Abstract
Background
Gliomas with mutant isocitrate dehydrogenase (IDH) produce high levels of 2-hydroxyglutarate (2HG) that can be quantitatively measured by 3D magnetic resonance spectroscopic imaging (MRSI). Current glioma MRI primarily relies upon fluid-attenuated inversion recovery (FLAIR) hyperintensity for treatment planning, although this lacks specificity for tumor cells. Here, we investigated the relationship between 2HG and FLAIR in mutant IDH glioma patients to determine whether 2HG mapping is valuable for radiotherapy planning.
Methods
Seventeen patients with mutant IDH1 gliomas were imaged by 3 T MRI. A 3D MRSI sequence was employed to specifically image 2HG. FLAIR imaging was performed using standard clinical protocol. Regions of interest (ROIs) were determined for FLAIR and optimally thresholded 2HG hyperintensities. The overlap, displacement, and volumes of 2HG and FLAIR ROIs were calculated.
Results
In 8 of 17 (47%) patients, the 2HG volume was larger than FLAIR volume. Across the entire cohort, the mean volume of 2HG was 35.3 cc (range, 5.3–92.7 cc), while the mean volume of FLAIR was 35.8 cc (range, 6.3–140.8 cc). FLAIR and 2HG ROIs had mean overlap of 0.28 (Dice coefficients range, 0.03–0.57) and mean displacement of 12.2 mm (range, 3.2–23.5 mm) between their centers of mass.
Conclusions
Our results indicate that for a substantial number of patients, the 2HG volumetric assessment of tumor burden is more extensive than FLAIR volume. In addition, there is only partial overlap and asymmetric displacement between the centers of FLAIR and 2HG ROIs. These results may have important implications for radiotherapy planning of IDH mutant glioma.
Keywords: glioma, isocitrate dehydrogenase (IDH) mutation, 2-hydroxyglutarate (2HG), magnetic resonance spectroscopic imaging (MRSI), radiotherapy
Cancer-associated mutations in the isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2, or collectively IDH) enzymes are frequently found in 50%–80% of grades II–III glioma and secondary glioblastoma, resulting in a distinct glioma subtype with unique molecular profile, clinical phenotype, radiological pattern, prognosis, and treatment response.1–4 These glioma IDH mutations, more often cytosolic IDH1 (90%) than mitochondrial IDH2 (10%), result not only in a loss of the enzyme's ability to catalyze the oxidation of isocitrate to α-ketoglutarate, but also in a new ability of the enzyme to catalyze the NADPH-dependent reduction of α-ketoglutarate to R-2-hydroxyglutarate (2HG).5 This neomorphic activity markedly increases the levels of the oncometabolite 2HG in human malignant gliomas harboring IDH1 mutations.5 The excess 2HG accumulated in vivo is thought to contribute to the formation and malignant progression of gliomas through epigenetic modifications of chromatin.6–10
Simultaneous with excess 2HG accumulation there is concomitant depletion of NADPH, which plays an important role in replenishing glutathione to maintain the redox balance in these cancer cells. Because IDH enzymes are the primary source of NADPH in the brain,11 mutation of IDH may significantly impair the overall ability of tumor cells to neutralize free radicals. As a consequence, it has been proposed that reactive oxygen species generated during radiotherapy could have a more deleterious effect on mutant IDH cells than on wild-type cells. Indeed, it has been shown in cell cultures that glioma cells transfected with mutant IDH alleles are more sensitive to radiation compared with their wild-type counterparts.12 Consistent with this proposal, analysis of anatomical MRI in glioblastoma patients has also shown that changes in postradiation 3D volumes of contrast-enhanced T1 and fluid attenuated inversion recovery (FLAIR) correlate with IDH mutation status, supporting increased radiosensitivity of mutant IDH glioblastoma.13
Optimizing radiation therapy planning is particularly important in the case of mutant IDH glioma patients and has been the main motivation in our current investigations. IDH glioma patients have prolonged overall survival (3–5 times longer) compared with wild-type patients,1,2 hence reducing adverse effects of radiation or neurotoxicity and improving quality of life in these patients is highly relevant.14–16 On the other hand, increased radiosensitivity of mutant IDH tumor cells underscores the importance of techniques to better target radiation dosing both spatially and temporally in this group of patients, to optimize the clinical benefit of treatment.
MRI T2-weighted (T2w) FLAIR hyperintensity has traditionally proven to be a neuroimaging method of primary importance for glioma target definition in surgical4 and radiotherapy planning and assessment.17 In particular, this has been the case for mutant IDH gliomas which are typically nonenhancing tumors.18 Of course, there are several important nuances that play a role in target definition for gliomas. First and foremost is the recognition that gliomas are infiltrative cancers, with tumor cells that extend well beyond the radiographic margin of disease, however defined or thresholded. Thus, radiation treatment plans are routinely extended beyond a delineated lesional volume by a defined margin. This therapeutic margin extension must be balanced against the normal brain irradiation and late toxicity risk in these more sparsely tumor-infiltrated areas. Furthermore, quantification of FLAIR hyperintensity can be problematic because the relation between edema probed by FLAIR and the density of cancer cells is ambiguous and can vary between individual cases. This variability limits the specificity of FLAIR to identify true tumor extent and to optimally guide treatment planning.19–21
As an opportunity to improve this situation, 2HG overproduction is highly specific to IDH mutation and therefore represents an ideal biomarker that can be imaged and quantified to assess tumor burden and monitor treatment response.22–27 The level of 2HG in the tumor correlates with the number of tumor cells producing 2HG, hence we hypothesized that 2HG imaging holds the potential to be more specific and quantitative than FLAIR for radiotherapy planning of mutant IDH glioma patients. The utility of magnetic resonance spectroscopic imaging (MRSI) for tumor target definition has been previously demonstrated for gliomas using common brain metabolites such as N-acetylaspartate, choline, and lactate.28–30 In the work described here, we extend this concept to the special case of 2HG.
