Abstract
Clustered apurinic/apyrimidinic (AP; abasic) DNA lesions produced by ionizing radiation are by far more cytotoxic than isolated AP lesion entities. The structure and dynamics of a series of seven 23-bp oligonucleotides featuring simple bistranded clustered damage sites, comprising of two AP sites, zero, one, three or five bases 3′ or 5′ apart from each other, were investigated through 400 ns explicit solvent molecular dynamics simulations. They provide representative structures of synthetically engineered multiply damage sites-containing oligonucleotides whose repair was investigated experimentally (Nucl. Acids Res. 2004, 32:5609-5620; Nucl. Acids Res. 2002, 30: 2800–2808). The inspection of extrahelical positioning of the AP sites, bulge and non Watson–Crick hydrogen bonding corroborates the experimental measurements of repair efficiencies by bacterial or human AP endonucleases Nfo and APE1, respectively. This study provides unprecedented knowledge into the structure and dynamics of clustered abasic DNA lesions, notably rationalizing the non-symmetry with respect to 3′ to 5′ position. In addition, it provides strong mechanistic insights and basis for future studies on the effects of clustered DNA damage on the recognition and processing of these lesions by bacterial or human DNA repair enzymes specialized in the processing of such lesions.
INTRODUCTION
Abasic DNA sites constitute one of the most common families of DNA lesions. An apurinic/apyrimidinic (AP) site is produced by the excision of one DNA constituent base leaving the uncapped deoxyribose moiety. Most notably AP lesions may be produced directly, as a consequence of exposure to ionizing radiation (IR) or as the products of monofunctional DNA N-glycosylases and base excision repair (BER) enzymes (1). In solution the AP sugar moiety experiences equilibrium between its cyclic and open forms (See Supplementary Data, Supplementary Figure S1). Although, the ring-opened form triggers dangerous interstrand cross-links (2) the equilibrium strongly favors the cyclic form that accounts for about 99% of the occurrence (3). NMR and molecular modeling agree in evidencing a strong local deformation of the DNA double-helix in response to the lesion (4). This distortion is also exacerbated by the fact that AP sites are prone to adopt an extra-helical position that may also correlate with the ease of repair. Indeed, the lesion is usually flipped out and inserted in a specific protein pocket during the excision reaction (5,6).
In the case of exposure to IR or even high oxidative stress, a specific pattern of highly localized ionizations and free radical attack is produced as also supported by various biophysical track structure models and Monte Carlo simulations. (7,8) The clustering of DNA damage may involve AP sites as well as double strand breaks (DSBs) and others non-DSB lesions like oxidized bases and it is currently accepted as one of the most important types of DNA damage with high biological significance due to its repair resistance as also reviewed in (9). Recently the repair resistance of clustered DNA damages comprising of bulky guanine adducts opposite to AP sites has also been considered(10). DNA lesion clustering induction, its role(s) in cell homeostasis, systemic effects and the pathways involved in its in vivo processing and repair, remain still an open question in current biology (11–13). Clustered abasic sites (14) (clustered AP) have been produced in oligonucleotides (15,16), plasmid, nucleosome (17–19) or found in irradiated DNA (20), cells and animals (21).
Repair processing of clustered AP has been investigated (17,21) and the available data show a clear dependence of the repair efficiency on the relative position of the clustered AP sites. Interestingly it appears that bacterial excision enzymes such as the Escherichia coli endonuclease IV (Nfo) behave quite differently than the corresponding human endonuclease APE1. However, the molecular bases of the observed specificity still need to be clarified. Earlier NMR and molecular dynamics studies have shown that when two bistranded AP sites are staggered 5′ to each other and 1–3 base pairs apart, they are located in the minor groove when adopting an extrahelical conformations. Therefore, major structural changes for APE1 incision are required. In contrast, when AP residues are staggered 3′ to each other and 1–3 bp apart they are extruded on the major groove, adopting a conformation that may favor enzyme binding by lowering kinetic energy barriers (4).
Numerical simulation techniques have nowadays reached a considerable maturity (22) and can complement experimental studies in tackling DNA damage (23) and repair problems, offering the possibility to visualize the interaction between DNA and enzymes at an atomistic resolution. Molecular modeling has been applied to damaged oligonucleotides, such as abasic sites (24–26), bulky adducts (10,27), cross-links (28) or photolesions, (23,29) and also to duplexes in interaction with repair enzymes (30). Molecular dynamics (MD) explorations of combined 8-oxoguanine and abasic site have been achieved independently (31,32), yet the combination of two AP to give clustered adducts is still missing. On the other hand the structural bases for the recognition of simple AP sites have been rationalized (33) and hence may offer a good comparison to understand the difference induced by the cluster lesion.
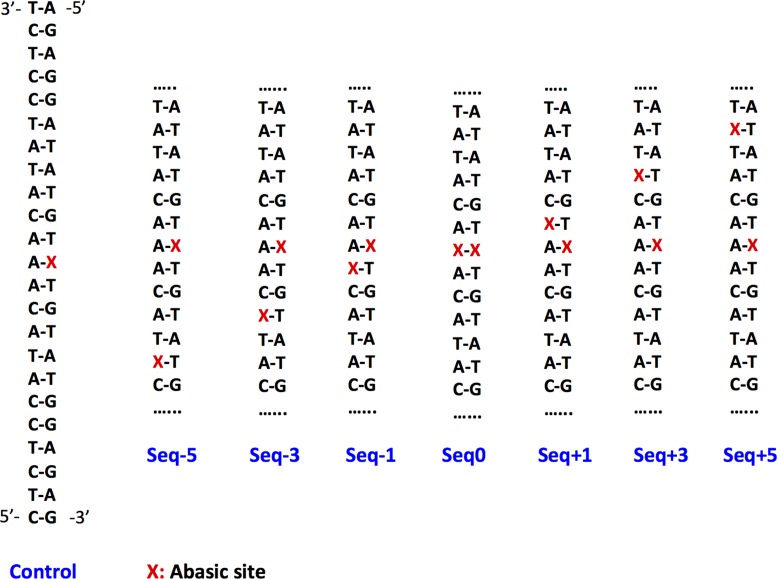
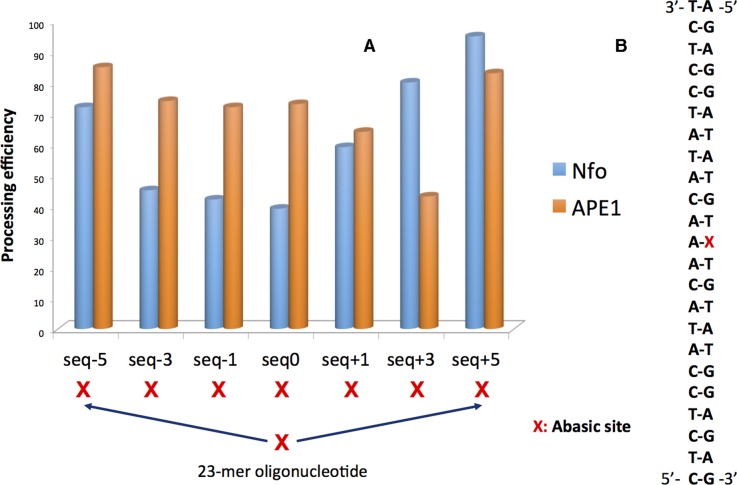
In this work, we explore by means of explicit solvent MD simulations, the structural evolution of the seven clustered AP oligonucleotides previously studied by Georgakilas et al. (17,21). They feature an AP site as the 35th nucleobase and a second one on the complementary strand. Their relative position is designed as seq ±0, 1, 3, 5 as shown in Scheme 1, in particular in seq0 the two AP sites are directly facing each other. Most notably a clear relative position dependence effect of cluster damages has been evidenced especially in the case of bacterial repair (21), as evidenced in Figure 1. MD simulations will allow us to unravel all the distinct structural deformations of clustered AP-containing strands as compared to the reference simple AP, providing a structural framework to rationalize the observed repair efficiency.
