Atp25 is a complex mitochondrial precursor that is cleaved three times by the matrix processing peptidase, resulting in two mature proteins. One of these, the ribosome-silencing factor Rsf, binds mitochondrial ribosomes. Cytosolic expression of Rsf alone (in yeast) is toxic and is prevented by the unconventional processing of Atp25.
Abstract
Whereas the structure and function of cytosolic ribosomes are well characterized, we only have a limited understanding of the mitochondrial translation apparatus. Using SILAC-based proteomic profiling, we identified 13 proteins that cofractionated with the mitochondrial ribosome, most of which play a role in translation or ribosomal biogenesis. One of these proteins is a homologue of the bacterial ribosome-silencing factor (Rsf). This protein is generated from the composite precursor protein Atp25 upon internal cleavage by the matrix processing peptidase MPP, and in this respect, it differs from all other characterized mitochondrial proteins of baker’s yeast. We observed that cytosolic expression of Rsf, but not of noncleaved Atp25 protein, is toxic. Our results suggest that eukaryotic cells face the challenge of avoiding negative interference from the biogenesis of their two distinct translation machineries.
INTRODUCTION
In their overall structure and function, mitochondrial ribosomes are similar to cytosolic 80S and bacterial 70S ribosomes (Greber and Ban, 2016; Ott et al., 2016). However, mitochondrial ribosomes were remodeled considerably during eukaryotic evolution and show significant differences compared with bacterial ribosomes (Smits et al., 2007; Beckmann and Herrmann, 2015). A common feature of all present-day eukaryotes is the highly reduced mitochondrial genome, which only encodes a small number of proteins, most of which are hydrophobic membrane proteins (Gray et al., 1999). Presumably, this specialization in synthesizing a very small number of hydrophobic membrane proteins explains the two major differences between mitochondrial ribosomes and those of bacteria and the eukaryotic cytosol:
Mitochondrial ribosomes of many organisms show tight physical connection with the inner membrane, which was reported from both biochemical fractionation studies (Liu and Spremulli, 2000; Jia et al., 2003; Szyrach et al., 2003) and in situ visualization by microscopy (Watson, 1972; Pfeffer et al., 2015). Membrane binding is mediated by a protein of the large subunit, called Mba1 in yeast and Mrpl45 in mammals, positioned adjacent to the polypeptide exit tunnel and directs nascent chains to the Oxa1 insertion machinery of the inner membrane (Ott et al., 2006; Jia et al., 2009; Greber et al., 2014b). The tight contact to the membrane presumably explains the substantial remodeling of the region around the exit tunnel of the mitochondrial ribosome (Sharma et al., 2003; Brown et al., 2014; Greber et al., 2014b).
The strongly degenerate rRNA core structure is another common feature of the mitochondrial ribosome (Amunts et al., 2014; Brown et al., 2014; Greber et al., 2014a). It is most obvious in animals (and some parasites), in which rRNA content of the ribosome is reduced close to what might be the minimum required for catalytic activity. Here many structural rRNA regions were lost, in some cases replaced by proteins (O’Brien, 2002). In fungi, reduction of the rRNA is less extreme, but its structure and base pairing–mediated stability strongly degenerates (van der Sluis et al., 2015).
In the past, the protein composition of high salt–treated, gradient-purified ribosomes under stringent conditions was identified by proteomics (Graack et al., 1999; Koc et al., 2001a, b; Suzuki et al., 2001). A recent study followed an interesting complementary approach to identify the proteome of mitochondrial ribosomes of Saccharomyces cerevisiae under very mild conditions (10 mM potassium acetate), using an affinity purification strategy of an epitope-tagged ribosome (Kehrein et al., 2015). A surprisingly large number of components copurified with mitochondrial ribosomes, including many factors involved in mitochondrial gene expression.
In this study, we used a third approach, relying on stable isotope labeling by amino acids in cell culture (SILAC)–based isotope labeling and comparative profiling of mitochondrial fractions containing or lacking ribosomes to reduce the number of false-positive interaction partners. We thereby identified 28 (85%) and 37 (84%) of the previously described proteins of the small and large subunit, respectively, plus 13 additional proteins, seven of which are essential for mitochondrial translation. We found that one of these additional proteins, mitochondrial ribosome-silencing factor (Rsf), was generated from a composite precursor protein of Atp25 (Zeng et al., 2008). The Atp25 precursor consisted of a matrix-targeting sequence (MTS), the Rsf domain, and a C-terminal region (M domain).
In bacteria, Rsf was shown to block protein synthesis under starvation conditions (Häuser et al., 2012), but the function of its mitochondrial counterpart is largely unknown. Three recent studies on the mammalian homologue indicate a function in biogenesis or functional maintenance of the large subunit (Rorbach et al., 2012; Wanschers et al., 2012; Fung et al., 2013). We observed that the expression of Rsf in the yeast cytosol is toxic, whereas the expression of Rsf-M domain fusion, such as present in the Atp25 precursor, was much better tolerated. Thus the unconventional organization of Atp25 as a tandem protein may serve as a strategy to prevent interference of the Rsf precursor with the cytosolic translation machinery.
RESULTS
Proteomic analysis of the mitochondrial ribosome
To isolate mitochondria devoid of assembled ribosomes, we used strains that lacked mitochondrial DNA (rho0) and hence mitochondrial rRNA. In addition, we generated a strain in which the expression of a central ribosomal protein (Mrp20, the homologue of bacterial L23) was under control of the GAL10 promoter. Mrp20 is essential for the assembly of the large ribosomal subunit (Fearon and Mason, 1992).
Mrp20 is also referred to as uL23m according to novel nomenclature suggested by Ban et al. (2014). For compatibility with protein names in published studies and the Saccharomyces genome database, we use the yeast nomenclature in this study. To avoid confusion with the nomenclature with protein names, we give a synonym list in Supplemental Table S1.
