Gene expression analysis indicates that p53-mediated suppression of proliferation of polyploid cells can be averted by increased levels of oncogenes such as cyclin D2. Tetraploid cells can adapt and continue to proliferate despite having increased genome content and a wild-type p53 signaling cascade.
Abstract
Tetraploidization, or genome doubling, is a prominent event in tumorigenesis, primarily because cell division in polyploid cells is error-prone and produces aneuploid cells. This study investigates changes in gene expression evoked in acute and adapted tetraploid cells and their effect on cell-cycle progression. Acute polyploidy was generated by knockdown of the essential regulator of cytokinesis anillin, which resulted in cytokinesis failure and formation of binucleate cells, or by chemical inhibition of Aurora kinases, causing abnormal mitotic exit with formation of single cells with aberrant nuclear morphology. Transcriptome analysis of these acute tetraploid cells revealed common signatures of activation of the tumor-suppressor protein p53. Suppression of proliferation in these cells was dependent on p53 and its transcriptional target, CDK inhibitor p21. Rare proliferating tetraploid cells can emerge from acute polyploid populations. Gene expression analysis of single cell–derived, adapted tetraploid clones showed up-regulation of several p53 target genes and cyclin D2, the activator of CDK4/6/2. Overexpression of cyclin D2 in diploid cells strongly potentiated the ability to proliferate with increased DNA content despite the presence of functional p53. These results indicate that p53-mediated suppression of proliferation of polyploid cells can be averted by increased levels of oncogenes such as cyclin D2, elucidating a possible route for tetraploidy-mediated genomic instability in carcinogenesis.
INTRODUCTION
Polyploidy refers to the exact multiplication of the haploid chromosomal number to a DNA content higher than the diploid (2N). Tetraploidy (4N) usually results from a doubling of the diploid chromosome number. Ordinarily, DNA and centrosome replication is limited to once per cell cycle. In events of failed mitosis, cytokinesis, or cell fusion, genome doublings are accompanied by acquisition of supernumerary centrosomes. Cell divisions in polyploid cells with extra centrosomes are error-prone and lead to the formation of aneuploid cells that are often genetically unstable and acquire further numerical and structural chromosomal aberrations (Nigg, 2006; Ganem et al., 2007, 2009; Godinho et al., 2009; Gisselsson et al., 2010). Proliferation of these cells can thus produce rapid and massive genome and proteome changes in populations of cells (Potapova et al., 2013; Giam and Rancati, 2015). Recurrent erroneous mitoses facilitate production of variable karyotypes, leading to genetic diversity within progeny (Holland and Cleveland, 2009; Gordon et al., 2012). Studies in eukaryotic microbes have shown that altered chromosome copy number can compensate for detrimental mutations or lead to resistance to environmental stress, promoting the survival of aneuploid cells (Selmecki et al., 2006, 2009; Thompson et al., 2006; Chen et al., 2012, 2015; Kaya et al., 2015; Liu et al., 2015).
Genome and centrosome doublings are recognized as prominent events in tumorigenesis. Cancer is an ever-evolving system whose adaptability is the main reason for failure in anticancer therapy and emergence of drug resistance. Cancer genome sequencing studies revealed that genome-doubling events are common throughout oncogenic progression in many cancers and are associated with elevated aggressiveness and risk of recurrence (Carter et al., 2012; Zack et al., 2013; de Bruin et al., 2014; Dewhurst et al., 2014). Tetraploidy has been detected in early stages of many cancers (Ornitz et al., 1987; Galipeau et al., 1996; Giannoudis et al., 2000; Olaharski et al., 2006). Experimental evidence also suggests that tetraploidy may be directly connected with tumor initiation (Fujiwara et al., 2005; Castillo et al., 2007; Davoli and de Lange, 2012). Moreover, in vitro studies have demonstrated that tetraploid cells can be more resistant to certain chemotherapeutic drugs than their diploid counterparts (Puig et al., 2008; Balsas et al., 2012; Sharma et al., 2013; Zhang et al., 2014; Kuznetsova et al., 2015). Overall tetraploidization can be viewed as an important culprit in oncogenesis.
To maintain genetic uniformity and functional coherence within a multicellular organism, mechanisms must exist to prevent deviant polyploid cells from dividing further. Indeed, many studies have shown that many types of nontransformed cells stop proliferation after the induction of polyploidy by various methods, such as failed mitotic cell division after pharmacological disruption of the actin or microtubule cytoskeleton, treatment with DNA-damaging agents, and viral-induced cell fusion. In all cases, the cessation of proliferation was found to depend on the tumor suppressor transcription factor p53 (Hirano and Kurimura, 1974; Minn et al., 1996; Khan and Wahl, 1998; Lanni and Jacks, 1998; Casenghi et al., 1999; Andreassen et al., 2001; Margolis et al., 2003; Yang et al., 2004; Duelli et al., 2005; Fujiwara et al., 2005; Incassati et al., 2006; Davoli et al., 2010; Vitale et al., 2010). Conversely, other studies postulated that nontransformed cells cannot arrest the cell cycle efficiently in response to tetraploidy per se, but instead the suppression of proliferation observed in some cases might have been a p53-dependent stress response unrelated to the increased DNA content (Guidotti et al., 2003; Uetake and Sluder, 2004; Wong and Stearns, 2005). Discerning the mechanism of p53 pathway activation in polyploid cells has been challenging because p53 can be activated by a wide variety of external and internal stressors (Vogelstein et al., 2000; Horn and Vousden, 2007; Vousden and lane, 2007). Certain instances of tetraploidization involving exposure to microtubule and actin polymerization inhibitors as well as DNA synthesis inhibitors may be accompanied by DNA damage and oxidative and other types of stress, all of which are potent p53 inducers. In binucleate cells with multiple centrosomes obtained by transient treatments with cytochalasin B, activation of the Hippo pathway kinase LATS2 was the key mechanism that triggered activation of p53 (Ganem et al., 2014). Another study of tetraploid arrest in cytochalasin B–treated cells concluded that p53 activation and cessation of proliferation depend on the function of the CDK inhibitor p16INK4a (Panopoulos et al., 2014). Piecing together the molecular circuitry of stress signals in different instances of polyploidization and the links to the cell cycle and activation of p53 is an ongoing effort.
This work presents the first systematic exploration of gene expression changes incited by polyploidization through different routes. We asked two main questions. 1) What changes in gene expression are evoked by acute (recent) tetraploidy? 2) What genetic changes underlie the ability of tetraploid cells to proliferate continuously? Transcriptome analysis by next-generation RNA sequencing (RNAseq) in Aurora kinase–inhibited and cytokinesis-failed cells indicated that despite morphological differences, gene expression changes in these cells had similar signatures, especially in the context of the p53 pathway activation. To address the mechanisms of adaptation to tetraploidy, we generated clonal proliferating tetraploid cells and analyzed their transcriptome by RNAseq. This analysis shows a large number of differentially expressed genes, indicating alterations in multiple signaling and metabolic pathways. Among the handful of commonly up-regulated transcripts, several genes were also up-regulated in acute p53-activated cells, indicating some degree of p53 activation in proliferating tetraploid cells. One of the commonly up-regulated genes unrelated to p53 activation was the G1 CDK-activating protein cyclin D2. We show that overexpression of cyclin D2 can potentiate adaptation to tetraploidy even in the presence of a functional p53.
RESULTS
Acute polyploidization activates p53-dependent transcriptional program
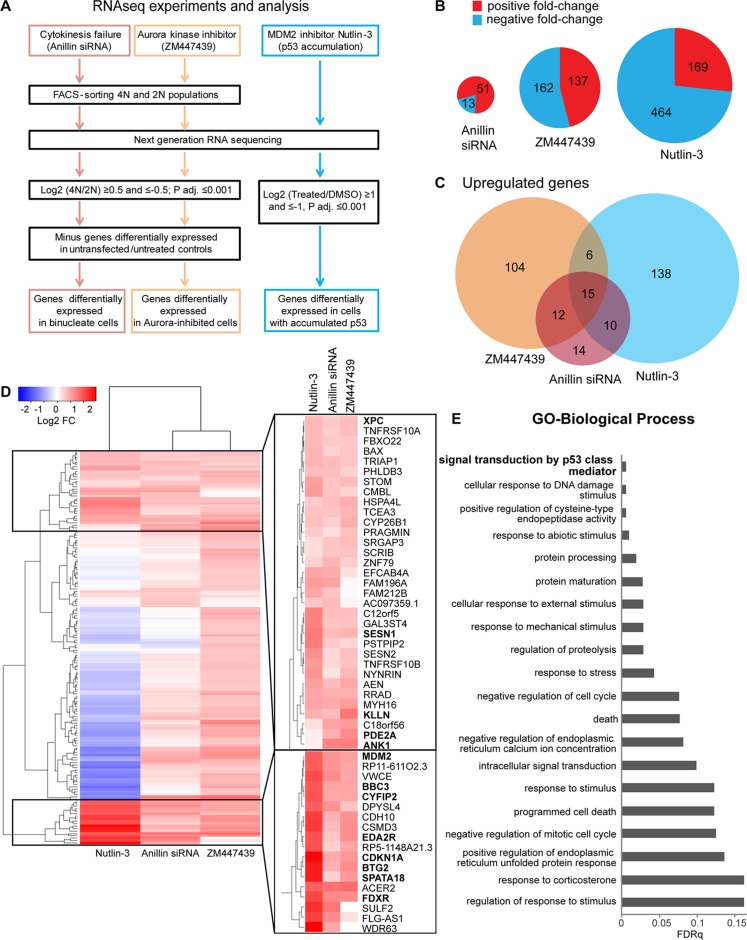
To explore changes in gene expression induced by acute tetraploidy, we investigated the transcriptional program activated in differentially induced, tetraploid hTERT-immortalized RPE1 cells. An important aspect of design of our study was that acute tetraploid cells were derived from the same diploid RPE1 cell line using two different methods: cytokinesis failure and aberrant mitotic exit (Supplemental Figure S1A). Cytokinesis failure in this study was induced by small interfering RNA (siRNA)–mediated knockdown of the scaffold protein anillin, an essential regulator of the contractile ring formation during cytokinesis (Piekny and Glotzer, 2008; Tse et al., 2011). Anillin knockdown results in the formation of binucleate tetraploid cells without global perturbation of the actin cytoskeleton. The fact that this protein is known for only its cytokinetic function makes the possibility of its knockdown inducing some non–tetraploidy-related stress less likely. Chemical inhibition of mitotic Aurora kinases was used to induce aberrant mitotic exit without chromosome segregation and cytokinesis, producing tetraploid cells with a single nucleus that is misshapen and/or fragmented. Aurora kinases are serine/threonine kinases that regulate several critical steps of mitotic progression, particularly stable attachment of kinetochores to microtubules, signaling of the mitotic spindle checkpoint, and cytokinesis (Taylor and Peters, 2008). A potent, well-characterized inhibitor called ZM447439, which is more specific toward Aurora B (Gadea and Ruderman, 2005), was used in most experiments. Inhibition of Aurora B activity in mitosis manifests in premature mitotic exit, chromosome missegregation, and cytokinesis failure, generating daughter cells with single tetraploid nuclei (Kallio et al., 2002; Wang et al., 2006). Of importance, inhibition of this kinase does not cause mitotic delay, which by itself can trigger G1 arrest in daughter cells (Uetake and Sluder, 2010; Orth et al., 2012). To study the transcriptional response to acute polyploidization, we designed a comprehensive RNAseq analysis of acute tetraploid cells (Figure 1A). Cells treated with Aurora kinase inhibitor and anillin siRNA for 24 h were labeled with Hoechst 33342 and sorted into 2N and 4N populations by fluorescence-activated cell sorting (FACS; Supplemental Figure S1, B and C). The 24-h treatment period, equal to the approximate duration of one cell cycle, was chosen because we were interested in early tetraploidy response genes.
FIGURE 1:
Transcriptome analysis of acute tetraploid cells and diploid p53-accumulated cells by RNAseq. (A) RNAseq experimental setup and analysis. Tetraploid Aurora-inhibited RPE1 cells were obtained by treatment with Aurora kinase inhibitor ZM447439 for 24 h, tetraploid binucleate RPE1 cells were obtained by siRNA-mediated knockdown of the essential cytokinesis regulator anillin for 24 h, and p53- activated cells were obtained by treatment of RPE1 cells with 2.5 μM nutlin-3 for 24 h. (B) Total number of differentially expressed genes. Red, positive fold change; blue, negative fold change. Only genes with log2 average reads per kilobase per million reads (RPKM) expression values >0 are included. Size of pie charts is proportional to total number of differentially expressed genes. (C) Venn diagram depicting genes up-regulated in tetraploid cells treated with Aurora inhibitor (orange) or anillin siRNA (maroon) and diploid cells treated with nutlin-3 (blue). Only genes with log2 average RPKM expression values >0 are included. (D) Hierarchical clustering of all genes up-regulated in tetraploid ZM447439-treated cells and all genes up-regulated in tetraploid anillin siRNA–treated binucleate cells based on 4N/2N fold change (log2 FC ≥ 0.5, p adjusted 0.001). Only genes with log2 average RPKM expression values >0 are included. For cells exposed to nutlin-3, FC is ratio of nutlin-3 over DMSO-treated cells. White, no change; red, up-regulated genes; blue, down-regulated genes. Two clusters of commonly up-regulated genes are expanded. Bold indicates genes chosen for subsequent validation. (E) Enriched biological process GO terms of selected clusters determined using the Gene Ontology Enrichment Analysis and Visualization tool (www.biomedcentral.com/1471-2105/10/48) and ranked by false discovery rate q value. Nonredundant GO terms were identified using REVIGO (journals.plos.org/plosone/article?id=10.1371/journal.pone.0021800). Top 20 nonredundant GO terms.
Three rounds of FACS sorting from different populations of cells were performed to generate three biological replicates. The 2N and 4N populations from the same rounds of FACS sorting were used for RNAseq to determine the ratio of transcripts in 4N over 2N cells. However, FACS sorting of 4N populations based on DNA content cannot distinguish tetraploid G1/G0 from diploid G2/M cells. For this reason, each experiment included vehicle and mock siRNA-treated controls that were also FACS sorted into 2N (G1/G0) and 4N (G2/M) populations. This allowed filtering out of differentially expressed genes commonly enriched in 2N or 4N populations in both treatment and controls, revealing transcripts selectively enriched or depleted in tetraploid G1/G0 populations. Finally, because previous studies showed that polyploidization activates the p53 signaling pathway (Coward and Harding, 2014), we also compared gene expression between acute tetraploid cells and cells in which p53 activation was induced without polyploidization, DNA damage, or other stresses using nutlin-3, which causes p53 accumulation by inhibition of the p53 E3 ubiquitin ligase MDM2 (Vassilev et al., 2004).
The total number of differentially expressed genes was lowest in anillin-knockdown binucleate tetraploid cells compared with diploid cells and highest in nutlin-treated cells compared with untreated cells (Figure 1B). In cells treated with anillin siRNA, nearly four times more transcripts were enriched in 4N than in 2N after removing genes differentially expressed in a mock-transfected 4N (G2/M) population. Imposing a stringent cutoff of adjusted p < 0.001, we found that most of the gene transcripts selectively up-regulated in anillin-knockdown tetraploid cells overlapped with genes up-regulated in Aurora kinase inhibitor–induced tetraploid cells, in nutlin-3–treated cells, or both (Figure 1C). There was almost no overlap among genes enriched in anillin siRNA or Aurora inhibitor–treated 2N population (Supplemental Figure S2A), possibly because these 2N populations were distinct: in anillin-knockdown cells, the 2N population may have comprised untransfected cells, whereas in Aurora inhibitor–treated cells, the 2N population may represent naturally existing quiescent cells.
