Abstract
Cyanobacterial regulation of gene expression must contend with a genome organization that lacks apparent functional context, as the majority of cellular processes and metabolic pathways are encoded by genes found at disparate locations across the genome and relatively few transcription factors exist. In this study, global transcript abundance data from the model cyanobacterium Synechococcus sp. PCC 7002 grown under 42 different conditions was analyzed using Context-Likelihood of Relatedness (CLR). The resulting network, organized into 11 modules, provided insight into transcriptional network topology as well as grouping genes by function and linking their response to specific environmental variables. When used in conjunction with genome sequences, the network allowed identification and expansion of novel potential targets of both DNA binding proteins and sRNA regulators. These results offer a new perspective into the multi-level regulation that governs cellular adaptations of the fast-growing physiologically robust cyanobacterium Synechococcus sp. PCC 7002 to changing environmental variables. It also provides a methodological high-throughput approach to studying multi-scale regulatory mechanisms that operate in cyanobacteria. Finally, it provides valuable context for integrating systems-level data to enhance gene grouping based on annotated function, especially in organisms where traditional context analyses cannot be implemented due to lack of operon-based functional organization.
INTRODUCTION
The cyanobacterial research community was one of the first scientific groups to enter the genomic era, when, in 1996, Synechocystis sp. PCC 6803 became only the third organism to be completely sequenced (1). Many subsequent genome sequences have been derived from other cyanobacterial species and have been assembled and deposited in the NCBI sequence database (http://www.ncbi.nlm.nih.gov/genomes). However, despite the early and continued activity across this extremely diverse bacterial phylum, the application of systems biology approaches in cyanobacterial research still lags behind other Eubacteria. This is likely due, at least in part, to the unique genomic and genetic properties of cyanobacteria, wherein functionally related genes are less frequently clustered in the genome (2) and the overall number of operons is significantly reduced (3). These properties render genome context analysis, which infers functional relatedness based on gene co-localization, relatively ineffective.
The use of regulon analysis to identify functionally related genes based on their regulation by a common transcription factor is also difficult due to the low number of transcription factors in cyanobacteria (4) compared to other bacteria. This may lead to a regulatory system that is comprised of larger regulons and increases the likelihood that different pathways are controlled by a single regulator responding to many different environmental stimuli (5). The lack of transcription factors also suggests that cyanobacteria may utilize alternative controls for gene expression including post-transcriptional mechanisms. Indeed, several studies have revealed a large number of small RNA (sRNA) species in cyanobacteria (6–10) as well as candidate targets of their regulation, predicted based on homologous base-pairing (11). Notably, the targets of post-transcriptional regulation include heat shock and light-induced proteins, which typically are parts of globally controlled pathways in other species (12–14).
The role of cyanobacteria as primary producers, a keystone function in many aquatic and terrestrial ecosystems, makes understanding the linkages between genomic content and regulatory strategies in these species extremely important. Global transcriptome analysis is an effective high-throughput means to identify coordinated gene expression and to infer functional relationships. RNA sequencing (RNA-seq) is rapidly becoming the standard for global transcriptome analysis because of its unparalleled resolution and its ability for de novo transcript identification. The increasing availability of transcriptional profiles collected under many different environmental conditions has the potential to yield new insights into the coordination of gene expression compared to what was previously concluded in smaller targeted studies. Gene association models based on co-expression are independent of genomic or operon structure and can be used to predict gene function (15), identify regulatory targets (16) or examine the importance and centrality of conserved genes across species (17).
To apply this gene organization methodology to cyanobacteria, we utilized the wealth of transcriptomic data available for Synechococcus sp. PCC 7002 (hereafter, Synechococcus 7002), a model unicellular marine cyanobacterium that was first isolated from mud flats associated with a fish farm in Puerto Rico (18). As an inhabitant of marine and freshwater interfacial systems (e.g. estuaries and tidal zones) (19), the ecophysiological success of this photoautotroph depends on its ability to adapt rapidly to drastic shifts in temperature, salinity, light and nutrient availability. Like other cyanobacteria, Synechococcus 7002 modulates its cellular responses using a genomic structure that places related genes at disparate sites in the genome and with a reduced number of transcription factors that govern the expression of specific cellular pathways. The available RNA-seq data sets used in this study, consisting of 42 distinct physiological conditions (20–23), were examined using the Context Likelihood of Relatedness program (CLR) (24) to develop a transcript-based gene association network of Synechococcus 7002. The resulting network was then grouped into modules of co-expressed genes. The outcomes of this study aptly illustrate the broad applicability of a CLR-based approach for integration of expression data to enhance correlation of genes, an important step toward the general principles underlying genomic organization and regulatory landscape of prokaryotic photoautotrophs.
MATERIALS AND METHODS
Strains, culture conditions and analysis of raw data
Expression data for Synechococcus 7002, representing 42 discrete growth conditions, were either generated from continuous culture experiments carried out for this study or sourced from previously reported experiments that examined growth of the organism under nutrient limitation, varying irradiance levels, extremes of cell density, temperature and salinity, as well as co-cultivation with a heterotrophic bacterium Shewanella W3-18 (20–23). Data were derived either from studies carried out at the Pacific Northwest National Laboratory (PNNL) (20) or from a series of studies carried out by Ludwig and Bryant at the Pennsylvania State University and deposited into the Gene Expression Omnibus (GEO) database (21–23). Experimental conditions that have not yet been reported but are included in this study include carbon-, light- and nitrogen-limited growth of Synechococcus 7002, growth of the organism under a range of irradiance levels, as well as growth of a high-light/high-O2 adapted strain of Synechococcus 7002 under variable oxygen levels. For these conditions that have not yet been reported the continuous cultivation of Synechococcus 7002, operated in chemostat or turbidostat modes, was carried out with A+ medium (25) in a photobioreactor operated at 30°C with a dilution rate of 0.1 h–1 as described previously (26). In carbon-, nitrogen- or light-limited chemostats, steady-state growth was supported at 7.7 mM NaHCO3, 0.9 mM NH4Cl or 140 μE m–2 s–1, respectively. To examine response to a range of irradiance levels Synechococcus 7002 was grown in turbidostat mode under six irradiance levels ranging from 33–760 μmol photons m–2 s–1. Finally, a high-light and high-O2 adapted strain of Synechococcus 7002 was grown under either 7.1% or 16.5% dissolved O2. Supplementary Table S1 summarizes the growth conditions used in this study. RNA from conditions 1–18 (Supplementary Table S1) was extracted and processed as described in Beliaev et al. 2014 (20) while RNA from conditions 19–42 was processed as described in Ludwig and Bryant (21). Sequencing was performed using SOLiD 5500XL protocol (20) (conditions 1–18) or with the SOLiD™ 3 or 3Plus protocol (21) (conditions 19–42). All raw RNA-seq files were aligned to the complete genome of Synechococcus sp. PCC 7002 (NCBI Accession # CP000951) and gene expression levels for all conditions were determined as reads per kilobase per million reads (RPKM), normalized to the upper quartile of expressed genes using the Rockhopper program as previously described (27).
Generation of the full co-expression network, module detection and functional enrichment
Using expression values for each gene in each condition (excluding uncharacterized non-coding transcripts such as sRNAs), a co-expression network was generated with the CLR method (24,28). To reduce the detection of spurious links between co-expressed genes, a resampling approach was used wherein CLR was run 500 times, each time with 38/42 randomly selected conditions and only gene (node) pairs with an edge Z-score of 4.5 (4.5 standard deviations above the mean of the mutual information score of a given gene pair with all other genes) in at least 375 (75%) of the runs were assigned edges between them in the final consensus network reported here. A Z-score of 4.5 was chosen based on the lack of structure in networks with lower Z-scores (Supplementary Figure S1). To determine how many runs of CLR were required for robust analysis, two separate resampling test analyses were carried out, each with 500 runs of CLR, with each analysis having a different random seed to insure that each resampling analysis contained a unique set of randomly sampled conditions for each run. We observed that the similarity of the final networks converged at ∼97% after 500 runs and that additional runs did little to increase convergence. The node-edge structure is provided as a .sif file (Supplementary Data File), that can be viewed in Cytoscape (http://www.cytoscape.org/, (29)).
To identify robust modules of co-expressed genes (30), we used a similar resampling approach to the one used for generating the consensus network. Using the same 500 networks inferred for the consensus network, we detected modules in each of them with the fastgreedy algorithm (as implemented in the R igraph package). If two genes were included in the same module in 75% of the networks, that gene pair was retained. The persistent gene pairs were then included in a ‘module network’, in which an edge between two genes meant that they were consistently found in the same module. Although this network representation had a clear modular structure (i.e. grouping of genes), there were still a subset of genes where their module membership was still ambiguous. To resolve such ambiguities, we used the fastgreedy algorithm again to assign genes to their final modules. We call a grouping of genes in this network a consensus module, and each module was required to have a minimum of 12 genes to be included in our final analysis. The clustering coefficient of each module was then calculated by averaging the local clustering coefficient of each node in a given module. A P-value was also assigned to each module by determining how often (among 1000 iterations) the clustering co-efficient of a random set of n genes (where n is equal to the number of genes in the module) exceeded the clustering co-efficient of the module as reported in the full network. Visualization of the network was carried out using Cytoscape (29). Functional enrichment was carried out on genes within modules using Fisher's exact test along with a curated Synechococcus 7002 genome annotation file.
Identification of transcription factor binding motifs within modules
Regulatory motifs were predicted in the intergenic regions of genes in the same module using the Gibbs recursive sampler (31). Intergenic regions of at least 20 bp in length and directly upstream of the genes in the co-expression modules were extracted and subjected to multiple runs of the Gibbs recursive sampler, using all combinations of the following parameters: prediction of one or two motif models, the model width specified as 14 or 16 bases (allowed to fragment to 22 or 24 bases, respectively), and the model defined as palindromic or non-palindromic. A maximum of three sites per intergenic was allowed for all Gibbs runs. A position-specific background model (32) was employed, sampling was performed with 20 random seeds and 1000 iterations were used with a plateau period of 200. From these parameter combinations, the most probable motifs were identified as those with positive maximum a posteriori probability (MAP) value; a positive MAP value indicates that the motif alignment is more likely than the unaligned random background.
