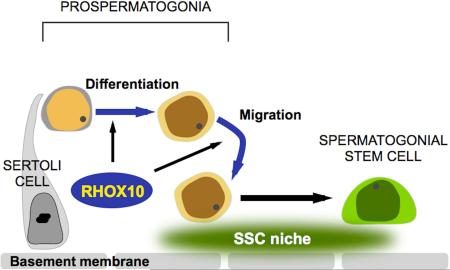
Summary
The developmental origins of most adult stem cells are poorly understood. Here, we report the identification of a transcription factor—RHOX10—that is critical for the initial establishment of spermatogonial stem cells (SSCs). Conditional loss of the entire 33-gene X-linked homeobox gene cluster that includes Rhox10 causes progressive spermatogenic decline, a phenotype indistinguishable from that caused by loss of only Rhox10. We demonstrate that this phenotype results from dramatically reduced SSC generation. Using a battery of approaches, including single cell-RNAseq (scRNAseq) analysis, we show that Rhox10 drives SSC generation by promoting Pro-spermatogonia differentiation. Rhox10 also regulates batteries of migration genes and promotes the migration of Pro-spermatogonia into the SSC niche. The identification of an X-linked homeobox gene that drives the initial generation of SSCs has implications for the evolution of X-linked gene clusters and sheds light on regulatory mechanisms influencing adult stem cell generation in general.
Graphical Abstract
Introduction
Spermatogonial stem cells (SSCs) support spermatogenesis throughout adult life. While considerable strides have been made in our understanding of SSCs, little is known about the process that initially generates these cells. The broadly defined cell type that gives rise to SSCs is called a “Pro-spermatogonium” (ProSG; also called a gonocyte), a finite and transient cell type that is poorly understood (Culty, 2009). Several stages of ProSG exist that are defined by their anatomical position and mitotic activity (Figure 1A). Multiplying (M) ProSG are derived from primordial germ cells (PGCs), the rapidly dividing cells responsible for initial germ cell seeding of the gonad. M-ProSG give rise to primary transitional (T1) ProSG, which are mitotically silent but molecularly active, as they reestablish DNA methylation marks erased in M-ProSG and PGCs. Upon resumption of mitosis, T1-ProSG become secondary transitional (T2) ProSG, which are regarded as the immediate precursor cells that give rise to SSCs (McCarrey, 2013). A unique feature of T2 ProSG is that they are migratory, a characteristic that allows them to traverse from the center of the seminiferous epithelial tubule to its periphery. This alters their local microenvironment, including signals that evidence suggests drive these precursor cells to differentiate into SSCs (Manku and Culty, 2015). An analogous migration step is thought to occur in humans (Guatelli-Steinberg and Boyce, 2012). To date, no transcription factors that drive this conserved migration event into the “SSC niche” have been identified.
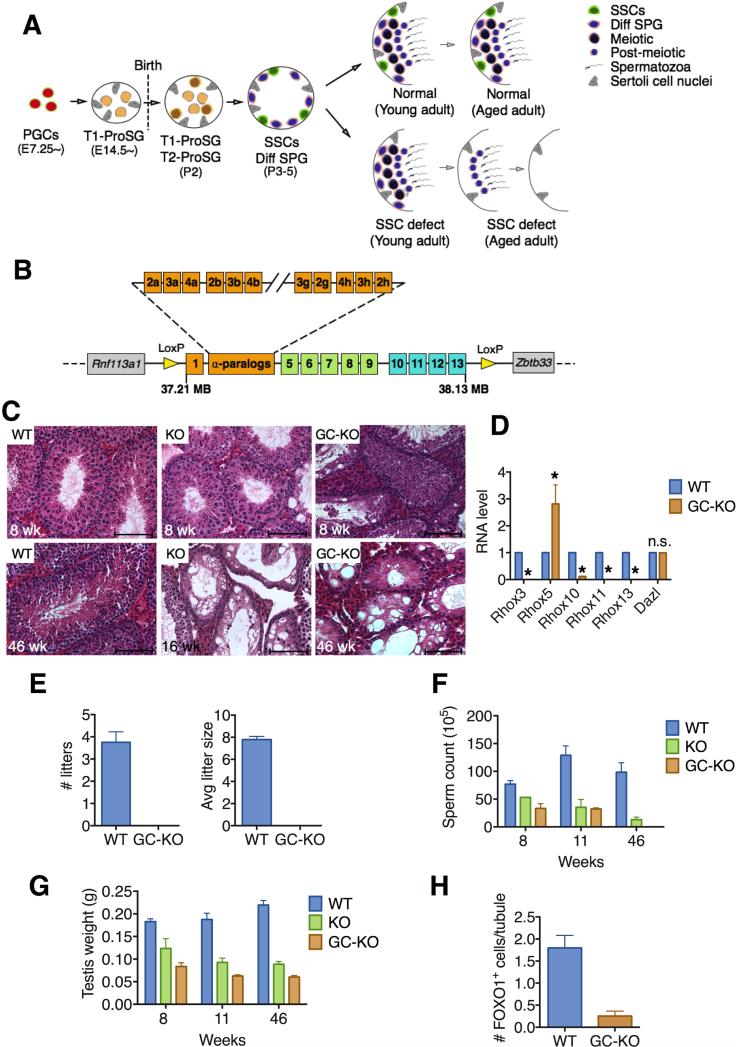
Figure 1. Loss of the Rhox Cluster Causes Progressive Spermatogenic Decline.
(A) Male germ cell development in normal and SSC-deficient mice. A SSC defect causes a progressive decline in spermatogenesis because the first wave of spermatogenesis is SSC independent. PGCs, primordial germ cells; ProSG, prospermatogonia; SSCs, spermatogonial stem cells; Diff SPG, differentiating spermatogonia.
(B) Strategy to conditionally delete the entire Rhox cluster: insertion of loxP sites (yellow arrows) at the beginning and end of the ~920-kb Rhox cluster (see Figure S1A for exact location of loxP sites).
(C) Hematoxylin and eosin staining of testes sections from Rhox-c-KO (labeled “KO”), Rhox-c fl/y;Vasa-Cre (labeled “GC-KO”), and control (WT) mice. Upper and lower rows are from 8- and 16-to-46-week-old mice, respectively. Scale bars = 100 μm.
(D) qRT-PCR analysis demonstrating selective defect in expression of germ cell-expressed Rhox genes in testes from 8 week-old Rhox-c fl/y;Vasa-Cre mice (GC-KO) relative to WT mice. Values were normalized to Rpl19 mRNA level and denote the mean fold change ± standard error of the mean (SEM). Asterisks indicate the difference is statistically significant (P<0.05).
(E) Testis weight of Rhox-c-KO (KO), Rhox-c fl/y; Vasa-Cre (GC-KO), and WT mice of the indicated ages.
(F) Epididymal sperm count of Rhox-c-KO (KO), Rhox-c fl/y; Vasa-Cre (GC-KO), and WT mice of the indicated ages.
(G) Fertility analysis of adult male Rhox-c fl/y;Vasa-Cre (GC-KO) and WT mice, each housed with two BL6 female mice (initially 8-weeks old) for 4 months. Values denote the mean ± standard error of the mean (SEM). Asterisks indicate that the difference is statistically significant (P<0.05).
(H) Quantification of FOXO1-positive spermatogonia in testes from 8 week-old Rhox-c fl/y;Vasa-Cre (GC-KO) and WT mice (n=3 per genotype). All values are mean ± SEM.
See also Figure S1
ProSG are regarded as the cellular precursors of not only SSCs but also differentiating A-spermatogonia, the cells responsible for the first wave of spermatogenesis (Culty, 2013; McCarrey, 2013). This first wave, which bypasses the SSC step, was likely selected for over evolutionary time because it permits rapid generation of sperm, thereby bestowing fertility to young males. Evidence suggests that the retinoic acid (RA) and NOTCH signaling pathways participate in the early differentiation events essential for this first wave of spermatogenesis (Manku and Culty, 2015). In contrast, the signals required to generate the SSCs responsible for the subsequent waves of spermatogenesis remain undefined.
In this communication, we fill this gap by identifying a transcription factor critical for this developmental step. This transcription factor—RHOX10—is encoded by a member of the X-linked reproductive homeobox (Rhox) gene cluster (Geyer and Eddy, 2008; MacLean and Wilkinson, 2010; Maclean et al., 2005). All members of the Rhox gene cluster are expressed in the reproductive tract, suggesting that the Rhox gene cluster encodes transcription factors devoted to regulating genes critical for the reproductive tract (Maclean et al., 2005). While much is known about the regulation of Rhox genes and some about their functions from in vitro experiments, little is known about their in vivo roles (MacLean and Wilkinson, 2010). Indeed, no germ cell-expressed Rhox genes have yet to be ascribed a function in vivo. In this report, we demonstrate that the germ cell-expressed gene, Rhox10, is essential for efficient SSC establishment through its ability to promote the differentiation of ProSG into SSCs. The discovery of a transcription factor important for both ProSG differentiation and SSC establishment provides an opportunity to gain insights into these critical steps of gametogenesis.
Results
Loss of the Rhox Homeobox Cluster Causes Progressive Spermatogenic Decline
The mouse X chromosome is highly enriched in large gene clusters exhibiting testis-biased expression (Mueller et al., 2008). The abundance of such testes-expressed gene clusters on the X chromosome strongly suggests their importance for fertility, but the function of these clusters is not known. To determine the biological roles of the X-linked Rhox homeobox gene cluster, we generated mutant mice designed to conditionally delete all 33 Rhox genes, which comprise a ~920-kb region of the X chromosome (Figures 1B and S1A). After breeding with EIIa-Cre mice, which express CRE ubiquitously, we obtained complete Rhox cluster KO (Rhox-c-KO) mice that lacked expression of all Rhox genes we tested (Figure S1B). We were surprised that these global Rhox-c-KO mice were viable given that all Rhox genes are expressed in placenta (Maclean et al., 2005). However, we note that few Rhox-c-KO mice appeared to be viable, as only 11 of 56 male progeny from Rhox-c+/− and Rhox-c+/y parents had a mutant (Rhox-c−/y) genotype, which is ~4 fold lower than the expected 1:1 Mendelian segregation ratio. Those that survived were runted (Figures S1C and S1D). Viability was further reduced when Rhox-c-heterozygous mice from a mixed genetic background (FVB/129/BL6) were backcrossed into a pure C57BL/6J (BL6) genetic background; mice with >93.75% BL6 genetic contribution did not give rise to any viable KO progeny.
The survival of mixed genetic background global Rhox-c-KO mice provided an opportunity to examine their male reproductive defects. Young adult Rhox-c-KO mice (6-weeks old) had only modestly reduced testes size and sperm number compared to control littermate mice (Figures S1E and S1F). However, they failed to exhibit the increase in testes size and sperm number that occurred in control mice after sexual maturity (Figures S1E and S1F). Likewise, histological analysis showed that while young Rhox-c-KO mice had generally normal seminiferous tubules, aged Rhox-c-KO mice had seminiferous tubules that were vacuolar, many of which lacked some or all germ cells (Figure 1C). When only some germ cell layers were missing, they tended to be immature cell layers. Indeed, even young sexually mature (8-week old) Rhox-c-KO mice had some tubules with missing layers of spermatogonia or both spermatogonia and spermatocytes (Figure 1C, upper, and see below). This phenotype—missing early germ cell layers coupled with progressive spermatogenic decline—is characteristic of mice defective in either the generation or maintenance of SSCs (Figure 1A) (Song and Wilkinson, 2014). The delayed timing of this phenotype follows from the considerable evidence that the first wave of spermatogenesis is independent on SSCs, while subsequent waves of spermatogenesis depend on SSCs (Figure 1A) (Yoshida et al., 2006).
