Abstract
Novel families of short interspersed nuclear element (SINE) sequences in the human pathogenic fungus Aspergillus fumigatus, clinical isolate Af293, were identified and categorised into tRNA-related and 5S rRNA-related SINEs. Eight predicted tRNA-related SINE families originating from different tRNAs, and nominated as AfuSINE2 sequences, contained target site duplications of short direct repeat sequences (4–14 bp) flanking the elements, an extended tRNA-unrelated region and typical features of RNA polymerase III promoter sequences. The elements ranged in size from 140–493 bp and were present in low copy number in the genome and five out of eight were actively transcribed. One putative tRNAArg-derived sequence, AfuSINE2-1a possessed a unique feature of repeated trinucleotide ACT residues at its 3’-terminus. This element was similar in sequence to the I-4_AO element found in A. oryzae and an I-1_AF long nuclear interspersed element-like sequence identified in A. fumigatus Af293. Families of 5S rRNA-related SINE sequences, nominated as AfuSINE3, were also identified and their 5'-5S rRNA-related regions show 50–65% and 60–75% similarity to respectively A. fumigatus 5S rRNAs and SINE3-1_AO found in A. oryzae. A. fumigatus Af293 contains five copies of AfuSINE3 sequences ranging in size from 259–343 bp and two out of five AfuSINE3 sequences were actively transcribed. Investigations on AfuSINE distribution in the fungal genome revealed that the elements are enriched in pericentromeric and subtelomeric regions and inserted within gene-rich regions. We also demonstrated that some, but not all, AfuSINE sequences are targeted by host RNA silencing mechanisms. Finally, we demonstrated that infection of the fungus with mycoviruses had no apparent effects on SINE activity.
Introduction
Short interspersed nuclear element (SINE) sequences are short repetitive, non-coding sequences ranging in size from 100–600 bp. SINE sequences are widely distributed in eukaryotic genomes and have crucial roles in genome organization, genome evolution and modulating gene expression. SINE sequences have been implicated as being involved in cell survival during physiological stresses including heat shock, DNA damage, irradiation, oxidative stress, low temperature, exposure to toxic agents, and infection by pathogens or protoplast isolation [1–5]. SINE sequences are referred to as non-autonomous retrotransposons because they usually depend on enzymes encoded by long nuclear interspersed element (LINE) sequences for reverse transcription and retrotransposition. SINE sequences are transcribed by RNA polymerase III (pol III). Subsequently RNA pol III-SINE transcripts are reverse transcribed by reverse transcriptase (RT) and then re-integrated by endonuclease (EN) into various sites in the genome [6]. Some SINE sequences, which possess an intact RNA pol III promoter, are functionally active but the majority are not actively transcribed [6–8]. Large numbers of SINE sequences in eukaryotes are derived from tRNAs while others are derived from 5S rRNAs or 7SL RNAs [9].
To date SINE sequences have been reported in several fungi. For example MgSINE, isolated from the rice blast fungus Magnaporthe grisea, is a 472 bp tRNA-derived SINE present in ca. 100 copies in the genome which possesses features similar to mammalian SINE sequences [10]. Another SINE identified in M. grisea is the mgsr1 SINE which is an 800 bp element present in ca. 40 copies in the genome [11]. Foxy is an active SINE family found in Fusarium oxysporum f. sp. lycopersici strain Fo1007 and is present in ca. 160 copies in the genome [7]. SINE sequences have also been identified in several other filamentous fungi including Egr1 (700 bp, ca. 50 copies; [12]) and egh1 (EGH24; 900 bp; [13]) both in Erysiphe graminis, nrs1 (500 bp, 11 copies) in Nectria haematococca [14], SINE2-1_BG (a tRNA-derived SINE) in barley powdery mildew, Blumeria graminis [15, 16] and fifteen families of infSINEs sequences in the oomycete Phytophthora infestans [17].
Aspergillus fumigatus is a saprophytic and thermotolerant filamentous fungus which produces large numbers of asexual spores. However, the production of functional sexual spores (cleistothecia and ascospores) and its teleomorph Neosartorya fumigata have been described [18]. Aspergillus fumigatus is an opportunistic, airborne fungal pathogen that causes pulmonary invasive aspergillosis and is responsible for 90% of fungal infections in immunocompromised patients [19]. A non-LTR (long terminal repeat) retrotransposon I-1_AF, which belongs to the Tad clade of LINE-like element (LLEs), has been found in several copies in the A. fumigatus Af293 genome. These elements encode a DNA/RNA-binding protein, EN, RT and RNase H and insert randomly in the genome or precisely at the same target site in Afut2_AF [20, 21]. Recently a full description of the LLEs in several clinical and environmental isolates of A. fumigatus has been presented [22]. However, the existence of SINE sequences in the A. fumigatus genome is as yet not recorded.
The aim of this study was to investigate the occurrence of SINE sequences in the genome of the prototype A. fumigatus Af293 isolate. The abundance and distribution of SINE sequences were identified by interrogating the genomic DNA sequence for the occurrence of well-characterized signature motif sequences using computational analyses prior to mapping on the fungus genome and assessing potential transcription activity and silencing. Additionally the insertion patterns and copy numbers of the elements were compared in isogenic virus-free and virus-infected A. fumigatus isolates.
Materials and Methods
Computational Identification of SINE Sequences in the A. fumigatus Af293 Genome
Sequence dataset
The A. fumigatus Af293 reference genome sequence was accessed through the NCBI database and was also retrieved from several other online sources including the Central Aspergillus Resource (CADRE; [23]), the BROAD INSTITUTE Aspergillus comparative database and the Aspergillus Genome Database (AspGD; [24]).
Search strategy and computational analyses
CENSOR software, developed by the Genetic Information Research Institute (GIRI; [25]) was used to interrogate the A. fumigatus Af293 genome sequences and screen for putative SINE sequences against a reference collection of repeats. Homologous portions of the query and reference sequences were masked and reports classifying all of the repeats that were discovered were generated. The RepeatMasker server (version: open-4.0) [26], which screens DNA sequences for interspersed repeats, was also used to screen the Af293 genome sequences against reference sequences in the Repbase libraries [27]. The reported sequences, which were classified as non-long terminal repeats (non-LTRs), SINE sequences, SINE2/tRNA and SINE3/5S were selected as SINE candidates and subjected to further analysis.
To identify tRNA-related SINE sequences, genomic sequences were examined using the tRNAScan-SE 1.21 program [28] to search for the presence of basic tRNA features such as RNA pol III promoter sequences and tRNA cloverleaf secondary structures using relaxed parameters and the EuFindtRNA algorithm. Subsequently selected sequences predicted from the program were resubmitted to tRNAScan-SE using the default settings. Sequences yielding positive reads were discarded as they were considered to be true tRNAs while the remaining sequences yielding negative reads and showed no support for a 5’-tRNA-related region, were considered for further analyses. The tRNA cloverleaf secondary structures and tRNA origin of the remaining sequences were also identified. In addition, the Genomic tRNA Database (GtRNAdb) was aligned with all the A. fumigatus tRNA sequences to separate tRNAs from candidate SINE sequences.
All predicted SINE sequences retrieved from Censor, RepeatMasker and tRNAScan-SE were submitted to BLAST analysis using accessible database sequences from the NCBI and A. fumigatus Genome Map Viewer to expand the boundaries of the masked sequences to 1,000 bp upstream and 2,000 bp downstream of both termini. Sequences with predicted tRNA-related structures were manually inspected for the presence of degenerate RNA pol III promoter A and B box sequences, extended 3’ tRNA-unrelated sequences upstream of an oligothymidine tract and also target site duplication (TSD) sequences flanking the SINE sequences. An additional check for the presence of the RNA pol III promoter sequence was performed by multiple sequence alignment of the predicted SINE sequences with the A. fumigatus Af293 tRNA gene sequences obtained from GtRNAdb. To identify 5S rRNA-related SINE sequences, sequences classified as SINE3/5S using Censor and RepeatMasker were aligned with the A. fumigatus 5S rRNAs and searched for the RNA pol III promoter (A, IE and C boxes) sequence. The process framework used for computational analysis of AfuSINEs is shown in S1 Fig.