A new 3D MRSI sequence has been recently developed to image 2HG signal in the brain to detect oncogenic IDH1 mutations and monitor treatment response.22,31 In this work, we investigated the potential value of 2HG imaging for tumor definition, compared with traditional FLAIR imaging. We analyzed the relationship between spatial extent and intensity landscape of 2HG and FLAIR images in a group of mutant IDH glioma patients. We describe a normalization technique for 2HG signal relative to the level of creatine in the healthy-appearing white matter of the contralateral hemisphere. Using histogram analysis, we derived a reproducible and optimal threshold for the ratio of 2HG over healthy creatine (2HG/hCr) that can be used to segment tumor region of interest (ROI). We quantified the agreement between ROIs obtained from 2HG and FLAIR images using 3 metrics: ROI volumes, ROI overlap, and ROI displacement. Strikingly, we found that in a substantial number of patients, the 2HG volume is larger than FLAIR volume, with only a partial overlap and an asymmetric displacement between centers of mass of 2HG and FLAIR ROIs. These results suggest that 2HG imaging can have a role in refining the definition of tumor extent with potential implications for radiotherapy planning and for subsequent treatment monitoring.
Materials and Methods
Patients
The institutional review board approved this study. Informed consent was obtained from all patients. We recruited 17 adult glioma patients (8 M, 9 F; age 23–87 y; mean 46) who had diagnosed gliomas of World Health Organization (WHO) grades II–IV (Table 1). All patients were recruited, consented, and scanned at Massachusetts General Hospital.
Table 1.
Demographic, histological and molecular profile of patients
| Patient # | Gender | Age | Grade | Type | IDH1 | Molecular Markers | Tumor Location |
|---|---|---|---|---|---|---|---|
| Pt 1 | F | 43 | 2 | OD | R132H | 1p/19q, MGMT | Frontal |
| Pt 2 | F | 33 | 3 | AA | R132H | PDGFR | Frontal/temporal |
| Pt 3 | M | 45 | 3 | AOD | R132H | 1p/19q, MGMT, PIK3CA | Frontal |
| Pt 4 | M | 34 | 4 | GBMO | R132H | – | Frontal |
| Pt 5 | M | 55 | 3 | AOD | R132H | – | Frontal |
| Pt 6 | F | 53 | 3 | AOA | R132H | 1p/19q, TP53 | Frontal |
| Pt 7 | F | 38 | 2 | OA | R132H | 19q | Occipital |
| Pt 8 | M | 23 | 2 | A | R132H | MGMT, TP53 | Frontal |
| Pt 9 | M | 41 | 3 | AA | R132H | – | Frontal/temporal |
| Pt 10 | F | 38 | 3 | AA | R132H | 19q | Parietal |
| Pt 11 | M | 43 | 3 | AOA | R132H | 19q, MGMT, TP53 | Frontal |
| Pt 12 | F | 87 | 3 | AOD | R132H | 1p/19q, MGMT | Frontal |
| Pt 13 | M | 33 | 2 | A | R132G | – | Parietal |
| Pt 14 | F | 42 | 3 | AA | R132S | – | Frontal |
| Pt 15 | F | 63 | 4 | GBM | R132H | – | Temporal/parietal |
| Pt 16 | F | 55 | 4 | GBMO | R132H | 1p/19q | Frontal |
| Pt 17 | M | 50 | 2 | A | R132H | – | Frontal |
Abbreviations: A, astrocytoma; OD, oligodendrogliomas; OA, oligoastrocytoma; AA, anaplastic astrocytoma; AOA, anaplastic oligoastrocytoma; AOD, anaplastic oligodendrogliomas; GBM, glioblastoma multiforme; GBMO, glioblastoma multiforme with oligodendroglial component; R132H/G/S, arginine 132 to histidine/glycine/serine; 1p/19q, codeletion of 1p and 19q chromosome arms; MGMT, O6-methylguanine DNA methyltransferase; PDGFR, platelet-derived growth factor receptor; PIK3CA, phosphatidylinositol-4,5-bisphosphate 3-kinase, catalytic subunit alpha; TP53, tumor protein p53.
Genomics and Molecular Diagnosis
IDH mutational status was confirmed by immunohistochemistry analysis using an antihuman R132H antibody (Dianova)32 or by genetic sequencing (SNaPshot).33 In addition, molecular markers such as 1p/19q codeletion,34 O6-DNA methylguanine-methyltransferase promoter methylation,35 mutation of tumor protein 53, and platelet derived growth factor receptor amplification were tested using fluorescence in-situ hybridization and PCR.33
MR Imaging
All images were acquired on a clinical 3 T MR scanner (Tim Trio, Siemens) with a 32-channel head coil. A robust 3D MRSI sequence for 2HG imaging was newly developed31 by integrating 3 highly optimized modules: (i) adiabatic J-difference spectral editing MEGA-LASER, (ii) spiral spectroscopic imaging, and (iii) real-time prospective motion correction, shim update with reacquisition. This sequence provides increased signal-to-noise ratio (SNR) for 2HG while minimizing subtraction errors caused by subject motion or scanner instability.
The acquisition parameters of the 3D MRSI sequence were: repetition time (TR) = 1600 ms, echo time (TE) = 68 ms, field of view (FOV) = 200 × 200 × 200 mm3, volume of interest = 100 × 80 × 50 mm3, 20 mm isotropic voxels, acquisition matrix 10 × 10 × 10 zero-filled to 16 × 16 × 16, number of acquisitions = 20, acquisition time = 9:55 min:s. The timing of the MEGA-LASER excitation was optimized for the maximum 2HG signal, and MEGA pulses of 65 Hz bandwidth were applied in an interleaved fashion at 1.9 ppm (ON) and 7.5 ppm (OFF) to edit the 2HG signal (Hα) at 4.02 ppm.