Spermatogenic failure can result from loss of germ cell- or somatic cell-expressed genes. To test which of these is responsible for the spermatogenic defects in global Rhox-c-KO mice, we mated floxed-Rhox cluster mice with Vasa-Cre transgenic mice, as they expresses CRE specifically in germ cells beginning at embryonic day (E) 15.5 (Gallardo et al., 2007). The Rhox cluster was specifically ablated in germ cells, as germ cell-expressed Rhox genes (Rhox3, Rhox10, Rhox11, and Rhox13) but not a Sertoli cell-expressed Rhox gene (Rhox5) (MacLean and Wilkinson, 2010; Song et al., 2012), exhibited dramatically reduced expression (Figure 1D). These germ cell-specific Rhox-c--KO mice were infertile (Figure 1E) and exhibited reproductive defects indistinguishable from those in global Rhox-c-KO mice (Figures 1C, 1F and 1G). Consistent with their having a SSC defect, Rhox-c-KO mice had dramatically reduced number of undifferentiated spermatogonia, based on FOXO1 marker analysis (Figure 1H). Together, these data suggest that one or more germ cell-expressed genes in the Rhox gene cluster are critical for the generation and/or maintenance of SSCs.
Identification of Germ Cell-Expressed Rhox Genes
To determine which genes in the Rhox cluster are candidates to be responsible for the putative SSC defect in Rhox-c-KO mice, we considered their different expression patterns. Our germ cell-specific conditional KO mice experiments indicated that only germ cell-expressed Rhox genes are viable candidates, which excluded Rhox5 and Rhox8, as both are expressed in Sertoli cells (MacLean and Wilkinson, 2010). To determine whether there are other germ cell-expressed Rhox genes, we performed irradiation (IR) experiments, as IR selectively ablates most spermatogonia, leading to the depletion of the more advanced types of germ cells, and leaving an enrichment of somatic cells (Zhang et al., 2006). As evidence of this method's specificity, the Rhox genes previously shown to be germ cell specific (Rhox3, Rhox10, and Rhox13), as well as the germ cell gene, Dazl, exhibited strongly reduced expression in IR testes (Figure S1G), while Sertoli cell-specific genes (Rhox5, Rhox8, and Gata1) displayed elevated expression (Figure S1H). IR analysis revealed that Rhox2, Rhox7, Rhox11, and Rhox12 all exhibited a germ-cell expression pattern (Figure S1G), while Rhox9 exhibited a somatic-cell expression pattern (Figure S1H). Rhox1, Rhox4, and Rhox6 displayed either modestly reduced or not significantly altered expression in IR testes (Figure S1G), suggesting that these 3 genes are expressed in both germ and somatic cell populations.
Together, the above analysis indicated that the Rhox2, Rhox3, Rhox7, Rhox10, Rhox11, Rhox12, and Rhox13 are expressed in germ cells and thus all have the potential to be responsible for the spermatogenic defects in mice lacking the Rhox cluster in male germ cells. We ruled out Rhox3 and Rhox13 as likely to be involved, as they are expressed in later stage germ cells than SSCs (Geyer et al., 2012; Song et al., 2015). Likewise, Rhox11 and Rhox12 are also unlikely to be responsible, as their postnatal expression pattern is consistent with primary expression in spermatids (Maclean et al., 2005). Among the three remaining genes—Rhox2, Rhox7, and Rhox10—we regarded Rhox10 as the best candidate because it is the only Rhox gene expressed at higher level in Id4-eGfp+ cells (Asingle [As] spermatogonia highly enriched in SSCs) than in Id4-eGfp− cells, (primarily spermatogonial progenitors and largely lack detectable SSC activity) (Supplemental Data Set S1 from Chan et al., 2014). The high expression of Rhox10 in undifferentiated spermatogonia was corroborated by our previous findings that Rhox10 is more highly expressed in the undifferentiated spermatogonial cell fraction than any other purified germ cell fraction (Song et al., 2012). Rhox10 mRNA is also abundant in adult male Jsd mice testes, which only contain undifferentiated A-spermatogonia (de Rooij et al., 1999), whereas it is undetectable in mutant mice testes lacking all germ cells (Song et al., 2012). Finally, RHOX10 protein is at abundant in germline stem (GS) cells (Song et al., 2012).
Loss of Rhox10 Causes an Analogous Phenotype as Loss of the Entire Rhox Cluster
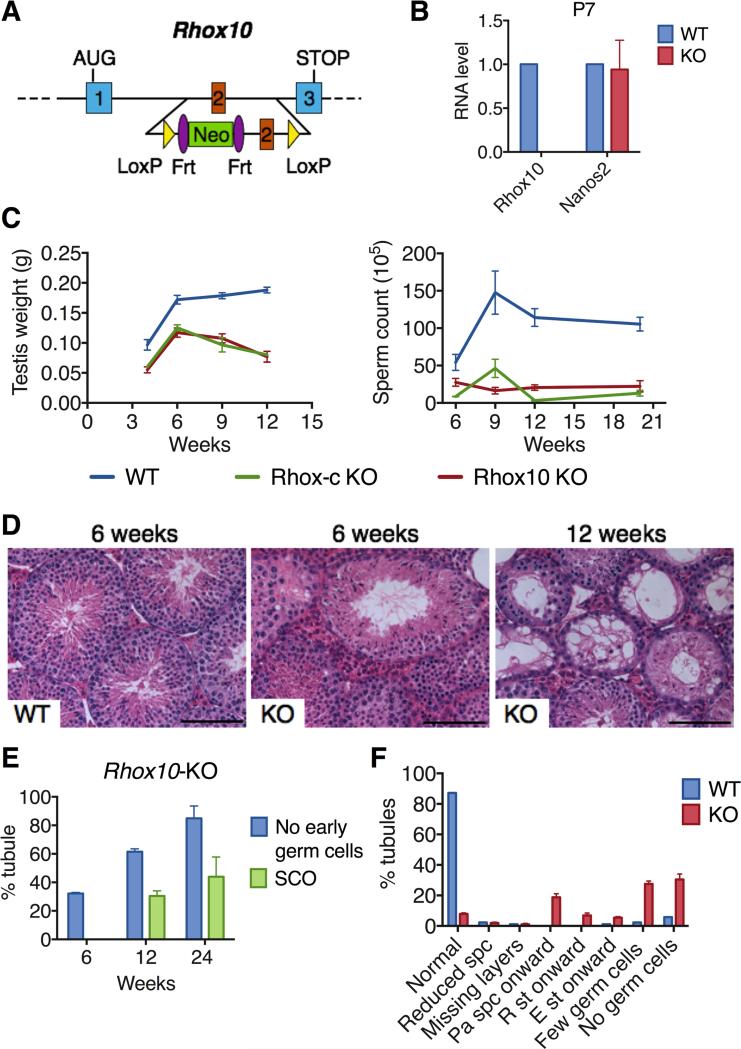
To elucidate whether loss of Rhox10 is responsible for the putative SSC defects in Rhox-c-KO mice, we generated Rhox10-floxed mice. We introduced loxP sites at positions designed to delete exon 2, which encodes an essential portion of the RHOX10 DNA-binding domain (Figure 2A). Using standard mouse gene targeting approaches, we obtained mice with a floxed-Rhox10 allele and mated these with EIIa-Cre mice to generate global Rhox10-KO mice (Figures S2A-B). These Rhox10-KO mice lacked Rhox10 expression (Figure 2B) and were normal in overall appearance and body weight. Rhox10-KO mice exhibited spermatogenic defects that worsened over time, with kinetics that largely mimicked that of Rhox-c-KO mice (Figures 2C, S2C and S2D). In contrast, seminal vesicle weight was normal in both Rhox10- and Rhox-c-KO mice (Figure S2E), suggesting that the spermatogenic defects in these mutant mice are unlikely to be secondary to an androgen or luteinizing hormone defect (Almenara et al., 2001). Histological analysis showed that, like Rhox-c-KO mice, Rhox10-KO mice exhibited a progressive decline in seminiferous tubule morphology consistent with a SSC defect (Figures 2D-F). Nonetheless, even at only 6 weeks of age, less than 50% of Rhox10-KO tubules looked normal (Figure S2F), and many tubules (~30%) had mature germ cells in the absence of immature germ cells (spermatogonia and early spermatocytes), indicative of a SSC defect (Figure 2E). By 12 weeks, less than 10% of the tubules in KO mice retained normal morphology (Figure 2F). Other histological defects were observed that were consistent with SSC defects, including missing layers of germ cells (Figures 2F and S2F). Many of tubule sections completely lacked detectable germ cells at both 12 and 24 weeks (Figure 2E), demonstrating a complete failure of spermatogenesis. We note that Rhox10 appears to act primarily in immature germ cells (ProSG or undifferentiated spermatogonia) since conditional deletion of Rhox10 at the spermatogonia stage using Stra8-Cre mice (Sadate-Ngatchou et al., 2008) did not cause a significant alteration in either testes weight or sperm number (Figure S2G).
Figure 2. Loss of Rhox10 Causes Progressive Spermatogenic Decline.
(A) Strategy to conditionally mutate Rhox10. LoxP sites were inserted on either side of exon 2, which encodes a critical part of the RHOX10 homeodomain.
(B) qRT-PCR analysis of Rhox10-KO (KO) and WT littermate control mice. Nanos2 is a germ cell marker. Values were normalized to Rpl19 mRNA level and denote the mean fold change ± standard error of the mean (SEM).
(C) Testis weight (left) and epididymal sperm count (right) of Rhox10-KO, Rhox-c-KO and control (WT) mice of the indicated ages.
(D) Hematoxylin and eosin staining of representative testis sections from Rhox10-KO (KO) and control (WT) mice of the indicated ages. Scale bars =100 μm.
(E) Quantification of percentage of seminiferous tubule sections without early germ cells or without any detectable germ cells (Sertoli cell only [SCO]) in testes sections from Rhox10-KO mice of the indicated ages. Littermate control mice at all ages had <5% abnormal tubules. Values denote the mean % tubules ± standard error of the mean (SEM). n=2-4, 50-100 tubules per each sections were counted.
(F) Seminiferous tubule abnormalities in 12 week-old Rhox10-KO (KO) and littermate control (WT) mice. spc, spermatocytes; pa spc, pachytene spermatocytes; R st, round spermatids; E st, elongating spermatids.
See also Figure S2
In addition to generating few sperm, Rhox10- and Rhox-c-KO mice had a high proportion of sperm with abnormal morphology (Figure S2H). The predominant phenotype in both mutants was perturbed head morphology and rolled back tails (Figure S2H). We also observed that Rhox10-KO male mice were subfertile (Figure S2I), which may result from both their decreased sperm count and their low frequency of sperm with normal morphology. Together, these results demonstrated that Rhox10 is an essential member of the Rhox gene cluster that is critical for spermatogenesis, sperm development, and normal fertility. The specific spermatogenesis defects in Rhox10-KO mice are consistent with a SSC defect, a possibility we examine in more detail below.
