragment, yielding the ligated RNA product (Figure 1D) (117–121). Thus, in contrast to the ends ligated by the ATP-grasp RNA ligases, the RtcB ligases utilize a cyclic 2′,3′ phosphate group at the 3′ end of the 5′ RNA fragment and a free 5′ OH group at the 5′ end of the 3′ RNA fragment (Figure 1B,D). This reaction mechanism also enables RtcB to repair ends with 3′ phosphates (122). This dichotomy in substrate prerequisites mirrors the distinct RNA ends produced by metal-dependent and -independent RNases (Figure 1A, Table 1). As an additional wrinkle, Shuman and colleagues have demonstrated a further activity for RtcB: at least some bacterial RtcB domains apparently cap broken DNA strands via direct transfer of GMP to the DNA 3′ phosphate group. These caps shield DNA from further exonucleolytic degradation, while acting as a primer for DNA synthesis by repair polymerases (123–127). RtcB ligases discussed in the systems below are likely primarily involved in RNA repair by merit of their genome contextual associations and we observe no persuasive contextual evidence linking RtcB to DNA repair; however, it is possible that RtcB domains found in genomes with no conserved context could additionally function in DNA end-protection.
Polymerase-type nucleotidyltransferase domains
Template-independent and -dependent nucleotidyltransferases (NTases) perform unique, and oftentimes poorly understood, roles in RNA repair. Such NTases are typically implicated in addition of nucleotides at RNA ends exposed by endonucleolytic cleavage. Nucleotide addition has, to date, been implicated in opposing roles: either the stabilization/protection of RNA from further exonucleolytic degradation or, conversely, in tagging damaged RNA for exonucleolytic degradation.
CCA-adding enzymes and other members of the DNA polymerase β (Polβ) NTase superfamily
CCA-adding enzymes are members of the Polβ NTase superfamily (Figure 2B) which catalyze the serial template-independent addition of the CCA trinucleotide to 3′ ends of tRNAs (Figure 1E). Such CCA-free ends are produced during tRNA maturation by cleavage by the metallo-beta-lactamase (MBL) fold endonuclease tRNase Z (128). Evolutionary reasoning supports a scenario wherein CCA-adding enzymes first emerged as repair enzymes acting to restore genomically-encoded CCA 3′ termini in tRNAs, the configuration required for aminoacylation (129). Such a role is still observed in many bacterial lineages with tRNA termini retaining genome-encoded CCA (130,131). This enzyme likely bestowed a selective advantage by preventing exonucleolytic degradation and extending tRNA life, perhaps even countering effectors with exonuclease activity in biological conflicts. Fixation of the CCA-adding enzyme relaxed the constraint for a genomic CCA and the addition of this trinucleotide became a required step for tRNA maturation in several lineages. Other representatives of the Polβ superfamily have been characterized in parallel repair/protective roles: the poly(A)polymerases (PAP) add a protective poly(A) tail to mRNAs following 3′ end endonucleolytic cleavage during transcript maturation again catalyzed by an MBL fold endonuclease. The TUT1 polymerases of the related TRF clade adds a poly(U) tail to protect the 3′ end of U6 snRNA (132). In contrast, polynucleotide tags added by other representatives of the superfamily, specifically the TRF clade, appear to route various classes of RNA for exonucleolytic degradation (14).
Thg1 family 5′-3′ polymerases
Thg1 catalyzes template-dependent or -independent addition of the distinctive guanine nucleotide at the 5′ end of tRNAHis, which is complementary to a cytosine just upstream of the CCA trinucleotide, during its maturation. This base is essential for its recognition by histidine aminoacyl-tRNA synthetase (HisRS) (133,134). Thg1 requires 5′ OH phosphorylation at exposed 5′ RNA ends to initiate nucleotide addition (Figure 1F) (135). Thg1 contains a RRM-fold palm domain, forming a higher-order clade of such domains with the minimal CRISPR polymerase (mCpol), CRISPR polymerase and GGDEF-like nucleotide cyclase domains (Figure 2B) (55,136,137). Like CCA-adding enzymes, Thg1 likely emerged ancestrally as a general 5′ repair enzyme in prokaryotes (138), later to be fixed as histidine 5′ adding enzymes in certain lineages like the eukaryotes.
Prim-pol domains
Prim-pol domains belong to the archaeo-eukaryotic primase superfamily which contains a derived version of the RRM fold polymerase palm domain, and is capable of catalyzing both polymerase and primase activities in the known context of oxidative lesion DNA repair (139,140). Despite their exclusive characterization to this point in DNA repair pathways, prim-pol enzymes generate RNA polynucleotides, bringing up the possibility that these could function in RNA-related contexts (see below for details).
Enzymes ‘cleaning’ or processing RNA termini
These domains are in part a specific evolutionary response to the distinctive 3′ and 5′ ends of RNA, especially the distinct products of metal-dependent and -independent RNases. Moreover, the specific biochemical requirements inherent to different ligases which ‘heal’ the cleaved RNA backbone or RNA polymerases which elongate them necessitates processing of the ends of RNAs before they can be acted upon by these enzymes. The primary end ‘cleaning’ actions are performed by enzymes that either add or remove phosphate groups. A further group of enzymes subject RNA termini to modifications which may complement the other cleaning enzymes or protect the ends from further attack.
Phosphate group-adding domains: P-loop kinases
A family of kinase domains belonging to the DxTN clade of the P-loop kinase superfamily of the P-loop NTPase fold is frequently coupled to the ATP-grasp ligases, and is commonly referred to as the polynucleotide kinase (PNK) family (Figure 2C) (141). The well-documented role of this enzyme is phosphorylation of the 5′ OH group of an RNA fragment with a phosphate group required for reactions such as the ATP-grasp ligase-catalyzed ligation and extension by Thg1 enzymes (Figure 1B) (142–144).
The distinct Clp1 family of kinases which have convergently evolved nucleic acid kinase activity belong to the SIMIBI clade of P-loop GTPases (145) and are related to other kinases within that clade such as the protein tyrosine kinase Etk. Members of the Clp1 family catalyze ATP-dependent 5′ phosphorylation during tRNA ligation following intron excision in certain eukaryotes (146–148). Clp1 and its paralogs have also been implicated in other RNA maturation pathways, including mRNA and rRNA (149–152). Clp1 displays a stark archaeo-eukaryotic phyletic distribution, with some bacteria likely acquiring it via horizontal gene transfer (HGT). While Clp1-mediated 5′ nucleic acid end phosphorylation is observed in archaea (153), endogenous substrates await further characterization. Prokaryotic genome context provides few clues to Clp1 function, although in a subset of crenarchaea Clp1-like domains are genomically linked to YqgF-like nucleases of the RNaseH fold (154,155) (Supplementary Material). YqgF nuclease activity has been linked to a range of nucleic acid-related functions (156–158); supporting a role for Clp1 in nucleic acid end processing in prokaryotes, perhaps interacting with a range of substrates mirroring the current experimental evidence for its eukaryotic counterparts.
Phosphate group-removing domains
A wide range of phosphoesterase domains displaying distinct structural folds, which catalyze either removal or alteration of the linkages of a phosphate group, have been identified as functionally associating with RNA ligases (Figure 2D). These domains display a characteristic interchangeability in the domain architectural network, suggesting repeated acquisition/displacement of phosphoesterase domains, even in instances where the RNA ligase with which it is associated belongs to the same family (Figure 3A). The most widespread of these is the 2H phosphoesterase domain (159), which was initially characterized as a cyclic phosphodiesterase (160,161) involved in tRNA ligation following intron removal (162,163). RNA ligase-associated 2H domains break 2′,3′-cyclic phosphates, yielding a free 3′ OH and a 2′ phosphate group which might then be processed further (see below; Figure 1C) (162,164).
An additional, well-studied phosphoesterase domain associating with RNA ligases belongs to the Rossmannoid fold HAD superfamily domain of phosphoesterases (Figure 2D) (165). Experimentally-characterized RNA ligase-associated HAD domains, in a reaction typical of the HAD superfamily (166,167), cleave the cyclic phosphate group from the 3′ end of a RNA fragment, yielding a free 3′ OH at the end for the ATP-grasp ligases to join the RNA fragments (Figure 1C) (168,169). A third phosphatase belongs to the calcineurin-like fold (Figure 2D), which contains various well-characterized phosphatases like the 2′-3′ cAMP phosphodiesterases, protein phosphoserine phosphatases, and sphingomyelin phosphodiesterases as well as the bacterial SbcD and yeast MRE11 nucleases (170,171). Calcineurin-like phosphatases mechanistically hew closely to HAD phosphatases, removing the cyclic phosphate group from the 3′ end of the RNA fragment (Figure 1C) (142).
A number of other phosphoesterase domains with distinct protein folds show propensity to act as phosphatases on nucleotide or nucleic acid substrates (Figures 1C, and 2D). These include the HD, synaptojanin-like (also termed ‘Exonuclease-Endonuclease-Phosphatase (EEP)’ superfamily), PTPase and HIT-like superfamilies (Figures 1C and 2D). HD domains (172) have been linked to a range of phosphoesterase activities including removal of phosphate groups by the SpoT enzyme (173), cNMP phosphodiester bond cleavage (174), and 5′-deoxyribonucleotide phosphoester bond cleavage (175). The synaptojanin-like superfamily, including the inositol polyphosphate 5-phosphatases (176), also contains members with nuclease and potential nucleotide phosphatase activity (177). Members of the PTPase superfamily, while primarily studied as protein tyrosine or dual-specificity protein serine/tyrosine phosphatases, also operate on other substrates including lipids and tRNAs (178–182). Finally, members of the HIT-like superfamily have been shown to be nucleotide phosphoesterases that hydrolyze the 5′-adenylated RNA that has been misincorporated into genomic DNA (183). Indeed such adducts can also conceivably form as a result of aborted ligation during RNA repair. While none of these phosphoesterase domains have been experimentally characterized in RNA-ligation-related end-processing systems, we present evidence via contextual inference for their potential involvement in RNA repair comparable to the previously characterized phosphoesterases described in the preceding paragraphs (see below).
Domains catalyzing modifications of RNA termini
KptA-like 2′-phosphotransferase
As noted above, action of end-processing enzymes such as those of the 2H superfamily result in termini with a phosphate group attached to the 2′ end. Such ends are cleaned by the KptA 2′-phosphotransferase (Figure 1C and 2E), which contains a catalytic domain of the ADP-ribosyltransferase (ART) superfamily fused to an N-terminal La-type winged helix-turn-helix (wHTH) domain (81,184–187).The ART domain uses NAD+ as a substrate to attack the 2′ phosphate group on tRNA with the ADP-ribose moiety. The phosphate thus leaves as the 1′,2′-cyclic phosphate ADP-ribose (Figure 1C) (188–190). This compound is then processed by another enzyme containing a catalytic Macro domain (Figure 2E), in conjunction with a 2H enzyme, to release ADP-ribose by removing the phosphate group from it (191).
RtcA cyclase
RtcA belongs to the same superfamily as the 5-enolpyruvylshikimate-3-phosphate (EPSP) synthases (Figure 2E), which catalyze the formation of EPSP from phosphoenolpyruvate and 3-phosphoshikimate in biosynthesis of aromatic amino acids (192). This superfamily contains a version of the IF3-C fold, an inferred ancestral RNA-binding fold within which enzymes acting on nucleic acids have repeatedly emerged (193). RtcA has been shown to catalyze cyclization of termini with 3′-phosphate or 2′-phosphate groups to 2′,3′ cyclic phosphate (194,195) (Figure 1D). Predominant genome associations of RtcA with RtcB suggest that they might function together in 3′-end processing during RNA ligation (see discussion below). Additional roles for RtcA could include cyclic termini recognition/cyclic phosphate end maintenance, a possibility for eukaryotic RtcA acting on U6 snRNA (196).
Hen1 methyltransferase
Hen1 is a two-domain protein combining a N-terminal Hen1-L domain which couples it to RNA ligase systems (142,197) and the S-adenosyl methionine-dependent Rossmann fold methyltransferase domain of the Hen1 clade, which methylates the 2′ OH on the 5′ RNA fragment prior to ligation (Figure 1B). This modification is thought to effectively ‘seal’ the re-ligated tRNA against further toxin attack by introduction of the methyl group which cannot be accommodated by RNase active sites (142,197–201). Notably, Hen1 methyltransferases appear to have been recruited to similar roles in eukaryotes: modification of free 2′ OH groups at 3′ ends of small RNA has been observed in animal piRNAs (202,203), ciliate scnRNAs (204), and plant miRNAs (205,206). In plant miRNAs, the protection afforded by Hen1-mediated methylation, at least in some contexts (207), competes with the degradation-stimulating polynucleotide tags added by TRF clade Polβ NTase enzymes (208,209).
Accessory domains involved in RNA repair
Ligand-sensing CARF and WYL domains
The RtcR protein is a σ54 transcriptional coactivator of the operon coding for RtcA and RtcB (Figure 4A, E–M, Supplementary Material). It displays the characteristic architecture of these coactivators with a central ATPase domain of the AAA+ superfamily (210,211) and a C-terminal HTH domain (212). The N-terminal region was long assumed to harbor the domain responsible for recognizing a signal which would activate expression of RtcA/RtcB (194): a recent study identified this region as harboring a CARF domain (213). In distinct conflict systems, including CRISPR/Cas, the CARF domain, like the WYL domain with which it is genomically linked, is predicted to specifically bind nucleotide and nucleotide-derived ligands in a range of regulatory roles (Figure 4A) (55,213). While the RtcR ligand remains to be experimentally identified, the presence of a CARF domain suggests that it might function as a sensor for cyclic termini of RNA to regulate RNA ligation-based repair systems (see below). Similar to other conflict systems (213), WYL domain proteins are also present in several RNA ligase systems and predicted to sense similar ligands and regulate transcription via their fused HTH domains (Figure 4E–M). Despite their shared function, the mechanism of regulation by transcription factors with the WYL and CARF domains is likely to be distinct, with the former being a repressor that relieves a transcriptional block on ligand sensing and the latter directly activating transcription (see below) (213).
Figure 4.
Genome context network of RtcB ligase-based and related RNA repair systems. (A) Contextual network; domains displaying mutual exclusivity wrapped together in dotted lines. (B and C) Structures of RtcB ligase-associating domains. Despite multimerization in crystal structures, archease exists as monomer in solution (216). Swapped strand in obligate dimer archease structure colored in cyan. (D–R) Representatives of RtcB RNA ligase-centered RNA repair and related systems. The system of labeling is as in Figure 3.
Archease domain
The archease domain displays a pronounced archaeo-eukaryotic phyletic distribution, although many bacteria have acquired it via horizontal gene transfer (HGT) (214). It contains a unique fold which appears to have emerged from a combination of two homologous ancestral 3-stranded units (Figure 4B) (215). Archease has been shown to function as a chaperone which promotes reaction turnover of tRNA ligation during intron excision in archaea and eukaryotes (216–218), possibly by enhancing formation of the RtcB-guanylate intermediate during the reaction (217). Archease has also been linked to a similar functional role during tRNA methylation in the archaeon Pyrococcus (219) and is also found in many bacteria which do not have tRNA introns (218), suggesting it functions as a general chaperone in distinct processes, including RNA ligation following attack by RNase effectors in bacteria (218).
Rot/TROVE proteins and their non-coding YRNA partners
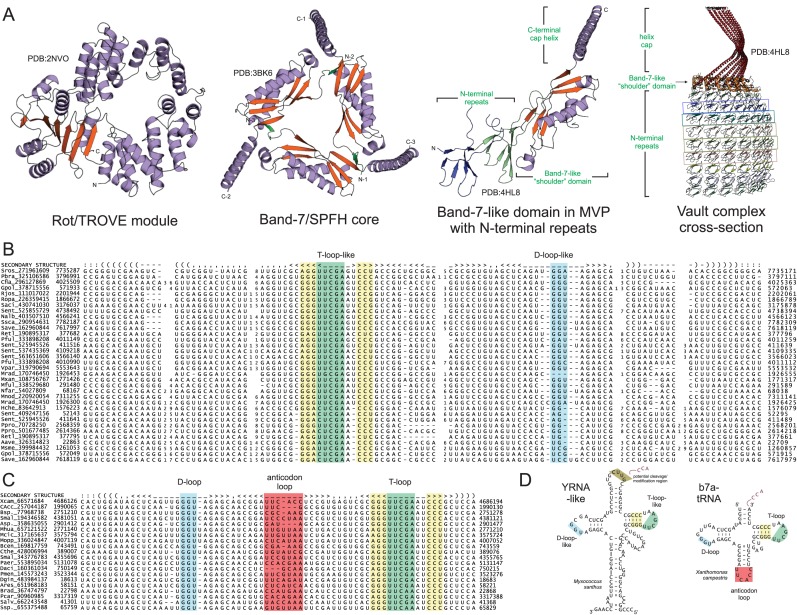
Proteins of the Rot/TROVE superfamily contain an eponymous module (220,221) comprised of multiple bihelical repeats fused to a C-terminal vWA domain (Figure 5A) (222,223), or more rarely, a TerD domain (52). Members of this superfamily are key components of several ribonucleoprotein complexes including the animal telomerase and vault complexes and the eukaryotic and bacterial Ro RNP complex (220,221) which had been linked to stress response (224), UV irradiation response (225), and non-coding RNA quality control (226–228). Additionally, recent studies have shown the bacterial Ro RNP to associate with the polynucleotide phosphorylase (PNPase), an exoRNase which plays a key role in degradation of misfolded RNA (226,229). Related proteins with the Rot/TROVE module and vWA proteins are found associated with RNA ligases (195,230), pointing to a further role in RNA repair. Interestingly, the Rot/TROVE protein associates with a non-coding RNA, the YRNA, which appears to ‘tether’ it and the PNPase and helps position the target single-stranded RNA in a ring-like structure formed by Rot/TROVE for degradation (Figure 5A) (230). A recent analysis revealed YRNAs to be present across diverse bacterial lineages (231). Our analysis, using nucleotide regions adjacent to the Rot/TROVE and vWA protein as starting points for iterative homology searches combining the BLASTN and Infernal programs (232), verified earlier findings but recovered a more expansive set of predicted YRNAs (Figure 5B, Supplementary Material). Notably, we observe that YRNAs almost always occur in tandem on the bacterial genome, with between 2–4 repeats present next to the Rot/TROVE gene (Figure 4G, L, Supplementary Material), thereby generalizing a previous observation in Salmonella enterica (230).
Figure 5.
Structures of RtcB ligase-associating and related domains. (A) Individual domains in multi-domain proteins are labeled in green. Swapped strands in obligate Band-7 core domain are colored green. Repeat domains found N-terminal to core Band-7-like domain in MVP are colored as individual units. Interacting Band-7-like N-terminal repeat domains found in structure of the Vault complex are colored as per repeat. (B and C) Multiple sequence alignments of YRNAs (B) and b7a-tRNAs (C). Genome sequence position is provided to the left and right of sequences. Predicted secondary structure features are given on the top line of alignment in WUSS notation. Poorly-conserved regions replaced by numbers. (D) Secondary structure depictions of YRNA and b7a-tRNAs. Key features are shaded to match (B and C). Potential modification/cleavage region for YRNA described in (231) shaded in brown. All domain and organism abbreviations are provided in Supplementary Material.
