Abstract
Transcription factors have been considered undruggable, but this paradigm has been recently challenged. DNA binding natural product mithramycin (MTM) is a potent antagonist of oncogenic transcription factor EWS–FLI1. Structural details of MTM recognition of DNA, including the FLI1 binding sequence GGA(A/T), are needed to understand how MTM interferes with EWS–FLI1. We report a crystal structure of an MTM analogue MTM SA–Trp bound to a DNA oligomer containing a site GGCC, and two structures of a novel analogue MTM SA–Phe in complex with DNA. MTM SA–Phe is bound to sites AGGG and GGGT on one DNA, and to AGGG and GGGA(T) (a FLI1 binding site) on the other, revealing how MTM recognizes different DNA sequences. Unexpectedly, at sub-micromolar concentrations MTMs stabilize FLI1–DNA complex on GGAA repeats, which are critical for the oncogenic function of EWS–FLI1. We also directly demonstrate by nuclear magnetic resonance formation of a ternary FLI1–DNA–MTM complex on a single GGAA FLI1/MTM binding site. These biochemical and structural data and a new FLI1–DNA structure suggest that MTM binds the minor groove and perturbs FLI1 bound nearby in the major groove. This ternary complex model may lead to development of novel MTM analogues that selectively target EWS–FLI1 or other oncogenic transcription factors, as anti-cancer therapeutics.
INTRODUCTION
Specific targeting of transcription factors by small molecules is an attractive concept that has been difficult to realize. Recently, mithramycin (MTM; Figure 1), a DNA binding aureolic acid natural product, was identified in high-throughput screening of 50 000 small molecules as a highly potent (low-nanomolar) and modestly selective inhibitor of abnormal oncogenic transcription factor EWS–FLI1 in Ewing sarcoma (ES) (1). ES is a rare devastating cancer affecting mostly children and young adults that is caused by EWS–FLI1 in the majority of ES patients. Because MTM antagonizes EWS–FLI1 without affecting its expression level (1), MTM likely directly interferes with the EWS–FLI1 function on DNA. Due to its anti-tumor activity caused by the potent inhibition of EWS–FLI1 in mouse ES xenografts, MTM recently underwent a phase I/II clinical trial as an ES therapeutic (National Cancer Institute clinical trial NCT01610570), where toxicity was observed at doses below efficacious doses of this agent. Because of the off-target toxicity, more selective anti-ES analogues of MTM are needed.
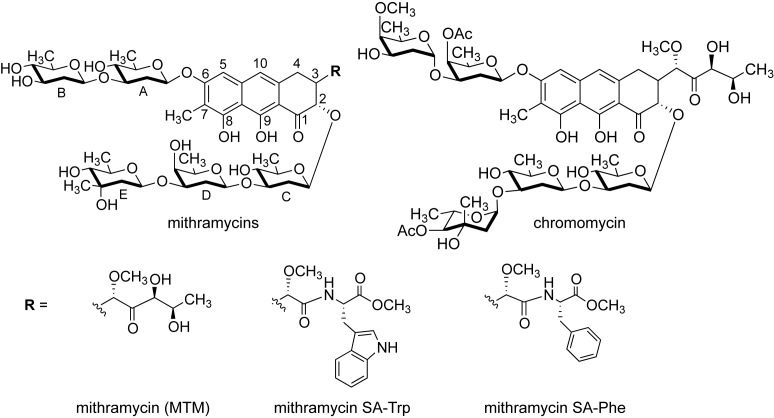
Figure 1.
Chemical structures of mithramycin, its analogues and chromomycin A3.
EWS–FLI1 is an abnormal fusion of parts of two proteins (EWS and FLI1), containing the DNA binding domain (DBD) of FLI1 (2,3), which is a result of a chromosomal translocation. The FLI1 DBD is critical for the transcription and oncogenic transformation functions (4) of EWS–FLI1. Because ES cells, but not other cells, contain EWS–FLI1, it is an attractive target for development of novel anti-cancer agents (5–7). MTM analogues with improved selectivity against this fusion would have a wider therapeutic window than MTM does. analogues of MTM with modified B- and E-sugar moieties and the 3-side chain (see the atom numbering and group nomenclature in Figure 1) were recently shown to have lower toxicity or higher anti-ES potency in vitro and in mouse ES xenografts (8). Recently, semi-synthetic MTM analogues of the 3-side chain were demonstrated to have improved potency against non-small lung cancer cells A549 (9) and improved anti-tumor activity and toxicity in colon and melanoma mouse xenograft models (10). This 3-side chain of MTM was previously proposed not to be involved in direct DNA contacts (11); therefore, its modifications may not interfere significantly with DNA recognition properties of MTM analogues. Indeed, we demonstrated that a recently developed MTM analogue, MTM SA–Trp, had a similar DNA sequence preference and binding affinity to those of MTM (12). Nevertheless, we also showed in the same study that 3-side chain substitutions could modulate the DNA binding affinity of the analogues by as much as ∼20-fold. Detailed mechanistic understanding of MTM–DNA binding and inhibition of EWS–FLI1 is necessary for development of highly selective MTM analogues that would specifically target EWS–FLI1. Principles of such design may then also be applied to design MTM analogues as chemical probes for selective targeting of other transcription factors.
MTM is a non-intercalating DNA binding molecule (11). We hypothesized that the disruption of EWS–FLI1 function by MTM is due to some effect of MTM on FLI1 DBD at oncogenic promoters. MTM binds in the minor groove of DNA as a dimer, in which the two MTM monomers are coordinated by a divalent metal ion (11,13–15). Pioneering solution structural studies of MTM bound to palindromic GGCC DNA in the Patel laboratory by a combination of nuclear magnetic resonance (NMR) and molecular dynamics elegantly demonstrated that an MTM dimer recognized the central GC, so that the 2-fold dyad symmetry of the MTM dimer matched that of the DNA (11,16). Due to the intrinsic limitations of isotopic labeling in this and similar methods, the DNA conformation cannot be unambiguously resolved and needs to be assumed. Another structurally characterized aureolic acid natural product, chromomycin A3, contains O-methylated or O-acetylated groups in three out of five sugar moieties as well as numerous differences from MTM in the stereochemistry of the sugars and its glycosylic linkages (Figure 1). A crystal structure of chromomycin in complex with a similar GGCC-containing DNA oligomer was consistent with the basic observations made from the MTM–DNA solution structure (17). This structure revealed for the first time the octahedral coordination of the divalent metal ion by the chromomycin monomers and two water molecules. However, the chromomycin–DNA crystal structure exhibited significant differences from the previously reported solution structures of both MTM– and chromomycin–DNA complexes (18), in DNA conformations and the dispositions of the chromophore rings, presumably due to the limitations of the NMR data (17) as well as chemical differences. Therefore, these aspects of MTM–DNA complexes remain unclear. We have recently established that a DNA motif X(G/C)(G/C)X is necessary and sufficient for high-affinity binding by MTM (12). Notably, the recognition sequence for FLI1 and other related (ETS-family) transcription factors GGA(A/T) is a strong MTM binding sequence, but is not a palindrome. We and others have demonstrated that XGGX is a particularly high-affinity sequence (12,19). Therefore, another critical question to be addressed in elucidating the MTM mode of action and DNA recognition is how MTM binds non-palindromic DNA sequences.
In this work, we harnessed the structural variation of novel semi-synthetic MTM analogues to perform a detailed structural investigation of MTM–DNA complex formation for different DNA sequences and probed the effect of MTMs on the stability of FLI1–DNA complex. Together with a new crystal structure of FLI1 DBD–DNA complex, these studies led us to propose a structural model of MTM antagonism of EWS–FLI1.
MATERIALS AND METHODS
Synthesis of MTM SA–Trp and MTM SA–Phe
MTM SA–Trp was produced and characterized as reported recently (12). To synthesize MTM SA–Phe, we used an analogous PyBop coupling reaction, except for the use of L-Phe methyl ester instead of L-Trp methyl ester in the synthesis. MTM SA–Phe was characterized by NMR spectroscopy and liquid chromatography–mass spectrometry (LCMS) analysis (Supplementary Figures S1–5 and Table S1). Prior to their use in the crystallization and binding assays, MTM and its analogueueues were dissolved in dimethyl sulfoxide (DMSO) at 20 mM.
Crystallization, data collection and structure determination for MTM analogue–DNA complexes
Single-stranded palindromic DNA oligonucleotides 5′-AGAGGCCTCT-3′, 5′-AGGGTACCCT-3′, 5′-AGGGATCCCT-3′ were purchased from IDT (Coralville, IA, USA). The double-stranded oligomers were formed by dissolving each DNA to 2 mM in 10 mM Tris–HCl pH 8.0 (pH adjusted at room temperature), heating the solutions to 95°C and slowly cooling them to 4°C. The crystals of MTM SA–Trp- AGGGTACCCT, MTM SA–Phe- AGGGTACCCT and MTM SA–Phe-AGGGATCCCT were grown by vapor diffusion in 1 μl hanging drops containing 10 mM sodium cacodylate pH 6.0, 4 mM zinc acetate dihydrate, 0.85 mM DNA, 1.8 mM MTM SA–Trp or MTM SA–Phe, 2 mM spermine incubated against 35% v/v 2-methyl-2,4-pentanediol at 21°C. The crystals were taken directly from these drops and frozen in liquid nitrogen by quick immersion.