Rhox10 Promotes SSC Establishment
To further address whether Rhox10-KO mice have a SSC defect, we used several markers selectively expressed in undifferentiated spermatogonia, the most immature germ cell population in the adult testis (Phillips et al., 2010). First, we performed immunofluorescence analysis with PLZF, a SSC maintenance factor that marks all stages of undifferentiated spermatogonia (Phillips et al., 2010; Song and Wilkinson, 2014) and observed fewer PLZF+ cells in testes from Rhox10-KO mice than in littermate control mice (Figures S3A and S3B). The deficit in PLZF+ cells was more prominent after 4 weeks of age, consistent with the progressive decline in spermatogenesis in Rhox10-KO seminiferous tubules (Figures 2C-E). Our experiments (described below) revealed that Rhox10 loss only affects a specific subset of PLZF+ cells (the SSCs), explaining why we only observed a modest reduction in total PLZF+ cell number. Double staining with the proliferation marker, KI67, showed that while Rhox10-KO mice had less KI67/PLZF double-positive cells than control mice, the ratio of KI67/PLZF double-positive cells per total PLZF+ cells was not affected by loss of Rhox10 (Figure S3C), suggesting that Rhox10 does not promote undifferentiated spermatogonia proliferation and that instead it acts by some other mechanism.
Using a more selective marker, GFRα1 (Phillips et al., 2010), coupled with whole mount staining (Figure 3A), we found that adult Rhox10-KO mice had significantly reduced numbers of GFRα1+ As spermatogonia (which is enriched in SSCs [de Rooij and Griswold, 2012]) and a modest reduction in Apr spermatogonia (which has some SSC activity [de Rooij and Griswold, 2012]) (Figure 3B). In contrast, there was no significant change in the number of longer chains of GFRα1+ spermatogonia, which are mainly spermatogonial progenitors (de Rooij and Griswold, 2012). To assess the timing of the defect, we examined young postnatal mice when SSCs are undergoing their initial expansion. Flow cytometric analysis showed that the number of cells marked with either GFRα1 or another marker widely used to enrich for SSCs, THY1 (Oatley and Brinster, 2008), were both reduced in P8 Rhox10-KO testes relative to control P8 testes (Figures S3D and S3E). In contrast, the number of cells positive for the broader undifferentiated spermatogonial marker, PLZF, was not significantly changed in early postnatal Rhox10-KO mice (Figure S3B). Together, these results demonstrated that loss of Rhox10 reduces the number of a subset of undifferentiated spermatogonia during the early postnatal period, when SSCs first form.
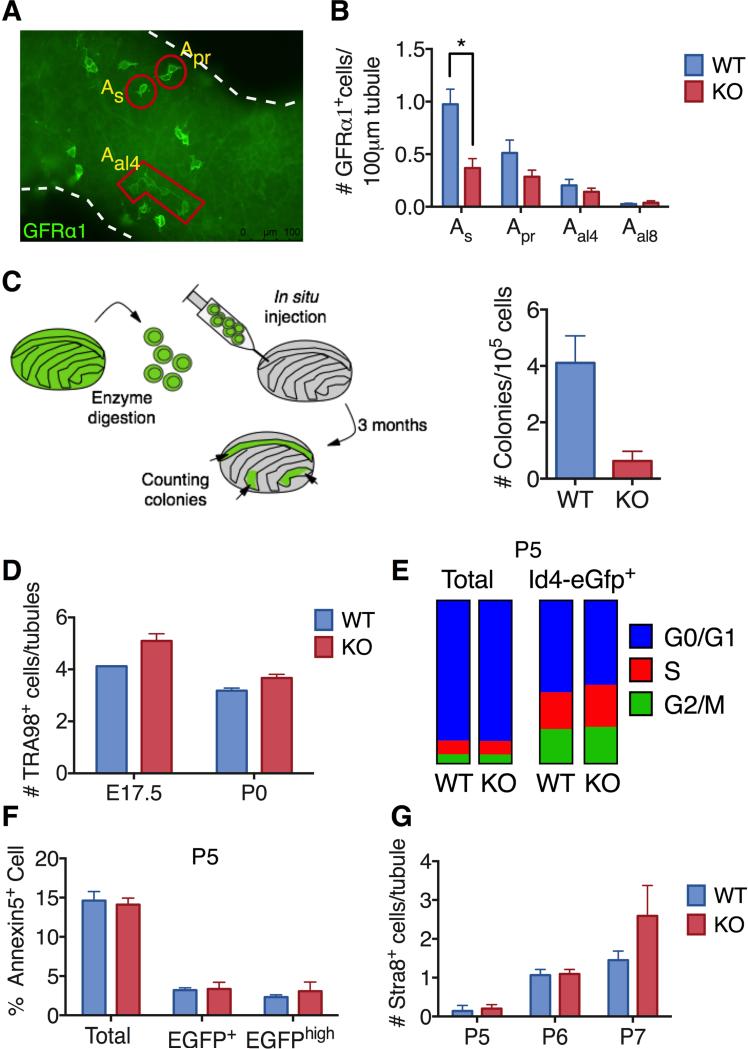
Figure 3. Loss of Rhox10 Causes SSC Defects.
(A) Representative image of whole mount staining of 6 month-old adult mice seminiferous tubules with GFRα1 antibody. As, Apr and Aal4 cells are positively stained with the GFRα1 antibody.
(B) Quantification of GFRα1+ spermatogonia in testes from 6- to 10-month-old Rhox10-KO and littermate control (WT) mice (n=4-5 per genotype). All values are mean ± SEM. Asterisks indicate differences that are statistically significant (P<0.05).
(C) Germ cell transplantation analysis. Donor-derived colonies in recipient testes were counted and normalized to 105 cells injected per testis 3 month after transplantation of testicular cells from P7-8 Rhox10-KO (Rhox10−/y;Ubc-Gfp) mice (n=4) and littermate control (WT) (Rhox10+/y;Ubc-Gfp) mice (n=5).
(D) Germ (TRA98+) cells per tubule in testes sections from Rhox10-KO and littermate control (WT) mice of the indicated ages (n=1-4, each time point).
(E) Cell cycle analysis of either total testicular cells or Id4-eGfp+ cells from P5 Rhox10-KO and littermate control (WT) mice (n=5).
(F) Flow cytometric analysis of Annexin V+ cells (among total, Id4-eGfp+ or Id4-eGfphigh testicular cells) from P5 Rhox10-KO mice (n=6) and littermate control (WT) mice (n=9).
(G) Quantification of STRA8+ cells in testes sections from Rhox10-KO and littermate control (WT) mice of the ages indicated (n=2-5, each time point). All values are mean ± SEM.
See also figure S3
To quantitatively determine the number of SSCs in Rhox10-KO mice, we used the “gold standard” for measuring SSC frequency – the germ cell transplantation assay (Figure 3C) (Oatley and Brinster, 2008). Germ cell transplantation analysis revealed that early postnatal (P7-8) Rhox10-KO testes yielded dramatically fewer (6-fold) colonies than did control littermate testes (Figure 3C). The few SSCs in Rhox10-KO mice at a postnatal time point when SSCs are normally first being generated, coupled with the other phenotypic defects in Rhox10-KO mice, strongly suggests that Rhox10 is critical for the initial establishment of SSCs.
One means by which Rhox10 could promote SSC establishment is by driving the generation of fetal germ cells. To test this, we stained embryonic (E17.5) and newborn (P0) testes sections with the general germ cell marker, TRA98. We did not observe a significant difference in the number of TRA98+ germ cells in Rhox10-KO mice compared to WT littermate controls at either time point (Figure 3D). This suggests that Rhox10 acts at some step after birth to promote the initial establishment of SSCs. To investigate the timing, we used reporter mice expressing EGFP from the Id4 promoter, as these mice mark a subset of As spermatogonia highly enriched for SSCs (Chan et al., 2014). Consistent with our germ cell transplantation analysis that showed that P7-8 Rhox10-KO mice have few SSCs (Figure 3C), FACS analysis demonstrated that P7 KO mice also had low numbers of Id4-eGfp+ cells (Figure S3G). Rhox10-KO mice also had a deficit of Id4-eGfp+ cells two days earlier (at P5, Figure S3F), soon after SSCs first emerge (McCarrey, 2013), providing more support for the notion that Rhox10 promotes the initial establishment of SSCs. While the reduction in the entire P5 Id4-eGfp+ cell population was only ~2 fold, we demonstrate below that loss of Rhox10 has a more dramatic effect on Id4-eGfp+ cells enriched for SSCs using a second marker.
Another recently described SSC marker is PAX7, which, like Id4-eGfp, marks a subset of As spermatogonia that are enriched for SSC activity (Aloisio et al., 2014). However, whether PAX7 and Id4-eGfp label the same subsets of As spermatogonia or SSCs has not been determined. To address how Rhox10 influences spermatogonial cell subsets labeled with these two markers, we performed double-staining analysis of P5 testis sections. Analysis of WT cells revealed that PAX7 was exclusively expressed in a subset of Id4-eGfp+ cells (Figure S3H). Loss of Rhox10 caused a significant reduction in only one subset – Id4-eGfpHI/PAX7− cells (Figure S3H). None of the cell subsets expressing PAX7 were significantly reduced in level in Rhox10-KO mice. Together with our germ cell transplantation analysis (Figure 3C), our results lead us to conclude that Rhox10 promotes the initial establishment of SSCs marked with Id4-eGfp but not PAX7.
We next addressed the underlying cellular mechanism by which Rhox10 promotes SSC establishment. To examine whether Rhox10 stimulates SSC proliferation, we co-stained with Id4-eGfp and the proliferation marker, KI67. This analysis showed that loss of Rhox10 did not significantly reduce the proportion of cells positive for KI67 among Id4-eGfp+ cells in P5 testes sections, suggesting there was no proliferative defect (Figure S3I). We also performed cell cycle analysis and found that loss of Rhox10 did not cause significant difference in the distribution of cells across the cell cycle in either Id4-eGfp+ cells or total testicular cells (Figure 3E). We next examined whether instead Rhox10 promotes the survival of Id4-eGfp+ cells. Using FACS analysis, we found that loss of Rhox10 did not have a significant effect on the proportion of cells positive for the apoptosis marker, Annexin 5, among Id4-eGfp+, Id4-eGfphigh, or total testicular cells at P5 (Figure 3F). Together, this analysis suggested that Rhox10 does not affect the survival or proliferation of early postnatal male germ cells. We note, however, that this does not rule out the possibility that Rhox10 promotes the proliferation and/or survival of Id4-eGfp-negative SSCs (which evidence suggests are a very minor population of the germ cells with SSC activity [Chan et al., 2014]) or a small subset of Id4-eGfp-positive SSCs. Another possibility is that Rhox10 promotes SSC establishment by suppressing the conversion of ProSG into differentiating A-spermatogonia, the germ cells responsible for generating the first wave of spermatogenesis. By suppressing the generation of differentiating A-spermatogonia, Rhox10 would instead favor the conversion of ProSG into SSCs. Using STRA8, a marker for differentiating A-spermatogonia, we found that the number of STRA8+ cells was not significantly increased in Rhox10-KO mice at any of the time points we tested (Figure 3G), suggesting that Rhox10 does not promote SSC establishment by inhibiting spermatogonial differentiation. This finding, coupled with our evidence that Rhox10 does not act by promoting SSC proliferation or survival (Figures 3E, 3F and S3I), led us to investigate whether, instead, Rhox10 promotes SSC establishment by directly driving the differentiation of ProSG to form SSCs, as described below.
