Abstract
SOX9 encodes a transcription factor that governs cell fate specification throughout development and tissue homeostasis. Elevated SOX9 is implicated in the genesis and progression of human tumors by increasing cell proliferation and epithelial-mesenchymal transition. We found that in response to UV irradiation or genotoxic chemotherapeutics, SOX9 is actively degraded in various cancer types and in normal epithelial cells, through a pathway independent of p53, ATM, ATR and DNA-PK. SOX9 is phosphorylated by GSK3β, facilitating the binding of SOX9 to the F-box protein FBW7α, an E3 ligase that functions in the DNA damage response pathway. The binding of FBW7α to the SOX9 K2 domain at T236-T240 targets SOX9 for subsequent ubiquitination and proteasomal destruction. Exogenous overexpression of SOX9 after genotoxic stress increases cell survival. Our findings reveal a novel regulatory mechanism for SOX9 stability and uncover a unique function of SOX9 in the cellular response to DNA damage. This new mechanism underlying a FBW7-SOX9 axis in cancer could have implications in therapy resistance.
INTRODUCTION
SOX9 is a member of the structurally related sex-determining region Y (Sry)-box-containing (Sox) family of transcription factors essential for carrying out diverse functions during development (1–3). In humans, heterozygous germline mutations of SOX9 cause Campomelic Dysplasia (CD), a severe form of dwarfism characterized by gross cartilage and bone abnormalities, as well as phenotypic sex reversal in most males (4). Although a small percentage survive to adulthood (5), the majority of patients with CD die in the neonatal period from respiratory distress caused by abnormal airway development or rib cage abnormalities (6). Similar to humans, homozygous loss of SOX9 in mice causes embryonic lethality at E11.5, and heterozygous SOX9 knockout mice die perinatally (7). Conditional SOX9 knockout mouse models have further revealed that SOX9 regulates osteochondrogenic differentiation, extracellular matrix production, cell proliferation and branching morphogenesis (8–11). In adult tissues, SOX9 expression is necessary for the maintenance of stem and progenitor cell populations (12–14).
SOX9 has been implicated in tumorigenesis or tumor progression in numerous organs and tissues (15–22). In the KRASG12D pancreas cancer mouse model, SOX9 expression is required for development of pancreatic intraepithelial neoplasia, and overexpression of SOX9 accelerates formation of premalignant lesions (20). Several oncogenic pathways, such as KRAS, Wnt/β-catenin, Sonic hedgehog, Notch, TGFβ, NF-κB and YAP1 regulate SOX9 transcription (20,23–30). Although the precise downstream mechanisms responsible for the pro-tumorigenic properties of SOX9 have not been fully uncovered, it is known that SOX9 enhances epithelial-mesenchymal transition and cell proliferation across multiple tumor types (21,31–33). Upregulated SOX9 in tumors has also been correlated with poor survival in cancer patients (21,34–35). SOX9 might contribute to poor survival by increasing resistance to therapy. Expression of SOX9 was shown to impart radiation-resistance to intestinal stem cells (36), and SOX9 increased resistance to tyrosine kinase inhibitors (37). Moreover, SOX9 overexpression was reported to participate in resistance to ultraviolet (UV) irradiation in keratinocytes (38). Further exploration of how SOX9 confers resistance to these external agents could lead to discovery of novel therapeutic strategies.
During the DNA damage response pathway, commonly initiated by genotoxic drugs or irradiation, many key proteins are actively degraded through the ubiquitin–proteasome system. The protein ubiquitination pathway is catalyzed by the ubiquitin-activating E1 enzyme, the ubiquitin-conjugating E2 enzyme and the ubiquitin–protein E3 ligase. Crucially, E3 ligases determine the substrate specificity for ubiquitination and subsequent degradation (39). Thus, E3 ligases are essential for regulating the specificity of the DNA damage response. A pivotal E3 ligase complex that regulates protein stability in development and cancer is the SKP1–cullin1–F-box protein (SCF) complex (39). The FBW7 tumor suppressor is a member of the SCF complex and is the most highly mutated F-box protein in human cancer (39,40). FBW7 targets numerous oncoproteins for degradation, including MYC, JUN, Cyclin E and Notch1 (39,40). FBW7 binds to its substrate at a conserved phosphodegron (CPD) motif within the substrate that must first be phosphorylated (40). FBW7 is also participates directly in the DNA damage response. FBW7 regulates the stability of MYC (41) and polo-like kinase 1 (PLK1) (42) in response to UV-induced DNA damage. Moreover, FBW7 is recruited to DNA double strand breaks and participates in DNA repair (43).
In the present study, we found that the developmental- and cancer-associated transcription factor, SOX9, is degraded in response to DNA damage caused by cytotoxic drugs or UV-irradiation. We identified a novel regulatory mechanism for the depletion of SOX9 induced by DNA damage that involves phosphorylation of SOX9 within its K2 transactivation domain by GSK3β, followed by the targeting of SOX9 by FBW7 for ubiquitination and proteasomal degradation (44). We also found that forced overexpression of SOX9 increased cell survival after exposure to UV irradiation, suggesting a potential mechanism driving the resistance of high SOX9-expressing cells to certain forms of genotoxic therapy. SOX9 can be added to the growing repertoire of proteins that are actively modulated by genotoxic stress and that participate in the DNA damage response.
MATERIALS AND METHODS
Cell lines
HCT116 FBXW7+/+ and FBXW7−/− colon cancer cell lines, as well as HCT116 and p53-null derivatives were kind gifts from Dr Bert Vogelstein (45). C8161 melanoma cells were provided by Karine Cohen–Solal (Rutgers Cancer Institute of New Jersey), H460 sh-control and H460-p53shRNA lung cancer cells were provided by Zhaohui Feng (Rutgers Cancer Institute of New Jersey) (46). All other cell lines, A549, H1299 and H460 lung cancer, DLD1 colon cancer, U2OS osteosarcoma and Beas2b immortalized human bronchial epithelial cells, were obtained from ATCC. Cell cultures were maintained in RPMI plus 10% fetal bovine serum.
In vitro drug treatment
Clinical-grade cisplatin and doxorubicin were obtained from the Rutgers Cancer Institute of New Jersey Pharmacy. Cells were treated with 30 μM cisplatin or 7.4 μM (4 μg/ml) doxorubicin for 24 h before collection.
Irradiation
Cells were irradiated with UVC in a UV Stratalinker1800 (Agilent) at 70–90% confluence in a petri dish without the lid or media. The fluence output was confirmed with a UVX digital radiometer and UVX-25 sensor (UVP). After UV irradiation, fresh media was added. Where indicated, cells were transiently transfected 24 or 48 h before irradiation. Inhibitors were added 1 h prior to UV irradiation and added back to the media immediately after irradiation. Half-life measurements were performed after UV or without UV in 20 μg/ml cycloheximide (Sigma-Aldrich) for the indicated time periods. For γ-irradiation, cells were irradiated with Cs-137 γ-rays (dose rate: 0.893 Gy/min) 18 h after the cells were plated and lysates were collected at the indicated times after irradiation. Inhibitors were obtained from Sigma-Aldrich: MG132 (20 μM), H89 (20 μM), SB202190 (10 μM), KU55933 (10 μM), PD98059 (10 μM). Other drug concentrations are indicated.
Plasmids and transfections
The HA-FBW7 and HA-GSK3 constructs were a gift of Wenyi Wei (47). GST-FBXL3a, GST-FBXL13, GST-FBXL18, GST-FBXL18, GST-FBW4, GST-FBW6 and GST-FBW7 were gifts of Wade Harper (Harvard Medical School). The SOX9-pCMV-Tag2 plasmid was validated and described previously (22,31). GFP-C1 was purchased from Clontech. The pDZ Flag USP28 plasmid was a gift from Martin Eilers (Addgene plasmid # 15665) (41). HA-GSK3β and HA-GSK3β S9A plasmids were a kind gift from Jim Woodgett (Addgene plasmid #14753, 14754) (48). SOX9 mutants were generated with the QuikChange II XL Site-Directed Mutagenesis kit (Stratagene) according to the manufacturer's instructions. Transfections were performed using Fugene (Promega) as described (31). siRNAs against ATR, ATM and DNA-PK were purchased from Santa Cruz and transfected with Lipofectimine RNAiMAX (Life Technologies), following the manufacturer's instructions. Cells were collected or experiments were performed 48 h after transfection, unless otherwise indicated.
Western blot
Standard immunoblots were performed as previously described (31). Antibodies against the following proteins were purchased from Cell Signaling: GST (8146), FLAG (8146), HA (2367), GFP (2559), GSK3β (9315), Phospho-histone H3 (9701), Notch1-ICD (4147), phospho-CHK1 (2341), phospho-CHK2 (2661), phospho-ERK (9101), phospho-AKT (4060), phospho-CREB (9198), phospho-MAPKAPK2 (3007), PARP (9542), ATM (2873), ATR (2790), DNA-PK (4602) and GSK3β (9315). Antibodies against SOX9 (Millipore, AB5535), FBW7 (R&D, MAB7776), p53 (Abcam, ab1101), GAPDH (Millipore, MAB374) and α-Tubulin (Sigma, T9026) were used. Rabbit polyclonal phospho-T236-SOX9 was custom-generated by Bethyl laboratories. Specificity was assessed by detection of wild-type but not T236-mutant SOX9, as well as loss of detection after treatment of lysates with λ-phosphatase.
Immunoprecipitation
Cells were transfected 48 h prior to cell collection, treated with 20 μM MG132 for the last 8 h, and lysed in lysis buffer [50 mM Tris.HCl (pH7.4), 150 mM NaCl, 10% Glycerol, 1 mM EDTA], 1% Triton-X-100, phosphatase inhibitors and protease inhibitor cocktails 2 and 3 (Sigma-Aldrich). The cell lysates were cleared by centrifugation and incubated with Flag M2 protein A/G (Cell Signaling) beads overnight. The beads were boiled after extensive washing and the proteins were detected by Western blotting, followed by quantification using Image J software (http://rsb.info.nih.gov/ij). For endogenous immunoprecipitation, SOX9 was immunoprecipitated from lysates that were prepared from two 10-cm dishes with a rabbit polyclonal antibody against SOX9 (Millipore, AB5535), using Protein A/G plus-Agarose IP Reagent (Santa Cruz), following the manufacturer's instructions. For immunoprecipitation under denatured conditions, cells were lysed in denaturing buffer (1% SDS, 5 mM EDTA, phosphatase and protease inhibitors), boiled for 5 min and passed through a 26-g needle several times. Lysates were diluted 1:10 in the non-denaturing lysis buffer as used for Western blotting. Immunoprecipitation was carried out as described above.