X-ray diffraction data for the crystals of the MTM SA–Trp–DNA complex were collected at 100 K at sector 22-ID (SER-CAT) of the Advanced Photon Source (APS) of the Argonne National Laboratory (Argonne, IL, USA). The data for the crystals of the MTM SA–Phe complexes were collected at sector 21-ID (Life Sciences–Collaborative Access Team; LS–CAT) at the same facility. Data indexing, integration and scaling were performed with HKL2000 (20). The crystal structures were determined by using the anomalous signal from bound Zn2+ ions by the single–wavelength anomalous dispersion (SAD) method with Phenix suite (21). The Zn2+ sites were located by the HySS module (22) and then used for phasing by Phaser (23). The periodic electron density for the DNA backbone and partial density for the bases and the MTM analogues was well discernible in the experimental map and allowed us to build an initial DNA model. Subsequent rounds of refinement with Refmac (24) and model building with Coot (25) improved the map quality and allowed us to complete the DNA models and build MTM SA–Trp and MTM–Phe. The data collection and refinement statistics are given in Supplementary Table S2. The structures of MTM SA–DNA, MTM SA–Phe-AGGGTACCCT and MTM SA–Phe–AGGGATCCCT were deposited in the Protein Data Bank with accession codes 5JVW, 5JW0 and 5JW2, respectively.
Cloning, expression and purification of the DNA binding domain of FLI1
A pET28a plasmid encoding a EWS–FLI1 fusion (26) was a kind gift from Dr Jeffrey Toretsky. This vector served as a template for polymerase chain reaction (PCR) amplification of the region encoding the FLI1 DBD (residues 276–375), by using primers 5′-ATCAACCACATATGCCTGGAAGCGGGCAGATCCAGC-3′ and 5′-ATCCTCGAGCTACTCGGTCGGATGTGGCTGCAG-3′. The PCR product, after appropriate processing, was inserted into a modified pET19b-pps vector (27) between NdeI and XhoI restriction sites. The resulting protein product contained an N-terminal decahistidine tag cleavable by Prescission protease (GE Healthcare). The protein was expressed and purified by using a recently published protocol (28) with minor modifications, as follows. The protein was expressed in BL21(DE3)-RIL cells at 13°C for 18 h upon induction with 1 mM IPTG and purified on Ni-IMAC HisTrap FF column (GE Healthcare) and washed thoroughly with 1 M NaCl to remove possible contaminating DNA prior to the elution. The tag was removed by overnight cleavage with PreScission protease at 4°C followed by further purification on a size-exclusion Sephacryl S-200 column (GE Healthcare) equilibrated in 40 mM Tris–HCl pH 8.0 (pH adjusted at room temperature), 0.15 M NaCl and 2 mM β-mercaptoethanol. The fractions containing protein were pooled and concentrated using an Amicon Ultra-15 centrifugal filter device (Millipore) to 2 mg/ml. This preparation was then used to form and further purify protein–DNA complexes for crystallization. For DNA binding studies, we used a previously reported construct of FLI1 DBD (residues 259–399), expressed and purified analogously (28).
Double-stranded 11-bp DNA oligomer consisting of complementary strands 5′-GACCGGAAGTG-3′ and 5′-CACTTCCGGTC-3′ (IDT) was annealed as described previously (28). The double-stranded oligomer was added to the purified FLI1 DBD at the 1.2:1 DNA: FLI1 molar ratio. This protein-DNA mixture was then diluted 5-fold with a buffer not containing NaCl (40 mM Tris–HCl pH 8.0 and 2 mM β-mercaptoethanol), incubated on ice for 15 min and then purified on a size-exclusion Sephacryl S-200 column equilibrated in 20 mM Tris–HCl pH 8.0. An elution peak at a position distinct from that of protein alone represented the protein–DNA complex. The peak fractions were pooled, concentrated to ∼20 mg/ml. This preparation was flash-frozen in liquid nitrogen, and stored at −80°C for crystallization.
Crystallization, data collection and crystal structure determination for the FLI1 DBD–DNA complex
The crystals of the FLI1 DBD–DNA complex were grown at 21°C by vapor diffusion in hanging drops containing a mixture of 1 μl of the concentrated protein–DNA complex and 1 μl of the reservoir solution (1.6 M ammonium sulfate, 0.1 M sodium acetate pH 4.6). The crystals were gradually transferred into the cryoprotectant solution (1.6 M ammonium sulfate, 0.1 M sodium acetate, pH 4.6 and 28% sucrose) and then frozen in liquid nitrogen by quick immersion. The X-ray diffraction data were collected at 100 K at the sector 22-ID (SER-CAT) of the APS and processed with HKL2000 (20). The structure was determined by molecular replacement with program MOLREP (29) with our previously published structure of FLI1 DBD–DNA (PDB accession code 5E8I; (28)) as a search model. The structure was then iteratively rebuilt and refined with COOT (25) and REFMAC (24), respectively. The data collection and refinement statistics are given in Supplementary Table S3. The crystal structure of the FLI1 DBD–DNA complex and the structure factor amplitudes were deposited in the Protein Data Bank with accession code 5JVT.
DNA binding assays
A single-stranded DNA oligomer labeled with a 6-carboxyfluorescein (6FAM) at the 5′-end 5′-6FAM-AA(GGAA)6-3′ was annealed with its complementary unlabeled oligomer (both from IDT) as described above. The binding mixture contained 50 nM of this double-stranded DNA oligomer, 500 nM FLI1 DBD (residues 259–399) and MTM or its analogue (at a specified concentration) in the binding buffer (20 mM Tris–HCl, pH 8.0 (adjusted at room temperature), 50 mM NaCl, 10 mM MgCl2). The mixture was incubated for 15 min at 22°C. Heparin was then added to the final concentration of 7.5 mg/ml and the mixture was incubated for 10 min before fluorescence measurements. The fluorescence polarization measurements were carried out in 96-well plates on a SpectraMax M5 microplate reader (Molecular Devices), at excitation and emission wavelengths of 495 and 520 nm, respectively.
NMR experiments
The 15N-labeled FLI1 DBD was expressed in M9 minimal media with 15N-labeled NH4Cl as the sole source of nitrogen. The double-stranded DNA 12-mer TTGAAGGAAGAG was prepared as described above. The HSQC spectra were acquired on a Bruker Avance III 500 spectrometer equipped with a 1H/13C/15N cryoprobe at the University of Texas Health Science Center at San Antonio Biomolecular NMR Core. NMR samples contained 1 mM 15N-labeled FLI1 alone or complexed with dsDNA (5′-TTGAAGGAAGAG-3′), in 20 mM Tris–HCl, pH 7.0, 5 mM MgCl2, 5% D2O and 3% DMSO-d6, in the presence and absence of 1 mM MTM SA–Trp.
RESULTS
Crystal structure of MTM SA–Trp in complex with oligomer AGAGGCCTCT
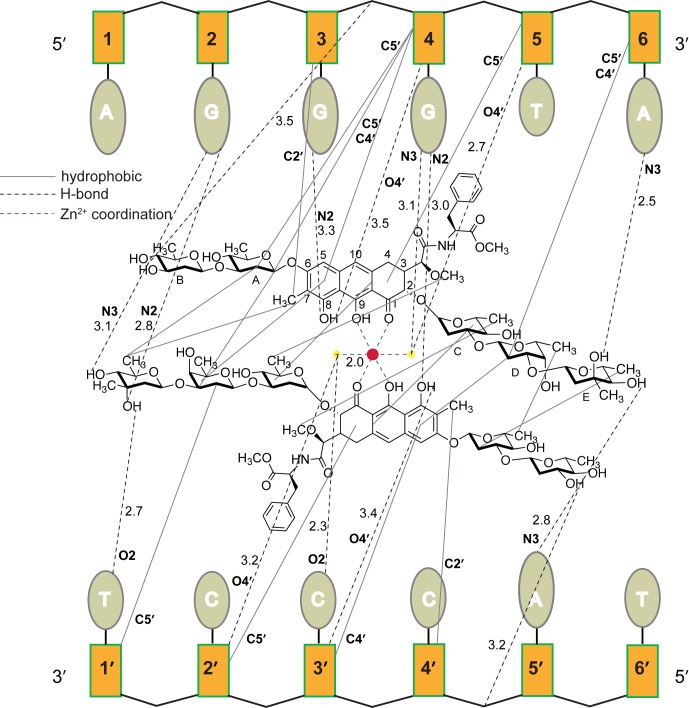
A crystal structure of MTM or an MTM–DNA complex has not been obtained to date. Crystallization experiments with MTM and DNA oligomers of different sequences and sizes did not yield diffracting crystals. Our recently reported analogue MTM SA–Trp and a novel analogue MTM SA–Phe (Figure 1) ultimately have yielded high-quality crystals, when in complex with DNA. We crystallized and determined a structure of a complex of MTM SA–Trp with a double-stranded palindromic 10-bp DNA oligomer 5′-AGAGGCCTCT-3′ containing a central MTM recognition sequence GGCC (Figure 2A). The crystal structure was refined at 2.0 Å resolution (Supplementary Table S2). Zn2+ was used as a divalent metal mediating the dimer formation of MTM SA–Trp (Figure 2A and B, Supplementary Figure S6), as Zn2+ was previously shown to stabilize the dimer of MTM better than Mg2+, without affecting other functions of MTM (30,31). Moreover, anomalous X-ray scattering by Zn2+ ions bound by MTM SA–Trp and DNA was used to determine the crystal structure by single-wavelength anomalous dispersion (SAD). The crystals contained two nearly identical complexes of MTM SA–Trp–DNA per asymmetric unit. Crystal packing interactions between complexes involved stacking of the indole ring of the Trp residue of one MTM SA–Trp in the dimer against a terminal A of the DNA of another complex in the crystal (Supplementary Figure S7), which explained why this analogue was critical for co-crystallization with DNA.