Expanding regulons of DNA binding transcription factors and non-coding RNAs
Well-characterized DNA binding transcription factors were analyzed further through generation of their 2nd order network neighborhood. This network neighborhood contains all genes that have an edge with the regulator as well as any genes that have an edge with a gene that has an edge with the regulator. For this more specific analysis a new consensus network was generated with a Z-score cutoff of 3.0 rather than 4.5. Reducing the Z-score in this way allowed for a larger pool of potential targets of regulators while still maintaining a strict Z-score cutoff. Well-characterized regulators with a significant number of their known targets, as determined by RegPrecise (33), within their 2nd order neighborhoods were examined further. First, a motif matrix for the binding site of each regulator was generated through analysis of the promoters of known targets, again from RegPrecise, using the MEME program (34). Except in the case of NrtR2 (SYNPCC7002_A2383) known binding sites were only drawn from Synechococcus 7002 genes. For NrtR2 there were only three known binding sites so the consensus binding site was made up of these three sites plus two additional sites from Cyanothece sp. PCC 7425. Promoter-containing regions (inclusive of 250 bp upstream of the ATG start codon) for all genes within the 2nd order neighborhood were then scanned using the Find Individual Motif Occurrences program to find matches with the motif matrix generated by MEME. Genes that (i) were within the 2nd order neighborhood of the regulator, (ii) contained the motif for the regulator under analysis in their promoter with a P-value of < 0.0001 and (iii) had not previously been named a target of the regulator by the RegPrecise database were considered putative new targets of the regulator. A similar analysis was carried out with non-coding transcripts (sRNAs) identified in the RNA-seq data set. The only difference was that a Z-score of 2.0 was used and instead of using motif analysis, the TargetRNA2 (35) program was used to generate lists of putative targets of sRNAs. These lists of putative targets were then cross-referenced against the list of mRNAs having edges with the sRNA under analysis to identify targets of high possibility.
RESULTS
Global analysis of the consensus Co-expression network
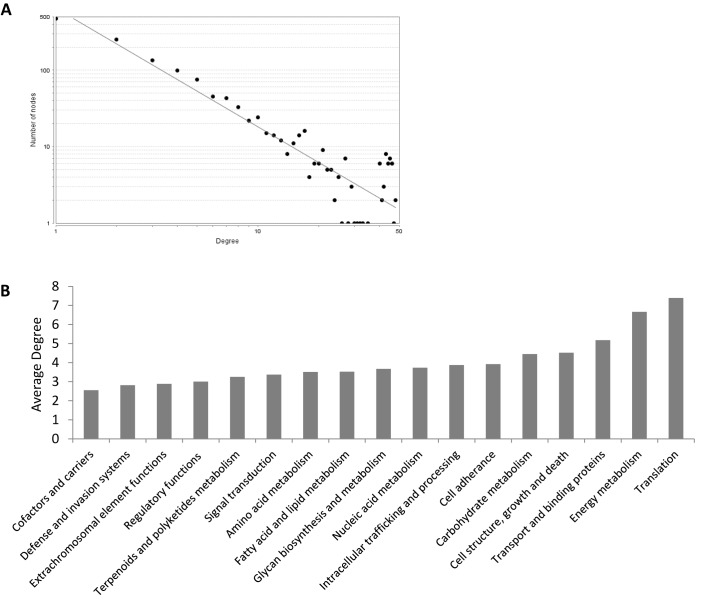
The 42 RNA-seq data sets used in this study were either generated de novo or collected from previous studies examining acclimation of Synechococcus 7002 to a broad range of environmental variables (see Supplementary Table S1; (20–23)). A co-expression network was inferred using CLR methodology in conjunction with a resampling approach, which significantly increased the robustness of the inferences. The final consensus network consisted of 1386 genes (nodes) with 3916 connections (edges) between them corresponding to ∼43% of the Synechococcus 7002 genome (Supplementary Table S2, Supplementary Data File). The consensus network was used to calculate the degree of each node (i.e. the number of edges a node has with other nodes), which can be used as a proxy for the centrality of a gene in a given pathway as well as its essentiality to the overall survival and fitness of the organism (15,36,37). The degree distribution of the network fit a power law (correlation = 0.98), a common feature of scale-free biological networks (Figure 1A) (38). Genes with the highest degree were involved in energy metabolism, metabolite transport and translation (Figure 1B; Supplementary Table S2). The latter were dominated by genes encoding ribosomal proteins associated with both the large and the small ribosome subunits; many of these had high degree values driven by their connections with other genes encoding ribosomal proteins. Genes involved in energy metabolism with high degree values included atpACHG and other genes encoding subunits of the F1/F0 ATP-ase complex. Other energy metabolism genes encoding the structural components of the photosystem II (psb) and the phycobilisome antenna complex (cpc) also had a large number of connections in the network, as did three genes encoding enzymes of the Carbon–Benson cycle (pgk, SYNPCC7002_A1585; glpX, SYNPCC7002_A1301; and tktA, SYNPCC7002_A1022). Transport of molecules into the cell is obviously crucial for growth and, as a result, genes with high degree values included iron uptake and carbon fixation genes, as well as the MRP-family of Na+/H+ antiporters (SYNPCC7002_A2373-2380), which establish a sodium gradient necessary for the transport of bicarbonate into the cyanobacterial cell (39,40). In addition to genes with annotated functions, network topology analysis also identified a number of completely uncharacterized genes with high degree values (Supplementary Table S2) likely pointing to their essentiality for growth and metabolism of Synechococcus 7002 as well as other cyanobacteria.
Figure 1.
Network Characteristics. (A) The node degree distribution is shown with degree on the x-axis and number of nodes on the y-axis. The power line fit is also shown (correlation = 0.98, grey line). Fitting a power law is a common feature of biological networks (38). (B) The average degree (number of edges a gene has with other genes) of genes comprising each of the main roles used in the annotation is shown on the y-axis with the name of the function on the x-axis. Genes with zero edges were removed before calculating averages.
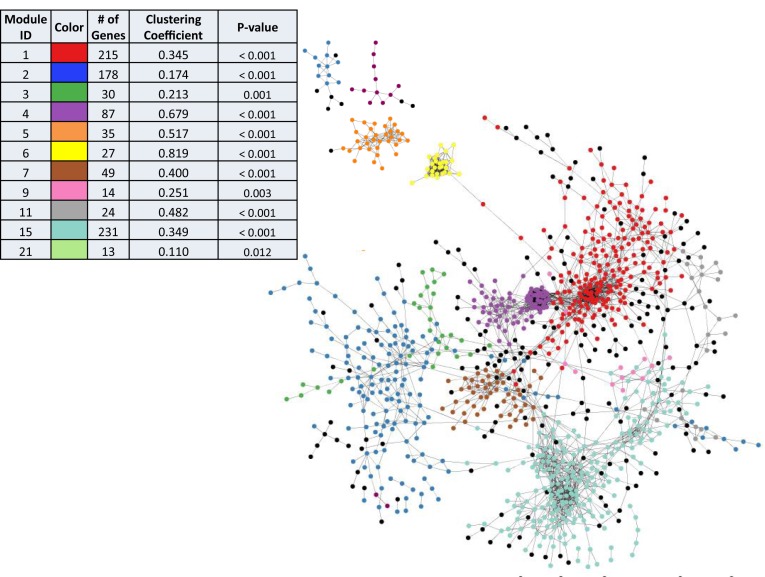
The obtained node-edge network can be organized further by grouping genes into modules, structured such that connections within a given module (intra-module edges) are dense, while connections between two distinct modules (inter-module edges) are sparse. As a result of this clustering, we were able to organize 903 genes into 11 distinct modules that contained between 13 and 231 genes each, representing ∼28% of the Synechococcus 7002 genome (Figure 2; Supplementary Table S2). Because the edges are highly concentrated within modules, this means that, on average, the genes within a given module have a higher concordance of co-expression across conditions compared to genes in another module. Module density, which was determined quantitatively by calculating the clustering coefficient of each module (Figure 2), was generally inversely related to the module size. The high clustering coefficient displayed by Modules 4 (n = 87 genes), 5 (n = 35 genes) and 6 (n = 27 genes), indicates that many genes within this module have high co-expression values across growth conditions. In contrast, Modules 1 (n = 215 genes), 2 (n = 178 genes) and 15 (n = 231 genes), which are much larger, contained less intra-module edges, indicating lower level of co-expression between genes within these modules. All modules were also statistically significant with a P-value < 0.05.
Figure 2.
Global Expression Map of Synechococcus 7002. Lines (edges) between genes (nodes), represented as circles, indicate co-expression between the two genes connected. Nodes are colored according to the module to which they were assigned to using the fastgreedy algorithm. A total of 903 genes (28% of genome) could be assigned to modules. Black nodes have a statistically significant edge with a node in the network but do not belong to a specific module. Other nodes that were not connected to the main network are not shown. Table in upper left shows module color scheme, size of modules, clustering coefficients and P-values.