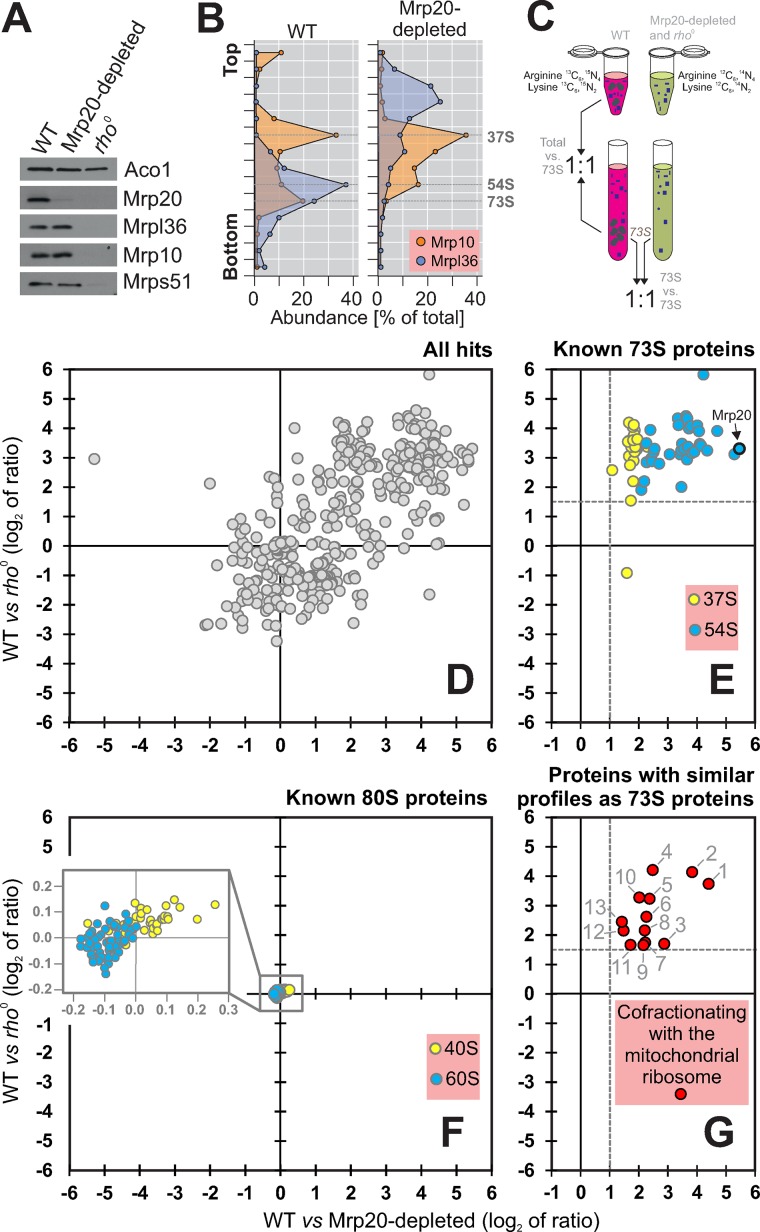
By shifting the culture for 24 h to glucose-based medium, we depleted Mrp20 (Figure 1A). Sucrose gradient centrifugation confirmed that Mrp20 depletion prevented assembly of the large (54S) but not of the small subunit (37S) of the mitochondrial ribosome (Figure 1B).
FIGURE 1:
Identification of proteins cofractionating with mitochondrial ribosomes. (A) Proteins of the large (Mrp20, Mrpl36) and small (Mrp10, Mrps51) ribosomal subunits, as well as of aconitase (Aco1), were analyzed by Western blotting of mitochondrial extracts isolated from the indicated strains. (B) Mitochondria were isolated from wild-type and Mrp20-depleted cells, lysed, and separated on a linear sucrose gradient. Proteins of 16 fractions were analyzed by Western blotting with Mrp10- and Mrpl36-specific antibodies. The signals were quantified. The graph shows their distribution in the gradient. (C) Schematic overview of the differential profiling strategy. (D–G) For the SILAC analysis, wild-type cells were grown in the presence of “heavy” (13C6, 15N4-arginine, 13C6, 15N2-lysine) medium. The rho0 cells and cells of the Mrp20-depleted mutant were grown in “light” (12C6, 14N4-arginine, 12C6, 14N2-lysine) medium. Mitochondrial extracts were separated on a linear sucrose gradient. The 73S-containing fractions were pooled and their protein profiles analyzed by mass spectrometry. (D) Plot of the log2 ratios of all 466 identified proteins obtained in these two experiments. (E, F) Proteins of the large (blue) and small (yellow) subunits of mitochondrial (E) and cytosolic (F) ribosomes. (G) Proteins that cofractionated with mitochondrial ribosomes. The proteins shown were all enriched at least threefold in the 73S fraction over the total extract of wild-type mitochondria.
SILAC is a powerful technique for quantitatively assessing differences in the protein composition of complex samples (Ong et al., 2002). We designed a SILAC-based strategy to identify proteins associated with mitochondrial ribosomes. To this end, we grew two yeast cultures in parallel, one containing and one lacking mitochondrial ribosomes, in the presence of “light” (12C6, 14N4-arginine, 12C6, 14N2-lysine) or “heavy” (13C6, 15N4-arginine, 13C6, 15N2-lysine) amino acids, respectively. Mitochondria were purified from these cultures and lysed under low-salt conditions (50 mM NH4Cl, 5 mM MgSO4) before the extract was subjected to sucrose gradient centrifugation (Figure 1C). Proteins of the total mitochondrial extracts and the 73S fractions of the gradients were mixed, digested with trypsin, and analyzed by mass spectrometry. Because each tryptic peptide contains either one lysine or one arginine residue, the ratios of each peptide could be determined accurately in the samples.
Next we compared the peptide ratios of the 73S fractions of wild type to rho0 with wild type to Mrp20-depleted cells (Figure 1D). We identified 466 proteins in both samples (Supplemental Tables S1 and S2), many of which were enriched in the wild type relative to the mutants. These proteins included ribosomal subunits but also many components of the respiratory chain whose stable accumulation in mitochondria depends on the function of the mitochondrial ribosome. However, when we removed all proteins that were not at least threefold enriched in the 73S fraction relative to the total, the remaining proteins fell into three categories (Figure 1, E–G). 1) Previously identified subunits of the mitochondrial ribosomes (Supplemental Table S2). We identified 28 (85%) and 37 (84%) of the previously described proteins of the large and small subunit, respectively. The few proteins we missed (Mrpl27, Mrpl37, Mrpl44, Mrp10, Rsm19, Mrps12, and Mrps18) were predominantly very small and positively charged, which explains why they did not provide enough unambiguous peptides. Of interest, the proteins of the small (Figure 1E, yellow) and large (Figure 1E, blue) subunit were clearly distinct in the segregation pattern due to the specific depletion of the large-subunit proteins in the Mrp20-depletion sample. 2) We identified many proteins of the 80S ribosome that were coisolated with the mitochondria. All of these proteins scattered around the intersection of the axes and hence were not influenced by the absence or presence of mitochondrial ribosomes (Figure 1F). 3) As a third group, we identified 13 additional proteins that exactly resembled in their fractionation profile the subunits of the mitochondrial ribosome (Figure 1G).