Cluster analysis of all transcripts up-regulated in anillin-knockdown or Aurora kinase inhibitor–induced tetraploid cells showed high similarity among these two groups, with some clusters of genes also up-regulated in nutlin-3–treated cells (Figure 1D). Enriched gene ontology (GO) terms for selected clusters of genes commonly up-regulated in all populations are shown in Figure 1E. The top GO category for these clusters was the p53 signaling pathway. For a separate cluster containing transcripts that overall behave more similarly in acute tetraploidy situations than in nutlin-3–treated cells, the GO term analysis pointed out enrichment of amino acid and nucleotide metabolic processes and response to stimulus (Supplemental Figure S2B). Supplemental Table S1 lists the transcripts enriched and depleted in acute tetraploid cells and in cells treated with nutlin-3.
Acute tetraploidization suppresses the cell cycle by activating the p53 signaling pathway
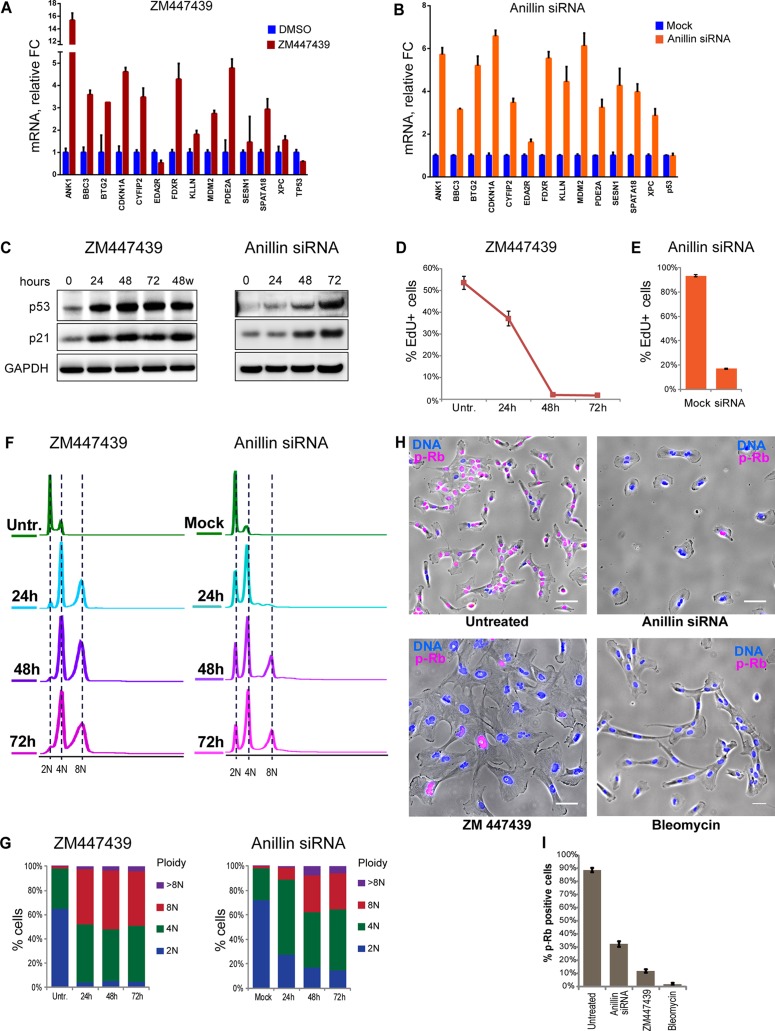
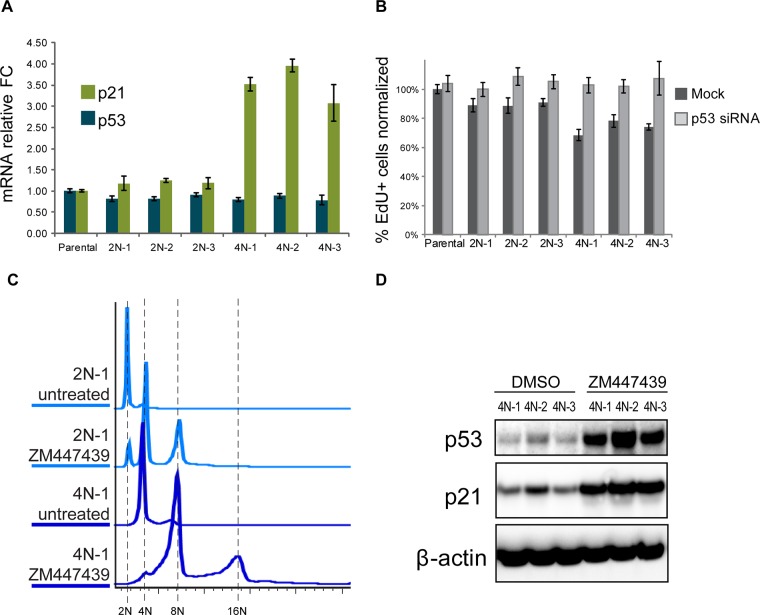
A group of genes commonly up-regulated in acute polyploid cells was chosen for validation by quantitative real-time PCR. Most of the genes examined showed increased expression in tetraploid cells induced by Aurora inhibition and anillin knockdown (Figure 2, A and B) but also in nutlin-3–treated cells (Supplemental Figure S3). These genes included the CDK inhibitor CDKN1A (p21), a well-studied direct transcriptional target of p53 (el-Deiry et al., 1993, 1995; Abbas and Dutta, 2009). Consistently, P53 and p21 protein levels accumulated overtime in both types of tetraploid cells (Figure 2C). ZM447439 is a reversible Aurora kinase inhibitor. Of importance, washing it out 48 h after induction of polyploidization did not reduce the amount of p21 and p53 accumulated in these cells, indicating that the activation of p53 in Aurora kinase inhibitor–treated cells was not a direct effect of the chemical on the p53 pathway.
FIGURE 2:
Validation of up-regulation of selected genes and cell cycle arrest in polyploid cells. (A, B) Real-time qPCR analysis of selected genes after 72-h treatment of RPE1 cells with Aurora inhibitor ZM447439 (A) and anillin siRNA (B). All values were normalized to vehicle (DMSO)- treated control (A) or mock-transfected control (B). Bar heights represent mean relative fold change (n = 3); error bars represent SEM. (C) Western blot analysis of p53 and p21 protein levels in cells treated with ZM447439 and anillin siRNA for 24, 48, and 72 h. In the last lane of the blot of Aurora inhibitor–treated cells, the drug was washed out after 48-h treatment and the sample was collected 24 h after washout. (D) Time-course treatment of RPE1 cells with Aurora inhibitor ZM447439 for 24, 48, and 72 h. The same cell cultures were split equally for indicated treatments, followed by EdU incorporation for 6 h. EdU was detected after fixation and counterstaining with Hoechst 33342, and percentage of EdU-positive cells was quantified by imaging. Error bars represent SD. (E) EdU incorporation in RPE1 cells treated with anillin siRNA for 48 h. EdU was allowed to incorporate for 24 h, followed by fixation and counterstaining with Hoechst 33342. EdU incorporation was quantified by imaging. Percentages of EdU-positive cells were normalized to the p53 siRNA. Error bars represent SD. (F) Flow cytometric profiles of time-course treatments with ZM447439 or anillin siRNA for 24, 48, and 72 h. Cells were fixed and stained with propidium iodide. (G) Quantification of ploidy distributions in F. (H, I) Immunofluorescence of proliferation marker phospho–Rb Ser-807/811 (red) after 48-h treatment with anillin siRNA (binucleate) or ZM447439 (aberrant multilobed nuclei) polyploid cells. Bleomycin (3 μg/ml) was used as a control for cell cycle arrest. Nuclei were counterstained with Hoechst 33342 (blue). Bar, 40 μm. (I) Quantification of cells positive for p-Rb based on the imaging; error bars denote SD.
The issue of whether tetraploidy itself arrests the cell cycle or whether the arrest is caused by some stress due to the treatment and unrelated to genome doubling has been controversial. Using two distinct methods of inducing tetraploidy allowed us to interpret phenotypic commonalities as outcomes of the unifying change in these cells—the increased DNA content. To assay DNA synthesis and proliferation rate quantitatively, we used the 5-ethynyl-2′-deoxyuridine (EdU) incorporation assay (Salic and Mitchison, 2008). In Aurora-inhibited tetraploid cells, EdU incorporation decreased over time and ceased 48 h after drug addition (Figure 2D). In binucleate cells resulting from anillin knockdown, the fraction of EdU-positive cells decrease by ∼80% 48 h after siRNA transfection (Figure 2E). Therefore, in both instances of acute polyploidization, suppression of proliferation was observed. Analysis of ploidy increase over time after polyploidization revealed that in a substantial fraction of cells, ploidy continued to increase after reaching tetraploid DNA content but stopped after reaching octoploid DNA content. In both methods of polyploidy induction, ploidy increase stopped concurrently with cessation of EdU incorporation (Figure 2, F and G).
During the course of this work, a number of nonmalignant hTERT-immortalized and primary cell lines were tested for their ability to arrest the cell cycle in response to polyploidization by Aurora inhibition and cytokinesis failure. All nonmalignant cells examined arrested the cell cycle at 4N and 8N DNA content, with variable proportion of 4N and 8N populations (unpublished data). This finding may reconcile some of the debate over the existence of the cell cycle arrest after tetraploidization: the cell cycle was not arrested in all cells after becoming tetraploid, but it was arrested after becoming octoploid.
To verify that the cell cycle in acute polyploid cells stopped at the G1/G0 phase of the cell cycle, we labeled polyploid cells with antibody against the retinoblastoma tumor suppressor protein Rb phosphorylated on Ser-807/811. Rb prevents the cell cycle progression from G0 to G1 and into S phase by inhibiting transcriptional activity of E2F family of transcription factors involved in transcription of proliferation-associated genes (Giacinti and Giordano, 2006). Phosphorylation of human Rb on Ser-807/811 releases E2F transcription factors and allows cell cycle progression (Zarkowska and Mittnacht, 1997; Macdonald and Dick, 2012). Immunofluorescence for Rb phosphorylated on Ser-807/811 showed that after 48 h of Aurora kinase inhibitor treatment or transfection of anillin siRNA, most cells were phospho-Rb negative (Figure 2, H and I). This confirms that the majority of acute induced polyploid cells arrest the cell cycle in G1/G0 stage with dephosphorylated Rb.
A previous study proposed that the cellular response to polyploidy might be linked to DNA damage (Hayashi and Karlseder, 2013). In support of this view, certain protocols used to induce polyploidy are accompanied by DNA damage, such as telomere deprotection during mitotic delay or arrest (Davoli et al., 2010; Davoli and de Lange, 2012) and tetraploidy induced with DNA-damaging agents (Puig et al., 2008; Ichijima et al., 2010; Shen et al., 2013). We therefore investigated the presence of DNA damage in our experimental systems by using immunofluorescent labeling of phospho–histone H2AX, an early DNA damage marker. Binucleate cells that resulted from anillin knockdown did not show accumulation of DNA damage foci, in contrast to positive control cells exposed to γ-irradiation. In Aurora kinase–inhibited tetraploid cells, pH2AX was localized almost exclusively to micronuclei (Supplemental Figure S4A). Studies have shown that DNA damage in micronuclei accumulates due to compromised nuclear envelope structure and aberrant DNA replication. Although micronuclei do accumulate DNA damage, they are unable to elicit an efficient downstream signaling cascade that leads to cell cycle arrest (Crasta et al., 2012; Hatch et al., 2013). Knocking down DNA-damage kinases ATM, ATR, and both in Aurora-inhibited cells had no effect on DNA content or EdU incorporation (Supplemental Figure S4, B and C). These observations suggest that it is unlikely that G1 arrest in tetraploid cells in our experiments was due primarily to DNA damage.
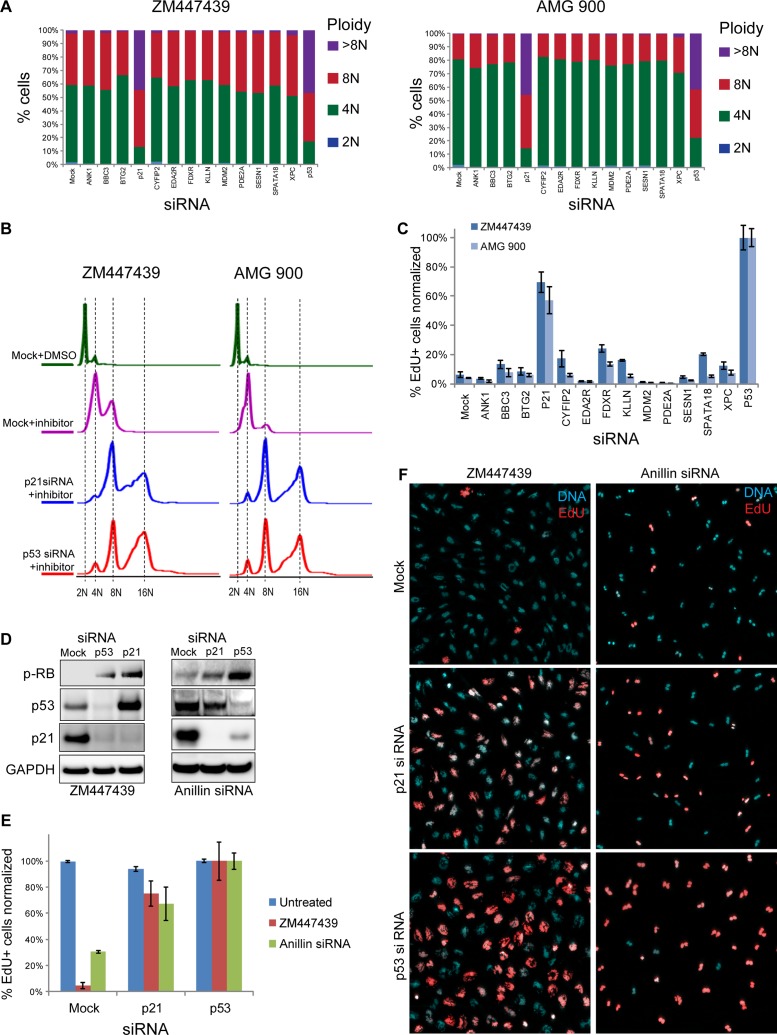
Next we used siRNA-mediated knockdown to test the functional role of genes that were validated by quantitative PCR (qPCR) as up-regulated in acute polyploidy. In addition to ZM447439, we used a structurally distinct pan-Aurora inhibitor, AMG900 (Payton et al., 2010; Geuns-Meyer et al., 2015), a chemotherapeutic agent that demonstrated promising antitumor activity in preclinical testing (Linardopoulos and Blagg, 2015). For these experiments, we collected samples 72 h after siRNA transfection, when control cells fully arrested the cell cycle, and the siRNA knockdown was maximal. siRNA-mediated knockdown of 13 genes that were up-regulated in tetraploid cells showed that only CDKN1A (p21) knockdown caused an increase in ploidy to the level comparable to that with knockdown of p53 (Figure 3, A and B). Consistent with ploidy increase, proliferation assay by EdU incorporation demonstrated an increase in proliferation only in p21- and p53-knockdown cells treated with ZM447439 or AMG900 (Figure 3C). Next we analyzed p53 and p21 protein levels in Aurora-inhibited and anillin-knockdown tetraploid cells. siRNA against CDKN1A (p21) causes a major decrease in protein level, and p53 siRNA causes a strong decrease of both p53 and p21 proteins. Phosphorylation of Rb on Ser-807/811 in both p53 and p21 siRNA–treated cells indicated an active cell cycle (Figure 3D). Cell cycle progression in p53 and p21 knockdowns in Aurora-inhibited or anillin-knockdown tetraploid cells was confirmed by EdU incorporation (Figure 3, E and F).