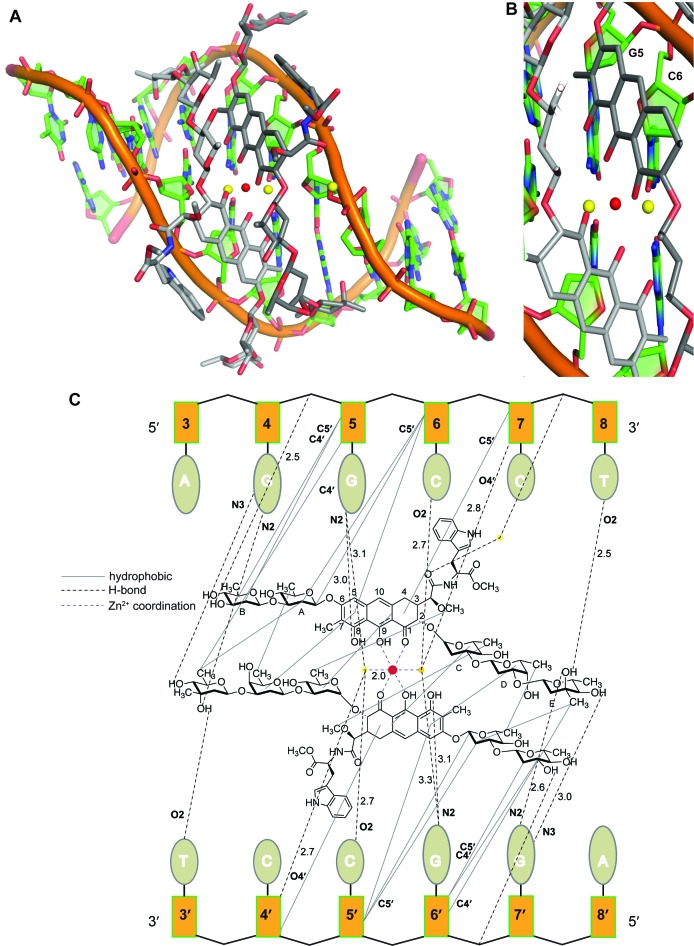
Figure 2.
Structure of MTM SA–Trp–DNA complex and the ligand–DNA interactions. (A) Crystal structure of MTM SA–Trp–DNA complex. The two molecules of MTM analogue are shown as light-gray and dark-gray sticks, the DNA is in orange, the Zn2+ ion and its coordinating waters are shown as red and yellow balls, respectively. (B) The zoomed-in view of the Zn2+ coordination and the GC motif recognition. (C) A schematic of the interactions in the MTM SA–Trp–DNA complex.
The dimer of MTM SA–Trp is bound to the DNA with the same dyad symmetry as the DNA palindrome (Figure 2A). The dimer lies in the minor groove of the DNA, with the sugar side chains in extended conformations. The monomers in the dimer are oriented in an approximately anti-parallel fashion relative to one another, so that the disaccharide side chain of each monomer points toward the 5′-end of the adjacent DNA strand. Even though the binding mode of MTM SA–Trp to DNA and MTM SA–Trp–DNA interactions are generally similar to those in the solution structure of MTM–DNA complex (where the same DNA sequence was used) (11), there are differences in the mutual disposition of the chromophore cores in the MTM dimers and very large differences between the conformations of the saccharide side chains of MTM SA–Trp and the DNA in our crystal structure and the respective conformations of MTM and the presumed A-form DNA (of the same sequence) in the solution structure. These differences reflect the inaccuracy of the stereochemistry assumptions in the solution structure necessitated by the intrinsic paucity of the NMR data, and not any deficiency of that elegant NMR study. Likewise, differences between the previous structure of chromomycin–DNA(TTGGCCAA) (17) and our structure of MTM SA–Trp–DNA complex are quite significant: atomic positional differences of DNA backbones reach ∼4 Å and angle differences between the respective bases are as large as ∼20–30°. Similar differences were observed in the natural product conformations. The DNA oligomers are different in sequence outside of the core GGCC sequence; therefore, some of the conformational differences outside of the core sequence may be due to the DNA sequence differences. Significant differences are present in the GGCC regions and in the MTM chromophore dispositions between the two structures. Most prominently, the minor groove in the core GGCC region is wider by ∼2 Å in the chromomycin–DNA structure and, if one superimposes one of the two chromophore rings of the chromomycin and MTM SA–Trp, the other two chromophore rings of the respective dimers in the two structures form ∼20° angle to one another, with the two chromomycin rings appearing as a more obtuse wedge in the wider minor groove. The wider minor groove in the chromomycin–DNA structure appears to be needed to accommodate the bulkier acetyl groups on the chromomycin sugar moieties, although the effects of other chemical and stereochemical differences between the two DNA binding molecules cannot be ruled out.
The 3-side chains bearing a Trp residue protrude out into the solvent and do not interact with the DNA, except for a water-mediated hydrogen bond between the carbonyl oxygen adjacent to cores of some of MTM SA–Trp molecules in the crystal and nearby DNA phosphate groups (Figure 2C). The sugar B moieties of each monomer also protrude out of the minor groove, propped by hydrophobic interactions with the deoxyribose moieties of G5/G6’ (Figure 2A and C). The two tricyclic cores form a dimer through a coordinated Zn2+ ion, which is bound to the O1 and O9 atoms of each core and two water molecules (Figure 2B and C). All six Zn2+-coordinating ligand atoms are located 2.0 A away from the Zn2+, forming a perfect octahedral coordination shell. The planes of the chromophore cores are at an 85° angle relative to one another, fitting snugly into the wide minor groove (with the width of ∼14 Å, as defined by 3DNA program (32)) of this G/C rich DNA oligomer. The MTM SA–Trp dimer interacts with a 6-base pair region AGGCCT of the 10-mer DNA, where the two nucleotides on the 5′-end of this region on either strand do not interact with the MTM SA–Trp (Figure 2C). With the exception of the E sugars, each MTM SA–Trp molecule interacts only with one DNA strand (Figure 2C). The chromophore and the di- and the tri-sugar moieties of each monomer of MTM SA–Trp make extensive hydrophobic contacts with the phosphodiester backbone of the DNA and with the other monomer. Specifically, the chromophore rings of each monomer interact with C and D sugar moieties of the other monomer as well as with the two deoxyribose moieties of the CC region of the DNA. The E sugars of each MTM SA–Trp molecule are in non-polar contacts only with the A and B sugars of the other MTM SA–Trp across the dimeric interface. In addition to non-polar interactions and Zn2+ coordination, there are nine hydrogen bonds between each DNA strand and an MTM SA–Trp or a bound water. Six of them are formed with DNA bases, one with a deoxyribose moiety and two with phosphate groups. There are three hydrogen bonds per strand with the bases of the central GC base pair, which is the core recognition sequence (12). Two of them are from a Zn2+-coordinated water and the 8-hydroxyl group of the chromophore to the N2 of the G base. The third hydrogen bond is made between the other Zn2+-coordinated water and the O2 of the C base (Figure 2C). Two other hydrogen bonds are made between the 3- and 4-hydroxyl groups of the E sugar and the N2 and the N3 of the G base immediately preceding the central GC, and the other base-directed hydrogen bond is between the 3-hydroxyl group of the E sugar as a donor of its hydrogen to the O2 of the 3′-terminal T8/T3’ base of the interacting 6-bp region. In contrast with the central (G/C)(G/C)-recognizing rigid chromophore-Zn2+-water scaffold, dictated by the Zn2+ coordination geometry, the flexibility of the saccharide side chains may allow MTM to adapt to different flanking DNA sequences, which would explains the lack of sequence preference of MTM to these flanking sequences (12,33).