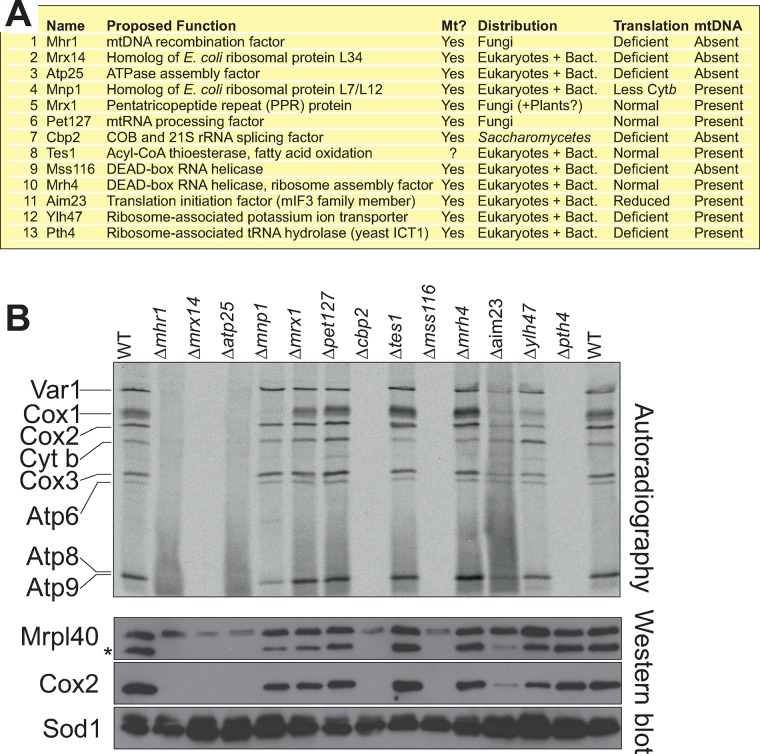
Aim23, Atp25, Cbp2, Mhr1, Mss11, Mrx14, and Pth4 are required for mitochondrial translation
With the exception of Tes1, all of the 13 proteins that cofractionated with mitochondrial ribosomes were previously shown to be present in mitochondria (Figure 2A). Tes1 is a peroxisomal acyl-CoA thioesterase involved in β-oxidation of oleate. We did not further follow up whether Tes1 is dually localized to mitochondria, but there is an increasing list of proteins found in both organelles, and the expression of TES1 mRNA is very similarly regulated to that of several mitochondrial proteins, including SDH8, OM14, OM45, PET10, and SDH4 (Hibbs et al., 2007). Most of the other proteins identified are either homologues of bacterial ribosomal proteins (Mnp1, Mrx14) or were previously linked to mitochondrial gene expression and/or assembly of the mitochondrial ribosome (Sato and Miyakawa, 2004; Frazier et al., 2006; Richter et al., 2010; De Silva et al., 2013; Kehrein et al., 2015; Kuzmenko et al., 2016). However, in several cases, a physical interaction with mitochondrial ribosomes was not detected before. We observed that seven of these mutants (Δaim23, Δatp25, Δcbp2, Δmhr1, Δmss11, Δmrx14, and Δpth4) showed no or strongly diminished mitochondrial protein synthesis, which coincided in most cases with the absence of mitochondrial DNA (Figure 2B and Supplemental Figure S1). This is explained by the fact that the presence of mitochondrial ribosomes is essential for the maintenance of the mitochondrial genome. Cells lacking the mitochondrial tRNA hydrolase Pth1 did not synthesize mitochondrial proteins despite the presence of mitochondrial DNA. In Δaim23, in which mitochondrial translation was not completely blocked, the mitochondrial DNA remained stable. The deletion of MNP1 caused a selective reduction of newly synthesized Cox1, which is indicative for mutants defective in the assembly of cytochrome c oxidase (Perez-Martinez et al., 2003; Barrientos et al., 2004).
FIGURE 2:
Mhr1, Mrx14, Atp25, Cbp2, Mss116, Aim23, and Pth4 are critical for mitochondrial translation. (A) Overview of the proteins that cofractionated with mitochondrial ribosomes. Mitochondrial localization (Mt?) was previously documented for all components except for Tes1. Distribution of potential homologues identified by BLAST searches. Bact., bacteria. The presence of mtDNA and the ability to synthesize mitochondrially encoded proteins was analyzed as described in Figure 2B and Supplemental Figure S1. (B) Cells of the indicated strains were grown in fermentative carbon source to logarithmic growth phase. Cytosolic translation was blocked with cycloheximide before mitochondrial translation products were labeled by addition of [35S]methionine for 20 min. The samples were analyzed by SDS–PAGE and autoradiography, as well by Western blotting with antibodies against the mitochondrially encoded protein Cox2 and the nuclear encoded proteins Mrpl40 and Sod1. The asterisk indicates the Cox2 signal.
The precursor of Atp25 gives rise to two mature proteins
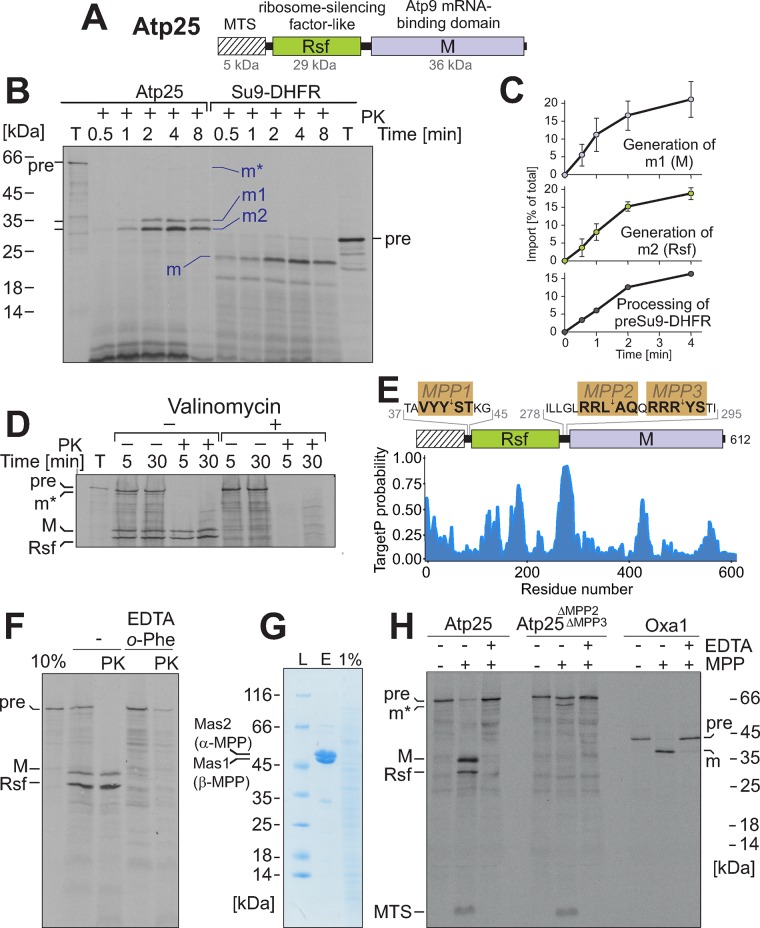
One of the proteins cofractionating with mitochondrial ribosomes was Atp25, which was identified as a factor critical for assembly of the ATPase complex (Zeng et al., 2008). The ATP25 gene codes for a protein that consists of three different parts (Figure 3A): an N-terminal MTS, a domain related to the Rsf of bacteria (Häuser et al., 2012; Li et al., 2015), and a domain previously shown to stabilize the ATP9 mRNA (Zeng et al., 2008), which we will refer to as the M domain. When we synthesized this precursor protein in vitro in the presence of [35S]methionine and incubated it with isolated mitochondria, the protein was efficiently imported and became protease resistant (Figure 3B). Of interest, the precursor protein, which had a calculated mass of 70 kDa but migrated at ∼65 kDa, gave rise to two mature fragments with apparent masses of 30 and 35 kDa (Figure 3B, m1 and m2). In addition, a very small amount of 62-kDa form was observed transiently at early time points (Figure 3B, m*). The m1 and m2 fragments were generated with comparable kinetics, suggesting that they are formed simultaneously rather than subsequently. The efficiency by which both fragments were generated was similar to that of the maturation of the well-characterized model substrate Su9-DHFR (Figure 3, B and C) and depended on the mitochondrial membrane potential (Figure 3D).
