Abstract
DNA segment exchange by site-specific serine recombinases (SRs) is thought to proceed by rigid-body rotation of the two halves of the synaptic complex, following the cleavages that create the two pairs of exchangeable ends. It remains unresolved how the amount of rotation occurring between cleavage and religation is controlled. We report single-DNA experiments for Bxb1 integrase, a model SR, where dynamics of individual synapses were observed, using relaxation of supercoiling to report on cleavage and rotation events. Relaxation events often consist of multiple rotations, with the number of rotations per relaxation event and rotation velocity sensitive to DNA sequence at the center of the recombination crossover site, torsional stress and salt concentration. Bulk and single-DNA experiments indicate that the thermodynamic stability of the annealed, but cleaved, crossover sites controls ligation efficiency of recombinant and parental synaptic complexes, regulating the number of rotations during a breakage-religation cycle. The outcome is consistent with a ‘controlled rotation’ model analogous to that observed for type IB topoisomerases, with religation probability varying in accord with DNA base-pairing free energies at the crossover site. Significantly, we find no evidence for a special regulatory mechanism favoring ligation and product release after a single 180° rotation.
INTRODUCTION
Serine integrases belong to a family of site-specific recombinases that exchange two segments of DNA and are named for their conserved active site residue that is critical for DNA cleavage. No net nucleotides are gained or lost during the reaction catalyzed by serine recombinases (SRs), and no high energy or metal cofactors are required for catalysis, making it an energetically conservative process (1). Instead, the energy of the cleaved phosphodiester bonds is stored in the form of covalent phosphoserine linkages between DNA and enzyme.
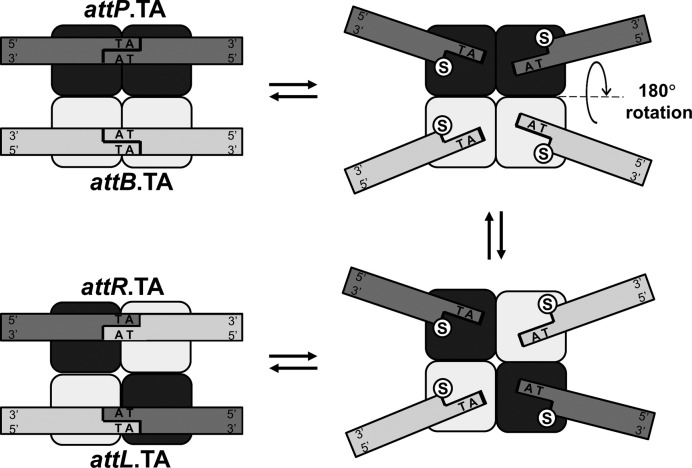
All serine integrases likely function by the same basic mechanism whereby two dimers of protein subunits bind two ∼50 bp double stranded DNA attachment sites (att). The two DNA-bound dimers associate to form a synaptic complex composed of a tetrameric core of protein subunits flanked by a set of two att sites (Figure 1). The recombinase binding (attachment) sites are denoted as attP (phage), attB (bacteria) and their recombination products as attL (left) and attR (right). Nucleophilic attack by the conserved serine residue in each protomer cleaves both duplexes resulting in covalent 5′-phosphoserine linkages and two nucleotide overhangs with free 3′ hydroxyls. Structural (2–6), topological (7–10) and biophysical (11) studies have examined the next step of the reaction, which has been proposed to be DNA half-site exchange via subunit rotation of half of the tetrameric complex along a planar hydrophobic dimer–dimer interface. Nucleophilic attack by the free 3′ hydroxyls reseals the nicks and completes the process that forms the recombinant products. While additional cofactors are not required, accessory proteins, supercoiling state and orientation and sequence of recombination sites control the outcome of the reaction pathways of SRs (12–16). Due to their simplicity and fidelity, large SRs are of interest for precision genome engineering and synthetic biology applications (17) and there is great interest in understanding their mechanism (18,19).
Figure 1.
Site-specific recombination by serine integrases. Cleavage leads to a state in which half-sites can be exchanged by rotation (upper right to lower right). Hybridization and religation after an odd number of half turns will result in recombination (lower left); religation after an even number of half turns returns to the parental state (upper left).
Bxb1 integrase (Int) is a large (500 amino acid) SR from a bacteriophage of Mycobacterium smegmatis with a ∼150 amino acid canonical N-terminal catalytic domain and extended C-terminal domain responsible for coordination of attachment sites (20,21). Bxb1 Int is able to recombine attP × attB to form attL and attR, but it cannot recombine attL × attR without an extra directionality factor, nor can it recombine alternative pairs of sites (22). Int requires neither regulatory DNA sites nor supercoiling, rendering it a simple and tractable model system. However, biochemical studies have shown that the central dinucleotide sequence regulates the outcome of an attP/attB synaptic complex, via complementarity of the two-nucleotide overhangs (13). After an initial 180° rotation, if the 3′ overhangs of the new partner half-sites cannot base-pair, religation cannot occur. In this case where the recombinant 2-base overhangs are incompatible, the parental strands are able to reform following an additional 180° rotation (Figure 1) (23). This limits prophage insertions to the correct orientation (13,14) and prevents inversions instead of prophage excisions.
A few distinct models for the SR strand exchange mechanism have been discussed. Given a rotational mechanism, ‘gated’ models propose that rotation is tightly regulated, where a single 180° or 360° rotation is the most heavily favored outcome (10). Alternately, one could envision a ‘controlled rotation’ model with a fixed ligation probability at each opportunity, with the possibility of multiple rotations occurring, depending on the probability of religation per turn; experimental data in favor of such a model include direct observation of multiple-turn events by Bxb1 Int (11). Finally, we note that a non-rotational model has been proposed, based on a type-II-topoisomerase-like mechanism to tightly restrict topology change during recombination (24) although such models are incompatible with experiments suggesting a rotational mechanism (11).
Within the rotational models, a key question is whether rotations within the synaptic complex of various SRs are limited to the first ligation-compatible opportunity (180° for recombination of parallel-aligned attB and attP sites, or 360° for rejoining the parental sites or for antiparallel attP and attB sites). For a rotational process that can undergo 180° rotations in synaptic complexes with parallel-aligned sites or 360° in those with antiparallel sites, it is difficult to imagine how such strict rotational control could occur. However, if the number of rotations is determined stochastically by the lifetime of the pre-ligation state and the rate of DNA annealing and religation, then a sufficiently efficient annealing and religation would give the appearance of a gating process, whereas inefficiency would lead to multiple rotations preceding ligation, similar to the ‘controlled rotation’ of type IB and type IC topoisomerases (25,26).
Ensemble experiments with Bxb1 Int (11,13) and ϕC31 Int (14), both members of the large SR family, have shown evidence of multiple 180° rotations when using supercoiled substrates, leading to a variety of topologically unique products, suggesting a controlled rotation mechanism. Evidence for the gating model was provided by a hybrid recombination system constructed of a ϕC31 Int att site adjacent to Tn3 resolvase regulatory sites, which showed highly limited rotations when the DNA substrates were preincubated with Tn3 resolvase (10). This system may suppress multiple rotations by artificial imposition of topological constraints; experiments with ϕC31 Int in the absence of Tn3 resolvase yielded a greater variety of topologically distinct products, consistent with this hypothesis. Similar results were observed when Bxb1 Int was used with its cognate att sites in place of ϕC31 Int. The presence of glycerol and absence of MgCl2 in those experiments also may have affected the stability of cleaved intermediates (27–29).
Single-molecule magnetic tweezers studies have provided a method to probe Bxb1 Int recombination (11) whereby the results of synaptic rotations are directly observed. Prior studies showed that most supercoils stored in the attP tethered substrate were removed during a single synapsis and strand exchange event with a recombinationally-incompatible attB oligonucleotide (11). Here we present ensemble and single molecule experiments that show that the balance between ligation and rotation depends on substrate sequence, DNA torsional stress, and reaction conditions. Specifically, we focused on investigating the role played by the thermodynamic stability of the central two base pairs of the crossover sites in controlling rotation during the recombination reaction.
