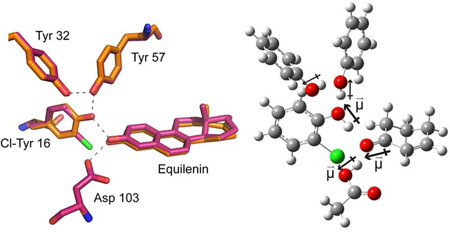
Abstract
The vibrational Stark effect (VSE) has been used to measure the electric field in the active site of ketosteroid isomerase (KSI). These measured fields correlate with ΔG‡ in a series of conventional mutants yielding an estimate for the electrostatic contribution to catalysis (Fried et al. Science, 2014, 346, 1510–1513). In this work we test this result with much more conservative variants in which individual Tyr residues in the active site are replaced by 3-chlorotyrosine via amber suppression. The electric fields sensed at the position of the carbonyl bond involved in charge displacement during catalysis were characterized using the VSE, where the field sensitivity has been calibrated by vibrational Stark spectroscopy, solvatochromism, and MD simulations. A linear relationship is observed between the electric field and ΔG‡ that interpolates between wild-type and more drastic conventional mutations, reinforcing the evaluation of the electrostatic contribution to catalysis in KSI. A simplified model and calculation are developed to estimate changes in the electric field accompanying changes in the extended hydrogen-bond network in the active site. The results are consistent with a model in which the O-H group of a key active site tyrosine functions by imposing a static electrostatic potential onto the carbonyl bond. The model suggests that the contribution to catalysis from the active site hydrogen bonds is of similar weight to the distal interactions from the rest of the protein. A similar linear correlation was also observed between the proton affinity of KSI’s active site and the catalytic rate, suggesting a direct connection between the strength of the H-bond and the electric field it exerts.
Graphical abstract
Introduction
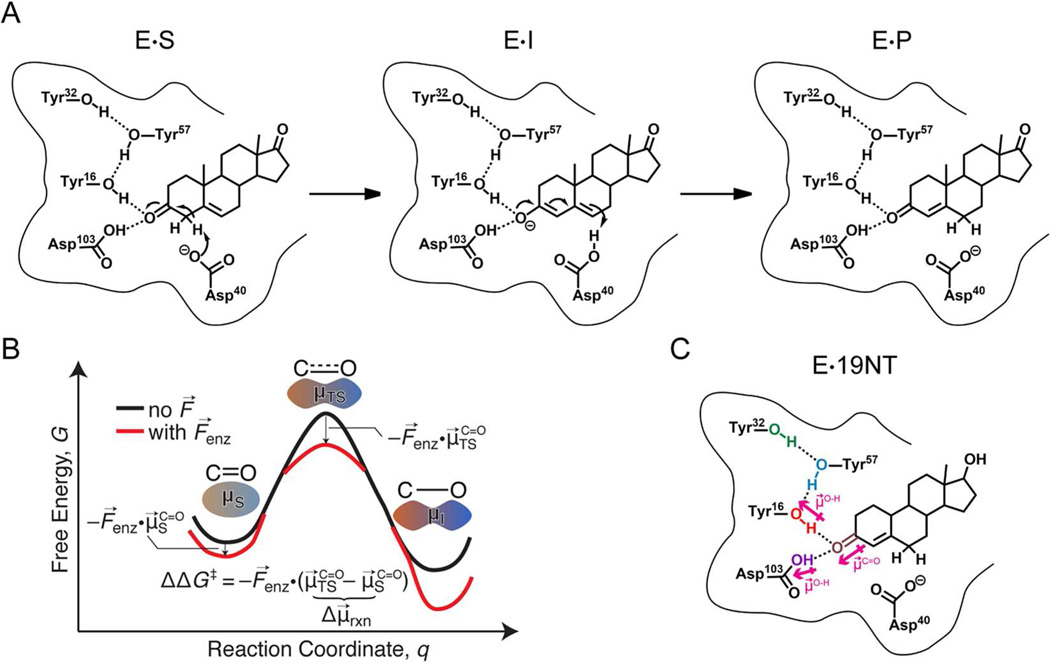
Electrostatic stabilization has been widely discussed as contributing to enzymes’ high catalytic proficiency.1 The essential concept is that enzymes create a pre-organized electrostatic environment in the active site that preferentially stabilizes the charge distribution of the transition state more than the substrate to accelerate the reaction.1,2 Recent work from our lab using the vibrational Stark effect (VSE) to probe the electric field exerted at the active site of the model enzyme ketosteroid isomerase (KSI) provided an experimental approach to quantifying the contribution of electrostatic stabilization to catalysis in this enzyme.3 Ketosteroid isomerase catalyzes the isomerization of its steroid substrate via formation of a dienolate intermediate with a reaction rate approximately 1011.5-fold faster than the uncatalyzed reaction (Figure. 1A).4,5 An extended H-bonding network is present in the active site composed of the tyrosine triad (Y16, Y32, and Y57) and aspartic acid 103 (Asp103). Fried et al.3 utilized a product analog, 19-nortestosterone (19NT, Figure 1C), whose C=O group is located in KSI’s active site in the same manner as the carbonyl bond of the substrate that undergoes a charge rearrangement in KSI’s rate-limiting step (Figure 1B), to probe the electric field sensed by this bond. The IR spectral shifts for this carbonyl group were evaluated using vibrational Stark spectroscopy, solvatochromic data and molecular dynamics simulations6 and demonstrated that a very large electric field is exerted onto the C=O group of 19NT. The magnitude of the electric field was found to be linearly correlated with the activation free energy in wild-type and a series of conventional mutants, and from this the electrostatic contribution to catalysis could be obtained.3
Figure 1. Catalysis by ketosteroid isomerase (KSI) and product-like inhibitor, 19-nortestosterone (19NT).