Band-7 domain proteins and their non-coding RNA partners
In several bacteria we found the genes coding for ROT/TROVE proteins and YRNAs to be missing in the genomic neighborhoods of RNA ligase systems. Instead these coded for a protein containing the Band-7 domain (also referred to as the SPFH or PHB domain) (233–236), mutually-exclusively to the former genes (Figure 4A). The Band-7 domain is further related to the shoulder domain of the Major Vault protein (MVP) of the small RNA associated Vault complex (Figure 5A) (237); it is also distantly-related to the Ribosomal S3AE superfamily of proteins, the bacterial flagellar basal body-associated protein FliL, and the Bacillus YqfA sublancin immunity protein (238) (Burroughs AM, Aravind L, personal observations). However, the functions of the Band-7 domain have long remained largely enigmatic though members of the superfamily have been implicated in membrane lipid recognition (239), facilitating protein-complex recruitment/assembly (240), serine protease chaperone activity (241), and cellular stress response (242). The Band-7 domain proteins associating with RNA ligase systems correspond to the SPFH9 family (234), which notably contains no transmembrane helices, an otherwise common feature in the Band-7/SPFH superfamily. Instead they show a large N-terminal region harboring at least two tandem repeats of a globular α+β domain (Figure 5A, Supplementary Material). Analysis of this N-terminal domain revealed a distant relationship to the repeats observed N-terminal to the Band-7 (MVP-shoulder) domain in the Major Vault Protein which forms the toroidal core of the Vault complex (237,243,244), for which bacterial homologs were also discovered (Supplementary Material). Thus, by analogy, members of the SPFH9 family found in RNA repair contexts could form a multimeric toroidal structure with parallels to the Vault and convergently similar to the ring-like structure of the ROT/TROVE proteins (Figure 5A) (227). Moreover, we detected a distant relationship between this N-terminal repeat domain and the N-terminal region found in all Band-7 superfamily members, suggesting that the repeat may have initially emerged in bacteria through partial duplication of that part of the Band-7 domain.
Analyzing the genomic regions immediately adjacent to Band-7 domain genes using iterative homology searches, we detected a previously-unidentified class of non-coding RNAs distinct from the YRNA family associated with the ROT/TROVE domain proteins (Figure 5C, Supplementary Material). Instead its sequence and predicted structure strongly resembled tRNAs. Accordingly, we named this noncoding RNA the b7a-tRNA, for band-7-associating tRNA. In relatively rare instances, we observe one of these ncRNAs associating with the protein which it does not usually associate, i.e. a YRNA associating with a band7/SPFH protein or b7a-tRNA associating with a ROT/TROVE or both together in the same gene-neighborhood (Figure 4H,L, Supplementary Material). This observation strongly supports a functional equivalence between the two classes of ncRNAs. Additionally, iterative searches with the YRNAs also recovered bona fide tRNAs, consistent with the shared sequence and predicted structural features between these two classes of RNA (Figure 5D) (231). These are strongly suggestive of a common evolutionary origin for the b7a-tRNA and the YRNAs involved in RNA repair from bona fide tRNAs. There is a direct structural equivalence between the tRNA T-loop and the T-loop-like sequence in these repair-related ncRNAs, both of which contain the highly-conserved UCGA motif bracketed by a stem containing the conserved CCC and GGG motifs (Figure 5B–D) (228,231,245,246). On the other side in the characteristic ‘clover-leaf’ architecture the two RNAs contain another equivalent loop corresponding to the tRNA D-loop (Figure 5B–D). The striking sequence and structural parallels between the YRNA and b7a-tRNA families suggests the T-loop-like structure could mediate important interactions in the context of RNA ligation (see below).
The predicted RNA-binding domain, MJ1316
The MJ1316 domain (also called Domain of Unknown Function 504 in pfam (247)), was originally predicted to be a RNA-binding domain based on analysis of its domain architectural contexts (159). Our updated survey of genome contexts revealed a striking tendency for MJ1316 to repeatedly occur in RNA-repair systems (see below, Figure 3). The MJ1316 superfamily contains several well-conserved positively-charged amino acid residues and also a near-absolutely conserved histidine residue in a predicted secondary structure comprised of a single α-helix followed by a β-meander of 3–5 strands (Supplementary Material). These new observations suggest it to be a key player in RNA systems and more specifically implicate it in recognition of RNA 3′ ends, most probably 2′,3′-cyclic phosphate groups (see below).
The PSα domain
Our analysis of the contextual connections of RNA repair system components revealed a poorly-characterized, all α-helical domain typified by the human PDCD5/TFAR19 and budding yeast Sdd2 proteins. Accordingly, we named it the PSα domain (Figure 4C). Previous studies had dubiously described the archaeal version of the PSα domain (MTH1615) from Methanobacterium thermoautotrophicum as a dsDNA-binding domain (248). However, this role is not supported by subsequent studies (249,250) and we recommend against using the name dsDNA-binding, as currently provided by the Pfam database. PSα domains show a strongly archaeo-eukaryotic phyletic pattern, which combined with its presence in archaeal ribosomal superoperons, points to a role in association with ribosomes (see below, Figure 4R, Supplementary Material).
THE DIVERSITY OF RNA REPAIR SYSTEMS
The above survey indicates that the chief domains involved in RNA repair are relatively few; however, they are combined in several distinct ways in diverse domain architectures and mobile gene-neighborhoods to spawn a considerable variety of RNA repair systems. At the highest level these systems can be divided into those which are centered on a ligase domain and those which are not. The former category is by far the most widespread across Life and can be further divided into those containing either an ATP-grasp or a RtcB ligase. All characterized RNA ligase families are present across several distinctive systems, many of which display similar syntactical themes of domain architectural and operonic linkages suggesting a certain functional equivalence between them. In the following sections, we discuss a unified ontology covering the syntactical diversity of these RNA-repair systems.
ATP-grasp RNA ligase systems
These systems can be further classified based on their complexity, which spans the entire range from single component standalone ligases to multicomponent systems where the ligases are combined with increasing numbers of additional components. In the multi-component systems, the basic set of additional components are P-loop kinases and phosphoesterases. More complex systems are marked by the persistent presence of additional enzymatic domains such a methylases and Polβ NTases (Figure 3A–Q).
Standalone ATP-grasp ligases
The simplest system consists of ligases lacking strong operonic linkages, but containing characteristic domain fusions or C-terminal extensions. The best studied of these systems are the Rnl2-like ligases with a distinctive C-terminal extension (Figure 3B) which specifically recognizes nicked duplex RNA (111,112). They have been shown to perform duplex RNA nick repair, both in the context of kinetoplastid RNA editing (REL1 and REL2 enzymes) (251,252) and in bacteria and their phages (77,253). These are also found in haloarchaea, large nucleo-cytoplasmic large DNA viruses (NCLDVs) (254), and diverse eukaryotic lineages (Supplementary Material). While the in vivo ligation targets of bacterial and phage Rnl2-like ligases are poorly-understood, one possible substrate are nicked RNAs generated by uncharacterized RNA toxins (Table 1). The so-called Rnl5-like ligases (76,109) are a subfamily of Rnl2-like ligases and appear to similarly act as self-contained, single protein systems. These lack any C-terminal helical extension, and are instead N-terminally fused to an RNA-binding OB fold domain (Figure 3C). Rnl5 has been shown to repair tRNAs with cleaved anticodon-loops, and are found across several disparate eukaryotic, bacterial, and phage lineages (76,109) (Figure 3C, Supplementary Material).
Systems combining Rnl1-like ligases with kinase and phosphoesterase components
These were first identified over four decades ago in the phage T4 and combine a ligase module of the Rnl1-like family (22) with a P-loop kinase and either a HAD domain phosphatase or a HD domain in at least two distinct systems (Figure 3D–E). Such enzymes are observed in phages, a limited set of actinobacteria, firmicutes, cyanobacteria, and NCLDVs; in NCLDVs all three domains are fused to the same polypeptide while elsewhere the ligase is a standalone protein with a C-terminal extension and the kinase N-terminally fused to the phosphoesterase (often referred to as a ‘polynucleotide kinase’) is encoded by an adjacent downstream gene (Figure 3D–E, Supplementary Material) (163). In phages, these systems repair host nuclease-mediated cleavage of host tRNALys to reinitiate translation of phage mRNAs (163,255). Similarly, the genome-encoded bacterial versions could be deployed against potential toxin attacks. Solo versions of the ligase seen in this system are present in a limited set of eukaryotes including the heterolobosean Naegleria, the mollusk endosymbiont Capsaspora, and certain amoebozoans (Supplementary Material). These eukaryotic versions could still associate with kinases and/or phosphoesterases, although cognate HAD family orthologs are not present in these genomes.
A similar, well-studied system combines a Rnl1 family ligase with the P-loop kinase and a phosphoesterase of the 2H superfamily in the same polypeptide, typified by yeast Trl1p (Figure 3F). This system has been studied in tRNA ligation following intron removal (164) in fungi and plants (256–259). Recent research identified comparable but divergent versions in ciliates (260,261), representatives of the SAR (Stramenopiles-Alveolates-Rhizarian) lineage, amoebozoans, and certain marine metazoans including the cephalochordate Branchiostoma (148). We further identified homologous versions in certain kinetoplastids (Supplementary Material). In Branchiostoma the Rnl1 ligase is a standalone protein (Figure 3G) which participates in tRNA intron splicing with cognate kinase and 2H domains encoded elsewhere in the genome (148). A comparable process could be active in several organisms where orthologous ligases occur as standalone proteins (Figure 3G, Supplementary Material). Variants of the basic three-domain architecture are observed in apicomplexans, which tend to combine the core with a second copy of either the ligase (Toxoplasma, Hammondia, and Neospora), the 2H domain (Eimeria), or the kinase (Perkinsus) domains (Figure 3F).
Beyond these, a poorly-understood system combines a Rnl1-like ligase with a calcineurin-like phosphoesterase and a P-loop kinase often in the same polypeptide (Figure 3H). These again span several bacterial and phage lineages while showing sporadic representation within any single lineage (Supplementary Material). Two major variations to the core system are observed: (i) in several lineages, the ligase is found fused directly to the kinase with the calcineurin-like phosphoesterase encoded as a solo protein; (ii) sporadic neighborhood association is observed with KptA and Thg1, a Rot/TROVE+vWA protein, and a Macro domain (Figure 3H, Supplementary Material).
Systems predominantly centered on Rnl2-like ligases with kinases and phosphoesterases
Two additional uncharacterized systems are predicted to act similarly to the above but predominantly contain a Rnl2-like family ligase. The first, sporadically observed across several bacterial lineages (Supplementary Material), combines two genes respectively coding for a Rnl2-like family ligase and a fusion of a N-terminal HD phosphoesterase and P-loop kinase domains (Figure 3I, Supplementary Material). Several variations to the core of this system are observed: (i) in firmicutes, the HD and kinase order is inverted, (ii) operonic linkage with a distinct ligase system is observed in some bacteriodetes and α/γ-proteobacteria, combining with the RtcB ligase and RF-1 domains (see below, Supplementary Material) and (iii) in some cases the system includes genes for an additional P-loop kinase and a Thg1 polymerase. The Thg1-kinase gene pair, which is also seen in at least one other system (Figure 3K), could specifically function in repairing 5′ ends via its polymerase action in combination with ligation of an endonucleolytic cleavage.
The second system, sporadically observed in several eukaryotic lineages including fungi, the nucleomorph Guillardia, kinetoplastid Angomonas, and oomycete Phytopthora, fuses Rnl2-like ligase modules to a N-terminal 2H fold phosphoesterase and an additional phosphoesterase domain of the PTPase superfamily, and a P-loop kinase (Figure 3J, Supplementary Material). This system is found in many of the same organisms containing one or more copies of the architecturally similar Rnl1-like ligase, 2H phosphoesterase, and P-loop kinase module described above (Figure 3F–G). This suggests a functional differentiation between these systems or a degree of backup between them. In the current system the PTPase domain could potentially remove 2′ phosphate groups from the RNA fragment after the 2′,3′-cyclic phosphate is cleaved by the 2H domain (see above). Another variation is observed in Emiliania, which lacks the 2H domain but contains a 3′→5′ exonuclease of the RNase H fold between the PTPase and kinase domains (Figure 3J).
ATP-grasp ligases associated with a Hen1 methylase and/or a novel Polβ NTase
The most common of these displays at its core a calcineurin phosphoesterase, a P-loop kinase and a Rnl4-like ligase, all fused together in the same polypeptide (Figure 3K) (171). While some representatives contain only this core configuration, it is mostly additionally found to associate with a Hen1 RNA methylase protein (Figures 1B, and 3K). In several gene neighborhoods, distributed broadly across bacteria, these are further coupled to a gene encoding a novel family of Polβ NTase proteins, often sandwiched between those coding for the Hen1 and the three-domain protein with the ligase component (Figure 3K). Due to its repeated association with RNA ligases, we hereafter refer to this family as the RlaP (RNA ligase-associating Polβ) NTase family (Figures 3A, K–M, and 4A). These systems less-frequently display additional associations which might potentially represent adaptations to further escalation of the arms-race in the conflict. These include combinations with P-loop kinase, Thg1, Macro, and Rot/TROVE+vWA domains (Figure 3K, Supplementary Material). We also observe that on a few occasions the Rnl4-like ligase is encoded as a solo domain, found adjacent to a further P-loop kinase and HAD phosphatase fusion protein as found in the above Rnl1-centric systems (Figure 3K, Supplementary Material). Additionally, if Hen1 is not operonically linked in these systems, it might be encoded elsewhere in the genome and has been shown to physically associate with the Rnl4, HAD and P-loop kinase domains via its Hen1-L domain (199).
A distinct system from baculoviruses and entomopoxviruses combines, in the same polypeptide, a member of the RlaP NTase family with a Rnl2-like ligase (Figure 3L, Supplementary Material). The same linkage, this time with a Rnl1-like ligase, is observed in some bacteria (operonic) and some eukaryotes (domain fusion in Acanthamoeba and Naegleria); however, in the eukaryotic proteins the ligase is predicted to be inactive based on the lack of conservation of catalytic residues (Figure 3M, Supplementary Material). This suggests that it might merely serve as a RNA-end-recognition module which localizes the action of the RlaP NTase. RlaP NTases are also coupled with RtcB RNA-ligases as discussed below. These multiple independent combinations between RNA ligases and RlaP NTases further emphasizes the importance of the functional interaction between these two modules.
ATP-grasp ligase systems containing polyA polymerase family NTases
A previously uncharacterized system with complex architecture found sporadically in bacteria combines genes coding for a Rnl2-like ligase, a P-loop kinase, and sometimes a HD phosphatase with an additional gene encoding a large, multi-domain protein. This protein, from N-terminus to C-terminus, combines a MJ1316 domain, a synaptojanin-like phosphoesterase, a 2H phosphoesterase, and a Polβ NTase module (Figure 3N, Supplementary Material). This Polβ NTase module belongs to the eukaryotic polyA polymerase (PAP) clade, and shares their characteristic C-terminal PβCD (262), and PAP RNA-binding domains (Figure 3N). The conserved presence of the three domain PAP module suggests a comparable function in adding terminal polynucleotide tails. Presence of this PAP module, distinct from the RlaP NTase from the above systems, is intriguing and suggests possible terminal polynucleotide elongation either competing with or acting in parallel to ligation. The strong coupling of the synaptojanin-like and the 2H phosphoesterase domains suggests a cooperative relationship, with 2H acting on the cyclic phosphate followed by the former hydrolyzing the 2′ phosphate (Figure 1C). As noted above, the MJ1316 domain might act in cyclic phosphate recognition to recruit these systems to damaged RNA.
Large eukaryotic ‘Swiss-army knife’-type ligase proteins
Parallel to the above bacterial proteins, we observed several previously-unreported large eukaryotic proteins combining the ligase module to multiple enzymatic domains related to RNA repair. These include a protein from the haptophyte Emiliania containing a Rnl1-ligase coupled to two HAD domains from distinct families, including one previously uncharacterized family (Supplementary Material), a RNase H fold 3′→5′ exonuclease, 2H, P-loop kinase, prim-pol, and Ras-type GTPase domains (Figure 3O). The stramenopile Aphanomyces fuses a Rnl2-like ligase module with a PAP-like Polβ NTase module, another previously-uncharacterized HAD family (Supplementary Material), 2H, P-loop kinase, MJ1316, and RWD domains in a single protein (Figure 3O). Another stramenopile, Phaeodactylum tricornutum, combines a Rnl1 ligase with MJ1316, 2H and P-loop kinase domains. Finally, the chlorophytes Volvox and Chlamydomonas contain proteins combining Rnl1-like ligases with a mRNA-capping guanylyltransferase module, a P-loop kinase, and sometimes an ubiquitin (Ub)-ligase recruiting F-Box domain (Figure 3O, Supplementary Material). These combinations suggest the proteins potentially coordinate multiple repair activities, with mRNA as one potential substrate. Importantly, multiple links to Ub-binding or conjugation related domains suggest, as noted previously in eukaryotes (263), RNA processing activities might be regulated via ubiquitination.
ATP-grasp ligase systems coupled with endonuclease domains
A unique system, belonging to a vast assemblage of previously-undescribed conflict systems centered on the presence of a distinct clade of P-loop NTPase domains of the ABC superfamily (Iyer LM, Burroughs AM, Aravind L, unpublished), combines a gene encoding the said ABC NTPase with those coding for a distinct lineage of the Rnl2-like clade of ligases, a HNH endonuclease, and a PIN domain RNase (Figure 3P, Supplementary Material). In a few organisms the PIN and ABC NTPase are absent and the ligase and HNH are fused together in the same protein. The system is highly mobile, sporadically present across several diverse bacteria and at least one archaea and archaeal virus (Figure 3P, Supplementary Material). By analogy to other conflict systems containing effector domains and ABC NTPases, like the HEPN domain RNase-containing RloC system (264), the ABC NTPase likely functions to sense nucleotide/nucleic acid-derived signals triggered by viral infection and regulate the activity of the associated effector domains. HNH and PIN domain nucleases are known to be deployed as effectors in multiple distinct conflict systems, the former usually against DNA and the latter against RNA (Table 1). However, the combination of a RNA-ligase domain with these nucleic acid-targeting effectors is, to our knowledge, without precedent. It is conceivable that this ligase helps repair cellular RNAs cleaved by the effectors of the invader even as the HNH and PIN domains target the invader directly or indirectly. It is also possible that the ligase helps heal cellular RNAs (e.g. tRNAs) that were cleaved to limit the infection of the invasive entity once the infection is overcome by action of the HNH and/or PIN effectors.
Another group of systems exclusive to both bacterial and archaeal extremophiles are typified by two linked genes respectively encoding a Rnl3-like ligase and a distinct PIN domain RNase (Figure 3Q). Unlike classical T-A systems, the two genes are in the opposite direction, typically tail-to-tail (Supplementary Material). The exclusive presence of this system in extremophile genomes and the decoupling of the Rnl3-ligases from the PIN nuclease in certain genomes suggest that it might perform specific regulatory functions as opposed to being a classic T-A system. Such a Rnl3-type ligase has been experimentally shown to catalyze circularization of single-stranded RNA in vitro (265,266); hence, one regulatory possibility might involve alternative circularization and linearization or cleavage and re-ligation of specific RNAs in response to environmental stress. Such regulation has been reported to stabilize the circularly-permutated signal recognition particle (SRP) RNA in the thermophile Thermoproteus; this system could similarly act in RNA stabilization (267).