In addition, structural images including 3D axial T2w (TR/TE = 3200/420 ms, resolution 1 mm × 1 mm × 1 mm) and 2D FLAIR images (TR/inversion time/TE = 10000/2500/70 ms, resolution 0.43 mm × 0.43 mm × 5 mm) were acquired. In 8 patients, diffusion tensor imaging was also acquired (TR/TE = 7980/84 ms, b-values = 0/700 s/mm2, gradient directions = 12, FOV = 236 × 236 × 118 mm3, matrix = 128 × 128 × 64).
Image Processing
MRSI data were fitted with LCModel36 software to quantify the levels of 2HG and creatine. Cramér-Rao lower bounds (CRLB) less than 25% (relative CRLB) as calculated by LCModel were considered acceptable for metabolic goodness of fit. Three-dimensional metabolic maps were reconstructed from the LCModel fits using a combination of MINC (Montreal Neurological Institute), FSL (FMRIB Software Library), and Matlab (Mathworks) software tools.22
The 2HG signal was standardized using the mean value of hCr. With the presumption that the level of hCr within healthy white matter is stable and comparable across patients, we outlined reference tissue containing normal-appearing white matter in the hemisphere contralateral to the tumor site, and the mean value of hCr was calculated within this reference tissue. All values in the 2HG image voxels were divided by the mean value of hCr, and the generated map is called 2HG/hCr throughout this paper. Metabolic maps were further coregistered to the T2w anatomical image using robust register tools of Freesurfer software.37 Because of the 3D coverage and isotropic 1 mm high resolution, the T2w image was chosen as the common space for analysis of both metabolic and FLAIR images.
Tumor ROIs were manually outlined on 2D FLAIR images and verified by 2 neuroradiologists (O.R. and G.R.G.). The 5-mm-thick 2D clinical FLAIR images were upsampled to 1 mm in the slice direction with the help of the T2w image using a feature-based nonlocal means technique38 and then coregistered to the T2w image by rigid coregistration using FSL. Upsampling considerably reduced the partial volume effects caused by large slice thickness and improved visual appearance. Likewise, the FLAIR ROIs were similarly upsampled and coregistered to the T2w image. Since the MRSI FOV did not cover the entire brain, we masked the FLAIR FOV by the MRSI FOV so that the comparison was performed within the same volume of interest.
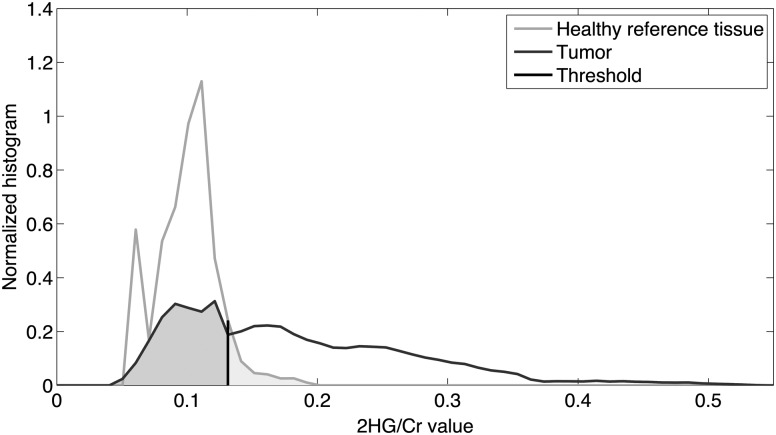
The next step was to threshold the 2HG/hCr image for tumor segmentation. The 2HG/hCr cutoff value was determined by minimizing the error rate of classifying tissues into tumor and healthy. To optimize this threshold across all patients, we generated normalized histograms for both tumor and healthy-appearing tissues, respectively. The total area under each histogram distribution was normalized to unity, rendering histograms independent from the volume of the ROI. We then averaged the histograms over all patients and obtained an average histogram for tumor and an average histogram for healthy-appearing reference tissues, respectively (Fig. 1). The optimal threshold was then calculated by minimizing the summation of the area under the curve for 2HG/hCr values below the threshold within the tumor and the area under the curve for 2HG/hCr values above the threshold within the healthy tissue, which corresponded to the average error when we classified the 2 tissue types by thresholding the 2HG/hCr values (shaded region in Fig. 1). Using this threshold, we segmented the tumor ROI to include all voxels that have 2HG/hCr values above the cutoff value.
Fig. 1.
Normalized histograms of 2HG/hCr values within tumor and healthy regions averaged over all patients. The optimal 2HG/hCr threshold of 0.13 for tumor classification has been determined by minimizing the overall error. Shaded areas under the curve correspond to the overall error for tissue classification.
To compare the 2HG/hCr and FLAIR ROIs and determine how similar they were, we calculated their Dice coefficient, as well as the distance between their centers of mass. A Dice coefficient39 is a measure for the similarity of 2 ROIs, ranging from 0 (no overlap) to 1 (perfect agreement) and is calculated by
where V2HG and VFLAIR are the 2HG/hCr and FLAIR ROIs and |·| is the size operator. Higher values of Dice coefficients correspond to more similarity and overlap of the 2 ROIs. We also compared the volumes of 2HG/hCr and FLAIR ROIs.
Radiation Treatment Plans
Volumes of 2HG/hCr and FLAIR were compared against clinical tumor volumes (CTVs) and planned target volumes defined in radiotherapy plans. For gliomas the CTV is obtained by expansion up to 20–25 mm (depending on the grade) from the gross tumor volume visible on diagnostic anatomical (MRI/CT) imaging, and the planned target volume is obtained by further extension of the CTV up to 5 mm.40 Typically the expansion is homogeneous and isotropic, with subsequent editing to avoid in some cases critical anatomical brain structures or account for anatomical barriers.
Results
Clinical and molecular profiles of the mutant IDH glioma patients are shown in Table 1. A total of 17 patients were included in this study. Patients were diagnosed with gliomas of WHO grades II (n = 5), III (n = 9), and IV (n = 3) and had confirmed IDH1 status of either canonical R132H mutation (n = 15) or noncanonical R132G (n = 1) and R132S (n = 1) mutations.