Mass spectrometry
Endogenous SOX9 immunoprecipitation products were separated by SDS-PAGE. The SOX9 bands were excised and digested with Asp-N using a standard in-gel digestion protocol. Nano-LC-MSMS was done as described (49). Peak lists in the format of MASCOT Generic Format was generated using the Proteome Discover 1.4 (ThermoFisher). Data were searched against SwissPort most updated database with species set to human using MASCOT 2.2 search engine. Carbamidomethylation on cysteine was set as static modification. Oxidation on methionine, phosphorylation on serine, threonine and tyrosine were set as dynamic modifications. The identified phosphorylated peptides of SOX9 were manually inspected.
In vitro phosphorylation
An empty GST vector or wild-type and mutant GST-SOX9 fragments (residues 186–318) were purified from Escherichia coli using a standard protocol. The kinase assay was performed at 30°C in a kinase buffer with recombinant human GSK3β, 200 μM ATP and 5 μCi[32P]-ATP. Reactions were terminated with Laemmli SDS sample buffer. The phosphorylation of SOX9 was detected by autoradiography. Presence of GST or GST-SOX9 was detected by Coomassie blue staining.
GST pull-down
HEK293T cells expressing HA-FBW7 were lysed and cleared by centrifugation. The GST proteins were purified using glutathione sepharose 4B (AmershamBiosciences), and the bound HA-FBW7 was detected by Western blot. Binding to immobilized GST proteins was performed as described previously (50). Where indicated, the GST-SOX9 fragments proteins were incubated with GSK3 in the presence of 200 μM ATP for 1 h prior to the binding assays.
Polymerase Chain Reaction (PCR)
RNA was extracted from HCT116 FBW7+/+ and FBW7−/− cells, cDNA was generated and GAPDH was amplified following standard procedures (31). Exons 4–6 of FBW7 was amplified using primers Forward, 5′AACGACGCCGAATTACATCT3′, and Reverse, 5′TGCTTGTAGCAGGTCTTTGG3′, to yield either a 251 bp polymerase chain reaction (PCR) product if exon 5 was intact, or no PCR product if exon 5 was deleted, as originally demonstrated (45).
Cell synchronization
For synchronization in early S phase, H1299 cells were incubated with 200 mM thymidine for 15 h, grown in the absence of thymidine for 8 h and incubated in thymidine again for 16 h. For synchronization in G0/G1 phase, cells were incubated in serum-free media containing 0.1% bovine serum albumin for 24 h. Synchronization was confirmed by cell cycle analysis of propidium iodide-stained cells.
Clonogenic assay
Cells were plated (50 000 or 200 cells) 24 h after transfection, and exposed to 40 J/m2 UVC 24 h later. Plates were fixed and stained with coomassie blue 14 days after irradiation, or 7 days after plating in the case of unirradiated cells. Colonies that were ≥200 μm in diameter were counted using ImageJ software (https://imagej.nih.gov/ij/).
Statistical analyses
Mean values and standard deviations are shown in graphs that were generated from several repeats of biological experiments, unless otherwise stated. The used statistical tests are indicated in the Figure legends. Calculations were performed by use of STATA version 12 software (STATA Corp). A P-value that was ≤0.05 was used as the criterion of statistical significance. All statistical tests were two-sided.
RESULTS
Drug- and UV-induced DNA damage depletes SOX9
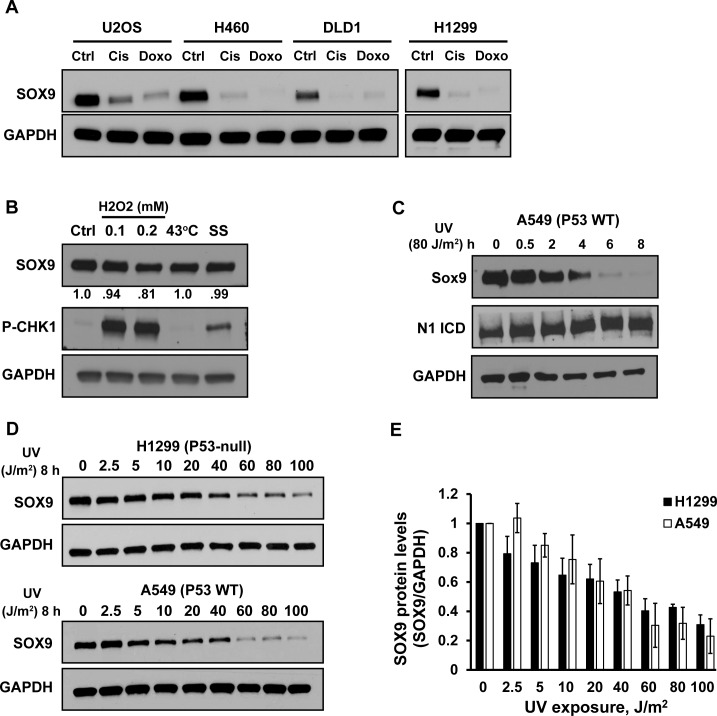
During our studies relating SOX9 expression to genotoxic drug sensitivity in cancer, we were surprised that both cisplatin and doxorubicin caused a reduction of SOX9 protein levels in several cancer cell lines (Figure 1A). Cisplatin forms inter-strand DNA crosslinks that interfere with DNA replication (51), whereas doxorubicin intercalates into DNA, causes DNA breaks and prevents DNA replication (52). To explore our findings further, we tested various cellular stressors. As expected, hydrogen peroxide, which causes DNA damage through oxidative stress, resulted in activation of the DNA damage response, evidenced by activation of CHK1, but it caused only a modest decrease in SOX9 (Figure 1B). In contrast, neither exposure of cells to 43°C, which induces a rapid heat stress response, nor serum starvation, which initiates a cellular stress response by depriving cells of growth factors, caused a loss of SOX9 (Figure 1B). We concluded that SOX9 depletion is likely a consequence of certain forms of DNA damage but not an effect of a general cellular stress response.
Figure 1.
Chemotherapeutic drugs and UV irradiation deplete SOX9. (A) Western blot of SOX9 protein levels in various cancer cell lines after 24 h of treatment with cisplatin (Cis) (30 μM), doxorubicin (Doxo) (7.4 μM) or untreated (Ctrl). The blots for H1299 were run in parallel with the other cell lines and exposed on the same film. GAPDH was used as a loading control. (B) Western blot of SOX9 protein levels in untreated (Ctrl) H1299 cells or cells after treatment with 0.1 or 0.2 mM hydrogen peroxide for 4 h, heat-shock (43°C) for 2 h or serum starvation (SS) for 24 h. Numbers below represent densitometric determination of the change in SOX9 protein levels compared to control, normalized to GAPDH. Phosphorylated CHK-1 served as a control for activation of the DNA damage response. (C) Western blot of SOX9 and Notch1 intracellular domain (N1 ICD) in A549 cells. Lysates were prepared at the indicated times (0–8 h) after UV irradiation (80 J/m2). GAPDH was used as a loading control. (D) Western blot of SOX9 protein levels in A549 and H1299 cells 8 h after varying doses of UV irradiation (0–100 J/m2). (E) The graph shows average SOX9 protein levels normalized to GAPDH, from the data in (D) in three independent experiments. Error bars indicate the standard deviation.
We next tested if SOX9 is depleted in response to other types of DNA damage. We exposed cells to UV irradiation, which causes pyrimidine dimers, induces DNA strand breaks and prevents DNA replication. UV exposure decreased SOX9 protein levels in a time- and dose-dependent manner. SOX9 was significantly depleted as early as 2–4 h after exposure, and at a UV fluence as low as 5 J/m2 (Figure 1C–E). UV-induced SOX9 depletion was also apparent across additional cell lines from lung cancer, colon cancer, osteosarcoma, melanoma and immortalized human bronchial epithelial cells (Supplementary Figure S1A and B). UV-induced loss of SOX9 was prolonged and recovery was not reached until 96 h after exposure (Supplementary Figure S1C). UV irradiation had no effect on Notch1 (Figure 1C), that transcriptionally regulates SOX9 expression (31,53), suggesting that the decrease in SOX9 is specific. We also confirmed that decreased SOX9 after UV exposure was not due to cell cycle-dependent expression of SOX9, as there were no changes in SOX9 protein levels in the different cell cycle phases (Supplementary Figure S1F). We were unable to detect any loss of SOX9 after varying doses of γ-irradiation, although there was a slight increase at higher doses (Supplementary Figure S1E). Thus, elimination of SOX9 is unique to certain forms of DNA damage. This observation is similar to p21, where is has been reported to be depleted after UV irradiation but up-regulated by γ-irradiation (54).
Mechanistic insight into SOX9 protein degradation after UV irradiation
We next sought to define the molecular mechanisms responsible for the depletion of SOX9 after UV-induced DNA damage. P53 coordinates the cellular response to DNA damage and is activated by UV (55). However, SOX9 down-regulation occurred after UV exposure in cell lines regardless of TP53 status or expression (Figure 1C, D and Supplementary Figure S1A–D). Thus, depletion of SOX9 after UV exposure is likely independent of p53.
Activation of the ATR, ATM and DNA-PK checkpoint pathways are major events in the cellular response to UV damage (56). Additional pathways are activated during the DNA damage response, including PKA/cAMP, PI3K and p38/MAPK (57–59). We surmised that at least one of these DNA damage response pathways would contribute to the loss of SOX9 after UV irradiation. However, experimental evidence failed to implicate any of these pathways in UV-induced elimination of SOX9. Specifically, inhibition of ATR with either VE821 or AZ20 prevented UV-induced CHK1 activation, but did not prevent SOX9 depletion (Supplementary Figure S2A). Likewise, inhibition of ATM with KU55933 prevented CHK2 activation caused by UV exposure but didn't abrogate SOX9 depletion (Supplementary Figure S2B). UV irradiation also induced loss of SOX9 in DNA-PK deficient cells (Supplementary Figure S2C). Moreover, UV irradiation caused the depletion of SOX9 when ATM, ATR and DNA-PK expressions were knocked by siRNA (Supplementary Figure S2D). Next, inhibition of PI3K by Wortmannin or LY294002 prevented AKT phosphorylation but did not abrogate UV-induced SOX9 loss (Supplementary Figure S2E). Similarly, inhibition of PKA with H89 prevented phosphorylation of CREB after UV irradiation, but had no effect on attenuating the loss of SOX9 (Supplementary Figure S2F). We also inhibited MEK with PD98059 and p38 MAPK with SB202-190, and verified that activation of ERK was attenuated and phosphorylation of MAP-KAPK2 was blocked after UV exposure, respectively, but UV still resulted in SOX9 degradation (Supplementary Figure S2B, S2F). In summary, UV-induced SOX9 depletion in cancer cells is independent of ATR/ATM, DNA-PK, as well as PKA/cAMP, PI3K and p38/MAPK.
