C9orf72 interacts strongly with SMCR8 and depends on this interaction for its stability. Lysosomes are major sites of C9orf72 subcellular localization, and abnormal lysosome morphology is seen in its absence. Defects are found in the regulation of the lysosome-localized mTORC1 signaling pathway in C9orf72 KO cells.
Abstract
Hexanucleotide expansion in an intron of the C9orf72 gene causes amyotrophic lateral sclerosis and frontotemporal dementia. However, beyond bioinformatics predictions that suggested structural similarity to folliculin, the Birt-Hogg-Dubé syndrome tumor suppressor, little is known about the normal functions of the C9orf72 protein. To address this problem, we used genome-editing strategies to investigate C9orf72 interactions, subcellular localization, and knockout (KO) phenotypes. We found that C9orf72 robustly interacts with SMCR8 (a protein of previously unknown function). We also observed that C9orf72 localizes to lysosomes and that such localization is negatively regulated by amino acid availability. Analysis of C9orf72 KO, SMCR8 KO, and double-KO cell lines revealed phenotypes that are consistent with a function for C9orf72 at lysosomes. These include abnormally swollen lysosomes in the absence of C9orf72 and impaired responses of mTORC1 signaling to changes in amino acid availability (a lysosome-dependent process) after depletion of either C9orf72 or SMCR8. Collectively these results identify strong physical and functional interactions between C9orf72 and SMCR8 and support a lysosomal site of action for this protein complex.
INTRODUCTION
The C9orf72 gene has attracted widespread attention due the contribution of an expanded hexanucleotide repeat within an intronic region as a major risk factor for both frontotemporal dementia (FTD) and amyotrophic lateral sclerosis (ALS; DeJesus-Hernandez et al., 2011; Renton et al., 2011; Freibaum et al., 2015; O’Rourke et al., 2015; Peters et al., 2015; Zhang et al., 2015). Research into potential disease-causing mechanisms has yielded evidence that toxic effects of the hexanucleotide expansions can arise from both the repeat-containing RNAs (Haeusler et al., 2014; Freibaum et al., 2015; Zhang et al., 2015) and the dipeptide repeats that are translated from them by non-ATG mechanisms (Kwon et al., 2014; Mizielinska et al., 2014). However, the possible disease contributions of the C9orf72 protein insufficiency reported in conjunction with these expanded hexanucleotide repeats (Waite et al., 2014) have been difficult to define due to the lack of knowledge concerning the normal functions of C9orf72.
Although little is known about the function of the C9orf72 protein, bioinformatic studies have predicted that the major structural feature of this protein is a DENN domain (Zhang et al., 2012; Levine et al., 2013). DENN-domain proteins are best characterized as regulators of intracellular membrane traffic through their actions as guanine nucleotide exchange factors (GEFs) for specific Rab GTPases (Yoshimura et al., 2010; Marat et al., 2011; Wu et al., 2011). This raises the possibility of important membrane-trafficking functions for C9orf72, and evidence has been presented that implicates C9ORF72 in the regulation of endosomal and autophagic membrane traffic (Farg et al., 2014; Sellier et al., 2016). However, folliculin (FLCN) and the FLCN-interacting proteins (FNIPs), the DENN-domain proteins with the closest predicted structural similarity to C9orf72 (Levine et al., 2013; Zhang et al., 2015), instead form a complex that interacts with the lysosome-localized Rag GTPases and is critical for regulating their responsiveness to changes in amino acid availability (Petit et al., 2013; Tsun et al., 2013). More specifically, the FLCN-FNIP complex has GTPase-activating protein (GAP) activity toward the RagC/D GTPases (Tsun et al., 2013). The major function for the Rag GTPases is not related to membrane traffic. Instead, their best-understood function is to recruit mechanistic target of rapamycin complex 1 (mTORC1) to the surface of lysosomes when intracellular amino acids are abundant. This recruitment of mTORC1 to lysosomes is critical for its activation by growth factor signals that act through the Rheb GTPase (Bar-Peled and Sabatini, 2014; Ferguson, 2015). Thus, whereas the predicted presence of a DENN domain in C9orf72 suggests possible roles in membrane traffic and/or nutrient sensing, more detailed investigation of the C9orf72 protein is required to determine a specific site of action and function.
To address this problem, we implemented a combination of genetic and cell biological approaches to identify cellular functions of C9orf72. Through these efforts, we discovered that C9orf72 interacts strongly and selectively with Smith–Magenis Chromosome Region gene 8 (SMCR8), a previously uncharacterized protein that is also predicted to contain an FLCN-like DENN domain (Zhang et al., 2012; Levine et al., 2013). This interaction with SMCR8 is critical for C9orf72 stability. With respect to subcellular localization, we observed that C9orf72 is recruited to the surface of lysosomes in a manner that is negatively regulated by amino acid availability. Analysis of C9orf72 and SMCR8 knockout (KO) cells revealed roles for C9orf72 and SMCR8 in the regulation of lysosome morphology and cellular response to amino acid availability as measured by changes in mTORC1 signaling. These findings identify the lysosome as an important subcellular site of C9orf72 protein function and provide a foundation for the future investigation of possible C9orf72 protein contributions to both neurodegenerative disease and nutrient-dependent signaling mediated by mTORC1. Given the major role played by mTORC1 signaling in the control of cell growth and the dysregulation of such signaling in many cancers (Zoncu et al., 2011), our results suggest that contributions of C9orf72 and SMCR8 to this set of diseases are also worthy of further investigation.
RESULTS
C9orf72 and SMCR8 proteins form a robust complex
On the basis of the lysosomal function for the FLCN-FNIP1/2 heterodimer in the regulation of Rag GTPases by amino acid availability (Petit et al., 2013; Tsun et al., 2013) and the prediction (Zhang et al., 2012; Levine et al., 2013) of their structural similarity to C9orf72 (Supplemental Figure S1A), we hypothesized that C9orf72 could function in a manner that is similar to FLCN and FNIP1/2. To unambiguously detect C9orf72 expressed from its endogenous locus, we took advantage of a clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 gene-editing strategy that we previously established for the study of FLCN (Petit et al., 2013) to modify the endogenous C9orf72 locus in HEK293FT cells so as to yield the expression of an N-terminally epitope-tagged, double-hemagglutinin (2xHA)–C9orf72 protein. After successful generation of a clonal population of gene-edited cells harboring the 2xHA tag in the C9orf72 locus (as confirmed by DNA sequencing), we confirmed translation of the 2XHA-C9orf72 protein by detection of a single band of ∼55 kDa after immunoblotting with an anti-HA antibody (Figure 1A). The specificity of this 2xHA-C9orf72 signal was further supported by its suppression with two different C9orf72 siRNAs (Figure 1A). These experiments support the production of a C9orf72 protein of the predicted molecular weight for full-length C9orf72 (54 382 Da). However, we did not detect the smaller, 25-kDa, form of C9orf72 that was previously reported (Atkinson et al., 2015). Because the 25-kDa form of C9orf72 is predicted to share the same amino terminus as the 55-kDa isoform, the lack of detection in our study does not reflect a limitation of our tagging strategy but could be explained by the lack of expression of this smaller isoform in HEK293FT cells, human versus mouse differences, or issues with antibody specificity in the previous studies of C9orf72 protein from the mouse brain (Atkinson et al., 2015).
FIGURE 1:
C9orf72 interacts robustly and directly with SMCR8. (A) Immunoblotting reveals the successful detection of 2xHA-C9orf72 expressed from its endogenous locus in HEK293FT cells. Suppression of this signal by two distinct C9orf72 siRNAs confirms specificity. (B) Anti-HA immunoprecipitation of 2xHA-C9orf72 from genome-edited HEK293FT cells, followed by immunoblotting for the indicated endogenously expressed proteins to assess their interaction with C9orf72. Nonspecific band on SMCR8 blot is indicated by an asterisk. (C) After their cotransfection, anti-GFP immunoprecipitation and subsequent immunoblotting experiments reveal the interaction of SMCR8-tdTomato with GFP-C9orf72 but not with GFP alone. (D) Coomassie staining of an SDS–PAGE gel with resolved anti-GFP immunoprecipitates from cells transfected with SMCR8-tdTomato and GFP or GFP-C9orf72. (E) Anti-HA immunoprecipitates from 2xHA-C9orf72 cells were washed with the indicated salt and detergent concentrations, followed by immunoblotting detection of 2xHA-C9orf72 and SMCR8. (F) Immunoblot analysis of siRNA-mediated depletion of SMCR8 and C9orf72 depletion in HEK293FT cells. Nonspecific band on SMCR8 blot is indicated by an asterisk.