Histogram analysis of 2HG/hCr values in patients is presented in Fig. 1. The 2HG/hCr values ranged from 0.05 to 0.53 in the tumor histogram, and from 0.06 to 0.19 in the healthy histogram. The mean values of the 2 histograms were 0.2 for tumor and 0.1 for healthy, respectively. The 2HG/hCr cutoff value of 0.13 was found to best separate tumor and healthy 2HG/hCr histograms. As can be seen, the healthy histogram falls sharply to 0.19 after the 0.13 threshold, while the tumor histogram has a significantly higher and longer tail extending to 0.53, beyond the 0.13 cutoff value. Typically, a z-score is used in radiotherapy planning to express the level of trust in mapping tumor volume, and the threshold of 0.13 corresponds to a z-score of 1.34 in the case of 2HG/hCr. To further demonstrate reliable editing of the 2HG signal and the localization accuracy of our 3D MRSI method, results from phantom measurements are shown in Supplementary Fig. S1. The agreement between tumor ROIs defined by 2HG/hCr > 0.13 and manually outlined on FLAIR is further analyzed in Figs 2–4 and Table 2.
Fig. 2.
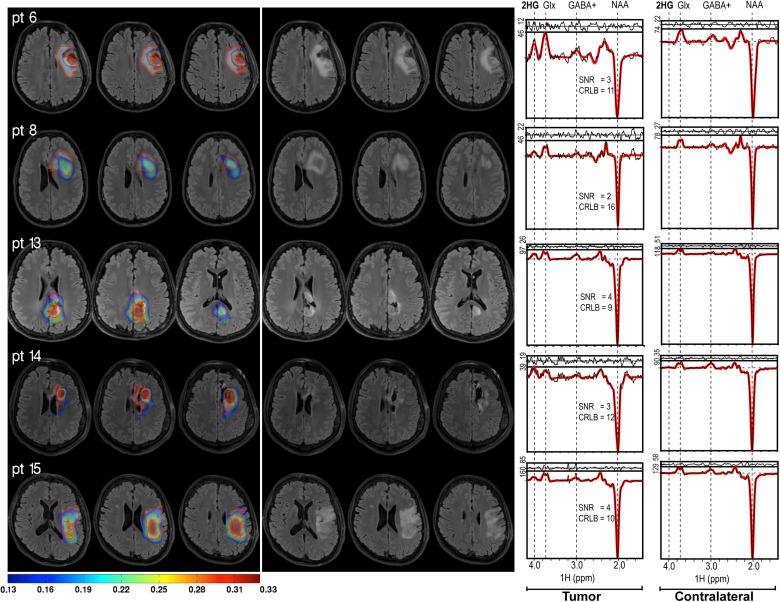
Example of representative 2HG/hCr maps overlaid on FLAIR images for 5 different patients. The outline of FLAIR ROIs is shown as red contour (left). The same 2HG/hCr intensity range (0.13–0.33) is shown for all patients. The corresponding FLAIR images are shown on the right. Examples of 2HG edited spectra from tumor and contralateral voxels are shown in the rightmost column, indicating the SNR and CRLB of 2HG.
Fig. 3.
Comparison of 2HG/hCr and FLAIR ROIs: (A) box plots of volumes, (B) scatter plot of volumes, (C) Dice coefficients, (D) distance between the centers of mass. Linear model fit (solid red line) with 95% CI (dashed red line) is shown in (B). Patients with volume of 2HG/hCr larger than volume of FLAIR are shown in red, and the opposite patients are shown in blue (C and D).
Fig. 4.
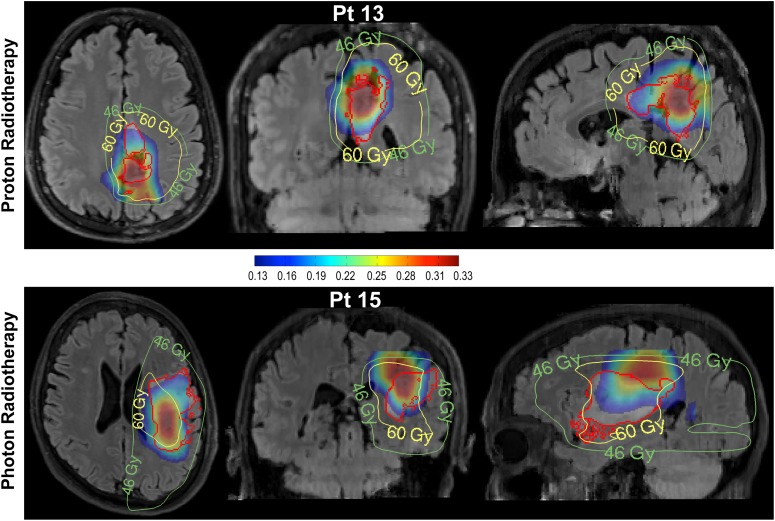
Radiation treatment plans and 2HG/hCr maps overlaid on the FLAIR image for one patient treated by proton beam therapy (upper row) and another patient treated by photon radiation therapy (lower row). The 2HG/hCr intensity scale is (0.13–0.33), radiation plans show the planned target volumes at 46 and 60 Gy. The FLAIR ROI is shown by the red contour.
Table 2.