RtcB RNA ligase systems
Despite the initial functional characterization for RtcB ligases in tRNA intron removal/ligation, the entire range of endogenous substrates ligated by these enzymes remains unclear. Genomic evidence suggests that RtcB is the core of diverse ligase systems, whose rapid divergence and mobility across distinct prokaryotic lineages indicate a role in RNA ligation during conflict (Figure 4A, D–L).
Archease-coupled systems
RtcB ligases associate frequently with the archease chaperone protein across a range of distinct prokaryotic lineages (Figure 4D, Supplementary Material) (216); this pair participates in tRNA intron splicing in eukaryotes and archaea (216–218). The archease-RtcB gene pair is frequently observed in diverse bacteria, even those lacking introns in their tRNAs (218). This supports the inference that ligation catalyzed by this pair is likely to be important for repair of RNA damaged in biological conflicts. In certain bacteria, the archease-RtcB gene pair is combined with further components (Figure 4D, Supplementary Material): (i) an association with phosphoribosyltransferase (PRTase) and a α/β hydrolase domain, which can be fused together on the same polypeptide or encoded in separate genes. In certain bacteria this gene neighborhood might include a further gene coding for a Rossmann-fold methylase which is distinct from Hen1. It is possible that the PRTase and the α/β hydrolase generate a nucleotide signal similar to what has been previously proposed in the Ter system that activates RNA repair (52). The additional methylase in some of these systems might function in end-protection akin to Hen1. (ii) A distinct association occurs with a 2H phosphoesterase, either in conjunction with the archease-RtcB dyad (e.g. Rubrobacter) or independent of it, with the 2H domain directly fused to the archease (e.g. Waddlia) (Figure 4D). (iii) An association with a RNA-base modifying Rossmann-fold methyltransferase and its PUA domain partner is found in certain members of the archaeal lineage of Thermococcales (216). This association is consistent with the previously reported chaperone-like role of archease with respect to the RNA-base methylases (219).
RtcB systems with transcription factors
Though comparably or even more mobile in bacteria, this class of operons shows largely mutually-exclusive phyletic profiles to the RtcB-archease operons (Figure 4A, E–L, Supplementary Material). This points to their functional equivalence in conflict-related RNA repair, albeit via different mechanisms as suggested by the distinctness of their components beyond RtcB. There is considerable variation in the additional components of this system across bacteria beyond the basic core formed by the RtcB (Figure 4A, E–L): (i) the previously characterized versions contain a gene for the RtcR transcription factor (TF) on the opposite coding strand (Figure 4E) (194). (ii) We noted that RtcR is frequently replaced by another TF combining a WYL domain (see above) with a c1-repressor-like HTH domain and occasionally a second distinct HTH domain or on rare occasions, in lieu of the HTH, a MetJ/Arc-like ribbon-helix-helix DNA-binding domain. Some of these neighborhoods might also encode a second such WYL fused to a distinct HTH domain (Figure 4E). The mutually exclusive phyletic patterns of the variants respectively with CARF (RtcR) and WYL domains as the sensors of the activating ligands likely reflect the distinct modes of transcriptional regulation of the two TFs (213) with RtcR primarily acting as a ligand-induced activator of RtcA-RtcB expression (194,195) and the WYL-cHTH protein as a repressor (see above, Figure 4A). (iii) The repeated loss of the RtcA domain yielding a two-gene operon (Figure 4F)—even in the proteobacterial lineage, where RtcA is widely found, it has been repeatedly lost (Supplementary Material). In this regard it should be noted that RtcA is never found in systems with a WYL TF. This suggests that it emerged relatively late in prokaryotic evolution and performs a more ancillary function in RNA repair. Given that the RtcA can generate 2′-3′ cyclic phosphates at RNA termini, it might function both to recognize cleaved ends and channel them for ligation by versions of RtcB ligases with either a preference for substrates with a cyclic end or under certain specific conditions (Figure 1A,D) (195).
Additional components accreted to the RtcB-TF systems
Versions of the above operons have often accreted additional functional modules. One such is a protein with ROT/TROVE and vWA domains (195,220,221). As noted above, a gene for this protein occurs in mutually exclusive gene-neighborhoods with one coding for a band-7/SPFH domain protein of the SPFH9 family. These systems further frequently include the above-described ncRNAs, YRNA and b7a-tRNA, typically adjacent to the ROT/TROVE+vWA and band-7/SPFH genes, respectively (Figure 4G, Rot/TROVE neighborhoods and 4H, band-7/SPFH neighborhoods). While the protein components are evolutionarily unrelated, their mutual exclusivity indicates that they together with the counterpart ncRNA are likely to be functionally equivalent (Figure 4A). The further mutual exclusivity observed between these systems and the archease-RtcB systems in bacterial genomes could point to a comparable chaperone-like role for these ribonucleoproteins (RNPs) in facilitating the assembly of RtcB-containing complexes, or potentially as a scaffold which augments the ligase activity. Moreover, their occasional combination with systems centered on Rnl1-like and Rnl4-like ligases suggests that they might more generally perform such a role even in other ligase systems (Figure 3H and K). This proposal is supported by recent experimental work which points to a general role for the Rot/TROVE+vWA-YRNA RNP in the correct positioning of a RNA substrate and its enzyme partner (230), paralleling the results from the archease systems (216–218).
Two further components are observed in these neighborhoods, although at a lower frequency (Supplementary Material): the first is a zinc ribbon protein which appears to occur specifically in those systems also containing the RtcR TF (Figure 4J). This zinc ribbon family is largely restricted to bacteria with occasional lateral transfers to eukaryotes. In bacteria it is potentially it is encoded by within multiple distinct gene-neighborhoods potentially coding for conflict systems (Burroughs AM, Aravind L, personal observations). These observations suggest the domain might function in recognizing specific macromolecules in multiple conflict-related contexts. The second is the KptA phosphotransferase domain which potentially helps cleanup 2′-phosphates in RNA that might have escaped ligation via a RtcB-dependent ligase (Figure 4K).
Coupling of the RlaP NTase to RtcB-TF systems
We frequently observed the RtcB-TF gene-neighborhoods include the RlaP NTase also found associating with the ATP-grasp RNA ligases (Figure 3K–M). The RlaP NTase coupling is observed across the various RtcB-TF systems described above but not in any of the RtcB-archease systems (Figure 4A, J–L). Less frequently, certain bacteria possess systems comparable to the RtcB systems described above where there is only a RlaP NTase component but no RtcB (Figure 4M). Further, the RlaP is found embedded in a subset of CRISPR/Cas operons (Figure 4N). Additionally, the RlaP NTase might also be found as a standalone protein in several caudate bacteriophages that infect most major bacterial lineages (Supplementary Material). The RlaP family shows several distinct features absent in other members of the Pol-β NTase superfamily, such as a characteristic C-terminal α-helical extension beyond the core NTase domain which has a nearly absolutely conserved lysine and arginine that might have a role in catalysis and/or substrate recognition (Supplementary Material). Given the distinctness of this family, it is likely to play a role in RNA repair different from what has been observed for the CCA-adding enzyme and polyA polymerase clade. Its strong association with mechanistically distinct RNA ligases utilizing different substrates suggests that the action of the RlaP enzymes is likely closely coupled with ligase activity. Hence, it probably operates on RNA ends to selectively modify them alongside repair through ligation.
RtcB-RFH systems
Prior genome analyses identified the coupling of RtcB with a distinctive paralog of class-I release factors (RFs), termed RFH in bacteria (Figure 4O, Supplementary Material) (268). Canonical RFs directly recognize and associate with stop codons in the ribosomal A-site leading to translation termination (269), and it has been persuasively argued that the RFH family retains the sequence and structural features necessary to act similarly (268). At the time of that study, RtcB function had yet to be elucidated. In light of what we currently know of RtcB, we can now predict that this two-gene system likely couples release of the nascent peptide chain with RNA repair. Given that RNA toxins are known to act by ‘jamming’ the ribosome, such a system might be useful in relieving the blocked ribosome while also healing damaged rRNA or tRNA (Table 1, see above). Unlike the above systems, RFH-coupled systems are not found in mutually exclusive genome contexts, supporting a non-redundant role with the other RtcB systems (Supplementary Material). At least two stramenopiles (Thalassiosira and Phaeodactylum) contain a RFH module N-terminally fused to the MJ1316 domain (Figure 4P). This might function analogous to the bacterial systems with MJ1316 sensing a damaged RNA-terminus while RFH acts to release peptide chains.
RtcB-PSα system
In thaumarchaeota we identified a previously-undescribed, conserved system linking RtcB with genes coding for proteins with a PSα domain and a further uncharacterized domain (Figure 4Q). In addition to its linkage to the ribosomal super-operon, in certain euryarchaea the PSα domain protein is also operonically-linked to the MJ0690-type PP-loop domain, an enzyme previously predicted to participate in RNA-modification pathways (Figure 4R, Supplementary Material) (270,271). Hence, it is possible that the PSα protein recruits RtcB to the ribosome to allow ribosome-linked RNA repair in thaumarchaeota. Additionally, it could also play a role as a chaperone or a scaffold as suggested for components in the above systems, consistent with characterized roles in eukaryotes (249,250).
Systems lacking a canonical ligase component
The available genome data points to systems containing components with several of the domains clearly characteristic of RNA repair but lacking a canonical ligase component. While some of these systems are likely to participate in RNA repair steps other than or separate from ligation, the remaining are likely to function in conjunction with any of the ligase systems that have been described above which are encoded elsewhere in the genome.
Potential RNA end-processing systems with the MJ1316 domain
Several systems defined by conserved gene-neighborhoods and multi-domain architectures combine the MJ1316 domain, predicted to play a role in damaged RNA end-recognition, with other partners potentially involved in RNA repair (Figure 3A, R–W). Given the above-noted conserved histidine in the MJ1316 domain (Supplementary Material), we cannot entirely rule out the possibility that it possesses some enzymatic activity of its own specific to certain RNA termini. In several proteobacterial lineages, MJ1316 combines with RtcA in its only conserved context outside of association with RtcB (Figure 3R, Supplementary Material). It is possible that here the MJ1316 domain and RtcA function together in discriminating cyclic from non-cyclic ends and helps channel the latter for cyclization. A context conserved in several euryarchaea combines MJ1316 with a MBL domain (Figure 3S, Supplementary Material). As several MBL families (e.g. tRNase Z (128)) possess RNase activity (272), this pairing could suggest recruitment by MJ1316 of RNase activity for end-processing.
Several bacteria contain a protein whose core comprises a MJ1316 domain fused to a C-terminal 2H domain (Figure 3T). Such a protein appears to have been transferred to the animal lineage prior to its radiation and is prototyped by the human protein Leng9 (Figure 3T) (159). It is possible that these proteins function in concert with ligases encoded elsewhere in the genome in processing RNAs with cyclic phosphate ends. Notably, this protein shows considerable variability across animals with numerous lineage-specific combinations of the above core to additional domains. Fusions seen in diverse animal lineages are to either a RNA-binding Zn-knuckle domain or to a Ub-system linked RWD domain or to both of them (Figure 3T, Supplementary Material). Notably, even in the vertebrate lineage, where the core architecture is largely fixed, subtle variations are detected; the core is further elaborated in certain organisms by fusions to the actin cytoskeleton-interacting PBD and BORG_CEP domains and on occasions there is loss of the 2H or Zn-knuckle domains (Figure 3T). Recent studies observed striking expression of mammalian Leng9 in lymphoid cells in response to rinderpest virus infection, suggesting a potential role for these proteins in coping with RNA damage occurring in response to virus infection (273,274).
‘Swiss-army knife’-type proteins with MJ1316 domains
These large proteins thematically resemble those described earlier in the context of ATP-grasp ligases (Figure 3O) but are distinguished from them in lacking a ligase domain and unlike them are found in both eukaryotes and diverse bacteria. However, they all contain a MJ1316 domain. The bacterial proteins are standalone versions of those found linked in conserved neighborhoods with Rnl2-like ligases (Figure 3N, Supplementary Material). These appear to have been laterally transferred to fungi while undergoing multiple circular permutations of the domain order with synaptojanin-like and 2H phosphoesterase domains flipped in the domain architecture and MJ1316 found at the extreme C-terminus (Figure 3U, Supplementary Material). Versions from the choanoflagellid Monosiga, the sponge Amphimedon, and the stramenopile Saprolegnia have thematically similar architectures: at their core, these proteins combine the MJ1316 domain with one or two 2H phosphoesterases, a HAD phosphoesterase domain of a previously-uncharacterized family, a P-loop kinase, and a PAP-like Polβ NTase module (Figure 3V, Supplementary Material). The RWD and potentially RNA-interacting R3H domains are also sometimes found fused to these proteins at the extreme N- and C-termini, respectively (Figure 3V, Supplementary Material). Diverse microbial eukaryotes contain one or more similar proteins, some with loss of the above domains in their architectures (Figure 3V, Supplementary Material). Given their thematic relationship to the above-described ligase systems, it is conceivable that they act as multipurpose end-processing enzymes that work along with ligases encoded elsewhere in the genome.
Prim-pol-centric systems with links to RNAi
Kinetoplastids, heteroloboseans, and haptophytes possess a remarkable set of proteins that contain a highly divergent version of the prim-pol domain. In the haptophyte Emiliania the prim-pol domain is combined with several other RNA-end-processing (e.g. P-loop kinase and multiple HAD phosphoesterases, and 2H), RNA-binding (R3H) and RNase (3′→5′ exonuclease) domains in a manner reminiscent of the above-described ‘Swiss-army knife’ proteins (Figure 3O,V,W). In the heterolobosean Naegleria gruberi, this prim-pol domain is combined with a N-terminal MJ1316 domain, whereas in the kinetoplastid Angomonas deanei, there are further synaptojanin-like and 2H phosphoesterase domains between those two domains (Figure 3W, Supplementary Material). Another kinetoplastid, Perkinsela, has a similar architecture to Angomonas deanei with a RWD domain instead of the MJ1316 domain (Figure 3W). In other kinetoplastids, this prim-pol domain occurs as part of the Dicer-like protein DCL1 (Figures 3X, and 6A). The kinetoplastid Dicer-like proteins include the cytoplasmic DCL1-like (275) and the paralogous nuclear DCL2-like proteins (276) (e.g. Trypanosoma brucei and Leishmania braziliensis), which have been previously noted for the rapid divergence of their RNase III domains, an uncharacteristically large gap between the two RNase III domains, and presence of globular regions in the C-terminal extension of DCL1 (14). Our identification of the prim-pol represents the first definitive domain assignment for any of these previously-uncharacterized regions (Figure 6A). Further analysis led to the identification of the KptA-like La (a RNA-binding winged HTH domain (277)) and KptA-ADP-ribosyltransferase domains (Figure 2E) immediately after the first of the two Dicer RNase III repeats in both the kinteoplastid Dicer-like proteins (Figure 6B). The KptA domain in both of the Dicer-like paralogues appears to have lost most residues required for catalyzing the phospho-transfer to NAD. However, several positively-charged residues specifically implicated in recognition of the 2′ phosphate group on RNA remain well-conserved (Figure 6B, Supplementary Material) (278,279), suggesting the domain retains its ability to recognize RNA ends with such phosphate groups.
Figure 6.
Alignments and evolutionary scenarios of RNA-repair enzymes. Multiple sequence alignment of prim-pol domains (A), KptA domain found in kinetoplastid DCL1 and DCL2 proteins (B). Alignments are labeled as described in Figure 5. Names of experimentally-characterized proteins: orange. Conserved positions corresponding to known substrate binding residues: ‘*’; conserved residues unique to a family, predicted to function similarly: ‘%’. Coloring scheme and abbreviations for organisms are provided in Supplementary Material. (C) Stylized phylogenetic tree depicting relationships between prim-pol families, broadly labeled at the top of the tree. Branches are collapsed at levels containing clearly-delineable and labeled monophyletic groups. Bootstrap values are shown for major branches only (complete tree available in Supplementary Material). (D and E) Major events in the evolutionary history of ATP-grasp-like ligases (D) and the CCA-adding enzyme-like polymerases of the DNA pol-β superfamily (E). Inferred functional and architectural shifts are marked/labeled with red lines/lettering. Dashed lines indicate uncertain origins for a lineage.
The prim-pol domain from these proteins belongs to a previously unidentified family (Figure 6A and C, Supplementary Material). Phylogenetic analysis placed them together with those encoded by the NCLDVs (139), suggesting origins distinct from most eukaryotic prim-pols, including previously-studied prim-pol families in kinetoplastids and some newly identified ones (Figure 6C) (280). While prim-pol domains were previously only known to function in DNA repair and replication, based on their domain architectures, these kinetoplastid-heterolobosean versions can be confidently linked to RNA-processing and repair, thus implicating the prim-pol domain in such activities for the first time. While prim-pol domains are known to generate RNA using a DNA template, the above contextual associations do not support such a role, especially given its presence in DCL1, a cytoplasmic protein (275). Hence, these prim-pol domains are predicted to possess RNA priming and RNA polymerase activity independently of a DNA template.
While the role of such a RNA polymerase activity remains unknown, some interesting possibilities are raised by the data available from kinetoplastid RNAi systems. The two Dicer-like enzymes are only present in kinetoplastids retaining core RNAi machinery, which additionally includes the following components: (i) at least one PIWI domain (281–283) which binds small RNA cargo; (ii) the Maelstrom-like small RNA maturation factor RIF4; (iii) the DCL1 cofactor RIF5 (284) which contains tandem copies of the Staphylococcus nuclease (SNase) domain (14). Notably, the RNA-dependent RNA Polymerase (RdRP), a component of many eukaryotic RNAi pathways catalyzing small RNA transcript amplification, is absent in kinetoplastids (14). Small RNA profiling in kinetoplastids reveals an endogenous affinity of DCL1 for small RNAs derived from diverse mobile elements and repeats (285,286). Hence, one interesting functional prediction, at least for the DCL1 prim-pol domains, could be as a polymerase effectively functioning as the RdRP component. However, the presence of the inactive KptA domain suggestive of 2′ phosphate recognition and the other domain fusions noted above (Figure 3W, Supplementary Material) suggest that the prim-pol domains might have an active role in trypanosome-specific RNA-repair. This could happen in the context of an uncharacterized cytoplasmic RNA-editing process parallel to what has been reported in the kinetoplast (287), or in response to as-yet uncharacterized conflicts. Given that prim-pol domains have previously been demonstrated to be capable of template-independent terminal nucleotide transferase activity (288), they could even play such a role in RNA repair, consistent with architectural similarities observed between the Emiliania and other large ‘Swiss army knife’ repair proteins, which could posit a functional equivalence between the prim-pol and PAP-like polymerase modules (Figure 3O, V and W). In this scenario they might act similar to the PAP-like polymerase modules found in several of the above-described RNA repair systems in the synthesis of terminal nucleotide extensions.