Because FLCN and FNIP proteins heterodimerize with one another (Baba et al., 2006), we predicted that C9orf72 might also interact with FLCN, FNIP1, or SMCR8. SMCR8 was named for its chromosomal localization near the RA1 gene that is responsible for Smith–Magenis syndrome (Slager et al., 2003). Therefore SMCR8’s name does not provide insight into its function. Although little is known about the cellular functions of SMCR8, it is believed to have arisen via duplication of the FLCN gene and is predicted to exhibit close structural similarity with FLCN (Zhang et al., 2012; Levine et al., 2013). We tested for interactions of C9orf72 with FLCN, FNIP1, and SMCR8 by performing an anti-HA immunoprecipitation of 2xHA-C9orf72 (expressed from the endogenous locus) followed by immunoblotting for the endogenously expressed FLCN, FNIP1, and SMCR8 proteins. These experiments revealed the copurification of SMCR8 but not FLCN or FNIP1 with C9orf72 (Figure 1B). This interaction between C9ORF72 and SMCR8 was also observed after C9orf72-GFP and SMCR8-tdTomato cotransfection and anti–green fluorescent protein (GFP) immunoprecipitation (Figure 1C). The direct nature of the C9orf72-SMCR8 interaction was revealed by Coomassie staining of SDS–PAGE gels containing such anti-GFP immunoprecipitates, which resulted in the detection of near-1:1 stoichiometry between C9orf72-GFP and SMCR8-tdTomato and no other bands of significant abundance (Figure 1D). Resistance to high-salt concentrations and detergent is an indication of the robustness of a protein complex (Dibble et al., 2012). To assess the strength of the C9orf72-SMCR8 interactions, we anti-HA immunoprecipitated the endogenous C9orf72-SMCR8 complex and subjected it to washes containing varying concentrations of NaCl or SDS. The C9orf72-SMCR8 complex was unperturbed by washes with salt concentrations as high as 1.5 M NaCl, as well as up to 0.1% SDS (Figure 1E). Interaction with SMCR8 is likely to be crucial for C9orf72 function because SMCR8 knockdown was accompanied by strong reduction in C9orf72 protein levels (Figure 1F).
Subunits of a protein complex are expected to share similar patterns of gain and loss through evolutionary history. We thus applied the clustering by inferred models of evolution method of phylogenetic profiling (Li et al., 2014) to C9orf72 and SMCR8. These analyses revealed that the gene with the most highly correlated evolutionary history with C9orf72 is SMCR8 and vice versa (Supplemental Figure S1, B and C). This shared pattern of evolutionary conservation further supports the likely functional significance of their interactions. Other genes that shared correlated evolutionary history with both C9orf72 and SMCR8 were cystatin A (CSTA) and COMMD1, proteins involved in regulation of lysosomal protease activity and endosomal membrane traffic, respectively (Phillips-Krawczak et al., 2015; Stoka et al., 2016).
Collectively, through analysis of coimmunoprecipitation of endogenous and overexpressed proteins, small interfering RNA (siRNA)–mediated knockdowns, and phylogenetic profiling, we identified a robust C9orf72-SMCR8 protein complex analogous to, but distinct from, the heterodimer formed by the structurally related FLCN and FNIP proteins.
Amino acid availability regulates the recruitment of C9orf72 to lysosomes
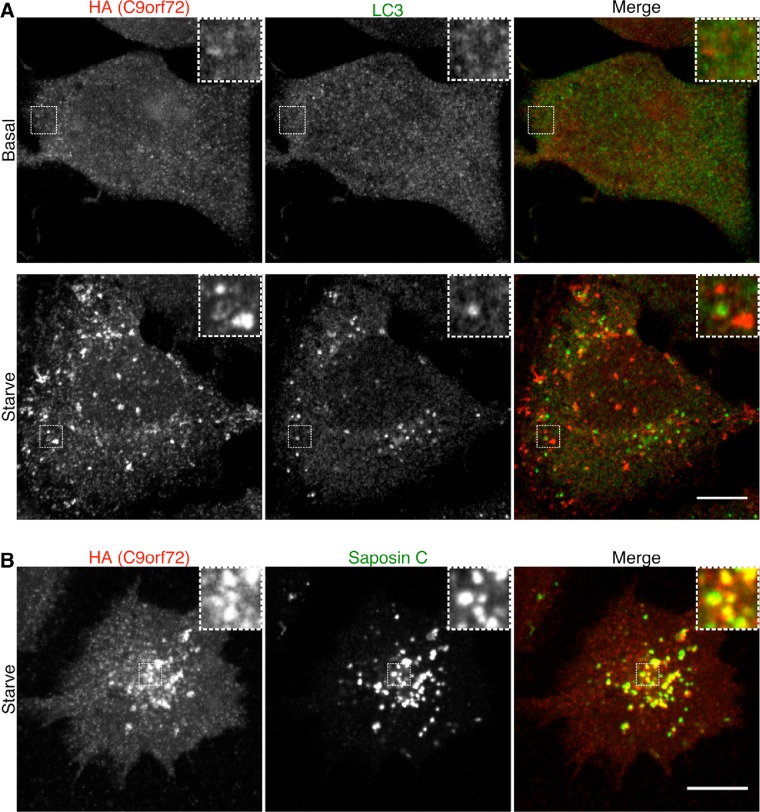
To gain further insight into C9orf72 function, we used the 2xHA tag to identify its subcellular localization. Owing to the low abundance of the C9orf72 protein, immunofluorescence detection of this protein was challenging and required the use of a tertiary antibody to amplify the signal beyond what is achieved with the traditional primary plus fluorescent secondary antibody approach of indirect immunofluorescence (similar to what we previously used for the detection of endogenous expression levels of HA-tagged FLCN protein; Petit et al., 2013). These immunofluorescence experiments revealed a predominantly diffuse cytoplasmic localization pattern for C9orf72 when cells were analyzed under basal cell culture conditions (Figure 2A). Having previously observed that FLCN recruitment to lysosomes was negatively regulated by amino acid availability (Petit et al., 2013), we investigated the effect of starvation (1 h without serum or amino acids) on C9orf72 localization and observed that it became concentrated on lysosomes (Figure 2B, as revealed by colocalization with lysosome-associated membrane protein 1 [LAMP1]). In support of the lysosomes as a major site of C9orf72 localization, 92% of C9orf72 puncta in starved cells colocalized with LAMP1 (17 cells, two independent experiments). The key role for amino acids in regulating C9orf72 recruitment to lysosomes is supported by the redistribution of C9orf72 back into the cytoplasm after just 15 min of amino acid refeeding (Figure 2C). The specificity of the punctate 2xHA-C9orf72 signal observed under starvation conditions was confirmed by its sensitivity to C9orf72 siRNA (Supplemental Figure S2A). We also investigated the effect of starvation on the interaction between C9orf72 and SMCR8 and found that it was not affected by this treatment (Figure 2D). The lysosomal localization of the C9orf72 puncta during starvation conditions was further supported by its colocalization (94% of C9orf72 puncta from 15 cells, two independent experiments) with Saposin C (a luminal protein of lysosomes; Figure 3B). Whereas starvation also induces the formation of autophagosomes (Figure 3A), only minimal colocalization was observed between C9orf72 and LC-3, a protein of autophagosomes (Figure 3A; 12% of C9orf72 puncta from 14 cells, two independent experiments). This low level of colocalization between C9orf72 and LC-3 may reflect the delivery of LC-3 to lysosomes via autophagosome-lysosome fusion. Whereas C9orf72 localization to lysosomes was regulated by amino acid availability, direct mTOR inhibition with torin 1 (Thoreen et al., 2009) did not elicit a similar change in C9orf72 localization (Supplemental Figure S2B). We thus conclude that C9orf72’s recruitment to lysosomes is not a downstream result of the mTORC1 inactivation that is triggered by amino acid starvation and may instead reflect a more direct response to changes in amino acid levels.
FIGURE 2:
Amino acid availability regulates C9orf72 recruitment to lysosomes. Immunofluorescence images showing 2xHA-tagged endogenous C9ORF72’s localization in (A) basal, (B) starved (1-h serum and amino acid–free RPMI), and (C) starvation followed by amino acid refeeding (15 min). C9ORF72’s recruitment to lysosomes is enhanced by amino acid starvation, as revealed by its colocalization with LAMP1 (a lysosomal integral membrane protein) in starved conditions. Insets, higher magnification of boxed regions of interest. Experiments were performed in HEK293FT cells, and images were obtained by spinning-disk confocal microscopy. Scale bar, 10 μm. (D) Anti-HA immunoprecipitations from 2xHA-C9orf72 cells harvested under starvation and amino acid–fed conditions revealed similar levels of SMCR8 coimmunoprecipitation. Analysis of S6 phosphorylation served as a positive control for assessing cellular responses to changes in amino acid availability via the mTORC1 signaling pathway.