Crystal structures of MTM SA–Phe in complex with AGGGATCCCT and AGGGTACCCT
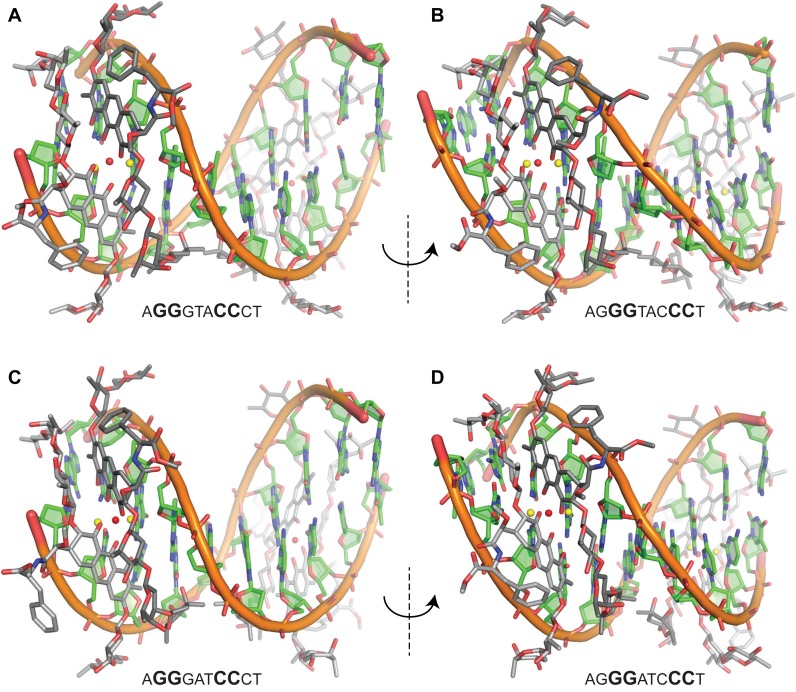
We recently demonstrated that both of these two palindromic DNA sequences are high-affinity MTM binding sequences, as they contain XGGX sites (12). We obtained crystal structures of these complexes, where Zn2+, as above, was used to form an MTM SA–Phe dimer and to serve as a source of the anomalous signal for crystal structure determination by SAD. The two complexes formed crystals in the same crystal form, with one MTM SA–Phe–DNA complex per asymmetric unit. In these crystal structures, each DNA oligomer was bound to two MTM dimers (Figure 3). Unexpectedly, in each structure one dimer was bound to the first GG register (i.e. to the AGGG site; Figure 3A and C) and the other dimer was bound to the second GG register (i.e. to the GGGT and GGGA on the complementary strand; Figure 3B and D).
Figure 3.
Crystal structures of MTM SA–Phe–DNA complexes. (A) The MTM SA–Phe bound to the first GG register of the AGGGTACCCT oligomer. (B) The MTM SA–Phe bound to the second GG register of the AGGGTACCCT oligomer in the same complex as in panel A. (C) The MTM SA–Phe bound to the first GG register of the AGGGATCCCT oligomer. (D) The MTM SA–Phe bound to the second GG register of the AGGGATCCCT oligomer in the same complex as in panel C.
MTM SA–Phe dimers are bound to the AGGG(TA) and AGGG(AT) sites on the two DNA oligomers very similarly (Figure 4 and Supplementary Figure S8), and only a few minor differences are observed, as follows. In the former case (Figure 4) a hydrogen bond is observed between the 3-hydroxyl group of the E-sugar of one of the MTM SA–Phe molecules as a hydrogen bond donor and the O2 of the T base of the AGGGTA region, as was also observed in the MTM SA–Trp–DNA complex (Figure 2C). In the complex with AGGG(AT) (Supplementary Figure S8), an A is in place of the T; here the hydrogen is instead donated to the O4’ atom of the deoxyribose moiety of the A nucleotide. Unlike the complex with AGGG(TA), this complex also contains a hydrogen bond between the 3-hydroxyl of the B-sugar of the other MTM SA–Phe monomer and a phosphate group of the bottom DNA strand. A similar hydrogen bond is observed in the MTM SA–Trp–DNA complex. The other interactions appear to be present in both complexes.
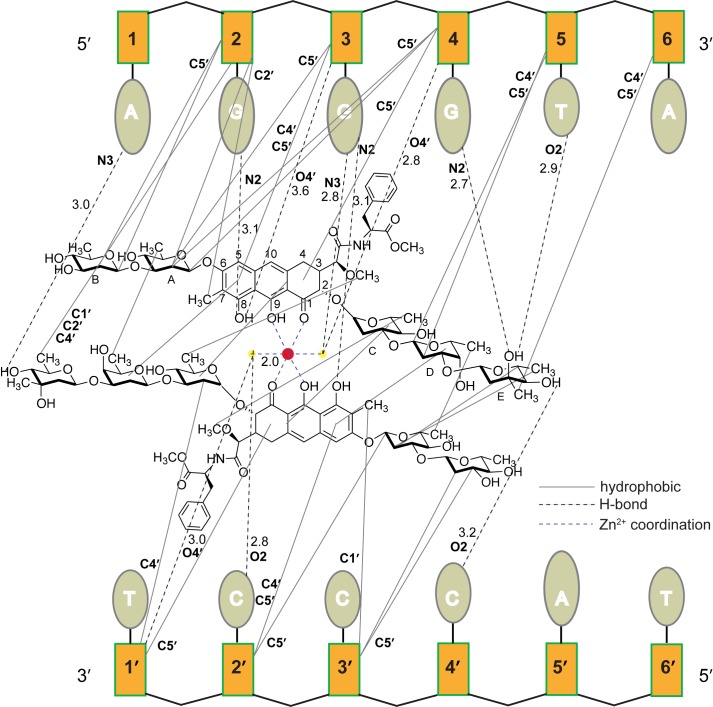
Figure 4.
A schematic of the interactions in the complex of MTM SA–Phe with the first GG register of AGGGTACCCT DNA.
As expected from the non-palindromic nature of sequences AGGGTA and AGGGAT, there is a pronounced interaction asymmetry in the MTM SA–Phe–DNA interactions. The top strand makes a more extensive set of contacts with the MTM analog than does the bottom strand. All six nucleotides of the AGGG(TA)/(AT) region are engaged in the interactions with the MTM analogue on that strand, including the A/T nucleotide on the 3′-end of the region. This interaction is a non-polar contact between the 3-methyl group of the E-sugar and the C4’ and C5’ deoxyribose atoms of the A/T nucleotide. Sugar D of one MTM SA–Phe molecule in the dimer is engaged in hydrophobic contacts with the deoxyribose group of the preceding nucleotide, whereas the sugar D groups of MTM SA–Trp are not engaged in interactions with the palindromic GGCC site. The core of MTM SA–Phe is also in a more intimate contact with the top strand than for the GGCC site. For example, the 7-methyl group of one of the MTM SA–Phe monomers is in non-polar contact with the deoxyribose moiety of the first G of the AGGG motif. The top strand bias is also observed for the hydrogen bonds: seven bonds are formed with the top strand and only three with the bottom one. Remarkably, one of the MTM analogue monomers in the dimer does not form any hydrogen bonds with DNA. The MTM SA–Phe core of the other monomer and the Zn2+−coordinating water molecules are engaged in five hydrogen bonds with the GGG region, whereas only two hydrogen bonds are observed between these structural elements of the MTM analogue and the complementary strand. The hydrogen bond formed by the 8-hydroxyl of one of the cores to the N2 amino group of the first G is the same as that in complex with the GGCC sequence (Figure 2C). Unlike the latter, the 8-hydroxyl of the other core forms a hydrogen bond with the N2 of the second G, an apparent consequence of the sequence difference. Another hydrogen bond not observed in the complex with GGCC is also from the 8-hydroxyl donating its hydrogen to O4′ of the deoxyribose of the second G. Notably, the N2 amino groups of all three G's in the AGGG sequence and the N2 amino groups of all the G bases of both strands of the GGCC sequence are engaged in hydrogen bonds with the MTM analogues. The propensity of MTM to satisfy the hydrogen bonding potential of the N2 amino group was demonstrated previously by binding studies with DNA containing 2,6-diaminopurine substitutions (34). Our structures provide an atomic view of this adaptation to the XGGX sequence by the MTM dimer core and, ultimately, the basis of the partial disengagement from the bottom strand and the resulting top strand bias. The two Zn2+ coordinating water molecules form two hydrogen bonds each. One water forms a bifurcated interaction with one strand and the other water forms a bifurcated interaction with the other strand in the same fashion as observed in the complex of MTM SA–Trp with GGCC (except for bonding to the N3 of G3 in the AGGG sequence in place of the bond to the O2 of the C6/C5’ in the GGCC sequence; Figure 2C). The hydrogen bonds to the other strand from either water molecule are no longer present in the complexes with AGGG. The E-sugar groups form some of the same or similar hydrogen bonds to those observed in the complex with GGCC. One prominent difference, also reflecting the bias toward the AGGG strand is that the 3-hydroxyl group of the E-sugar forms a hydrogen bond with the N2 of G4 instead of the N2 of G7’ on the complementary strand in the GGCC sequence (Figure 2C).
The binding to the second GG register by the other MTM SA–Phe dimer in each structure (Figure 5 and Supplementary Figure S9) is best described as a shift of all interactions relative to those with the first GG register, with a few relatively minor exceptions. Even though more hydrogen bonds are formed between the backbone of the bottom DNA strand than for complexes with the other register, the overall top strand bias is still present. For example, eight hydrogen bonds are formed with the top strand and six with the bottom one. As expected, all core interactions all shifted by one base pair, maintaining the top strand bias. An additional weak hydrogen bond is formed between the 8-hydroxyl group and the O4’ of the deoxyribose of C3’. The E-sugar of the same MTM SA–Phe monomer forms a hydrogen bond both with N2 and N3 of G2, in keeping with the previous observations that the N2 of all the G bases are engaged in the hydrogen bonds with the MTM analogue. The differences between binding to the GGGTA (Figure 5) and GGGAT (Supplementary Figure S9) sequences are mainly in the patterns of hydrogen bonding from an E-sugar to the variable bases. For example, in each case the base at the 5′ position is engaged in interactions with the E-sugar moiety (O2 of T5’ or N3 of A5’, respectively).
Figure 5.
A schematic of the interactions in the complex of MTM SA–Phe with the second GG register of AGGGTACCCT DNA.