EVOLUTIONARY HISTORY OF COUNTER-EFFECTOR RNA REPAIR
The provenance of RNA repair enzymes
In extant biological systems mRNAs and rRNAs are often produced in considerable abundance and are present in several times the copy number of the genomic coding sequence. Hence, targeting them requires highly efficient effector mechanisms or those that exploit vulnerabilities, such as ‘jamming’ of ribosomes in course of translation. Since the ribosome and core translation apparatus in a form comparable to what is found in extant organisms can be confidently traced back to a period before the last universal common ancestor (LUCA), it is conceivable that such RNA-targeting mechanisms arose early in evolution. Several independent lines of inference suggest that RNA might have also once played a larger role as the primary genetic material alongside DNA or prior to the emergence of DNA as genomic material. If this inference were correct, then it would mean that both RNA-targeting and RNA repair mechanisms might be of great antiquity. However, direct evidence available from comparative genomics does not allow us to infer a complex RNA repair apparatus as being directly inherited from the LUCA. This is in sharp contrast to other RNA-base modifying and RNA-processing enzymes for which a sizeable and diverse repertoire can be traced back to the LUCA (271). Moreover, this is also unlike what is known for DNA repair systems, where certain major components such as recombination-based repair centered on the RecA-family enzymes, double-strand break repair by Mre11-Rad50-like proteins, and the common precursor of the UvrC-Endonuclease V nuclease domain go back to the LUCA (289,290). Further, several DNA repair systems are deeply conserved within each or at least two of Life's superkingdoms and show a reasonably strong signal for vertical inheritance even in prokaryotes (290,291). In distinct contrast, major RNA repair systems described here are considerably more mobile and patchy in their distributions, especially in the prokaryotic superkingdoms.
Due to the above reasons, the early evolutionary history of the RNA repair systems remains difficult to reconstruct. Of the above-described components, the RtcB RNA ligase appears to be one component that can be reasonably confidently reconstructed as being in the LUCA: first, it is found widely in both the prokaryotic superkingdoms. Second, despite the evidence for inter-superkingdom lateral transfers, there are distinct archaeal and bacterial branches which cover several deep lineages. Third, it displays a distinct protein fold with no evidence for more recent derivation from any other domain inferred as being in the LUCA (116). In contrast, its partner the archease shows a strong archaeo-eukaryotic phyletic pattern, suggesting that its bacterial representatives might have been secondarily derived via horizontal transfer. It is also possible that a representative of the CARF domain family was present in the LUCA. However, it seems to have been recruited for distinct sensor roles in different conflict-related contexts: as a nucleotide sensor in CRISPR/CAS systems on one hand and as the sensor of an as-yet-undiscovered signal of RNA damage in the RtcB systems (213). Unlike RtcB, ATP-grasp RNA-ligases cannot be reconstructed as being in the LUCA. Both sequence and structural evidence favors the deepest split among nucleic acid ligases as being between the ATP-dependent and NAD-dependent DNA ligases (Figure 6D), which act as the primary DNA ligase respectively in the archaeo-eukaryotic and bacterial clades (292). Hence, it appears that the RNA ligases were derived from them on two independent occasions: (1) the ancestor of the Rnl1, Rnl2 (including Rnl5 and PIN-associated ligases) and Rnl3 and (2) the precursor of the Rnl4 ligases (Figure 6D). In a similar vein, the mRNA-capping guanylyltransferase of eukaryotes appears to have also been independently derived from ATP-dependent DNA ligases to operate on RNA (Figure 6D).
Another enzyme which can be reconstructed as being in the LUCA based on phyletic pattern analysis is a Polβ superfamily nucleotidyltransferase that functioned as the CCA-adding enzyme and probably also doubled as the polyA polymerase (Figure 6E). However, the post-LUCA evolutionary history of these enzymes is rather complex with multiple diversification and lateral transfer events (Figure 6E) (293,294). Notably, the eukaryotic CCA-adding enzyme was derived from the bacterial rather than the archaeal version (Figure 6E), suggesting displacement of the ancestral version by a version from a bacterial source (perhaps the primary endosymbiont) during eukaryogenesis. Finally, the above recovery of eukaryotic PolyA-polymerase like modules in bacterial RNA repair contexts suggests that the eukaryote-type PolyA polymerase clade appears to have emerged during the diversification of the CCA-adding enzyme-like clade in a bacterial RNA-repair context (Figure 6E). It was subsequently acquired by the lineage leading to eukaryotes prior to the last eukaryotic common ancestor (LECA) and diversified greatly within eukaryotes (293,294).
The ‘prokaryotic phase’ of diversification of RNA repair systems
In prokaryotes the evolution of RNA repair systems is marked by three major evolutionary trends: (i) extensive inter-organismal mobility including transfer between superkingdoms and gene-loss; (ii) repeated recruitment of biochemically equivalent but non-orthologous enzymatic, scaffolding/chaperone modules and non-coding RNAs for comparable functions in RNA repair; (iii) extensive domain shuffling along with repeated re-formulation of similar domain architectures and operonic structures. In a broad sense these evolutionary trends have been previously observed in other prokaryotic systems including CRISPR/Cas, nucleotide second-messenger-dependent conflict systems, R-M systems and prokaryotic Ub-systems (15,55,295–298). However, only a few DNA-repair systems, like the Ku-DNA ligase and prim-pol-containing DNA repair systems, show comparable tendencies (139,299). This is consistent with all of these systems sharing a role in biological conflicts: the direct and unrelenting consequences of negative fitness outcomes in such conflicts impose a strong selective pressure that can account for the above features. Indeed, they also display the hallmarks of an ‘arms race’ situation because similar evolutionary trends have been reported for effectors which are deployed in an ‘offensive’ role in biological conflicts (300,301).
In both prokaryotes and eukaryotes, components from both the offensive and defensive systems appear to have been domesticated to perform roles in tRNA maturation. Thus, we interpret the tRNA splicing enzyme with a restriction endonuclease-like catalytic domain as probably arising from the domestication of an ancient effector endonuclease of a conflict system. This in turn probably facilitated the fixation of the tRNA intron which itself probably emerged as a defensive mechanism against tRNA-targeting effectors. Notably, the tendency of repair systems to associate with tRNAs which are frequently targeted by effectors might have also favored the recruitment of tRNA-like ncRNAs as structural scaffolds in some of these systems with ROT/TROVE an SPFH9/band-7 proteins. Certain components might also switch from a conserved role in tRNA maturation to conflict-related repair, particularly upon undergoing trans-superkingdom lateral transfers. We observe that certain innovations, such as the archease, MJ1316 and Thg1, appear to have been emerged first in the archaeo-eukaryotic lineage and were subsequently transferred to bacteria (136,159,214,215). In contrast, KptA was likely innovated in bacteria and transferred to archaea (81). At least the former set of domains play a likely role in processing of conserved RNAs in the archaeo-eukaryotic lineage, while in bacteria they are considerably more mobile, suggesting a role primarily in conflict related to RNA-damage.
We also note that despite the rampant tendencies for repeated displacement and operonic or domain-architectural mixing-and-matching, certain strong syntactical rules are discernable, pointing to biochemical restrictions. For example, while RtcB is not fused to other domains and rarely occurs in the same gene contexts with phosphoesterase modules (Figure 4A, D–L, N, P), the ATP-grasp ligases show the opposite trend (Figure 3A–Q). This is consistent with the biochemistry of RtcB, which suggests that it is a self-contained enzyme capable of repairing cyclic 2′-3′ or 3′ phosphate ends by itself. In contrast, the linkage of ATP-grasp ligases with end-processing nucleic acid kinase and diverse phosphatase domains allowed them to extend their repair capacity from ends primarily produced by metal-dependent RNases to those produced by metal-independent ones. In the context of ‘end-cleaning’, we also noted that in addition to KptA, other ADP-ribose-derivative processing enzymes such as the Macro domain and 2H domains are often combined with RNA repair systems (Figures 3H, and 4K, Supplementary Material). This suggests that the previously-observed coupling of processing of ADP-ribose derivatives during tRNA splicing (190) is likely to have a more widespread role in efficient RNA repair (80).
Repeated acquisition of prokaryotic RNA repair systems by eukaryotes
The structure of the eukaryotic cell, with the sequestering of genomic and functional nucleic acids inside of organellar membranes like the nuclear membrane, limits their exposure to nuclease toxins to a certain degree. However, existence of extensive counter-RNA conflict strategies in eukaryotes and recent research into the eukaryotic Crinkler and related toxin delivery systems (302) shows that such protection is hardly foolproof (303,304). Consistent with observations on other conflict systems, even in the case of RNA repair, eukaryotes have repeatedly acquired components which have been innovated in the prokaryotic world. The most striking examples of these are seen in the large ‘Swiss-army-knife’-type proteins from phylogenetically distant microbial eukaryotic lineages combining diverse sets of domains (Figure 3O, V and W). This suggests that they are the functional equivalents of the multi-component prokaryotic systems, with coupling of diverse domains in a single polypeptide being the direct consequence of the absence of operons in eukaryotic genomes. Not only have domains in these proteins been acquired via lateral transfer from prokaryotes, they also show signs of transfer between eukaryotic lineages, as suggested by their patchy phyletic patterns (Figure 3O, V and W). Further, given that in several cases direct architectural cognates are not presently observed in prokaryotes, it is likely that they have emerged via accretion of distinct domains from various sources into a single polypeptide in eukaryotes. These might involve sources beyond bacterial RNA-repair systems as seen in the case of the acquisition of the prim-pol domain from a likely NCLDV source (Figures 3W and X and 6A).
Most of these large multi-domain RNA repair proteins are observed in microbial eukaryotes or those which pass through a distinct unicellular phase in their lifecycle. This is consistent with attacks on their RNA potentially completely nullifying their fitness as in the case of bacteria. One possible corollary is that the emergence of multicellularity in multiple eukaryotic lineages was a further defense against such attacks, which allows for included fitness in the context of a multicellular colony that is then favored by kin-selection (300,301). However, even in multicellular forms viral infections pose a potential selective pressure for evolution of conflict-related RNA repair, especially given that multicellular organisms deploy their own counter-RNA effectors of several types during viral infection (e.g. interferon pathway) (305). Many of the Leng9-like MJ1316 domain-containing proteins, which across several eukaryotic lineages display domain shuffling reminiscent of prokaryotic RNA repair systems, appear to have evolved in such contexts (Figure 3T).
The origin of eukaryotes was also accompanied by the extensive expansion of cellular RNA-centric systems. This occurred both in terms of complexification of the systems for rRNA and tRNA maturation inherited from archaea and also development of entirely new systems (271). In this process, both effector-derived RNase and RNA-repair domains were taken up at various stages of eukaryotic evolution and ‘institutionalized’ into these emerging systems. PIN- and HEPN-domain RNases ultimately originating in prokaryotic conflict systems have been incorporated into roles such as rRNA processing, nonsense-mediated decay (NMD) and NMD-like processes (68), the misfolded protein response, and other stress-related responses in the endoplasmic reticulum (44). The RtcB ligase was ‘institutionalized’ as the primary ligase in tRNA maturation via intron-excision. However, on several occasions (e.g. fungi and land plants) it was displaced by the ATP-grasp ligases (306). Similarly, they were also incorporated into lineage-specific processes such as kinetoplastid RNA-editing (252). The previously-documented antagonistic regulatory roles for the Hen1-like methylases and certain polyA polymerase-related TRF clade Polβ NTases in eukaryotic RNAi systems (207–209) might again represent a eukaryote-specific regulatory adaptation of components ultimately drawn from mobile bacterial RNA repair pathways. The discovery reported here of prim-pol domains and inactive KptA phosphatases in kinetoplastid Dicer-like proteins suggests that such recruitment in lineage-specific eukaryotic RNAi contexts might be more extensive (Figures 3X and 6A and B). Finally, as part of this study we uncovered mobile bacterial vault systems which code for orthologs of the eukaryotic Major Vault Protein (MVP) with SPFH9/band-7 and multiple N-terminal repeats (Supplementary Material). Given that they exactly mirror the architecture of their eukaryotic counterparts we propose that they are likely to constitute a toroidal complex similar to the eukaryotic vault and bind ncRNAs. While these bacterial versions are the likely precursors of their eukaryotic cognates, their exact roles in RNA biology still remain obscure.
Viral acquisitions of RNA repair systems
Given that host counter-viral responses in both eukaryotes and prokaryotes involve deployment of RNA-targeting effectors that limit viral protein synthesis or replication by different means, it is not surprising that viruses have repeatedly acquired RNA repair components. Distinct viral lineages, including the caudate bacteriophages, NCLDVs, and baculoviruses have acquired similar ligase systems (Figure 3B–D, H, L, M and P). All of these systems are likely to function similarly in restoring host or viral RNAs which have been damaged by the deployment of a RNase effector. In the case of multiple eukaryotic RNA viruses, 2H phosphoesterase domains have been incorporated into the viral proteome and might serve an important function in processing viral RNAs (159). Several bacteriophages, NCLDVs and baculoviruses also contain the RlaP NTase found associated with both ATP-grasp and RtcB RNA ligases in cellular systems (Supplementary Material). Since it might occur either as a standalone protein or in association with ligase domains, we suspect that this protein might have an important role in RNA repair on its own. Finally, the viral RNA repair systems might have been the progenitors of some of the enzymes, such as the Rnl1-like ATP-grasp ligases. Indeed, it is even possible that such viral sources contributed comparable enzymes to eukaryotes on more than one occasion.
CONCLUSIONS
As the current survey indicates, there are several notable directions that remain unexplored both in terms of effectors and the repair mechanisms deployed against them. On the effector side, the past several decades have yielded a vast increase in the knowledge of the mechanisms, structure, and mode of delivery of RNA toxins. However, a better understanding of the targets of specific toxins, particularly those which do not appear to target tRNA and are likely to be active against rRNA, mRNA, or some other RNA class, need further work. At the same time, little is known of the constraints governing the timing, cellular context, and strength (i.e. environment, stress conditions, and absolute expression) of the deployment of many of these RNA toxins. Finally, while much research has been devoted to understanding RNase and ribodeglycosylating toxins, fundamental questions regarding the biochemistry of other more recently-described classes, like the RNA deaminase and possibly ADP-ribosylating toxins that modify RNA, remain as yet unanswered.
Recognition of their diversity and elucidation of the distinct mechanisms utilized by RNA-repair systems has resulted in a profound shift in the understanding of how the cell copes with toxin-induced RNA damage (307). However, several fundamental questions relating to both the biochemistry and ecological significance of these systems await further exploration. In terms of biochemistry, we have outlined several domains in this survey that necessarily need further investigation in terms of their activities, such as the MJ1316, PSα, the RlaP NTase, band-7/SPFH, and prim-pol domains. Likewise, several whole systems with these and other components, such as the novel ligase systems combined with PIN domain nuclease, are in need of further investigation. We also have very little knowledge regarding what repair systems might be deployed against ribodeglycosylating and deaminase toxins and whether effector-independent RNA damage plays a major role in eliciting a repair response. Further, issues such as the coordination between multiple domains in the large proteins, such as the eukaryotic Swiss army-knife proteins, and the functional interface between RNAi and RNA repair remains unexplored. At a higher level of function, the arms race between the classes of toxins effecting RNA damage and the classes of repair systems countering them remains in need of more detailed exploration in terms of functional correspondence between the various effector and repair systems. Among the most pressing ecological questions are the inter-relationship between factors such as inter-organismal conflicts and intracellular conflicts with invasive entities and the role of RNA repair as a mechanism of surviving damage caused by effectors in such conflicts.
Hence, we hope that the survey provided helps guide future wet-lab studies investigating such unanswered questions pertaining to RNA repair.
AVAILABILITY
Supplementary Material will also be made available from internally-hosted FTP site: ftp://ftp.ncbi.nih.gov/pub/aravind/RNA_REPAIR/Supplementary_Material.html.
Description of Materials and Methods used in the paper is provided in Supplementary Material.
Supplementary Material
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, National Library of Medicine. We thank Raven Huang for helpful discussions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the National Institutes of Health (NIH); National Library of Medicine. The open access publication charge for this paper has been waived by Oxford University Press - NAR.
Conflict of interest statement. None declared.