Requirement of GSK3β for SOX9 protein depletion after UV irradiation
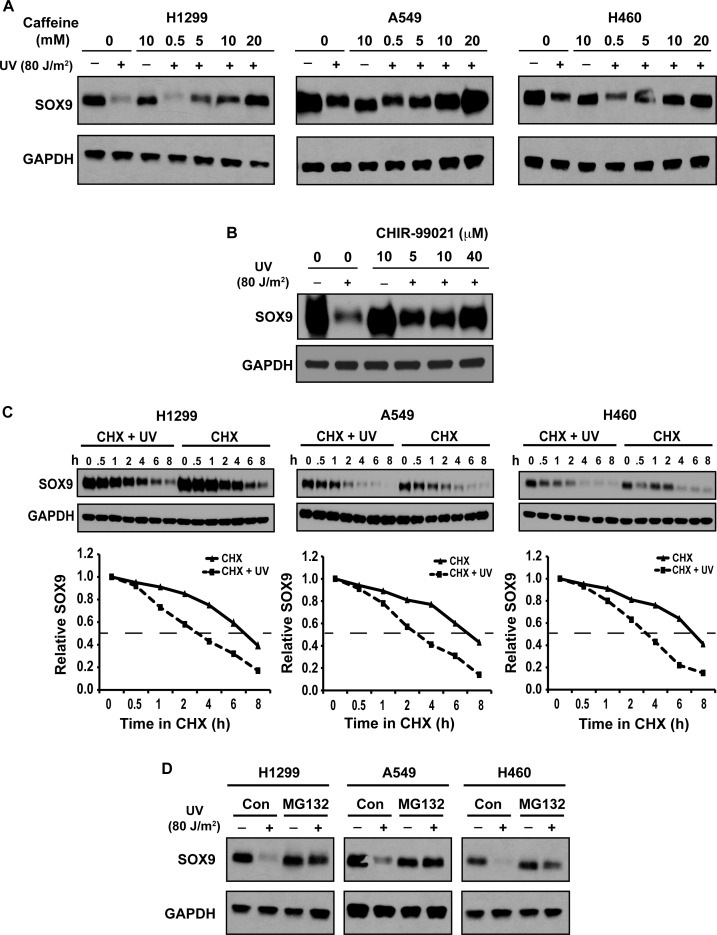
We had noted that pretreatment of cells with a low concentration of caffeine (5 mM) attenuated the reduction of SOX9 protein in A549 cells caused by UV exposure (Supplementary Figure S2B). We next confirmed this in multiple cell lines. Caffeine completely prevented UV-induced SOX9 degradation in a dose-dependent manner (Figure 2A). There was a slight reduction of SOX9 protein gel mobility in UV-irradiated cells as compared to unirradiated cells that was prevented by caffeine treatment (Figure 2A), suggesting that UV-irradiation induces a SOX9 protein modification inhibited by caffeine. Although caffeine is a widely-used inhibitor of ATM and ATR pathways, it is also a potent indirect inhibitor of GSK3β, either through inhibition of ATM/ATR (60) or activation of PKA (61). There had been no prior reports that GSK3β phosphorylates SOX9 or regulates SOX9 stability, but GSK3β is involved in the regulation of p21 and MYC degradation after UV irradiation (41,60). Thus, we asked whether GSK3β regulates UV-induced SOX9 destabilization. To test this hypothesis, we pretreated cells with the GSK3 inhibitor CHIR99021 prior to UV irradiation. Inhibition of GSK3 abrogated UV-induced SOX9 degradation in a dose-dependent manner (Figure 2B), suggesting that GSK3β plays a role in regulating SOX9 stability after DNA damage.
Figure 2.
SOX9 depletion after UV irradiation is inhibited by caffeine or GSK3 inhibition and is mediated by the proteasome pathway. (A) Dose-dependent inhibition of SOX9 depletion after UV by caffeine. Western blot of SOX9 levels in H1299, A549 and H460 cells 8 h after UV irradiation (80 J/m2) that were pre-treated with escalating doses of caffeine (0–20 mM). GAPDH was used as a loading control. (B) UV-induced SOX9 degradation is prevented by inhibition of GSK3. Western blot of SOX9 levels 8 h after UV irradiation (80 J/m2) in H1299 cells that were pre-treated with escalating doses of the GSK3 inhibitor CHIR-99021 (0–40 μM). GAPDH was used as a loading control. (C) Cycloheximide (CHX) chase assay on UV irradiated (80 J/m2) or unirradiated H1299, A549 and H460 cells. SOX9 protein levels were detected by Western blot at the indicated times (0–8 h). The half-life of SOX9 was determined by densitometric quantitation. In the graphical representation, the signal at 0 h was set to 1.0 and used as a standard for subsequent times, after normalization by the GAPDH loading control. (D) UV-induced proteasome-dependent degradation of SOX9. Western blot of SOX9 protein levels in unirradiated or UV-irradiated (8 h, 80 J/m2) H1299, A549 and H460 cells, either with or without a 6-h treatment with the proteasome inhibitor MG132. GAPDH was used as a loading control.
FBW7 targets SOX9 for destruction after UV-induced DNA damage
Because the data thus far suggested that the regulation of SOX9 depletion after UV irradiation is post-translational, we next tested if UV-induced SOX9 loss was due to reduced protein stability. Cycloheximide (CHX) was used to block de novo protein synthesis. We determined the half-life of SOX9 was about 7 h in unirradiated cells, but it was reduced to about 3 h in cells exposed to UV (Figure 2C). Moreover, the reduction of SOX9 protein levels after exposure to UV was completely abrogated by a 6 h treatment with the 26S proteasome inhibitor, MG132, (Figure 2D), suggesting that SOX9 was degraded through a ubiquitin–proteasome pathway in response to DNA damage. We also noted that under normal conditions, a 6 h treatment with MG132 did not increase SOX9 protein levels, similar to a recent report (62).
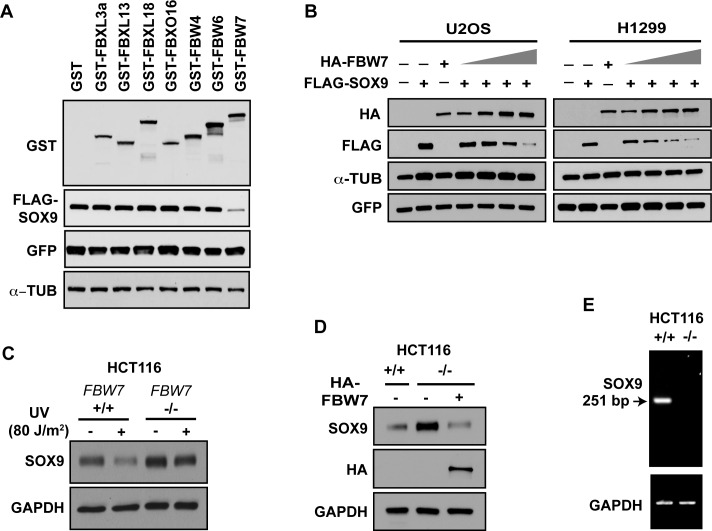
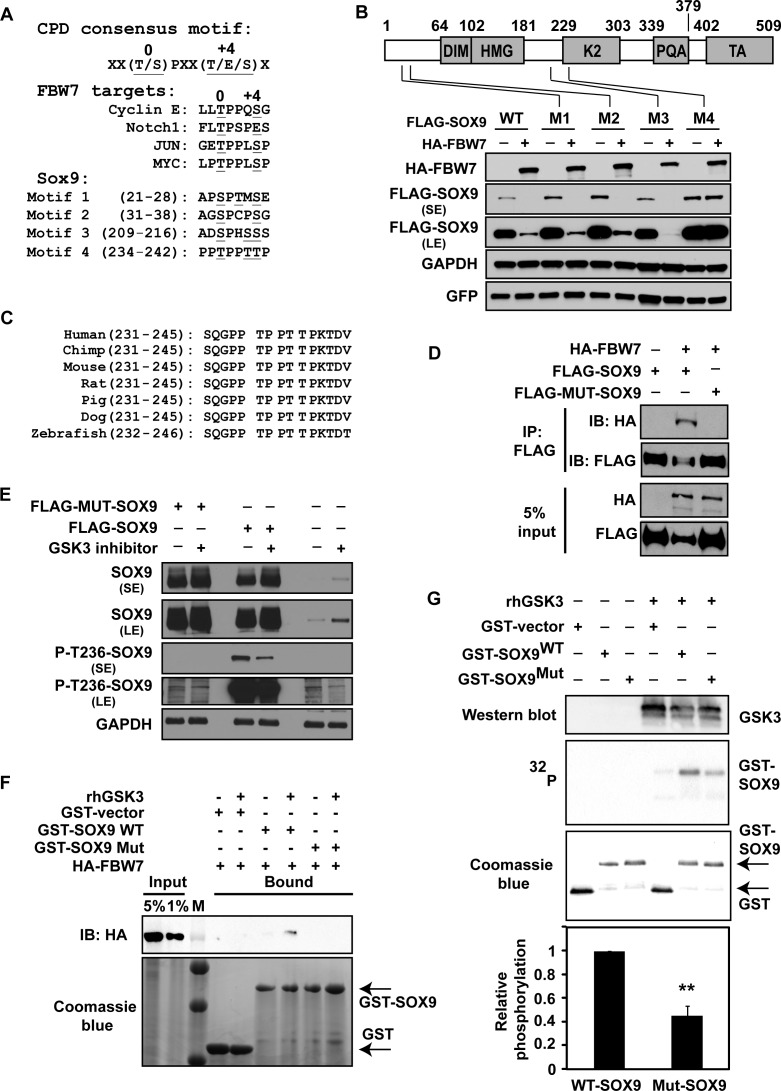
In order to identify the E3 ligase that may meditate the ubiquitination of SOX9 leading to its proteasomal degradation after DNA damage, we screened the F-box protein family of E3 ligases, which typically require their target proteins to first be phosphorylated by GSK3β (39). We co-expressed SOX9 with empty GST vector, or GST-tagged FBXL3a, FBXL13, FBXL18, FBXO16, FBW4, FBW6 or FBW7. As shown in Figure 3A, the protein levels of SOX9 were decreased when it was co-expressed with FBW7, but none of the other E3 ligases were able to alter SOX9 levels, suggesting that SOX9 can be targeted for proteasomal degradation through FBW7.
Figure 3.