Rhox10 Promotes ProSG-to-SSC Progression
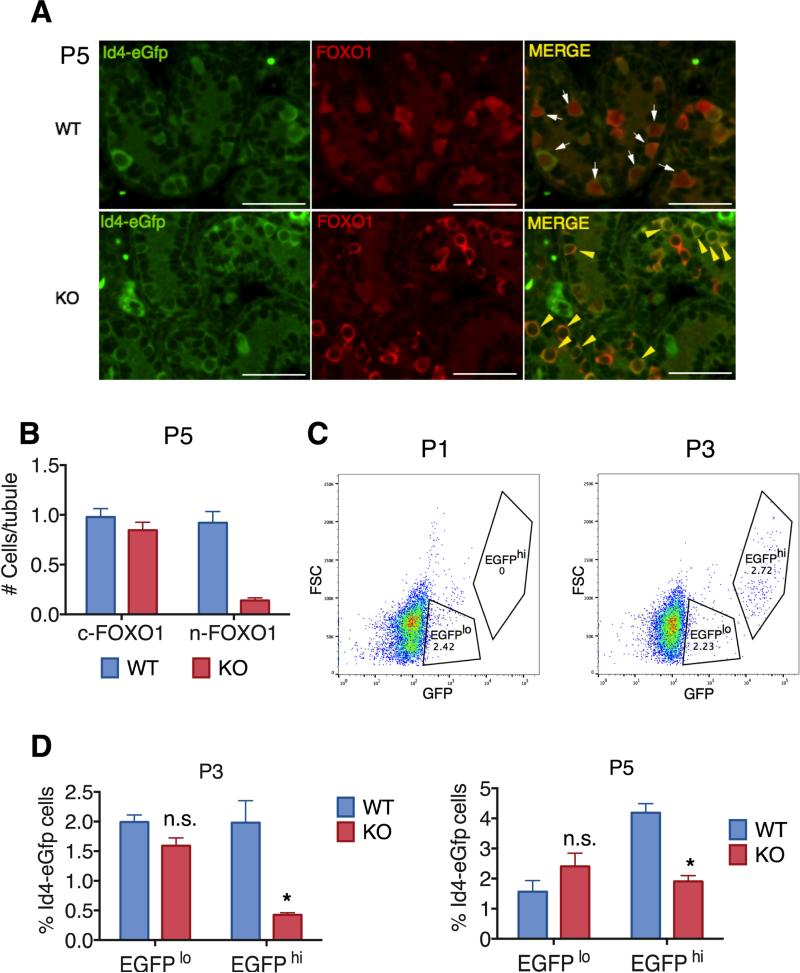
As a first approach to test whether RHOX10 promotes the differentiation of ProSG into SSCs, we made use of the discovery that the transcription factor, FOXO1, translocates from the cytoplasm to the nucleus when ProSG differentiate into SSCs and A-spermatogonia during the early postnatal period (Goertz et al., 2011). Thus, the location of FOXO1 serves as a marker to distinguish between ProSG and SSCs, when combined with the Id4-eGfp marker, which labels both ProSG and SSCs (Chan et al., 2014). Using this double-labeling approach, we observed that P5 Rhox10-KO mice had dramatically fewer nuclear (n)-FOXO1+/Id4-eGfp+ cells than control mice (Figures 4A and 4B), consistent with our many lines of evidence (above) that these KO mice have few SSCs (Figures 3C and S3D-G). Indeed, the decrease in nFOXO1+/Id4-eGfp+ cells (6.5 fold) was similar to the reduction in SSCs (6 fold), as measured by germ cell transplantation (Figure 3C). In striking contrast, the number of cytoplasmic (c)-FOXO1+/Id4-eGfp+ cells was not significantly reduced in the KO mice, consistent with suppressed differentiation of these ProSG-enriched cells. Similar results were observed when we analyzed ProSG and SSCs using FOXO1/PLZF double staining (Figures S4A and S4B).
Figure 4. Rhox10 Promotes ProSG-to-SSC Progression.
(A) Representative images of testes sections from P5 Rhox10-KO (Rhox10−/y;Id4-eGfp) mice and littermate control (Rhox10+/y;Id4-eGfp) mice stained for the indicated markers. Mice with the same genotype were used for panels B-D. Scale bars = 60 μm.
(B) Quantification of cytoplasmic c-FOXO1+/Id4-eGfp+ and nuclear n-FOXO1+/Id4-eGfp+ cells in testes sections from P5 Rhox10-KO mice (n=3) and littermate control (WT) mice (n=2).
(C) Flow cytometric analysis of GFP intensity (x-axis) and forward scatter (size, y-axis) of testicular cells from P1 and P3 Id4-eGfp WT mice. Cells were judged as exhibiting high (hi) or low (lo) expression of EGFP.
(D) Id4-eGfp+ cell populations identified by FACS (analyzed as in panel C) derived from P3 (n=3) and P5 (n=6-8) Rhox10-KO mice and littermate control (WT) mice. All values are mean ± SEM. Asterisks indicate statistically significant differences (P<0.05).
See also Figure S4
As an independent means to assess the possibility that Rhox10-KO mice have a ProSG-to-SSC progression defect, we used FACS analysis. Id4-eGfp+ cells were analyzed at two time points: P1, when ProSG are the only germ cells present, and at P3, when SSCs begin to differentiate from ProSG (Mclean et al., 2003). We observed that WT mice had a single major population of Id4-eGfp+ cells at P1, whereas, at P3, another cell population emerged that expressed higher levels of EGFP (Figure 4C). This suggested that the EGFPlo and EGFPhi cells are ProSG and SSCs, respectively. The EGFPhi cells that emerged at P3 expanded in number, such that by P5, they represented 4% of total testicular cells (Figure 4D), in agreement with previous evidence that SSCs are fully established by P5 (McCarrey, 2013). Rhox10-KO mice had a normal number of the EGFPlo “ProSG” subset but largely failed to generate the EGFPhi “SSC” subset, based on analysis at both P3 and P5 (Figure 4D). This data verified that Rhox10-KO mice have a defect in SSC generation and assigned the timing of this defect as occurring at ~P3, when SSCs first differentiate from ProSG (McCarrey, 2013). This progression defect appeared to be specific, as the total number of germ cells was not significantly reduced in KO mice relative to control mice between P3 and P7 (Figure S4C).
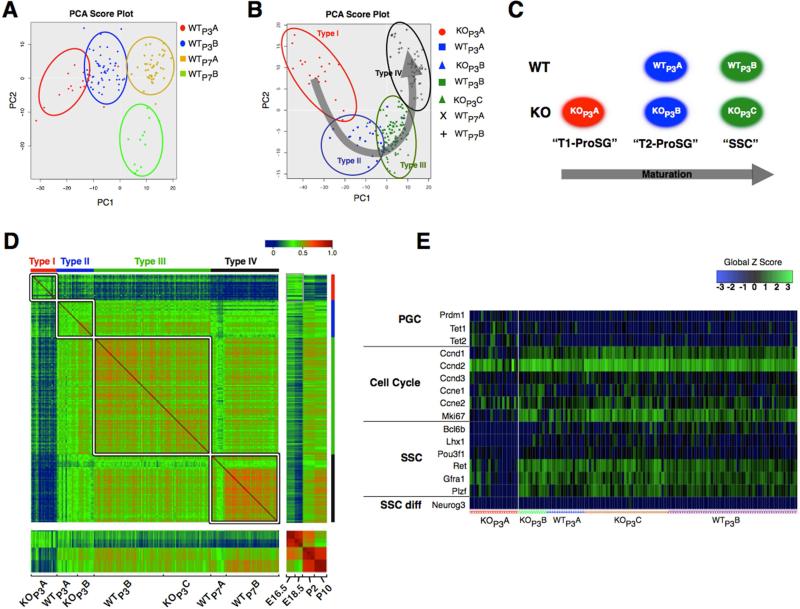
To analyze the ProSG-to-SSC progression defect in depth, we employed scRNAseq analysis, which provides an unbiased means to distinguish between cell subsets. This is particularly critical for analysis of ProSG and SSCs, as there are no known markers that label either of these germ cell subsets exclusively. To enrich for these two cell subsets, we used FACS-purified Id4-eGfp+ cells at different time points (P3 testes have both ProSG and SSCs, while P7 testes have SSCs without contaminating ProSG). Principal component analysis (PCA) was used to identify the most divergently expressed genes to allow us to define cell subsets in P3 and P7 Id4-eGfp+ cells by hierarchical clustering. PCA identified two distinct P3 Id4-eGfp+ subsets (WTP3A and WTP3B) and two distinct P7 Id4-eGfp+ subsets (WTP7A and WTP7B) in WT mice (Figure 5A). We defined WTP3A as ProSG, based on the large PCA distance between them and the P7 cell subsets, which lack ProSG (Culty, 2009). We defined WTP3B and WTP7A as being enriched in SSCs, based on their close proximity in the PCA plot and the fact that SSCs are the only known germ cell subset present at both P3 and P7 in Id4-eGfp+ cells (McCarrey, 2013). The final cell subset—WTP7B—had a PCA score that suggested they are the differentiated progeny of SSCs – spermatogonial progenitors. All these cell subset assignments were further supported by comparison with RNAseq data from male germ cells purified from specific embryonic and postnatal time points (see below).
Figure 5. Single-Cell RNAseq Analysis of Id4-eGfp+ Cell Subsets.
(A) PCA of Id4-eGfp+ cells from P3 and P7 WT Id4-eGfp mice, analyzed by SC-RNAseq, using the 1009 most differentially expressed genes.
(B) PCA-defined Id4-eGfp+ cell clusters containing cells from P3 Rhox10-KO (Rhox10−/y; Id4-eGfp) and littermate control (Rhox10+/y; Id4-eGfp) mice. The four clusters (Type I through Type IV) were defined based on the gene expression profiles described in panel A. The arrow represents the direction of germ cell maturation.
(C) The P3 Id4-eGfp+ cell subsets are enriched in the indicated cell subsets, based on the evidence described in the text.
(D) Pairwise correlations of gene expression profiles of purified germ cells from the time points indicated (Pastor et al., 2014) and the single cells from our study. The genes examined were the 1009 differentially expressed genes described in panel A.
(E) Expression of the indicated genes in individual Id4-eGfp+ cells. Of note, neither KOP3A cells nor any of the other P3 Id4-eGfp-positive cell subsets expressed the SSC progenitor marker, Neurog3.
To elucidate the nature of the cell subset defects in Rhox10-KO mice, we chose to analyze the P3 time point, as this is when defects were first observed in these mutant mice by FACS analysis (Figure 4D). Three distinct KO Id4-eGfp+ subsets were defined by PCA at this time point. When we plotted these 3 KO cell subsets together with the 4 WT subsets defined above, 4 clusters of related cells could easily be discerned, which we named Type I to IV, in order of increasing maturity (Figure 5B). To discriminate between these clusters, we identified genes expressed in a larger proportion of cells in a given cluster than in the other clusters. The type-I cluster, which contains only a single cell subset—KOP3A—is enriched for genes expressed in FACS-purified E16.5 and E18.5 germ cells defined using RNAseq analysis by Pastor et al. (Pastor et al., 2014) (Figure S5A). Because the vast majority of germ cells in E16.5 and 18.5 testes are T1-ProSG (McCarrey, 2013), we assigned this KO cell subset as T1-ProSG. The type-II cluster, which are comprised of WTP3A cells and a related KO cell subset, KOP3B, is enriched in genes expressed in FACS-purified P2 germ cells defined by Pastor et al. (Pastor et al., 2014) (Figure S5A), indicating that the cells in this cluster are likely to be predominantly T2-ProSG, the major germ cell type at P2 (McCarrey, 2013). The type-III cluster, which contains WTP3B cells and a related KO cell subset, KOP3C, are enriched in newly emerging SSCs, based on their PCA-defined relationship to the other cell subsets. These P3 cell subset assignments are summarized in Figure 5C. The type-IV cluster, which contains the two WT Id4-eGfp+ subsets from P7 testes, is enriched in genes expressed in FACS-purified P10 germ cells (Pastor et al., 2014) (Figure S5A), implying that these cells are the most mature cell subset and thus are mainly established SSCs and spermatogonial progenitors.