MATERIALS AND METHODS
DNA constructs for ensemble experiments
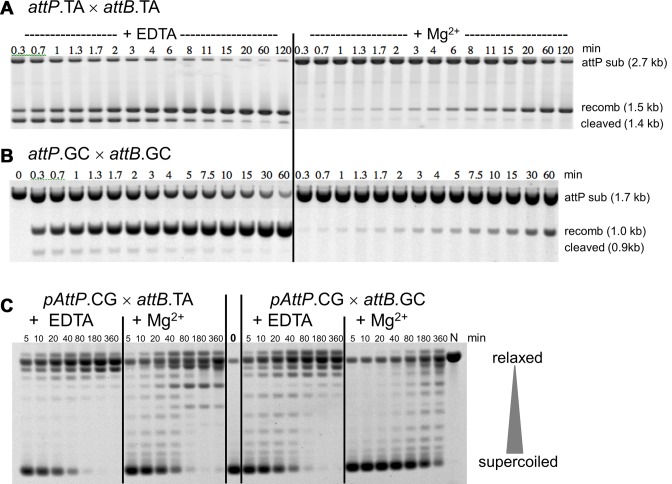
We used 2.7 kb plasmids: pAttP.TA, pAttB.TA and pAttP.CG; 3.1 kb plasmids: pAttP+ ( = pAttP.GT, the wild-type attP+ site), pAttP.CT and pAttP.GC; and the 3.5 kb plasmids: pAttB+ ( = pAttB.GT, the wild-type attB+ site), pAttB.CT and pAttB.GC. Long linear attP DNAs (Figure 2A and B) were made by restriction digestion. Short linear partners with various attB crossover sequences were 312 bp and were made by polymerase chain reaction (PCR) and purified by QIAquick spin columns (Qiagen).
Figure 2.
Kinetic analysis of substrate cleavage and supercoil relaxation by Bxb1 Int in ensemble reactions. (A) A 2.7 kb linear substrate containing attP.TA at its center was reacted with Int and a 312 bp attB.TA in the absence or presence of Mg2+ for the times indicated and the products were analyzed by agarose gel electrophoresis. The two attP cleavage products (both 1.36 kb) migrate as a single band, as do the two recombinant products (1.52 kb). The fast-migrating attB bands have been omitted. (B) Similar experiments with a 1.7 kb linear attP.GC and a 312 bp attB.GC, showing the cleavage and recombinant products of 860 bp and 1.0 kb respectively. (C) A supercoiled plasmid with attP.CG was reacted with mismatched (and recombination-incompatible) 312 bp attB substrates containing either attB.TA or attB.GC for the times indicated, in the absence or presence of Mg2+. Agarose gel electrophoresis reveals the extent of plasmid relaxation: more relaxed topological isomers migrate slower than those with more supercoils. The lanes marked 0 and N show the unreacted and nicked forms of the pAttP.GC substrate; the position of linearized substrate is indicated. As supercoils are relaxed their migration rate is reduced, with the most relaxed forms running close to the nicked marker. However, because the precise helical parameters of DNA are influenced by presence or absence of Mg2+ and univalent ionic conditions, the most relaxed (but covalently closed) species migrate at positions that differ from each other and from that of the nicked marker (N). Thus, the most relaxed species formed in the presence of Mg2+ behave as slightly positively supercoiled in the gel and form the 2–3 bands clearly visible in the later lanes of the reactions with Mg2+ that migrate ahead of the nicked marker. See Supplementary Figure S1 for an alternative image of these data.
Ensemble experiments
Bxb1 Int was purified as described previously (20). For ensemble kinetics experiments with linear DNAs, Int was incubated with each individual substrate for 15 min (to ensure cysteine reduction and maximum att site occupancy); the reactions were initiated by rapidly mixing the two solutions. Reactions were in 20 mM Tris HCl, pH 8.0, 5 mM dithiothreitol (DTT) and 50 mM NaCl, with either 5 mM ethylenediaminetetraacetic acid (EDTA) or 10 mM MgCl2 as indicated and were incubated at 37°C. For the reactions in Figure 2, attP plasmids (linear or supercoiled) were at 40 nM and the attB sites (all 312 bp) were at 330 nM to maximize synapsis; Int was 2 μM. For recombination reactions between supercoiled attP and attB plasmids, each parental DNA was at 50 nM, Int was 1 μM and these reactions also contained spermidine at 10 mM. All reactions were stopped by addition of SDS (to 0.1%) and treated with proteinase K (0.1 mg/ml). Bxb1 Int was stored in 50 mM Tris, pH 8.0, 300 mM NaCl, 1 mM EDTA and 50% glycerol and concentrated to >10 μM, such that only a few microliters were used for each experiment depending on the batch.
DNA constructs for single-DNA experiments
Linear DNA fragments used in single-molecule supercoiling relaxation experiments were derived from the plasmid pNG1175 (9702 bp) (11) which was constructed from pFOS-1 (9691 bp, New England Biolabs) by insertion of a single attP site between MfeI and BstXI restriction sites. pNG1175 was linearized by cutting at nearby SpeI and ApaI restriction sites; the resulting linear molecule was ligated to ∼900 bp PCR products carrying either biotinylated or digoxigenin-labeled nucleotides, and prepared with SpeI and ApaI-compatible ends, respectively. The resulting linear constructs were 11.4 kb in length, with roughly 900-bp of biotin- and digoxigenin-labeled DNA at their ends, allowing multiple tethering of the ends to streptavidin- or anti-digoxigenin-coated surfaces. The multiple tethers constrain the two DNA strands sufficiently that they may be supercoiled by rotation of the magnetic particle.
Single-DNA experiments
Flow cells were assembled for each experiment and contained 2.8 μm streptavidin coated paramagnetic beads (Invitrogen Dynabeads, M280) tethered to the surface of an anti-digoxigenin coated glass coverslip via a linear pNG1175 DNA molecule with biotinylated and digoxigenin-labeled ends. Flow cell contents were viewed with a bright field microscope and a 100× 1.3 NA immersion oil objective (Olympus). Translation in the z direction of a permanent magnet under the objective stage controlled the force on the bead while 360° rotations of the magnet controlled the linking number of the tethered DNA molecule. Bead position in three dimensions was tracked with custom lab-written software, which uses an untethered bead non-specifically bound to the glass surface as a reference point. Position fluctuations in the x-y plane were used to calibrate the force on the tethered beads while changes in the z direction relative to the reference bead were used to measure the tether extension (30).
Calibration measurements of extension as a function of force and ΔLk were performed on each tether used in relaxation experiments. This ensured that a magnetic bead was only tethered by a single DNA molecule of the correct length (Supplementary Figure S2). In the extension versus ΔLk measurement, as ΔLk is changed from 0, the extension of the DNA decreases slightly from ∼2.4 for the first ∼12 turns and then the extension decreases linearly by ∼55 nm/ΔLk. Since calibration slopes varied from tether to tether over the range 55 ± 5 nm/ΔLk, linking number-extension calibration was required for each tether to ensure accurate measurements.
Single-molecule experiments were carried out in buffer conditions similar to (20): 50 or 100 mM potassium glutamate (KGlu), 10 mM HEPES, 1 mM DTT and pH = 7.5, at temperature of 37°C, with either 10 mM EDTA or a combination of 5 mM MgCl2 and 2 mM EDTA added as indicated (3 mM excess MgCl2). A total of 2 mM EDTA was required in the buffer to prevent nonspecific collapse of tethers. Bxb1 Int (300 nM) was added to flow cells after 30 s preincubation with 100 nM of attB oligo.
RESULTS
Half-life of the cleaved state is reduced by strengthening base-pairing of the central dinucleotide
Before the DNA half-sites within the cleaved intermediate can be rejoined, we hypothesize that the complementary 2-base 3′ overhangs must base-pair. If so, we would expect that once crossover site cleavages have occurred, thermodynamic stability of the pre-ligation state(s) may affect re-joining kinetics. Ensemble experiments have previously observed a surprisingly long (1.8 min) half-life for the cleaved state for att.TA sites (denotes att site with a 5′-TA central dinucleotide) with no Mg2+, absence of which weakens base-pairing (11,29,31) (Figure 2C of (11)). To test our hypothesis, we examined the effect of increasing the strength of the base-pairing interaction on the kinetic profile of the reaction either by adding Mg2+ or by changing the central dinucleotide to the thermodynamically more stable sequence 5′-GC (See Supplementary Table S1 for Gibbs free energies of crossover sequences used based on Refs. (32,33)).
As shown in Figure 2A, addition of 10 mM Mg2+ to the reaction with att.TA sites dramatically reduces the half-life of the cleaved intermediate; this intermediate never accumulates but is rapidly chased into recombinant products. Interestingly, the overall rate of the reaction (as measured by substrate reacted or by recombinant formed) is reduced; in the presence of Mg2+ the halfway point is at about 60 min, while in its absence the substrate is more than 50% depleted by 20 s and recombinant formation achieves 50% in 3 min.