Comparison of 2HG/Cre and FLAIR ROIs by distance between their centers of mass, Dice coefficient, and their volumes
| Patient # | Distance (mm) | Dice Coefficient | 2HG/Cre Volume (cc) | FLAIR* Volume (cc) | FLAIR** Volume (cc) |
|---|---|---|---|---|---|
| Pt 1 | 17.8 | 0.03 | 6.8 | 11.9 | 12.5 |
| Pt 2 | 18.8 | 0.29 | 30.2 | 55.9 | 111.2 |
| Pt 3 | 11.6 | 0.13 | 5.4 | 13.1 | 16.8 |
| Pt 4 | 8.1 | 0.26 | 9.3 | 27.5 | 45.5 |
| Pt 5 | 17.9 | 0.18 | 11.2 | 25.8 | 35.6 |
| Pt 6 | 8.3 | 0.49 | 37.3 | 52.8 | 68.6 |
| Pt 7 | 14.2 | 0.15 | 5.3 | 27.3 | 43.8 |
| Pt 8 | 15.8 | 0.42 | 44.3 | 101.4 | 155.7 |
| Pt 9 | 7.6 | 0.32 | 32.0 | 140.8 | 354.5 |
| Pt 10 | 12.4 | 0.18 | 10.4 | 7.6 | 9.7 |
| Pt 11 | 13.7 | 0.27 | 33.5 | 10.2 | 26.5 |
| Pt 12 | 23.5 | 0.23 | 92.7 | 38.9 | 58.5 |
| Pt 13 | 5.4 | 0.38 | 77.0 | 18.6 | 19.1 |
| Pt 14 | 3.2 | 0.37 | 40.8 | 11.9 | 12.5 |
| Pt 15 | 12.8 | 0.57 | 80.1 | 43.1 | 51.8 |
| Pt 16 | 6.7 | 0.31 | 67.5 | 14.8 | 25.4 |
| Pt 17 | 9.4 | 0.27 | 16.3 | 6.3 | 8.1 |
FLAIR volume: *limited to the field of view of 2HG MRSI; **total FLAIR volume.
Figure 2 shows representative 2HG/hCr maps thresholded above 0.13 and overlaid on the FLAIR images for several patients. For qualitative comparison, the outline of FLAIR ROI is highlighted. Although there is partial agreement on the approximate location of the tumors, the 2 ROIs are generally divergent in their shape and size, showing varying degrees of agreement. In some cases, the 2HG/hCr maps show foci of high signal at the edge of the FLAIR ROI. This scenario was seen in patients with small residual tumor after surgery (eg, patients 6 and 14 in Fig. 2). Across all patients, the distribution of 2HG/hCr is heterogeneous and asymmetric with respect to the FLAIR ROI. Example of 2HG edited spectra, together with spectral fitting results, SNR, and CRLB estimations are shown in the rightmost column.
Table 2 presents the quantitative comparison of 2HG/hCr and FLAIR ROIs for all subjects. The 2HG/hCr and FLAIR volumes had similar mean values (35.3 cc and 35.8 cc, respectively) but different ranges (5.3 cc–92.7 cc, and 6.3 cc–140.8 cc, respectively). Almost half of the patients (8 of 17) had 2HG/hCr volumes larger than FLAIR volumes. Note that this was true for both FLAIR volume restricted to the MRSI FOV as well as total FLAIR volume. There was a trend for weak correlation between the volumes of 2HG/hCr and FLAIR (r = 0.3, P = .24 by Spearman correlation). However, besides the global agreement that volumes provide, regional differences are also important. The Dice coefficients and distances between centers of mass of the 2 ROIs are a measure of the regional agreement. The distance between the centers of 2HG/hCr and FLAIR ROIs ranged from 3.2 mm to 23.5 mm with a mean value of 12.2 mm, while their Dice coefficients ranged from 0.03 to 0.57 with a mean value of 0.28, indicating an asymmetric displacement of the 2 ROIs. The MRSI restricted FLAIR volume ranged from 39% to 98% of the total FLAIR volume, with a smaller displacement between their centers from 0 to 12.49 mm (mean 5.25 mm).
Figure 3A–D compares graphically the volumes of 2HG/hCr and FLAIR ROIs. As shown by box plot in Fig. 3A, there is a larger difference between the median volumes of the 2 ROIs compared with their mean volumes, with a larger median volume for 2HG/hCr (32 cm3) than FLAIR (25.8 cm3). The scatter plot in Fig. 3B shows results of a linear model fit, with a moderate correlation between the 2 volumes and 2 large outliers where FLAIR volume exceeds by 2- to 3-fold the 2HG/hCr volumes. Figure 3C reports the Dice coefficients and the distances between the centers of mass of the 2HG/hCr and FLAIR ROIs. In general the regional agreement of 2 ROIs is dominated by the size of the 2HG/hCr ROI. Thus, higher Dice coefficients and lower intercenter distances were found for patients that had the volume of 2HG/hCr larger than the volume of FLAIR. The mean Dice coefficient for patients who had 2HG/hCr volume larger than FLAIR volume was 0.34 versus 0.24 (P = .09) for the opposite patients, while the intercenter distances were 10.8 mm versus 13.3 mm (P = .3), respectively, for the same 2 groups of patients. There was more variation (58%) for Dice coefficients when FLAIR volume was larger than 2HG/hCr volume versus variation (37%) for the opposite scenario. In addition, we investigated the potential relation between SNR of 2HG, tumor grade, tumor volume, and mismatch of 2HG and FLAIR volumes. Results presented in Supplementary Fig. S2 indicate a trend for correlation between SNR and tumor volume (Supplementary Fig. S2A) or tumor grade (Supplementary Fig. S2B). A trend for increased Dice coefficients for higher-grade tumors is also shown by our data (Supplementary Fig. S2C); however, no correlation reached statistical significance.
Figure 4 illustrates the type of additional information that may be available from 2HG/hCr volumetric assessment, using 2 opposing scenarios from one patient treated with proton beam therapy (upper row) and another patient treated with photon radiation therapy (lower row), respectively. In the case of the proton beam therapy patient with a grade II glioma, it can be seen that high 2HG/hCr is limited to only 1 of the 2 FLAIR hyperintense areas, while high radiation dose covers the entire FLAIR ROI, including areas of low 2HG/hCr. On the other hand, in the case of the patient treated with photon radiation therapy who had a large glioblastoma tumor, it can be seen that areas of high 2HG/hCr extend superiorly in the coronal/sagittal views into regions of low radiation dose (Fig. 4, lower row). Note that 2HG/hCr levels are comparable between the low-grade (patient 13) and high-grade (patient 15) glioma patients. Although this seems to contradict the trend and expectation that 2HG levels may be larger in higher-grade tumors due to increased cell density, in this case the similar results are explained by the fact that noncanonical mutations such as IDH1-R132G in patient 13 are more efficient41 in producing 2HG compared with canonical IDH1-R132H mutations in patient 15, hence compensating for cell density.