FIGURE 3:
siRNA-mediated knockdown of p53 and p21 overrides cell cycle arrest in polyploid cells. (A) Quantification of ploidy distributions in RPE1 cells exposed to two different Aurora kinase inhibitors—ZM447439 and AMG 900—and treated with siRNAs for indicated genes for 72 h. Only p53 and p21 siRNA–treated cells demonstrated shifts toward increased ploidy content, consistent with continuous cell cycle progression. (B) Flow cytometric profiles of cells treated with Aurora kinase inhibitors AZM447439 and AMG 900 and siRNAs for p21 and p53. Cells were fixed 72 h after siRNA transfection and stained with propidium iodide. (C) EdU incorporation in RPE1 cells exposed to ZM447439 and AMG 900 and treated with siRNAs for indicated genes for 72 h. Cells were allowed to incorporate EdU for 24 h, followed by fixation and counterstaining with Hoechst 33342. Percentages of EdU-positive cells were normalized to the p53 siRNA (the average number of EdU-positive cells in p53 siRNA–treated samples was assigned 100% value). Bar heights represent the mean (n = 4); error bars represent SD. (D) Western blot analysis of p53- and p21-knockdown cells treated with ZM447439 and anillin siRNA for 72 h. Note increased levels of phospho-Rb (Ser-807/811) in p53 and p21 siRNAs, indicating active cell cycle. (E) EdU incorporation in Aurora kinase–inhibited and anillin-knockdown RPE1 cells treated with siRNAs to p53 and p21. Percentages of the EdU-positive cells were normalized to the p53 siRNA as in C. Bar heights represent the mean (n = 4); error bars represent SD. (F) Nuclear morphology and EdU incorporation in Aurora-inhibited and anillin siRNA polyploid cells treated with siRNAs to p21 and p53. Note aberrant multilobed nuclei in Aurora inhibitor ZM447439–treated cells and binucleate cells in anillin siRNA. EdU was detected with fluorescent azide (red); DNA is counterstained with Hoechst 33342 (cyan). Note the increase in EdU incorporation in p21- and p53-knockdown cells.
Together these data support the notion that the cell cycle arrest in acute polyploid cells depends on p53 and its transcriptional target CDK inhibitor p21 and that this genome surveillance mechanism is distinct from the DNA damage response.
RNAseq analysis of proliferating tetraploid clones adapted after acute polyploidy induction
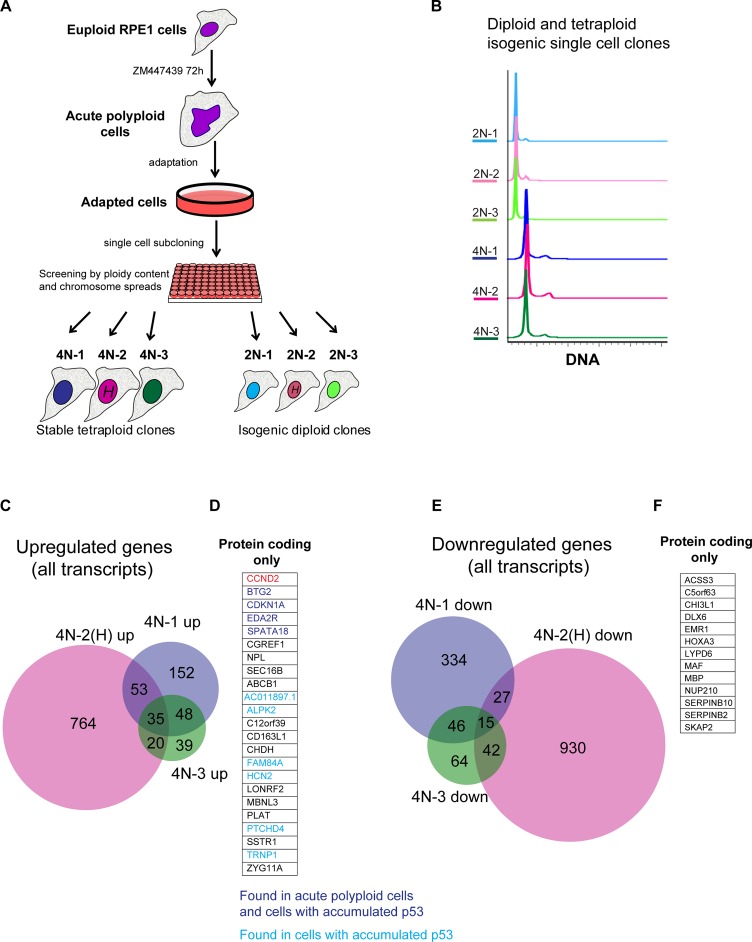
The p53 pathway is dysfunctional in most cancers, and many cancers undergo genome doubling(s) during their evolution. Whereas acute polyploid cells arrest the cell cycle in a p53-dependent manner, there are many reports of proliferating cells with tetraploid genomic content (Puig et al., 2008; Balsas et al., 2012; Ohshima and Seyama, 2013; Sharma et al., 2013; Ganem et al., 2014; Zhang et al., 2014; Kuznetsova et al., 2015). It is unclear whether suppression of the p53 pathway is required for cancer cells to escape tetraploid arrest or proliferating tetraploid cells can emerge without the concurrent loss of functional p53. To explore mechanisms of adaptation to tetraploidy, we performed an evolution experiment in which a large population of acute polyploid cells obtained by inhibition of Aurora B was allowed to survive for several weeks until proliferating cells emerged (Materials and Methods). During the adaptation period, most of the polyploid cells appeared quiescent. After several weeks, proliferating zones of varying densities appeared among quiescent cells. At that point, cells were lifted off the tissue culture flask and subcloned using a flow cytometer at a density of one cell per well, yielding single-cell clones. The DNA content of these single-cell–derived cell lines was analyzed by flow cytometry and chromosome counting. Of 169 single-cell clones obtained by this method, most had a FACS profile consistent with diploid DNA content, but many had shifted profiles, indicating possible aneuploidy, and only 3, designated 4N number 1 (4N-1), 4N number 2 (4N-2), and 4N number 1 (4N-3), were tetraploid (Figure 4, A and B). For each of these tetraploid single cell clones, we picked an isogenic diploid clone generated in parallel from the same plate. In one of the single-cell subcloning experiment, we labeled cells with Hoechst 33342 in the hope of enriching the number of tetraploid cells by flow cytometry. The tetraploid clone 4N-2 and corresponding diploid 2N-2 were produced from this experiment.
FIGURE 4:
Generation of proliferating tetraploid single-cell clones from acute polyploid Aurora-inhibited cells and analysis of their transcriptome by RNAseq. (A) Generation of proliferating tetraploid RPE1 single cell clones (4N 1–3) from euploid RPE1 cell culture treated with Aurora inhibitor ZM447439. Asynchronously growing cells were treated with ZM447439 for 72 h and allowed to survive for several weeks, until proliferating populations emerged. Proliferating cells were plated in 96-well plates at a density of one cell per well. The ploidy content of each single-cell clone was analyzed by FACS. Isogenic diploid single-cell clones (2N 1–3) were picked from the same 96-well plates as the corresponding tetraploid clones. Single-cell clones 4N-2 and 2N-2 underwent Hoechst 33342 labeling (H) before subcloning. (B) Ploidy analysis of generated tetraploid and isogenic diploid single-cell clones. Cells were fixed and stained with propidium iodide. FACS profiles indicate doubling of the DNA content. (C) Venn diagram of genes up-regulated in stable tetraploid single-cell clones in comparison to isogenic diploid single-cell clones. Only genes with log2 FC ≥ 1, p adjusted ≤ 0.001, and expression values log2 average RPKM > 0 are included. (D) Expanded list of genes from C, showing names of overlapping genes up-regulated in all three clones (protein-coding transcripts only). Genes that were previously found to be up-regulated in acute polyploid cells (Aurora inhibited and anillin knockdown) are highlighted in dark blue. Genes that were previously found to be up-regulated only in p53-accumulated cells treated with nutlin-3 are highlighted in light blue. (E) Venn diagram of genes down-regulated in stable tetraploid single-cell clones in comparison to isogenic diploid single-cell clones. Includes genes with log2 FC ≤ −1, p adjusted ≤ 0.001, log2 average RPKM > 0. (F) Expanded list of overlapping genes found in E, showing names of genes down-regulated in all three tetraploid single-cell clones (protein-coding transcripts only).
We examined karyotypes of these cells using the spectral karyotyping technology recently developed in our lab (Potapova et al., 2015). The 4N-1 and 4N-3 tetraploid clones had a tetraploid karyotype with two extra copies of chromosomes 12, and the clone 4N-2 in addition lost a copy of chromosome 1. One extra copy of chromosome 12 was also found in all isogenic diploid clones (Supplemental Figure S5A). The RPE1 cell line has 46 chromosomes according to the American Type Culture Collection (ATCC), but we found that it tends to acquire an extra copy of chromosome 12 with passages (Supplemental Figure S5B). Nonetheless, although gaining an extra copy of chromosome 12 may provide some selective advantage for the RPE1 cell line in long-term culture, it cannot be considered a selective advantage specific for tetraploidy because diploid RPE1 cells also gain it.
Analysis of the transcriptome of the three tetraploid single cell clones by RNAseq revealed that they had more unique differentially expressed transcripts than they had common (Figure 4, C and E). This suggests that these tetraploid single-cell clones, although subjected to the same treatment (with the exception of Hoechst 33342 staining in the isolation of clone 4N-2), undertook distinct evolutionary routes. Clone 4N-2 had much more differentially expressed genes than did clones that were never exposed to Hoechst. These cells also became trisomic for chromosome 1. Hoechst is a widely used DNA intercalating dye, and our results indicate that even though Hoechst did not impede cell proliferation, it had an effect on transcriptome and karyotype evolution. Nevertheless, there was a group of commonly up- or down-regulated transcripts in all three tetraploid clones. Of 23 significantly up-regulated protein-encoding transcripts, 4, including CDKN1A (p21), were previously found to be up-regulated in acute tetraploid cells and p53-activated cells (Figure 4D, purple), and 6 transcripts were identified in p53-activated cells only (Figure 4D, blue). This indicated that the only common gene expression signature in proliferating tetraploid cells was activation of the p53 pathway. However, in proliferating tetraploid cells, the p53-mediated downstream response is slighter and different from that in acute tetraploidy situations. Outside of the p53-dependent group of genes, there were 13 commonly up-regulated protein-coding genes (Figure 4D) and 13 commonly down-regulated protein-coding genes (Figure 4F). GO term and pathway analysis found no particular pathway enrichment among these genes. One of the most up-regulated protein-coding genes in all three tetraploid single cell clones that was not part of the p53 signaling network was cyclin D2 (CCND2). Supplemental Table S2 lists the transcripts differentially expressed in proliferating tetraploid cells.
Adapted tetraploid single-cell clones retain the ability to activate p53 upon further polyploidization
The foregoing RNAseq result suggested that in all three tetraploid single-cell clones, p53 signaling is activated but perhaps not fully. This prompted us to measure the mRNA levels of p53 transcriptional target CDKN1A (p21; Figure 5A). Aligned RNAseq reads were checked for variants in TP53 transcript using SAMtools, and no variants segregating specifically to the 4N group were found (unpublished data), indicating that the p53 in these cells was likely not mutated. Consistent with RNAseq results, transcript levels of CDKN1A (p21) were increased threefold to fourfold, which was lower than in acute tetraploidy (sixfold to eightfold increase; Figure 2, A and B) but considerably higher than in corresponding diploid cells. Evidently, this dampened p53 activity had some effect on the proliferation rate—EdU incorporation in tetraploid cells was slightly slower than in diploid cells, which was effectively compensated by knocking down the p53 (Figure 5B). To test whether the proliferating tetraploid clones were capable of mounting the p53 response to ploidy increase, we induced acute polyploidy with Aurora kinase inhibitor ZM447439 in these already tetraploid cells. The pattern of DNA content increase in these cells was similar to that of isogenic euploid clones (Figure 5C), and p53 and p21 proteins accumulated to high levels (Figure 5D). These results indicated that, paradoxically, the adapted tetraploid clones were able to proliferate continuously despite their ongoing weak p53 signaling and were capable of triggering full-scale p53 activation upon further increase in ploidy content. This suggests that the adaptation process essentially resets the tolerance of the p53 network to the doubled-genome content.
FIGURE 5:
p53-dependent transcriptional and cell cycle effects in proliferating tetraploid cells. (A) Real-time qPCR analysis of p21 and p53 genes in parental RPE1 cells and proliferating tetraploid and isogenic diploid single-cell clones. p21 mRNA levels were increased threefold to fourfold in tetraploid cell lines, whereas p53 mRNA levels were unchanged. All values were normalized to the parental RPE1. Bar heights represent average relative fold change (n = 3); error bars represent SEM. (B) Effect of p53 knockdown on proliferation of tetraploid single-cell clones. Cells were treated with siRNA for 72 h and allowed to incorporate EdU for 8 h. Percentages of EdU-positive cells were normalized to parental cell line not transfected with p53 siRNA (the number of EdU-positive cells in the parental untransfected sample was assigned 100% value). Bar heights represent the mean (n = 4); error bars represent SD. (C) Flow cytometric profiles of diploid and tetraploid isogenic cell lines treated with ZM447439 for 72 h. Like the diploid cell lines, tetraploid cells showed doubled or quadrupled DNA content. Cells were fixed and stained with propidium iodide. (D) Western blot analysis of p53 and p21 protein levels in proliferating tetraploid and isogenic diploid single-cell clones treated with Aurora inhibitor ZM447439 for 72 h.