Inferring regulatory patterns through functional enrichment of network modules
To determine the association of genes with metabolic and regulatory processes in each module, we carried out functional enrichment analysis using a highly comprehensive Synechococcus 7002 genome annotation, in which genes were classified based on a main role, subrole and subsystem category assignment. With the exception of Module 4, which was comprised mainly of non-coding tRNA genes, all modules were enriched for multiple functions (Table 1; Supplementary Table S3), indicating the presence of functional and transcriptomic relatedness within each grouping. Interestingly, modules displaying the highest density also showed some of the most significantly enriched functions. All enrichments in reported in Table 1 had a P-value of < 0.05
Table 1. Functional Enrichment of Modules.
| Module ID | Subrole | Percentage in Module | Ratio* | Subsystem | Percentage in Module | Ratio* |
|---|---|---|---|---|---|---|
| 1 | Purine metabolism | 3.72 | 4.24 | Pyruvate dehydrogenase | 1.86 | 14.86 |
| Ribosomal proteins: synthesis and modification | 18.14 | 8.16 | Ribosome large subunit | 10.70 | 10.68 | |
| Ribosome small subunit | 7.44 | 11.32 | ||||
| RNA polymerase RpoABCEZ | 1.86 | 11.88 | ||||
| 2 | Polysaccharide and lipopolysaccharide metabolism | 6.74 | 5.98 | Lipopolysaccharide biosynthesis | 1.12 | 11.96 |
| Polysaccharide biosynthesis | 8.43 | 7.92 | ||||
| 5 | Cations and iron carrying compounds | 31.43 | 12.24 | Iron (III) transport system AfuABC | 5.71 | 60.84 |
| Biosynthesis of siderophore group | 14.29 | 57.04 | Iron uptake FhuBCD2 | 5.71 | 60.84 | |
| Iron uptake FhuBCD3 | 11.43 | 73.01 | ||||
| 6 | CO2 fixation | 18.52 | 53.77 | |||
| NAD(P)H:quinone oxidoreductase | 11.11 | 14.79 | ||||
| NADH dehydrogenase I | 3.70 | 118.30 | ||||
| Cations and iron carrying compounds | 33.33 | 12.98 | Sodium:hydrogen antiport Mrp | 29.63 | 118.30 | |
| 7 | Porphyrin and chlorophyll metabolism | 8.16 | 5.79 | Photosystem I main subunits | 8.16 | 43.46 |
| Photosynthesis | 30.61 | 16.57 | Photosystem I other common subunits | 6.12 | 48.89 | |
| Photosystem II main subunits | 6.12 | 21.73 | ||||
| Photosystem II other common subunits | 4.08 | 10.03 | ||||
| Phycobilisome | 2.04 | 32.59 | ||||
| Photosynthesis – antenna proteins | 14.29 | 19.01 | Phycocyanin biosynthesis | 8.16 | 37.25 | |
| 15 | Toxin-antitoxin systems | 6.93 | 2.35 | Toxin-antitoxin system 16 | 0.87 | 13.83 |
| Restriction-modification systems | 3.90 | 5.41 | Toxin-antitoxin system 18 | 0.87 | 13.83 | |
| Toxin-antitoxin system 8 | 0.87 | 13.83 | ||||
| Type II restriction-modification systems | 0.87 | 9.22 | ||||
| Type III restriction-modification system pAQ5 | 0.87 | 13.83 | ||||
| Type IV restriction system LlaI | 0.87 | 13.83 | ||||
*Ratio refers to the percentage of genes in a given category in the module/the percentage of genes in the same category in the genome as a whole.
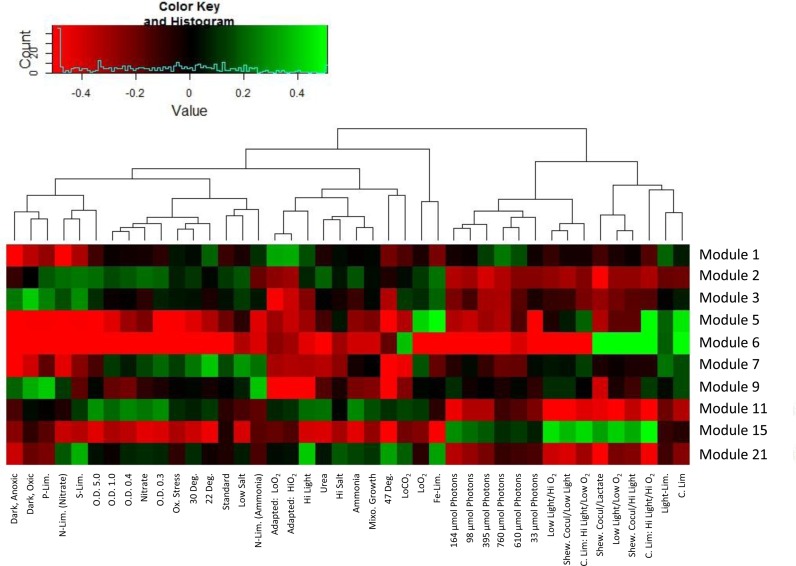
The vast majority of growth-related, biosynthetic and energy metabolism genes were found in Module 1 (215 genes). The latter was highly enriched for translation processes containing 23 out of 31 genes encoding subunits of the large ribosomal subunit and 16 out of 21 genes encoding subunits of the small ribosomal subunit. With a single exception, no other ribosomal protein genes were assigned to any other module. Nucleotide metabolism, pyruvate metabolism and transcription processes were also enriched within Module 1, which contained eight purine metabolism genes, including purAQLHTE, all of the pyruvate dehydrogenase complex genes, pdhABCD as well as a pyruvate kinase gene pyk and 4/5 RNA polymerase genes rpoAC1C2B. The relative transcript abundances of genes in Module 1 were unchanged across the majority of experimental conditions with the exception of those treatments associated with Synechococcus 7002 after it has acclimated to high light and high oxygen (Figure 3). An appreciable decrease in transcript abundances across Module 1 was observed when Synechococcus 7002 was cultivated in the absence of key nutrients such as light, N, P and S; in contrast, broad upregulation of growth-related genes was seen in cultures acclimated to high irradiance conditions.
Figure 3.
Response of Modules to Growth Conditions. Conditions are shown at the bottom of the heat map and the modules on the right. Dendogram depicts clustering of conditions. Module response was determined by taking the log2 value of the median of the expression level of all genes in the module under each condition normalized to the mean of the medians across all conditions.
The other two large groupings were represented by Modules 2 (178 genes) and 15 (231 genes); notably, nearly half of the genes in these modules encoded proteins of unknown function. In Module 2, the enriched categories included genes encoding glycan and polysaccharides biosynthesis pathways as well as cell division and defense systems (Table 1; Supplementary Table S3). The average transcript levels did not display significant responses to any of the experimental conditions, although a general decrease in expression was seen during co-cultivation with Shewanella and lactate and a general increase during changes in cell density and nitrogen levels. Genes in Module 15 showed a significant increase in expression during co-cultivation of Synechococcus 7002 with Shewanella (Figure 3). Strong enrichment for genes involved in defense against invasion by phage and restriction modification systems was observed in this module (Table 1; Supplementary Table S3). In addition, three CRISPR genes, cas2, cas6 and cas10, as well as several toxin-antitoxin gene pairs, were also present in Module 15, and all of them encoded functions involved in microbial competition and ‘warfare’.
Among smaller groupings, Module 5 (35 genes) displayed one of the highest clustering coefficients and was highly enriched for genes involved in iron uptake (31% of genes in this module were involved in uptake of iron and other compounds) and siderophore biosynthesis (14%) (Table 1; Supplementary Table S3). Consistent with their putative function, genes in Module 5 were iron-responsive as relative transcript abundances in this module were 31- to 35-fold higher under iron-limiting conditions when compared to similar experiments examining P, S or N limitations (Figure 3). Furthermore, genes in Module 5 displayed significantly elevated transcript abundances in steady state chemostat cultures grown under light-, carbon- or oxygen-limiting conditions.
Module 6 (27 genes) also showed tight clustering and contained a functionally diverse group of genes, with most encoding components of the carbon capture and fixation machinery in Synechococcus 7002. Among those, the largest enriched categories were comprised of sodium/hydrogen antiporters (29%) and CO2 fixation (18%) genes, which encoded putative SbtAB and BicA bicarbonate symporters as well MRP and NapA antiporters. While these two Na+/H+ antiporters can function to confer salt tolerance, their role here is most likely linked to developing a charge gradient necessary for a sodium-dependent bicarbonate transport (39,40). Other genes in this module included the NAD(P)H:quinone oxidoreductase subunits that facilitate carbon dioxide uptake (ndhD3 and ndhJ), the CO2 hydration protein (cupA) and the CO2 fixation regulator (ccmR). Module 6 also contained seven genes of unknown function and their assignment to Module 6 suggests that they may also be involved in CO2 acquisition (Supplementary Table S2). Transcripts for genes in this module increased their expression up to 30-fold under carbon, light and O2-limiting conditions (Figure 3) when bicarbonate concentrations were limiting growth. Aside from these changes, however, transcript levels of genes in Module 6 were largely unchanged across the rest of the experimental conditions.
Finally, Module 7 (49 genes) contained another essential group of genes, those involved in photosynthetic functions, displaying enrichment for genes encoding photosystems (31%) and antenna proteins (14%). Specifically, these included genes encoding subunits of photosystem I (psaABDFIKL), photosystem II (psbCOU, psbD1, psbD2 and psb27) reaction center components, as well as phycobilisome antenna genes (cpcABCG). The transcript abundances in Module 7 were relatively unchanged across most of the tested conditions, with the exception of nitrogen-limitation, heat shock (47°C) and dark conditions, when the abundances decreased and lower temperatures (22°C) where expression increased (Figure 3).
Network topology analysis reveals metabolic coordination
Even though modules were formed by minimizing edges between genes in different modules, the remaining intra-module connections can indicate specific functional interactions between cellular processes or pathways. For example, Modules 1 and 4 were both involved in translation processes in Synechococcus 7002, being enriched for ribosomal protein and tRNA genes, respectively. Because of the close functional association between tRNAs and ribosomal structures, there were a large number of edges connecting genes in Module 4 to those in Module 1 (Supplementary Figure S2A). The large number of connections between Modules 4 and 1 were, in fact, almost exclusively limited to four genes in Module 1 that had connections with genes in Module 4: an ATPase gene atpC, two ribosomal protein genes, rplO and rplF and an RNA polymerase gene, rpoC2. The ribosomal, ATPase and translation genes that link Modules 1 and 4 are all intimately tied to growth, as are the tRNA genes of Module 4. Nodes with edges that link two different modules are examples of bottlenecks, and such genes are believed to be important in bacterial growth and replication (15).
