Abstract
In 1992, the Brugada syndrome (BrS) was recognized as a disease responsible for sudden cardiac death, characterized by a right bundle-branch block with ST segment elevation in the leads V1 and V2. This syndrome is highly associated with sudden cardiac death, especially in young males. BrS is currently diagnosed in patients with ST-segment elevation showing type 1 morphology ≥ 2 mm in ≥1 leads among the right precordial leads V1 or V2 positioned in the 2nd, 3rd, or 4th intercostal space, and occurring either spontaneously or after a provocative drug test by the intravenous administration of Class I antiarrhythmic drugs. With accumulated findings, the BrS inheritance model is believed to be an autosomal dominant inheritable model with incomplete penetrance, although most patients with BrS were sporadic cases. SCN5A, which was identified as the first BrS-associated gene in 1998, has emerged as the most common gene associated with BrS, and more than 10 BrS-associated genes have been identified thereafter. Mutation-specific genetic testing is recommended for the family members and appropriate relatives following the identification of BrS-causative mutations in an index patient. In addition, comprehensive or BrS1 (SCN5A) targeted genetic testing could be useful for patients in whom a cardiologist has established a clinical index of suspicion for BrS based on the patient׳s clinical history, family history, and the expressed electrocardiographic (resting 12-lead ECGs and/or provocative drug challenge testing) phenotype.
Over the past 20 years, extensive research in this field has allowed better understanding of the pathophysiology, genetic background, and management of BrS even though controversies still exist. In this review article, a background of genetics, the genetic background of BrS, the genotype and phenotype relationship, the role of genetic screening in clinical practice, and the interpretation of the identified genetic variants have been addressed based on this understanding.
Keywords: Genetics, Brugada syndrome
1. Background of basic genetics
The human genome contains approximately 3 billion nucleotide base pairs contained in 23 chromosome pairs. Each chromosome contains hundreds to thousands of genes and the estimated 30,000 genes in the human genome express approximately 100,000 proteins [1]. The Human Genome Project (HGP) was proposed in the 1980s and was completed in 2003. The 1000 Genomes Project was conducted between 2008 and 2015, generating the largest public catalogue of human variations and genotype data. The completion of these projects provides a source book for biology, medicine, and genetic research.
Sanger sequencing was used for the genetic screening of BrS and was considered the gold standard for DNA sequencing in the subsequent two and a half decades [2]. In the past few years, new technologies (large-insert clone arrays, oligonucleotide arrays, target-gene sequencing, whole-exome sequencing (WES), and whole-genome sequencing (WGS)) have been developed, thus allowing the detection of medium- to large-sized genomic regions at a single nucleotide resolution. These technological genomic advancements are able to provide high-throughput screening and can detect genetic variations in patients with high accuracy and reduced cost. Although scientists and policy advisers deal intensively with the interpretation and handling of the onslaught and ambiguity of genome-wide data, we are rapidly moving toward a new era of genetic research for BrS, which is full of opportunities as well as a mountain of challenges.
2. Terminology
2.1. Polymorphism vs. mutation
The genome of any person is more than 99% similar to that of an unrelated individual. This tiny variability allows individuals to be distinguished by means of genetic testing. When a nucleotide change occurs in more than 1% of the general population, it is called a “polymorphism.” In contrast, a mutation occurs in less than 0.5% of the population and is defined as a permanent change in the nucleotide sequence that results in altered amino acids. The terms “mutation” and “polymorphism” have been used widely but often lead to confusion because of incorrect assumptions regarding their respective pathogenic and benign effects. In 2015, the American College of Medical Genetics and Genomics (ACMG) recommended that both terms be replaced by the term “variant” with the following modifiers: (i) pathogenic, (ii) likely pathogenic, (iii) uncertain significance, (iv) likely benign, or (v) benign [3].
2.2. Penetrance and expressivity
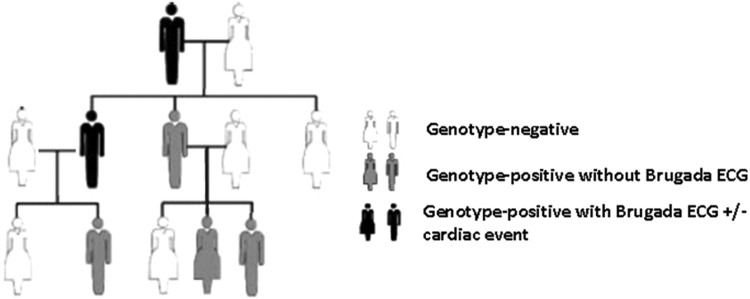
In medical genetics, penetrance is the proportion of individuals with the mutation who exhibit clinical symptoms. For example, in a family with 10 members, if 4 out of 10 are carriers of a pathogenic variant in the SCN5A gene but only 2 of the 4 carriers have type 1 BrS ECG, the penetrance in this family is 50%. The penetrance of BrS is lower than that of the congenital long QT syndrome. In a study conducted in 2000, Priori et al. estimated that the overall disease penetrance across 4 small BrS families harboring mutations in the SCN5A gene was 16% based on their ECG analysis (range 12.5–50%) [4]. In contrast, the mean penetrance across multiple long QT syndrome subtypes in a population-based study was shown to be ~40% (range 25–100%) [5], [6] (Fig. 1).
Fig. 1.
An example of a representative multi-generation pedigree displaying incomplete penetrance (33%) and variable expressivity as some individuals display Brugada ECG without any cardiac events.
Expressivity is used to describe the variations in a phenotype among individuals carrying the same pathogenic variants. Different degrees of expression in different individuals may be due to variation in the allelic constitution of the remaining genome or due to environmental factors. For example, individuals in the same BrS family who carry the same SCN5A pathogenic variant could show different electrocardiographic patterns ranging from Brugada type I ECG to conduction disturbance, or even long QT.
3. Clear clinical diagnosis of Brugada syndrome before genetic testing
Many clinical conditions may lead to ST-segment elevation in the right precordial leads, which mimic the BrS ECG patterns. For example, exposure to some drugs and ionic imbalance may produce a Brugada-like ST-segment elevation. Previous reports have described this as an acquired Brugada syndrome or Brugada phenocopy. It presents with an ECG pattern identical to type I (Coved), type 2, or type 3 (Saddleback) Brugada patterns but differs etiologically from true BrS. Prior to diagnosis of the true or congenital Brugada syndrome, all of the following conditions should be ruled out (Table 1).
Table 1.
