Actin stimulates oligomerization and mitochondrial accumulation of Drp1, a mitochondrial fission protein. Drp1 binds actin filaments in an unusually dynamic manner that is strongly influenced by guanine nucleotide.
Abstract
Drp1 is a dynamin-family GTPase recruited to mitochondria and peroxisomes, where it oligomerizes and drives membrane fission. Regulation of mitochondrial Drp1 recruitment is not fully understood. We previously showed that Drp1 binds actin filaments directly, and actin polymerization is necessary for mitochondrial Drp1 oligomerization in mammals. Here we show the Drp1/actin interaction displays unusual properties that are influenced by several factors. At saturation, only a fraction Drp1 binds actin filaments, and the off-rate of actin-bound Drp1 is significantly increased by unbound Drp1. GDP and GTP accelerate and decelerate Drp1/actin binding dynamics, respectively. Actin has a biphasic effect on Drp1 GTP hydrolysis, increasing at low actin:Drp1 ratio but returning to baseline at high ratio. Drp1 also bundles filaments. Bundles have reduced dynamics but follow the same trends as single filaments. Drp1 preferentially incorporates into bundles at higher ionic strength. We measure Drp1 concentration to be ∼0.5 μM in U2OS cell cytosol, suggesting the actin-binding affinity measured here (Kd = 0.6 μM) is in the physiologically relevant range. The ability of Drp1 to bind actin filaments in a highly dynamic manner provides potential for actin filaments to serve as reservoirs of oligomerization-competent Drp1 that can be accessed for mitochondrial fission.
INTRODUCTION
Mitochondria routinely undergo fission and fusion events, helping to maintain cellular distribution and homeostasis. In neurons, fission allows for efficient mitochondrial transport, by which mitochondria travel long distances between the cell body and synapse (Schwarz, 2013). During mitosis, fission helps to partition the mitochondrial network between daughter cells (Taguchi et al., 2007; Kashatus et al., 2011). In addition, fission acts as quality control by removing damaged mitochondrial segments during mitophagy (Youle and van der Bliek, 2012). Defective mitochondrial dynamics are associated with several neurological disorders including Alzheimer’s, Huntington’s, Parkinson’s, and Charcot Marie Tooth diseases (Chen and Chan, 2009; Chang and Blackstone, 2010; Nunnari and Suomalainen, 2012).
The dynamin-family GTPase Drp1 provides a driving force during mitochondrial fission through its oligomerization and constriction capabilities (Bui and Shaw, 2013; Friedman and Nunnari, 2014; Hatch et al., 2014). Drp1 oligomerization and mitochondrial binding are in dynamic equilibrium even in the absence of fission, but fission signals target the maturation of mitochondrially bound Drp1 oligomers to fission sites (Ji et al., 2015). Even after Drp1 is stably recruited to fission sites, there is still a significant lag before fission, suggesting that additional maturation of the fission complex occurs.
The molecules that recruit Drp1 to fission sites are still being elucidated. Several proteins on the outer mitochondrial membrane bind Drp1, including Mff, MiD49, MiD51, and Fis1 (Gandre-Babbe and van der Bliek, 2008; Otera et al., 2010; Palmer et al., 2011). Depletion of some of these receptors in mammalian cells decreases Drp1 enrichment on mitochondria and increases mitochondrial length (Palmer et al., 2011, 2013; Loson et al., 2013; Shen et al., 2014; Osellame et al., 2016; Otera et al., 2016). Drp1 receptors perform a similar role in yeast, in which the Drp1 homologue, Dnm1, interacts with its mitochondrial receptor, Fis1, through adaptor protein Mdv1, which stimulates Dnm1 oligomerization and GTP hydrolysis (Lackner et al., 2009). The mitochondrial-specific lipid cardiolipin may also play a role in Drp1 assembly at fission sites due to cardiolipin's potent stimulatory effect on Drp1 GTPase activity (Bustillo-Zabalbeitia et al., 2014; Macdonald et al., 2014; Stepanyants et al., 2015). Finally, mitochondrial fission occurs preferentially at contact sites with the endoplasmic reticulum (ER; Friedman et al., 2011), possibly activating Drp1 recruitment molecules.