Discussion
A mainstay of treatment for gliomas is radiation therapy, which in the modern era is administered using carefully modulated techniques to optimize target coverage and minimize normal brain exposure. However, target definition is a challenge for radiation planning and there are known limitations to the specificity of anatomical MRI such as FLAIR for characterizing the extent of these infiltrative tumors. Hence molecular imaging methods are needed to more specifically evaluate gliomas for improved tumor definition and to facilitate better treatment planning and monitoring. To investigate this potential, we compared 2HG and FLAIR imaging and evaluated their regional and global agreement in mutant IDH glioma patients.
In the case of mutant IDH gliomas, a potential imaging biomarker is the 2HG/hCr ratio, for which we determined an optimal normalization and thresholding. The normalized 2HG values to the mean value of healthy creatine (2HG/hCr) may be used to distinguish mutant versus nonmutant IDH brain tissue intrasubject and segment mutant IDH tumor volume. Using the normalized 2HG images, we obtained the cutoff value of 0.13 for 2HG/hCr above which the tumor classification error was minimized compared with healthy tissues. Assuming a total creatine (creatine + phosphocreatine) level of 5–10 mM in the healthy brain,42 the 0.13 threshold for the 2HG/hCr ratio corresponds to a minimum 2HG level that is in good agreement with the minimum concentration detectable by in vivo MRSI. The 0.13 threshold corresponded to a z-score of 1.34 for 2HG/hCr tumor volume mapping.
Importantly, we observed that 2HG/hCr and FLAIR do not completely agree on the spatial distribution and characteristics of the tumor. In general, there was more agreement in the global measures such as mean tumor volumes. There was more disagreement in the regional measures of tumors, suggesting that there are substantial opportunities for improvement in target definition. This disagreement was evident from the relatively small values of Dice coefficients and comparatively large values of distance between the centers of masses of the 2 ROIs. There was a trend for more disagreement when FLAIR volume was larger than 2HG/hCr volume, which may be explained by the fact that FLAIR probes a mixture of tumor and peritumoral edema. There was also a trend for higher SNR of 2HG for larger and higher-grade tumors, with a trend for more overlap between 2HG/hCr and FLAIR volumes for higher-grade tumors. Although not statistically significant, these trends suggest increased 2HG production throughout the entire FLAIR volume for higher-grade tumors. Visual analysis revealed also that the level of 2HG/hCr was not uniform within the FLAIR ROI, often with a high-intensity area in an eccentric position and extending beyond the FLAIR margin. Similar results have been recently reported for choline hyperintensity extending outside FLAIR volume in an MRSI study on glioblastoma patients.30
These findings have significant implications and may be relevant for treatment planning by radiotherapy as well as for treatment monitoring. Conventionally, radiation therapy is planned with substantial reliance on FLAIR images. Areas of hyperintensity are outlined in the FLAIR images and radiation plans are designed in such a way that the FLAIR ROI and an isotropic expansion margin around it receive uniform and high levels of radiation. The use of FLAIR images for radiation planning may not be ideal because the intensity of FLAIR images may not correspond to the concentration of cancer cells. On the other hand, recent results have been reported showing that 2HG levels detected by single-voxel MRS correlate with tumor cell density determined by histopathology on biopsies.43 To investigate this possibility, we analyzed the relation between 2HG levels in our multivoxel MRSI and apparent diffusion coefficient (ADC) as a surrogate imaging biomarker for cell density (Supplementary Fig. S3). We found an inverse correlation between 2HG levels and ADC values, with tumor areas that have low ADC (hence higher cell density) showing larger 2HG levels. With nonuniform levels of cancer cells within a tumor, uniform irradiation may not be an optimal approach. On the other hand, multiple-region biopsy sampling of the brain tissues in order to specify the concentration of tumor cells within and around the tumor is not practical for brain tumors. Thus, other imaging biomarkers are needed for radiation therapy planning.
In particular, on closer inspection it appears that for some patients (Fig. 4), molecular imaging by 2HG/hCr may be valuable in refining radiotherapy planning. Assuming the 2HG/hCr map represents the most metabolically active component of the tumor, these cases suggest that the radiation therapy target may be suboptimally defined by FLAIR alone and could be improved by incorporating the 2HG/hCr map. Hence, the 2HG/hCr maps have the potential to guide modulation of the radiation dose according to the underlying molecular biology of the tumor, in addition to anatomical imaging sequences. Conversely, there is clearly a threshold effect with 2HG/hCr identifying areas of higher tumor cell density, and the optimal contribution of each sequence for tumor target delineation remains to be determined.
Mutant IDH glioma are nonetheless an important test case for the convergence of radiation genomics44 and imaging genomics.45 Integrating 2HG/hCr and FLAIR maps into radiation therapy has 2 potential benefits: (i) radiation dose can be increased to regions of greater tumor cell density as determined by areas of high 2HG/hCr, and (ii) regions of FLAIR hyperintensity having low 2HG/hCr signal may be spared high-dose radiation and thereby reduce the potential adverse effects of radiation therapy. The increased radiosensitivity12 and particular imaging phenotype3 of mutant IDH glioma might also suggest different radiation strategies46–48 compared with wild-type patients.