(A) Reaction mechanism for KSI catalyzed isomerization of 5-androstene-3,17-dione (substrate) to 4-androstene-3,17-dione (product). In the intermediate state, the H-bond network formed by the tyrosine triad and Asp103 (oxyanion hole) stabilizes the negative charge of the enolate. The dipole moment along the C=O bond increases as the reaction proceeds from substrate to intermediate, and the possible effect of the electric field, F⃗enz, from the protein on the free energy of the transition state compared with the starting state is illustrated in the simplified reaction coordinate diagram in (B). (C) Complex between KSI and inhibitor 19NT that closely mimics the natural substrate.
While site-directed mutagenesis is widely used, a legitimate criticism of this approach, particularly for active site residues in enzymes, is that the changes in chemical functionality using natural amino acids are intrinsically large (e.g. Tyr to Phe or Asp to Asn) and can lead to unexpected structural rearrangements or water binding pockets which can confound the interpretation of the results. More specifically in the case of KSI (and a number of other enzymes), the removal of specific H-bond interactions prevents a direct comparison of electrostatic stabilization with a widely-discussed alternative proposal that KSI’s active site preferentially forms a strong, short H-bond with the intermediate relative to the substrate to provide additional stabilization energy.7,8 Therefore, in this work we introduce more subtle and conservative changes by site-specifically replacing each of the key Tyr residues with 3-Cl-Tyr (Cl-Y)9 in the KSI active site10 to provide a critical test of the earlier analysis on electric field/activation free energy correlations. We then probe the physical basis of the large electric field and demonstrate the functional connection between the preferential H-bond interactions and electrostatic stabilization.
Results and Discussion
The electrostatic contribution to catalysis in conservative KSI-Cl-Y mutants
The enzymatic activities of the wild type and the KSI-Cl-Y variants on the natural substrate 5-androstenedione (5-AND) were determined with high accuracy (Table 1). Similar to the effect on the catalysis of the slow substrate 5(10)-estrene-3,17-dione (Table S1)10, small but systematic changes were observed in these conservative variants, making them ideal candidates to critically test the electric field/activation free energy correlations mapped by vibrational Stark spectroscopy.3
