Summary
A major hurdle for in vitro culturing of primary endothelial cells (ECs) is that they readily dedifferentiate, hampering their use for therapeutic applications. Human embryonic stem cells (hESCs) may provide an unlimited cell source; however, most current protocols deriving endothelial progenitor cells (EPCs) from hESCs use direct differentiation approaches albeit on undefined matrices, yet final yields are insufficient. We developed a method to culture monolayer hESCs on stem cell niche laminin (LN) LN511 or LN521 matrix. Here, we report a chemically defined, xeno-free protocol for differentiation of hESCs to EPCs using LN521 as the main culture substrate. We were able to generate ∼95% functional EPCs defined as VEGFR2+CD34+CD31+VE-Cadherin+. RNA-sequencing analyses of hESCs, EPCs, and primary human umbilical vein endothelial cells showed differentiation-related EC expression signatures, regarding basement membrane composition, cell-matrix interactions, and changes in endothelial lineage markers. Our results may facilitate production of stable ECs for the treatment of vascular diseases and in vitro cell modeling.
Highlights
-
•
Differentiation of hESCs to EPCs in chemically defined and xeno-free systems
-
•
Laminin 521 as the main substrate for differentiation
-
•
EPCs are functional and express both progenitor and mature endothelial markers
-
•
Six-week continuous culturing allows them to mature further, relative to HUVECs
Tryggvason and colleagues have developed a protocol to differentiate hESCs to endothelial progenitor cells (EPCs) on human recombinant laminin matrices in a chemically defined and xeno-free environment. These hESC-derived cells are functional and express specific markers in the endothelial lineage, holding promise to provide a pure, unlimited source of EPCs for therapeutic purposes and in vitro disease modeling.
Introduction
Human pluripotent stem cells (hPSCs) have the ability to proliferate indefinitely and the potential to differentiate into any somatic cell type in the human body (Takahashi et al., 2007, Thomson et al., 1998). They can be a great resource for the development of cell transfer-based therapy of diseases and injuries, such as myocardial infarction and heart failure (reviewed in Sanganalmath and Bolli, 2013), Parkinson's disease (Kriks et al., 2011), and hepatic injury (Woo et al., 2012). In addition, many in vitro models mimicking human systems, such as the blood-brain barrier or glomerular filtration barrier, would require a large quantity of cells with stable phenotypes for testing novel drugs and to understand their mechanisms of action (Kinney et al., 2014). In this regard, hPSC-derived cells are a preferable source for cell therapies compared with primary cells isolated directly from human tissues, as they readily dedifferentiate and senesce in vitro. It is, therefore, of utmost importance to develop chemically defined, xeno-free, and reproducible strategies for directing differentiation of hPSCs to various somatic cell lineages with a high yield of pure populations.
As methods for derivation and expansion of hPSCs have largely become established, there has been a growing interest in the development of differentiation protocols to generate endothelial cell lineages that could be used for clinical applications, such as engineered blood vessels (Wang et al., 2007), vascular graft coatings (Campagnolo et al., 2015), or co-implantation with cardiomyocytes to treat myocardial infarction (Sahara et al., 2014). As far as human cell therapy applications are concerned, it is important to generate such cells in defined and xeno-free conditions. The three most common methods used to differentiate hPSCs into endothelial lineage involve the formation of embryoid bodies (Adams et al., 2013, Goldman et al., 2009), irradiated mouse embryonic fibroblasts (MEFs) or human foreskin fibroblast feeder cells (Sahara et al., 2014, Wang et al., 2007), and mouse tumor-derived Matrigel as a supporting matrix for maintenance and differentiation (Lian et al., 2014, Patsch et al., 2015). Embryoid bodies exhibit heterogeneous patterns of differentiated cell lineages and their viability is influenced by size, resulting in a slow and inconsistent differentiation process (Van Winkle et al., 2012). Importantly, maintenance of hPSCs dissociated from embryoid bodies often requires Rho-associated kinase (ROCK) inhibitor to minimize apoptosis (Watanabe et al., 2007). This may potentially enrich for tumorigenic cells and, thus, limit their applications in human therapies. In contrast, the maintenance and differentiation of hPSCs in homogeneous monolayer cultures can overcome drawbacks of the embryoid body system. Nevertheless, culturing hPSCs on the Matrigel matrix, MEFs, or human feeder cells introduces xeno-products and undefined substances. In addition, such systems are prone to batch-to-batch differences and lack of consistency among differentiation protocols.
Our laboratory, together with others, has developed methods that are void of animal components and completely chemically defined for human embryonic stem cell (hESC) derivation, expansion, and maintenance of their self-renewal capacity (Melkoumian et al., 2010, Rodin et al., 2010, Rodin et al., 2014a). We have demonstrated that stem cell niche-specific human laminin (LN) LN511 or LN521 substratum can support pluripotency of hESCs in the absence of ROCK inhibitor (Rodin et al., 2010). Importantly, a combination matrix of LN521 and E-Cadherin fully supports clonal derivation and clonal survival of hESCs (Rodin et al., 2014b). Here, we have explored whether such a system could be applied to the generation of endothelial cell lineage from hESCs.
Immediately beneath all endothelia is a specialized basement membrane (BM) matrix that contains highly tissue- and cell-type-specific trimeric isoforms of laminins that significantly influence cell behaviors, such as cell adhesion, differentiation, and phenotype stability. Each laminin molecule comprises three chains, α, β, and γ, that exist in five, four, and three genetically distinct forms, respectively (Domogatskaya et al., 2012). The laminin isoforms are named after the chain composition (e.g., LN521 consists of α5, β2, and γ1 chains). In mammalian tissues there exist at least 16 laminin isoforms with various degrees of cell specificity. For example, LN511 and LN521 are dominant in embryonic BM and the inner cell mass of the blastocyst (Miner et al., 1998), while LN111 (the Matrigel isoform) is mainly present in Reichert's membrane (Klaffky et al., 2001). Indeed, we have previously shown that human recombinant LN511 or LN521 alone can support long-term self-renewal of mouse ESCs (Domogatskaya et al., 2008) and hESCs in completely defined and xeno-free culture conditions (Rodin et al., 2010, Rodin et al., 2014a). In subendothelial BM of most vessels, the laminins contain α4 and α5 chains, with α4 being expressed in various vessel types independent of developmental stages, while α5, a ubiquitous laminin chain, is expressed in placental vasculature and most capillary BM postnatally (Sorokin et al., 1997). We have shown that mice lacking the α4 chain exhibit hemorrhages during embryonic and neonatal stages as well as impaired microvessel maturation (Thyboll et al., 2002). These mice exhibited excessive filopodia and tip cell formation in the retina, indicating that α4 chain plays a crucial role in regulating tip cell numbers and vascular density (Stenzel et al., 2011). On the other hand, deletion of the α5 chain gene causes embryonic lethality at the later stage, possibly due to placental dysfunction (Miner et al., 1998). This emphasizes the critical developmental roles of laminin chains during embryogenesis and the maintenance of structural integrity in adult tissues.
In this study, we have developed a three-step protocol to differentiate hESCs to endothelial progenitor cells (EPCs) using human recombinant laminins that are biologically relevant (LN521, LN511, and LN421) to mimic the in vivo endothelial substrata. First, vascular cell lineages emerge from mesoderm during embryogenesis (Huber et al., 2004). The next phase directs these mesoderm-committed cells toward the endothelial lineage (CD34+vascular endothelial growth factor receptor [VEGFR2]+CD31+VE-Cadherin+) (Sahara et al., 2014). At the last stage, to obtain a pure population of ECs, a CD31+ population is purified using CD31-coupled magnetic beads. With this three-step protocol, we were able to obtain a 95% CD34+VEGFR2+CD31+VE-Cadherin+ population after 15 days. Using RNA-sequencing (RNA-seq) technology, we compared transcriptomes of our hESC-derived EPCs with that of human umbilical vein endothelial cells (HUVECs), which are fully mature and the most commonly used human primary ECs. The results confirmed expression of markers in the early endothelial lineage, as well as the presence of some mature markers. In short, our differentiation protocol allows efficient generation of pure EPCs in a chemically defined and xeno-free system, which can be applicable for therapeutic purposes as well as modeling human diseases.
