The mechanisms that mediate the temporal and spatial recruitment of cell division factors to the spindle midzone and midbody remain unclear. Cell division is regulated by the conserved RNA-binding protein, ATX-2/Ataxin-2, which facilitates the targeting of ZEN-4 to the spindle midzone by mediating PAR-5.
Abstract
The spindle midzone harbors both microtubules and proteins necessary for furrow formation and the completion of cytokinesis. However, the mechanisms that mediate the temporal and spatial recruitment of cell division factors to the spindle midzone and midbody remain unclear. Here we describe a mechanism governed by the conserved RNA-binding protein ATX-2/Ataxin-2, which targets and maintains ZEN-4 at the spindle midzone. ATX-2 does this by regulating the amount of PAR-5 at mitotic structures, particularly the spindle, centrosomes, and midbody. Preventing ATX-2 function leads to elevated levels of PAR-5, enhanced chromatin and centrosome localization of PAR-5–GFP, and ultimately a reduction of ZEN-4–GFP at the spindle midzone. Codepletion of ATX-2 and PAR-5 rescued the localization of ZEN-4 at the spindle midzone, indicating that ATX-2 mediates the localization of ZEN-4 upstream of PAR-5. We provide the first direct evidence that ATX-2 is necessary for cytokinesis and suggest a model in which ATX-2 facilitates the targeting of ZEN-4 to the spindle midzone by mediating the posttranscriptional regulation of PAR-5.
INTRODUCTION
In animal cells, cytokinesis requires the dynamic interplay of microtubules, membrane, and actin to coordinate the positioning and formation of the cleavage furrow (Knoblich, 2010; Green et al., 2012; Mierzwa and Gerlich, 2014; D’Avino et al., 2015). The cleavage plane in many animal systems is dependent on a structure called the spindle midzone, which is formed from overlapping spindle microtubules in anaphase (Wheatley and Wang, 1996; Glotzer, 2004; Galli and van den Heuvel, 2008; Pacquelet et al., 2015; Landino and Ohi, 2016). The assembly and stability of the spindle midzone are regulated by a conserved complex called centralspindlin, which consists of KIF23/ZEN-4 and RacGAP/CYK-4. KIF23/ZEN-4 is a kinesin-6 family member that binds to a nonmotor subunit, RacGAP/CYK-4, with which it works in concert to bundle and anchor the midzone to the cleavage furrow (Mishima et al., 2002; Lekomtsev et al., 2012; White and Glotzer, 2012; Glotzer, 2013; Davies et al., 2015). Despite recent data on how centralspindlin is initially targeted (Douglas et al., 2010; Basant et al., 2015), the regulation of this complex and associated factors remains unclear. Although several genomic and proteomic screens have identified factors required for spindle assembly and cytokinesis (Goshima and Vale, 2003; Echard et al., 2004; Eggert et al., 2004; Skop et al., 2004; Neumann et al., 2010; Bonner et al., 2011; Moutinho-Pereira et al., 2013), many of these factors have not been fully characterized. One such factor, Ataxin-2/ATX-2, identified in a proteomic screen of isolated mammalian midbodies, is necessary for cytokinesis (Skop et al., 2004) and is associated with two important human diseases, spinocerebellar ataxia type II (SCA2) and amyotrophic lateral sclerosis (ALS; Pulst et al., 1996; Elden et al., 2010).
SCA2 and ALS are neurodegenerative diseases associated with varying numbers of polyglutamine (CAG) repeats in the RNA-binding protein Ataxin-2 (Pulst et al., 1996; Elden et al., 2010; Fischbeck and Pulst, 2011; Van Damme et al., 2011; Fittschen et al., 2015). SCA2 belongs to a family of diseases that includes Huntington’s disease, dentatorubral-pallidolusian atrophy, and spinal and bulbar muscular atrophy (Shao and Diamond, 2007). Individuals with SCA2 suffer degeneration and atrophy of Purkinje cells in the cerebellum, leading to defects in muscle coordination and balance (ataxia), as well as a shortened lifespan (Scherzed et al., 2012). Although human Ataxin-2 has been widely studied as a regulator of stress granules and processing bodies (Nonhoff et al., 2007; van de Loo et al., 2009; Kaehler et al., 2012; Yokoshi et al., 2014), its precise cellular role is unclear. Work in several model organisms has begun to shed light on our understanding of Ataxin-2 function in the cell (Satterfield et al., 2002; Ciosk et al., 2004; Maine et al., 2004; Kasumu et al., 2012), yet many questions remain.
Ataxin-2 belongs to a conserved family of cytoplasmic Like-SM (Lsm) proteins widely expressed in eukaryotes (Albrecht et al., 2004; Jimenez-Lopez and Guzman, 2014). Ataxin-2 contains three conserved RNA-binding domains: SM-ATX, LsmAD, and PAM2 (Albrecht et al., 2004; Yokoshi et al., 2014). Ataxin-2 binds and stabilizes mRNA directly or through a conserved association with poly(A)-binding protein cytoplasmic 1 (PABPC1), mediating the stability, transport, and translation of target mRNAs (Kozlov et al., 2001; Goss and Kleiman, 2013). Ataxin-2 positively regulates translation by directly binding to the cis-regulatory elements in the 3′ untranslated regions (UTRs) of its target mRNAs. Of the proteins encoded by the Ataxin-2 mRNA targets, ∼40% are involved in intracellular and protein transport, and >21% are associated with the cell cycle, mitosis, and meiosis, including Cyclin D1, KIF23/MKLP1, KIF4, 14-3-3σ/Stratifin, Rab-11, Cdc42, Par-6, RhoA, PRC1, and the entire ESCRT I-II-III complex (Yokoshi et al., 2014), suggesting that Ataxin-2 may be particularly critical during the cell cycle. However, the role of Ataxin-2 in mitosis has not been described.
On the cellular level, Ataxin-2 is necessary for the assembly of stress granules and P-bodies, both of which regulate mRNA expression, signaling, and cell fate (Leung and Sharp, 2006; Anderson and Kedersha, 2007; Nonhoff et al., 2007; Thomas et al., 2011; Kaehler et al., 2012; Ohshima et al., 2015). Ataxin-2 has been shown to localize to the Golgi and rough endoplasmic reticulum (RER; Huynh et al., 2003; van de Loo et al., 2009), yet the function of these associations is unclear. In Drosophila, DAtx2 binds directly to polyribosomes, and DAtx2 mutants exhibit neuronal and germline defects (Satterfield et al., 2002; Satterfield and Pallanck, 2006). In Caenorhabditis elegans, ATX-2 also functions in the germline by mediating the translation of germline-specific genes (Ciosk et al., 2004; Maine et al., 2004). Although these findings have provided insight into the cellular function of Ataxin-2/DAtx2/ATX-2, the precise molecular and cellular role of ATX-2 during mitosis has not been described.
In this study, we characterized the role of the homologue of Ataxin-2, ATX-2, in the early C. elegans embryo. Using genetics and live-cell imaging, we determined that cytokinesis requires ATX-2 to regulate a molecular mechanism necessary to target and maintain ZEN-4 to the spindle midzone. ATX-2 orchestrates the amount of PAR-5 on the mitotic spindle, centrosomes, chromatin, and midbody, and loss of ATX-2 leads to elevated PAR-5 protein levels. Elevated PAR-5 levels cause defects in the targeting of ZEN-4–green fluorescent protein (GFP) to the spindle midzone. When ATX-2 and PAR-5 were codepleted, the targeting of ZEN-4–GFP to the midzone was similar to that for control embryos, suggesting that ATX-2 functions upstream of PAR-5 in the spindle midzone assembly pathway (Douglas et al., 2010; Basant et al., 2015). Our data suggest a novel function for the RNA-binding protein, ATX-2, in mitosis by regulating PAR-5 and spindle midzone assembly.
RESULTS
ATX-2 is necessary for cytokinesis, the anaphase/telophase transition, and pronuclear size
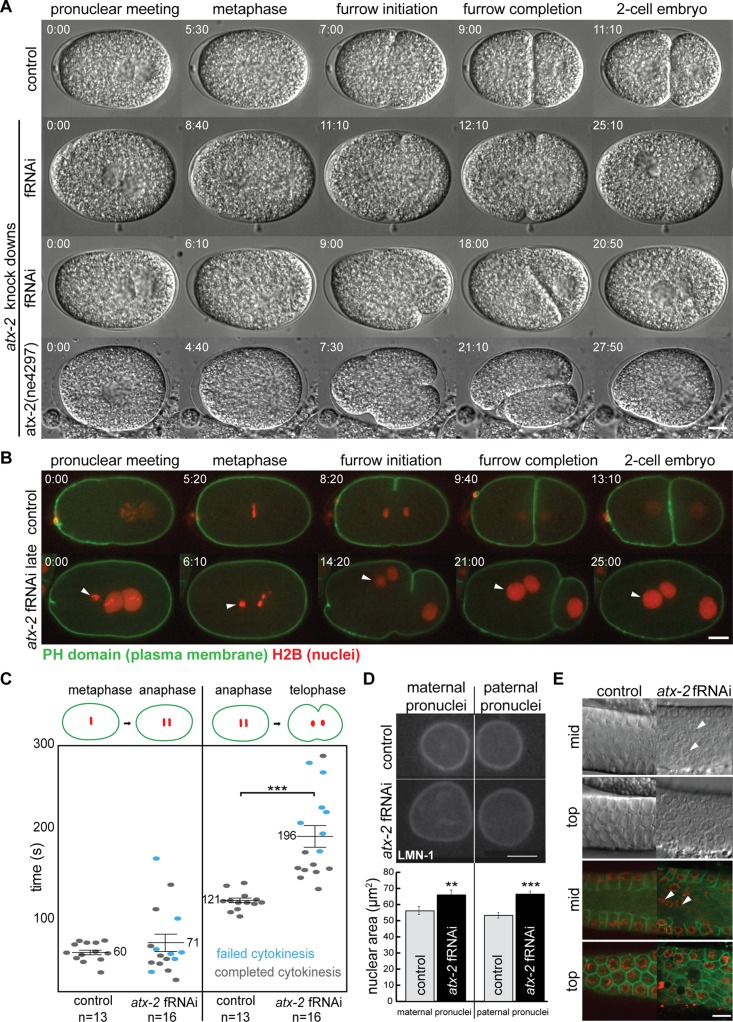
Ataxin-2 was identified in a proteomic screen of isolated mammalian midbodies, and depletion of the C. elegans orthologue, ATX-2, indicated a role in cell division (Skop et al., 2004), yet many questions remain. To determine the specific role for ATX-2 in mitosis, we compared atx-2 feeding RNA interference (fRNAi)–treated embryos and an atx-2 temperature-sensitive (ts) mutant strain (ne4297; Q919stop) with control embryos and observed the early embryonic divisions using in vivo videomicroscopy (Figure 1A and Supplemental Figure S1C). In control embryos, the cleavage furrow initiated and completed to form two blastomeres of unequal size (Figure 1A, control, and Supplemental Movie S1), whereas atx-2 fRNAi–treated embryos exhibited early (17%; n = 3 of 18) and late (28%; n = 5 of 18) cytokinesis failures (Figure 1A and Supplemental Movies S2 and S3). Cytokinesis failures occurred well into the second and third divisions in atx-2 fRNAi–treated embryos, resulting in multinucleate embryos (unpublished data). Similar embryonic phenotypes were observed in atx-2(ne4297) embryos at the restrictive temperature (22 or 24°C; n = 8 of 24; Figure 1A, atx-2(ne4297), and Supplemental Movie S4). ATX-2 protein levels were reduced in both atx-2 fRNAi and atx-2(ne4297) embryos (Supplemental Figure S1, A and B), and for the rest of our experiments, we used both RNAi feeding and atx-2 ts mutants to assay the loss of ATX-2.