Other bottlenecks present in the Synechococcus 7002 network included a single link between Module 6 and Module 1 that passes through two genes, the NAD(P)H gene ndhB and the RuBisCo chaperone, rbcX (Supplementary Figure S2B). Although both of these genes are assigned to Module 1, Module 6 is strongly enriched for carbon acquisition processes. As the RuBisCo complex is associated with both carbon metabolism and growth, it is expected that it would form a link between the growth and replication genes in Module 1 and the carbon acquisition genes in Module 6. Indeed, the rbcX gene has an edge with the ndhB, which in turn has an edge with ndhF3 in Module 6. On the other side, rbcX has an edge with the carboxysome structural protein gene ccmM in Module 1. The observation that ndhB is also found linking Module 6 and Module 1 is of interest as previous studies have shown that, in addition to its role in respiration, NdhB is intimately involved in CO2 uptake in cyanobacterial species (41–43). Finally, the dependence of cyanobacterial growth on light acquisition was highlighted by two bottlenecks linking Module 1 and Module 7 (Supplementary Figure S2C) – one of the bottlenecks passed through a photosystem II gene (psbB) while the other passed through a gene for the alpha subunit of allophycocyanin (apcA). The psbB gene in Module 1 was connected with three Module 7 genes, psbC, psbD1 and psbD2, all encoding proteins of photosystem II. Similarly, the apcA gene (also of Module 1) has eleven edges with Module 7 genes. The structure of this network clearly shows the central importance of bottleneck genes, which connect distinct, but functionally related, processes in Synechococcus 7002.
Delineating transcriptional subnetworks through condition-specific responses
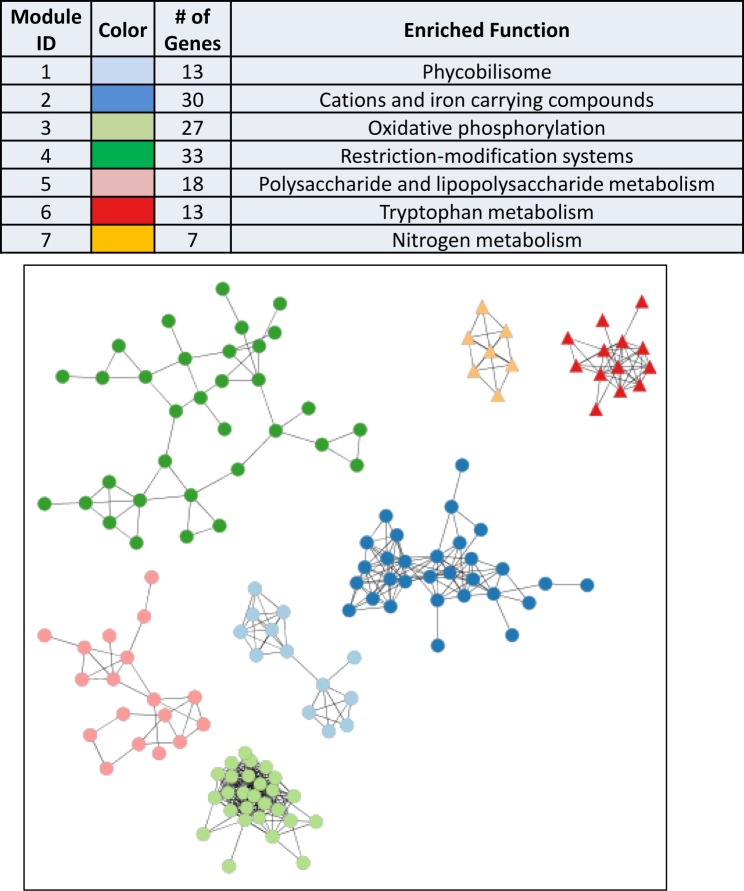
The highly compartmentalized co-expression network was also exploited as an organizational and data mining tool to examine the response of Synechococcus 7002 to specific conditions. When examining the entire data set (Supplementary Table S1), a large number of differentially expressed genes were identified when comparing carbon-limited (CL) to nitrogen-limited (NL) chemostat cultures, with a total of 437 genes displaying >2-fold change in transcript levels (q-value < 0.05). A subnetwork reconstruction, using only these genes and the structure of the full network identified, was then carried out. Modularization of this subnetwork identified seven modules, which separated cleanly based on the relative expression levels of the genes comprising them (Figure 4). Genes displaying decreased transcript abundances under NL were organized into five discrete clusters, which were enriched in iron regulation and oxidative phosphorylation, as well as defense and invasion functions (Supplementary Table S4). Genes showing increased expression under NL were grouped into the final two modules and were enriched in formate utilization and nitrate uptake functions (Supplementary Table S4).
Figure 4.
Network Analysis of Carbon and Nitrogen Regulated Genes. Identified subnetworks of genes change in expression by at least 2-fold when comparing carbon-limited (CL) conditions to nitrogen-limited (NL) conditions. Colors indicate genes categorized into new modules based on the edge-node structure of the subnetwork. Table indicates color and number of genes in each module, a representative enriched function is also shown. Genes that exhibited decreased or increased expression under NL conditions compared to CL conditions are indicated by circles and triangles, respectively.
It is important to note that modularization of Synechococcus 7002 genes showing differential expression under NL versus CL conditions was able to group only 32% of these genes into modules (141/437). Despite this, a greater number of functional roles were enriched with modularization compared to when the entire data set was grouped into only two large groups comprised of genes showing increased (169 genes) or decreased (268 genes) expression (Supplementary Table S5). In addition, some functional roles were identified only after modularization of regulated genes (oxidative phosphorylation, nitrate uptake, defense systems). These observations suggest that modularization as a tool to better analyze specific conditions is best used in conjunction with more traditional categorization of genes (such as by increased or decreased expression) rather than as a strict replacement. Moreover, because edges within the full network are structured to be condition independent, this approach could be applied to the analysis of environmental conditions that were not used in the construction of the full network presented here, demonstrating the extensive utility of gene co-expression networks built from RNA-seq data.
Modularization and network neighborhood structure analyses enhance regulon predictions
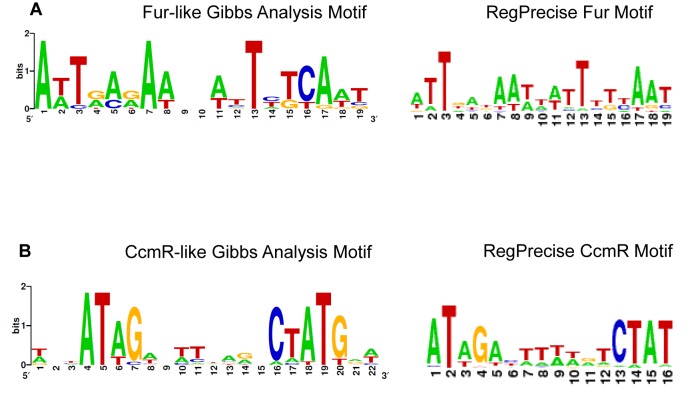
Grouping of genes into modules is carried out based on the co-expression similarity. Because of this, genes in the same module are likely to be enriched for genes co-regulated by the same DNA-binding transcription factors. To examine this further, the intergenic regions within each module were examined using the Gibbs sampler (31) for common binding motifs, with significant motifs being found in Modules 5 and 6 (Figure 5). As expected, the intergenic regions upstream of genes in Module 5 were strongly enriched for putative ferric uptake regulator (Fur) binding sites, whose activity is regulated by Fe2+ (44,45). A total of 12 Fur binding sites were detected in Module 5 (Table 2, Figure 5A), and of these, 5 were previously predicted in RegPrecise, including the putative iron transporters (futC, fecB), the iron responsive regulator (pchR) and the siderophore receptor (schT). In addition to these five, we detected putative Fur binding sites in the intergenic regions of a bacterioferritin-like gene and two genes of unknown function (SYNPCC7002_A1857 and SYNPCC7002_A2659), which have not been previously associated with the Fur regulon.
Figure 5.
Binding Site Motifs Identified in Promoter Regions. Examination of the promoter regions of genes in modules revealed a Fur-like binding site in (A) Module 5 and a CcmR-like binding site in (B) Module 6. Nucleotide numbering is shown in the x-axis and bit scores on the y-axis. Height of nucleotides indicates enrichment at that position. The motif derived through Gibbs analysis for each module is shown on the left and the corresponding motif defined in RegPrecise for either Fur or CcmR is shown on the right.
Table 2. Gibbs Analysis of Modules 5 and 6.