Results
Step 1: Mesoderm Induction
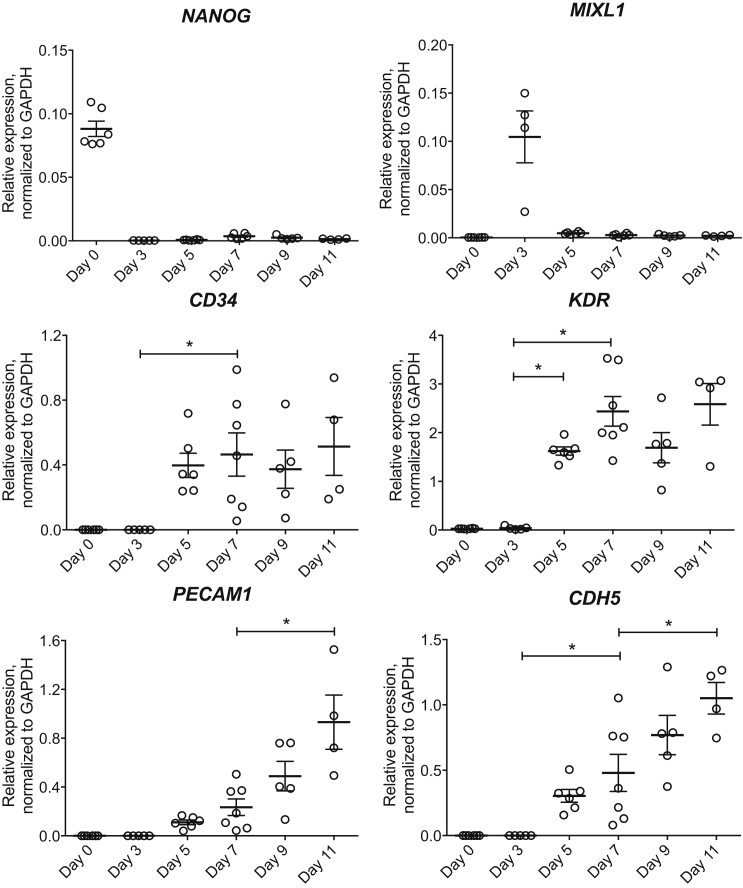
We have previously reported that human recombinant LN511 or LN521 alone can support monolayer culture and self-renewal of hESCs (Rodin et al., 2010, Rodin et al., 2014a). In the present study, hESCs were seeded onto LN521- or LN511-coated plates 2–3 days prior to the start of differentiation (day 0) (Figure 1). Cells collected on day 0 were characterized and their pluripotency confirmed by high mRNA expression of NANOG (Figure 2), 99% Oct3/4+, and 100% TRA-1-60+ and weak expression of VEGFR2 in total cell population (Figure 3A), as reported previously (Sahara et al., 2014). During embryogenesis, endothelial progenitors are derived from a subpopulation of mesoderm (Huber et al., 2004). Several studies have emphasized critical roles of bone morphogenetic protein 4 (BMP4), Activin/Nodal, and Wnt signaling activation via glycogen synthase kinase 3 (GSK3) inhibition in directing hESCs toward the mesodermal lineage (Goldman et al., 2009, Lian et al., 2014, Patsch et al., 2015, Sahara et al., 2014, Sumi et al., 2008). When the cultures reached 40%–50% confluency (Figure 1A), NutriStem was replaced by mesoderm induction medium containing 20 ng/mL BMP4, 10 ng/mL activin A, and 6 μM GSK3β-specific inhibitor CHIR99021. These concentrations were previously optimized to derive ECs from hESCs (James et al., 2010, Lian et al., 2014, Patsch et al., 2015, Sahara et al., 2014). Indeed, after 3 days of mesoderm induction, we observed a drastic change in cell morphology (Figure 1B) and the peak expression of MIXL1 (Figure 2), a transcription factor expressed during mesodermal development. Similar cell morphological changes were observed with HS1001 and HS983A hESC lines (Figure S1).
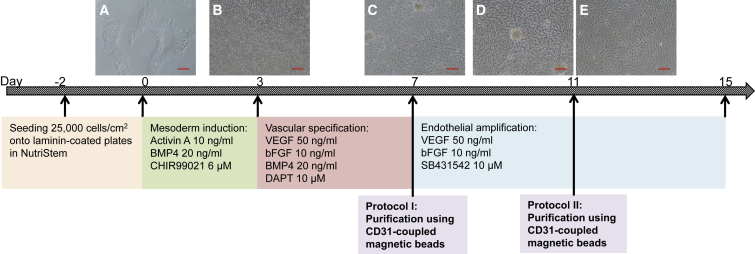
Figure 1.
Differentiation Protocol
(A) hESCs (H1) were seeded on LN521- or LN511-coated plates. Differentiation started on day 0.
(B) Phase 1 (from day 0 to day 3) directed hESCs into mesodermal lineage.
(C) Phase 2 (from day 3 to day 7) induced vascular specification. CD31+ population was purified by MACS beads on day 7 (protocol I) or day 11 (protocol II), and plated onto new LN521- or LN511-coated plates (with and without LN421).
(D) Cell morphology on day 11 of differentiation without purification (phase 3).
(E) Cells were purified on day 7 and continued to expand until day 11 (protocol I). For both protocols, cells were harvested for analysis 4 days after purification (day 11 and day 15, respectively).
Scale bars, 100 μm. See also Figure S1.
Figure 2.
Gene Expression during Differentiation
Gene expression of pluripotency marker (NANOG), mesoderm marker (MIXL1), and endothelial lineage markers (CD34, KDR, PECAM1, and CDH5) was analyzed on days 0, 3, 5, 7, 9, and 11 without purification, normalized to GAPDH. Means ± SEM; each dot represents one independent differentiation batch (n = 4–7). ∗p < 0.05.
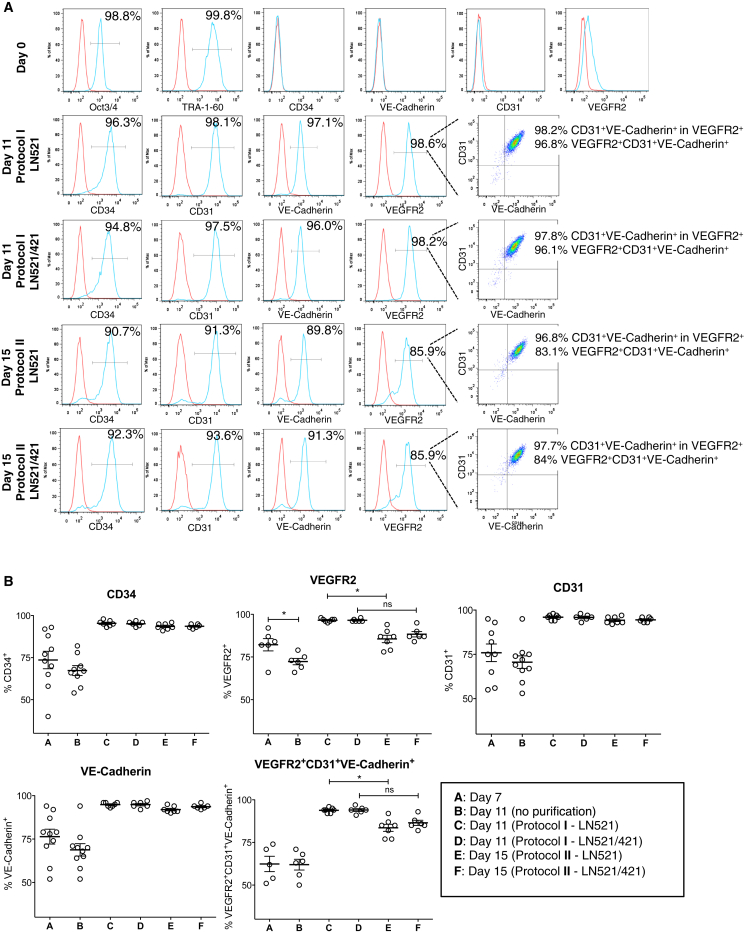
Figure 3.
Flow Cytometry Analysis of Specific Markers during Differentiation
(A) hESCs were immunostained with pluripotency markers Oct3/4 and TRA-1-60, and endothelial lineage markers for antibody specificity testing. In protocol I, we obtained almost 100% pure population of EPCs (CD34+ and VEGFR2+CD31+VE-Cadherin+) on either LN521 or LN521/421 substrate. In protocol II, we observed a decrease in VEGFR2 expression 4 days later while CD31 and VE-Cadherin expression still maintained almost 100%. Red trace, isotype control; blue trace, positive population. y axis represents percentage of maximum count.