FBW7α targets SOX9 for ubiquitination and destruction after UV-induced DNA damage. (A) Screen of F-box proteins. H293T cells were co-transfected with FLAG-SOX9 and an empty vector or the GST-tagged F-box proteins FBXL3a, FBXL13, FBXL18, FBXO16, FBW4, FBW6 or FBW7, as well as green fluorescent protein (GFP) for a transfection control. Lysates were immunoblotted for FLAG and GST. α-Tubulin (α-TUB) was used as a loading control. (B) Western blot of FLAG-SOX9, HA-FBW7 and GFP of lysates from U2OS and H1299 cells that were co-transfected with FLAG-SOX9, GFP and increasing amounts of HA-FBW7α. α-tubulin was used as a loading control. (C) FBW7 mediates UV-induced SOX9 degradation. FBW7+/+ or FBW7−/− HCT116 colon cancer cells UV irradiated (8 h, 80 J/m2), or unirradiated, were immunoblotted for SOX9. GAPDH was used as a loading control. (D) FBW7 decreases endogenous SOX9. FBW7−/− HCT116 were transfected with HA-FBW7. FBW7+/+ and FBW7−/− HCT116 cells were immunoblotted for SOX9 and HA-FBW7. GAPDH was used as a loading control. (E) RNA from FBW7+/+ and FBW7−/− HCT116 cells were examined for deletion of exon 5 in FBW7, utilizing primers that span exons 4 and 6 and that yield a 251 bp product. The absence of an RT-PCR product in lane 2 signifies deletion of exon 5 and de-stabilization of the mRNA. GAPDH was used as a control for mRNA quality.
FBW7 is a key component of the SCF E3 ubiquitin ligase complex that has been reported to regulate MYC stability in response to UV-induced DNA damage (41). Thus, we set out to further test whether FBW7 regulates the destruction of SOX9. Co-expression of SOX9 with increasing amounts of FBW7α, an isoform of FBW7 that is localized primarily in the nucleoplasm, resulted in incrementally reduced SOX9 levels (Figure 3B). To assess if degradation of endogenous SOX9 is downstream of and specific to FBW7, we examined HCT116 isogenic cell lines that were either FBW7-wild-type or -null (by deletion of exon 5) (45). Endogenous SOX9 levels were higher in the FBW7-null cells (Figure 3C). Moreover, after UV-induced DNA damage, SOX9 was depleted in FBW7-wild-type cells, but not in FBW7-null cells (Figure 3C). In order to exclude clonal selection, and to confirm specificity, we re-introduced FBW7 into HCT116 FBW7-null cells, and this fully restored SOX9 degradation (Figure 3D). We also confirmed the deletion of exon 5 FBW7 (Figure 3E), which results in FBW7 mRNA degradation (45). These data in total confirm that FBW7 targets SOX9 for degradation and accelerates SOX9 loss after UV-induced DNA damage.
Phosphorylation by GSK3β triggers FBW7-mediated SOX9 depletion
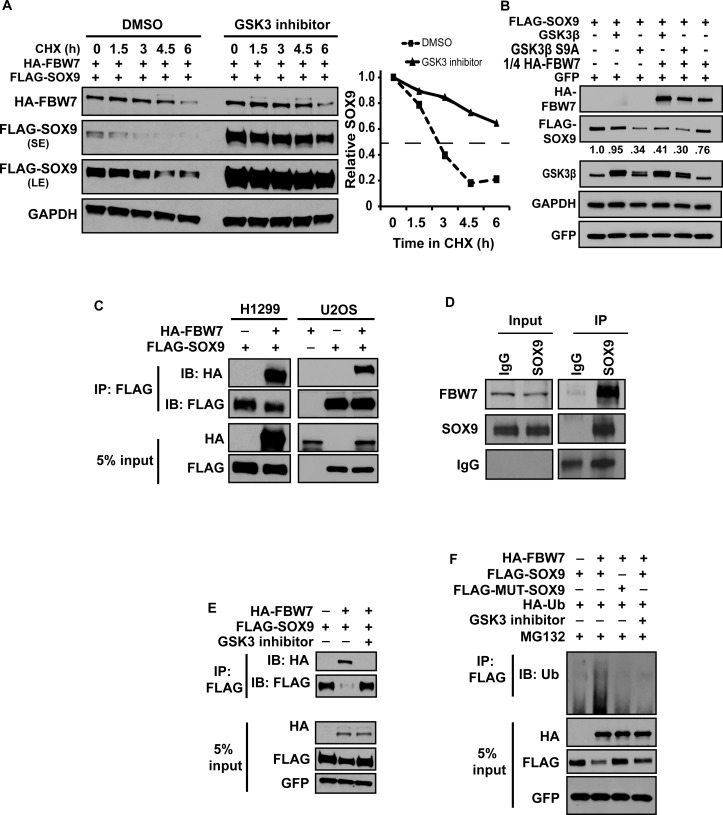
Based on our findings that inhibition of GSK3β prevents DNA damage-induced SOX9 degradation, and that FBW7 targets SOX9 for degradation after DNA damage, we reasoned that phosphorylation of SOX9 by GSK3β is required for FBW7-mediated SOX9 degradation. When SOX9 and FBW7 were transiently co-overexpressed, the half-life of SOX9 was about 2 h, but it was increased to more than 6 h when GSK3 was inhibited (Figure 4A). In line with this, SOX9 was decreased when constitutively active mutant (S9A) GSK3β was expressed, in comparison to wild-type GSK3β (Figure 4B). Moreover, when cells were co-transfected with a reduced molar ratio of FBW7 to SOX9 (1:4) that did not deplete SOX9 levels substantially, overexpression of either wild-type GSK3β or constitutively active GSK3β caused a further reduction in SOX9 (Figure 4B), demonstrating that SOX9 destabilization mediated by FBW7 is enhanced by GSK3β.
Figure 4.
UV-induced SOX9 degradation requires GSK3 activity. (A) GSK3 enhances FBW7α-mediated SOX9 degradation. H1299 cells were co-transfected with HA-FBW7α and FLAG-SOX9, and were either untreated or treated with CHIR-99021 in the presence of CHX. Lysates were collected at varying times and probed for FLAG-SOX9 and HA-FBW7. The graph on the right depicts the half-life of SOX9, determined by densitometric quantitation. GAPDH was used as a normalization control. SE, short film exposure; LE, long film exposure. (B) H1299 cells were transfected with FLAG-SOX9 with or without wild-type (WT) GSK3β or constitutively active S9A GSK3β and with or without HA-FBW7α, which was transfected at a 1:4 molar ratio to the other plasmids. GFP was transfected as a control. Lysates were immunoblotted for FLAG-SOX9, HA-FBW7, GFP and GSK3β. Levels of FLAG-SOX9 were quantified by densitometry, normalized to GAPDH. (C) FBW7α interacts with SOX9. H1299 and U2OS cells were transfected with HA-FBW7α along with FLAG-SOX9. Lysates were immunoprecipitated with anti-FLAG and immunoblotted for FLAG-SOX9 and HA-FBW7. A Western blot of the 5% input lysates is also shown (D) Endogenous FBW7 interacts with endogenous SOX9. SOX9 was immunoprecipitated (IP) from H1299 lysates, and proteins were immunoblotted for FBW7 and SOX9, as well as IgG. A Western blot of the 5% input lysates is also shown. (E) Interaction between FBW7α and SOX9 is dependent on GSK3. H1299 cells were transfected with HA-FBW7α along with FLAG-SOX9 and GFP as a transfection control, either untreated or treated with the GSK3 inhibitor, CHIR99021. Lysates were immunoprecipitated with anti-FLAG and immunoblotted for FLAG-SOX9 and HA-FBW7. A Western blot of the 5% input lysates is also shown. (F) FBW7 targets SOX9 for ubiquitination. H1299 cells were co-transfected with either FLAG-SOX9 or FLAG-MUT-SOX9 (CPD-mutant), and HA-Ub with or without HA-FBW7α. Cells were treated with MG132 with or without CHIR99021 for 6 h before collection of lysates. GFP was transfected as a control. Denatured lysates were immunoprecipitated with anti-FLAG and immunoblotted for Ubiquitin. The 5% input lysates were also immunoblotted for HA, FLAG and GFP.
FBW7 interacts with SOX9 in vivo
Direct binding to FBW7 is necessary for the destruction of its substrates (40,63). Thus, the correlation between FBW7 and SOX9 prompted us to investigate whether FBW7 interacts with SOX9. We performed a co-immunoprecipitation on lysates from cells in which FLAG-SOX9 was co-transfected with or without HA-FBW7α in the presence of MG132. FBW7 was shown to be pulled down by SOX9 (Figure 4C). Moreover, endogenous FBW7 was also pulled down by immunoprecipitation of endogenous SOX9 (Figure 4D), further confirming the interaction between FBW7 and SOX9. Additionally, inhibition of GSK3 abolished the interaction between SOX9 and FBW7α (Figure 4E). To test whether the interaction of FBW7 with SOX9 results in ubiquitination of SOX9, we co-overexpressed ubiquitin with FLAG-SOX9 in the presence of MG132, and we immunoprecipitated SOX9 under denatured conditions. Poly-ubiquitinated SOX9 was increased when cells were co-transfected with FBW7α, and moreover, ubiquitination of SOX9 was abrogated after the addition of the GSK3 inhibitor (Figure 4F). These data confirmed that FBW7α targets SOX9 for ubiquitination and proteasomal destruction in a GSK3β-dependent manner.
FBW7 binds to the K2 transactivation domain of SOX9
We next mapped the regions of SOX9 that are required for its interaction with FBW7. We scanned the SOX9 amino acid sequence for CDC4 phosphodegron (CPD) motifs that are characteristic of FBW7α substrates, where the ‘0’ and ‘+4’ sites are most frequently a phosphorylated threonine or serine, and the ‘0’ site is typically phosphorylated by GSK3β (40). We identified four potential motifs (Figure 5A). To ascertain if one or more of the identified motifs are functional CPDs, we mutated the threonine and serine residues within each motif to alanine (underlined in Figure 5A). We then co-expressed wild-type or mutant SOX9 (CPD motifs 1–4) along with FBW7. Only mutation of one putative CPD motif, comprising amino acids 236–240, prevented the decrease in SOX9 levels caused by FBW7α (Figure 5B). The CPD is conserved across vertebrates (Figure 5C) and is located in the K2 domain of SOX9 that has potential transactivation and protein binding functions (64). As expected, the CPD-mutant SOX9 protein was more stable than CPD-WT SOX9 when co-overexpressed with FBW7 (Supplementary Figure S3). In a co-immunoprecipitation assay, wild-type SOX9 but not the CPD-mutant SOX9 was able to interact with FBW7 in vivo (Figure 5D), confirming that FBW7 interacts with SOX9 within SOX9's K2 domain, at T236-T240. To test if the CPD motif is phosphorylated in vivo, we immunoprecipitated endogenous SOX9 and performed liquid chromatography-tandem mass spectrometry, which indicated that T236, as well as either T239 or T240, are phosphorylated (Supplementary Figure S4A). We next individually mutated T236, T239 and T240. Mutation of T236 to Ala substantially attenuated, and mutation of T240 to Ala partly attenuated the reduction of SOX9 by FBW7; however, mutation of T239 to Ala had essentially no effect (Supplementary Figure S4B), demonstrating that phosphorylation of T236 and T240 are key steps facilitating the recognition of SOX9 by FBW7. Moreover, P-T236 was decreased after the addition of the GSK3β inhibitor, CHIR-99021 (Figure 5E). We could also detect P-T236 in overexpressed wild-type SOX9 that was decreased by GSK3β inhibition. However, as expected, P-T236 was not detected in the CPD-mutant SOX9 (Figure 5E). Last, a database search of PhosphoSitePlus (http://www.phosphosite.org/) revealed that phosphorylation at T236-SOX9 was previously reported (65).