REFERENCES
- 1.Crick F.H. On protein synthesis. Symp. Soc. Exp. Biol. 1958;12:138–163. [PubMed] [Google Scholar]
- 2.Alberts B., Johnson A., Lewis J., Morgan D., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. sixth edn. Garland Science; 2014. [Google Scholar]
- 3.Willingham A.T., Gingeras T.R. TUF love for “junk" DNA. Cell. 2006;125:1215–1220. doi: 10.1016/j.cell.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 4.Mattick J.S., Makunin I.V. Non-coding RNA. Hum. Mol. Genet. 2006;15:R17–R29. doi: 10.1093/hmg/ddl046. [DOI] [PubMed] [Google Scholar]
- 5.Saxena A., Carninci P. Whole transcriptome analysis: what are we still missing? Wiley Interdisc. Rev. Syst. Biol. Med. 2011;3:527–543. doi: 10.1002/wsbm.135. [DOI] [PubMed] [Google Scholar]
- 6.Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdisc. Rev. RNA. 2010;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rutenberg-Schoenberg M., Sexton A.N., Simon M.D. The properties of long noncoding RNAs that regulate chromatin. Annu. Rev. Genomics Hum. Genet. 2016 doi: 10.1146/annurev-genom-090314-024939. doi:10.1146/annurev-genom-090314-024939. [DOI] [PubMed] [Google Scholar]
- 8.Breaker R.R. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kruger K., Grabowski P.J., Zaug A.J., Sands J., Gottschling D.E., Cech T.R. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 10.Guerrier-Takada C., Gardiner K., Marsh T., Pace N., Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 11.Prody G.A., Bakos J.T., Buzayan J.M., Schneider I.R., Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 12.Nissen P., Hansen J., Ban N., Moore P.B., Steitz T.A. The structural basis of ribosome activity in peptide bond synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 13.Birnstiel M.L. Structure and Function of Major and Minor Small Nuclear Ribonucleoprotein Particles. 1 edn. Berlin, Heidelberg: Springer-Verlag; 1988. [Google Scholar]
- 14.Burroughs A.M., Ando Y., Aravind L. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdisc. Rev. RNA. 2014;5:141–181. doi: 10.1002/wrna.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Makarova K.S., Aravind L., Wolf Y.I., Koonin E.V. Unification of Cas protein families and a simple scenario for the origin and evolution of CRISPR-Cas systems. Biol. Direct. 2011;6:38. doi: 10.1186/1745-6150-6-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Masaki H., Ogawa T. The modes of action of colicins E5 and D, and related cytotoxic tRNases. Biochimie. 2002;84:433–438. doi: 10.1016/s0300-9084(02)01425-6. [DOI] [PubMed] [Google Scholar]
- 17.Lacadena J., Alvarez-Garcia E., Carreras-Sangra N., Herrero-Galan E., Alegre-Cebollada J., Garcia-Ortega L., Onaderra M., Gavilanes J.G., Martinez del Pozo A. Fungal ribotoxins: molecular dissection of a family of natural killers. FEMS Microbiol. Rev. 2007;31:212–237. doi: 10.1111/j.1574-6976.2006.00063.x. [DOI] [PubMed] [Google Scholar]
- 18.Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014;5:3538. doi: 10.1038/ncomms4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peach S.E., York K., Hesselberth J.R. Global analysis of RNA cleavage by 5′-hydroxyl RNA sequencing. Nucleic Acids Res. 2015;43:e108. doi: 10.1093/nar/gkv536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma C.M., Vogel J. Differential RNA-seq: the approach behind and the biological insight gained. Curr. Opin. Microbiol. 2014;19:97–105. doi: 10.1016/j.mib.2014.06.010. [DOI] [PubMed] [Google Scholar]
- 21.Huang R.H. Unique 2′-O-methylation by Hen1 in eukaryotic RNA interference and bacterial RNA repair. Biochemistry. 2012;51:4087–4095. doi: 10.1021/bi300497x. [DOI] [PubMed] [Google Scholar]
- 22.Silber R., Malathi V.G., Hurwitz J. Purification and properties of bacteriophage T4-induced RNA ligase. Proc. Natl. Acad. Sci. U.S.A. 1972;69:3009–3013. doi: 10.1073/pnas.69.10.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tanaka N., Shuman S. RtcB is the RNA ligase component of an Escherichia coli RNA repair operon. J. Biol. Chem. 2011;286:7727–7731. doi: 10.1074/jbc.C111.219022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith J.M.P.G.R. The logic of animal conflict. Nature. 1973:15–18. [Google Scholar]
- 25.Hurst L.D., Atlan A., Bengtsson B.O. Genetic conflicts. Q. Rev. Biol. 1996;71:317–364. doi: 10.1086/419442. [DOI] [PubMed] [Google Scholar]
- 26.Burt A., Trivers R. Genes in Conflict: The Biology of Selfish Genetic Elements. Cambridge: The Belknap Press of Harvard University Press; 2006. [Google Scholar]
- 27.Werren J.H. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. U.S.A. 2011;108(Suppl. 2):10863–10870. doi: 10.1073/pnas.1102343108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jablonowski D., Schaffrath R. Zymocin, a composite chitinase and tRNase killer toxin from yeast. Biochem. Soc. Trans. 2007;35:1533–1537. doi: 10.1042/BST0351533. [DOI] [PubMed] [Google Scholar]
- 29.Klassen R., Paluszynski J.P., Wemhoff S., Pfeiffer A., Fricke J., Meinhardt F. The primary target of the killer toxin from Pichia acaciae is tRNA(Gln) Mol. Microbiol. 2008;69:681–697. doi: 10.1111/j.1365-2958.2008.06319.x. [DOI] [PubMed] [Google Scholar]
- 30.Lu J., Huang B., Esberg A., Johansson M.J., Bystrom A.S. The Kluyveromyces lactis gamma-toxin targets tRNA anticodons. RNA. 2005;11:1648–1654. doi: 10.1261/rna.2172105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chakravarty A.K., Smith P., Jalan R., Shuman S. Structure, mechanism, and specificity of a eukaryal tRNA restriction enzyme involved in self-nonself discrimination. Cell Rep. 2014;7:339–347. doi: 10.1016/j.celrep.2014.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olombrada M., Martinez-del-Pozo A., Medina P., Budia F., Gavilanes J.G., Garcia-Ortega L. Fungal ribotoxins: natural protein-based weapons against insects. Toxicon. 2014;83:69–74. doi: 10.1016/j.toxicon.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 33.Brandhorst T., Dowd P.F., Kenealy W.R. The ribosome-inactivating protein restrictocin deters insect feeding on Aspergillus restrictus. Microbiology. 1996;142:1551–1556. doi: 10.1099/13500872-142-6-1551. [DOI] [PubMed] [Google Scholar]
- 34.Stirpe F., Battelli M.G. Ribosome-inactivating proteins: progress and problems. Cell. Mol. Life Sci.: CMLS. 2006;63:1850–1866. doi: 10.1007/s00018-006-6078-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lainhart W., Stolfa G., Koudelka G.B. Shiga toxin as a bacterial defense against a eukaryotic predator, Tetrahymena thermophila. J. Bacteriol. 2009;191:5116–5122. doi: 10.1128/JB.00508-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Asakura H., Makino S., Kobori H., Watarai M., Shirahata T., Ikeda T., Takeshi K. Phylogenetic diversity and similarity of active sites of Shiga toxin (stx) in Shiga toxin-producing Escherichia coli (STEC) isolates from humans and animals. Epidemiol. Infect. 2001;127:27–36. doi: 10.1017/s0950268801005635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dao D.N., Kessin R.H., Ennis H.L. Developmental cheating and the evolutionary biology of Dictyostelium and Myxococcus. Microbiology. 2000;146:1505–1512. doi: 10.1099/00221287-146-7-1505. [DOI] [PubMed] [Google Scholar]
- 38.Zhang D., Iyer L.M., Aravind L. A novel immunity system for bacterial nucleic acid degrading toxins and its recruitment in various eukaryotic and DNA viral systems. Nucleic Acids Res. 2011;39:4532–4552. doi: 10.1093/nar/gkr036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang D., de Souza R.F., Anantharaman V., Iyer L.M., Aravind L. Polymorphic toxin systems: comprehensive characterization of trafficking modes, processing, mechanisms of action, immunity and ecology using comparative genomics. Biol. Direct. 2012;7:18. doi: 10.1186/1745-6150-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang D., Iyer L.M., Burroughs A.M., Aravind L. Resilience of biochemical activity in protein domains in the face of structural divergence. Curr. Opin. Struct. Biol. 2014;26:92–103. doi: 10.1016/j.sbi.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Aoki S.K., Diner E.J., de Roodenbeke C.T., Burgess B.R., Poole S.J., Braaten B.A., Jones A.M., Webb J.S., Hayes C.S., Cotter P.A., et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature. 2010;468:439–442. doi: 10.1038/nature09490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hale C.R., Zhao P., Olson S., Duff M.O., Graveley B.R., Wells L., Terns R.M., Terns M.P. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Staals R.H., Agari Y., Maki-Yonekura S., Zhu Y., Taylor D.W., van Duijn E., Barendregt A., Vlot M., Koehorst J.J., Sakamoto K., et al. Structure and activity of the RNA-targeting Type III-B CRISPR-Cas complex of Thermus thermophilus. Mol. Cell. 2013;52:135–145. doi: 10.1016/j.molcel.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anantharaman V., Makarova K.S., Burroughs A.M., Koonin E.V., Aravind L. Comprehensive analysis of the HEPN superfamily: identification of novel roles in intra-genomic conflicts, defense, pathogenesis and RNA processing. Biol. Direct. 2013;8:15. doi: 10.1186/1745-6150-8-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shmakov S., Abudayyeh O.O., Makarova K.S., Wolf Y.I., Gootenberg J.S., Semenova E., Minakhin L., Joung J., Konermann S., Severinov K., et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hoskisson P.A., Sumby P., Smith M.C. The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology. 2015;477:100–109. doi: 10.1016/j.virol.2014.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bickle T.A., Kruger D.H. Biology of DNA restriction. Microbiol. Rev. 1993;57:434–450. doi: 10.1128/mr.57.2.434-450.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uzan M., Miller E.S. Post-transcriptional control by bacteriophage T4: mRNA decay and inhibition of translation initiation. Virol. J. 2010;7:360. doi: 10.1186/1743-422X-7-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meineke B., Shuman S. Determinants of the cytotoxicity of PrrC anticodon nuclease and its amelioration by tRNA repair. RNA. 2012;18:145–154. doi: 10.1261/rna.030171.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bedford D.J., Laity C., Buttner M.J. Two genes involved in the phase-variable phi C31 resistance mechanism of Streptomyces coelicolor A3(2) J. Bacteriol. 1995;177:4681–4689. doi: 10.1128/jb.177.16.4681-4689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sumby P., Smith M.C. Genetics of the phage growth limitation (Pgl) system of Streptomyces coelicolor A3(2) Mol. Microbiol. 2002;44:489–500. doi: 10.1046/j.1365-2958.2002.02896.x. [DOI] [PubMed] [Google Scholar]
- 52.Anantharaman V., Iyer L.M., Aravind L. Ter-dependent stress response systems: novel pathways related to metal sensing, production of a nucleoside-like metabolite, and DNA-processing. Mol. bioSyst. 2012;8:3142–3165. doi: 10.1039/c2mb25239b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parreira R., Ehrlich S.D., Chopin M.C. Dramatic decay of phage transcripts in lactococcal cells carrying the abortive infection determinant AbiB. Mol. Microbiol. 1996;19:221–230. doi: 10.1046/j.1365-2958.1996.371896.x. [DOI] [PubMed] [Google Scholar]
- 54.Dai G., Su P., Allison G.E., Geller B.L., Zhu P., Kim W.S., Dunn N.W. Molecular characterization of a new abortive infection system (AbiU) from Lactococcus lactis LL51-1. Appl. Environ. Microbiol. 2001;67:5225–5232. doi: 10.1128/AEM.67.11.5225-5232.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burroughs A.M., Zhang D., Schaffer D.E., Iyer L.M., Aravind L. Comparative genomic analyses reveal a vast, novel network of nucleotide-centric systems in biological conflicts, immunity and signaling. Nucleic Acids Res. 2015;43:10633–10654. doi: 10.1093/nar/gkv1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schroder H.C., Suhadolnik R.J., Pfleiderer W., Charubala R., Muller W.E. (2′-5′)Oligoadenylate and intracellular immunity against retrovirus infection. Int. J. Biochem. 1992;24:55–63. doi: 10.1016/0020-711x(92)90229-t. [DOI] [PubMed] [Google Scholar]
- 57.Choi U.Y., Kang J.S., Hwang Y.S., Kim Y.J. Oligoadenylate synthase-like (OASL) proteins: dual functions and associations with diseases. Exp. Mol. Med. 2015;47:e144. doi: 10.1038/emm.2014.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gupta S.K., Haigh B.J., Griffin F.J., Wheeler T.T. The mammalian secreted RNases: mechanisms of action in host defence. Innate Immun. 2013;19:86–97. doi: 10.1177/1753425912446955. [DOI] [PubMed] [Google Scholar]
- 59.Blin J., Fitzgerald K.A. Perspective: The RNA exosome, cytokine gene regulation and links to autoimmunity. Cytokine. 2015;74:175–180. doi: 10.1016/j.cyto.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 60.Irvin J.D. Purification and partial characterization of the antiviral protein from Phytolacca americana which inhibits eukaryotic protein synthesis. Arch. Biochem. Biophys. 1975;169:522–528. doi: 10.1016/0003-9861(75)90195-2. [DOI] [PubMed] [Google Scholar]
- 61.Obrig T.G., Irvin J.D., Hardesty B. The effect of an antiviral peptide on the ribosomal reactions of the peptide elongation enzymes, EF-I and EF-II. Arch. Biochem. Biophys. 1973;155:278–289. doi: 10.1016/0003-9861(73)90116-1. [DOI] [PubMed] [Google Scholar]
- 62.Cook G.M., Robson J.R., Frampton R.A., McKenzie J., Przybilski R., Fineran P.C., Arcus V.L. Ribonucleases in bacterial toxin-antitoxin systems. Biochim. Biophys. Acta. 2013;1829:523–531. doi: 10.1016/j.bbagrm.2013.02.007. [DOI] [PubMed] [Google Scholar]
- 63.Beloglazova N., Brown G., Zimmerman M.D., Proudfoot M., Makarova K.S., Kudritska M., Kochinyan S., Wang S., Chruszcz M., Minor W., et al. A novel family of sequence-specific endoribonucleases associated with the clustered regularly interspaced short palindromic repeats. J. Biol. Chem. 2008;283:20361–20371. doi: 10.1074/jbc.M803225200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gunderson F.F., Mallama C.A., Fairbairn S.G., Cianciotto N.P. Nuclease activity of Legionella pneumophila Cas2 promotes intracellular infection of amoebal host cells. Infect. Immun. 2015;83:1008–1018. doi: 10.1128/IAI.03102-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dixit B., Ghosh K.K., Fernandes G., Kumar P., Gogoi P., Kumar M. Dual nuclease activity of a Cas2 protein in CRISPR-Cas subtype I-B of Leptospira interrogans. FEBS Lett. 2016;590:1002–1016. doi: 10.1002/1873-3468.12124. [DOI] [PubMed] [Google Scholar]
- 66.Abudayyeh O.O., Gootenberg J.S., Konermann S., Joung J., Slaymaker I.M., Cox D.B., Shmakov S., Makarova K.S., Semenova E., Minakhin L., et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:557–566. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.James R., Penfold C.N., Moore G.R., Kleanthous C. Killing of E coli cells by E group nuclease colicins. Biochimie. 2002;84:381–389. doi: 10.1016/s0300-9084(02)01450-5. [DOI] [PubMed] [Google Scholar]
- 68.Anantharaman V., Aravind L. New connections in the prokaryotic toxin-antitoxin network: relationship with the eukaryotic nonsense-mediated RNA decay system. Genome Biol. 2003;4:R81. doi: 10.1186/gb-2003-4-12-r81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Winther K.S., Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Das U., Pogenberg V., Subhramanyam U.K., Wilmanns M., Gourinath S., Srinivasan A. Crystal structure of the VapBC-15 complex from Mycobacterium tuberculosis reveals a two-metal ion dependent PIN-domain ribonuclease and a variable mode of toxin-antitoxin assembly. J. Struct. Biol. 2014;188:249–258. doi: 10.1016/j.jsb.2014.10.002. [DOI] [PubMed] [Google Scholar]
- 71.Olovnikov I., Chan K., Sachidanandam R., Newman D.K., Aravin A.A. Bacterial argonaute samples the transcriptome to identify foreign DNA. Mol. Cell. 2013;51:594–605. doi: 10.1016/j.molcel.2013.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yuan Y.R., Pei Y., Ma J.B., Kuryavyi V., Zhadina M., Meister G., Chen H.Y., Dauter Z., Tuschl T., Patel D.J. Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Mol. Cell. 2005;19:405–419. doi: 10.1016/j.molcel.2005.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Y., Juranek S., Li H., Sheng G., Tuschl T., Patel D.J. Structure of an argonaute silencing complex with a seed-containing guide DNA and target RNA duplex. Nature. 2008;456:921–926. doi: 10.1038/nature07666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yan X., Guo W., Yuan Y.A. Crystal structures of CRISPR-associated Csx3 reveal a manganese-dependent deadenylation exoribonuclease. RNA Biol. 2015;12:749–760. doi: 10.1080/15476286.2015.1051300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Schmier B.J., Shuman S. Effects of 3′-OH and 5′-PO4 base mispairs and damaged base lesions on the fidelity of nick sealing by Deinococcus radiodurans RNA ligase. J. Bacteriol. 2014;196:1704–1712. doi: 10.1128/JB.00020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Unciuleac M.C., Shuman S. Characterization of a novel eukaryal nick-sealing RNA ligase from Naegleria gruberi. RNA. 2015;21:824–832. doi: 10.1261/rna.049197.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nandakumar J., Ho C.K., Lima C.D., Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J. Biol. Chem. 2004;279:31337–31347. doi: 10.1074/jbc.M402394200. [DOI] [PubMed] [Google Scholar]
- 78.Walsh M.J., Dodd J.E., Hautbergue G.M. Ribosome-inactivating proteins: potent poisons and molecular tools. Virulence. 2013;4:774–784. doi: 10.4161/viru.26399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iyer L.M., Zhang D., Rogozin I.B., Aravind L. Evolution of the deaminase fold and multiple origins of eukaryotic editing and mutagenic nucleic acid deaminases from bacterial toxin systems. Nucleic Acids Res. 2011;39:9473–9497. doi: 10.1093/nar/gkr691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Aravind L., Zhang D., de Souza R.F., Anand S., Iyer L.M. The natural history of ADP-ribosyltransferases and the ADP-ribosylation system. Curr. Top. Microbiol. Immunol. 2015;384:3–32. doi: 10.1007/82_2014_414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.de Souza R.F., Aravind L. Identification of novel components of NAD-utilizing metabolic pathways and prediction of their biochemical functions. Mol. bioSyst. 2012;8:1661–1677. doi: 10.1039/c2mb05487f. [DOI] [PubMed] [Google Scholar]
