Xenopus TACC2 is a microtubule plus end–tracking protein that localizes in front of EB1 and overlaps with TACC1 and TACC3 in cultured embryonic mesenchymal cells and neuronal growth cones. TACC2 OE can promote increased microtubule polymerization in mesenchymal cells but not growth cones, suggesting cell-type specificity to its function.
Abstract
Microtubule dynamics is regulated by plus end–tracking proteins (+TIPs), which localize to the plus ends of microtubules (MTs). We previously showed that TACC1 and TACC3, members of the transforming acidic coiled-coil protein family, can act as +TIPs to regulate MT dynamics in Xenopus laevis. Here we characterize TACC2 as a +TIP that localizes to MT plus ends in front of EB1 and overlapping with TACC1 and TACC3 in multiple embryonic cell types. We also show that TACC2 can promote MT polymerization in mesenchymal cells but not neuronal growth cones, thus displaying cell-type specificity. Structure–function analysis demonstrates that the C-terminal region of TACC2 is both necessary and sufficient to localize to MT plus ends and promote increased rates of MT polymerization, whereas the N-terminal region cannot bind to MT plus ends but can act in a dominant-negative capacity to reduce polymerization rates. Finally, we analyze mRNA expression patterns in Xenopus embryos for each TACC protein and observe neural enrichment of TACC3 expression compared with TACC1 and TACC2, which are also expressed in mesodermal tissues, including somites. Overall these data provide a novel assessment of all three TACC proteins as a family of +TIPs by highlighting the unique attributes of each, as well as their collective characteristics.
INTRODUCTION
Plus end–tracking proteins (+TIPs) are a diverse family of proteins that regulate microtubule (MT) plus-end dynamics (Akhmanova and Steinmetz, 2008). Although many studies have identified specific functions for the various +TIPs, much is unknown regarding how +TIPs interact with MT plus ends and with each other to control MT dynamics in living cells, particularly during embryonic development. The transforming acidic coiled-coil (TACC) domain family of proteins (consisting of TACC1, TACC2, and TACC3) represents one important group of +TIPs that regulate MT dynamics (Peset and Vernos, 2008), yet only a handful of studies examined their potential MT plus end–regulatory roles (Lee et al., 2001; Samereier et al., 2010; Nwagbara et al., 2014; Gutierrez-Caballero et al., 2015; Lucaj et al., 2015; Hussmann et al., 2016). Previous immunostaining approaches demonstrated that all three TACC family members interact with MTs and concentrate at centrosomes and mitotic spindles (Gergely et al., 2000). We found that both TACC1 and TACC3 can act as +TIPs that promote MT polymerization in many Xenopus embryonic cell types (Lucaj et al., 2015; Nwagbara et al., 2014). However, no previous studies examined a possible MT plus end–regulatory function for TACC2.
TACC2 was first identified as a centrosomal protein with weak localization to the mitotic spindle (Gergely et al., 2000). Early studies showed that TACC2 is expressed throughout murine embryonic development (Lauffart et al., 2003). However, TACC2-deficient mice were viable and displayed no obvious developmental abnormalities (Schuendeln et al., 2004). In contrast, in HeLa cells, loss of TACC2 centrosomal localization led to defects in mitotic spindle function (Dou et al., 2004). In addition, later studies of TACC2 suggested that it can function as both a tumor suppressor and an oncogenic protein (Lauffart et al., 2003; Cheng et al., 2010; Takayama et al., 2012), suggesting that its requirement and function might vary between cell types and model systems. Furthermore, it was suggested that the three TACC family proteins might exhibit differing degrees of functional redundancy (Gergely et al., 2000; Sadek et al., 2003).
In this study, we examined the localization and function of TACC2 in Xenopus embryonic cells. First, we used live imaging of green fluorescent protein (GFP)–TACC2 to determine its subcellular localization in two embryonic cell types and investigated its role in MT dynamics. Then we performed structure–function analysis of TACC2 in the context of MT localization and dynamics. Finally, we compared mRNA expression patterns of TACC2 to those of other TACC proteins during embryonic development. Taken together, our findings give new insights into TACC2 function, how it regulates MT plus-end dynamics, and how it may display unique regulatory roles compared with the other TACC members.