(B) Quantification of flow cytometry analysis of endothelial lineage markers. Both protocols significantly improved the yields of CD34+, VEGFR2+, CD31+, and VE-Cadherin+ populations. VEGFR2 expression was significantly reduced from days 7 to 11 (without purification). Similar reduction was observed when CD31+ cells were purified using protocol I versus protocol II on LN521. Means ± SEM; each dot represents one independent differentiation batch (n = 6–10). ∗p < 0.05.
See also Figures S2 and S3.
Step 2: Vascular Specification
To drive these mesoderm-committed cells toward the endothelial lineage, we switched to vascular specification medium on day 3. Sahara et al. (2014) reported that the combination of Notch signaling inhibitor DAPT and VEGF promoted efficient generation of EPCs, defined as VEGFR2highVE-Cadherin+CD34+CD14−. The presence of BMP4 throughout this stage also facilitated the generation of immature ECs (Goldman et al., 2009), while basic fibroblast growth factor (bFGF) was reported to be necessary during vascular specification and endothelial amplification stages (James et al., 2010). After 2 days of incubation in vascular specification medium (day 5), we observed a significant increase in KDR (encoding VEGFR2) mRNA expression versus day 3 (p < 0.001, Figure 2). On day 7, KDR mRNA expression increased further together with significant increases in CD34 and CDH5 (encoding VE-Cadherin) expression, compared with day 3 (p < 0.0001, p < 0.05, and p < 0.05, respectively; Figure 2), suggesting that the majority of the culture contained endothelial progenitors. Flow cytometry analyses of hESC-derived cells collected on day 7 showed 74% CD34+, 82% VEGFR2+, 76% CD31+, 76% VE-Cadherin+, and overall 62% VEGFR2+CD31+VE-Cadherin+ in the total cell population (Figure 3B), indicating that there was a significant population of CD31-expressing cells at this time point with relatively low mRNA expression.
Step 3: Endothelial Amplification
From day 7 on, hESC-derived cells were maintained in endothelial amplification medium, containing SB431542, an inhibitor of transforming growth factor β (TGF-β) signaling, together with VEGF and bFGF. TGF-β inhibition has been suggested to maintain the vascular-committed state following specification and to prevent the loss of endothelial identity, thus promoting the expansion of pure populations of hESC-derived ECs (James et al., 2010). To obtain a pure population of ECs and remove cell clumps (Figures 1C and 1D), we tested two different protocols: purification of CD31+ cells on day 7 (protocol I) and on day 11 (protocol II). As suggested from the mRNA expression profile, cells on day 11 (without purification) expressed significantly higher EC markers PECAM1 (encoding CD31) and CDH5 mRNA levels, compared with day 7 (p < 0.0001 and p < 0.01, respectively; Figure 2). Therefore, we purified cells at these two time points to compare the final yields between purified cultures from endothelial progenitors (protocol I) and from more mature cells (protocol II). Four days after purification, 4.25 EPCs from protocol I and 5.03 EPCs from protocol II were generated from every one hESC plated. To study the role of laminin α4 subunit in maintaining the phenotypes of the derived EPCs, we plated CD31+-purified cells on LN521 (or LN511) with or without LN421 substrata in both protocols. Figure 3A demonstrated typical fluorescence-activated cell sorting (FACS) analysis data on comparing protocol I (LN521 and LN521/421) with protocol II (LN521 and LN521/421). Figure 3B summarizes final yields of each protocol by FACS analyses. The VEGFR2+ population was reduced significantly after 4 days in endothelial amplification medium (days 7 through 11, without purification), suggesting the maturation progress in endothelial lineage. We did not observe significant differences in CD31+ or VE-Cadherin+ populations by FACS between days 7 and 11 even though their mRNA levels were increased during endothelial amplification, also indicating the progression toward maturity of hESC-derived cells in our cultures.
Both purification protocols have significantly improved the final yield of EPCs. Specifically, in protocol I, on both LN521 and LN521/421, we have consistently obtained 97% VEGFR2+, 95% CD34+, 96% CD31+, and 95% VE-Cadherin+ populations, whereby 94% of the final culture was triple-positive for VEGFR2, CD31, and VE-Cadherin (Figures 3A and 3B). In protocol II, 4 days after purification, the hESC-derived cultures contained 94% CD34+, 95% CD31+, and 93% VE-Cadherin+ populations, without any significant differences compared with those in protocol I. Interestingly, the VEGFR2+ population decreased to 86% (on LN521) and 88% (on LN521/421). As a result, the triple-positive populations of VEGFR2, CD31, and VE-Cadherin were 84% (on LN521) and 87% (on LN521/421). One-way ANOVA analysis revealed a significantly lower VEGFR2+ population in protocol II compared with protocol I on LN521 alone; in consequence, the VEGFR2+CD31+VE-Cadherin+ population from protocol II was also lower than that from protocol I. However, with the addition of LN421 in the coating substrata, the differences in VEGFR2+ and VEGFR2+CD31+VE-Cadherin+ populations were not significant between the two protocols. The lower yield of VEGFR2+ population in protocol II could again be attributed to the maturity of EPCs when cells were cultured for 4 more days before purification. Overall, with either protocol, we were able to achieve almost pure populations of EPCs that highly expressed endothelial lineage markers VEGFR2, CD31, and VE-Cadherin. Similar results were obtained with HS1001 and HS983A cell lines (Figure S2).
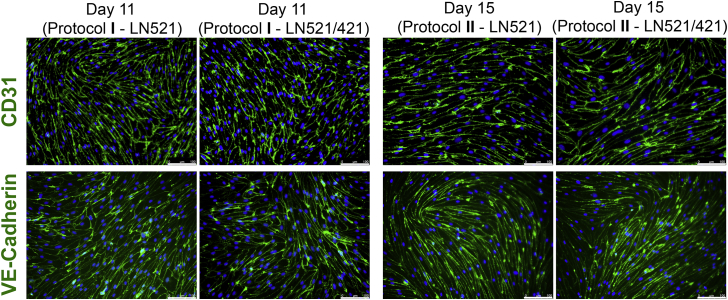
To confirm the cellular localization of CD31, we performed immunofluorescence analysis at the endpoints of both protocols (Figure 4). We observed similar membrane localization of CD31 and of VE-Cadherin on either LN521 or LN521/421 in the entire cultures, regardless of whether protocol I or II was used for EPC purification.
Figure 4.
Immunofluorescent Analysis of CD31 and VE-Cadherin after Purification
Cells were purified on day 7 (protocol I) and day 11 (protocol II), replated on either LN521 or LN521/421, and immunostained with anti-CD31 and anti-VE-Cadherin antibodies. Green, CD31 or VE-Cadherin; blue, DAPI. Scale bars, 100 μm.
We also obtained a 98% culture of EPCs when using LN511 and LN511/421 as coating substrates (Figure S3). However, we did not observe significant differences in VEGFR2+ or VEGFR2+CD31+VE-Cadherin+ populations between protocols on either substrate. As both LN521 and LN511 have been shown to support long-term self-renewal of hESCs, we suggested that LN511 could be used as an alternative coating substrate.