Table 1.
Kinetic Parameters for Wild-type (WT) and Cl-Y-KSI variants (mutant) with 5-AND
|
kcat (103 s−1) |
KM (µM) |
kcat / KM (107M−1 s−1) |
kcat ratio (mutant/WT) |
kcat / KM ratio (mutant/WT) |
ΔG‡
[a] (kcal mol−1) |
|
|---|---|---|---|---|---|---|
| WT | 17.3 ± 0.5 | 78 ± 5 | 22.2 ± 2.1 | 1 | 1 | 11.68 ± 0.02 |
| Cl-Y57 | 4.08 ± 0.26 | 41 ± 6 | 9.9 ± 2.0 | 0.24 ± 0.02 | 0.45 ± 0.13 | 12.54 ± 0.04 |
| Cl-Y32 | 13.8 ± 1.1 | 79 ± 12 | 17.5 ± 4.0 | 0.80 ± 0.09 | 0.79 ± 0.25 | 11.82 ± 0.05 |
| Cl-Y16 | 4.51 ± 0.30 | 48 ± 7 | 9.3 ± 1.9 | 0.26 ± 0.02 | 0.42 ± 0.12 | 12.48 ± 0.04 |
| Cl-Y57 D103N |
0.336 ± 0.012 | 70 ± 5 | 0.56 ± 0.06 | 0.019 ± 0.001 | 0.025 ± 0.005 | 14.02 ± 0.02 |
ΔG‡ = −R·T· ln[kcat/(kBT/h)]; T = 293K; R = gas constant; kB = Bolzmann’s constant; h = Planck’s constant
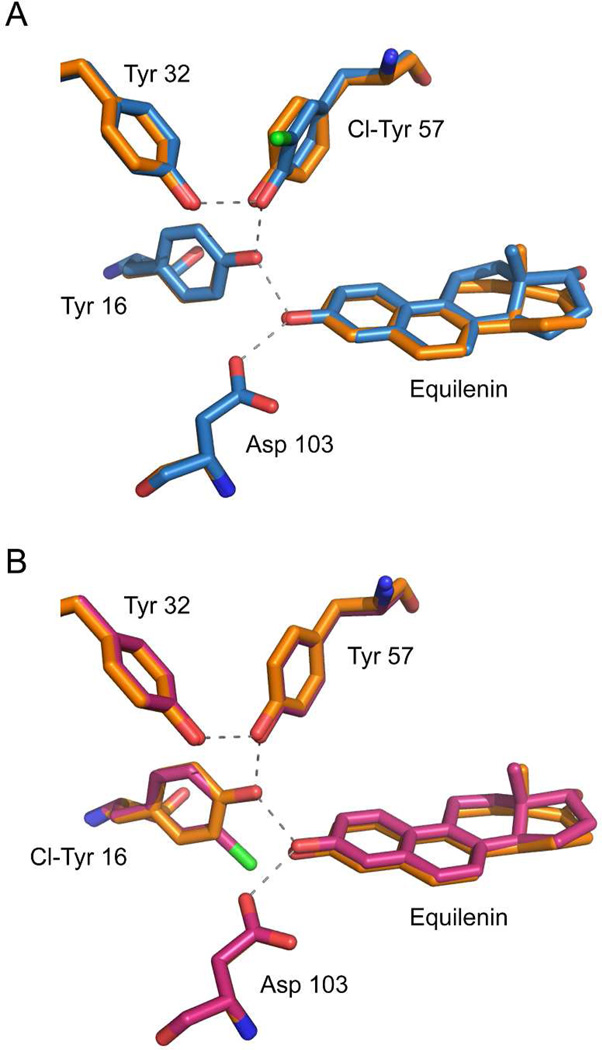
The crystal structures of KSI-Cl-Y16 and KSI-Cl-Y57 bound with the transition state analog equilenin (PDB ID: 5KP1, 5KP3) have an overall RMS deviation of 0.20 and 0.23 Å when superimposed onto the structure of wild type KSI (PDB: 1OH0), suggesting little change in the overall enzyme architecture within the error of the structures (Figure 2, statistics in Table S2). Equilenin was chosen as the bound substrate for crystallization because of its tight binding and its similar binding mode as that of 19NT (Figure S1, PDB ID: 5KP4). Focusing on the H-bond network in the active site, in KSI-Cl-Y57, the plane of the benzene ring of Cl-Y57 rotates about 27° from that of Y57 in wild type and the O-O distance between Cl-Y57 and Y16 is shortened by 0.15 Å. The rest of the active site residues are unperturbed within the uncertainty of the X-ray structures (Table S3). In KSI-Cl-Y16, the plane of the benzene ring of Cl-Y16 rotates about 10° from that of Y16 in the wild type, the O-O distance between Y57 and Cl-Y16 is shortened by 0.07 Å on average, and that between Asp103 and the ligand is lengthened by 0.10 Å with the rest unperturbed. We note that although the benzene rings of the Cl-Y residues are not co-planar with their tyrosine counterparts