FIGURE 3:
Atp25 is a composite precursor protein. (A) Structure of the Atp25 precursor. The MTS and the Rsf and M domains are indicated. (B) Radiolabeled Atp25 and Su9-DHFR were incubated with mitochondria for the times indicated. Mitochondria were incubated with proteinase K (PK) to remove nonimported material and analyzed by SDS–PAGE and autoradiography. Ten percent of the total (T) lysate used per import lane was loaded for control. Positions of the precursor (pre) and matured (m) species are indicated. (C) The import of Atp25 was performed three times, as described. The signals were quantified. Mean values and SDs. (D) Import experiment with radiolabeled Atp25 in the presence or absence of valinomycin, which depletes the mitochondrial membrane potential. (E) Positions of the three sequences that match the consensus for the MPP processing site. The primary sequence of the protein of interest was truncated sequentially residue by residue from the N-terminus. The resulting sequences were submitted to the TargetP server (Emanuelsson et al., 2000), and the mTP value for each sequence was obtained and plotted along the residue number of the protein of interest. (F) Mitochondria were preincubated with 20 mM EDTA and 2 mM o-phenanthroline to inhibit MPP activity before radiolabeled Atp25 was imported for 20 min as described for C. (G) Recombinant hexahistidine-tagged MPP (Luciano et al., 1997) was purified (E, eluate) from an E. coli extract (1% shown for control) on Ni-NTA beads and analyzed by SDS–PAGE and Coomassie staining. (H) Radiolabeled Atp25 precursor, an Atp25 variant lacking the linker between the Rsf and M domains (i.e., the predicted MPP2 and MPP3 cleavage sites), and the well-characterized protein Oxa1 were incubated with purified MPP in the absence or presence of the MPP inhibitor EDTA. Positions of the different fragments generated.
The role of mitochondrial processing peptidase (MPP) in the proteolytic removal of MTSs is well established, but in baker’s yeast, examples in which MPP generates different mature proteins from one composite precursor are not known. In the sequence of the Atp25 precursor, we identified three potential MPP cleavage sites that resemble the previously published consensus (Vögtle et al., 2009). Two of these sites (MPP2 and MPP3) are positioned between the Rsf and the M domains in a stretch that, according to the TargetP algorithm, forms an MTS-like structure (Figure 3E). Indeed, inhibition of MPP prevented processing of Atp25 (Figure 3F). Moreover, in vitro incubation of Atp25 with purified MPP resulted in three fragments resembling the MTS, the Rsf, and the M domain (Figure 3G,H). Deletion of the MPP2 and MPP3 sequences abolished the internal cleavage by MPP. Thus we conclude that Atp25 represents a composite precursor protein from which two mature fragments are generated by internal processing by MPP.
Atp25 contains an internal MTS
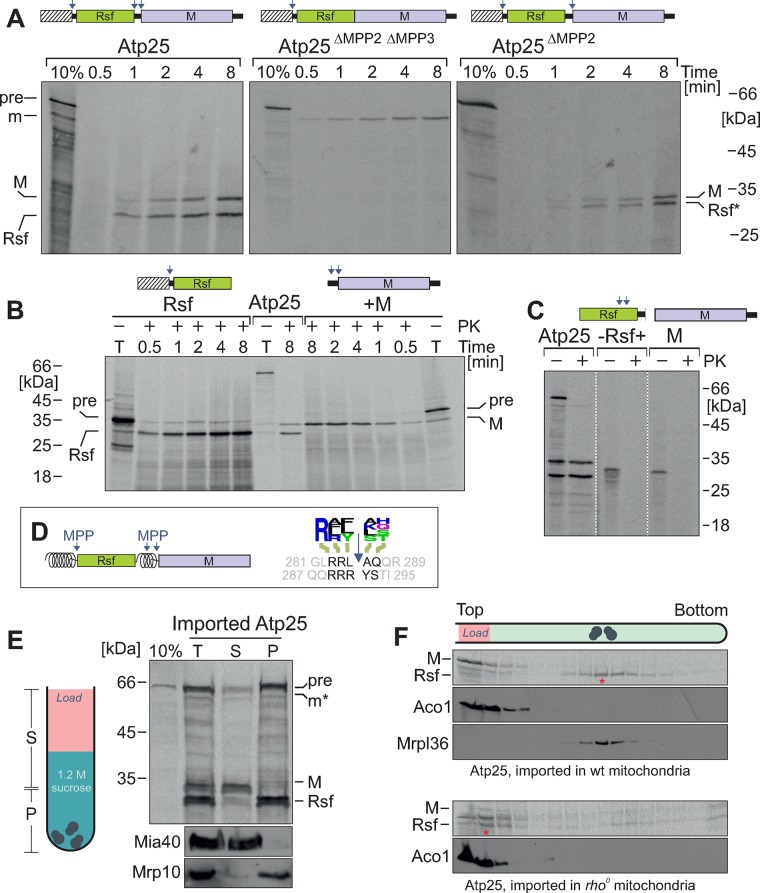
To identify the sequences that serve as targeting and processing signals in Atp25, we constructed a series of deletion variants. First, we synthesized a variant of Atp25 that lacked the region between the Rsf and M domains containing the MPP2 and MPP3 processing sites and incubated it with isolated mitochondria (Figure 3A). This version was still imported into mitochondria, giving rise to a protease-protected mature form of ∼60 kDa. MPP cleavage at MPP1 obviously matured this precursor into an Rsf-M fusion protein that was not further cleaved. If only the MPP2 site was mutagenized, the resulting Atp25ΔMPP2 protein was internally cleaved into two matured species in the mitochondria. Here the Rsf-containing fragment also included the stretch between the MPP2 and MPP3 cleavage sites (Figure 4A, Rsf*). This proved that both internal cleavage sites are used in the endogenous Atp25 precursor protein.
FIGURE 4:
Atp25 is processed by three different MPP sites. (A–C) The indicated Atp25 variants were synthesized in reticulocyte lysate in the presence of [35S]methionine and incubated with wild-type mitochondria for the times indicated. Import and processing of these radiolabeled proteins were monitored by SDS–PAGE and autoradiography. (D) Schematic representation of the MTSs in Atp25 (shown as spirals) and the two internal MPP sites that adhere to the MPP consensus sequence identified on the basis of the mitochondrial N-proteome (Vögtle et al., 2009; shown as logo plot). (E) After import of radiolabeled Atp25 into wild-type mitochondria for 20 min, mitochondria were reisolated, washed, and lysed. From the total extract (T), ribosomes (P) were separated from other proteins (S) by centrifugation through a 1.2 M sucrose cushion. Western blot signals for nonribosomal protein Mia40 and ribosomal protein Mrp10 are shown for control. (F) Cofractionation experiment as described for E; however, here a linear sucrose gradient (10–34% sucrose) was used. Bottom, Atp25 was imported in rho0 mitochondria, which lack rRNA and, hence, assembled ribosomes.
When we generated truncated variants that represented a version in which Atp25 was split at the MPP2 site into two halves, both species were efficiently imported into mitochondria (Figure 4B). Thus the stretch between the Rsf and M domain obviously represents a functional MTS that can on its own efficiently drive the translocation of the M domain into mitochondria. In the absence of any matrix-targeting sequences, the Rsf and the M domain were not imported (Figure 4C).
From this, we conclude that the Atp25 precursor is a composite preprotein that represents two fully functional mitochondrial precursors fused in tandem (Figure 4D). During or directly after the import reaction, the internal MTS is spliced out by MPP cleavage at the MPP2 and MPP3 sites.