RESULTS AND DISCUSSION
TACC2 can act as a +TIP that localizes in front of end-binding protein 1 in many embryonic cell types
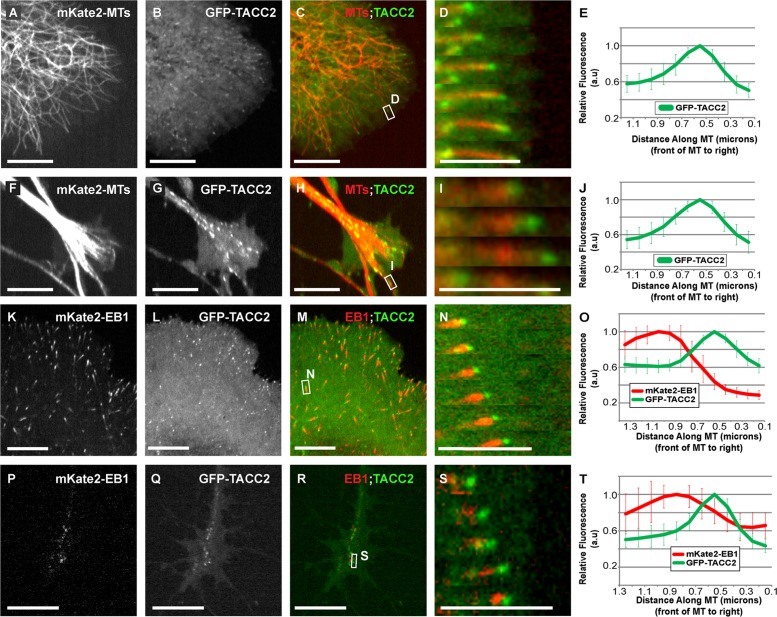
We first determined whether TACC2 could act as a +TIP. We observed that GFP-TACC2 strongly localized as puncta that tracked along growing MT plus ends in embryonic mesenchymal cells (Figure 1, A–E, and Supplemental Movie S1) and in neuronal growth cones (Figure 1, F–J, and Supplemental Movie S2). Colocalization analysis of GFP-TACC2 with mKate2–end-binding protein 1 (EB1), which binds all growing MT plus ends (Stepanova et al., 2003), demonstrated that the peak of GFP-TACC2 intensity was slightly distal to mKate2-EB1 (Figure 1, K–N, and Supplemental Movie S3), occurring ∼0.5 μm in front of mKate2-EB1 in mesenchymal cells (Figure 1O). This was similar to our findings for TACC1 and TACC3, which were ∼0.4 and ∼0.5 μm in front of EB1, respectively (Nwagbara et al., 2014; Lucaj et al., 2015). We determined that the distance in peak intensity of EB1 versus TACC2 matches that of EB1/TACC3 and is significantly different from that of EB1/TACC1 (Supplemental Figure S1A) in mesenchymal cells. However, in neuronal growth cones, the average distance between peak intensities of GFP-TACC2 and mKate2-EB1 was slightly less, with GFP-TACC2 ∼0.3 μm distal to mKate2-EB1 (Figure 1P-T). This suggests that the MT localization of TACC2 varies slightly between these two cell types. However, we cannot exclude the possibility that this difference in mesenchymal cells versus neuronal growth cones could be due to the imprecise nature of fluorescence intensity plot profiles when measuring distances so close to the diffraction limit.
FIGURE 1:
TACC2 can act as a +TIP that localizes in front of EB1 in many embryonic cell types. (A–J) Representative images of mKate2-tubulin, GFP-TACC2, and merged in cultured embryonic mesenchymal cells derived from neural tube (A–C) and neuronal growth cone (F–H). See Supplemental Movies S1 and S2. (D, I) Magnified time-lapse montage of the boxed regions in C and H. The time interval between frames is ∼2–3 s. (E, J) Fluorescence intensity profile of GFP-TACC2. (K–T) Images of mKate2-EB1, GFP-TACC2, and merged in cultured embryonic mesenchymal cell (K–M) and neuronal growth cone (P–R). See Supplemental Movie S3. (N, S) Magnified time-lapse montage of the boxed region in M and R. For N, the green channel was translated left by ∼0.2 μm to account for the different acquisition times of the red and green channels (MT plus-end velocity ∼0.24 μm/s in this series, and the green channel was imaged 600 ms after the red channel). This allows for a more accurate visualization of +TIP colocalization. For S, the green channel was translated left by ∼0.1 μm (plus end was moving at 0.085 μm/s, and the green channel was imaged 600 ms after the red channel). For all fluorescence intensity profiles, ∼20 MTs for each condition were quantified by intensity line scans. The MT plus end is toward the right, and error bars represent SDs. Scale bars, 5 μm (montages), 10 μm (all others).
TACC2 overlaps with both TACC1 and TACC3 and localizes to the plus end through its C-terminal domain
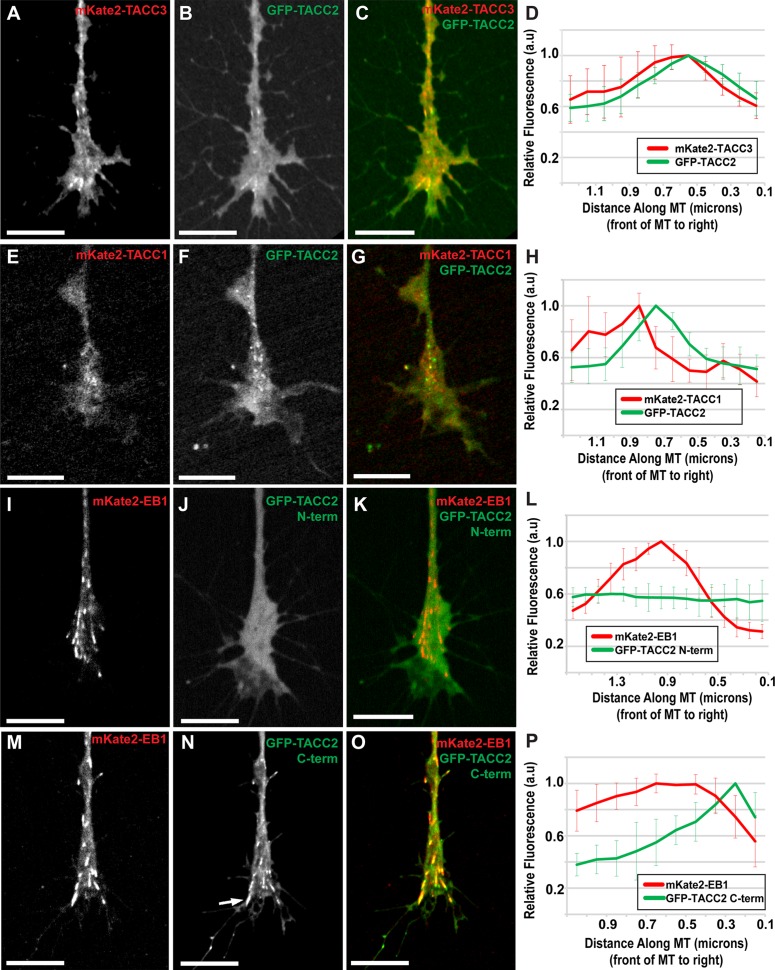
We next performed colocalization analysis of TACC2 with either TACC1 or TACC3. Consistent with our analyses of the TACC proteins with EB1, we observed that GFP-TACC2 closely overlaps with mKate2-TACC3 (Figure 2, A–D), whereas it localizes just in front of mKate2-TACC1 (Figure 2, E–H). These findings align well with our previous measurements showing TACC3 localizing slightly distal to TACC1 on the MT plus end (Nwagbara et al., 2014; Lucaj et al., 2015), with TACC2 appearing to occupy the same MT sublocalization as TACC3.