FIGURE 3:
C9orf72 localizes more robustly to lysosomes than autophagosomes under starvation conditions. (A) Immunofluorescence images of endogenously expressed 2xHA-C9orf72 and LC-3 (autophagosome marker) in basal cell growth conditions and starved (1-h serum and amino acid–free RPMI) conditions. (B) Immunofluorescence images of 2xHA-C9orf72 and Saposin C (protein of the lysosome lumen) in starved conditions. Scale bar, 10 μm.
Characterization of C9orf72 and SMCR8 KO cells
To gain insight into the functions of C9orf72 and SMCR8, we used CRISPR/Cas9 genome editing to generate C9orf72 and SMCR8 KO cell lines. A C9orf72 plus SMCR8 double-KO cell line was also made by retargeting the C9orf72 KO cell line with an SMCR8 sgRNA. In addition to genomic DNA sequencing, which confirmed the presence of frameshift mutations in the respective KO cell lines (Supplemental Figure S3A), Western blotting confirmed the loss of the C9orf72 and SMCR8 proteins in their respective KO cell lines (Figure 4A). As observed with SMCR8 siRNA, depletion of SMCR8 was accompanied by a strong reduction in the levels of C9orf72. In contrast to the siRNA knockdown experiments (Figure 1F), C9orf72 KO was accompanied by a modest reduction in SMCR8 protein levels (Figure 4A). This difference between effects of C9orf72 siRNA knockdown and CRISPR KO on SMCR8 abundance may reflect either complete C9orf72 depletion in the KO or the transient nature of the siRNA experiments. The reciprocal dependence of C9orf72 and SMCR8 protein levels on the presence of each other supports the likely functional importance of this interaction. The less complete depletion of SMCR8 protein levels in the C9orf72 KO could reflect an intrinsic difference in the stability of these two proteins or the existence of an additional, as-yet-undiscovered protein complex that contains and stabilizes SMCR8 but not C9orf72.
FIGURE 4:
Analysis of C9orf72 and SMCR8 KO cell lines. (A) Immunoblot analysis of SMCR8 and C9orf72 abundance in KO cell lines. Asterisk indicates nonspecific band in SMCR8 immunoblots. (B) LAMP1 immunofluorescence in cells of the indicated genotypes (maximum intensity projections of confocal image z-stacks acquired through full cell thickness). Cell edges are indicated with a dotted white line. Boxes indicate regions of interest displayed in higher-magnification insets. Scale bar, 10 μm.
The recruitment of C9orf72 to lysosomes under starvation conditions (Figure 2) suggested a lysosomal function for C9orf72. Consistent with this prediction, late endosomes and lysosomes (as revealed by LAMP1 staining) were enlarged and more compactly clustered in the perinuclear region in C9orf72 KO cells than in either wild-type (WT) or SMCR8 KO cells (Figure 4B). The effects of C9orf72 KO on lysosome appearance (size and subcellular distribution), in combination with the localization of C9orf72 to lysosomes, support the lysosome as the site of action for C9orf72.
Although not showing the lysosome clustering that was evident in the C9orf72 KO cells, the SMCR8 KO cells appeared significantly larger than WT or C9orf72 KOs (Figures 4B and 5A), and this difference was robustly supported by quantification of cellular footprint area (Figure 5B). Furthermore, measurement of nuclear size, a parameter that scales with cell volume and thus represents a readily measurable indicator of overall cell size (Snijder et al., 2009; Levy and Heald, 2012), revealed an increase in SMCR8 KOs that was similar to what was observed for analysis of cell footprints (Figure 5C). Analysis of cells in suspension was previously used to quantify changes in cell size (Sancak et al., 2008; Thoreen et al., 2009). This strategy allows the efficient analysis of large numbers of cells and is not affected by potential differences in cell spreading or substrate interactions. When applied to our KO cells, flow cytometry analysis of cells in suspension yielded results that paralleled our measurements of adherent cells (Figure 5D). Of interest, C9orf72-SMCR8 double-KO cells reversed the lysosome clustering and enlargement of C9orf72 single KOs (Figure 4B). Similarly, double KO of C9orf72 and SMCR8 did not exhibit the increased-cell-size phenotype of the SMCR8 single KO (Figures 4B and 5, A–D).
FIGURE 5:
SMCR8 KO causes an increase in cell size. (A) F-actin (phalloidin) and nuclear (DAPI) labeling of cells of the indicated genotypes reveals the enlargement of SMCR8 KO cells. Scale bar, 10 μm. (B) Scatterplot of cell areas as determined by measuring the perimeter of phalloidin-stained cells (>30 cells measured/experiment, three experiments, mean ± SEM summarized by lines and whiskers; ****p < 0.0001, analysis of variance (ANOVA) with Tukey’s multiple comparisons posttest). (C) Scatterplot of nuclear areas as determined by measuring the outline of DAPI-stained nuclei (>30 cells measured/experiment, three experiments, mean ± SEM summarized by lines and whiskers; ****p < 0.0001, ANOVA with Tukey’s multiple comparisons posttest). (D) Cell diameter distributions of WT, C9orf72 KO, SMCR8 KO, and C9orf72/SMCR8 double-KO cell lines. Cell diameters were measured in suspension by flow cytometry (data represent the average of results from two independent experiments with >1800 cells measured for each genotype per experiment).
Altered mTORC1 signaling in C9orf72 and SMCR8 KO cells
mTORC1 signaling is tightly coupled to lysosomal amino acid–sensing machinery (Bar-Peled and Sabatini, 2014; Ferguson, 2015). Amino acid–regulated recruitment of C9orf72 to lysosomes (Figure 2) suggested a potential role for C9orf72 in coordinating the response of mTORC1 to changes in amino acid availability. Because mTORC1 is a major regulator of cell size (Kim et al., 2002; Eltschinger and Loewith, 2016), altered mTORC1 signaling could also contribute to the increased size of SMCR8 KO cells. We therefore measured mTORC1 signaling under basal cell growth conditions by assessing the phosphorylation state of ribosomal protein S6 (S235/S236 phosphorylation sites), a downstream readout that we previously established to be a robust reporter of mTORC1 signaling in HeLa cells (Petit et al., 2013). Whereas basal S6 phosphorylation in C9orf72 KO and C9orf72-SMCR8 double-KO cells was similar to that in WT cells, it increased by approximately twofold in SMCR8 single KOs (Figure 6, A and B). To explore further the link between elevated mTORC1 signaling and increased cell size after SMCR8 depletion, we also knocked down SMCR8 with siRNA. This alternative method of SMCR8 depletion also caused an increase in cell size and an increase in S6 phosphorylation (Figure 6, C and D). The role of elevated mTORC1 signaling in driving this increase in cell size was supported by the observation that mTORC1 inhibition (Torin1 treatment) blocked the cell size difference between control and SMCR8 siRNA–transfected cells (Figure 6, C and D).
FIGURE 6:
Impaired regulation of mTORC1 in C9orf72 and SMCR8 KO cells. (A) Immunoblot analysis of ribosomal protein S6 phosphorylation (S235/S236) under normal growth conditions. (B) Summary of S6 phosphorylation levels. Mean ± SEM; ****p < 0.0001 (ANOVA with Dunnett’s posttest); three to seven experiments per genotype (three for the double-KO line). (C) Increased cell size after SMCR8 depletion is mTOR dependent. Flow cytometry analysis of HeLa cell diameter after treatment with the indicated siRNAs ± 200 nM torin 1 (1300 cells measured/condition). (D) Immunoblot analysis of HeLa cells treated with indicated siRNAs and/or 200 nM torin 1 confirms the effectiveness of SMCR8 depletion and mTORC1 inhibition. (E) Immunoblot analysis of S6 phosphorylation after starvation (1.5 h) and subsequent amino acid refeeding (15 min). (F) Summary of S6 phosphorylation levels after starvation and amino acid refeeding (WT refed condition was normalized to 1, mean ± SEM; **p < 0.01, ****p < 0.0001, ANOVA with Dunnett’s posttest; four to eight experiments, four for double-KO cell line).
