Spatial and temporal control of MT dynamics is important for proper spindle assembly and chromosome segregation. The kinesin-8 Kif18B spatially regulates astral MT dynamics. Not all members of a single kinesin superfamily control MT dynamics in a similar fashion.
Abstract
The spatial and temporal control of microtubule dynamics is fundamentally important for proper spindle assembly and chromosome segregation. This is achieved, in part, by the multitude of proteins that bind to and regulate spindle microtubules, including kinesin superfamily members, which act as microtubule-destabilizing enzymes. These fall into two general classes: the kinesin-13 proteins, which directly depolymerize microtubules, and the kinesin-8 proteins, which are plus end–directed motors that either destabilize microtubules or cap the microtubule plus ends. Here we analyze the contribution of a PtK kinesin-8 protein, Kif18B, in the control of mitotic microtubule dynamics. Knockdown of Kif18B causes defects in spindle microtubule organization and a dramatic increase in astral microtubules. Kif18B-knockdown cells had defects in chromosome alignment, but there were no defects in chromosome segregation. The long astral microtubules that occur in the absence of Kif18B are limited in length by the cell cortex. Using EB1 tracking, we show that Kif18B activity is spatially controlled, as loss of Kif18B has the most dramatic effect on the lifetimes of astral microtubules that extend toward the cell cortex. Together our studies provide new insight into how diverse kinesins contribute to spatial microtubule organization in the spindle.
INTRODUCTION
One fundamental problem in biology is to understand how cells spatially regulate so many dynamic cellular events. A quintessential example of this is the complex regulation of mitotic spindle dynamics that occurs during cell division to govern the accurate segregation of chromosomes. The spindle is composed of three distinct functional classes of microtubules (MTs), which differ in both location and stability. Although all MTs are composed of the same α/β-tubulin heterodimers, the different dynamics of the MT subclasses suggest that cellular proteins must spatially and temporally control their dynamics.
A key class of MT dynamics regulators includes members of the kinesin superfamily (Walczak et al., 2013). Kinesin-13 proteins, including MCAK, Kif2A, and Kif2B, use the energy of ATP hydrolysis to directly depolymerize MTs from both ends (Desai et al., 1999; Hunter et al., 2003). Kinesin-13 family members are involved in spindle assembly, spindle MT flux, astral MT dynamics and organization, and kinetochore–MT attachments (Maney et al., 1998; Kline-Smith et al., 2004; Rogers et al., 2004; Ganem et al., 2005; Mennella et al., 2005; Moore et al., 2005; Manning et al., 2007; Ohi et al., 2007; Bakhoum et al., 2009; Rizk et al., 2009; Ems-McClung and Walczak, 2010; Domnitz et al., 2012). In contrast, the kinesin-8 proteins, including Kip3 in yeast and Kif18A, Kif18B, and Kif19 in mammalian cells, are both MT plus-end motor proteins and plus end-destabilizing enzymes (Gupta et al., 2006; Varga et al., 2006; Du et al., 2010; Stumpff et al., 2011; Su et al., 2011), but their mechanism of action is not fully understood. Functionally, the kinesin-8 proteins are also involved in the regulation of spindle, astral, and kinetochore-fiber MTs, but much less is known about how they control MT dynamics in each of these processes (DeZwaan et al., 1997; Cottingham et al., 1999; West et al., 2001, 2002; Garcia et al., 2002; Gatt et al., 2005; Mayr et al., 2007; Stumpff et al., 2008; Stout et al., 2011; Tanenbaum et al., 2011).
The best-studied kinesin-8 protein is Kip3. Kip3 is a plus end–directed MT depolymerase (Gupta et al., 2006; Varga et al., 2006) that preferentially acts on longer MTs (Varga et al., 2006, 2009). Mutations in Kip3 are associated with long astral MTs and increased benomyl sensitivity (Cottingham et al., 1999; Gupta et al., 2006), supporting a role in controlling MT dynamics. Its function in cells is more complex, as it was recently shown to act both as a catastrophe factor and a rescue factor on MTs in the bud tip, in which the motor domain is required for catastrophe induction and the tail domain is needed for MT rescue (Fukuda et al., 2014). It has also been shown to cross-link and slide MTs to control spindle length (Su et al., 2013; Rizk et al., 2014). These studies highlight the complex modes of action of this protein.
Mammalian Kif18A is a plus-end motor and a plus end–capping protein (Mayr et al., 2007; Du et al., 2010; Stumpff et al., 2011). It has a well-established role in controlling chromosome oscillations by its association with kinetochore-fiber MT plus ends and by inducing MT pausing, consistent with its proposed role as a capping protein (Stumpff et al., 2008, 2012). Kif18A targets to MT plus ends with its motor domain but uses its tail domain both to maintain MT association and to enhance processivity (Stumpff et al., 2011; Weaver et al., 2011). In contrast to Kif18A, much less is known about Kif18B in terms of both its activity and its function. Kif18B is also a plus end–directed motor, although it is far less processive than Kif18A (Shin et al., 2015). Kif18B has a second MT-binding site in the tail, which promotes diffusion along the MT lattice and decreases dissociation from the MT lattice in vitro (Shin et al., 2015). There is no evidence as to whether Kif18B directly destabilizes MTs. In cells, it binds to EB1 both in vitro and in vivo, and this association is required for its localization to MT plus ends (Stout et al., 2011; Tanenbaum et al., 2011). Furthermore, Kif18B can associate with the kinesin-13 MCAK in cells, where it may control astral MT dynamics (Stout et al., 2011; Tanenbaum et al., 2011). Knockdown of Kif18B resulted in an increase in both the length and number of astral MTs (Stout et al., 2011; Tanenbaum et al., 2011).
While the foregoing studies propose an important role for Kif18B in organizing astral MTs, it is not known whether Kif18B affects other aspects of mitotic progression or how Kif18B controls MT dynamics in cells. Here we show that knockdown of Kif18B perturbs chromosome alignment but does not affect the timing of mitotic progression. Kif18B acts preferentially on astral MTs to limit MT lifetime. In addition, loss of Kif18B causes a dramatic increase in MT polymer that extends toward the cell cortex and is exacerbated in the absence of the cortical actin cytoskeleton. Our studies provide new insight into the complexities of MT dynamics regulation in mitosis and highlight how individual members of the same kinesin subfamily can have different physiological activities.