Characterization, classification, distribution and location of SINE sequences was achieved by comparative analysis of the sequences. All predicted AfuSINE sequences were compared against the complete genome sequence of Af293 available on AspGD and CADRE by BLAST to search for locations of each element on the chromosomes. Multiple sequence alignments were performed using the Clustal Omega package [29] and MAFFT multiple sequence alignment software version 7 [30] and manual edition of the sequences. Phylogenetic trees were constructed using the neighbor-joining (NJ) method on MAFFT alignments [30].
Fungal Strains and Culture Conditions
A. fumigatus Af293, which is a clinical strain, was used throughout the study. Af293 is naturally infected with a dsRNA mycovirus, Aspergillus fumigatus tetramycovirus-1 (AfuTmV-1), and NK125 is an isogenic, cured virus-free strain of the fungus [31]. A 20 μl suspension of fungal conidia was inoculated on Aspergillus Complete Media (ACM; [32]) agar plates and incubated for 4 days at 7C. Ampicillin (100 μg/ml) was added to the growth medium when required. For liquid culture cultivation, a 10 ml suspension of ca. 5x108 conidia/ml was inoculated into 300 ml ACM broth and incubated at 37°C for 7 days on a rotary shaker at 150 rpm.
Nucleic Acid Preparations
A. fumigatus genomic DNA was prepared using the DNeasy® Plant Mini Kit (QIAGEN, UK) using the mini protocol as described by the manufacturer. Total RNA extracts were prepared using the RNeasy® Plant Mini Kit (QIAGEN, UK) with the small scale protocol or Trizol as described by the manufacturers. DNA contaminants were removed from total RNA preparations using TURBOTM DNase (Ambion, UK) prior to cDNA synthesis.
PCR and RT-PCR Amplification of A. fumigatus SINE Sequences
Sequence specific primers were designed based on the sequences of candidate SINE sequences to cover the promoter regions, yielding amplicons ca. 100–150 bp in size (S1 Table). PCR amplification was performed under high stringency PCR conditions described as follows; 94°C, 5 min; 35 cycles of 94°C 30 sec, 55 or 60°C 30 sec and 72°C 30 sec; followed by final extension at 72°C for 5 min. RT-PCR amplification of A. fumigatus Af293 total RNA was performed using the same PCR cycling procedure and primer sets. Complementary DNA was synthesised using 10 μl of total RNA (ca. 50 ng/μl) using either random hexamers or sequence specific primers and Superscript III reverse transcriptase (Invitrogen, UK) following the manufacturer’s protocol. PCR amplicons were separated by gel electrophoresis and visualized by SYBR safe DNA staining.Southern Blot Hybridization of A. fumigatus SINE Sequence Amplicons.
Aliquots of 10 μg of genomic DNA were individually digested overnight with 1 μl of HindIII (New England Biolabs) at 37°C. Genomic DNA was also used as a DNA template for probe preparation and was labelled using the PCR DIG probe synthesis kit (Roche, UK) according to the manufacturer’s instructions. Restriction fragments of genomic DNA were separated by electrophoresis on 1% w/v agarose gels containing SYBR safe DNA stain in 1XTAE. Gels were denatured in 0.25 N HCl and 0.5M NaOH + 1.5 M NaCl, followed by neutralization in 0.5 M Tris-HCl + 1.5 M NaCl. The DNA was then transferred onto a positively charged Amersham HybondTM-N membrane (GE healthcare) and nucleic acids were subsequently fixed by UV-cross linking prior to probing.
Detection of Small RNA Molecules Homologous to A. fumigatus SINE Sequences
Small, low molecular weight (LMW) RNAs were isolated using the procedure of Lu et al. [33] with some minor modifications. Total RNA, high molecular weight RNA (HMW RNA) and LMW RNA fractions were analyzed on 1.5% agarose and 15% (w/v) polyacrylamide Tris-borate-EDTA-urea gels (Bio-Rad, Sweden) (Fig 1A–1C). For the detection of homologous SINE small RNAs, 100 μg of the small RNA fraction was separated on polyacrylamide gels. For northern blot hybridization of small RNAs, nucleic acids were transferred to nylon membranes and fixed using 1-ethyl-3-(3-dimethylaminopropryl carbodiimide; EDC)-cross linking [34]. Prior to hybridization, probes were cleaved to an average length of 50 bp by alkaline hydrolysis as described by Kreuze et al. [35].
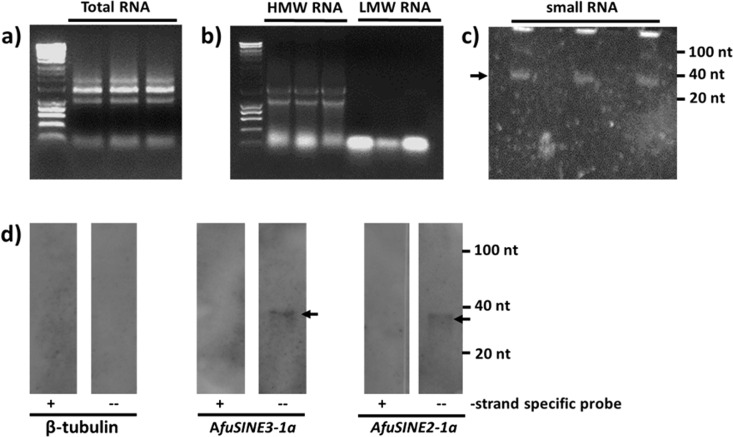
Fig 1. Detection of small RNA molecules homologous to AfuSINE sequences.
Total RNAs isolated with Trizol were electrophoresed on 1.5% agarose gels (a). After separation by PEG precipitation, high molecular weight RNAs (HMW RNAs) and low molecular weight RNAs (LMW RNAs) were electrophoresed on 1.5% agarose gels (b). The LMW fractions from 400 ng/μl of total RNA were resolved on 15% (w/v) polyacrylamide Tris-borate-EDTA-urea gels and stained with SYBR Gold Nucleic Acid Gel Stain (c). Northern blot hybridization of small RNAs homologous to AfuSINE sequences in A. fumigatus Af293 (d). LMW RNA fractions were isolated using TRIzol and 5 μg RNA sample was loaded into each well for northern blot analysis. Only small RNAs (<40 nt; arrowed) from the antisense strand were detected.
Secondary Structure Prediction of A. fumigatus SINE Transcripts
The secondary structures of SINEs were predicted using Mfold [36] using default settings with a folding temperature set to 37°C, which is the optimal growth temperature of A. fumigatus Af293. RNA structures showing the lowest free energies were accepted as the most likely structures for the AfuSINE sequences.
Results and Discussion
Search Strategy and Identification of AfuSINE Sequences by Computational Analyses
Here we report for the first time the presence and distribution of SINE sequences in the A. fumigatus Af293 genome by exploiting the availability of its complete genome sequence to investigate their incidence. In this study, a computational search strategy was developed to predict the occurrence of A. fumigatus SINE sequences (designated as AfuSINE sequences) using several bioinformatic software packages. The Af293 genomic sequences were initially screened for the interspersed repeat consensus of SINE sequences using Censor and RepeatMasker against the available sequences from the reference collection of repeats in the Repbase database. The tRNAscan-SE search server was also used to screen for tRNA-related SINEs using relaxed parameters. In total 17,269 candidate sequences were retrieved. These include simple repeats, small RNAs, tRNAs, 5S rRNAs, 7SL RNAs, tRNA- and 5S rRNA-related sequences, sequences homologous to SINE and LINE sequences as well as other transposons. However, since these programs identify exact sequence matches, any data retrieved required further screening with the tRNAscan-SE program using default parameters to eliminate true tRNA sequences. The 5S rRNA sequences were eliminated by BLAST analysis against Af293 5S rRNAs on the NCBI database. Subsequently, the sequences that do not exhibit SINE characteristics were discarded, leaving 145 candidate sequences nominated SINE2 sequences (tRNA-derived SINE sequences) and SINE3 sequences (5S rRNA-derived SINE sequences) for further investigations including the extension of SINE boundaries, BLAST analysis, and inspection of 5S rRNA-related SINE and tRNA-related SINE features.