MTM analogues can bind DNA together with the FLI1 DBD and affect the stability of FLI1 DBD–DNA complex
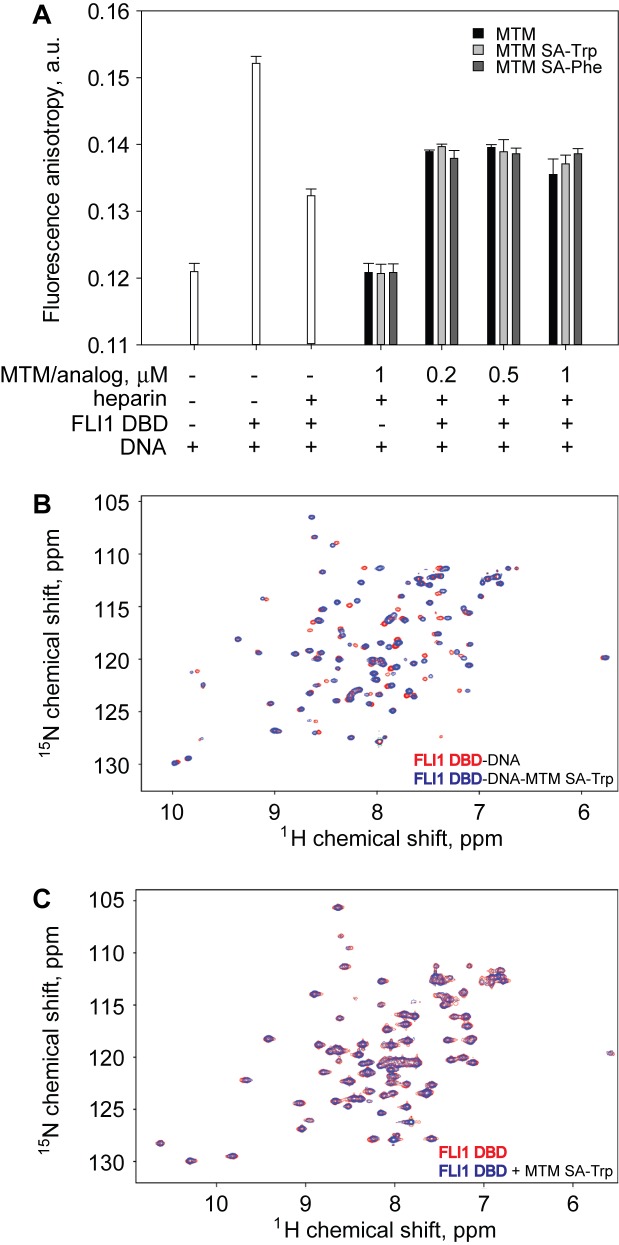
Because MTM and its analogues bind GG sites on the DNA in the minor groove whereas FLI1 DBD binds its recognition site GGA(A/T) in the major groove (28), we hypothesized that a molecule of an MTM or its analogue may bind a FLI1 DBD recognition site non-competitively with the FLI1 DBD and perturb the stability of the FLI1 DBD–DNA complex. To test this hypothesis, we formed a FLI1 DBD complex on a 6FAM-labeled DNA containing six GGAA repeats (AA(GGAA)6) and investigated the stability of this complex upon titrating in MTM and its analogues, by measuring fluorescence polarization of the 6FAM probe. This DNA sequence was designed based on the previous finding that a minimum of five GGAA repeats was necessary and sufficient for the oncogenic transformation by EWS–FLI1, driven by binding to these repeats (35). Binding of the FLI1 DBD to this DNA resulted in an increase in fluorescence anisotropy (Figure 6A). At these conditions ([DNA] = 50 nM, [FLI1 DBD] = 500 nM), DNA is nearly completely saturated with bound FLI1 DBD. We then probed the lifetime of these complexes by a heparin challenge. Heparin is a polyanionic competitor that binds free FLI1 DBD and prevents it from binding DNA. Therefore, at high enough concentration heparin drives dissociation of FLI1 DBD the DNA. Indeed, upon a 10 min challenge with heparin (at 7.5 μg/ml), approximately half of FLI1–DNA complexes dissociated from the DNA (Figure 6A). We carried out an analogous heparin challenge in the presence of increasing concentrations of MTM, MTM SA–Trp and MTM SA–Phe. At 1 μM, none of these compounds had an effect on fluorescence anisotropy in the absence of FLI1 (Figure 6A). We observed that at concentrations of 0.2–1 μM, these compounds resulted in a significant increase in fluorescence anisotropy in the heparin challenge experiment, stabilizing about half of FLI1–DNA complexes that would have dissociated without MTM. Above 0.2 μM this stabilizing effect was progressively weaker with increasing concentration of MTM, and this trend continued at higher concentrations. These data demonstrate that not only do MTM and its analogues bind DNA together with FLI DBD, but at sub-micromolar concentrations they can have a stabilizing effect on FLI1–DNA complex formation.
Figure 6.
Formation of a ternary FLI1 DBD–DNA–MTM complex. (A) The stabilizing effect of MTM and its analogues at 1 μM and lower concentrations on the FLI DBD complexes with AA(GGAA)6 DNA upon heparin challenge. (B) The HSQC NMR spectra of FLI1–DBD–DNA complex in the absence (red) and in the presence (blue) of MTM SA–Trp. (C) The HSQC NMR spectra of FLI1–DBD in the absence (red) and in the presence (blue) of MTM SA–Trp.
To probe directly whether a FLI1–DNA–MTM ternary complex can form on a short DNA oligomer with a single FLI1/MTM binding site GGAA site and no other MTM or FLI1 binding sequences, we acquired HSQC NMR spectra for an 15N-labeled FLI1 DBD in complex with a double-stranded DNA 12-mer TTGAAGGAAGAG in the absence and in the presence of the stoichiometric amount of MTM SA–Trp (Figure 6B). Additionally, to test a possibility for interactions of FLI1 DBD with MTM in the absence of DNA and to consider a potential for MTM to compete with FLI1 DBD for DNA binding, we collected HSQC NMR spectra with the 15N-labeled FLI1 DBD alone and in the presence of MTM SA–Trp (Figure 6C). The resonance signals the FLI1 DBD are well dispersed in all cases, indicating a stable structure, in agreement with our crystallographic structural studies (28). Upon addition of the MTM analogue, chemical shifts of the FLI1 DBD bound to DNA undergo significant changes (Figure 6B). The resulting spectral pattern (blue resonances in Figure 6B) is different from that of the FLI1 DBD alone (red resonances in Figure 6C), indicating formation of the ternary FLI1 DBD–DNA–MTM SA–Trp complex, rather than displacement of the FLI1 DBD from DNA. These spectral differences show that the FLI1 DBD MTM SA–Trp binding perturbs the structure of the FLI1 DBD bound to DNA, consistent with the ternary complex formation observed in the fluorescence anisotropy experiments. Furthermore, the spectral patterns of the free FLI1 DBD in the absence and in the presence of the MTM analogue (Figure 6C) are highly similar, demonstrating that MTM does not interact with the FLI1 DBD in the absence of DNA. These data are the first direct demonstration of the formation of ternary FLI1–DNA–MTM complex.
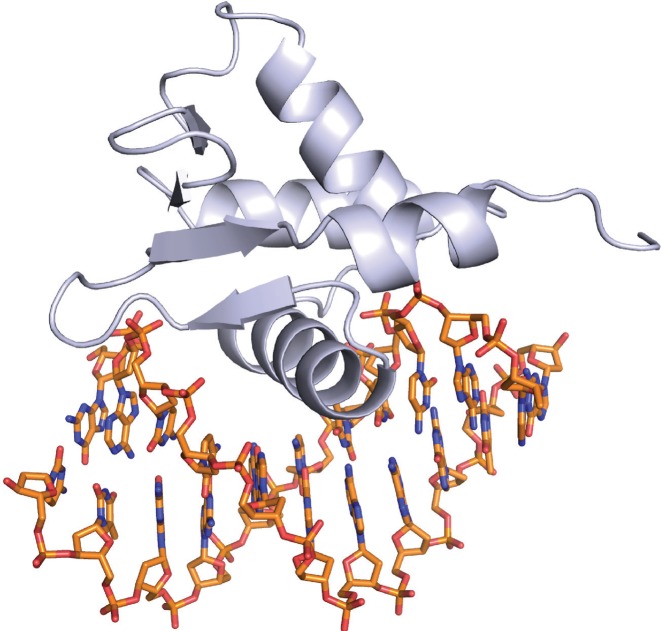
DNA conformations in the structures of MTM analogue–DNA complexes and in a structure of FLI1 DBD in complex with 11-mer DNA GACCGGAAGTG DNA
To analyze whether MTM binding perturbs the DNA conformation and whether MTM has a potential to affect FLI1–DNA binding through these DNA conformational changes, we analyzed the DNA conformations in our structures of MTM analogue–DNA complexes and in a new crystal structure of FLI1 DBD in complex with an 11-mer DNA containing a single FLI1 binding site GGAA, by using 3DNA software (32). The crystals of this FLI1 DBD–DNA complex were in a new crystal form, yielding a higher resolution structure (Figure 7) than our previously reported structure of FLI1 DBD with a one base pair shorter 10-mer ACCGGAAGTG (28). The DNA binding and dimerization properties of the FLI1 DBD were described in that study. These properties are very similar in our new crystal structure; therefore, here we will consider only the conformation of the DNA in complex with FLI1 DBD.
Figure 7.
The crystal structure of FLI1 DBD in complex with the 11-mer DNA GACCGGAAGTG.