RESULTS AND DISCUSSION
Kif18B regulates astral MTs to control proper chromosome alignment
Previous studies have shown that loss of Kif18B causes an increase in astral MT number and density in HeLa cells (Stout et al., 2011). To understand further how Kif18B affected the dynamics of the MTs and how those changes affected mitotic progression, we sought to examine Kif18B function in PtK cells, a cell type more amenable to imaging of the MT array. We used degenerate PCR to identify cDNA sequence of the motor domain of PtK Kif18B. Additional sequence from the 3′ end of the gene was obtained from a small-scale sequencing project of PtK cells (Jagesh Shah, personal communication). These sequences were used to generate siRNAs for Kif18B knockdown. Knockdown of Kif18B caused ∼90% reduction at the mRNA level using either of two independent small interfering RNAs (siRNAs; 88 ± 4% for Kif18B-1 and 87 ± 4% for Kif18B-2). Antibodies to human Kif18B did not cross-react in PtK cells, and our attempts to generate antibodies specific to the tail of PtK Kif18B were unsuccessful; therefore we were unable to assess the extent of knockdown at the protein level.
Cells in which Kif18B was knocked down had an increase in astral MTs (Figure 1, A–C), which appeared even more dramatic than observed previously in HeLa cells (Stout et al., 2011). The effects were most dramatic in prometaphase, where MTs in these cells appeared quite rigid as they extended toward the cell cortex, and cells often appeared to have shorter or distorted spindles (Figure 1, B and C). Quantification of the mitotic stage distribution showed that both siRNAs resulted in a decrease in percentage of cells in metaphase, and Kif18B-2 siRNA also caused a statistically significant increase in percentage of cells in prometaphase (Figure 1D). These results are distinct from what was observed with Kif18B knockdown in HeLa cells (Stout et al., 2011), but this may be because PtK cells have far fewer chromosomes than HeLa cells and so it is easier to score chromosome alignment. Our results here suggest that changes in astral MT organization and density could affect the ability of cells to properly align chromosomes at the spindle equator.
FIGURE 1:
Kif18B knockdown causes a decrease in the percentage of metaphase cells. Cells transfected with (A) control, (B) Kif18B-1, or (C) Kif18B-2 siRNAs were fixed and stained for MTs (red) and DNA (blue). Representative deconvolved images from the indicated mitotic stages. Scale bar, 10 μm. (D) Distribution of mitotic stages quantified from six experiments in which 100 cells were counted per experiment, plotted as mean ± SD. *p < 0.05.
To address whether loss of Kif18B causes defects in mitotic progression, we used time-lapse imaging of cells from prophase through cytokinesis and scored movies for qualitative defects in chromosome behavior (Figure 2, A and B, and Supplemental Movie S1). Kif18B knockdown caused an increase in percentage of cells in which not all chromosomes congressed or had a poor alignment of chromosomes, where chromosomes were not tightly aligned at the metaphase plate (Figure 2C), consistent with our observation in fixed cells that there is a reduction in percentage of cells in metaphase. The defects in chromosome alignment were not due to off-target effects of the Kif18B siRNAs knocking down Kif18A, as the siRNAs did not have sufficient homology with Kif18A sequence (Udy et al., 2015) to cause knockdown (58% identity with Kif18B-1 and 75% identity with Kif18B-2). Consistent with this sequence analysis, there was no significant reduction of Kif18A mRNA by quantitative PCR (qPCR) with either oligo (p = 0.32 for Kif18B-1 and p = 0.20 for Kif18B-2).
FIGURE 2:
Kif18B knockdown does not affect mitotic progression but causes poor chromosome alignment in metaphase. Selected frames from phase-contrast time-lapse images of cells transfected with (A) control or (B) Kif18B-2 siRNAs and then imaged for 2 h at 30-s intervals. Scale bar, 10 μm. (C) Percentage of cells with indicated phenotype in control (20 cells) or Kif18B RNAi (30 cells). (D) Average timing between mitotic stages. Dots represent individual measurements, and bar and whiskers represent mean ± SD.
One potential model for how Kif18B knockdown might perturb chromosome alignment is that excess MT polymerization resulted in an increase in the polar ejection force on chromosome arms and a disruption in chromosome arm positioning (Kapoor and Compton, 2002). Computational modeling predicted that changes in chromosome arm orientation might increase merotelic attachments, which result in defects in chromosome segregation (Cimini et al., 2001; Cimini, 2008; Paul et al., 2009). To test this idea, we examined how Kif18B RNA interference (RNAi) cells progressed through anaphase. Of the cells imaged, only one of 20 control cells failed to enter anaphase, whereas five of 30 Kif18B-knockdown cells failed to enter anaphase during the 2-h imaging time window. There was no effect on the fidelity of chromosome segregation, as Kif18B knockdown did not increase percentage of cells with lagging chromosomes during anaphase. This suggests that altered forces on chromosome arms in the absence of Kif18B do not cause errors in kinetochore–MT attachments and distinguishes the phenotype of Kif18B knockdown from that of MCAK knockdown, which causes an increase in percentage of cells with defects in chromosome segregation (Stout et al., 2006). In support of this finding, MCAK localization was not disrupted in the Kif18B-knockdown cells (Supplemental Figure S1), and MCAK levels were not reduced, but instead there was a twofold increase in the amount of MCAK mRNA in Kif18B-knockdown cells (p < 0.05 for Kif18B-1 and Kif18B-2). Although there was no difference in the average time of mitotic progression between any stages, there was a statistically significant increase in the variance of the time from nuclear envelope breakdown to metaphase alignment (Figure 2D). These results suggest that loss of Kif18B causes a defect in spindle MT organization that hinders the ability of chromosomes to properly align on the spindle equator without affecting the fidelity of chromosome segregation.
Kif18B knockdown increases astral MT length but does not affect kinetochore MT length
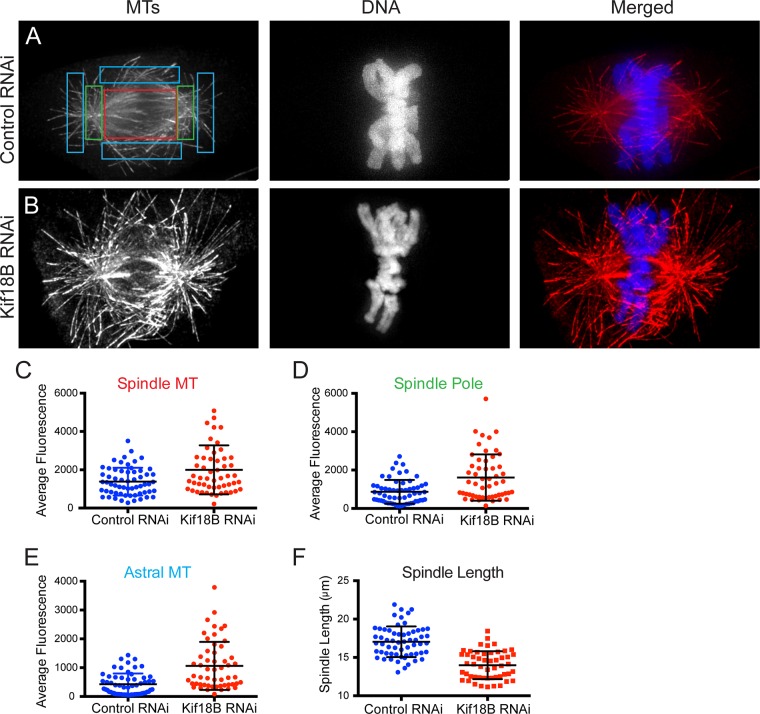
The foregoing results show that Kif18B knockdown causes defects in MT organization that affect chromosome alignment. To ask which populations of MTs were affected by the depletion of Kif18B, we used quantification of MT fluorescence to measure the relative changes in MT density in the spindle, near the poles and of the astral MTs (Figure 3, A and B; Rizk et al., 2009). There was a 45% increase in MT fluorescence in the spindle (Figure 3C; p < 0.01), an 85% increase in the region of the poles (Figure 3D; p < 0.001), and a 150% increase in the astral MT region (Figure 3E; p < 0.001). These results show that whereas Kif18B contributes to the dynamics of all MTs in the spindle, its effects on astral MTs are most prominent, consistent with the observation that Kif18 is localized primarily to the plus tips of astral MTs (Stout et al., 2011; Tanenbaum et al., 2011). Furthermore, these changes in MT polymer amounts were also correlated with reduced spindle length by 18% (p < 0.001), consistent with our qualitative analysis of the spindles (Figure 1).