Following up on the observation that C9orf72 recruitment to lysosomes is regulated by amino acid availability, we next assessed the effect of C9orf72 and SMCR8 KOs on the acute responsiveness of mTORC1 signaling to changes in amino acid availability. These experiments revealed that the responsiveness of mTORC1 to amino acid refeeding was impaired in both the C9orf72 and SMCR8 single-KO cell lines (Figure 6, E and F). C9orf72 KO cells starved efficiently but were impaired in their ability to rephosphorylate S6 upon amino acid refeeding (Figure 6, E and F). Meanwhile, whereas the SMCR8 KO cells were more resistant to the effects of starvation (perhaps due to their greater size and higher basal levels of mTORC1 activity), they were also acutely insensitive to amino acid refeeding (Figure 6, E and F). Remarkably, the C9orf72-SMCR8 double-KO cells were indistinguishable from WT in these assays. Such results could reflect dominant-negative effects of the low levels of C9orf72 and SMCR8 that persist in the absence of their binding partner (Figure 4A). Although more detailed insight into the mechanisms that support distinct functions and interactions of these proteins would be required to thoroughly resolve this matter, our observations of amino acid availability regulating the localization of C9orf72 to lysosomes, the effects of C9orf72 and SMCR8 KOs on lysosome appearance, and the defective mTORC1 signaling pathway response of C9orf72 and SMCR8 KO cells to changes in amino acid availability strongly suggest an important function for these proteins on lysosomes.
Intact amino acid–regulated recruitment of mTOR to lysosomes in C9orf72 and SMCR8 KO cells
Although the C9orf72-SMCR8 complex is similar to FLCN-FNIP1/2 with respect to predicted DENN-domain structures, lysosomal site of action, and role in coordinating cellular responses to amino acid availability, the specific functions of each protein complex appear to be distinct. FLCN-FNIP1/2 heterodimers are required for mTOR localization to lysosomes and activation by amino acids. FLCN-FNIP1/2 exerts these effects through direct regulation of the Rag GTPases that control mTORC1’s recruitment to lysosomes (Petit et al., 2013; Tsun et al., 2013). In contrast, even though C9orf72 and SMCR8 KO cells also show defects in the regulation of mTORC1 by amino acids (Figure 6, E and F), they are not defective with respect to the regulation of mTOR localization by changes in amino acid levels (Figure 7, A–D). In particular, mTOR recruitment to lysosomes still occurs in response to amino acid refeeding in each of these KO cell lines (Figure 7, B–D). The intact regulation of mTOR localization by changes in amino acid availability in both C9orf72 and SMCR8 KO cells suggests that the C9orf72-SMCR8 heterodimer is not required for controlling the Rag GTPase-mediated recruitment of mTORC1 to lysosomes. This interpretation is supported by observations that FLCN but not SMCR8 interacts with the Rags (Supplemental Figure S4). To rule out the possibility that C9orf72 and/or SMCR8 KO cells have adapted in a manner the renders mTOR recruitment to lysosomes independent of the Rags, we took advantage of a previously validated RagC siRNA (Roczniak-Ferguson et al., 2012) to examine the effects of RagC depletion on mTOR recruitment to lysosomes and found that it is highly RagC dependent in each of the WT, C9orf72 KO, SMCR8 KO, and C9orf72-SMCR8 double-KO cell lines (Supplemental Figure S5). These results indicate that C9orf72 and SMCR8 contribute to regulation of mTORC1 signaling by amino acid availability in a Rag-independent manner and provide a foundation for future elucidation of underlying mechanisms.
FIGURE 7:
Intact regulation of mTOR localization in C9orf72 KO, SMCR8 KO, and double-KO cells. mTOR localization was observed under basal growth conditions and starvation (1.5 h) and amino acid refed (15 min) conditions in WT (A), C9orf72 (B), SMCR8 (C), and double-KO (D) cells. The lysosomal colocalization of this punctate mTOR staining has been thoroughly established in previous studies (Petit et al., 2013; Ferguson, 2015). Scale bar, 10 μm.
DISCUSSION
Our discovery of a C9orf72-SMCR8 interaction builds on our previous efforts to elucidate lysosomal functions of the FLCN-FNIP protein complex (Petit et al., 2013) and the predictions of structurally similar DENN domains in FLCN, FNIP1 and 2, SMCR8, and C9orf72 (Zhang et al., 2012; Levine et al., 2013). FLCN and FNIP proteins form heterodimers (Baba et al., 2006) that localize to lysosomes, where they support the activation of mTORC1 signaling by intracellular amino acids (Petit et al., 2013; Tsun et al., 2013). Such actions of FLCN-FNIP1/2 are mediated by direct interactions between FLCN-FNIP and the Rag GTPases. This enables the Rag-dependent recruitment of mTORC1 to the surface of lysosomes and mTORC1’s subsequent activation (Sancak et al., 2010; Petit et al., 2013; Tsun et al., 2013).
The bioinformatics studies (Zhang et al., 2012; Levine et al., 2013) that predicted a similar DENN-like domain as the major folded domain within FLCN, FNIP1, FNIP2, SMCR8, and C9orf72 (Supplemental Figure S1A) also suggested a subdivision within this group of proteins in which FLCN and SMCR8 are most similar to one another and C9orf72 most closely resembles FNIP1 and FNIP2. On the basis of these predictions, we initially suspected that C9orf72 might interact with FLCN in a manner similar to what was shown for FNIP1 (Baba et al., 2006) but instead found that C9orf72 interacts selectively with SMCR8 (Figure 1). This family of DENN-like proteins can thus be divided into distinct protein complexes comprising FLCN-FNIP1/2 and SMCR8-C9orf72, respectively. Our investigation of C9orf72 and SMCR8 and their comparison to the FLCN-FNIP heterodimer thus generated new insights into important similarities and differences between cellular functions of this subfamily of DENN domain–containing proteins.
Whereas SMCR8-C9orf72 form a distinct complex, C9orf72 shares with FLCN-FNIP1 the ability to be recruited to lysosomes in a manner that is negatively regulated by amino acid availability (Petit et al., 2013). Several pieces of evidence additionally support the likely localization of SMCR8 to lysosomes. First, the interaction between C9orf72 and SMCR8 is strong and influences the stability of both C9orf72 and SMCR8 (Figures 1 and 4A). Second, the C9orf72-SMCR8 interaction was not disassembled during amino acid starvation (Figure 2D). Third, SMCR8 was previously identified as a component of purified lysosomes via proteomic methods (Schroder et al., 2007). Although the recruitment of FLCN-FNIP to lysosomes is believed to be mediated by interactions with Rag GTPase heterodimers (Petit et al., 2013; Tsun et al., 2013), we were unable to detect similar Rag interactions for the C9orf72-SMCR8 complex (Supplemental Figure S4). The mechanism that supports recruitment of C9orf72-SMCR8 to lysosomes thus represents an important question that needs to be addressed in future studies.
The presence of DENN domains in FLCN and FNIP proteins, combined with established roles for other DENN domain–containing proteins as Rab GEFs (Yoshimura et al., 2010; Marat et al., 2011), suggested a possible function for FLCN and FNIP1/2 as Rab GEFs. However, whereas in vitro studies implicated FLCN as a Rab35 GEF (Nookala et al., 2012), the in vivo significance of this finding remains uncertain. In fact, the best-characterized function for the FLCN-FNIP complex is to act as a GAP for RagC (Tsun et al., 2013). In line with this finding, LST7, the budding yeast homologue of FLCN, encodes a much smaller protein, which does not contain a complete DENN domain. However, it still interacts with Lst4 (homologous to FNIP1/2) and regulates Tor signaling by acting as a GAP for Gtr2, the yeast RagC homologue (Pacitto et al., 2015; Peli-Gulli et al., 2015). Based on the lessons learned from these studies of the FLCN-FNIP complex, it is possible that C9orf72-SMCR8 has non-Rab GTPase targets or that their major molecular function is to serve as a GAP rather than a GEF. Nonetheless, C9orf72 was recently reported to act as a GEF toward Rab8a and Rab39b (Sellier et al., 2016). Because these particular Rabs have not previously been localized to lysosomes, it remains to be determined how their regulation relates to the regulated lysosomal localization of C9orf72 (Figures 2 and 3), the lysosome-related phenotypes that we observed in C9orf72 and SMCR8 KO cells (Figures 4 and 5), and the occurrence of abnormally enlarged lysosomes that was recently reported in macrophages and microglia from C9orf72 KO mice (O’Rourke et al., 2016).
Of interest, while our manuscript was under revision, C9orf72 and SMCR8 interactions were independently identified in conjunction with a third protein, WDR41, which robustly associates with this complex (Sellier et al., 2016; Sullivan et al., 2016; Xiao et al., 2016). Although little else is known concerning WDR41 functions, like SMCR8, it was previously detected in a proteomic analysis of lysosomal membranes (Schroder et al., 2007), consistent with the possibility that it forms a complex with C9orf72 and SMCR8 on lysosomes. These new findings highlight the importance of future studies to define both the structure of the C9orf72-SMCR8-WDR41 protein complex and the specific mechanisms that control its subcellular localization and molecular functions.