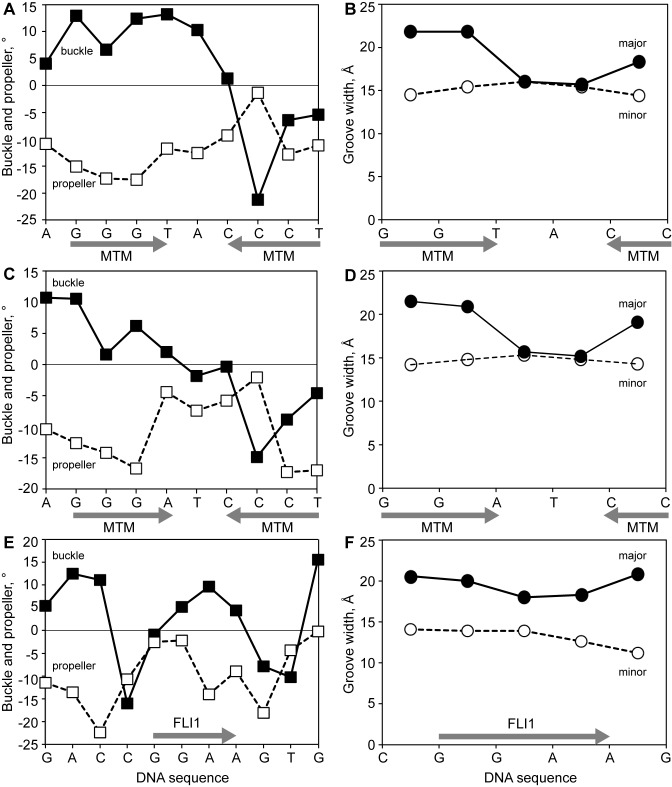
Binding of MTM SA–Phe to two different registers of the same DNA sequence allows us to observe potential effects of binding of this MTM analogue on DNA conformation (Figure 8). The DNA conformations of AGGGTACCCT and AGGGATCCCT oligomers in complex with MTM SA–Phe differ, at least in part, due to the different sequences of these two oligomers. Nevertheless, the general behavior of several parameters that exhibit the largest variation of values along the DNA is similar for the two DNA sequences. Several parameters, such as buckle, propeller and the widths of the major and minor grooves, vary significantly along the DNA and exhibit a pronounced asymmetry with respect to the middle of the DNA. This asymmetry indicates the two MTMs exert different effects on DNA conformation by binding to different registers of the oligomers. Binding of MTM SA–Phe to the second GG register (GGG(T/A)) is associated with positive buckle values, whereas binding to the first GG register (AGGG), close to the DNA end, is associated with negative buckle values. Propeller values are negative, but increase toward the zero value asymmetrically, also likely as a result of the asymmetry of MTM analog binding. The DNA major groove is compressed asymmetrically with respect to the middle of the DNA oligomer. On the other hand, the minor groove is widened throughout the DNA, which could be either a result of MTM binding or an intrinsic property of the sequence. In assessing whether these potential conformational effects of the MTM analogue binding to a GGA(A/T) site could favor or disfavor FLI1 binding to the same site, we compared structural parameter values for binding of the MTM analog to the second DNA register, in other words, the GG of the GG(T/A) sequences, to the values for the GGAA sequence in the FLI1 DBD–DNA structure (Figure 8E and F). The buckle values of the GGAA region bound to FLI1 are positive, as with the GG(AT)/(TA) conformation in the MTM SA–Phe–DNA complex. Again, for both FLI1 DBD and MTM analogue binding the DNA major groove is compressed. This effect is stronger as a result of MTM analog binding, as the major groove has the same width as the minor groove in the MTM analogue DNA complexes. In both FLI1 DBD– and MTM analog–DNA complexes the minor grooves of the binding sites are widened. In summary, conformational perturbations that appear to be caused by MTM SA–Phe binding to DNA are consistent with FLI1 DBD binding to the same site.
Figure 8.
DNA structural parameters for MTM SA–Phe–DNA and FLI1 DBD–DNA complexes. The sites of the bound MTM SA–Phe and FLI1 DBD are marked by gray arrows. Panels A and B display buckle and propeller (A) and major and minor groove widths (B) for the complex of MTM SA–Phe with AGGGTACCCT. Panels C and D display buckle and propeller (C) and major and minor groove widths (D) for the complex of MTM SA–Phe with AGGGATCCCT. Panels E and F display buckle and propeller (E) and major and minor groove widths (F) for the complex of FLI1 DBD with GACCGGAAGTG.
MTM conformations in the structures of MTM analogue–DNA complexes
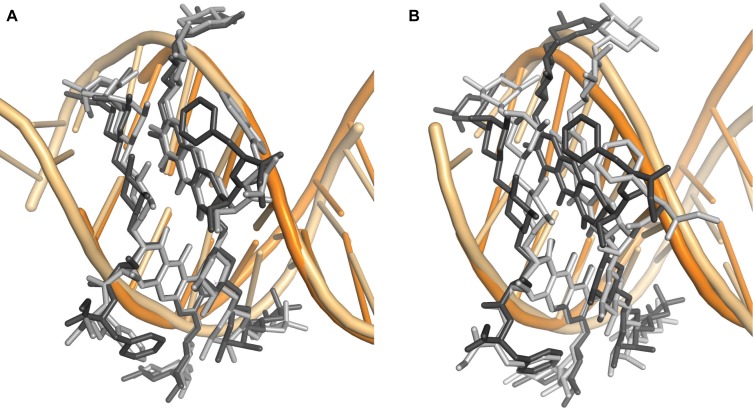
To establish if MTM analogues also undergo DNA sequence-dependent structural adaptation upon binding DNA, we superimposed cores of one of MTM analogue monomers in different complexes and compared the conformations of the cores and the side chains in different dimers. The conformations of the dimeric cores of the MTM analogues in the complexes with the GGCC and AGGG sites (the first GG register in either AGGGT or AGGGA sequences) are nearly identical. The disaccharide and the trisaccharide appendages exhibit greater conformational plasticity (Figure 9A). The largest atomic positional differences of ∼2.5 Å are observed for the E-sugars, the most distant groups from the cores. These conformational differences of the disaccharide and the trisaccharide tails are consistent with the minor differences in the DNA backbone conformations of these regions, especially for the middle GC and GG in the GGCC and AGGG sequences, respectively. The 3-side chains are either solvent-exposed (in MTM SA–Phe) or are involved in crystal packing interactions (in MTM SA–Trp). These side chains adopt somewhat different conformations dictated by their environment in the respective crystals.
Figure 9.
Superimposition of MTM analogues in different complexes with DNA. (A) Superimposition of MTM SA–Trp (light-gray)-DNA and MTM SA–Phe (dark-gray)-AGGGTACCCT (first GG register). (B) Superimposition of MTM SA–Phe (dark-gray)-AGGGTACCCT (first GG register) with MTM SA–Phe (light-gray)-AGGGTACCCT (second GG register).
The MTM SA–Phe bound to the GGG(T/A) sites, in other words, to the second GG register exhibits much larger conformational differences from that bound to the first GG register or from MTM SA–Trp bound to the GGCC site (Figure 9B). These differences appear to follow the large DNA conformational differences between the two halves of the DNA oligomers described in the previous section. While the positions of the Zn2+ and the coordinating water and oxygen atoms are essentially unchanged, one of the cores is rotated toward the other core by ∼18°. This rotation is a superposition of a rotation around the axis going through the Zn2+ perpendicular to the chromophore plane and a rotation around the axis parallel to the Zn2+-coordinating oxygens. The conformations of the tri- and di-saccharide tails of the two MTM monomers point toward the center DNA end are quite similar, with atomic position shifts not exceeding ∼1.5 Å. In contrast, significant conformational differences between the tails are observed on the other side of the cores. For example, these trisaccharide tails are shifted relative to one another by ∼3.9 Å, and the disaccharide tails are shifted by ∼3.1 Å. These observations indicate that MTM analogues, like DNA, undergo conformational changes upon formation of MTM–DNA complexes.
DISCUSSION
Small molecules that could regulate transcription of specific genes in therapeutic or research applications are highly attractive, but they have not yet been developed. Initiation of transcription has been recognized to be a kinetic bottleneck in the entire gene expression pathway (36); therefore, transcription initiation represents both a highly regulated process and an excellent target for chemical inhibition. Transcription in bacteria and human is targeted by clinically used drugs, such as rifampicin and trabectedin, respectively. While rifampicin interacts directly with the bacterial RNA polymerase, trabectedin forms a covalent adduct with DNA, which disrupts proper function of RNA polymerase II and DNA binding transcription factors. An individual transcription factor generally regulates a relatively small set of promoters and, in that regard, it appears to be a promising target for small molecules. However, the DNA or protein binding surfaces of transcription factors typically lack small hydrophobic cavities suitable for binding drug-like compounds and, for this reason, transcription factors have been viewed as undruggable. On the other hand, small molecules that bind DNA do not recognize a long enough DNA sequence to be specific, and binding to multiple sites leads to off-target effects and toxicity. Synthetic polymeric molecules, such as polyamides, that would bind in the minor groove of the DNA over a sufficiently long DNA sequence to ensure specificity to compete off transcription factors are being developed (37). However, sequence recognition from the minor groove is limited by poor accessibility of DNA bases from that groove. A molecule that would combine chemical features responsible for DNA binding and transcription factor binding would be very desirable. DNA binding natural products are potential candidates for development of such bifunctional molecules, as they generally have favorable pharmaceutical properties, they are large enough to have some DNA sequence preference and they can contain groups that could be chemically elaborated to install protein interacting functionalities.