FIGURE 3:
Kif18B knockdown increases MT polymer levels. Cells transfected with (A) control or (B) Kif18B-1 siRNAs were fixed and stained for MTs (red) and DNA (blue). Color-coded boxes were drawn to indicate the quantification of spindle (red), spindle pole (green), and astral MTs (cyan). Scale bar, 10 μm. (C–E) Average fluorescence intensity measurements calculated for control and Kif18B RNAi cells in regions corresponding to (C) the spindle, (D) the spindle poles, and (E) the astral MTs. (F) Spindle length from the same cells analyzed for MT fluorescence intensity in A–E. Dot plots for a total of 60 control spindles and 54 Kif18B RNAi spindles, with bars and whiskers corresponding to mean ± SD. Three experiments.
One possibility for the defects in chromosome alignment seen with Kif18B RNAi would be if Kif18B disrupted kinetochore-fiber MT dynamics and in turn kinetochore-fiber length. Because chromosome position at the spindle equator is balanced by forces from MTs coming from opposite spindle poles, we used monopolar spindles to look at kinetochore-fiber MT lengths in the absence of these opposing forces. Cells were transfected with control or Kif18B siRNAs, incubated for 72 h, and then treated with 100 μM monastrol during the last 3 h of the transfection to arrest mitotic cells in a monopolar spindle configuration. Kif18B knockdown resulted in a dramatic increase in the astral-cortical MTs (Figure 4, A and B), which resulted in a 70% increase in the area of the monopolar spindle (Figure 4C; p < 0.0001). This increase in area was due to an increase in the average length of the astral-cortical MTs by 40% (Figure 4D; p < 0.0001). There was no change in the length of the kinetochore-fiber MTs (Figure 4E; p = 0.32). These results suggest that whereas Kif18B affects chromosome positioning, these defects are due to excess MT polymerization and to changes in spindle MT organization rather than to a direct effect on kinetochore-fiber MTs. Furthermore, because there is no effect on kinetochore-fiber length, this suggests that the shorter spindles are due to changes in other MT subclasses.
FIGURE 4:
Kif18B knockdown increases astral MT length but not kinetochore-fiber MT length. Cells were transfected with (A) control or (B) Kif18B-1 siRNAs and treated with monastrol 3 h before fixation. Cells were stained for MTs (red), DNA (blue), and kinetochores (green). Scale bar, 10 μm. (C) Monopolar spindle area from 55 control and 56 Kif18B RNAi spindles in three independent experiments. Dots represent individual measurements, and bar and whiskers represent mean ± SD. Frequency distribution of (D) astral MT length and (E) kinetochore MT length from three independent experiments in which eight length measurements were averaged per spindle.
The long-MT defect is enhanced in the absence of the actin cytoskeleton
When analyzing the morphology of the spindles in Kif18B RNAi cells, we often noticed that the astral MTs appeared long and rigid as they extended toward the cortex. Because some members of the kinesin-8 family are MT-destabilizing enzymes and Kif18B localizes to the plus ends of astral MTs, loss of Kif18B may prevent MT catastrophe near the cortex. The MTs may then become anchored in the cortex by interactions with the cortical cytoskeletal network. To test this idea, we first asked whether depolymerization of the actin cytoskeleton cortex abrogated the long-MT phenotype seen after Kif18B knockdown. We treated cells with 0.1 μg/ml cytochalasin D for 30 min, a condition that was sufficient to depolymerize the actin but that did not cause so much cell distortion that the cells de-adhered from the coverslip. The long and rigid astral MTs seen after Kif18B RNAi were readily apparent in cytochalasin-treated cells (Figure 5A). However, many of the cells were distorted, preventing accurate quantification of the MT polymer levels. To overcome this problem, we treated cells with monastrol for 3–5 h before cytochalasin D treatment, resulting in monopolar spindles with much more uniform morphology (Figure 5B). We found that in control dimethyl sulfoxide (DMSO)–treated cells, loss of Kif18B caused an 89% increase in astral MT area (p < 0.001), similar to what we reported earlier. Cytochalasin D treatment had no effect on control transfected cells (p = 0.91). In contrast, there was an additional 32% increase in astral MT area (p < 0.001) when Kif18B-knockdown cells were treated with cytochalasin D. Together these studies support the idea that Kif18B regulates astral MT dynamics to constrain the growth of astral MTs. Removal of the actin cytoskeletal cortex in addition to Kif18B depletion allows the MTs to grow even longer than with removal of Kif18B alone, suggesting that MT length is also constrained by the cortex.
FIGURE 5:
Depolymerization of the actin cytoskeleton does not suppress the long astral MTs caused by Kif18B knockdown. (A) Cells transfected with control or Kif18B-2 siRNAs were treated with DMSO or cytochalasin D for 30 min before being fixed and stained for MTs (green), actin (red), and DNA (blue). Images were collected on a GE Healthcare OMX 3D-SIM microscope. (B) Cells treated as in A, except that they were treated with monastrol for 3 h before cytochalasin D treatment and fixation. (C) Monopolar spindle area from 45 spindles in three independent experiments. Dots represent individual measurements, and bar and whiskers represent mean ± SD. ***p < 0.001. Scale bar, 10 μm.
