Abstract
Inherited arrhythmias, such as cardiomyopathies and cardiac ion channelopathies, along with coronary heart disease (CHD) are three most common disorders that predispose adults to sudden cardiac death. In the last three decades, causal genes in inherited arrhythmias have been successfully identified. At the same time, it has become evident that the genetic architectures are more complex than previously known. Recent advancements in DNA sequencing technology (next generation sequencing) have enabled us to study such complex genetic traits.
This article discusses indications for genetic testing of patients with inherited arrhythmias. Further, it describes the benefits and challenges that we face in the era of next generation sequencing. Finally, it briefly discusses genetic counseling, in which a multidisciplinary approach is required due to the increased complexity of the genetic information related to inherited arrhythmias.
Abbreviations: ARVD/C, arrhythmogenic right ventricular dysplasia/cardiomyopathy; BrS, Brugada syndrome; CHD, coronary heart disease; CPVT, catecholaminergic polymorphic ventricular tachycardia; DCM, dilated cardiomyopathy; GWAS, genome wide association study; HCM, hypertrophic cardiomyopathy; HF, heart failure; ICD, implantable cardioverter defibrillator; NGS, next generation sequencing; LQTS, long QT syndrome; SCD, sudden cardiac death; VA, ventricular arrhythmia; VF, ventricular fibrillation; WES, whole exome sequencing
Keywords: Inherited arrhythmias, Genetic testing, Cardiac ion channelopathies, Cardiomyopathies, Genetic counseling
1. Introduction
Sudden cardiac death (SCD) is a tragic event and the aftermath is devastating to the surviving family and community. SCD occurs in nearly 400,000 cases every year in the US [1]. In adult populations, cardiomyopathies and cardiac ion channelopathies, along with coronary heart disease (CHD), are the most common conditions that predispose patients to SCD. According to recent epidemiological studies of SCD in Western countries, CHD, cardiomyopathies, and ion channelopathies are diagnosed in ~75%, 10–15%, and 1–2% of the SCD cases, respectively [2]. In Japan, approximately 50,000 new cases of SCD are diagnosed every year [3]. CHD is less common in Japan than in Western countries: CHD, cardiomyopathies, and ion channelopathies account for 50–60%, 30–35% and 10% of the SCD cases, respectively [2]. In children or adults younger than 35 years of age, cardiomyopathies and ion channelopathies account for significant proportions of SCD [4]. Because of the low survival rate in aborted SCD cases (≤10%) [2], identification of patients at risk of arrhythmia is very important for the prevention of SCD.
Inherited arrhythmia-susceptibility genes have been successfully identified in the last three decades. This has broadened both our understanding of the mechanisms and the clinical management of inherited arrhythmias. At the same time, it has become evident that the genetic architectures of inherited arrhythmias are actually more complex than previously known, involving more genetic components than a single gene locus. A recent advance in sequencing technology (next generation sequencing [NGS]) has facilitated the search for such genetic components spread over the genome.
This article provides a concise overview of genetic testing for inherited arrhythmias used in current clinical practice. It also provides a short introduction to NGS technology. Further, it briefly discusses genetic counseling, in which a multidisciplinary approach is required due to the increased complexity of genetic information regarding inherited arrhythmias. Because this article aims to provide basic knowledge regarding inherited arrhythmias for readers who are unfamiliar with this field, more detailed clinical and genetic characteristics of inherited arrhythmias are not within the scope of this article and can be found elsewhere [5], [6], [7], [8], [9], [10], [11].
2. Genetic testing for inherited arrhythmias
2.1. Classic genetic testing: candidate gene approach
Disease-causing genes for inherited arrhythmias have been successfully identified in the last three decades, resulting in a large impact on patient care. This success has largely been through the use of classical linkage mapping. Linkage mapping is performed in affected family members, followed by candidate gene sequencing within an identified disease susceptibility locus. Such analysis was performed with the assumption that inherited arrhythmias are Mendelian (monogenic) disorders and that a single mutation contributes substantially to the risk. In Table 1, the most commonly identified genes in inherited arrhythmias are shown.
Table 1.
Major genes associated with inherited arrhythmias.
| Disease | Genes (yield, %) |
|---|---|
| Long QT syndrome | KCNQ1 (30–35), KCNH2 (25–30), SCN5A (5–10) [25] |
| Catecholaminergic Polymorphic Ventricular Tachycardia | RYR2 (60–65),CASQ2 (<5) [47], [49] |
| Brugada Syndrome | SCN5A (20) [60] |
| Hypertrophic cardiomyopathy | MYBPC3 (30–40), MYH7 (20–30), TNNT2 (10), TNNI3 (7) [66] |
| Dilated cardiomyopathy | TTN (~25) [80] |
| Arrhythmogenic right ventricular dysplasia/cardiomyopathy | PKP2 (46), PLN (5) [62] |
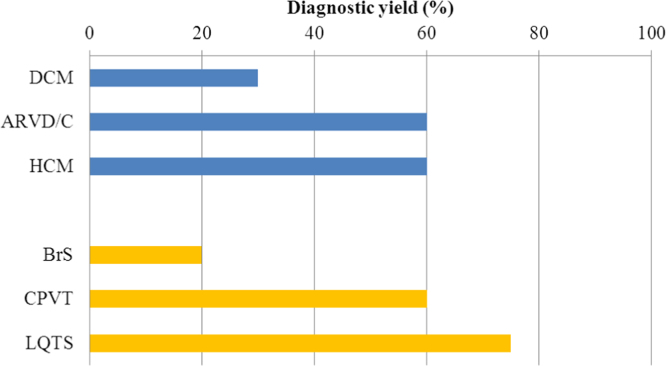
Presently, genetic testing for causal genes in a proband is used to confirm diagnosis. Genetic testing can also be used to perform cascade screening (mutation-specific testing) in family members of genotype-positive probands to exclude a diagnosis (in the case of a negative test) or to allow targeted testing (in the case of a positive test) [11]. Diagnostic, therapeutic, and prognostic values of genetic testing are most relevant in long QT syndrome (LQTS) wherein genotype–phenotype correlation (i.e. the association between a certain mutation [genotype] and clinical characteristics [phenotypes]) is robust. In catecholaminergic polymorphic ventricular tachycardia (CPVT), genetic testing plays an important role in diagnosis. The clinical utility of genetic testing is limited in the remaining inherited arrhythmias [11], because of as yet unknown genotype–phenotype correlation or because of the low yield of genetic testing (Fig. 1).
Fig. 1.
Diagnostic yields of genetic testing in inherited arrhythmias. Diagnostic yields are variable among cardiomyopathies and channelopathies. Data shown was taken from [11].
Genotype–phenotype correlation is complicated when some phenotypes, such as electrocardiographic abnormalities and arrhythmias, do not manifest in all individuals carrying the same gene mutation (incomplete penetrance), and when the type and severity of the phenotypes vary between genotype-positive individuals (variable expressivity) [12]. For example, in families with Brugada syndrome (BrS) that carry the familial SCN5A mutation, not all affected family members (mutation carriers) show diagnostic type 1 BrS ECG or other symptoms [13]. Such puzzling phenomena are the focus of ongoing genetic research. Meanwhile, consensus reports provide recommendations to guide clinicians in the appropriate use of genetic testing for inherited arrhythmias [11]. It is important to keep in mind that in any inherited arrhythmia, clinical examinations are critical for accurate diagnosis before genetic testing is implemented.