FIGURE 2:
TACC2 localizes to the plus end through its C-terminal domain and overlaps with both TACC1 and TACC3. (A–C) Representative images of mKate2-TACC3, GFP-TACC2, and merged in cultured neuronal growth cones. (D) Fluorescence intensity profiles of GFP-TACC2 and mKate2-TACC3. (E–G) mKate2-TACC1, GFP-TACC2, and merge with GFP-TACC2 fluorescence intensity plot profile (H). (I–K) Images of mKate2-EB1, GFP-TACC2 N-term (aa 1–540), and merged with the plot of fluorescence intensity profiles (L). (M–O) Images of mKate2-EB1, GFP-TACC2 C-term (aa 923-1168), and merged with the plot showing their fluorescence intensity profiles (P). Arrow in N points to lattice binding of TACC protein. Time interval between all frames is ∼2 s. For D, the green plot profile was translated ∼0.2 μm left (MT plus-end velocity ∼0.124 μm/s, green channel imaged 1.5 s after red) to accurately represent relative localization. For H, the green profile was translated left based on the velocity of each comet measured to account for difference in acquisition time points between channels (green channel imaged 1.5 s after red). In P, the green profile was translated 0.1 μm (MT plus-end velocity ∼0.148 μm/s, red channel imaged 0.96 s after green). For all fluorescence intensity profiles, ∼10 MTs for each condition were quantified by intensity line scans, and SDs are shown in error bars. MT plus end is at the right. Scale bars, 10 μm.
The TACC2 protein contains two structural elements—SxIP motifs and a TACC domain—that could mediate its plus-end localization (Supplemental Figure S1B). TACC2 has two SxIP motifs known to facilitate interaction with EB proteins and recruit +TIPs to growing MT plus ends (Honnappa et al., 2009). In addition, the C-terminus consists primarily of a TACC domain, which is highly conserved across all three TACC family members (Supplemental Figure S1, C and D; Gergely et al., 2000; Peset and Vernos, 2008) and is required for plus-end tracking of TACC1 and TACC3 (Nwagbara et al., 2014; Lucaj et al., 2015). Of note, however, the SxIP motifs in TACC2 set it apart from TACC1 and TACC3, which have only “SxIP-like” motifs (Supplemental Figure S1, C and D). To investigate the structural mechanism by which TACC2 interacts with the MT plus end, we examined the localization of several N- or C-terminally truncated proteins (Supplemental Figure S1B and Figure 2, I–P).
Despite the fact that SxIP motifs are known to mediate plus-end tracking of other +TIPs (Honnappa et al., 2009), neither N-terminal TACC2 region (amino acids [aa] 1–540 or 1–724, containing one or two SxIP motifs, respectively) was sufficient to bind to MT plus ends (Figure 2, I–L, and unpublished data). However, TACC2 proteins lacking the N-terminal regions containing either just the TACC domain (aa 923–1168; Figure 2, M–P) or the TACC domain and the second SxIP motif (aa 639–1168; unpublished data) were sufficient to drive MT plus-end tracking in a manner similar to the full-length protein, albeit with some lattice binding (Figure 2N, arrow). This observation is in line with our past deletion construct analysis of GFP-TACC1 and GFP-TACC3 (Nwagbara et al., 2014; Lucaj et al., 2015), suggesting that the N-terminus of TACC proteins might help to constrain plus-end tracking to the very MT tip. Together these results show that the C-terminal amino acids 923–1168, encompassing the TACC domain, are both necessary and sufficient for TACC2 plus-end tracking. Given that the C-termini for TACC1, 2, and 3 are all necessary to track MT plus ends and exhibit strong sequence similarity (Supplemental Figure S1, C and D), this conserved TACC domain appears to be the structural element that mediates the shared +TIP behavior of the TACC family.