MTM is a highly potent antagonist of oncogenic transcription factor EWS–FLI1 in ES. Off-target toxicity of MTM, likely due to its low DNA sequence specificity (an MTM-box X(G/C)(G/C)X is sufficient for tight MTM binding (12)), poses a barrier in developing MTM as a clinically useful anti-cancer agent. However, MTM is chemically complex, containing groups that can be modified by semi-synthetic methods to yield analogues with superior anti-cancer or toxicity properties (8–10,12,38–41). Basic understanding of the molecular mechanism of MTM binding to DNA and its interference with transcription is necessary to improve MTM analogues in a rational fashion. A set of crystal structures of novel MTM analogues, MTM SA–Trp and MTM SA–Phe bound to four different MTM-boxes reported here provides a rationale for and detailed structural view of the MTM-box recognition by MTM. For example, non-palindromic sequences are recognized through the bias toward G bases. We also demonstrate that both MTM and DNA alter their conformations upon complex formation, a phenomenon that is common in protein–DNA recognition. MTM binds DNA in the minor groove, whereas ETS-family transcription factors including FLI1 (28) or its close relative ERG (42) bind DNA in the major groove, which has prompted us to hypothesize that MTM can modulate functions of these transcription factors by binding DNA together with them, instead of competing with them by occluding some of the protein binding surface on the DNA. Then the effect of MTM or its analog on the transcription function could be allosteric, through the DNA conformational changes, such as those observed in this study by NMR, or through direct interactions between an MTM analogue and the transcription factor. Indeed, we found that MTM and its analogues had a stabilizing effect on the FLI1 DBD bound to a repeating GGAA sequence, which is critical for the oncogenic function of EWS–FLI1 at some promoters. One of the MTM analogue–DNA complexes that we structurally characterized was formed on the GGAT sequence (Figure 3D), an ETS-family recognition sequence. Even though the conformations of the GGAT site are generally comparable in the MTM SA–Phe and FLI1 DBD complexes, they are not the same (Figure 8): there are differences both in the backbone and base conformations. Therefore, the conformation of the DNA in the ternary complex is likely a compromise between these two conformations. The conformations of the DNA bound MTM and FLI1 in the ternary complex with DNA must then also be somewhat different from those in the respective binary complexes. These changes or even direct interactions between the MTM analog and FLI1 may account for the chemical shift changes observed in the NMR experiments (Figure 6). Future determination of the structure of the ternary complex will shed light on the exact nature of these conformational changes.
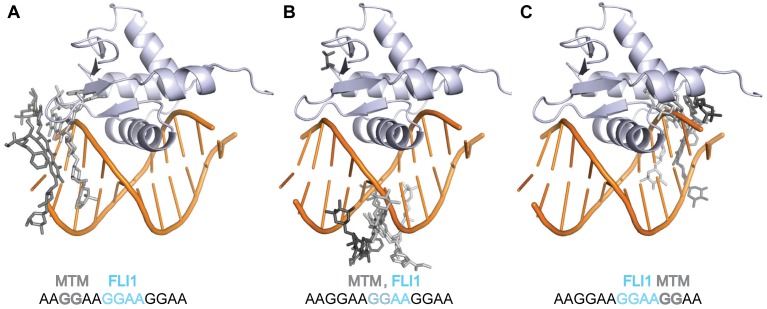
In order to understand how MTM and FLI1 may interact on DNA, by using the structure of MTM SA–Phe in complex with the GGAT site and the structure of FLI1 DBD–DNA complex, we constructed structural models of MTM–DNA–FLI1 complexes. We made structural models of three ternary complexes MTM SA–Phe–DNA–FLI1 DBD by superimposing the DNA in these two structures in different DNA registers, which would correspond to sequence GGAAGGAAGG (Figure 10). In these structures the FLI1 DBD is bound to the interior GGAA site, and DNA is superimposed in three different registers so that MTM SA–Phe is bound to the upstream GG (Figure 10A), the middle GG (Figure 10B) and the downstream GG (Figure 10C). The MTM analogue interacts with FLI1 DBD differently in these three registers. When bound to the same GG as the FLI1–DBD (Figure 10B), the MTM analogue makes no direct contacts with the protein; any interactions would be transmitted through DNA conformational changes. These interactions, in principle, can be favorable and stabilize the protein–DNA complex. One of such stabilizing conformational changes may be the major groove narrowing that we observed in both FLI1 DBD–DNA and MTM analogue–DNA complex. The upstream bound MTM analogue does not exhibit severe steric clashes with the protein (Figure 10A) and can interact with the protein directly through its 3-side chain or its B-sugar moiety. Such interactions, in principle, can either stabilize or destabilize the protein–DNA complex, depending on the chemical nature of the interacting groups on the MTM analogue. The 3-side chain stands out as an antenna-like structural feature that could be optimized for specific interactions with transcription factors such as FLI1 or ERG, in addition to the effect of this side chain on DNA binding (12). In contrast, the MTM analogue bound downstream (Figure 10C) severely clashes with the protein and is likely to act as a competitor. The last mode of binding can serve as an explanation of the observed destabilizing effect of MTM analogues on FLI1 DBD–DNA complexes at concentrations above 1 μM. Detailed structural and biochemical studies of ternary complexes of MTM analogues, FLI1 DBD and DNA are needed to test these models and obtain a more accurate picture of MTM effects on transcription factors.
Figure 10.
Structural models of the MTM SA–Phe–DNA–FLI1 DBD complex with GGAA repeats. (A) The MTM analogue bound to the upstream GG site. (B) The MTM analogue bound to the same GG site as the FLI1 DBD. (C) The MTM analogue bound to the downstream GG site. The respective binding sites for the MTM analogue and the FLI1 DBD are shown schematically on the bottom.
In summary, this study clarified the structural basis of MTM analogue–DNA recognition and demonstrated complex effects of MTM binding to DNA on the stability of FLI1–DNA complexes. The structural models of ternary complexes of MTM analogue–DNA–FLI1 DBD provided a potential explanation for these effects. These models and future experiments testing them will be invaluable in developing transcription factor-selective MTM analogues for their use as chemical probes and anti-neoplastic agents in clinic.
Supplementary Material
Acknowledgments
We thank Jhong-Min Chen for initial preparations of MTM SA–Trp and MTM SA–Phe, Samantha Hancock for preliminary analysis of the MTM SA–Trp–DNA structure and the staff of sector LS-CAT at the Advanced Photon Source at the Argonne National Laboratory for the assistance with the synchrotron data collection.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
University of Kentucky Markey Cancer Center Pilot Award (to O.V.T., J.R.); National Institutes of Health (NIH) [CA091901 to J.R.]; US Department of Defense Prostate Cancer Research Program [PC150300 to O.V.T and J.R.]; NIH P30 to NMR Core Facility at the University of Texas Health Science Center at San Antonio and Cancer Therapy and Research Center [CA054174]. Funding for open access charge: NIH.
Conflict of interest statement. None declared.