Kif18B controls lifetime and growth rate of astral MTs
To look at MT dynamics more directly, we did time-lapse microscopy to visualize the plus tip–tracking protein EB1 in spindles of PtK cells transfected with control or Kif18B siRNAs (Figure 6, A and B; Tirnauer et al., 2002). Green fluorescent protein (GFP)–EB1 tracks were automatically detected using plusTipTracker (Applegate et al., 2011). We found a 1.5-fold increase in the average number of EB1 tracks per spindle when Kif18B was knocked down (Supplemental Figure S2A and Supplemental Movie S2). Because Kif18B has been proposed to be a regulator of MT dynamics, we first asked whether Kif18B knockdown changes the MT lifetime of all MTs in a spindle. Indeed, we found a 15% increase in the average MT lifetime in Kif18B siRNA cells compared with controls (p < 0.0001; Supplemental Figure S2B).
FIGURE 6:
Kif18B knockdown causes an increase in the lifetime of astral MTs and a decrease in MT growth rate in general. (A, B) First frame of EB1-GFP image series of spindles in PtK1 cells transfected with (A) control or (B) Kif18B-2 siRNA. Scale bar, 10 μm. (C, D) The plusTipTracker software was used to detect and link EB1 comets from time-lapse images of spindles in A and B. Tracked comets were divided into three subspindle regions and color coded accordingly: astral MTs to the left and right of spindle poles (aster_lr) are in magenta, astral MTs in the top and bottom region of the spindle body (aster_tb) are in green, and MTs in the central region of the spindle body are in cyan (Spindle_midzone). (E) Lifetime of EB1 tracks in the three subspindle regions was fit with a one-phase exponential decay to determine the rate constant K (s−1), and the average lifetime (τ) was determined from K−1. Data represent mean ± 95% confidence interval from the fit (see also Supplemental Figure S2) for 11 spindles for the control with 1045 tracks in aster_lr region, 1063 tracks in aster_tb region, and 4030 tracks in spindle_midzone region and 16 Kif18B RNAi spindles with 3605 tracks in aster_lr region, 2295 tracks in aster_tb region, and 7828 tracks in spindle_midzone region. (F) Histograms of EB1 track velocity in the entire spindle of control and Kif18B knockdowns graphed with the fitting curve of a Gaussian distribution. The mean and SD derived from each fit is indicated. Control RNAi, 11 spindles with 7782 tracks; Kif18B RNAi, 16 spindles with 16414 tracks.
To address whether knocking down Kif18B specifically affected the dynamics of different populations of MTs, we looked at the lifetime of EB1 tracks in three distinct subspindle regions: aster_lr (astral MTs that extend from the left and right of each spindle pole toward the cortex), aster_tb (astral MTs that extend toward the cortex on the top and bottom of the spindle), and spindle_midzone (MTs that are within the center of the spindle body; Figure 6, C–D). Analysis of MT lifetimes in the three subspindle regions revealed that there was a more dramatic increase in the astral regions in Kif18B siRNA cells but only a relatively small increase in the lifetime of the MTs in the spindle midzone (Figure 6E). In particular, the average MT lifetime in the aster_lr region increased by 25% in Kif18B RNAi relative to control (p < 0.0001; Figure 6E and Supplemental Figure S2C). The average MT lifetime in the aster_tb region increased even more dramatically by 37% in Kif18B RNAi (p < 0.0001; Figure 6E and Supplemental Figure S2D). In contrast, there was only a 6% increase in the average MT lifetime in the spindle_midzone region in Kif18B RNAi cells, which was not statistically different from controls (p = 0.07; Figure 6E and Supplemental Figure S2E). These results show that loss of Kif18B specifically affects the dynamics of astral MTs near the cortex but not of MTs in the spindle.
Because knocking down MT dynamic regulators affects MT assembly rates (Wordeman et al., 2016), we also measured the velocity of the movement of EB1 tracks as a readout for MT growth rate. We found a 6% decrease in the mean velocity of MTs in Kif18B RNAi cells, which was not statistically different from control (p = 0.09; Figure 6F). We also noticed a 20% decrease in the variance of velocity in Kif18B RNAi spindles (p = 0.0001), suggesting that there are likely subpopulations of MTs that are differentially affected by knockdown of Kif18B. Analysis of MT growth rate in the subspindle regions revealed a bigger change of the mean and variance of velocity in the astral regions of Kif18 RNAi cells compared with controls. Specifically, we found a 5% decrease in the mean velocity in the aster_lr region in Kif18B-knockdown relative to control (p < 0.0001; Supplemental Figure S2F). However, the decrease was much larger (15%) in the aster_tb region (p < 0.0001; Supplemental Figure S2G). We also found a modest 6% decrease in the spindle_midzone region (p < 0.0001; Supplemental Figure S2H). There was a decrease in the variance of relative distribution of velocity in the three subspindle regions, with 22, 15, and 18% decrease in aster_lr, aster_tb, and spindle_midzone, respectively (F test, p < 0.0001 for all three regions; Supplemental Figure S2, F–H). Overall our results are consistent with the idea that Kif18B is a MT-destabilizing enzyme that differentially regulates astral MT dynamics.
Differential regulation of MT dynamics by kinesin-8 and kinesin-13 proteins
Our data provide new insights into the complexities by which kinesins can regulate MT dynamics in cells and provide the first report of how loss of Kif18B affects MT dynamics in cells. For the kinesin-8 family in vertebrates, focus has largely been on Kif18A, which dampens chromosome oscillations by suppressing MT dynamics on kinetochore MTs (Du et al., 2010; Stumpff et al., 2012). Consistent with this idea, purified GFP-Kif18A also dampens the dynamics of purified MTs in vitro (Stumpff et al., 2012). Our results show that knockdown of Kif18B reduces MT growth velocities, similar to the phenotype of knockdown of MCAK but opposite to knockdown of Kif18A (Wordeman et al., 2016), suggesting that different members of the kinesin-8 superfamily can have opposing effects on MT dynamics. This finding is somewhat surprising because it was shown that the catalytic domain of Kif18B can functionally substitute for the catalytic domain of Kif18A in terms of chromosome positioning (Kim et al., 2014), implying that these catalytic domains are equivalent. This suggests that the differences in regulation of MT dynamics come either from the populations of MTs on which the proteins act or from their tail domains, which regulate association with MTs. Indeed, the tails of Kif18A and Kif18B have differing effects on their association with MTs. For Kif18A, the tail domain has a second MT-binding site, which helps increase processivity of the motor to reach the long ends of MTs (Stumpff et al., 2011; Weaver et al., 2011). In contrast, although the tail of Kif18B also has a second MT-binding site, Kif18B is much less processive than Kif18A, and the second MT-binding site contributes to diffusive behavior by decreasing the off rate from the MT lattice (Shin et al., 2015). These results suggest that the tail domains of kinesin-8 proteins play important roles in spatially regulating MT dynamics.