Adapted tetraploid cells regain diploid-like nucleus–cell volume ratio and centrosome number
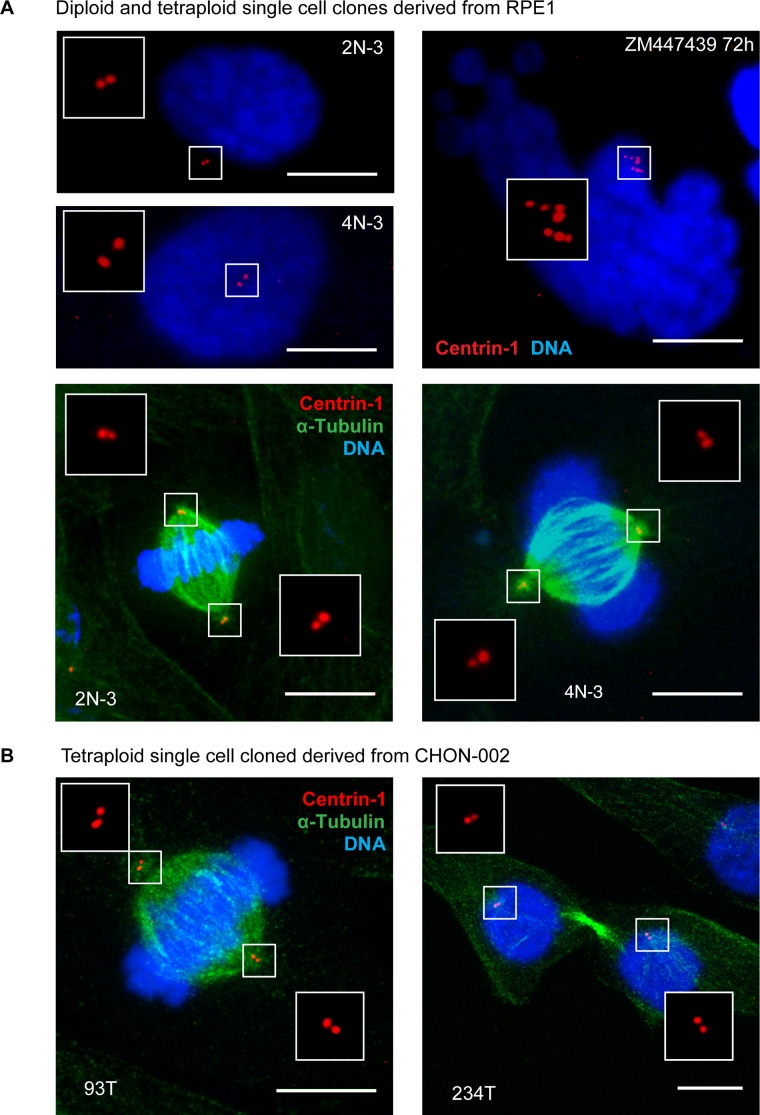
Proliferating tetraploid cells maintained the original fibroblast-like morphology with a single round nucleus. The cellular volume in proliferating tetraploid cells, as measured using the Coulter counter, approximately doubled compared with isogenic diploid cells, with some variability among the three clones (Supplemental Figure S6A). Therefore the cell volume in tetraploid RPE1 cells scaled roughly proportionally with the ploidy. Estimating the nuclear size by measuring the area of fluorescence DNA signal showed increased nuclear area in tetraploid compared with isogenic diploid cells (Supplemental Figure S6, B and C). An unexpected feature of these tetraploid clones was the normal number of centrioles, identical to that of diploid RPE1 cells (Figure 6A). All three tetraploid clones had two centrioles in the G1 phase of the cell cycle and four centrioles in mitosis. This is different from G1-arrested acute polyploid cells, which can have four to eight centrioles (Figure 6A, top right) and build multipolar mitotic spindles if dividing (Nigg, 2002; Holland and Cleveland, 2009). We confirmed this observation in two other proliferating tetraploid clonal cell lines generated from the hTERT-immortalized human diploid fibroblast cell line CHON-002 by repeated nocodazole washout. Like RPE1-derived tetraploid clones, tetraploid cells generated from CHON-002 by a different method have two centrioles in interphase and build bipolar mitotic spindles (Figure 6B). The normal number of centrioles allows tetraploid cells to build a bipolar mitotic spindle and segregate chromosomes equally between the two daughter cells during mitosis. Losing extra centrioles during evolution appears to be a very essential nongenetic adaptation that allows tetraploid cells to maintain a relatively stable genomic content.
FIGURE 6:
Established proliferating tetraploid cells have normal number of centrioles. (A) Immunofluorescence of a centriole component centrin-1 (red). Top left, diploid and established tetraploid cells in interphase. Note two centrin-1 dots in both cells. Top right, acute polyploid cell in interphase. This cell was treated with Aurora kinase inhibitor ZM447439. It displays characteristic nuclear morphology and has multiple centrioles. Bottom, diploid and proliferating tetraploid cells have equal centrosome number and form bipolar mitotic spindles in metaphase. DNA was counterstained with DAPI (blue); α-tubulin was labeled by immunostaining (green). At least 30 cells were examined for every image. Maximum projections of representative confocal stacks. (B) Centrin-1 labeling of proliferating tetraploid CHON-002 single-cell clones. Immunofluorescence labeling of centrin-1 (red) and α-tubulin (green) of proliferating tetraploid single-cell clones designated 93T (metaphase cell) and 234T (telophase cell). Nuclei were counterstained with DAPI (blue). Maximum projections of confocal stacks. Like proliferating tetraploid RPE1, established tetraploid cells derived from CHON-002 have centrosome number equal to diploid and form bipolar mitotic spindles. Bar, 10 μm.
The role of cyclin D2 in potentiation of adaptation to the tetraploid genome
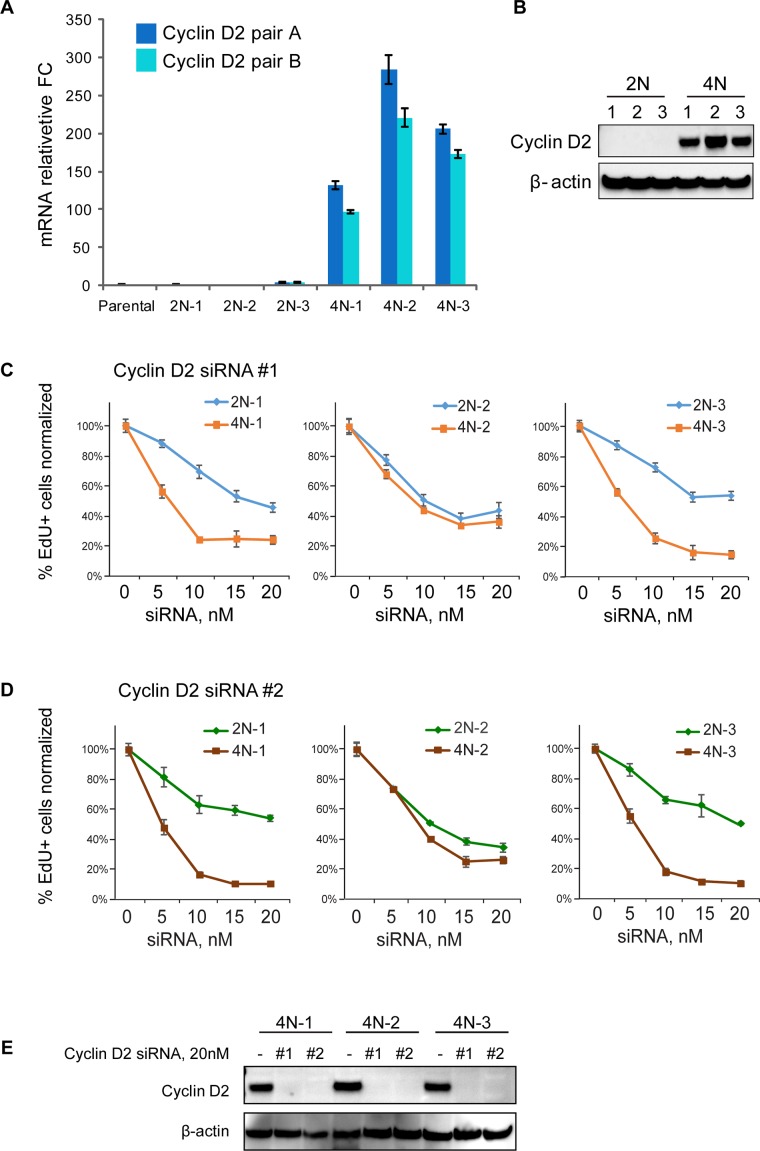
Cyclin D2 was one of the highest commonly up-regulated protein encoding genes outside of the network of p53 target genes in adapted tetraploid clones. This D-type cyclin is normally expressed during embryonic development (Sicinski et al., 1996) and forms complexes mainly with CDK 4, 6, and 2 and CDK inhibitor proteins. Cyclin D2-CDK 4/6 complexes are capable of phosphorylation and inhibition of the Rb protein and are known for promoting G1–S transition (Giacinti and Giordano, 2006). Analysis of cyclin D2 mRNA and protein levels confirmed that it was expressed at very low levels in diploid cells but became greatly up-regulated in adapted tetraploid cells (Figure 7, A and B). We also found increased expression of cyclin D2 in one of the two tetraploid single cell clones derived from CHON-002 fibroblasts (Supplemental Figure S7, A and B), demonstrating that increased endogenous expression of this gene in tetraploid cells is not limited to just one cell type. To examine the functional importance of cyclin D2 in proliferating tetraploid RPE1 cells, we studied the effect of gradual reduction of cyclin D2 levels on proliferation. We used two different siRNAs against cyclin D2 at increasing concentrations and measured EdU incorporation (Figure 7, C and D). Knockdown efficiency of the highest siRNA concentration is shown in Figure 7E. Both siRNAs resulted in dosage-dependent reduction of EdU incorporation, and the dosage response was significantly sharper in tetraploid clones 4N-1 and 4N-3 than in their isogenic diploid 2N-1 and 2N-3 sister clones. This difference was not as pronounced as in clone 4N-2, which was exposed to Hoechst 33342 during its evolution and had the greatest gene expression changes overall. This clone also had highest levels of cyclin D2, and because cyclin D2 knockdown also caused some reduction of EdU incorporation in diploid cells, there may have been enough residual protein left to support proliferation. However, since exposure to DNA intercalating agents is mutagenic, we cannot exclude the possibility that this cell line may have carried additional proliferation drivers.
FIGURE 7:
Dependence of tetraploid RPE1 cells on cyclin D2 for proliferation. (A) Real-time qPCR analysis of cyclin D2 mRNA levels in proliferating tetraploid and isogenic diploid single-cell clones. Two different primer pairs yield similar results. All values were normalized to the parental RPE1. Bar heights represent the mean relative fold change (n = 3); error bars represent SEM. (B) Western blot analysis of endogenous cyclin D2 protein levels in in proliferating tetraploid and isogenic diploid single-cell clones. (C, D) Concentration-dependent effect on proliferation of two different cyclin D2 siRNAs in established tetraploid and isogenic diploid cell lines. The proliferation was assessed by EdU incorporation. Cells were treated with indicated siRNAs at 5, 10, 15, and 20 nM concentration for 72 h and allowed to incorporate EdU for 8 h. Percentages of the EdU-positive cells were normalized to the parental cell line not transfected with cyclin D2 siRNA (the number of EdU-positive cells in 0 nM siRNA samples was assigned 100% value). Each data point represents an average of four images. Error bars represent SD. (E) Western blot analysis of the cyclin D2 knockdown efficiency by two cyclin D2 siRNAs used in C and D. Cells were treated with 20 nM siRNA for 72 h.
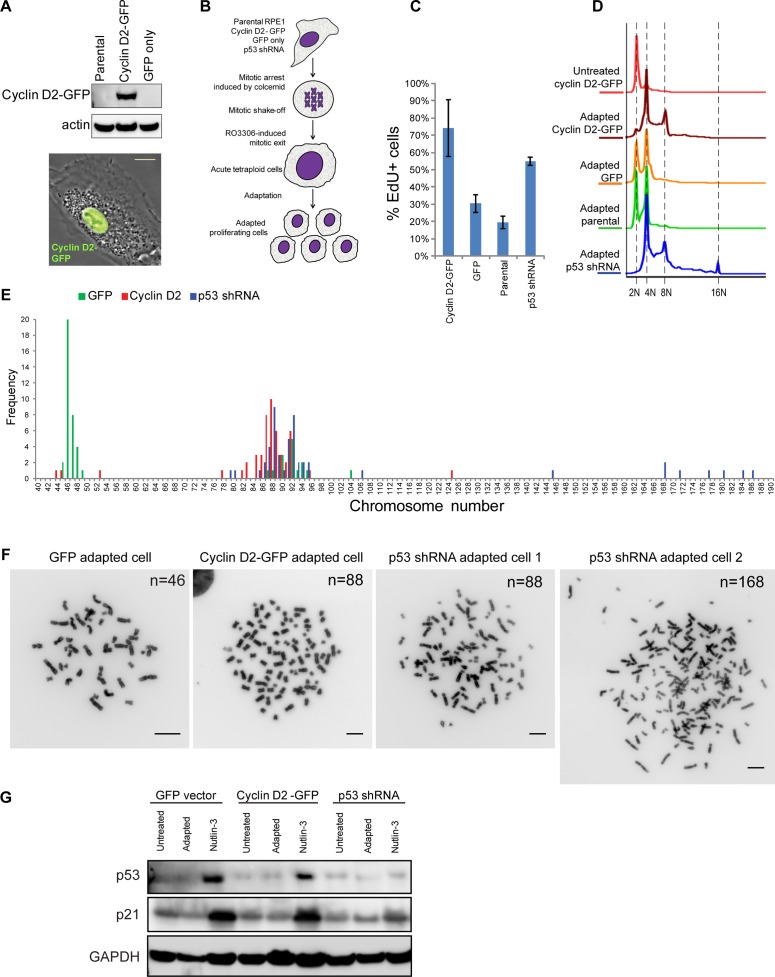
We next tested whether increased expression of cyclin D2 can potentiate long-term adaptation to the doubling of the genome. For this, we cloned human cyclin D2 into a green fluorescent protein (GFP)–tagging mammalian expression vector and generated a stable RPE1 cell line overexpressing cyclin D2–GFP (Figure 8A). For a negative control, we generated a cell line expressing empty GFP vector in parallel. For a positive control that can sustain proliferation after tetraploidization, we generated a stable cell line in which expression of p53 was silenced by a short hairpin RNA (shRNA). To ensure that the ability of cyclin D2 to potentiate adaptation to increased DNA content was not limited to instances in which tetraploidy was induced by Aurora kinase inhibitors, we used a different method of tetraploidy induction. Instead of using the Aurora inhibitor, we induced tetraploidy in these cell lines by mitotic exit in the presence of microtubule poison Colcemid, similar to the protocol used in Ohshima and Seyama (2013). Specifically, we collected mitotic cells after Colcemid treatment and induced mitotic exit with the CDK1 inhibitor RO-3306 (Vassilev et al., 2006). G1-arrested tetraploid cells were cultured until proliferating, adapted populations emerged, typically for 2–3 wk (Figure 8B). We observed that proliferating cells emerged first in cyclin D2–overexpressing cells and last in GFP-expressing and parental RPE1 cells. Among adapted cells that were generally slow growing, cyclin D2–overexpressing cells exhibited the greatest EdU incorporation rate (Figure 8C), indicating high proliferation. Ploidy analysis of adapted cyclin D2-GFP–overexpressing cells showed peaks at 4N and 8N DNA content (Figure 8D). Cyclin D2–overexpressing adapted cells had ploidy profile similar to that of p53-knockdown cells, with the note that cells expressing p53 shRNA also showed a small peak at 16N. By contrast, adapted parental and GFP-expressing cells showed nearly equivalent peaks of 2N and 4N populations. These data indicated that adapted cyclin D2–overexpressing cells can proliferate with increased ploidy similarly to cells lacking p53.
FIGURE 8:
Experimental adaptation to tetraploidy after forced mitotic exit. (A) Western blot (top) and phase contrast/fluorescence image (bottom) of RPE1 cells overexpressing cyclin D2–GFP. Bar, 10 μm. (B) Design of experimental adaptation to tetraploidy after forced mitotic exit. Adaptation experiments included cells overexpressing cyclin D2-GFP, GFP empty vector, p53 shRNA, and parental RPE1 cell lines of early passage. Mitotic cells were collected after incubation in Colcemid for 6 h and treated with 10 μM CDK1 inhibitor RO-3306 to induce mitotic exit. After drug washout, cells were left to survive until proliferating populations emerged. Each experiment was performed in triplicate. (C) EdU incorporation assay of adapted cells overexpressing cyclin D2-GFP, cells expressing GFP empty vector, parental cells, and cells expressing p53 shRNA. Cells were allowed to incorporate EdU for 24 h. Bar heights represent the mean (n = 4); error bars denote SD. (D) Ploidy distributions of adapted cells overexpressing cyclin D2-GFP, cells expressing GFP empty vector, parental cells, and cells expressing p53 shRNA. Cells were fixed and stained with propidium iodide. Histograms indicate doubling of the DNA content predominantly in cells overexpressing cyclin D2-GFP and cells expressing p53 shRNA. (E) Chromosome counts from adapted populations of cells overexpressing cyclin D2–GFP or GFP alone and expressing p53 shRNA. Fifty spreads were counted from each group. (F) Representative mitotic chromosome spreads from every adapted population; n, chromosome number. Bar, 10 μm. (G) Western blot analysis of p53 and p21 protein levels in cells expressing GFP, cyclin D2–GFP, and p53 shRNA before adaptation (untreated), after adaptation, and treated with 2.5 μM nutlin-3 for 24 h.