Scheme 1.
Representation of the sequence of the studied oligonucleotides as taken from (17,21).
Figure 1.
(A) Repair rate of the different clustered AP oligomer using Nfo (Escherichia coli) and APE1 (human) enzymes, respectively. (B) The original DNA double strand featuring the position of the AP marked as X. The clustered AP containing lesions have been produced inserting a secondary AP site in the complementary strand and up to five residues apart from the original lesion, see also Scheme 1. Data are adapted from references (17,21).
MATERIALS AND METHODS
Oligonucleotide structure
The choice for the specific oligonucleotide sequence presented in Figure 1 was based on the analytical existing results for AP DNA cluster processing for both Nfo and APE1 enzymes as described in the original publications by Georgakilas et al. (17,21). One of the reason behind the choice of this specific sequence is that it did form no hairpin loops and was thermally stable in solution even after the induction of the AP site. Current theoretical calculations agree with this initial finding and additionally using BLAST (blast.ncbi.nlm.nih.gov) sequence alignment analysis it was found that the specific sequence shared great homology with human chromosomal regions (see Supplementary Data).
Computational protocol
The Amber12 software (34) package and the Amber ff99 force field (35,36) including bsc0 corrections (37) were used for all classical MD simulations. Seven oligonucleotides were built on top of canonical B-DNA sequences produced with the Nucleic Acids Builder module of Amber (34). Abasic sites were inserted at specific positions to generate the seven sequences corresponding to the synthetized oligonucleotides of reference (17), with the xleap module. The parameters for the AP site (see Supplementary Data) were generated with antechamber and parmcheck subprograms, and atom point charges were computed using the RESP protocol (38), see Supplementary Table S1 and Supplementary Figure S2. Sodium cations (Na+) were added in order to neutralize the systems, which were immersed in a truncated octahedral TIP3P (39) water box, counting between 18 272 and 22 405 water molecules depending on the system. Each AP-containing oligonucleotide structure was first minimized in a 10 000 steps simulation, including 5000 steps of steepest descent. Then, a thermalization step was performed to heat each system from 0 to 300 K in 20 ps. The temperature was kept constant during the following steps using Langevin thermostat with a collision frequency γln of 1 ps−1. A first 100 ps equilibration run was performed in NPT conditions, followed by a second one in NVT conditions. Finally, a 400 ns production was executed with constant pressure.
In order to check how the cluster lesions affect the global B-DNA structure, we also computed the overall bending of the whole clustered AP oligomers and compared it to the control, i.e. the strand containing only one AP site. We used the bending measurement method as defined within the Curves+ program (40). To assess residency times for hydrogen bonds, we fix a cut-off on the H…O of H…N distances of 2.4 Å. We adopt the criterion that extrahelical positions correspond to a distance C1′…C1′ greater than 14 Å, as proposed by Lavery et al. (25). This criterion was used to infer the percentages of extrahelicity for abasic sites and other pyrimidines. Furthermore, we also calculated the minor groove occupancy by the extrahelical AP; details are given in Supplementary Data (Supplementary Table S2 and Supplementary Figure S3).
Note that due to the equilibrium between the closed and open form AP sites can exist as α- or β-anomers. However, as pointed out both theoretically (41) and via NMR determinations (32) the two isomers give rise to duplexes having the same structural characteristics. This aspect will be computationally too demanding to analyze systematically, yet preliminary MDs on seq0 and seq+1 show a conservation of the structural and dynamical properties.
RESULTS
MD simulations were performed over 400 ns, for seven sequences and a reference one as depicted in Figure 1 and Scheme 1. We described hereafter the main structural outcomes obtained for these oligonucleotides. Particular attention will also be given to the global bending and extra-helical position of the AP sites as reported in Table 1.
Table 1. Average structural descriptors for the damaged oligonucleotides over 400 ns and the latest 40 ns along MD simulations. Please note that the global parameter such has bending is not strongly affected by the presence of the cluster lesion. On the other hand the extrahelicity of the AP site, which is usually considered a signature of this class of lesions, is strongly altered and shows a remarkable clustered AP position dependency that can partially correlate with the processing rate.
| Sequence | Bend (400 ns) | Bend (40 ns) | AP extrahelicity |
|---|---|---|---|
| seq-5 | 34.1 ± 18.7° | 25.2 ± 13.4° | AP7: 12.4% ; AP35: 12.5 % |
| seq-3 | 38.3 ± 19.3° | 24.9 ± 19.3° | A1P12: 36.6% ; AP35: 27.7 % |
| seq-1 | 31.1 ± 16.5° | 23.6 ± 14.1° | AP11: 2.5% ; AP35: 3.9 % |
| seq0 | 34.3 ± 17.8° | 35.5 ± 18.2° | AP12 and AP35: 81.2 % |
| seq+1 | 27.0 ± 14.4° | 27.4 ± 13.5° | AP13: 2.1% ; AP35: 4.2 % |
| seq+3 | 35.3 ± 19.4° | 33.7 ± 18.1° | AP15: 13.2% ; AP35: 3.9 % |
| seq+5 | 31.5 ± 16.5° | 27.3 ± 13.6° | AP17: 30.3% ; AP35: 28.0 % |
| Reference | 32.3 ± 17.2° | 25.3 ± 11.8° | AP35: 35.9 % |
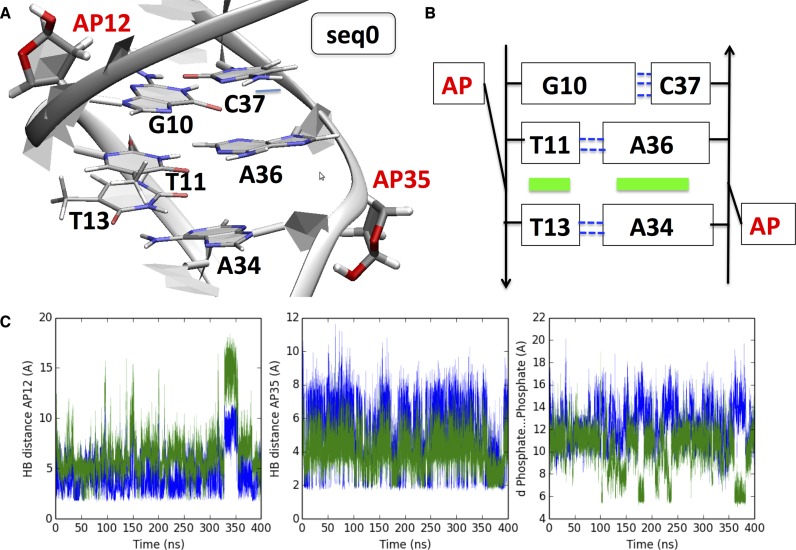
We first describe the situation where the two AP sites face each other (seq0). Because of the constraints imposed by the DNA backbone, the damaged sites are too distant to associate together by HB as the B-helix evolves in time. Instead, the oligonucleotide deploys an alternative strategy of stabilization by excluding the two AP sites (i.e. forcing their extrahelical residency), this in turn enables to stack T11 and T13, and A34 and A36. The former conformation is easily accessed and the structural reorganization occurs in less than 2 ns. The two Watson–Crick base pairs T11:A36 and T13:A34, hence maintain their pairing all along the simulation. As a result of the peculiar stabilization strategy the two AP sites present the same high extrahelical residency of 80%. Yet the two AP sites are still directed toward the core of the helix due to transient HB interactions with O4′ and N3 of proximal adenines (see Figure 2C). The 23-bp oligonucleotide presents a locally narrower minor groove in correspondence of the AP sites due to the B helix compression subsequent to the ejection of the lesions, but no marked deviation of the backbone. In contrast, backbone deformation leading to bulge have been observed in the case of single AP site and extensively studied (42–45). The absence of bulge is an important feature that one can relate to the lack of repair of seq0: the duplex structural reorganization, acts as such to make the damaged oligonucleotide extremely close to a non-damaged 22-bp B-DNA strand with, in its center, two dangling extrahelical AP sites. Consequently, not only recognition is made harder but a repair would imply the disruption of the newly formed and stable T11…T13 and A34…A36 stacking (Figure 2), i.e. an energetic penalty corresponding to ∼10 kcal/mol. However, even though the structural factors are certainly important the corresponding barrier is not too high and indeed seq0 is only barely less efficiently excised than seq-1 or seq-3 by Nfo. Concerning the evolution of the strand one may also note that the bending assumes an average value of 35.5 ± 18.2° over the last 40 ns, which is only slightly more pronounced than the single-AP-containing control sequence (25.3 ± 11.8°).
