Abstract
A truly universal nucleobase enables a host of novel applications such as simplified templates for PCR primers, randomized sequencing and DNA based devices. A universal base must pair indiscriminately to each of the canonical bases with little or preferably no destabilization of the overall duplex. In reality, many candidates either destabilize the duplex or do not base pair indiscriminatingly. The novel base 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin- 4-amine) N8-(2′deoxyribonucleoside), a deoxyadenosine analog (UB), pairs with each of the natural DNA bases with little sequence preference. We have utilized NMR complemented with molecular dynamic calculations to characterize the structure and dynamics of a UB incorporated into a DNA duplex. The UB participates in base stacking with little to no perturbation of the local structure yet forms an unusual base pair that samples multiple conformations. These local dynamics result in the complete disappearance of a single UB proton resonance under native conditions. Accommodation of the UB is additionally stabilized via heightened backbone conformational sampling. NMR combined with various computational techniques has allowed for a comprehensive characterization of both structural and dynamic effects of the UB in a DNA duplex and underlines that the UB as a strong candidate for universal base applications.
INTRODUCTION
The genetic code relies on the precise base pairing of the four natural bases. However, an objective in synthetic biology is the expansion of the ‘genetic alphabet’ with the development of unnatural bases. These efforts have shown promise in a variety of roles including polymerase chain reaction (PCR) amplification, RNA transcription and DNA primers and probes (1–3). In contrast to novel synthetic base pairs that feature specific pairing, a universal base would be indiscriminate in its base pairing and ideally pair with all four natural bases uniformly. In the biotechnology field, a truly universal base could be implemented in probes and primers and would be highly useful (4–11).
Ideally, a universal base should be incorporated into a DNA duplex environment with little perturbation to the duplex structure. Natural DNA base pairs are formed via canonical base stacking and Watson–Crick hydrogen bonding patterns generating a regular B-type DNA. Thus, regular base stacking, hydrogen bonding and steric interactions are important design features for the stable incorporation of any synthetic nucleobase analog within a duplex environment. Examples of universal base designs range from single and fused aromatic ring systems to aliphatic and even abasic designs; each of these schemes undermine either structure and/or stability of the DNA duplex (12–15). Recent examples of a prominent family of universal bases include the non-hydrogen bonding 3-nitropyrrol and 5-nitrolindole analogs. NMR and thermostability studies of theses analogs reveal a unique intercalating base stacking scheme, which aids in its ambiguous base pairing scheme while maintaining an overall B-type conformation with a reduced thermostability (ΔTm ∼6°C per synthetic nucleobase in a 16er oligonucleotide sequence) (16). Although novel, this intercalation presents an altered local conformation as compared to canonical B-type DNA, which may or may not be desirable.
The Seela lab has developed a series of N8 linked 2′ deoxyadenosine analogs that exhibit universal base properties (17–22). The 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin-4- amine) N8-(2′deoxyribonucleoside) deoxyadenosine analog (UB) base pairs indiscriminately with all four natural base pairs despite the unique N8 glycosidic linkage (Figure 1) (18,20–22). In dodecamer duplexes where UB is paired with all four natural bases, the Tm values are within 2°C of one another. Compared to an A:T base pair, the UB-(A,C,G,T) base pair results in a drop in thermal stability with ΔTm of −4 to −6°C. Additionally, it exhibits less fluctuations than mismatches (mismatch ΔTm values of −2 to −12°C), and is more stable than abasic sites (abasic ΔTm values of −10 to −17°C) (18).
Figure 1.

UB schematic. Schematic of 8-aza-7-deazaadenine (pyrazolo[3,4-d]pyrimidin-4-amine) N8-(2′deoxyribonucleoside) (UB) in the “anti” orientation.
Understanding how the UB is incorporated into a duplex environment will aid in a refined design of universal bases and shed light on its interaction with DNA servicing proteins and enzymes (16). Solution state NMR was utilized to determine a high-resolution structure of a DNA duplex containing a UB opposite a dT; the duplex was also probed for regions of unusual dynamics and chemical exchange. NMR data complemented with extended molecular dynamics, molecular dynamics with time averaged restraints and chemical shift calculations reveal that the UB is well accommodated in the duplex with very little impact to global and local geometry. Nevertheless, UB is also the source of unusual dynamics on the high μs to low ms timescale, which results in the disappearance of the UB base H2 proton in all NMR spectra under native conditions but with little impact on other resonances.
MATERIALS AND METHODS
Oligonucleotides
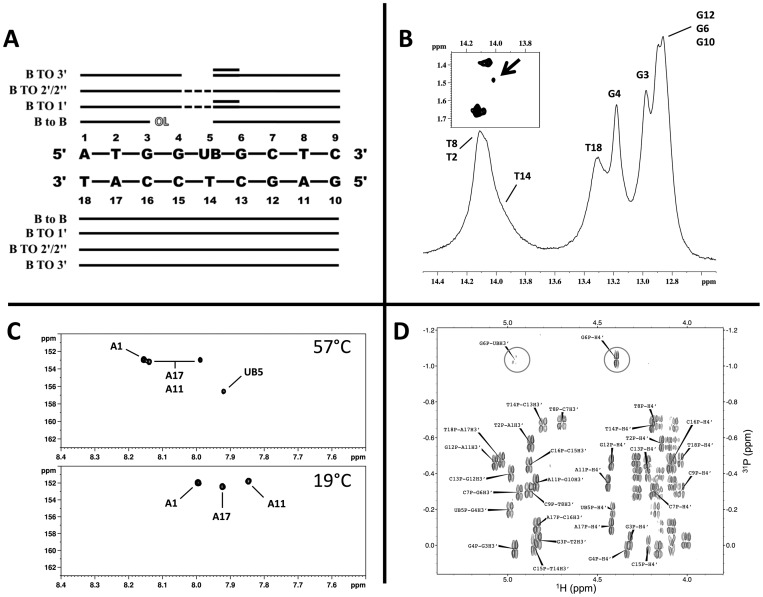
UB containing oligonucleotides were synthesized and purified as previously described (18,19). All other oligonucleotides were purchased from University of Calgary Core DNA Services and purified via ion exchange (Resource 15Q column) and size exclusion liquid chromatography. Purity was assessed via denaturing polyacrylamide gels. Concentrations were calculated using extinction coefficients from the sum of mononucleotides (absorbance at 260 nm, 80°C) (23). The extinction coefficient for the UB is 6600 M−1cm−1 (18). Sequences used for NMR are as follows (X = UB or dA):

Melting temperature studies
DNA duplexes for Tm measurements are detailed in Table 1. Samples for Tm measurements were in 100 mM NaCl, 10 mM MgCl2 and 10 mM sodium cacodylate with a pH of 7.0. Total DNA strand concentration was 10 μM. UV absorbance was measured on a Cary-1/1E UV/VIS spectrophotometer. Thermodynamic data were calculated using the MeltWin 3.0 package. Tm measurements in 100 mM NaCl with 10 mM sodium phosphate at a pH of 6.8 were also completed on the NMR control sample (A:T) and UB sample (UB:T) and analyzed as described previously (23).
Table 1. (A) Thermodynamic properties of UB containing oligonucleotides in the same sequence context but with varying base pair partners for the UB. Also included for comparison are the same sequences with an A:T base pair and a A:C mismatch. The X represents the UB. Sample conditions are as follows: 10 μM total strand concentration, 100 mM NaCl, 10 mM MgCl2 and 10 mM sodium cacodylate with a pH of 7.0. aMelting curve is not fitable. (B) Thermodynamic properties of the UB containing oligonucleotide and control sequence using NMR buffer conditions (100 mM NaCl, 10 mM sodium phosphate with a pH of 6.8). The X represents the UB. For both sections, ΔG was calculated at 21°C using ΔG = ΔH − TΔS.