The most striking germ cell subset defect in Rhox10-KO mice was the accumulation of KOP3A cells, a cell subset not present in control P3 mice (Figures 5B and 5C). These KOP3A cells are highly enriched in T1-ProSG, based on 4 lines of evidence. First, as mentioned above, genes enriched in KOP3A cells closely match those in purified E16.5 and E18.5 germ cells (Figure S5A). Second, pairwise comparison of gene expression profiles showed that KOP3A cells have a transcriptome more related to E18.5 germ cells than that of purified germ cells from any other time point or the 4 other Id4-eGfp+ cell subsets from the same time point (Figures 5D and S5B). Third, KOP3A cells tended to express genes known to mark PGCs but not SSCs (Figure 5E). Finally, KOP3A cells poorly expressed cell cycle genes (Figure 5E), consistent with the fact that T1-ProSG are, by definition, non-proliferative.
The accumulation of T1-ProSG cells in P3 Rhox10-KO mice indicates that loss ofRhox10 impairs the progression of T1-ProSG. Coupled with our evidence that Rhox10-KO mice have fewer SSCs when they are normally first generated (Figures 3C and 4), this strongly suggests that Rhox10 drives the differentiation of T1-ProSG to ultimately form SSCs, a model we elaborate on below.
Rhox10 Regulates Cell Migration Genes and Promotes ProSG Migration
RHOX10 is a member of the homeobox superfamily (Maclean et al., 2005) and thus presumably is a transcription factor. To identify genes regulated by RHOX10 in specific germ cell subsets, we compared the transcriptomes of Rhox10-KO Id4-eGfp+ cell subsets with corresponding WT Id4-eGfp+ cell subsets using the Single-Cell Differential Expression (SCDE) software (see Experimental Procedures). This analysis identified both statistically up- and down-regulated genes in two distinct Rhox10-KO cell subsets (Figure 6A). Interestingly, more genes were downregulated than upregulated in both Rhox10-KO cell subsets, suggesting that RHOX10 primarily serves as a positive regulator of transcription.
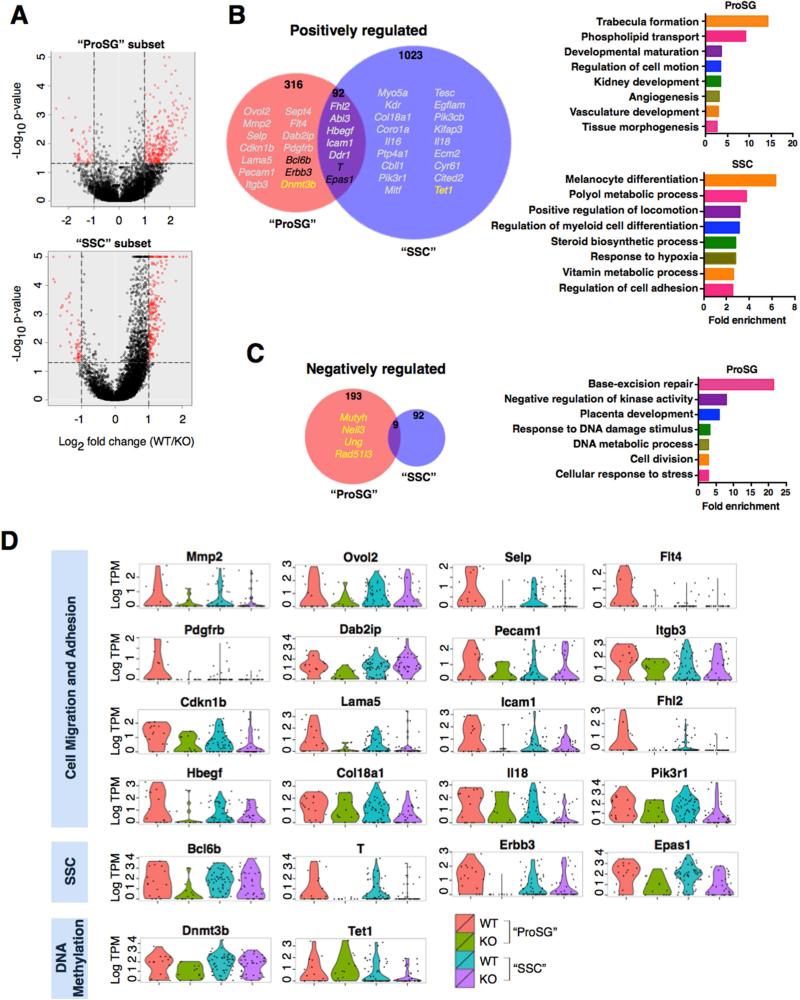
Figure 6. Rhox10-Regulated Genes in Germ Cell Subsets.
(A) SC-RNAseq coupled with SCDE analysis was used to identify genes differentially expressed in the “ProSG subset” (KOP3B vs. WTP3A; left) and “SSC subset” (KOP3C vs. WTP3B; right), as defined in Figure 5. Red points are genes >2-fold differentially expressed (P<0.05).
(B-C) (Left) Selected Rhox10-regulated genes (defined in panel A) are indicated with different colors: grey, Cell migration and adhesion; black, SSC genes; yellow, DNA methylation (>2 fold, P<0.05). Note that “Positively regulated” refers to genes upregulated by Rhox10 (i.e., downregulated in Rhox10-KO cells) and “Negatively regulated” refers to the converse. (Right) Selected highly enriched GO categories are shown (>2.5 fold enrichment, P<0.02). Note that there is no significantly enriched GO category for Rhox10-negatively regulated genes in the SSC subset.
(D) The level of RHOX10-positively regulated genes in single cells in the “ProSG” and “SSC” subsets.
To identify classes of genes regulated by RHOX10, we performed DAVID gene ontology analysis (Figures 6B, 6C and S6). Among the most prominently enriched (>2.5-fold enrichment) categories of RHOX10-positively regulated genes in both cell subsets were those encoding proteins involved in “Regulation of Cell Motion”, “Regulation of Cell Adhesion” and “Positive Regulation of Locomotion.” This suggested that genes encoding proteins involved in cell migration are major targets of RHOX10, a notion supported by the fact that the most statistically enriched (14.3 fold) functional category in the “ProSG subset”—“Trabecular formation”—consists of only cell migration genes: Ovol2, Fhl1, and Mmp2. In addition, other genes encoding cell migration and adhesion protein were regulated by RHOX10 in these two cell subsets (cell migration-related genes are marked in grey in Figure 6B; their expression profiles—depicted as a violin plot to depict expression in individual cells—is shown in Figure 6D). All told, we found that at least 33 known “migration genes” are regulated by Rhox10. This was intriguing given that the migration of ProSG to the periphery of seminiferous tubules accompanies their differentiation into SSCs (Culty, 2009; Payne, 2013). The linkage of these two events, coupled with the finding that Rhox10 regulates batteries of migration genes and promotes SSC generation, led us to test whether Rhox10 has a role in ProSG migration. To address this, we quantified the number of germ cells in the center and periphery of KO and WT tubules during the developmental window when this migration event occurs. In support of a migration defect, we observed that Rhox10-KO mice had a statistically higher proportion of TRA98+ (total) germ cells than control mice in the tubule central region at P3, P5 and P7 (Figures 7A, 7B and S7A). This was also observed for germ cells defined by morphological criteria at P3 and those defined with another germ cell marker (MVH) at P5 (Figures S7B and S7C).
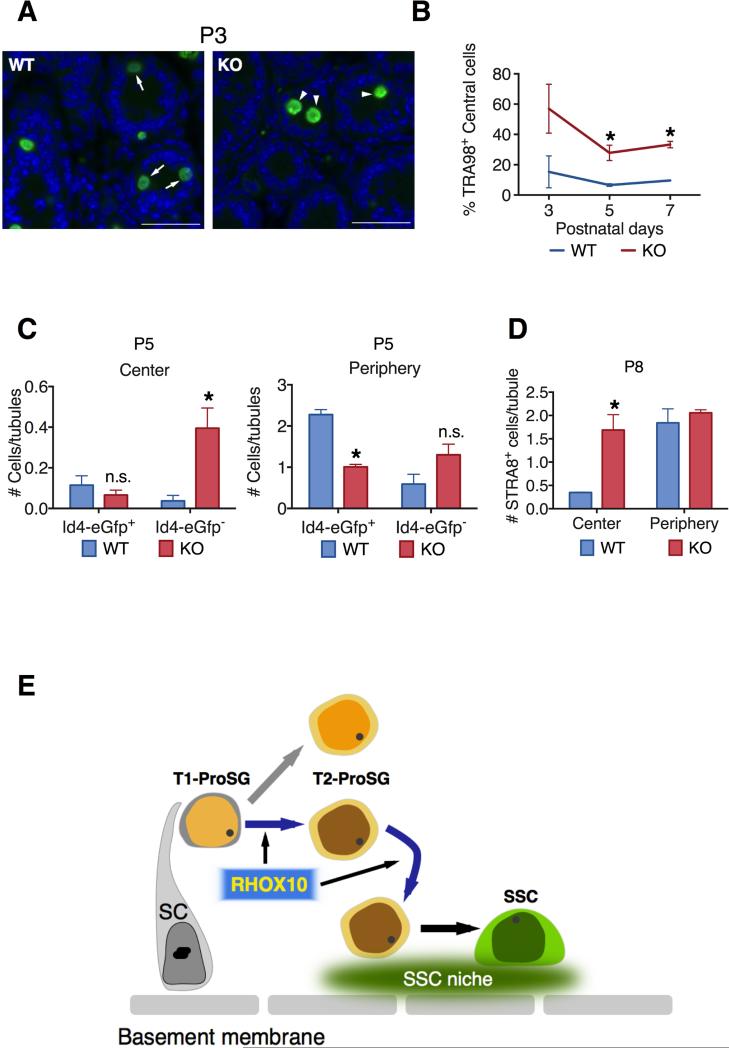
Figure 7. Evidence that Rhox10 Promotes ProSG Migration into the SSC Niche.
(A) Representative images of germ cells (stained with TRA98) in testes cross-sections from P3 Rhox10-KO mice and littermate control (WT) mice. Arrows, cells located in the periphery; Arrow heads, cells located in the center. Scale bars = 20 μm.
(B) Percentage of TRA98+ germ cells located in the center of seminiferous tubules as compared to total TRA98+ germ cells in Rhox10-KO mice and littermate control (WT) mice of the indicated age (n= 2-5 each time point). All values are mean ± SEM. Asterisks indicate statistically significant differences (P<0.05).