Common causes of acquired Brugada syndrome or Brugada phenocopy.
| Category | Examples |
|---|---|
| Metabolic factors or electrolytes | Hypothermia, adrenal crisis/insufficiency, hyper or hypokalemia, hypercalcemia |
| Neuromuscular Diseases | Duchenne׳s muscular dystrophy, spinal and bulbar muscular atrophy (SBMA), Friedreich ataxia, myotonic dystrophy [68] |
| Toxin or poison | Cannabis, cocaine, ethanol, heroin, ketamine, aluminium phosphide fatal poisoning |
| Mechanical compression | Rhabdomyoma of the interventricular septum/mediastinal lipoma/mass lesions, pectus excavatum, post-esophageal reconstruction, hemopericardium |
| Vascular factors | RV infarction, pulmonary embolism, Dissecting aortic aneurysm |
| Myocardial and Pericardial Disease | Chagasic cardiomyopathy, cardiac amyloidosis, myocarditis, pericarditis, Arrhythmogenic right ventricular dysplasia |
| ECG modulation | Improper application of a high pass filter (ex. 0.5-Hz filter) |
| Miscellaneous | Repaired pentalogy of Fallot, electrocution |
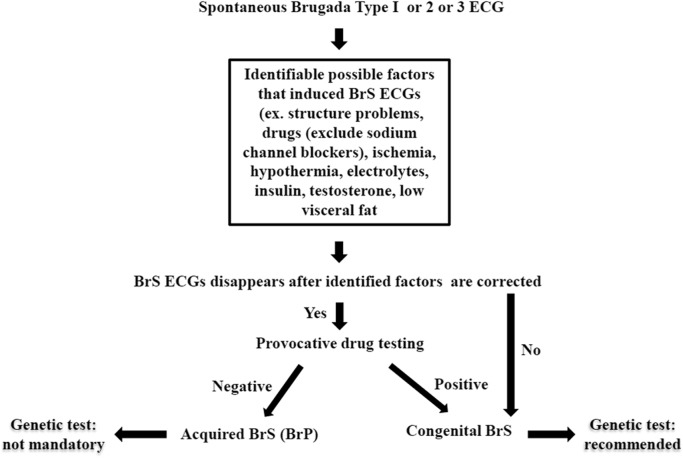
Acquired BrS usually has an identifiable underlying condition that elicits a BrS ECG pattern. Once the underlying condition is resolved, the ECG normalizes completely. Provocative testing with flecainide, ajmaline, pilsicainide, procainamide, or other sodium channel blockers is an important method to differentiate congenital BrS from acquired BrS. The result of these tests should be negative in patients with acquired BrS. The flowchart for such testing is shown in Fig. 2.
Fig. 2.
The flowchart for differentiating congenital BrS from acquired BrS before conducting genetic tests.
After clinical confirmation of congenital BrS, genetic testing is recommended for patients with congenital BrS but is not mandatory for those with acquired BrS. Thus, it is important to make a clear BrS diagnosis before conducting genetic tests.
4. Genetic background of Brugada syndrome
In 1996, the term “Brugada syndrome” was used to describe what was known as “right bundle branch block, persistent ST segment elevation, and sudden death syndrome”[7]. In 1998, the first BrS-associated gene, SCN5A, which encodes the alpha-subunit of the voltage-gated Nav1.5 cardiac sodium channel, was reported to be associated with the disease [8]. In the past 2 decades, several BrS-associated genes and modifier genes were reported and most of these primarily encode sodium, potassium, and calcium channels or the proteins associated with these channels (Table 2 and Fig. 3). Despite these genetic studies, only 30–35% of clinically diagnosed cases can be genetically diagnosed, indicating that 65–70% of BrS patients remain genetically unresolved.
Table 2.
Identified genes and modifier genes linked to Brugada syndrome.
| BrS subtype | Ion channel | BrS-associated Gene | Locus | Protein | Functional Effect | % of Disease | Reference |
|---|---|---|---|---|---|---|---|
| BrS1 | Sodium channel | SCN5A | 3p21 | Nav1.5 | LOF | 20–25 (Caucasian); 10-15 (Asian) | [8], [17] |
| BrS5 | SCN1B | 19q13.1 | Navβ1 | LOF | 1–2 | [25], [38] | |
| BrS17 | SCN2B | 11q23 | Navβ2 | LOF | Rare | [26] | |
| BrS7 | SCN3B | 11q23.2 | Navβ3 | LOF | Rare | [27] | |
| BrS18 | SCN10A | 3p22.2 | Nav1.8 | LOF | 2.5–16 | [28], [29], [30], [31], [46] | |
| BrS11 | Sodium channel-associated | RANGRF | 17p13.1 | MOG1 | LOF | Rare | [32] |
| BrS2 | GPD1-L | 3p24 | G3PD1L | LOF | Rare | [33], [38] | |
| BrS15 | SLMAP | 3p21.2-p14.3 | SLMAP | LOF | Rare | [34] | |
| BrS20 | PKP2 | 12p11.21 | plakophilin-2 | plakophilin-2 cause sodium current deficit | 2.5 | [35] | |
| BrS16 | TRPM4 | 19q13.33 | NSCCa | LOF | 8 | [36] | |
| BrS3 | Calcium channel | CACNA1C | 12p13.3 | Cav1.2 | LOF | 6–7 | [37], [38] |
| BrS4 | CACNB2b | 10p12.33 | Cavβ2 | LOF | 4–5 | [37], [38] | |
| BrS10 | CACNA2D1 | 7q21-22 | Cavα2δ−1 | LOF | Rare | [39] | |
| BrS21 | Potassium channel | ABCC9 | 12p12.1 | SUR2A (sulfonylurea receptor subunit 2 A), IK-ATP | GOF | 4–5 | [44] |
| BrS13 | KCND3 | 1p13.2 | Kv4.3, Ito | GOF | Rare | [40] | |
| BrS6 | KCNE3 | 11q13-14 | MiRP2, Ito/Iks | GOF | <1 | [38], [41] | |
| BrS9 | KCNJ8 | 12p12.1 | Kir6.1, IK-ATP | GOF | Rare | [43], [69] | |
| BrS8 | KCNH2 | 7q35 | Kv11.1, Ikr | GOF | 1–2 | [45] | |
| BrS-modifier Gene | |||||||
| BrS19 | Sodium channel | HEY2 | 6q22 | Nav1.5 | LOF | ? | [46] |
| BrS14 | Potassium channel | HCN4 | 15q24.1 | If | LOF | Rare | [47], [70] |
| BrS12 | KCNE5 | Xq22.3 | MiRP4, Kv4.3, Ito | GOF | Rare | [42] | |
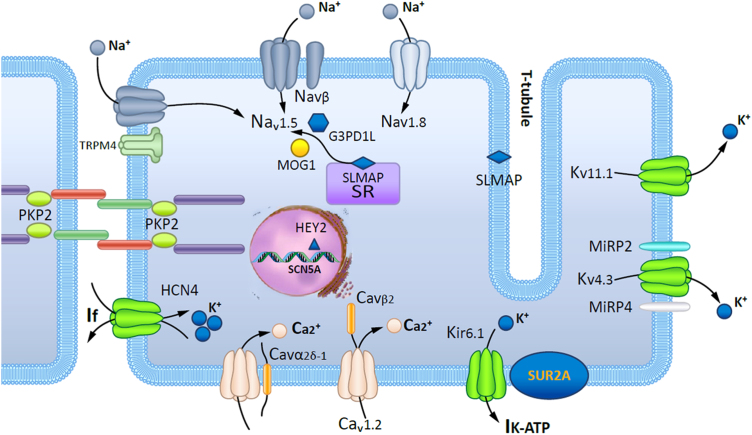
Fig. 3.