Following this analysis, thirteen candidate SINE sequences were identified which were divided into AfuSINE2 and AfuSINE3 types. Most of these sequences possessed features in common with generic SINE sequences but some lacked TSD and intact internal RNA pol III sequences. However some did contain masked sequences similar to SINE sequences from a broad spectrum of reference organisms including A. oryzae, A. nidulans, B. graminis, Cryptococcus neoformans, arabidopsis, hedgehogs, mosquitoes and humans. The organization and structure of representative AfuSINE sequences are shown in Table 1 and S2–S5 Figs.
Table 1. List of 5 putative 5S rRNA-related SINE sequences (AfuSINE3) and 8 putative tRNA-related SINE sequences (AfuSINE2).
| Chr.# | AfuSINEs | AfuSINE sequences | Origin | Length (bp) | Activity | Estimated copy no. |
|---|---|---|---|---|---|---|
| 1 | AfuSINE3-1a | TAGGCTCCAACCAAGCACATACGACCATAGGGTGTGGAAAACAGGGCTTCCCGTCCGCTCAACCGTACTTAAGCCAGGTGTAGAATGTGGTCAATCCATAGAAAGCCCTTTTGCCTCCAAAGCAGCGGCGGGATGCTTTTCGTATGAGTGCTGGTGATGTGGGTGAGAACTCCAGGGGGCCCCATCACCCCTGGTGATGATGGCTCTAGGGGTTTGATGAAGGCTAACTTAACAGCTGACTGCAGAAGAAAACAGTGATTCAGCTGCTTTTAATAGGCTC | 5S rRNA | 280 | Active | 5 |
| 3 | AfuSINE3-3a | AGCTTGAAAACAAGCACATACGACCATAGGGTGTGGAAAACAGGGCTTCCCGTCCGCTCAGCCGTACTTAAGCCACATGTATCAACGTGGTCGGAAGTTATATGCGCCCTTAATGGCCTGCTGCTTTACGGGTGAGAGACGCTAAACCATAGGGTGAGTGTGGGTGAGTGAAGGTGAGCTGAGTGATGCTATCCAACGCCATGGGTGAGCAAGGGTGATCAGTGGGTGACAGTGTGATTCTATAATGACTAGACTCCGTCTATCGCTAGCTTAAA | 5S rRNA | 275 | Inactive | NA |
| 3 | AfuSINE3-3c | ATTCAGTTATCTTTCTCCCAGCACATACGACCATAGGGTGTGGAAAACAGGGCTTCCCGTCCGCTCAGCCGTACTTAAGCCACACGTAGAACGTGGTCAATCCCTAGAAAGCCTTTTTGCTTCTAAAGCAGCGGCGTAATTCTTTTAGTATGAGTGCTGGTGATACGGGTGAGGACCCCGTCACCCCTGGTGATGATGGCTTACTGGTGGTGACGAGGGCTCCAGGGGGGTGACGAAGGCGTACTCAGAAGTCAACCACAGAAGATACAGTGATTGAGCTACTTTTAATGGGCTCTGCGCCCCGCAGAAGTAATGCACTTGATTATTCCTGAGTCCATACAGT | 5S rRNA | 343 | Active | 5 |
| 4 | AfuSINE3-4a | CGTCAGATTGCACAATGACCATAGGGTGTGGAAAACAGGGCTTCCTGTCCGCTCAGCCGTATTCAAACCAGTATTATTAATTTGGTCGGAAGTTATATGCGTCCCTAAAGGCCTGCCGCTCTACGGATGAGAGACACTGAACCATAGGGTGAGTGTGGTGAGTGAAGGTGAGCAGAACGATGCTATCTATTGAATTATTCACATCGGCGATCCGTAGAACAGGTTGGTCCCACGTCCTGGACACGCCCATACTCCGTCA | 5S rRNA | 259 | Inactive | NA |
| 5 | AfuSINE3-5c | AGCTTCTATATGAATTTCTTGCACGAATATCCTAGCACATACGACCATAGGGTGTGGAAAACAGGGCCTCCTGGCTGCTCGGCCATACTTAAGCCAGTATTATTAATATAGTCAAAAGTTATATGCACCCTTAAAGGCCTGCTACTTTACAGATAAGAGATACTAAACCATAGGGTGAGTATAGGTGAGTAAAGGTGAGCTAAGTAATGCTATCTAACAGCATACTTGAGCATGGTGATTAGTAGATAACAGAGTGATTCTATCATGACTAGACTCCTCTATCACTAGCTTCAATAT | 5S rRNA | 297 | Inactive | NA |
| 1 | AfuSINE2-1a | AAGTGTACATAGAGCCGGTAGCGGTAGCGTAGTGGTAAGCGCTCCGAGGCAGCCTCTAGAGAAGTAGGATTGGTGACCCTGCTTTTTTGGAGGTTATGGGTTCGATTCCCGTCGCTGGCACAACATTTACCACCACAATGGAAGATCACTTCCCACAATGGTATCAAGGCCACTCCCTTATCGCAAGGTGGTGGGGAAGTTGGAACAATCACAGGCCTGTAAGGCGAGGCTCTAAATTCGCCCTCATGTAATGGAACAAAATGTAACTAGACACACAAGGATTAGCTATAGTCGATACCTGCATATCGCCCAAGGCGAGGGGTCAGCGTATGAGTACTACTACTACTACTACTACTAAGTGTACATAGAGA | tRNA/Arg | 370 | Active | 4 |
| 3 | AfuSINE2-3a | AGGGAGTTTGATATCCCATTTTGGAGGGACTGGCCGGCTCTGGGGTCGGTCGTTAAGGCGCTCCGCCATTCATCTGCAGGTTGGATAACAGCCGGGCAATTTTCGTGACTGAAAATCCCATCCATCTGAAACTCTTGTGAGTATGTTGAAAGCCGACTTTTCAAGACGCCAATCTCAGGGCCATCCGCAGCTGAAAGATAACTCTCGAAGCAATGCCGGCACTGTAGAAAGCAGCAACGGAGAAGAGGCGATTGTTGGTGTCCGGCCAACTTGACATAACGATCGAATAGATCGAACATTCGTTGCGAGGTTTCGACGGCCAGATCAATCATATCGGCACTGGGAGACCGACAGTGGCGGACAAGTGCGAGAATCACAAAGACGGCTGTTAGATGCAAGGTAAGAAGAGGCGGCGTAGTAGAGTCGGACCTGGGATTTGAGTATTGTGCTAGAAGAATCGCCTGTTGTTCCTGGATCTGGAGCAGGAGGGAGTB | tRNA/Ser | 493 | Active | 1 |
| 4 | AfuSINE2-4a | ATGTACATCAGGTGTATGCGGCCTGGGCAGAAATATGATGGGTAGATATCAAGATATCGTAAGTATTCCAGCTCCTCCACTGATTGACGATCCTTTTGTTATCTTGAGCCAAGGAATTTGGTGCCTTTGGGCCTGAGGTTCCAGATCCTCCCCCACATCCGCGAACCTCATTTCGTGGTTAAGGACAACATTAAATAACCGTGAATTTACTCCGTAGTGTCTCGACCCTTTGGGGGGTGATCTTGTAAAGATTAGGAAAGAATGAGAGAAATAAATCACAGATTTTTTTTTTTTTTTTTTCGTAAGAAAGAAAAGGCAACTCCCTCGTCACAGCAGGATTTATGTACATC | tRNA/Ser | 349 | Active | 1 |
| 7 | AfuSINE2-7a | TCAAAACGCTGGAACTGGGTTTATTTCCATTTGGCGGAATGGAAAGGTTCGAAACAGCCCTGGAGGTGTTCGAATAGTAGAGTATGGTCAGCTTCGCTTCCTGCTCGTGTGACTATGATGTAAAGATGTTGGGTTATTTTATTTCAATCAATCAGTGGGCCGCATTCTATTCTTACAAGTCCCTTCAATGGCAATCATGGATATCAGATGGAGAGATTTGCGATGCTTCTGTGTCTCTAATAATAAGCAGTCTTCAGATGAGTGGAGGCGGCAGAGAAGGGGAGCAATGGGGACCAATGGGGAGCAATGGGGAGTAATAAGGAGCAACAAGGATGAAGAGGATTGGTGTGACGACCCCTGCATACAACACTTGTAGGAAACGACATCTTATCAATCTCAAA | tRNA/Gly | 401 | Inactive | NA |
| 5 | AfuSINE2-5d | TTTTTAGTTGCTACGTAGGCTTAATGGAAGGAACATGTTCGAGGATGGTCGGAAAATCGCTGGAGGTGTTCAATCCCCGAGGTACGTCATCTGGAACTGAATCAAACCTCTCCACTCGAGGGAGAGCTGAGAGTATTAGGTGGTTTTCGTACTTATATCCAAGGCTCATTATCACGAATGCCGTACAAACAAACACAATCGAGATACTCCATACTTGAATTATTTTCAGT | tRNA/Ser | 230 | Active | 2 |