Figure 2.
(A) Cartoon representation of the final structure obtained after 400 ns for seq0 and (B) scheme of the non-covalent interactions reshaping the oligonucleotide: green boxes indicate new π-stacking, and dashed blue lines indicate Watson–Crick hydrogen bonds. (C) Time evolution of HB and phosphate–phosphate distances. Left panel: blue line AP12H1…T11O4′ distance, green line AP11H1…A36N1. Middle panel: AP35H1…A12N1 distance blue line, AP35H1…A34N3 distance green line. Right panel: blue line A12P…C37P distance, green line T15P…A36P; see also Supplementary Figure S4 for explicative diagrams.
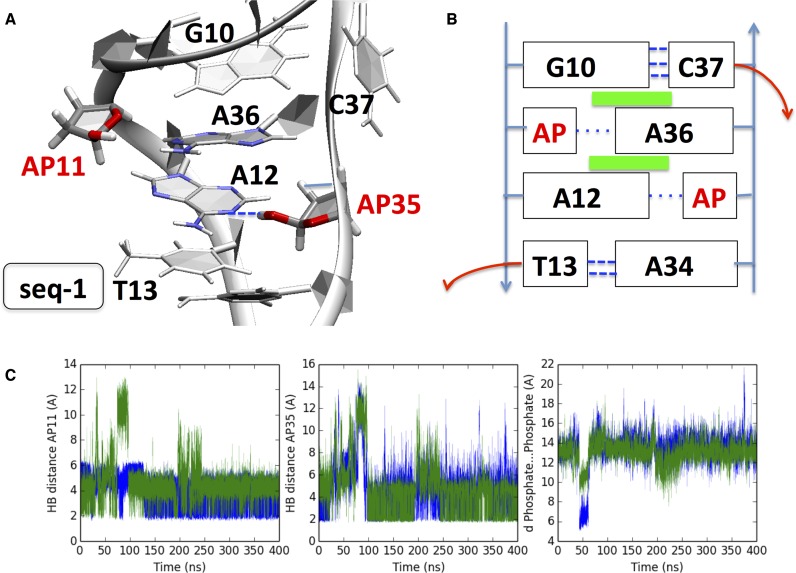
As the two AP sites are shifted by one base pair upstream (staggered 3′), i.e. for seq-1 a very distinct stabilization scheme is evidenced (Figure 3) with the two AP sites now occupying only scarcely extrahelical positions (∼3%, see Table 1). Figure 3C reveals that AP11 first pairs with A36:N1, through the terminal hydroxyl hydrogen H1, until a most stable HB with the vicinal O5′ atom occurs ∼330 ns. AP35 also involves a stable HB with A12:N1 up to 60 ns, and reforms interactions at 250 ns, also implying A34:N3. As a consequence all along the MD trajectory one can observe a swapping between the two HB patterns. The presence of the two AP sites initially isolates the orphan nucleobases A12 and A36 that are stabilized partly by HB formation. However, A36 reinstate favorable π-stacking interactions by intercalating between G10 and A12. This interstrand stacking of three purines triggers stabilization (compared to a situation where pyrimidines would be involved) and is accompanied by a noticeable local narrowing of the double helix (C1′…C1′ distances of 11.2 ± 1.3 Å, versus 18.2 ± 0.5 Å in absence of lesions). The geometrical effects induced by this peculiar arrangement are propagated to the nearby bases: C37 adopts an extrahelical position at 136 ns up to the end of the simulation, the former configuration is moreover locked by one strong hydrogen bond with one phosphate oxygen A36:O2P (see Supplementary Figure S7). Furthermore, we see that seq-1 does not exhibit a significant deviation of the overall bending when compared to the control sequence.
Figure 3.
(A) Cartoon representation of the final structure obtained after 400 ns for seq-1. (B) Diagram showing the interaction pattern: Green rectangles indicate transverse π-stacking, and dashed blue lines new hydrogen bonds (thick ones for Watson–Crick pairing). (C) Left panel: blue line AP11H1…AP11O5′ distance, green line AP11H1…A36N1. Middle panel: blue line AP35H1…A12N1distance, green line AP35H1…A34N3 distance. Right panel: blue line A12P…C37P distance, green line T13P…A36P; see also Supplementary Figure S5 for explicative diagrams. The two AP sites are mostly directed within the helix, owing to HB with vicinal nucleobases: the cytosine C37 adopts an extrahelical position. A36 develops interstrand stacking with G10 and A12.