Figure 5.
Phosphorylation of SOX9 in the K2 domain by GSK3β triggers SOX9 destruction by FBW7α. (A) Alignment of the putative SOX9 consensus phosphodegron (CPD) sites (Motifs 1–4) with the FBW7 CPD sequence known for other FBW7 target proteins. The SOX9 serine and threonine residues that were mutated to alanine are underlined. (B) The FBW7α CPD binding site on SOX9 comprises amino acids 236–240. The top schematic depicts the locations of the DIM, HMG, K2, PQA and TA domains of SOX9, as well as the locations of the putative CPD motifs. Below, H1299 cells were transfected with WT FLAG-SOX9 or the indicated FLAG-SOX9 mutants (M1–M4) with GFP and with or without HA-FBW7α. Lysates were immunoblotted for GFP, FLAG and HA. GAPDH was used as a normalization control. SE, short film exposure; LE, long film exposure. (C) The SOX9 CPD sequence targeted by FBW7α is conserved across different species. The sequence shown for zebrafish is the SOX9a ortholog. (D) SOX9 interacts with FBW7α at the K2 domain CPD motif. H1299 cells were co-transfected with HA-FBW7α and either wild-type FLAG-SOX9 or CPD-mutant FLAG-MUT-SOX9. Lysates were immunoprecipitated with anti-FLAG and immunoblotted for HA and FLAG. A Western blot of lysates comprising 5% of input is also shown for HA and FLAG. (E) The SOX9 K2 domain is phosphorylated by GSK3β at T236. Beas2b cells were transfected with the FLAG-control, wild-type FLAG-SOX9 or CPD-mutant FLAG-MUT-SOX9. Cells were treated with 40 μM CHIR-99021 for 8 h, 48 h after transfection. A Western blot of lysates is shown for SOX9, P-T236-SOX9 and GAPDH. SE, short film exposure; LE, long film exposure. (F) FBW7α directly interacts with SOX9 in vitro. A binding assay was performed on HA-FBW7α protein and a GST-tagged WT SOX9 fragment comprising amino acids 186–318, or a GST-tagged SOX9 mutated at the CPD motif, with or without human recombinant GSK3β (rhGSK3β). The input and GST-immunoprecipitated proteins were immunoblotted for HA-FBW7. Total GST and GST-SOX9 were visualized by a coomassie blue stain. M, protein marker. (G) GSK3β directly phosphorylates SOX9. An in vitro kinase assay was performed using human recombinant GSK3β and purified GST, wild-type GST-SOX9 or a CPD-mutant GST-SOX9. 32P GST-SOX9 is shown. Total GST and GST-SOX9 were visualized by a coomassie blue stain. Presence of GSK3β was confirmed by immunoblotting. Graph represents the average phosphorylation of CPD-mutant SOX relative to wild-type SOX9 in three independent experiments. Error bars represent the standard deviation. A two-tailed Student's t-test was used to compare groups. **, P < 0.01.
Because alterations in protein stability in vivo could reflect indirect effects of FBW7, we performed an in vitro binding assay. Purified FBW7α and a fragment of WT SOX9 (comprising amino acids 181–320) were able to directly interact; and this interaction was lost when the CPD site was mutated (Figure 5F). To test if GSK3β directly phosphorylates SOX9 within the FBW7 consensus binding motif, we performed an in vitro kinase assay using a purified fragment of WT or CPD-mutant SOX9 and recombinant human GSK3. Wild-type SOX9 was phosphorylated by GSK3, and the level of phosphorylation was reduced by 50% in the CPD mutant SOX9 (Figure 5G), demonstrating that GSK3 phosphorylates SOX9 within the FBW7 binding motif, although it is unclear if GSK3 phosphorylates T236, T240 or both. Diminished phosphorylation of the CPD-mutant protein suggested the presence of an additional GSK3β phosphorylation site(s) on SOX9. Last, the CPD-mutant form of SOX9 was unable to be ubiquitinated when co-expressed with FBW7 (Figure 4F). These data in total confirmed that the FBW7 CPD motif in SOX9 spans amino acids 236–240, that T236 is phosphorylated by GSK3β, that FBW7 targets GSK3-phosphorylated SOX9 for degradation, and that FBW7 regulates SOX9 stability under normal physiological conditions.
SOX9 depletion after DNA damage contributes to cell death
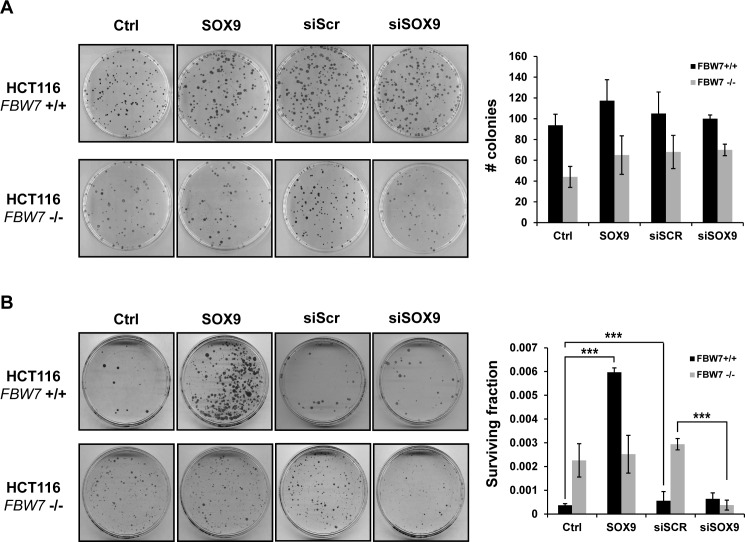
SOX9 upregulation has been shown to bypass cellular senescence or prevent apoptosis in non-tumor cells (36,66–68), but there has been no established link between SOX9 and cell survival in cancer. We surmised that the active elimination of SOX9 contributes to cell death during the DNA damage response. Exogenously introduced SOX9 driven by a strong CMV promoter was not depleted after UV exposure (Supplementary Figure S1C), which allowed us to functionally test the role of persistent SOX9 levels after UV irradiation. Under basal conditions, transient SOX9 overexpression or siRNA-mediated knock-down of SOX9 had no effect on altering colony-formation ability (Figure 6A). However, overexpression of SOX9 increased the fraction of HCT116 FBW7-wild-type cells that survived after UV irradiation by 10-fold (Figure 6B). Moreover, HCT116 FBW7-null cells had an increased colony-forming ability as compared to HCT116 FBW7-wild-type cells, though the colony sizes were smaller (Figure 6B). In HCT116 FBW7-null cells in which SOX9 protein levels are unaltered after UV irradiation (Figure 3C), transient upregulation of SOX9 had no effect on cell survival, as evidenced by no increase in the surviving fraction after UV irradiation (Figure 6B). It is important to note that wild-type SOX9 was overexpressed, and perhaps overexpression of un-degradable SOX9 that is mutated at the FBW7 CPD site would have a different effect on cell survival. We next transiently knocked down SOX9 prior to UV irradiation to rescue the inability to deplete SOX9 caused by the loss of FBW7, and this completely abolished the increased colony-forming ability after UV irradiation. Transient knock-down of SOX9 in HCT116 FBW7-wild-type cells that deplete SOX9 after UV exposure had no significant effect on the surviving fraction after UV exposure (Figure 6B). This demonstrates that SOX9 is likely a participant in the role of FBW7 in mediating UV-induced cell death.
Figure 6.
Loss of SOX9 mediated by FBW7 after DNA damage mediates long-term cell survival. Cell clonogenic assays were performed on HCT116 FBW7 wild-type (+/+) and null (-/-) cells with (A) no treatment or (B) after exposure to UV irradiation at 40 J/m2. Cells were transiently transfected with SOX9 or vector control (Ctrl), or with siRNA against SOX9 (siSOX9) or a scrambled siRNA (siSCR), 24 h before irradiation. Plates were stained with coomassie blue. Graphs to the right represent the average number of colonies out of (A) 200 cells per plate, or the average fraction of surviving cells out of (B) 50 000 cells in three independent experiments. A two-tailed Student's t-test was used to compare groups. Error bars show standard deviation; ***P < 0.0001.
DISCUSSION
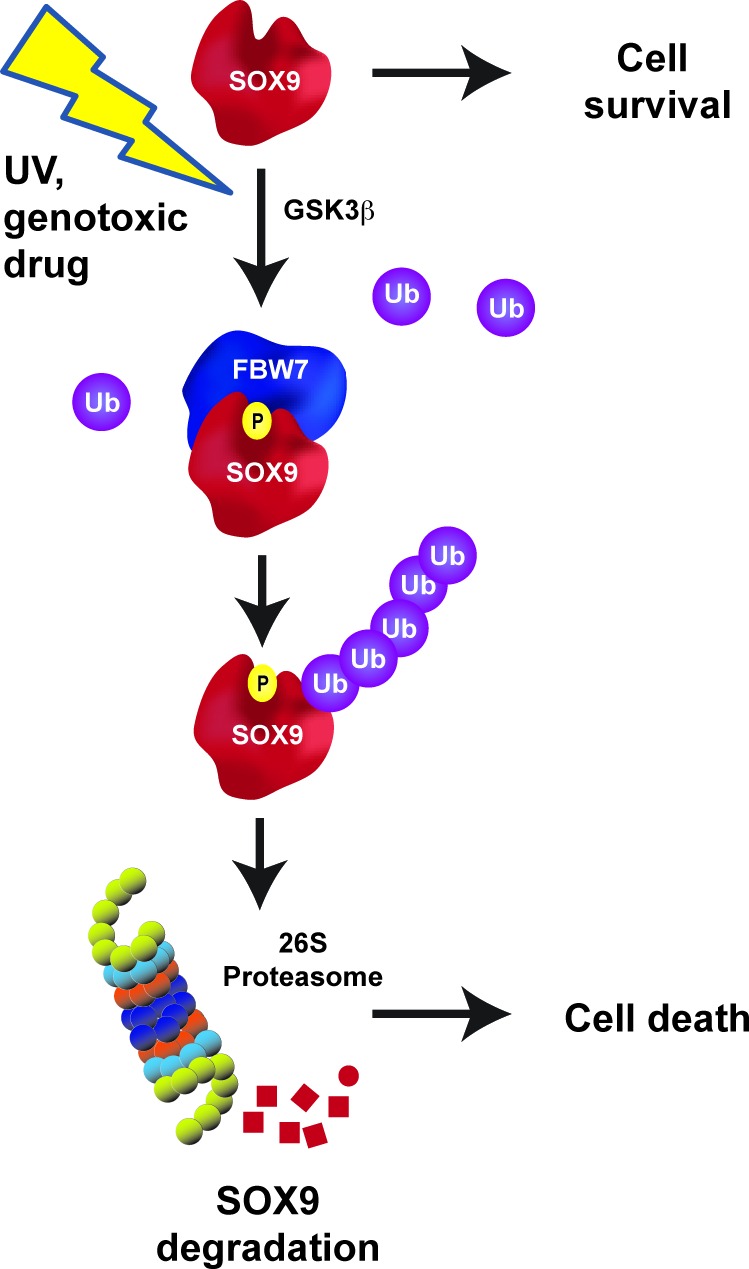
In this study we identified an E3 ligase, FBW7, as a key regulator of SOX9 depletion after genotoxic stress. We present evidence that the induction of DNA damage by cytotoxic chemotherapeutic agents or UV-irradiation causes a rapid and robust loss of SOX9 across multiple cancer cell types and in immortalized human epithelial cells. We also show that SOX9 depletion after genotoxic stress is independent of many classical DNA damage response pathways including p53, ATR and ATM. We propose a model in which UV-induced SOX9 ubiquitination and proteasomal degradation requires phosphorylation by GSK3β. Failure of GSK3β to phosphorylate SOX9 prevents the binding of FBW7 to the consensus CPD site on SOX9, located within the K2 transactivation domain, and prevents the degradation of SOX9 both under normal conditions and after DNA damage. Indeed, Sangfelt and colleagues also observed that SOX9 protein is stabilized by FBW7 repression in meduloblastoma under normal conditions (69). In our study, loss of SOX9 after DNA damage participates in UV damage-induced cell death, because overexpressed SOX9 increases cell survival even after genotoxic stress (Figure 7). These results suggest that SOX9 is a new signaling link between the FBW7 tumor suppressor pathway and the cellular response to DNA damage.