Rsf binds to mitochondrial ribosomes
We next asked whether both Atp25 domains bind to ribosomes. For this, we imported radiolabeled Atp25 precursor into mitochondria, lysed the mitochondria, and isolated a ribosome-containing pellet by centrifugation through a sucrose cushion (Figure 4E). Whereas the Rsf domain was exclusively found in the ribosome pellet, the M domain remained in the supernatant. The cofractionation with ribosomes was even more obvious when, subsequent to the import reaction, the mitochondrial extract was loaded onto a continuous sucrose gradient (Figure 4F). Here all of the Rsf domain cofractionated with mitochondrial ribosomes (Figure 4F, asterisk) whereas all of the M domain remained at the top of the gradient. When Atp25 was imported into rho0 mitochondria, which lack ribosomes, the Rsf domain also remained at the top, confirming that the Rsf domain is pulled into the sucrose gradient due to its affinity to ribosomes (Figure 5B, bottom).
FIGURE 5:
Expression of Rsf, but not of Rsf-M, in the cytosol reduces cellular fitness. (A) Rsf-HA (corresponding to residues 1–278 of Atp25) was expressed in yeast cells. A cell extract was loaded onto a linear 10–34% sucrose gradient. The gradient was harvested by pumping it through a photometer recording the distribution of ribosomes at 260 nm. Proteins from 14 fractions were precipitated and analyzed by Western blotting. (B) Tenfold serial dilutions were dropped either on glucose (noninducing) or galactose (inducing) plates. The plates were incubated at 35°C. Expression of Rsf but not of Rsf-M interfered with cell growth. (C) Growth curves of the same strains as in B in glucose- and galactose-containing media. Expression of Rsf retarded cell growth (arrowhead). Mean values of four replicates. SDs are plotted as gray bars. (D) Extracts from the stains used in B were loaded onto linear 10–34% sucrose gradients. The distribution of ribosomes was analyzed spectroscopically. Relative area volumes of the monosome peaks. (E, F) Yeast cells expressing Rsf-M, Rsf, or M in the cytosol were grown to log phase, and [35S]methionine was added for the times indicated. Cells were harvested and lysed. Proteins were resolved either by SDS–PAGE or dot blot analysis and visualized by autoradiography. (G) A C-terminally hexahistidine-tagged version of Rsf (residues 43–292) was expressed in E. coli, purified from inclusion bodies, refolded, and added to reticulocyte lysate (Promega) for an in vitro translation reaction. Bovine serum albumin (BSA) was used for control. Addition of Rsf prevented protein synthesis of this cytosolic translation system. (H) Model for the import and processing of Atp25. In mitochondria, Rsf associates with the mitochondrial ribosome in the matrix. Our observations suggest that Rsf can interfere with the cytosolic translation machinery, which is prevented by its synthesis as a composite Atp25 precursor protein.
Is the tandem organization of the Atp25 precursor relevant for the ribosome binding of Rsf? To address this question, we expressed a variant that represented only the first half of the protein (comprising the MTS and Rsf). When cellular extracts were analyzed on a sucrose gradient, the mature Rsf domain produced from this version comigrated with ribosomes (Figure 5A), showing that the tandem organization is not critical for binding of Rsf to the mitochondrial ribosome. On expression of this MTS-Rsf variant, but not of the normal Atp25 protein, we observed a considerable accumulation of nonimported precursor, which might suggest that this form is trapped in the cytosol (Figure 5A, pre). This precursor species sedimented deep in the sucrose gradient and comigrated with cytosolic polysomes, suggesting that the cytosolic Rsf associates with 80S ribosomes or other large cytosolic protein complexes.
We therefore tested whether cytosolic expression of the Rsf domain had an effect on cell growth (Figure 5, B and C). Whereas cytosolic expression of Rsf strongly reduced cell growth on plates as well as in liquid cultures, expression of the Rsf-M tandem variant was much better tolerated. This growth inhibition was observed only upon overexpression of Rsf, but it points to a toxic effect of the protein that is not observed when Rsf is part of the Rsf-M fusion. To test whether the Rsf domain has the potential to interfere with the cytosolic translation system, we analyzed the number of 80S ribosomes in extracts of these cells (Figure 5D). Expression of Rsf in the cytosol strongly reduced the number of monosomes and cytosols observed on sucrose gradients, whereas cytosolic expression of the Rsf-M fusion had a much smaller effect. This was consistent with the efficiency of cytosolic protein synthesis in these cells, which was analyzed by incubating cells with [35S]methionine: the expression of Rsf considerably inhibited cytosolic translation (Figure 5, E and F), which explains the strong growth defect of this strain. To test whether the strong effect of Rsf on translation in directly or indirectly caused, we purified the Rsf domain after bacterial expression in inclusion bodies, refolded the protein, and added it to an in vitro translation system (Figure 5G). The addition of Rsf, but not that of bovine serum albumin, strongly inhibited the translation reaction, suggesting that Rsf interferes not only with bacterial but also with eukaryotic translation. In summary, we observe that the mitochondrial Rsf, when present in the cytosol, is highly toxic because it interferes with 80S ribosomes and blocks protein synthesis. The Rsf-M fusion protein was much better tolerated, suggesting that the complex structure of the precursor makes it much better tolerated in the cytosol. Thus the organization of Atp25 as a composite precursor might be used by eukaryotic cells to avoid the interference of the Rsf domain with the cytosolic translation system (Figure 5H).
DISCUSSION
In this study, we present a differential proteomics approach to identify components that associated with ribosomes in a highly specific manner but with lower affinity. Most of these components represented RNA-processing factors and helicases, translation enzymes, or homologues of constituents of the bacterial ribosome. Several of these proteins were also identified in a recent, less-stringent proteomic study that proposed that at least 196 proteins are loosely associated with the mitochondrial ribosome in a large structure referred to as MIOREX complex (Kehrein et al., 2015). The number of interactors identified in our study is much smaller, despite similarly mild lysis conditions (Supplemental Tables S1 and S2). This is most likely explained in part by the fact that we filtered out components that were not enriched in the 73S fraction to reduce the number of false-positive hits. However, this might also have removed those ribosomal interactors that have a considerable nonribosomal fraction. One of the factors we identified, Aim23, is a homologue of the bacterial initiation factor IF3, which was shown very recently to be ribosome associated and relevant, although not essential, for mitochondrial protein synthesis (Kuzmenko et al., 2016), which is consistent with our findings. The differential proteomic profiling that we used here will serve as a very good starting point to address the dynamic association of factors with ribosomes during their assembly process.
One of the factors that we found to be associated with mitochondrial ribosomes was the Rsf part of the Atp25 preprotein. Because we detected only the Rsf domain and not the M domain with isolated ribosomes, we realized that the Atp25 precursor gives rise to two distinct mature proteins. This composite organization was already suggested in the initial study on Atp25 based on the small mass that was observed with an epitope-tagged Atp25 version (Zeng et al., 2008). We identified MPP as the processing protease for the internal Atp25 cleavages. To our knowledge, this is the first example of a mitochondrial protein in baker’s yeast with such a tandem organization, but similar mitochondrial tandem precursors were described in Neurospora crassa, Schizosaccharomyces pombe, and plants (Gessert et al., 1994; Oshima et al., 2005; Abdelnoor et al., 2006; Khalimonchuk et al., 2006). Intriguingly, most of these tandem precursors give rise to proteins of the mitochondrial ribosome.