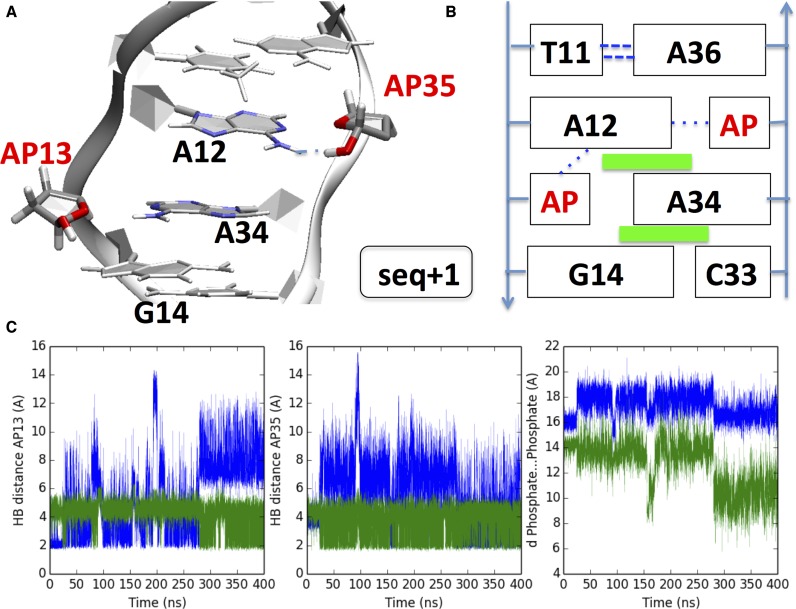
In the case of seq+1, i.e. the reciprocal downstream duplex featuring the two abasic sites at 13th and 35th positions, we observe very different structural features, with notably no extrusion of pyrimidines (Figure 4). AP13 and AP35 sites rapidly seek to form hydrogen bonds with the orphan A12 and A34 bases, and most notably the two AP sites do not adopt extrahelical position. AP13 is hydrogen bonded to A12:N3, but this interaction is disrupted after around 280 ns as AP13 favors a pairing with A34:N1 and also a more transient interaction with a proximal O5′ oxygen of the sugar moiety. A12, through its nitrogen N1, becomes pivotal in favoring a rearrangement ultimately ending up in the formation of a HB for AP35. Also G14 ends forming a hydrogen bond with the AP13 residue further stabilizing its position (see Figure 4). One identifies a stacking between A12, A34 and G14, but with only a limited impact on the C1′…C1′ distances compared to seq-1, that correlates with the non-exclusion of the vicinal pyrimidines. The duplex exhibits a bend angle of 27.0 ± 14.4°, almost constant along the trajectory.
Figure 4.
(A) Cartoon representation of the final structure obtained after 400 ns for seq+1 and (B) diagram illustrating the non-covalent interactions and the position of the bases. Dispersion interaction are represented as green rectangles, while dotted blue lines represent hydrogen bonds. (C) Left panel: blue line AP13H1…A12N3 distance, green line AP13H1…AP13:O5′. Middle panel: blue line A12N1…AP35H1 distance, green line AP35O5′…AP35H1distance. Right panel: blue line A12:P – A36:P distance, green line AP13 – AP35; see also Supplementary Figure S6 for explicative diagrams. The two AP sites are mostly directed within the helix, owing to HB with vicinal nucleobases: however, an interstrand stacking between A12…A34…G14 is observed.
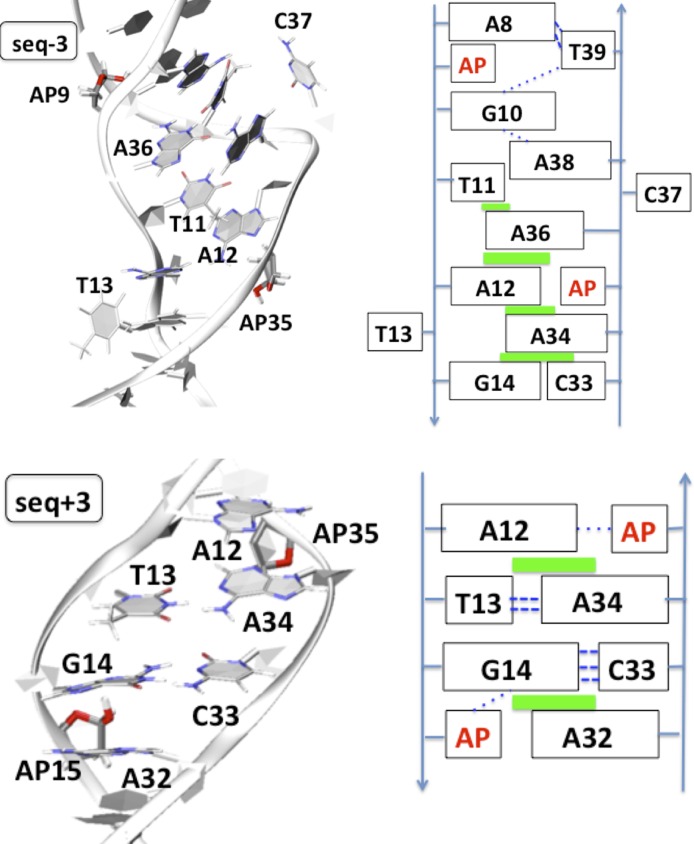
As the AP sites are more widely separated, like in seq+3, the duplex still evolves to develop interstrand purine stacking, for A12 and A34 as shown in Figure 5 (bottom). This implies a moderate constriction of the B-helix reflected by C1′…C1′ distances up to ∼16.3 ± 1.1 Å. We also note that the Watson–Crick pairing of the two base pairs in between the defects, T13…A34 and G14…C33 is maintained. The AP15 and AP35 damages adopt transiently extrahelical positions accounting for 13.2% versus 3.9%: hence they are only barely solvent exposed, and their –OH group transiently points toward an HB partner available in the surrounding (G14 and A12, respectively). On the other hand the orphan A32 base adopts an intra-helical position and develops attractive stacking interactions with G14. The DNA backbone is flexible enough to accommodate interstrand stacking, and the two central base pairs are left unaltered in their Watson–Crick pairing. Interestingly the structure is slightly more bended than the reference sequence by more than 8°.
Figure 5.
Cartoon representation of the final structure obtained after 400 ns for seq−3 (top) and seq+3 (bottom). A schematic representation is given at the right where green squares indicate π-stacking, and dashed blue lines new hydrogen bonds (thick ones for Watson–Crick pairing).
The structural evolution of the 23-bp duplex seq-3 (Figure 5 top) featuring AP sites as 9th and 35th residues is also driven by the need to fill up the space left unoccupied by the excised thymines. Differently from seq+3 the pairing between the central non-damaged base pairs (T11…A36 and G10…C37) rapidly experience a disruption at ∼34 and 64 ns ending up to the exclusion of C37 at ∼280 ns and T13 at ∼320 ns. This conformation constitute a very distinctive feature of seq-3 and present specifically complex features, such as the stabilization of the two ejected pyrimidines between 76 and 244 ns via a single HB and extended intra- and inter-strand π-stacking developing from T11 to C33 base. Not surprisingly, due to the complex structural rearrangements the duplex exhibits polymorphism as evidenced by the bending that can reach values up to 75°.
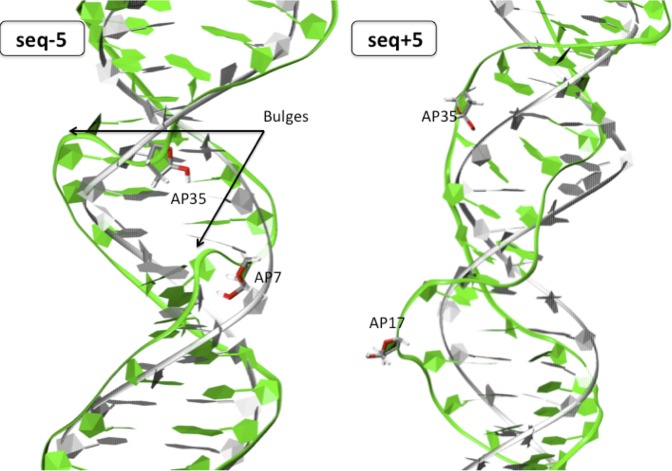
Further increasing the separation between the two AP sites leads to a situation where they begin to behave independently and partially loose the correlation between the AP sites. The structure of seq-5 (Figure 6 left panel) follows the rather well established behavior of isolated AP sites although we observe an extra-helical occupation of ∼12.5%, i.e. a noticeably lower extrahelicity than for isolated AP sites. The AP sugar on the other hand explores a broad range of sugar puckers angles giving rise to a well established DNA bulge typical of AP strands (42). AP35 exhibits many HB partners along the MD trajectory, yet mostly interacts with the oxygen of A34. A similar interaction pattern is found for AP7, which interacts with its O5′ despite the steric constraint it induces to the backbone. The stacking remains identical to the one of the reference structure, with no deviation for the orphan A12 and A40 and no interstrand stacking. The DNA bending is only weakly perturbed (25.2 ± 13.4°) and assumes values close to the ones experienced by single AP sites.