TACC2 can regulate microtubule plus-end dynamics through its C-terminal domain in a cell type–specific manner
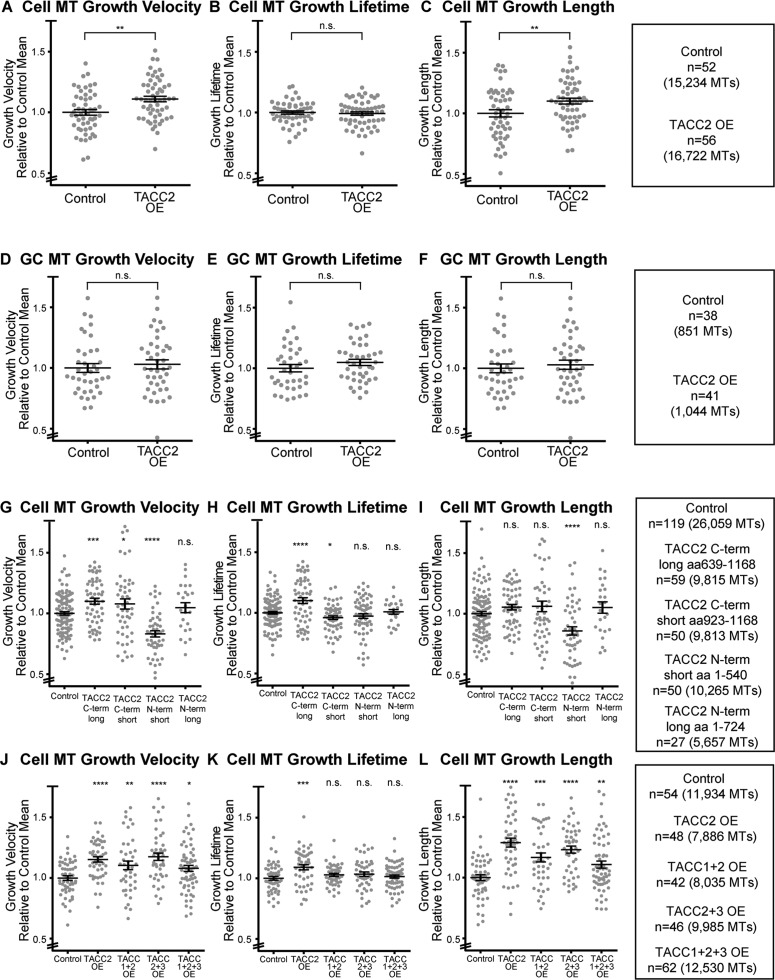
In light of our characterization of TACC2 as a +TIP, we next assessed whether TACC2 could modulate MT plus-end dynamics. We overexpressed GFP-TACC2, acquired time-lapse images of mKate2-EB1, which is a marker of MT polymerization, and quantified parameters of MT polymerization dynamics. In mesenchymal cells derived from the neural tube, mean MT growth velocity was 11% faster with TACC2 overexpression (OE; Figure 3A), mean MT growth-track lifetime was not statistically significantly different (Figure 3B), and mean MT growth-track length was 10% longer (Figure 3C). These data suggest that TACC2 can promote MT polymerization either directly or indirectly. These findings are similar to our previous MT dynamics analyses after TACC1 OE and TACC3 OE, which showed faster velocities, longer tracks, and no change in lifetime (Nwagbara et al., 2014; Lucaj et al., 2015). However, the magnitude of these differences varies, depending on the TACC protein, with TACC1 OE showing the greatest increase in mean MT growth velocities and growth track lengths (∼25% greater). Thus the entire TACC family appears to promote MT polymerization rates, but variations may exist in how each TACC protein exerts its effect.
FIGURE 3:
TACC2 can regulate microtubule plus-end dynamics through its C-terminal domain in a cell type–specific manner. All raw data of automated tracking of mKate2-EB1 comets were normalized to the same-day control means, and all reported values are relative ones combined over at least three separate experiments. (A–C) Quantification of mean values of MT parameters (velocity, lifetime, length) upon TACC2 OE in embryonic mesenchymal cells. (D–F) MT parameters in growth cones (GC). (G–I) MT parameters in mesenchymal cells on overexpression of C- or N-terminal-truncated TACC2. (J–L) Quantification of MT parameters in mesenchymal cells when multiple combinations of TACC family members were expressed. Bars on dot plots show mean and SEM. To assess statistical significance, ANOVA tests were first performed (for multiple conditions), followed by an unpaired t test. ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, n.s., not significant.
To assess possible cell-type specificity in the effect of TACC2 on MT dynamics, we performed the same OE analysis in neuronal growth cones. We observed no significant increase in mean MT growth velocity, MT growth-track lifetime, or MT growth-track length (Figure 3, D–F). These findings not only stand in contrast to our findings for TACC2 OE in mesenchymal cells (Figure 3, A–C), but they also differ from previous data for MT dynamics with TACC3 OE in growth cones, which showed faster mean growth velocities and longer growth-track lengths (Nwagbara et al., 2014). These data suggest cell-type specificity to TACC2 function, in contrast to TACC3. It is intriguing that the TACC3-interacting MT polymerase XMAP215 also has a different effect on MT dynamics when levels are manipulated in the growth cone compared with mesenchymal cells (Lowery et al., 2013). Thus the exact role of TACC2 compared with that of other TACC proteins and possibly XMAP215 in regulating MT plus-end dynamics in various embryonic cell types will need to be explored further.
We also examined whether OE of the GFP-TACC2 deletion proteins would affect MT plus-end dynamics. Mesenchymal cells overexpressing the C-terminal amino acids 639–1168 of GFP-TACC2 showed a 10% increase in mean MT growth velocity and lifetime (Figure 3, G–I), suggesting that TACC2 may promote MT polymerization through its conserved C-terminal TACC domain. However, OE of the C-terminal amino acids 923–1168, while sufficient for plus-end tracking, was still able to promote increased MT growth velocity but led to a slight decrease in MT lifetime (Figure 3, G–I). Of interest, overexpressing the GFP-tagged N-terminal region (amino acids 1–540, containing only one SxIP motif) of TACC2 resulted in a 17% decrease in mean MT growth velocity and 14% shorter MT growth-track lengths (Figure 3, G–I). The longer N-terminal region (amino acids 1–724, with both SxIP motifs) did not show any effect on MT dynamics. Thus, although the shorter N-terminal region was not able to track MT plus ends (Figure 2, I–K), it was still able to indirectly affect MT plus-end dynamics, perhaps acting in a dominant-negative capacity. However, the mechanism by which this occurs needs to be investigated further. These systematic deletion analyses data for TACC2 differ from our previous findings for TACC1, which showed no effect on MT dynamics upon OE of either the N- or C-terminal domain of GFP-TACC1 (Lucaj et al., 2015). Thus, whereas the behavior of full-length TACC2 generally mirrors that of its family members, TACC2 also appears to modulate MT dynamics in a unique, domain-specific way.