2.2. New sequencing technology: next generation sequencing (NGS)
As suggested by the incomplete penetrance and variable expressivity seen in mutant genotypes, the genetic architecture of inherited arrhythmias is more complex than initially thought. It is hypothesized that in addition to a mutation in a single disease-susceptibility gene, as seen in pathogenic variants found in <1% of the general population, other genetic (and environmental) factors are involved in the ultimate manifestation of inherited arrhythmias [10]. In recent years, it became possible, using NGS technology, to screen for such common or rare variants (frequency in the general population >1–5% and <1%, respectively) [14]. These variants have a smaller impact on disease risk than mutations, but have an aggregated influence on disease manifestation.
NGS is a sequencing technology that enables sequencing at a massive scale across the genome to find additional genetic loci involved in a disease. In the last decade, genome wide association studies (GWAS) were conducted to identify common single nucleotide polymorphisms (SNPs) associated with SCD or ECG parameters, because SCD/ECG parameters were shown to have heritable components [14]. With regard to rare inherited arrhythmias, Bezzina et al. was the first to report an association between common SNPs and BrS [15]. These exciting findings using GWAS pave the way for functional analyses and for better understanding of the mechanisms of inherited arrhythmias.
NGS is used in gene panels to sequence multiple genes simultaneously (e.g. 20–80 comprehensive arrhythmia or cardiomyopathy-associated genes) or to sequence whole exomes/genomes in order to find variants with intermediate to large effect sizes on phenotypes [14]. In the last couple of years, the successful application of NGS for the discovery of rare variants in heritable arrhythmia syndromes has been reported. Intriguing examples are cases of early onset and recurrent episodes of ventricular fibrillation (VF) without identification of pathogenic mutation when using classical genetic testing [16], [17]. VF was refractory to all medical treatments in these cases. Furthermore, clinical or familial history did not provide clues for effective therapy, except for the use of an implantable cardioverter defibrillator [ICD]. Whole exome sequencing (WES) was performed in symptomatic siblings [16] or in parents and a child (parent–child trios) [17], which led to discovery of mutations in the calmodulin genes. As just described, WES is becoming a promising approach to uncover pathogenic variants associated with heritable conditions.
NGS is not a perfect tool, but is a developing method to uncover new variants/mutations that may be involved in disease conditions. The technique is expected to allow improved diagnosis, better prognostic evaluation, or enhanced individual therapy. However, we are currently facing an increased number of rare variants of uncertain significance (VUS), that is, variants whose association with disease risk is unknown, as a result of massive sequence data. To confirm pathogenicity in VUS, a number of criteria need to be carefully evaluated by experts in the field, such as geneticists, cardiologists, and genetic counselors, to allow the findings to be interpreted in the context of clinical practice. These criteria include cosegregation of the unraveled variant with disease, absence or extreme rarity of the variant in control cohorts, types of mutation (missense mutation vs. nonsense, frameshift, insertion/deletion mutations that lead to truncated protein products), and results of functional studies.
3. Inherited arrhythmias with structurally normal hearts: cardiac ion channelopathies
Cardiac ion channelopathies, such as LQTS, CPVT and BrS, are heritable conditions with an increased risk of SCD in otherwise normal hearts. An ion channelopathy manifests when ion channel function is disrupted by a mutation in a gene encoding a cardiac ion channel or an ion channel-related protein. Clinical and genetic characteristics of each condition are described below.
3.1. Long QT syndrome (LQTS)
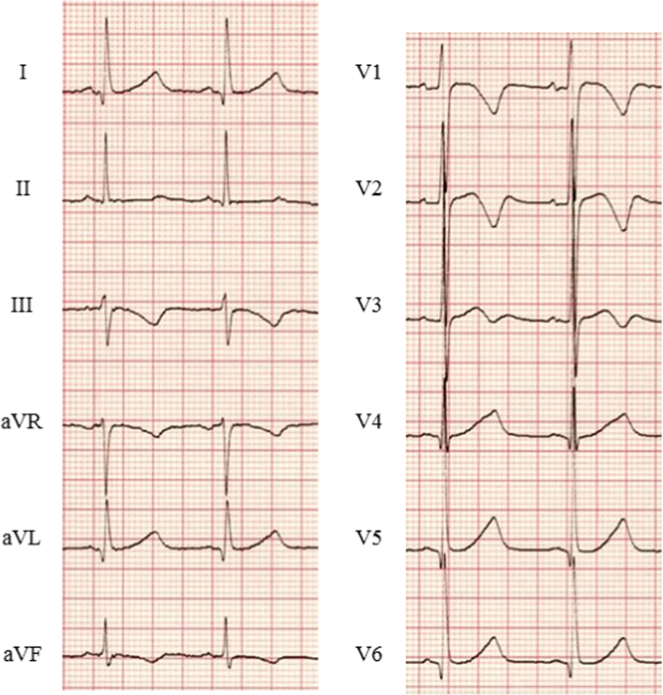
Congenital LQTS is characterized by a prolonged QT interval (Fig. 2) and syncope/VF secondary to peculiar polymorphic ventricular tachycardia, called torsades de pointes (TdP). The prevalence of LQTS is estimated to be as high as 1 in 2000 [18].
Fig. 2.
ECG in a patient with LQTS type 2. Baseline ECG of a patient with LQTS type 2 is shown. QTc interval was prolonged and abnormal T waves (negative in V1–V3) were observed (HR 71 bpm, QTc 480 ms). LQTS=long QT syndrome; QTc=corrected QT interval (Bazett׳s formula).
The first cardiac event occurs in teenagers (13±9, 18±10, and 16±10 years in LQT1, LQT2, and LQT3, respectively) [19]. Syncope is the most common, first symptom, while aborted sudden cardiac arrest (ACA)/SCD is rare (1–3%) [20]. The mainstay of LQTS diagnostics is the evaluation of clinical characteristics, including symptoms, family history and electrocardiographic findings, using the diagnostic scoring system (Schwartz score) [21]. When the diagnosis is established, beta-blockers are introduced as the first-line of therapy. ICD is indicated in aborted SCD cases [22], [23].
3.1.1. Genetic testing in LQTS
LQTS segregates in a Mendelian fashion; in the majority of cases of LQTS, it is transmitted in an autosomal dominant pattern (Romano–Ward syndrome), and in the minority of cases, in an autosomal recessive pattern (Jervell and Lange–Nielsen syndrome) [11]. To date, mutations in 15 different genes have been implicated in LQTS [5]. A causal mutation is found in approximately 75% of LQTS patients with a Schwartz score ≥4 [24]. The KCNQ1 gene, encoding the Kv7.1 potassium channel (LQT1, 30–35%), the KCNH2 gene, encoding theKv11.1 potassium channel (LQT2, 25–30%), and the SCN5A gene, encoding the Nav1.5 sodium channel (LQT3, 5–10%) make up more than 90% of the cases of genotype-positive LQTS (Table 1) [25]. Testing the remaining minor LQTS-associated genes increases the diagnostic yield by only <5%, while it also increases the chance of encountering a false-positive result [26]. Therefore, comprehensive screening of LQTS genes is not currently a common practice.
Because genotype–phenotype correlation is robust in the major LQTS subtypes (LQT1–LQT3), genotype can be inferred by thorough clinical evaluation. For example, in LQT1, cardiac events occur during physical exertion or emotional stress, typically during swimming [27]. Auditorial stimulation, such as an alarm clock or a telephone bell, is a typical trigger for ventricular arrhythmia (VA) in LQT2 [28]. In LQT3, symptoms are generally observed during rest or at night [27], [29]. T wave morphology may also help to distinguish different LQTS subtypes (LQT1 [broad T wave], LQT2 [biphasic T wave] and LQT3 [late-appearing T wave]) [30].