- 82.Makarova K.S., Anantharaman V., Aravind L., Koonin E.V. Live virus-free or die: coupling of antivirus immunity and programmed suicide or dormancy in prokaryotes. Biol. Direct. 2012;7:40. doi: 10.1186/1745-6150-7-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chopin M.C., Chopin A., Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr. Opin. Microbiol. 2005;8:473–479. doi: 10.1016/j.mib.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 84.Blanga-Kanfi S., Amitsur M., Azem A., Kaufmann G. PrrC-anticodon nuclease: functional organization of a prototypical bacterial restriction RNase. Nucleic Acids Res. 2006;34:3209–3219. doi: 10.1093/nar/gkl415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Linder P., Doelz R., Gubler M., Bickle T.A. An anticodon nuclease gene inserted into a hsd region encoding a type I DNA restriction system. Nucleic Acids Res. 1990;18:7170. doi: 10.1093/nar/18.23.7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Szewczak A.A., Moore P.B., Chang Y.L., Wool I.G. The conformation of the sarcin/ricin loop from 28S ribosomal RNA. Proc. Natl. Acad. Sci. U.S.A. 1993;90:9581–9585. doi: 10.1073/pnas.90.20.9581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hausner T.P., Atmadja J., Nierhaus K.H. Evidence that the G2661 region of 23S rRNA is located at the ribosomal binding sites of both elongation factors. Biochimie. 1987;69:911–923. doi: 10.1016/0300-9084(87)90225-2. [DOI] [PubMed] [Google Scholar]
- 88.Montanaro L., Sperti S., Mattioli A., Testoni G., Stirpe F. Inhibition by ricin of protein synthesis in vitro. Inhibition of the binding of elongation factor 2 and of adenosine diphosphate-ribosylated elongation factor 2 to ribosomes. Biochem. J. 1975;146:127–131. doi: 10.1042/bj1460127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fernandez-Puentes C., Vazquez D. Effects of some proteins that inactivate the eukaryotic ribosome. FEBS Lett. 1977;78:143–146. doi: 10.1016/0014-5793(77)80292-5. [DOI] [PubMed] [Google Scholar]
- 90.Winther K.S., Brodersen D.E., Brown A.K., Gerdes K. VapC20 of Mycobacterium tuberculosis cleaves the sarcin-ricin loop of 23S rRNA. Nat. Commun. 2013;4:2796. doi: 10.1038/ncomms3796. [DOI] [PubMed] [Google Scholar]
- 91.Cruz J.W., Sharp J.D., Hoffer E.D., Maehigashi T., Vvedenskaya I.O., Konkimalla A., Husson R.N., Nickels B.E., Dunham C.M., Woychik N.A. Growth-regulating Mycobacterium tuberculosis VapC-mt4 toxin is an isoacceptor-specific tRNase. Nat. Commun. 2015;6:7480. doi: 10.1038/ncomms8480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ramisetty B.C., Natarajan B., Santhosh R.S. mazEF-mediated programmed cell death in bacteria: “what is this?". Crit. Rev. Microbiol. 2015;41:89–100. doi: 10.3109/1040841X.2013.804030. [DOI] [PubMed] [Google Scholar]
- 93.Koga M., Otsuka Y., Lemire S., Yonesaki T. Escherichia coli rnlA and rnlB compose a novel toxin-antitoxin system. Genetics. 2011;187:123–130. doi: 10.1534/genetics.110.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Otsuka Y., Yonesaki T. Dmd of bacteriophage T4 functions as an antitoxin against Escherichia coli LsoA and RnlA toxins. Mol. Microbiol. 2012;83:669–681. doi: 10.1111/j.1365-2958.2012.07975.x. [DOI] [PubMed] [Google Scholar]
- 95.Neubauer C., Gao Y.G., Andersen K.R., Dunham C.M., Kelley A.C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D.E. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kawano M., Aravind L., Storz G. An antisense RNA controls synthesis of an SOS-induced toxin evolved from an antitoxin. Mol. Microbiol. 2007;64:738–754. doi: 10.1111/j.1365-2958.2007.05688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aguilera A., Garcia-Muse T. R loops: from transcription byproducts to threats to genome stability. Mol. Cell. 2012;46:115–124. doi: 10.1016/j.molcel.2012.04.009. [DOI] [PubMed] [Google Scholar]
- 98.Burroughs A.M., Iyer L.M., Aravind L. Two novel PIWI families: roles in inter-genomic conflicts in bacteria and Mediator-dependent modulation of transcription in eukaryotes. Biol. Direct. 2013;8:13. doi: 10.1186/1745-6150-8-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ng C.L., Lang K., Meenan N.A., Sharma A., Kelley A.C., Kleanthous C., Ramakrishnan V. Structural basis for 16S ribosomal RNA cleavage by the cytotoxic domain of colicin E3. Nat. Struct. Mol. Biol. 2010;17:1241–1246. doi: 10.1038/nsmb.1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Aravind L. Guilt by association: contextual information in genome analysis. Genome Res. 2000;10:1074–1077. doi: 10.1101/gr.10.8.1074. [DOI] [PubMed] [Google Scholar]
- 101.Overbeek R., Fonstein M., D'Souza M., Pusch G.D., Maltsev N. The use of gene clusters to infer functional coupling. Proc. Natl. Acad. Sci. U.S.A. 1999;96:2896–2901. doi: 10.1073/pnas.96.6.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Galperin M.Y., Koonin E.V. A diverse superfamily of enzymes with ATP-dependent carboxylate-amine/thiol ligase activity. Protein Sci. 1997;6:2639–2643. doi: 10.1002/pro.5560061218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Iyer L.M., Abhiman S., Maxwell Burroughs A., Aravind L. Amidoligases with ATP-grasp, glutamine synthetase-like and acetyltransferase-like domains: synthesis of novel metabolites and peptide modifications of proteins. Mol. bioSyst. 2009;5:1636–1660. doi: 10.1039/b917682a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Burroughs A.M., Zhang D., Aravind L. The eukaryotic translation initiation regulator CDC123 defines a divergent clade of ATP-grasp enzymes with a predicted role in novel protein modifications. Biol. Direct. 2015;10:21. doi: 10.1186/s13062-015-0053-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Balaji S., Aravind L. The RAGNYA fold: a novel fold with multiple topological variants found in functionally diverse nucleic acid, nucleotide and peptide-binding proteins. Nucleic Acids Res. 2007;35:5658–5671. doi: 10.1093/nar/gkm558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Shuman S., Schwer B. RNA capping enzyme and DNA ligase: a superfamily of covalent nucleotidyl transferases. Mol. Microbiol. 1995;17:405–410. doi: 10.1111/j.1365-2958.1995.mmi_17030405.x. [DOI] [PubMed] [Google Scholar]
- 107.Aravind L., Koonin E.V. Gleaning non-trivial structural, functional and evolutionary information about proteins by iterative database searches. J. Mol. Biol. 1999;287:1023–1040. doi: 10.1006/jmbi.1999.2653. [DOI] [PubMed] [Google Scholar]
- 108.Shuman S., Lima C.D. The polynucleotide ligase and RNA capping enzyme superfamily of covalent nucleotidyltransferases. Curr. Opin. Struct. Biol. 2004;14:757–764. doi: 10.1016/j.sbi.2004.10.006. [DOI] [PubMed] [Google Scholar]
- 109.Unciuleac M.C., Goldgur Y., Shuman S. Structure and two-metal mechanism of a eukaryal nick-sealing RNA ligase. Proc. Natl. Acad. Sci. U.S.A. 2015;112:13868–13873. doi: 10.1073/pnas.1516536112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wang L.K., Nandakumar J., Schwer B., Shuman S. The C-terminal domain of T4 RNA ligase 1 confers specificity for tRNA repair. RNA. 2007;13:1235–1244. doi: 10.1261/rna.591807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ho C.K., Wang L.K., Lima C.D., Shuman S. Structure and mechanism of RNA ligase. Structure. 2004;12:327–339. doi: 10.1016/j.str.2004.01.011. [DOI] [PubMed] [Google Scholar]
- 112.Nandakumar J., Shuman S. How an RNA ligase discriminates RNA versus DNA damage. Mol. Cell. 2004;16:211–221. doi: 10.1016/j.molcel.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 113.Tanaka N., Meineke B., Shuman S. RtcB, a novel RNA ligase, can catalyze tRNA splicing and HAC1 mRNA splicing in vivo. J. Biol. Chem. 2011;286:30253–30257. doi: 10.1074/jbc.C111.274597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Englert M., Sheppard K., Aslanian A., Yates J.R., 3rd, Soll D. Archaeal 3′-phosphate RNA splicing ligase characterization identifies the missing component in tRNA maturation. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1290–1295. doi: 10.1073/pnas.1018307108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Popow J., Englert M., Weitzer S., Schleiffer A., Mierzwa B., Mechtler K., Trowitzsch S., Will C.L., Luhrmann R., Soll D., et al. HSPC117 is the essential subunit of a human tRNA splicing ligase complex. Science. 2011;331:760–764. doi: 10.1126/science.1197847. [DOI] [PubMed] [Google Scholar]
- 116.Okada C., Maegawa Y., Yao M., Tanaka I. Crystal structure of an RtcB homolog protein (PH1602-extein protein) from Pyrococcus horikoshii reveals a novel fold. Proteins. 2006;63:1119–1122. doi: 10.1002/prot.20912. [DOI] [PubMed] [Google Scholar]
- 117.Desai K.K., Bingman C.A., Phillips G.N., Jr, Raines R.T. Structures of the noncanonical RNA ligase RtcB reveal the mechanism of histidine guanylylation. Biochemistry. 2013;52:2518–2525. doi: 10.1021/bi4002375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Tanaka N., Chakravarty A.K., Maughan B., Shuman S. Novel mechanism of RNA repair by RtcB via sequential 2′,3′-cyclic phosphodiesterase and 3′-Phosphate/5′-hydroxyl ligation reactions. J. Biol. Chem. 2011;286:43134–43143. doi: 10.1074/jbc.M111.302133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chakravarty A.K., Subbotin R., Chait B.T., Shuman S. RNA ligase RtcB splices 3′-phosphate and 5′-OH ends via covalent RtcB-(histidinyl)-GMP and polynucleotide-(3′)pp(5′)G intermediates. Proc. Natl. Acad. Sci. U.S.A. 2012;109:6072–6077. doi: 10.1073/pnas.1201207109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Chakravarty A.K., Shuman S. The sequential 2′,3′-cyclic phosphodiesterase and 3′-phosphate/5′-OH ligation steps of the RtcB RNA splicing pathway are GTP-dependent. Nucleic Acids Res. 2012;40:8558–8567. doi: 10.1093/nar/gks558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Englert M., Xia S., Okada C., Nakamura A., Tanavde V., Yao M., Eom S.H., Konigsberg W.H., Soll D., Wang J. Structural and mechanistic insights into guanylylation of RNA-splicing ligase RtcB joining RNA between 3′-terminal phosphate and 5′-OH. Proc. Natl. Acad. Sci. U.S.A. 2012;109:15235–15240. doi: 10.1073/pnas.1213795109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Chakravarty A.K., Shuman S. RNA 3′-phosphate cyclase (RtcA) catalyzes ligase-like adenylylation of DNA and RNA 5′-monophosphate ends. J. Biol. Chem. 2011;286:4117–4122. doi: 10.1074/jbc.M110.196766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Das U., Chakravarty A.K., Remus B.S., Shuman S. Rewriting the rules for end joining via enzymatic splicing of DNA 3′-PO4 and 5′-OH ends. Proc. Natl. Acad. Sci. U.S.A. 2013;110:20437–20442. doi: 10.1073/pnas.1314289110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Das U., Chauleau M., Ordonez H., Shuman S. Impact of DNA3′pp5′G capping on repair reactions at DNA 3′ ends. Proc. Natl. Acad. Sci. U.S.A. 2014;111:11317–11322. doi: 10.1073/pnas.1409203111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Chauleau M., Jacewicz A., Shuman S. DNA3′pp5′G de-capping activity of aprataxin: effect of cap nucleoside analogs and structural basis for guanosine recognition. Nucleic Acids Res. 2015;43:6075–6083. doi: 10.1093/nar/gkv501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Maughan W.P., Shuman S. Characterization of 3′-phosphate RNA ligase paralogs RtcB1, RtcB2, and RtcB3 from Myxococcus xanthus highlights DNA and RNA 5′-phosphate capping activity of RtcB3. J. Bacteriol. 2015;197:3616–3624. doi: 10.1128/JB.00631-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Chauleau M., Das U., Shuman S. Effects of DNA3′pp5′G capping on 3′ end repair reactions and of an embedded pyrophosphate-linked guanylate on ribonucleotide surveillance. Nucleic Acids Res. 2015;43:3197–3207. doi: 10.1093/nar/gkv179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Vogel A., Schilling O., Spath B., Marchfelder A. The tRNase Z family of proteins: physiological functions, substrate specificity and structural properties. Biol. Chem. 2005;386:1253–1264. doi: 10.1515/BC.2005.142. [DOI] [PubMed] [Google Scholar]
- 129.Sprinzl M., Cramer F. The -C-C-A end of tRNA and its role in protein biosynthesis. Progress Nucleic Res. Mol. Biol. 1979;22:1–69. doi: 10.1016/s0079-6603(08)60798-9. [DOI] [PubMed] [Google Scholar]
- 130.Zhu L., Deutscher M.P. tRNA nucleotidyltransferase is not essential for Escherichia coli viability. EMBO J. 1987;6:2473–2477. doi: 10.1002/j.1460-2075.1987.tb02528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Marck C., Grosjean H. tRNomics: analysis of tRNA genes from 50 genomes of Eukarya, Archaea, and Bacteria reveals anticodon-sparing strategies and domain-specific features. RNA. 2002;8:1189–1232. doi: 10.1017/s1355838202022021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Trippe R., Sandrock B., Benecke B.J. A highly specific terminal uridylyl transferase modifies the 3′-end of U6 small nuclear RNA. Nucleic Acids Res. 1998;26:3119–3126. doi: 10.1093/nar/26.13.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Himeno H., Hasegawa T., Ueda T., Watanabe K., Miura K., Shimizu M. Role of the extra G-C pair at the end of the acceptor stem of tRNA(His) in aminoacylation. Nucleic Acids Res. 1989;17:7855–7863. doi: 10.1093/nar/17.19.7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Rosen A.E., Brooks B.S., Guth E., Francklyn C.S., Musier-Forsyth K. Evolutionary conservation of a functionally important backbone phosphate group critical for aminoacylation of histidine tRNAs. RNA. 2006;12:1315–1322. doi: 10.1261/rna.78606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jahn D., Pande S. Histidine tRNA guanylyltransferase from Saccharomyces cerevisiae. II. Catalytic mechanism. J. Biol. Chem. 1991;266:22832–22836. [PubMed] [Google Scholar]
- 136.Anantharaman V., Iyer L.M., Aravind L. Presence of a classical RRM-fold palm domain in Thg1-type 3′- 5′ nucleic acid polymerases and the origin of the GGDEF and CRISPR polymerase domains. Biol. Direct. 2010;5:43. doi: 10.1186/1745-6150-5-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hyde S.J., Eckenroth B.E., Smith B.A., Eberley W.A., Heintz N.H., Jackman J.E., Doublie S. tRNA(His) guanylyltransferase (THG1), a unique 3′-5′ nucleotidyl transferase, shares unexpected structural homology with canonical 5′-3′ DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 2010;107:20305–20310. doi: 10.1073/pnas.1010436107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Rao B.S., Maris E.L., Jackman J.E. tRNA 5′-end repair activities of tRNAHis guanylyltransferase (Thg1)-like proteins from Bacteria and Archaea. Nucleic Acids Res. 2011;39:1833–1842. doi: 10.1093/nar/gkq976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Iyer L.M., Koonin E.V., Leipe D.D., Aravind L. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 2005;33:3875–3896. doi: 10.1093/nar/gki702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lipps G., Weinzierl A.O., von Scheven G., Buchen C., Cramer P. Structure of a bifunctional DNA primase-polymerase. Nat. Struct. Mol. Biol. 2004;11:157–162. doi: 10.1038/nsmb723. [DOI] [PubMed] [Google Scholar]
- 141.Leipe D.D., Koonin E.V., Aravind L. Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 2003;333:781–815. doi: 10.1016/j.jmb.2003.08.040. [DOI] [PubMed] [Google Scholar]
- 142.Wang L.K., Das U., Smith P., Shuman S. Structure and mechanism of the polynucleotide kinase component of the bacterial Pnkp-Hen1 RNA repair system. RNA. 2012;18:2277–2286. doi: 10.1261/rna.036061.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Das U., Wang L.K., Smith P., Jacewicz A., Shuman S. Structures of bacterial polynucleotide kinase in a Michaelis complex with GTP*Mg2+ and 5′-OH oligonucleotide and a product complex with GDP*Mg2+ and 5′-PO4 oligonucleotide reveal a mechanism of general acid-base catalysis and the determinants of phosphoacceptor recognition. Nucleic Acids Res. 2014;42:1152–1161. doi: 10.1093/nar/gkt936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Das U., Wang L.K., Smith P., Munir A., Shuman S. Structures of bacterial polynucleotide kinase in a michaelis complex with nucleoside triphosphate (NTP)-Mg2+ and 5′-OH RNA and a mixed substrate-product complex with NTP-Mg2+ and a 5′-phosphorylated oligonucleotide. J. Bacteriol. 2014;196:4285–4292. doi: 10.1128/JB.02197-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Leipe D.D., Wolf Y.I., Koonin E.V., Aravind L. Classification and evolution of P-loop GTPases and related ATPases. J. Mol. Biol. 2002;317:41–72. doi: 10.1006/jmbi.2001.5378. [DOI] [PubMed] [Google Scholar]
- 146.Weitzer S., Martinez J. The human RNA kinase hClp1 is active on 3′ transfer RNA exons and short interfering RNAs. Nature. 2007;447:222–226. doi: 10.1038/nature05777. [DOI] [PubMed] [Google Scholar]
- 147.Dikfidan A., Loll B., Zeymer C., Magler I., Clausen T., Meinhart A. RNA specificity and regulation of catalysis in the eukaryotic polynucleotide kinase Clp1. Mol. Cell. 2014;54:975–986. doi: 10.1016/j.molcel.2014.04.005. [DOI] [PubMed] [Google Scholar]
- 148.Englert M., Sheppard K., Gundllapalli S., Beier H., Soll D. Branchiostoma floridae has separate healing and sealing enzymes for 5′-phosphate RNA ligation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:16834–16839. doi: 10.1073/pnas.1011703107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Minvielle-Sebastia L., Preker P.J., Wiederkehr T., Strahm Y., Keller W. The major yeast poly(A)-binding protein is associated with cleavage factor IA and functions in premessenger RNA 3′-end formation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7897–7902. doi: 10.1073/pnas.94.15.7897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Heindl K., Martinez J. Nol9 is a novel polynucleotide 5′-kinase involved in ribosomal RNA processing. EMBO J. 2010;29:4161–4171. doi: 10.1038/emboj.2010.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Braglia P., Heindl K., Schleiffer A., Martinez J., Proudfoot N.J. Role of the RNA/DNA kinase Grc3 in transcription termination by RNA polymerase I. EMBO Rep. 2010;11:758–764. doi: 10.1038/embor.2010.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Kitano E., Hayashi A., Kanai D., Shinmyozu K., Nakayama J. Roles of fission yeast Grc3 protein in ribosomal RNA processing and heterochromatic gene silencing. J. Biol. Chem. 2011;286:15391–15402. doi: 10.1074/jbc.M110.201343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Jain R., Shuman S. Characterization of a thermostable archaeal polynucleotide kinase homologous to human Clp1. RNA. 2009;15:923–931. doi: 10.1261/rna.1492809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Aravind L., Makarova K.S., Koonin E.V. SURVEY AND SUMMARY: holliday junction resolvases and related nucleases: identification of new families, phyletic distribution and evolutionary trajectories. Nucleic Acids Res. 2000;28:3417–3432. doi: 10.1093/nar/28.18.3417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Liu D., Wang Y.S., Wyss D.F. Solution structure of the hypothetical protein YqgF from Escherichia coli reveals an RNAse H fold. J. Biomol. NMR. 2003;27:389–392. doi: 10.1023/a:1025840121177. [DOI] [PubMed] [Google Scholar]
- 156.Nautiyal A., Sandhya Rani P., Sharples G.J., Muniyappa K. Mycobacterium tuberculosis RuvX is a Holliday junction resolvase formed by dimerisation of the monomeric YqgF nuclease domain. Mol. Microbiol. 2016;101:182. doi: 10.1111/mmi.13338. [DOI] [PubMed] [Google Scholar]