Recent studies have yielded tremendous insight into the mechanisms by which lysosomes serve as a major platform for integrating signals that are critical for control of the mTORC1 signaling pathway (Bar-Peled and Sabatini, 2014; Ferguson, 2015). Chief among these signals are amino acid availability and growth factor signaling via the phosphoinositide 3-kinase–AKT pathway. Amino acids are best characterized by their ability to regulate mTORC1 via several distinct mechanisms that converge on the Rag GTPases and thus determine their ability to recruit mTORC1 to the surface of lysosomes (Bar-Peled and Sabatini, 2014; Efeyan et al., 2014; Ferguson, 2015; Saxton et al., 2016; Wolfson et al., 2016). For this reason, as well as the previously established lysosomal role of the FLCN-FNIP complex in Rag regulation (Petit et al., 2013; Tsun et al., 2013), we investigated the possibility that the Rag GTPases represent targets of C9orf72-SMCR8, which could explain defects in the regulation of mTORC1 signaling by amino acids in C9orf72 and SMCR8 KO cells. However, in contrast to the previously established model for FLCN-FNIP–dependent Rag regulation, our immunoprecipitation experiments did not detect interactions between Rags and C9orf72-SMCR8 (Supplemental Figure S4), nor was C9orf72 or SMCR8 essential for acute amino acid–regulated recruitment of mTOR to lysosomes (Figure 7). These observations suggest the existence of additional factors, beyond the Rag GTPases, that communicate amino acid availability to the regulation of mTORC1.
Our analysis of C9orf72 and SMCR8 single- and double-KO cell lines revealed a complex pattern with respect to changes in lysosome appearance, cell size, and mTORC1 signaling. Lysosomes were enlarged and clustered in the C9orf72 KO cells. However, this phenotype was reversed by the subsequent disruption of the SMCR8 gene. Similarly, the increase in both basal mTORC1 signaling and cell size in the single SMCR8 KO was not observed after mutation of both C9orf72 and SMCR8. Furthermore, whereas both the C9orf72 and SMCR8 single-KO cells were unable to efficiently activate mTORC1 in response to acute amino acid refeeding, the double-KO cells responded as robustly as WT cells to this stimulus. The lysosome-related phenotypes of the C9orf72 and SMCR8 single-KO cell lines, combined with evidence supporting the localization of both C9orf72 and SMCR8 to lysosomes, strongly point to the lysosome as a major intracellular site of action for these proteins.
The reduced levels of C9orf72 and SMCR8 after KO of their binding partner (Figures 1F and 4A) demonstrate the importance of C9orf72-SMCR8 interactions in controlling their respective stabilities. It is possible that unstable C9orf72 and SMCR8 proteins that remain in the respective single-KO cells exert a dominant-negative effect on the lysosomal machinery that represents the natural target of the C9orf72-SMCR8 protein complex. Alternatively, SMCR8 could play a role in a different protein complex that acts as a brake on mTORC1 activity and is negatively regulated by the C9orf72-SMCR8 complex. This issue remains to be resolved.
Our analysis of both C9orf72 and SMCR8 single- and double-KO cell lines sheds important new light on recent reports of phenotypes, including lysosomal defects, in C9orf72 single-KO mice and cell lines (Atanasio et al., 2016; O’Rourke et al., 2016; Sellier et al., 2016; Sullivan et al., 2016). For example, the overabundance of enlarged lysosomes in macrophages and microglia from C9orf72 KO mice (O’Rourke et al., 2016) parallels the lysosome changes that we uncovered in C9orf72 KO HeLa cells. However, reversal of the C9orf72 KO phenotype that we observed in C9orf72-SMCR8 double-KO cells suggests an SMCR8-related gain of function rather than simple loss of C9orf72 as the cause of the C9orf72 KO lysosome defects that we and others have observed in diverse cell types. SMCR8 was previously identified as a target of mTORC1-dependent phosphorylation (Hsu et al., 2011). Therefore, SMCR8-dependent phenotypes related to lysosomes and mTORC1 signaling could also involve complex feedback loops. Furthermore, given the broad regulation of numerous aspects of cell physiology and metabolism by mTORC1 signaling, changes arising due to loss of C9orf72-SMCR8 functions at lysosomes could have pleiotropic indirect effects.
The localization of C9orf72 to lysosomes sheds light on neurodegenerative diseases that extend beyond the strong genetic link between C9orf72 hexanucleotide expansions and ALS-FTD. For example, C9orf72 accumulations have been observed within swollen axons surrounding Alzheimer’s disease amyloid plaques (Satoh et al., 2012). Such axonal swellings also contain high levels of progranulin (Gowrishankar et al., 2015), another protein with FTD-linked mutations (Baker et al., 2006; Cruts et al., 2006). The underlying cell biological mechanism that explains the enrichment of such proteins at amyloid plaques is the high abundance of lysosomes within the swollen axons that surround each amyloid plaque (Gowrishankar et al., 2015). Of interest, the possibility of altered C9orf72-dependent lysosome functions in ALS-FTD parallels the identification of other genes with lysosomal functions, including progranulin (Baker et al., 2006; Cruts et al., 2006; Gowrishankar et al., 2015), TMEM106B (Van Deerlin et al., 2010; Finch et al., 2011), and Rab38 and/or cathepsin C (Ferrari et al., 2014) as modulators of FTD risk. Although it remains to be established whether such genes define a pathway of general relevance to ALS and/or FTD susceptibility and, if so, what cell type is most relevant for such disease risk, it is interesting that C9orf72 and progranulin have recently been linked to microglial lysosome function (Lui et al., 2016; O’Rourke et al., 2016). While it can be speculated that the high phagocytic activity of microglia places a strong demand on lysosomes that unmasks subtle defects that can be tolerated by most other cell types, the specific molecular mechanisms that underlie such defects remain unclear. Our new findings indicate the importance of taking SMCR8 into account while dissecting such mechanisms and furthermore provide a highly experimentally tractable cell culture model for pursuing such studies of the relationship between C9orf72 and lysosome cell biology.
In this study, we define robust interactions between C9orf72 and SMCR8 and demonstrate a role for this protein complex in the regulation of lysosomal responses to changes in amino acid availability. More specifically, amino acid availability acts acutely (within minutes) to negatively regulate the recruitment of C9orf72 to lysosomes. Lysosome-related functions for C9orf72 and SMCR8 are further supported by observations of altered lysosome subcellular position, morphology, and responsiveness of mTORC1 signaling to changes in amino acid availability in cells depleted of these proteins by CRISPR/Cas9 KO strategies. The observations presented in this study will support multiple lines of future investigation. These include elucidation of the structural basis for C9orf72-SMCR8 interactions; identification of the mechanisms by which amino acid availability regulates C9orf72 recruitment to lysosomes, dissection of the mechanisms that link C9orf72 to regulation of lysosome size and subcellular positioning, and dissection of the role played by the C9orf72-SMCR8 complex in the regulation of mTORC1 signaling by amino acid availability. The new cell biological understanding of the C9orf72 protein provided by this study will also support investigation of the potential contributions of reduced C9orf72 protein levels to neurodegenerative diseases arising from C9orf72 hexanucleotide expansions. Regardless of whether reduced C9orf72 protein levels contribute to ALS-FTD disease mechanisms in C9orf72 hexanucleotide expansion carriers, understanding the C9orf72 protein will be critical for evaluating the risks associated with therapeutic strategies, such as antisense oligonucleotide approaches (Donnelly et al., 2013), that aim to suppress expression of the repeat expansion RNA.
In addition to interest in physiological and pathophysiological functions of C9orf72 that has been generated by insights from ALS-FTD, the role played by C9orf72 and SMCR8 in the regulation of the mTORC1 signaling pathway suggests that understanding their functions may also have relevance to cancer biology. Given the recent reports of monocytic cell dysfunction and autoimmunity in C9orf72 KO mice (Atanasio et al., 2016; Burberry et al., 2016; O’Rourke et al., 2016), understanding the contributions of C9orf72 to hematopoietic cell function will also benefit from further elucidation of direct biochemical actions of C9orf72.