Schematic representation of a cardiomyocyte exhibiting the proteins involved in Brugada syndrome pathogenesis.
4.1. Differing ethnic background of Brugada syndrome
As early as 1917 sudden unexpected nocturnal death (SUND) in young, apparently healthy, adults has been reported in Manila, Guam, and Hawaii [9], [10]. Although the exact prevalence of BrS is still uncertain, it presents marked geographical differences. It was estimated to affect 5/10,000 individuals in Western countries and 12/10,000 individuals in Southeast Asia [11], [12], especially in Thailand and the Philippines where Brugada syndrome is considered to be the leading cause of sudden death in young men. After pooling all the community-based and hospital-based population studies in the world, type 1 ECG was found to appear more frequently in Asia (0–0.36%) and Europe (0–0.25%) than in the United States (0.03%). Type 2 and type 3 ECG is more prevalent in Asia (0.12–2.23%) than in Europe (0–0.6%) or the United States (0.02%) [13]. Data from subjects of African origin are highly scarce. Confining the analyses to community-based studies, which may be more representative of the general population, Asia still shows the highest prevalence of type 1 ECG globally (0–0.67%). There is a greater burden of sudden cardiac death due to BrS in Asian regions than that in Europe or America.
In 1998, Chen et al. reported that BrS has a genetic abnormality linked to mutations in SCN5A, which are present in 20–25% of BrS cases in Caucasian populations [8], [14]. In previous Japanese studies, the mutation rate of SCN5A was around 11–14% (personal communications) whereas in the Han Chinese population, <10% BrS patients SCN5Ain Taiwan had SCN5A mutations [15]. This implied that the distribution of disease-causing genes among BrS patients in the Asian population might differ from that in the Caucasian population. Furthermore, certain SCN5A promoter polymorphisms in a haplotype variant with a relatively high prevalence in Asians, were reported to not only reduced transcriptional activity in vitro, but also modulate variability in cardiac conduction as assessed by PR and QRS durations [16]. These observations implied the existence of variations in the genetic architecture of Asian BrS and Caucasian, which requires further research.
4.2. Sodium channel
SCN5A, the most common BrS-associated gene, was identified in approximately 20-25% of BrS patients. Until 2010, almost 300 SCN5A mutations have been reported in BrS, which include missense mutations, nonsense mutations, nucleotide insertion/deletions, and splice site mutations [17]. The number of SCN5A mutations is still increasing. SCN5A is responsible for the fast upstroke in phase 0 of the cardiac action potential, and pathogenic variations result in sodium channel dysfunction, which leads to slowing of conduction in the heart. Functional studies of these mutations revealed loss of function in the sodium channels that was affected through different mechanisms including decreased expression of the sodium channel protein (Nav1.5) in the sarcolemma [18], expression of non-functional channels [19], or altered gating properties (delayed activation, earlier inactivation, faster inactivation, enhanced slow inactivation, and delayed recovery from inactivation) [20], [21], [22], [23], [24].
Besides SCN5A genes, many BrS-related genes play a role in regulating sodium channel function. Several pathogenic variations in three genes (SCN1B, SCN2B, and SCN3B) encoding the β subunits of the Nav1.5 sodium channel, have been discovered to modify the channel function. SCN1B encodes the β1- and β1b- subunits, which are auxiliary function-modifying subunits of the cardiac Na+ channel, and decreases the INa current by affecting Na+ channel trafficking [25]. SCN2B encodes the β2-subunit of the Na+ cardiac channel. The D211G mutation in SCN2B decreases Nav1.5 expression on CHO cells [26]. Mutations in SCN3B, which encodes the β3-subunit of the Na+ cardiac channel, and leads to a loss of function in the Na+ cardiac channel, is also reported to cause BrS [27]. SCN10A, encodes the neuronal sodium channel Nav1.8, and modulates SCN5A expression and the electrical function of the heart; it is reported that mutations in SCN10A account for 16% of patients with the typical BrS identified in a cohort of 150 BrS affected Caucasian individuals [28]. However, three more studies have reported a lower percentage of SCN10A mutation in the SCN5A mutation negative BrS in the Caucasian (3.8%) and Japanese BrS patients (2.5%) [29], [30], [31]. Determining the true prevalence of SCN10A mutations in patients with BrS requires more data from different ethnicities.
4.3. Sodium channel-associated
RAN guanine nucleotide release factor (RANGRF) gene, encoding MOG1, was reported to impair the trafficking of Nav1.5 to the membrane, leading to INa reduction [32]. Mutations in the glycerol-3-phosphate dehydrogenase 1-like gene (GPD1L) may also affect the trafficking of cardiac Na+ channels to the cell surface and cause ~50% reduction in the inward Na+ current [33]. The gene for sarcolemmal membrane-associated protein (SLMAP) that is found in T-tubules and in the sarcoplasmic reticulum, which has an unknown function, causes BrS by modulating the intracellular trafficking of the Nav1.5 channel [34]. The gene for Plakophilin-2 (PKP2) was the primary gene responsible for arrhythmogenic right ventricular cardiomyopathy (ARVC). Recently, a correlation was identified between the loss of PKP2 expression PKP2 and reduced INa in BrS patients [35]. The transient receptor potential melastatin protein number 4 (TRPM4) is a calcium-activated nonselective cation channel, that is a member of a large family of transient receptor potential genes; of 248 BrS cases without SCN5A mutations, 8% SCN5Acarry mutations in TRPM4. Because of its effect on the resting membrane potential, both reduction and increase in TRPM4 channel function, may reduce the availability of Na+ channels and thus lead to BrS [36].