(C) Quantification of FOXO1+ cells co-stained for Id4-eGfp located in the center (Left) or periphery (Right) of seminiferous tubules in P5 Rhox10-KO mice (n=3) and littermate control (WT) mice (n=2). All values are mean ± SEM. Asterisks indicate statistically significant differences (P<0.05).
(D) Quantification of STRA8+ cells (differentiating A-spermatogonia) located in the center or periphery of seminiferous tubules in P8 Rhox10-KO mice and littermate control (WT) mice (n=2). All values are mean ± SEM. Asterisks indicate statistically significant differences (P<0.05).
(E) Model: Rhox10 promotes ProSG progression into SSCs. The model posits that Rhox10 drives both the T1-ProSG-to-T2-ProSG differentiation step and the ProSG migration step, which temporal studies suggest may occur simultaneously. Depicted are two kinds of T2-ProSG, each with a different fate, a possibility that is consistent with published studies (Kluin and de Rooij, 1981; Murphey et al., 2013), but has not been directly demonstrated. SC, Sertoli cell.
See also Figure S7.
To test if the migration event is critical for SSC establishment, we stained for Id4-eGfp, which marks late-stage ProSG, some SSCs, and progenitors at P5. We co-stained with FOXO1, as it marks all stages of ProSG, SSCs, and progenitors, but excludes differentiating A-spermatogonia. This analysis revealed that Rhox10 KO mice had dramatically more Id4-eGfp−/FOXO1+ cells (immature ProSG) in the tubule center than control mice (Figure 7C; labeled only “Id4-eGfp−” for simplicity). In contrast, the number of Id4-eGfp−/FOXO1+ cells in the tubule periphery was not statistically different between the KO and control. These data indicated that there is a buildup of immature ProSG in the tubule center of KO mice, consistent with these mice possessing a migration defect. In control mice, most FOXO1+ cells are Id4-eGfp+ and localized in the tubule periphery (Figure 7C), consistent with previously defined SSC establishment time line and location (McCarrey, 2013). KO mice had Id4-eGfp+ cells predominantly in the tubule periphery, albeit their frequency was much lower than in control mice, consistent with the reduced number of SSCs in KO mice (Figure 3C). Together, these data strongly suggest that Rhox10 KO mice have a defect in the migration of ProSG into the tubule periphery, but this block is not complete and thus some ProSG reach the tubule periphery and differentiate into SSCs and progenitors.
Like SSCs, differentiating A-spermatogonia are generated from ProSG after they migrate to the seminiferous tubule periphery (Culty, 2009), but this is a later event that does not begin in earnest until after P5 (Figure 3G). As described above, we found that loss of Rhox10 did not measurably affect the generation of differentiating A-spermatogonia (as measured using the STRA8 marker; Figure 3G). We examined whether this is because the ProSG that give rise to differentiating A-spermatogonia migrate to the tubule periphery independently of Rhox10. Contrary to this hypothesis, we found that Rhox10-KO mice accumulated STRA8+ cells in the center of seminiferous tubules (Figure 7D). Indeed, the majority of germ cells in the central region (marked with general germ-cell markers) were stained with STRA8 (compare Figures 7D with S7A). This data suggested that the conversion of ProSG into differentiating A-spermatogonia does not absolutely require the migration of ProSG to the tubule periphery. Together, these data support a model in which RHOX10 selectively drives the differentiation of ProSG into SSCs because this transcription factor promotes ProSG migration, an event that is required for them to generate SSCs, but not differentiating A-spermatogonia (Figure 7E).
Discussion
While much has been learned about the self-renewal and differentiation of SSCs, how SSCs are initially generated is poorly understood. In this communication, we report the first identification of a transcription factor that promotes the initial establishment of SSCs. We were led to the identification of RHOX10 as an SSC establishment factor by our discovery that loss of the large homeobox gene cluster encoding this factor caused a phenotype consistent with a deficiency in SSC generation or maintenance. We subsequently selectively knocked out only the Rhox10 gene and found that this led to SSC-associated defects largely indistinguishable from those caused by loss of the entire Rhox gene cluster. Using a battery of markers and different approaches, we obtained several lines of evidence that Rhox10 acts by promoting SSC establishment through its ability to drive ProSG differentiation. To address how Rhox10 might promote the differentiation of ProSG into SSCs, it was necessary for us to distinguish between different ProSG subsets. This was a challenge given that, to our knowledge, no markers (other than proliferation markers) distinguish between ProSG subsets (Culty, 2009; Manku and Culty, 2015). Indeed, very few markers have been found that distinguish ProSG from SSCs, probably because these two cell stages are highly related (Culty, 2009; Manku and Culty, 2015) and SSCs have not yet been purified to homogeneity (Oatley and Brinster, 2008). To overcome this limitation, we used scRNAseq, which revealed that T1-ProSG accumulate in Rhox10-KO mice at a stage in which they normally differentiate into T2-ProSG, demonstrating that Rhox10 drives the progression of T1-ProSG. Together, these results indicated that Rhox10 drives the initial establishment of SSCs by promoting a specific step in ProSG differentiation.
Our results have implications for understanding how germ cells normally develop in vivo. In mice, a critical step occurs between P0.5 and P3, when non-proliferative T1-ProSG convert to mitotically active T2-ProSG and migrate to the periphery of the seminiferous tubule (Culty, 2009). Coincident with this migration event, some T2-ProSG differentiate into SSCs, a process that continues for the next several days (Culty, 2013; McCarrey, 2013). During the later phases of this SSC generation period (between P5 and P7), T2-ProSG also give rise to differentiating A-spermatogonia, the precursor cells responsible for the first wave of spermatogenesis. It is not known what determines whether a ProSG will become a SSC or a differentiating A-spermatogonium. One possibility is that there are different subsets of T2-ProSG, each with different potentials for generating these 2 cell types. In support, two morphologically distinct ProSG generated at birth have been identified that exhibit morphological similarities with undifferentiated spermatogonia and differentiating A-spermatogonia, respectively (Kluin and de Rooij, 1981). Furthermore, a recent study showed that germ cell subsets differ considerably in their mutation frequencies in a manner consistent with the notion that a subset of ProSG with enhanced genome fidelity (low mutation rate) are predestined to become SSCs (Murphey et al., 2013). Our results also support the existence of ProSG heterogeneity. In particular, our finding that Rhox10 preferentially promoted the generation of SSCs, not differentiating A-spermatogonia, coupled with our finding that Rhox10-KO mice accumulated T1-ProSG that fail to progress, support a model in which Rhox10 drives the conversion of T1-ProSG into a specific subset of T2-ProSG that give rise to SSCs, not differentiating A-spermatogonia (Figure 7E). An intriguing alternative possibility is that Rhox10 promotes the differentiation of T1-ProSG directly into SSCs, thereby bypassing the T2-ProSG step entirely.
We obtained evidence that loss of Rhox10 causes partial retention of germ cells in the center of the seminiferous tubules at time points when germ cells normally migrate to their periphery. This migratory event is critical, as evidence suggests it places ProSG in the appropriate niche that allows them to become self-renewing SSCs (Payne, 2013). We postulate that by promoting the migration of cells to this stem cell niche, RHOX10 serves as a critical transcription factor that not only places SSCs in the appropriate environment for their long-term function as stem cells, but also drives their initial generation. Supporting this, the majority of Id4-eGfp+ cells are found only in the periphery of tubules in both WT and Rhox10 KO testes. In striking contrast, most centrally located germ cells in Rhox10 KO testes become differentiating A-spermatogonia, as judged using the marker STRA8. Given the evidence that germ cells that remain in the center of tubules undergo apoptosis (Roosen-Runge and Leik, 1968), it is possible that the migration-deficient STRA8+ cells in Rhox10 KO mice eventually die and do not contribute to the first wave of spermatogenesis. However, we did not observe significantly increased germ cell apoptosis in Rhox10 KO compared to control mice at either P12 or P21 (as measured by TUNEL; H.W.S., unpublished observations) and sperm count was only modestly reduced in young mature Rhox10 KO compared to control mice (Figure 2C), indicative of a largely normal first wave of spermatogenesis. Together, we believe the weight of the evidence suggests that migration of ProSG to the seminiferous tubule periphery is not necessary for at least the initial differentiation of A-spermatogonia. In agreement with this, it was recently shown that RA injection into testes prior to the ProSG migration step (at P1) is able to trigger the conversion of ProSG into differentiating A-spermatogonia (Busada et al., 2014). Together, our results support a model in which Rhox10 serves to promote the migration of ProSG into the SSC niche in order to drive them to specifically differentiate into SSCs, not differentiating A-spermatogonia (Figure 7E).
To understand how Rhox10 drives SSC establishment, it is critical to identify genes regulated by Rhox10. A challenge in identifying genes regulated by transcription factors in complex cell populations in vivo is that it is typically not possible to distinguish between genes exhibiting differential expression because of altered expression in a given cell type (i.e., real targets) versus changes in cell subset composition (i.e., increased or decreased numbers of cells in which such genes are highly expressed). This is particularly problematic in the testis, which harbors many cell types, including germ cells at many stages of differentiation. Even highly purified undifferentiated spermatogonia enriched for SSCs have recently been shown to exhibit considerable heterogeneity (Hermann et al., 2015). We therefore elected to employ scRNAseq analysis to identify genes regulated by the RHOX10 transcription factor in specific germ cell subsets. From this analysis, we identified RHOX10-regulated genes in two specific germ cell subsets.
Most strikingly, we found that a large number of RHOX10-regulated genes encode cell migration proteins. Among them, Pdgfrb and Dab2ip, have been directly shown to drive ProSG migration in neonatal testes (Basciani et al., 2008; Xu et al., 2015). Of note, Pdgfrb not only promotes ProSG migration but also the accompanying step, resumption of ProSG proliferation (Basciani et al., 2008; Payne, 2013; Thuillier et al., 2010). Given that we obtained evidence that Rhox10 also acts on the same two developmental steps—promoting the differentiation of mitotically inactive ProSG (T1-ProSG) into mitotically active ProSG (T2-ProSG) and driving the migration of T2-ProSG into the SSC niche—it is tempting to speculate that Pdgfrb is a key effector gene downstream of RHOX10. In addition, two of the three known VEGF receptor genes are positively regulated by RHOX10 - Flt4 (which encodes VEGFR-3) and Kdr (which encodes VEGFR-2). Although best known for its role in vascular development, the VEGF pathway also has non-vascular roles, including cell fate decision of SSCs and cell migration events independent of vascular development (Breen, 2007; Nalbandian et al., 2003; Sargent et al., 2016). We also identified 10 RHOX10-regulated genes that belong to the ADAM, integrin, and tetraspanin families, all of which have been implicated in ProSG migration (Tres and Kierszenbaum, 2005). Thus, RHOX10 regulates a large set of genes that have the potential to act downstream of RHOX10 to drive the migration of ProSG to the SSC niche.
Another class of RHOX10-regulated genes identified by our scRNAseq analysis is those previously shown to have ProSG- and SSC-associated functions. For example, RHOX10 regulates genes encoding transcription factors previously shown to function in SSCs. RHOX10-regulated genes in this category include Brachyury (T), which encodes a transcription factor that appears to promote SSC self-renewal (Wu et al., 2011), and Bcl6b, which encodes a BTB/POZ zinc finger transcription factor in the same subfamily as PLZF that shares with PLZF the ability to drive SSC maintenance (Oatley et al., 2006). Both T and BCL6B are candidates to also function in ProSG-to-SSC differentiation, as we showed both are expressed in ProSG and they were both previously shown to promote SSC frequency in undifferentiated spermatogonia cultures, as assessed by germ-cell transplantation. Indeed, T is known to promote the generation of PGCs (Aramaki et al., 2013). Other Rhox10-regulated genes encode factors driving ProSG/SSC proliferation and/or survival, including Erbb3, which encodes a receptor tyrosine kinase that has been implicated in driving ProSG proliferation (Toyoda-Ohno et al., 1999) and Epas1, which encodes a hypoxia-induced transcription factor that supports proliferation of germline stem cell cultures (Huang et al., 2014).