| 3 | AfuSINE2-3c | TGTGATGCCTTTCCACACTGCATGGTTGTCTCGGCACAGATGGCAAAGCCGTTCCTGGCAACAGTTAATTGTTCAGTTCAAATCTGTTAAAACGGGGTCGTACTTACCATAGACTGTCGTCGAGCATCTGCCGCAGCTCTAGCCAAAAGACCCAGTGATTTCTGCCTATGAATGACAGGTCAGCTTTGTGGGGGCTTTGCA | tRNA/Ala | 201 | Active | 5 |
| 4 | AfuSINE2-4c | CTTGGAGATTACATACGTATCCTGCAATGGGCAGAATCAACAACGTGTACATTTTACGCGTAAATGGATCGATTCCATATGTACCACTCTCTAAGCCAACGCAACCATGAAAGGGTGAGAAGACCCTGACGGCCACCGAAACACTTACCAACCAGTTCTTGGTTGTGAACATAGTATGGATAATTTGGTAGCTATAGTTGATTTGAATAATGGTGCTGTCGCACCTCGCAATGTCTTTTTAGCATCTTGGAGA | tRNA/Thr | 253 | Inactive | NA |
| 7 | AfuSINE2-7e | AATTACTTATATAGCAGAGTGGTAGAGTCATAGGCTTCATATTTTTACAGCAATGGAGATGAGATAAGGATTCGATTTCTATATATTTTAGTACTAGATAATATTAACTTTAAGGGAAGCTATAGGATTTTCCCCTAATT | tRNA/Met | 140 | Inactive | NA |
Annotations: NNNN = RNA pol III promoter sequences; NNNN = target site duplication (TSD);NNNN = short repetitive sequence
NNNN = 5' tRNA-related CCA end; NNNN = ACATT at 3’ end
A Sequence underlined by dotted line is similar to I-4_AO#LINE/Tad1 of A. oryzae (Galagan et. al., 2005; Kapitonov and Jurka, 2006) and I-1_AF A. fumigatus Af293 LINE.
B The 5’ tRNA-related region is similar to a 481 bp LFSINE_Vert (SINE2/tRNA#His) identified in Latimeria menadoensis (Bejerano et al., 2006).
C The 5’ tRNA-related region is similar to a 268 bp MIRc (SINE2/tRNA#Pro) identified in mammals (Smit and Riggs, 1995).
5S rRNA-related SINE sequences (AfuSINE3)
Families of five 5S rRNA-derived SINE-like sequences (AfuSINE3) were discovered which are homologous to the 5’-terminal region of fungal 5S rRNA sequences. The elements are present in five related copies following in silico analysis (nominated AfuSINE3-1a, AfuSINE3-3a, AfuSINE3-3c, AfuSINE3-4a and AfuSINE3-5c on chromosomes 1, 3, 4 and 5, respectively) with an average length of 259–343 bp. All five AfuSINE3 sequences are related to SINE3-1_OA, which is a 5S rRNA-derived A. oryzae SINE (Table 1 and S4 Fig).
Pairwise alignment of the AfuSINE3 sequences with 5S rRNAs revealed that the 5’-terminal head regions (positions 1 to 64) are well-preserved and are very similar to the sequences of A. fumigatus 5S rRNA (S4 Fig). Significant similarity was also observed when the 5’-terminal head together with portions of the body and tail of the AfuSINE3 sequences were aligned with A. oryzae SINE3-1_AO (S4 Fig) which in turn is 50–65% homologous and 60–75%% similar to 5S rRNA gene sequences. With at least 60% similarity to the 5S rRNA species and a 60-nucleotide overlap, these observations suggests that all AfuSINE3 sequences are indeed derived from 5S rRNA.
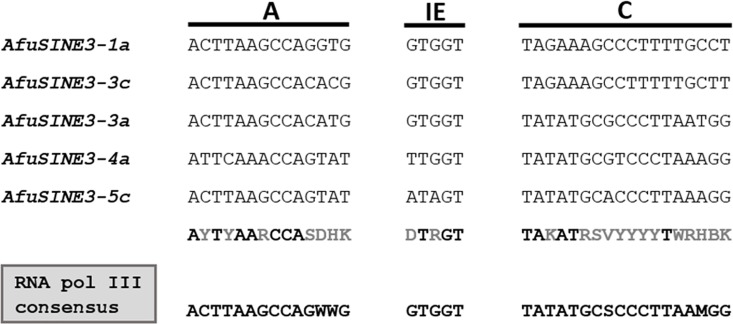
A comparison of the five AfuSINE3 sequences (positions 1 to 119) revealed similar conservation at the 5’-terminal head sequences when some parts of the body regions were included (S6 Fig). However excluding the upstream sequence resulted in significant similarity only being found at the beginning (24 nt) of the 3’-terminal divergent body sequence of the five AfuSINE3 candidates (S7 Fig). Regions of three type I RNA pol III domains (A, IE and C boxes) were poorly conserved in AfuSINE3 as compared to typical 5S rRNAs (Fig 2 and S8 Fig). Additionally, the AfuSINE3 sequences lack a termination signal for RNA pol III transcription (GCTTTTCG) apart from AfuSINE3-1a. This feature may result in continuous transcription activity extending the 5S rRNA-related region towards the 3’ termini of the AfuSINE3 sequences.
Fig 2. Consensus sequences of A, IE and C box promoters of type I RNA pol III in 5S rRNA-derived AfuSINE sequences (AfuSINE3s).
Phylogenetic analysis showed that AfuSINE3-1a and AfuSINE3-3c cluster in the same clade as SINE3-1_OA (S9 Fig). Two main considerations used to define an AfuSINE family in this study are 1) their same common origin and 2) their same sequence module/structure. However, the terminal (tail) sequences of some familial SINE sequences were variable and thus these were omitted from the AfuSINE phylogram. From these criteria and the alignment of the AfuSINE3 candidate sequences were considered to have a common origin since the alignment showed similarities in the 5’-terminal head and some parts of the body regions. Phylogenetic analysis showed similar results, revealing all five predicted AfuSINE3 sequences grouped into two closely related, distinct lineages. Thus, it can be inferred that AfuSINE3 sequences originate from the same gene.
tRNA-related SINE sequences (AfuSINE2)
Eight tRNA-derived SINE-like sequences (AfuSINE2) originating from different tRNAs were identified which ranged in size from 140–493 bp. In order to identify internal RNA pol III promoter sequences (A and B boxes) with a 10–11 bp consensus sequence, the 5’-terminal regions of the AfuSINE2 sequences were aligned with A. fumigatus Af293 tRNA sequences. However, both the AfuSINE2 RNA pol III A and B box promoter sequences were degenerate with the former more degenerate than the latter.