Figure 7.
Schematic model of the FBW7-SOX9 pathway under basal conditions and after DNA damage. Under basal conditions, SOX9 protein stability is tightly controlled by a phosphorylation by GSK3β within the CPD motif located within the K2 domain of SOX9. Phosphorylated SOX9 is recognized by FBW7 that targets SOX9 for proteasomal degradation.
Exposure to UV irradiation or genotoxic chemicals causes a wide range of changes that serve to protect the cell from permanent DNA damage. The outcome of these changes is generally either DNA repair or apoptosis, depending on the degree of damage. DNA-damaged cells induce a dose-dependent transcriptional regulation of cell cycle proteins, and numerous cell cycle genes are suppressed after 12–24 h, including DNA polymerase, Fen1, primase, CDK7, cyclin B1, cyclin A and CDC6, among others (70). The activity of cell cycle-related proteins would be inhibited faster by active proteolysis. UV-irradiation causes degradation of Myc, p21 (41,54), as well as the replication proteins HBO1, PLK1, CDT1, CDC6 and MCM10 (42,71–74). Protein depletion is highly specific; levels of other cell cycle proteins are unchanged after UV exposure, such as other MCM family members, or the replication initiators ORC1–ORC5 (74), and we did not observe any changes in levels of activated Notch1 (Figure 1D). Due to the high selectivity of proteins that are degraded, it is highly plausible that the active depletion of SOX9 after UV- or drug-induced DNA damage serves a crucial purpose for the cellular response to genotoxic stress.
The kinetics of SOX9 degradation raises the possibility of the involvement of the apoptotic pathway. There are some indications in the literature that SOX9 overexpression may prevent apoptosis after DNA damage. For example, microarray data from murine cells with gut-derived intestinal epithelial stem cell characteristics, and high SOX9 expression, express genes associated with cell cycle re-entry, DNA repair and anti-apoptosis (67). In a more recent study, knock-out of SOX9 within the intestinal crypt increased the incidence of apoptosis after irradiation (36). Moreover, in the developing limb buds, conditional knockout of SOX9 caused extensive apoptosis (68). Additional studies would be needed to determine if apoptosis is a functional consequence of SOX9 depletion after DNA damage. In addition, it would be worthwhile to examine what role, if any, activation of apoptosis has on the regulation of SOX9.
The cellular response to UVC exposure, best observed by activation of the S-phase checkpoint, occurs at fluences as low as 1 J/m2, much lower than the fluence needed to degrade SOX9. This raises the question of why are higher levels of UV (>5 J/m2) required for SOX9 degradation? The observation we report for SOX9 has some similarities to p21, which is also degraded in response to UV irradiation (54,60,75–77). The amount of DNA damage required for the detection of p21 degradation is incompatible with long-term cell survival (78), yet it was determined that failure to degrade p21 facilitates the replication of damaged DNA (79). The authors speculated that the amount of UV exposure needed to degrade p21 total protein levels may be much higher than what is needed for p21's function at the molecular level. Thus, processes restricted to subcellular molecular processes at lower UV fluences may not be evident at the global level, and unaffected protein levels may not imply a lack of activation of a cellular response. Likewise, we predict that although relatively high levels of UV are required to observe depletion of total SOX9 protein levels, SOX9 might actively participate in the cellular response to sub-lethal amounts of DNA damage even when SOX9 degradation is not detectable globally.
This study raises numerous additional questions. For example, what is the mechanistic role of SOX9 loss in the context of the DNA damage response? If the FBW7 axis targets SOX9 for degradation under basal conditions, what is the triggering event that accelerates the loss of SOX9 after genotoxic stress? Why is SOX9 depleted after exposure to UV, or treatment with doxorubicin or cisplatin, but increased after γ-irradiation, when all of these DNA damaging agents induce stalled replication forks? All of the aforementioned genotoxic agents elicit different types of DNA damage that converge on replication fork stalling and collapse. Could SOX9 participate in a cellular response that is upstream of that convergence? Does SOX9 have cellular response functions in addition to its transcriptional activity? Why would a stem cell and developmental gene be actively degraded after DNA damage when the DNA damage response is a universal mechanism? Because SOX9 is only expressed in certain cell types, perhaps a function of SOX9 is to participate in the DNA damage response only within the stem and progenitor cell pool, or during development, that is utilized by cancer cells for their survival advantage. It is known that progression through the cell cycle after genotoxic stress causes unperturbed DNA replication, prevents DNA break repair and leads to genomic instability. One explanation for the active and rapid degradation of SOX9 after exposure to DNA damaging agents could be to prevent the incorporation of permanent DNA replication errors into the genome. SOX9 has been reported to transcriptionally upregulate p21 expression (18,80). It is possible that the rapid depletion of SOX9 facilitates a gene expression profile that enhances the DNA damage response. It would also be interesting to test if the inability to degrade SOX9 after DNA damage, caused either by overexpression of CPD-mutant SOX9 or by the loss of functional FBW7, enhances genomic instability or resistance to chemotherapy. Given the prevalence of FBW7 mutations and SOX9 overexpression in multiple tumor types, such a scenario would suggest that targeting the FBW7-SOX9 axis could have therapeutic implications. Moreover, failure to deplete SOX9 in tumor cells during chemotherapy could potentially contribute to tumor growth and progression.
Supplementary Material
Acknowledgments
The authors thank Bert Vogelstein for FBW7 and HCT116 knockout cell lines and Zhaohui Feng for the H460 sh-control and H460-p53shRNA cells. The authors also thank Wade Harper, Wenyi Wei, Martin Eilers and Jim Woodgett for providing plasmids.
Author contribution statement: X.H., W.L. and S.R.P. designed the study. X.H., W.L., R.S., J.J.S., X.F., C.K.T., K.M.M., H.E.S. and S.R.P. performed the research and analyzed the data. S.F.B., H.I., X.F.S.Z., Z.S., H.E.S., L.X.L. and S.R.P. directed and supervised the studies. S.R.P. wrote the manuscript and all co-authors edited the manuscript.
Footnotes
Present address: Xuehui Hong, Department of Gastrointestinal Surgery, Institute of Gastrointestinal Oncology, Zhongshan Hospital, Xiamen University, Xiamen, China.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Cancer Institute (NCI), National Institutes of Health (NIH) [R01CA190578 to S.R.P., R01CA156706, R01CA195612 to Z.S. and R01CA123391, R01CA166575, R01CA173519 to X.F.S.Z.]; Lung Cancer Research Foundation [to S.R.P.]; Shared Resource of The Cancer Institute of New Jersey [P30CA072720]; NCI Ruth L. Kirschstein National Research Service F30 Award [to K.M.M.]. Funding for open access charge: NCI [R01CA190578 to S.R.P.].
Conflict of interest statement. None declared.