4.4. Calcium channel
BrS-susceptibility genes were also found among calcium channels (CACNA1C, CACNB2b, and CACNA2D1). Mutations in CACNA1C and CACNB2b are associated with 11.5% of BrS cases, wherein patients generally present with shorter-than normal QT intervals [17]. Putative mutations in the genes encoding the calcium channel, the voltage-dependent, L-type, α1c subunit (CACNA1C), and the β−2b subunits (CACNB2b) of the L-type cardiac Ca2+ channel caused a decrease in ICa current, resulting in a combined BrS/short QT syndrome [37], [38]. CACNA2D1, encoding the calcium channel, voltage-dependent, L-type, α−2/δ subunit 1, regulates the current density and activation/inactivation kinetics of the Ca2+ channel and is associated with BrS [39].
4.5. Potassium channel
Apart from sodium and calcium channels, putative gain of function mutations in genes encoding channels that conduct outward potassium currents (KCND3, KCNE3, KCNE5KCNE5, and KCNJ8) have also been reported in a few BrS cases. Mutations in KCND3 genes, encoding a Kv4.3 potassium voltage-gated channel, Shal-related subfamily, member 3, lead to an increase of the Ito current in the right ventricle, and have been linked to BrS [40]. KCNE3 (encoding MiRP2) is a protein that decreases the potassium (K+) transient outward current (Ito) current by interacting with the channel Kv4.3, and results in increased Ito magnitude and density in the human heart [38], [41]. KCNE5 (KCNE1L) is located on the X chromosome and encodes an auxiliary β-subunit for K channels (potassium voltage-gated channel, Isk-related family, member 5). Mutations in KCNE5 modified potassium channels that lead to an increase in the Ito current have been linked to BrS [42]. Potassium inwardly rectifying channel, subfamily J, member 8 (KCNJ8) gene encodes Kir6.1 [43]. The gene for ATP-binding cassette, subfamily C member 9 (ABCC9) encodes the sulfonylurea receptor subunit 2 A (SUR2A) [44]. Functional KATP channels have an octameric subunit structure with four pore-forming subunits (Kir6.1) and four sulfonylurea receptors (SUR2A). Gain-of-function mutations in KATP channels induced by pathogenic variants in KCNJ8 or ABCC9, accompanied with loss-of-function pathogenic variants in SCN5A may result in a severe arrhythmic phenotype of BrS [43]. Four KCNH2 mutations, T152I, R164C, W927G, and R1135H were identified in 236 Japanese consecutive probands with BrS or Brugada-like ECG. Three of these mutation carriers showed QTc intervals shorter than 360 milliseconds and one experienced VF. Patch-clamp analyses showed that all KCNH2 mutations exerted gain-of-function effects on IKr channels in CHO cells [45].
4.6. Potassium channel-associated
In a transgenic mouse study, HEY2, which encodes the transcriptional repressor hairy/enhancer-of-split related with YRPW motif protein, was found to play a role in the regulation of SCN5A expression and conduction velocity in the heart, suggesting that BrS may originate from altered transcriptional programming during cardiac development. One SNP, rs9388451 near the HEY2 gene was significantly associated with BrS through a genome-wide association study in 312 individuals with BrS [46]. HCN4 encodes the hyperpolarization activated cyclic nucleotide-gated potassium channel 4, which is a voltage-gated ion channel mediating the pacemaker (If) current in the heart. It has been reported in a few BrS patients, but its causative role is still not clear [47], [48].
Until now, GPD1L is the only gene identified by a classical genetic linkage study, whereas all other genes were identified only in a single patient, in a few unrelated patients, or in small families through candidate gene analysis [17]. This is an important limitation that deserves further study before genes are implicated in the pathogenesis of BrS or any other disease entity [49].
4.7. Inherited model of BrS
The inheritance of BrS was believed to present an autosomal dominant mode of transmission with incomplete penetrance [50]. The penetrance of BrS is age- and sex-dependent and most lethal events occur in men after the fourth decade of life [51], [52]. In a long-term follow-up study, age-related depolarization abnormalities (e.g. slowing of conduction) in the heart modulate the clinical ECG phenotype [53], especially in patients with SCN5A-positive mutations. Gender is also a modifier of both ECG phenotype and the risk of SCD in BrS [54]. The relative risk for cardiac events in men is 3.34 times that in women. These sex-specific differences may be linked to the differential effect of sex hormones on cardiac ion channel current densities and functional cardiac repolarization reserves [6].
In the past decades, more observations suggest that BrS has a heterogeneous genetic basis and is a disease with a more complex inheritance. Overall, <40% of BrS cases are familial, whereas other cases are sporadic. Despite its symptomatic or asymptomatic status, a positive family history of sudden cardiac death at a young age (<45 years) in BrS patients was quite low (10–30%). In addition, low disease penetrance was observed based on ECG analysis in family members carrying SCN5A mutations (12.5–50%, average 16%) [4]. In a study with large families that had at least 5 family members carrying SCN5A mutations, 8 SCN5A mutation-negative BrS individuals were identified in 5 families (3 belonging to 3 different families) [55]. Furthermore, familial linkage analyses have largely been unsuccessful in uncovering new disease-causing genes because most of the BrS families are rather small for powerful linkage analysis, low penetrance, and variable expressivity of diseases. In a GWAS study, Bezzina et al. identified three common genetic variants that contributed to BrS susceptibility [46]. Disease susceptibility increased consistently with increasing numbers of risk alleles, when these loci were considered in aggregate [16]. The above observations provide evidence that the genetic architecture of BrS could be more complicated than we have understood until date and it is more like an oligogenic disease.
4.8. Overlapping syndromes and genetic heterogeneity of BrS
After two decades, the SCN5A gene has become the most common BrS-causing gene [56]. However, several reports have demonstrated increasing clinical and biophysical overlap among various types of SCN5A mutations, referred to as an “overlap syndrome” of cardiac sodium channel disease [57]. A single mutation in SCN5A can cause several phenotypes in the same family or in a single patient, such as BrS, long QT syndrome type 3 (LQTS3), sick sinus syndrome, and cardiac conduction diseases [58]. A pathogenic variant in the SCN5A gene has been identified in patients affected by the early repolarization syndrome (ERS) [59]. The presence of progressive cardiac conduction disease (PCCD) in BrS families is frequent and has been described as a different expression of the BrS genetic phenotype. Several pathogenic variations in the SCN5A gene are associated with both PCCD and BrS [60]. One gene could result in multiple phenotypes, even in one family. The BrS and sick sinus syndrome (SSS) were both identified within a single family to be caused by a SCN5A mutation causing loss-of-function in INa [61]. One particular mutation may cause a mixed clinical phenotype in one carrier, and to a single arrhythmia syndrome in another. The reasons for this remain largely elusive. Clinical and genetic diagnoses of SCN5A-related overlap syndromes are often complicated further by reduced penetrance and variable disease expressivity, even within a single family. Large cohorts of patients carrying the same overlap syndrome with an SCN5A mutation are needed to gain more insight into the different factors that determine disease severity, expressivity, and outcome.