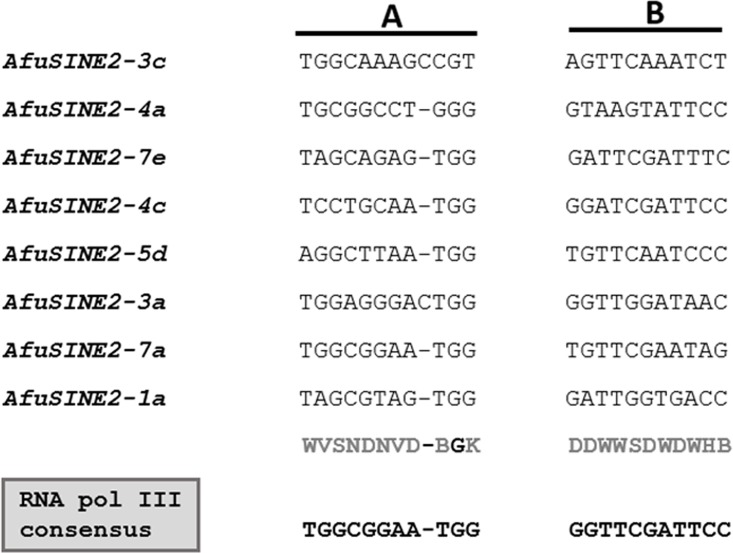
Consensus sequences of the RNA pol III promoter A and B boxes of tRNA-derived AfuSINE sequences are shown in Fig 3. The 4–14 bp long TSD repeat sequences (a characteristic feature of SINE genomic insertions) were partially degraded in some AfuSINE2 sequences with 1 or 2 residues missing e.g. in AfuSINE2-3c and AfuSINE2-5d. This degradation phenomenon might reflect the age of the SINE and concern nucleotide substitution over many generations, over an extended period of time [17]. As a consequence it’s possible that AfuSINE2-1a might have emerged more recently that the other AfuSINE2 sequences since it possessed intact direct repeats. It has been documented that SINE insertion into the genome usually produces short 5–8 bp TSDs at the 5’- and 3’-termini of the sequence. However if SINE insertion is very ancient, TSD identification might be difficult due to mutations in the sequence. Also a TC motif located immediately upstream of the A/T-rich tail and an additional B box (B’ box) downstream of the RNA pol III promoter were absent from most of the AfuSINE2 sequences except AfuSINE2-1a which contained an additional B’ box GGTTCGATTCC sequences 20 bp downstream of the B box. Interestingly this feature has been reported in some tRNA-related VES SINEs, P.k. SINEs in bats and SINE B1 in rodents [37, 38, 39].
Fig 3. Consensus A and B box promoter sequences of RNA pol III in tRNA-derived AfuSINE sequences (AfuSINE2s).
Additionally, none of the 5’-terminal regions of the AfuSINE2 sequences could be folded into tRNA-like cloverleaf structures, which is in contrast to the MgSINE and the MIR SINE sequences [10, 40].
No significant similarity was observed among the predicted AfuSINE2 sequences and they appear to originate from different tRNA-related sequences (S10 Fig). The ancestral tRNA of each AfuSINE2 was identified using the tRNAscan-SE and multiple sequence alignment of the predicted tRNA sequences. The results indicated that the five AfuSINE2 sequences are similar in sequence to the 5’-tRNA-related regions of respectively tRNAAla, tRNAArg, tRNAGly, tRNAMet, tRNAThrand tRNASer. Phylogenetic analysis revealed that the predicted AfuSINE2 sequences do not group into distinct lineages as the bootstrap supports are too low to statistically support any of the nodes (S11 Fig).
Similarity of AfuSINE Sequences to Retrotransposons
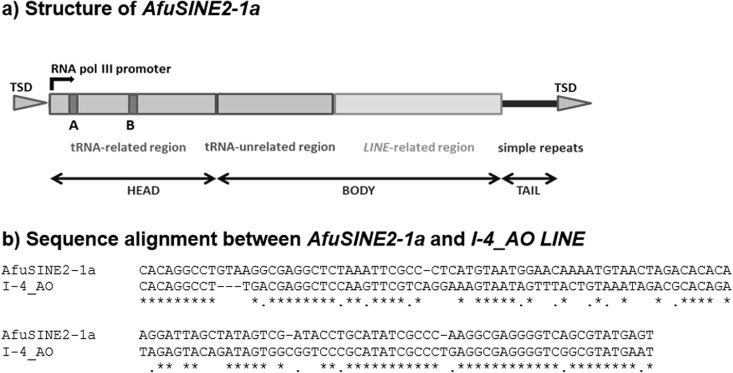
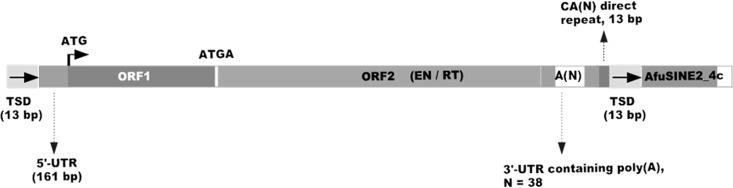
Interestingly the AfuSINE2-1a 3’-terminal sequence was similar to that of LINE, I-4_AO#LINE/Tad1, identified in A. oryzae [20, 21] following CENSOR analysis (Fig 4). Subsequently, the element was aligned manually with retrotransposons found in the A. fumigatus Af293 genome such as a retrotransposon-like element (Afut1-LTR; [41]) and a non-LTR retrotransposon (I-1_AF; [20, 21]). The analysis revealed that the 3’-terminus of the AfuSINE2-1a at positions 210–335 appears similar to the 3’-untranslated region (UTR) of the I-1_AF LINE-like sequence which terminates the RT gene in ORF2. It has been proposed that sequence and structural similarity of the 3’-terminal region of tRNA-derived SINE sequence with a corresponding LINE sequence is crucial for its retrotransposition [42]. Since AfuSINEs are non-autonomous retrotransposons with no gene coding capability, the elements may need to exploit the action of other retrotransposons (such as LINEs) for their amplification and insertion into the genome. Thus, the presence and transcription activity of retrotransposons such as I-1_AF LINE sequences could signify the occurrence and activity of AfuSINE2-1a. Additionally, it was noted that the 3’-terminus of AfuSINE2-1a terminates with a series of short repetitive ACT trinucleotide sequences.
Fig 4. Structure of the AfuSINE2-1a tRNA-derived SINE.
Structure of the AfuSINE2-1a tRNA-derived SINE found in A. fumigatus Af293 (a) and the alignment of its 3'-terminus which is related to I-4_AO LINE (b).
Distribution and Location of AfuSINE Sequences
The distribution and location of AfuSINE sequences on the eight chromosomes of the A. fumigatus Af293 genome were investigated by a comparative analysis of the sequences. Computational analyses revealed that AfuSINE sequences are not abundant in the fungal genome (Fig 5). AfuSINE sequences are dispersed on chromosomes 1, 3, 4, 5 and 7, and are more abundant on chromosomes 3 and 4. The elements are randomly dispersed in pericentromeric and subtelomeric regions on the chromosomes and inserted within gene-rich regions, normally in intergenic regions or close to coding regions similar to LLEs [22]. These observations suggest that the insertion of SINE sequences and LLE sequences in A. fumigatus chromosomes may be linked.
Fig 5. Mapping of SINE-like sequences of the A. fumigatus Af293 genome.
Mapping of SINE-like sequences on eight chromosomes of the A. fumigatus Af293 genome including previously described LINE-like sequences (LLEs; [22]). Chromosome numbers are shown on the left and the size of each chromosome is shown on the right.
Interestingly, AfuSINE2_4c on chromosome 4 appears to be located very close to a LINE-like retrotransposable sequence (LLE#4_3.0); a subclass previously identified in the A. fumigatus Af293 genome [22]. The LLE#4_3.0 is a non-LTR retrotransposon from I clade which is flanked by 13 bp TSDs and contains two overlapping ORFs (open reading frames) ORF1 and ORF2. ORF1 encodes a 413-aa DNA/RNA-binding protein (pos. 175–1669) and ORF2 encodes a 1273-aa polyprotein (pos. 1666–5487) encoding an EN and a RT. The insertion of AfuSINE2_4c next to the RT of LLE#4_3.0 LINE-like element (Fig 6) suggests that activity of AfuSINE2_4c possibly relies on this LINE. However, AfuSINE2_4c is transcriptionally inactive possibly because of inactivation of the LLE#4_3.0 RT domain by mutation. Since most of the LLE sequences identified in the genome of A. fumigatus Af293 are not intact, this could potentially contribute to loss of transcription and retrotransposition activities for some AfuSINE sequences.
Fig 6. Structure of LINE-like element.