Given that all three full-length TACC proteins can increase MT growth velocities when overexpressed in mesenchymal cells, we next investigated whether combinations of TACC1, TACC2, and TACC3 would demonstrate a synergistic effect on MT dynamics. We overexpressed each TACC protein by injecting only one-fourth of the amount of mRNA that we used for our initial TACC2 OE experiments. Cells with this lower amount of GFP-TACC2 still exhibited significantly faster MT growth velocity than did controls (Figure 3J). However, there did not appear to be a combinatorial effect on MT growth velocity, as overexpressing the TACC members in combination did not show increased velocity, lifetime, or length beyond that of TACC2 alone (Figure 3, J–L), and an analysis of variance (ANOVA) test among the four TACC OE combinations did not show statistical significance in MT growth velocity across those conditions. Only TACC2 OE alone showed a slight increase in MT growth lifetime. All four OE combinations resulted in longer mean MT growth-track lengths, but again, no synergistic interactions were apparent. These data are in agreement with our previous findings that showed no synergistic effect between TACC1 and TACC3 (Lucaj et al., 2015), suggesting that there may be an upper threshold rate of MT polymerization that is reached with OE of any single TACC member in this cell type.
TACC family members are differentially expressed during embryonic development
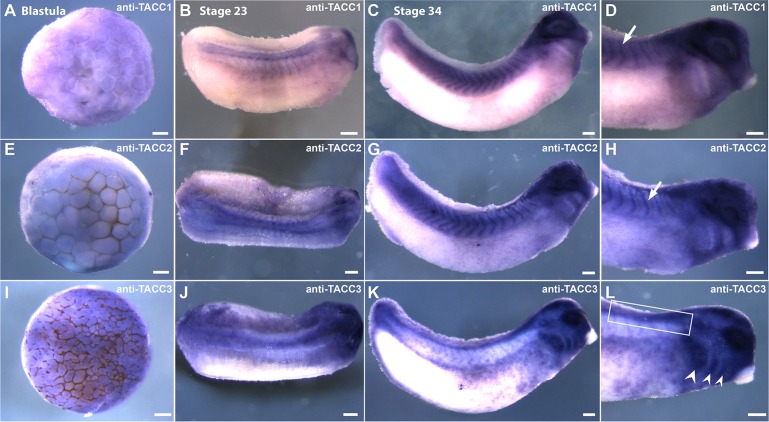
To further explore possible cell type–specific roles of the TACC family in embryonic development, we assessed expression of TACC mRNA transcripts in Xenopus laevis embryos using whole-mount in situ hybridization (Figure 4). Uniform, diffuse expression of all three TACC transcripts was evident at the blastula stage (Figure 4, A, E, and I), but some differences in expression emerged at later stages. At stage 23 (neurula), all three TACCs were expressed in the neural tube (Figure 4, B, F, and J), but by stage 34 (tadpole stage), TACC1 and TACC2 expression appeared to be restricted to the anterior regions of the neural tube and the hindbrain, whereas TACC3 was present along the entire length of the neural tube (Figure 4, C, G, and K). Consistent expression of TACC1 and TACC2, but not TACC3, was also evident in early somite tissue and was strongest at stage 34 (Figure 4, C and G), when TACC3 showed strong labeling of the pharyngeal arches (Figure 4L, arrows). TACC1 and TACC2 were not expressed in these regions, instead exhibiting diffuse staining in surrounding tissue with a notable lack of signal in the pharyngeal arches.
FIGURE 4:
TACC family members are differentially expressed during embryonic development. Cell type–specific expression of TACC mRNAs shown by whole-mount in situ hybridization using full-length antisense probes to TACC1, TACC2, and TACC3. TACC1 is expressed uniformly at the late blastula stage (A), in somites, neural tube, and hindbrain at stage 23 (B), and in somites, eye, and anterior neural tube at stage 34 (C, D). TACC2 expression is similar to that of TACC1 at the blastula stage (E) and also increases in a tissue-specific manner after neurulation, apparent in the neural tube, early brain, and presomitic mesoderm at stage 23 (F). At stage 34, TACC2 is expressed in the somitic mesoderm, the ocular lens, and the anterior neural tube (G, H). Arrows in D and H indicate somitic mesoderm. TACC3 expression resembles that of TACC1 and TACC2 at the blastula stage (I), but its expression becomes more neural specific by stage 23, showing expression along the entire length of the neural tube, although strongest anteriorly near the brain (J). At stage 34, TACC3 is strongly expressed in the pharyngeal arches (L, arrows), otic vesicle, and trunk neural crest (K, L). Neural tube and trunk neural crest are indicated by the box in L. Scale bars, 150 μm.