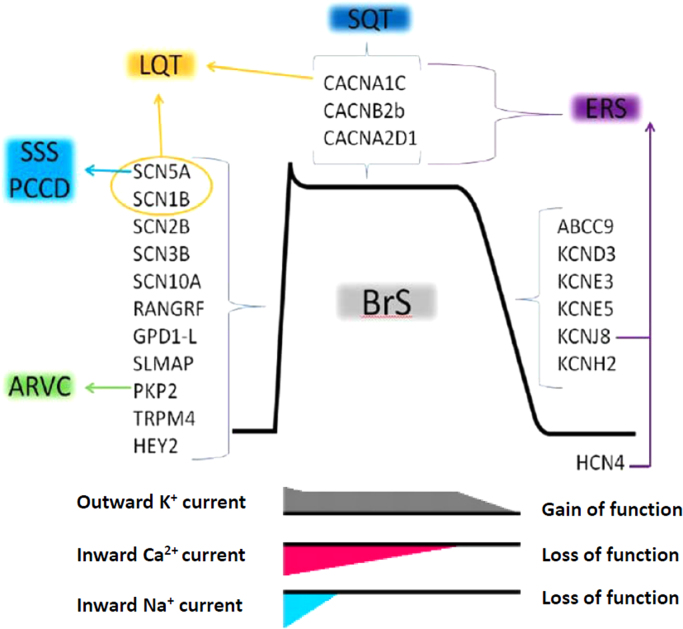
The genetic heterogeneity of BrS showed overlapping genetic features with other inherited arrhythmia diseases (ERS, PCCD, SSS, short QT syndrome, LQTS3, and arrhythmogenic right ventricular cardiomyopathy). The genes associated with both BrS and other inherited arrhythmia diseases are shown in Fig. 4.
Fig. 4.
Genes associated with Brugada syndrome (BrS) and other inherited arrhythmia diseases.
5. The role of genetic testing in the clinical practice for Brugada syndrome
BrS is partly a cardiac disorder, with an autosomal dominant inheritable model showing incomplete penetrance. The current expert consensus statement recommends mutation-specific genetic testing for family members and appropriate relatives following identification of the BrS-causative mutation in an index patient (Class I recommendation) [62]. The primary role of genetic testing in BrS is to confirm the disorder in the index patient, and cascade testing in relatives to distinguish those who need clinical follow-up (for development of conduction disease or syncopal episodes) and preventive measures (e.g., avoiding specific drugs, prompt management of fever) from those who are both clinically and genetically unaffected [56], [63].
5.1. Genetic testing for diagnostic implications
BrS is diagnosed clinically (type 1 Brugada ECG pattern) at this point. The result of genetic testing is not considered one of the criteria for BrS diagnosis. Genetic testing is not recommended in the absence of a diagnostic ECG. Nevertheless, comprehensive or SCN5A targeted genetic testing can be useful for any patient in whom a cardiologist has established a clinical index of suspicion for BrS based on clinical examination (patient׳s clinical history, family history, and ECG phenotype) [56]. In addition, mutation-specific genetic testing in family members of successfully genotyped probands may play a decisive role in determining who should take precautions in certain conditions and who should be followed-up. Both clinically diagnosed BrS patients and asymptomatic SCN5A mutation-positive subjects are advised to treat fever with antipyretic drugs immediately, and to avoid intake of excessive alcohol and drugs that decrease sodium channel availability and functionality [56].
5.2. Genetic testing for prognosis
The data linking genetic information to clinical prognosis are being accumulated but are still limited at this moment. In a previous meta-analysis and results from the FINGER Brugada Syndrome registry, the status of SCN5A mutations does not seem to be an independent risk factor for future cardiac events (syncope or SCD) [54], [64]. However, in a subset analysis for mutation types, patients carrying an SCN5A mutation that causes a prematurely truncated protein (stop codon or frameshift), were more likely to present with syncope and develop prolonged PR and QRS intervals (increased conduction delay at different levels in the heart) compared to patients harboring missense SCN5A mutations [65]. In 2010, Nishii et al. reported that the shock-free survival rate was significantly lower in BrS patients with SCN5A mutations than that in those without SCN5A mutations in the Japanese population [66]. In the Han Chinese population, we observed that SCN5A mutations may not be a predictor of recurrent arrhythmic events in symptomatic BrS patients, but may predict the timing of the onset of the first cardiac event in a study from Taiwan (unpublished data).
5.3. Genetic testing for treatment
Compared to the long QT syndrome, the genetic evidence of BrS for adjusting treatment is scarce at this point. The result of genetic testing does not have an impact on the decision for choosing medications or for ICD implantation.
6. Interpretation of identified genetic variants and genetic counseling
Although the current expert consensus statement recommends that comprehensive or BrS1 (SCN5A) targeted BrS genetic testing can be useful for genetic testing [63], not all the published SCN5A mutations in BrS have been subjected to functional analysis, and 2–5% background rate of rare variants was reported in healthy subjects [17]. Before the exome data from the NHLI GO Exome Sequencing Project (ESP) was published in 2012, the distribution of genetic variations in BrS-associated genes from the large general population was still elusive. Upon reviewing the published variants associated with BrS, ESP harbored 22 of 303 (7%) variants in SCN5A [67]. Many variants were originally thought to be disease causing based on their absence in modestly sized healthy controls, but they could possibly be rare or low frequency benign variants. In addition, SCN5A mutations in BrS are also known to have incomplete penetrance and variable expressivity. As a result, careful interpretation of identified variants is extremely important for BrS patients or family members, especially for single probands or for single small families. Genetic counseling that outlines the possible explanations of genetic results and multidisciplinary clinics are essential to unravel the complicated questions from patients and family members.