Our work also reveals that the effect of loss of Kif18B on MT lifetime is not uniform in all parts of the spindle. The effect of Kif18B knockdown is seen more prominently on astral MTs near the cortex, suggesting that the behavior of Kif18B is more similar to that of the yeast kinesin-8, Kip3, which has a strong action on cortical MTs (Gupta et al., 2006), or to Klp 5/6 in Schizosaccharomyces pombe (Tischer et al., 2009). Also important in this light is the observation that knockdown of Kif18B has a more prominent effect on those MTs that interacted with the cortex above and below the spindle rather than the astral MTs that extended to the left and right of each spindle pole. The MTs that extend to the cortex above and below the spindle are on average longer than the ones that interact with the cortex closer to the poles. This may suggest that Kif18B shares another property with Kip3, in that it acts preferentially on longer MTs (Varga et al., 2006), an idea that remains to be tested.
There is strong evidence that Kip3 activity is spatially controlled, and it has multiple effects on MT dynamics, depending on where it is acting in the cell (Fukuda et al., 2014), further supporting the idea of spatial regulation of activity. Our finding that actin depolymerization allows for a further increase in MT length in the absence of Kif18B suggests that Kif18B normally acts at MT plus ends rather than at the cortex, and without Kif18B, the MTs will persist in a state of unbounded growth, consistent with the increase in MT lifetimes seen in the absence of Kif18B. An important avenue for further study will be to understand how Kif18B affects the dynamics of purified MTs and whether its effects on dynamics are more similar to those of Kip3, Kif18A, or MCAK.
One recent study presented the surprising result that long-term knockdown of MCAK or Kif18A had opposite effects on MT growth velocities compared with short-term perturbation of the protein, which was due to tubulin autoregulation (Wordeman et al., 2016). Our data on Kif18B knockdown are more similar to those seen with long-term knockdown of MCAK, suggesting that these two proteins will have similar effects on MT dynamics. This is consistent with the morphological changes to the spindle and astral MTs seen after knockdown, which are very similar between Kif18B and MCAK knockdown but distinct from changes with Kif18A knockdown (Mayr et al., 2007; Stumpff et al., 2008). One explanation for the similar phenotypes comes from studies proposing that Kif18B directly targets MCAK to the MT plus ends (Tanenbaum et al., 2011), such that knockdown of Kif18B would also disrupt MCAK localization. We do not favor this idea because we saw no obvious disruption of MCAK localization in cells with Kif18B knockdown. We also did not find defects in chromosome segregation, which occur after MCAK knockdown (Stout et al., 2006). We cannot rule out the possibility that Kif18B and MCAK only function together at the ends of astral MTs and not on kinetochore–MTs; however, MCAK still localized to MT plus ends in Kif18B RNAi cells (Supplemental Figure S1). Furthermore, immunoprecipitation of MCAK pulls down no detectable Kif18B in Xenopus egg extracts, and in fact, the levels of Kif18B are at least 100-fold less than MCAK in the extract (Yount and Walczak, unpublished results). These results suggest that MCAK depletion alone is sufficient to cause the long-MT phenotype, even in systems with little or no Kif18B, suggesting that Kif18B has the ability to perturb MT dynamics in an MCAK-independent manner. Looking to the future, it will be important to understand the biochemical mechanisms by which these individual proteins regulate dynamics and then understand how these activities are coordinated near the cell cortex.
MATERIALS AND METHODS
Cell culture and RNAi
PtK2 cells were cultured at 37°C in a complete medium composed of Opti-MEM, 10% fetal bovine serum, GlutaMAX, and penicillin/streptomycin. GFP-EB1–expressing PtK1 cells were similarly cultured except using Ham’s F-12 instead of Opti-MEM. For RNAi, cells were plated at a density of 25,000 cells/ml in six-well culture dishes onto poly-l-lysine–coated coverslips or into MatTek dishes. After 24 h, the cells were transfected with 20 μM siRNA using Oligofectamine or 10 nM siRNA using Lipofectamine RNAiMAX according to the manufacturer’s instructions. At 24 h posttransfection, the medium was removed from each well and replaced with 2 ml of fresh medium. Cells were analyzed at 72 h posttransfection. The following siRNAs (Dharmacon, Lafayette, CO) were used: negative control #2 (UGGUUUAUGUUGUGUGA), PtK2 Kif18B-1 (GAACCAGAAUCGUAAGCAAUU), and PtK2 Kif18B-2 RNAi oligonucleotide (CAGCCCAGCUAGCGGAAUAUU).
To form monopolar spindles, cells were incubated at 37°C in medium containing 100 μM monastrol for 3 h before fixation or imaging. To depolymerize the actin, cells were incubated for 30 min at 37°C in medium containing 1 μg/ml cytochalasin D in DMSO. All controls contained an equivalent amount of DMSO in medium.
qPCR analysis
The extent of knockdown was analyzed by qPCR using Sybr Green (Agilent Technologies, Santa Clara, CA) on a Stratagene Mx3000P quantitative PCR thermal cycler (Agilent Technologies). RNA was extracted from 2 ml of cells at 72 h posttransfection using the Qiagen RNAEasy isolation kit. cDNA was synthesized using the AffinitySCRIPT qPCR cDNA synthesis kit. All reactions were run in triplicate per sample, and each sample was run in duplicate. All knockdown values presented are from four independent siRNA transfections. The following primers were used in qPCR: PtK–glyceraldehyde-3-phosphate dehydrogenase (forward, GTGT-TCCTACTCCCAACGTATC); reverse, TGCCTGCTTCACCACTTT), PtK-Kif18B (forward, AGAGGGATGAAAGCACCATAAA; reverse, TAGCCTCTTGGTCTTGTCTTTG), PtK-MCAK (forward, TGACTTTGAGGATGTGGCAG; reverse, AGGGATTTTGGAGAGGGTTG), and PtK-Kif18A (forward, CTCGGAGGAAACTGTCAAACT; reverse, TTGCACGGTTGGCATACT)
Phase contrast imaging
Time-lapse phase contrast microscopy was carried out on a Nikon TE-300 inverted microscope (Nikon Instruments, Melville, NY) with a 40×, 0.6 numerical aperture Plan Fluor objective at 68-76 h posttransfection. Samples were maintained at 37°C using a 400 ASI airstream incubator (Nevtek, Williamsville, VA). Images were collected onto a CoolSnap CF camera (Photometrics, Tucson, AZ) at 30-s intervals using 100-ms exposure times. The microscope shutter and camera were controlled with MetaMorph software (Molecular Devices, Sunnyvale, CA). Images were processed in FIJI for contrast enhancement and preparation of movies. All timing and phenotype analysis was done manually using frame-by-frame analysis and recording of digital time stamps.