When the central dinucleotide is switched to GC on both linear substrates while omitting Mg2+ (Figure 2B, left panel), the half-life of the cleaved intermediate is again dramatically reduced compared to the corresponding att.TA reaction (compare Figure 2A, left). The apparent reaction rate is also slower than the TA reaction with depletion of the substrate reaching 50% by about 2 min. Addition of Mg2+ (Figure 2B, right panel) to the att.GC reaction further reduces the reaction rate (the amount of recombinant formed in 60 min in the presence of Mg2+ is the same as that formed in 20 s in its absence), and the cleaved intermediate becomes essentially undetectable, consistent with an extremely short half-life. We conclude that for recombination reactions between linear att-carrying DNAs, increasing the strength of the base-pairing interaction that helps to constrain the unjoined half-sites in a pre-ligation state reduces the half-life of the cleaved state, while at the same time reducing the overall rate of substrate depletion and recombinant formation. We caution that is possible that varying the dinucleotide sequence may alter the reaction kinetics by affecting the Int–DNA interaction, but the correlation of both sequence and Mg2+ results to DNA thermodynamic stability most simply suggests that strength of the dinucleotide base pairing interactions modulates the rejoining rate.
Strengthening base-pairing of the central dinucleotide sequence suppresses synaptic rotation
In a reaction between a supercoiled substrate and a linear partner with mismatched central dinucleotides, recombination (following a 180° rotation) is prevented by the mismatch but a subsequent 180° rotation, re-establishing the parental configuration, relaxes substrate supercoils; each 360° rotation relaxes one unit of DNA linking number and only integer multiples of 360° rotations can occur. We performed such reactions using the plasmid pAttP.CG as shown in Figure 2C, which indicates the cleavage-rotation-religation reaction via the rate of depletion of the supercoiled substrate (lowest gel band) and accumulation of relaxed topoisomers (upper bands running near the nicked plasmid in lane N). When reacted with attB.TA in the absence of Mg2+ the portion of the plasmid substrate that is neither nicked nor linearized is converted to circles that are mostly fully relaxed even at the earliest time point when the majority of the substrate is unreacted. Therefore, most relaxed circles are likely to be products from single synaptic complexes. In the presence of 10 mM Mg2+, however, the initial (5 min) circular products retain substantial superhelicity (Figure 2C, bands between supercoiled and relaxed in second panel from the left) and only become fully relaxed at 80 min after several synaptic encounters between the two substrates. When the partner substrate is the relatively stable attB.GC (Figure 2C, two rightmost panels), even in the absence of Mg2+, the reaction rate is substantially slower and most circular products formed in the initial 5 min retain substantial superhelicity.
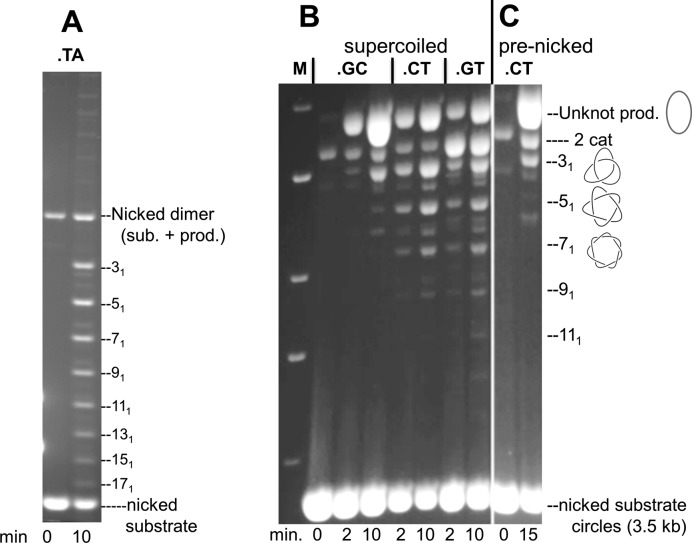
We also analyzed the topology of products from reactions in which both partners were on separate supercoiled plasmids and had identical central dinucleotides, a situation analogous to the normal integration of Bxb1 phage (Figure 3 and Supplementary Figure S1). As we had seen before (11), if the partners both contained the ‘weak’ att.TA sites, the majority of recombinant products (>90%) form a ladder of knots indicative of multiple rotations within a single synapse prior to the final ligation; the unknotted form resulting from a single 180° rotation accounts for <10% of these products (Figure 3A). Changing the central dinucleotide to GC reduces the proportion of knotted products; the unknotted product accounts for about 75% of the total products, although the 31 and 51 knots (indicating 1 and 2 extra 360° rotations) can readily be detected, as can catenated parental circles (products of one or more 360° rotations). With CT as the central dinucleotide, results are complicated by the fact that, with a non-palindromic central sequence, only half of the initial synapses are appropriately aligned for recombination. Nevertheless the characteristic ladder of knotted recombinants is readily seen and less than 50% of the recombinants are unknotted. Interestingly, pre-nicking the CT substrates largely eliminates the multiple rotations and the distribution of products looks very similar to the GC reaction (compare Figure 3C with B). These data clearly show that in reactions that can lead directly to recombination, multiple rotations before the final ligation are common, but the number of these extra 360° rotations is sharply reduced by dinucleotide sequences that stabilize the pre-ligation intermediate, or by lack of superhelical torque.
Figure 3.
Bxb1 Int recombination of two supercoiled plasmids. Integrase reactions using supercoiled substrates with attP and attB produce recombinant products with multiple knot nodes. Int reactions were performed at 37°C with ∼50 nM pAttP and pAttB supercoiled plasmids in standard reaction buffer containing spermidine (10 mM) to increase reaction efficiency and product yield. After the times indicated, samples were treated with the nicking enzyme Nb.BsrNDI (New England Biolabs) at 65°C for 60 min and then with SDS and proteinase K. Products were analyzed by electrophoresis (45 V, 20 h) on a 1% agarose-E buffer gel. The gel was subsequently stained with ethidium bromide to visualize the DNA. (A) Reaction contained pAttP.TA and pAttB.TA, both 2.73 kb. (B) Reactions contained pAttP.GC (3.1 kb) and pAttB.GC (3.5 kb) (lanes 2–4, counting from left), pAttP.CT (3.1 kb) and pAttB.CT (3.5 kb) (lanes 5–6), or pAttB.GT (3.5 kb) and pAttP.GT (3.1 kb) (lanes 7–8), and were run for the times indicated. (C) A reaction of pAttP.CT and pAttB.CT in which both parents were pre-nicked to remove supercoiling before Int addition. M indicates 1 kb ladder. The unknotted recombinants, the recombinant torus knots (31, 51, 71, 91, 111, …) and the 2-noded catenane of the parental plasmids are shown. Note that the 6.2 kb dimeric form of the 3.1 kb pAttP plasmids migrates at the same position as the 2-noded catenane.
Single-molecule detection of cleavage and strand exchange by the Bxb1 synapse
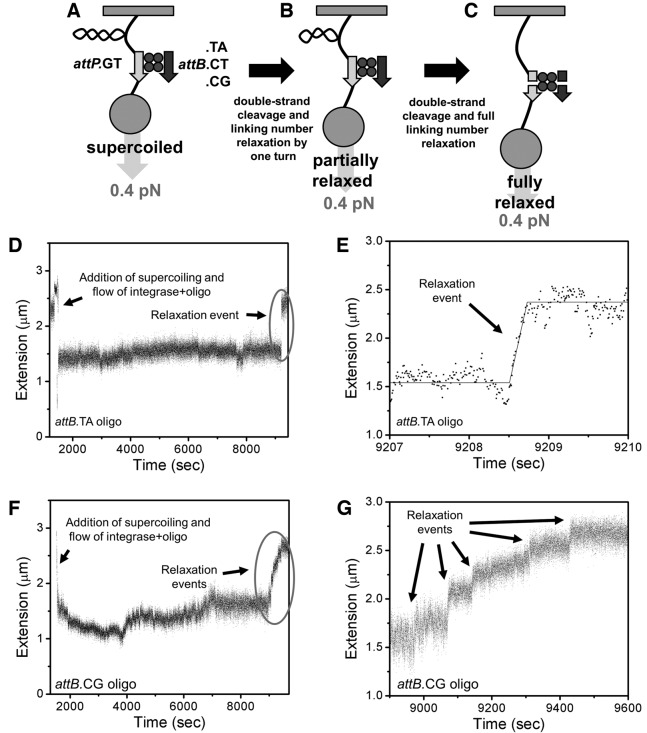
We used a single-DNA supercoil-relaxation magnetic-tweezers-based approach (11,34) to obtain information about individual cleavage-rotation events (Figure 4A–C). This experiment is the single-molecule analog of the supercoil relaxation assay used to probe synapse rotation in Figure 2C (the one difference being use of attP.GT in single-molecule experiments versus attP.CG in the bulk experiment of Figure 2C). Experiments used a 9.7 kb tethered DNA containing a single internal WT attP+ with GT crossover site (11). None of the attB oligos used in any experiments contained a GT, forcing religation to occur after returning to the parental DNA sequences. We emphasize that all experiments used the same attP+-carrying tethered DNA, and thus all changes in synapse properties are solely due to changes in the partner attB sequence on the oligo.