Another prominent group of RHOX10-regulated genes is those encoding factors involved in DNA methylation. In the “ProSG subset,” we identified the de novo DNA methyltransferase enzyme gene, Dnmt3b, as being positively regulated by Rhox10, which raises the possibility that Rhox10 promotes the genome-wide DNA methylation that occurs in ProSG (McCarrey, 2013). Further support for this notion is our finding that RHOX10 negatively regulates genes involved in base-excision repair (Mutyh, Neil3, Ung, and Rad51l3), a pathway that works together with the ten-eleven translocation (TET) family proteins to elicit genome-wide DNA demethylation during PGC reprograming (Yamaguchi et al., 2013). In the “SSC subset,” we found that RHOX10 positively regulates the Tet1 gene, raising the possibility that Rhox10 stimulates the demethylation and subsequent transcriptional activation of genes in SSCs (Hammoud et al., 2014). The notion that Rhox10 influences DNA demethylation/methylation events in early germ cells is intriguing given the many studies that have shown that Rhox genes are themselves regulated by DNA methylation, both in vitro and in vivo (MacLean and Wilkinson, 2010; Maeder et al., 2013; Richardson et al., 2014). This raises the possibility that RHOX10 participates in a DNA methylation feedback network in undifferentiated germ cells.
While we found that Rhox10-KO mice have a strong SSC establishment defect, it is worth noting that the generation of SSCs is not completely abolished in Rhox10-KO mice. Thus, even though most seminiferous tubules in these mutant mice are abnormal, spermatogenesis does take place and some sperm are generated, even in older mice that depend on SSCs for spermatogenesis. One explanation for why a small number of SSCs (~15% of normal) are still capable of being generated in Rhox10-KO mice is that these residual SSCs are generated by a minor pathway different than the one acted upon by Rhox10. Indeed, a recent study provided evidence for the existence of at least two different populations of SSCs that self renew using different growth factors (Takashima et al., 2015). Given that only a single round of replication can allow a single SSC to give rise of 4000 spermatids (Yoshida, 2008), it is not surprising that the small number of remaining SSCs in Rhox10-KO mice can support spermatogenesis to some degree.
RHOX10 may have functions in addition to SSC establishment. For example, it may also be involved in SSC maintenance. However, it is unlikely to have an essential role in SSC maintenance, as we found that aged Rhox10-KO mice (up to 48 weeks old) had comparable testes weight and sperm number as to 6- to 12-week Rhox10-KO mice (Figures S2C and S2D). Since RHOX10 is expressed in all stages of spermatogonia and even early meiotic spermatocytes (Song et al., 2012), it is also possible that RHOX10 has functions in post-SSC stages of germ cells. However, against this possibility, we found that Rhox10Fl/y;Stra8-Cre mice, which conditionally ablate Rhox10 at the spermatogonia stage (Sadate-Ngatchou et al., 2008), did not exhibit a statistically significant alteration in either testes weight or sperm number compared to Rhox10Fl/y littermate mice. Thus, either Rhox10 does not function after the SSC stage or its function is compensated by other genes when it is absent.
Our finding that Rhox10-KO mice had SSC-related defects strikingly similar to those of whole Rhox cluster-KO mice suggests that Rhox10 is the only gene in the Rhox cluster with significant SSC-promoting activity. Other members of this 33-gene cluster are likely to function in other aspects of spermatogenesis. The Sertoli cell-expressed Rhox genes, Rhox5 and Rhox8, have been shown to promote germ cell survival, the generation of normal numbers of motile sperm, and male fertility, in part through regulation of androgen-dependent events in Sertoli cells (Hu et al., 2010; Maclean et al., 2005; Welborn et al., 2015). Germ cell-expressed Rhox genes in addition to Rhox10 are likely to also have roles in spermatogenesis, based on our finding that mice lacking the entire Rhox cluster in germ cells have post-SSC defects not observed in Rhox10-KO mice (H.W.S., unpublished observation). Candidate Rhox genes responsible for these defects are Rhox11 and the Rhox3 paralogs, as these Rhox genes are highly expressed in round and elongating spermatids (Maclean et al., 2005; Song et al., 2015).
Our discovery of a transcription factor that drives the initial establishment of stem cells in the testis has potential clinical implications. For example, a block in the ProSG differentiation step is likely to cause a subset of testicular tumors (Sonne et al., 2009) and thus aberrations in Rhox10 and other SSC establishment genes may contribute to such germ cell tumors. Understanding the ProSG-to-SSC transition also has the potential to lead to diagnostic and treatment approaches for many infertility cases, as there is an established correlation between defects in this step and human infertility (Hadziselimovic and Herzog, 2001). Finally, through its ability to drive the formation of SSCs, RHOX10 has the potential to be used for reprogramming strategies to cure infertility in humans.
Experimental Procedures
Animals
Mouse colonies were maintained in agreement with protocols approved by the Institutional Animal Care and Use Committee at the University of California, San Diego.
Generation of Rhox-c-KO mice and Rhox10-KO mice
Rhox-c- and Rhox10-floxed mice were generated by conventional conditional gene KO procedures involving the use of targeting vectors to introduce loxP sites. See Supplemental Experimental Procedures for details.
Histology and immunofluorescence
Histological analysis and immunostaining were performed as conventional procedures. See Supplemental Experimental Procedures for details and antibodies used.
SC-RNAseq analysis
Single testicular cells from Id4-eGfp mice were prepared from P7 WT, P3 WT, and P3 Rhox10-KO mice. Id4-eGfp+ cells were sorted using a FACSAria II machine (BD Biosciences). Individual GFP+ cells were captured on a C1 chip using the C1 Single-Cell Microfluidic Auto Prep System (Fluidigm) and reverse transcription and cDNA amplification was performed on the C1 chip using the SMARTer Ultra Low RNA Kit for Illumina Sequencing (Clontech Inc.). Sequencing was performed using Illumina's HiSeq2500 (La Jolla Institute for Allergy and Immunology and University of California, San Diego). See Supplemental Experimental Procedures for details.
Transcriptome data analysis
RNA sequencing data was analyzed using the Singular 3.5.2 software. Principal component analysis was used for each sample condition to identify the top 200 genes that best account for variance in the first three principal components for that sample set. These variant genes were used in hierarchical clustering analysis using the R package gplots to split each sample into subgroups. To identify potential Rhox10 target genes, pairwise SCDE analysis was performed within similar type subgroups (Type II: WTP3A versus KOP3B; Type III: WTP3B versus KOP3C). See Supplemental Experimental Procedures for detail analysis.
Germ cell transplantation analysis
Donor testicular cells for germ cell transplantation analysis were isolated from P7-8 male mice. Four Rhox10-KO pups and five WT littermate control pups were used for transplantation as described previously (Ogawa et al., 1997). See Supplemental Experimental Procedures for details.
Statistical Methods
Unpaired t test or two-way ANOVA was used when applicable to determine significance. Data were analyzed using Prism 6. Significant values are considered as p<0.05.
Supplementary Material
Acknowledgments
The authors thank Jon Oatley (Washington State University) for generously providing Id4-eGfp transgenic mice. The authors also thank Gina Aloisio and Diego Castrillon (University of Texas Southwestern Medical Center) for technical advice in performing PAX7 staining. This work was supported by the National Institutes of Health grant R01-GM119128-01 and California Institute for Regenerative Medicine grant RB5-07210.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
Most experiments were conceived, performed, and analyzed by H.S., with help from A.B., A.Z., D.S., E.B., E.S., M.P, T.P, M.R, M.R., and V.S. Germ cell transplantation experiment was performed by M.S. and K.O. sc-RNAseq analysis was performed by B.L and K.Z. Help with testicular phenotypic analysis was provided by D.D. The manuscript was written by H.S. and M.W.
Accession Numbers
The sc-RNAseq data are submitted to NIH Gene Expression Omnibus (GEO) under accession number GSE82174.