Despite the potential advantages, there are several important practical limitations for 2HG imaging that are related to MRSI data acquisition, such as brain coverage, data quality, spatial resolution, and sensitivity. MRSI protocols often use limited brain coverage to avoid large artifacts from subcutaneous and bone marrow lipids of the head. Current 2HG imaging protocols using this type of acquisition may not cover the entire tumor, especially if the tumor is peripheral in the brain. The quality of MRSI data is not homogeneous across the brain, in particular spectral resolution and SNR are reduced in regions with inhomogeneous B0 magnetic field from air–tissue interfaces around sinuses, bullae, and ear canals. Because of the generally lower concentration of metabolites, the spatial resolution in MRSI is lower compared with anatomical MRI, and typically the minimum concentration of metabolites that can be detected by in vivo MRSI in humans is around 1 mM. Thus, patients who have tumors with unfavorable location, small size, lower tumor cell density, and less efficient 2HG production may not be adequately imaged with 2HG MRSI. However, technical improvements are possible for data quality and spatial resolution to expand the usage of 2HG imaging into these patients. One other limitation today is that our real-time 3D MRSI methodology for 2HG editing was implemented and operational only on Siemens scanners, though porting it to other platforms is certainly possible. Thus, at the moment, 2HG imaging clearly cannot replace anatomical imaging such as FLAIR for radiotherapy planning, but it may provide valuable further information that could guide spatiotemporal modulation of the radiation dose. Further investigation is required to determine how treatment planning can be optimally modified having the 2HG/hCr maps.
In summary, we demonstrated that a 2HG/hCr map provides molecular assessment of mutant IDH tumors, which is complementary to FLAIR, and may provide significant additional utility for tumor definition in treatment planning and subsequent monitoring. We anticipate that this approach may have a significant impact on the management and outcome of mutant IDH glioma patients.
Supplementary Material
Funding
This work was supported by the National Cancer Institute (1K22CA178269-01 to O.CA., 1P50CA165962-01 to T.T.B and D.P.C), Harvard-MIT Bridge Project (D.P.C. and O.C.A.), Burroughs-Wellcome Career Award (D.P.C.), and MGH-MIT Grand Challenge 3: Neuroscience (O.C.A.).
Supplementary Material
Acknowledgments
We acknowledge Drs Borjan Gagoski (Boston Children Hospital), Andrew van der Kouwe and Dylan Tisdall (Massachusetts General Hospital), and Himanshu Bhat (Siemens Medical Solutions) for help with MRSI sequence development, and Dr Malgorzata Marjanska (CMRR, University of Minnesota) for providing the LCModel basis set for 2HG fitting.
Conflict of interests statement. There are no conflicts of interest to disclose.
References
- 1.Parsons DW, Jones S, Zhang XS et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yan H, Parsons DW, Jin GL et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carrillo JA, Lai A, Nghiemphu PL et al. Relationship between tumor enhancement, edema, IDH1 mutational status, MGMT promoter methylation, and survival in glioblastoma. AJNR Am J Neuroradiol. 2012;33(7):1349–1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beiko J, Suki D, Hess KR et al. IDH1 mutant malignant astrocytomas are more amenable to surgical resection and have a survival benefit associated with maximal surgical resection. Neuro Oncol. 2014;16(1):81–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dang L, White DW, Gross S et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462(7274):739–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Noushmehr H, Weisenberger DJ, Diefes K et al. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turcan S, Rohle D, Goenka A et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu C, Ward PS, Kapoor GS et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483(7390):474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu W, Yang H, Liu Y et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chowdhury R, Yeoh KK, Tian Y-M et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12(5):463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bleeker FE, Atai NA, Lamba S et al. The prognostic IDH1 (R132) mutation is associated with reduced NADP(+)-dependent IDH activity in glioblastoma. Acta Neuropathol. 2011;119(4):487–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li S, Chou AP, Chen W et al. Overexpression of isocitrate dehydrogenase mutant proteins renders glioma cells more sensitive to radiation. Neuro Oncol. 2013;15(1):57–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tran AN, Lai A, Li S et al. Increased sensitivity to radiochemotherapy in IDH1 mutant glioblastoma as demonstrated by serial quantitative MR volumetry. Neuro Oncol. 2014;16(3):414–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Bent MJ, Afra D, de Witte O et al. Long-term efficacy of early versus delayed radiotherapy for low-grade astrocytoma and oligodendroglioma in adults: the EORTC 22845 randomised trial. Lancet. 2005;366(9490):985–990. [DOI] [PubMed] [Google Scholar]
- 15.Douw L, Klein M, Fagel SS et al. Cognitive and radiological effects of radiotherapy in patients with low-grade glioma: long-term follow-up. Lancet Neurol. 2009;8(9):810–818. [DOI] [PubMed] [Google Scholar]
- 16.Forst DA, Nahed BV, Loeffler JS et al. Low-grade gliomas. Oncologist. 2014;19(4):403–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stall B, Zach L, Ning H et al. Comparison of T2 and FLAIR imaging for target delineation in high grade gliomas. Radiat Oncol. 2010;5(5) doi:10.1186/1748-1717X-1185-1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carrillo JA, Lai A, Nghiemphu P et al. Non-contrast enhancement in glioblastoma is associated with IDH-1 mutation and multifocality. Neuro Oncol. 2010;12:113–113. [Google Scholar]
- 19.Wen PY, Macdonald DR, Reardon DA et al. Updated response assessment criteria for high-grade gliomas: Response Assessment in Neuro-Oncology working group. J Clin Oncol. 2010;28(11):1963–1972. [DOI] [PubMed] [Google Scholar]