MATERIALS AND METHODS
Cell culture and transfection
HEK293FT (Life Technologies, Carlsbad, CA) and HeLa M cells (provided by P. De Camilli, Yale University, New Haven, CT) were grown in high-glucose DMEM (+l-glutamine), 10% fetal bovine serum, and 1% penicillin/streptomycin supplement (Invitrogen, Carlsbad, CA). Where indicated, cells were starved for 90 min by incubation with amino acid–free RPMI media (US Biological, Swampscott, MA). Amino acid refeeding was performed with 1× MEM amino acid supplement (Invitrogen) added to starvation medium for 15 min. Transfections were performed with 500 ng of DNA, 100 μl of OptiMEM (Invitrogen), and 1.5 μl of FuGENE 6 transfection reagent (Promega, Madison, WI) per 35-mm dish and scaled up proportionately for larger and smaller dishes. siRNA transfections were performed with 5 μl of RNAiMAX (Invitrogen), 500 μl of OptiMEM (Invitrogen), and 7.5 μl of 20 μM siRNA stock suspended in 100 mM KAc and 30 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid, pH 7.5. We used 250,000 HEK 293FT cells per 35-mm dish for siRNA transfection and incubated them for 2 d posttransfection before experiments. siRNA sequences purchased from Integrated DNA Technologies (IDT, Coralville, IA) are summarized in Supplemental Table S1.
Plasmids
Human C9orf72 (residues 1–481; Uniprot identifier Q96LT7-1) was PCR amplified from cDNA (ThermoFisher Scientific, Waltham MA) and cloned into the pEGFP-C2 vector (Clontech, Mountain View, CA) to generate an N-terminally GFP-tagged C9orf72 protein. Human SMCR8 (residues 1–937; Uniprot identifier Q8TEV9-1) was also PCR amplified from cDNA (Clone 40148405; Thermo Fisher Scientific) and cloned into the ptDTomato-N1 plasmid (Clontech) to generate an SMCR8 protein with tdTomato at its C-terminus. Plasmids encoding HA–glutathione S-transferase (GST)–tagged human Rag GTPases in pRK5 vector and mutants thereof (RagA T21L, RagA Q66L, RagC Q120L, and RagC S75L) were obtained from D. Sabatini (Massachusetts Institute of Technology, Cambridge, MA) via Addgene (Sancak et al., 2008).
Antibodies
Antibodies used in this study are described in Supplemental Table S2.
Immunoblotting and immunoprecipitation
Cells were lysed in Tris-buffered saline (TBS) plus 1% Triton X-100 plus protease and phosphatase inhibitor cocktails (Roche Diagnostics, Indianapolis IN), and insoluble material was cleared by centrifugation for 10 min at 20,000 × g. Immunoprecipitations were performed on the resulting lysates. For anti-GFP immunoprecipitations, GFP-Trap resin (ChromoTek, Planegg-Martinsried, Germany) was incubated with lysates for 1 h with rotation at 4°C, followed by five washes with lysis buffer and elution in 2× Laemmli buffer. For pulldown of HA-GST Rag proteins, the same procedure was used with glutathione Sepharose 4B (GE Healthcare, Pittsburgh PA) and a 2-h incubation. For anti-HA immunoprecipitations, we used anti-HA Affinity Matrix (Roche Diagnostics) and a 3-h incubation. Immunoblotting was performed with 4–15% gradient Mini-PROTEAN TGX precast polyacrylamide gels and nitrocellulose membranes (Bio-Rad, Hercules CA). Where indicated, gels were stained with Imperial Coomassie protein stain (ThermoFisher Scientific). Blots were blocked with 5% milk, and antibodies were incubated with 5% milk or bovine serum albumin in TBS with 0.1% Tween 20. Chemiluminescence detection of horseradish peroxidase signals was performed on a VersaDoc imaging station (Bio-Rad), and ImageJ (National Institutes of Health, Bethesda, MD) was used to quantify band intensities.
CRISPR/Cas9 genome editing
Use of the CRISPR/Cas9 genome-editing system for the insertion of epitope tags at endogenous loci was described previously (Petit et al., 2013). The specific guide RNA and single-stranded DNA (ssDNA) repair template (IDT Ultramer format) sequences used to insert the 2xHA epitope tag in the endogenous C9orf72 locus are given in Supplemental Table S3. Guide RNA–encoding DNA oligonucleotides were ligated into Bbs1-digested PX459 vector (acquired from Feng Zhang [Massachusetts Institute of Technology] via Addgene) and transformed into Stabl3-competent Escherichia coli cells (Ran et al., 2013). Successful insertion of the guide RNA–encoding DNA sequence was confirmed by sequencing. All oligonucleotides were purchased from IDT.
For 2xHA-tag insertion into the endogenous C9orf72 locus, 3 μg of PX459 vector containing C9orf72 guide RNA and 3 μl of 100 μM ssDNA repair template were electroporated into 1 million HEK 293FT cells (Cell line kit V, electroporation program Q01; Lonza, Basel, Switzerland). Clonal cell lines were subsequently isolated from this pool of cells and screened for HA signal by immunoblotting and immunofluorescence. To confirm further the correct in-frame insertion of the 2xHA tag in these cells, genomic DNA surrounding the C9orf72 start codon was sequenced. To this end, genomic DNA was extracted (QuickExtract DNA extraction solution; Epicentre Biotechnologies, Madison, WI), the region of interest was amplified by PCR (primers described in Supplemental Table S4), cloned into the pCR-Blunt TOPO vector (Zero Blunt TOPO PCR cloning kit; ThermoFisher Scientific), and transformed into TOP10-competent E. coli cells, and plasmid DNA isolated from multiple colonies was sequenced.
Guide RNA sequences used to generate C9orf72 and SMCR8 KO cell lines are summarized in Supplemental Table S3. Annealed oligonucleotides were cloned into the px459 vector, and 0.4 μg of such plasmids was transfected with FuGENE 6 into 35,000 HeLa cells per/well of a 24-well dish. Transfected cells were selected with 2 μg/ml puromycin for 2 d, and surviving cells were subsequently plated at clonal density. After selection and expansion of clonal populations, KOs were first identified by Western blotting and subsequently confirmed by sequencing of PCR-amplified genomic DNA (primers defined in Supplemental Table S4). C9orf72-SMCR8 double-KO cells were generated by transfection of the C9orf72 KO line with the SMCR8 sgRNA plasmid, puromycin selection, and the isolation and validation of a clonal double KO cell population.
Immunofluorescence staining and microscopy
Immunofluorescence staining and confocal microscopy were largely performed as described previously (Petit et al., 2013). Optimal preservation of lysosome morphology and detection of 2x-HA C9orf72 was achieved by direct dropwise addition of one volume of 8% paraformaldehyde (PFA) in 0.1 M sodium phosphate buffer to cells growing on class coverslips in growth medium. After fixing for 30 min at room temperature, cells were washed with phosphate-buffered saline (PBS), blocked, and permeabilized with 5% normal donkey serum (Jackson ImmunoResearch, West Grove, PA)/0.1% saponin/PBS for 1 h. Subsequent antibody incubations were performed in this buffer.
For measurements of nuclear and cell footprint area, spinning-disk confocal images were acquired with a 40× CFI Plan Apo, oil immersion objective (numerical aperture 1.0). Nuclei were identified by 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen) staining, and cell edges were identified by phalloidin (Alexa 594–phalloidin; Invitrogen) staining of PFA-fixed cells. Cell Profiler software (Carpenter et al., 2006) was used for area measurements. More than 30 cells were measured per experiment in three experiments per cell line. GraphPad Prism was used for all statistical analysis. Colocalization of C9orf72 with LAMP1, Saposin C, and LC3 under starvation conditions was determined by visual identification of 2xHA-C9orf72 puncta followed by determination of the coenrichment of the respective organelle markers on a spot-by-spot basis.
Flow cytometry analysis of cell diameter
One million cells were plated in 150-mm dishes, incubated for 24 h, and harvested by trypsinization. Cells were then washed with PBS and fixed with 2% PFA in 0.1 M sodium phosphate buffer for 20 min at room temperature. Samples were washed to remove fixative and filtered using 40-μm nylon cell strainers (BD Falcon, Durham, NC). For analysis, cells were suspended to a final volume of 100 μl in PBS. To determine cell diameter, bright-field images of individual cells were acquired using an Amnis ImageStreamX mkII imaging flow cytometer and analyzed with INSPIRE software. More than 1800 cells were measured per genotype per experiment.
For cell diameter measurement of SMCR8 siRNA/torin1–treated cells, 500,000 WT HeLa M cells were transfected with control or SMCR8 siRNA and plated in a 150-mm dish. After 24 h, 200 nM torin1 was added to cells receiving this treatment, followed by a 48-h incubation. Cells were then prepared for cell diameter measurement as described.