| Fur Binding Sites in Module 5 | ||||||
|---|---|---|---|---|---|---|
| Motif | Motif Start Site* | Motif Stop Site** | Upstream Gene*** | Downstream Gene | Probability | Motif Identified in RegPrecise+ |
| 1 | 902085 | 902103 | futC A0871; Iron transport | ybeB A0872; Translation | 0.97 | Yes |
| 2 | 1946476 | 1946494 | A1857; Transport | A1858; Unknown | 0.81 | No |
| 3 | 2613428 | 2613446 | dprA A2506; DNA uptake | futA1 A2507; Iron transporter | 0.78 | No |
| 4++ | 2792428 | 2792446 | A2659; Unknown | A2660; Multicopper oxidase | 0.97 | No |
| 5++ | 2793956 | 2793974 | bfd A2661; Iron homeostasis | bfr A2663; Iron homeostasis | 0.95 | No |
| 6 | 7367 | 7385 | G0007; Iron Homeostasis | G0008; Unknown | 0.77 | No |
| 7 | 102222 | 102240 | tonB G0090; Transport | fecB G0091; Iron homeostasis | 0.90 | Yes |
| 8 | 112330 | 112348 | G0099; AraC Regulator | G0100; Transport | 0.62 | Yes |
| 9 | 119454 | 119472 | pchR G0104; Iron homeostasis | G0105; Unknown | 0.96 | Yes |
| 10 | 157500 | 157518 | exbB G0137; Transport | schT G0138; Iron homeostasis | 0.59 | Yes |
| 11 | 157651 | 157669 | exbB G0137; Transport | schT G0138; Iron homeostasis | 0.69 | Yes |
| 12 | 157686 | 157704 | exbB G0137; Transport | schT G0138; Iron homeostasis | 0.97 | Yes |
| CcmR Binding Sites in Module 6 | ||||||
| Motif | Motif Start Site | Motif Stop Site | Upstream Gene | Downstream Gene | Probability | Motif Identified in RegPrecise |
| 1 | 178199 | 178220 | A0170; Unknown | ccmR A0171; Regulator | 0.99 | Yes |
| 2 | 178231 | 178252 | A0170; Unknown | ccmR A0171; Regulator | 0.99 | Yes |
| 3 | 502490 | 502511 | sbtA A0470; Transport | A0471; Unknown | 0.98 | Yes |
| 4 | 503129 | 503150 | sbtA A0470; Transport | A0471; Unknown | 1 | Yes |
| 5 | 503544 | 503565 | A0471; Unknown | sbtB A0472; Unknown | 0.8 | Yes |
| 6 | 503576 | 503597 | A0471; Unknown | sbtB A0472; Unknown | 0.94 | Yes |
| 7 | 503615 | 503636 | A0471; Unknown | sbtB A0472; Unknown | 0.59 | Yes |
| 8 | 2370407 | 2370428 | A2287; Unknown | A2288; Unknown | 0.98 | No |
| 9 | 2370439 | 2370460 | A2287; Unknown | A2288; Unknown | 0.99 | No |
| 10 | 2452657 | 2452678 | mltA A2370; Metabolism | bicA A2371; Transport | 1 | Yes |
| 11 | 2452707 | 2452728 | mltA A2370; Metabolism | bicA A2371; Transport | 1 | Yes |
| 12 | 11474 | 11495 | G0009; Unknown | G0010; Unknown | 0.97 | No |
| 13 | 11506 | 11527 | G0009; Unknown | G0010; Unknown | 0.85 | No |
| 14 | 14517 | 14538 | porB G0011; Transport | pacL G0012; Transport | 0.83 | Yes |
| 15 | 14549 | 14570 | porB G0011; Transport | pacL G0012; Transport | 0.77 | Yes |
*Genomic location of the motif start site is shown.
**Genomic location of the motif stop site is shown.
***Gene names are indicated along with locus tags (‘SYNPCC7002_’ has been removed for ease of viewing) and functional roles.
+Genes in bold are those with motif sites identified in this study that had not previously been associated with the regulator under analysis.
++At the time of analysis these motifs were within intergenic regions, they now overlap slightly with annotated ORFs.
Similarly, analysis of the intergenic regions in Module 6 resulted in the identification of putative motifs for the carbon concentrating mechanism regulator (CcmR; Figure 5B). A total of 15 putative CcmR binding sites were detected upstream of seven different genes (Table 2), which included ccmR itself, sbtA/sbtB bicarbonate transporter genes, as well as the bicarbonate porin porB gene (40). Of the five genes predicted to have CcmR binding sites according to RegPrecise (33), all were detected in Module 6 and we detected CcmR-like sites in the intergenic regions upstream of all five. In addition to the known members of the CcmR regulon (40), several other genes within Module 6 were predicted to contain CcmR-binding sites within their putative promoter regions. These encoded uncharacterized proteins, including the periplasmic protein SYNPCC7002_G0009 located near the porB bicarbonate transporter (SYNPCC7002_G0011).
Results from previous studies (24), as well as those described above, show that regulators and their targets are often associated with each other in a co-expression network. To expand the regulons of well-characterized transcription factors, we next used regulator topology to detect new putative targets of four well-characterized transcriptional regulators in Synechococcus 7002: ccmR (controlling carbon uptake), ntcB (controlling nitrogen assimilation), sufR (controlling iron-sulfur biogenesis) and nrtR2 (controlling genes responsible for NAD biosynthesis). We collected the 2nd order network neighborhood of these four transcriptional regulators, extracted from our CLR-derived global network and examined genes within this neighborhood to identify those that also had sequences in their upstream regions (defined as 250 bp upstream of the ATG start codon) that match the binding motif for the given regulator. To increase the chances of finding genes with sites for a given regulator, we lowered the Z-score from 4.5 to 3.0 before making the 2nd order network neighborhood of each regulator. In this way, several new putative targets were found for this subset of regulators in Synechococcus 7002.
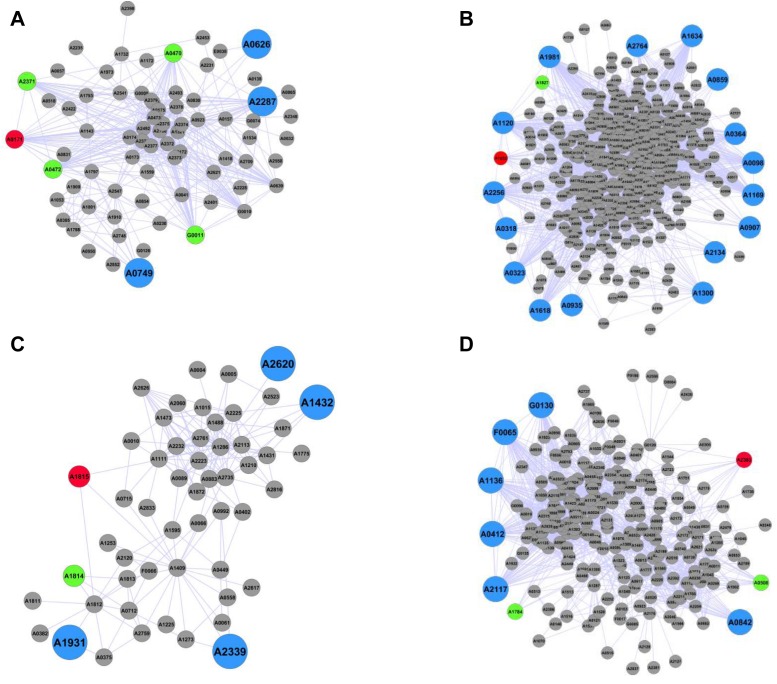
The 2nd order network neighborhood of ccmR contained 74 genes, including all of those previously identified as part of the CcmR regulon (33), as well three new target genes that contained a putative CcmR binding site in the upstream region (Figure 6A, Supplementary Table S6). One of them, SYNPCC7002_A0626, encodes an ABC-type transporter, similar to many of the other genes controlled by CcmR involved with the transport of carbon sources into the cell. We also compared the gene composition in each of the subnetworks surrounding the four regulators to the gene composition of the modules in the full network (Figure 2, Supplementary Table S2). For most of the subnetworks there was significant overlap with the genes in particular modules, suggesting that genes of some of the most enriched functions in each module are controlled by a single regulator. The subnetwork surrounding CcmR contained all genes of Module 6, a module that also contains CcmR itself and was strongly enriched for the carbon transport mechanisms that CcmR regulates. The network neighborhood of the ntcB regulator contained 453 genes, including two that were previously predicted to be in the NtcB regulon (33) and 16 additional genes that contain an NtcB-binding site in the upstream region but have not been previously reported as members of the NtcB regulon (Figure 6B, Supplementary Table S6). This subnetwork was also contained many genes in Modules 2 (comprised of genes involved in glycan metabolism) as well as Modules 11 and 21 (every gene of Module 21 was found in this subnetwork). Both of these latter two modules contain genes involved with amino acid metabolism. Similarly, neighborhoods of sufR and nrtR2 (33) contained previously predicted as well as unknown members of their respective regulons that contained putative DNA binding sites in their upstream regions (Figure 6C, D and Supplementary Table S6). While every one of these predictions may not be true targets of their regulators, this approach provides specific hypotheses for further analysis and regulon expansion for known DNA-binding proteins. Finally, while the sufR subnetwork did not show strong enrichment for any particular module, the nrtR2 subnetwork contained a large number of genes in Module 7, containing 29/49 of the genes in this module which is enriched for genes involved in photosynthesis.
Figure 6.
Second-Order Network Neighborhoods of Individual Regulators. Figures (A–D) depict the 2nd order network neighborhoods associated with transcriptional regulator genes, shown as red circles; (A) ccmR (SYNPCC7002_A0171), (B) ntcB (SYNPCC7002_A1632), (C) sufR (SYNPCC7002_A1815) and (D) nrtR2 (SYNPCC7002_A2383). Green circles denote connected genes previously recognized as belonging to the indicated regulon and containing a binding site as determined by RegPrecise, while large blue circles denote genes that are not reported as targets of the regulator in RegPrecise and contain a binding site of the given regulator as determined by Find Individual Motif Occurrences.