Our results support past findings that established a role for the TACC proteins in the differentiation of early embryonic tissues. The more neural-specific expression pattern of TACC3 that we observed merits special consideration. Up-regulation of TACC3 in response to neural differentiation cues, followed by a decrease in expression in terminally differentiated cells, was reported in rat PC12 cells treated with nerve growth factor (Sadek et al., 2003). Moreover, TACC3 expression, which is prominent in multipotent neural progenitors, such as the neural crest and in undefined cell types of the early brain, was previously shown in Xenopus between stages 17 and 24 (Tessmar et al., 2002). Our findings that show persistent TACC3 expression after stage 23 implies a putative role in MT-dependent processes that are especially active around stages 28–34, such as neurite outgrowth, neural crest cell migration, and cell proliferation. If increased TACC3 levels were exclusively involved in neural specification at the early tadpole stage, such high expression in the pharyngeal arches and otic vesicle would not be observed, as most of these cells are fated to develop into craniofacial cartilage. Higher expression here could be a sign of the need for specific cytoskeletal regulation during the dynamic processes of embryogenesis, which require large-scale cytoskeletal rearrangements. For example, TACC3 has been shown to promote axon outgrowth in Xenopus neurites, likely by regulating MT polymerization dynamics (Nwagbara et al., 2014).
In conclusion, we used live imaging in cultured embryonic cells to provide the first evidence that TACC2 can function as a +TIP to regulate MT dynamics. Moreover, we show that TACC2 localizes to the MT plus end through its C-terminal domain, similar to other TACC members, indicating a conserved function of this domain for MT plus-end localization. Finally, our data suggest that although all three TACC members can promote MT polymerization, each displays cell-type specificity in its expression and function: TACC3 is more highly expressed in neural tissues and cranial neural crest cells, whereas TACC1 and TACC2 are expressed in somitic mesoderm. In addition, although our previous work showed that TACC3 OE resulted in increased MT growth velocities in embryonic neuronal growth cones, TACC2 OE did not increase MT growth velocities in this cell type. Future studies will continue to explore the unique and collective functions of the TACC proteins in regulating MT dynamics during embryonic development.
MATERIALS AND METHODS
Embryos
Eggs obtained from female X. laevis frogs (NASCO, Fort Atkinson, WI) were fertilized in vitro, dejellied, and cultured at 13–22°C in 0.1X Marc’s modified Ringer’s (MMR) using standard methods (Sive et al., 2010). Embryos were staged according to Nieuwkoop and Faber (1994). All experiments were approved by the Boston College Institutional Animal Care and Use Committee and performed according to national regulatory standards.
Culture of Xenopus embryonic explants
Embryos were cultured in 0.1× MMR at 22°C to stages 22–24, and embryonic explants were dissected and cultured on poly-l-lysine (100 μg/ml)– and laminin (20 μg/ml)-coated coverslips, as described previously (Nwagbara et al., 2014; Lucaj et al., 2015). Cells were imaged at room temperature 18–24 h after plating.
Cloning of TACC2
The TACC2 sequence was designed based on the TACC2 annotated sequence from the laevis genome, version 7.1 (www.xenbase.org, RRID: SCR_003280), using the UTA World Cup 2014 transcript model of Scaffold16863:9073321…9127841 (+ strand), which was predicted to contain the X. laevis TACC2 genomic sequence. The TACC2 sequence was synthesized, cloned into the pUC57 plasmid (GenScript, Piscataway, NJ), and then subcloned into the GFP-pCS2+ vector to tag GFP in the N-terminus of TACC2. The Xenopus TACC2 sequence was compared with different mammalian isoforms of TACC2, as well as with Xenopus TACC1 and TACC3, using Clustal Omega (www.ebi.ac.uk/Tools/msa/clustalo/; RRID: SCR_001591).
Constructs and RNA
Capped mRNA was transcribed in vitro using the SP6 mMessage mMachine Kit (Life Technologies, Grand Island, NY). RNA was purified with LiCl precipitation and resuspended in nuclease-free water. Constructs used were GFP-TACC2, GFP-TACC2-Nterm short, GFP-tACC2-Nterm long, GFP-TACC2-Cterm short, GFP-TACC2-Cterm long, GFP-TACC3, mKate2-TACC3 (TACC3 pET30a was gift from the Richter lab [University of Massachusetts Medical, Worcester, MA]), GFP-TACC1, mKate2-TACC1 (all TACC constructs subcloned into pCS2+ vector), mKate2-tubulin (Shcherbo et al., 2009) in pT7TS, and mKate2-EB1 in pCS2+. The dorsal blastomeres of embryos were injected four times at the two-to-four-cell stage (in 0.1× MMR containing 5% Ficoll) with total mRNA amount per embryo (variations were specific to each experiment) of 500–3000 pg of GFP-TACC2, 1500 pg of GFP-TACC2 N-term constructs, 1500 pg of GFP-TACC2 C-term constructs, 500 pg of GFP-TACC3, 2000–3000 pg of mKate2-TACC3, 500 pg of GFP-TACC1, 2000–4000 pg of mKate2-TACC1, 1000 pg of mKate2-tubulin, and 200–300 pg of mKate2-EB1.
Confocal microscopy
Live images were collected with a CSU-X1M 5000 spinning-disk confocal (Yokogawa, Tokyo, Japan) on a Zeiss Axio Observer inverted motorized microscope with a Zeiss 63× Plan Apo 1.4 numerical aperture lens (Zeiss, Thornwood, NY). Images were acquired with an ORCA R2 charge-coupled device camera (Hamamatsu, Hamamatsu, Japan) controlled with Zen software. For time lapse, images were collected about every 2 s for 1 min. For two-color colocalizations in Figures 1 and 2, the red and green channels were imaged sequentially. Thus the true extent of potential overlap between plus-end accumulations could not be examined from these image time series without correcting for plus-end movement. Figures 1, A–D, F–I, K–M, and P–R, and 2, A–C, E–G, I–K, and M–O, show the original acquisitions without correction. However, for the single-MT montages (Figure 1, N and S), we performed x-axis translation on the green channel to correct for the time displacement. For this analysis, the green channel was translated in the x-axis, after calculating the frame-to-frame velocity of the growing MT plus end, to account for the time delay between channels for each examined MT (using the Translate function in ImageJ [National Institutes of Health, Bethesda, MD]). Details of displacement measurements are given in the legends for Figures 1 and 2.