Immunofluorescence staining
Cells were fixed in −20°C methanol, a combination of formaldehyde and glutaraldehyde, or glutaraldehyde alone. Cells were rinsed in phosphate-buffered saline (PBS; 12 mM PO42−, 137 mM NaCl, and 3 mM KCl, pH 7.4) and then fixed for 20 min in 4% formaldehyde and 0.1% glutaraldehyde in PHEM (60 mM K–1,4-piperazinediethanesulfonic acid [PIPES], 25 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, 10 mM ethylene glycol tetraacetic acid [EGTA], and 2 mM MgCl2, pH 6.9). The fixative was quenched with sodium borohydride in TBS (20 mM Tris, 150 mM NaCl, pH 7.5) for 5 min. The fixed cells were rinsed three times with TBS-Tx (TBS with 0.1% Triton X-100) before proceeding with immunofluorescence. Alternatively, cells were washed with room temperature MTSB (4 M glycerol, 100 mM PIPES, pH 6.8, 1 mM EGTA, 5 mM MgCl2) for 2 min, extracted with MTSB plus 0.5% Triton for 2 min, and then washed again with MTSB for 2 min before fixation in 1% glutaraldehyde in PBS for 10 min. Glutaraldehyde was quenched with three 10-min washes of 0.5 mg/ml NaBH4 in PBS. After fixation, the cells were rinsed in TBS-Tx before processing for immunofluorescence.
Cells were blocked for 30 min in Ab Dil (2% bovine serum albumin in TBS-Tx) at room temperature. Primary antibodies included DM1α (1/500; Sigma-Aldrich, St. Louis, MO), ACA (1/10; Antibodies Inc., Davis, CA), or 5 μg/ml anti–PtK-MCAK (Rizk et al., 2009), which were diluted in Ab Dil, and cells were stained for 30 min each. Cells were rinsed three times in TBS-Tx and then incubated with DyLight or Alexa Fluor secondary antibodies at 1–2 μg/ml final concentration in Ab Dil and incubated for 30 min at room temperature. For labeling of the actin cytoskeleton, cells were washed three times in TBS-Tx and incubated with 1/1000 rhodamine-phalloidin for 30 min at room temperature. All coverslips were washed three times with TBS-Tx, incubated with Hoechst (2–10 μg/ml) for 5 min, washed three times with TBS-Tx, and then mounted in mounting medium (0.5% phenylenediamine in 20 mM Tris, pH 8.8, in 90% glycerol) or ProLong Diamond/Gold (Invitrogen, Carlsbad, CA) and sealed with nail polish.
Microscopy
Immunofluorescence imaging was performed with a Nikon Eclipse 90i microscope using a 100× Apochromatic PLAN objective (numerical aperture [NA] 1.4) and a CoolSnap HQ charge-coupled device (CCD) camera (Photometrics). MetaMorph was used to control the camera and emission/excitation filters. The z-stacks of cells were taken at 0.2- or 0.5-μm steps through the cell volume. Images were deconvolved with AutoQuant X 3D deconvolution software (Media Cybernetics, Rockville, MD). Superresolution imaging was performed using the OMX 3D-SIM Super-Resolution system controlled by the DV-OMX software (GE Healthcare Life Sciences, Pittsburgh, PA). Images were acquired using a UNIPLANAPO 100×/1.4 NA objective and immersion oil with a refractive index of 1.514. The z-stacks of cells were taken at 0.125-μm steps through the cell volume. Images were processed using softWoRx and IMARIS three-dimensional (3D) imaging software (Bitplane, Concord, MA). All micrographs were processed in FIJI for contrast enhancement, and figures were assembled in Adobe Illustrator (Adobe, San Jose, CA).
Image analysis and quantification
MT subgroup measurements were made on deconvolved 3D reconstructed images essentially as described previously (Rizk et al., 2009). In MetaMorph, images were rotated so that the spindle was horizontal. Boxes were drawn over each MT population with corresponding boxes in regions outside the spindle for background measurements. For astral-cortical MT intensity, four 30 × 150–pixel boxes were positioned around the spindle (two horizontal and two vertical). For pole intensity, two 30 × 100–pixel boxes were placed over the centrosomes. For spindle MT intensity, two 100 × 150–pixel boxes were placed in the spindle between the poles and spindle equator. Fluorescence values were collected from at least 15–20 cells per condition for each of three independent experiments. Spindle lengths were calculated as pole-to-pole distances on the deconvolved reconstructed images in FIJI. For monopolar cells, astral MT length and pole–kinetochore distances were measured on deconvolved 3D reconstructed images using the line tool and ACA as a kinetochore marker.
For quantification of astral MT area in monopolar spindles, z-stack images were processed for maximal projection and contrast enhancement using a custom-written macro in FIJI. Monopolar spindle area was then measured using Cell Profiler (Broad Institute, Cambridge, MA) by a macro in which each image was sequentially opened, and the spindle area was determined by a user-drawn circle around the spindle. Area measurements were automatically exported to Excel (Microsoft, Redmond, WA) and then converted into micrometer units. All graphs were assembled with GraphPad Prism (La Jolla, CA), which was also used for statistical analysis.
EB1 imaging and analysis
EB1-GFP expressing PtK1 cells (Tirnauer et al., 2002) were plated onto MatTek dishes and transfected as described. Cells were imaged with a custom-built spinning-disk confocal microscope. A Nikon TE2000U inverted microscope was equipped with a CSU-10 spinning-disk confocal head (Yokogawa, Sugarland, TX) and illuminated with a 10-mW, 488-nm laser. Images were captured with a 60×/1.4 PlanApo objective and collected on a Cascade-II electron-multiplying CCD camera (Photometrics) using a 200-ms exposure and 2-s time interval for a total of 2 min. The microscope was controlled using Nikon Elements software.
EB1 comets were automatically detected and frame-to-frame linked using the plusTipTracker MATLAB software package (MathWorks, Natick, MA; Applegate et al., 2011). A region of interest was drawn along the cell boundary to exclude tracks from adjacent cells. The parameters used were maximum gap length, 0; minimum track length, three frames; backward angle, 10; forward angle, 30; search radius range, 1–5 pixels; fluctuation radius (pixels), 1; and maximum shrinkage factor, 1.5. After detection, the x, y-coordinates of EB1 position in the tracks were extracted from plusTipTracker and further analyzed in MATLAB.
For positional analysis of EB1 tracks, the coordinates of the two spindle poles and the upper and lower boundaries of a spindle body were input manually. For the ease of analysis, spindles were rotated to an orientation with the long axis in the horizontal position using a matrix rotation function, and tracks were divided into subspindle regions based on the location of the first comet in each track (Zong et al., 2016). Because astral MTs emanating from a spindle pole toward the cell cortex can move farther away toward the left or the right of the respective poles or toward the cortex near the spindle equator, we defined two astral MT populations based on their location. 1) aster_lr is the combined area to the left of the left pole and to the right of the right pole. MTs in this region were color coded in magenta. 2) aster_tb is the combined area containing astral MTs that emanate toward the top and toward the bottom cortex, respectively, of the spindle body. MTs in this region were color coded in green. To get a relatively pure population of nonastral MTs—in other words, to reduce the number of astral MTs—we chose the spindle midzone as a centered rectangle with a length equal to 60% of the spindle length (determined by the pole positions) and a width equal to 60% of the spindle width (determined by the upper and lower boundaries of the spindle body) for the analysis of nonastral MT dynamics. MTs in this region were color coded in cyan.