in WT, the hydroxyl group of the residue and the carbonyl of the substrate (the chemical group directly responsible for the H-bond) retain the same relative orientation as in WT. Also, the position of the general base Asp40 is preserved for both variants (Figure S2), minimizing the entropic perturbation to catalysis from misplacing the residue directly involved in the reaction (i.e. the breaking and formation of covalent bonds, Figure 1A).11,12
Figure 2. Structures of the tyrosine clusters in KSI-Cl-Y variants bound with transition state analog equilenin, superimposed onto a high-resolution structure of KSI bound with equilenin (1OH0), shown in orange.
(A) KSI-Cl-Y57 (chain a) aligned with wild type (chain a) with an overall RMS of 0.23. (B) KSI-Cl-Y16 (chain a) aligned with wild type (chain a) with an overall RMS of 0.20.
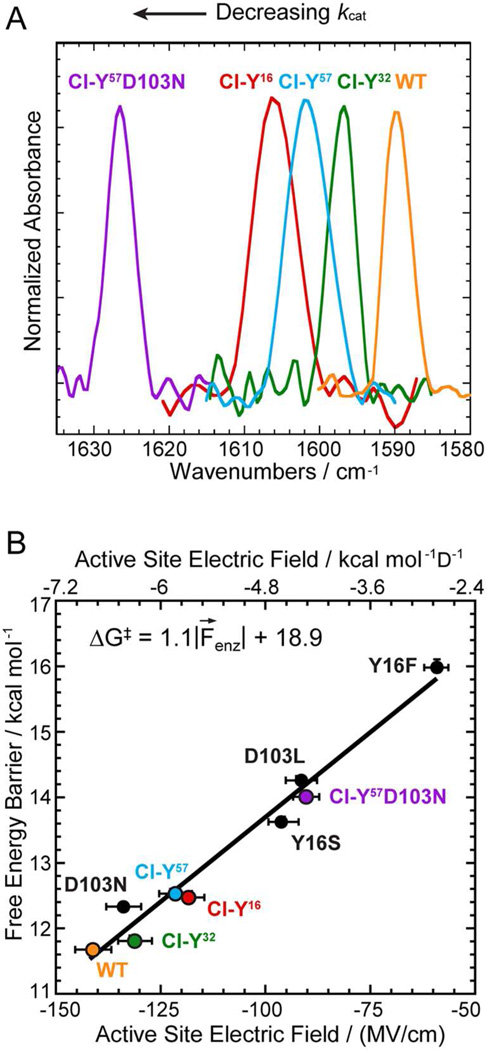
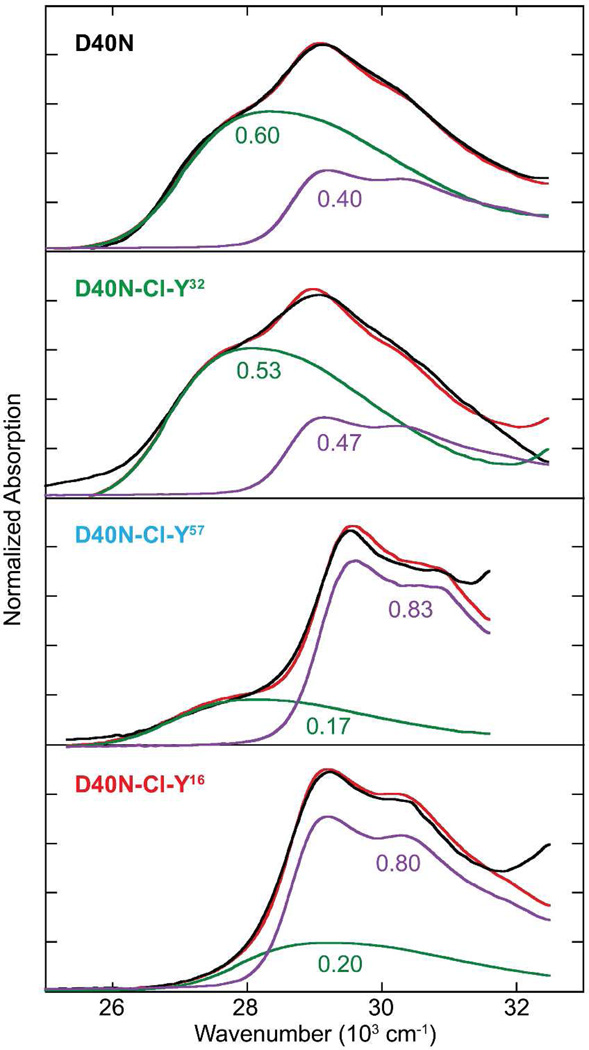
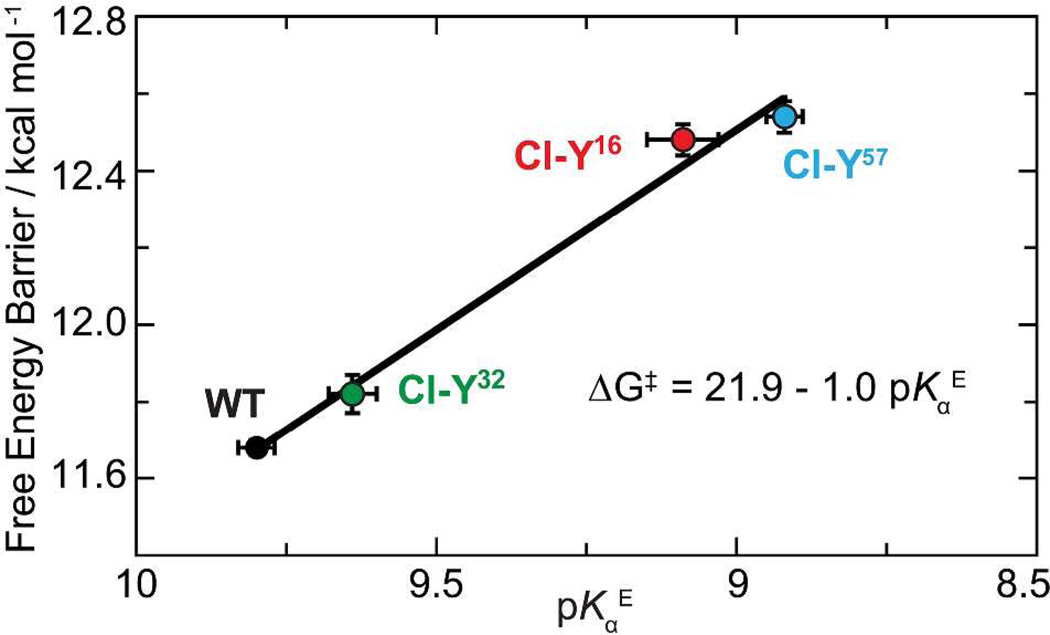
The preservation of the active site H-bond network in these conservative variants allows us to relate the activation free energy to the frequency shift of the IR stretch of the carbonyl group of bound 19NT, and from this to the electric field felt by the carbonyl group. As shown in Figure 3A, there is a systematic shift to the blue of the C=O stretch band as Y is replaced by Cl-Y at different positions. The electric field corresponding to these frequency shifts in the KSI variants was assigned to an absolute scale from 19NT’s solvatochromism calibration curve (Table S4).3,13,14 When the apparent activation energies of these conservative variants and their corresponding electric fields were plotted together with the conventional mutants, a robust linear correlation was observed (Figure 3B; R2 = 0.98). The correlation between the activation free energy and the electric field is highly similar to the one derived from the conventional mutants (Figure S3),3 demonstrating that changes in electrostatics associated with small perturbations on the H-bond interactions affect catalysis by the same physical mechanism as changes in electrostatics caused by large perturbations.
Figure 3. Preservation of the linear correlation between electric field and activation free energy in the conservative variant series using Cl-Y substitutions.