Figure 6.
Cartoon representation of seq−5 (left) and seq+5 (right) after 400 ns of classical simulations. Both structures exhibit backbone deformation.
The symmetrical sequence seq+5 (Figure 6 right panel) shares common overall features, with no marked reorganization such as the ones observed for closer AP sites. The extrahelical occupancy accounts for roughly 30% along our simulations, this value being extremely close to the one of the control double-strand. As for the case of seq-5 it also shows a distinctive bulge all along the trajectory due to the backbone deformation in correspondence to the two AP sites. The Watson–Crick pairing of the four separating base pair is maintained all along the 400 ns trajectory, and the bending of the helix remains limited (27.3 ± 13.6°).
DISCUSSION
AP sites can occur either as a result of regular oxidative stress or IR, or as an intermediate product resulting from the removal of an oxidized base lesion by a DNA glycosylase. Unrepaired AP sites are highly toxic and can carry mutagenic potential since they are non-coding and they can result in transversions due to the erroneous incorporation of DNA bases opposite to the AP site in replication or repair (46,47). Whereas the occurrence of one isolated AP site has been more widely studied and is now well characterized, quantifying bistranded clustered DNA damage induction and repair turns out to be more difficult. In addition, it is very important to address the relationship between clustered AP sites and DSBs. Accumulating experimental but also theoretical evidences, suggest that, the processing of a DSB can be impeded or inhibited by the presence of non-DSB neighboring lesions and the same stands for the processing of an oxidized base or AP site due to the presence of breaks or non-DSB lesions. (48–50) Understanding the processing of an AP site within a cluster lesion is considered of major importance. Most notably, different in vitro repair studies have shown that processing of complex lesions follows a sequential established hierarchy in the case of an AP site(s) and an 8-oxodG or SSB etc. Specifically, it is suggested that the AP sites and the resulting SSB(s) are processed and repaired before any DNA glycosylase binds and excises the neighboring base lesions (13,46,51–53), thus leading to the so-called ‘DSB avoidance’ and repair retardation of the lesions belonging to a cluster (9). However, ‘DSB avoidance’ is not universal and is mostly likely to occur for cluster composed of oxidative damaged bases it is not always happening in case of bistranded AP site or complex, uracil-containing lesions (54,55). Hence, elucidating at a molecular level the structural deformations induced on DNA by complex lesions patterns, as well as the interplay with repair enzymes recognition, is of clear importance and relevance.
To examine the effects of such complex damages on DNA structure, we have analyzed a series of cluster abasic lesions-containing oligonucleotides differing by the number of separating base pairs (up to four) and the strand orientation. We stress out the need for hundreds of ns dynamics to capture the inherent complex reorganization of the duplex and the multiple conformations spanned by these complex systems. In contrast with experimental techniques such as NMR, our simulations allow probing transient interactions such as weak hydrogen bonds, which may ultimately dictate the global structure rearrangements, as well as the conformational flexibility of the systems. We evidence the following general trends: (i) the duplex evolves, as much as possible, to restore an ideal B-helicity with a global bending close to 30°, this aspect is also coherent with the NMR results recently found by Zalesak et al. on DNA strand featuring clustered AP and 8-oxoguanine damages (56). However (ii) interstrand dispersive interactions take place and play a major role in stabilizing the AP sites (iii) the local constriction of the helix, i.e. a narrower minor groove, correlates with the number of separating base pairs, (iv) AP positions can be locked by strong hydrogen bonding, and (v), when separated by 4 base pairs, the AP sites behave almost independently.
Clusters of closely spaced oxidative DNA lesions present challenges to the cellular repair machinery, especially in the case of bacterial organisms (17,21,57). As expected the closest the AP sites the more difficult the repair-enzyme processing with seq0 being the less efficiently repaired one. However, a stunning low repair ratio has been evidenced for some sequences such as seq-3 and to a lesser extent seq-1 suggesting the emergence of peculiar interactions and structural deformations strongly affecting the recognition patterns between damaged DNA strands and proteins. On the contrary, human endonuclease experiences a globally higher processing rate and a less pronounced dependency on the secondary AP position. Our results provide a set of conformations constituting a preliminary step to reveal the interactions of clustered AP sites with repair enzymes. Yet, the relation between the repair ratio and the structure is not entirely straightforward. For instance, formation of a complex with a repair enzyme could induce a constraint on DNA (57) that may trigger new conformations of the duplex, whose flexibility may be increased by the clustered AP. Furthermore, clustered AP sites embrace a combinatorial biochemistry, and their complex structural, dynamical and thermodynamical (58,59) properties may be strongly sequence dependent.
Human APE1 and its counterpart the bacterial enzyme Nfo exhibit a striking difference in their repair capacity of clustered AP sites. With the exception of only one particular sequence (seq+3), APE1 has a higher processing rate than the Nfo protein (Figure 1). This suggests different evolutionary pathways (33), which are probably rooted in different interaction patterns between the oligonucleotides and the repair proteins. As revealed by the crystal structures (6), APE1 has an extended contact area between the enzyme and the DNA, furthermore strong electrostatic interactions between positively-charged residues and the negatively-charged DNA backbone induces rather large structural deformations of the helix. This fact was also elegantly revealed by Beloglazova et al. (60) in their study of thermodynamic, kinetic and structural basis for recognition of AP site by APE1. Hence, in the case of APE1 specific interaction and DNA deformation dominates, and hence the repair ratio cannot be correlated with the duplex deformation, this aspect and notably the surprisingly low repair of seq+3 by APE1 will be the object of a forthcoming contribution. On the other hand interaction with Nfo (61) is much more local and, as a consequence, the DNA structure is almost not constrained by the protein. Hence, in the case of Nfo, the structural modifications of the DNA duplex will directly influence the stability of the protein/nucleic acid aggregate and hence will largely correlate with the observed process efficiency. However, one should be aware of the fact that the process rate depends on many complex factors, comprising the extrusion of the lesioned base, its accommodation in the active pocket and the subsequent interaction developing with adjacent bases, therefore an absolute quantitative correlation cannot be drawn on the basis of structural factors, only.
Consistently with previous observations and computational studies, our data clearly show that the distortion induced by a single AP site is not inducing a permanent gap or hole in the strongly coupled and organized double helix. Instead, and also taking advantage of the well-known flexibility of the DNA backbone, the macromolecule experiences a considerable local rearrangement to restore the maximal number of attractive non-covalent interactions. Two of the most paradigmatic modifications are the significant extrahelical position of the AP site and the induction of an important kink and bulge in the helix. Normally those two features are recognized as flags triggering the repair process (28).