In addition to these factors, several laboratories have shown that actin polymerization at or near fission sites can stimulate oligomeric maturation of Drp1 on mitochondria in mammalian cells (De Vos et al., 2005; DuBoff et al., 2012; Korobova et al., 2013; Ji et al., 2015; Li et al., 2015; Manor et al., 2015). This actin polymerization is stimulated by a protein on the ER, INF2 (Korobova et al., 2013), in conjunction with a mitochondrially bound protein, Spire1C (Manor et al., 2015). Actin polymerization precedes Drp1 recruitment to mitochondria, and suppression of actin polymerization blocks Drp1 maturation into stable oligomers on mitochondria (Korobova et al., 2013; Ji et al., 2015). Biochemically, Drp1 binds actin filaments directly, and actin stimulates Drp1 GTPase activity (Ji et al., 2015). Other dynamin-family proteins also bind directly to actin filaments or actin-binding proteins (Schafer et al., 2002; Mooren et al., 2009; Gu et al., 2010; Palmer et al., 2015), and actin filaments precede recruitment of dynamin at clathrin-mediated endocytosis sites (Grassart et al., 2014).
An important question concerns how the multiple Drp1-interacting molecules coordinate to trigger effective Drp1 oligomeric maturation at fission sites. In this work, we investigate the unusual binding characteristics between Drp1 and actin filaments. We find that the interaction is extremely dynamic, with several factors dramatically influencing Drp1 turnover on actin filaments, including unbound Drp1, GTP, and GDP. Combined with our previous cellular findings that actin accumulates before Drp1 during mitochondrial fission (Ji et al., 2015), these results suggest that actin filaments may serve as a dynamic reservoir for oligomerization-competent Drp1.
RESULTS
Drp1 displays noncanonical actin filament–binding properties
Previously we reported that the human Drp1 000 isoform directly binds to actin filaments with an apparent Kd of 0.8 μM (Ji et al., 2015). We asked whether this affinity was in the physiological range by determining the cellular concentration of Drp1 in U2OS human osteosarcoma cells. By quantitative Western blotting, Drp1 concentration is 0.51 ± 0.09 μM in the cytoplasm, based on our calculation of cytoplasmic volume (see Materials and Methods; 6.31 × 105 ± 0.5 × 105 molecules/cell; Supplemental Figure S1, A–C). Drp1 migrates as three distinct bands, presumably reflecting splice variation or posttranslational modification (Chang and Blackstone, 2010; Otera et al., 2013; Strack et al., 2013). We determined the concentration of cytoplasmic actin to be 208 ± 41 μM (2.57 × 108 ± 0.36 × 108 molecules/cell; Supplemental Figure S1, D and E). Because 40–75% of actin is estimated to be polymerized in unstimulated mammalian cells (Pollard et al., 2000), an appreciable portion of Drp1 is predicted to interact with actin filaments in principle.
A peculiarity of the Drp1/actin filament–binding reaction in vitro is that less than half of the total Drp1 is bound to filaments at saturating actin concentration, as judged by high-speed cosedimentation assay (Ji et al., 2015). We conducted a series of sedimentation assays to determine the percentage Drp1 bound at saturation with higher accuracy and found that this value, as well as binding affinity, varies within the following ranges under identical conditions of 1.3 μM Drp1 and 65 mM NaCl (Supplemental Figure S2A): Kdapp from 0.25 to 0.87 μM (mean 0.59 ± 0.20 μM) and maximum percentage Drp1 bound from 31.1 to 51.2% (mean 42.3 ± 6.7%). Representative binding assays are shown in Figure 1A and Supplemental Figure S2B. Despite this variability, this assay allows testing of factors that contribute to the peculiar subsaturating binding of Drp1 to actin filaments.