FIGURE 1:
ATX-2 is necessary during mitosis. (A) Early and late cytokinesis defects are observed in atx-2 fRNAi–treated and atx-2(ne4297) embryos. DIC time-lapse images of control, atx-2 fRNAi-treated, and atx-2(ne4297) (24°C) embryos throughout the first cell division. In control embryos, the cleavage furrow initiates (7:00) and subsequently completes (9:00), resulting in a two-cell embryo (11:10). In atx-2 fRNAi–treated embryos, an early cytokinesis failure occurs when the cleavage furrow ingresses one-fourth of the way into the cell (fRNAi, top montage; 12:10) and subsequently retracts (fRNAi early; 25:10), and a late cytokinesis failure occurs when the furrow ingresses three-fourths or more into the embryo (fRNAi late; 18:00) before retraction (fRNAi, bottom montage; 20:50). In atx-2(ne4297) embryos, the cleavage furrow initiates (7:30) and subsequently retracts, resulting in a multinucleate embryo (27:50). Time in minutes:seconds is given relative to pronuclear meeting. Scale bar, 10 μm. (B) Membrane and DNA dynamics in control and atx-2 fRNAi–treated embryos coexpressing GFP–PH domain and mCherry–histone H2B. In control embryos, the daughter set of chromosomes segregates, the furrow initiates in anaphase (8:20), and the cleavage furrow completes, yielding a two-cell embryo (13:10). The extra nucleus in the atx-2 fRNAi embryo (0:00) indicates a polar body extrusion failure (arrowhead). A delay is observed between metaphase (6:10) and furrow initiation (14:20) as quantified in C. At 25:00 min, the furrow retracts, resulting in a multinucleate cell. Maternal and paternal pronuclei are enlarged in ATX-2–depleted embryos (quantified in D). (C) Quantification of the cell cycle delay observed in control and ATX-2 depleted embryos (B). Each point represents a measurement from a single embryo. Gray points represent embryos in which cytokinesis successfully completes; blue points represent embryos in which cytokinesis failed to complete. Solid lines denote the mean. Results are mean ± SEM. Asterisks indicate statistical significance for compared data (p < 0.001; Welch’s t test). (D) Graph quantifies pronuclear size (area in μm2) in control (n = 10) and atx-2 fRNAi (n = 14) embryos. Control (top) and atx-2 fRNA–treated (bottom) embryos coexpressing mCherry–histone H2B and YFP–LMN-1 just before pronuclear meeting. This size increase is seen in both maternal (left) and paternal (right) pronuclei. Results are mean ± SEM. Asterisks represent level of significance for compared data. **p < 0.01, ***p < 0.001 (Welch’s t test). Scale bars, 5 μm. (E) GFP–PH domain, mCherry–histone H2B, and control or atx-2 fRNAi–treated germlines. Top and mid focal plane germline images taken by DIC and confocal microscopy in N2 worms and worms coexpressing GFP–PH domain and mCherry–histone H2B. Control germlines have T-shaped membranes at the mid focal plane and hexagonal cells at the top focal plane, with each cell housing one nucleus within. In atx-2 fRNAi–treated germlines, cytokinesis defects disrupt the membrane morphology, causing nuclei to be displaced into the rachis (arrowheads). Scale bar, 10 μm.
To monitor plasma membrane dynamics during cell division, we depleted ATX-2 using fRNAi in worms coexpressing GFP–PH domain and mCherry–histone H2B (Green et al., 2008). Under control conditions, two polar bodies were extruded to the anterior of the embryo after meiosis II (Figure 1B and Supplemental Movie S5). After pronuclear meeting, the chromosomes aligned at the metaphase plate, segregated to opposite poles in anaphase, and remained condensed until telophase (Figure 1B and Supplemental Movie S5). The cleavage furrow initiated in anaphase and completed in cytokinesis, yielding two blastomeres of unequal size (Figure 1B). In 33% of the atx-2 fRNAi–treated embryos, an extra body of histone H2B–GFP was observed in the anterior (n = 6 of 18; Figure 1B and Supplemental Movie S6), likely from polar body extrusion failures in meiosis II. This compacted DNA migrated along with the maternal pronuclei yet never mixed with the maternal or paternal pronuclei during mitosis (Figure 1B and Supplemental Movie S6). These polar body extrusion defects were similar to those observed in ZEN-4 and CAR-1 mutant embryos (Raich et al., 1998; Audhya et al., 2005; Squirrell et al., 2006). After metaphase, the chromosomes segregated in anaphase, and the cleavage furrow initiated and appeared to complete before subsequently retracting, leading to blastomeres with multiple nuclei in 44% of the ATX-2–depleted embryos (n = 7 of 16; Figure 1B). In some embryos, the chromatin decondensed before furrow initiation, suggesting a delay in furrow initiation, precocious decondensation of chromosomes, or a cell cycle defect (Figure 1B). The furrow initiation delay mimics phenotypes observed in cells treated with Taxol, in which inhibition of microtubule dynamics resulted in defects in the timing and completion of the furrow (Shannon et al., 2005; Strickland et al., 2005). No obvious plasma membrane defects were observed, except for failures in meiotic and mitotic cytokinesis.
Given that we observed a cell cycle delay in atx-2 fRNAi–treated embryos, we sought to determine and quantify the time between the chromatin phases in mitosis in control and atx-2 fRNAi–treated embryos (Figure 1C). In control embryos, the transition from metaphase to anaphase onset took an average of 60 ± 3 s, and the transition from anaphase to telophase took an average of 121 ± 3 s (n = 13). We defined the metaphase-to-anaphase transition as the period when the metaphase plate first forms until anaphase onset, and we defined the anaphase-to-telophase transition as the period from anaphase onset until the initial decondensation of the chromosomes. In atx-2 fRNAi–treated embryos, the metaphase-to-anaphase transition averaged 71 ± 10 s, which was not statistically significant (values are mean ± SEM; p > 0.05; Figure 1C). However, atx-2 fRNAi–treated embryos averaged 196 ± 13 s to complete the anaphase-to-telophase transition, which was statistically significant (n = 16; p < 0.00005; Figure 1C). The delay during the anaphase-to-telophase transition was significant regardless of a successful completion (gray points) or cytokinesis failure (blue points), suggesting a specific role for ATX-2 during the anaphase-to-telophase transition. This specific cell cycle delay is similar to those observed in Staufen-depleted U2OS cells (Boulay et al., 2014), suggesting a role in mediating cell division factors during this time. Of note, both Staufen and Ataxin-2 bind mRNAs encoding proteins that mediate aspects of the cell cycle (LeGendre et al., 2013; Boulay et al., 2014; Yokoshi et al., 2014), suggesting that the function of ATX-2 may be temporally coordinated.
In atx-2 fRNAi–treated embryos, we also observed that the pronuclei appeared larger (Figure 1, B and D). To quantify pronuclear size, we used a strain that coexpressed mCherry–histone H2B and yellow fluorescent protein (YFP)–LMN-1 (Lamin; Portier et al., 2007) to visualize the DNA and nuclear envelope, respectively (Figure 1D). We then compared the pronuclear area (in μm2) of the maternal and paternal pronuclei in control (n = 10) and atx-2 fRNAi–treated (n = 14) embryos. On average, the maternal pronuclei in atx-2 fRNAi–treated embryos were 9.82 μm2 larger (p = 0.009; control, 56.12 μm2; atx-2 fRNAi, 65.94μm2), and the paternal pronuclei were 13.18 μm2 larger (p = 0.00009; control, 53.25 μm2, atx-2 fRNAi, 66.43 μm2), than controls. These results support observations that ATX-2 functions in meiosis (Ciosk et al., 2004; Maine et al., 2004) by contributing to chromatin remodeling and nuclear envelope assembly in some way.
Depletion of ATX-2 perturbs germline cytokinesis
ATX-2 is necessary for proper germline development, including germline proliferation and oogenesis. As a result, when ATX-2 is depleted, the C. elegans germline shrinks and becomes masculinized, causing sterility (Ciosk et al., 2004; Maine et al., 2004). To determine whether defective germline cytokinesis failures are the cause of the sterility observed, we monitored membrane dynamics in control and atx-2 fRNAi–treated worms coexpressing GFP–PH domain and mCherry–histone H2B (Green et al., 2008). At the mid focal plane in control animals, the germline nuclei were separated by a T-shaped membrane surrounding the rachis (common cytoplasm). At the top focal plane, the germline is organized into hexagonal cells with one nucleus each (Skop et al., 2001; Green et al., 2008; Pazdernik and Schedl, 2013; Figure 1E). At the mid focal plane in atx-2 fRNAi–treated worms, the T-shaped membranes were replaced with Y-shaped, I-shaped, or looped membranes, with stray nuclei localized within the rachis (Figure 1E, white arrowheads). At the top focal plane, the hexagonal membrane pattern was abnormal (Figure 1E). ATX-2 plays an important role in germline membrane structure and may contribute to the sterility observed (Ciosk et al., 2004; Maine et al., 2004).
ATX-2 localizes to cytoplasmic granules, centrosomes, the spindle, the spindle midzone, and midbody
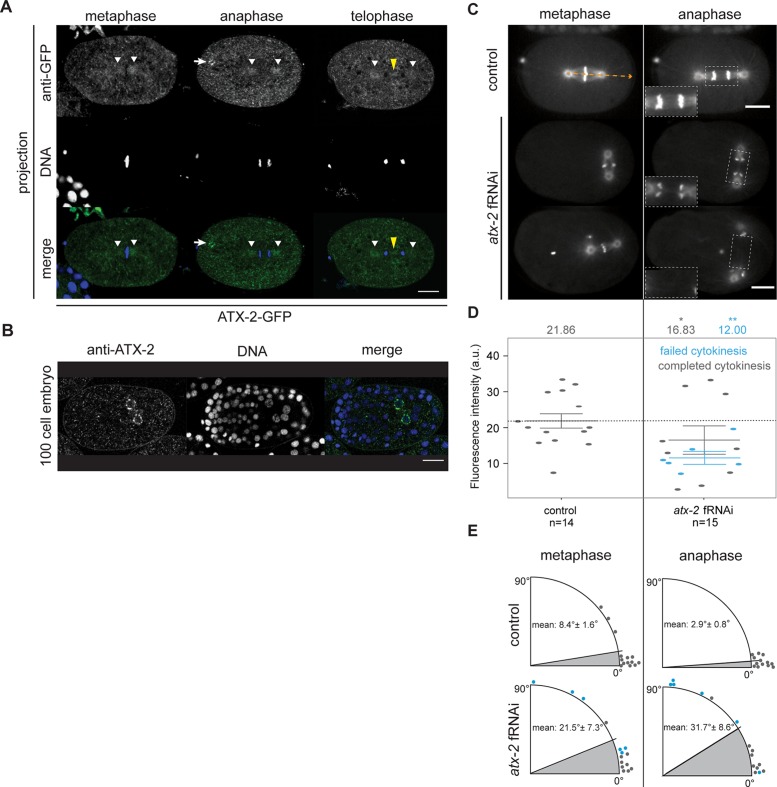
To determine the temporal and spatial localization of ATX-2 in the C. elegans embryo, we constructed an ATX-2-GFP strain (MAD63; see Materials and Methods). The GFP signal was specific, as we detected a reduction of the ATX-2-GFP signal in atx-2 fRNAi–treated embryos (Supplemental Figure S2A). In mitosis, ATX-2-GFP decorated dynamic puncta of various sizes throughout the cytoplasm of early embryos (Supplemental Figure S2A and Supplemental Movie S7). These puncta were similar in size to P-granules, except that they did not polarize to the posterior pole after pronuclear meeting, suggesting that they may be RNA stress granules, a previously attributed function for Ataxin-2 in mammalian cells (Nonhoff et al., 2007; Kaehler et al., 2012). To determine the precise localization pattern during mitosis, we fixed, immunostained, and projected 15 optical 0.1-μm sections per embryo of ATX-2-GFP–expressing embryos with anti-GFP antibody. In metaphase-stage embryos, we detected an enrichment of ATX-2 puncta adjacent to the metaphase plate (white arrowheads), around the centrosomes in anaphase (white arrowheads), on the spindle midzone, and at the midbody during telophase (yellow arrowheads; Figure 2A). During anaphase, we observed enrichment of ATX-2-GFP on a round structure in the anterior of the embryo (white arrow), likely an organelle or vesicle-like structure 2 μm in size. In late-stage (∼100 cell) embryos, ATX-2 localized to cytoplasmic puncta, as well as around the periphery of the germline precursor nuclei, Z2 and Z3 cells (Figure 2B). Overall we determined that ATX-2 localizes to nonpolarizing cytoplasmic granules, around germline precursor nuclei, and to spindle-associated structures. These data indicate that ATX-2 may function not only in RNA-associated puncta or granules as previously reported (Nonhoff et al., 2007; Kaehler et al., 2012; Figley et al., 2014), but also specifically on the spindle during mitosis.