Supplementary Material
Acknowledgments
We appreciate the contributions of Matthew Schrag to initiating early aspects of this research. This work was supported in part by grants from the National Institutes of Health (GM105718 and AG047270), the Consortium for Frontotemporal Dementia Research, and the Ellison Medical Foundation to S.M.F. J.A. was supported by National Institutes of Health training grants (T32GM007223 and F31GM119249). Flow cytometry studies were supported by Shared Instrument Grant 1-S10-RR-026526-01. Microscopy studies were supported by the Yale University Program in Cellular Neuroscience, Neurodegeneration and Repair Imaging Facility.
Abbreviations used:
- ALS
amyotrophic lateral sclerosis
- C9orf72
chromosome 9 open reading frame 72
- CRISPR
clustered regularly interspaced short palindromic repeats
- DAPI
4’,6-diamidino-2-phenylindole
- DENN
differentially expressed in normal and neoplastic cells
- FLCN
folliculin
- FNIP
FLCN-interacting protein
- FTD
frontotemporal dementia
- GAP
GTPase-activating protein
- GEF
guanine nucleotide exchange factor
- GFP
green fluorescent protein
- HA
hemagglutinin
- IP
immunoprecipitation
- KO
knockout
- LAMP1
lysosome-associated membrane protein 1
- LC3
microtubule-associated proteins 1A/1B light chain 3B
- mTORC1
mechanistic target of rapamycin complex 1
- sgRNA
single-guide RNA
- siRNA
small interfering RNA
- SMCR8
Smith-Magenis Chromosome Region gene 8
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-01-0003) on August 24, 2016.
REFERENCES
- Atanasio A, Decman V, White D, Ramos M, Ikiz B, Lee HC, Siao CJ, Brydges S, LaRosa E, Bai Y, et al. C9orf72 ablation causes immune dysregulation characterized by leukocyte expansion, autoantibody production, and glomerulonephropathy in mice. Sci Rep. 2016;6:23204. doi: 10.1038/srep23204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson RA, Fernandez-Martos CM, Atkin JD, Vickers JC, King AE. C9ORF72 expression and cellular localization over mouse development. Acta Neuropathol Commun. 2015;3:59. doi: 10.1186/s40478-015-0238-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Hong SB, Sharma N, Warren MB, Nickerson ML, Iwamatsu A, Esposito D, Gillette WK, Hopkins RF, 3rd, Hartley JL, et al. Folliculin encoded by the BHD gene interacts with a binding protein, FNIP1, and AMPK, and is involved in AMPK and mTOR signaling. Proc Natl Acad Sci USA. 2006;103:15552–15557. doi: 10.1073/pnas.0603781103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker M, Mackenzie IR, Pickering-Brown SM, Gass J, Rademakers R, Lindholm C, Snowden J, Adamson J, Sadovnick AD, Rollinson S, et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- Bar-Peled L, Sabatini DM. Regulation of mTORC1 by amino acids. Trends Cell Biol. 2014;24:400–406. doi: 10.1016/j.tcb.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burberry A, Suzuki N, Wang J, Moccia R, Mordes DA, Stewart MH, Suzuki-Uematsu S, Ghosh S, Singh A, Merkle FT, et al. Loss-of-function mutations in the C9ORF72 mouse ortholog cause fatal autoimmune disease. Sci Transl Med. 2016;8:347ra393. doi: 10.1126/scitranslmed.aaf6038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruts M, Gijselinck I, van der Zee J, Engelborghs S, Wils H, Pirici D, Rademakers R, Vandenberghe R, Dermaut B, Martin JJ, et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibble CC, Elis W, Menon S, Qin W, Klekota J, Asara JM, Finan PM, Kwiatkowski DJ, Murphy LO, Manning BD. TBC1D7 is a third subunit of the TSC1-TSC2 complex upstream of mTORC1. Mol Cell. 2012;47:535–546. doi: 10.1016/j.molcel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Haeusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efeyan A, Schweitzer LD, Bilate AM, Chang S, Kirak O, Lamming DW, Sabatini DM. RagA, but not RagB, is essential for embryonic development and adult mice. Dev Cell. 2014;29:321–329. doi: 10.1016/j.devcel.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eltschinger S, Loewith R. TOR complexes and the maintenance of cellular homeostasis. Trends Cell Biol. 2016;26:148–159. doi: 10.1016/j.tcb.2015.10.003. [DOI] [PubMed] [Google Scholar]
- Farg MA, Sundaramoorthy V, Sultana JM, Yang S, Atkinson RA, Levina V, Halloran MA, Gleeson PA, Blair IP, Soo KY, et al. C9ORF72, implicated in amytrophic lateral sclerosis and frontotemporal dementia, regulates endosomal trafficking. Hum Mol Genet. 2014;23:3579–3595. doi: 10.1093/hmg/ddu068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson SM. Beyond indigestion: emerging roles for lysosome-based signaling in human disease. Curr Opin Cell Biol. 2015;35:59–68. doi: 10.1016/j.ceb.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JB, Dobson-Stone C, Brooks WS, Schofield PR, Halliday GM, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. 2014;13:686–699. doi: 10.1016/S1474-4422(14)70065-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch N, Carrasquillo MM, Baker M, Rutherford NJ, Coppola G, Dejesus-Hernandez M, Crook R, Hunter T, Ghidoni R, Benussi L, et al. TMEM106B regulates progranulin levels and the penetrance of FTLD in GRN mutation carriers. Neurology. 2011;76:467–474. doi: 10.1212/WNL.0b013e31820a0e3b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freibaum BD, Lu Y, Lopez-Gonzalez R, Kim NC, Almeida S, Lee KH, Badders N, Valentine M, Miller BL, Wong PC, et al. GGGGCC repeat expansion in C9orf72 compromises nucleocytoplasmic transport. Nature. 2015;525:129–133. doi: 10.1038/nature14974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gowrishankar S, Yuan P, Wu Y, Schrag M, Paradise S, Grutzendler J, De Camilli P, Ferguson SM. Massive accumulation of luminal protease-deficient axonal lysosomes at Alzheimer’s disease amyloid plaques. Proc Natl Acad Sci USA. 2015;112:E3699–E3708. doi: 10.1073/pnas.1510329112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeusler AR, Donnelly CJ, Periz G, Simko EA, Shaw PG, Kim MS, Maragakis NJ, Troncoso JC, Pandey A, Sattler R, et al. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, et al. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science. 2011;332:1317–1322. doi: 10.1126/science.1199498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell. 2002;110:163–175. doi: 10.1016/s0092-8674(02)00808-5. [DOI] [PubMed] [Google Scholar]
- Kwon I, Xiang S, Kato M, Wu L, Theodoropoulos P, Wang T, Kim J, Yun J, Xie Y, McKnight SL. Poly-dipeptides encoded by the C9orf72 repeats bind nucleoli, impede RNA biogenesis, and kill cells. Science. 2014;345:1139–1145. doi: 10.1126/science.1254917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine TP, Daniels RD, Gatta AT, Wong LH, Hayes MJ. The product of C9orf72, a gene strongly implicated in neurodegeneration, is structurally related to DENN Rab-GEFs. Bioinformatics. 2013;29:499–503. doi: 10.1093/bioinformatics/bts725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy DL, Heald R. Mechanisms of intracellular scaling. Annu Rev Cell Dev Biol. 2012;28:113–135. doi: 10.1146/annurev-cellbio-092910-154158. [DOI] [PubMed] [Google Scholar]