|
NMR sample conditions
NMR samples were completed in a buffer containing 10 mM sodium phosphate, 100 mM NaCl, with 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS) added as an internal reference. The pH* for D2O samples was 6.36; the pH for H2O samples was 6.79. NMR samples for imino proton spectra collected at supercooled conditions were transferred to 1 mm borate glass capillaries with the ends flame sealed; capillaries were combined in a standard 8 mm NMR tube (24,25). Temperatures below −5°C were reduced at a rate of 1°C per hour (26).
NMR experiments and structure calculations
Detailed methods for NMR experiments and structure calculations are provided in Supplementary Data. Briefly, NMR experiments were conducted on a Bruker Avance 600 MHz NMR equipped with a 5 mm or 8 mm QXI probe, 1H {13C, 15N, 31P}; HSQC and 13C T1 experiments were conducted on a 500 MHz Avance systems equipped with a 5 mm TXI cryoprobe 1H{13C, 15N}. Standard protocols were utilized for the collection of 1D 1H and 31P and 2D COSY, TOCSY, CTNOESY, HPCOR, HSQC, and NOESY spectra as previously described (27–30). D2O NOESY spectra were collected with mixing times of 75, 150 and 250 ms and an 8 s interscan delay. RDC values were determined for NMR signals from the sugar and base regions using f2 coupled HSQC spectra collected in the presence and absence of pf1 phage (ASLA) (2H splitting of 18.0 and 32.2 Hz giving concentrations of 20.3 and 36.3 mg/ml for the UB and control samples respectively) (31). Structure calculation utilized an iterative RANDMARDI process with MARDIGRAS, CORMA and AMBER 9.0 cycles as described previously (27,28,32–39). The B-type backbone torsion angles (except for the experimentally determined ϵ torsion angles) and Watson–Crick hydrogen bond restraints were not included for the UB5 residue.
The AMBER parm99 force-field was appended to include specific parameters for the UB (40). (Protocols for the AMBER force-field paramaterization can be found in Supplementary Data). Starting structures were canonical B-type DNA with the incorporation of UB at position 5. Fully restrained molecular dynamics were conducted in explicit water (TIP3P) and charges were neutralized with the addition of 16 Na+ counterions; simulation times were 10 ns. The final bundle of structures was selected from the last 100 ps at a rate of 1 structure per 10 ps. Structural and helical parameters were determined using the Curves+ algorithm (41). Heavy atom RMSD calculations were completed in VMD 1.9 (42). Molecular visualization was completed in VMD 1.9 and UCSF Chimera (42–44).
Computational methods
For a detailed description of the molecular dynamics simulations, see Supplementary Data. Briefly, molecular dynamics simulations using time-averaged restraints (MDtar) and AMBER 9.0 with a τ = 20 ps were completed as described previously (45). The PDQPRO algorithm was used in conjunction with CORMA to select high probability structures from MDtar trajectory of structures (46). Unconstrained extended molecular dynamic simulations were completed on the UB and control oligonucleotides with additional GC base pairs at the terminal ends to stabilize the ends and avoid artifacts (47). The sequence used for unconstrained extended molecular dynamics simulations is:

where X is either the UB or dA. NMR chemical shift calculations were completed with Gaussian 03 (revision C.02) using a DFT B3LYP/6-31g(d,p) basis set and the GIAO method in vacuum.
RESULTS
Thermal stability
The thermal stability for the four UB containing oligonucleotides are similar with Tm and ΔG values all within 1°C and 0.5 kcal/mol of one another with the UB opposite dT having a slightly higher Tm than the others (Table 1, section A). Although the UB thermal stabilities are compromised compared to the canonical A:T base pairing (a reduction from 42°C for the A:T duplex to 33.2–34°C for the UB oligonucleotides), all four canonical bases opposite the UB exhibit a similar stability underlining its universal base pairing capabilities (Table 1). Differences in ΔG as compared to the control are ∼2.2-2.7 kcal/mol lower than the control potentially indicating different stacking or hydrogen bonding pattern for the UB containing duplexes. Additionally, thermal stability studies were conducted on the UB:T and control samples using NMR buffer conditions (Supplementary Figure S17). As anticipated, the thermal stabilities for the UB and control sequence were lower in the absence of Mg2+ with the difference in Tm and ΔG between the two samples being higher as compared to the Mg2+ containing samples (UB:dT and control Tm 31.4 ± 0.4°C and 41.7 ± 0.4°C, respectively; ΔΔG 2.6 kcal/mol) (Table 1, section B). Based on these results and the UB being an adenosine analog, NMR and dynamics studies were limited to the slightly more stable UB opposite the dT.
UB glycosidic bond orientation
The unique UB glycosidic bond orientation will be defined using the same naming convention as originally reported where χ = O4′-C1′-N8-C7 and the minimum distance between H7 and H1′ is defined as ‘anti’ (18). The ‘syn’ and ‘anti’ conformations yield quite different H7-H1′ distances (‘syn’ 3.8 Å and ‘anti’ 2.5 Å) and was used to characterize the glycosidic bond torsion angle (Figure 1). A NOESY spectra with a 75 ms mixing time was used to estimate the distance between the UB5 H7 and H1′ protons at 2.3 Å. This establishes the base orientation as ‘anti’ with the H7 proton pointing into the minor groove and the H2 proton in the major groove. These results were verified with H7 to H2′, H2″ and H3′ distances. The residues surrounding the UB are all in canonical anti conformations.
NMR structure data
All anticipated NOE base to H1′ contacts were identified, with the only unusual features being a reduction in intensity between G4 H8 and UB5 H1′ and the previously described high intensity UB5 H7 to UB5 H1′. All base to base, H2′, H2″ and H3′ pathways are accounted for with the exception of the base to base and base to H3′ pathways between G4 to UB5 (Figure 4A). In addition, several unique crosspeaks were observed: G6 H1′ to UB5 H7 and G6 H3′ to UB5 H7 as well as UB5 H1′ to G6 H3′ and UB5 H1′ to G6 H4′ (Figure 4A). Collectively, these unique contacts along with the G4 H8 to UB5 H1′ NOESY crosspeaks, confirm an intrahelical orientation of the UB base (Supplementary Figure S2B). The measured NOEs and intensities for the rest of the oligonucleotide are consistent with an overall B type helix.
Figure 4.
NOE connectivity and NMR spectra indicating unusual dynamics. (A) NOE pathways. Solid line signifies normal NOE contacts; dotted line = weak NOE, double line = unanticipated NOE contacts; OL = signal overlap. See text for details of unanticipated NOE contacts. (B) Supercooled aqueous NMR spectra of the imino proton region at −12°C showing the appearance of a ‘shoulder’ peak at 14 ppm. Inset: NOESY spectrum collected at −12°C showing crosspeaks between the imino proton region and methyl region. Arrow indicates the appearance of a new crosspeak. (C) 1H-13C HSQC spectra focused on the adenosine C2–H2 region; top spectrum is collected at 57°C, bottom spectrum is at 19°C. (D) 1H–31P HETCOR spectrum showing crosspeaks for H3′–P (4.6–5.1 ppm, 1H) and P–H4′ (3.9–4.5 ppm, 1H) (22°C). Circled are the weakened intensity for the UB5 H3′– G6 P and normal intensity G6 P–G6 H4′ crosspeaks.