FIGURE 2:
ATX-2 localizes to mitotic structures and is required for spindle orientation and midzone dynamics. (A) Fixed ATX-2–GFP embryos in metaphase, anaphase, and telophase and stained with anti-GFP antibody (green) and DAPI (blue). ATX-2–GFP localized to cytoplasmic puncta, 2-μm spherical aggregates (white arrows), centrosomes (white arrowheads), and the spindle midzone (yellow arrowheads). Images are average projections of 0.1-μm Z-stacks spanning 1.5 μm. Scale bar, 10 μm. (B) Fixed 100-cell stage N2 embryo stained with anti–ATX-2 antibody (green) and DAPI (blue). ATX-2 localized to cytoplasmic puncta and Z2 and Z3 cells. Image is a single focal plane. Scale bar, 10 μm. (C) Microtubule dynamics in control and atx-2 fRNAi–treated embryos coexpressing TBB-2–GFP and GFP–HIS-11 in metaphase and anaphase. Insets, 1.5×-magnified view of spindle midzone microtubules in dashed boxes. Control embryos contain discrete midzone microtubules, whereas atx-2 fRNAi–treated embryos show faint or no visible midzone microtubules. Orange arrow shows angle measurement relative to horizontal (0º). (D) Quantification of TBB-2–GFP fluorescence intensity in control and atx-2 fRNAi–treated embryos. Each point represents a fluorescence intensity measurement from a single embryo. Gray points represent embryos in which cytokinesis successfully completed; blue points represent embryos in which cytokinesis failed to complete. Reduced TBB-2–GFP fluorescence intensity at the spindle midzone was observed in ATX-2–depleted embryos. Solid lines denote the mean. Results are mean ± SEM. Asterisks represent level of significance for compared data. *p < 0.05, **p < 0.01 (Welch’s t test). (E) Quantification of spindle displacement from 0º measured at metaphase and anaphase in control (n = 14) and atx-2 fRNAi–treated (n = 15) embryos. Gray points represent embryos in which cytokinesis successfully completed; blue dots represent embryos in which cytokinesis failed to complete. In control embryos, the spindle is oriented along the anterior–posterior axis, with average displacement varying between 3 and 8º. In atx-2 fRNAi–treated embryos, the average displacement increases to between 21 and 32º. Extreme spindle orientation defects correlate with cytokinesis defects in atx-2 fRNAi–treated embryos.
ATX-2 is necessary for P-granule localization
Ataxin-2 is important for the assembly and dynamics of RNA stress granules and P-bodies (Nonhoff et al., 2007; Kaehler et al., 2012). To determine whether ATX-2 regulates RNA granules in a similar way, we monitored the dynamics of RNA-rich P-granules, using PGL-1–GFP (Cheeks et al., 2004). In control embryos, by pronuclear meeting, P-granules of various sizes segregated to the posterior of the cell (Supplemental Figure S2D, 0:00 min) and at the two-cell stage remained in the P1 cell (Supplemental Figure S2D, 14:00 min, and Supplemental Movie S15; Strome, 2005). Any P-granules remaining in the AB cell of the embryo were rapidly degraded (Strome and Wood, 1983; Hird et al., 1996). P-granules in atx-2 fRNAi–treated embryos did not segregate properly to the posterior of the embryo by pronuclear meeting and were larger, and several granules persisted in the AB cell at the two-cell stage when cell division completed (n = 12 of 17; Supplemental Figure S2D, middle, and Supplemental Movie S16) or in the anterior of the cell when cytokinesis failed (n = 3 of 3; Supplemental Figure S2D, bottom, and Supplemental Movie S17). These data suggest a failure in the segregation to the posterior, a failure to degrade P-granules in the anterior, or a defect in RNA granule dynamics. This observation is consistent with the loss of known ATX-2 interacting proteins PAB-1 and CGH-1 (Updike and Strome, 2009; Ko et al., 2013). This observation is consistent with a disruption to cell polarity (Watts et al., 1996; Gotta et al., 2001) that we observed in ATX-2–depleted embryos (Supplemental Figure S2C). Here, atx-2 fRNAi–treated embryos coexpressing the opposing polarity domains of PAR-6 (Supplemental Figure S2C, green) and PAR-2 (red) failed to occupy their normal cortical localization. Compared to control embryos, PAR-2 occupied 8% more of the cortical cell length (Supplemental Figure S2C and Supplemental Movies S13 and S14). Taken together, the results show that ATX-2 appears to be important for regulating P-granule dynamics; further study is necessary, however, to determine the specific relationship between ATX-2, PGL-1, and P-granule dynamics.
ATX-2 depletion leads to spindle orientation and midzone defects
Successful asymmetric cell division requires proper spindle midzone microtubule dynamics and alignment of the mitotic spindle along the axis of polarity (Hyman and White, 1987; Lu and Johnston, 2013; Landino and Ohi, 2016). Given that we observed cytokinesis defects, we sought to determine whether ATX-2 is necessary for microtubule dynamics during the cell cycle. To do this, we monitored microtubules in the early embryo using a strain coexpressing GFP-tagged tubulin and histones (TBB-2–GFP; HIS-11–GFP; Skop et al., 2004). In control embryos, the spindle assembled along the anterior–posterior axis and, during anaphase, elongated to form the spindle midzone (Figure 2C and Supplemental Movie S8). In some ATX-2–depleted embryos, the spindle aligned properly and the midzones appeared similar to control embryos (n = 5 of 15; unpublished data). However, in most cases when ATX-2 was depleted, we observed spindle orientation defects in both metaphase (n = 4 of 15) and anaphase (n = 9 of 15), as well as a reduction or loss of spindle midzone microtubules in late anaphase (n = 6 of 15; Figure 2, C–E, insets, and Supplemental Movies S9 and S10). In control embryos, TBB-2–GFP fluorescence intensity during anaphase was on average 21.86 arbitrary units (a.u.; n = 14; measured by drawing a region of interest [ROI] around the spindle midzone in anaphase). In ATX-2–depleted embryos, TBB-2–GFP fluorescence intensity at the spindle midzone was significantly reduced to 12.00 a.u. (p = 0.002) when cytokinesis failed, and there was no significant reduction in midzone fluorescence intensity (16.83 a.u.) when cytokinesis completed (n = 15; Figure 2, C and D). Most of the ATX-2–depleted embryos that failed in cytokinesis had reduced spindle midzone fluorescence (n = 5 of 6), but not all ATX-2–depleted embryos that had reduced spindle midzone fluorescence failed in cytokinesis. The reduced spindle midzone microtubules were not rescued after codepletion of ATX-2 and PAR-5 (Supplemental Figure S2, E and F). The midzone defects were similar to those observed in car-1 fRNAi–treated embryos (Audhya et al., 2005; Squirrell et al., 2006). Cytokinesis failures in atx-2 fRNAi–treated embryos were not the result of a disruption in actin at the furrow or in the cortex (Supplemental Figure S2B and Supplemental Movies S11 and S12), suggesting that the primary roles of ATX-2 may be in spindle alignment and midzone assembly.
ATX-2 targets ZEN-4 to the spindle midzone but does not regulate ZEN-4 RNA or protein levels
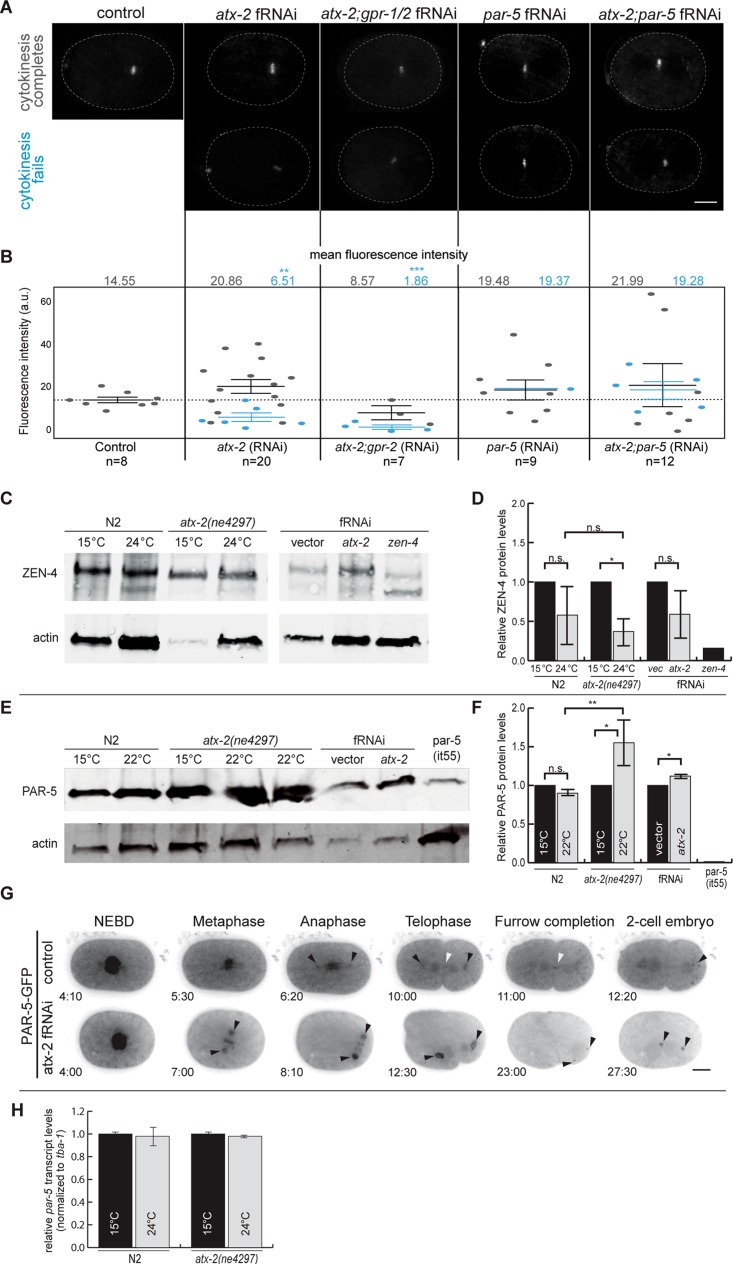
Given that the spindle midzone was weak or absent in many of our ATX-2–depleted embryos, we sought to determine whether ZEN-4 properly localized during mitosis. In control embryos, ZEN-4–GFP first appeared at the spindle midzone in anaphase and persisted until the end of cytokinesis (Figure 3A). In control embryos, ZEN-4–GFP fluorescence intensity during anaphase was on average 14.55 a.u. (n = 8; measured by drawing an ROI around the spindle midzone in anaphase). In ATX-2–depleted embryos, ZEN-4–GFP fluorescence intensity ranged between 1.43 and 40.72 a.u. at the spindle midzone (n = 20; Figure 3, A and B). Ectopic ZEN-4–GFP puncta (yellow boxes) were also observed in the posterior of the embryo near or associated with the centrosomes (Supplemental Figure S3A). When cytokinesis failed, ZEN-4–GFP intensity levels were reduced to 6.51 a.u., compared with control levels of 14.55 a.u. (n = 6 of 20; Figure 3B). When cytokinesis did not fail, ZEN-4–GFP intensity levels varied greatly (3.82–40.72 a.u.) and averaged 20.86 a.u. (n = 14 of 20; Figure 3B), suggesting that ATX-2 may function to modulate the levels of ZEN-4 protein at the spindle midzone. Furthermore, these data suggest that in the absence of ATX-2, ZEN-4–GFP localization to the spindle midzone was disrupted. Expression of CYK-4–GFP, the C. elegans homologue of RacGAP and a member of the centralspindlin complex, at the spindle midzone was not affected by ATX-2 depletion (Supplemental Figure S3, B and C). In atx-2(ne4297) germlines, ZEN-4 also failed to localize to the T-shaped membrane structure, often extending into or completely across the rachis (Supplemental Figure S3D). Our data suggest that ATX-2 is necessary to maintain proper ZEN-4 at the spindle midzone and germline furrows and could also mediate ZEN-4 levels during mitosis.