Topology of regulatory interactions suggests a multilayered strategy to gene regulation
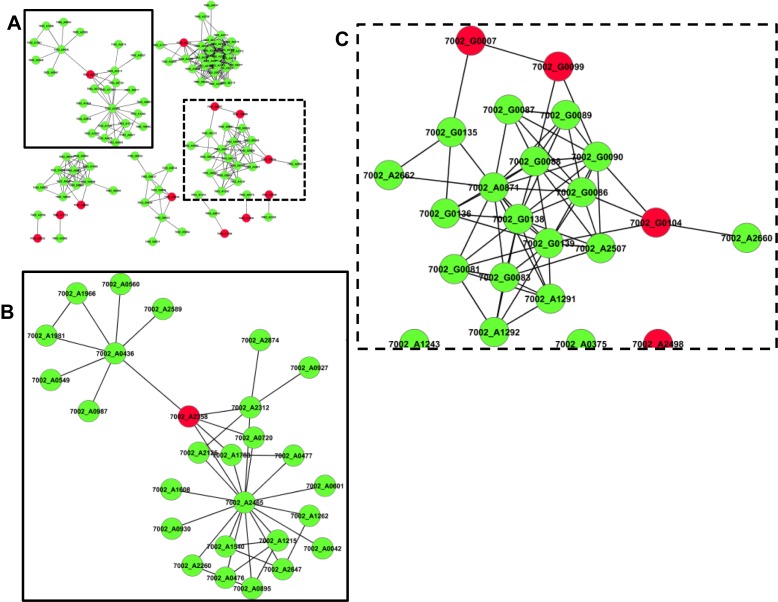
The relatively low number of transcription factors in Synechococcus 7002 may indicate different regulatory strategies that involve multi-regulator interactions with the RNA polymerase to control gene expression (4). To gain insight into the mechanistic aspects of transcriptional regulation in cyanobacteria, we examined the 2nd order network of each regulator that was connected to at least one gene, a requirement that only 26% (12/46) of the known regulators in Synechococcus 7002 satisfied. When compared to the full network, in which ∼43% of all genes were assigned edges, this number was significantly lower, indicating that regulators are less likely to have edges compared to the average gene. Regulators were also less likely to be organized into modules, with only 15% (7/46) of regulators assigned to a module. This is in contrast to the global network, where 28% of genes could be assigned to modules. Analysis of resulting regulator subnetworks revealed that a majority of regulators have edges with multiple genes (Figure 7A). Furthermore, two genes controlled by a single regulator may not have an edge between them, as is the case with an Xre-family regulator (SYNPCC7002_A2358), which controls two separate groups of genes (Figure 7B). The lack of edges between targets of the same transcription factor suggests that these genes are putatively involved in separate cellular processes or are responding to different environmental stimuli. Furthermore, edges linking regulators, such as those found between the two AraC-family regulators (SYNPCC7002_G0007 and SYNPCC7002_G0099) suggests potential co-expression mechanisms (Figure 7C) or that regulators may control additional regulators. Finally, we also identified instances in which the same gene displayed edges with multiple regulators as exemplified by tonB (SYNPCC7002_G0090), which had an edge with both SYNPCC7002_G0099 and pchR (SYNPCC7002_G0104) (Figure 7C).
Figure 7.
Network Topology of Regulators. (A) Subnetwork of transcriptional regulators (green) that contained an edge with at least one gene (red) and their 2nd order network neighborhood. (B) Inset shows a regulator that spans two distinct groups of genes. (C) Inset shows regulators controlling other regulators and non-regulators responding to the input from more than one regulator.
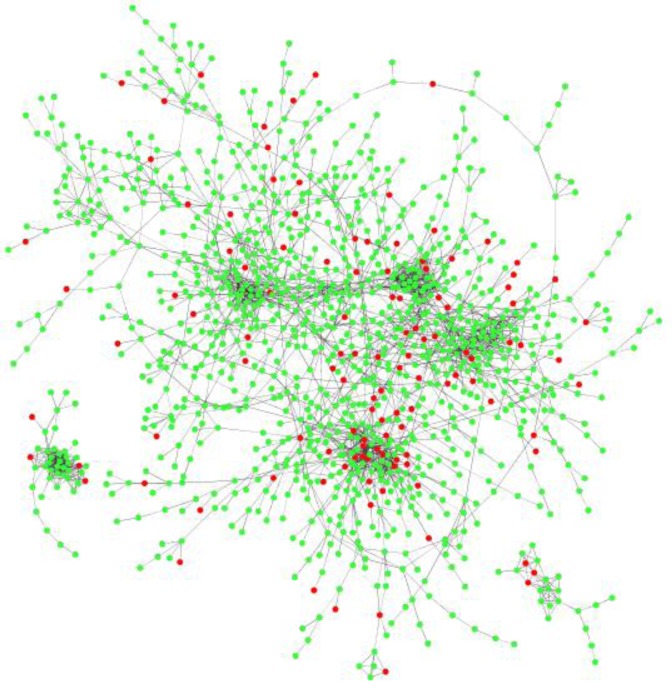
The relative lack of classical DNA-binding regulators in Synechococcus 7002 also suggests the presence of other mechanisms controlling gene expression, such as regulatory sRNAs. Using samples in which the short RNA transcripts were not depleted during isolation (conditions 1–18; Supplementary Table S1), we identified a total of 346 short RNA transcripts that were expressed either within intergenic regions or opposite protein coding regions. The identified short transcripts were used to reconstruct a new network, in which edges between sRNA and mRNA may indicate the putative targets of a given sRNA regulator (Figure 8). We next used the TargetRNA2 program (46) to generate lists of putative targets for the sRNAs in our data set. TargetRNA2 takes an input sRNA in fasta format and returns a list of possible targets based on several criteria. These include conserved regions of the sRNA as well as exposed regions of the sRNA in a predicted folded structure. Such regions are more likely to participate in binding with putative targets. Folding structure of the possible mRNA targets is also considered. Finally, the binding energy of several possible regions of interactions is calculated and the mRNA transcripts with the most energetically favorable interactions with the input sRNA are listed as possible targets. For a given sRNA in our Synechococcus 7002 data set, this list of putative targets was then cross-referenced against the list of mRNAs having edges with the sRNA. Several sRNAs that had edges with mRNAs in our network were also predicted by TargetRNA2 to be putative targets of the sRNA. sRNA_303, a 111-nucleotide transcript expressed from at 1866513–1866623, had edges with 189 mRNA genes, and TargetRNA2 provided 61 possible targets for this sRNA. The intersection of these two lists identified four high-quality targets for sRNA_303: the tRNA modifier, mnmE (SYNPCC7002_A1170), DNA polymerase 1 (polA: SYNPCC7002_A1280), SYNPCC7002_A1877, a bacteriophage related gene and SYNPCC7002_A2158, an endonuclease toxin. sRNA_332, a 160-nucleotide transcript at 2008442–2008601, had edges with 205 mRNA genes and was linked to 23 potential targets and the intersection of these two lists identified two high-quality targets of sRNA_332, SYNPCC7002_A0706, a monophosphatase family protein and SYNPCC7002_A0721, a hypothetical protein. This approach highlights how regulator-target pairings can extend to post-transcriptional mechanisms, and indeed, other studies have also successfully linked sRNAs and their targets using gene co-expression networks (47,48).
Figure 8.
Small RNAs of Synechococcus 7002. A network of sRNAs and mRNAs is shown. Red nodes are sRNAs and green nodes are mRNAs. The network shown was built from a higher Z-value (3.5) then that used for mRNA-sRNA target prediction.
DISCUSSION
Compared to some well-studied heterotrophic microorganisms like Escherichia coli or Bacillus subtilis, cyanobacteria are still relatively understudied. Furthermore, because cyanobacteria are photoautotrophs and respond to important environmental signals including light, inorganic carbon (CO2 and bicarbonate), as well as other nutrients that are not sensed by most heterotrophs, their regulatory networks are likely to differ significantly from those that have been previously characterized. The Synechococcus 7002 genome encodes about 3200 proteins, but a large percentage (35%) of the predicted proteins are of unknown function. Moreover, families of paralogous genes are fairly common in cyanobacterial genomes, and the functions of many of the paralogous gene products are still unknown, although this is slowly changing. For example, the 2-oxoglutarate decarboxylase that functions in the TCA cycle in cyanobacteria was originally annotated as acetolactate synthase (49), and a divergent paralog (ChlF) of a core subunit of Photosystem II (PsbA) was recently shown to encode a light-dependent oxidoreductase that converts chlorophyll a (or chlorophyllide a) into chlorophyll f (or chlorophyllide f) (50). Cyanobacteria contain many light-responsive phytochromes and cyanobacteriochromes (51–54), but the regulatory networks associated with these light-sensing proteins are known in only a few cases (52,55). Nostoc punctiforme has 21 different photoreceptors that account for more than 40 different chromophore-binding, light-sensing domains (54), yet the functions of most of these proteins remain unknown.
The large number of predicted proteins with unidentified functions, as well as differences in the types of environmental signals sensed, places some limitations on the conclusions that can be drawn from global network studies such as those reported here. Furthermore, it is likely that some of the numerous hypothetical proteins might be required to control the complex interactions that occur in bacterial communities, but detailed studies on this subject have only very recently begun for cyanobacteria. Recent examples of co-cultivation studies include Synechococcus sp. PCC 7002 and Shewanella oneidensis (20), Prochlorococcus sp. NATL2A and Alteromonas macleodii MIT1002 (56) and Thermosynechococcus sp. and Meiothermus sp. (Bernstein, et al., unpublished results). Network analyses will become more meaningful as functions are assigned to additional proteins. In spite of these limitations, the analyses reported here shed new light on global regulatory networks in Synechococcus 7002 that can probably be extended in at least some cases to other cyanobacteria. In addition, networks such as these not only allow for an examination of central processes and expansion of known regulons, as shown in this study, but they can be a powerful way to globally organize genes that are functionally related but physically separated in the genome. This can lead to better functional annotation of unknown genes, a processes termed ‘guilt-by-association’ (57–61), and can be a powerful tool to annotate genes in species, such as cyanobacteria, where genomic context is lacking and hypothetical proteins are numerous.
Computational analysis of biological systems has been historically driven by comparative approaches that use sequence homology and genome context to draw functional inferences. However, in many biological systems, including cyanobacteria, sequence context-based methodology is not easy to apply due to unique genomic organization, complex regulatory landscape and other fundamental differences between cyanobacteria and other species such as those described above (3,4). To circumvent these challenges, we examined a compendium of global gene expression data to specifically probe linkages of functionally related processes and the regulatory aspects of Synechococcus 7002 biology by network analysis through grouping genes based on co-expression. The resulting 1386-gene network reconstruction was further organized into 11 modules, which were highly enriched in specific functions that represent key metabolic and cellular pathways. The densest modules, or portions of modules, were enriched for genes involved in growth and replication (photosynthesis, central metabolism and translation), indicating a tightly coordinated response for these genes across all conditions. Less dense modules (e.g. polysaccharide metabolism, defense systems) generally showed less coordination and lack of a uniform response to specific conditions. The structure of specific modules, or portions of modules associated with different functional categories likely reflects a specific regulatory strategy employed by Synechococcus 7002, whereby pathways involved in resource acquisition are tightly linked to changing nutrient levels, diel cycling and alterations in carbon concentrations. Module organization also allows for the grouping of sets of genes whose expression is controlled by a single regulator. This allows for the possibility of identifying new targets of regulators based on their position in the network with respect to known targets of a regulator. In the case of Module 5 we identified several genes whose promoters contained binding sites for the Fur protein, including genes that had not yet been identified as members of the Fur regulon by RegPrecise. Recently, a study examining the effects of a fur deletion in Synechococcus 7002 was published (62) and 33/35 (94%) of the genes in Module 5 showed differential regulation in a fur mutant compared to a wild-type strain, including several of the genes that we identified as Fur regulated but were not yet reported in RegPrecise. Similarly, in the case of Module 6 we show that several of the genes are likely regulated by CcmR. A case study was also recently published examining the regulon of this protein (63) and showed that 26/27 of the genes in Module 6 show differential expression in a ccmR mutant compared to a wild-type strain. The identification of new targets of regulators that were then confirmed through generation of knockouts speaks to the biological relevance of the network presented here and shows how it can be used to gain information on regulator-target pairing.