Structure of LINE-like element (LLE#4_3.0; [22]) on A. fumigatus Af293 chromosome 4 showing insertion of the AfuSINE2_4c sequence next to the LLE RT.
Transcription Activity of AfuSINE Sequences in the Genome by RT-PCR
Initially PCR amplification was performed using genomic DNA as a template under high stringency PCR conditions. The results of these experiments showed the expected amplicons for the thirteen candidate AfuSINE sequences (S12 Fig, lane 1 throughout) proving the existence of the sequences on the A. fumigatus Af293 genome and confirming that the genomic information in the database is correct. Additionally, a ladder of DNA amplicons of increasing size was not found following PCR amplification indicating that AfuSINE sequences are not present as an array on the genome.
Subsequently, RT-PCR amplification was performed using total RNA with the same sets of primers to investigate the production of AfuSINE transcripts. The results illustrated that seven out of thirteen AfuSINEs are transcriptionally active (viz. AfuSINE3-1a, AfuSINE3-3c, AfuSINE2-1a, AfuSINE2-3a, AfuSINE2-3c, AfuSINE2-4a and AfuSINE2-5d; S12 Fig, lane 2 throughout). No amplicons were observed from the cDNA (-RT) samples, indicating that contaminating genomic DNA was absent from the reaction mixtures for cDNA synthesis (S12 Fig, lane 3 throughout). All RT-PCR amplicons were sequenced to confirm their identity. These results illustrated that the levels of transcription of the AfuSINEs are very low as compared to the β-tubulin gene which is a constitutively expressed house-keeping gene. Additionally RT-PCR analysis clearly demonstrated that not all of the AfuSINE sequences in the A. fumigatus Af293 genome are transcribed.
Estimation of AfuSINE Copy Number in the A. fumigatus Af293 Genome
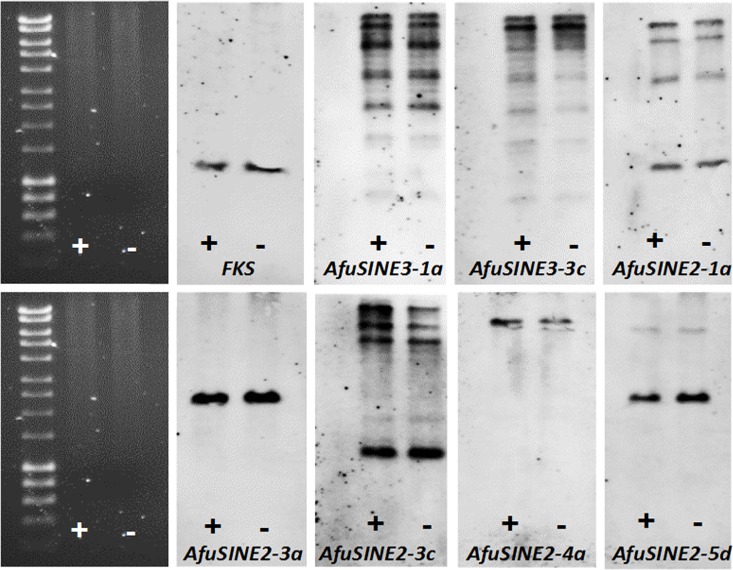
Only actively transcribed SINE sequences viz. AfuSINE3-1a, AfuSINE3-3c, AfuSINE2-1a, AfuSINE2-3a, AfuSINE2-3c, AfuSINE2-4a and AfuSINE2-5d were investigated in this study. The copy numbers and transposition of the seven AfuSINE sequences above per genome of two isogenic A. fumigatus Af293 lines, one infected with AfuTmV-1 and one virus-free (NK125; [31]) were examined by Southern blot hybridization. A. fumigatus Af293 genomic DNA was digested with HindIII which does not restrict the AfuSINE sequences, thus it can be assumed that any single hybridizing band observed corresponds to one copy of each AfuSINE. Southern analysis of the AfuSINE2 sequences showed strong, single hybridization signals for the AfuSINE2-3a, AfuSINE2-4a and AfuSINE2-5d sequences indicating that these elements are present as single copies in the genome (Fig 7). Four or more strong hybridization signals were detected with AfuSINE2-1a and AfuSINE2-3c, indicating that at least four copies of these elements are present in the genome. At least five strong hybridization signals were detected for both AfuSINE3-1a and AfuSINE3-3c, indicating several copies of each AfuSINE3 sequence dispersed in the genome. The A. fumigatus fks gene, which is present as a single copy, was used as a control in these experiments.
Fig 7. Southern blot hybridization of AfuSINE sequences in the genome of A. fumigatus Af293.
Southern blot hybridization of AfuSINE sequences in the genome of A. fumigatus Af293 (virus-infected strain; indicated by plus sign) and A. fumigatus NK125 (virus-free strain; indicated by minus sign). HindIII-digested DNA of each strain was separated in 1% agarose gels in 1xTAE, denatured and blotted onto nylon membrane. Hyperladder 1 (M; 10 kbp; Bioline) was used as marker.
These results indicate that SINE sequences in A. fumigatus are present in very low copy number. For instance AfuSINE3 sequences are present as ca. five copies which is in contrast to other eukaryotic SINE families where ca. 104 copies of SINE3 types are present in the zebra fish genome [8] and in M. grisea the Mg-SINE where the copy number was estimated to be ca. 100 [10]. Because of the known and significant divergence in the Af293 SINE sequences and potential insertions and/or deletions within them it is likely that the copy numbers estimated here are underestimates and represent the minimum copy numbers for each element. It is also likely that A. fumigatus SINE RNA transcripts are silenced since Aspergilli encode all of the enzymes required for the RNA silencing process and are active in vivo [43]. As the number of repetitive elements in A. fumigatus are limited, this could contribute to the small size of the A. fumigatus genome (29.4 Megabases) as compared to for instance the oomycete P. infestans genome which consists of a large proportion of TEs and repeats contributing to a genome size of 240 Megabases [17]. Thus, SINE abundance is clearly diverse and variable between different fungal and oomycete species. Low copy number non-LTR retrotransposons with degenerate sequences are likely to be lost from the genome as a result of genetic drift and natural selection [44].
Infection of A. fumigatus with mycoviruses causes some effects on retrotransposon activity. For instance in A. fumigatus strain A56, it has been demonstrated recently that chrysovirus infection stimulates LLE mobilization [22]. However infection of A. fumigatus strain Af293 with AfuTmV-1 had no obvious effects on AfuSINE transposition or copy number since both virus-free and virus-infected isogenic lines showed identical hybridization patterns (Fig 7).
Detection of Small RNA Molecules Homologous to AfuSINE Sequences
To confirm AfuSINE regulation and gene silencing in A. fumigatus Af293, small RNAs homologous to AfuSINE sequences were identified following northern blot hybridization. DIG-labeled probes for AfuSINE sequences were in vitro transcribed in both sense and antisense orientation to detect strand-specific small RNAs. In this study, only four representative actively transcribed AfuSINEs (AfuSINE3-1a, AfuSINE2-1a, AfuSINE2-3a and AfuSINE2-4a) were selected for detection of small RNAs.
Autoradiographic analysis showed weak hybridization signals of a band corresponding to <40 nt for the antisense (-) strands of AfuSINE3-1a and AfuSINE2-1a (Fig 1D) while small RNAs homologous to AfuSINE2-3a and AfuSINE2-4a were not detected. Constitutively expressed control β-tubulin probes were also used to detect small RNAs homologous to the β-tubulin gene. No hybridization signals were found in the controls, suggesting that hybridization of small RNAs with AfuSINE probes was not attributable to mRNA degradation.
The presence of small RNAs (<40 nt) homologous to AfuSINE3-1a and AfuSINE2-1a sequences suggests that these elements may be targeted for degradation and silencing in the fungus. However, small RNAs were not detected with AfuSINE2-3a and AfuSINE2-4a suggesting that not all AfuSINEs are targeted by host RNA silencing. In addition, hybridization signals observed from the northern blot analysis are very weak possibly attributable to low transcription activity of the elements resulting in low abundance of siRNAs.