Concerning the dynamic behavior of clustered AP sites, our results suggest a complex situation, associated with serious consequences on the efficiency of repair, especially in the case of Nfo. Furthermore, the induced structural deformations are strongly entangled with the specificity inherent to a single DNA sequence and hence cannot be captured by using only one simple descriptor. As an interesting example, and in agreement with previous NMR analysis (4), we found that the extrahelical behavior is strongly dependent on the specific clustered AP. Furthermore, while in seq0 and seq−3 the extruded AP sites occupy preferentially the minor groove, in the case of seq±1 and seq±5 the major groove occupancy predominates, seq+3 has a particular behavior with only one AP site preferentially occupying the minor groove. Consistently with the experimental observation (17, 21), we may safely conclude that the presence of four separating base pairs represents the limit under which two AP sites can no longer be considered as independent. Consequently, the repair rate for such well separated cluster sites is quite high and, even in the case of Nfo, close to 100% for seq+5, since no peculiar structural distortion hamper the recognition by the repair enzyme (Figure 1). Even if a general conclusion can be drawn, it is however important to note, that some sequence effects still exist, and seq-5 has a slightly lower repair ratio, which we relate to a lower percentage of extrahelicity for both AP sites (∼12 %, versus ∼30% for seq+5 and ∼33% for single AP sites).
Seq0 is the oligomer showing the less efficient repair by Nfo, yet the extrahelicity of the two AP sites is extremely high, close to 80%. The ejection of the AP sites is accompanied by a strong deformation of the DNA that consistently shrinks in order to reinstate π-stacking between the vicinal base pairs surrounding the two APs. This deformation is accompanied by a narrowing of the minor groove, in absence of any important kinking. Hence, seq0 is characterized by a global structure significantly different from the one shown by a conventional AP site, thus precluding the recognition by the bacterial repair enzyme that indeed necessitates interaction with the minor groove.
Low extrahelicities and important local deformations characterize the duplexes seq±1. However, once again the seq-1 is somehow peculiar since non-damaged nucleobases too assume extrahelical position. This fact, also reported based on NMR structures (4,32,62), once again imposes structural modifications in the groove region, i.e. in the regions developing interactions with the Nfo enzyme, hence the repair ratio is even lower than for the corresponding seq+1.
However, the most striking position dependence is the one happening between seq+3 and seq−3. Indeed, seq+3 is almost perfectly repaired by Nfo and its efficiency is comparable with seq−5. Interestingly enough, all the structural parameters, particularly the extrahelicity and the kink, are extremely similar for the two cases. On the other hand the strong structural deformation, evidenced for seq−3 with the ejection of non-damaged bases, and, the development of an extended interstrand stacking between different base pairs, result in a very low repair ratio, which could be the consequence of a weak interaction of the altered DNA with Nfo.
The strong asymmetry in Nfo repair efficiency observed experimentally is dictated by the various local environments experienced by the AP sites and the nearby nucleobases. In particular, an extremely complex pattern of non-covalent interactions between the double-helix constituents comes into play, going up to the development of transverse stacking. Even if it is not the sole parameter playing an important role, extrahelicity propensity and conversely the presence of hydrogen bonds or transverse stacking locking the AP site, is certainly strongly correlated to the Nfo repair ratio of a given cluster lesion. While the local embedding and structural deformations around a cluster site are fundamental to understand its repair propensity, global deformations such as DNA bend angle do not have any strong influence. All these effects ultimately sum up to give a clear relation between the Nfo repair efficiency and the separation of the AP sites. The repair efficiency increases with the distance separating the residues bearing lesions. For AP sites distant by 5 bp apart to each other, i.e. at a distance larger than the enzyme recognition area (around 17 Å), repair is almost as efficient as for simple lesions. However, in some cases clustered AP position may severely alter the previous trend, or even almost reverse it, so that seq-3 has a dramatic low repair.
Supplementary Material
Acknowledgments
Calculations were performed using the local HPC resources of PSMN at ENS-Lyon and at Université de Lorraine – Nancy. The support by the COST Action CM1201 “Biomimetic Radical Chemistry" is gratefully acknowledged. This work was performed within the framework of the LABEX PRIMES (ANR-11-LABX0063) of Université de Lyon, within the program “Investissements d'Avenir" (ANR-11-IDEX0007) operated by the French National Research Agency (ANR). Dr Georgakilas was supported by an EU grant MC-CIG-303514.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
COST Action CM1201 “Biomimetic Radical Chemistry"; this work was performed within the framework of the LABEX PRIMES (ANR-11-LABX0063) of Université de Lyon, within the program “Investissements d'Avenir" (ANR-11-IDEX0007) operated by the French National Research Agency (ANR); EU grant [MC-CIG-303514 to Dr Georgakilas]. Funding for open access charge: University of Lorraine and CNRS.
Conflict of interest statement. None declared.
REFERENCES
- 1.Demple B., Sung J.-S. Molecular and biological roles of Ape1 protein in mammalian base excision repair. DNA Repair (Amst) 2005;4:1442–1449. doi: 10.1016/j.dnarep.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Clauson C., Schärer O.D., Niedernhofer L. Advances in understanding the complex mechanisms of DNA interstrand cross-link repair. Cold Spring Harb. Perspect. Biol. 2013;5:a012732. doi: 10.1101/cshperspect.a012732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gates K.S. An overview of chemical processes that damage cellular DNA: spontaneous hydrolysis, alkylation, and reactions with radicals. Chem. Res. Toxicol. 2009;22:1747–1760. doi: 10.1021/tx900242k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hazel R.D., Tian K., de Los Santos C. NMR solution structures of bistranded abasic site lesions in DNA. Biochemistry. 2008;47:11909–11919. doi: 10.1021/bi800950t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mol C.D., Izumi T., Mitra S., Tainer J.A. DNA-bound structures and mutants reveal abasic DNA binding by APE1 and DNA repair coordination [corrected] Nature. 2000;403:451–456. doi: 10.1038/35000249. [DOI] [PubMed] [Google Scholar]