- 157.Kurata T., Nakanishi S., Hashimoto M., Taoka M., Yamazaki Y., Isobe T., Kato J. Novel essential gene Involved in 16S rRNA processing in Escherichia coli. J. Mol. Biol. 2015;427:955–965. doi: 10.1016/j.jmb.2014.12.013. [DOI] [PubMed] [Google Scholar]
- 158.Iwamoto A., Osawa A., Kawai M., Honda H., Yoshida S., Furuya N., Kato J. Mutations in the essential Escherichia coli gene, yqgF, and their effects on transcription. J. Mol. Microbiol. Biotechnol. 2012;22:17–23. doi: 10.1159/000336517. [DOI] [PubMed] [Google Scholar]
- 159.Mazumder R., Iyer L.M., Vasudevan S., Aravind L. Detection of novel members, structure-function analysis and evolutionary classification of the 2H phosphoesterase superfamily. Nucleic Acids Res. 2002;30:5229–5243. doi: 10.1093/nar/gkf645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Culver G.M., Consaul S.A., Tycowski K.T., Filipowicz W., Phizicky E.M. tRNA splicing in yeast and wheat germ. A cyclic phosphodiesterase implicated in the metabolism of ADP-ribose 1“,2"-cyclic phosphate. J. Biol. Chem. 1994;269:24928–24934. [PubMed] [Google Scholar]
- 161.Tyc K., Kellenberger C., Filipowicz W. Purification and characterization of wheat germ 2′,3′-cyclic nucleotide 3′-phosphodiesterase. J. Biol. Chem. 1987;262:12994–13000. [PubMed] [Google Scholar]
- 162.Xu Q., Teplow D., Lee T.D., Abelson J. Domain structure in yeast tRNA ligase. Biochemistry. 1990;29:6132–6138. doi: 10.1021/bi00478a004. [DOI] [PubMed] [Google Scholar]
- 163.Amitsur M., Levitz R., Kaufmann G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Greer C.L., Peebles C.L., Gegenheimer P., Abelson J. Mechanism of action of a yeast RNA ligase in tRNA splicing. Cell. 1983;32:537–546. doi: 10.1016/0092-8674(83)90473-7. [DOI] [PubMed] [Google Scholar]
- 165.Burroughs A.M., Allen K.N., Dunaway-Mariano D., Aravind L. Evolutionary genomics of the HAD superfamily: understanding the structural adaptations and catalytic diversity in a superfamily of phosphoesterases and allied enzymes. J. Mol. Biol. 2006;361:1003–1034. doi: 10.1016/j.jmb.2006.06.049. [DOI] [PubMed] [Google Scholar]
- 166.Morais M.C., Zhang W., Baker A.S., Zhang G., Dunaway-Mariano D., Allen K.N. The crystal structure of bacillus cereus phosphonoacetaldehyde hydrolase: insight into catalysis of phosphorus bond cleavage and catalytic diversification within the HAD enzyme superfamily. Biochemistry. 2000;39:10385–10396. doi: 10.1021/bi001171j. [DOI] [PubMed] [Google Scholar]
- 167.Allen K.N., Dunaway-Mariano D. Phosphoryl group transfer: evolution of a catalytic scaffold. Trends Biochem. Sci. 2004;29:495–503. doi: 10.1016/j.tibs.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 168.Soltis D.A., Uhlenbeck O.C. Isolation and characterization of two mutant forms of T4 polynucleotide kinase. J. Biol. Chem. 1982;257:11332–11339. [PubMed] [Google Scholar]
- 169.Wang L.K., Lima C.D., Shuman S. Structure and mechanism of T4 polynucleotide kinase: an RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Sharples G.J., Leach D.R. Structural and functional similarities between the SbcCD proteins of Escherichia coli and the RAD50 and MRE11 (RAD32) recombination and repair proteins of yeast. Mol. Microbiol. 1995;17:1215–1217. doi: 10.1111/j.1365-2958.1995.mmi_17061215_1.x. [DOI] [PubMed] [Google Scholar]
- 171.Aravind L., Koonin E.V. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Aravind L., Koonin E.V. The HD domain defines a new superfamily of metal-dependent phosphohydrolases. Trends Biochem. Sci. 1998;23:469–472. doi: 10.1016/s0968-0004(98)01293-6. [DOI] [PubMed] [Google Scholar]
- 173.Hogg T., Mechold U., Malke H., Cashel M., Hilgenfeld R. Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response [corrected] Cell. 2004;117:57–68. doi: 10.1016/s0092-8674(04)00260-0. [DOI] [PubMed] [Google Scholar]
- 174.Francis S.H., Colbran J.L., McAllister-Lucas L.M., Corbin J.D. Zinc interactions and conserved motifs of the cGMP-binding cGMP-specific phosphodiesterase suggest that it is a zinc hydrolase. J. Biol. Chem. 1994;269:22477–22480. [PubMed] [Google Scholar]
- 175.Zimmerman M.D., Proudfoot M., Yakunin A., Minor W. Structural insight into the mechanism of substrate specificity and catalytic activity of an HD-domain phosphohydrolase: the 5′-deoxyribonucleotidase YfbR from Escherichia coli. J. Mol. Biol. 2008;378:215–226. doi: 10.1016/j.jmb.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Guo S., Stolz L.E., Lemrow S.M., York J.D. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J. Biol. Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- 177.Whisstock J.C., Romero S., Gurung R., Nandurkar H., Ooms L.M., Bottomley S.P., Mitchell C.A. The inositol polyphosphate 5-phosphatases and the apurinic/apyrimidinic base excision repair endonucleases share a common mechanism for catalysis. J. Biol. Chem. 2000;275:37055–37061. doi: 10.1074/jbc.M006244200. [DOI] [PubMed] [Google Scholar]
- 178.Barford D., Das A.K., Egloff M.P. The structure and mechanism of protein phosphatases: insights into catalysis and regulation. Annu. Rev. Biophys. Biomol. Struct. 1998;27:133–164. doi: 10.1146/annurev.biophys.27.1.133. [DOI] [PubMed] [Google Scholar]
- 179.Nandurkar H.H., Layton M., Laporte J., Selan C., Corcoran L., Caldwell K.K., Mochizuki Y., Majerus P.W., Mitchell C.A. Identification of myotubularin as the lipid phosphatase catalytic subunit associated with the 3-phosphatase adapter protein, 3-PAP. Proc. Natl. Acad. Sci. U.S.A. 2003;100:8660–8665. doi: 10.1073/pnas.1033097100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Berger P., Bonneick S., Willi S., Wymann M., Suter U. Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum. Mol. Genet. 2002;11:1569–1579. doi: 10.1093/hmg/11.13.1569. [DOI] [PubMed] [Google Scholar]
- 181.Begley M.J., Taylor G.S., Kim S.A., Veine D.M., Dixon J.E., Stuckey J.A. Crystal structure of a phosphoinositide phosphatase, MTMR2: insights into myotubular myopathy and Charcot-Marie-Tooth syndrome. Mol. Cell. 2003;12:1391–1402. doi: 10.1016/s1097-2765(03)00486-6. [DOI] [PubMed] [Google Scholar]
- 182.Czerwoniec A., Bujnicki J.M. Identification and modeling of a phosphatase-like domain in a tRNA 2′-O-ribosyl phosphate transferase Rit1p. Cell Cycle. 2011;10:3566–3570. doi: 10.4161/cc.10.20.17857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Tumbale P., Williams J.S., Schellenberg M.J., Kunkel T.A., Williams R.S. Aprataxin resolves adenylated RNA-DNA junctions to maintain genome integrity. Nature. 2014;506:111–115. doi: 10.1038/nature12824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Hottiger M.O., Hassa P.O., Luscher B., Schuler H., Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem. Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 185.Han S., Tainer J.A. The ARTT motif and a unified structural understanding of substrate recognition in ADP-ribosylating bacterial toxins and eukaryotic ADP-ribosyltransferases. Int. J. Med. Microbiol.: IJMM. 2002;291:523–529. doi: 10.1078/1438-4221-00162. [DOI] [PubMed] [Google Scholar]
- 186.Ame J.C., Spenlehauer C., de Murcia G. The PARP superfamily. BioEssays. 2004;26:882–893. doi: 10.1002/bies.20085. [DOI] [PubMed] [Google Scholar]
- 187.Kato-Murayama M., Bessho Y., Shirouzu M., Yokoyama S. Crystal structure of the RNA 2′-phosphotransferase from Aeropyrum pernix K1. J. Mol. Biol. 2005;348:295–305. doi: 10.1016/j.jmb.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 188.Spinelli S.L., Kierzek R., Turner D.H., Phizicky E.M. Transient ADP-ribosylation of a 2′-phosphate implicated in its removal from ligated tRNA during splicing in yeast. J. Biol. Chem. 1999;274:2637–2644. doi: 10.1074/jbc.274.5.2637. [DOI] [PubMed] [Google Scholar]
- 189.McCraith S.M., Phizicky E.M. An enzyme from Saccharomyces cerevisiae uses NAD+ to transfer the splice junction 2′-phosphate from ligated tRNA to an acceptor molecule. J. Biol. Chem. 1991;266:11986–11992. [PubMed] [Google Scholar]
- 190.Culver G.M., McCraith S.M., Zillmann M., Kierzek R., Michaud N., LaReau R.D., Turner D.H., Phizicky E.M. An NAD derivative produced during transfer RNA splicing: ADP-ribose 1“-2" cyclic phosphate. Science. 1993;261:206–208. doi: 10.1126/science.8392224. [DOI] [PubMed] [Google Scholar]
- 191.Shull N.P., Spinelli S.L., Phizicky E.M. A highly specific phosphatase that acts on ADP-ribose 1″-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res. 2005;33:650–660. doi: 10.1093/nar/gki211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Brown K.A., Carpenter E.P., Watson K.A., Coggins J.R., Hawkins A.R., Koch M.H., Svergun D.I. Twists and turns: a tale of two shikimate-pathway enzymes. Biochem. Soc. Trans. 2003;31:543–547. doi: 10.1042/bst0310543. [DOI] [PubMed] [Google Scholar]
- 193.Aravind L., Iyer L.M., Anantharaman V. The two faces of Alba: the evolutionary connection between proteins participating in chromatin structure and RNA metabolism. Genome Biol. 2003;4:R64. doi: 10.1186/gb-2003-4-10-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Genschik P., Drabikowski K., Filipowicz W. Characterization of the Escherichia coli RNA 3′-terminal phosphate cyclase and its sigma54-regulated operon. J. Biol. Chem. 1998;273:25516–25526. doi: 10.1074/jbc.273.39.25516. [DOI] [PubMed] [Google Scholar]
- 195.Das U., Shuman S. 2′-Phosphate cyclase activity of RtcA: a potential rationale for the operon organization of RtcA with an RNA repair ligase RtcB in Escherichia coli and other bacterial taxa. RNA. 2013;19:1355–1362. doi: 10.1261/rna.039917.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Desai K.K., Bingman C.A., Cheng C.L., Phillips G.N., Jr, Raines R.T. Structure of RNA 3′-phosphate cyclase bound to substrate RNA. RNA. 2014;20:1560–1566. doi: 10.1261/rna.045823.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Mui Chan C., Zhou C., Brunzelle J.S., Huang R.H. Structural and biochemical insights into 2′-O-methylation at the 3′-terminal nucleotide of RNA by Hen1. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17699–17704. doi: 10.1073/pnas.0907540106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Martins A., Shuman S. An end-healing enzyme from Clostridium thermocellum with 5′ kinase, 2′,3′ phosphatase, and adenylyltransferase activities. RNA. 2005;11:1271–1280. doi: 10.1261/rna.2690505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wang P., Selvadurai K., Huang R.H. Reconstitution and structure of a bacterial Pnkp1-Rnl-Hen1 RNA repair complex. Nat. Commun. 2015;6:6876. doi: 10.1038/ncomms7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Zhang C., Chan C.M., Wang P., Huang R.H. Probing the substrate specificity of the bacterial Pnkp/Hen1 RNA repair system using synthetic RNAs. RNA. 2012;18:335–344. doi: 10.1261/rna.030502.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Chan C.M., Zhou C., Huang R.H. Reconstituting bacterial RNA repair and modification in vitro. Science. 2009;326:247. doi: 10.1126/science.1179480. [DOI] [PubMed] [Google Scholar]
- 202.Horwich M.D., Li C., Matranga C., Vagin V., Farley G., Wang P., Zamore P.D. The Drosophila RNA methyltransferase, DmHen1, modifies germline piRNAs and single-stranded siRNAs in RISC. Curr. Biol.: CB. 2007;17:1265–1272. doi: 10.1016/j.cub.2007.06.030. [DOI] [PubMed] [Google Scholar]
- 203.Saito K., Sakaguchi Y., Suzuki T., Siomi H., Siomi M.C. Pimet, the Drosophila homolog of HEN1, mediates 2′-O-methylation of Piwi- interacting RNAs at their 3′ ends. Genes Dev. 2007;21:1603–1608. doi: 10.1101/gad.1563607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Kurth H.M., Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Li J., Yang Z., Yu B., Liu J., Chen X. Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr. Biol.: CB. 2005;15:1501–1507. doi: 10.1016/j.cub.2005.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Yu B., Yang Z., Li J., Minakhina S., Yang M., Padgett R.W., Steward R., Chen X. Methylation as a crucial step in plant microRNA biogenesis. Science. 2005;307:932–935. doi: 10.1126/science.1107130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Tu B., Liu L., Xu C., Zhai J., Li S., Lopez M.A., Zhao Y., Yu Y., Ramachandran V., Ren G., et al. Distinct and cooperative activities of HESO1 and URT1 nucleotidyl transferases in microRNA turnover in Arabidopsis. PLoS Genet. 2015;11:e1005119. doi: 10.1371/journal.pgen.1005119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ren G., Chen X., Yu B. Uridylation of miRNAs by hen1 suppressor1 in Arabidopsis. Curr. Biol.: CB. 2012;22:695–700. doi: 10.1016/j.cub.2012.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Zhao Y., Yu Y., Zhai J., Ramachandran V., Dinh T.T., Meyers B.C., Mo B., Chen X. The Arabidopsis nucleotidyl transferase HESO1 uridylates unmethylated small RNAs to trigger their degradation. Curr. Biol.: CB. 2012;22:689–694. doi: 10.1016/j.cub.2012.02.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Neuwald A.F., Aravind L., Spouge J.L., Koonin E.V. AAA+: A class of chaperone-like ATPases associated with the assembly, operation, and disassembly of protein complexes. Genome Res. 1999;9:27–43. [PubMed] [Google Scholar]
- 211.Iyer L.M., Leipe D.D., Koonin E.V., Aravind L. Evolutionary history and higher order classification of AAA+ ATPases. J. Struct. Biol. 2004;146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 212.Morett E., Segovia L. The sigma 54 bacterial enhancer-binding protein family: mechanism of action and phylogenetic relationship of their functional domains. J. Bacteriol. 1993;175:6067–6074. doi: 10.1128/jb.175.19.6067-6074.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Makarova K.S., Anantharaman V., Grishin N.V., Koonin E.V., Aravind L. CARF and WYL domains: ligand-binding regulators of prokaryotic defense systems. Front. Genet. 2014;5:102. doi: 10.3389/fgene.2014.00102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Canaves J.M. Predicted role for the archease protein family based on structural and sequence analysis of TM1083 and MTH1598, two proteins structurally characterized through structural genomics efforts. Proteins. 2004;56:19–27. doi: 10.1002/prot.20141. [DOI] [PubMed] [Google Scholar]
- 215.Anantharaman V., Aravind L. The SHS2 module is a common structural theme in functionally diverse protein groups, like Rpb7p, FtsA, GyrI, and MTH1598/TM1083 superfamilies. Proteins. 2004;56:795–807. doi: 10.1002/prot.20140. [DOI] [PubMed] [Google Scholar]
- 216.Desai K.K., Cheng C.L., Bingman C.A., Phillips G.N., Jr, Raines R.T. A tRNA splicing operon: Archease endows RtcB with dual GTP/ATP cofactor specificity and accelerates RNA ligation. Nucleic Acids Res. 2014;42:3931–3942. doi: 10.1093/nar/gkt1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Popow J., Jurkin J., Schleiffer A., Martinez J. Analysis of orthologous groups reveals archease and DDX1 as tRNA splicing factors. Nature. 2014;511:104–107. doi: 10.1038/nature13284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Desai K.K., Beltrame A.L., Raines R.T. Coevolution of RtcB and Archease created a multiple-turnover RNA ligase. RNA. 2015;21:1866–1872. doi: 10.1261/rna.052639.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Auxilien S., El Khadali F., Rasmussen A., Douthwaite S., Grosjean H. Archease from Pyrococcus abyssi improves substrate specificity and solubility of a tRNA m5C methyltransferase. J. Biol. Chem. 2007;282:18711–18721. doi: 10.1074/jbc.M607459200. [DOI] [PubMed] [Google Scholar]
- 220.Makarova K.S., Aravind L., Wolf Y.I., Tatusov R.L., Minton K.W., Koonin E.V., Daly M.J. Genome of the extremely radiation-resistant bacterium Deinococcus radiodurans viewed from the perspective of comparative genomics. Microbiol. Mol. Biol. Rev.: MMBR. 2001;65:44–79. doi: 10.1128/MMBR.65.1.44-79.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Bateman A., Kickhoefer V. The TROVE module: a common element in Telomerase, Ro and Vault ribonucleoproteins. BMC Bioinformatics. 2003;4:49. doi: 10.1186/1471-2105-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Ruggeri Z.M., Ware J. von Willebrand factor. FASEB J. 1993;7:308–316. doi: 10.1096/fasebj.7.2.8440408. [DOI] [PubMed] [Google Scholar]
- 223.Bork P., Rohde K. More von Willebrand factor type A domains? Sequence similarities with malaria thrombospondin-related anonymous protein, dihydropyridine-sensitive calcium channel and inter-alpha-trypsin inhibitor. Biochem. J. 1991;279:908–910. doi: 10.1042/bj2790908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Purcell A.W., Todd A., Kinoshita G., Lynch T.A., Keech C.L., Gething M.J., Gordon T.P. Association of stress proteins with autoantigens: a possible mechanism for triggering autoimmunity? Clin. Exp. Immunol. 2003;132:193–200. doi: 10.1046/j.1365-2249.2003.02153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Chen X., Quinn A.M., Wolin S.L. Ro ribonucleoproteins contribute to the resistance of Deinococcus radiodurans to ultraviolet irradiation. Genes Dev. 2000;14:777–782. [PMC free article] [PubMed] [Google Scholar]
- 226.Chen X., Wurtmann E.J., Van Batavia J., Zybailov B., Washburn M.P., Wolin S.L. An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans. Genes Dev. 2007;21:1328–1339. doi: 10.1101/gad.1548207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Stein A.J., Fuchs G., Fu C., Wolin S.L., Reinisch K.M. Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell. 2005;121:529–539. doi: 10.1016/j.cell.2005.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Sim S., Wolin S.L. Emerging roles for the Ro 60-kDa autoantigen in noncoding RNA metabolism. Wiley Interdiscipl. Rev. RNA. 2011;2:686–699. doi: 10.1002/wrna.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Wurtmann E.J., Wolin S.L. A role for a bacterial ortholog of the Ro autoantigen in starvation-induced rRNA degradation. Proc. Natl. Acad. Sci. U.S.A. 2010;107:4022–4027. doi: 10.1073/pnas.1000307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Chen X., Taylor D.W., Fowler C.C., Galan J.E., Wang H.W., Wolin S.L. An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell. 2013;153:166–177. doi: 10.1016/j.cell.2013.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Chen X., Sim S., Wurtmann E.J., Feke A., Wolin S.L. Bacterial noncoding Y RNAs are widespread and mimic tRNAs. RNA. 2014;20:1715–1724. doi: 10.1261/rna.047241.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Nawrocki E.P., Eddy S.R. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29:2933–2935. doi: 10.1093/bioinformatics/btt509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Tavernarakis N., Driscoll M., Kyrpides N.C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 1999;24:425–427. doi: 10.1016/s0968-0004(99)01467-x. [DOI] [PubMed] [Google Scholar]