Functional Analyses of hESC-Derived EPCs
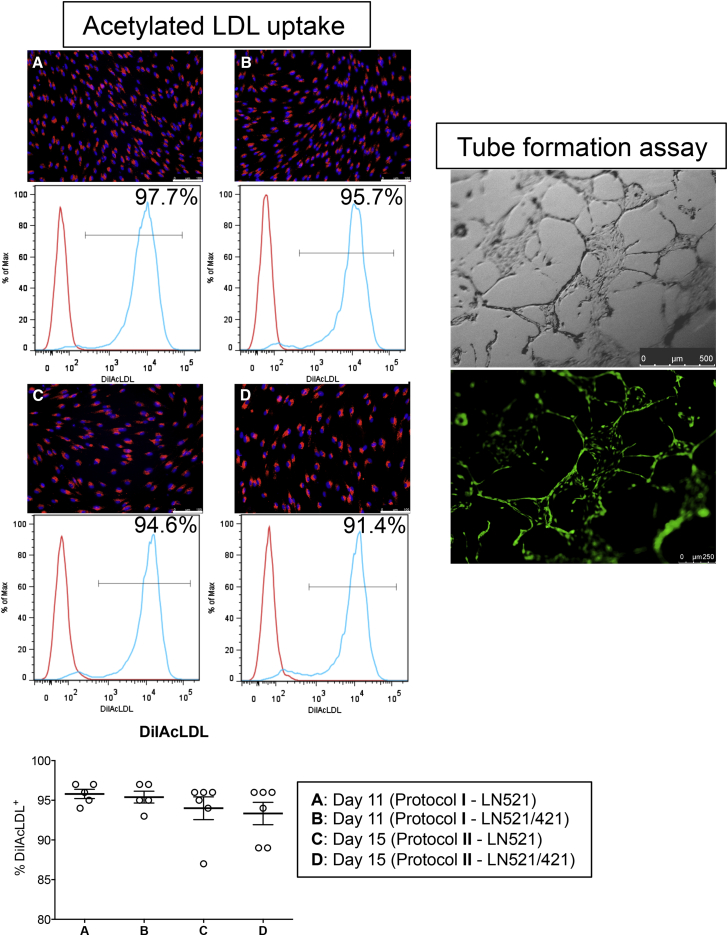
One of the hallmarks of ECs is their ability to take up acetylated low-density lipoprotein (AcLDL) via the “scavenger cell pathway” of LDL metabolism (Voyta et al., 1984). At the endpoints of each protocol, we incubated our endothelial cultures with 1 μg/mL AcLDL conjugated with DiI dye. After incubation, cells were either harvested for FACS analysis or visualized by fluorescence microscopy (Figure 5). In protocol I, 96% of EPCs in both LN521 and LN521/421 cultures were positive for DiIAcLDL, while 94% were positive from protocol II, suggesting our cultures consisted of nearly homogeneous EPCs. The presence of perinuclear fluorescent puncta also confirmed that cells had taken up the red dye. Similar results were seen when EPCs were derived from HS1001 and HS983A lines (Figure S4) and from H1 cultured on LN511 (Figure S5).
Figure 5.
Functional Assays
Acetylated LDL uptake: cells were purified on day 7 (A and B; protocol I) and day 11 (C and D; protocol II), replated on either LN521 or LN521/421, and incubated with DiIAcLDL 4 days after purification. In all conditions, 93%–96% of DiIAcLDL+ population was obtained from the culture. The uptake was confirmed by perinuclear staining of DiI dye (red). Blue, DAPI. Scale bars, 100 μm. Means ± SEM; each dot represents one independent differentiation batch (n = 5–6). See also Figures S4 and S5.
Tube formation assay: On day 11, cells were dissociated into single-cell suspension and plated onto Matrigel. Tube-like structures were observed 4 days later. Calcein-AM dye (green) stained live cells in the culture.
We then carried out a tube formation assay on Matrigel (protocol adapted from Patsch et al., 2015) to determine the angiogenic potential of our hESC-derived cells. We observed tube-like structures 4 days after plating purified EPCs at a low density. Figure 5 shows typical tube formation observed when replating EPCs in protocol I onto LN521. To assess the viability of the cells in this assay, we added calcein-AM dye to the culture and were able to detect strong green fluorescent signals emanating from the vascular network-like structures.
Gene Expression Profiles of hESC-Derived EPCs
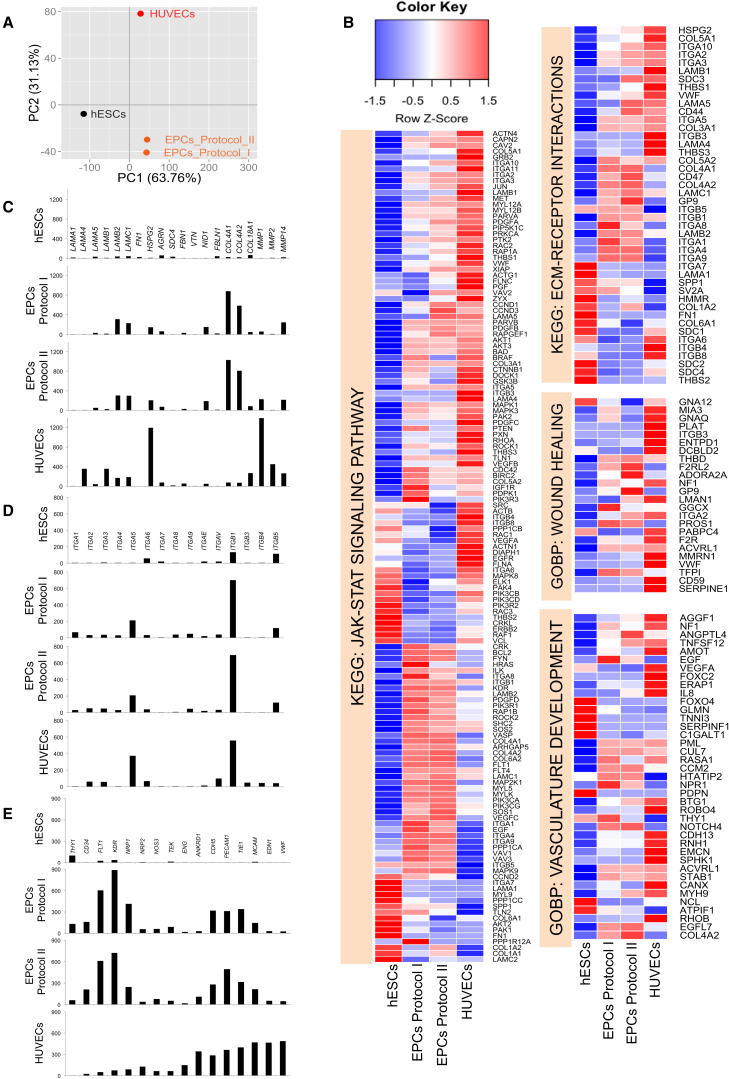
We did not observe significant differences in VEGFR2, CD31, and VE-Cadherin protein expression via FACS analyses, CD31 membrane localization, or AcLDL uptake when purified cells were replated onto LN521/421, compared with LN521 alone (Figures 3, 4, and 5). Therefore, we focused on characterizing EPCs derived from either protocol on LN521 alone, with references to hESCs and fully mature HUVECs that were also cultured on LN521. To identify global transcriptomic changes across these cell states, we carried out RNA-seq analysis and analyzed the transcriptome abundance data for the enrichment of biological processes in different cell types, compared with hESCs. Principal component analysis, based on the overall transcriptome signatures from expressed genes (fragments per kilobase of exon per million reads [FPKM] >5 in at least one sample), revealed 63.7% of the total expression variability in the first principal component due to the separation of hESCs from the other cell types. The second principal component explained 31.3% of the variability originating from the separation of EPCs samples from the mature HUVECs (Figure 6A). We next performed gene set enrichment analysis (GSEA) on the transcriptomic data to identify biological pathways that are enriched for up- and downregulated genes in EPC and HUVEC samples, compared with hESCs. Using the KEGG and Gene Ontology Biological Process (GOBP) pathway repositories, we identified several biological mechanisms that were significantly altered at a false discovery rate (FDR) of <5% (data not shown). From this list, a selection of key KEGG and GOBP pathways related to extracellular matrix (ECM) biology and endothelial lineage was further investigated to identify the pattern of changes in their constituent genes across hESC, EPC, and HUVEC samples. Specifically, from their gene-level expression patterns via heatmaps, we highlighted changes in the GOBP pathways “wound healing” and “vasculature development,” and in the KEGG pathways “JAK-STAT signaling” and “ECM-receptor interactions” (Figure 6B). For each of these pathways, we observed subsets of genes that displayed cell-stage-specific maximal expression, suggesting that different segments of the pathways were operative at different points of cellular differentiation. For example, considering the “ECM-receptor interactions” pathway, we identified a subset of genes that show lowest expression in the hESC state and highest expression in the HUVECs (e.g., ITGA2, VWF, HSPG2, etc.). Another set of genes in the same pathway showed highest levels of expression in hESCs and most reduced expression in the HUVECs (e.g., ITGA7, SPP1, COL1A2, etc.). A third category of genes showed maximal expression in EPC samples compared with both hESCs and HUVECs (e.g., COL4A1, COL4A2, ITGA4, etc.).
Figure 6.
Global Transcriptome Analyses of hESCs, hESC-Derived EPCs, and HUVECs
(A) Principal component analysis of 10,370 genes with FPKM >5 in at least one sample revealed 63.76% variability of the dataset along component 1 and 31.13% along component 2.