To assess ploidy content in individual adapted cells, we counted chromosomes in mitotic spreads from these cells (Figure 8, E and F). In cells expressing the empty GFP vector, chromosome counts peaked around 46–48, with a small proportion of spreads in the near-tetraploid (86–94) range. Of note, only few cells were perfectly tetraploid, confirming that proliferating tetraploid cells exhibit chromosome losses and genomic instability. Chromosome counts from cyclin D2–overexpressing cells peaked in the range 80–96, specifying that most of these cells were near-tetraploid aneuploid. Most cells expressing p53 shRNA also showed near-tetraploid aneuploid chromosomal content, but in addition some spreads had >140 chromosomes, sometimes ≥180, indicating that lack of p53 in some cases can permit even near-octoploid chromosome number.
Finally, we confirmed that the cyclin D2-GFP–overexpressing cells, which have the propensity to adapt to increased ploidy content, can activate p53. Examining protein levels of p53 and its transcription target p21 in cells before adaptation, after adaptation, and after treatment with nutlin-3 showed that when treated with nutlin, cyclin D2-GFP–overexpressing cells can accumulate p53 as robustly as cells that express GFP only (Figure 8G). Accumulation of p21 indicated that p53 is functional in these cells. P53 shRNA–expressing cells did not show accumulation of p53 or p21. Adapted tetraploid cells do not show a significant accumulation of p53, suggesting again that in proliferating cells already adapted to increased ploidy, p53 signaling is somehow abolished. Together these data indicate that cyclin D2 potentiates long-term adaptation to increased ploidy content in the presence of the functional p53 protein.
DISCUSSION
Polyploidy can lead to formation of aneuploid cells and catalyze vast genomic instability if polyploid cells have the capacity to proliferate. This work investigated consequences of acute (recent) tetraploidy and potential mechanisms of adaptation to the suppression of proliferation associated with it. Acute tetraploidy can result from many mitotic defects. Particularly common causes are cytokinesis failure and impaired mitotic exit. Morphologically, these tetraploid cells are distinct, and the first aim of our study was to compare physiological responses elicited in distinct instances of tetraploidization. We used two ways to induce acute tetraploidy. Inhibition of Aurora kinases induced formation of cells with aberrant multilobed nuclei—a consequence of defective mitotic exit—and knockdown of the cytokinesis regulator anillin resulted in formation of binucleate cells. Many previous studies obtained tetraploid binucleate cells by transient treatment with actin polymerization inhibitor cytochalasin B or D. Previous work (Uetake and Sluder, 2004) showed that treatment with high concentrations of cytochalasins (≤10 μM) causes enough stress to trigger cell cycle arrest, but lower concentrations (<5 μM) do not. We decided to avoid using cytochalasins in this study after finding that cytochalasin D treatment even at 2.5 μM causes some transcriptional activation of p53 regardless of ploidy (unpublished data).
The gene expression analysis of differentially induced tetraploid RPE1 cells showed a high degree of similarity among up-regulated genes, although overall Aurora kinase–inhibited cells had more differentially expressed transcripts than did anillin-knockdown cells (Figure 1D). Moreover, there was considerable overlap between genes enriched in tetraploid populations with genes induced in diploid cells by MDM2 inhibitor nutlin-3. Because nutlin-3 causes p53 accumulation in the absence of stresses such as DNA damage, this result indicated a close connection between acute tetraploidization and induction of p53. Genes up-regulated in tetraploid cells but not in diploid nutlin-3–treated cells were mostly metabolic (Supplemental Figure S2) and did not reveal an obvious linkage to the cell cycle arrest. RNAseq study detected only 12 genes that were significantly up-regulated in both Aurora-inhibited and binucleate tetraploid cells but did not show up in nutlin-3–treated cells (Figure 1C): ANK1, C18ORF56, CTSL2, FBXO22, KLLN, NAPRT1, PDE2A, RRAD, SCRIB, SRGAP3, TCEA3, and XPC. Nine of these genes (ANK1, C18ORF56, FBXO22, KLLN, PDE2A, RRAD, SCRIB, TCEA3, XPC) also tended to be up-regulated in nutlin-3–treated cells but did not meet our stringent fold-change and/or p-value criteria. Some of these genes (ANK1, KLLN, PDE2A, XPC) were confirmed to be up-regulated in nutlin-3–treated cells by the qPCR (Supplemental Figure S3), indicating that they may also be p53 targets. Together these data can imply that the category of genes we were looking for—up-regulated only in tetraploid cells but not in p53-activated diploid cells—may not exist. We can further speculate that the mechanism(s) triggering p53 activation in tetraploid cells may not rely on an upstream genetic network in our experimental paradigm.
A long-standing debate with regard to ploidy regulation concerned whether tetraploid cells with functional p53 arrest the cell cycle, because many tetraploid cells can divide again (Guidotti et al., 2003; Uetake and Sluder, 2004; Wong and Stearns, 2005). Our time-course analysis of ploidy increase showed that in both binucleate and Aurora kinase–inhibited populations, a considerable portion of cells stopped proliferating with tetraploid DNA content, whereas the rest of the cells arrested after reaching octoploid DNA content (Figure 2, F and G). Our finding that in some cells, the cell cycle blockage occurs at 4N DNA content and in others at 8N might reconcile this controversy. We also confirmed that the suppression of proliferation in these polyploid cells is principally mediated by the p53 pathway and relies on p21, a direct target of p53-dependent transcription.
Why do some cells arrest at 4N DNA content and others at 8N? The p53 protein levels vary and oscillate at different rates even in unperturbed cells (Loewer et al., 2010). It is possible that due to this endogenous variation, the level of p53 transcriptional activity required to trigger the G1/G0 arrest can be reached after becoming tetraploid, or after one more cell cycle. The amounts of G1/S cyclin/CDK complexes subject to inhibition by p21 may also vary from cell to cell. In rare events of cellular adaptation to tetraploidy, p21/CDK balance may shift toward CDK activation, leading to cell cycle progression. This is consistent with the finding that all adapted tetraploid single-cell clones overexpressed cyclin D2, which can activate CDKs 4, 6, and 2.
A recent study searching for genes involved in cessation of proliferation of binucleate tetraploid cells induced by transient incubation with cytochalasin B postulated that p53 in this experimental system may be activated via the Hippo tumor suppressor pathway (Ganem et al., 2014). Our RNAseq data did not show evidence of Hippo pathway activation in acute tetraploid cells. For instance, the expression of YAP/TAZ target genes such as INHBA, EDN1, and FSTL1 was not significantly decreased (unpublished data). These genes would be expected to show down-regulation if their transcriptional regulators YAP/TAZ were inactivated by the Hippo pathway kinases LATS1 and/or LATS2 (Mo et al., 2012; Yu et al., 2012; Lv et al., 2015). The aforementioned study also reported down-regulation of LATS2 transcript in evolved tetraploids generated from cytochalasin B–treated cells. In proliferating tetraploid single-cell clones generated in this study, the amount of transcripts of Hippo pathway kinases LATS1 and LATS2 was not reduced. Another recent study suggested that the tetraploid arrest was elicited by the CDK inhibitor p16INK4 (Panopoulos et al., 2014). Our experimental system, however, did not detect accumulation of the p16/INK4. The use of different experimental models and approaches may account for these differences, the reconciliation of which requires further exploration.
Cyclin D2 is recognized as an oncogene in many cancers. Copy-number gains of cyclin D2 are frequently detected in intracranial germ cell tumors (Terashima et al., 2014). Cyclin D2 was shown to be essential for proliferation of glioblastoma stem cells (Koyama-Nasu et al., 2013). Overexpression of cyclin D2 was reported in chronic lymphocytic leukemia, small lymphocytic lymphoma (Decker et al., 2002; Igawa et al., 2011), and T-cell acute lymphoblastic leukemia (Karrman et al., 2006). Increased expression of cyclin D2 was also associated with cancer invasiveness, specifically in human squamous carcinoma (Liu et al., 2002) and colorectal cancer (Sarkar et al., 2010). Search for genetic alterations of cyclin D2 using The Cancer Genome Atlas via the cBioPortal for Cancer Genomics (Cerami et al., 2012; Gao et al., 2013) revealed increased copy number of this gene in testicular, breast, and ovarian cancer (Supplemental Figure S8). To the best of our knowledge, there are no studies exploring cyclin D2 expression in the context of ploidy doubling and karyotype changes during tumor evolution.
In our study, the >100-fold increase in cyclin D2 mRNA and protein levels in proliferating tetraploid cells is not likely to result from gene copy-number gain. Cyclin D2 was expressed in diploid cells at a very low level, indicating that its transcription was suppressed (Figure 7, A and B). The expression of cyclin D2 may be regulated by various mechanisms. For instance, a subset of breast cancers silenced cyclin D2 by promoter methylation (Evron et al., 2001), whereas a study of gastric carcinoma showed that overexpression of cyclin D2 was correlated with undermethylation at its promoter (Oshimo et al., 2003). Cyclin D2 promoter was reported to be under control of various transcription factors in different experimental systems: Stat5 in T-cells (Moon et al., 2004), MYCN in developing neurons (Knoepfler et al., 2002; Alvarez-Rodriguez and Pons, 2009) and cardiomyocytes (Zhong et al., 2006), GATA4 in cardiomyocytes (Rojas et al., 2008; Yamak et al., 2012, 2014), and Gli in developing hair follicles (Mill et al., 2003). However, other mechanisms of controlling cyclin D2 gene expression may exist. For instance, a study exploring a superenhancer mechanism of regulating oncogenes detected formation of superenhancers associated with cyclin D2 in multiple myeloma and colorectal cancer samples (Hnisz et al., 2013). More research is needed to understand the molecular mechanism of activating cyclin D2 transcription during tetraploid adaptation.
Activation of the p53 tumor suppressor pathway is a powerful safeguard against the massive chromosome instability that would arise if polyploid cells—a frequent product of failure of several individual steps of mitotic cell division—did not halt further proliferation. The importance of this genome surveillance mechanism in prevention of tumorigenesis was highlighted by the finding that in a xenograft tumor model, tetraploid cells were tumorigenic only if derived from p53−/− cells (Fujiwara et al., 2005). Indeed, the p53 pathway is inactivated in most human cancers (Brown et al., 2009; Olivier et al., 2010). Acute polyploidization selects for mutations that have compromised p53 pathway because, as we and others have shown, p53-deficient tetraploid cells can continue to proliferate and give rise to karyotypically heterogeneous aneuploid population. Here, we also demonstrated that p53-dependent suppression of proliferation can be circumvented more easily by increasing levels of oncogenic drivers such as cyclin D2. This raises the possibility that abnormal increase in cyclin D2 expression due to promoter hyperactivation or copy-number gain might be an early driver of cancer development.
MATERIALS AND METHODS
Cell culture, plasmid, and siRNA transfection
hTERT RPE1 and hTERT CHON-002 (ATCC, Manassas, VA) were grown at 37°C in 5% CO2 in DMEM supplemented with 10% fetal bovine serum (FBS).
Plasmid encoding wild-type human cyclin D2 was obtained from Addgene (plasmid 8958). The cyclin D2 gene was inserted into pAcGFP1-N1 vector (Clontech, Mountain View, CA) using a Gibson assembly strategy and Gibson Assembly Master Mix (E2611S; NEB, Ipswich, MA) to generate cyclin D2-GFP.
To generate stable cell lines overexpressing human cyclin D2–GFP or expressing GFP only, RPE1 cells were transfected using X-tremeGENE 9 DNA Transfection Reagent (Roche, Basel, Switzerland) according to the manufacturer’s directions. Transfected cells were selected with 1 mg/ml G418 (A.G. Scientific, San Diego, CA). Emerged drug-resistant cells were FACS sorted for GFP expression. To generate RPE1 cells stably expressing p53 shRNA, cells were transduced with lentiviral particles (V3LHS_333920; GE Dharmacon, Lafayette, CO) in the presence of 10 μg/ml Polybrene (EMD Millipore, Darmstadt, Germany).
For siRNA-mediated knockdowns, RPE1 cells were transfected with siRNA using Lipofectamine RNAiMAX (Thermo Fisher Scientific, Waltham, MA) according to the manufacturer’s directions. siRNAs targeting indicated genes were obtained from GE Dharmacon. Table 1 lists siRNA sequences used. Knockdown of each mRNA is shown in Supplemental Figure S9.
TABLE 1:
siRNA sequences used in this study.