As for clinical symptoms of minor LQTS subtypes, severe phenotypes, such as longer QTc or early onset of arrhythmic events [31], [32], suggest the presence of compound mutations in approximately 10% of the genotype-positive patients [33]. The presence of extracardiac phenotypes point to two rare subtypes: LQT7 (Andersen–Tawil syndrome) and LQT8 (Timothy syndrome). LQT7 is caused by mutations in the KCNJ2 gene, which results in loss-of-function of the inward rectifying potassium channel Kir2.1 [34], and may manifest periodic paralysis and dysmorphic features as extracardiac symptoms. LQT8 is an extremely rare and highly malignant form of LQTS, carrying mutations in the CACNA1c gene [35], [36], [37], [38]. Typical extracardiac manifestations are dysmorphic facial features, immune deficiency, developmental delay, and syndactyly; however, not all LQT8 patients develop these extracardiac symptoms [35], [36], [37], [38].
Prognosis of LQTS is influenced by the location and type of mutation. In LQT1, missense mutations and transmembrane mutations cause a longer QT interval and an increased risk of cardiac events [39]. In LQT2, the risk of SCD is higher in females than in males. Among LQT2 females, the risk of SCD is not significantly different between patients with and without pore–loop mutations [40]. Among LQT2 males, patients with pore–loop mutations [40] and missense mutations [41] showed higher chances of SCD than patients with non-pore–loop mutations.
In LQTS therapeutics, genotypes are particularly helpful in predicting the efficacy of therapy using beta-blockers. For the LQTS subtypes LQT1–LQT3, therapy using beta-blockers is extremely effective for patients with LQT1 but less protective for those with LQT2 and LQT3 [42]. In patients with LQT3, mexiletine may be used in addition to beta-blockers because it shortens QTc by blocking sodium channel currents [43]. However, caution is warranted when using mexiletine to treat LQT3 patients because its effect is dependent on the type of mutation; it can be beneficial in some LQT3 patients, but harmful in others [44], [45].
3.2. Catecholaminergic polymorphic ventricular tachycardia (CPVT)
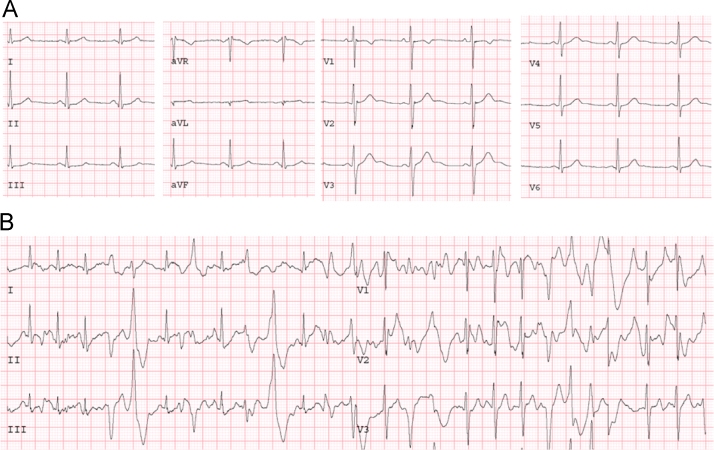
CPVT is characterized by polymorphic ventricular tachycardia (bidirectional VT) triggered by adrenergic stimulation, such as exercise or emotional stress. The prevalence of CPVT in the general population is estimated to be 1:10,000 [7]. Patients are typically admitted to a hospital with syncopal episodes caused by polymorphic VT, which may degenerate to VF and SCD. Cardiac examinations reveal no structural abnormality in the heart and normal baseline ECG. Therefore, establishing patient history, such as symptoms after physical/emotional stress, family history of SCD, is important in the diagnosis of CPVT. Exercise testing is the gold standard for diagnosis: during an exercise test, ventricular premature beats and non-sustained VT are reproducibly induced (Fig. 3). The mean age for the onset of symptoms is 8 years. Approximately 30% of patients with CPVT develop symptoms before 10 years of age, and 60% before 40 years [11].
Fig. 3.
Exercise stress testing of CPVT. Baseline ECG (A) and an ECG during exercise stress testing (B) in a patient with CPVT are shown. The ECG during exercise testing shows VES and NSVT. CPVT=catecholaminergic polymorphic ventricular tachycardia; NSVT=non-sustained ventricular tachycardia; VES=ventricular extra systoles.
Beta-blockers, combined with exercise restriction, are the first-line of therapy for CPVT. Among several beta-blockers, Nadolol, a long-acting, non-selective beta-blocker, seems to be more effective than others [46]. When patients are under treatment with beta-blockers, fatal/near-fatal events at 4 year- and 8 year-follow up occur in 1–7% and 11–14% of the patients, respectively [7], mostly because of poor drug compliance. Therefore, it is crucial to inform CVPT patients about the potential fatal consequences of non-compliance to medication. For those refractory to beta-blockers, flecainide, in addition to the beta-blockers, seems effective [7].
3.2.1. Genetic testing in CPVT
In CPVT, 60–65% of the patients carry a mutation in the RYR2 gene, which is transmitted in an autosomal dominant pattern, whereas, a mutation in the CASQ2 gene, which is transmitted in an autosomal recessive pattern, is found in only small percentage of index cases (Table 1) [47], [48], [49]. Mutations in these genes trigger arrhythmias as a result of increased calcium release from the sarcoplasmic reticulum. Rarely, CPVT is diagnosed with mutations in the calmodulin genes, CALM1 [50] and CALM2 [51]. A CPVT phenotype also develops in Andersen–Tawil syndrome (LQT7), which is caused by mutations in the KCNJ2 gene [52]. Thus, it is particularly relevant for RYR2/CASQ2 mutation-negative patients to consider being screened for mutations in these genes [52].
Because SCD can be the first manifestation of CPVT, genetic testing is clinically relevant, especially for the management of family members. The most recent HRS/EHRA guidelines recommend screening for CPVT susceptibility genes in index cases with clinically established/suspected CPVT, and cascade screening in family members of index cases with a putative, pathogenic mutation [11]. At present, there are no genotype-based risk stratification or therapeutic approaches in CPVT [11].
3.3. Brugada syndrome (BrS)
BrS is diagnosed by the presence of its characteristic ECG pattern: type1 BrS ECG (Fig. 4). It is defined as ≥2 mm ST-segment elevation with coved-type morphology in ≥1 lead of the right precordial leads (V1–V3 in 4th–2nd intercostal spaces) [23]. The typical ECG pattern may manifest spontaneously, either after a sodium channel blocker is administered during drug-challenge testing or during fever, and it may fluctuate over time. The prevalence of BrS is estimated to be 5:10,000 in the general population [53].
Fig. 4.
ECG in a patient with BrS. Spontaneously appearing type 1 ECG was observed in lead V2. BrS=Brugada syndrome.
In BrS, phenotypes are heterogeneous. Symptoms, such as VF or syncope due to VA, typically occur at rest or during sleep in adults that are in their 40s. Meanwhile, some individuals with diagnostic ECG pattern remain asymptomatic for life. Therefore, it is critical to accurately identify patients at risk, although the risk stratification in BrS is still ill defined. As expected, patients with a history of VF are at high risk for recurrent VF (approximately 8–10% of patients/year) [54], [55]. It is generally agreed that syncope in the presence of spontaneous type 1 ECG identifies high-risk patients [23]. The outcome of asymptomatic cases are much more benign compared to that in symptomatic cases, even in the presence of spontaneous type 1 ECG (approximately 0.5–0.8% of patients/year) [54], [55].
The only effective therapy for BrS is implantation of ICD. However, because ICD implants have a high complication rate (approximately 9% of patients/year) [56], ICD should be indicated only in those with documented, or strongly suspected, VAs [23]. In cases with repetitive VF attacks, isoproterenol infusion is effective in acute settings to suppress VF [57]. In chronic settings, quinidine, a Class Ia antiarrhythmic drug, is used as an adjunctive therapy to treat recurrent VAs [58].