7. Perspectives
Genetics is one of the links between basic science and clinical medicine. Advances in genetics could probably improve the future diagnosis, identification of possibly affected individuals, risk stratification, treatment, and prevention of BrS. Research in genetics is emerging at this point with many hopes and challenges. BrS has been recognized for more than two decades and many studies on BrS have disclosed part of its genetic underpinning, pathophysiological mechanisms, clinical characteristics, and electrophysiological findings. However, many questions remain unresolved with regard to the exact mechanisms, the complete genetic architecture, the role of polymorphisms and gene modifiers, and the environmental factors (e.g., hormones, fever) involved. Using new molecular biotechnological techniques like next-generation sequencing (exome or whole genome sequencing), we expect that more BrS-associated genes would be detected in the near future. With more genetic data, personalized treatment may be available in the future although we are just at the beginning of the molecular genetics era. Finally, the field of genetics is moving extremely faster than expected. Validating the present data periodically with the most up-to-date information regarding BrS is therefore necessary.
Funding
This review article received no grants from any funding agency in the public, commercial, or not-for-profit sectors.
Conflict of Interest
The authors declare that there are no conflicts of interest related to this study.
Acknowledgments
JMJ Juang is partially supported by research grants from NTUH-104-S2649, NTUH-104-S2671, NTUH104-2640, NTUH104-UN001, NTUH104L005, MOST 103-2314-B-002-189, and MOST-104-2314-B-002-193-MY3. Dr. Juang also thanks the staff members of the Sixth Core Lab, Department of Medical Research, National Taiwan University Hospital (NTUH) for providing technical support. MH is supported by research grants from the Ministry of Education, Culture, Science, and Technology of Japan; health science research grants from the Ministry of Health, Labour and Welfare of Japan for Clinical Research on Measures for Intractable Diseases (H24-033, H26-040, H27-032); and Translational Research Funds from the Japan Circulation Society.
References
- 1.Mitchel CR. Biology concept and connections. California; 1997.
- 2.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Richards S., Aziz N., Bale S. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Priori S.G., Napolitano C., Gasparini M. Clinical and genetic heterogeneity of right bundle branch block and ST-segment elevation syndrome: a prospective evaluation of 52 families. Circulation. 2000;102(20):2509–2515. doi: 10.1161/01.cir.102.20.2509. [DOI] [PubMed] [Google Scholar]
- 5.Berge K.E., Haugaa K.H., Fruh A. Molecular genetic analysis of long QT syndrome in Norway indicating a high prevalence of heterozygous mutation carriers. Scand J Clin Lab Invest. 2008;68(5):362–368. doi: 10.1080/00365510701765643. [DOI] [PubMed] [Google Scholar]
- 6.Giudicessi J.R., Ackerman M.J. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res. 2013;161(1):1–14. doi: 10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miyazaki T., Mitamura H., Miyoshi S. Autonomic and antiarrhythmic drug modulation of ST segment elevation in patients with Brugada syndrome. J Am Coll Cardiol. 1996;27(5):1061–1070. doi: 10.1016/0735-1097(95)00613-3. [DOI] [PubMed] [Google Scholar]
- 8.Chen Q., Kirsch G.E., Zhang D. Genetic basis and molecular mechanism for idiopathic ventricular fibrillation. Nature. 1998;392(6673):293–296. doi: 10.1038/32675. [DOI] [PubMed] [Google Scholar]
- 9.Algunas H.G.M.P. notas sobre bangungut. Rev Filip Med Farm. 1917;8:437–442. [Google Scholar]
- 10.Cruz J.Z. The pathology of "bangungut". J Philipp Med Assoc. 1951;27(7):476–481. [PubMed] [Google Scholar]
- 11.Mizusawa Y., Wilde A.A. Brugada syndrome. Circ Arrhythm Electrophysiol. 2012;5(3):606–616. doi: 10.1161/CIRCEP.111.964577. [DOI] [PubMed] [Google Scholar]
- 12.Berne P., Brugada J. Brugada syndrome 2012. Circ J. 2012;76(7):1563–1571. doi: 10.1253/circj.cj-12-0717. [DOI] [PubMed] [Google Scholar]
- 13.Juang J.M., Chen C.Y., Chen Y.H. Prevalence and prognosis of Brugada electrocardiogram patterns in an elderly Han Chinese population: a nation-wide community-based study (HALST cohort) Europace. 2015;17(Suppl 2):ii54–ii62. doi: 10.1093/europace/euv141. [DOI] [PubMed] [Google Scholar]
- 14.Alings M., Wilde A. "Brugada" syndrome: clinical data and suggested pathophysiological mechanism. Circulation. 1999;99(5):666–673. doi: 10.1161/01.cir.99.5.666. [DOI] [PubMed] [Google Scholar]
- 15.Juang J.M., Tsai C.T., Lin L.Y. Unique clinical characteristics and SCN5A mutations in patients with Brugada syndrome in Taiwan. J Formos Med Assoc. 2015;114(7):620–626. doi: 10.1016/j.jfma.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 16.Bezzina C.R., Shimizu W., Yang P. Common sodium channel promoter haplotype in asian subjects underlies variability in cardiac conduction. Circulation. 2006;113(3):338–344. doi: 10.1161/CIRCULATIONAHA.105.580811. [DOI] [PubMed] [Google Scholar]
- 17.Kapplinger J.D., Tester D.J., Alders M. An international compendium of mutations in the SCN5A-encoded cardiac sodium channel in patients referred for Brugada syndrome genetic testing. Heart Rhythm. 2010;7(1):33–46. doi: 10.1016/j.hrthm.2009.09.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valdivia C.R., Tester D.J., Rok B.A. A trafficking defective, Brugada syndrome-causing SCN5A mutation rescued by drugs. Cardiovasc Res. 2004;62(1):53–62. doi: 10.1016/j.cardiores.2004.01.022. [DOI] [PubMed] [Google Scholar]
- 19.Kyndt F., Probst V., Potet F. Novel SCN5A mutation leading either to isolated cardiac conduction defect or Brugada syndrome in a large French family. Circulation. 2001;104(25):3081–3086. doi: 10.1161/hc5001.100834. [DOI] [PubMed] [Google Scholar]
- 20.Bezzina C., Veldkamp M.W., van Den Berg M.P. A single Na(+) channel mutation causing both long-QT and Brugada syndromes. Circ Res. 1999;85(12):1206–1213. doi: 10.1161/01.res.85.12.1206. [DOI] [PubMed] [Google Scholar]