1D and 2D imino proton spectra were used to detect and assign base pairs. All imino proton resonance except the T14 imino proton were observed under normal conditions (7°C) and exhibit chemical shifts indicative of normal A:T and G:C Watson–Crick base pairing. Of particular note, the bases flanking the UB5 (G4 and G6) exhibit sharp line shapes indicating stable base pair formation. All imino protons resonances were detected for the control sequence (Figure 2A). NOESY spectra collected in H2O were used to identify the imino/amino pathways. The UB5 amino group could not be identified in NOESY spectra collected in H2O. Finally, a weak NOESY crosspeak between G4's imino proton and UB5 H7 was observed, which further confirms the UB5 intrahelical orientation. Interstrand NOESY crosspeaks involving UB5 or T14 were not observed.
Figure 2.
Imino proton and 31P NMR spectra. (A) 1H imino proton spectrum of the control and UB sequences at 7°C (top and bottom, respectively). Imino protons for residues G4 and G6 of the UB sequence (which flank UB5) are labeled in grey. (B) 31P spectra for the control and UB sequences at 22°C. Arrows and circles indicate the G6 and C15 phosphorous resonances, which exhibit significant perturbations in chemical shifts as compared to the control.
Absolute values of differences in chemical shifts (|UB – control| = Δ ppm) for sugar protons and phosphorus resonances were calculated. As shown in Supplementary Figure S2A, the largest chemical shift perturbations for the sugar protons were localized near the UB5:T14 base pair with the T14 sugar protons containing the largest overall chemical shift changes and the largest changes in individual proton resonances. Base proton chemical shift changes were mainly localized to C15 (Supplementary Tables S2 and S3). As compared to the control sequence, 31P chemical shifts for UB5-P-G6 and T14-P-C15 are shifted +0.52 and −0.56 ppm from their control values, respectively (Figure 2B, Supplementary Tables S2 and S3).
All measured three bond sugar coupling values for UB5 were consistent with a 2′ endo / 3′ endo equilibrium and place the UB5 sugar in an S conformation with a pseudorotation value of 125–170° and fraction south of 80%. With the exception of C7 (50% fs), all other residues are predominately in an S conformation (Supplementary Table S4). For the control sequence, all residues measured were found to be in S conformation. Coupling values for control C7 could not be measured due to signal overlap, however, examination of the COSY coupling patterns suggest a similar fraction south range as the UB sequence (50% fs). Experimentally determined epsilon (ϵ) torsion angles did not reveal a unique perturbation for any of the residues including UB5 and flanking bases (ϵ torsion angle range of 163° to 171°) (Supplementary Table S5). Interestingly, the 1H – 31P HETCOR experiments did show a weak intensity for the H3′ to 31P crosspeak in the UB5-P-G6 sequence (Figure 4D).
Structure calculation
The starting conformation for both duplexes was a B-type geometry with the UB5:T14 base pair in a reverse Watson–Crick-like base pair. For both sequences, the rMD trajectory was analyzed for variations in structure conformation through the monitoring of key torsion angles and base pair distances (torsion angles presented in Figure 5; base pair distance data not shown). Little variation was observed allowing for sampling from the final 100 ps of the trajectory. The UB containing duplex is highly restrained containing a total of 480 restraints giving an average 27 restraints per residue (Table 2). The average heavy atom R.M.S.D. for the final bundle of structures is 0.42 Å, and the structure with the overall lowest CORMA RX and AMBER restraint violations was chosen as the final representative structure (Supplementary Figure S3). The structure shows excellent agreement with NOESY data with total CORMA RX values ranging from 4.4 to 5.5 and AMBER distance and RDC violations of 36.6 and 12.9 kcal/mol respectively (RX values multiplied by 100). The control structure has fewer restraints than the UB structure due to signal overlap (∼22 restraints per residue); it too exhibits excellent agreement with NOESY data with total CORMA RX values ranging from 3.9 to 5.4 and AMBER distance and RDC violations of 14.3 and 3.0 kcal/mol for all restraints (Table 2). Both the UB5 and the A5 residues are well restrained in each of the structures with the UB5 H7 proton having a total of 7 distance restraints and the A5 H8 proton having 8 restraints (Supplementary Figure S2B).
Figure 5.
(A) G4 and UB5 backbone torsion angles versus MD trajectory time for rMD, MDtar and long term unconstrained MD simulations for the UB sequence. From top to bottom: G4 epsilon, G4 zeta, UB5 alpha, UB5 beta and UB5 gamma backbone torsion angles. From left to right: 9 ns of restrained MD, 1 ns of MD using time averaged restraints and 400 ns of unconstrained MD. Occurrences of populations A, B and C are labeled and separated with dashed lines in the MDtar graphs. For all graphs: X-axis is the trajectory time in ns and the Y-axis is torsion angle in degrees. Gauche− (g−) = −60° (i.e. 300°), gauche+ (g+) = +60° and trans (t) = 180°. (B) Scatter plots of torsion angles versus UB5 alpha torsion angle from the unconstrained MD trajectory. For all plots, the X axis is UB5 alpha torsion angle. Side projections are histograms for the respective torsion angle. Populations are outlined as follows: blue box = population A, orange box = population B, green box = population C.
Table 2. Summary of NMR restraints used for the structure development of the UB and control structures. Qualitative distance restraints involving exchangeable protons were obtained from a H2O NOESY spectrum with a mixing time of 150 ms. The structure rMD calculations were run for 10 ns. Charges were neutralized with 16 Na+ ions and simulations were conducted in explicit TIP3P water. Final structures underwent fully restrained energy minimization (2000 steps steepest decent). Heavy atom R.M.S.D. values were calculated in VMD version 1.9. Of note, residue A1 for both the control and UB sequences exhibit a syn/anti equilibrium with <25% syn population, which was evident in the integrated NOESY crosspeaks (Supplementary Figure S4). Because this observation was present and similar for both sequences and obviously not a result of the UB incorporation, the A1 residue was constrained to an anti conformation for both sequences.

|
Structure analysis
All bases including both the UB5 and T14 bases are intrahelical and the overall global geometry is B-type (Supplementary Figure S3). The UB5:T14 base pair does not form a hydrogen bond involving the T14 imino proton, which is consistent with the absence of the lack of the T14 imino proton. Instead, the amino group of UB5 is hydrogen bonded to the T14 O4 carbonyl (Figure 3A). This unusual base pairing results in heightened stretch and opening (0.75 Å and −23°, respectively) manifesting in an increase of 1 Å in the UB5:T14 anomeric carbon distance as compared to the control structure (Figure 3A, Supplementary Figure S3). As a consequence of the unique N8 UB base connectivity and orientation, the local stacking of G4-UB5-G6 is slightly disrupted with somewhat G4 poorly stacked on top of UB5. Interestingly, the perturbation is confined to the UB5; G4 and G6 remain in a similar conformation as the control sequence (Figure 3B). This unusual orientation was persistent with repeated structure calculations indicating that the energetically unfavorable base pair configuration is represented in all of the UB5:T14 base pair restraints. The overall solved UB and control structures are quite comparable with a total heavy atom R.M.S.D. of 0.98 Å (due to different connectivity and atom placement, UB5/A5 were excluded from the calculation).
Figure 3.
UB5:T14 and A5:T14 base conformations. (A) Base pair for UB5:T14 (top) and the control A5:T14 (bottom) (both are from the solved rMD structures). Anomeric carbon distances are referenced. Sugar and backbone are in grey; base colors: blue = nitrogen, grey = carbon, red = oxygen, white = hydrogen. (B) Base stacking for G4 (yellow), UB5/A5 (red) and G6 (blue). UB sequence on left, control on right (both are from the solved rMD structures). (C) Predicted UB-T base pair hydrogen bonding pattern determined from previous thermal stability studies (Seela F. & Debelak, H., NAR, 2000).