FIGURE 3:
ATX-2 regulates ZEN-4 and PAR-5 localization during mitosis and posttranscriptionally regulates PAR-5 expression. (A) Visualization of ZEN-4-GFP localization in control and atx-2 fRNAi–, atx-2; gpr-2 fRNAi–, par-5 fRNAi–, and atx-2; par-5 fRNAi–treated embryos. In control embryos, ZEN-4 localizes to the spindle midzone. In atx-2 fRNAi–and atx-2; gpr-2 fRNAi–treated embryos, ZEN-4 expression at the midzone is reduced. Images are maximum projections of 0.5-μm Z-stacks spanning 1.5 μm. Scale bars, 10 μm. (B) Quantification of ZEN-4–GFP fluorescence intensity in control and atx-2 fRNAi–, and atx-2; gpr-2 fRNAi–, par-5 fRNAi–, and atx-2; par-5 fRNAi–treated embryos. Each point represents a fluorescence intensity measurement from a single embryo. Gray points represent embryos in which cytokinesis successfully completed; blue points represent embryos in which cytokinesis failed to complete. When cytokinesis fails in atx-2 fRNAi– and atx-2; gpr-2 fRNAi–treated embryos, ZEN-4–GFP expression is reduced at the spindle midzone. Codepletion of ATX-2 and PAR-5 rescues the ATX-2 single-depletion phenotype. Solid lines denote the mean. Results are mean ± SEM. (C, D) Quantitative Western blot analysis of relative protein expression levels of ZEN-4 in control and atx-2(ne4297) worms grown at 15 or 24°C and atx-2 fRNAi–treated embryos from N2 at similar conditions. Protein levels were normalized to actin. ZEN-4 protein expression levels were inconsistent in control temperature shifts from 15 to 24°C, and we cannot conclude that ZEN-4 expression is altered with ATX-2 depletion. As an antibody control, zen-4 fRNAi–treated embryos were probed with ZEN-4 antibody. Results are mean ± SEM. Asterisks represent level of significance for compared data. *p < 0.05. (E, F) Quantitative Western blot analysis of relative protein expression levels of PAR-5 in control and atx-2(ne4297) worms grown at 15 or 22°C and atx-2 fRNAi–treated embryos. Protein levels were normalized to actin. In ATX-2–depleted embryos, PAR-5 protein expression levels were increased 1.12- to 1.55-fold above controls. As an antibody control, par-5(it55) mutant embryos were probed with PAR-5 antibody. Results are mean ± SEM. Asterisks represent level of significance for compared data. *p < 0.05, **p < 0.01. (G) PAR-5-GFP dynamics in control and atx-2 fRNAi–treated embryos. In control embryos, PAR-5-GFP localizes to nuclei (4:10), the metaphase plate (5:30), centrosomes (10:00, black arrowheads), the spindle midzone, faintly along the cleavage furrow (11:00, white arrowhead), and posterior cytoplasmic granules (12:20). In atx-2 fRNAi–treated embryos, PAR-5–GFP accumulates at the metaphase plate (7:00) and the centrosomes (7:00, black arrowheads). PAR-5–GFP then associates with the spindle midzone and midbody and faintly at the cleavage furrow (22:30). During mitosis, PAR-5–GFP also associates with cytoplasmic granules in the anterior and posterior of the embryos (27:30). Time in minutes:seconds is given relative to pronuclear meeting. Scale bar, 10 μm. (H) qPCR analysis of the relative transcript expression levels of par-5 in control and atx-2(ne4297) worms grown at 15 or 24°C. mRNA levels were normalized to tba-1 (α-tubulin), and 24°C samples were compared with their respective 15°C control sample. par-5 mRNA expression is unchanged when ATX-2 is depleted. Results are mean ± SEM.
To determine whether this might be the case, we quantified ZEN-4 using a quantitative Western blot. We loaded equal amounts of embryo lysate from control (fRNAi empty vector) and atx-2 fRNAi–treated embryos. In addition, we compared lysates from N2 and atx-2(ne4297) embryos at permissive (15°C) and restrictive (24°C) temperatures. We then probed with an anti–ZEN-4 antibody (gift from MIchael Glotzer, University of Chicago; Figure 3C) and measured ZEN-4 protein expression. In both N2 and atx-2(ne4297) embryos at the restrictive temperature (24°C), ZEN-4 protein levels were reduced twofold to threefold compared with embryos at the permissive temperature (15°C), suggesting that ZEN-4 protein expression is unstable when the temperature is increased (Figure 3D). We observed a slight decrease in ZEN-4 protein expression between the atx-2(ne4297) embryos at 15 and 24°C (p = 0.2; n = 3). However, there was no significant difference between N2 and atx-2(ne4297) embryos at 24°C. Furthermore, atx-2 fRNAi–treated embryos did not exhibit a significant decrease in ZEN-4 protein expression compared with control (Figure 3D). Taking the results together, we cannot conclude that ATX-2 depletion causes a decrease in ZEN-4 protein expression. To determine whether ZEN-4 transcript levels were affected, we performed real-time quantitative PCR (RT-qPCR). Here, zen-4 transcript levels were unaffected after ATX-2 depletion (Supplemental Figure S3E), suggesting that ATX-2 does not regulate either ZEN-4 mRNA or protein directly. The reduced levels of ZEN-4-GFP at the spindle midzone could be a secondary phenotype resulting from decreased midzone microtubules or premature lengthening of the mitotic spindle. To address the latter possibility, we inhibited premature lengthening of the mitotic spindle by depleting GPR-1/2, a protein necessary for the spindle-pulling forces (Srinivasan et al., 2003; Audhya et al., 2005). We codepleted GPR-1/2 and ATX-2 using fRNAi and measured ZEN-4–GFP fluorescence intensity in live embryos. Codepletion limited the premature spindle elongation phenotype observed in ATX-2–depleted embryos but did not rescue the reduction of ZEN-4–GFP fluorescence at the spindle midzone (Figure 3, A and B), indicating that ATX-2 functions to modulate ZEN-4 targeting to the spindle midzone.
ATX-2 regulates PAR-5 posttranscriptionally
To determine how ATX-2 might function in targeting of ZEN-4 to the spindle midzone, we selected PAR-5, the homologue of mammalian 14-3-3σ, which has been shown to coordinate mitotic translation and cytokinesis (Wilker et al., 2007). PAR-5 is also a negative regulator of ZEN-4 and plays a role in the activation of ZEN-4 during mitosis (Douglas et al., 2010; Basant et al., 2015). Finally, mammalian 14-3-3σ (PAR-5) and KIF23 (ZEN-4) mRNAs are direct Ataxin-2 targets (Yokoshi et al., 2014). Given these previous data and our work, we hypothesized that ATX-2 could regulate the expression of PAR-5 in some way. To monitor PAR-5 protein expression, we quantified PAR-5 protein levels using a quantitative Western blot. Lysates from control (empty vector), atx-2 fRNAi–treated, N2, and atx-2(ne4297) (at permissive [15°C] and restrictive temperatures [22°C]) embryos were probed with anti–PAR-5 antibody (gift from Andy Golden, National Institutes of Health; Morton et al., 2002). We determined that depletion of ATX-2, either by fRNAi or in the atx-2(ne4297) mutant embryos at 22°C, led to a 1.12- to 1.55-fold increase in PAR-5 protein expression above controls (p = 0.047 [RNAi] and 0.022 [atx-2(ne4297)]; n = 3; Figure 3, E and F). These data revealed that PAR-5 protein levels are regulated by ATX-2 and may explain the unstable targeting of ZEN-4-GFP to the midzone in ATX-2–depleted embryos (Figure 3A).
If ATX-2 regulates PAR-5 protein levels necessary for targeting ZEN-4 to the midzone, we predict that ZEN-4 localization could be rescued by depleting both ATX-2 and PAR-5. To test this, we codepleted ATX-2 and PAR-5 using fRNAi in ZEN-4-GFP–expressing embryos and measured ZEN-4–GFP fluorescence intensity levels at the spindle midzone (Figure 3, A and B). As a control, ZEN-4–GFP fluorescence intensity levels were measured in ATX-2– and PAR-5–depleted embryos. Here, ZEN-4–GFP expression was similar to that for control embryos (Figure 3, A and B). These data suggest that in the absence of both ATX-2 and PAR-5, ZEN-4–GFP is able to localize to the midzone, revealing that ATX-2 likely functions to modulate PAR-5 during mitosis in some way.
Given that Ataxin-2 directly binds to 14-3-3σ mRNA in mammalian cells (Yokoshi et al., 2014), we sought to determine whether par-5 mRNA levels are regulated by ATX-2. To monitor par-5 expression, we quantified par-5 mRNA levels using RT-qPCR on cDNA from control (N2), atx-2(ne4297) (permissive, 15°C), and atx-2(ne4297) (restrictive, 24°C) worms. The par-5 expression levels were normalized to tba-1(tubulin). We determined that par-5 expression levels were unchanged after ATX-2 depletion compared with control (Figure 3H), confirming that the loss of ATX-2 specifically alters PAR-5 protein levels (Figure 3, E and F). We conclude that ATX-2 modulates PAR-5 posttranscriptionally.
To investigate how ATX-2 might regulate PAR-5, we sought to determine whether PAR-5 and ATX-2 shared similar localization patterns during mitosis. PAR-5 localization was originally described as cytoplasmic in early embryos (Morton et al., 2002), but the cell cycle pattern had not been described. Given this, we used PAR-5-GFP–expressing embryos (Mikl and Cowan, 2014). To our surprise, PAR-5-GFP localized around the metaphase plate, to the centrosomes (black arrowheads), to the spindle midzone, to the midbody (white arrowheads), and faintly along the cleavage furrow in telophase (Figure 3G and Supplemental Movie S18), very similarly to ATX-2. In ATX-2–depleted embryos, we observed a decrease in PAR-5–GFP around the metaphase plate and a dramatic increase in PAR-5:GFP at the centrosomes (black arrowheads; Figure 3G and Supplemental Movie S19). During anaphase, PAR-5 maintained its localization to the centrosomes and chromatin, and in late anaphase, PAR-5 puncta dispersed into the cytoplasm, appearing to come from the spindle midzone (Supplemental Movie S19). In telophase, PAR-5–GFP accumulated at the periphery of the centrosomes and less was observed around the chromatin. In sum, our data suggest that during mitosis, ATX-2 mediates the levels and the temporal and spatial localization of PAR-5 (Figures 3 and 4). Ultimately, our data place ATX-2 upstream of PAR-5 in a molecular pathway necessary to target and maintain ZEN-4 to the spindle midzone. We provided the first direct evidence that ATX-2 is necessary for cytokinesis by mediating the posttranscriptional regulation of PAR-5.