References
- Almenara A, Escalante G, Gazzo E, Gonzales GF. Transillumination to evaluate spermatogenesis: effect of testosterone enanthate in adult male rats. Arch. Androl. 2001;46:21–27. doi: 10.1080/01485010150211119. [DOI] [PubMed] [Google Scholar]
- Aloisio GM, Nakada Y, Saatcioglu HD, Peña CG, Baker MD, Tarnawa ED, Mukherjee J, Manjunath H, Bugde A, Sengupta AL, et al. PAX7 expression defines germline stem cells in the adult testis. J. Clin. Invest. 2014;124:3929–3944. doi: 10.1172/JCI75943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aramaki S, Hayashi K, Kurimoto K, Ohta H, Yabuta Y, Iwanari H, Mochizuki Y, Hamakubo T, Kato Y, Shirahige K, et al. A mesodermal factor, T, specifies mouse germ cell fate by directly activating germline determinants. Dev. Cell. 2013;27:516–529. doi: 10.1016/j.devcel.2013.11.001. [DOI] [PubMed] [Google Scholar]
- Basciani S, De Luca G, Dolci S, Brama M, Arizzi M, Mariani S, Rosano G, Spera G, Gnessi L. Platelet-derived growth factor receptor beta-subtype regulates proliferation and migration of gonocytes. Endocrinology. 2008;149:6226–6235. doi: 10.1210/en.2008-0349. [DOI] [PubMed] [Google Scholar]
- Breen EC. VEGF in biological control. J. Cell. Biochem. 2007;102:1358–1367. doi: 10.1002/jcb.21579. [DOI] [PubMed] [Google Scholar]
- Busada JT, Kaye EP, Renegar RH, Geyer CB. Retinoic acid induces multiple hallmarks of the prospermatogonia-to-spermatogonia transition in the neonatal mouse. Biol. Reprod. 2014;90:64. doi: 10.1095/biolreprod.113.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan F, Oatley MJ, Kaucher AV, Yang Q-E, Bieberich CJ, Shashikant CS, Oatley JM. Functional and molecular features of the Id4+ germline stem cell population in mouse testes. Genes Dev. 2014;28:1351–1362. doi: 10.1101/gad.240465.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culty M. Gonocytes, the forgotten cells of the germ cell lineage. Birth Defects Res. C. Embryo Today. 2009;87:1–26. doi: 10.1002/bdrc.20142. [DOI] [PubMed] [Google Scholar]
- Culty M. Gonocytes, from the fifties to the present: is there a reason to change the name? Biol. Reprod. 2013;89:46. doi: 10.1095/biolreprod.113.110544. [DOI] [PubMed] [Google Scholar]
- Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line, Vasa-Cre. Genesis. 2007;45:413–417. doi: 10.1002/dvg.20310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Eddy EM. Identification and characterization of Rhox13, a novel X-linked mouse homeobox gene. Gene. 2008;423:194–200. doi: 10.1016/j.gene.2008.06.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer CB, Saba R, Kato Y, Anderson AJ, Chappell VK, Saga Y, Eddy EM. Rhox13 is translated in premeiotic germ cells in male and female mice and is regulated by NANOS2 in the male. Biol. Reprod. 2012;86:127. doi: 10.1095/biolreprod.111.094938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J. Clin. Invest. 2011;121:3456–3466. doi: 10.1172/JCI57984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guatelli-Steinberg D, Boyce J. The Postnatal Endocrine Surge and Its Effects on Subsequent Sexual Growth. Handbook of Growth and Growth Monitoring in Health and Disease (Springer Science) 2012 [Google Scholar]
- Hadziselimovic F, Herzog B. The importance of both an early orchidopexy and germ cell maturation for fertility. Lancet. 2001;358:1156–1157. doi: 10.1016/S0140-6736(01)06274-2. [DOI] [PubMed] [Google Scholar]
- Hammoud SS, Low DHP, Yi C, Carrell DT, Guccione E, Cairns BR. Chromatin and transcription transitions of mammalian adult germline stem cells and spermatogenesis. Cell Stem Cell. 2014;15:239–253. doi: 10.1016/j.stem.2014.04.006. [DOI] [PubMed] [Google Scholar]
- Hermann BP, Mutoji KN, Velte EK, Ko D, Oatley JM, Geyer CB, McCarrey JR. Transcriptional and translational heterogeneity among neonatal mouse spermatogonia. Biol. Reprod. 2015;92:54. doi: 10.1095/biolreprod.114.125757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z, Dandekar D, O'Shaughnessy PJ, De Gendt K, Verhoeven G, Wilkinson MF. Androgen-induced Rhox homeobox genes modulate the expression of AR- regulated genes. Mol. Endocrinol. 2010;24:60–75. doi: 10.1210/me.2009-0303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y-H, Lin M-H, Wang P-C, Wu Y-C, Chiang H-L, Wang Y-L, Chang J-H, Huang Y-K, Gu S-Y, Ho H-N, et al. Hypoxia inducible factor 2α/insulin-like growth factor receptor signal loop supports the proliferation and Oct-4 maintenance of mouse germline stem cells. Mol. Hum. Reprod. 2014;20:526–537. doi: 10.1093/molehr/gau016. [DOI] [PubMed] [Google Scholar]
- Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int. J. Androl. 1981;4:475–493. doi: 10.1111/j.1365-2605.1981.tb00732.x. [DOI] [PubMed] [Google Scholar]
- MacLean JA, Wilkinson MF. The Rhox genes. Reproduction. 2010;140:195–213. doi: 10.1530/REP-10-0100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maclean J. a, Chen M. a, Wayne CM, Bruce SR, Rao M, Meistrich ML, Macleod C, Wilkinson MF. Rhox: a new homeobox gene cluster. Cell. 2005;120:369–382. doi: 10.1016/j.cell.2004.12.022. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Angstman JF, Richardson ME, Linder SJ, Cascio VM, Tsai SQ, Ho QH, Sander JD, Reyon D, Bernstein BE, et al. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manku G, Culty M. Mammalian gonocyte and spermatogonia differentiation: recent advances and remaining challenges. Reproduction. 2015;149:R139–R157. doi: 10.1530/REP-14-0431. [DOI] [PubMed] [Google Scholar]
- McCarrey JR. Toward a more precise and informative nomenclature describing fetal and neonatal male germ cells in rodents. Biol. Reprod. 2013;89:47. doi: 10.1095/biolreprod.113.110502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mclean DJ, Friel PJ, Johnston DS, Griswold MD. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol. Reprod. 2003;69:2085–2091. doi: 10.1095/biolreprod.103.017020. [DOI] [PubMed] [Google Scholar]
- Mueller JL, Mahadevaiah SK, Park PJ, Warburton PE, Page DC, Turner JMA. The mouse X chromosome is enriched for multicopy testis genes showing postmeiotic expression. Nat. Genet. 2008;40:794–799. doi: 10.1038/ng.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphey P, McLean DJ, McMahan CA, Walter CA, McCarrey JR. Enhanced genetic integrity in mouse germ cells. Biol. Reprod. 2013;88:6. doi: 10.1095/biolreprod.112.103481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbandian A, Dettin L, Dym M, Ravindranath N. Expression of vascular endothelial growth factor receptors during male germ cell differentiation in the mouse. Biol. Reprod. 2003;69:985–994. doi: 10.1095/biolreprod.102.013581. [DOI] [PubMed] [Google Scholar]
- Oatley JM, Brinster RL. Regulation of spermatogonial stem cell self-renewal in mammals. Annu. Rev. Cell Dev. Biol. 2008;24:263–286. doi: 10.1146/annurev.cellbio.24.110707.175355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc. Natl. Acad. Sci. U. S. A. 2006;103:9524–9529. doi: 10.1073/pnas.0603332103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa T, Aréchaga JM, Avarbock MR, Brinster RL. Transplantation of testis germinal cells into mouse seminiferous tubules. Int. J. Dev. Biol. 1997;41:111–122. [PubMed] [Google Scholar]
- Ohbo K, Yoshida S, Ohmura M, Ohneda O, Ogawa T, Tsuchiya H, Kuwana T, Kehler J, Abe K, Schöler HR, et al. Identification and characterization of stem cells in prepubertal spermatogenesis in mice. Dev. Biol. 2003;258:209–225. doi: 10.1016/s0012-1606(03)00111-8. [DOI] [PubMed] [Google Scholar]
- Pastor WA, Stroud H, Nee K, Liu W, Pezic D, Manakov S, Lee SA, Moissiard G, Zamudio N, Bourc'his D, et al. MORC1 represses transposable elements in the mouse male germline. Nat. Commun. 2014;5:5795. doi: 10.1038/ncomms6795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payne CJ. Cycling to and from a stem cell niche: the temporal and spatial odyssey of mitotic male germ cells. Int. J. Dev. Biol. 2013;57:169–177. doi: 10.1387/ijdb.130006cp. [DOI] [PubMed] [Google Scholar]
- Phillips BT, Gassei K, Orwig KE. Spermatogonial stem cell regulation and spermatogenesis. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2010;365:1663–1678. doi: 10.1098/rstb.2010.0026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson ME, Bleiziffer A, Tüttelmann F, Gromoll J, Wilkinson MF. Epigenetic regulation of the RHOX homeobox gene cluster and its association with human male infertility. Hum. Mol. Genet. 2014;23:12–23. doi: 10.1093/hmg/ddt392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Rooij DG, Griswold MD. Questions about spermatogonia posed and answered since 2000. J. Androl. 2012;33:1085–1095. doi: 10.2164/jandrol.112.016832. [DOI] [PubMed] [Google Scholar]
- De Rooij DG, Okabe M, Nishimune Y. Arrest of spermatogonial differentiation in jsd/jsd, Sl17H/Sl17H, and cryptorchid mice. Biol. Reprod. 1999;61:842–847. doi: 10.1095/biolreprod61.3.842. [DOI] [PubMed] [Google Scholar]
- Roosen-Runge EC, Leik J. Gonocyte degeneration in the postnatal male rat. Am. J. Anat. 1968;122:275–299. doi: 10.1002/aja.1001220208. [DOI] [PubMed] [Google Scholar]
- Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis. 2008;46:738–742. doi: 10.1002/dvg.20437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent KM, Clopton DT, Lu N, Pohlmeier WE, Cupp AS. VEGFA splicing: divergent isoforms regulate spermatogonial stem cell maintenance. Cell Tissue Res. 2016;363:31–45. doi: 10.1007/s00441-015-2297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H-W, Wilkinson MF. Transcriptional control of spermatogonial maintenance and differentiation. Semin. Cell Dev. Biol. 2014;30:14–26. doi: 10.1016/j.semcdb.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H-W, Dann CT, McCarrey JR, Meistrich ML, Cornwall GA, Wilkinson MF. Dynamic expression pattern and subcellular localization of the Rhox10 homeobox transcription factor during early germ cell development. Reproduction. 2012;143:611–624. doi: 10.1530/REP-11-0479. [DOI] [PubMed] [Google Scholar]
- Song H-W, Bettegowda A, Oliver D, Yan W, Phan MH, de Rooij DG, Corbett MA, Wilkinson MF. shRNA off-target effects in vivo: impaired endogenous siRNA expression and spermatogenic defects. PLoS One. 2015;10:e0118549. doi: 10.1371/journal.pone.0118549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, Harrison NJ, Schwager C, Abdollahi A, Huber PE, et al. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res. 2009;69:5241–5250. doi: 10.1158/0008-5472.CAN-08-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takashima S, Kanatsu-Shinohara M, Tanaka T, Morimoto H, Inoue K, Ogonuki N, Jijiwa M, Takahashi M, Ogura A, Shinohara T. Functional differences between GDNF-dependent and FGF2-dependent mouse spermatogonial stem cell self-renewal. Stem Cell Reports. 2015;4:489–502. doi: 10.1016/j.stemcr.2015.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuillier R, Mazer M, Manku G, Boisvert A, Wang Y, Culty M. Interdependence of platelet-derived growth factor and estrogen-signaling pathways in inducing neonatal rat testicular gonocytes proliferation. Biol. Reprod. 2010;82:825–836. doi: 10.1095/biolreprod.109.081729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toyoda-Ohno H, Obinata M, Matsui Y. Members of the ErbB receptor tyrosine kinases are involved in germ cell development in fetal mouse gonads. Dev. Biol. 1999;215:399–406. doi: 10.1006/dbio.1999.9482. [DOI] [PubMed] [Google Scholar]
- Tres LL, Kierszenbaum AL. The ADAM-integrin-tetraspanin complex in fetal and postnatal testicular cords. Birth Defects Res. C. Embryo Today. 2005;75:130–141. doi: 10.1002/bdrc.20041. [DOI] [PubMed] [Google Scholar]
- Welborn JP, Davis MG, Ebers SD, Stodden GR, Hayashi K, Cheatwood JC, Rao MK, MacLean JA. Rhox8 Ablation in the Sertoli Cells Using a Tissue-Specific RNAi Approach Results in Impaired Male Fertility in Mice. Biol. Reprod. 2015 doi: 10.1095/biolreprod.114.124834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Goodyear SM, Tobias JW, Avarbock MR, Brinster RL. Spermatogonial stem cell self-renewal requires ETV5-mediated downstream activation of Brachyury in mice. Biol. Reprod. 2011;85:1114–1123. doi: 10.1095/biolreprod.111.091793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Wan P, Wang M, Zhang J, Gao X, Hu B, Han J, Chen L, Sun K, Wu J, et al. AIP1-mediated actin disassembly is required for postnatal germ cell migration and spermatogonial stem cell niche establishment. Cell Death Dis. 2015;6:e1818. doi: 10.1038/cddis.2015.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi S, Shen L, Liu Y, Sendler D, Zhang Y. Role of Tet1 in erasure of genomic imprinting. Nature. 2013;504:460–464. doi: 10.1038/nature12805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S. Spermatogenic stem cell system in the mouse testis. Cold Spring Harb. Symp. Quant. Biol. 2008;73:25–32. doi: 10.1101/sqb.2008.73.046. [DOI] [PubMed] [Google Scholar]
- Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development. 2006;133:1495–1505. doi: 10.1242/dev.02316. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Shao S, Meistrich ML. Irradiated mouse testes efficiently support spermatogenesis derived from donor germ cells of mice and rats. J. Androl. 2006;27:365–375. doi: 10.2164/jandrol.05179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.