- 20.van den Bent MJ, Wefel JS, Schiff D et al. Response assessment in neuro-oncology (a report of the RANO group): assessment of outcome in trials of diffuse low-grade gliomas. Lancet Oncol. 2011;12(6):583–593. [DOI] [PubMed] [Google Scholar]
- 21.Brandsma D, van den Bent MJ, Chang JH et al. Pseudoprogression and pseudoresponse in the management of high-grade glioma: optimal decision timing according to the response assessment of the neuro-oncology working group. Curr Opin Neurol. 2009;22(6):633–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andronesi OC, Loebel F, Bogner W et al. Treatment response assessment in IDH-mutant glioma patients by non-invasive 3D functional spectroscopic mapping of 2-hydroxyglutarate. Clin Cancer Res. 2016;22(7):1632–1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Andronesi OC, Kim GS, Gerstner E et al. Detection of 2-hydroxyglutarate in IDH-mutated glioma patients by in vivo spectral-editing and 2D correlation magnetic resonance spectroscopy. Sci Transl Med. 2012;4(116):116ra114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi C, Ganji SK, DeBerardinis RJ et al. 2-Hydroxyglutarate detection by magnetic resonance spectroscopy in subjects with IDH-mutated gliomas. Nat Med. 2012;18(4):624–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pope WB, Prins RM, Thomas MA et al. Non-invasive detection of 2-hydroxyglutarate and other metabolites in IDH1 mutant glioma patients using magnetic resonance spectroscopy. J Neurooncol. 2012;107(1):197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Elkhaled A, Jalbert LE, Phillips JJ et al. Magnetic resonance of 2-hydroxyglutarate in IDH1-mutated low-grade gliomas. Sci Transl Med. 2012;4(116):116ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kalinina J, Carroll A, Wang L et al. Detection of "oncometabolite" 2-hydroxyglutarate by magnetic resonance analysis as a biomarker of IDH1/2 mutations in glioma. J Mol Med (Berl). 2012;90(10):1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang CF, Chan AA, Larson D et al. Potential value of MR spectroscopic imaging for the radiosurgical management of patients with recurrent high-grade gliomas. Technol Cancer Res Treat. 2007;6(5):375–382. [DOI] [PubMed] [Google Scholar]
- 29.Ken S, Vieillevigne L, Franceries X et al. Integration method of 3D MR spectroscopy into treatment planning system for glioblastoma IMRT dose painting with integrated simultaneous boost. Radiat Oncol. 2013;8(1) doi:10.1186/1748-1717X-1188-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parra NA, Maudsley AA, Gupta RK et al. Volumetric spectroscopic imaging of glioblastoma multiforme radiation treatment volumes. Int J Radiat Oncol Biol Phys. 2014;90(2):376–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bogner W, Gagoski B, Hess AT et al. 3D GABA imaging with real-time motion correction, shim update and reacquisition of adiabatic spiral MRSI. Neuroimage. 2014;103:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Capper D, Zentgraf H, Balss J et al. Monoclonal antibody specific for IDH1 R132H mutation. Acta Neuropathol. 2009;118(5):599–601. [DOI] [PubMed] [Google Scholar]
- 33.Chi AS, Batchelor TT, Dias-Santagata D et al. Prospective, high-throughput molecular profiling of human gliomas. J Neurooncol. 2012;110(1):89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cairncross JG, Ueki K, Zlatescu MC et al. Specific genetic predictors of chemotherapeutic response and survival in patients with anaplastic oligodendrogliomas. J Natl Cancer Inst. 1998;90(19):1473–1479. [DOI] [PubMed] [Google Scholar]
- 35.Hegi ME, Diserens AC, Gorlia T et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. [DOI] [PubMed] [Google Scholar]
- 36.Provencher SW. Estimation of metabolite concentrations from localized in-vivo proton NMR-spectra. Magn Reson Med. 1993;30(6):672–679. [DOI] [PubMed] [Google Scholar]
- 37.Reuter M, Rosas HD, Fischl B. Highly accurate inverse consistent registration: a robust approach. Neuroimage. 2010;53(4):1181–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jafari-Khouzani K. MRI upsampling using feature-based nonlocal means approach. IEEE Trans Med Imaging. 2014;33(10):1969–1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zou KH, Warfield SK, Bharatha A et al. Statistical validation of image segmentation quality based on a spatial overlap index. Acad Radiol. 2004;11(2):178–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitfield GA, Kennedy SR, Djoukhadar IK et al. Imaging and target volume delineation in glioma. Clin Oncol (R Coll Radiol). 2014;26(7):364–376. [DOI] [PubMed] [Google Scholar]
- 41.Ward PS, Cross JR, Lu C et al. Identification of additional IDH mutations associated with oncometabolite R(-)-2-hydroxyglutarate production. Oncogene. 2012;31(19):2491–2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Govindaraju V, Young K, Maudsley AA. Proton NMR chemical shifts and coupling constants for brain metabolites. NMR in Biomedicine. 2000;13(3):129–153. [DOI] [PubMed] [Google Scholar]
- 43.de la Fuente MI, Young RJ, Rubel J et al. Integration of 2-hydroxyglutarate-proton magnetic resonance spectroscopy into clinical practice for disease monitoring in isocitrate dehydrogenase-mutant glioma. Neuro Oncol. 2016;18(2):283–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kerns SL, West CM, Andreassen CN et al. Radiogenomics: the search for genetic predictors of radiotherapy response. Future Oncol. 2014;10(15):2391–2406. [DOI] [PubMed] [Google Scholar]
- 45.ElBanan MG, Amer AM, Zinn PO et al. Imaging genomics of glioblastoma: state of the art bridge between genomics and neuroradiology. Neuroimaging Clin N Am. 2015;25(1):141–153. [DOI] [PubMed] [Google Scholar]
- 46.Shih HA, Sherman JC, Nachtigall LB et al. Proton therapy for low-grade gliomas: results from a prospective trial. Cancer. 2015;121(10):1712–1719. [DOI] [PubMed] [Google Scholar]
- 47.Unkelbach J, Menze BH, Konukoglu E et al. Radiotherapy planning for glioblastoma based on a tumor growth model: implications for spatial dose redistribution. Phys Med Biol. 2014;59(3):771–789. [DOI] [PubMed] [Google Scholar]
- 48.Dennis ER, Bussiere MR, Niemierko A et al. A comparison of critical structure dose and toxicity risks in patients with low grade gliomas treated with IMRT versus proton radiation therapy. Technol Cancer Res Treat. 2013;12(1):1–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.