Supercooled aqueous NMR yields a T14 imino proton signal
Previous thermal stability studies with UB containing oligonucleotides and other UB-like analogs predicted a reverse Watson–Crick like UB-T base pair and would exhibit a hydrogen bonding pattern with the involvement of the thymine imino proton (17,18) (Figure 3C). Due to the quite unusual UB5-T14 hydrogen bonding pattern found in the solved structure, further probing of the imino proton spectra was conducted via supercooled aqueous NMR utilizing 1 mm capillaries. At temperatures >4°C, no T14 imino proton signal is observed. When temperatures are dropped to −8°C, a shoulder begins to emerge at 13.6 ppm suggestive of the formation of an additional A:T–like base pair (Supplementary Figure S8). A NOESY spectrum collected at −12°C was used to confirm the identity of the new resonance as T14 through the appearance of a cross peak between the new imino proton resonance and a methyl resonance similar to T14's methyl chemical shift at 21°C (Figure 4B, Supplementary Table S2). All observed imino proton resonances shifted as anticipated with decreasing temperatures indicating that an overall global conformation change in the oligonucleotide did not occur (Supplementary Figure S9).
Base opening rates
Base pair lifetimes were calculated for non-overlapped imino proton peaks of the UB and control sequences through the addition of NH4OH as a catalyst (Supplementary Figure S18). For the control sequence, the A5:T14 base pair exhibits a lifetime of 3 to 5 ms; because no T14 imino proton signal is observed under standard conditions for the UB sequence, a corresponding base opening for the UB5:T14 base pair could not determined. Significant overlap of the control sequence limited quantitative analysis to G6:C13 which was found to have a base pair lifetime of 40 to 50 ms. Based on findings from other studies, these results are consistent with canonical A:T and G:C Watson–Crick base pair lifetimes (48–50). Similar to the control sequence, the base pair lifetimes for many of the UB sequence residues could not be quantitatively determined due to signal overlap. G4-C15 exhibits a lifetime of ∼3 ms. The G6 and G12 imino proton signals are overlapped and lifetimes could not be determined individually. Therefore, a qualitative approach was taken with some overlapped peaks where the imino proton resonance's recovery was compared to other peaks in the same sequence. Under high concentrations of catalyst, stable base pairs recover more slowly than short lived ones. Inspection of the UB sequence's G6/G12 overlapped peaks reveal that the signal recovers more slowly than the G4 peak suggesting that G6 and G12 have a longer base pair lifetime (Supplementary Figure S19). Additionally, although the control G3, G4 and G12 imino proton peaks are overlapped, the relative position of the G4 resonance within the overlapped peak is known to be on the downfield side (Figure 2A). With this, G6 appears to recover comparably with G4 suggesting that G4 and G6 have similar base pair lifetimes (Supplementary Figure S20). For both sequences, the imino proton resonance for T2:A17 and T8:A11 disappear upon the first addition of NH4OH (∼3 mM NH3).
‘The case of the disappearing H2 proton’
The UB5-H2 proton has a similar chemical environment as compared to an adenosine H2 and is anticipated to have similar NMR and chemical traits. Under denaturing conditions, all 4 H2 protons in the UB sequence are readily identified by both 1D 1H NMR and 1H–13C HSQC spectra. A spectrum recorded for the single strand identified the UB5 H2 and C2 resonances at 7.92 and 156 ppm, respectively. Unexpectedly, when annealed, the UB5-H2 proton vanishes from all spectra; only the three adenosine H2 protons can be identified in 1D 1H, HSQC and NOESY spectra (Figure 4C, Supplementary Figure S1). The UB H2 proton reappears upon subsequent denaturing of the duplex; this process is readily reversible. Careful examination and integration of the base region ruled out resonance overlap. This anomaly is restricted to the UB5 H2 proton; the UB5 H7 proton, UB5 sugar protons as well as protons of other neighboring residues including the T14 protons were readily identified and exhibit sharp peaks in 1D spectra.
Reduced crosspeak intensities
A number of 2D NMR crosspeaks in the HSQC, low-flip angle COSY and 1H–31P HETCOR spectra have lower intensities than anticipated. These include the HSQC T14 H1′/C1′, G6 H1′/C1′ and UB5 base H7/C7 crosspeaks, COSY UB5 and T14 H1′–H2′ and H1′–H2″ crosspeaks, and the UB5 H3′–G6 P HETCOR crosspeak (Figure 4D, Supplementary Figures S5, S6 and S7). In all instances, the underlying coupling values responsible for the crosspeaks were identified and measured. Measured 1JC-H values were within anticipated ranges with the UB5 1JC7-H7 at 197 Hz. COSY 3JH-H couplings for UB5 and T14 sugar protons exhibit values anticipated for a predominant fs conformation, and the 3JH3′-P for UB5 H3′–G6 P was measured at 3.85 Hz from ctNOESY experiments (Supplementary Tables S4 and S5). Taken together, these peak anomalies are suggestive of unusual dynamics.
13C T1 measurements
To gain insight into motions on the ps–ns time frame, natural abundance 13C T1 relaxation rates were measured for the UB sequence (Supplementary Figure S21). For this size molecule, lower T1 values correlate with increased mobility. To avoid artifacts from terminal residues, the analysis was limited to the immediate bases surrounding the UB residue. At 19°C, the anomeric carbons for G4, UB5, G6 and T14 are comparable with only the G4 showing a slightly reduced value (0.34 ± .03 s compared to a range of 0.38 to 0.41 s) (C13 and C15 not determined due to signal overlap). However, at 9°C, G4's anomeric T1 value is markedly lower than the other values (0.38 ± .05 s compared to the others, range of 0.47 to 0.52 s). Data for the G4 and G6 base carbons could not be determined, however, C13 and C15 base carbons were evaluated revealing that C15 exhibits a somewhat lower T1 relaxation compared to C13 (C13 = 0.35 and 0.42 s, C15 = 0.25 and 0.21 s for C5 and C6 carbons, respectively). This is consistent with the reduced anomeric T1 value observed for G4 (C15's base pair) indicating a slight increase in mobility on the 5′ side of UB5. Of note, UB5's C7 relaxation was measured at 0.33 ± 0.03 s, which is comparable to A11's C8 (0.31 ± 0.03 s).
Molecular dynamics with time averaged restraints
In order to evaluate possible conformational sampling while still using the NMR restraints, molecular dynamics calculations using time-averaged restraints (MDtar) were performed where distance restraints are enforced over a period of time as opposed to constant enforcement (51,52). This treatment allows for the evolution of dynamics when they are not readily apparent in the NMR data (implementation detail are in Supplementary Data Materials and Methods). For the control structure, the MDtar simulations do not yield any significant changes in conformations or torsion angles for A5 or the surrounding residues. In contrast, the UB MDtar simulation reveals dynamic flexibility of the backbone torsion angles at the UB5 residue with no significant changes in base orientations (Supplementary Figure S10A and B). As shown in Figure 5A, the UB5 α/γ torsion angles transition from a canonical g−/g+ (gauche−/gauche+) through a g−/t (gauche−/ trans) and ultimately to a g+/t (gauche+/ trans) conformation (labeled as populations A, B and C on Figure 5A, respectively). Concurrent with these transitions, the β torsion angle transitions from trans to gauche+ back to trans over the course of the simulation. The ϵ and ζ torsion angles were used to analyze BI/BII transitions. For the G4 residue, populations A and B are in a BI like conformation. However, once the α/γ torsion angles adopt a g+/t geometry (population C), G4 samples the entire BI and BII conformational space. All other torsion angles surrounding the UB residue including its base pair T14 exhibit stable, canonical torsion angles. As anticipated, the AMBER restraint penalties over the course of the MDtar trajectory drop to nearly half (∼18 kcal/mol and 8 kcal/mol for the UB and control trajectories, respectively).