Figure 4.
Single-DNA experiments show that relaxation behavior of an attP substrate is influenced by the crossover sequence of the partner attB site. (A) Experimental design. A supercoiled single DNA containing a WT attP.GT site is tethered to a surface and pulled and twisted via a magnetic particle at its end. The attB site is supplied on a 52 bp DNA oligomer, along with Bxb1 Int enzyme. Force is set at 0.4 pN, which generates DNA torque similar to that present in a supercoiled bacterial plasmid or chromosome (σ ≈ −0.05 (35)). (B) Cleavage and rotation at the synapse leads to partial relaxation of supercoiling, and extension of the DNA which is detected in real time. (C) Further cleavage and rotation removes all stored supercoils, allowing full extension of the tether. (D) Example of a relaxation event obtained in a reaction with attB.TA. In this experiment, enzyme and attB were supplied at time ≈1500 s. (E) Same data as in C but with magnified timescale showing the 3 s period within which the complete relaxation event occurs. (F) Example of a relaxation ‘event’ obtained in a reaction with attB.CG. (G) Same data as in E but with magnified timescale showing a run of small relaxation steps.
Forces <0.5 pN imparted nearly symmetrical extension versus ΔLk behavior in the positive and negative supercoil regimes, whereas DNA melting occurred in the negative ΔLk regime at forces >0.5 pN (Supplementary Figure S2), in accord with prior studies (30). All Bxb1 experiments were performed at 0.4 ± 0.05 pN. This level of force generates nearly constant DNA torsional stress in plectonemic supercoils (formed in the nearly straight ‘wings’ of Supplementary Figure S2) corresponding to that found in a physiologically supercoiled plasmid (σ = −0.05) (35). This type of calibration data allows DNA extension to be converted to linking number, and therefore for changes in extension to be converted to changes in linking number.
We first carried out experiments with attB.TA; after verification of a tether as a single supercoilable 9.7 kb DNA, a mix of Bxb1 Int protein (300 nM) pre-bound with the attB.TA oligomer (100 nM) in assay buffer (50 mM KGlu, no MgCl2) was added to the flow cell and ΔLk was set to −30. We note that due to the flattening out of the extension versus ΔLk response (Supplementary Figure S2, squares, |ΔLk| < 8) the maximum relaxation step size observable is a linking number change from 20 to 22; the experiment cannot detect relaxation of the final ∼10 turns due to the small extension change that occurs in that range. A typical attB.TA experiment (Figure 4D) showed a long initial period with little change in extension (some slow variations occur due to nonspecific protein–DNA interactions and slow dynamics of the initially supercoiled DNA), terminated by an abrupt jump in extension >500 nm, initiated by cleavage of the DNA. In general we refer to ‘steps’ as events or individual jumps in extension (or corresponding numbers of turns n relaxed); multiple steps occurring in succession will be referred to as a ‘run’ of steps or a series of events.
Tether extensions in periods bracketing a typical relaxation event show Brownian fluctuations with standard deviation 100 nm and mean duration of 0.1 s (Figure 4E). Extension jumps were identified from data of this type, and then fit with custom step-finding software that provided an estimate of step size (μm) and duration (s), as well as relaxation velocity (during the step) in μm/s. In some events, we could obtain a step size but not a velocity due to limitations in time resolution (10 ms) and the large amplitude of Brownian noise. Extension versus ΔLk data (Supplementary Figure S2) were used to determine the number of turns removed per step (n) and the velocity (turns/s; one turn refers to a 360° rotation, or equivalently |ΔLk| = 1). As in the bulk experiments, single 180° rotations are not expected due to the mismatched central dinucleotide sequences.
The noise amplitude is large enough that it interferes with detection of small (n < 3) events. The experimental extension fluctuations are well described as Gaussian distributed noise with standard deviation σ = 110 nm and a correlation time of 0.08 s. We carried out simulations of the relaxation process using this noise amplitude and correlation time and concluded that steps with n = 1 (extension change of ∼50 nm) are undetectable, while steps with n = 2 (∼ 100 nm) can only be reliably detected if they are well separated from other step events (Supplementary Figure S3). We reliably detect larger steps (n ≥ 3).
In a set of nine experiments (i.e. nine single molecules) with the attB.TA oligomer, we observed 10 events, which were primarily relaxations involving single, relatively large steps, with numbers of turns removed ranging from n = 3 to 21 (Figure 5A, dark bars). Note that the n = 22 cutoff on our measurements is well above the peak in the event size distribution of Figure 5A. Runs predominantly consisted of single steps with n = 10 to 15 (only one run of two steps was observed). The bulk of the steps were smaller than the maximum step size measurable (corresponding to n = 22 turns) and were predominantly relaxation events following the initial setting of DNA linking number to −30. We note that the step size distribution is dominated by quite large events of more than 10 turns.
Figure 5.
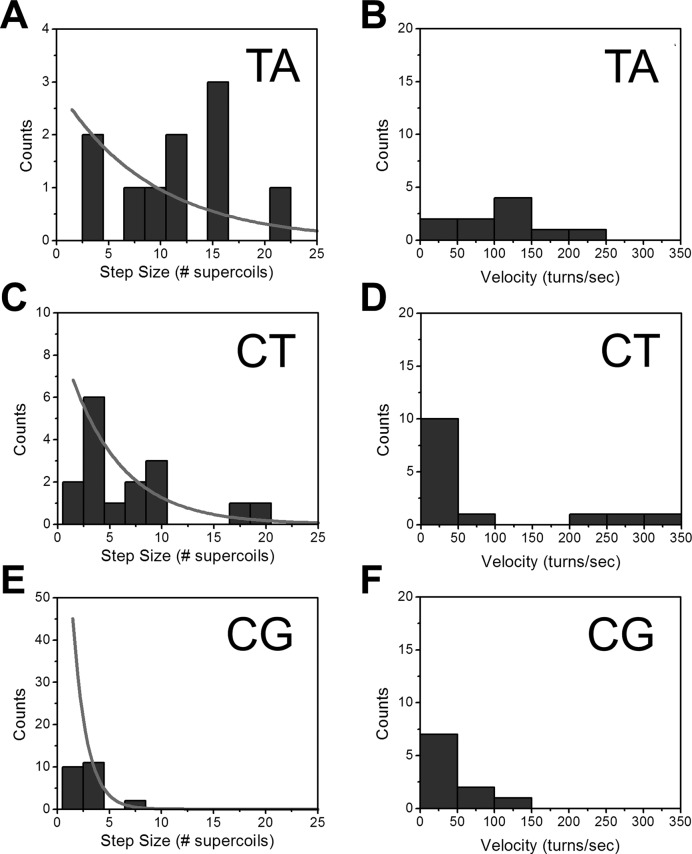
Step size and rotational velocity distributions. For the three attB sequence oligos studied (A, B: attB.TA; C, D; attB.CT; E, F: attB.CG), the distributions of step sizes and step velocities are shown (solid dark gray bars). Step size distributions bins have width 2, and are centered at n = 1.5, 2.5,… . With increasing base pairing stability the widths of the step size and the velocity distributions are both gradually reduced. The n > 2 step-size distributions are consistent with the controlled rotation model (solid gray curves in A, C, E), except in the leftmost bin (n = 1 and 2), corresponding to events obscured by Brownian noise.
Based on the observed steps, the mean number of turns released per step was  = 11.5 ± 1.8 (Figure 6A), and the average rotational velocity was 106 ± 16 turns/s (5.7 ± 0.7 μm/s; Figures 5B and 6B). Velocities were computed only for steps where n = 3 or more turns were released; velocities for smaller steps could not be reliably estimated due to Brownian noise and time resolution obscuring step start and end times.