(A) Infrared spectra of 19NT sulfate bound to the active site of wild-type and KSI-Cl-Y variants blue shift as kcat decreases. (B) Plot of enzymatic unimolecular free energy barrier, ΔG‡, against the electric field |F⃗enz| experienced by 19NT sulfate’s C=O group in the active site of each KSI variant obtained from the IR frequency/field correlation for 19NT.
A simple model to relate changes in the electric field to changes in the extended hydrogen-bond network
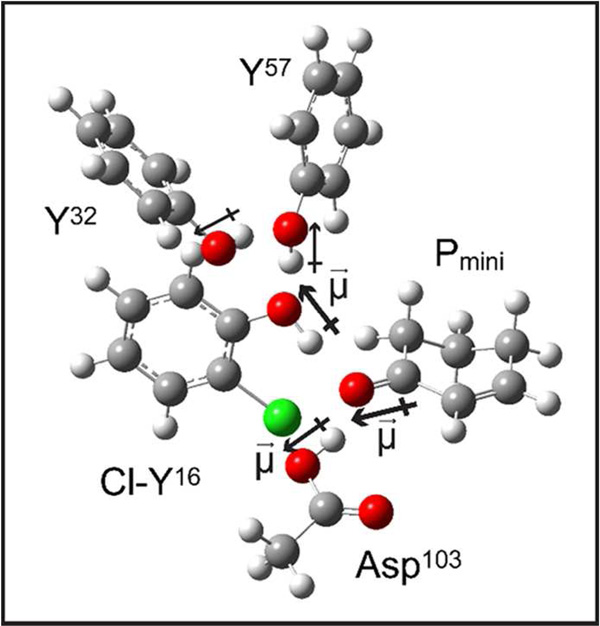
The KSI-Cl-Y variants offer a relatively simple system to examine the origins of changes in the electric field and their connection with the H-bond interactions. The most straightforward model to relate the resulting electric field to the H-bond network is the dominant dipole moments of the O-H group of tyrosine and Asp103 imposing a static electrostatic potential onto the carbonyl of 19NT (Figure 1C). The carbonyl group would experience a smaller electric field when H-bonded with Cl-Tyr than Tyr as the inductive effect of Cl decreases its O-H dipole moment. Such an effect was indeed observed in a simple solution mixture mimicking this particular aspect of KSI’s active site interaction: placing acetophenone in an excess amount of 2-chlorophenol from phenol blue-shifts the vibrational frequency of its carbonyl group by 3 cm−1, corresponding to a ~3 MV/cm decrease in electric field (Figure S4). While conceptually similar, the actual effect upon Cl-Tyr substitution at each site in the Tyr H-bond network of KSI is more profound and complex, and a proper treatment would require a full quantum simulation of the cluster as described earlier for the apo-enzyme15; here we provide a semi-quantitative estimate for the change in electric field in each mutant based on a simplified computational model of the active site consisting of phenol, 2-chlorophenol and acetic acid to represent the active site residues Y32/Y57, Cl-Y16, and Asp103, respectively, and 2-cyclohexen-1-one to mimic the A-ring of 19NT that is directly involved in the H-bond network (Figure 4). The change in the electric field on the carbonyl of 19NT can be the result of a smaller O-H dipole moment of the modified Tyr or/and a shift in the relative distances and orientations of the active site residues’ hydroxyl groups and the carbonyl. Therefore, we first performed electric field and partial charges calculations on the simplified active site complex with fixed geometry, where the heavy atom coordinates were directly taken from the crystal structures, H atoms were added, and their coordinates were optimized for the entire complex by energy minimization at the B3LYP/6-31G(d,p) level. Next, we investigated the sensitivity of the calculated electric field to the O-O distance of a given geometry, which allowed us to dissect the calculated electric field shift into the changes in the magnitude or/and the relative positioning of the dipole moments, and to understand the bias in our modeling scheme.
Figure 4. A simplified molecular complex mimicking the active site geometry of KSI used for DFT calculations.
To construct the complex for KSI-Cl-Y16, phenol, 2-Chlorophenol and acetic acid are used to represent the active site residues Y32/Y57, Cl-Y16, and Asp 103, respectively and 2-Cyclohexen-1-one (Pmini) mimics the A-ring of 19NT that is directly involved in the H-bond network.16 The heavy atom coordinates were taken from the crystal structures and fixed. The hydrogen atoms were added and their positions were optimized by energy minimization at the B3LYP/6-31G(d,p) level. The same strategy is applied to KSI-Cl-Y32 and KSI-Cl-Y57, as shown in Figure S5.