3.3.1. Genetic testing in BrS
BrS is transmitted in an autosomal dominant pattern. Loss of function mutations in SCN5A, encoding the Nav1.5 sodium channel, account for approximately 20% of BrS cases (Table 1) [59]. Other genes reported in BrS are found in <5% [60] of cases, and the majority of BrS patients remain genetically elusive. Minor BrS-associated genes include: sodium channel β-subunit genes (SCN1B, SCN3B); Glycereol-3 phosphate dehydrogenase 1-like enzyme (GPD1L); MOG1, which affects trafficking of sodium channels; calcium channel genes (CACNA1C, CACNB2b, CACNA2D1); and genes that affect the Ito (KCNE3, KCND3), IK-ATP (KCNJ8), and the If (HCN4) currents [6].
Thorough clinical evaluation, but not genetic testing, is the standard for diagnosis of BrS. The recent HRS/EHRA consensus guidelines recommend cascade screening in family members of genotype-positive index cases [11]. Screening SCN5A can be useful in clinically suspected BrS index cases. At present, genotype is not relevant for prognostic or therapeutic measures [11].
4. Inherited arrhythmias with structural abnormality: cardiomyopathies
To date, thousands of variants have been associated with inherited cardiomyopathies. A major challenge is to determine whether these variants are truly pathogenic or benign polymorphisms. In relation to arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C), a recent clinical study involving a large number of patients has provided valuable information regarding the genotype–phenotype correlation [61], [62].
4.1. Hypertrophic cardiomyopathy (HCM)
HCM is defined by the presence of increased left ventricular wall thickness that is not solely explained by abnormal (pressure or volume) loading conditions [63]. HCM includes both familial and non-familial subtypes, regardless of the presence of extra-cardiac traits. HCM is relatively common disease: its prevalence is estimated to be approximately 1:500 in the general population [64], which is similar among different ethnic groups [63]. HCM is the most common cause of SCD in young, otherwise healthy individuals [65]. Symptoms, such as heart failure (HF), may occur in HCM patients due to diastolic dysfunction. A small proportion of HCM patients eventually develop systolic dysfunction.
4.1.1. Genetic testing in HCM
HCM shows predominantly an autosomal dominant transmission with age-related penetrance, and >50% of HCM patients are estimated to have familial disease [66]. HCM is known as a “disease of the sarcomere.” Approximately 60% of HCM cases harbor mutations in sarcomere genes, which encode for components of the contractile apparatus of the heart. Most mutations have been identified in the β-myosin heavy chain gene, MYH7 (20–30%), and in the cardiac myosin-binding protein C gene, MYBPC3, (30–40%). Less frequent mutations have been identified in the cardiac troponin T gene, TNNT2 (10%), and in the cardiac troponin I gene, TNNI3 (7%) (Table 1). Mutations in other sarcomere genes are rare. In an Asian familial HCM cohort, the yield of MYBPC3 gene mutations was less than in Western countries (MYH7 19%, MYBPC3 11%, TNNT2 12%, TNNI3 3%) [9].
Carriers of pathogenic mutations typically harbor a single mutation. Five to seven percent of cases carry ≥2 mutations. These can be compound heterozygous mutations, which are heterozygous mutations in each allele of the same gene, digenic heterozygous mutations, which are heterozygous mutations in two different genes, or homozygous mutations [66]. In rare cases, left ventricular hypertrophy may be caused by non-sarcomeric gene mutations. For example, in 25% of children with HCM, metabolic disease (e.g. Pompe disease), malformation syndromes (e.g. Noonan syndrome), and neuromuscular disorders (e.g. Friedreich ataxia) are diagnosed [67]. In adults, non-sarcomeric gene mutations are rarely observed, but can include Fabry disease, Danon disease, familial amyloidosis, or mitochondrial cardiomyopathies [66].
Based on the 2014 ESC guidelines for HCM, genetic testing is recommended in definite HCM index cases (a wall thickness ≥15 mm in one or more LV myocardial segments measured by any imaging modality) and in family members of genotype-positive index cases [63]. Genetic testing for clinical management, including diagnosis, risk stratification, and therapy, is currently of limited value [63].
4.2. Dilated cardiomyopathy (DCM)
DCM is characterized by left ventricular enlargement and systolic dysfunction with a prevalence of 1:2500 in the general population [68], [69]. Clinical manifestations include HF, thromboembolism, and SCD. Most patients are diagnosed with DCM between the ages of 20 and 50 [70]. While different causes may lead to DCM, 20–50% of DCM cases are estimated to carry a genetic component based on the presence of family history [11]. These patients are often referred as ‘idiopathic’ or ‘inherited’ DCM.
4.2.1. Genetic testing in DCM
Inherited DCM is predominantly transmitted as an autosomal dominant trait. In a minority of cases, X-linked, recessive, or mitochondrial inheritance patterns occur [71]. To date, more than 40 genes that encode cytoskeletal, sarcomeric or Z-disk proteins have been associated with DCM or DCM in combination with early-onset conduction system disease. Mutations in ion channel genes and desmosome genes have also been associated with DCM. Variants associated with DCM are found most frequently in TTN (up to 25%) [9], [66]. Variants in other genes are rarely found. Familial DCM is observed in 20–35% of cases when first-degree relatives are screened, underscoring the need for cascade screening in family members. Currently, no prognostic or therapeutic implications of genetic testing exist in DCM except for LMNA and DES variants, which carry an increased risk of SCD [11].
4.3. Arrhythmogenic right ventricular dysplasia/cardiomyopathy (ARVD/C)
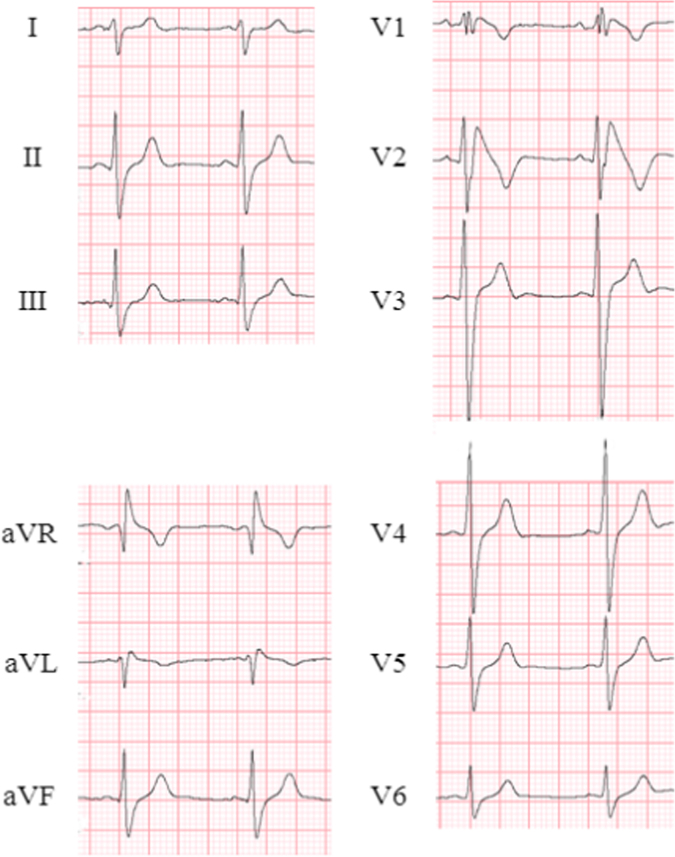
ARVD/C is a heritable cardiomyopathy characterized by fibrofatty replacement in the right ventricle. The prevalence in the general population is 1:5000 [8]. Patients typically present with arrhythmia-related symptoms, including palpitations, syncope, or SCD, between the ages of 20 and 59 years [62]. ECG of a patient with ARVD/C is shown in Fig. 5. A recent cohort study reported that in 439 unrelated probands with ARVD/C who fulfilled the 2010 diagnostic task force criteria [72], sustained VA including aborted SCD was the 1st symptom in 61% of the cases [62]. At a median of 7 years follow-up, 72% developed sustained VA. Appropriate risk stratification, including indication for ICD [73], led to a good prognosis, which included cardiac mortality in 7% [62]. HF manifests less commonly and usually develops at a later stage.