- Li Y, Calvo SE, Gutman R, Liu JS, Mootha VK. Expansion of biological pathways based on evolutionary inference. Cell. 2014;158:213–225. doi: 10.1016/j.cell.2014.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui H, Zhang J, Makinson SR, Cahill MK, Kelley KW, Huang HY, Shang Y, Oldham MC, Martens LH, Gao F, et al. Progranulin deficiency promotes circuit-specific synaptic pruning by microglia via complement activation. Cell. 2016;165:921–935. doi: 10.1016/j.cell.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marat AL, Dokainish H, McPherson PS. DENN domain proteins: regulators of Rab GTPases. J Biol Chem. 2011;286:13791–13800. doi: 10.1074/jbc.R110.217067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizielinska S, Gronke S, Niccoli T, Ridler CE, Clayton EL, Devoy A, Moens T, Norona FE, Woollacott IO, Pietrzyk J, et al. C9orf72 repeat expansions cause neurodegeneration in Drosophila through arginine-rich proteins. Science. 2014;345:1192–1194. doi: 10.1126/science.1256800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nookala RK, Langemeyer L, Pacitto A, Ochoa-Montano B, Donaldson JC, Blaszczyk BK, Chirgadze DY, Barr FA, Bazan JF, Blundell TL. Crystal structure of folliculin reveals a hidDENN function in genetically inherited renal cancer. Open Biol. 2012;2:120071. doi: 10.1098/rsob.120071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Muhammad AK, Gendron TF, Kim KJ, Austin A, Cady J, Liu EY, Zarrow J, Grant S, et al. C9orf72 BAC transgenic mice display typical pathologic features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Rourke JG, Bogdanik L, Yanez A, Lall D, Wolf AJ, Muhammad AK, Ho R, Carmona S, Vit JP, Zarrow J, et al. C9orf72 is required for proper macrophage and microglial function in mice. Science. 2016;351:1324–1329. doi: 10.1126/science.aaf1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacitto A, Ascher DB, Wong LH, Blaszczyk BK, Nookala RK, Zhang N, Dokudovskaya S, Levine TP, Blundell TL. Lst4, the yeast Fnip1/2 orthologue, is a DENN-family protein. Open Biol. 2015;5:150174. doi: 10.1098/rsob.150174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peli-Gulli MP, Sardu A, Panchaud N, Raucci S, De Virgilio C. Amino acids stimulate TORC1 through Lst4-Lst7, a GTPase-activating protein complex for the Rag family GTPase Gtr2. Cell Rep. 2015;13:1–7. doi: 10.1016/j.celrep.2015.08.059. [DOI] [PubMed] [Google Scholar]
- Peters OM, Cabrera GT, Tran H, Gendron TF, McKeon JE, Metterville J, Weiss A, Wightman N, Salameh J, Kim J, et al. Human C9ORF72 hexanucleotide expansion reproduces RNA foci and dipeptide repeat proteins but not neurodegeneration in BAC transgenic mice. Neuron. 2015;88:902–909. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit CS, Roczniak-Ferguson A, Ferguson SM. Recruitment of folliculin to lysosomes supports the amino acid-dependent activation of Rag GTPases. J Cell Biol. 2013;202:1107–1122. doi: 10.1083/jcb.201307084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips-Krawczak CA, Singla A, Starokadomskyy P, Deng Z, Osborne DG, Li H, Dick CJ, Gomez TS, Koenecke M, Zhang JS, et al. COMMD1 is linked to the WASH complex and regulates endosomal trafficking of the copper transporter ATP7A. Mol Biol Cell. 2015;26:91–103. doi: 10.1091/mbc.E14-06-1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roczniak-Ferguson A, Petit CS, Froehlich F, Qian S, Ky J, Angarola B, Walther TC, Ferguson SM. The transcription factor TFEB links mTORC1 signaling to transcriptional control of lysosome homeostasis. Sci Signal. 2012;5:ra42. doi: 10.1126/scisignal.2002790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Bar-Peled L, Zoncu R, Markhard AL, Nada S, Sabatini DM. Ragulator-Rag complex targets mTORC1 to the lysosomal surface and is necessary for its activation by amino acids. Cell. 2010;141:290–303. doi: 10.1016/j.cell.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science. 2008;320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh J, Tabunoki H, Ishida T, Saito Y, Arima K. Dystrophic neurites express C9orf72 in Alzheimer’s disease brains. Alzheimers Res Ther. 2012;4:33. doi: 10.1186/alzrt136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxton RA, Knockenhauer KE, Wolfson RL, Chantranupong L, Pacold ME, Wang T, Schwartz TU, Sabatini DM. Structural basis for leucine sensing by the Sestrin2-mTORC1 pathway. Science. 2016;351:53–58. doi: 10.1126/science.aad2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder B, Wrocklage C, Pan C, Jager R, Kosters B, Schafer H, Elsasser HP, Mann M, Hasilik A. Integral and associated lysosomal membrane proteins. Traffic. 2007;8:1676–1686. doi: 10.1111/j.1600-0854.2007.00643.x. [DOI] [PubMed] [Google Scholar]
- Sellier C, Campanari ML, Corbier CJ, Gaucherot A, Kolb-Cheynel I, Oulad-Abdelghani M, Ruffenach F, Page A, Ciura S, Kabashi E, et al. Loss of C9ORF72 impairs autophagy and synergizes with polyQ Ataxin-2 to induce motor neuron dysfunction and cell death. EMBO J. 2016;35:1276–1297. doi: 10.15252/embj.201593350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slager RE, Newton TL, Vlangos CN, Finucane B, Elsea SH. Mutations in RAI1 associated with Smith-Magenis syndrome. Nat Genet. 2003;33:466–468. doi: 10.1038/ng1126. [DOI] [PubMed] [Google Scholar]
- Snijder B, Sacher R, Ramo P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520–523. doi: 10.1038/nature08282. [DOI] [PubMed] [Google Scholar]
- Stoka V, Turk V, Turk B. Lysosomal cathepsins and their regulation in aging and neurodegeneration. Ageing Res Rev. 2016 doi: 10.1016/j.arr.2016.04.010. 2016(Apr 26), S1568-1637(16)30067-8. [DOI] [PubMed] [Google Scholar]
- Sullivan PM, Zhou X, Robins AM, Paushter DH, Kim D, Smolka MB, Hu F. The ALS/FTLD associated protein C9orf72 associates with SMCR8 and WDR41 to regulate the autophagy-lysosome pathway. Acta Neuropathol Commun. 2016;4:51. doi: 10.1186/s40478-016-0324-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen CC, Kang SA, Chang JW, Liu Q, Zhang J, Gao Y, Reichling LJ, Sim T, Sabatini DM, Gray NS. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284:8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsun ZY, Bar-Peled L, Chantranupong L, Zoncu R, Wang T, Kim C, Spooner E, Sabatini DM. The folliculin tumor suppressor is a GAP for the RagC/D GTPases that signal amino acid levels to mTORC1. Mol Cell. 2013;52:495–505. doi: 10.1016/j.molcel.2013.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Deerlin VM, Sleiman PM, Martinez-Lage M, Chen-Plotkin A, Wang LS, Graff-Radford NR, Dickson DW, Rademakers R, Boeve BF, Grossman M, et al. Common variants at 7p21 are associated with frontotemporal lobar degeneration with TDP-43 inclusions. Nat Genet. 2010;42:234–239. doi: 10.1038/ng.536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waite AJ, Baumer D, East S, Neal J, Morris HR, Ansorge O, Blake DJ. Reduced C9orf72 protein levels in frontal cortex of amyotrophic lateral sclerosis and frontotemporal degeneration brain with the C9ORF72 hexanucleotide repeat expansion. Neurobiol Aging. 2014;35 doi: 10.1016/j.neurobiolaging.2014.01.016. 1779.e1775–1779.e1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfson RL, Chantranupong L, Saxton RA, Shen K, Scaria SM, Cantor JR, Sabatini DM. Sestrin2 is a leucine sensor for the mTORC1 pathway. Science. 2016;351:43–48. doi: 10.1126/science.aab2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Bradley MJ, Cai Y, Kummel D, De La Cruz EM, Barr FA, Reinisch KM. Insights regarding guanine nucleotide exchange from the structure of a DENN-domain protein complexed with its Rab GTPase substrate. Proc Natl Acad Sci USA. 2011;108:18672–18677. doi: 10.1073/pnas.1110415108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, MacNair L, McLean J, McGoldrick P, McKeever P, Soleimani S, Keith J, Zinman L, Rogaeva E, Robertson J. C9orf72 isoforms in amyotrophic lateral sclerosis and frontotemporal lobar degeneration. Brain Res. 2016 doi: 10.1016/j.brainres.2016.04.062. 2016(Apr 29), S0006-8993(16)30311-0. [DOI] [PubMed] [Google Scholar]
- Yoshimura S, Gerondopoulos A, Linford A, Rigden DJ, Barr FA. Family-wide characterization of the DENN domain Rab GDP-GTP exchange factors. J Cell Biol. 2010;191:367–381. doi: 10.1083/jcb.201008051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Iyer LM, He F, Aravind L. Discovery of novel DENN proteins: implications for the evolution of eukaryotic intracellular membrane structures and human disease. Front Genet. 2012;3:283. doi: 10.3389/fgene.2012.00283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Donnelly CJ, Haeusler AR, Grima JC, Machamer JB, Steinwald P, Daley EL, Miller SJ, Cunningham KM, Vidensky S, et al. The C9orf72 repeat expansion disrupts nucleocytoplasmic transport. Nature. 2015;525:56–61. doi: 10.1038/nature14973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoncu R, Efeyan A, Sabatini DM. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol. 2011;12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.