REFERENCES
- 1.Gubbay J., Collignon J., Koopman P., Capel B., Economou A., Munsterberg A., Vivian N., Goodfellow P., Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- 2.Soullier S., Jay P., Poulat F., Vanacker J.M., Berta P., Laudet V. Diversification pattern of the HMG and SOX family members during evolution. J. Mol. Evol. 1999;48:517–527. doi: 10.1007/pl00006495. [DOI] [PubMed] [Google Scholar]
- 3.Kamachi Y., Kondoh H. Sox proteins: Regulators of cell fate specification and differentiation. Development. 2013;140:4129–4144. doi: 10.1242/dev.091793. [DOI] [PubMed] [Google Scholar]
- 4.Schafer A.J., Foster J.W., Kwok C., Weller P.A., Guioli S., Goodfellow P.N. Campomelic dysplasia with XY sex reversal: diverse phenotypes resulting from mutations in a single gene. Ann. N. Y. Acad. Sci. 1996;785:137–149. doi: 10.1111/j.1749-6632.1996.tb56252.x. [DOI] [PubMed] [Google Scholar]
- 5.Mansour S., Offiah A.C., McDowall S., Sim P., Tolmie J., Hall C. The phenotype of survivors of campomelic dysplasia. J. Med. Genet. 2002;39:597–602. doi: 10.1136/jmg.39.8.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mansour S., Hall C.M., Pembrey M.E., Young I.D. A clinical and genetic study of campomelic dysplasia. J. Med. Genet. 1995;32:415–420. doi: 10.1136/jmg.32.6.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., van de Kant H.J., Wegner M., de Rooij D.G., Behringer R.R., Schedl A. Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development. 2004;131:1891–1901. doi: 10.1242/dev.01087. [DOI] [PubMed] [Google Scholar]
- 8.Akiyama H. Control of chondrogenesis by the transcription factor Sox9. Mod. Rheumatol. 2008;18:213–219. doi: 10.1007/s10165-008-0048-x. [DOI] [PubMed] [Google Scholar]
- 9.Lefebvre V., Dumitriu B., Penzo-Mendez A., Han Y., Pallavi B. Control of cell fate and differentiation by Sry-related high-mobility-group box (Sox) transcription factors. Int. J. Biochem. Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rockich B.E., Hrycaj S.M., Shih H.P., Nagy M.S., Ferguson M.A., Kopp J.L., Sander M., Wellik D.M., Spence J.R. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Seymour P.A. Sox9: A master regulator of the pancreatic program. Rev. Diabet. Stud. 2014;11:51–83. doi: 10.1900/RDS.2014.11.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Blache P., van de Wetering M., Duluc I., Domon C., Berta P., Freund J.N., Clevers H., Jay P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004;166:37–47. doi: 10.1083/jcb.200311021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vidal V.P., Chaboissier M.C., Lutzkendorf S., Cotsarelis G., Mill P., Hui C.C., Ortonne N., Ortonne J.P., Schedl A. Sox9 is essential for outer root sheath differentiation and the formation of the hair stem cell compartment. Curr. Biol. 2005;15:1340–1351. doi: 10.1016/j.cub.2005.06.064. [DOI] [PubMed] [Google Scholar]
- 14.Kadaja M., Keyes B.E., Lin M., Pasolli H.A., Genander M., Polak L., Stokes N., Zheng D., Fuchs E. SOX9: a stem cell transcriptional regulator of secreted niche signaling factors. Genes Dev. 2014;28:328–341. doi: 10.1101/gad.233247.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang G., Lunardi A., Zhang J., Chen Z., Ala U., Webster K.A., Tay Y., Gonzalez-Billalabeitia E., Egia A., Shaffer D.R., et al. Zbtb7a suppresses prostate cancer through repression of a Sox9-dependent pathway for cellular senescence bypass and tumor invasion. Nat. Genet. 2013;45:739–746. doi: 10.1038/ng.2654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Drivdahl R., Haugk K.H., Sprenger C.C., Nelson P.S., Tennant M.K., Plymate S.R. Suppression of growth and tumorigenicity in the prostate tumor cell line M12 by overexpression of the transcription factor SOX9. Oncogene. 2004;23:4584–4593. doi: 10.1038/sj.onc.1207603. [DOI] [PubMed] [Google Scholar]
- 17.Kato N., Fukase M., Motoyama T. Expression of a transcription factor, SOX9, in Sertoli-stromal cell tumors of the ovary. Int. J. Gynecol. Pathol. 2004;23:180–181. doi: 10.1097/00004347-200404000-00014. [DOI] [PubMed] [Google Scholar]
- 18.Passeron T., Valencia J.C., Namiki T., Vieira W.D., Passeron H., Miyamura Y., Hearing V.J. Upregulation of SOX9 inhibits the growth of human and mouse melanomas and restores their sensitivity to retinoic acid. J. Clin. Invest. 2009;119:954–963. doi: 10.1172/JCI34015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Swartling F.J., Ferletta M., Kastemar M., Weiss W.A., Westermark B. Cyclic GMP-dependent protein kinase II inhibits cell proliferation, Sox9 expression and Akt phosphorylation in human glioma cell lines. Oncogene. 2009;28:3121–3131. doi: 10.1038/onc.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kopp J.L., von F.G., Mayes E., Liu F.F., Dubois C.L., Morris J.P., Pan F.C., Akiyama H., Wright C.V., Jensen K., et al. Identification of Sox9-dependent acinar-to-ductal reprogramming as the principal mechanism for initiation of pancreatic ductal adenocarcinoma. Cancer Cell. 2012;22:737–750. doi: 10.1016/j.ccr.2012.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo W., Keckesova Z., Donaher J.L., Shibue T., Tischler V., Reinhardt F., Itzkovitz S., Noske A., Zurrer-Hardi U., Bell G., et al. Slug and Sox9 cooperatively determine the mammary stem cell state. Cell. 2012;148:1015–1028. doi: 10.1016/j.cell.2012.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jiang S.S., Fang W.T., Hou Y.H., Huang S.F., Yen B.L., Chang J.L., Li S.M., Liu H.P., Liu Y.L., Huang C.T., et al. Upregulation of SOX9 in lung adenocarcinoma and its involvement in the regulation of cell growth and tumorigenicity. Clin. Cancer Res. 2010;16:4363–4373. doi: 10.1158/1078-0432.CCR-10-0138. [DOI] [PubMed] [Google Scholar]
- 23.Mead T.J., Wang Q., Bhattaram P., Dy P., Afelik S., Jensen J., Lefebvre V. A far-upstream (-70 kb) enhancer mediates Sox9 auto-regulation in somatic tissues during development and adult regeneration. Nucleic Acids Res. 2013;41:4459–4469. doi: 10.1093/nar/gkt140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ling S., Chang X., Schultz L., Lee T.K., Chaux A., Marchionni L., Netto G.J., Sidransky D., Berman D.M. An EGFR-ERK-SOX9 signaling cascade links urothelial development and regeneration to cancer. Cancer Res. 2011;71:3812–3821. doi: 10.1158/0008-5472.CAN-10-3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Es J.H., van Gijn M.E., Riccio O., van den Born M., Vooijs M., Begthel H., Cozijnsen M., Robine S., Winton D.J., Radtke F., et al. Notch/gamma-secretase inhibition turns proliferative cells in intestinal crypts and adenomas into goblet cells. Nature. 2005;435:959–963. doi: 10.1038/nature03659. [DOI] [PubMed] [Google Scholar]
- 26.Pritchett J., Harvey E., Athwal V., Berry A., Rowe C., Oakley F., Moles A., Mann D.A., Bobola N., Sharrocks A.D., et al. Osteopontin is a novel downstream target of SOX9 with diagnostic implications for progression of liver fibrosis in humans. Hepatology. 2012;56:1108–1116. doi: 10.1002/hep.25758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paganelli M., Nyabi O., Sid B., Evraerts J., El Malmi I., Heremans Y., Dolle L., Benton C., Calderon P.B., van Grunsven L., et al. Downregulation of Sox9 expression associates with hepatogenic differentiation of human liver mesenchymal stem/progenitor cells. Stem Cells Dev. 2014;23:1377–1391. doi: 10.1089/scd.2013.0169. [DOI] [PubMed] [Google Scholar]
- 28.Sun L., Mathews L.A., Cabarcas S.M., Zhang X., Yang A., Zhang Y., Young M.R., Klarmann K.D., Keller J.R., Farrar W.L. Epigenetic regulation of SOX9 by the NF-kappaB signaling pathway in pancreatic cancer stem cells. Stem Cells. 2013;31:1454–1466. doi: 10.1002/stem.1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Song S., Ajani J.A., Honjo S., Maru D.M., Chen Q., Scott A.W., Heallen T.R., Xiao L., Hofstetter W.L., Weston B., et al. Hippo coactivator YAP1 upregulates SOX9 and endows esophageal cancer cells with stem-like properties. Cancer Res. 2014;74:4170–4182. doi: 10.1158/0008-5472.CAN-13-3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larsimont J.C., Youssef K.K., Sanchez-Danes A., Sukumaran V., Defrance M., Delatte B., Liagre M., Baatsen P., Marine J.C., Lippens S., et al. Sox9 controls self-renewal of oncogene targeted cells and links tumor initiation and invasion. Cell Stem Cell. 2015;17:60–73. doi: 10.1016/j.stem.2015.05.008. [DOI] [PubMed] [Google Scholar]
- 31.Capaccione K.M., Hong X., Morgan K.M., Liu W., Bishop J.M., Liu L., Markert E., Deen M., Minerowicz C., Bertino J.R., et al. Sox9 mediates Notch1-induced mesenchymal features in lung adenocarcinoma. Oncotarget. 2014;5:3636–3650. doi: 10.18632/oncotarget.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jo A., Denduluri S., Zhang B., Wang Z., Yin L., Yan Z., Kang R., Shi L.L., Mok J., Lee M.J., et al. The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis. 2014;1:149–161. doi: 10.1016/j.gendis.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma F., Ye H., He H.H., Gerrin S.J., Chen S., Tanenbaum B.A., Cai C., Sowalsky A.G., He L., Wang H., et al. SOX9 drives WNT pathway activation in prostate cancer. J. Clin. Invest. 2016;126:1745–1758. doi: 10.1172/JCI78815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou C.H., Ye L.P., Ye S.X., Li Y., Zhang X.Y., Xu X.Y., Gong L.Y. Clinical significance of SOX9 in human non-small cell lung cancer progression and overall patient survival. J. Exp. Clin. Cancer Res. 2012;31:1–9. doi: 10.1186/1756-9966-31-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruun J., Kolberg M., Nesland J.M., Svindland A., Nesbakken A., Lothe R.A. Prognostic Significance of beta-Catenin, E-Cadherin, and SOX9 in Colorectal Cancer: Results from a Large Population-Representative Series. Front Oncol. 2014;4:1–16. doi: 10.3389/fonc.2014.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roche K.C., Gracz A.D., Liu X.F., Newton V., Akiyama H., Magness S.T. SOX9 Maintains Reserve Stem Cells and Preserves Radioresistance in Mouse Small Intestine. Gastroenterology. 2015;149:1553–1563. doi: 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li X.L., Chen X.Q., Zhang M.N., Chen N., Nie L., Xu M., Gong J., Shen P.F., Su Z.Z., Weng X., et al. SOX9 was involved in TKIs resistance in renal cell carcinoma via Raf/MEK/ERK signaling pathway. Int. J. Clin. Exp. Pathol. 2015;8:3871–3881. [PMC free article] [PubMed] [Google Scholar]