We identified MPP as the processing peptidase that separates Rsf and the M domain. The consensus sequence for MPP cleavage was comprehensively analyzed by a proteome-wide analysis (Vögtle et al., 2009). The internal MPP cleavage sites that we identified in Atp25 matched this consensus [R][AFR][FLY]↓[AKLS][HSTQ], a sequence that can be found also internally in other mitochondrial proteins, such as Nam2 (after residue 742), Mrp7 (after residue 367), and Nca2 (after residue 277). Because many MPP substrates deviate from this consensus (Vögtle et al., 2009), there might be even more tandem precursors encoded in the yeast genome.
It is not clear why proteins form tandem precursors. This may just be by chance, for example, as the result of a genome rearrangement that caused the in-frame fusion of two sequences coding for mitochondrial precursors. In the case of Atp25, this possibility appears to be less likely, given the strong conservation of the tandem organization among organisms that harbor the subunit 9 of the ATPase on the mitochondrial genome and therefore express an M domain (Supplemental Figure S2). Gene fusions are frequently observed in genomes, and those that are fixed during evolution often link two proteins that cooperate functionally or need to be expressed at equal levels (Yanai et al., 2002; Leonard and Richards, 2012). This is not obvious for most of the mitochondrial composite precursors identified so far, which often fuse proteins of very different function. However, in many cases, one partner is a protein of the mitochondrial ribosome. Our results presented here suggest that these ribosomal proteins might negatively interfere with the cytosolic translation machinery in some way. Their insertion into a larger composite precursor might reduce the risk of their misincorporation into the 80S ribosome. The assembly of two distinct ribosomes might be a critical challenge for eukaryotic cells that has not been studied so far. This might have considerable consequences for the evolution of eukaryotic cells and might even explain why the assembly of the 80S ribosome was relocated from the cytosol into the nucleus or why many mitochondrial genomes still contain genes for ribosomal subunits. It will be interesting in the future to analyze in more detail the strategies by which eukaryotic cells ensure the immaculate assembly of their two ribosomal particles.
MATERIALS AND METHODS
Cell culture, strains, and preparation of mitochondria
For SILAC experiments, the strain YPH499 ARG4::URA3 with or without mitochondrial genome (rho0) was used (Sikorski and Hieter, 1989). To regulate Mrp20 levels, a GAL promoter was integrated in front of the chromosomal MRP20 gene. See the Supplemental Materials for details on the construction of Atp25 variants and mutants. Cells were grown in minimal medium supplemented with 2% glucose or galactose at 30°C (Peleh et al., 2016). Isolation of mitochondria, in vitro import experiments with radiolabeled proteins, detection of mitochondrial protein synthesis, SILAC labeling, and mass spectrometry were performed as described (Bode et al., 2015; Peleh et al., 2016).
Construction of Atp25 variants
The Atp25-coding region or a fragment of it was amplified by PCR and cloned into pGEM4 (Promega, Madison, WI) using the EcoRI and HindIII restriction sites. For construction of the Atp25ΔMPP2 ΔMPP3 version, the sequence encoding for amino acid residues 279–293 were deleted and replaced by the restriction sites of Xba1 and SalI. These sites were used to insert a linker to restore the MPP3 deletion site, giving rise to the Atp25ΔMPP2 variant. For expression of Rsf-HA in yeast cells, the sequence corresponding to residues 1–278 was cloned into the expression plasmid pYX143 under control of a GAL10 promoter fused to the sequence of a hemagglutinin (HA) tag. For expression of Rsf and Rsf-M, pYX223 vectors were used; for these constructs, sequences were used corresponding to residues 43–292 and 43–612 of Atp25, respectively. The protein variants used consisted of the following residues of Atp25: Atp25 (1–612); Atp25ΔMPP2 ΔMPP3 (1–278,VDGSR, 294–612); Atp25ΔMPP2 (1–278, VDNTPQQNKTQQRRRYSTSR, 294–612); Rsf (1–278); +M (M, 272–612); -Rsf+ (M, 43–292); and M (M, 312–612).
Growth curves
The growth curves were obtained by an ELx808 absorbance microplate reader (BioTek, Winooski, VT). Precultures of 100 μl were inoculated at an OD of 0.1 in microtiter plates and sealed with an air-permeable membrane (Breathe-Easy; Sigma-Aldrich, St. Louis, MO). Growth was determined at maximum shaking for 35 h at 35°C.
Sucrose gradient analysis of mitochondrial ribosomal proteins
Isolated mitochondria (3 mg) were incubated with translation buffer for 10 min at 30°C. Translation was stopped by addition of 10 μg/ml chloramphenicol. Mitochondria were pelleted and resuspended in 250 μl of 1% Triton X-100, 50 mM NH4Cl, 5 mM MgSO4, 1× complete protease inhibitor (Roche, Basel, Switzerland), and 20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4. After a clarifying spin for 10 min at 25,000 × g, 4°C, the lysate was loaded onto a linear sucrose gradient (12 ml; 10–34% sucrose, 0.1% Triton X-100, 50 mM NH4Cl, 5 mM MgSO4, 1× complete protease inhibitor (Roche), 20 mM HEPES, pH 7.4) and centrifuged in an SW41 rotor (Beckman, Brea, CA) at 33,000 rpm for 5.5 h at 4°C. The gradient was fractionated, and the containing proteins were precipitated with trichloroacetic acid. For SILAC analysis fractions 11 of 16 of the different samples were mixed 1:1 before proteins were precipitated.
MPP in vitro cleavage assay
The in vitro cleavage reactions were performed in 150 mM NaCl, 10% glycerol, 100 μM MnCl2, and 50 mM Tris, pH 7.4, in the presence of 250 μg of purified MPP and 5% reticulocyte lysate containing the substrate. If not otherwise stated, the incubation time was 1 h at 30°C. To inhibit MPP, the same reaction was performed in the presence of 2.5 mM EDTA.
Purification and refolding of Rsf
Escherichia coli cells harboring the pET19b-MTS-RsfHis7 plasmid were grown to an OD600 of 0.8 at 37°C and 1 mM isopropyl-β-d-thiogalactoside (IPTG) was added. After incubation for 16 h, 50 ml of the culture was harvested. The cells were resuspended in phosphate-buffered saline analyzed in three freeze–thaw cycles and 15 cycles of sonification for 1 s at 60% duty level, and then the lysate was cleared at 16,000 × g for 15 min at 4°C. The pellet was resuspended in denaturing buffer (2 M urea, 500 mM NaCl, 2% Triton X-100, 20 mM Tris-HCl, pH 8), sonified 15 times, and centrifuged (16,000 × g for 10 min at 20°C). The obtained pellet was resuspended in resolving buffer (6 M guanidine hydrochloride, 500 mM NaCl, 1 mM β-mercaptoethanol, 20 mM Tris-HCl, pH 8) and tumbled for 1 h at 25°C. After a clarifying spin (10 min, 25,000 × g, 25°C), the supernatant was passed through a 0.45-μm filter and loaded on a 1-ml Ni–nitriloacetic acid (NTA) column (Qiagen, Venlo, Netherlands) equilibrated with resolving buffer. The column was washed with five column volumes of resolving buffer followed by seven column volumes of refolding buffer A (6 M urea, 500 mM NaCl, 1 mM β-mercaptoethanol, 20 mM imidazole, 20 mM Tris-HCl, pH 8). The bound protein was then refolded on the column using five column volumes of refolding buffer B (like refolding buffer A without urea). Subsequently the protein was eluted in increasing amounts of imidazole in the elution buffer (500 mM NaCl, 1 mM β-mercaptoethanol, 20 mM Tris-HCl, pH 8) starting from 20 mM imidazole to final 500 mM imidazole. The purified protein was soluble in the aqueous solution, and no denatured protein was detected by dynamic light scattering.