| Gene | SiRNA sequence |
|---|---|
| ANK1 | GAGUAGGGCUAGUGAAUGU |
| GUUAGGGACUUAAGAAUCU | |
| GAGUGAACUUGGUGCUCUA | |
| UAUCAUAGUCCGUGUUCAA | |
| BBC3 | CGGACGACCUCAACGCACA |
| CCGAGAUGGAGCCCAAUUA | |
| CCUGGAGGGUCCUGUACAA | |
| GUAGAUACCGGAAUGAAUU | |
| BTG2 | GAACCGACAUGCUCCCGGA |
| GCAUUCGCAUCAACCACAA | |
| GGUCAUAGAGCUACCGUAU | |
| AGACAAAGGUUACUAAUUG | |
| CYFIP2 | GCUGUUGGGUAGAUCAAUU |
| GACUUGAACAGACUCAUUA | |
| AGAUAGAGCUGGCCAGAUA | |
| GAGCUGAGUGUAAGAAUUA | |
| EDA2R | CAAUGUGCCUUCCAGUUGA |
| UUUCAGACCCAGCCACUUA | |
| GCCCUUAACUCUAAUGAGG | |
| GCAGUUUGAGGCUGAUAAA | |
| FDXR | UCAGCAGCAUUGGGUAUAA |
| UCACUAGACUGGAGGGUGU | |
| CAAGUGGCCUUCACCAUUA | |
| GAACGGACAUCACGAAGGC | |
| KLLN | CAAGAGACCUAGCGCAGAA |
| CCAAAAUAAUGUCGAAAGA | |
| UGAGAGAGCUGAUUCGGAA | |
| UGGAAAGUACGGAACGGUA | |
| MDM2 | GCCAGUAUAUUAUGACUAA |
| GAACAAGAGACCCUGGUUA | |
| GAAUUUAGACAACCUGAAA | |
| GAUGAGAAGCAACAACAUA | |
| PDE2A | GGAAAGUCCGGGAGGCUAU |
| CGACGGCCUUCUCCAUCUA | |
| CCCAUUCUCUCCUAUACAA | |
| GGCCAUGGUUCAGCAAGUU | |
| SESN1 | GAACUUGGCAUUAGAAUUC |
| GCUCAAUGGUUUAGAGAAU | |
| CAGCAGAGAUUCAACUACU | |
| UUAGUGAACCUGCAUGUAA | |
| SPATA18 | GCAGCUACGACUCGGAUUU |
| GGAGCUUGGAGGAGCGGAA | |
| GUUUGCAGAUGGAUCGUAA | |
| AUAAGAAACAGCUCCGAAA | |
| XPC | GCAAAUGGCUUCUAUCGAA |
| UGAAAUAUGAGGCCAUCUA | |
| GAGAAGUACCCUACAAGAU | |
| GGAGGGCGAUGAAACGUUU | |
| ATM | GCAAAGCCCUAGUAACAUA |
| GGUGUGAUCUUCAGUAUAU | |
| GAGAGGAGACAGCUUGUUA | |
| GAUGGGAGGCCUAGGAUUU | |
| ATR | GAGAAAGGAUUGUAGACUA |
| GCAACUCGCCUAACAGAUA | |
| CCACGAAUGUUAACUCUAU | |
| CCGCUAAUCUUCUAACAUU | |
| ANLN | GCAAACAACUAGAAACCAA |
| GGCGAUGCCUCUUUGAAUA | |
| GAUCAAGCAUUAGCAGAAA | |
| ACGCAACACUUUUGAAUUA | |
| CCND2 | GAUCGCAACUGGAAGUGUG |
| UGACUGAGCUGCUGGCUAA | |
| GCUCAGACCUUCAUUGCUC | |
| UCUCAAAGCUUGCCAGGAG | |
| CDKN1A | CGACUGUGAUGCGCUAAUG |
| CCUAAUCCGCCCACAGGAA | |
| CGUCAGAACCCAUGCGGCA | |
| AGACCAGCAUGACAGAUUU | |
| TP53 | GAAAUUUGCGUGUGGAGUA |
| GUGCAGCUGUGGGUUGAUU | |
| GCAGUCAGAUCCUAGCGUC | |
| GGAGAAUAUUUCACCCUUC |
Quantitative real-time PCR
RNA isolation was performed using a Maxwell 16 LEV simplyRNA Tissue kit on a Maxwell 16 Research Instrument (Promega, Madison, WI). Reverse transcription reactions were carried out using 2000 ng of each RNA template in 20 μl of total volume reactions using SuperScript VILO Master Mix (Thermo Fisher Scientific). Reactions containing cDNA were diluted for qPCR 10–15 times in 1× PerfeCTa SYBR Green FastMix (Quanta Biosciences, Beverly, MA) and cycled on the ABI 7900HT fast real-time PCR system (Applied Biosystems, Foster City, CA) according to ABI’s standard protocol. Each sample was measured in triplicate. Analysis of the fluorescence curves was done using ABI’s SDS2.4 software. All curves that showed errors as determined by the SDS2.4 software were excluded. The remaining Ct values were exported and analyzed using Biogazelle qBase plus, version 2.4, software to generate normalized relative quantities. Each primer pair was confirmed for linearity of amplification by analyzing standard curves. Primers used in this study are listed later. Hypoxanthine phosphoribosyltransferase (HPRT), ATP5B, ACTB, TBL1, B2M, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and ELF1 were used as endogenous controls. Each measurement was performed in triplicate. Table 2 lists primer sequences used for qPCR.
TABLE 2:
Primers used for qPCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Housekeeping genes | ||
| HPRT | TGCTGAGGATTTGGAAAGGG | ACAGAGGGCTACAATGTGATG |
| ATP5B | GATCCTCTAGACTCCACCTCTC | AGAAAGTTCATCCATACCCAGG |
| ACTB | ACCTTCTACAATGAGCTGCG | CCTGGATAGCAACGTACATGG |
| TBL1 | AATAGACCTCATGCCAGTTACG | GCAACAGCCTTTACATTGGG |
| B2M | GGCATTCCTGAAGCTGACAG | TGGATGACGTGAGTAAACCTG |
| GAPDH | ACATCGCTCAGACACCATG | TGTAGTTGAGGTCAATGAAGGG |
| ELF1 | CCTCAATATGGATTCCCCTGG | CTCTTAGGCTGTTCTGGTGATG |
| Genes of interest | ||
| ANK1 | CAATTCACGGATGAGCAGGG | CCTCTACAGTCACCTCCTCG |
| ATM | TGCTGACAATCATCACCAAGTTC | TCTCCCTTCGTGTCCTGGAA |
| ATR | AACTGGGTAGCTCGACTAGAAATGA | TTCATTGTAATCTGGTCTTTTGTTGAGA |
| BBC3 | CTCTCGGTGCTCCTTCACTC | GTGGTCACGTTTGGCTCATT |
| BTG2 | GAGGCACTCACAGAGCACTA | ATGGGGTCCATCTTGTGGTT |
| CDKN1A | TGTCACTGTCTTGTACCCTTG | GCGTTTGGAGTGGTAGAAATC |
| CCND2 (1) | AACCTGCTCACCATCGAGGA | CTGCGCATGTAGGGTTGGAT |
| CCND2 (2) | GCTGGGGAAGTTGAAGTGGA | TGCTTGCGGATCAGAGACAG |
| EDA2R | AGCTTACCGTGAAAGGAGGG | AATCCATGGTGGGAAGGAGTA |
| FDXR | TCCTACTGACCCCACCTG | CACTAGCCACACTGTCTTCAC |
| KLLN | GTTACACAAGCACCCACATC | TTTGGCTTGCTCTTAGGGTAG |
| MDM2 | TGCCAAGCTTCTCTGTGAAAG | TCCTTTTGATCACTCCCACC |
| PDE2A | ACCACAGACTTCTCCTCTGC | TTGTAGATCAGCTCCGCGAT |
| SESN1 | AGATGAGGCAGTTACAGGAATG | ATGACGAGATACAGCTCTTGC |
| SPATA18 | ACGAACGTTGCAGGAAAAGC | TTACGTCCTCCTTCTTGGGC |
| TP53 | TGTGACTTGCACGTACTCCC | ACCATCGCTATCTGAGCAGC |
| XPC | AGAACCACCCTCTGTATGCC | TGCACACAATCCCTGGAGTA |
Flow cytometry and cell volume measurements
For DNA content analysis, cells were trypsinized, fixed in 70% ethanol, and stained with FxCycle PI/RNase Staining Solution (Thermo Fisher Scientific) according to manufacturer’s directions. Data from labeled cells were collected on the MACSQuant flow cytometer (Miltenyi Biotec, Bergisch Gladbach, Germany) with 488-nm laser and analyzed using Flowjo software.
Cell size measurements were conducted on a Sony EC800 cytometer (Sony Biotechnology, San Jose, CA) equipped with a Coulter volume detector. A standard curve was generated using National Institute of Standards and Technology–certified size standard calibration particles, and cell samples were run using the same instrument settings as size standards. Forward scatter (FCS) files from the instrument were analyzed in FlowJo, and mean electronic volume (Coulter volume) data for the cell and bead populations was exported for analysis and plotting in Excel. Events were gated on light scatter (FSC and side scatter) to exclude small debris and large aggregates from analysis before exporting size data.
FACS sorting live cells for RNAseq
Cell sorting for diploid and tetraploid DNA content was performed on a BD (Franklin Lakes, NJ) Influx sorter running Sortware v1.0.1.6. Fluidics was set up at 20 psi with a 100-μm nozzle tip and 1× Leinco clear-sort sheath fluid (S632). Hoechst 33342 (Sigma-Aldrich, St. Louis, MO) was excited using 100-mW laser power at 351 nm and detected on detector using a 450/20 emission filter. Cells were gated first on FSC pulse width versus FSC pulse height to exclude debris, doubles, and aggregates before gating on DNA content by plotting 450/20 detector intensity histograms. Trypsinized cells were stained for 60 min at 37°C with 2 μg/ml Hoechst 33342 before sorting and then kept cold for the duration of sorting. Cells were sorted on doublet discriminator (FSC pulse width vs. FSC pulse height) and Hoechst intensity (pulse height) gates. Sorted cells were spun down and flash-frozen in liquid nitrogen to be used for RNA isolation. Each experiment was performed in triplicate—that is, three different populations of cells were FACS sorted on different days to generate biological replicates for RNAseq experiments. To access the quality of sorting, we plated a small number of sorted cells on glass-bottom dishes (MatTek), allowed them to attach for several hours, and imaged them live on a Nikon (Tokyo, Japan) TE2000 wide-field microscope equipped with an environmental chamber using the 20× air objective.
RNAseq library preparation, sequencing, and analysis
RNA isolation was performed using a Maxwell 16 LEV simplyRNA Tissue kit on a Maxwell 16 Research Instrument (Promega). The quality of RNA was analyzed by an Agilent 2100 Bioanalyzer. Total RNA was normalized to 100, 250, or 500 ng, and libraries were prepared according to manufacturer’s protocol using TruSeq RNA Sample Prep Kit v2 or TruSeq Stranded mRNA LT Kit (RS-122-2101 and RS-122-2001, Illumina). Resulting short-fragment libraries were checked for quality and quantity using an Agilent 2100 Bioanalyzer or LabChip GX (PerkinElmer, Waltham, MA) and the Qubit Fluorometer (Life Technologies, Carlsbad, CA). Libraries were pooled, requantified, and sequenced as 50–base pair single read on the Illumina (San Diego, CA) HiSeq 2500 instrument using HiSeq Control Software. After sequencing, Illumina Primary Analysis and Secondary Analysis were run to demultiplex reads for all libraries and generate FASTQ files. Sequencing data analysis was carried out by mapping sequence reads to version hg19 of the human genome (hgdownload.cse.ucsc.edu/downloads.html) using TopHat, version 2. Three biological replicates were assessed for gene expression differences. Data were analyzed in the R environment using the edgeR library (Robinson et al., 2010) for normalization and calculation of p values between treatments. The p values were adjusted for multiple hypothesis testing by the method of Benjamini and Hochberg (1995). Differentially expressed transcripts in anillin siRNA– and ZM447439-treated samples were specified by applying log2 4N/2N fold change (FC) ≥ 0.5 for enriched in 4N and log2 FC ≤ −0.5 for enriched in 2N; p adjusted ≤ 0.001. For 4N/2N sorted cells, analysis was performed by taking pairing information into account; that is, 4N and 2N samples from each round of sorting were treated as pairs. To select transcripts changed only in tetraploid G1 cells and not in diploid G2/M cells, we excluded transcripts with log2 FC ≥0.5 and log2 FC ≤ −0.5, p adjusted ≤ 0.001, which were also found in untreated controls. To find genes differentially expressed in nutlin-3–treated samples, we applied log2[treated/dimethyl sulfoxide (DMSO; vehicle)] ≥1 and ≤ −1, p adjusted ≤0.001, criteria. To find transcripts differentially expressed in proliferating tetraploid cells, we applied log2(4N clone/2N isogenic clone) ≥1 and ≤ −1, p adjusted ≤ 0.001, criteria and examined each isolate independently.
Data access
The raw data from this study have been submitted to the National Center for Biotechnology Information Gene Expression Omnibus (GSE86105; www.ncbi.nlm.nih.gov/geo/).
Generation of proliferating tetraploid cells
Proliferating tetraploid single-cell clones from acute polyploid Aurora-inhibited cells.
Asynchronously growing euploid RPE1 cells were seeded in a T75 flask and treated with 2.5 μM ZM447439 for 72 h. After the treatment, drug was washed out, and cells were cultured in the same vessel for several weeks. Growth medium was refreshed regularly, and cells were examined under the microscope every few days. Once proliferating populations in the vessel became apparent, cells were trypsinized and plated in 96-well plates at one cell per well density using a MoFlo flow cytometer. After single-cell plating, cells were cultured in regular growth medium until they reached confluency, generating proliferating cultures from single cells. Each well that contained proliferating cells was treated with trypsin to lift cells off the well. Part of the cell culture from each well was fixed for ploidy analysis and another part was cryopreserved. DNA content of each single-cell clone was determined by FACS. Tetraploid and isogenic diploid single-cell clones were expanded and subjected to further analysis.
Tetraploidy after forced mitotic exit and adaptation.
RPE1 cells expressing cyclin D2–GFP, GFP empty vector, p53 shRNA, and parental cells were incubated in Karyomax Colcemid solution (1:100; Life Technologies) for 6 h. Mitotic cells were collected by shake-off, seeded on plates, and treated with 10 μM CDK1 inhibitor RO-3306 overnight to induce mitotic exit. After drug washout, cells remained in culture and were examined under the microscope regularly until adapted, proliferating cell populations emerged. Adapted cells were trypsinized, expanded for one or two passages and subjected to experimental protocols.
Proliferating tetraploid CHON-002 single-cell clones.
Asynchronously growing hTERT CHON-002 cells were synchronized by double thymidine (2 mM) block, followed by three cycles of 12-h incubation with 0.1 μg/ml of nocodazole. Mitotic shake-off was performed at the last nocodazole treatment to enrich for mitotic cells. At 22 h after the last nocodazole treatment, single cells were sorted into 96-well plates and allowed to grow for 3–4 wk. DNA content analysis by FACS was performed to assess ploidy. Tetraploidy was confirmed by analyzing chromosome spreads.
Chromosome spreads, counting, and karyotyping
Chromosome spreads were obtained by arresting cells in mitosis in Karyomax Colcemid solution (1:100; Life Technologies) for 4–8 h, followed by harvesting using trypsin. Trypsinized cells were collected by centrifugation for 5 min at 300 × g and gently resuspended in a small amount of medium (∼1 ml). Resuspended cells were allowed to swell for 7–10 min in 0.4% KCl solution at room temperature and prefixed by addition of freshly prepared methanol: acetic acid (3:1) fixative solution (∼100 μl per 10 ml of total volume). Prefixed cells were collected by centrifugation and fixed in methanol:acetic acid (3:1) fixative solution. Spreads were dropped on a glass slide and incubated at 65°C for at least 2 h.
For counting, spreads were mounted in Vectashield containing 4′,6-diamidino-2-phenylindole (DAPI; Vector Laboratories, Burlingame, CA), imaged on a Nikon TE-2000 wide-field fluorescence microscope (60× oil immersion objective) equipped with a CoolSNAP HQ2 charge-coupled device (CCD) camera (Photometrics, Tucson, AZ), and counted using the Cell Counter plug-in in ImageJ (National Institutes of Health, Bethesda, MD). Spectral karyotyping was performed using in-house–generated chromosome paints and the open source Karyotype Identification via Spectral Separation analysis package for ImageJ. For a detailed description of probe generation, hybridization, and analysis procedures, see Potapova et al. (2015).
EdU labeling
For EdU incorporation assays, cells were typically seeded in multiwell black, optically clear bottom, tissue culture–treated plates (PerkinElmer) and treated with 10 μM EdU (Thermo Fisher Scientific) for the indicated times. Cells were fixed in 4% paraformaldehyde (PFA)/phosphate-buffered saline (PBS) for 15 min and then permeabilized with 0.5% Triton X-100. Fixed cells were washed with PBS and stained with 1 μM Alexa Fluor 488 or Alexa Fluor 555–conjugated azide diluted in PBS containing 2 mM CuSO4 and 50 mM ascorbic acid. To counterstain the DNA, Hoechst 33342 was added to 2 μg/ml. Cells were incubated for several hours or overnight at room temperature protected from light and evaporation and then washed three times with PBS.
High-throughput imaging, analysis, and quantification
Multiwell plates with labeled cells were imaged on a Celigo Imaging Cytometer (Nexcelom) or Nikon TE-2000 wide-field fluorescence microscope equipped with 10× air objective. Images were quantified with Celigo or MetaMorph software. Each image typically contained several hundred cells; three or four images were analyzed to determine the mean number of positive cells and SD.