(B) Detailed gene-level heatmaps for key KEGG and GOBP biological pathways that are significantly upregulated in EPCs and HUVECs, compared with hESCs (FDR <5). Rows represent genes and columns represent samples. Row z-score transformation was performed on log2 of FPKM values with blue denoting a lower and red a higher expression level compared with the average expression level.
(C–E) Relative expression of extracellular matrix proteins (C), different α and β integrin subunits (D), and endothelial lineage markers (E) were plotted in their respective scales. y axis represents normalized FPKM values from RNA-seq.
These findings are further elaborated in the gene-level plots (Figures 6C–6E). Here, we extracted relevant genes within each pathway and plotted their expression profiles (in normalized FPKM) on the same scale. First, we examined the expression of some major ECM genes during maturation of hESC-derived endothelial lineage (Figure 6C). Our primary HUVECs exhibited high expression of laminin α4, β1, β2, and γ1 chains, corresponding to LN411 and LN421, revealing that mature ECs express high levels of those isoforms that are specific for subendothelial BM. In comparison, α5, β2, and γ1 were the predominant laminin chains found in the hESC-derived EPCs. HUVECs also expressed high levels of HSPG2 (encoding Perlecan) as well as matrix metallopeptidases MMP1 and MMP2, while hESC-derived EPCs significantly exhibited higher levels of collagen IV α1 and α2 subunits (COL4A1 and COL4A2) compared with those in HUVECs.
Next, expression analysis of major receptors for ECM proteins, e.g., integrin subunits, provided insight into cell-matrix interactions (Figure 6D). The data demonstrated a shift from α6β1 integrin (ITGA6, ITGB1) in hESCs (Toya et al., 2015) to predominantly α5β1 (ITGA5) integrin in the EPCs (Francis et al., 2002), and to a lesser extent αVβ5 (ITGAV, ITGB5), an integrin required for VEGF- or TGF-α-induced angiogenesis (Friedlander et al., 1995).
Lastly, the expression profile of the EPCs revealed upregulation of specific genes along the endothelial lineage (Figure 6E). Compared with mature HUVECs, the hESC-derived EPCs from both protocols highly expressed early endothelial markers, such as CD34, FLT1, KDR, and NRP1 (encoding Neuropilin 1, which together with CD31 identified a population of hESC-derived cells capable of giving rise to stable cord-blood endothelial colony-forming cells [Prasain et al., 2014]). The expression of some mature endothelial markers, such as CDH5, TIE1, TEK, MCAM, and PECAM1, was comparable between EPCs and HUVECs. These results suggested that hESC-derived EPCs from both protocols were still at a progenitor stage.
Maturation of hESC-Derived EPCs
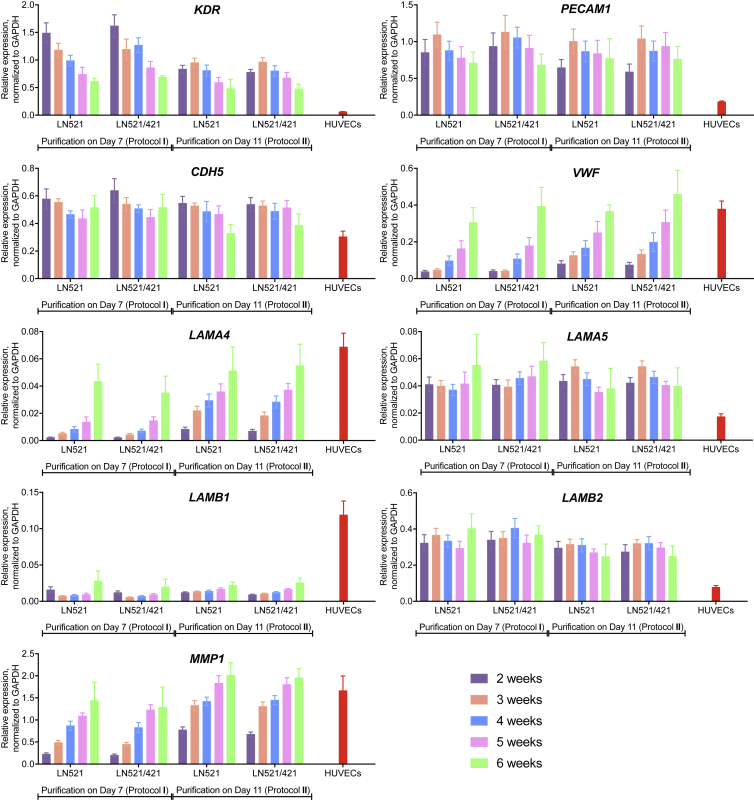
To determine whether our hESC-derived EPCs had the potential to reach a fully mature phenotype, we allowed cells to mature further in endothelial amplification medium and analyzed their mRNA expression profiles at 2, 3, 4, 5, and 6 weeks after purification (Figure 7). During the maturation, cell numbers did not decline and were similar in both protocols (Figure S6). We observed a decrease in mRNA level of KDR (immature marker) and increases in the expression of VWF and LAMA4 (encoding laminin α4 chain) (mature endothelial markers) over time, comparable with those of HUVECs. Importantly, high PECAM1 and CDH5 expression levels were maintained over long-term culturing. The major laminin β chain expressed by our EPCs was β2 (LAMB2), whereas HUVECs showed a much higher β1 mRNA level (LAMB1). Interestingly, one of the key matrix metallopeptidases, MMP1, exhibited an increase in gene expression over time to a similar extent as seen in HUVECs, indicating a faster turnover rate once the cells become more mature. Overall, we have demonstrated that hESC-derived EPCs from protocol II were more mature than those from protocol I, as evidenced by lower KDR and higher VWF, LAMA4, and MMP1 expression levels. We also showed that the presence of a TGF-β inhibitor allowed hESC-derived EPCs to mature to ECs.
Figure 7.
Expression Profiles of hESC-Derived Cells during Long-Term Culturing
Gene expression of EC-specific markers and BM components were analyzed 2, 3, 4, 5, and 6 weeks after purification in each protocol, compared with those of HUVECs, normalized to GAPDH. Means ± SEM (n = 3–10 independent batches; n = 3 HUVEC lines from three separate cords).
Discussion
Our laboratory has produced multiple laminin isoforms as recombinant proteins, and developed chemically defined and xeno-free hESC derivation and culture systems using biologically relevant substrata LN511 or LN521. Both of these laminins, which are developmentally regulated, are also present in other BMs that, additionally, may contain other tissue-specific laminin isoforms (Domogatskaya et al., 2012, Miner et al., 1998). The results of this study demonstrate the possibility to produce high-quality, homogeneous cultures of EPCs from pluripotent hESCs that are cultured on these two laminins found in all subendothelial BM. Our hESC-derived cells were functionally demonstrated by tube formation and DiIAcLDL uptake assays, and expressed specific markers for the endothelial lineage and vasculogenesis.
Global transcriptome analysis shed light on the expression of genes involved in cell-cell and cell-matrix interactions, as well as expression signatures during differentiation toward maturity of the endothelial cell lineage (Figure 6). The data revealed that pluripotent hESCs do not produce high amounts of BM components, emphasizing the importance of providing the initial relevant supporting matrix, such as LN521, for hESCs to survive and maintain their pluripotency (Rodin et al., 2014a). hESCs, EPCs, and mature HUVECs did not produce Fibronectin (FN1), Fibulin-1 (FBLN1), Fibrillin-1 (FBN1), or Vitronectin (VTN), suggesting that these proteins are not biologically relevant coating substrata when culturing these cells in vitro. Interestingly, the subendothelium-specific laminin α4 chain did not appear until cells became fully mature. Along the endothelial lineage, our EPCs expressed collagen type IV as the main structural BM component, which is likely to provide structural support, while laminin α5 chain expression was low, possibly because it was added exogenously. When we allowed our cells to mature further after purification, important endothelial-specific BM markers, such as von Willebrand factor, laminin α4 chain, and matrix metallopeptidase MMP1 had an increase in mRNA levels over the 6-week period, reaching levels similar to those of HUVECs. Our results suggested that a switch in ECM composition might occur alongside a faster turnover as the cells progress toward maturation. For example, Perlecan was the main BM component expressed in HUVECs, indicating that this proteoglycan bearing a high affinity for growth factors is needed for EC proliferation during vascular development (Zoeller et al., 2009). Both laminin α4 chain and matrix metallopeptidases MMP1, MMP2, and MMP14 were highly expressed, suggesting that the main laminins LN411 and LN421 in the HUVEC BM undergo a constant turnover.