The lifetime and velocity of EB1 tracks were calculated from the x, y-coordinates of each EB1 comet as in Applegate et al. (2011). The lifetime of a track is determined as the amount of time a comet persists in frames before it disappears. The velocity of a track was averaged from all of the frame-to-frame velocities. The frequency distribution of lifetime and velocity were fitted to an exponential and Gaussian distribution, respectively, using the MATLAB curve-fitting app. The average lifetime (τ) was calculated as the reciprocal of the rate constant K from the exponential fit. All graphs were plotted in MATLAB, and data analysis was done with MATLAB and GraphPad Prism.
Supplementary Material
Acknowledgments
We thank Rania Rizk, Laura Wetzel, and Yuxi Li for early experiments and help in designing the image quantification protocols. We also thank J. Tirnauer for the GFP-EB1–expressing cells, J. Shah for unpublished PtK sequences for Kif18B, and M. Gupta for suggesting the actin depolymerization experiment. We further thank Jim Powers of the Light Microscopy Imaging Center for assistance with imaging and offer a special thanks to Amber Yount and Sanjay Shrestha for comments on the manuscript. This work was supported by National Institutes of Health Grant GM059618 to C.E.W. The Indiana University Light Microscopy Imaging Center is supported in part by the Office of Vice Provost for Research at Indiana University.
Abbreviations used:
- aster_lr
aster left right
- aster_tb
aster top bottom
- GFP
green fluorescent protein
- MT
microtubule
- RNAi
RNA interference
- siRNA
short interfering RNA.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0254) on August 24, 2016.
REFERENCES
- Applegate KT, Besson S, Matov A, Bagonis MH, Jaqaman K, Danuser G. plusTipTracker: quantitative image analysis software for the measurement of microtubule dynamics. J Struc Biol. 2011;176:168–184. doi: 10.1016/j.jsb.2011.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimini D. Merotelic kinetochore orientation, aneuploidy, and cancer. Biochim Biophys Acta. 2008;1786:32–40. doi: 10.1016/j.bbcan.2008.05.003. [DOI] [PubMed] [Google Scholar]
- Cimini D, Howell B, Maddox P, Khodjakov A, Degrassi F, Salmon ED. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J Cell Biol. 2001;153:517–527. doi: 10.1083/jcb.153.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottingham FR, Gheber L, Miller DL, Hoyt MA. Novel roles for saccharomyces cerevisiae mitotic spindle motors. J Cell Biol. 1999;147:335–350. doi: 10.1083/jcb.147.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/s0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- DeZwaan TM, Ellingson E, Pellman D, Roof DM. Kinesin-related KIP3 of Saccharomyces cerevisiae is required for a distinct step in nuclear migration. J Cell Biol. 1997;138:1023–1040. doi: 10.1083/jcb.138.5.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domnitz SB, Wagenbach M, Decarreau J, Wordeman L. MCAK activity at microtubule tips regulates spindle microtubule length to promote robust kinetochore attachment. J Cell Biol. 2012;197:231–237. doi: 10.1083/jcb.201108147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, English CA, Ohi R. The kinesin-8 Kif18A dampens microtubule plus-end dynamics. Curr Biol. 2010;20:374–380. doi: 10.1016/j.cub.2009.12.049. [DOI] [PubMed] [Google Scholar]
- Ems-McClung SC, Walczak CE. Kinesin-13s in mitosis: key players in the spatial and temporal organization of spindle microtubules. Semin Cell Dev Biol. 2010;21:276–282. doi: 10.1016/j.semcdb.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Luchniak A, Murphy ER, Gupta ML., Jr Spatial control of microtubule length and lifetime by opposing stabilizing and destabilizing functions of Kinesin-8. Curr Biol. 2014;24:1826–1835. doi: 10.1016/j.cub.2014.06.069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Upton K, Compton DA. Efficient mitosis in human cells lacking poleward microtubule flux. Curr Biol. 2005;15:1827–1832. doi: 10.1016/j.cub.2005.08.065. [DOI] [PubMed] [Google Scholar]
- Garcia MA, Koonrugsa N, Toda T. Spindle-kinetochore attachment requires the combined action of Kin I-like Klp5/6 and Alp14/Dis1-MAPs in fission yeast. EMBO J. 2002;21:6015–6024. doi: 10.1093/emboj/cdf611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatt MK, Savoian MS, Riparbelli MG, Massarelli C, Callaini G, Glover DM. Klp67A destabilises pre-anaphase microtubules but subsequently is required to stabilise the central spindle. J Cell Sci. 2005;118:2671–2682. doi: 10.1242/jcs.02410. [DOI] [PubMed] [Google Scholar]
- Gupta ML, Jr, Carvalho P, Roof DM, Pellman D. Plus end-specific depolymerase activity of Kip3, a kinesin-8 protein, explains its role in positioning the yeast mitotic spindle. Nat Cell Biol. 2006;8:913–923. doi: 10.1038/ncb1457. [DOI] [PubMed] [Google Scholar]
- Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The Kin I kinesin MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing comlex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/s1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapoor TM, Compton DA. Searching for the middle ground: mechanisms of chromosome alignment during mitosis. J Cell Biol. 2002;157:551–556. doi: 10.1083/jcb.200202073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Fonseca C, Stumpff J. A unique kinesin-8 surface loop provides specificity for chromosome alignment. Mol Biol Cell. 2014;25:3319–3329. doi: 10.1091/mbc.E14-06-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maney T, Hunter AW, Wagenbach M, Wordeman L. Mitotic centromere-associated kinesin is important for anaphase chromosome segregation. J Cell Biol. 1998;142:787–801. doi: 10.1083/jcb.142.3.787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning AL, Ganem NJ, Bakhoum SF, Wagenbach M, Wordeman L, Compton DA. The kinesin-13 proteins Kif2a, Kif2b, and Kif2c/MCAK have distinct roles during mitosis in human cells. Mol Biol Cell. 2007;18:2970–2979. doi: 10.1091/mbc.E07-02-0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr MI, Hummer S, Bormann J, Gruner T, Adio S, Woehlke G, Mayer TU. The human kinesin Kif18A is a motile microtubule depolymerase essential for chromosome congression. Curr Biol. 2007;17:488–498. doi: 10.1016/j.cub.2007.02.036. [DOI] [PubMed] [Google Scholar]
- Mennella V, Rogers GC, Rogers SL, Buster DW, Vale RD, Sharp DJ. Functionally distinct kinesin-13 family members cooperate to regulate microtubule dynamics during interphase. Nat Cell Biol. 2005;7:235–245. doi: 10.1038/ncb1222. [DOI] [PubMed] [Google Scholar]