Immunofluorescence and nuclear size measurement
For p-Rb and pH2AX immunofluorescence, cells were fixed in 4% PFA/PBS for 15 min and permeabilized with 0.5% Triton X-100. For centrin labeling, cells were grown on glass coverslips and fixed with ice-cold methanol for 10 min. Blocking was done with 5% boiled normal goat serum in PBS/0.5% Triton X-100. Primary and secondary antibodies were diluted in 2.5% (wt/vol) bovine serum albumin (BSA)/PBS/0.5% Triton X-100. Specimens were incubated with primary antibodies overnight, washed three times for 5–10 min, and incubated with fluorescently conjugated secondary antibodies for 2–4 h. All washes were performed with PBS/0.5% Triton X-100. DNA was counterstained with DAPI or Hoechst 33342 (Thermo Fisher Scientific). Vectashield (Vector Laboratories) was used for mounting. p-Rb and pH2AX images were acquired with the Nikon TE-2000 wide-field fluorescence microscope (20× air and 60× oil immersion objectives) equipped with a CoolSNAP HQ2 CCD camera. For p-Rb immunofluorescence, three or four tiled images containing hundreds of cells were analyzed to determine the mean number of positive cells and the SD. Centrin and tubulin images were acquired using an LSM710 confocal microscope (Zeiss, Oberkochen, Germany) using the 63×/1.40 numerical aperture oil objective. At least 30 cells were imaged for each condition. Data were visualized and analyzed using ImageJ. To measure nuclear size, fixed cells stained with DAPI and imaged on a Nikon TE-2000 microscope equipped with a 20× extralong–working distance air objective. After background subtraction, images were thresholded and the area of each object quantified using MetaMorph software.
Western blotting
Cells were collected by spinning down trypsinized cultures at ∼200 × g for 5 min at 4°C; trypsin was neutralized by addition of fetal bovine serum before centrifugation. Cell pellets were washed with ice-cold PBS and lysed in ice-cold RIPA buffer supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific) for 30 min. Lysates were cleared by centrifugation at 16, 000 × g for 10 min. at 4°C and boiled in NuPAGE protein sample buffer (Thermo Fisher Scientific) containing 5% 2-mercaptoethanol (Sigma-Aldrich). Protein samples were separated by SDS–PAGE in 4–12% Bis-Tris gels (Thermo Fisher Scientific), transferred to polyvinylidene fluoride membrane, blocked in SuperBlock (TBS) Blocking Buffer (Thermo Fisher Scientific). and washed with Tris-buffered saline/Tween-20. Primary antibodies were detected using horseradish peroxidase (HRP)–conjugated secondary antibodies and developed using the ECL 2 (Thermo Fisher Scientific) or WesternBright (Advansta, Menlo Park, CA) detection kit. Chemiluminescence was detected using G:Box Chemi XT4 (Syngene, Cambridge, UK).
Antibodies used in this study
We used the following antibodies:
p53 (7F5) rabbit monoclonal antibody (mAb), 2527 (1:500; Cell Signaling Technology, Danvers, MA)
p21 Waf1/Cip1 (12D1) rabbit mAb, 2947 (1:1000; Cell Signaling Technology)
Phospho-Rb (Ser-807/811; D20B12) rabbit mAb, 8516 (1:500; Cell Signaling Technology)
GAPDH (D16H11) rabbit mAb, 5174 (1:1000; Cell Signaling Technology)
Cyclin D2 (D52F9) rabbit mAb, 3741 (1:500; Cell Signaling Technology)
Anti-centrin, clone 20H5, 04-1624 (1:500; EMD Millipore)
Anti–α-tubulin antibody, ab15246 (1:500; Abcam, Cambridge, UK)
Phospho–histone H2AX (Ser-139; 20E3) rabbit mAb, 9718 (1:500; Cell Signaling Technology)
Anti–β-actin antibody, ab8227 (1:5000; Abcam)
Secondary antibodies for immunofluorescence (Alexa 488 and Alexa 594 conjugates) were obtained from Life Technologies and used at 1:500 dilution. Secondary HRP-conjugated antibodies for Western blotting were from Cell Signaling Technology and typically used at 1:5000 dilution.
Chemical inhibitors used in this study
We used the following chemical inhibitors: ZM447439 (Selleckchem, Houston, TX) at 2.5 μM, AMG 900 (Selleckchem) at 2 μM, Nutlin-3 (Sigma-Aldrich) at 2.5 μM, Bleomycin (BLEOCIN, EMD Millipore) at 3 μg/ml, and RO-3306 (Tocris, Bristol, UK) at 10 μM.
Supplementary Material
Acknowledgments
We thank Jennifer Gerton, Richard Shrock, and Gary Gorbsky for critical review of the manuscript and Jay Unruh and Mark Mattingly for constructive comments. We are grateful to the Molecular Biology, Flow Cytometry, and Imaging core facilities at the Stowers Institute for enabling many of our experiments. We also thank members of the Rong Li lab for discussions. This work was supported by a William Neaves Award from the Stowers Institute for Medical Research, Grant R35-GM-118172 from the National Institutes of Health to R.L., A*STAR Investigatorship Award 1437a00119 to G.R., and Postdoctoral Fellowship PF-12-129-01-CCG from the American Cancer Society to T.P.
Abbreviations used:
- CDK
cyclin-dependent kinase
- DAPI
4’,6-diamidino-2-phenylindole
- DMSO
dimethyl sulfoxide
- EdU
5-ethynyl-2’-deoxyuridine
- FACS
fluorescence-activated cell sorting
- GFP
green fluorescent protein
- GO
gene ontology.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0268) on August 24, 2016.
REFERENCES
- Abbas T, Dutta A. p21 in cancer: intricate networks and multiple activities. Nat Rev Cancer. 2009;9:400–414. doi: 10.1038/nrc2657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Rodriguez R, Pons S. Expression of the proneural gene encoding Mash1 suppresses MYCN mitotic activity. J Cell Sci. 2009;122:595–599. doi: 10.1242/jcs.037556. [DOI] [PubMed] [Google Scholar]
- Andreassen PR, Lohez OD, Lacroix FB, Margolis RL. Tetraploid state induces p53-dependent arrest of nontransformed mammalian cells in G1. Mol Biol Cell. 2001;12:1315–1328. doi: 10.1091/mbc.12.5.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balsas P, Galan-Malo P, Marzo I, Naval J. Bortezomib resistance in a myeloma cell line is associated to PSMbeta5 overexpression and polyploidy. Leuk Res. 2012;36:212–218. doi: 10.1016/j.leukres.2011.09.011. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- Brown CJ, Lain S, Verma CS, Fersht AR, Lane DP. Awakening guardian angels: drugging the p53 pathway. Nat Rev Cancer. 2009;9:862–873. doi: 10.1038/nrc2763. [DOI] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casenghi M, Mangiacasale R, Tuynder M, Caillet-Fauquet P, Elhajouji A, Lavia P, Mousset S, Kirsch-Volders M, Cundari E. p53-independent apoptosis and p53-dependent block of DNA rereplication following mitotic spindle inhibition in human cells. Exp Cell Res. 1999;250:339–350. doi: 10.1006/excr.1999.4554. [DOI] [PubMed] [Google Scholar]
- Castillo A, Morse HC, 3rd, Godfrey VL, Naeem R, Justice MJ. Overexpression of Eg5 causes genomic instability and tumor formation in mice. Cancer Res. 2007;67:10138–10147. doi: 10.1158/0008-5472.CAN-07-0326. [DOI] [PubMed] [Google Scholar]
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Bradford WD, Seidel CW, Li R. Hsp90 stress potentiates rapid cellular adaptation through induction of aneuploidy. Nature. 2012;482:246–250. doi: 10.1038/nature10795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Mulla WA, Kucharavy A, Tsai HJ, Rubinstein B, Conkright J, McCroskey S, Bradford WD, Weems L, Haug JS, et al. Targeting the adaptability of heterogeneous aneuploids. Cell. 2015;160:771–784. doi: 10.1016/j.cell.2015.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coward J, Harding A. Size does matter: why polyploid tumor cells are critical drug targets in the war on cancer. Front Oncol. 2014;4:123. doi: 10.3389/fonc.2014.00123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, de Lange T. Telomere-driven tetraploidization occurs in human cells undergoing crisis and promotes transformation of mouse cells. Cancer Cell. 2012;21:765–776. doi: 10.1016/j.ccr.2012.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davoli T, Denchi EL, de Lange T. Persistent telomere damage induces bypass of mitosis and tetraploidy. Cell. 2010;141:81–93. doi: 10.1016/j.cell.2010.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruin EC, McGranahan N, Mitter R, Salm M, Wedge DC, Yates L, Jamal-Hanjani M, Shafi S, Murugaesu N, Rowan AJ, et al. Spatial and temporal diversity in genomic instability processes defines lung cancer evolution. Science. 2014;346:251–256. doi: 10.1126/science.1253462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker T, Schneller F, Hipp S, Miething C, Jahn T, Duyster J, Peschel C. Cell cycle progression of chronic lymphocytic leukemia cells is controlled by cyclin D2, cyclin D3, cyclin-dependent kinase (cdk) 4 and the cdk inhibitor p27. Leukemia. 2002;16:327–334. doi: 10.1038/sj.leu.2402389. [DOI] [PubMed] [Google Scholar]
- Dewhurst SM, McGranahan N, Burrell RA, Rowan AJ, Gronroos E, Endesfelder D, Joshi T, Mouradov D, Gibbs P, Ward RL, et al. Tolerance of whole-genome doubling propagates chromosomal instability and accelerates cancer genome evolution. Cancer Discov. 2014;4:175–185. doi: 10.1158/2159-8290.CD-13-0285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duelli DM, Hearn S, Myers MP, Lazebnik Y. A primate virus generates transformed human cells by fusion. J Cell Biol. 2005;171:493–503. doi: 10.1083/jcb.200507069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- el-Deiry WS, Tokino T, Waldman T, Oliner JD, Velculescu VE, Burrell M, Hill DE, Healy E, Rees JL, Hamilton SR, et al. Topological control of p21WAF1/CIP1 expression in normal and neoplastic tissues. Cancer Res. 1995;55:2910–2919. [PubMed] [Google Scholar]
- Evron E, Umbricht CB, Korz D, Raman V, Loeb DM, Niranjan B, Buluwela L, Weitzman SA, Marks J, Sukumar S. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61:2782–2787. [PubMed] [Google Scholar]
- Fujiwara T, Bandi M, Nitta M, Ivanova EV, Bronson RT, Pellman D. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 2005;437:1043–1047. doi: 10.1038/nature04217. [DOI] [PubMed] [Google Scholar]
- Gadea BB, Ruderman JV. Aurora kinase inhibitor ZM447439 blocks chromosome-induced spindle assembly, the completion of chromosome condensation, and the establishment of the spindle integrity checkpoint in Xenopus egg extracts. Mol Biol Cell. 2005;16:1305–1318. doi: 10.1091/mbc.E04-10-0891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galipeau PC, Cowan DS, Sanchez CA, Barrett MT, Emond MJ, Levine DS, Rabinovitch PS, Reid BJ. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci USA. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Cornils H, Chiu SY, O’Rourke KP, Arnaud J, Yimlamai D, Thery M, Camargo FD, Pellman D. Cytokinesis failure triggers hippo tumor suppressor pathway activation. Cell. 2014;158:833–848. doi: 10.1016/j.cell.2014.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem NJ, Storchova Z, Pellman D. Tetraploidy, aneuploidy and cancer. Curr Opin Genet Dev. 2007;17:157–162. doi: 10.1016/j.gde.2007.02.011. [DOI] [PubMed] [Google Scholar]
- Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geuns-Meyer S, Cee VJ, Deak HL, Du B, Hodous BL, Nguyen HN, Olivieri PR, Schenkel LB, Vaida KR, Andrews P, et al. Discovery of N-(4-(3-(2-aminopyrimidin-4-yl)pyridin-2-yloxy)phenyl)-4-(4-methylthiophen-2-yl)p hthalazin-1-amine (AMG 900), a highly selective, orally bioavailable inhibitor of aurora kinases with activity against multidrug-resistant cancer cell lines. J Med Chem. 2015;58:5189–5207. doi: 10.1021/acs.jmedchem.5b00183. [DOI] [PubMed] [Google Scholar]
- Giacinti C, Giordano A. RB and cell cycle progression. Oncogene. 2006;25:5220–5227. doi: 10.1038/sj.onc.1209615. [DOI] [PubMed] [Google Scholar]
- Giam M, Rancati G. Aneuploidy and chromosomal instability in cancer: a jackpot to chaos. Cell Div. 2015;10:3. doi: 10.1186/s13008-015-0009-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannoudis A, Evans MF, Southern SA, Herrington CS. Basal keratinocyte tetrasomy in low-grade squamous intra-epithelial lesions of the cervix is restricted to high and intermediate risk HPV infection but is not type-specific. Br J Cancer. 2000;82:424–428. doi: 10.1054/bjoc.1999.0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gisselsson D, Jin Y, Lindgren D, Persson J, Gisselsson L, Hanks S, Sehic D, Mengelbier LH, Ora I, Rahman N, et al. Generation of trisomies in cancer cells by multipolar mitosis and incomplete cytokinesis. Proc Natl Acad Sci USA. 2010;107:20489–20493. doi: 10.1073/pnas.1006829107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- Gordon DJ, Resio B, Pellman D. Causes and consequences of aneuploidy in cancer. Nat Rev Genet. 2012;13:189–203. doi: 10.1038/nrg3123. [DOI] [PubMed] [Google Scholar]
- Guidotti JE, Bregerie O, Robert A, Debey P, Brechot C, Desdouets C. Liver cell polyploidization: a pivotal role for binuclear hepatocytes. J Biol Chem. 2003;278:19095–19101. doi: 10.1074/jbc.M300982200. [DOI] [PubMed] [Google Scholar]
- Hatch EM, Fischer AH, Deerinck TJ, Hetzer MW. Catastrophic nuclear envelope collapse in cancer cell micronuclei. Cell. 2013;154:47–60. doi: 10.1016/j.cell.2013.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MT, Karlseder J. DNA damage associated with mitosis and cytokinesis failure. Oncogene. 2013;32:4593–4601. doi: 10.1038/onc.2012.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano A, Kurimura T. Virally transformed cells and cytochalasin B. I. The effect of cytochalasin B on cytokinesis, karyokinesis and DNA synthesis in cells. Exp Cell Res. 1974;89:111–120. doi: 10.1016/0014-4827(74)90193-1. [DOI] [PubMed] [Google Scholar]
- Hnisz D, Abraham BJ, Lee TI, Lau A, Saint-Andre V, Sigova AA, Hoke HA, Young RA. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Boveri revisited: chromosomal instability, aneuploidy and tumorigenesis. Nat Rev Mol Cell Biol. 2009;10:478–487. doi: 10.1038/nrm2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horn HF, Vousden KH. Coping with stress: multiple ways to activate p53. Oncogene. 2007;26:1306–1316. doi: 10.1038/sj.onc.1210263. [DOI] [PubMed] [Google Scholar]
- Ichijima Y, Yoshioka K, Yoshioka Y, Shinohe K, Fujimori H, Unno J, Takagi M, Goto H, Inagaki M, Mizutani S, et al. DNA lesions induced by replication stress trigger mitotic aberration and tetraploidy development. PLoS One. 2010;5:e8821. doi: 10.1371/journal.pone.0008821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Sato Y, Takata K, Fushimi S, Tamura M, Nakamura N, Maeda Y, Orita Y, Tanimoto M, Yoshino T. Cyclin D2 is overexpressed in proliferation centers of chronic lymphocytic leukemia/small lymphocytic lymphoma. Cancer Sci. 2011;102:2103–2107. doi: 10.1111/j.1349-7006.2011.02046.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Incassati A, Patel D, McCance DJ. Induction of tetraploidy through loss of p53 and upregulation of Plk1 by human papillomavirus type-16 E6. Oncogene. 2006;25:2444–2451. doi: 10.1038/sj.onc.1209276. [DOI] [PubMed] [Google Scholar]