In summary, our study has demonstrated the first computational search for SINE sequences in the A. fumigatus Af293 genomic DNA. Distribution, copy number, transcription activity and silencing of these elements have been described. Further research is required to investigate SINE distribution among other clinical and environmental A. fumigatus isolates and to assess the effects of AfuSINE sequences on adaptability and pathogenicity. Potentially active AfuSINE sequences, for example AfuSINE2-1a, might be further developed as a reverse genetic tool. The fact that this element was actively transcribed, possessed intact 14 bp TSDs, a 5’-tRNA-related region corresponding to tRNAArg as reported for most active tRNA-derived SINE [38, 45, 46], sequence similarity to the I-1_AF LINE-like element and was a target for host silencing illustrates its potential.
Supporting Information
(TIF)
Predicted secondary structure was performed using Mfold program (http://mfold.rna.albany.edu/?q=mfold) (Zuker, 2003).
(TIF)
Predicted secondary structure was performed using Mfold program (http://mfold.rna.albany.edu/?q=mfold) (Zuker, 2003).
(TIF)
The alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The 5S rRNA-related and part of the body regions of each sequence were selected for the alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The 5S rRNA-unrelated regions of each sequence were selected for the alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The A, IE, and C boxes, which constitute the type 1 pol III promoter, are aligned and highlighted in grey.
(TIF)
The 5S rRNA-related regions (nt 1–119) of each sequence were selected for the alignment. A phylogenetic tree was constructed using the fast Fourier transform MAFFT program L9INS-1(2). A bootstrap test was conducted with 1,000 resamplings for the neighbor-joining trees. Numbers on the nodes indicate percentage of bootstrap support from 1,000 replicates with branch lengths indicated.
(TIF)
The tRNA-related region (nt 1–72) of each sequence was selected for alignment which was performed using the MAFFT with L-INS-i parameter. Potential A and B boxes are highlighted in grey.
(TIF)
The tRNA-related region (nt 1–72) of each sequence was selected for the alignment. A phylogenetic tree was constructed using the fast Fourier transform MAFFT program L9INS-1(2). A bootstrap test was conducted with 1,000 resamplings for the neighbor-joining trees. Numbers on the nodes indicate percentage of bootstrap support from 1,000 replicates with branch lengths indicated. MIRc is a SINE2/tRNA (Pro) from mammals, Foxy is a tRNA-derived SINE from Fusarium oxysporum f.sp. lycopersici, SINE2-1_BG is a SINE2/tRNA (Gly) from barley powdery mildew Blumeria graminis, MgSINE is a tRNA-derive SINE from Magnaporthe grisea, and LFSINE_Vert is a SINE2/tRNA (His) from Latimeria.
(TIF)
Agarose gel electrophoresis showing the PCR and RT-PCR products of the 13 candidate AfuSINE sequences; Lane 1 for each AfuSINE shows PCR amplicons generated from genomic DNA; Lane 2 shows for each AfuSINE, amplicons generated following RT-PCR; Lane 3 shows for each AfuSINE, amplicons generated from (-RT) negative controls RT-PCR. Hyperladder 1 (M; 10 kbp; Bioline) and Quick-Load® 100 bp DNA Ladder (NEB) were used as markers. Electrophoretic analysis was performed in 2.5% agarose gels for 3 h at 80 V.
(TIF)
(PDF)
Acknowledgments
We would like to thank Dr Elaine Bignell for supplying us with the Aspergillus fumigatus Af293 isolate. We also thank Dr Stephen C. Whisson at the James Hutton Institute and Dr Michael Tristem at the Department of Life Sciences, Imperial College London for their valuable advice, comments and helpful discussions. LK thanks the Royal Thai Government for supporting her PhD program and RHAC thanks The Leverhulme Trust for an Emeritus Fellowship.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The authors received no specific funding for this work.
References
- 1.Bradshaw VA and McEntee K. DNA damage activates transcription and transposition of yeast Ty retrotransposons. Mol Gen Genet. 1989; 218: 465–474. [DOI] [PubMed] [Google Scholar]
- 2.Paquin CE and Williams VM. In: Eukaryotic transposable elements as mutagenic agents, Banbury Report 30, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY; 1988.
- 3.Wessler SR. Turned on by stress. Plant retrotransposons. Curr Biol. 1996; 6: 959–961. [DOI] [PubMed] [Google Scholar]
- 4.Grandbastien MA. Activation of plant retrotransposons under stress conditions. Trends Plant Sci. 1998; 3: 181–187. [Google Scholar]
- 5.Capy P, Gasperi G, Biémont C and Bazin C. Stress and transposable elements: Co-evolution or useful parasites? Heredity. 2000; 85: 101–106. [DOI] [PubMed] [Google Scholar]
- 6.Kramerov DA and Vassetzky NS. SINEs. WIRs:RNA. 2011; 2(6): 772–786. [DOI] [PubMed] [Google Scholar]
- 7.Mes JJ, Haring MA and Cornelissen BJ. Foxy: an active family of short interspersed nuclear elements from Fusarium oxysporum. Mol Gen Genet. 2000; 263: 271–280. [DOI] [PubMed] [Google Scholar]
- 8.Kapitonov VV and Jurka J. A novel class of SINE elements derived from 5S RNA. Mol Biol Evol. 2003; 20: 694–702. 10.1093/molbev/msg075 [DOI] [PubMed] [Google Scholar]
- 9.Kramerov DA and Vassetzky NS. Origin and evolution of SINEs in eukaryotic genomes. Heredity. 2011; 107: 487–495. 10.1038/hdy.2011.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kachroo P, Leong SA and Chatto BB. Mg-SINE: A short interspersed nuclear element from the rice blast fungus, Magnaporthe grisea. Proc Natl Acad Sci USA. 1995; 92: 11125–11129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sone T, Suto M and Tomita F. Host species-specific repetitive DNA sequence in the genome of Magnaporthe grisea, the rice blast fungus. Biosci Biotechnol Biochem. 1993; 57: 1228–1230. 10.1271/bbb.57.1228 [DOI] [PubMed] [Google Scholar]
- 12.Wei YD, Collinge DB, Smeregaard-Peterson V and Thordal-Christensen H. Characterization of the transcript of a new class of retroposon-type repetitive element cloned from the powdery mildew fungus Erysiphe graminis. Mol Gen Genet. 1996; 250: 477–482. [DOI] [PubMed] [Google Scholar]
- 13.Rasmussen M, Rossen L and Giese H. SINE-like properties of a highly repetitive element in the genome of the obligateparasitic fungus Erysiphe graminis f. sp. hordei. Mol Gen Genet. 1993; 239: 298–303. [DOI] [PubMed] [Google Scholar]
- 14.Kim HG, Meinhardt LW, Benny U and Kistler HC. NrsI, a repetitive element linked to pisatin demethylase genes on a dispensable chromosome of Nectria haematococca. Mol Plant Microbe Interactions. 1995; 8: 524–531. [DOI] [PubMed] [Google Scholar]
- 15.Spanu PD, Abbott JC, Amsalem J, Burgis TA, Soanes DM, Stuber K, et al. Genome expansion and gene loss in powdery mildew fungi reveal trade-offs in extreme parasitism. Science. 2010; 330: 1543–1546. 10.1126/science.1194573 [DOI] [PubMed] [Google Scholar]
- 16.Bao W and Jurka J. SINEs from barley powdery mildew. Repbase Reports. 2011; 11(9): 2582–2582. [Google Scholar]
- 17.Whisson SC, Avrova AO, Lavrova O and Pritchard L. Families of short interspersed elements in the genome of the oomycete plant pathogen, Phytophthora infestans. Fungal Genet Biol. 2005; 42: 351–365. 10.1016/j.fgb.2005.01.004 [DOI] [PubMed] [Google Scholar]
- 18.O’Gorman CM, Fuller HT and Dyer PS. Discovery of a sexual cycle in the opportunistic fungal pathogen Aspergillus fumigatus. Nature. 2009; 457: 471–474. 10.1038/nature07528 [DOI] [PubMed] [Google Scholar]
- 19.Latgé JP. Aspergillus fumigatus and Aspergillosis. Clin Microbio. Rev. 1999; 12(2): 310–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Galagan JE, Calvo SE, Cuomo C, Ma LJ, Wortman JR, Batzoglou S, et al. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature. 2005; 438: 1105–1115. 10.1038/nature04341 [DOI] [PubMed] [Google Scholar]