FIGURE 4:
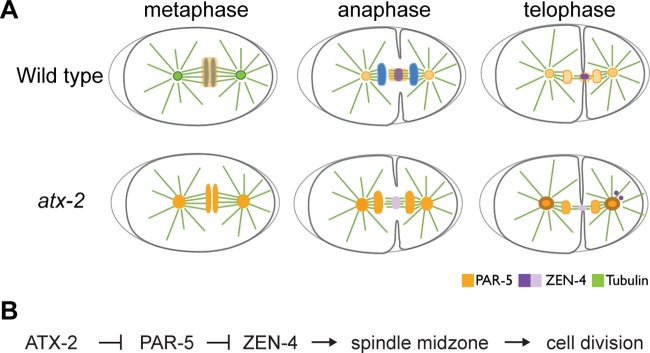
Mitosis requires ATX-2. (A) Model summarizing the role of ATX-2 during mitosis. In control embryos, PAR-5 (orange) localizes around the metaphase plate and faintly to the centrosomes. During anaphase and telophase, PAR-5 localizes to the spindle midzone, where it maintains ZEN-4 (purple). In ATX-2–depleted embryos, PAR-5 localization accumulates at the metaphase plate and centrosomes. During anaphase and telophase, PAR-5 is lost from the spindle midzone, which leads to the loss of ZEN-4 from the midzone. ZEN-4 puncta localize ectopically near the centrosomes in the posterior of the embryo. (B) Our data suggest that ATX-2 functions upstream of PAR-5 in the targeting and maintaining of ZEN-4 at the spindle midzone.
DISCUSSION
The work presented here provides a novel function for the conserved RNA-binding protein ATX-2 in mitosis. We show that cytokinesis requires ATX-2 by regulating a novel pathway necessary to target ZEN-4 to the spindle midzone. ATX-2 does this by properly targeting PAR-5 to mitotic structures and functioning as a molecular rheostat for PAR-5, a known inhibitor of ZEN-4 (Douglas et al., 2010; Basant et al., 2015). Loss of ATX-2 leads to increased levels of PAR-5, which lead to a reduction of ZEN-4 at the midzone. This loss of ZEN-4 at the midzone is rescued upon codepletion of ATX-2 and PAR-5, suggesting that ATX-2 functions upstream of PAR-5. Our results suggest that ATX-2 promotes cell division by facilitating the targeting of ZEN-4 to the spindle midzone through the posttranscriptional regulation of PAR-5.
ATX-2 plays an important role in cytokinesis
Mammalian Ataxin-2 localizes to the Golgi, RER, and RNA stress granules and plays a role in RNA transport and processing (Huynh et al., 2003; van de Loo et al., 2009; Yokoshi et al., 2014), yet the precise cellular role for Ataxin-2 is unclear. To further understand the cell biology of Ataxin-2, we uncovered additional roles for ATX-2 by studying its role in developing C. elegans embryos. Here loss of ATX-2 led to cell cycle delays, cytokinesis failures, and disrupted spindle midzone microtubule bundles. Furthermore, ATX-2 localized to cytoplasmic puncta (similar to RNA granules) and aggregated around mitotic structures, including the spindle, centrosomes, the spindle midzone, and the midbody (Figure 2A). Although human Ataxin-2 has been widely studied as a regulator of stress granules and processing bodies (Nonhoff et al., 2007; van de Loo et al., 2009; Kaehler et al., 2012; Yokoshi et al., 2014), we found a new and important role for ATX-2 in mitosis.
Stable targeting of ZEN-4–GFP to the midzone requires ATX-2
Cytokinesis and spindle midzone assembly require centralspindlin to bundle and stabilize microtubules within the midzone (Raich et al., 1998; Severson et al., 2000; Mishima et al., 2002; Zhu et al., 2005). Our data show, for the first time, that ATX-2 is important for maintaining midzone function. The loss of ATX-2 mimics ZEN-4 mutants in which there is either reduced or loss of midzone microtubules (Raich et al., 1998; Mishima et al., 2002). ATX-2 does not directly regulate the expression of ZEN-4 protein or zen-4 mRNA expression (Figure 3D and Supplemental Figure S3E), and ZEN-4-GFP is often ectopically localized at or near centrosomes (Supplemental Figure S3A). These data indicate that ATX-2 mediates a mechanism that targets and maintains ZEN-4 at the midzone.
Targeting of ZEN-4 to the spindle midzone is mediated by AIR-2 and PAR-5, which activate or repress ZEN-4 localization to the midzone, respectively (Douglas et al., 2010; Basant et al., 2015). We found a new mechanism mediated by ATX-2 that regulates ZEN-4 localization to the midzone by modulating PAR-5 protein levels and activity (Figures 3 and 4). Given that mammalian Ataxin-2 also directly binds to the transcript of the zen-4 homologue KIF23 (Yokoshi et al., 2014), we hypothesize that ATX-2 mediates ZEN-4 localization by proper targeting or transport of its mRNA to midzone microtubules, where it is locally activated. This mechanism for ZEN-4 activity would be novel for any kinesin (Nislow et al., 1992; Hirokawa, 1998, 2009; Mishima et al., 2002; Miki et al., 2005). In neurons, mRNAs are often transported out to microtubule bundles at synapses, where they are locally activated and function (Sutton and Schuman, 2006; Jung and Holt, 2011). This transport coupled with local translation enables neurons to selectively alter the protein composition of each postsynaptic site (Falley et al., 2009; Puthanveettil, 2013) and may be analogous to the events that occur during cell division, given the many similarities and functions of the cytoskeletal structures involved. Known asymmetric cell division genes, including RhoA, Par3, and β-actin, are transported as mRNAs out to the synapse, where they are translated (Kanai et al., 2004; Wu et al., 2005; Leung et al., 2006; Hengst et al., 2009), suggesting a possibility that other cell division genes could be regulated in a similar manner during mitosis. Understanding how KIF23/ZEN-4 mRNAs and associated RNA-binding proteins like Ataxin-2/ATX-2 are regulated during mitosis will likely reveal mechanisms that mediate the temporal and spatial patterning of cell division factors and provide insight into the biological function of Ataxin-2 that is relevant to spinocerebellar ataxia and neurodegenerative disease.
ATX-2 regulates PAR-5 expression and localization during mitosis
Successful cell division requires a stable spindle midzone (Wheatley and Wang, 1996; Bringmann and Hyman, 2005; Khmelinskii and Schiebel, 2008; Lewellyn et al., 2011; White and Glotzer, 2012; Landino and Ohi, 2016). Recent work from the Glotzer lab suggests that PAR-5 may play an important role in midzone assembly (Basant et al., 2015); however, many questions remain. Here our study found that PAR-5 is not only important for division but also is temporally and spatially regulated during mitosis. Once believed to be cytoplasmically localized (Morton et al., 2002; Mikl and Cowan, 2014), we found a novel pattern for PAR-5 in mitosis, in which PAR-5 is enriched around the centrosomes, metaphase plate, midzone, and midbody (Figure 3G). Loss of ATX-2 leads to a dramatic shift in PAR-5–GFP localization primarily to the centrosomes and around the metaphase plate. In mammalian tissue culture cells, the PAR-5 orthologue, 14-3-3σ, is necessary for cytokinesis by regulating the switch between cap-dependent and -independent translation, and yet the localization and regulation of 14-3-3σ was not described (Wilker et al., 2007). We found that the mitotic localization of PAR-5 is not only dependent on ATX-2 but also is important for its expression and activity.
The role of RNA-binding proteins in mitosis
It has become evident that RNA-binding proteins play an important role in the regulation of target mRNAs throughout development (Bandziulis et al., 1989; Colegrove-Otero et al., 2005; Gerstberger et al., 2014; Ye and Blelloch, 2014; Brinegar and Cooper, 2016). However, how mRNAs are regulated during mitosis remains unclear. Once believed to cease in mitosis, cap-independent translation plays a significant role (Wilker et al., 2007). How translation is both spatially and temporally regulated during spindle assembly and cytokinesis is an important step in understanding how cells coordinate the activity and dynamics of proteins necessary for cell division. It remains to be determined how ATX-2/Ataxin-2 regulates the function and dynamics of mRNA targets during cell division. Uncovering these mechanisms might contribute to our understanding of neurodegenerative disease.
MATERIALS AND METHODS
Worm strains and ATX-2–GFP construction
Strains used in this study, include N2 (control; Brenner, 1974), OD95 (GFP-PHPLC1delta1;mCherry-HIS-58; Green et al., 2008), OD139 (mCherry–histone H2B;YFP-LMN-1; Portier et al., 2007), TY3558 (TBB-2-GFP;GFP-HIS-11; Skop et al., 2004), WM210 (atx-2(ts); ne4297; Q919stop; isolated by ENU mutagenesis), SS629 (GFP-PGL-1; Cheeks et al., 2004), TH120 (GFP-PAR-2;mCherry-PAR-6; Schonegg et al., 2007), KK299 (par-5(it55); Morton et al., 2002), MG170 (ZEN-4-GFP; Kaitna et al., 2000), UE50 (GFP-PAR-5; Mikl and Cowan, 2014), PF100 (MOE-1-GFP; Motegi et al., 2006), and WH279 (CYK-4-GFP; Verbrugghe and White, 2004). All strains were maintained at 24°C, with the exception of, atx-2(ne4297), which was maintained at 15°C and shifted to 22 or 24°C to deplete ATX-2 (see Western blot, Supplemental Figure S1B). Some strains were provided by the Caenorhabditis Genetics Center (St. Paul, MN). The ATX-2–GFP strain MAD63 (dqSi1[Pmex-5::ATX-2a::GFP::tbb-2 3′ UTR + unc-119(+)] II; unc-119(ed3) III) was generated as follows: cDNA of the a isoform of atx-2 (atx-2a) was amplified from pT7β (generously provided by Rafal Ciosk, Friedrich Miescher Institute for Biomedical Research) and then inserted into pCR®8/GW/TOPO (Invitrogen, Carlsbad, CA). The primers used to amplify the atx-2a were sense primer, 5′-ATGTCAACACCAACGGGTCTTCCTGCG-3′, and antisense primer, 5′-CGGGGGGCTCTGAGAGTGCTG-3′. The resulting atx-2a vector was then combined with pJA252 (Pmex-5 w/o stop; Addgene plasmid 21512; Zeiser et al., 2011), pJA256 ((Gly)5Ala::gfp::tbb-2 3′UTR; Addgene plasmid 21509; Zeiser et al., 2011), and pCFJ150 (pDESTttTi5605[R4-R3], II; Addgene plasmid 19329; Frokjaer-Jensen et al., 2008) in a Gateway LR Clonase reaction (Invitrogen) to make the final construct, pMMG07, Pmex-5-/atx-2a/(Gly)5Ala::gfp::tbb-2 3′UTR. The resulting construct was injected into the Mos1 insertion strain EG6699 (Frokjaer-Jensen et al., 2008). The injection mix contained pMMG07 (22.5 ng/μl), pCFJ601 (50 ng/μl; Addgene plasmid 34874; Frokjaer-Jensen et al., 2012), pMA122 (10 ng/μl; Addgene plasmid 34873; Frokjaer-Jensen et al., 2012), and the coinjection markers pGH8 (10 ng/μl; Addgene plasmid 19359; Frokjaer-Jensen et al., 2008), pCFJ90 (2.5 ng/μl; Addgene plasmid 19327; Frokjaer-Jensen et al., 2008), and pCFJ104 (5 ng/μl; Addgene plasmid 19328; Frokjaer-Jensen et al., 2008). Rescued worms were screened for fluorescence as described previously (Frokjaer-Jensen et al., 2008, 2012). MAD63 rescued atx-2(ne4297).
RNA interference
To knock down ATX-2, ZEN-4, GPR-1/2, and PAR-5 in vivo, the pL4440 feeding vectors specific to these genes were obtained from the Ahringer feeding RNAi library and sequence verified as previously described (Timmons and Fire, 1998; Kamath et al., 2001, 2003; Kamath and Ahringer, 2003; Ahringer, 2006). RNAi experiments were performed at 24°C for 16–22 h.