Given that the ECM becomes more complex as ECs differentiate, it is of particular interest to understand the outside-in signaling that might modulate their behavior. Similar to other cell types, ECs interact with their environment in large part via the integrin family of adhesion receptors that consists of α and β subunits. The pairing of these two subunits confers specificity of binding to one or more ligands. ECs have been shown to express different integrin heterodimers, including α1β1, α2β1, α3β1, α5β1, αVβ1, αVβ5, α6β1, and αVβ3 (reviewed in Luscinskas and Lawler, 1994). Our analyses showed the highest expression of β1 integrin subunit in both EPCs and mature HUVECs, explaining its indispensable role for cell-cell junction integrity in growing and maturing vasculature (Yamamoto et al., 2015). Among the integrin heterodimers, α5β1 integrin interacts with fibronectin to regulate early vasculogenesis and angiogenesis (Francis et al., 2002). Our results suggested α5β1 integrin as a potential receptor for LN411 and LN421 as well.
Compared with other protocols developed for differentiation of hPSCs to EPCs, our approach offers major advantages from the point of view of both clinical and research applications. First, it emphasizes a crucial role for specific biologically relevant laminin isoforms at different stages of EC differentiation and phenotype maintenance. Since laminins are tissue specific, they are likely to play a significant role in transducing signals in cell-matrix interaction. By differentiating hESCs on their relevant laminin molecules, this approach allows the mimicking of the in vivo cell niche. This, in turn, enables detailed analyses of cell-type-specific molecular signal transduction pathways. Second, without any animal products and undefined components in the system, we can confidently provide a reproducible system to generate a large amount of functional ECs that may be used in regenerative medicine for the treatment of vascular diseases, myocardial infarction, heart failure, and diabetes. Lastly, the use of specific recombinant matrix proteins such as laminins has enabled the development of reproducible differentiation protocols for obtaining a homogeneous population of EPCs. These cells may have the potential to mature further into particular endothelia depending upon the specific cues provided by other cell types in the microenvironment, for instance, the tight endothelium in the blood-brain barrier or the fenestrated endothelium in the glomerular filtration barrier. With an unlimited source of pure EPCs, such in vitro models can now be studied in molecular detail and are most appropriate for the screening of potential therapeutic drugs.
Experimental Procedures
Differentiation Protocol
Tissue culture plates (24-well; Costar) were coated overnight at 4°C with sterile LN521, LN511, LN521/421, and LN511/421 (1:1 ratio) combinations at 10 μg/mL according to the manufacturer's instructions (LN521 and LN421 were purchased from BioLamina, LN511 was generated in our laboratory). hESC line H1 (WiCell Research Institute) was cultured as monolayer on pre-coated plates and maintained in NutriStem hESC XF medium (Biological Industries). We also tested our protocol on two other cell lines, HS1001 and HS983A, derived at the Karolinska Institute, and obtained results similar to those obtained with the H1 cell line.
Figure 1 illustrates our three-step differentiation protocol. Specifically, hESCs were plated on LN521 or LN511 pre-coated 24-well tissue culture plates at a density of 50,000–100,000 cells/well (depending on the cell line) and refreshed daily with NutriStem. The basal medium used throughout the differentiation process consisted of DMEM/F12 medium, 1× chemically defined lipid concentrate, 0.1× insulin-transferrin-selenium-X, 2 mM GlutaMAX (Gibco), 450 μM mono-thio glycerol (Sigma), and 50 μg/mL L-ascorbic acid 2-phosphate (Sigma). When hESC cultures reached 40%–50% confluency (after 2–3 days depending on the cell line), on day 0 of differentiation NutriStem was replaced with mesoderm induction medium, consisting of basal medium supplemented with 10 ng/mL activin A, 20 ng/mL BMP4 (R&D Systems), and 6 μM CHIR99021 (Tocris Bioscience). After 3 days, the medium was replaced by vascular specification medium, consisting of basal medium supplemented with 50 ng/mL VEGF165 (Gibco), 10 ng/mL bFGF, 20 ng/mL BMP4 (R&D Systems), and 10 μM DAPT, a Notch signaling inhibitor (Tocris Bioscience). For protocol I, on day 7 of differentiation adherent differentiated cells were dissociated with TrypLESelect (Gibco) and MACS purified using a CD31 Microbead Kit according to the manufacturer's instructions (Miltenyi Biotec). CD31+ cells were replated on LN521- or LN511-coated plates (with or without LN421) at the density of 32,500 cells/cm2 in endothelial amplification medium, consisting of basal medium supplemented with 50 ng/mL VEGF165, 10 ng/mL bFGF, and 10 μM SB431542, a TGF-β signaling inhibitor (Tocris Bioscience), expanded for 4 more days, and characterized on day 11. For protocol II, vascular lineage committed cells were allowed to grow in endothelial amplification medium until day 11, then CD31+ population was purified and hESC-derived cells were harvested on day 15 for characterization. All media were refreshed every other day.
HUVEC Isolation
Human umbilical cords were freshly obtained from the KK Women's and Children's Hospital, Singapore, with the approval from the Singhealth Centralized Institutional Review Board (ref. no: CIRB Ref: 2014/323/D). The protocol to isolate and culture HUVECs was adapted from Baudin et al. (2007). In brief, umbilical vein was flushed with 1× PBS to remove all red blood cells and digested with 2 mg/mL collagenase (Roche) solution for 10 min in 37°C saline bath. Cells were detached from the cord vein, plated onto LN521-coated plates, and cultured in M199 medium supplemented with 1% L-GlutaMAX, 1% penicillin-streptomycin, 15 mM HEPES (Gibco), 0.135% NaHCO3 (Lonza), 30 μg/mL endothelial cell growth supplement (Sigma), 10 U/mL heparin (Merck), and 20% fetal bovine serum (Gibco). HUVECs were passaged onto new LN521-coated plates upon confluency. In this study, we compared the expression profiles of our hESC-derived cells with HUVECs at passage 1 to ensure the maintenance of their phenotypic features.
qPCR Analysis
Total RNA from HUVECs and hESC-derived cells at different time points was purified using an RNeasy Micro Kit (Qiagen) according to the manufacturer's instructions. The yield was determined by NanoDrop ND-2000 spectrophotometer (NanoDrop Technologies). For qRT-PCR analysis, cDNA was synthesized from 500 ng of total RNA in a 20-μL reaction mixture using a TaqMan Reverse Transcription Reagents Kit (Applied BioSystems) according to the manufacturer's instructions. Real-time qRT-PCR was performed with synthesized cDNA in assay mix containing iQ SYBR Green Super mix (Bio-Rad) and primers for genes of interest. GAPDH was used as the normalizing control. Primer sequences are listed in Table S1.
FACS Analysis
Cells were collected at different time points during differentiation process and single-cell suspensions were fixed with Fixation Reagent (medium A; Life Technologies) for 15 min at room temperature, washed with FACS buffer (0.5% BSA, 2 mM EDTA in 1× PBS), blocked with 5% goat serum in FACS buffer, immunostained with primary antibodies in Permeabilization Reagent (medium B; Life Technologies) for 15 min at room temperature, detected with secondary antibodies diluted in 1% goat serum in FACS buffer. For fluorophore-conjugated antibodies, fixed cells were incubated with antibodies diluted in medium B and human FcR blocking reagent (Miltenyi Biotec, 1:50) for 30 min at room temperature. Stained cells were resuspended in FACS buffer and subjected to FACS analysis (MACSQuant VYB, Miltenyi Biotec) to detect the expression of specific cell-surface markers and transcription factors. Antibodies used in this study are summarized in Table S2. CD34 was assessed via single staining while VEGFR2, CD31, and VE-Cadherin were assessed together via triple staining, i.e., the VEGFR2+ population was first determined via histogram analysis, followed by scatterplot analysis of both CD31 and VE-Cadherin within the VEGFR2+ population. CD31 and VE-Cadherin were analyzed individually, overlaid with their respective isotype controls to determine gate settings for the scatterplot. Data were analyzed using MACSQuantify (Miltenyi Biotec) and FlowJo software.