Cyanobacteria have fewer DNA-binding regulators compared to other prokaryotes (4). To apply our network to the study of cyanobacterial regulators we examined the topology of genes in their local neighborhood. This analysis identified two related observations about the topology of regulators: (i) they are less likely to have edges with other genes and (ii) are less likely to be organized into modules. There are likely several reasons for this observation, but it may be related to the relatively small number of regulators in Synechococcus 7002 and other cyanobacteria. It is possible that with a small number of regulators, many may respond to more than one environmental condition or stimuli. Response(s) to multiple inputs may result in associations between a single regulator and mRNAs of many different processes, making identification of specific regulatory pathways difficult. If a regulator responds to multiple inputs, it may also act in multiple pathways, allowing for tight transcriptional control with fewer DNA binding proteins. The topological analysis shown here already suggests that DNA binding regulators can span groups of genes responsible for different processes (Figure 7). The observation that regulators have fewer edges compared to other genes may also be because the RNA expression level of DNA-binding regulators is often very static, with the presence or absence of a small molecule effector, rather than regulator abundance at the mRNA level, leading to increased or decreased function of the regulator. This would reduce the chances of a regulator sharing an edge with its target.
As was done above with the examination of carbon- and nitrogen-limiting conditions, the linkages and grouping of genes represented here can also be applied to experimental conditions that have not yet been studied or have not been specifically incorporated into this network. This reflects the translational nature of this work and its application to cyanobacteria in general, a powerful approach considering that cyanobacteria are often the primary producers of several types of terrestrial and aquatic microbial communities. Analysis of such communities in regards to coordination of process between species is also a problem of linking related genes across physical space as different species occupy different spatial locations in the community. When applied to these microbial communities, the co-expression network approach presented here becomes a powerful way to link genes located in different species that are transcriptionally, and thus perhaps functionally, related. Such approaches can help decipher complex community interactions and lead to greater understanding of principles guiding community behavior.
ACCESSION NUMBERS
The data sets that have not yet been published that support the results of this article are available in the Gene Expression Omnibus repository, with accession numbers GSE72691 and GSE72880.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the help of Margaret Romine and Jim Fredrickson of the PNNL for help with the manuscript. A significant portion of the research was performed using the Environmental Molecular Sciences Laboratory (EMSL), a national scientific user facility sponsored by DOE BER and located at PNNL. PNNL is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO 1830.
Author contributions: R.M. collected and analyzed the data and wrote the manuscript, C.O. analyzed the data and contributed to the manuscript, E.H. performed the bacterial growth experiments, L.M.M. carried out the RNA sequencing, L.A.M. analyzed the data regarding Gibbs analysis, J.M. analyzed the data and contributed to the manuscript, R.T. analyzed raw sequencing files, M.L. performed the bacterial growth and RNA seq experiments, D.A.B. designed some of the transcription experiments and contributed to writing the manuscript, and A.B. guided the study and contributed to the manuscript. All authors read and approved the final manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genomic Science Program (GSP); Office of Biological and Environmental Research (BER); U.S. Department of Energy (DOE); PNNL Foundational and Biofuels Scientific Focus Area [to A.B]; Air Force Office of Scientific Research Support [FA9550-05-1-0365 (MURI), FA9550-11-1-0148]; National Science Foundation [MCB-1021725 to D. A. B]; PNNL is operated for the DOE by Battelle Memorial Institute under Contract DE-AC05-76RLO 1830. Funding for open access charge: PNNL Foundational and Biofuels Scientific Focus Area [to A.B].
Conflict of interest statement. None declared.
REFERENCES
- 1.Kaneko T., Sato S., Kotani H., Tanaka A., Asamizu E., Nakamura Y., Miyajima N., Hisosawa M., Sugiura M., Sasamoto S., et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 2.Murata N., Suzuki I. Exploitation of genomic sequences in a systematic analysis to access how cyanobacteria sense environmental stress. J. Exp. Bot. 2006;57:235–247. doi: 10.1093/jxb/erj005. [DOI] [PubMed] [Google Scholar]
- 3.Itoh T., Takemoto K., Mori H., Gojobori T. Evolutionary instability of operon structures disclosed by sequence comparisons of complete microbial genomes. Mol. Biol. Evol. 1999;16:332–346. doi: 10.1093/oxfordjournals.molbev.a026114. [DOI] [PubMed] [Google Scholar]
- 4.Minezaki Y., Homma K., Nishikawa K. Genome-wide survey of transcription factors in prokaryotes reveals many bacteria-specific families not found in archaea. DNA Res. 2005;12:269–280. doi: 10.1093/dnares/dsi016. [DOI] [PubMed] [Google Scholar]
- 5.Wagner R. Transcription Regulation in Prokaryotes. NY: Oxford University Press Inc; 2000. [Google Scholar]
- 6.Axmann I.M., Kensche P., Vogel J., Kohl S., Herzel H., Hess W.R. Identification of cyanobacterial non-coding RNAs by comparative genome analysis. Genome Biol. 2005;6:R73. doi: 10.1186/gb-2005-6-9-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beck C., Hertel S., Rediger A., Lehmann R., Wiegard A., Kolsch A., Heilmann B., Georg J., Hess W.R., Axmann I.M. Daily expression pattern of protein-encoding genes and small noncoding RNAs in Synechocystis sp. strain PCC 6803. Appl. Environ. Microbiol. 2014;80:5195–5206. doi: 10.1128/AEM.01086-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kopf M., Moke F., Bauwe H., Hess W.R., Hagemann M. Expression profiling of the bloom-forming cyanobacterium Nodularia CCY9414 under light and oxidative stress conditions. ISME J. 2015;9:2139–2152. doi: 10.1038/ismej.2015.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Steglich C., Futschik M.E., Lindell D., Voss B., Chisholm S.W., Hess W.R. The challenge of regulation in a minimal photoautotroph: non-coding RNAs in Prochlorococcus. PLoS Genet. 2008;4:e1000173. doi: 10.1371/journal.pgen.1000173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W., Chen H., He C.L., Wang Q. Deep sequencing-based identification of small regulatory RNAs in Synechocystis sp. PCC 6803. PloS One. 2014;9:e92711. doi: 10.1371/journal.pone.0092711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gierga G., Voss B., Hess W.R. Non-coding RNAs in marine Synechococcus and their regulation under environmentally relevant stress conditions. ISME J. 2012;6:1544–1557. doi: 10.1038/ismej.2011.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kojima K., Nakamoto H. Post-transcriptional control of the cyanobacterial hspA heat-shock induction. Biochem. Biophys. Res. Commun. 2005;331:583–588. doi: 10.1016/j.bbrc.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 13.Samartzidou H., Widger W.R. Transcriptional and posttranscriptional control of mRNA from lrtA, a light-repressed transcript in Synechococcus sp. PCC 7002. Plant Physiol. 1998;117:225–234. doi: 10.1104/pp.117.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tan X., Varughese M., Widger W.R. A light-repressed transcript found in Synechococcus PCC 7002 is similar to a chloroplast-specific small subunit ribosomal protein and to a transcription modulator protein associated with sigma 54. J. Biol. Chem. 1994;269:20905–20912. [PubMed] [Google Scholar]
- 15.McDermott J.E., Taylor R.C., Yoon H., Heffron F. Bottlenecks and hubs in inferred networks are important for virulence in Salmonella typhimurium. J. Comput. Biol. 2009;16:169–180. doi: 10.1089/cmb.2008.04TT. [DOI] [PubMed] [Google Scholar]
- 16.Ishchukov I., Wu Y., Van Puyvelde S., Vanderleyden J., Marchal K. Inferring the relation between transcriptional and posttranscriptional regulation from expression compendia. BMC Microbiol. 2014;14:14. doi: 10.1186/1471-2180-14-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Netotea S., Sundell D., Street N.R., Hvidsten T.R. ComPlEx: conservation and divergence of co-expression networks in A. thaliana, Populus and O. sativa. BMC Genomics. 2014;15:106. doi: 10.1186/1471-2164-15-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Baleen C. Studies on Marine Blue Green Algae. Bot. Mar. 1962;4:129–139. [Google Scholar]
- 19.Rippka R., Deruelles J., Waterbury J.B., Herdman M., Stanier R.Y. Generic assignments, strain histories and properties of pure cultures of Cyanobacteria. Microbiology. 1979;111:1–61. [Google Scholar]