Live imaging
Embryo dissection was performed in Shelton’s Growth Medium, and embryos were mounted on 2% agar pads (made with egg salt buffer) in a drop of Shelton’s Growth Medium (Shelton and Bowerman, 1996). The pads were covered with a 22 mm × 22 mm coverslip and sealed in place with Vaseline. We found that this modified mount abrogates the osmotic defect observed in atx-2 fRNAi or atx-2(ne4297) embryos while allowing for optimal imaging during the first two cell divisions. Time-lapse images were taken every 10 s using a 200M inverted Axioscope microscope (Carl Zeiss, Oberkochen, Germany) equipped with a spinning-disk confocal scan head (QLC100; VisiTech, Sunderland, United Kingdom), an Orca 285 differential interference contrast (DIC) camera (Hamamatsu, Japan), and an Orca ER camera (Hamamatsu, Japan). The DIC and Orca ER cameras were operated through MetaMorph software (version 7.7.11.0; Molecular Devices, Sunnyvale, CA). Image rotating and cropping was performed using Fiji (ImageJ) software (Schindelin et al., 2012). Fluorescence intensity of ZEN-4–GFP was obtained by drawing a region of interest of equal area around the spindle midzone of each embryo. These regions of interest were then used to measure the average pixel intensity. Background fluorescence obtained from a nonfluorescent region in the embryo anterior was subtracted.
Immunostaining
Immunostaining was performed using a methanol–4% formaldehyde fix as described previously (Skop and White, 1998) using the primary antibodies anti–ATX-2 mouse monoclonal (1:100; Ciosk et al., 2004), anti-GFP rabbit polyclonal (1:100; ab6556; Abcam, Cambridge, United Kingdom), anti–ZEN-4 rabbit serum (1:150; gift from Michael Glotzer, University of Chicago), and anti-actin [C4] mouse monoclonal (1:200; 691002; MP Biomedicals, Santa Ana, CA) diluted in PBSB (1× phosphate-buffered saline [PBS], 1% bovine serum albumin) and incubated overnight at 4°C. Unbound primary antibodies were removed by three washes for 5 min each with 1× PBS plus 0.5% Tween-20. Secondary antibodies used over the course of several different experiments were applied as follows: Alexa Fluor 488 anti-mouse (A-11001; Molecular Probes, Eugene, OR), Alexa Fluor 568 anti-mouse (A-11004; Molecular Probes), and Alexa Fluor 488 anti-rabbit (A-11008; Molecular Probes), with all secondary antibodies diluted 1:200 in PBSB. After 2 h of incubation in the dark at ambient conditions, unbound secondary antibodies were removed as described. The fixed and stained embryos were mounted with 8 μl of Vectashield with 4′,6-diamidino-2-phenylindole (DAPI; H-1200; Vector Laboratories, Burlingame, CA). Confocal imaging was performed on a departmental Zeiss 510 Confocal LSM operated with ZEN software (Carl Zeiss).
Western blot
Total SDS-soluble protein was extracted from control RNAi, atx-2 RNAi, zen-4 RNAi, N2 15°C, N2 22°C, N2 24°C, atx-2(ne4297) 15°C, atx-2(ne4297) 22°C, atx-2(ne4297) 24°C, and par-5(it55) embryos. Protein concentration was determined by a modified Lowry assay (Dulley and Grieve, 1975). For the horseradish peroxidase (HRP) Western blot, 60 μg of protein per lane was separated on a 4–15% SDS–PAGE minigel (TGX; Bio-Rad, Hercules, CA) and then transferred to an Immun-Blot polyvinylidene fluoride membrane (Bio-Rad) using standard conditions for semidry blotting. The blots were blocked with PBS plus 0.1% Tween-20 containing 4% skim milk powder for 1 h at ambient conditions. Membranes were probed with primary anti–ATX-2 antibody (mAbP1G12; Ciosk et al., 2004) at a dilution of 1:150 and mouse anti-actin monoclonal Clone 4 (MP Biomedicals) antibody at a dilution of 1:20,000 for 16–18 h at 4°C. Primary antibody binding was detected with anti-mouse HRP-conjugated antibody (1:5000; GE Amersham, Little Chalfont, United Kingdom). After washing with PBST three times (5 min each) at ambient conditions, blots were developed with ECL Plus (Amersham). For the quantitative Western blot, 20 μg of protein per lane was separated on a 4–15% SDS–PAGE minigel (TGX) and transferred to a Millipore Immobilon-FL membrane using standard conditions for semidry blotting. The blot was blocked in Odyssey Tris buffered saline–based blocking solution (LI-COR Biosciences, Lincoln, NE) for 1 h at room temperature before being probed with primary antibodies for 16–18 h at 4°C. The immunoblot was visualized with infrared fluorescence IRDye secondary antibodies at a dilution of 1:15,000 (LI-COR Biosciences) using an Odyssey imager (LI-COR Biosciences). Polyclonal rabbit anti–ZEN-4 serum (a gift from M. Glotzer) at a dilution of 1:1000 or polyclonal rabbit anti–PAR-5 antibody (a gift from A. Golden) at a dilution of 1:2000 and mouse anti-actin monoclonal Clone 4 antibody (MP Biomedicals) at a dilution of 1:750 were used as the primary antibodies. Blots were repeated three times. Quantification of protein levels, after normalization to the actin loading control, was performed on Image Studio Software (LI-COR Biosciences).
RNA isolation and cDNA synthesis for RT-qPCR
Total RNA was isolated from four independent worm samples of N2 15°C, N2 24°C, atx-2(ne4297) 15°C, and atx-2(ne4297) 24°C synchronized adult populations using TRIzol Reagent (15596-026; Invitrogen). RNA samples were treated with TURBO DNase (AM2238; Life Technologies, Carlsbad, CA). RNA quantity and purity were measured using a NanoDrop spectrophotometer. cDNA was made from 5 μg of total RNA using the SuperScript III First-Strand Synthesis System for RT-PCR (Invitrogen) kit and the Oligo(dT)20 primer. To confirm whether there was DNA contamination in each RNA sample, a no-transcriptase control was made and verified by RT-qPCR. For use in RT-qPCRs, cDNA was diluted 100-fold. cDNA preparations for each RNA sample were stored at −20°C.
RT-qPCR
Reference gene (act-1, cdc-42, tba-1) primers were obtained from previously published data (Zhang et al., 2012). The zen-4 (AGACGCCAGAAATGGAGTATTG forward, CGCTTGACGAGACTTCAACA reverse), atx-2 (AGCAGCCTCAGCAAGTATTC forward, CAGATCGCTCGCCCATAAG reverse), and par-5 (CCGTTCATCGTGGAGAGTTATC forward, TCTTGGCAGATGTCGTTGAG reverse) primers were designed using the RealTime PCR design tool (IDT). tba-1 was chosen as the reference gene using NormFinder. RT-qPCR was performed in triplicate 10 μl/well reactions (5 μl of SYBR Green [Roche, Basel, Switzerland], 2 μl of cDNA, 0.5 μl of forward primer, 0.5 μl of reverse primer, and 2 μl of RNase-free water) on the cDNA samples prepared as described. All reactions were analyzed with a Roche Light Cycler 480. Target transcripts were normalized to tba-1 to obtain a relative expression ratio of each target mRNA, and each 24°C sample was compared with its respective 15°C control. RT-qPCR protocols followed the Minimum Information for Publication of Quantitative Real-Time PCR Experiments Guidelines (Bustin et al., 2009).
Statistical analysis
To determine the statistical significance of quantified values, control and ATX-2–depleted embryos were compared using unpaired t tests. First, the variances of the two sample values were compared using an F test. In our case, all variances were found to be unequal between control and ATX-2–depleted embryos. Therefore we compared the groups using a Welch’s t test (unpaired, two-tailed). The statistical calculations were performed using Excel (Microsoft, Redmond, WA). The statistical significance cutoff was p < 0.05.
Supplementary Material
Acknowledgments
We thank Craig Mello, Rafal Ciosk, Keith Blackwell, James Priess, Michael Glotzer, Ken Kemphues, Carrie Cowan, Martin Mikl, Timothy Gomez, and Andy Golden for the generous gifts of reagents and guidance. We thank Robert D. White, Kevin O’Connell, William Sullivan, Judith Kimble, Phil Anderson, Mi Hye Song, Masanori Mishima, Christopher Nicchitta, Eleanor Maine, Kevin Eliceiri, Bill Bement, Mark Burkard, Kate O’Connor-Giles, Francisco Pelegri, Aki Ikeda, the Xin Sun lab, Gregory Fischer, Celeste Eno, and Kathryn VanDenHeuvel for suggestions and advice. This work was supported by grants to A.R.S. from the National Science Foundation (MCB 115800), the National Institutes of Health (K01 HL092583), and the Caenorhabditis Genetics Center (St. Paul, MN), which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440).
Abbreviations used:
- fRNAi
feeding RNA interference
- GFP
green fluorescent protein
- SCA2
spinocerebellar ataxia type II.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-04-0219) on August 24, 2016.