In order to obtain representative structures, the PDQPRO algorithm was used to compare the MDtar ensemble of structures with the NMR NOESY data (45,46). The analysis yields 5 high probability structures (individual structure probabilities are 11%, 10%, 20%, 13% and 14%) for the UB sequence; one each from clusters A and B, and 3 from cluster C. All PDQPRO selected structures have the UB5:T14 base pair in the same configuration as the solved structure with little variation in orientation (Supplementary Figure S10A and B). For the three PDQPRO structures from cluster C, none were in full canonical BI or BII conformations, but 2 exhibit BII type characteristics (ϵ – ζ: 39.5° and 26.6°), and one with BI characteristics (ϵ – ζ: −40.7°). The two other structures representing clusters A and B were in the canonical BI range (−83.0° and −86.9°). Despite the time-averaged enforcement of distance restraints, the five structures overlay well with most conformational changes occurring in the backbone geometries (Supplementary Figure S10A, B and C). These results indicate that the enhanced backbone flexibility is unique to the UB5 residue, is not exhibited by the control A5 residue, and is supported by the NMR experimental data.
Long-term unconstrained Molecular Dynamics
Following MDtar analysis, long-term unconstrained dynamics simulations were run to predict whether the backbone conformational changes are consistently concerted, if they are long lived, and if there are other changes. The heavy atom RMSD values reveal a bimodal distribution of populations for the UB5-T14 base pair only; this is not observed for other base pairs in the UB duplex or the control duplex (Supplementary Figure S11). At the onset of the unconstrained molecular dynamics simulation, the UB5–T14 base pair moves to the predicted reverse Watson–Crick like base pair and does not sample another configuration (Figure 3C, Supplementary Figure S14B). Other than this initial shift in the UB5–T14 hydrogen bonding pattern, no other prominent changes were evident in the base orientation of the surrounding residues. Remarkably, the shift in base pair hydrogen bonding patterns only affects the conformational sampling of the backbone geometry at the UB5 residue as observed in the MDtar calculations.
The backbone at UB5 exhibits 3 distinct populations similar to those observed in the MDtar trajectory where the UB5 α, β and γ torsion angles exhibit concerted transitions (Figure 5A, Supplementary Figure S12). Correlating the UB5 α torsion angle versus other torsion angles confirms concerted backbone transitions and allows the populations to be culled into three separate populations with torsion angles similar to the MDtar results (i.e. A, B and C) (Figure 5A and B, Supplementary Figure S22, Supplementary Table S1). Interestingly, only the UB5 β torsion angle exhibits a bimodal distribution with heavy atom RMSD for the UB5:T14 base pair (Supplementary Figures S11C and S12). The α, β and γ torsion angles for the residues flanking the UB as well as T14 and flanking residues remain in essentially static canonical torsion angles (Supplementary Figure S12). Population B is the most frequently sampled conformation in the MD simulation representing ∼50% of the total trajectory (Supplementary Table S1). Unconstrained MD simulations with the control sequence do not reveal any significant changes in conformation at the A5 residue emphasizing that the observed results for the UB sequence are unique to the UB5 (Supplementary Figure S12).
Analysis of the UB sequence G4's ϵ and ζ torsion angles reveals that populations A and B are in a BI conformation, while C samples both BI and BII space with BII sampling occurring 14.1% of the trajectory. Within the C population, lower values of UB5 α (α < 100°) tend favor a BII like conformation while higher values favors a BI like conformation (Figure 5B, Supplementary Figure S13). Analysis of T14's ϵ and ζ torsion angles reveal that it is in a BII conformation 8.6% of the trajectory. However, unlike G4, the sampling appears to be independent from the UB5 torsion angle changes (Supplementary Figure S13). Surprisingly, despite the heightened G4 ϵ and ζ and UB5 α, β and γ flexibility, the UB5 ϵ and ζ torsion angles are quite static and remain in the BI conformation (UB5 = 0% BII) (Supplementary Figures S12 and S13).
DISCUSSION
To determine how the UB is situated in duplex DNA, solution NMR and MD calculations were used to probe the structure and dynamics of the UB in an oligonucleotide. The standard restrained MD NMR procedure resulted in a single high-resolution solution structure that maintains a remarkably unaltered B-type helical geometry with the UB base well accommodated, intrahelical, involved in base stacking, yet exhibiting an unusual base pair conformation. Further probing with supercooled aqueous NMR provided evidence of an alternative UB5:T14 base pair conformation correlating to the previously predicted and energetically favorable configuration. Additionally, several diminished 2D NMR crosspeak intensities provide evidence that the UB duplex exhibits unusual dynamics on the NMR time-scale (Figure 4B, D, Supplementary Figures S5, S6 and S7). Notwithstanding these indications of dynamics, all observed 1D resonances that could be identified exhibit sharp peaks (the only exceptions being the missing UB H2 and T14 imino proton resonances), all NOESY and TOCSY crosspeaks were assigned, and no exchange peaks were observed (Supplementary Figure S1). Although quite low CORMA Rx values were obtained for both structures, the higher AMBER distance restraint penalties and deviations for the UB structure compare to the control indicate an increase in dynamics (Table 2).
Of the weak 2D crosspeaks intensities, the low intensity of UB5 H3′–G6 P HETCOR crosspeak is insightful. A reduced H3′–P crosspeak intensity can be the result of small 3JH3′-P values, yet for the UB sequence, the 3JH3′-P coupling values were measured and are in the anticipated values for B-type DNA (UB5 H3′–G6 P 3JH3′-P = 3.8 Hz). Thus, the weak crosspeak is not a result of a small 3JH3′-P coupling, but due to backbone dynamics (Supplementary Table S5).
The rMD NMR solved structure conforms to the NMR data and does not exhibit altered backbone torsion angles for the UB or any of the surrounding residues despite the fact that the backbone torsion angles for the UB (aside from the experimentally determined ϵ and sugar puckering) were unconstrained during structure calculations (Supplementary Figure S3). MDtar calculations were utilized to probe for conformational changes that may be contained in the NMR data but not readily apparent. The calculations reveal concerted α/γ and BI/BII sampling in the backbone torsion angles around G4 and UB5 that did not manifest themselves in the conventional structure calculation protocol. This sampling was also observed in the long term unconstrained MD simulations. In contrast, the control sequence does not show any significant deviations in the backbone configuration in either simulation indicating the effect originated from the UB.
These predicted motions are supportive of the diminished UB5 COSY sugar proton and UB5 H3′–P HETCOR crosspeaks. Additionally, the BII torsion angle populations for UB5 and T14 predicted by the long-term molecular dynamics simulations are consistent with the 31P chemical shift resonances. 31P resonances are partially dependent on BI/BII populations with downfield shifts generally attributed to a higher BII population (53–56). In the molecular dynamics simulation, UB5-P-G6 exhibits 0% BII population whereas the control sequence's A5-P-G6 sampled the BII conformational space 4% of the trajectory. This trend is consistent with the upfield shifted 31P resonance of −1.04 ppm (Δppm = −0.52 ppm as compared to the control sequence). Similarly, the T14-P-C15 of the UB sequence samples the BII population 8.6% compared to 0.7% in the control sequence; this again is consistent with the downfield shifted signal (Δppm = +0.56 ppm as compared to the control. Although the 31P resonance for G4-P-UB5 does not deviate much from the control value despite the enhanced G4 BII sampling found in the molecular dynamics simulations, the complex motions exhibited in the UB5 α, β and γ torsion angles may offset the impact on the 31P data) (Figure 2B, Supplementary Figure S13).