- 20.Beliaev A.S., Romine M.F., Serres M., Bernstein H.C., Linggi B.E., Markillie L.M., Isern N.G., Chrisler W.B., Kucek L.A., Hill E.A., et al. Inference of interactions in cyanobacterial-heterotrophic co-cultures via transcriptome sequencing. ISME J. 2014;8:2243–2255. doi: 10.1038/ismej.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig M., Bryant D.A. Transcription profiling of the model cyanobacterium synechococcus sp. strain PCC 7002 by Next-Gen (SOLiD) sequencing of cDNA. Front. Microbiol. 2011;2:41. doi: 10.3389/fmicb.2011.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig M., Bryant D.A. Acclimation of the global transcriptome of the cyanobacterium synechococcus sp. strain PCC 7002 to nutrient limitations and different nitrogen sources. Front. Microbiol. 2012;3:145. doi: 10.3389/fmicb.2012.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ludwig M., Bryant D.A. Synechococcus sp. strain PCC 7002 transcriptome: Acclimation to temperature, salinity, oxidative stress, and mixotrophic growth conditions. Front. Microbiol. 2012;3:354. doi: 10.3389/fmicb.2012.00354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Faith J.J., Hayete B., Thaden J.T., Mogno I., Wierzbowski J., Cottarel G., Kasif S., Collins J.J., Gardner T.S. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevens S.E., Porter R.D. Transformation in Agmenellum quadruplicatum. Proc. Natl. Acad. Sci. U.S.A. 1980;77:6052–6056. doi: 10.1073/pnas.77.10.6052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Melnicki M.R., Pinchuk G.E., Hill E.A., Kucek L.A., Stolyar S.M., Fredrickson J.K., Konopka A.E., Beliaev A.S. Feedback-controlled LED photobioreactor for photophysiological studies of cyanobacteria. Bioresour. Technol. 2013;134:127–133. doi: 10.1016/j.biortech.2013.01.079. [DOI] [PubMed] [Google Scholar]
- 27.McClure R., Balasubramanian D., Sun Y., Bobrovskyy M., Sumby P., Genco C.A., Vanderpool C.K., Tjaden B. Computational analysis of bacterial RNA-Seq data. Nucleic Acids Res. 2013;41:e140. doi: 10.1093/nar/gkt444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trevino S., 3rd, Sun Y., Cooper T.F., Bassler K.E. Robust detection of hierarchical communities from Escherichia coli gene expression data. PLoS Comput. Biol. 2012;8:e1002391. doi: 10.1371/journal.pcbi.1002391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Csardi G., Nepusz T. The igraph software package for complex network research. InterJournal. 2006:1695–1704. [Google Scholar]
- 31.Thompson W., Rouchka E.C., Lawrence C.E. Gibbs recursive sampler: Finding transcription factor binding sites. Nucleic Acids Res. 2003;31:3580–3585. doi: 10.1093/nar/gkg608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J.S., Lawrence C.E. Bayesian inference on biopolymer models. Bioinformatics. 1999;15:38–52. doi: 10.1093/bioinformatics/15.1.38. [DOI] [PubMed] [Google Scholar]
- 33.Novichkov P.S., Kazakov A.E., Ravcheev D.A., Leyn S.A., Kovaleva G.Y., Sutormin R.A., Kazanov M.D., Riehl W., Arkin A.P., Dubchak I., et al. RegPrecise 3.0–a resource for genome-scale exploration of transcriptional regulation in bacteria. BMC Genomics. 2013;14:745. doi: 10.1186/1471-2164-14-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 35.Kery M.B., Feldman M., Livny J., Tjaden B. TargetRNA2: identifying targets of small regulatory RNAs in bacteria. Nucleic Acids Res. 2014;42:W124–W129. doi: 10.1093/nar/gku317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McDermott J.E., Costa M., Janszen D., Singhal M., Tilton S.C. Separating the drivers from the driven: Integrative network and pathway approaches aid identification of disease biomarkers from high-throughput data. Dis. Markers. 2010;28:253–266. doi: 10.3233/DMA-2010-0695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song H.S., McClure R.S., Bernstein H.C., Overall C.C., Hill E.A., Beliaev A.S. Integrated in silico analyses of regulatory and metabolic networks of synechococcus sp. PCC 7002 reveal relationships between gene centrality and essentiality. Life. 2015;5:1127–1140. doi: 10.3390/life5021127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barabasi A.L., Oltvai Z.N. Network biology: understanding the cell's functional organization. Nat. Rev. Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 39.Burnap R.L., Hagemann M., Kaplan A. Regulation of CO2 concentrating mechanism in cyanobacteria. Life. 2015;5:348–371. doi: 10.3390/life5010348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Woodger F.J., Bryant D.A., Price G.D. Transcriptional regulation of the CO2-concentrating mechanism in a euryhaline, coastal marine cyanobacterium, Synechococcus sp. Strain PCC 7002: role of NdhR/CcmR. J. Bacteriol. 2007;189:3335–3347. doi: 10.1128/JB.01745-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Battchikova N., Eisenhut M., Aro E.M. Cyanobacterial NDH-1 complexes: Novel insights and remaining puzzles. Biochim. Et Biophys. Acta. 2011;1807:935–944. doi: 10.1016/j.bbabio.2010.10.017. [DOI] [PubMed] [Google Scholar]
- 42.Ogawa T. A gene homologous to the subunit-2 gene of NADH dehydrogenase is essential to inorganic carbon transport of Synechocystis PCC6803. Proc. Natl. Acad. Sci. U.S.A. 1991;88:4275–4279. doi: 10.1073/pnas.88.10.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ogawa T. Cloning and inactivation of a gene essential to inorganic carbon transport of synechocystis PCC6803. Plant Physiol. 1991;96:280–284. doi: 10.1104/pp.96.1.280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fillat M.F. The FUR (ferric uptake regulator) superfamily: diversity and versatility of key transcriptional regulators. Arch. Biochem. Biophys. 2014;546:41–52. doi: 10.1016/j.abb.2014.01.029. [DOI] [PubMed] [Google Scholar]
- 45.Troxell B., Fink R.C., Porwollik S., McClelland M., Hassan H.M. The Fur regulon in anaerobically grown Salmonella enterica sv. typhimurium: identification of new Fur targets. BMC Microbiol. 2011;11:236. doi: 10.1186/1471-2180-11-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tjaden B. Computational identification of sRNA targets. Methods Mol. Biol. 2012;905:227–234. doi: 10.1007/978-1-61779-949-5_14. [DOI] [PubMed] [Google Scholar]
- 47.Haning K., Cho S.H., Contreras L.M. Small RNAs in mycobacteria: an unfolding story. Front. Cell. Infect. Microbiol. 2014;4:96. doi: 10.3389/fcimb.2014.00096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Modi S.R., Camacho D.M., Kohanski M.A., Walker G.C., Collins J.J. Functional characterization of bacterial sRNAs using a network biology approach. Proc. Natl. Acad. Sci. U.S.A. 2011;108:15522–15527. doi: 10.1073/pnas.1104318108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang S., Bryant D.A. The tricarboxylic acid cycle in cyanobacteria. Science. 2011;334:1551–1553. doi: 10.1126/science.1210858. [DOI] [PubMed] [Google Scholar]
- 50.Ho M.Y., Shen G., Canniffe D.P., Zhao C., Bryant D.A. Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science. 2016:aaf9178. doi: 10.1126/science.aaf9178. [DOI] [PubMed] [Google Scholar]
- 51.Anders K., Essen L.O. The family of phytochrome-like photoreceptors: Diverse, complex and multi-colored, but very useful. Curr. Opin. Struct. Biol. 2015;35:7–16. doi: 10.1016/j.sbi.2015.07.005. [DOI] [PubMed] [Google Scholar]
- 52.Bhaya D. In the Limelight: Photoreceptors in Cyanobacteria. mBio. 2016;7 doi: 10.1128/mBio.00741-16. doi:10.1128/mBio.00741-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ikeuchi M., Ishizuka T. Cyanobacteriochromes: A new superfamily of tetrapyrrole-binding photoreceptors in cyanobacteria. Photochem. Photobiol. Sci. 2008;7:1159–1167. doi: 10.1039/b802660m. [DOI] [PubMed] [Google Scholar]
- 54.Rockwell N.C., Martin S.S., Gan F., Bryant D.A., Lagarias J.C. NpR3784 is the prototype for a distinctive group of red/green cyanobacteriochromes using alternative Phe residues for photoproduct tuning. Photochem. Photobiol. Sci. 2015;14:258–269. doi: 10.1039/c4pp00336e. [DOI] [PubMed] [Google Scholar]
- 55.Gutu A., Kehoe D.M. Emerging perspectives on the mechanisms, regulation, and distribution of light color acclimation in cyanobacteria. Mol. Plant. 2012;5:1–13. doi: 10.1093/mp/ssr054. [DOI] [PubMed] [Google Scholar]
- 56.Biller S.J., Coe A., Chisholm S.W. Torn apart and reunited: impact of a heterotroph on the transcriptome of Prochlorococcus. ISME J. 2016 doi: 10.1038/ismej.2016.82. doi:10.1038/ismej.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gillis J., Pavlidis P. The impact of multifunctional genes on ‘guilt by association’ analysis. PloS one. 2011;6:e17258. doi: 10.1371/journal.pone.0017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gillis J., Pavlidis P. ‘Guilt by association’ is the exception rather than the rule in gene networks. PLoS Comput. Biol. 2012;8:e1002444. doi: 10.1371/journal.pcbi.1002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.van Dam S., Cordeiro R., Craig T., van Dam J., Wood S.H., de Magalhaes J.P. GeneFriends: an online co-expression analysis tool to identify novel gene targets for aging and complex diseases. BMC Genomics. 2012;13:535. doi: 10.1186/1471-2164-13-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wolfe C.J., Kohane I.S., Butte A.J. Systematic survey reveals general applicability of ‘guilt-by-association’ within gene coexpression networks. BMC Bioinformatics. 2005;6:227. doi: 10.1186/1471-2105-6-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yoon H., Ansong C., McDermott J.E., Gritsenko M., Smith R.D., Heffron F., Adkins J.N. Systems analysis of multiple regulator perturbations allows discovery of virulence factors in Salmonella. BMC Syst. Biol. 2011;5:100. doi: 10.1186/1752-0509-5-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ludwig M., Chua T.T., Chew C.Y., Bryant D.A. Fur-type transcriptional repressors and metal homeostasis in the cyanobacterium Synechococcus sp. PCC 7002. Front. Microbiol. 2015;6:1217. doi: 10.3389/fmicb.2015.01217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krishnan A., Zhang S., Liu Y., Tadmori K.A., Bryant D.A., Dismukes G.C. Consequences of ccmR deletion on respiration, fermentation and H metabolism in cyanobacterium Synechococcus sp. PCC 7002. Biotechnol. Bioeng. 2015;113:1448–1459. doi: 10.1002/bit.25913. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.