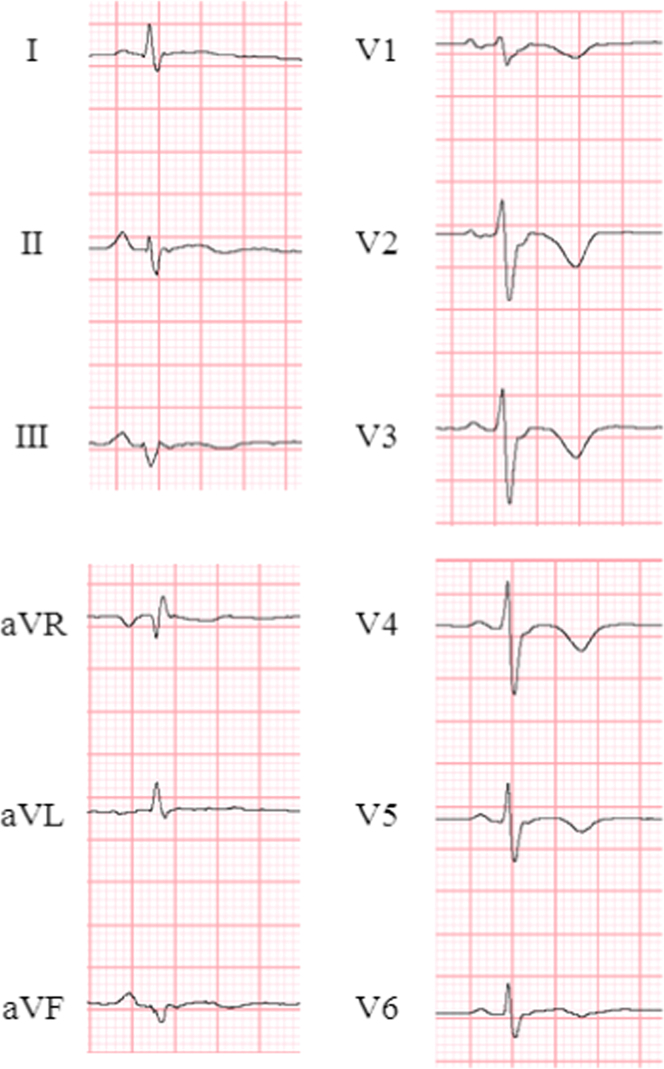
Fig. 5.
ECG in a patient with ARVD/C. Epsilon waves and inverted T waves (V1–V6) were detected. ARVD/C=arrhythmogenic right ventricular dysplasia/cardiomyopathy.
4.3.1. Genetic testing in ARVD/C
ARVD/C is inherited predominantly as an autosomal dominant trait, although it is also transmitted as an autosomal recessive trait (Naxos disease or Carvajal syndrome) in a minority of cases [74]. It is widely considered to be a desmosomal disease. Desmosomes are one of the junctional complexes at the intercalated disk that are essential for structural and functional integrity of cardiac tissue. Desmosomal gene mutations lead to defective desmosomal components and to the development of structural abnormalities typical of ARVD/C. Mutations in five desmosomal genes, including junction plakoglobin [JUP], DSP, PKP2, desmoglein-2 [DSG2], and desmocollin-2 [DSC2], cause ARVD/C. More recently, the extradesmosomal genes transforming growth factor beta3 (TGFB3), transmembrane protein 43 (TMEM43), desmin (DES), titin (TTN), phospholamban (PLN), lamin A/C (LMNA), and αT-catenin (CTNNA3) have been associated with atypical forms of ARVD/C [75].
ARVD/C susceptibility gene mutations are found in approximately 60% of ARVD/C index cases, of which the majority are desmosomal gene mutations (PKP2 [46%], PLN [5%], DSG2 [4%], DSP [3%], and DSC2 [1%]) [62]. A recent large cohort study collected from the Johns Hopkins and the Dutch ARVD/C registries [61] reported that of 555 ARVD/C index patients and family members who carry a pathogenic mutation, 80% carried a single PKP2 mutation. Other desmosomal gene mutations (JUP, DSG2, DSC2, DSP) were found in 10% of the study population. Compound mutations were found in 4%, which led to earlier onset of symptoms including VA and HF. Among single mutation carriers, premature truncating, splice site, and missense mutations were identified in 60%, 23%, and 14%, respectively. However, outcomes, including arrhythmic events, HF, and death, were not significantly different between different types of mutations. Among Asians, PKP2 gene mutations were most commonly found (54%) in a Chinese ARVD/C cohort consisting of 100 ARVD/C probands [76], whereas, in a small series of ARVD/C in Japan (35 patients), mutations in PKP2, DSP, and DSG2 were found in 28.6%, 20%, and 8.6%, respectively [77]. This implies diverse proportions of AVRD/C causal genes among different ethnicities. Of note, pathogenicity of an identified missense variant requires careful evaluation because missense variants in ARVD/C susceptibility genes are commonly found (16%) as background noise in the general population [78]. The ARVD/C Genetic Variants Database is currently available online (http://www.arvcdatabase.info/), and summarizes ARVD/C-associated variants and their pathogenesis [75].
While the 2010 diagnostic task force criteria include identification of a putative ARVD/C susceptibility gene mutation as a new major diagnostic criterion [72], genetic testing should be used as a confirmatory tool for diagnosis of ARVD/C in index cases. Cascade screening is recommended in family members [74]. A recent large cohort study reported that carriers of mutations in desmoplakin (DSP) are more likely to present with SCD and more prone to left ventricular dysfunction and HF than carriers with mutations in Plakophilin-2 (PKP2) [61]. Prognostic and therapeutic implications of genetic testing in ARVD/C await further analyses of large-scale clinical studies.
5. Genetic counseling in inherited arrhythmias
Since genetic testing was implemented in clinical practice, the importance of genetic counseling in probands as well as in family members with inherited arrhythmias has been recognized. When a patient is confronted with the risk of sudden death and a possibility of transmitting a potentially fatal disease to his/her children, psychological consequences may naturally occur. Such psychological distress can, in fact, be addressed and improved by pre- and post-genetic test counseling [79]. A major problem currently faced in the era of NGS is that rare VUS are increasingly identified in disease-susceptibility genes, while there is no standard platform that allows proper processing and integration of raw massive genomic data with clinical data. Furthermore, NGS provides a chance to incidentally uncover pathogenic mutations in other, non-inherited arrhythmia-related conditions.
It is of paramount importance to assemble a team of experts to interpret the complex results of genetic testing. Therefore, a multidisciplinary approach involving the cooperation of cardiologists, geneticists, genetic counselors, and psychologists, and the interaction between experienced academic and local hospitals is strongly recommended [11]. Meanwhile, in research settings, collaboration and data sharing between research groups is encouraged to advance our understanding of the intricate genetic architectures of inherited arrhythmias.