- 21.Dumaine R., Towbin J.A., Brugada P. Ionic mechanisms responsible for the electrocardiographic phenotype of the Brugada syndrome are temperature dependent. Circ Res. 1999;85(9):803–809. doi: 10.1161/01.res.85.9.803. [DOI] [PubMed] [Google Scholar]
- 22.Akai J., Makita N., Sakurada H. A novel SCN5A mutation associated with idiopathic ventricular fibrillation without typical ECG findings of Brugada syndrome. FEBS Lett. 2000;479(1–2):29–34. doi: 10.1016/s0014-5793(00)01875-5. [DOI] [PubMed] [Google Scholar]
- 23.Amin A.S., Verkerk A.O., Bhuiyan Z.A. Novel Brugada syndrome-causing mutation in ion-conducting pore of cardiac Na+ channel does not affect ion selectivity properties. Acta Physiol Scand. 2005;185(4):291–301. doi: 10.1111/j.1365-201X.2005.01496.x. [DOI] [PubMed] [Google Scholar]
- 24.Lei M., Huang C.L., Zhang Y. Genetic Na+ channelopathies and sinus node dysfunction. Prog Biophys Mol Biol. 2008;98(2-3):171–178. doi: 10.1016/j.pbiomolbio.2008.10.003. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe H., Koopmann T.T., Le Scouarnec S. Sodium channel beta1 subunit mutations associated with Brugada syndrome and cardiac conduction disease in humans. J Clin Invest. 2008;118(6):2260–2268. doi: 10.1172/JCI33891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riuro H., Beltran-Alvarez P., Tarradas A. A missense mutation in the sodium channel beta2 subunit reveals SCN2B as a new candidate gene for Brugada syndrome. Hum Mutat. 2013;34(7):961–966. doi: 10.1002/humu.22328. [DOI] [PubMed] [Google Scholar]
- 27.Hu D., Barajas-Martinez H., Burashnikov E. A mutation in the beta 3 subunit of the cardiac sodium channel associated with Brugada ECG phenotype. Circ Cardiovasc Genet. 2009;2(3):270–278. doi: 10.1161/CIRCGENETICS.108.829192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu D., Barajas-Martinez H., Pfeiffer R. Mutations in SCN10A are responsible for a large fraction of cases of Brugada syndrome. J Am Coll Cardiol. 2014;64(1):66–79. doi: 10.1016/j.jacc.2014.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Behr E.R., Savio-Galimberti E., Barc J. Role of common and rare variants in SCN10A: results from the Brugada syndrome QRS locus gene discovery collaborative study. Cardiovasc Res. 2015;106(3):520–529. doi: 10.1093/cvr/cvv042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Scouarnec S., Karakachoff M., Gourraud J.B. Testing the burden of rare variation in arrhythmia-susceptibility genes provides new insights into molecular diagnosis for Brugada syndrome. Hum Mol Genet. 2015;24(10):2757–2763. doi: 10.1093/hmg/ddv036. [DOI] [PubMed] [Google Scholar]
- 31.Fukuyama M., Ohno S., Makiyama T. Novel SCN10A variants associated with Brugada syndrome. Europace. 2015;18(6):905–911. doi: 10.1093/europace/euv078. [DOI] [PubMed] [Google Scholar]
- 32.Kattygnarath D., Maugenre S., Neyroud N. MOG1: a new susceptibility gene for Brugada syndrome. Circ Cardiovasc Genet. 2011;4(3):261–268. doi: 10.1161/CIRCGENETICS.110.959130. [DOI] [PubMed] [Google Scholar]
- 33.London B., Michalec M., Mehdi H. Mutation in glycerol-3-phosphate dehydrogenase 1 like gene (GPD1-L) decreases cardiac Na+ current and causes inherited arrhythmias. Circulation. 2007;116(20):2260–2268. doi: 10.1161/CIRCULATIONAHA.107.703330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ishikawa T., Sato A., Marcou C.A. A novel disease gene for Brugada syndrome: sarcolemmal membrane-associated protein gene mutations impair intracellular trafficking of hNav1.5. Circ Arrhythm Electrophysiol. 2012;5(6):1098–1107. doi: 10.1161/CIRCEP.111.969972. [DOI] [PubMed] [Google Scholar]
- 35.Cerrone M., Lin X., Zhang M. Missense mutations in plakophilin-2 cause sodium current deficit and associate with a Brugada syndrome phenotype. Circulation. 2014;129(10):1092–1103. doi: 10.1161/CIRCULATIONAHA.113.003077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu H., Chatel S., Simard C. Molecular genetics and functional anomalies in a series of 248 Brugada cases with 11 mutations in the TRPM4 channel. PLoS One. 2013;8(1):e54131. doi: 10.1371/journal.pone.0054131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Antzelevitch C., Pollevick G.D., Cordeiro J.M. Loss-of-function mutations in the cardiac calcium channel underlie a new clinical entity characterized by ST-segment elevation, short QT intervals, and sudden cardiac death. Circulation. 2007;115(4):442–449. doi: 10.1161/CIRCULATIONAHA.106.668392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Antzelevitch C., Nof E. Brugada syndrome: recent advances and controversies. Curr Cardiol Rep. 2008;10(5):376–383. doi: 10.1007/s11886-008-0060-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burashnikov E., Pfeiffer R., Barajas-Martinez H. Mutations in the cardiac L-type calcium channel associated with inherited J-wave syndromes and sudden cardiac death. Heart Rhythm. 2010;7(12):1872–1882. doi: 10.1016/j.hrthm.2010.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Giudicessi J.R., Ye D., Tester D.J. Transient outward current (I(to)) gain-of-function mutations in the KCND3-encoded Kv4.3 potassium channel and Brugada syndrome. Heart Rhythm. 2011;8(7):1024–1032. doi: 10.1016/j.hrthm.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Delpon E., Cordeiro J.M., Nunez L. Functional effects of KCNE3 mutation and its role in the development of Brugada syndrome. Circ Arrhythmia Electrophysiol. 2008;1(3):209–218. doi: 10.1161/CIRCEP.107.748103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ohno S., Zankov D.P., Ding W.G. KCNE5 (KCNE1L) variants are novel modulators of Brugada syndrome and idiopathic ventricular fibrillation. Circ Arrhythm Electrophysiol. 2011;4(3):352–361. doi: 10.1161/CIRCEP.110.959619. [DOI] [PubMed] [Google Scholar]