REFERENCES
- Ahringer J. Reverse genetics. 2006. In: WormBook, ed. The C. elegans Research Community, WormBook, doi/10.1895/wormbook.1.47.1, http://www.wormbook.org/ (accessed January 2012)
- Albrecht M, Golatta M, Wullner U, Lengauer T. Structural and functional analysis of ataxin-2 and ataxin-3. Eur J Biochem. 2004;271:3155–3170. doi: 10.1111/j.1432-1033.2004.04245.x. [DOI] [PubMed] [Google Scholar]
- Anderson P, Kedersha N. On again, off again: the SRC-3 transcriptional coactivator moonlights as a translational corepressor. Mol Cell. 2007;25:796–797. doi: 10.1016/j.molcel.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Audhya A, Hyndman F, McLeod IX, Maddox AS, Yates JR, 3rd, Desai A, Oegema K. A complex containing the Sm protein CAR-1 and the RNA helicase CGH-1 is required for embryonic cytokinesis in Caenorhabditis elegans. J Cell Biol. 2005;171:267–279. doi: 10.1083/jcb.200506124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandziulis RJ, Swanson MS, Dreyfuss G. RNA-binding proteins as developmental regulators. Genes Dev. 1989;3:431–437. doi: 10.1101/gad.3.4.431. [DOI] [PubMed] [Google Scholar]
- Basant A, Lekomtsev S, Tse YC, Zhang D, Longhini KM, Petronczki M, Glotzer M. Aurora B kinase promotes cytokinesis by inducing centralspindlin oligomers that associate with the plasma membrane. Dev Cell. 2015;33:204–215. doi: 10.1016/j.devcel.2015.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner MK, Poole DS, Xu T, Sarkeshik A, Yates JR, 3rd, Skop AR. Mitotic spindle proteomics in Chinese hamster ovary cells. PLoS One. 2011;6:e20489. doi: 10.1371/journal.pone.0020489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulay K, Ghram M, Viranaicken W, Trepanier V, Mollet S, Frechina C, DesGroseillers L. Cell cycle-dependent regulation of the RNA-binding protein Staufen1. Nucleic Acids Res. 2014;42:7867–7883. doi: 10.1093/nar/gku506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. The genetics of Caenorhabditis elegans. Genetics. 1974;77:71–94. doi: 10.1093/genetics/77.1.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinegar AE, Cooper TA. Roles for RNA-binding proteins in development and disease. Brain Res. 2016;1647 doi: 10.1016/j.brainres.2016.02.050. doi: 10.1016/j.brainres.2016.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- Bustin SA, Benes V, Garson JA, Hellemans J, Huggett J, Kubista M, Mueller R, Nolan T, Pfaffl MW, Shipley GL, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Cheeks RJ, Canman JC, Gabriel WN, Meyer N, Strome S, Goldstein B. C. elegans PAR proteins function by mobilizing and stabilizing asymmetrically localized protein complexes. Curr Biol. 2004;14:851–862. doi: 10.1016/j.cub.2004.05.022. [DOI] [PubMed] [Google Scholar]
- Ciosk R, DePalma M, Priess JR. ATX-2, the C. elegans ortholog of ataxin 2, functions in translational regulation in the germline. Development. 2004;131:4831–4841. doi: 10.1242/dev.01352. [DOI] [PubMed] [Google Scholar]
- Colegrove-Otero LJ, Minshall N, Standart N. RNA-binding proteins in early development. Crit Rev Biochem Mol Biol. 2005;40:21–73. doi: 10.1080/10409230590918612. [DOI] [PubMed] [Google Scholar]
- Davies T, Kodera N, Kaminski Schierle GS, Rees E, Erdelyi M, Kaminski CF, Ando T, Mishima M. CYK4 promotes antiparallel microtubule bundling by optimizing MKLP1 neck conformation. PLoS Biol. 2015;13:e1002121. doi: 10.1371/journal.pbio.1002121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Avino PP, Giansanti MG, Petronczki M. Cytokinesis in animal cells. Cold Spring Harb Perspect Biol. 2015;7:a015834. doi: 10.1101/cshperspect.a015834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas ME, Davies T, Joseph N, Mishima M. Aurora B and 14-3-3 coordinately regulate clustering of centralspindlin during cytokinesis. Curr Biol. 2010;20:927–933. doi: 10.1016/j.cub.2010.03.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulley JR, Grieve PA. A simple technique for eliminating interference by detergents in the Lowry method of protein determination. Anal Biochem. 1975;64:136–141. doi: 10.1016/0003-2697(75)90415-7. [DOI] [PubMed] [Google Scholar]
- Echard A, Hickson GR, Foley E, O’Farrell PH. Terminal cytokinesis events uncovered after an RNAi screen. Curr Biol. 2004;14:1685–1693. doi: 10.1016/j.cub.2004.08.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elden AC, Kim HJ, Hart MP, Chen-Plotkin AS, Johnson BS, Fang X, Armakola M, Geser F, Greene R, Lu MM, et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature. 2010;466:1069–1075. doi: 10.1038/nature09320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falley K, Schutt J, Iglauer P, Menke K, Maas C, Kneussel M, Kindler S, Wouters FS, Richter D, Kreienkamp HJ. Shank1 mRNA: dendritic transport by kinesin and translational control by the 5’untranslated region. Traffic. 2009;10:844–857. doi: 10.1111/j.1600-0854.2009.00912.x. [DOI] [PubMed] [Google Scholar]
- Figley MD, Bieri G, Kolaitis RM, Taylor JP, Gitler AD. Profilin 1 associates with stress granules and ALS-linked mutations alter stress granule dynamics. J Neurosci. 2014;34:8083–8097. doi: 10.1523/JNEUROSCI.0543-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischbeck KH, Pulst SM. Amyotrophic lateral sclerosis and spinocerebellar ataxia 2. Neurology. 2011;76:2050–2051. doi: 10.1212/WNL.0b013e31821f4498. [DOI] [PubMed] [Google Scholar]
- Fittschen M, Lastres-Becker I, Halbach MV, Damrath E, Gispert S, Azizov M, Walter M, Muller S, Auburger G. Genetic ablation of ataxin-2 increases several global translation factors in their transcript abundance but decreases translation rate. Neurogenetics. 2015;16:181–192. doi: 10.1007/s10048-015-0441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Ailion M, Jorgensen EM. Improved Mos1-mediated transgenesis in C. elegans. Nat Methods. 2012;9:117–118. doi: 10.1038/nmeth.1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frokjaer-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet. 2008;40:1375–1383. doi: 10.1038/ng.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli M, van den Heuvel S. Determination of the cleavage plane in early C. elegans embryos. Annu Rev Genet. 2008;42:389–411. doi: 10.1146/annurev.genet.40.110405.090523. [DOI] [PubMed] [Google Scholar]
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15:829–845. doi: 10.1038/nrg3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. Cleavage furrow positioning. J Cell Biol. 2004;164:347–351. doi: 10.1083/jcb.200310112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M. Cytokinesis: centralspindlin moonlights as a membrane anchor. Curr Biol. 2013;23:R145–R147. doi: 10.1016/j.cub.2013.01.006. [DOI] [PubMed] [Google Scholar]
- Goshima G, Vale RD. The roles of microtubule-based motor proteins in mitosis: comprehensive RNAi analysis in the Drosophila S2 cell line. J Cell Biol. 2003;162:1003–1016. doi: 10.1083/jcb.200303022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goss DJ, Kleiman FE. Poly(A) binding proteins: are they all created equal. Wiley Interdiscip Rev RNA. 2013;4:167–179. doi: 10.1002/wrna.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gotta M, Abraham MC, Ahringer J. CDC-42 controls early cell polarity and spindle orientation in C. elegans. Cur Biol. 2001;11:482–488. doi: 10.1016/s0960-9822(01)00142-7. [DOI] [PubMed] [Google Scholar]
- Green RA, Audhya A, Pozniakovsky A, Dammermann A, Pemble H, Monen J, Portier N, Hyman A, Desai A, Oegema K. Expression and imaging of fluorescent proteins in the C. elegans gonad and early embryo. Methods Cell Biol. 2008;85:179–218. doi: 10.1016/S0091-679X(08)85009-1. [DOI] [PubMed] [Google Scholar]
- Green RA, Paluch E, Oegema K. Cytokinesis in animal cells. Annu Rev Cell Dev Biol. 2012;28:29–58. doi: 10.1146/annurev-cellbio-101011-155718. [DOI] [PubMed] [Google Scholar]
- Hengst U, Deglincerti A, Kim HJ, Jeon NL, Jaffrey SR. Axonal elongation triggered by stimulus-induced local translation of a polarity complex protein. Nat Cell Biol. 2009;11:1024–1030. doi: 10.1038/ncb1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hird SN, Paulsen JE, Strome S. Segregation of germ granules in living Caenorhabditis elegans embryos: cell-type-specific mechanisms for cytoplasmic localisation. Development. 1996;122:1303–1312. doi: 10.1242/dev.122.4.1303. [DOI] [PubMed] [Google Scholar]
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. doi: 10.1126/science.279.5350.519. [DOI] [PubMed] [Google Scholar]
- Hirokawa N, Noda Y, Tanaka Y, Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. doi: 10.1038/nrm2774. [DOI] [PubMed] [Google Scholar]
- Huynh DP, Yang HT, Vakharia H, Nguyen D, Pulst SM. Expansion of the polyQ repeat in ataxin-2 alters its Golgi localization, disrupts the Golgi complex and causes cell death. Hum Mol Genet. 2003;12:1485–1496. doi: 10.1093/hmg/ddg175. [DOI] [PubMed] [Google Scholar]
- Hyman AA, White JG. Determination of cell division axes in the early embryogenesis of Caenorhabditis elegans. J Cell Biol. 1987;105:2123–2135. doi: 10.1083/jcb.105.5.2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Lopez D, Guzman P. Insights into the evolution and domain structure of Ataxin-2 proteins across eukaryotes. BMC Res Notes. 2014;7:453. doi: 10.1186/1756-0500-7-453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H, Holt CE. Local translation of mRNAs in neural development. Wiley Interdiscip Rev RNA. 2011;2:153–165. doi: 10.1002/wrna.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaehler C, Isensee J, Nonhoff U, Terrey M, Hucho T, Lehrach H, Krobitsch S. Ataxin-2-like is a regulator of stress granules and processing bodies. PLoS One. 2012;7:e50134. doi: 10.1371/journal.pone.0050134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaitna S, Mendoza M, Jantsch-Plunger V, Glotzer M. Incenp and an aurora-like kinase form a complex essential for chromosome segregation and efficient completion of cytokinesis. Curr Biol. 2000;10:1172–1181. doi: 10.1016/s0960-9822(00)00721-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2:RESEARCH0002. doi: 10.1186/gb-2000-2-1-research0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- Kasumu AW, Liang X, Egorova P, Vorontsova D, Bezprozvanny I. Chronic suppression of inositol 1,4,5-triphosphate receptor-mediated calcium signaling in cerebellar purkinje cells alleviates pathological phenotype in spinocerebellar ataxia 2 mice. J Neurosci. 2012;32:12786–12796. doi: 10.1523/JNEUROSCI.1643-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A, Schiebel E. Assembling the spindle midzone in the right place at the right time. Cell Cycle. 2008;7:283–286. doi: 10.4161/cc.7.3.5349. [DOI] [PubMed] [Google Scholar]
- Knoblich JA. Asymmetric cell division: recent developments and their implications for tumour biology. Nat Rev Mol Cell Biol. 2010;11:849–860. doi: 10.1038/nrm3010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko S, Kawasaki I, Shim YH. PAB-1, a Caenorhabditis elegans poly(A)-binding protein, regulates mRNA metabolism in germline by interacting with CGH-1 and CAR-1. PLoS One. 2013;8:e84798. doi: 10.1371/journal.pone.0084798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlov G, Trempe JF, Khaleghpour K, Kahvejian A, Ekiel I, Gehring K. Structure and function of the C-terminal PABC domain of human poly(A)-binding protein. Proc Natl Acad Sci USA. 2001;98:4409–4413. doi: 10.1073/pnas.071024998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landino J, Ohi R. The timing of midzone stabilization during cytokinesis depends on myosin II activity and an interaction between INCENP and actin. Curr Biol. 2016;26:698–706. doi: 10.1016/j.cub.2016.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LeGendre JB, Campbell ZT, Kroll-Conner P, Anderson P, Kimble J, Wickens M. RNA targets and specificity of Staufen, a double-stranded RNA-binding protein in Caenorhabditis elegans. J Biol Chem. 2013;288:2532–2545. doi: 10.1074/jbc.M112.397349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lekomtsev S, Su KC, Pye VE, Blight K, Sundaramoorthy S, Takaki T, Collinson LM, Cherepanov P, Divecha N, Petronczki M. Centralspindlin links the mitotic spindle to the plasma membrane during cytokinesis. Nature. 2012;492:276–279. doi: 10.1038/nature11773. [DOI] [PubMed] [Google Scholar]
- Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb Symp Quant Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