- Moore AT, Rankin KE, von Dassow G, Peris L, Wagenbach M, Ovechkina Y, Andrieux A, Job D, Wordeman L. MCAK associates with the tips of polymerizing microtubules. J Cell Biol. 2005;169:391–397. doi: 10.1083/jcb.200411089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohi R, Burbank K, Liu Q, Mitchison TJ. Nonredundant functions of kinesin-13s during meiotic spindle assembly. Curr Biol. 2007;17:935–939. doi: 10.1016/j.cub.2007.04.057. [DOI] [PubMed] [Google Scholar]
- Paul R, Wollman R, Silkworth WT, Nardi IK, Cimini D, Mogilner A. Computer simulations predict that chromosome movements and rotations accelerate mitotic spindle assembly without compromising accuracy. Proc Natl Acad Sci USA. 2009;106:15708–15713. doi: 10.1073/pnas.0908261106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Bohannon K, Wetzel L, Powers JA, Shaw SL, Walczak CE. MCAK and paclitaxel have differential effects on spindle organization and microtubule dynamics. Mol Biol Cell. 2009;20:1639–1651. doi: 10.1091/mbc.E08-09-0985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk RS, Discipio KA, Proudfoot KG, Gupta ML., Jr The kinesin-8 Kip3 scales anaphase spindle length by suppression of midzone microtubule polymerization. J Cell Biol. 2014;204:965–975. doi: 10.1083/jcb.201312039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- Shin Y, Du Y, Collier SE, Ohi MD, Lang MJ, Ohi R. Biased Brownian motion as a mechanism to facilitate nanometer-scale exploration of the microtubule plus end by a kinesin-8. Proc Natl Acad Sci USA. 2015;112:E3826–E3835. doi: 10.1073/pnas.1500272112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JR, Rizk RS, Kline SL, Walczak CE. Deciphering protein function during mitosis in PtK cells using RNAi. BMC Cell Biol. 2006;7:26. doi: 10.1186/1471-2121-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stout JR, Yount AL, Powers JA, Leblanc C, Ems-McClung SC, Walczak CE. Kif18B interacts with EB1 and controls astral microtubule length during mitosis. Mol Biol Cell. 2011;22:3070–3080. doi: 10.1091/mbc.E11-04-0363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Du Y, English CA, Maliga Z, Wagenbach M, Asbury CL, Wordeman L, Ohi R. A tethering mechanism controls the processivity and kinetochore-microtubule plus-end enrichment of the Kinesin-8 Kif18A. Mol Cell. 2011;43:764–775. doi: 10.1016/j.molcel.2011.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, von Dassow G, Wagenbach M, Asbury C, Wordeman L. The kinesin-8 motor Kif18A suppresses kinetochore movements to control mitotic chromosome alignment. Dev Cell. 2008;14:252–262. doi: 10.1016/j.devcel.2007.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpff J, Wagenbach M, Franck A, Asbury CL, Wordeman L. Kif18A and chromokinesins confine centromere movements via microtubule growth suppression and spatial control of kinetochore tension. Dev Cell. 2012;22:1017–1029. doi: 10.1016/j.devcel.2012.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Arellano-Santoyo H, Portran D, Gaillard J, Vantard M, Thery M, Pellman D. Microtubule-sliding activity of a kinesin-8 promotes spindle assembly and spindle-length control. Nat Cell Biol. 2013;15:948–957. doi: 10.1038/ncb2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Qiu W, Gupta ML, Jr, Pereira-Leal JB, Reck-Peterson SL, Pellman D. Mechanisms underlying the dual-mode regulation of microtubule dynamics by kip3/kinesin-8. Mol Cell. 2011;43:751–763. doi: 10.1016/j.molcel.2011.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, van der Vaart B, Galli M, Akhmanova A, Medema RH. A Complex of Kif18b and MCAK promotes microtubule depolymerization and is negatively regulated by Aurora kinases. Curr Biol. 2011;21:1356–1365. doi: 10.1016/j.cub.2011.07.017. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Canman JC, Salmon ED, Mitchison TJ. EB1 targets to kinetochores with attached, polymerizing microtubules. Mol Biol Cell. 2002;13:4308–4316. doi: 10.1091/mbc.E02-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tischer C, Brunner D, Dogterom M. Force- and kinesin-8-dependent effects in the spatial regulation of fission yeast microtubule dynamics. Mol Syst Biol. 2009;5:250. doi: 10.1038/msb.2009.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udy DB, Voorhies M, Chan PP, Lowe TM, Dumont S. Draft de novo transcriptome of the rat kangaroo Potorous tridactylus as a tool for cell biology. PLoS One. 2015;10:e0134738. doi: 10.1371/journal.pone.0134738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varga V, Helenius J, Tanaka K, Hyman AA, Tanaka TU, Howard J. Yeast kinesin-8 depolymerizes microtubules in a length-dependent manner. Nat Cell Biol. 2006;8:957–962. doi: 10.1038/ncb1462. [DOI] [PubMed] [Google Scholar]
- Varga V, Leduc C, Bormuth V, Diez S, Howard J. Kinesin-8 motors act cooperatively to mediate length-dependent microtubule depolymerization. Cell. 2009;138:1174–1183. doi: 10.1016/j.cell.2009.07.032. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Gayek S, Ohi R. Microtubule-depolymerizing kinesins. Annu Rev Cell Dev Biol. 2013;29:417–441. doi: 10.1146/annurev-cellbio-101512-122345. [DOI] [PubMed] [Google Scholar]
- Weaver LN, Ems-McClung SC, Stout JR, Leblanc C, Shaw SL, Gardner MK, Walczak CE. Kif18A uses a microtubule binding site in the tail for plus-end localization and spindle length regulation. Curr Biol. 2011;21:1500–1506. doi: 10.1016/j.cub.2011.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West RR, Malmstrom T, McIntosh JR. Kinesins klp5+ and klp6+ are required for normal chromosome movement in mitosis. J Cell Sci. 2002;115:931–940. doi: 10.1242/jcs.115.5.931. [DOI] [PubMed] [Google Scholar]
- West RR, Malmstrom T, Troxell CL, McIntosh JR. Two related kinesins, klp5+ and klp6+, foster microtubule disassembly and are required for meiosis in fission yeast. Mol Biol Cell. 2001;12:3919–3932. doi: 10.1091/mbc.12.12.3919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wordeman L, Decarreau J, Vicente JJ, Wagenbach M. Divergent microtubule assembly rates after short- versus long-term loss of end-modulating kinesins. Mol Biol Cell. 2016;27:1300–1309. doi: 10.1091/mbc.E15-11-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zong H, Carnes SK, Moe C, Walczak CE, Ems-McClung SC. The far C-terminus of MCAK regulates its conformation and spindle pole focusing. Mol Biol Cell. 2016;27:1451–1464. doi: 10.1091/mbc.E15-10-0699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.