- 43.Barajas-Martinez H., Hu D., Ferrer T. Molecular genetic and functional association of Brugada and early repolarization syndromes with S422L missense mutation in KCNJ8. Heart Rhythm. 2012;9(4):548–555. doi: 10.1016/j.hrthm.2011.10.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hu D., Barajas-Martinez H., Terzic A. ABCC9 is a novel Brugada and early repolarization syndrome susceptibility gene. Int J Cardiol. 2014;171(3):431–442. doi: 10.1016/j.ijcard.2013.12.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Q., Ohno S., Ding W.G. Gain-of-function KCNH2 mutations in patients with Brugada syndrome. J Cardiovasc Electrophysiol. 2014;25(5):522–530. doi: 10.1111/jce.12361. [DOI] [PubMed] [Google Scholar]
- 46.Bezzina C.R., Barc J., Mizusawa Y. Common variants at SCN5A-SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45(9):1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ueda K., Hirano Y., Higashiuesato Y. Role of HCN4 channel in preventing ventricular arrhythmia. J Hum Genet. 2009;54(2):115–121. doi: 10.1038/jhg.2008.16. [DOI] [PubMed] [Google Scholar]
- 48.Wilde A.A., Behr E.R. Genetic testing for inherited cardiac disease. Nat Rev Cardiol. 2013;10(10):571–583. doi: 10.1038/nrcardio.2013.108. [DOI] [PubMed] [Google Scholar]
- 49.Wilde A.A., Ackerman M.J. Exercise extreme caution when calling rare genetic variants novel arrhythmia syndrome susceptibility mutations. Heart Rhythm. 2010;7(12):1883–1885. doi: 10.1016/j.hrthm.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 50.Priori S.G., Napolitano C., Giordano U. Brugada syndrome and sudden cardiac death in children. Lancet. 2000;355(9206):808–809. doi: 10.1016/S0140-6736(99)05277-0. [DOI] [PubMed] [Google Scholar]
- 51.Nademanee K., Veerakul G., Nimmannit S. Arrhythmogenic marker for the sudden unexplained death syndrome in Thai men. Circulation. 1997;96(8):2595–2600. doi: 10.1161/01.cir.96.8.2595. [DOI] [PubMed] [Google Scholar]
- 52.Benito B., Sarkozy A., Mont L. Gender differences in clinical manifestations of Brugada syndrome. J Am Coll Cardiol. 2008;52(19):1567–1573. doi: 10.1016/j.jacc.2008.07.052. [DOI] [PubMed] [Google Scholar]
- 53.Yokokawa M., Noda T., Okamura H. Comparison of long-term follow-up of electrocardiographic features in Brugada syndrome between the SCN5A-positive probands and the SCN5A-negative probands. Am J Cardiol. 2007;100(4):649–655. doi: 10.1016/j.amjcard.2007.03.078. [DOI] [PubMed] [Google Scholar]
- 54.Gehi A.K., Duong T.D., Metz L.D. Risk stratification of individuals with the Brugada electrocardiogram: a meta-analysis. J Cardiovasc Electrophysiol. 2006;17(6):577–583. doi: 10.1111/j.1540-8167.2006.00455.x. [DOI] [PubMed] [Google Scholar]
- 55.Probst V., Wilde A.A., Barc J. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet. 2009;2(6):552–557. doi: 10.1161/CIRCGENETICS.109.853374. [DOI] [PubMed] [Google Scholar]
- 56.Ackerman M.J., Priori S.G., Willems S. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13(8):1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 57.Remme C.A., Wilde A.A. SCN5A overlap syndromes: no end to disease complexity? Europace. 2008;10(11):1253–1255. doi: 10.1093/europace/eun267. [DOI] [PubMed] [Google Scholar]
- 58.Makiyama T., Akao M., Tsuji K. High risk for bradyarrhythmic complications in patients with Brugada syndrome caused by SCN5A gene mutations. J Am Coll Cardiol. 2005;46(11):2100–2106. doi: 10.1016/j.jacc.2005.08.043. [DOI] [PubMed] [Google Scholar]
- 59.Li A., Behr E.R. Brugada syndrome: an update. Future Cardiol. 2013;9(2):253–271. doi: 10.2217/fca.12.82. [DOI] [PubMed] [Google Scholar]
- 60.Probst V., Allouis M., Sacher F. Progressive cardiac conduction defect is the prevailing phenotype in carriers of a Brugada syndrome SCN5A mutation. J Cardiovasc Electrophysiol. 2006;17(3):270–275. doi: 10.1111/j.1540-8167.2006.00349.x. [DOI] [PubMed] [Google Scholar]
- 61.Shimizu N., Iwamoto M., Nakano Y. Long-term electrocardiographic follow-up from childhood of an adult patient with Brugada syndrome associated with sick sinus syndrome. Circ J. 2009;73(3):575–579. doi: 10.1253/circj.cj-07-0659. [DOI] [PubMed] [Google Scholar]
- 62.Priori S.G., Wilde A.A., Horie M. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10(12):1932–1963. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 63.Priori S.G., Wilde A.A., Horie M. Executive summary: HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes. Europace. 2013;15(10):1389–1406. doi: 10.1093/europace/eut272. [DOI] [PubMed] [Google Scholar]
- 64.Probst V., Veltmann C., Eckardt L. Long-term prognosis of patients diagnosed with Brugada syndrome: Results from the FINGER Brugada Syndrome Registry. Circulation. 2010;121(5):635–643. doi: 10.1161/CIRCULATIONAHA.109.887026. [DOI] [PubMed] [Google Scholar]
- 65.Meregalli P.G., Tan H.L., Probst V. Type of SCN5A mutation determines clinical severity and degree of conduction slowing in loss-of-function sodium channelopathies. Heart Rhythm. 2009;6(3):341–348. doi: 10.1016/j.hrthm.2008.11.009. [DOI] [PubMed] [Google Scholar]
- 66.Nishii N., Ogawa M., Morita H. SCN5A mutation is associated with early and frequent recurrence of ventricular fibrillation in patients with Brugada syndrome. Circ J. 2010;74(12):2572–2578. doi: 10.1253/circj.cj-10-0445. [DOI] [PubMed] [Google Scholar]
- 67.Risgaard B., Jabbari R., Refsgaard L. High prevalence of genetic variants previously associated with Brugada syndrome in new exome data. Clin Genet. 2013;84(5):489–495. doi: 10.1111/cge.12126. [DOI] [PubMed] [Google Scholar]
- 68.Freyermuth F., Rau F., Kokunai Y. Splicing misregulation of SCN5A contributes to cardiac-conduction delay and heart arrhythmia in myotonic dystrophy. Nat Commun. 2016;7(11067):1–14. doi: 10.1038/ncomms11067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Medeiros-Domingo A., Tan B.H., Crotti L. Gain-of-function mutation S422L in the KCNJ8-encoded cardiac K(ATP) channel Kir6.1 as a pathogenic substrate for J-wave syndromes. Heart Rhythm. 2010;7(10):1466–1471. doi: 10.1016/j.hrthm.2010.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Crotti L., Marcou C.A., Tester D.J. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60(15):1410–1418. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]