6. Conclusions
Remarkable advances have been achieved in genetic testing of inherited arrhythmias in the last three decades. Technological advances in DNA sequencing have further increased the ability to analyze complex architectures of inherited arrhythmias. Yet there are many challenges to implementing NGS into clinical practice. Ceaseless efforts to assemble massive genetic data and to build standard platforms for integrating genetic data into clinical data will further expand our understanding of inherited arrhythmias and will improve patient care.
Conflict of interest
The author declares no conflicts of interest related to this study.
References
- 1.Mozaffarian D., Benjamin E.J., Go A.S. Heart disease and stroke statistics—2015 update: a report from the American Heart Association. Circulation. 2015;131:e29–322. doi: 10.1161/CIR.0000000000000152. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi M., Shimizu W., Albert C.M. The spectrum of epidemiology underlying sudden cardiac death. Circ Res. 2015;116:1887–1906. doi: 10.1161/CIRCRESAHA.116.304521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guidelines for risks and prevention of sudden cardiac death (JCS 2010): – digest version. Circ J. 2012;76:489–507. doi: 10.1253/circj.cj-88-0022. [DOI] [PubMed] [Google Scholar]
- 4.Kaltman J.R., Thompson P.D., Lantos J. Screening for sudden cardiac death in the young: report from a national heart, lung, and blood institute working group. Circulation. 2011;123:1911–1918. doi: 10.1161/CIRCULATIONAHA.110.017228. [DOI] [PubMed] [Google Scholar]
- 5.Mizusawa Y., Horie M., Wilde A.A. Genetic and clinical advances in congenital long QT syndrome. Circ J. 2014;78:2827–2833. doi: 10.1253/circj.cj-14-0905. [DOI] [PubMed] [Google Scholar]
- 6.Mizusawa Y., Wilde A.A. Brugada syndrome. Circ Arrhythm Electrophysiol. 2012;5:606–616. doi: 10.1161/CIRCEP.111.964577. [DOI] [PubMed] [Google Scholar]
- 7.van der Werf C., Wilde A.A. Catecholaminergic polymorphic ventricular tachycardia: from bench to bedside. Heart. 2013;99:497–504. doi: 10.1136/heartjnl-2012-302033. [DOI] [PubMed] [Google Scholar]
- 8.Calkins H. Arrhythmogenic right ventricular dysplasia/cardiomyopathy-three decades of progress. Circ J. 2015;79:901–913. doi: 10.1253/circj.CJ-15-0288. [DOI] [PubMed] [Google Scholar]
- 9.Kimura A. Molecular etiology and pathogenesis of hereditary cardiomyopathy. Circ J. 2008;72(Suppl. A):A38–A48. doi: 10.1253/circj.cj-08-0050. [DOI] [PubMed] [Google Scholar]
- 10.Bezzina C.R., Lahrouchi N., Priori S.G. Genetics of sudden cardiac death. Circ Res. 2015;116:1919–1936. doi: 10.1161/CIRCRESAHA.116.304030. [DOI] [PubMed] [Google Scholar]
- 11.Ackerman M.J., Priori S.G., Willems S. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Heart Rhythm. 2011;8:1308–1339. doi: 10.1016/j.hrthm.2011.05.020. [DOI] [PubMed] [Google Scholar]
- 12.Giudicessi J.R., Ackerman M.J. Determinants of incomplete penetrance and variable expressivity in heritable cardiac arrhythmia syndromes. Transl Res. 2013;161:1–14. doi: 10.1016/j.trsl.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Probst V., Wilde A.A., Barc J. SCN5A mutations and the role of genetic background in the pathophysiology of Brugada syndrome. Circ Cardiovasc Genet. 2009;2:552–557. doi: 10.1161/CIRCGENETICS.109.853374. [DOI] [PubMed] [Google Scholar]
- 14.Kolder I.C., Tanck M.W., Bezzina C.R. Common genetic variation modulating cardiac ECG parameters and susceptibility to sudden cardiac death. J Mol Cell Cardiol. 2012;52:620–629. doi: 10.1016/j.yjmcc.2011.12.014. [DOI] [PubMed] [Google Scholar]
- 15.Bezzina C.R., Barc J., Mizusawa Y. Common variants at SCN5A–SCN10A and HEY2 are associated with Brugada syndrome, a rare disease with high risk of sudden cardiac death. Nat Genet. 2013;45:1044–1049. doi: 10.1038/ng.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marsman R.F., Barc J., Beekman L. A mutation in CALM1 encoding calmodulin in familial idiopathic ventricular fibrillation in childhood and adolescence. J Am Coll Cardiol. 2014;63:259–266. doi: 10.1016/j.jacc.2013.07.091. [DOI] [PubMed] [Google Scholar]
- 17.Crotti L., Johnson C.N., Graf E. Calmodulin mutations associated with recurrent cardiac arrest in infants. Circulation. 2013;127:1009–1017. doi: 10.1161/CIRCULATIONAHA.112.001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwartz P.J., Stramba-Badiale M., Crotti L. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Priori S.G., Schwartz P.J., Napolitano C. Risk stratification in the long-QT syndrome. N Engl J Med. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 20.Zareba W., Moss A.J., Schwartz P.J. Influence of genotype on the clinical course of the long-QT syndrome. International Long-QT Syndrome Registry Research Group. N Engl J Med. 1998;339:960–965. doi: 10.1056/NEJM199810013391404. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz P.J., Crotti L. QTc behavior during exercise and genetic testing for the long-QT syndrome. Circulation. 2011;124:2181–2184. doi: 10.1161/CIRCULATIONAHA.111.062182. [DOI] [PubMed] [Google Scholar]
- 22.Zareba W., Moss A.J., Daubert J.P. Implantable cardioverter defibrillator in high-risk long QT syndrome patients. J Cardiovasc Electrophysiol. 2003;14:337–341. doi: 10.1046/j.1540-8167.2003.02545.x. [DOI] [PubMed] [Google Scholar]
- 23.Priori S.G., Wilde A.A., Horie M. HRS/EHRA/APHRS expert consensus statement on the diagnosis and management of patients with inherited primary arrhythmia syndromes: document endorsed by HRS, EHRA, and APHRS in May 2013 and by ACCF, AHA, PACES, and AEPC in June 2013. Heart Rhythm. 2013;10:1932–1963. doi: 10.1016/j.hrthm.2013.05.014. [DOI] [PubMed] [Google Scholar]
- 24.Tester D.J., Will M.L., Haglund C.M. Effect of clinical phenotype on yield of long QT syndrome genetic testing. J Am Coll Cardiol. 2006;47:764–768. doi: 10.1016/j.jacc.2005.09.056. [DOI] [PubMed] [Google Scholar]
- 25.Goldenberg I., Zareba W., Moss A.J. Long QT syndrome. Curr Probl Cardiol. 2008;33:629–694. doi: 10.1016/j.cpcardiol.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Giudicessi J.R., Ackerman M.J. Genetic testing in heritable cardiac arrhythmia syndromes: differentiating pathogenic mutations from background genetic noise. Curr Opin Cardiol. 2013;28:63–71. doi: 10.1097/HCO.0b013e32835b0a41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwartz P.J., Priori S.G., Spazzolini C. Genotype–phenotype correlation in the long-QT syndrome: gene-specific triggers for life-threatening arrhythmias. Circulation. 2001;103:89–95. doi: 10.1161/01.cir.103.1.89. [DOI] [PubMed] [Google Scholar]