plusTipTracker software analysis
MT dynamics was analyzed from mKate2-EB1 movies using plusTipTracker (Applegate et al., 2011; Lowery et al., 2013; Stout et al., 2014). The same parameters were used for all movies: maximum gap length, eight frames; minimum track length, three frames; search radius range, 5–12 pixels; maximum forward angle, 50°; maximum backward angle, 10°; maximum shrinkage factor, 0.8; fluctuation radius, 2.5 pixels; and time interval 2 s. Only cells with a minimum number of 10 MT tracks in a 1-min time lapse were included for analysis. Data collected from the analysis included mean MT growth velocity, mean MT growth-track lifetime, and mean MT growth-track length. MT growth-track lifetime is defined as the number of seconds of MT polymerization before pauses and catastrophe. MT growth-track length is the distance of persistent MT polymerization before pause or catastrophe. Because MT dynamics parameters were compiled from many individual experiments and there can be significant day-to-day fluctuations in control MT dynamics (in part, based on room temperature fluctuations), the final compiled data were normalized relative to the mean of the control data for each experiment. Dot plots were made using GraphPad Prism Version 6.00 for Windows (GraphPad Software, La Jolla, CA). To determine statistical differences, unpaired two-tailed t tests were used for comparing two conditions, and ANOVA tests were used to compare multiple conditions, followed by unpaired two-tailed t tests (GraphPad).
Image analysis and statistics
Phenotypic quantification was performed from multiple experiments to ensure reproducibility. Images were processed using Fiji, the image processing package that contains ImageJ (http://fiji.sc; RRID: SCR_002285). MT plus-end accumulations for fluorescently tagged TACC2, TACC2 deletion proteins, TACC1, TACC3, and EB1 were measured with the Line tool in ImageJ along their longest axis, which was approximated from their trajectory in the previous and subsequent frames. Line intensities were gathered using Plot Profile, and puncta lengths were determined where fluorescence signal became visually and statistically higher than noise. Graphs were made in Excel (Microsoft, Redmond, WA).
Whole-mount in situ hybridization
Embryos were fixed overnight at 4°C in a solution of 4% paraformaldehyde in phosphate-buffered saline (PBS), dehydrated using solutions of methanol in PBS, and stored in methanol at −20°C before in situ hybridization, which was performed on fixed embryos using a multibasket system, as previously described (Sive et al., 2007). The proteinase K treatment was shortened to 3–5 min, and embryos were bleached under a fluorescent light in 1.8× saline–sodium citrate, 1.5% H2O2, and 5% (vol/vol) formamide for 1 h before prehybridization. During hybridization, probe concentration was 0.5 μg/ml. All TACC constructs—TACC1, TACC2, and TACC3—used for the hybridization probes were subcloned into the pGEM T-easy vector (Promega, Madison, WI). The full-length antisense digoxigenin-labeled TACC hybridization probes were transcribed in vitro using the SP6 MAXIscript kit (Life Technologies) and the pGEM constructs. Probes were purified using LiCl precipitation and resuspended in nuclease-free water. Embryos were then imaged using a Zeiss AxioCam MRc attached to a Zeiss SteREO Discovery.V8 light microscope. Images were processed in Photoshop (Adobe, San Jose, CA) to superimpose multiple images of the same embryo in order to visualize several focal planes. Stained embryos for each condition were compared with other negative control embryos that proceeded through the entire in situ hybridization protocol but did not incubate with any hybridization probe.
Supplementary Material
Acknowledgments
We thank Aleksandra Ostojic, Claire Stauffer, and Kelly Hawkins for technical assistance with experiments and Amanda Dickinson for experimental suggestions. We also thank other members of the Lowery lab for manuscript critiques and Nancy McGilloway and Todd Gaines for excellent Xenopus husbandry. L.A.L. is funded by National Institutes of Health Grants R00 MH095768, R01 MH109651, and R03 DE025824 and March of Dimes Foundation Grant FY 16-220.
Abbreviations used:
- aa
amino acids
- EB1
end-binding protein 1
- GFP
green fluorescent protein
- MT
microtubule
- OE
overexpression
- TACC
transforming acidic coiled-coil
- +TIP
plus end–tracking protein.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0198) on August 24, 2016.