= 11.5 ± 1.8 (Figure 6A), and the average rotational velocity was 106 ± 16 turns/s (5.7 ± 0.7 μm/s; Figures 5B and 6B). Velocities were computed only for steps where n = 3 or more turns were released; velocities for smaller steps could not be reliably estimated due to Brownian noise and time resolution obscuring step start and end times.
Figure 6.
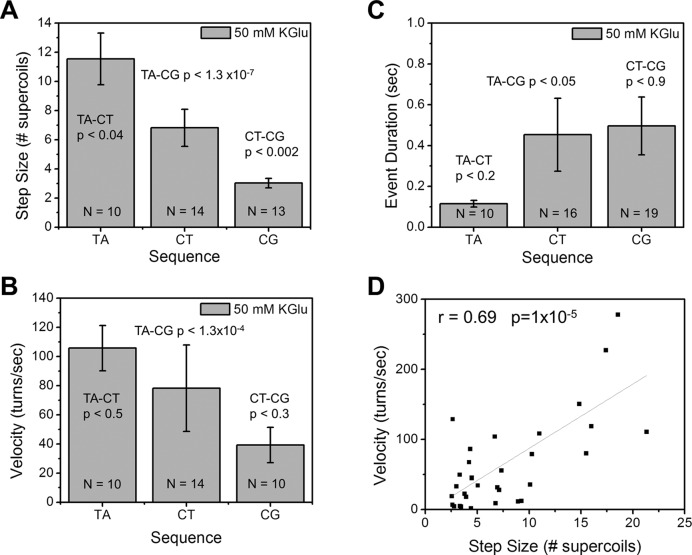
The number of rotations per relaxation event (the step size) and their velocity are dependent on the thermodynamic stability of the partner attB crossover sequence. (A) From a series of experiments of the form in Figure 4, average step sizes were computed. When comparing attB.TA and attB.CG data, P < 3 × 10−7. (B) Rotational velocity also is gradually reduced as crossover-base-pairing stability is increased. Comparing attB.TA and attB.CG data, P < 1.3 × 10−4. (C) The duration of the step events increases with base-pairing stability. Comparing .TA and .CG data, P < 0.05. (D) Rotational velocity and step size are strongly correlated. Fit line has slope 9.2±1.7 turns/sec.
In experiments with non-attB DNA oligomers in place of attB we found that cleavages were extremely rare; quantification of this was presented in Ref. (11). In a series of five additional control experiments, we verified that the cleavage rate in the presence of a non-attB oligo was consistent with that observed in Ref. (11), with one cleavage event occurring in a total of 19 h of incubation time, giving a cleavage rate of 1.5 × 10−5 s−1. We note that the single-molecule rates are much slower than those observed in the ensemble experiments; the latter are carried out at the highest concentration of reactants so that binding and synapsis were not rate limiting. In contrast, the single-molecule experiments were carried out at lower reactant concentrations. However, this does not fully explain the much lower rates observed in single-molecule versus ensemble experiments.
Single molecule relaxation experiments with attP+ show that increased base-pairing stability of the partner crossover reduces the number of rotations per cleavage event
The attB.TA oligo used above has the weakest base-pairing free energy (Supplementary Table S1). To look at the effect of strengthening this, we did similar experiments using attB.CT or attB.CG as the partner sites; these dinucleotide changes decrease the ΔGTA (denotes the free energy change ΔG of the base pairing interaction of the att-central tetranucleotide with a TA central dinucleotide) by 1.36 and 3.26 kcal/mol, respectively (unified nearest-neighbor energies of Supplementary Table S1).
With the attB.CT oligo, 9 experiments yielded 16 events; unlike the attB.TA case, multiple-step runs of relaxation (typically less than three steps) were common. The mean number of turns relaxed per observable step event was 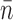 = 6.8 ± 1.3 (Figure 6A), with a distribution peaked at lower n than for the attB.TA case (Figure 5C, dark bars). The average velocity during attB.CT relaxations was 78 ± 27 turns/s (4.2 ± 1.5 μm/s, Figure 6B), similar within error to that previously reported for the .GT x .CT reaction (54 ± 5 turns/s) (11) and significantly slower than the velocity for .TA (Figure 5D). We note that there is a larger error in the estimation of velocities for smaller events.
= 6.8 ± 1.3 (Figure 6A), with a distribution peaked at lower n than for the attB.TA case (Figure 5C, dark bars). The average velocity during attB.CT relaxations was 78 ± 27 turns/s (4.2 ± 1.5 μm/s, Figure 6B), similar within error to that previously reported for the .GT x .CT reaction (54 ± 5 turns/s) (11) and significantly slower than the velocity for .TA (Figure 5D). We note that there is a larger error in the estimation of velocities for smaller events.
For attB.CG, smaller steps were observed, with more steps per relaxation run than in the attB.CT case (see Figure 4F and G). Six separate experiments with the attB.CG oligo yielded 23 events with relaxation runs consisting of three to nine steps with a narrower distribution of turns released per step than for the attB.CT case (Figure 5E, dark bars). The mean number of turns per step was 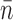 = 3.0 ± 0.3 and the mean velocity was 39 ± 12 turns/s (Figure 6B), both significantly smaller than the mean attB.CT step size and velocity.
= 3.0 ± 0.3 and the mean velocity was 39 ± 12 turns/s (Figure 6B), both significantly smaller than the mean attB.CT step size and velocity.
Results for the three oligos are shown in Figure 6 in order of increasing base-pairing stability (TA < CT < CG) and indicate that increased base-pairing stability decreases step size and rotational velocity, and increases event duration (Figure 6C). Plotting the observed step sizes and rotational velocities across our data set (34 steps) shows a strong correlation (Figure 6D).
In light of the preceding results for attB oligos of varied central dinucleotide sequence (all for 50 mM KGlu, 0 mM MgCl2), we hypothesized that increasing salt, which stabilizes DNA base pairing (i.e. increases DNA melting temperature; for univalent salt this occurs over a wide concentration range, while for divalent salt in 50–100 mM monovalent salt this occurs for divalent concentrations up to ∼10 mM (29,36,37)), would lead to a reduction in step size and rotation velocity. While there was a restricted range of univalent and divalent salt over which we observed single-molecule relaxation events, we did observe that increasing concentration of univalent salt (KGlu) from 50 to 100 mM led to a statistically significant reduction in step size for the attB.CT reaction (Supplementary Figure S4A). Effect of increased univalent salt on step velocity was less statistically significant (Supplementary Figure S4B), as were effects of increased divalent ions (MgCl2 increased from 0 to 3 mM, Supplementary Figure S4C and D), but all of these changes did lead to the expected trends, i.e. decreased step size and rotation velocity with increased salt concentration. However, when using the attB.TA oligo, addition of divalent salt produced a significant effect on both step size (P < 0.02) and velocity (P < 0.13) (Supplementary Figure S4E and F).
Although the simplest explanation for the observed supercoil relaxation is subunit rotation within the Int synapse, it is hard to exclude the possibility that a type-I topoisomerase activity of Bxb1 Int is responsible, for example due to cleavage followed by release and rotation of a free 3′-OH DNA strand end around the other strand (38). Several observations strongly argue against this, however. First, the dependence of the rotation rate and step size on dinucleotide sequence suggests that base pairing interactions are involved in limiting the rate of rotations. Second, the rates of Bxb1 Int-mediated rotation are essentially the same for either a supercoiled duplex or a pair of ‘braided’ DNAs which absolutely require double-stranded breaks and rotations within the Int-att synaptic complex (11), and are significantly slower than the rotation rates obtained with a DNA-nicking enzyme (11,39). Finally, and perhaps most significantly, we have previously carried out in vitro experiments on supercoiled plasmids showing that recombination proceeds without relaxing supercoiling: Supplementary Figure S2 of (11) shows that un-nicked recombination products migrate on an agarose gel slightly ahead of the supercoiled dimers of the parental plasmids (compare lanes 1 and 4); only when the products are nicked are they converted to the much more slowly migrating ‘ladder’ of knot types expected from multi-turn recombination reactions (lane 2).