Although most of the imino proton resonances were overlapped and prevented a direct comparison of residues, the base pair opening analysis does suggest a heightened base opening rate on the 5′ side of the UB (G4) compared to the 3′ side: ∼3 ms for the UB sequence G4:C16 compared to 40–50 ms for the control sequence G6:C13. For the control sequence, the G4 imino proton signal recovers similarly to G6 (Supplementary Figure S20) suggesting similar base pair lifetimes, however, this was not observed for the UB sequence where G4 appears to recover more quickly than the G6/G12 peak (Supplementary Figure S19) suggesting that G4 has a shorter base pair lifetime. This trend is also in agreement with the 13C T1 analysis revealing faster dynamics on the 5′ side. Backbone flexibility on the 5′ side of the UB may in part be lending to the accommodation of the UB insertion into the duplex environment.
The most glaring anomaly exhibited in the NMR data, however, is the missing UB H2 resonance, which defied detection in all NMR spectra under annealed conditions, yet could be readily observed under denaturing conditions and for the single strand (Figure 4C). The UB5 H2 proton is the only signal that exhibits this behavior. Because only a single base proton is affected, this behavior cannot be explained solely by the heightened backbone torsion angle sampling.
The supercooled aqueous NMR data provide evidence of a weak base pair with corresponding higher opening rates. The ‘missing’ UB H2 proton resonance can then be rationalized in context of the surrounding structure and chemical exchange. The ‘anti’ conformation of the UB and the sequence context of G4-UB5-G6 places the H2 proton in the major groove flanked by two carbonyl groups. Carbonyl shielding effects can cause quite dramatic chemical shift changes of protons (57).
With this, the data support at least 2 different UB5:T14 base pair configurations (Figures 3A, C, 4B and Supplementary Figure S14B). These expose the UB5 H2 proton to different environments from the adjacent carbonyls’ shielding cones resulting in chemical exchange for the UB5 H2 proton (Supplementary Figure S14A). The appearance of resonances in chemical exchange is dependent on the difference in chemical shifts and the time-scale of the motion. The fact that the H2 peak disappears from NMR spectra can provide limits on the time frame of the motion.
The broadest a NMR resonance can be due to chemical exchange is at the coalescence point, which is determined by the chemical shifts difference of the proton in the two environments:
 |
(1) |
where Δν is the difference in chemical shift (Hz) and kc is the exchange rate (s−1) at the coalescence point. The difference in chemical shifts must be relatively large as smaller differences would have resulted in an observable broad peak. Because there are no observable NMR resonances under annealed conditions for the UB H2 proton and our NMR and MD calculations indicate two possible orientations, a simple two state model (Equation 1) was used to estimate the timescale of the coalescence point. A symmetrical exchange was assumed since the absence of any signals did not allow us to estimate populations (Supplementary Figures S15 and S16).
Chemical shift differences were calculated using the NMR solved structure and the unconstrained MD model, which was supported by imino proton data collected under supercooled aqueous conditions. In addition, a third theoretical conformation where UB5 was flipped out from the flanking G4 and G6 bases was also used representing an extreme structural limit (57,58).
The difference in calculated UB H2 chemical shifts from the two NMR and MD base conformations is estimated to be 450 Hz resulting in a lifetime (1/kc) of ∼1 ms. Considering the completely flipped out model the difference in chemical shifts is ∼700 Hz that reduces the lifetime to ∼0.6 ms. Compared to the experimentally determined A5:T14 base pair lifetime of 3 to 5 ms, these theoretical values are not unreasonable given the inability to determine the T2 and T11 base opening rates and the ability to slow dynamics of the UB5–T14 base pair under supercooled aqueous conditions (48–50,59). Further support for the predicted timescale comes from the reduced HSQC intensities of UB5, G6 and T14 (Supplementary Figures S6 and S7). Comparison of HSQC peak intensities have been found to give qualitative information of local dynamics on the μs to ms time frame and has been used to characterize broad protein motions and protein–protein interactions (60,61). Additionally, spectra obtained under different NMR field strengths (400, 500 and 600 MHz) did reveal any signals for the UB H2; this was in line with our simulated spectra as well as our estimates above (Supplementary Figures S15 and S16).
The UB's impact on the dynamics and geometry of the DNA duplex are somewhat similar to that of DNA mismatches. As with many mismatches and other lesions, the DNA B-type conformation is robust and accommodates the UB:dT inclusion with little impact on the global geometry showing some similarity to mismatches with an observed increased base opening. The presence of a mismatch in a DNA duplex enhances the dynamics of the mismatch and surrounding residues similar to our findings. However, unlike many mismatches, there is not an observed transferred distortion of helical parameters to other residues outside of the UB5:T14 base pair; the UB disruption to the conformation of the duplex is confined to that base pair with flanking residues quite comparable to the control structure (Figure 3B) (48,62).
Additionally, the UB presents a similar reduction in thermostability as that of the 3-nitropyrrol and 5-nitrolindole universal bases. Both the UB and the nitropyrrol and nitrolindole gain thermal stability through conformational sampling yet achieve this in different ways. The nitropyrrol and nitrolindole bases are not involved in hydrogen bonding with their base pairs, but stabilize via a dynamic intercalation with the base pair causing an increase in rise between both the 3′ and 5′ flanking residues (16). In contrast, the UB presents a less disruptive effect on local base conformation, yet exhibits a heightened breathing rate and dynamic sampling in the phosphodiester backbone.
Taken together, the UB:dT base pair is accommodated within the duplex via entropic freedom in the backbone and base pairing dynamics, yet remarkably does not overly impact the rest of the DNA duplex or the overall B-type conformation about the UB. With the insight of the UB:dT duplex, the ΔH and ΔS data implies a similar stabilization scheme for the UB opposite the dG and dA. Intriguingly, the UB opposite dC is more enthalpically favored suggesting a more rigid structure, and should be characterized in future studies. Moreover, based on our solved structure, the UB's unique N8 connection and ‘anti’ orientation presents a quite different landscape of major and minor groove binding determinants with respects to canonical G-C and A-T base pairs. This unique presentation may be an attribute that could be exploited with custom groove binders. Our data show that the UB remains a viable universal base candidate.
ACCESSION CODES
Coordinates and NMR data have been deposited in the PDB and BMRB under accession codes 2n5o and 2n5p for the UB and control structures, respectively (BRMB codes: 25723 and 25724 for the UB and control, respectively).
Supplementary Material
Acknowledgments
The authors thank Christopher N. Johnson for his help with the NMR calculations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Georgia State University Molecular Basis of Disease Area of Focus (to A.M.S.C.); Georgia State University Brains and Behaviour Program (to M.E.); Georgia Cancer Coalition and a GSU research initiation grant (for M.W.G). Funding for open access charge: Georgia State Department of Chemistry (NMR account).
Conflict of interest statement. None declared.