- 6.Freudenthal B.D., Beard W.A., Cuneo M.J., Dyrkheeva N.S., Wilson S.H. Capturing snapshots of APE1 processing DNA damage. Nat. Struct. Mol. Biol. 2015;22:924–931. doi: 10.1038/nsmb.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe R., Rahmanian S., Nikjoo H. Spectrum of radiation-induced clustered non-DSB damage – A Monte Carlo track structure modeling and calculations. Radiat. Res. 2015;183:525–540. doi: 10.1667/RR13902.1. [DOI] [PubMed] [Google Scholar]
- 8.Nikjoo H., O'Neill P., Terrissol M., Goodhead D.T. Quantitative modelling of DNA damage using Monte Carlo track structure method. Radiat. Environ. Biophys. 1999;38:31–38. doi: 10.1007/s004110050135. [DOI] [PubMed] [Google Scholar]
- 9.Georgakilas A.G., O'Neill P., Stewart R.D. Induction and repair of clustered DNA lesions: What do we know so far? Radiat. Res. 2013;180:100–109. doi: 10.1667/RR3041.1. [DOI] [PubMed] [Google Scholar]
- 10.Liu Z., Ding S., Kropachev K., Jia L., Amin S., Broyde S., Geacintov N.E. Resistance to nucleotide excision repair of bulky guanine adducts opposite abasic sites in DNA duplexes and relationships between structure and function. PLoS One. 2015;10:e0137124. doi: 10.1371/journal.pone.0137124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikitaki Z., Hellweg C.E., Georkakilas A.G., Ravanat J.L. Stress-induced DNA damage biomarkers: Applications and limitations. Front. Chem. 2015;3:35. doi: 10.3389/fchem.2015.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shikazono N., Akamatsu K., Takahashi M., Noguchi M., Urushibara A., O'Neill P., Yokoya A. Significance of DNA polymerase I in in vivo processing of clustered DNA damage. Mutat. Res. Mol. Mech. Mutagen. 2013;749:9–15. doi: 10.1016/j.mrfmmm.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 13.Lomax M.E., Cunniffe S., O'Neill P. Efficiency of repair of an abasic site within DNA clustered damage sites by mammalian cell nuclear extracts. Biochemistry. 2004;43:11017–11026. doi: 10.1021/bi049560r. [DOI] [PubMed] [Google Scholar]
- 14.Sutherland B.M., Bennett P.V., Sidorkina O., Laval J. Clustered DNA damages induced in isolated DNA and in human cells by low doses of ionizing radiation. Proc. Natl. Acad. Sci. U.S.A. 2000;97:103–108. doi: 10.1073/pnas.97.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chastain P.D., Nakamura J., Rao S., Chu H., Ibrahim J.G., Swenberg J.A., Kaufman D.G. Abasic sites preferentially form at regions undergoing DNA replication. FASEB J. 2010;24:3674–3680. doi: 10.1096/fj.09-145276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chastain P.D., Nakamura J., Swenberg J., Kaufman D. Nonrandom AP site distribution in highly proliferative cells. FASEB J. 2006;20:2612–2614. doi: 10.1096/fj.06-6145fje. [DOI] [PubMed] [Google Scholar]
- 17.Georgakilas A.G., Bennett P.V., Sutherland B.M. High efficiency detection of bi-stranded abasic clusters in gamma-irradiated DNA by putrescine. Nucleic Acids Res. 2002;30:2800–2808. doi: 10.1093/nar/gkf393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Singh V., Das P. Condensation of DNA–a putative obstruction for repair process in abasic clustered DNA damage. DNA Repair (Amst) 2013;12:450–457. doi: 10.1016/j.dnarep.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 19.Akamatsu K., Shikazono N., Saito T. Localization estimation of ionizing radiation-induced abasic sites in DNA in the solid state using fluorescence resonance energy transfer. Radiat. Res. 2015;183:105–113. doi: 10.1667/RR13780.1. [DOI] [PubMed] [Google Scholar]
- 20.Eccles L.J., Menoni H., Angelov D., Lomax M.E., O'Neill P. Efficient cleavage of single and clustered AP site lesions within mono-nucleosome templates by CHO-K1 nuclear extract contrasts with retardation of incision by purified APE1. DNA Repair (Amst) 2015;35:27–36. doi: 10.1016/j.dnarep.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Georgakilas A.G., Bennett P.V., Wilson D.M., 3rd, Sutherland B.M. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 2004;32:5609–5620. doi: 10.1093/nar/gkh871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pérez A., Luque F.J., Orozco M. Frontiers in molecular dynamics simulations of DNA. Acc. Chem. Res. 2012;45:196–205. doi: 10.1021/ar2001217. [DOI] [PubMed] [Google Scholar]
- 23.Dumont E., Monari A. Understanding DNA under oxidative stress and sensitization: The role of molecular modeling. Front. Chem. 2015;3:43. doi: 10.3389/fchem.2015.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ayadi L., Coulombeau C., Lavery R. The impact of abasic sites on DNA flexibility. J. Biomol. Struct. Dyn. 2000;17:645–653. doi: 10.1080/07391102.2000.10506555. [DOI] [PubMed] [Google Scholar]
- 25.Ayadi L., Coulombeau C., Lavery R. Abasic sites in duplex DNA: Molecular modeling of sequence-dependent effects on conformation. Biophys J. 1999;77:3218–3226. doi: 10.1016/S0006-3495(99)77152-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barsky D., Foloppe N., Ahmadia S., Wilson D.M., 3rd, MacKerell A.D. Jr. New insights into the structure of abasic DNA from molecular dynamics simulations. Nucleic Acids Res. 2000;28:2613–2626. doi: 10.1093/nar/28.13.2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mu H., Kropachev K., Wang L., Zhang L., Kolbanovskiy A., Kolbanovskiy M., Geacintov N.E., Broyde S. Nucleotide excision repair of 2-acetylaminofluorene- and 2-aminofluorene-(C8)-guanine adducts: Molecular dynamics simulations elucidate how lesion structure and base sequence context impact repair efficiencies. Nucleic Acids Res. 2012;40:9675–9690. doi: 10.1093/nar/gks788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Churchill C.D.M., Eriksson L.A., Wetmore S.D. DNA distortion caused by uracil-containing intrastrand cross-links. J. Phys. Chem. B. 2016;120:1195–1204. doi: 10.1021/acs.jpcb.5b10381. [DOI] [PubMed] [Google Scholar]
- 29.Bignon E., Gattuso H., Morell C., Dumont E., Monari A. DNA photosensitization by an insider: Photophysics and triplet energy transfer of 5-Methyl-2-pyrimidone deoxyribonucleoside. Chemistry. 2015;21:11509–11516. doi: 10.1002/chem.201501212. [DOI] [PubMed] [Google Scholar]
- 30.Faraji S., Dreuw A. Physicochemical mechanism of light-driven DNA repair by (6-4) photolyases. Annu. Rev. Phys. Chem. 2014;65:275–292. doi: 10.1146/annurev-physchem-040513-103626. [DOI] [PubMed] [Google Scholar]
- 31.Fujimoto H., Pinak M., Nemoto T., O'Neill P., Kume E., Saito K., Maekawa H. Molecular dynamics simulation of clustered DNA damage sites containing 8-oxoguanine and abasic site. J. Comput. Chem. 2005;26:788–798. doi: 10.1002/jcc.20184. [DOI] [PubMed] [Google Scholar]
- 32.Zálešák J., Lourdin M., Krejčί L., Constant J.F., Jourdan M. Structure and dynamics of DNA duplexes containing a cluster of mutagenic 8-oxoguanine and abasic site lesions. J. Mol. Biol. 2014;426:1524–1538. doi: 10.1016/j.jmb.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 33.Lu D., Silhan J., MacDonald J.T. Structural basis for the recognition and cleavage of abasic DNA in Neisseria meningitidis. Proc. Natl. Acad. Sci. U.S.A. 2012;109:16852–16857. doi: 10.1073/pnas.1206563109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Case D.A., Berryman J.T., Betz R.M., Cerutti D.S., Chatham T.E. III, Darden T.A., Duke R.E., Giese T.J., Gohlke H., Goetz A.W., et al. AMBER. 2015.