Increased force causes larger steps and faster velocities
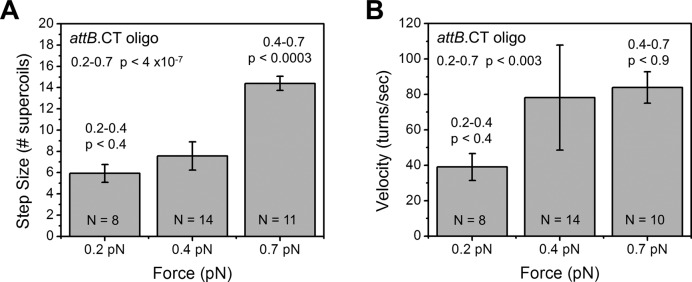
Given that in a magnetic tweezers DNA stretching-twisting experiment, torsional stress, or torque, in the DNA is controlled by applied force (35), and that for Bxb1 Int, rotation rate has been hypothesized to be controlled in part by enzymatic ‘friction’ (11), we reasoned that the velocity and step size might be sensitive to applied force. Previous experiments with attB.CT performed at 0.5 pN showed nearly complete relaxation of all supercoils in single step events (∼18 turns/event, as outlined in Figure 4, supercoil transitions from A to C, and bypasses state B), in contrast with the experiments at 0.4 pN presented above that generated an average step size of 6.8 turns/event (supercoil goes through states A, B and C). The experiments of this paper discussed above were carried out at 0.4 pN, which generates torsional stress in plectonemic DNA comparable to that found in DNA in Escherichia coli (σ = −0.05). We therefore carried out attB.CT × attP experiments at forces of 0.2 and 0.7 pN to compare with our results for 0.4 pN, and in order to examine differences between the 0.4 and 0.5 pN datasets.
For 0.2 pN, five experiments yielded eight steps of average size of 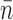 = 5.9 ± 0.8 (Figure 7A), the same as that obtained with 0.4 pN within measurement uncertainty. While the average velocity was smaller for 0.2 pN than for 0.4 pN, the large statistical error in the latter makes this effect not highly significant (Figure 7B). However, for 0.7 pN 11 experiments yielded 11 steps that were significantly larger and with rotations faster than for 0.2 pN (Figure 7). The 0.7 pN experiments were carried out for initially positive supercoiling (ΔLk = +30 turns) to avoid unwinding (torque-melting) the DNA; however, we have observed events to have similar size and speed at 0.4 pN for initial ΔLk = +30 and -30 (data not shown). Overall, the data indicate that increasing force (and therefore supercoil torque) from 0.2 to 0.7 pN causes larger steps and faster rotation.
= 5.9 ± 0.8 (Figure 7A), the same as that obtained with 0.4 pN within measurement uncertainty. While the average velocity was smaller for 0.2 pN than for 0.4 pN, the large statistical error in the latter makes this effect not highly significant (Figure 7B). However, for 0.7 pN 11 experiments yielded 11 steps that were significantly larger and with rotations faster than for 0.2 pN (Figure 7). The 0.7 pN experiments were carried out for initially positive supercoiling (ΔLk = +30 turns) to avoid unwinding (torque-melting) the DNA; however, we have observed events to have similar size and speed at 0.4 pN for initial ΔLk = +30 and -30 (data not shown). Overall, the data indicate that increasing force (and therefore supercoil torque) from 0.2 to 0.7 pN causes larger steps and faster rotation.
Figure 7.
Step size and rotational velocity are dependent on DNA torque. Experiments as in Figure 4 but with varied force (and DNA torque) show that as force (torque) is increased, average step size (A) and rotational velocity (B) both increase. P-values for differences between 0.2 and 0.7 pN data for step size and velocity are 4.0 × 10−7 and 0.003, respectively.
Step size distribution is consistent with the ‘controlled rotation’ model
The simplest model for the religation kinetics during a relaxation event is that there is a fixed probability 1-p of religation per turn, versus a probability p per turn of not ligating and continuing to rotate. This ‘controlled rotation’ model predicts an exponentially decaying form of the event size distribution, P(n) = (1−p) pn-1, The controlled rotation model has been found to describe relaxation of supercoiling by type IB and IC topoisomerases (25,26). The peak in the event-size distribution for this model is at n = 1, and the average event size is 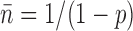 .
.
Our inability to detect events with a step size of n = 1 and our unreliable detection of n = 2 events complicate our data analysis. Nevertheless, we can ask whether our data for the larger steps (n > 2) are consistent with the controlled rotation model. We have used the maximum likelihood method to determine the relegation probability (1-p) for the steps with n > 2: this relates p to the mean of the observed event sizes via
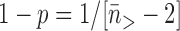 where
where 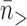 is the average size of events with n > 2. For the attB.TA data we find 1−p = 0.10±0.02; for attB.CT we find 1-p = 0.18 ± 0.04; for attB.CG we find p = 0.53 ± 0.14. The resulting event size distributions are consistent with the data for n > 2 (Figure 5A, C and E, solid curves; the curves in Figure 5A, C and E take on the value predicted for each bin at the bin centers of 1.5, 2.5, 3.5,… .). Since the statistical variance for each bin is approximately the square root of the counts within it, for n > 2 our observed step size distributions are consistent with the simple controlled rotation model.
is the average size of events with n > 2. For the attB.TA data we find 1−p = 0.10±0.02; for attB.CT we find 1-p = 0.18 ± 0.04; for attB.CG we find p = 0.53 ± 0.14. The resulting event size distributions are consistent with the data for n > 2 (Figure 5A, C and E, solid curves; the curves in Figure 5A, C and E take on the value predicted for each bin at the bin centers of 1.5, 2.5, 3.5,… .). Since the statistical variance for each bin is approximately the square root of the counts within it, for n > 2 our observed step size distributions are consistent with the simple controlled rotation model.
Small events with n = 1 and 2 can be explained by non-step relaxation
Although we fail to observe individual n = 1 and n = 2 steps, they will still contribute to overall relaxation of the substrate supercoils.To estimate the maximum number of undetected n = 1 and n = 2 events, we partitioned the recovery of DNA extension into a fraction 1-f contributed by the observed steps, and a fraction f that cannot be attributed to discrete observable events. For attB.TA, we find f = 0.14 ± 0.06: this is done by determining the total length change in individual relaxation experiments (e.g. Figure 4D or F), and then determining what fraction of that is in the form of identified steps (circled regions of Figure 4D and F), versus non-step relaxation.
The average f can be used to estimate an upper bound on the number of turns that were not detected, i.e. that were of size n = 1 or 2, via Nundetected = Ndetected f / (1-f), where Ndetected is the total number of turns that were detected in step events. For the attB.TA data (Ndetected = 115, f = 0.14) we have Nundetected = 19. Because of the broad distribution we expect these turns to be in roughly equal numbers of n = 1 and 2 events, indicating that the non-step relaxation could contain 12.5 step events, well in excess of the ∼3 predicted by the controlled rotation model fit to the events with n > 2 (Figure 5A, difference between leftmost bin and solid curve). We conclude that the attB.TA data are consistent with the controlled rotation model.
For the attB.CT data (Ndetected = 110, f = 0.34) we have Nundetected = 56, easily explaining the ∼6 missing n = 1 and 2 events predicted by the controlled rotation model (Figure 5C). For the attB.GC data (Ndetected = 70, f = 0.37) we have Nundetected = 41, again explaining the ∼35 missing n = 1 and 2 events in Figure 5E. For each case, the apparent deficit in small n = 1 and 2 events is consistent with the controlled rotation model, given that we are unable to observe all n = 1 and most n = 2 events.
DISCUSSION
We have reported a series of experiments that analyze the cleavage and rotation dynamics of the SR, Bxb1 Int. Together, both ensemble and single-molecule experiments indicate that the core enzyme tetramer does not, by itself, limit strand exchanges to a single rotation. Rather, our results suggest that a stochastic process determines whether each 180° rotation is followed by a ligation event or by another rotation (or, in the case of mis-matched or mis-aligned attP and attB sites, each 360° rotation; note that this situation occurs for roughly 50% of synapses that form in vivo). The choice between these two possibilities is determined primarily by the thermodynamic stability of the base pairing interactions that stabilize the pre-ligation state. Strong interactions favor the ligation process, limiting the number of rotations, whereas weak interactions result in inefficient ligation and more rapid rotation (less molecular ‘friction’), favoring multiple rotations.