REFERENCES
- 1.Nakano S., Fujii M., Sugimoto N. Use of nucleic Acid analogs for the study of nucleic Acid interactions. J. Nucleic Acids. 2011;2011:967098. doi: 10.4061/2011/967098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hirao I., Kimoto M., Yamashige R. Natural versus artificial creation of base pairs in DNA: Origin of nucleobases from the perspectives of unnatural base pair studies. Acc. Chem. Res. 2012;45:2055–2065. doi: 10.1021/ar200257x. [DOI] [PubMed] [Google Scholar]
- 3.Kimoto M., Kawai R., Mitsui T., Yokoyama S., Hirao I. Efficient PCR amplification by an unnatural base pair system. Nucleic Acids Symp. Ser. (Oxf) 2008:469–470. doi: 10.1093/nass/nrn238. [DOI] [PubMed] [Google Scholar]
- 4.Chiaramonte M., Moore C.L., Kincaid K., Kuchta R.D. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase α and Klenow fragment (DNA polymerase I) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- 5.Loakes D., Van Aerschotl A., Brown D.M., Hill F. Enzymatic recognition of acyclic universal base analogues in oligonucleotides. Nucleosides Nucleotides Nucleic Acids. 1996;15:1891–1904. [Google Scholar]
- 6.Too K., Loakes D. Universal base analogues and their applications to biotechnology. In: Herdewijn P, editor. Modified Nucleosides and Medicine. Weinheim: Wiley-VCH Verlag GmbH Co. KGaA; 2008. pp. 277–303. [Google Scholar]
- 7.Smith C.L., Simmonds A.C., Felix I.R., Hamilton A.L., Kumar S., Nampalli S., Loakes D., Hill F., Brown D.M. DNA polymerase incorporation of universal base triphosphates†. Nucleosides Nucleotides. 1998;17:541–554. [Google Scholar]
- 8.Sengupta S., Onodera K., Lai A., Melcher U. Molecular detection and identification of influenza viruses by oligonucleotide microarray hybridization. J. Clin. Microbiol. 2003;41:4542–4550. doi: 10.1128/JCM.41.10.4542-4550.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guschin D.Y., Mobarry B.K., Proudnikov D., Stahl D.A., Rittmann B.E., Mirzabekov A.D. Oligonucleotide microchips as genosensors for determinative and environmental studies in microbiology. Appl. Environ. Microbiol. 1997;63:2397–2402. doi: 10.1128/aem.63.6.2397-2402.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ryabinin V.A., Kostina E.V., Maksakova G.A., Neverov A.A., Chumakov K.M., Sinyakov A.N. Universal oligonucleotide microarray for sub-typing of Influenza A virus. PLoS One. 2011;6:e17529. doi: 10.1371/journal.pone.0017529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinheiro A.V., Han D., Shih W.M., Yan H. Challenges and opportunities for structural DNA nanotechnology. Nat. Nanotechnol. 2011;6:763–772. doi: 10.1038/nnano.2011.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loakes D. Survey and summary: The applications of universal DNA base analogues. Nucleic Acids Res. 2001;29:2437–2447. doi: 10.1093/nar/29.12.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liang F., Liu Y.-Z., Zhang P. Universal base analogues and their applications in DNA sequencing technology. RSC Adv. 2013;3:14910–14928. [Google Scholar]
- 14.Loakes D., Hill F., Brown D.M., Salisbury S.A. Stability and structure of DNA oligonucleotides containing non-specific base analogues. J. Mol. Biol. 1997;270:426–435. doi: 10.1006/jmbi.1997.1129. [DOI] [PubMed] [Google Scholar]
- 15.Martin F.H., Castro M.M., Aboul-ela F., Tinoco I., Jr Base pairing involving deoxyinosine: Implications for probe design. Nucleic Acids Res. 1985;13:8927–8938. doi: 10.1093/nar/13.24.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gallego J., Loakes D. Solution structure and dynamics of DNA duplexes containing the universal base analogues 5-nitroindole and 5-nitroindole 3-carboxamide. Nucleic Acids Res. 2007;35:2904–2912. doi: 10.1093/nar/gkm074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He J., Seela F. Oligonucleotides incorporating 8-aza-7-deazapurines: Synthesis and base pairing of nucleosides with nitrogen-8 as a glycosylation position. Org. Biomol. Chem. 2003;1:1873–1883. doi: 10.1039/b301608k. [DOI] [PubMed] [Google Scholar]
- 18.Seela F., Debelak H. The N(8)-(2′-deoxyribofuranoside) of 8-aza-7-deazaadenine: a universal nucleoside forming specific hydrogen bonds with the four canonical DNA constituents. Nucleic Acids Res. 2000;28:3224–3232. doi: 10.1093/nar/28.17.3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seela F., Kaiser K. 8-Aza-7-deazaadenine N8- and N8-(β-D-2′-Deoxyribofuranosides): Building blocks for automated DNA synthesis and properties of oligodeoxyribonucleotides. Helv. Chim. Acta. 1988;71:1813–1823. [Google Scholar]
- 20.Seela F., Jawalekar A.M., Sun L., Leonard P. Oligonucleotides containing pyrazolo[3,4-d]pyrimidines: 8-Aza-7-deazaadenines with bulky substituents in the 2- or 7-position. Nucleosides Nucleotides Nucleic Acids. 2005;24:1485–1505. doi: 10.1080/15257770500265745. [DOI] [PubMed] [Google Scholar]
- 21.Seela F., Zulauf M., Debelak H. Base-Pairing Properties of 8-aza-7-deazaadenine Linked via the 8-Position to the DNA Backbone. Helv. Chim. Acta. 2000;83:1437–1453. [Google Scholar]
- 22.Zhang X., Budow S., Leonard P., Eickmeier H., Seela F. N8-(2′-O-methylribofuranosyl)-8-aza-7-deazaadenine monohydrate. Acta Crystallogr. C. 2006;62:o79–o81. doi: 10.1107/S0108270105041983. [DOI] [PubMed] [Google Scholar]
- 23.Aramini J.M., Germann M. Solution structure of a DNA⊙ RNA hybrid containing an α-anomeric thymidine and polarity reversals: d (ATGG-3′-3′-αT-5′-5′-GCTC)⊙ r (gagcaccau) Biochemistry. 1999;38:15448–15458. doi: 10.1021/bi9915418. [DOI] [PubMed] [Google Scholar]
- 24.Plateau P., Gueron M. Exchangeable proton NMR without base-line distortion, using new strong-pulse sequences. J. Am. Chem. Soc. 1982;104:7310–7311. [Google Scholar]
- 25.Schroeder K.T., Skalicky J.J., Greenbaum N.L. NMR spectroscopy of RNA duplexes containing pseudouridine in supercooled water. RNA. 2005;11:1012–1016. doi: 10.1261/rna.2270205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szyperski T., Mills J.L. NMR-based structural biology of proteins in supercooled water. J. Struct. Funct. Genomics. 2011;12:1–7. doi: 10.1007/s10969-011-9111-5. [DOI] [PubMed] [Google Scholar]
- 27.Aramini J.M., Cleaver S., Pon R., Cunningham R., Germann M. Solution structure of a DNA duplex containing an [alpha]-anomeric adenosine: Insights into substrate recognition by endonuclease IV. J. Mol. Biol. 2004;338:77–91. doi: 10.1016/j.jmb.2004.02.035. [DOI] [PubMed] [Google Scholar]
- 28.Johnson C.N., Spring A.M., Sergueev D., Shaw B.R., Germann M.W. Structural basis of the RNase H1 activity on stereo regular borano phosphonate DNA/RNA hybrids. Biochemistry. 2011;50:3903–3912. doi: 10.1021/bi200083d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson C.N., Spring A.M., Desai S., Cunningham R.P., Germann M.W. DNA sequence context conceals alpha-anomeric lesions. J. Mol. Biol. 2012;416:425–437. doi: 10.1016/j.jmb.2011.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu Z., Tjandra N., Bax A. Measurement of 1H3′-31P dipolar couplings in a DNA oligonucleotide by constant-time NOESY difference spectroscopy. J. Biomol. NMR. 2001;19:367–370. doi: 10.1023/a:1011292803363. [DOI] [PubMed] [Google Scholar]