- 28.Wilde A.A., Jongbloed R.J., Doevendans P.A. Auditory stimuli as a trigger for arrhythmic events differentiate HERG-related (LQTS2) patients from KVLQT1-related patients (LQTS1) J Am Coll Cardiol. 1999;33:327–332. doi: 10.1016/s0735-1097(98)00578-6. [DOI] [PubMed] [Google Scholar]
- 29.Takigawa M., Kawamura M., Noda T. Seasonal and circadian distributions of cardiac events in genotyped patients with congenital long QT syndrome. Circ J. 2012;76:2112–2118. doi: 10.1253/circj.cj-12-0213. [DOI] [PubMed] [Google Scholar]
- 30.Moss A.J., Zareba W., Benhorin J. ECG T-wave patterns in genetically distinct forms of the hereditary long QT syndrome. Circulation. 1995;92:2929–2934. doi: 10.1161/01.cir.92.10.2929. [DOI] [PubMed] [Google Scholar]
- 31.Westenskow P., Splawski I., Timothy K.W. Compound mutations: a common cause of severe long-QT syndrome. Circulation. 2004;109:1834–1841. doi: 10.1161/01.CIR.0000125524.34234.13. [DOI] [PubMed] [Google Scholar]
- 32.Itoh H., Shimizu W., Hayashi K. Long QT syndrome with compound mutations is associated with a more severe phenotype: a Japanese multicenter study. Heart Rhythm. 2010;7:1411–1418. doi: 10.1016/j.hrthm.2010.06.013. [DOI] [PubMed] [Google Scholar]
- 33.Tester D.J., Will M.L., Haglund C.M. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 34.Plaster N.M., Tawil R., Tristani-Firouzi M. Mutations in Kir2.1 cause the developmental and episodic electrical phenotypes of Andersen׳s syndrome. Cell. 2001;105:511–519. doi: 10.1016/s0092-8674(01)00342-7. [DOI] [PubMed] [Google Scholar]
- 35.Splawski I., Timothy K.W., Sharpe L.M. Ca(V)1.2 calcium channel dysfunction causes a multisystem disorder including arrhythmia and autism. Cell. 2004;119:19–31. doi: 10.1016/j.cell.2004.09.011. [DOI] [PubMed] [Google Scholar]
- 36.Splawski I., Timothy K.W., Decher N. Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations. Proc Natl Acad Sci U S A. 2005;102:8089–8096. doi: 10.1073/pnas.0502506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gillis J., Burashnikov E., Antzelevitch C. Long QT, syndactyly, joint contractures, stroke and novel CACNA1C mutation: expanding the spectrum of Timothy syndrome. Am J Med Genet A. 2012;158A:182–187. doi: 10.1002/ajmg.a.34355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boczek N.J., Miller E.M., Ye D. Novel Timothy syndrome mutation leading to increase in CACNA1C window current. Heart Rhythm. 2015;12:211–219. doi: 10.1016/j.hrthm.2014.09.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moss A.J., Shimizu W., Wilde A.A. Clinical aspects of type-1 long-QT syndrome by location, coding type, and biophysical function of mutations involving the KCNQ1 gene. Circulation. 2007;115:2481–2489. doi: 10.1161/CIRCULATIONAHA.106.665406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Migdalovich D., Moss A.J., Lopes C.M. Mutation and gender-specific risk in type 2 long QT syndrome: implications for risk stratification for life-threatening cardiac events in patients with long QT syndrome. Heart Rhythm. 2011;8:1537–1543. doi: 10.1016/j.hrthm.2011.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shimizu W., Moss A.J., Wilde A.A. Genotype-phenotype aspects of type 2 long QT syndrome. J Am Coll Cardiol. 2009;54:2052–2062. doi: 10.1016/j.jacc.2009.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Priori S.G., Napolitano C., Schwartz P.J. Association of long QT syndrome loci and cardiac events among patients treated with beta-blockers. J Am Med Assoc. 2004;292:1341–1344. doi: 10.1001/jama.292.11.1341. [DOI] [PubMed] [Google Scholar]
- 43.Schwartz P.J., Priori S.G., Locati E.H. Long QT syndrome patients with mutations of the SCN5A and HERG genes have differential responses to Na+ channel blockade and to increases in heart rate. Implications for gene-specific therapy. Circulation. 1995;92:3381–3386. doi: 10.1161/01.cir.92.12.3381. [DOI] [PubMed] [Google Scholar]
- 44.Ruan Y., Liu N., Bloise R. Gating properties of SCN5A mutations and the response to mexiletine in long-QT syndrome type 3 patients. Circulation. 2007;116:1137–1144. doi: 10.1161/CIRCULATIONAHA.107.707877. [DOI] [PubMed] [Google Scholar]
- 45.Ruan Y., Denegri M., Liu N. Trafficking defects and gating abnormalities of a novel SCN5A mutation question gene-specific therapy in long QT syndrome type 3. Circ Res. 2010;106:1374–1383. doi: 10.1161/CIRCRESAHA.110.218891. [DOI] [PubMed] [Google Scholar]
- 46.Hayashi M., Denjoy I., Extramiana F. Incidence and risk factors of arrhythmic events in catecholaminergic polymorphic ventricular tachycardia. Circulation. 2009;119:2426–2434. doi: 10.1161/CIRCULATIONAHA.108.829267. [DOI] [PubMed] [Google Scholar]
- 47.Priori S.G., Napolitano C., Memmi M. Clinical and molecular characterization of patients with catecholaminergic polymorphic ventricular tachycardia. Circulation. 2002;106:69–74. doi: 10.1161/01.cir.0000020013.73106.d8. [DOI] [PubMed] [Google Scholar]
- 48.Medeiros-Domingo A., Bhuiyan Z.A., Tester D.J. The RYR2-encoded ryanodine receptor/calcium release channel in patients diagnosed previously with either catecholaminergic polymorphic ventricular tachycardia or genotype negative, exercise-induced long QT syndrome: a comprehensive open reading frame mutational analysis. J Am Coll Cardiol. 2009;54:2065–2074. doi: 10.1016/j.jacc.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bai R., Napolitano C., Bloise R. Yield of genetic screening in inherited cardiac channelopathies: how to prioritize access to genetic testing. Circ Arrhythm Electrophysiol. 2009;2:6–15. doi: 10.1161/CIRCEP.108.782888. [DOI] [PubMed] [Google Scholar]
- 50.Nyegaard M., Overgaard M.T., Sondergaard M.T. Mutations in calmodulin cause ventricular tachycardia and sudden cardiac death. Am J Hum Genet. 2012;91:703–712. doi: 10.1016/j.ajhg.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Makita N., Yagihara N., Crotti L. Novel calmodulin mutations associated with congenital arrhythmia susceptibility. Circ Cardiovasc Genet. 2014;7:466–474. doi: 10.1161/CIRCGENETICS.113.000459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sansone V., Tawil R. Management and treatment of Andersen–Tawil syndrome (ATS) Neurotherapeutics. 2007;4:233–237. doi: 10.1016/j.nurt.2007.01.005. [DOI] [PubMed] [Google Scholar]
- 53.Postema P.G. About Brugada syndrome and its prevalence. Europace. 2012;14:925–928. doi: 10.1093/europace/eus042. [DOI] [PubMed] [Google Scholar]
- 54.Kamakura S., Ohe T., Nakazawa K. Long-term prognosis of probands with Brugada-pattern ST-elevation in leads V1–V3. Circ Arrhythm Electrophysiol. 2009;2:495–503. doi: 10.1161/CIRCEP.108.816892. [DOI] [PubMed] [Google Scholar]
- 55.Probst V., Veltmann C., Eckardt L. Long-term prognosis of patients diagnosed with Brugada syndrome: results from the FINGER Brugada Syndrome Registry. Circulation. 2010;121:635–643. doi: 10.1161/CIRCULATIONAHA.109.887026. [DOI] [PubMed] [Google Scholar]