We suggest that the final outcome of a synaptic event is controlled by thermodynamics. The overall process is multi-step. Following formation of the parental synapse the joined and cleaved states are in equilibrium. Only while both sites are in the cleaved state can the complex escape from this paired configuration to initiate rotation; the rate of this escape will depend on the cleaved/joined equilibrium, and will be assisted by high superhelical torque but inhibited by higher thermodynamic stability of the paired-but-cleaved state. Following initiation of rotation, a chance for pairs of half-sites to re-anneal occurs once every 180°; the likelihood of this strongly depends on the stability of the paired state. The rates of joining (and re-cleavage) and synapse dissociation determine whether paired half-sites result in a joined product or are subjected to further rotations and repeat cycles of the process.
In this overall process, the only stage that is influenced by the half-site base-pairing interaction is the post-cleavage/pre-joined conformation of the synapse formed at positions 0/180°/360° of the rotating complex. When pairing interactions are strong, it is reasonable to expect that the lifetimes of these discrete conformational states increase, increasing the probability of religation while slowing synaptic rotation. We emphasize that during a multi-turn rotational event, the base-pairing interactions are transient; indeed the resolvase crystal structure traps a cleaved state in which base-pairing interactions are disrupted (2).
In the single DNA experiments with attB.CG, individual steps within a run are separated by pauses that may be several minutes in duration. It is likely that the synaptic complex remains intact during these periods and we suggest that for most of the pause one or both crossover sites within the parental synapse are in the joined state.
In addition to strengthening the base-pairing interaction, Mg2+ may also reduce rotations by shifting the equilibrium between cleaved and joined states towards the latter. This could explain the reduced rates of recombination and low yield of cleaved intermediates seen in the right-hand panels of Figure 2A and B.
One might ask to what degree the number of rotations following a DNA cleavage by a Bxb1 synapse is affected by whether the DNAs being reacted are recombination-competent or not. For the serine integrases, the attP and attB sites (and the Int–attP and Int–attB complexes) are functionally distinct. A parental synapse must structurally differ from a recombinant synapse, perhaps as proposed in a recent structural analysis (40). In principle, this might influence the outcome of a synaptic event between attP and attB sites. For example, the recombinant synapse (formed by 180° rotation of the cleaved parental synapse) might have a cleaved/joined equilibrium that favors the joined state, or once joined might have a rapid dissociation rate. Either of these scenarios would then favor recombinant formation over restoring the parental configuration and might result in an apparent ‘gating’ of the rotations specifically when recombination is assayed. However, we observe multiple rotations not only when we prevent recombination (as in the single DNA experiments, or as occurs in vivo for misaligned sites) but also when we allow recombination following the first 180° rotation (Figure 3), so if there is a bias towards forming a recombinant rather than a parental joint, it does not appear to be very strong. Recombination is ultimately favored because, once the recombinant synapse dissociates, Int alone cannot reconstitute it.
An analogous stochastic process has been proposed for the controlled relaxation of DNA supercoiling by type IB and IC topoisomerases. For these topoisomerases it is thought that there is a certain probability 1−p per rotation of religation occurring via a single rate-limiting step, which gives rise to an exponential distribution of step sizes (P(n) ≈ (1−p) pn-1), characterized by the smallest steps being the most probable. The ensemble recombination data shown in Figure 3 are, with the exception of the .TA data (see below), generally in accord with this expectation. And, while Brownian noise prevents detection of the anticipated n = 1 and 2 steps in the single molecule experiments, the size distributions of the larger steps (n > 2) are also consistent with a fixed probability of ligation that depends on the crossover sequence (Figure 5).
From our data we can estimate the ligation probability at each step. The ensemble recombination data using pairs of sites with identical crossover sequences give probabilities for ligation at the first opportunity (after 180°) of about 0.8 for att.GC sites, 0.5 for att.CT (Figure 3B), 0.4 for .GT and <0.1 for att.TA. In the single-DNA relaxation experiments the probabilities for ligation at the first opportunity (after 360°) are ∼0.53 for attB.CG, 0.18 for attB.CT and about 0.10 for attB.TA (all in combination with an attP.GT site). These data are consistent with synapse rotation properties being sensitive to the sequence composition at the central dinucleotide, suggesting that transient base-pairing plays a role. This conclusion is backed up by consistent effects of changes in salt concentration; changes in solution conditions strengthening DNA base-pairing lead to smaller steps and slower rotation (Supplementary Figure S4).
The att.TA ensemble recombination data show obvious deviations from the predictions of the simple controlled-rotation (fixed probability of ligation per turn) model. We found here (Figure 3A) and in our earlier experiments (Supplementary Figure S2 of (11)) that the amount of each recombinant knot species actually increased slightly over the first three or four cycles, with peak yields at the 51 and 71 knots respectively: why might this be? We suggest that with the very weak .TA interaction, the lifetime of the pre-ligation state, and thus its probability of ligation, is particularly sensitive to superhelical tension. After the first 180°, further rotations not only reduce the superhelical tension but also create more complex topological tangles that become energetically more costly to further rotations.
We do not believe there is any contradiction between our results and those of (10) who observed with the serine integrases from ϕC31 and Bxb1 a preponderance of products formed from the minimum number of rotations (180° for recombination) and concluded that the serine integrases performed a ‘gated’ rotation. Those authors, however, also observed products consistent with multiple rotations. We suggest that their adoption of a topologically constrained and intramolecular recombination system is largely responsible for the substantial enhancement of ligation and product release at the first opportunity. In that system, the torque experienced by the cleaved synapse may be largely, if only temporarily, eliminated. This would also explain why the resolvases such as Tn3 and γδ, which use a similarly constrained synaptic complex, only rarely produce products of processive cycles that have bypassed the initial ligation opportunity (10). Our own experiments on Bxb1 Int show no evidence that ligation is specifically favored following the first 180° rotation that would form a recombinant product.
In conclusion, we suggest that it may not be appropriate to use the term ‘gated’ to describe a rotational mechanism that can, in principle, undergo multiple rounds of rotation even if the chance of iterations is small. In the controlled rotation model, a high religation probability after a single rotation can give the impression of tight gating, even though there remains a nonzero probability of multiple rotations occurring between cleavage and religation. Bxb1 Int appears to exchange DNA half-sites by this type of mechanism as, most likely, do the other SRs.
Supplementary Material
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Northwestern University NSF Grants [MCB-1022117, DMR-1206868]; Northwestern University NIH Grant [GM105847], Northwestern University NCI Grant [CA193419]; University of Pittsburgh NIH Grant [AI059114]; Yale University NIH Grant [GM028470]; Yale University research funds. Funding for open access charge: Northwestern University NIH Grant [GM105847].
Conflict of interest statement. None declared.