- 31.Zweckstetter M., Bax A. Characterization of molecular alignment in aqueous suspensions of Pf1 bacteriophage. J. Biomol. NMR. 2001;20:365–377. doi: 10.1023/a:1011263920003. [DOI] [PubMed] [Google Scholar]
- 32.Case D.A., Cheatham T.E., 3rd, Darden T., Gohlke H., Luo R., Merz K.M., Jr, Onufriev A., Simmerling C., Wang B., Woods R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005;26:1668–1688. doi: 10.1002/jcc.20290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pearlman D.A., Case D.A., Caldwell J.W., Ross W.S., Cheatham T.E. III, DeBolt S., Ferguson G., Seibel G., Kollman P. AMBER, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules. Comp. Phys. Commun. 1995;91:1–41. [Google Scholar]
- 34.Borgias B.A., James T.L. COMATOSE, a method for constrained refinement of macromolecular structure based on two-dimensional nuclear Overhauser effect spectra. J. Magn. Reson. 1988;79:493–512. [Google Scholar]
- 35.Borgias B.A., James T.L. MARDIGRAS-A procedure for matrix analysis of relaxation for discerning geometry of an aqueous structure. J. Magn. Reson. 1990;87:475–487. [Google Scholar]
- 36.Keepers J.W., James T.L. A theoretical study of distance determinations from NMR. Two-dimensional nuclear overhauser effect spectra. J. Magn. Reson. 1984;57:404–426. [Google Scholar]
- 37.Thomas P.D., Basus V.J., James T.L. Protein solution structure determination using distances from two-dimensional nuclear Overhauser effect experiments: effect of approximations on the accuracy of derived structures. Proc. Natl. Acad. Sci. U.S.A. 1991;88:1237–1241. doi: 10.1073/pnas.88.4.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.James T.L. Relaxation matrix analysis of two-dimensional nuclear Overhauser effect spectra. Curr. Opin. Struct. Biol. 1991;1:1042–1053. [Google Scholar]
- 39.Liu H., Spielmann H.P., Ulyanov N.B., Wemmer D.E., James T.L. Interproton distance bounds from 2D NOE intensities: Effect of experimental noise and peak integration errors. J. Biomol. NMR. 1995;6:390–402. doi: 10.1007/BF00197638. [DOI] [PubMed] [Google Scholar]
- 40.Cheatham T.E., 3rd, Cieplak P., Kollman P.A. A modified version of the Cornell et al. force field with improved sugar pucker phases and helical repeat. J. Biomol. Struct. Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 41.Lavery R., Moakher M., Maddocks J.H., Petkeviciute D., Zakrzewska K. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Humphrey W., Dalke A., Schulten K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 43.Couch G.S., Hendrix D.K., Ferrin T.E. Nucleic acid visualization with UCSF Chimera. Nucleic Acids Res. 2006;34:e29. doi: 10.1093/nar/gnj031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 45.Aramini J.M., Mujeeb A., Ulyanov N.B., Germann M.W. Conformational dynamics in mixed alpha/beta-oligonucleotides containing polarity reversals: a molecular dynamics study using time-averaged restraints. J. Biomol. NMR. 2000;18:287–302. doi: 10.1023/a:1026798010342. [DOI] [PubMed] [Google Scholar]
- 46.Ulyanov N.B., Schmitz U., Kumar A., James T.L. Probability assessment of conformational ensembles: sugar repuckering in a DNA duplex in solution. Biophys J. 1995;68:13–24. doi: 10.1016/S0006-3495(95)80181-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zgarbová M., Otyepka M., Šponer J., Lankaš F., Jurečka P. Base pair fraying in molecular dynamics simulations of DNA and RNA. J. Chem. Theory Comput. 2014;10:3177–3189. doi: 10.1021/ct500120v. [DOI] [PubMed] [Google Scholar]
- 48.Mazurek A., Johnson C., Germann M., Fishel R. Sequence context effect for hMSH2-hMSH6 mismatch-dependent activation. Proc. Natl. Acad. Sci. U.S.A. 2009;106:4177–4182. doi: 10.1073/pnas.0808572106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gueron M., Leroy J.L. Studies of base pair kinetics by NMR measurement of proton exchange. Methods Enzymol. 1995;261:383–413. doi: 10.1016/s0076-6879(95)61018-9. [DOI] [PubMed] [Google Scholar]
- 50.Moe J.G., Russu I.M. Kinetics and energetics of base-pair opening in 5′-d(CGCGAATTCGCG)-3′ and a substituted dodecamer containing G.T mismatches. Biochemistry. 1992;31:8421–8428. doi: 10.1021/bi00151a005. [DOI] [PubMed] [Google Scholar]
- 51.Pearlman D.A., Kollman P.A. Are time-averaged restraints necessary for nuclear magnetic resonance refinement? A model study for DNA. J. Mol. Biol. 1991;220:457–479. doi: 10.1016/0022-2836(91)90024-z. [DOI] [PubMed] [Google Scholar]
- 52.Torda A.E., Scheek R.M., van Gunsteren W.F. Time-averaged nuclear Overhauser effect distance restraints applied to tendamistat. J. Mol. Biol. 1990;214:223–235. doi: 10.1016/0022-2836(90)90157-H. [DOI] [PubMed] [Google Scholar]
- 53.Gorenstein D.G. 31P NMR of DNA. Methods Enzymol. 1992;211:254–286. doi: 10.1016/0076-6879(92)11016-c. [DOI] [PubMed] [Google Scholar]
- 54.Gorenstein D.G., Lai K., Shah D.O. 31P and two-dimensional 31P/1H correlated NMR spectra of Duplex d(Ap[17O]Gp[18O]Cp[16O]T) and assignment of 31P signals in d(ApGpCpT)2-actinomycin D complex. Biochemistry. 1984;23:6717–6723. doi: 10.1021/bi00321a067. [DOI] [PubMed] [Google Scholar]
- 55.Precechtelova J., Novak P., Munzarova M.L., Kaupp M., Sklenar V. Phosphorus chemical shifts in a nucleic acid backbone from combined molecular dynamics and density functional calculations. J. Am. Chem. Soc. 2010;132:17139–17148. doi: 10.1021/ja104564g. [DOI] [PubMed] [Google Scholar]
- 56.Heddi B., Foloppe N., Bouchemal N., Hantz E., Hartmann B. Quantification of DNA BI/BII backbone states in solution. Implications for DNA overall structure and recognition. J. Am. Chem. Soc. 2006;128:9170–9177. doi: 10.1021/ja061686j. [DOI] [PubMed] [Google Scholar]
- 57.Martin N.H., Allen N.W., 3rd, Brown J.D., Kmiec D.M., Jr, Vo L. An NMR shielding model for protons above the plane of a carbonyl group. J. Mol. Graph. Model. 2003;22:127–131. doi: 10.1016/S1093-3263(03)00152-9. [DOI] [PubMed] [Google Scholar]
- 58.Martin N.H., Loveless D.M., Main K.L., Pyles A.K. Computation of through-space NMR shielding effects by functional groups common to peptides. J. Mol. Graph. Model. 2006;25:1–9. doi: 10.1016/j.jmgm.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 59.Nonin S., Leroy J.L., Gueron M. Terminal base pairs of oligodeoxynucleotides: imino proton exchange and fraying. Biochemistry. 1995;34:10652–10659. doi: 10.1021/bi00033a041. [DOI] [PubMed] [Google Scholar]
- 60.Granovsky A.E., Clark M.C., McElheny D., Heil G., Hong J., Liu X., Kim Y., Joachimiak G., Joachimiak A., Koide S., et al. Raf kinase inhibitory protein function is regulated via a flexible pocket and novel phosphorylation-dependent mechanism. Mol. Cell. Biol. 2009;29:1306–1320. doi: 10.1128/MCB.01271-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smrcka A.V., Kichik N., Tarrago T., Burroughs M., Park M.S., Itoga N.K., Stern H.A., Willardson B.M., Giralt E. NMR analysis of G-protein betagamma subunit complexes reveals a dynamic G(alpha)-Gbetagamma subunit interface and multiple protein recognition modes. Proc. Natl. Acad. Sci. U.S.A. 2010;107:639–644. doi: 10.1073/pnas.0909503107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rossetti G., Dans P.D., Gomez-Pinto I., Ivani I., Gonzalez C., Orozco M. The structural impact of DNA mismatches. Nucleic Acids Res. 2015;43:4309–4321. doi: 10.1093/nar/gkv254. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.