Immunocytochemistry
On day 11 (protocol I) or day 15 (protocol II), adherent cells were fixed with 4% paraformaldehyde in 1× PBS for 20 min at 4°C, permeabilized and blocked in 0.1% Triton X-100, 5% goat serum, and 1% BSA in 1× PBS for 15 min at room temperature. Cells were immunostained with primary antibodies, followed by secondary antibodies, 30 min each at room temperature (Table S2). The antibodies were diluted in 5% goat serum and 1% BSA in 1× PBS. Samples were preserved in ProLong Gold Antifade Reagent with DAPI (Life Technologies) and visualized under a Leica DMi8 fluorescent microscope.
DiIAcLDL Uptake Assay
On day 11 (protocol I) or day 15 (protocol II), cells were incubated with 1 μg/mL AcLDL labeled with the fluorescent probe DiI (DiIAcLDL; Alfa Aesar) in endothelial amplification medium for 5 hr at 37°C. Thereafter, cells were washed with Dulbecco’s PBS, dissociated in single-cell suspensions, fixed with medium A, and permeabilized with medium B. After a final wash with FACS buffer, cells were subjected to FACS analysis. To confirm the localization of DiIAcLDL taken up by hESC-derived cells, after 5 hr of incubation we fixed the cells with 4% paraformaldehyde for 20 min at 4°C and preserved them using ProLong Gold Antifade Reagent with DAPI. Fluorescent signals were detected using rhodamine filter of the Leica DMi8 fluorescent microscope.
Tube Formation Assay
Growth factor reduced Matrigel (Corning) was thawed on ice overnight, and 300 μL added in each well of a pre-chilled 24-well plate. The plate was incubated at 37°C for 45 min for gel formation. On day 11 of differentiation (protocol I), cells were dissociated by TrypLESelect and 7 × 104 cells were added onto Matrigel in 500 μL of endothelial amplification medium. After 4 days, tube formation was visualized under a light microscope. To determine the cell viability on Matrigel, we incubated cells with 500 μL of calcein-AM dye diluted in Hank’s balanced salt solution (8 μg/mL; Life Technologies) for 30 min at 37°C according to the manufacturer's instructions. Fluorescent images were obtained by a Leica DMi8 fluorescent microscope.
RNA-Seq and Bioinformatics Studies
RNA-seq libraries were prepared using Tru-Seq Stranded Total RNA with Ribo-Zero Gold kit protocol, according to the manufacturer's instructions (Illumina). Libraries were validated with an Agilent Bioanalyzer (Agilent Technologies), diluted, and applied to an Illumina flow cell using the Illumina Cluster Station. Sequencing was performed on an Illumina HiSeq2000 sequencer at the Duke-NUS Genome Biology Facility with the paired-end 100- bp read option.
The quality of the paired sequencing reads was ascertained via the FASTQC tool (Andrews, 2010). The average sequencing depth was 213.14 million reads per sample and the per-sequence quality scores were >30 for the majority of the reads. No further trimming of the bases was performed. Sequencing reads were then mapped to the human reference genome (hg19) using the TopHat-2.0.9 alignment tool (Kim et al., 2013). The mean mapping rate was 80.78%. Transcript/gene assembly and abundance estimation were performed using Cufflinks-2.1.1 (Trapnell et al., 2012), resulting in the generation of counts, normalized for transcript length and library size (FPKM). Pathway enrichment analysis was conducted using a ranked list of genes (ranked by their log2 fold change between two sample groups), and by using the pre-ranked option in the GSEA tool (Subramanian et al., 2005).
Statistical Analysis
Data are presented as means ± SEM from independent differentiation batches. Differences in relative mRNA expression and surface marker protein expression at different time points were assessed by one-way ANOVA, corrected for multiple comparisons using Tukey’s post hoc test. All graphs and statistical analyses were generated by Prism Software 7.0 (GraphPad). Differences were regarded as significant at p < 0.05.
Author Contributions
M.T.X.N. designed and performed experiments, analyzed data, and drafted the manuscript. E.O. carried out differentiation experiments on HS983A cell line. X.C. and S.G. analyzed RNA-seq data. K.H.T. supervised umbilical cord collection. O.H. provided HS1001 and HS983A lines. K.T. led the conception, supervised the project, and drafted the manuscript.
Acknowledgments
We thank Ms. Zuhua Cai for producing LN511 in our laboratory, and Ms. Su Ting Tay from Duke-NUS Genome Biology Facility for generating cDNA libraries and running RNA-seq. This work was funded in part by grant NMRC/STaR/0010/2012 from the National Medical Research Council of Singapore. K.T. is a cofounder and shareholder of BioLamina AB.
Published: September 29, 2016
Footnotes
Supplemental Information includes six figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.08.017.
Accession Numbers
The accession number for the RNA-seq data reported in this paper is GEO: GSE85953.
Supplemental Information
References
- Adams W.J., Zhang Y., Cloutier J., Kuchimanchi P., Newton G., Sehrawat S., Aird W.C., Mayadas T.N., Luscinskas F.W., Garcia-Cardena G. Functional vascular endothelium derived from human induced pluripotent stem cells. Stem Cell Rep. 2013;1:105–113. doi: 10.1016/j.stemcr.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews, S.. (2010). FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc.
- Baudin B., Bruneel A., Bosselut N., Vaubourdolle M. A protocol for isolation and culture of human umbilical vein endothelial cells. Nat. Protoc. 2007;2:481–485. doi: 10.1038/nprot.2007.54. [DOI] [PubMed] [Google Scholar]
- Campagnolo P., Tsai T.N., Hong X., Kirton J.P., So P.W., Margariti A., Di Bernardini E., Wong M.M., Hu Y., Stevens M.M. c-Kit+ progenitors generate vascular cells for tissue-engineered grafts through modulation of the Wnt/Klf4 pathway. Biomaterials. 2015;60:53–61. doi: 10.1016/j.biomaterials.2015.04.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domogatskaya A., Rodin S., Boutaud A., Tryggvason K. Laminin-511 but not -332, -111, or -411 enables mouse embryonic stem cell self-renewal in vitro. Stem Cells. 2008;26:2800–2809. doi: 10.1634/stemcells.2007-0389. [DOI] [PubMed] [Google Scholar]
- Domogatskaya A., Rodin S., Tryggvason K. Functional diversity of laminins. Annu. Rev. Cell Dev. Biol. 2012;28:523–553. doi: 10.1146/annurev-cellbio-101011-155750. [DOI] [PubMed] [Google Scholar]
- Francis S.E., Goh K.L., Hodivala-Dilke K., Bader B.L., Stark M., Davidson D., Hynes R.O. Central roles of alpha5beta1 integrin and fibronectin in vascular development in mouse embryos and embryoid bodies. Arterioscler. Thromb. Vasc. Biol. 2002;22:927–933. doi: 10.1161/01.atv.0000016045.93313.f2. [DOI] [PubMed] [Google Scholar]
- Friedlander M., Brooks P.C., Shaffer R.W., Kincaid C.M., Varner J.A., Cheresh D.A. Definition of two angiogenic pathways by distinct alpha v integrins. Science. 1995;270:1500–1502. doi: 10.1126/science.270.5241.1500. [DOI] [PubMed] [Google Scholar]
- Goldman O., Feraud O., Boyer-Di Ponio J., Driancourt C., Clay D., Le Bousse-Kerdiles M.C., Bennaceur-Griscelli A., Uzan G. A boost of BMP4 accelerates the commitment of human embryonic stem cells to the endothelial lineage. Stem Cells. 2009;27:1750–1759. doi: 10.1002/stem.100. [DOI] [PubMed] [Google Scholar]
- Huber T.L., Kouskoff V., Fehling H.J., Palis J., Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- James D., Nam H.S., Seandel M., Nolan D., Janovitz T., Tomishima M., Studer L., Lee G., Lyden D., Benezra R. Expansion and maintenance of human embryonic stem cell-derived endothelial cells by TGFbeta inhibition is Id1 dependent. Nat. Biotechnol. 2010;28:161–166. doi: 10.1038/nbt.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinney M.A., Hookway T.A., Wang Y., McDevitt T.C. Engineering three-dimensional stem cell morphogenesis for the development of tissue models and scalable regenerative therapeutics. Ann. Biomed. Eng. 2014;42:352–367. doi: 10.1007/s10439-013-0953-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaffky E., Williams R., Yao C.C., Ziober B., Kramer R., Sutherland A. Trophoblast-specific expression and function of the integrin alpha 7 subunit in the peri-implantation mouse embryo. Dev. Biol. 2001;239:161–175. doi: 10.1006/dbio.2001.0404. [DOI] [PubMed] [Google Scholar]