- Leung KM, van Horck FP, Lin AC, Allison R, Standart N, Holt CE. Asymmetrical beta-actin mRNA translation in growth cones mediates attractive turning to netrin-1. Nat Neurosci. 2006;9:1247–1256. doi: 10.1038/nn1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewellyn L, Carvalho A, Desai A, Maddox AS, Oegema K. The chromosomal passenger complex and centralspindlin independently contribute to contractile ring assembly. J Cell Biol. 2011;193:155–169. doi: 10.1083/jcb.201008138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu MS, Johnston CA. Molecular pathways regulating mitotic spindle orientation in animal cells. Development. 2013;140:1843–1856. doi: 10.1242/dev.087627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maine EM, Hansen D, Springer D, Vought VE. Caenorhabditis elegans atx-2 promotes germline proliferation and the oocyte fate. Genetics. 2004;168:817–830. doi: 10.1534/genetics.104.029355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mierzwa B, Gerlich DW. Cytokinetic abscission: molecular mechanisms and temporal control. Dev Cell. 2014;31:525–538. doi: 10.1016/j.devcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Miki H, Okada Y, Hirokawa N. Analysis of the kinesin superfamily: insights into structure and function. Trends Cell Biol. 2005;15:467–476. doi: 10.1016/j.tcb.2005.07.006. [DOI] [PubMed] [Google Scholar]
- Mikl M, Cowan CR. Alternative 3’ UTR selection controls PAR-5 homeostasis and cell polarity in C. elegans embryos. Cell Rep. 2014;8:1380–1390. doi: 10.1016/j.celrep.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Mishima M, Kaitna S, Glotzer M. Central spindle assembly and cytokinesis require a kinesin-like protein/RhoGAP complex with microtubule bundling activity. Dev Cell. 2002;2:41–54. doi: 10.1016/s1534-5807(01)00110-1. [DOI] [PubMed] [Google Scholar]
- Morton DG, Shakes DC, Nugent S, Dichoso D, Wang W, Golden A, Kemphues KJ. The Caenorhabditis elegans par-5 gene encodes a 14–3-3 protein required for cellular asymmetry in the early embryo. Dev Biol. 2002;241:47–58. doi: 10.1006/dbio.2001.0489. [DOI] [PubMed] [Google Scholar]
- Motegi F, Velarde NV, Piano F, Sugimoto A. Two phases of astral microtubule activity during cytokinesis in C. elegans embryos. Dev Cell. 2006;10:509–520. doi: 10.1016/j.devcel.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Moutinho-Pereira S, Stuurman N, Afonso O, Hornsveld M, Aguiar P, Goshima G, Vale RD, Maiato H. Genes involved in centrosome-independent mitotic spindle assembly in Drosophila S2 cells. Proc Natl Acad Sci USA. 2013;110:19808–19813. doi: 10.1073/pnas.1320013110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nislow C, Lombillo VA, Kuriyama R, McIntosh JR. A plus-end-directed motor enzyme that moves antiparallel microtubules in vitro localizes to the interzone of mitotic spindles. Nature. 1992;359:543–547. doi: 10.1038/359543a0. [DOI] [PubMed] [Google Scholar]
- Nonhoff U, Ralser M, Welzel F, Piccini I, Balzereit D, Yaspo ML, Lehrach H, Krobitsch S. Ataxin-2 interacts with the DEAD/H-box RNA helicase DDX6 and interferes with P-bodies and stress granules. Mol Biol Cell. 2007;18:1385–1396. doi: 10.1091/mbc.E06-12-1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohshima D, Arimoto-Matsuzaki K, Tomida T, Takekawa M, Ichikawa K. Spatio-temporal dynamics and mechanisms of stress granule assembly. PLoS Comput Biol. 2015;11:e1004326. doi: 10.1371/journal.pcbi.1004326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pacquelet A, Uhart P, Tassan JP, Michaux G. PAR-4 and anillin regulate myosin to coordinate spindle and furrow position during asymmetric division. J Cell Biol. 2015;210:1085–1099. doi: 10.1083/jcb.201503006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pazdernik N, Schedl T. Introduction to germ cell development in Caenorhabditis elegans. Adv Exp Med Biol. 2013;757:1–16. doi: 10.1007/978-1-4614-4015-4_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulst SM, Nechiporuk A, Nechiporuk T, Gispert S, Chen XN, Lopes-Cendes I, Pearlman S, Starkman S, Orozco-Diaz G, Lunkes A, et al. Moderate expansion of a normally biallelic trinucleotide repeat in spinocerebellar ataxia type 2. Nat Genet. 1996;14:269–276. doi: 10.1038/ng1196-269. [DOI] [PubMed] [Google Scholar]
- Puthanveettil SV. RNA transport and long-term memory storage. RNA Biol. 2013;10:1765–1770. doi: 10.4161/rna.27391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raich WB, Moran AN, Rothman JH, Hardin J. Cytokinesis and midzone microtubule organization in Caenorhabditis elegans require the kinesin-like protein ZEN-4. Mol Biol Cell. 1998;9:2037–2049. doi: 10.1091/mbc.9.8.2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield TF, Jackson SM, Pallanck LJ. A Drosophila homolog of the polyglutamine disease gene SCA2 is a dosage-sensitive regulator of actin filament formation. Genetics. 2002;162:1687–1702. doi: 10.1093/genetics/162.4.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satterfield TF, Pallanck LJ. Ataxin-2 and its Drosophila homolog, ATX2, physically assemble with polyribosomes. Hum Mol Genet. 2006;15:2523–2532. doi: 10.1093/hmg/ddl173. [DOI] [PubMed] [Google Scholar]
- Scherzed W, Brunt ER, Heinsen H, de Vos RA, Seidel K, Burk K, Schols L, Auburger G, Del Turco D, Deller T, et al. Pathoanatomy of cerebellar degeneration in spinocerebellar ataxia type 2 (SCA2) and type 3 (SCA3) Cerebellum. 2012;11:749–760. doi: 10.1007/s12311-011-0340-8. [DOI] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonegg S, Constantinescu AT, Hoege C, Hyman AA. The Rho GTPase-activating proteins RGA-3 and RGA-4 are required to set the initial size of PAR domains in Caenorhabditis elegans one-cell embryos. Proc Natl Acad Sci USA. 2007;104:14976–14981. doi: 10.1073/pnas.0706941104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, Hamill DR, Carter JC, Schumacher J, Bowerman B. The aurora-related kinase AIR-2 recruits ZEN-4/CeMKLP1 to the mitotic spindle at metaphase and is required for cytokinesis. Curr Biol. 2000;10:1162–1171. doi: 10.1016/s0960-9822(00)00715-6. [DOI] [PubMed] [Google Scholar]
- Shannon KB, Canman JC, Ben Moree C, Tirnauer JS, Salmon ED. Taxol-stabilized microtubules can position the cytokinetic furrow in mammalian cells. Mol Biol Cell. 2005;16:4423–4436. doi: 10.1091/mbc.E04-11-0974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao J, Diamond MI. Polyglutamine diseases: emerging concepts in pathogenesis and therapy. Hum Mol Genet. 2007;16(Spec No. 2):R115–R123. doi: 10.1093/hmg/ddm213. [DOI] [PubMed] [Google Scholar]
- Shelton CA, Bowerman B. Time-dependent responses to glp-1-mediated inductions in early C. elegans embryos. Development. 1996;122:2043–2050. doi: 10.1242/dev.122.7.2043. [DOI] [PubMed] [Google Scholar]
- Skop AR, Bergmann D, Mohler WA, White JG. Completion of cytokinesis in C. elegans requires a brefeldin A-sensitive membrane accumulation at the cleavage furrow apex. Curr Biol. 2001;11:735–746. doi: 10.1016/s0960-9822(01)00231-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, Liu H, Yates J, 3rd, Meyer BJ, Heald R. Dissection of the mammalian midbody proteome reveals conserved cytokinesis mechanisms. Science. 2004;305:61–66. doi: 10.1126/science.1097931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skop AR, White JG. The dynactin complex is required for cleavage plane specification in early Caenorhabditis elegans embryos. Curr Biol. 1998;8:1110–1116. doi: 10.1016/s0960-9822(98)70465-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squirrell JM, Eggers ZT, Luedke N, Saari B, Grimson A, Lyons GE, Anderson P, White JG. CAR-1, a protein that localizes with the mRNA decapping component DCAP-1, is required for cytokinesis and ER organization in Caenorhabditis elegans embryos. Mol Biol Cell. 2006;17:336–344. doi: 10.1091/mbc.E05-09-0874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan DG, Fisk RM, Xu H, van den Heuvel S. A complex of LIN-5 and GPR proteins regulates G protein signaling and spindle function in C elegans. Genes Dev. 2003;17:1225–1239. doi: 10.1101/gad.1081203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strickland LI, Wen Y, Gundersen GG, Burgess DR. Interaction between EB1 and p150(glued) is required for anaphase astral microtubule elongation and stimulation of cytokinesis. Curr Biol. 2005;15:2249–2255. doi: 10.1016/j.cub.2005.10.073. [DOI] [PubMed] [Google Scholar]
- Strome S. Specification of the germ line. WormBook. 2005;2005(Jul 28):1–10. doi: 10.1895/wormbook.1.9.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strome S, Wood WB. Generation of asymmetry and segregation of germ-line granules in early C. elegans embryos. Cell. 1983;35:15–25. doi: 10.1016/0092-8674(83)90203-9. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schuman EM. Dendritic protein synthesis, synaptic plasticity, and memory. Cell. 2006;127:49–58. doi: 10.1016/j.cell.2006.09.014. [DOI] [PubMed] [Google Scholar]
- Thomas MG, Loschi M, Desbats MA, Boccaccio GL. RNA granules: the good, the bad and the ugly. Cell Signal. 2011;23:324–334. doi: 10.1016/j.cellsig.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. doi: 10.1038/27579. [DOI] [PubMed] [Google Scholar]
- Updike DL, Strome S. A genomewide RNAi screen for genes that affect the stability, distribution and function of P granules in Caenorhabditis elegans. Genetics. 2009;183:1397–1419. doi: 10.1534/genetics.109.110171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme P, Veldink JH, van Blitterswijk M, Corveleyn A, van Vught PW, Thijs V, Dubois B, Matthijs G, van den Berg LH, Robberecht W. Expanded ATXN2 CAG repeat size in ALS identifies genetic overlap between ALS and SCA2. Neurology. 2011;76:2066–2072. doi: 10.1212/WNL.0b013e31821f445b. [DOI] [PubMed] [Google Scholar]
- van de Loo S, Eich F, Nonis D, Auburger G, Nowock J. Ataxin-2 associates with rough endoplasmic reticulum. Exp Neurol. 2009;215:110–118. doi: 10.1016/j.expneurol.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Verbrugghe KJ, White JG. SPD-1 is required for the formation of the spindle midzone but is not essential for the completion of cytokinesis in C. elegans embryos. Curr Biol. 2004;14:1755–1760. doi: 10.1016/j.cub.2004.09.055. [DOI] [PubMed] [Google Scholar]
- Watts JL, Etemad-Moghadam B, Guo S, Boyd L, Draper BW, Mello CC, Priess JR, Kemphues KJ. par-6, a gene involved in the establishment of asymmetry in early C. elegans embryos, mediates the asymmetric localization of PAR-3. Development. 1996;122:3133–3140. doi: 10.1242/dev.122.10.3133. [DOI] [PubMed] [Google Scholar]
- Wheatley SP, Wang Y. Midzone microtubule bundles are continuously required for cytokinesis in cultured epithelial cells. J Cell Biol. 1996;135:981–989. doi: 10.1083/jcb.135.4.981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White EA, Glotzer M. Centralspindlin: at the heart of cytokinesis. Cytoskeleton. 2012;69:882–892. doi: 10.1002/cm.21065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilker EW, van Vugt MA, Artim SA, Huang PH, Petersen CP, Reinhardt HC, Feng Y, Sharp PA, Sonenberg N, White FM, et al. 14-3-3sigma controls mitotic translation to facilitate cytokinesis. Nature. 2007;446:329–332. doi: 10.1038/nature05584. [DOI] [PubMed] [Google Scholar]
- Wu KY, Hengst U, Cox LJ, Macosko EZ, Jeromin A, Urquhart ER, Jaffrey SR. Local translation of RhoA regulates growth cone collapse. Nature. 2005;436:1020–1024. doi: 10.1038/nature03885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Blelloch R. Regulation of pluripotency by RNA binding proteins. Cell Stem Cell. 2014;15:271–280. doi: 10.1016/j.stem.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoshi M, Li Q, Yamamoto M, Okada H, Suzuki Y, Kawahara Y. Direct binding of Ataxin-2 to distinct elements in 3’ UTRs promotes mRNA stability and protein expression. Mol Cell. 2014;55:186–198. doi: 10.1016/j.molcel.2014.05.022. [DOI] [PubMed] [Google Scholar]
- Zeiser E, Frokjaer-Jensen C, Jorgensen E, Ahringer J. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS One. 2011;6:e20082. doi: 10.1371/journal.pone.0020082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Chen D, Smith MA, Zhang B, Pan X. Selection of reliable reference genes in Caenorhabditis elegans for analysis of nanotoxicity. PLoS One. 2012;7:e31849. doi: 10.1371/journal.pone.0031849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu C, Bossy-Wetzel E, Jiang W. Recruitment of MKLP1 to the spindle midzone/midbody by INCENP is essential for midbody formation and completion of cytokinesis in human cells. Biochem J. 2005;389:373–381. doi: 10.1042/BJ20050097. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.