MPP purification
The E. coli strain expressing histidine-tagged MPP plasmid was a kindly provided by Vincent Géli. The cells were grown at 37°C to an OD600 of 0.7 and induced with 1 mM IPTG for 4 h at 37°C. The bacteria were harvested and resuspended in buffer A (150 mM NaCl, 10 mM imidazole, 2 mM β-mercaptoethanol, 0.2% Nonidet NP-40, 50 mM Tris-HCl, pH 7.4) and then sonified 15 times for 1 s at 60% duty level. The cleared lysate was loaded on buffer A–equilibrated Ni-NTA Sepharose resin (Aminitra; Expedeon, San Diego, CA) and washed with buffer A without detergent. A second wash was performed with buffer A adjusted to 1 M NaCl. The enzyme was eluted in buffer C (150 mM NaCl, 300 mM imidazole, 10% glycerol, 50 mM Tris-HCl, pH 7.4).
Supplementary Material
Acknowledgments
We thank Sabine Knaus, Julia Malaika Maier, Eli van der Sluis, and Chantal Lang for help with some experiments, Doron Rapaport, Vincent Géli, and Tom Fox for reagents, and Bruce Morgan, Michael Wollin, and Nicolas Rouhier for critical comments on the manuscript. This work was funded by grants from the Deutsche Forschungsgemeinschaft (He2803/9-1) and the Landesforschungsinitiative Rheinland-Pfalz BioComp.
Abbreviations used:
- MPP
mitochondrial processing peptidase
- MTS
matrix-targeting sequence
- PK
proteinase K
- Rsf
ribosome-silencing factor
- SILAC
stable isotope labeling by amino acids in cell culture.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-07-0513) on August 31, 2016.
REFERENCES
- Abdelnoor RV, Christensen AC, Mohammed S, Munoz-Castillo B, Moriyama H, Mackenzie SA. Mitochondrial genome dynamics in plants and animals: convergent gene fusions of a MutS homologue. J Mol Evol. 2006;63:165–173. doi: 10.1007/s00239-005-0226-9. [DOI] [PubMed] [Google Scholar]
- Amunts A, Brown A, Bai XC, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the yeast mitochondrial large ribosomal subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban N, Beckmann R, Cate JH, Dinman JD, Dragon F, Ellis SR, Lafontaine DL, Lindahl L, Liljas A, Lipton JM, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos A, Zambrano A, Tzagoloff A. Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 2004;23:3472–3482. doi: 10.1038/sj.emboj.7600358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann R, Herrmann JM. Structural biology. Mitoribosome oddities. Science. 2015;348:288–289. doi: 10.1126/science.aab1054. [DOI] [PubMed] [Google Scholar]
- Bode M, Woellhaf MW, Bohnert M, van der Laan M, Sommer F, Jung M, Zimmermann R, Schroda M, Herrmann JM. Redox-regulated dynamic interplay between Cox19 and the copper-binding protein Cox11 in the intermembrane space of mitochondria facilitates biogenesis of cytochrome c oxidase. Mol Biol Cell. 2015;26:2385–2401. doi: 10.1091/mbc.E14-11-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown A, Amunts A, Bai XC, Sugimoto Y, Edwards PC, Murshudov G, Scheres SH, Ramakrishnan V. Structure of the large ribosomal subunit from human mitochondria. Science. 2014;346:718–722. doi: 10.1126/science.1258026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Silva D, Fontanesi F, Barrientos A. The DEAD box protein Mrh4 functions in the assembly of the mitochondrial large ribosomal subunit. Cell Metab. 2013;18:712–725. doi: 10.1016/j.cmet.2013.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G. Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol. 2000;300:1005–1016. doi: 10.1006/jmbi.2000.3903. [DOI] [PubMed] [Google Scholar]
- Fearon K, Mason TL. Structure and function of MRP20 and MRP49, the nuclear genes for two proteins of the 54 S subunit of the yeast mitochondrial ribosome. J Biol Chem. 1992;267:5162–5170. [PubMed] [Google Scholar]
- Frazier AE, Taylor RD, Mick DU, Warscheid B, Stoepel N, Meyer HE, Ryan MT, Guiard B, Rehling P. Mdm38 interacts with ribosomes and is a component of the mitochondrial protein export machinery. J Cell Biol. 2006;172:553–564. doi: 10.1083/jcb.200505060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung S, Nishimura T, Sasarman F, Shoubridge EA. The conserved interaction of C7orf30 with MRPL14 promotes biogenesis of the mitochondrial large ribosomal subunit and mitochondrial translation. Mol Biol Cell. 2013;24:184–193. doi: 10.1091/mbc.E12-09-0651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert SF, Kim JH, Nargang FE, Weiss RL. A polyprotein precursor of two mitochondrial enzymes in Neurospora crassa. Gene structure and precursor processing. J Biol Chem. 1994;269:8189–8203. [PubMed] [Google Scholar]
- Graack HR, Bryant ML, O’Brien TW. Identification of mammalian mitochondrial ribosomal proteins (MRPs) by N-terminal sequencing of purified bovine MRPs and comparison to data bank sequences: the large subribosomal particle. Biochemistry. 1999;38:16569–16577. doi: 10.1021/bi991543s. [DOI] [PubMed] [Google Scholar]
- Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Ban N. Structure and function of the mitochondrial ribosome. Annu Rev Biochem 85, 103–132. 2016 doi: 10.1146/annurev-biochem-060815-014343. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014a;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- Greber BJ, Boehringer D, Leitner A, Bieri P, Voigts-Hoffmann F, Erzberger JP, Leibundgut M, Aebersold R, Ban N. Architecture of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014b;505:515–519. doi: 10.1038/nature12890. [DOI] [PubMed] [Google Scholar]
- Häuser R, Pech M, Kijek J, Yamamoto H, Titz B, Naeve F, Tovchigrechko A, Yamamoto K, Szaflarski W, Takeuchi N, et al. RsfA (YbeB) proteins are conserved ribosomal silencing factors. PLoS Genet. 2012;8:e1002815. doi: 10.1371/journal.pgen.1002815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibbs MA, Hess DC, Myers CL, Huttenhower C, Li K, Troyanskaya OG. Exploring the functional landscape of gene expression: directed search of large microarray compendia. Bioinformatics. 2007;23:2692–2699. doi: 10.1093/bioinformatics/btm403. [DOI] [PubMed] [Google Scholar]
- Jia L, Dienhart M, Schramp M, McCauley M, Hell K, Stuart RA. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal hydrophilic region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia L, Kaur J, Stuart RA. Mapping of the Saccharomyces cerevisiae Oxa1-mitochondrial ribosome interface and identification of MrpL40, a ribosomal protein in close proximity to Oxa1 and critical for oxidative phosphorylation complex assembly. Eukaryot Cell. 2009;8:1792–1802. doi: 10.1128/EC.00219-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrein K, Schilling R, Moller-Hergt BV, Wurm CA, Jakobs S, Lamkemeyer T, Langer T, Ott M. Organization of mitochondrial gene expression in two distinct ribosome-containing assemblies. Cell Rep. 2015;10:843–853. doi: 10.1016/j.celrep.2015.01.012. [DOI] [PubMed] [Google Scholar]