- 234.Hinderhofer M., Walker C.A., Friemel A., Stuermer C.A., Moller H.M., Reuter A. Evolution of prokaryotic SPFH proteins. BMC Evol. Biol. 2009;9:10. doi: 10.1186/1471-2148-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Green J.B., Young J.P. Slipins: ancient origin, duplication and diversification of the stomatin protein family. BMC Evol. Biol. 2008;8:44. doi: 10.1186/1471-2148-8-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Nijtmans L.G., Artal S.M., Grivell L.A., Coates P.J. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell. Mol. Life Sci.: CMLS. 2002;59:143–155. doi: 10.1007/s00018-002-8411-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Tanaka H., Kato K., Yamashita E., Sumizawa T., Zhou Y., Yao M., Iwasaki K., Yoshimura M., Tsukihara T. The structure of rat liver vault at 3.5 angstrom resolution. Science. 2009;323:384–388. doi: 10.1126/science.1164975. [DOI] [PubMed] [Google Scholar]
- 238.Butcher B.G., Helmann J.D. Identification of Bacillus subtilis sigma-dependent genes that provide intrinsic resistance to antimicrobial compounds produced by Bacilli. Mol. Microbiol. 2006;60:765–782. doi: 10.1111/j.1365-2958.2006.05131.x. [DOI] [PubMed] [Google Scholar]
- 239.Bickel P.E., Scherer P.E., Schnitzer J.E., Oh P., Lisanti M.P., Lodish H.F. Flotillin and epidermal surface antigen define a new family of caveolae-associated integral membrane proteins. J. Biol. Chem. 1997;272:13793–13802. doi: 10.1074/jbc.272.21.13793. [DOI] [PubMed] [Google Scholar]
- 240.Langhorst M.F., Reuter A., Stuermer C.A. Scaffolding microdomains and beyond: the function of reggie/flotillin proteins. Cell. Mol. Life Sci.: CMLS. 2005;62:2228–2240. doi: 10.1007/s00018-005-5166-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Yokoyama H., Matsui I. A novel thermostable membrane protease forming an operon with a stomatin homolog from the hyperthermophilic archaebacterium Pyrococcus horikoshii. J. Biol. Chem. 2005;280:6588–6594. doi: 10.1074/jbc.M411748200. [DOI] [PubMed] [Google Scholar]
- 242.Chiba S., Ito K., Akiyama Y. The Escherichia coli plasma membrane contains two PHB (prohibitin homology) domain protein complexes of opposite orientations. Mol. Microbiol. 2006;60:448–457. doi: 10.1111/j.1365-2958.2006.05104.x. [DOI] [PubMed] [Google Scholar]
- 243.Anderson D.H., Kickhoefer V.A., Sievers S.A., Rome L.H., Eisenberg D. Draft crystal structure of the vault shell at 9-A resolution. PLoS Biol. 2007;5:e318. doi: 10.1371/journal.pbio.0050318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Casanas A., Querol-Audi J., Guerra P., Pous J., Tanaka H., Tsukihara T., Verdaguer N., Fita I. New features of vault architecture and dynamics revealed by novel refinement using the deformable elastic network approach. Acta Crystallogr. Sect. D, Biol. Crystallogr. 2013;69:1054–1061. doi: 10.1107/S0907444913004472. [DOI] [PubMed] [Google Scholar]
- 245.Kowalski M.P., Krude T. Functional roles of non-coding Y RNAs. Int. J. Biochem. Cell Biol. 2015;66:20–29. doi: 10.1016/j.biocel.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Wolin S.L., Belair C., Boccitto M., Chen X., Sim S., Taylor D.W., Wang H.W. Non-coding Y RNAs as tethers and gates: Insights from bacteria. RNA Biol. 2013;10:1602–1608. doi: 10.4161/rna.26166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Bateman A., Coggill P., Finn R.D. DUFs: families in search of function. Acta Crystallogr. Sect. F, Struct. Biol. Crystall. Commun. 2010;66:1148–1152. doi: 10.1107/S1744309110001685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Christendat D., Yee A., Dharamsi A., Kluger Y., Savchenko A., Cort J.R., Booth V., Mackereth C.D., Saridakis V., Ekiel I., et al. Structural proteomics of an archaeon. Nat. Struct. Biol. 2000;7:903–909. doi: 10.1038/82823. [DOI] [PubMed] [Google Scholar]
- 249.Park S.Y., Choi H.K., Jo S.H., Seo J., Han E.J., Choi K.C., Jeong J.W., Choi Y., Yoon H.G. YAF2 promotes TP53-mediated genotoxic stress response via stabilization of PDCD5. Biochim. Biophys. Acta. 2015;1853:1060–1072. doi: 10.1016/j.bbamcr.2015.01.006. [DOI] [PubMed] [Google Scholar]
- 250.Bock F.J., Tanzer M.C., Haschka M.D., Krumschnabel G., Sohm B., Goetsch K., Kofler R., Villunger A. The p53 binding protein PDCD5 is not rate-limiting in DNA damage induced cell death. Sci. Rep. 2015;5:11268. doi: 10.1038/srep11268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Blanc V., Alfonzo J.D., Aphasizhev R., Simpson L. The mitochondrial RNA ligase from Leishmania tarentolae can join RNA molecules bridged by a complementary RNA. J. Biol. Chem. 1999;274:24289–24296. doi: 10.1074/jbc.274.34.24289. [DOI] [PubMed] [Google Scholar]
- 252.Palazzo S.S., Panigrahi A.K., Igo R.P., Salavati R., Stuart K. Kinetoplastid RNA editing ligases: complex association, characterization, and substrate requirements. Mol. Biochem. Parasitol. 2003;127:161–167. doi: 10.1016/s0166-6851(02)00333-x. [DOI] [PubMed] [Google Scholar]
- 253.Nandakumar J., Shuman S., Lima C.D. RNA ligase structures reveal the basis for RNA specificity and conformational changes that drive ligation forward. Cell. 2006;127:71–84. doi: 10.1016/j.cell.2006.08.038. [DOI] [PubMed] [Google Scholar]
- 254.Iyer L.M., Balaji S., Koonin E.V., Aravind L. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 255.Jabbar M.A., Snyder L. Genetic and physiological studies of an Escherichia coli locus that restricts polynucleotide kinase- and RNA ligase-deficient mutants of bacteriophage T4. J. Virol. 1984;51:522–529. doi: 10.1128/jvi.51.2.522-529.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Konarska M., Filipowicz W., Gross H.J. RNA ligation via 2′-phosphomonoester, 3′5′-phosphodiester linkage: requirement of 2′,3′-cyclic phosphate termini and involvement of a 5′-hydroxyl polynucleotide kinase. Proc. Natl. Acad. Sci. U.S.A. 1982;79:1474–1478. doi: 10.1073/pnas.79.5.1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Pick L., Furneaux H., Hurwitz J. Purification of wheat germ RNA ligase. II. Mechanism of action of wheat germ RNA ligase. J. Biol. Chem. 1986;261:6694–6704. [PubMed] [Google Scholar]
- 258.Schwartz R.C., Greer C.L., Gegenheimer P., Abelson J. Enzymatic mechanism of an RNA ligase from wheat germ. J. Biol. Chem. 1983;258:8374–8383. [PubMed] [Google Scholar]
- 259.Pick L., Hurwitz J. Purification of wheat germ RNA ligase. I. Characterization of a ligase-associated 5′-hydroxyl polynucleotide kinase activity. J. Biol. Chem. 1986;261:6684–6693. [PubMed] [Google Scholar]
- 260.Englert M., Beier H. Plant tRNA ligases are multifunctional enzymes that have diverged in sequence and substrate specificity from RNA ligases of other phylogenetic origins. Nucleic Acids Res. 2005;33:388–399. doi: 10.1093/nar/gki174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Sabaliauskienė V., Jonušienė V., Labeikytė D., Juodka B. Isolation and partial purification of tRNA ligase from Tetrahymena pyriformis. Biologija. 2003:7–11. [Google Scholar]
- 262.Rogozin I.B., Aravind L., Koonin E.V. Differential action of natural selection on the N and C-terminal domains of 2′-5′ oligoadenylate synthetases and the potential nuclease function of the C-terminal domain. J. Mol. Biol. 2003;326:1449–1461. doi: 10.1016/s0022-2836(03)00055-x. [DOI] [PubMed] [Google Scholar]
- 263.Venancio T.M., Balaji S., Iyer L.M., Aravind L. Reconstructing the ubiquitin network: cross-talk with other systems and identification of novel functions. Genome Biol. 2009;10:R33. doi: 10.1186/gb-2009-10-3-r33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Klaiman D., Steinfels-Kohn E., Krutkina E., Davidov E., Kaufmann G. The wobble nucleotide-excising anticodon nuclease RloC is governed by the zinc-hook and DNA-dependent ATPase of its Rad50-like region. Nucleic Acids Res. 2012;40:8568–8578. doi: 10.1093/nar/gks593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Brooks M.A., Meslet-Cladiere L., Graille M., Kuhn J., Blondeau K., Myllykallio H., van Tilbeurgh H. The structure of an archaeal homodimeric ligase which has RNA circularization activity. Protein Sci. 2008;17:1336–1345. doi: 10.1110/ps.035493.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Torchia C., Takagi Y., Ho C.K. Archaeal RNA ligase is a homodimeric protein that catalyzes intramolecular ligation of single-stranded RNA and DNA. Nucleic Acids Res. 2008;36:6218–6227. doi: 10.1093/nar/gkn602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Plagens A., Daume M., Wiegel J., Randau L. Circularization restores signal recognition particle RNA functionality in Thermoproteus. eLife. 2015;4:e11623. doi: 10.7554/eLife.11623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Baranov P.V., Vestergaard B., Hamelryck T., Gesteland R.F., Nyborg J., Atkins J.F. Diverse bacterial genomes encode an operon of two genes, one of which is an unusual class-I release factor that potentially recognizes atypical mRNA signals other than normal stop codons. Biol. Direct. 2006;1:28. doi: 10.1186/1745-6150-1-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Kisselev L.L., Buckingham R.H. Translational termination comes of age. Trends Biochem. Sci. 2000;25:561–566. doi: 10.1016/s0968-0004(00)01669-8. [DOI] [PubMed] [Google Scholar]
- 270.Aravind L., Anantharaman V., Koonin E.V. Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: implications for protein evolution in the RNA. Proteins. 2002;48:1–14. doi: 10.1002/prot.10064. [DOI] [PubMed] [Google Scholar]
- 271.Anantharaman V., Koonin E.V., Aravind L. Comparative genomics and evolution of proteins involved in RNA metabolism. Nucleic Acids Res. 2002;30:1427–1464. doi: 10.1093/nar/30.7.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Aravind L. An evolutionary classification of the metallo-beta-lactamase fold proteins. In Silico Biol. 1999;1:69–91. [PubMed] [Google Scholar]
- 273.Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drablos F., Lennartsson A., Ronnerblad M., Hrydziuszko O., Vitezic M., et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Lizio M., Harshbarger J., Shimoji H., Severin J., Kasukawa T., Sahin S., Abugessaisa I., Fukuda S., Hori F., Ishikawa-Kato S., et al. Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 2015;16:22. doi: 10.1186/s13059-014-0560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Shi H., Tschudi C., Ullu E. An unusual Dicer-like1 protein fuels the RNA interference pathway in Trypanosoma brucei. RNA. 2006;12:2063–2072. doi: 10.1261/rna.246906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Patrick K.L., Shi H., Kolev N.G., Ersfeld K., Tschudi C., Ullu E. Distinct and overlapping roles for two Dicer-like proteins in the RNA interference pathways of the ancient eukaryote Trypanosoma brucei. Proc. Natl. Acad. Sci. U.S.A. 2009;106:17933–17938. doi: 10.1073/pnas.0907766106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Dong G., Chakshusmathi G., Wolin S.L., Reinisch K.M. Structure of the La motif: a winged helix domain mediates RNA binding via a conserved aromatic patch. EMBO J. 2004;23:1000–1007. doi: 10.1038/sj.emboj.7600115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Steiger M.A., Jackman J.E., Phizicky E.M. Analysis of 2′-phosphotransferase (Tpt1p) from Saccharomyces cerevisiae: evidence for a conserved two-step reaction mechanism. RNA. 2005;11:99–106. doi: 10.1261/rna.7194605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Sawaya R., Schwer B., Shuman S. Structure-function analysis of the yeast NAD+-dependent tRNA 2′-phosphotransferase Tpt1. RNA. 2005;11:107–113. doi: 10.1261/rna.7193705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Rudd S.G., Glover L., Jozwiakowski S.K., Horn D., Doherty A.J. PPL2 translesion polymerase is essential for the completion of chromosomal DNA replication in the African trypanosome. Mol. Cell. 2013;52:554–565. doi: 10.1016/j.molcel.2013.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Durand-Dubief M., Bastin P. TbAGO1, an argonaute protein required for RNA interference, is involved in mitosis and chromosome segregation in Trypanosoma brucei. BMC Biol. 2003;1:2. doi: 10.1186/1741-7007-1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Shi H., Djikeng A., Tschudi C., Ullu E. Argonaute protein in the early divergent eukaryote Trypanosoma brucei: control of small interfering RNA accumulation and retroposon transcript abundance. Mol. Cell. Biol. 2004;24:420–427. doi: 10.1128/MCB.24.1.420-427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Garcia Silva M.R., Tosar J.P., Frugier M., Pantano S., Bonilla B., Esteban L., Serra E., Rovira C., Robello C., Cayota A. Cloning, characterization and subcellular localization of a Trypanosoma cruzi argonaute protein defining a new subfamily distinctive of trypanosomatids. Gene. 2010;466:26–35. doi: 10.1016/j.gene.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 284.Barnes R.L., Shi H., Kolev N.G., Tschudi C., Ullu E. Comparative genomics reveals two novel RNAi factors in Trypanosoma brucei and provides insight into the core machinery. PLoS Pathogens. 2012;8:e1002678. doi: 10.1371/journal.ppat.1002678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 285.Tschudi C., Shi H., Franklin J.B., Ullu E. Small interfering RNA-producing loci in the ancient parasitic eukaryote Trypanosoma brucei. BMC Genomics. 2012;13:427. doi: 10.1186/1471-2164-13-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 286.Atayde V.D., Shi H., Franklin J.B., Carriero N., Notton T., Lye L.F., Owens K., Beverley S.M., Tschudi C., Ullu E. The structure and repertoire of small interfering RNAs in Leishmania (Viannia) braziliensis reveal diversification in the trypanosomatid RNAi pathway. Mol. Microbiol. 2013;87:580–593. doi: 10.1111/mmi.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Aphasizhev R., Aphasizheva I. Mitochondrial RNA editing in trypanosomes: small RNAs in control. Biochimie. 2014;100:125–131. doi: 10.1016/j.biochi.2014.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Keen B.A., Jozwiakowski S.K., Bailey L.J., Bianchi J., Doherty A.J. Molecular dissection of the domain architecture and catalytic activities of human PrimPol. Nucleic Acids Res. 2014;42:5830–5845. doi: 10.1093/nar/gku214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Iyer L.M., Makarova K.S., Koonin E.V., Aravind L. Comparative genomics of the FtsK-HerA superfamily of pumping ATPases: implications for the origins of chromosome segregation, cell division and viral capsid packaging. Nucleic Acids Res. 2004;32:5260–5279. doi: 10.1093/nar/gkh828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Aravind L., Walker D.R., Koonin E.V. Conserved domains in DNA repair proteins and evolution of repair systems. Nucleic Acids Res. 1999;27:1223–1242. doi: 10.1093/nar/27.5.1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Aravind L., Koonin E.V. The DNA-repair protein AlkB, EGL-9, and leprecan define new families of 2-oxoglutarate- and iron-dependent dioxygenases. Genome Biol. 2001;2 doi: 10.1186/gb-2001-2-3-research0007. RESEARCH0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Leipe D.D., Aravind L., Koonin E.V. Did DNA replication evolve twice independently? Nucleic Acids Res. 1999;27:3389–3401. doi: 10.1093/nar/27.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Aravind L., Koonin E.V. DNA polymerase beta-like nucleotidyltransferase superfamily: identification of three new families, classification and evolutionary history. Nucleic Acids Res. 1999;27:1609–1618. doi: 10.1093/nar/27.7.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Martin G., Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Burroughs A.M., Iyer L.M., Aravind L. Functional diversification of the RING finger and other binuclear treble clef domains in prokaryotes and the early evolution of the ubiquitin system. Mol. bioSyst. 2011;7:2261–2277. doi: 10.1039/c1mb05061c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Burroughs A.M., Iyer L.M., Aravind L. The natural history of ubiquitin and ubiquitin-related domains. Front Biosci. (Landmark Ed.) 2012;17:1433–1460. doi: 10.2741/3996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Rankin D.J., Rocha E.P., Brown S.P. What traits are carried on mobile genetic elements, and why? Heredity. 2011;106:1–10. doi: 10.1038/hdy.2010.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Soucy S.M., Huang J., Gogarten J.P. Horizontal gene transfer: building the web of life. Nat. Rev. Genet. 2015;16:472–482. doi: 10.1038/nrg3962. [DOI] [PubMed] [Google Scholar]
- 299.Aravind L., Koonin E.V. Prokaryotic homologs of the eukaryotic DNA-end-binding protein Ku, novel domains in the Ku protein and prediction of a prokaryotic double-strand break repair system. Genome Res. 2001;11:1365–1374. doi: 10.1101/gr.181001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Maynard Smith J.S.E. The Major Transitions in Evolution. NY: W. H. Freeman Spektrum; 1995. [Google Scholar]
- 301.Aravind L., Anantharaman V., Zhang D., de Souza R.F., Iyer L.M. Gene flow and biological conflict systems in the origin and evolution of eukaryotes. Front. Cell. Infect. Microbiol. 2012;2:89. doi: 10.3389/fcimb.2012.00089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302.Zhang D., Burroughs A.M., Vidal N.D., Iyer L.M., Aravind L. Transposons to toxins: the provenance, architecture and diversification of a widespread class of eukaryotic effectors. Nucleic Acids Res. 2016;44:3513–3533. doi: 10.1093/nar/gkw221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Ramirez-Garces D., Camborde L., Pel M.J., Jauneau A., Martinez Y., Neant I., Leclerc C., Moreau M., Dumas B., Gaulin E. CRN13 candidate effectors from plant and animal eukaryotic pathogens are DNA-binding proteins which trigger host DNA damage response. New Phytologist. 2016;210:602–617. doi: 10.1111/nph.13774. [DOI] [PubMed] [Google Scholar]
- 304.Stam R., Jupe J., Howden A.J., Morris J.A., Boevink P.C., Hedley P.E., Huitema E. Identification and Characterisation CRN Effectors in Phytophthora capsici Shows Modularity and Functional Diversity. PLoS One. 2013;8:e59517. doi: 10.1371/journal.pone.0059517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Hornung V., Hartmann R., Ablasser A., Hopfner K.P. OAS proteins and cGAS: unifying concepts in sensing and responding to cytosolic nucleic acids. Nat. Rev. Immunol. 2014;14:521–528. doi: 10.1038/nri3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 306.Popow J., Schleiffer A., Martinez J. Diversity and roles of (t)RNA ligases. Cell. Mol. Life Sci.: CMLS. 2012;69:2657–2670. doi: 10.1007/s00018-012-0944-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Phizicky E.M. Waste not, want not: a case for tRNA repair. Nat. Struct. Mol. Biol. 2008;15:783–784. doi: 10.1038/nsmb0808-783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.