- Kriks S., Shim J.W., Piao J., Ganat Y.M., Wakeman D.R., Xie Z., Carrillo-Reid L., Auyeung G., Antonacci C., Buch A. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson's disease. Nature. 2011;480:547–551. doi: 10.1038/nature10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian X., Bao X., Al-Ahmad A., Liu J., Wu Y., Dong W., Dunn K.K., Shusta E.V., Palecek S.P. Efficient differentiation of human pluripotent stem cells to endothelial progenitors via small-molecule activation of WNT signaling. Stem Cell Rep. 2014;3:804–816. doi: 10.1016/j.stemcr.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscinskas F.W., Lawler J. Integrins as dynamic regulators of vascular function. FASEB J. 1994;8:929–938. doi: 10.1096/fasebj.8.12.7522194. [DOI] [PubMed] [Google Scholar]
- Melkoumian Z., Weber J.L., Weber D.M., Fadeev A.G., Zhou Y., Dolley-Sonneville P., Yang J., Qiu L., Priest C.A., Shogbon C. Synthetic peptide-acrylate surfaces for long-term self-renewal and cardiomyocyte differentiation of human embryonic stem cells. Nat. Biotechnol. 2010;28:606–610. doi: 10.1038/nbt.1629. [DOI] [PubMed] [Google Scholar]
- Miner J.H., Cunningham J., Sanes J.R. Roles for laminin in embryogenesis: exencephaly, syndactyly, and placentopathy in mice lacking the laminin alpha5 chain. J. Cell Biol. 1998;143:1713–1723. doi: 10.1083/jcb.143.6.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patsch C., Challet-Meylan L., Thoma E.C., Urich E., Heckel T., O'Sullivan J.F., Grainger S.J., Kapp F.G., Sun L., Christensen K. Generation of vascular endothelial and smooth muscle cells from human pluripotent stem cells. Nat. Cell Biol. 2015;17:994–1003. doi: 10.1038/ncb3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasain N., Lee M.R., Vemula S., Meador J.L., Yoshimoto M., Ferkowicz M.J., Fett A., Gupta M., Rapp B.M., Saadatzadeh M.R. Differentiation of human pluripotent stem cells to cells similar to cord-blood endothelial colony-forming cells. Nat. Biotechnol. 2014;32:1151–1157. doi: 10.1038/nbt.3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodin S., Domogatskaya A., Strom S., Hansson E.M., Chien K.R., Inzunza J., Hovatta O., Tryggvason K. Long-term self-renewal of human pluripotent stem cells on human recombinant laminin-511. Nat. Biotechnol. 2010;28:611–615. doi: 10.1038/nbt.1620. [DOI] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Hovatta O., Tryggvason K. Monolayer culturing and cloning of human pluripotent stem cells on laminin-521-based matrices under xeno-free and chemically defined conditions. Nat. Protoc. 2014;9:2354–2368. doi: 10.1038/nprot.2014.159. [DOI] [PubMed] [Google Scholar]
- Rodin S., Antonsson L., Niaudet C., Simonson O.E., Salmela E., Hansson E.M., Domogatskaya A., Xiao Z., Damdimopoulou P., Sheikhi M. Clonal culturing of human embryonic stem cells on laminin-521/E-cadherin matrix in defined and xeno-free environment. Nat. Commun. 2014;5:3195. doi: 10.1038/ncomms4195. [DOI] [PubMed] [Google Scholar]
- Sahara M., Hansson E.M., Wernet O., Lui K.O., Spater D., Chien K.R. Manipulation of a VEGF-Notch signaling circuit drives formation of functional vascular endothelial progenitors from human pluripotent stem cells. Cell Res. 2014;24:820–841. doi: 10.1038/cr.2014.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanganalmath S.K., Bolli R. Cell therapy for heart failure: a comprehensive overview of experimental and clinical studies, current challenges, and future directions. Circ. Res. 2013;113:810–834. doi: 10.1161/CIRCRESAHA.113.300219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorokin L.M., Pausch F., Frieser M., Kroger S., Ohage E., Deutzmann R. Developmental regulation of the laminin alpha5 chain suggests a role in epithelial and endothelial cell maturation. Dev. Biol. 1997;189:285–300. doi: 10.1006/dbio.1997.8668. [DOI] [PubMed] [Google Scholar]
- Stenzel D., Franco C.A., Estrach S., Mettouchi A., Sauvaget D., Rosewell I., Schertel A., Armer H., Domogatskaya A., Rodin S. Endothelial basement membrane limits tip cell formation by inducing Dll4/Notch signalling in vivo. EMBO Rep. 2011;12:1135–1143. doi: 10.1038/embor.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumi T., Tsuneyoshi N., Nakatsuji N., Suemori H. Defining early lineage specification of human embryonic stem cells by the orchestrated balance of canonical Wnt/beta-catenin, Activin/Nodal and BMP signaling. Development. 2008;135:2969–2979. doi: 10.1242/dev.021121. [DOI] [PubMed] [Google Scholar]
- Takahashi K., Tanabe K., Ohnuki M., Narita M., Ichisaka T., Tomoda K., Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Thomson J.A., Itskovitz-Eldor J., Shapiro S.S., Waknitz M.A., Swiergiel J.J., Marshall V.S., Jones J.M. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- Thyboll J., Kortesmaa J., Cao R., Soininen R., Wang L., Iivanainen A., Sorokin L., Risling M., Cao Y., Tryggvason K. Deletion of the laminin alpha4 chain leads to impaired microvessel maturation. Mol. Cell Biol. 2002;22:1194–1202. doi: 10.1128/MCB.22.4.1194-1202.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toya S.P., Wary K.K., Mittal M., Li F., Toth P.T., Park C., Rehman J., Malik A.B. Integrin alpha6beta1 expressed in ESCs instructs the differentiation to endothelial cells. Stem Cells. 2015;33:1719–1729. doi: 10.1002/stem.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012;7:562–578. doi: 10.1038/nprot.2012.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Winkle A.P., Gates I.D., Kallos M.S. Mass transfer limitations in embryoid bodies during human embryonic stem cell differentiation. Cells Tissues Organs. 2012;196:34–47. doi: 10.1159/000330691. [DOI] [PubMed] [Google Scholar]
- Voyta J.C., Via D.P., Butterfield C.E., Zetter B.R. Identification and isolation of endothelial cells based on their increased uptake of acetylated-low density lipoprotein. J. Cell Biol. 1984;99:2034–2040. doi: 10.1083/jcb.99.6.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z.Z., Au P., Chen T., Shao Y., Daheron L.M., Bai H., Arzigian M., Fukumura D., Jain R.K., Scadden D.T. Endothelial cells derived from human embryonic stem cells form durable blood vessels in vivo. Nat. Biotechnol. 2007;25:317–318. doi: 10.1038/nbt1287. [DOI] [PubMed] [Google Scholar]
- Watanabe K., Ueno M., Kamiya D., Nishiyama A., Matsumura M., Wataya T., Takahashi J.B., Nishikawa S., Nishikawa S., Muguruma K. A ROCK inhibitor permits survival of dissociated human embryonic stem cells. Nat. Biotechnol. 2007;25:681–686. doi: 10.1038/nbt1310. [DOI] [PubMed] [Google Scholar]
- Woo D.H., Kim S.K., Lim H.J., Heo J., Park H.S., Kang G.Y., Kim S.E., You H.J., Hoeppner D.J., Kim Y. Direct and indirect contribution of human embryonic stem cell-derived hepatocyte-like cells to liver repair in mice. Gastroenterology. 2012;142:602–611. doi: 10.1053/j.gastro.2011.11.030. [DOI] [PubMed] [Google Scholar]
- Yamamoto H., Ehling M., Kato K., Kanai K., van Lessen M., Frye M., Zeuschner D., Nakayama M., Vestweber D., Adams R.H. Integrin beta1 controls VE-cadherin localization and blood vessel stability. Nat. Commun. 2015;6:6429. doi: 10.1038/ncomms7429. [DOI] [PubMed] [Google Scholar]
- Zoeller J.J., Whitelock J.M., Iozzo R.V. Perlecan regulates developmental angiogenesis by modulating the VEGF-VEGFR2 axis. Matrix Biol. 2009;28:284–291. doi: 10.1016/j.matbio.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.