- Kallio MJ, McCleland ML, Stukenberg PT, Gorbsky GJ. Inhibition of aurora B kinase blocks chromosome segregation, overrides the spindle checkpoint, and perturbs microtubule dynamics in mitosis. Curr Biol. 2002;12:900–905. doi: 10.1016/s0960-9822(02)00887-4. [DOI] [PubMed] [Google Scholar]
- Karrman K, Andersson A, Bjorgvinsdottir H, Strombeck B, Lassen C, Olofsson T, Nguyen-Khac F, Berger R, Bernard O, Fioretos T, et al. Deregulation of cyclin D2 by juxtaposition with T-cell receptor alpha/delta locus in t(12;14)(p13;q11)-positive childhood T-cell acute lymphoblastic leukemia. Eur J Haematol. 2006;77:27–34. doi: 10.1111/j.0902-4441.2006.t01-1-EJH2553.x. [DOI] [PubMed] [Google Scholar]
- Kaya A, Gerashchenko MV, Seim I, Labarre J, Toledano MB, Gladyshev VN. Adaptive aneuploidy protects against thiol peroxidase deficiency by increasing respiration via key mitochondrial proteins. Proc Natl Acad Sci USA. 2015;112:10685–10690. doi: 10.1073/pnas.1505315112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SH, Wahl GM. p53 and pRb prevent rereplication in response to microtubule inhibitors by mediating a reversible G1 arrest. Cancer Res. 1998;58:396–401. [PubMed] [Google Scholar]
- Knoepfler PS, Cheng PF, Eisenman RN. N-myc is essential during neurogenesis for the rapid expansion of progenitor cell populations and the inhibition of neuronal differentiation. Genes Dev. 2002;16:2699–2712. doi: 10.1101/gad.1021202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama-Nasu R, Nasu-Nishimura Y, Todo T, Ino Y, Saito N, Aburatani H, Funato K, Echizen K, Sugano H, Haruta R, et al. The critical role of cyclin D2 in cell cycle progression and tumorigenicity of glioblastoma stem cells. Oncogene. 2013;32:3840–3845. doi: 10.1038/onc.2012.399. [DOI] [PubMed] [Google Scholar]
- Kuznetsova AY, Seget K, Moeller GK, de Pagter MS, de Roos JA, Durrbaum M, Kuffer C, Muller S, Zaman GJ, Kloosterman WP, et al. Chromosomal instability, tolerance of mitotic errors and multidrug resistance are promoted by tetraploidization in human cells. Cell Cycle. 2015;14:2810–2820. doi: 10.1080/15384101.2015.1068482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanni JS, Jacks T. Characterization of the p53-dependent postmitotic checkpoint following spindle disruption. Mol Cell Biol. 1998;18:1055–1064. doi: 10.1128/mcb.18.2.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linardopoulos S, Blagg J. Aurora kinase inhibition: a new light in the sky? J Med Chem. 2015;58:5186–5188. doi: 10.1021/acs.jmedchem.5b00918. [DOI] [PubMed] [Google Scholar]
- Liu SC, Bassi DE, Zhang SY, Holoran D, Conti CJ, Klein-Szanto AJ. Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol Carcinog. 2002;34:131–139. doi: 10.1002/mc.10057. [DOI] [PubMed] [Google Scholar]
- Liu G, Yong MY, Yurieva M, Srinivasan KG, Liu J, Lim JS, Poidinger M, Wright GD, Zolezzi F, Choi H, et al. Gene essentiality is a quantitative property linked to cellular evolvability. Cell. 2015;163:1388–1399. doi: 10.1016/j.cell.2015.10.069. [DOI] [PubMed] [Google Scholar]
- Loewer A, Batchelor E, Gaglia G, Lahav G. Basal dynamics of p53 reveal transcriptionally attenuated pulses in cycling cells. Cell. 2010;142:89–100. doi: 10.1016/j.cell.2010.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv XB, Liu CY, Wang Z, Sun YP, Xiong Y, Lei QY, Guan KL. PARD3 induces TAZ activation and cell growth by promoting LATS1 and PP1 interaction. EMBO Rep. 2015;16:975–985. doi: 10.15252/embr.201439951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald JI, Dick FA. Posttranslational modifications of the retinoblastoma tumor suppressor protein as determinants of function. Genes Cancer. 2012;3:619–633. doi: 10.1177/1947601912473305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis RL, Lohez OD, Andreassen PR. G1 tetraploidy checkpoint and the suppression of tumorigenesis. J Cell Biochem. 2003;88:673–683. doi: 10.1002/jcb.10411. [DOI] [PubMed] [Google Scholar]
- Mill P, Mo R, Fu H, Grachtchouk M, Kim PC, Dlugosz AA, Hui CC. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 2003;17:282–294. doi: 10.1101/gad.1038103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minn AJ, Boise LH, Thompson CB. Expression of Bcl-xL and loss of p53 can cooperate to overcome a cell cycle checkpoint induced by mitotic spindle damage. Genes Dev. 1996;10:2621–2631. doi: 10.1101/gad.10.20.2621. [DOI] [PubMed] [Google Scholar]
- Mo JS, Yu FX, Gong R, Brown JH, Guan KL. Regulation of the Hippo-YAP pathway by protease-activated receptors (PARs) Genes Dev. 2012;26:2138–2143. doi: 10.1101/gad.197582.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon JJ, Rubio ED, Martino A, Krumm A, Nelson BH. A permissive role for phosphatidylinositol 3-kinase in the Stat5-mediated expression of cyclin D2 by the interleukin-2 receptor. J Biol Chem. 2004;279:5520–5527. doi: 10.1074/jbc.M308998200. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- Ohshima S, Seyama A. Establishment of proliferative tetraploid cells from normal human fibroblasts. Front Oncol. 2013;3:198. doi: 10.3389/fonc.2013.00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaharski AJ, Sotelo R, Solorza-Luna G, Gonsebatt ME, Guzman P, Mohar A, Eastmond DA. Tetraploidy and chromosomal instability are early events during cervical carcinogenesis. Carcinogenesis. 2006;27:337–343. doi: 10.1093/carcin/bgi218. [DOI] [PubMed] [Google Scholar]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: origins, consequences, and clinical use. Cold Spring Harb Perspect Biol. 2010;2:a001008. doi: 10.1101/cshperspect.a001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ornitz DM, Hammer RE, Messing A, Palmiter RD, Brinster RL. Pancreatic neoplasia induced by SV40 T-antigen expression in acinar cells of transgenic mice. Science. 1987;238:188–193. doi: 10.1126/science.2821617. [DOI] [PubMed] [Google Scholar]
- Orth JD, Loewer A, Lahav G, Mitchison TJ. Prolonged mitotic arrest triggers partial activation of apoptosis, resulting in DNA damage and p53 induction. Mol Biol Cell. 2012;23:567–576. doi: 10.1091/mbc.E11-09-0781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshimo Y, Nakayama H, Ito R, Kitadai Y, Yoshida K, Chayama K, Yasui W. Promoter methylation of cyclin D2 gene in gastric carcinoma. Int J Oncol. 2003;23:1663–1670. [PubMed] [Google Scholar]
- Panopoulos A, Pacios-Bras C, Choi J, Yenjerla M, Sussman MA, Fotedar R, Margolis RL. Failure of cell cleavage induces senescence in tetraploid primary cells. Mol Biol Cell. 2014;25:3105–3118. doi: 10.1091/mbc.E14-03-0844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payton M, Bush TL, Chung G, Ziegler B, Eden P, McElroy P, Ross S, Cee VJ, Deak HL, Hodous BL, et al. Preclinical evaluation of AMG 900, a novel potent and highly selective pan-aurora kinase inhibitor with activity in taxane-resistant tumor cell lines. Cancer Res. 2010;70:9846–9854. doi: 10.1158/0008-5472.CAN-10-3001. [DOI] [PubMed] [Google Scholar]
- Piekny AJ, Glotzer M. Anillin is a scaffold protein that links RhoA, actin, and myosin during cytokinesis. Curr Biol. 2008;18:30–36. doi: 10.1016/j.cub.2007.11.068. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Unruh JR, Box AC, Bradford WD, Seidel CW, Slaughter BD, Sivagnanam S, Wu Y, Li R. Karyotyping human and mouse cells using probes from single-sorted chromosomes and open source software. Biotechniques. 2015;59:335–346. doi: 10.2144/000114362. [DOI] [PubMed] [Google Scholar]
- Potapova TA, Zhu J, Li R. Aneuploidy and chromosomal instability: a vicious cycle driving cellular evolution and cancer genome chaos. Cancer Metastasis Rev. 2013;32:377–389. doi: 10.1007/s10555-013-9436-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig PE, Guilly MN, Bouchot A, Droin N, Cathelin D, Bouyer F, Favier L, Ghiringhelli F, Kroemer G, Solary E, et al. Tumor cells can escape DNA-damaging cisplatin through DNA endoreduplication and reversible polyploidy. Cell Biol Int. 2008;32:1031–1043. doi: 10.1016/j.cellbi.2008.04.021. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Kong SW, Agarwal P, Gilliss B, Pu WT, Black BL. GATA4 is a direct transcriptional activator of cyclin D2 and Cdk4 and is required for cardiomyocyte proliferation in anterior heart field-derived myocardium. Mol Cell Biol. 2008;28:5420–5431. doi: 10.1128/MCB.00717-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salic A, Mitchison TJ. A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc Natl Acad Sci USA. 2008;105:2415–2420. doi: 10.1073/pnas.0712168105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar R, Hunter IA, Rajaganeshan R, Perry SL, Guillou P, Jayne DG. Expression of cyclin D2 is an independent predictor of the development of hepatic metastasis in colorectal cancer. Colorectal Dis. 2010;12:316–323. doi: 10.1111/j.1463-1318.2009.01829.x. [DOI] [PubMed] [Google Scholar]
- Selmecki AM, Dulmage K, Cowen LE, Anderson JB, Berman J. Acquisition of aneuploidy provides increased fitness during the evolution of antifungal drug resistance. PLoS Genet. 2009;5:e1000705. doi: 10.1371/journal.pgen.1000705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selmecki A, Forche A, Berman J. Aneuploidy and isochromosome formation in drug-resistant Candida albicans. Science. 2006;313:367–370. doi: 10.1126/science.1128242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Zeng JY, Zhuang CM, Zhou YQ, Yao HP, Hu X, Zhang R, Wang MH. Small-molecule inhibitor BMS-777607 induces breast cancer cell polyploidy with increased resistance to cytotoxic chemotherapy agents. Mol Cancer Ther. 2013;12:725–736. doi: 10.1158/1535-7163.MCT-12-1079. [DOI] [PubMed] [Google Scholar]
- Shen H, Perez RE, Davaadelger B, Maki CG. Two 4N cell-cycle arrests contribute to cisplatin-resistance. PLoS One. 2013;8:e59848. doi: 10.1371/journal.pone.0059848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sicinski P, Donaher JL, Geng Y, Parker SB, Gardner H, Park MY, Robker RL, Richards JS, McGinnis LK, Biggers JD, et al. Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384:470–474. doi: 10.1038/384470a0. [DOI] [PubMed] [Google Scholar]
- Taylor S, Peters JM. Polo and Aurora kinases: lessons derived from chemical biology. Curr Opin Cell Biol. 2008;20:77–84. doi: 10.1016/j.ceb.2007.11.008. [DOI] [PubMed] [Google Scholar]
- Terashima K, Yu A, Chow WY, Hsu WC, Chen P, Wong S, Hung YS, Suzuki T, Nishikawa R, Matsutani M, et al. Genome-wide analysis of DNA copy number alterations and loss of heterozygosity in intracranial germ cell tumors. Pediatr Blood Cancer. 2014;61:593–600. doi: 10.1002/pbc.24833. [DOI] [PubMed] [Google Scholar]
- Thompson DA, Desai MM, Murray AW. Ploidy controls the success of mutators and nature of mutations during budding yeast evolution. Curr Biol. 2006;16:1581–1590. doi: 10.1016/j.cub.2006.06.070. [DOI] [PubMed] [Google Scholar]
- Tse YC, Piekny A, Glotzer M. Anillin promotes astral microtubule-directed cortical myosin polarization. Mol Biol Cell. 2011;22:3165–3175. doi: 10.1091/mbc.E11-05-0399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint.”. J Cell Biol. 2004;165:609–615. doi: 10.1083/jcb.200403014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Tovar C, Chen S, Knezevic D, Zhao X, Sun H, Heimbrook DC, Chen L. Selective small-molecule inhibitor reveals critical mitotic functions of human CDK1. Proc Natl Acad Sci USA. 2006;103:10660–10665. doi: 10.1073/pnas.0600447103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vitale I, Senovilla L, Jemaa M, Michaud M, Galluzzi L, Kepp O, Nanty L, Criollo A, Rello-Varona S, Manic G, et al. Multipolar mitosis of tetraploid cells: inhibition by p53 and dependency on Mos. EMBO J. 2010;29:1272–1284. doi: 10.1038/emboj.2010.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- Vousden KH, Lane DP. p53 in health and disease. Nat Rev Mol Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- Wang Y, Toppari J, Parvinen M, Kallio MJ. Inhibition of Aurora kinases perturbs chromosome alignment and spindle checkpoint signaling in rat spermatocytes. Exp Cell Res. 2006;312:3459–3470. doi: 10.1016/j.yexcr.2006.04.026. [DOI] [PubMed] [Google Scholar]
- Wong C, Stearns T. Mammalian cells lack checkpoints for tetraploidy, aberrant centrosome number, and cytokinesis failure. BMC Cell Biol. 2005;6:6. doi: 10.1186/1471-2121-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamak A, Latinkic BV, Dali R, Temsah R, Nemer M. Cyclin D2 is a GATA4 cofactor in cardiogenesis. Proc Natl Acad Sci USA. 2014;111:1415–1420. doi: 10.1073/pnas.1312993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamak A, Temsah R, Maharsy W, Caron S, Paradis P, Aries A, Nemer M. Cyclin D2 rescues size and function of GATA4 haplo-insufficient hearts. Am J Physiol Heart Circ Physiol. 2012;303:H1057–H1066. doi: 10.1152/ajpheart.00250.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D, Welm A, Bishop JM. Cell division and cell survival in the absence of survivin. Proc Natl Acad Sci USA. 2004;101:15100–15105. doi: 10.1073/pnas.0406665101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu FX, Zhao B, Panupinthu N, Jewell JL, Lian I, Wang LH, Zhao J, Yuan H, Tumaneng K, Li H, Fu XD, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150:780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zack TI, Schumacher SE, Carter SL, Cherniack AD, Saksena G, Tabak B, Lawrence MS, Zhsng CZ, Wala J, Mermel CH, et al. Pan-cancer patterns of somatic copy number alteration. Nat Genet. 2013;45:1134–1140. doi: 10.1038/ng.2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarkowska T, Mittnacht S. Differential phosphorylation of the retinoblastoma protein by G1/S cyclin-dependent kinases. J Biol Chem. 1997;272:12738–12746. doi: 10.1074/jbc.272.19.12738. [DOI] [PubMed] [Google Scholar]
- Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33:116–128. doi: 10.1038/onc.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong W, Mao S, Tobis S, Angelis E, Jordan MC, Roos KP, Fishbein MC, de Alboran IM, MacLellan WR. Hypertrophic growth in cardiac myocytes is mediated by Myc through a Cyclin D2-dependent pathway. EMBO J. 2006;25:3869–3879. doi: 10.1038/sj.emboj.7601252. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.