REFERENCES
- Akhmanova A, Steinmetz MO. Tracking the ends: a dynamic protein network controls the fate of microtubule tips. Nat Rev Mol Cell Biol. 2008;9:309–322. doi: 10.1038/nrm2369. [DOI] [PubMed] [Google Scholar]
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J Struct Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Douglas-Jones A, Yang X, Mansel RE, Jiang WG. Transforming acidic coiled-coil-containing protein 2 (TACC2) in human breast cancer, expression pattern and clinical/prognostic relevance. Cancer Genomics Proteomics. 2010;7:67–73. [PubMed] [Google Scholar]
- Dou Z, Ding X, Zereshki A, Zhang Y, Zhang J, Wang F, Sun J, Huang H, Yao X. TTK kinase is essential for the centrosomal localization of TACC2. FEBS Lett. 2004;572:51–56. doi: 10.1016/j.febslet.2004.06.092. [DOI] [PubMed] [Google Scholar]
- Gergely F, Karlsson C, Still I, Cowell J, Kilmartin J, Raff JW. The TACC domain identifies a family of centrosomal proteins that can interact with microtubules. Proc Natl Acad Sci USA. 2000;97:14352–14357. doi: 10.1073/pnas.97.26.14352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez-Caballero C, Burgess SG, Bayliss R, Royle SJ. TACC3-ch-TOG track the growing tips of microtubules independently of clathrin and Aurora-A phosphorylation. Biol Open 4, 170–179. 2015. [DOI] [PMC free article] [PubMed]
- Honnappa S, Gouveia SM, Weisbrich A, Damberger FF, Bhavesh NS, Jawhari H, Grigoriev I, van Rijssel FJ, Buey RM, Lawera A, et al. An EB1-binding motif acts as a microtubule tip localization signal. Cell. 2009;138:366–376. doi: 10.1016/j.cell.2009.04.065. [DOI] [PubMed] [Google Scholar]
- Hussmann F, Drummond DR, Peet DR, Martin DS, Cross RA. Alp7/TACC-Alp14/TOG generates long-lived, fast-growing MTs by an unconventional mechanism. Sci Rep. 2016;6:20653. doi: 10.1038/srep20653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffart B, Gangisetty O, Still IH. Molecular cloning, genomic structure and interactions of the putative breast tumor suppressor TACC2. Genomics. 2003;81:192–201. doi: 10.1016/s0888-7543(02)00039-3. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Lowery LA, Stout A, Faris AE, Ding L, Baird MA, Davidson MW, Danuser G, Van Vactor D. Growth cone-specific functions of XMAP215 in restricting microtubule dynamics and promoting axonal outgrowth. Neural Dev. 2013;8:22. doi: 10.1186/1749-8104-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucaj CM, Evans MF, Nwagbara BU, Ebbert PT, Baker CC, Volk JG, Francl AF, Ruvolo SP, Lowery LA. Xenopus TACC1 is a microtubule plus-end tracking protein that can regulate microtubule dynamics during embryonic development. Cytoskeleton (Hoboken) 2015;72:225–234. doi: 10.1002/cm.21224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwkoop PD, Faber J. Normal Table of Xenopus laevis (Daudin) New York: Garland; 1994. [Google Scholar]
- Nwagbara BU, Faris AE, Bearce EA, Erdogan B, Ebbert PT, Evans MF, Rutherford EL, Enzenbacher TB, Lowery LA. TACC3 is a microtubule plus end-tracking protein that promotes axon elongation and also regulates microtubule plus end dynamics in multiple embryonic cell types. Mol Biol Cell. 2014;25:3350–3362. doi: 10.1091/mbc.E14-06-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peset I, Vernos I. The TACC proteins: TACC-ling microtubule dynamics and centrosome function. Trends Cell Biol. 2008;18:379–388. doi: 10.1016/j.tcb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Sadek CM, Pelto-Huikko M, Tujague M, Steffensen KR, Wennerholm M, Gustafsson JA. TACC3 expression is tightly regulated during early differentiation. Gene Expr Patterns. 2003;3:203–211. doi: 10.1016/s1567-133x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- Samereier M, Meyer I, Koonce MP, Graf R. Live cell-imaging techniques for analyses of microtubules in Dictyostelium. Methods Cell Biol. 2010;97:341-357. doi: 10.1016/S0091-679X(10)97018-0. [DOI] [PubMed] [Google Scholar]
- Schuendeln MM, Piekorz RP, Wichmann C, Lee Y, McKinnon PJ, Boyd K, Takahashi Y, Ihle JN. The centrosomal, putative tumor suppressor protein TACC2 is dispensable for normal development, and deficiency does not lead to cancer. Mol Cell Biol. 2004;24:64036409. doi: 10.1128/MCB.24.14.6403-6409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbo D, Murphy CS, Ermakova GV, Solovieva EA, Chepurnykh TV, Shcheglov AS, Verkhusha VV, Pletnev VZ, Hazelwood KL, Roche PM, et al. Far-red fluorescent tags for protein imaging in living tissues. Biochem J. 2009;418:567–574. doi: 10.1042/BJ20081949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Baskets for in situ hybridization and immunohistochemistry. CSH Protoc. 2007;2007:pdb.prot4777. doi: 10.1101/pdb.prot4777. [DOI] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Microinjection of Xenopus embryos. Cold Spring Harb Protoc. 2010;2010:pdb.ip81. doi: 10.1101/pdb.ip81. [DOI] [PubMed] [Google Scholar]
- Stepanova T, Slemmer J, Hoogenraad CC, Lansbergen G, Dortland B, De Zeeuw CI, Grosveld F, van Cappellen G, Akhmanova A, Galjart N. Visualization of microtubule growth in cultured neurons via the use of EB3-GFP (end-binding protein 3-green fluorescent protein) J Neurosci. 2003;23:2655–2664. doi: 10.1523/JNEUROSCI.23-07-02655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout A, D’Amico S, Enzenbacher T, Ebbert P, Lowery LA. Using plusTipTracker software to measure microtubule dynamics in Xenopus laevis growth cones. J Vis Exp. 2014;2014(91):e52138. doi: 10.3791/52138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takayama K, Horie-Inoue K, Suzuki T, Urano T, Ikeda K, Fujimura T, Takahashi S, Homma Y, Ouchi Y, Inoue S. TACC2 is an androgen-responsive cell cycle regulator promoting androgen-mediated and castration-resistant growth of prostate cancer. Mol Endocrinol. 2012;26:748–761. doi: 10.1210/me.2011-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tessmar K, Loosli F, Wittbrodt J. A screen for co-factors of Six3. Mech Dev. 2002;117:103–113. doi: 10.1016/s0925-4773(02)00185-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.