The final optimized structures possess the extended H-bond network in the active site of KSI (Figure S5). The electric fields were calculated with several methodologies and compared with the experimental data (Table 2 and S6). The estimated total field from these specific H-bond interactions is about 65 MV/cm smaller than that observed from VSE for all KSI variants, indicating that the rest of the protein architecture contributes approximately half of the total electric field experienced by the C=O bond of the substrate, as suggested earlier.3 The predicted change in the electric field of KSI-Cl-Y16 relative to WT (21 MV/cm, Table 2) is consistent with the observed shift from the VSE data (23 ± 6 MV/cm). A smaller magnitude of the O-H dipole moment in Cl-Y16 is predicted and estimated to account for 70% of the change, while the lengthening of the O-O distance between residue 16 and the ligand and the perturbations to the O-H dipole of Asp103 contribute the rest (Table 2 and SI discussion 1). A dominant contribution to the electric field from the dipole-dipole interaction between the O-H of Tyr16 and the C=O of the ligand is also consistent with the kinetic data where the Tyr16 to Phe mutation has a much more detrimental effect on the catalytic rate than mutating Asp103 to Asn.17,18 On the other hand, the simple model did not reproduce the observed changes in the electric field from the more remote residues Cl-Y32 and Cl-Y57 (Table 2 and S6). Experimentally, substitution of Cl-Y at position 57 exhibits similar electrostatic effects on 19NT as Cl-Y16, indicating a strong interaction within the H-bond network of KSI that is capable of propagating secondary perturbations to the very site of the reaction center.10 The highly simplified model is clearly insufficient to capture such subtly coupled interactions as it predicted little or even opposite changes in the magnitude of the electric field upon distal substitutions. The discrepancy can be partially accounted for by the possible differences between the x-ray geometry modeled and the actual equilibrium structure of each variant in solution. An uncertainty of 0.10 Å in the O-O distance between Tyr16 and 19NT from the structural data could introduce an error of 8 MV/cm in the calculated electric field (Table S5), limiting the model’s ability to predict an observed shift of similar magnitude (for instance, 10 MV/cm for KSI-Cl-Y32, Table 2). Much more advanced theoretical methods, such as those developed in ref 15, should provide further insight.
Table 2.
Electric field projected along the carbonyl of 19NT from DFT calculations
| WT | KSI-Cl-Y16 | KSI-Cl-Y57 | KSI-Cl-Y32 | |
|---|---|---|---|---|
| O-H16 dipole (D)a | 2.74 | 2.62 | 2.89 | 2.71 |
| O16 – OL (Å) | 2.48 | 2.58 | 2.49 | 2.49 |
| Electric field calculated (MV/cm)b |
−74.5 (−50.8, −23.7) |
−53.4 (−33.6, −19.8) |
−80.9 (−55.0, −25.9) |
−75.5 (−51.8, −23.7) |
| Observed from VSE (MV/cm) |
−141.3 ± 4.3 | −118.5 ± 3.7 | −121.6 ± 3.9 | −131.3 ± 4.0 |
The dipole moments are calculated from ESP charges derived using Merz-Kollman scheme.
The electric field is calculated via Coulomb’s law from the O-H dipoles of the two residues (16, 103) that are directly H-bonded with 19NT. The first number in the parenthesis refers to the contribution to the electric field from residue 16 and the second to residue 103.
Direct comparison between electrostatic stabilization and preferential H-bond interaction
There has been considerable discussion of a catalytic contribution of KSI’s H-bond network due to the formation of a strong, short H-bond (SSHB) between Tyr16 and the intermediate along the reaction coordinate.7,8 In this formulation, differential stabilization of the intermediate (and transition state) by KSI happens because the intermediate is a better H-bond acceptor to the active site, rather than because it has a greater dipole moment. This putative increase in H-bond strength is proposed to occur due to the matched proton affinity between the oxyanion hole19 and the intermediate and the hydrophobic nature of the active site.18 The primary difference between such an SSHB and electrostatic stabilization is the additional energetic benefit that might arise from the hypothesized covalent (delocalized) nature of a strong H-bond.1,20,21 However, the consistent linear correlation between the electric field and the activation free energy across the full spectrum of KSI mutants, regardless of the presence of specific H-bonds, strongly suggests that the specific H-bond interactions within the active site of KSI play the same functional role in catalysis as the distal interactions from the enzyme’s overall architecture. If additional (covalent) stabilization did occur, we would observe a different slope for KSI-Cl-Y mutants because the barrier would be lowered more for the same measured electric field (see SI discussion 2).
While a matched proton affinity between the active site of wild type KSI and equilenin, a model for the intermediate, has been demonstrated previously19, the conservative KSI-Cl-Y variants allowed us to show next that the enzyme’s preferential H-bond interaction with the intermediate over the substrate provides similar energetic benefits as that proposed from the electrostatic model. We note that the pKa perturbations introduced by Cl-Y incorporation in these mutants are sufficient to impact the formation energy of a presumably strong H-bond interaction between the ligand and the enzyme, as suggested by a systematic change in their rate constants.23,24 We first rationalize our strategy to quantify the catalytic benefit from the differential proton affinity match. The proton affinity of each KSI variant’s active site (pKαE) is defined in terms of the solution pKa of a ligand that upon binding forms a 50:50 mixture of protonated and deprotonates states (this is denoted differently from the active site’s pKa because the active site cannot exchange protons with solvent when a ligand is bound).25 By deconvolving the electronic absorption spectrum of equilenin (or other naphthols) bound to the enzyme into protonated and deprotonated contributions, the fraction ionization of the bound substrate can be established (this can also be probed in the IR as shown in ref26). This is shown for WT and the Cl-Y variants in Figure 5 and Figure S7, and the extracted values of the proton affinities are listed in Table 3. Empirically, the free energy of H-bond formation (ΔfG°, which defines the strength of the H-bond) varies linearly with the degree of proton affinity match (ΔpKα) between the H-bond partners.20,27 Therefore, the difference in the H-bond strength between the intermediate and the enzyme (ΔfG°(E•I)) vs. the substrate and the enzyme (ΔfG°(E•S)) is proportional to the difference in the ΔpKα between the two pairs (|pKaI − pKαE| − |pKaS − pKαE|). The pKa of the intermediate was determined to be 10.0 ± 0.1 and that of the substrate’s conjugate acid is approximately −2,19,22 therefore, this model anticipates that the enzyme’s active site H-bond network provides maximal activation barrier reduction when its pKαE matches the intermediate, and no energetic benefit when its pKαE equals 4 (where the H-bonds with the intermediate and the substrate have the same strength).