- 21.Kapitonov VV and Jurka J. SINE3-1_AO, a family of 5S rRNA-derived nonautonomous non-LTR retrotransposons in the Aspergillus oryzae genome. Repbase Reports. 2006; 6: 45–45. [Google Scholar]
- 22.Huber F and Bignell E. Distribution, expression and expansion of Aspergillus fumigatus LINE-like retrotransposon populations in clinical and environmental isolates. Fungal Genet Biol. 2014; 64: 36–44. 10.1016/j.fgb.2014.01.002 [DOI] [PubMed] [Google Scholar]
- 23.Mabey JE, Anderson MJ, Giles PF, Miller CJ, Attwood TK, Paton NW, et al. CADRE: the central Aspergillus data Repository 2012. Nucleic Acids Res. 2012; 40: D660–6. 10.1093/nar/gkr971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnaud MB, Chibucos MC, Costanzo MC, Crabtree J, Inglis DO, Lotia A, et al. The Aspergillus Genome Database, a curated comparative genomics resource for gene, protein and sequence information for the Aspergillus research community. Nucleic Acids Res. 2010; 38: D420–7. 10.1093/nar/gkp751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohany O, Gentles AJ, Hankus L and Jurka J. Annotation, submission and screening of repetitive elements in Repbase: Repbase Submittor and Censor. BMC Bioinformatics. 2006; 7: 474 10.1186/1471-2105-7-474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smit AFA, Hubley R and Green P. RepeatMasker Open-4.0. 2013–2015. Available: www.repeatmasker.org
- 27.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends in Genetics. 2000; 16: 418–420. [DOI] [PubMed] [Google Scholar]
- 28.Lawe TM and Eddy SR. tRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1997; 25: 955–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McWilliam H, Li W, Uludag M, Squizzato S, Park YM, Buso N, et al. Analysis tool web services from the EMBL-EBI. Nucleic Acids Res. 2013; 41 (Web Server issue):W597–600. 10.1093/nar/gkt376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K and Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013; 30(4): 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kanhayuwa L, Kotta-Luizou I, Ozkan S, Gunning AP and Coutts RHA. A novel mycovirus from Aspergillus fumigatus contains four unique dsRNAs as its genome and is infectious as dsRNA. Proc Natl Acad Sci USA. 2015; 112: 9100–9105. 10.1073/pnas.1419225112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pontecorvo G, Roper JA, Hemmons LM, Macdonald KD and Bufton AW. The genetics of Aspergillus nidulans. Adv Genet. 1953; 5: 141–238. [DOI] [PubMed] [Google Scholar]
- 33.Lu C, Meyers BC and Green PJ. Construction of small RNA cDNA libraries for deep sequencing. Methods. 2007; 43: 110–117. 10.1016/j.ymeth.2007.05.002 [DOI] [PubMed] [Google Scholar]
- 34.Pall GS, Codony-Servat C, Byrne J, Ritchie L and Hamilton A. Carbodiimide-mediated cross-linking of RNA to nylon membrane improves the detection of siRNA, miRNA and piRNA by northern blot. Nucleic Acids Res. 2007; 35: e60, 10.1093/nar/gkm112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kreuze JF, Savenkov EI, Cuellar W, Li X and Valkonen JPT. Viral class 1 RNase III involved in suppression of RNA silencing. J Virol. 2005; 79: 7227–7238. 10.1128/JVI.79.11.7227-7238.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003; 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borodulina OR and Kramerov D A. Wide distribution of short interspersed elements among eukaryotic genomes. FEBS Letters. 1999; 457: 409–413. [DOI] [PubMed] [Google Scholar]
- 38.Fantaccione S, Woodrow P and Pontecorvo G. Identification of a family of SINEs and LINEs in the Pipistrellus kuhli genome: A new structure and functional symbiotic relationship. Genomics. 2008; 91: 178–185. 10.1016/j.ygeno.2007.10.008 [DOI] [PubMed] [Google Scholar]
- 39.Koval AP, Veniaminova NA and Kramerov DA. Additional B box of RNA polymerase III promoter in SINE B1 can be functional. Gene. 2011; 487(2): 113–117. 10.1016/j.gene.2011.08.001 [DOI] [PubMed] [Google Scholar]
- 40.Smit AF and Riggs AD. MIRs are classic, tRNA‐derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995; 23: 98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Neuveglise C, Sarfati J, Latge JP and Paris S. Afut1, a retrotransposon-like element from Aspergillus fumigatus. Nucleic Acids Res. 1996; 24: 1428–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ogiwara I, Miya M, Ohshima K and Okada N. Retropositional parasitism of SINEs on LINEs: Identification of SINEs and LINEs in Elasmobranchs.Mol Biol Evol. 1999; 16(9): 1238–1250. [DOI] [PubMed] [Google Scholar]
- 43.Hammond TM, Andrewski MD, Roossinck MJ and Keller NP. Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryot Cell. 2008; 7(2): 350–357. 10.1128/EC.00356-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brookfield JF and Badge RM. Population genetics models of transposable elements. Genetica. 1997; 100: 281–294. [PubMed] [Google Scholar]
- 45.Daniels GR and Deininger PL. A second major class of Alu family repeated DNA sequences in a primate genome. Nucleic Acids Res. 1983; 11: 7595–7610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fantaccione S, Russo C, Palomba P, Rienzo M and Pontecorvo G. A new pair of CR1-like LINE and tRNA-derived SINE in Podarcis sicula genome. Gene. 2004; 339: 189–198. 10.1016/j.gene.2004.06.051 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TIF)
Predicted secondary structure was performed using Mfold program (http://mfold.rna.albany.edu/?q=mfold) (Zuker, 2003).
(TIF)
Predicted secondary structure was performed using Mfold program (http://mfold.rna.albany.edu/?q=mfold) (Zuker, 2003).
(TIF)
The alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The 5S rRNA-related and part of the body regions of each sequence were selected for the alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The 5S rRNA-unrelated regions of each sequence were selected for the alignment which was performed using the Clustal Omega program available at the EMBL-EBI website.
(TIF)
The A, IE, and C boxes, which constitute the type 1 pol III promoter, are aligned and highlighted in grey.
(TIF)
The 5S rRNA-related regions (nt 1–119) of each sequence were selected for the alignment. A phylogenetic tree was constructed using the fast Fourier transform MAFFT program L9INS-1(2). A bootstrap test was conducted with 1,000 resamplings for the neighbor-joining trees. Numbers on the nodes indicate percentage of bootstrap support from 1,000 replicates with branch lengths indicated.
(TIF)
The tRNA-related region (nt 1–72) of each sequence was selected for alignment which was performed using the MAFFT with L-INS-i parameter. Potential A and B boxes are highlighted in grey.
(TIF)
The tRNA-related region (nt 1–72) of each sequence was selected for the alignment. A phylogenetic tree was constructed using the fast Fourier transform MAFFT program L9INS-1(2). A bootstrap test was conducted with 1,000 resamplings for the neighbor-joining trees. Numbers on the nodes indicate percentage of bootstrap support from 1,000 replicates with branch lengths indicated. MIRc is a SINE2/tRNA (Pro) from mammals, Foxy is a tRNA-derived SINE from Fusarium oxysporum f.sp. lycopersici, SINE2-1_BG is a SINE2/tRNA (Gly) from barley powdery mildew Blumeria graminis, MgSINE is a tRNA-derive SINE from Magnaporthe grisea, and LFSINE_Vert is a SINE2/tRNA (His) from Latimeria.
(TIF)
Agarose gel electrophoresis showing the PCR and RT-PCR products of the 13 candidate AfuSINE sequences; Lane 1 for each AfuSINE shows PCR amplicons generated from genomic DNA; Lane 2 shows for each AfuSINE, amplicons generated following RT-PCR; Lane 3 shows for each AfuSINE, amplicons generated from (-RT) negative controls RT-PCR. Hyperladder 1 (M; 10 kbp; Bioline) and Quick-Load® 100 bp DNA Ladder (NEB) were used as markers. Electrophoretic analysis was performed in 2.5% agarose gels for 3 h at 80 V.
(TIF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.