- Khalimonchuk O, Ott M, Funes S, Ostermann K, Rodel G, Herrmann JM. Sequential processing of a mitochondrial tandem protein: insights into protein import in Schizosaccharomyces pombe. Eukaryot Cell. 2006;5:997–1006. doi: 10.1128/EC.00092-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moseley A, Spremulli LL. The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem. 2001a;276:19363–19374. doi: 10.1074/jbc.M100727200. [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL. The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem. 2001b;276:43958–43969. doi: 10.1074/jbc.M106510200. [DOI] [PubMed] [Google Scholar]
- Kuzmenko A, Derbikova K, Salvatori R, Tankov S, Atkinson GC, Tenson T, Ott M, Kamenski P, Hauryliuk V. Aim-less translation: loss of Saccharomyces cerevisiae mitochondrial translation initiation factor mIF3/Aim23 leads to unbalanced protein synthesis. Sci Rep. 2016;6:18749. doi: 10.1038/srep18749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard G, Richards TA. Genome-scale comparative analysis of gene fusions, gene fissions, and the fungal tree of life. Proc Natl Acad Sci USA. 2012;109:21402–21407. doi: 10.1073/pnas.1210909110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Sun Q, Jiang C, Yang K, Hung LW, Zhang J, Sacchettini JC. Structure of ribosomal silencing factor bound to mycobacterium tuberculosis ribosome. Structure. 2015;23:1858–1865. doi: 10.1016/j.str.2015.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M, Spremulli L. Interaction of mammalian mitochondrial ribosomes with the inner membrane. J Biol Chem. 2000;275:29400–29406. doi: 10.1074/jbc.M002173200. [DOI] [PubMed] [Google Scholar]
- Luciano P, Geoffroy S, Brandt A, Hernandez JF, Geli V. Functional cooperation of the mitochondrial processing peptidase subunits. J Mol Biol. 1997;19:213–225. doi: 10.1006/jmbi.1997.1231. [DOI] [PubMed] [Google Scholar]
- O’Brien TW. Evolution of a protein-rich mitochondrial ribosome: implications for human genetic disease. Gene. 2002;286:73–79. doi: 10.1016/s0378-1119(01)00808-3. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Oshima T, Yamasaki E, Ogishima T, Kadowaki K, Ito A, Kitada S. Recognition and processing of a nuclear-encoded polyprotein precursor by mitochondrial processing peptidase. Biochem J. 2005;385:755–761. doi: 10.1042/BJ20041396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M, Amunts A, Brown A. Organization and regulation of mitochondrial protein synthesis. Annu Rev Biochem. 2016;85:77–101. doi: 10.1146/annurev-biochem-060815-014334. [DOI] [PubMed] [Google Scholar]
- Ott M, Prestele M, Bauerschmitt H, Funes S, Bonnefoy N, Herrmann JM. Mba1, a membrane-associated ribosome receptor in mitochondria. EMBO J. 2006;25:1603–1610. doi: 10.1038/sj.emboj.7601070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleh V, Cordat E, Herrmann JM. Mia40 is a trans-site receptor that drives protein import into the mitochondrial intermembrane space by hydrophobic substrate binding. Elife. 2016;5:e16177. doi: 10.7554/eLife.16177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Martinez X, Broadley SA, Fox TD. Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 2003;22:5951–5961. doi: 10.1093/emboj/cdg566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer S, Woellhaf MW, Herrmann JM, Forster F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat Commun. 2015;6:6019. doi: 10.1038/ncomms7019. [DOI] [PubMed] [Google Scholar]
- Richter R, Rorbach J, Pajak A, Smith PM, Wessels HJ, Huynen MA, Smeitink JA, Lightowlers RN, Chrzanowska-Lightowlers ZM. A functional peptidyl-tRNA hydrolase, ICT1, has been recruited into the human mitochondrial ribosome. EMBO J. 2010;29:1116–1125. doi: 10.1038/emboj.2010.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rorbach J, Gammage PA, Minczuk M. C7orf30 is necessary for biogenesis of the large subunit of the mitochondrial ribosome. Nucleic Acids Res. 2012;40:4097–4109. doi: 10.1093/nar/gkr1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato H, Miyakawa I. A 22 kDa protein specific for yeast mitochondrial nucleoids is an unidentified putative ribosomal protein encoded in open reading frame YGL068W. Protoplasma. 2004;223:175–182. doi: 10.1007/s00709-004-0040-z. [DOI] [PubMed] [Google Scholar]
- Sharma MR, Koc EC, Datta PP, Booth TM, Spremulli LL, Agrawal RK. Structure of the mammalian mitochondrial ribosome reveals an expanded functional role for its component proteins. Cell. 2003;115:97–108. doi: 10.1016/s0092-8674(03)00762-1. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smits P, Smeitink JA, van den Heuvel LP, Huynen MA, Ettema TJ. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Terasaki M, Takemoto-Hori C, Hanada T, Ueda T, Wada A, Watanabe K. Proteomic analysis of the mammalian mitochondrial ribosome. Identification of protein components in the 28 S small subunit. J Biol Chem. 2001;276:33181–33195. doi: 10.1074/jbc.M103236200. [DOI] [PubMed] [Google Scholar]
- Szyrach G, Ott M, Bonnefoy N, Neupert W, Herrmann JM. Ribosome binding to the Oxa1 complex facilitates cotranslational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, Frauenfeld J, Berninghausen O, Neupert W, Herrmann JM, Beckmann R. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol. 2015;7:1235–1251. doi: 10.1093/gbe/evv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vögtle FN, Wortelkamp S, Zahedi RP, Becker D, Leidhold C, Gevaert K, Kellermann J, Voos W, Sickmann A, Pfanner N, Meisinger C. Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell. 2009;139:428–439. doi: 10.1016/j.cell.2009.07.045. [DOI] [PubMed] [Google Scholar]
- Wanschers BF, Szklarczyk R, Pajak A, van den Brand MA, Gloerich J, Rodenburg RJ, Lightowlers RN, Nijtmans LG, Huynen MA. C7orf30 specifically associates with the large subunit of the mitochondrial ribosome and is involved in translation. Nucleic Acids Res. 2012;40:4040–4051. doi: 10.1093/nar/gkr1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson K. The organization of ribosomal granules within mitochondrial structures of aerobic and anaerobic cells of Saccharomyces cerevisae. J Cell Biol. 1972;55:721–726. doi: 10.1083/jcb.55.3.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanai I, Wolf YI, Koonin EV. Evolution of gene fusions: horizontal transfer versus independent events. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-5-research0024. research0024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng X, Barros MH, Shulman T, Tzagoloff A. ATP25, a new nuclear gene of Saccharomyces cerevisiae required for expression and assembly of the Atp9p subunit of mitochondrial ATPase. Mol Biol Cell. 2008;19:1366–1377. doi: 10.1091/mbc.E07-08-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.