- 56.Sacher F., Probst V., Iesaka Y. Outcome after implantation of a cardioverter-defibrillator in patients with Brugada syndrome: a multicenter study. Circulation. 2006;114:2317–2324. doi: 10.1161/CIRCULATIONAHA.106.628537. [DOI] [PubMed] [Google Scholar]
- 57.Suzuki H., Torigoe K., Numata O. Infant case with a malignant form of Brugada syndrome. J Cardiovasc Electrophysiol. 2000;11:1277–1280. doi: 10.1046/j.1540-8167.2000.01277.x. [DOI] [PubMed] [Google Scholar]
- 58.Belhassen B., Viskin S., Fish R. Effects of electrophysiologic-guided therapy with Class IA antiarrhythmic drugs on the long-term outcome of patients with idiopathic ventricular fibrillation with or without the Brugada syndrome. J Cardiovasc Electrophysiol. 1999;10:1301–1312. doi: 10.1111/j.1540-8167.1999.tb00183.x. [DOI] [PubMed] [Google Scholar]
- 59.Antzelevitch C., Brugada P., Borggrefe M. Brugada syndrome: report of the second consensus conference. Heart Rhythm. 2005;2:429–440. doi: 10.1016/j.hrthm.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 60.Crotti L., Marcou C.A., Tester D.J. Spectrum and prevalence of mutations involving BrS1- through BrS12-susceptibility genes in a cohort of unrelated patients referred for Brugada syndrome genetic testing: implications for genetic testing. J Am Coll Cardiol. 2012;60:1410–1418. doi: 10.1016/j.jacc.2012.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhonsale A., Groeneweg J.A., James C.A. Impact of genotype on clinical course in arrhythmogenic right ventricular dysplasia/cardiomyopathy-associated mutation carriers. Eur Heart J. 2015;36:847–855. doi: 10.1093/eurheartj/ehu509. [DOI] [PubMed] [Google Scholar]
- 62.Groeneweg J.A., Bhonsale A., James C.A. Clinical presentation, long-term follow-up, and outcomes of 1001 arrhythmogenic right ventricular dysplasia/cardiomyopathy patients and family members. Circ Cardiovasc Genet. 2015;8:437–446. doi: 10.1161/CIRCGENETICS.114.001003. [DOI] [PubMed] [Google Scholar]
- 63.Elliott P.M., Anastasakis A., Borger M.A. ESC Guidelines on diagnosis and management of hypertrophic cardiomyopathy: the Task Force for the Diagnosis and Management of Hypertrophic Cardiomyopathy of the European Society of Cardiology (ESC) Eur Heart J. 2014;2014(35):2733–2779. doi: 10.1093/eurheartj/ehu284. [DOI] [PubMed] [Google Scholar]
- 64.Maron B.J., Gardin J.M., Flack J.M. Prevalence of hypertrophic cardiomyopathy in a general population of young adults. Echocardiographic analysis of 4111 subjects in the CARDIA Study. Coronary Artery Risk Development in (Young) Adults. Circulation. 1995;92:785–789. doi: 10.1161/01.cir.92.4.785. [DOI] [PubMed] [Google Scholar]
- 65.Maron B.J. Sudden death in young athletes. N Engl J Med. 2003;349:1064–1075. doi: 10.1056/NEJMra022783. [DOI] [PubMed] [Google Scholar]
- 66.Ho C.Y., Charron P., Richard P. Genetic advances in sarcomeric cardiomyopathies: state of the art. Cardiovasc Res. 2015;105:397–408. doi: 10.1093/cvr/cvv025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Colan S.D., Lipshultz S.E., Lowe A.M. Epidemiology and cause-specific outcome of hypertrophic cardiomyopathy in children: findings from the Pediatric Cardiomyopathy Registry. Circulation. 2007;115:773–781. doi: 10.1161/CIRCULATIONAHA.106.621185. [DOI] [PubMed] [Google Scholar]
- 68.Maron B.J., Towbin J.A., Thiene G. Contemporary definitions and classification of the cardiomyopathies: an American Heart Association Scientific Statement from the Council on Clinical Cardiology, Heart Failure and Transplantation Committee; Quality of Care and Outcomes Research and Functional Genomics and Translational Biology Interdisciplinary Working Groups; and Council on Epidemiology and Prevention. Circulation. 2006;113:1807–1816. doi: 10.1161/CIRCULATIONAHA.106.174287. [DOI] [PubMed] [Google Scholar]
- 69.Towbin J.A., Lowe A.M., Colan S.D. Incidence, causes, and outcomes of dilated cardiomyopathy in children. J Am Med Assoc. 2006;296:1867–1876. doi: 10.1001/jama.296.15.1867. [DOI] [PubMed] [Google Scholar]
- 70.Dec G.W., Fuster V. Idiopathic dilated cardiomyopathy. N Engl J Med. 1994;331:1564–1575. doi: 10.1056/NEJM199412083312307. [DOI] [PubMed] [Google Scholar]
- 71.Towbin J.A. Inherited cardiomyopathies. Circ J. 2014;78:2347–2356. doi: 10.1253/circj.cj-14-0893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Marcus F.I., McKenna W.J., Sherrill D. Diagnosis of arrhythmogenic right ventricular cardiomyopathy/dysplasia: proposed modification of the Task Force Criteria. Eur Heart J. 2010;31:806–814. doi: 10.1093/eurheartj/ehq025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Corrado D., Wichter T., Link M.S. Treatment of arrhythmogenic right ventricular cardiomyopathy/dysplasia: an international task force consensus statement. Eur Heart J. 2015;36:3227–3237. doi: 10.1093/eurheartj/ehv162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ackerman M.J., Priori S.G., Willems S. HRS/EHRA expert consensus statement on the state of genetic testing for the channelopathies and cardiomyopathies: this document was developed as a partnership between the Heart Rhythm Society (HRS) and the European Heart Rhythm Association (EHRA) Europace. 2011;13:1077–1109. doi: 10.1093/europace/eur245. [DOI] [PubMed] [Google Scholar]
- 75.Lazzarini E., Jongbloed J.D., Pilichou K. The ARVD/C genetic variants database: 2014 update. Hum Mutat. 2015;36:403–410. doi: 10.1002/humu.22765. [DOI] [PubMed] [Google Scholar]
- 76.Bao J.R., Wang J.Z., Yao Y. Screening of pathogenic genes in Chinese patients with arrhythmogenic right ventricular cardiomyopathy. Chin Med J. 2013;126:4238–4241. [PubMed] [Google Scholar]
- 77.Ohno S., Nagaoka I., Fukuyama M. Age-dependent clinical and genetic characteristics in Japanese patients with arrhythmogenic right ventricular cardiomyopathy/dysplasia. Circ J. 2013;77:1534–1542. doi: 10.1253/circj.cj-12-1446. [DOI] [PubMed] [Google Scholar]
- 78.Kapplinger J.D., Landstrom A.P., Salisbury B.A. Distinguishing arrhythmogenic right ventricular cardiomyopathy/dysplasia-associated mutations from background genetic noise. J Am Coll Cardiol. 2011;57:2317–2327. doi: 10.1016/j.jacc.2010.12.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Christiaans I., van Langen I.M., Birnie E. Quality of life and psychological distress in hypertrophic cardiomyopathy mutation carriers: a cross-sectional cohort study. Am J Med Genet A. 2009;149A:602–612. doi: 10.1002/ajmg.a.32710. [DOI] [PubMed] [Google Scholar]
- 80.Herman D.S., Lam L., Taylor M.R. Truncations of titin causing dilated cardiomyopathy. N Engl J Med. 2012;366:619–628. doi: 10.1056/NEJMoa1110186. [DOI] [PMC free article] [PubMed] [Google Scholar]