REFERENCES
- 1.Grindley N.D.F., Whiteson K.L., Rice P.A. Mechanisms of site-specific recombination. Annu. Rev. Biochem. 2006;75:567–605. doi: 10.1146/annurev.biochem.73.011303.073908. [DOI] [PubMed] [Google Scholar]
- 2.Li W., Kamtekar S., Xiong Y., Sarkis G.J., Grindley N.D.F., Steitz T.A. Structure of a synaptic γδ resolvase tetramer covalently linked to two cleaved DNAs. Science. 2005;309:1210–1215. doi: 10.1126/science.1112064. [DOI] [PubMed] [Google Scholar]
- 3.Keenholtz R.A., Rowland S.J., Boocock M.R., Stark W.M., Rice P.A. Structural basis for catalytic activation of a serine recombinase. Structure. 2011;19:799–809. doi: 10.1016/j.str.2011.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan P., Gupta K., Van Duyne G.D. Tetrameric structure of a serine integrase catalytic domain. Structure. 2008;16:1275–1286. doi: 10.1016/j.str.2008.04.018. [DOI] [PubMed] [Google Scholar]
- 5.Ritacco C.J., Kamtekar S., Wang J., Steitz T.A. Crystal structure of an intermediate of rotating dimers within the synaptic tetramer of the G-segment invertase. Nucleic Acids Res. 2013;41:2673–2682. doi: 10.1093/nar/gks1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamtekar S., Ho R.S., Cocco M.J., Li W., Wenwieser S.V., Boocock M.R., Grindley N.D.F., Steitz T.A. Implications of structures of synaptic tetramers of γδ resolvase for the mechanism of recombination. Proc. Natl. Acad. Sci. U.S.A. 2006;103:10642–10647. doi: 10.1073/pnas.0604062103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stark W.M., Sherratt D.J., Boocock M.R. Site-specific recombination by Tn3 resolvase: topological changes in the forward and reverse reactions. Cell. 1989;58:779–790. doi: 10.1016/0092-8674(89)90111-6. [DOI] [PubMed] [Google Scholar]
- 8.Wasserman S.A., Dungan J.M., Cozzarelli N.R. Discovery of a predicted DNA knot substantiates a model for site-specific recombination. Science. 1985;229:171–174. doi: 10.1126/science.2990045. [DOI] [PubMed] [Google Scholar]
- 9.Stark W.M., Grindley N.D.F., Hatfull G.F., Boocock M.R. Resolvase-catalysed reactions between res sites differing in the central dinucleotide of subsite I. EMBO J. 1991;10:3541–3548. doi: 10.1002/j.1460-2075.1991.tb04918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olorunniji F.J., Buck D.E., Colloms S.D., McEwan A.R., Smith M.C., Stark W.M., Rosser S.J. Gated rotation mechanism of site-specific recombination by φC31 integrase. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19661–19666. doi: 10.1073/pnas.1210964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai H., Sun M., Ghosh P., Hatfull G.F., Grindley N.D.F., Marko J.F. Single-molecule analysis reveals the molecular bearing mechanism of DNA strand exchange by a serine recombinase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7419–7424. doi: 10.1073/pnas.1018436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rowland S.J., Stark W.M., Boocock M.R. Sin recombinase from Staphylococcus aureus: synaptic complex architecture and transposon targeting. Mol. Microbiol. 2002;44:607–619. doi: 10.1046/j.1365-2958.2002.02897.x. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh P., Kim A.I., Hatfull G.F. The orientation of mycobacteriophage Bxb1 integration is solely dependent on the central dinucleotide of attP and attB. Mol. Cell. 2003;12:1101–1111. doi: 10.1016/s1097-2765(03)00444-1. [DOI] [PubMed] [Google Scholar]
- 14.Smith M.C., Till R., Smith M.C. Switching the polarity of a bacteriophage integration system. Mol. Microbiol. 2004;51:1719–1728. doi: 10.1111/j.1365-2958.2003.03942.x. [DOI] [PubMed] [Google Scholar]
- 15.Johnson R.C., Glasgow A.C., Simon M.I. Spatial relationship of the Fis binding sites for Hin recombinational enhancer activity. Nature. 1987;329:462–465. doi: 10.1038/329462a0. [DOI] [PubMed] [Google Scholar]
- 16.Koch C., Kahmann R. Purification and properties of the Escherichia coli host factor required for inversion of the G segment in bacteriophage Mu. J. Biol. Chem. 1986;261:15673–15678. [PubMed] [Google Scholar]
- 17.Fogg P.C., Colloms S., Rosser S., Stark M., Smith M.C. New applications for phage integrases. J. Mol. Biol. 2014;426:2703–2716. doi: 10.1016/j.jmb.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bonnet J., Subsoontorn P., Endy D. Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. U.S.A. 2012;109:8884–8889. doi: 10.1073/pnas.1202344109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bonnet J., Endy D. Switches, switches, every where, in any drop we drink. Mol. Cell. 2013;49:232–233. doi: 10.1016/j.molcel.2013.01.005. [DOI] [PubMed] [Google Scholar]
- 20.Kim A.I., Ghosh P., Aaron M.A., Bibb L.A., Jain S., Hatfull G.F. Mycobacteriophage Bxb1 integrates into the Mycobacterium smegmatis groEL1 gene. Mol. Microbiol. 2003;50:463–473. doi: 10.1046/j.1365-2958.2003.03723.x. [DOI] [PubMed] [Google Scholar]
- 21.Van Duyne G.D., Rutherford K. Large serine recombinase domain structure and attachment site binding. Crit. Rev. Biochem. Mol. Biol. 2013;48:476–491. doi: 10.3109/10409238.2013.831807. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh P., Wasil L.R., Hatfull G.F. Control of phage Bxb1 excision by a novel recombination directionality factor. PLoS Biol. 2006;4:e186. doi: 10.1371/journal.pbio.0040186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stark W.M. The Serine Recombinases. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0046-2014. MDNA3-0046-2014. [DOI] [PubMed] [Google Scholar]
- 24.Yang W. Topoisomerases and site-specific recombinases: similarities in structure and mechanism. Crit. Rev. Biochem. Mol. Biol. 2010;45:520–534. doi: 10.3109/10409238.2010.513375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koster D.A., Croquette V., Dekker C., Shuman S., Dekker N.H. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature. 2005;434:671–674. doi: 10.1038/nature03395. [DOI] [PubMed] [Google Scholar]
- 26.Taneja B., Schnurr B., Slesarev A., Marko J.F., Mondragon A. Topoisomerase V relaxes supercoiled DNA by a constrained swiveling mechanism. Proc. Natl. Acad. Sci. U.S.A. 2007;104:14670–14675. doi: 10.1073/pnas.0701989104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson R.C., Bruist M.F. Intermediates in Hin-mediated DNA inversion: a role for Fis and the recombinational enhancer in the strand exchange reaction. EMBO J. 1989;8:1581–1590. doi: 10.1002/j.1460-2075.1989.tb03542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed R.R., Grindley N.D.F. Transposon-mediated site-specific recombination in vitro: DNA cleavage and protein-DNA linkage at the recombination site. Cell. 1981;25:721–728. doi: 10.1016/0092-8674(81)90179-3. [DOI] [PubMed] [Google Scholar]
- 29.Owczarzy R., Moreira B.G., You Y., Behlke M.A., Walder J.A. Predicting stability of DNA duplexes in solutions containing magnesium and monovalent cations. Biochemistry. 2008;47:5336–5353. doi: 10.1021/bi702363u. [DOI] [PubMed] [Google Scholar]
- 30.Strick T.R., Allemand J.F., Bensimon D., Bensimon A., Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 31.Ott G.S., Ziegler R., Bauer W.R. The DNA melting transition in aqueous magnesium salt solutions. Biochemistry. 1975;14:3431–3438. doi: 10.1021/bi00686a022. [DOI] [PubMed] [Google Scholar]
- 32.SantaLucia J., Jr A unified view of polymer, dumbbell, and oligonucleotide DNA nearest-neighbor thermodynamics. Proc. Natl. Acad. Sci. U.S.A. 1998;95:1460–1465. doi: 10.1073/pnas.95.4.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huguet J.M., Bizarro C.V., Forns N., Smith S.B., Bustamante C., Ritort F. Single-molecule derivation of salt dependent base-pair free energies in DNA. Proc. Natl. Acad. Sci. U.S.A. 2010;107:15431–15436. doi: 10.1073/pnas.1001454107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skoko D., Wong B., Johnson R.C., Marko J.F. Micromechanical analysis of the binding of DNA-bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA-protein complexes. Biochemistry. 2004;43:13867–13874. doi: 10.1021/bi048428o. [DOI] [PubMed] [Google Scholar]
- 35.Marko J.F. Torque and dynamics of linking number relaxation in stretched supercoiled DNA. Phys. Rev. E. 2007;76:021926. doi: 10.1103/PhysRevE.76.021926. [DOI] [PubMed] [Google Scholar]
- 36.Eichhorn G.L., Shin Y.A. Interaction of metal ions with polynucleotides and related compounds. XII. The relative effect of various metal ions on DNA helicity. J. Am. Chem. Soc. 1968;90:7323–7328. doi: 10.1021/ja01028a024. [DOI] [PubMed] [Google Scholar]
- 37.Bond J.P., Anderson C.F., Record M.T., Jr Conformational transitions of duplex and triplex nucleic acid helices: thermodynamic analysis of effects of salt concentration on stability using preferential interaction coefficients. Biophys. J. 1994;67:825–836. doi: 10.1016/S0006-3495(94)80542-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Falvey E., Hatfull G.F., Grindley N.D. Uncoupling of the recombination and topoisomerase activities of the γδ resolvase by a mutation at the crossover point. Nature. 1988;332:861–863. doi: 10.1038/332861a0. [DOI] [PubMed] [Google Scholar]
- 39.Bai H., Kath J.E., Zorgiebel F.M., Sun M., Ghosh P., Hatfull G.F., Grindley N.D.F., Marko J.F. Remote control of DNA-acting enzymes by varying the Brownian dynamics of a distant DNA end. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16546–16551. doi: 10.1073/pnas.1203118109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rutherford K., Yuan P., Perry K., Sharp R., Van Duyne G.D. Attachment site recognition and regulation of directionality by the serine integrases. Nucleic Acids Res. 2013;41:8341–8356. doi: 10.1093/nar/gkt580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.