Figure 5. Proton affinities of the oxyanion hole of the KSI-Cl-Y variants.
Absorption spectra of equilenin when bound to the active site of WT and KSI-Cl-Y variants fitted to a linear combination (red) of the protonated (purple) and deprotonated (green) species.
Table 3.
Proton affinity of KSI-Cl-Y variantsa
| Proton affinity (pKαE) |
ΔpKα(E•I) |pKaI − pKαE| |
ΔpKα(E•S) |pKaS − pKαE| |
|
|---|---|---|---|
| WT | 9.80 ± 0.03 | 0.20 ± 0.10 | 11.80 ± 0.10 |
| Cl-Y32 | 9.64 ± 0.04 | 0.36 ± 0.11 | 11.64 ± 0.11 |
| Cl-Y57 | 8.92 ± 0.03 | 1.08 ± 0.10 | 10.92 ± 0.10 |
| Cl-Y16 | 9.09 ± 0.06 | 0.91 ± 0.12 | 11.09 ± 0.12 |
As shown in Figure 6, a linear correlation is observed between the free energy barrier and the proton affinity of KSI. The linear trend indicates that changes in the enzyme’s additional stabilization of the intermediate by a stronger H-bond (ΔfG°(E•I) − ΔfG°(E•S)) can explain changes in barrier reduction. Extrapolating the correlation line to pKαE = 4 yields the activation barrier without the preferential H-bonding towards the intermediate (17.7 kcal/mol). Compared to wild type KSI, the preferential H-bond interaction is estimated to provide a stabilization energy of 6.1 ± 0.2 kcal/mol, which accounts for 60% of KSI’s total rate acceleration and agrees well with the partial contribution to barrier reduction from intermediate stabilization by computational studies.28 The value is also comparable to the estimated barrier reduction due to the electric field in the active site (7.3 ± 0.4 kcal/mol),3 providing an independent piece of evidence that the energetic contribution from the H-bond network in KSI is electrostatic and not due to special covalency.29 In addition, the slope of the correlation suggests that the stabilization energy becomes 1.0 kcal/mol smaller for every pKα unit mismatch between the enzyme and the intermediate, suggesting a noticeable sensitivity of KSI’s catalytic rate to preferential H-bond interactions. The linear slope is highly reminiscent to the Bronsted β coefficient which describes the dependence of the strength of the H-bond upon the basicity of the H-bond acceptor (in our case, the proton affinity of the protein) and is larger for more nonpolar environment.27,30 Analogous analysis yields β = 0.37 for the H-bond interaction between KSI and the ligand, reflecting a rather hydrophobic nature of KSI’s active site (calculation details in SI discussion 3).
Figure 6. Linear correlation between the proton affinity of KSI’s active site and the catalytic proficiency.
The catalytic proficiency improves linearly as the proton affinity of the enzyme (pKαE) approaches that of the intermediate (pKaI =10) (R2 = 0.99). The extra stabilization energy for barrier reduction is presumably due to the increased H-bond strength between the enzyme and the intermediate, which is estimated to be 6.3 ± 0.2 kcal/mol.
Conclusions
In summary, by utilizing amber suppression, we test and extend the correlation between the electric field in the active site of KSI and the activation free energy for catalysis by adding small but significant changes to the electric field and catalytic rate. The preserved linear trend demonstrates that the functional relevance of the specific H-bond interactions in KSI’s extended H-bond network is of electrostatic origin, stabilizing the intermediate to about the same extent as the rest of the protein skeleton. A similar linear correlation was also observed between the proton affinity of KSI’s active site and the catalytic rate, suggesting a direct connection between the strength of the H-bond and the electric field it exerts. The set of conservative variants whose H-bond network is preserved while the electrostatic properties are incrementally perturbed provides a well-controlled platform for high-level simulations to further address the functional role of H-bonds in an enzyme’s active site.15
Supplementary Material
Acknowledgments
This work was supported in part by grants from the NIH (GM27738 and GM118044 to SGB). We thank Professor Jiangyun Wang from the Institute of Biophysics, Chinese Academy of Sciences, for making available plasmids pEVOL-CLYRS. We thank staff at SSRL and Dr. Marc Deller for support with x-ray diffraction data collection and analysis. We also thank Dr. Stephen Fried, Dr. Lu Wang and Sam Schneider for useful discussions about this work.