- 38.Shi G., Sohn K.C., Li Z., Choi D.K., Park Y.M., Kim J.H., Fan Y.M., Nam Y.H., Kim S., Im M., et al. Expression and functional role of Sox9 in human epidermal keratinocytes. PLoS. One. 2013;8:e54355. doi: 10.1371/journal.pone.0054355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang Z., Liu P., Inuzuka H., Wei W. Roles of F-box proteins in cancer. Nat. Rev. Cancer. 2014;14:233–247. doi: 10.1038/nrc3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davis R.J., Welcker M., Clurman B.E. Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell. 2014;26:455–464. doi: 10.1016/j.ccell.2014.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Popov N., Herold S., Llamazares M., Schulein C., Eilers M. Fbw7 and Usp28 regulate myc protein stability in response to DNA damage. Cell Cycle. 2007;6:2327–2331. doi: 10.4161/cc.6.19.4804. [DOI] [PubMed] [Google Scholar]
- 42.Giraldez S., Herrero-Ruiz J., Mora-Santos M., Japon M.A., Tortolero M., Romero F. SCF(FBXW7alpha) modulates the intra-S-phase DNA-damage checkpoint by regulating Polo like kinase-1 stability. Oncotarget. 2014;5:4370–4383. doi: 10.18632/oncotarget.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Q., Karnak D., Tan M., Lawrence T.S., Morgan M.A., Sun Y. FBXW7 facilitates nonhomologous end-joining via K63-linked polyubiquitylation of XRCC4. Mol. Cell. 2016;61:419–433. doi: 10.1016/j.molcel.2015.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hong X., Liu W., Inuzuka H., Liu L., Pine S.R. Abstract 1957: Negative regulation of Sox9 by glycogen synthase kinase 3 beta phosphorylation and SCFFbw7-dependent ubiquitination in cancer. Cancer Res. 2015;75:1957. [Google Scholar]
- 45.Rajagopalan H., Jallepalli P.V., Rago C., Velculescu V.E., Kinzler K.W., Vogelstein B., Lengauer C. Inactivation of hCDC4 can cause chromosomal instability. Nature. 2004;428:77–81. doi: 10.1038/nature02313. [DOI] [PubMed] [Google Scholar]
- 46.Zhang C., Lin M., Wu R., Wang X., Yang B., Levine A.J., Hu W., Feng Z. Parkin, a p53 target gene, mediates the role of p53 in glucose metabolism and the Warburg effect. Proc. Natl. Acad. Sci. U.S.A. 2011;108:16259–16264. doi: 10.1073/pnas.1113884108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R.S., Zhai B., Wan L., Gutierrez A., Lau A.W., et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature. 2011;471:104–109. doi: 10.1038/nature09732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.He X., Saint-Jeannet J.P., Woodgett J.R., Varmus H.E., Dawid I.B. Glycogen synthase kinase-3 and dorsoventral patterning in Xenopus embryos. Nature. 1995;374:617–622. doi: 10.1038/374617a0. [DOI] [PubMed] [Google Scholar]
- 49.Xie X.J., Hsu F.N., Gao X., Xu W., Ni J.Q., Xing Y., Huang L., Hsiao H.C., Zheng H., Wang C., et al. CDK8-Cyclin C mediates nutritional regulation of developmental transitions through the ecdysone receptor in drosophila. PLoS Biol. 2015;13:e1002207. doi: 10.1371/journal.pbio.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fukushima H., Matsumoto A., Inuzuka H., Zhai B., Lau A.W., Wan L., Gao D., Shaik S., Yuan M., Gygi S.P., et al. SCF(Fbw7) modulates the NFkB signaling pathway by targeting NFkB2 for ubiquitination and destruction. Cell Rep. 2012;1:434–443. doi: 10.1016/j.celrep.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Siddik Z.H. Cisplatin: mode of cytotoxic action and molecular basis of resistance. Oncogene. 2003;22:7265–7279. doi: 10.1038/sj.onc.1206933. [DOI] [PubMed] [Google Scholar]
- 52.Yang F., Teves S.S., Kemp C.J., Henikoff S. Doxorubicin, DNA torsion and chromatin dynamics. Biochim. Biophys. Acta. 2014;1845:84–89. doi: 10.1016/j.bbcan.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martini S., Bernoth K., Main H., Ortega G.D., Lendahl U., Just U., Schwanbeck R. A critical role for Sox9 in notch-induced astrogliogenesis and stem cell maintenance. Stem Cells. 2013;31:741–751. doi: 10.1002/stem.1320. [DOI] [PubMed] [Google Scholar]
- 54.Bendjennat M., Boulaire J., Jascur T., Brickner H., Barbier V., Sarasin A., Fotedar A., Fotedar R. UV irradiation triggers ubiquitin-dependent degradation of p21(WAF1) to promote DNA repair. Cell. 2003;114:599–610. doi: 10.1016/j.cell.2003.08.001. [DOI] [PubMed] [Google Scholar]
- 55.Lakin N.D., Hann B.C., Jackson S.P. The ataxia-telangiectasia related protein ATR mediates DNA-dependent phosphorylation of p53. Oncogene. 1999;18:3989–3995. doi: 10.1038/sj.onc.1202973. [DOI] [PubMed] [Google Scholar]
- 56.Goldstein M., Kastan M.B. The DNA damage response: Implications for tumor responses to radiation and chemotherapy. Annu. Rev. Med. 2015;66:129–143. doi: 10.1146/annurev-med-081313-121208. [DOI] [PubMed] [Google Scholar]
- 57.Stokes M.P., Rush J., Macneill J., Ren J.M., Sprott K., Nardone J., Yang V., Beausoleil S.A., Gygi S.P., Livingstone M., et al. Profiling of UV-induced ATM/ATR signaling pathways. Proc. Natl. Acad. Sci. U.S.A. 2007;104:19855–19860. doi: 10.1073/pnas.0707579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kirkpatrick D.S., Bustos D.J., Dogan T., Chan J., Phu L., Young A., Friedman L.S., Belvin M., Song Q., Bakalarski C.E., et al. Phosphoproteomic characterization of DNA damage response in melanoma cells following MEK/PI3K dual inhibition. Proc. Natl. Acad. Sci. U.S.A. 2013;110:19426–19431. doi: 10.1073/pnas.1309473110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Searle J.S., Schollaert K.L., Wilkins B.J., Sanchez Y. The DNA damage checkpoint and PKA pathways converge on APC substrates and Cdc20 to regulate mitotic progression. Nat. Cell Biol. 2004;6:138–145. doi: 10.1038/ncb1092. [DOI] [PubMed] [Google Scholar]
- 60.Lee J.Y., Yu S.J., Park Y.G., Kim J., Sohn J. Glycogen synthase kinase 3beta phosphorylates p21WAF1/CIP1 for proteasomal degradation after UV irradiation. Mol. Cell. Biol. 2007;27:3187–3198. doi: 10.1128/MCB.01461-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ku B.M., Lee Y.K., Jeong J.Y., Ryu J., Choi J., Kim J.S., Cho Y.W., Roh G.S., Kim H.J., Cho G.J., et al. Caffeine inhibits cell proliferation and regulates PKA/GSK3beta pathways in U87MG human glioma cells. Mol. Cells. 2011;31:275–279. doi: 10.1007/s10059-011-0027-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Peck B.C., Sincavage J., Feinstein S., Mah A.T., Simmons J.G., Lund P.K., Sethupathy P. miR-30 family controls proliferation and differentiation of intestinal epithelial cell models by directing a broad gene expression program that includes SOX9 and the ubiquitin ligase pathway. J. Biol. Chem. 2016;291:15975–15984. doi: 10.1074/jbc.M116.733733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wang Z., Inuzuka H., Fukushima H., Wan L., Gao D., Shaik S., Sarkar F.H., Wei W. Emerging roles of the FBW7 tumour suppressor in stem cell differentiation. EMBO Rep. 2012;13:36–43. doi: 10.1038/embor.2011.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Schreiner S., Cossais F., Fischer K., Scholz S., Bosl M.R., Holtmann B., Sendtner M., Wegner M. Hypomorphic Sox10 alleles reveal novel protein functions and unravel developmental differences in glial lineages. Development. 2007;134:3271–3281. doi: 10.1242/dev.003350. [DOI] [PubMed] [Google Scholar]
- 65.Yi T., Zhai B., Yu Y., Kiyotsugu Y., Raschle T., Etzkorn M., Seo H.C., Nagiec M., Luna R.E., Reinherz E.L., et al. Quantitative phosphoproteomic analysis reveals system-wide signaling pathways downstream of SDF-1/CXCR4 in breast cancer stem cells. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E2182–E2190. doi: 10.1073/pnas.1404943111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Matheu A., Collado M., Wise C., Manterola L., Cekaite L., Tye A.J., Canamero M., Bujanda L., Schedl A., Cheah K.S., et al. Oncogenicity of the developmental transcription factor Sox9. Cancer Res. 2012;72:1301–1315. doi: 10.1158/0008-5472.CAN-11-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van L.L., Santoro M.A., Krebs A.E., Mah A.T., Dehmer J.J., Gracz A.D., Scull B.P., McNaughton K., Magness S.T., Lund P.K. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am. J. Physiol. Gastrointest. Liver Physiol. 2012;302:G1111–G1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Akiyama H., Chaboissier M.C., Martin J.F., Schedl A., de C.B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahmanto A.S., Savov V., Brunner A., Bolin S., Weishaupt H., Malyukova A., Rosén G., Čančer M., Hutter S., Sundström A., et al. FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO. J. 2016 doi: 10.15252/embj.201693889. doi:10.15252/embj.201693889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bertoli C., Skotheim J.M., de Bruin R.A. Control of cell cycle transcription during G1 and S phases. Nat. Rev. Mol. Cell Biol. 2013;14:518–528. doi: 10.1038/nrm3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Matsunuma R., Niida H., Ohhata T., Kitagawa K., Sakai S., Uchida C., Shiotani B., Matsumoto M., Nakayama K.I., Ogura H., et al. UV damage-induced phosphorylation of HBO1 triggers CRL4DDB2-mediated degradation to regulate cell proliferation. Mol. Cell. Biol. 2015;36:394–406. doi: 10.1128/MCB.00809-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Higa L.A., Mihaylov I.S., Banks D.P., Zheng J., Zhang H. Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 2003;5:1008–1015. doi: 10.1038/ncb1061. [DOI] [PubMed] [Google Scholar]
- 73.Hall J.R., Kow E., Nevis K.R., Lu C.K., Luce K.S., Zhong Q., Cook J.G. Cdc6 stability is regulated by the Huwe1 ubiquitin ligase after DNA damage. Mol. Biol. Cell. 2007;18:3340–3350. doi: 10.1091/mbc.E07-02-0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma A., Kaur M., Kar A., Ranade S.M., Saxena S. Ultraviolet radiation stress triggers the down-regulation of essential replication factor Mcm10. J. Biol. Chem. 2010;285:8352–8362. doi: 10.1074/jbc.M109.041129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H., Zeng S.X., Lu H. UV Induces p21 rapid turnover independently of ubiquitin and Skp2. J. Biol. Chem. 2006;281:26876–26883. doi: 10.1074/jbc.M605366200. [DOI] [PubMed] [Google Scholar]
- 76.Soria G., Podhajcer O., Prives C., Gottifredi V. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 2006;25:2829–2838. doi: 10.1038/sj.onc.1209315. [DOI] [PubMed] [Google Scholar]
- 77.Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 2008;22:2496–2506. doi: 10.1101/gad.1676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soria G., Gottifredi V. PCNA-coupled p21 degradation after DNA damage: The exception that confirms the rule? DNA Repair (Amst) 2010;9:358–364. doi: 10.1016/j.dnarep.2009.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mansilla S.F., Soria G., Vallerga M.B., Habif M., Martinez-Lopez W., Prives C., Gottifredi V. UV-triggered p21 degradation facilitates damaged-DNA replication and preserves genomic stability. Nucleic Acids Res. 2013;41:6942–6951. doi: 10.1093/nar/gkt475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wang H.Y., Lian P., Zheng P.S. SOX9, a potential tumor suppressor in cervical cancer, transactivates p21WAF1/CIP1 and suppresses cervical tumor growth. Oncotarget. 2015;6:20711–20722. doi: 10.18632/oncotarget.4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.