REFERENCES
- 1.Grohar P.J., Woldemichael G.M., Griffin L.B., Mendoza A., Chen Q.R., Yeung C., Currier D.G., Davis S., Khanna C., Khan J., et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J. NCI. 2011;103:962–978. doi: 10.1093/jnci/djr156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delattre O., Zucman J., Plougastel B., Desmaze C., Melot T., Peter M., Kovar H., Joubert I., de Jong P., Rouleau G., et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359:162–165. doi: 10.1038/359162a0. [DOI] [PubMed] [Google Scholar]
- 3.May W.A., Gishizky M.L., Lessnick S.L., Lunsford L.B., Lewis B.C., Delattre O., Zucman J., Thomas G., Denny C.T. Ewing sarcoma 11;22 translocation produces a chimeric transcription factor that requires the DNA-binding domain encoded by FLI1 for transformation. Proc. Natl. Acad. Sci. U.S.A. 1993;90:5752–5756. doi: 10.1073/pnas.90.12.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arvand A., Welford S.M., Teitell M.A., Denny C.T. The COOH-terminal domain of FLI-1 is necessary for full tumorigenesis and transcriptional modulation by EWS/FLI-1. Cancer Res. 2001;61:5311–5317. [PubMed] [Google Scholar]
- 5.Erkizan H.V., Scher L.J., Gamble S.E., Barber-Rotenberg J.S., Sajwan K.P., Uren A., Toretsky J.A. Novel peptide binds EWS-FLI1 and reduces the oncogenic potential in Ewing tumors. Cell Cycle. 2011;10:3397–3408. doi: 10.4161/cc.10.19.17734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minas T.Z., Han J., Javaheri T., Hong S.H., Schlederer M., Saygideger-Kont Y., Celik H., Mueller K.M., Temel I., Ozdemirli M., et al. YK-4-279 effectively antagonizes EWS-FLI1 induced leukemia in a transgenic mouse model. Oncotarget. 2015;6:37678–37694. doi: 10.18632/oncotarget.5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lamhamedi-Cherradi S.E., Menegaz B.A., Ramamoorthy V., Aiyer R.A., Maywald R.L., Buford A.S., Doolittle D.K., Culotta K.S., O'Dorisio J.E., Ludwig J.A. An oral formulation of YK-4-279: preclinical efficacy and acquired resistance patterns in Ewing sarcoma. Mol. Cancer. Ther. 2015;14:1591–1604. doi: 10.1158/1535-7163.MCT-14-0334. [DOI] [PubMed] [Google Scholar]
- 8.Osgood C.L., Maloney N., Kidd C.G., Kitchen-Goosen S.M., Segars L.E., Gebregiorgis M., Woldemichael G.M., He M., Sankar S., Lessnick S.L., et al. Identification of mithramycin analogues with improved targeting of the EWS-FLI1 transcription factor. Clin. Cancer. Res. 2016;22:4105–4118. doi: 10.1158/1078-0432.CCR-15-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scott D., Chen J.M., Bae Y., Rohr J. Semi-synthetic mithramycin SA derivatives with improved anticancer activity. Chem. Biol. Drug Des. 2013;81:615–624. doi: 10.1111/cbdd.12107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nunez L.E., Nybo S.E., Gonzalez-Sabin J., Perez M., Menendez N., Brana A.F., Shaaban K.A., He M., Moris F., Salas J.A., et al. A novel mithramycin analogue with high antitumor activity and less toxicity generated by combinatorial biosynthesis. J. Med. Chem. 2012;55:5813–5825. doi: 10.1021/jm300234t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sastry M., Patel D.J. Solution structure of the mithramycin dimer-DNA complex. Biochemistry. 1993;32:6588–6604. doi: 10.1021/bi00077a012. [DOI] [PubMed] [Google Scholar]
- 12.Weidenbach S., Hou C., Chen J.M., Tsodikov O.V., Rohr J. Dimerization and DNA recognition rules of mithramycin and its analogues. J. Inorg. Biochem. 2016;156:40–47. doi: 10.1016/j.jinorgbio.2015.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cons B.M., Fox K.R. Interaction of mithramycin with metal ions and DNA. Biochem. Biophys. Res. Commun. 1989;160:517–524. doi: 10.1016/0006-291x(89)92463-7. [DOI] [PubMed] [Google Scholar]
- 14.Aich P., Dasgupta D. Role of Mg++ in the mithramycin-DNA interaction: evidence for two types of mithramycin-Mg++ complex. Biochem. Biophys. Res. Commun. 1990;173:689–696. doi: 10.1016/s0006-291x(05)80090-7. [DOI] [PubMed] [Google Scholar]
- 15.Demicheli C., Albertini J.P., Garnier-Suillerot A. Interaction of mithramycin with DNA. Evidence that mithramycin binds to DNA as a dimer in a right-handed screw conformation. Eur. J. Biochem. 1991;198:333–338. doi: 10.1111/j.1432-1033.1991.tb16020.x. [DOI] [PubMed] [Google Scholar]
- 16.Sastry M., Fiala R., Patel D.J. Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J. Mol. Biol. 1995;251:674–689. doi: 10.1006/jmbi.1995.0464. [DOI] [PubMed] [Google Scholar]
- 17.Hou M.H., Robinson H., Gao Y.G., Wang A.H. Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion. Nucleic Acids Res. 2004;32:2214–2222. doi: 10.1093/nar/gkh549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gao X.L., Mirau P., Patel D.J. Structure refinement of the chromomycin dimer-DNA oligomer complex in solution. J. Mol. Biol. 1992;223:259–279. doi: 10.1016/0022-2836(92)90730-8. [DOI] [PubMed] [Google Scholar]
- 19.Fox K.R., Howarth N.R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucleic Acids Res. 1985;13:8695–8714. doi: 10.1093/nar/13.24.8695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Otwinowski Z., Minor W. Processing of X-ray diffraction data collected in oscillation mode. Macromol. Cryst. A. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 21.Adams P.D., Afonine P.V., Bunkoczi G., Chen V.B., Davis I.W., Echols N., Headd J.J., Hung L.W., Kapral G.J., Grosse-Kunstleve R.W., et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Cryst. D. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grosse-Kunstleve R.W., Adams P.D. Substructure search procedures for macromolecular structures. Acta Cryst. D. 2003;59:1966–1973. doi: 10.1107/s0907444903018043. [DOI] [PubMed] [Google Scholar]
- 23.McCoy A.J., Storoni L.C., Read R.J. Simple algorithm for a maximum-likelihood SAD function. Acta Cryst. D. 2004;60:1220–1228. doi: 10.1107/S0907444904009990. [DOI] [PubMed] [Google Scholar]
- 24.Murshudov G.N., Vagin A.A., Dodson E.J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 25.Emsley P., Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. D. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 26.Uren A., Tcherkasskaya O., Toretsky J.A. Recombinant EWS-FLI1 oncoprotein activates transcription. Biochemistry. 2004;43:13579–13589. doi: 10.1021/bi048776q. [DOI] [PubMed] [Google Scholar]
- 27.Biswas T., Tsodikov O.V. Hexameric ring structure of the N-terminal domain of Mycobacterium tuberculosis DnaB helicase. FEBS J. 2008;275:3064–3071. doi: 10.1111/j.1742-4658.2008.06460.x. [DOI] [PubMed] [Google Scholar]
- 28.Hou C., Tsodikov O.V. Structural basis for dimerization and DNA binding of transcription factor FLI1. Biochemistry. 2015;54:7365–7374. doi: 10.1021/acs.biochem.5b01121. [DOI] [PubMed] [Google Scholar]
- 29.Vagin A., Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Cryst. 1997;30:1022–1025. [Google Scholar]
- 30.Reyzer M.L., Brodbelt J.S., Kerwin S.M., Kumar D. Evaluation of complexation of metal-mediated DNA-binding drugs to oligonucleotides via electrospray ionization mass spectrometry. Nucleic Acids Res. 2001;29:E103–E103. doi: 10.1093/nar/29.21.e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Devi P.G., Pal S., Banerjee R., Dasgupta D. Association of antitumor antibiotics, mithramycin and chromomycin, with Zn(II) J. Inorg. Biochem. 2007;101:127–137. doi: 10.1016/j.jinorgbio.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 32.Lu X.J., Olson W.K. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hampshire A.J., Fox K.R. The effects of local DNA sequence on the interaction of ligands with their preferred binding sites. Biochimie. 2008;90:988–998. doi: 10.1016/j.biochi.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 34.Bailly C., Waring M.J. Transferring the purine 2-amino group from guanines to adenines in DNA changes the sequence-specific binding of antibiotics. Nucleic Acids Res. 1995;23:885–892. doi: 10.1093/nar/23.6.885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gangwal K., Sankar S., Hollenhorst P.C., Kinsey M., Haroldsen S.C., Shah A.A., Boucher K.M., Watkins W.S., Jorde L.B., Graves B.J., et al. Microsatellites as EWS/FLI response elements in Ewing's sarcoma. Proc. Natl. Acad. Sci. U.S.A. 2008;105:10149–10154. doi: 10.1073/pnas.0801073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.von Hippel P.H., Rees W.A., Rippe K., Wilson K.S. Specificity mechanisms in the control of transcription. Biophys. Chem. 1996;59:231–246. doi: 10.1016/0301-4622(96)00006-3. [DOI] [PubMed] [Google Scholar]
- 37.Trauger J.W., Baird E.E., Dervan P.B. Recognition of DNA by designed ligands at subnanomolar concentrations. Nature. 1996;382:559–561. doi: 10.1038/382559a0. [DOI] [PubMed] [Google Scholar]
- 38.Perez M., Baig I., Brana A.F., Salas J.A., Rohr J., Mendez C. Generation of new derivatives of the antitumor antibiotic mithramycin by altering the glycosylation pattern through combinatorial biosynthesis. Chembiochem. 2008;9:2295–2304. doi: 10.1002/cbic.200800299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Albertini V., Jain A., Vignati S., Napoli S., Rinaldi A., Kwee I., Nur-e-Alam M., Bergant J., Bertoni F., Carbone G.M., et al. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer Streptomyces argillaceus exhibits improved transcriptional repressor activity: implications for cancer therapy. Nucleic Acids Res. 2006;34:1721–1734. doi: 10.1093/nar/gkl063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Remsing L.L., Gonzalez A.M., Nur-e-Alam M., Fernandez-Lozano M.J., Brana A.F., Rix U., Oliveira M.A., Mendez C., Salas J.A., Rohr J. Mithramycin SK, a novel antitumor drug with improved therapeutic index, mithramycin SA, and demycarosyl-mithramycin SK: three new products generated in the mithramycin producer Streptomyces argillaceus through combinatorial biosynthesis. J. Am. Chem. Soc. 2003;125:5745–5753. doi: 10.1021/ja034162h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tornin J., Martinez-Cruzado L., Santos L., Rodriguez A., Nunez L.E., Oro P., Hermosilla M.A., Allonca E., Fernandez-Garcia M.T., Astudillo A., et al. Inhibition of Sp1 by the mithramycin analogue EC-8042 efficiently targets tumor initiating cells in sarcoma. Oncotarget. 2016;7:30935–30950. doi: 10.18632/oncotarget.8817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Regan M.C., Horanyi P.S., Pryor E.E., Jr, Sarver J.L., Cafiso D.S., Bushweller J.H. Structural and dynamic studies of the transcription factor ERG reveal DNA binding is allosterically autoinhibited. Proc. Natl. Acad. Sci. U.S.A. 2013;110:13374–13379. doi: 10.1073/pnas.1301726110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.