Footnotes
Supporting Information
Experimental details including protein preparation, crystallization, enzymatic assay, IR and electric field characterization and DFT calculations. Further discussion on the catalytic contributions of H-bonds and modeling bias are also presented. This material is available free of charge via the Internet as http://pubs.acs.org
Reference
- 1.Warshel A, Sharma PK, Kato M, Xiang Y, Liu H, Olsson MHM. Chem. Rev. 2006;106:3210–3235. doi: 10.1021/cr0503106. [DOI] [PubMed] [Google Scholar]
- 2.Warshel A. J. Biol. Chem. 1998;273:27035–27038. doi: 10.1074/jbc.273.42.27035. [DOI] [PubMed] [Google Scholar]
- 3.Fried SD, Bagchi S, Boxer SG. Science. 2014;346:1510–1513. doi: 10.1126/science.1259802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pollack RM. Bioorg. Chem. 2004;32:341–353. doi: 10.1016/j.bioorg.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 5.Radzicka A, Wolfenden R. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 6.Fried SD, Bagchi S, Boxer SG. J. Am. Chem. Soc. 2013;135:11181–11192. doi: 10.1021/ja403917z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cleland WW, Frey PA, Gerlt JA. J. Biol. Chem. 1998;273:25529–25532. doi: 10.1074/jbc.273.40.25529. [DOI] [PubMed] [Google Scholar]
- 8.Gerlt JA, Gassman PG. Biochemistry. 1993;115:11552–11568. doi: 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- 9.Liu X, Jiang L, Li J, Wang L, Yu Y, Zhou Q, Lv X, Gong W, Lu Y, Wang J. J. Am. Chem. Soc. 2014;136:13094–13097. doi: 10.1021/ja505219r. [DOI] [PubMed] [Google Scholar]
- 10.Wu Y, Fried SD, Boxer SG. Biochemistry. 2015;54:7110–7119. doi: 10.1021/acs.biochem.5b00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hawkinson DC, Pollack RM, Ambulos NP. Biochemistry. 1994;33:12172–12183. doi: 10.1021/bi00206a021. [DOI] [PubMed] [Google Scholar]
- 12.Schwans JP, Hanoian P, Lengerich BJ, Sunden F, Gonzalez A, Tsai Y, Hammes-Schiffer S, Herschlag D. Biochemistry. 2014;53:2541–2555. doi: 10.1021/bi401671t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fried SD, Boxer SG. Acc. Chem. Res. 2015;48:998–1006. doi: 10.1021/ar500464j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagchi S, Fried SD, Boxer SG. J. Am. Chem. Soc. 2012;134:10373–10376. doi: 10.1021/ja303895k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang L, Fried SD, Boxer SG, Markland TE. Proc. Natl. Acad. Sci. U. S. A. 2014;111:18454–18459. doi: 10.1073/pnas.1417923111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwans JP, Kraut DA, Herschlag D. Proc. Natl. Acad. Sci. U. S. A. 2009;106:14271–14275. doi: 10.1073/pnas.0901032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Choi G, Ha N, Kim M, Hong B, Oh B, Choi KY. Biochemistry. 2001;99:6828–6835. doi: 10.1021/bi002767+. [DOI] [PubMed] [Google Scholar]
- 18.Kim SW, Choi KY. J. Bacteriol. 1995;177:2602–2605. doi: 10.1128/jb.177.9.2602-2605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Childs W, Boxer SG. Biochemistry. 2010;49:2725–2731. doi: 10.1021/bi100074s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shan SO, Loh S, Herschlag D. Science. 1996;272:97–101. doi: 10.1126/science.272.5258.97. [DOI] [PubMed] [Google Scholar]
- 21.Oltrogge LM, Boxer SG. ACS Cent. Sci. 2015;1:148–156. doi: 10.1021/acscentsci.5b00160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zeng B, Pollack RM. J. Am. Chem. Soc. 1991;113:3838–3842. [Google Scholar]
- 23.Mckenzie RH. 2014 http://condensedconcepts.blogspot.com.au/2014/11/strong-hydrogen-bonds-can-be.html. [Google Scholar]
- 24.Natarajan A, Schwans JP, Herschlag D. J. Am. Chem. Soc. 2014;136:7643–7654. doi: 10.1021/ja413174b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fried SD, Boxer SG. Proc. Natl. Acad. Sci. U. S. A. 2013;110:12271–12276. doi: 10.1073/pnas.1310964110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fried SD, Boxer SG. J. Phys. Chem. B. 2012;116:690–697. doi: 10.1021/jp210544w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shan SO, Herschlag D. Proc. Natl. Acad. Sci. U. S. A. 1996;93:14474–14479. doi: 10.1073/pnas.93.25.14474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Feierberg I, Aqvist J. Biochemistry. 2002;41:15728–15735. doi: 10.1021/bi026873i. [DOI] [PubMed] [Google Scholar]
- 29.We note that energetic benefit estimated from the extrapolation to pKαE = 4 is not a reflection of the sole contribution from the active site H-bond, as the electrostatics from the protein architecture certainly affect the proton affinity, thus are encompassed in the measurements (see SI discussion 3).
- 30.Stahl N, Jencks WP. J. Am. Chem. Soc. 1986;108:4196–4205. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.