- 35.Cornell W.D., Cieplak P., Bayly C.I., Gould I.R., Merz K.M., Ferguson D.M., Spellmeyer D.C., Fox T., Caldwell J.W., Kollman P.A. A second generation force field for the simulation of proteins, nucleic acids, and organic molecules. J. Am. Chem. Soc. 1995;117:5179–5197. [Google Scholar]
- 36.Wang J., Wolf R.M., Caldwell J.W., Kollman P.A., Case D.A. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 37.Pérez A., Marchán I., Svozil D., Sponer J., Cheatham T.E., 3rd, Laughton C.A., Orozco M. Refinement of the AMBER force field for nucleic acids: Improving the description of alphaγConformers. Biophys. J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang J., Cieplak P., Kollman P.A. How well does a restrained electrostatic potential (RESP) model perform in calculating conformational energies of organic and biological molecules? J. Comput. Chem. 2000;21:1049–1074. [Google Scholar]
- 39.Mark P., Nilsson L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A. 2001;105:9954–9960. [Google Scholar]
- 40.Lavery R., Moakher M., Maddocks J.H. Conformational analysis of nucleic acids revisited: Curves+ Nucleic Acids Res. 2009;37:5917–5929. doi: 10.1093/nar/gkp608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.de los Santos C., El-khateeb M., Rege P., Tian K., Johnson F. Impact of the C1′ configuration of abasic sites on DNA duplex structure. Biochemistry. 2004;43:15349–15357. doi: 10.1021/bi048400c. [DOI] [PubMed] [Google Scholar]
- 42.Zeglis B.M., Boland J.A., Barton J.K. Recognition of abasic sites and single base bulges in DNA by a metalloinsertor. Biochemistry. 2009;48:839–849. doi: 10.1021/bi801885w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shi X., Beauchamp K.A., Harbury P.B., Herschlag D. From a structural average to the conformational ensemble of a DNA bulge. Proc. Natl. Acad. Sci. U.S.A. 2014;111:E1473–E1480. doi: 10.1073/pnas.1317032111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li M., Völker J., Breslauer K.J., Wilson D.M., 3rd APE1 incision activity at abasic sites in tandem repeat sequences. J. Mol. Biol. 2014;426:2183–2198. doi: 10.1016/j.jmb.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chiba J., Aoki S., Yamamoto J. Deformable nature of various damaged DNA duplexes estimated by an electrochemical analysis on electrodes. Chem. Commun. (Camb) 2014;50:11126–11128. doi: 10.1039/c4cc04513k. [DOI] [PubMed] [Google Scholar]
- 46.Cunniffe S., O'Neill P., Greenberg M.M., Lomax M.E. Reduced repair capacity of a DNA clustered damage site comprised of 8-oxo-7,8-dihydro-2′-deoxyguanosine and 2-deoxyribonolactone results in an increased mutagenic potential of these lesions. Mutat. Res. Mol. Mech. Mutagen. 2014;762:32–39. doi: 10.1016/j.mrfmmm.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kunkel T.A., Schaaper R.M., Loeb L.A. Depurination-induced infidelity of deoxyribonucleic acid synthesis with purified deoxyribonucleic acid replication proteins in vitro. Biochemistry. 1983;22:2378–2384. doi: 10.1021/bi00279a012. [DOI] [PubMed] [Google Scholar]
- 48.Lomax M.E., Folkes L.K., O'Neill P. Biological consequences of radiation-induced DNA damage: Relevance to radiotherapy. Clin. Oncol. (R. Coll. Radiol.) 2013;25:578–585. doi: 10.1016/j.clon.2013.06.007. [DOI] [PubMed] [Google Scholar]
- 49.Sage E., Harrison L. Clustered DNA lesion repair in eukaryotes: Relevance to mutagenesis and cell survival. Mutat. Res. Mol. Mech. Mutagen. 2011;711:123–133. doi: 10.1016/j.mrfmmm.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Eccles L.J., O'Neill P., Lomax M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat. Res. Mol. Mech. Mutagen. 2011;711:134–141. doi: 10.1016/j.mrfmmm.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malyarchuk S., Castore R., Harrison L. Apex1 can cleave complex clustered DNA lesions in cells. DNA Repair (Amst) 2009;8:1343–1354. doi: 10.1016/j.dnarep.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eccles L.J., Lomax M.E., O'Neill P. Hierarchy of lesion processing governs the repair, double-strand break formation and mutability of three-lesion clustered DNA damage. Nucleic Acids Res. 2010;38:1123–1134. doi: 10.1093/nar/gkp1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eot-Houllier G., Eon-Marchais S., Gasparutto D., Sage E. Processing of a complex multiply damaged DNA site by human cell extracts and purified repair proteins. Nucleic Acids Res. 2005;33:260–271. doi: 10.1093/nar/gki165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sedletska Y., Radicella J.P., Sage E. Replication fork collapse is a major cause of the high mutation frequency at three-base lesion clusters. Nucleic Acids Res. 2013;41:9339–9348. doi: 10.1093/nar/gkt731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kozmin S.G., Sedletska Y., Reynaud-Angelin A., Gasparutto D., Sage E. The formation of double-strand breaks at multiply damaged sites is driven by the kinetics of excision/incision at base damage in eukaryotic cells. Nucleic Acids Res. 2009;37:1767–1777. doi: 10.1093/nar/gkp010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zalesak J., Constant J.-F., Jourdan M. NMR solution structure of DNA featuring clustered 2′-deoxyribonolactone and 8-oxoguanine lesions. Biochemistry. 2016;55:3899–3906. doi: 10.1021/acs.biochem.6b00396. [DOI] [PubMed] [Google Scholar]
- 57.Hosfield D.J., Guan Y., Haas B.J., Cunningham R.P., Tainer J.A. Structure of the DNA repair enzyme endonuclease IV and its DNA complex: Double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell. 1999;98:397–408. doi: 10.1016/s0092-8674(00)81968-6. [DOI] [PubMed] [Google Scholar]
- 58.Ghosh S., Greenberg M.M. Correlation of thermal stability and structural distortion of DNA interstrand cross-links produced from oxidized abasic sites with their selective formation and repair. Biochemistry. 2015;54:6274–6283. doi: 10.1021/acs.biochem.5b00860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gelfand C.A., Plum G.E., Grollman A.P., Johnson F., Breslauer K.J. Thermodynamic consequences of an abasic lesion in duplex DNA are strongly dependent on base sequence. Biochemistry. 1998;37:7321–7327. doi: 10.1021/bi9803372. [DOI] [PubMed] [Google Scholar]
- 60.Beloglazova N.G., Kirpota O.O., Starostin K.V., Ishchenko A.A., Yamkovoy V.I., Zharkov D.O., Douglas K.T., Nevinsky G.A. Thermodynamic, kinetic and structural basis for recognition and repair of abasic sites in DNA by apurinic/apyrimidinic endonuclease from human placenta. Nucleic Acids Res. 2004;32:5134–5146. doi: 10.1093/nar/gkh846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mazouzi A., Vigouroux A., Aikeshev B., Brooks P.J., Saparbaev M.K., Morera S., Ishchenko A.A. Insight into mechanisms of 3′-5′ exonuclease activity and removal of bulky 8,5′-cyclopurine adducts by apurinic/apyrimidinic endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2013;110:E3071–E3080. doi: 10.1073/pnas.1305281110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lin Z., Hung K.N., Grollman A.P., de los Santos C. Solution structure of duplex DNA containing an extrahelical abasic site analog determined by NMR spectroscopy and molecular dynamics. Nucleic Acids Res. 1998;26:2385–2391. doi: 10.1093/nar/26.10.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.