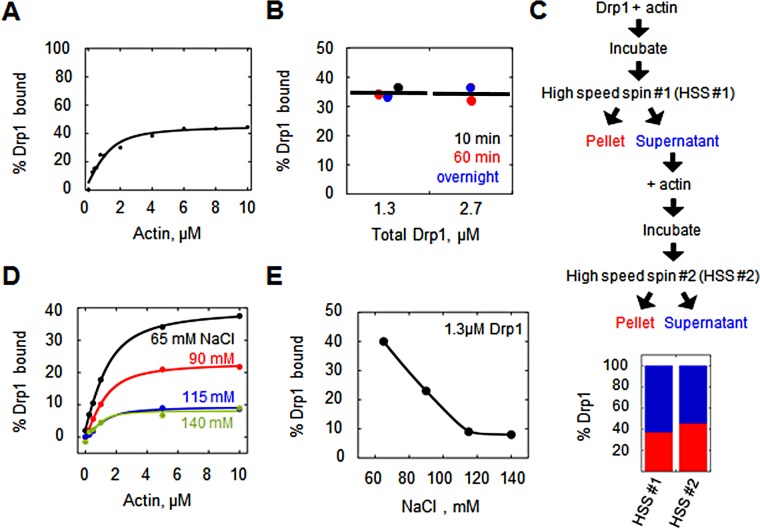
FIGURE 1:
Drp1 displays incomplete binding to actin filaments. (A) Percentage Drp1 bound vs. actin concentration from cosedimentation assays (raw data in Supplemental Figure S2B). Assay used 1.3 μM Drp1 and 65 mM NaCl. (B) Percentage Drp1 bound vs. Drp1 concentration at three incubation times (10 min, 60 min, and overnight) and two Drp1 concentrations (1.3 and 2.7 μM). Assay used 5 μM actin and 65 mM NaCl. (C) Sequential actin-binding assay containing 1.3 μM Drp1 and 65 mM NaCl, in which the supernatant from the first cosedimentation assay was added to fresh actin filaments for a second cosedimentation assay. Top, schematic; bottom, bar graph of results (red, Drp1 in pellet; blue, Drp1 in supernatant). Raw data in Supplemental Figure S2D. (D) Effect of ionic strength on Drp1/actin interaction. Cosedimentation assays containing 1.3 μM Drp1 and varying actin at varying ionic strength. (E) Percentage Drp1 bound at saturation of the cosedimentation assay in D.
One possible factor is a slow Drp1/actin binding equilibrium. However, varying the incubation time does not change the proportion of copelleting Drp1 (Figure 1B). This effect also is not due to Drp1-induced changes in the amount of polymerized actin, because this value does not change with changing Drp1 concentration (Supplemental Figure S2C), and the polymerization kinetics of actin are not altered by excess Drp1 (Ji et al., 2015).
Another possible explanation for the low percentage of Drp1 binding at saturation is that a significant proportion of the purified Drp1 is inactive. Arguing against this possibility, we previously showed that addition of a nonhydrolyzable GTP analogue (GMPPCP) causes near-complete oligomerization of Drp1 in solution (Ji et al., 2015). As an additional verification, we incubated our unbound Drp1 (the fraction remaining in the supernatant after incubation with saturating actin) with fresh actin filaments. A similar percentage of Drp1 binds actin in the second incubation (Figure 1C and Supplemental Figure S2D), indicating that the unbound Drp1 is capable of actin binding.
We also asked whether specific features of the actin filament could influence Drp1 binding. Drp1 binds throughout the length of the filament (Ji et al., 2015; our results), and so the availability of barbed or pointed ends does not appear to be a crucial factor. Another possibility is nucleotide state of the actin, because the affinity of another actin-binding protein, cofilin, is sensitive to this parameter, binding with lower affinity to ATP or ADP-Pi actin than to ADP actin (Pollard et al., 2000). In our assays, actin is predominantly in the ADP-bound state. We tested the effect of adding phosphate to the assays, which pushes actin to the ADP-Pi state (Carlier and Pantaloni, 1988). Phosphate does not change the maximal amount of Drp1 bound at saturation (Supplemental Figure S2E). In addition, the presence of rhodamine-phalloidin, which binds tightly to actin filaments, also has no apparent effect on Drp1 binding (Supplemental Figure S2F).
We also tested the effect of ionic strength on the Drp1/actin interaction, postulating that increasing ionic strength might reduce the affinity of the interaction. Surprisingly, apparent Drp1 binding affinity is similar or even slightly higher in these assays (0.7, 0.5, 0.4, and 0.2 μM for 65, 90, 115, and 140 mM NaCl, respectively; Figure 1D). However, the percentage of bound Drp1 at saturating actin is inversely proportional to ionic strength (Figure 1E).
These results suggest the existence of mechanisms that prevent complete Drp1 binding to actin filaments. Four possible explanations involve 1) Drp1 nucleotide state, 2) differential effect of Drp1 oligomeric state on actin-binding affinity, 3) noncanonical Drp1 binding kinetics to actin, and 4) differential binding to actin bundles versus single actin filaments. We test these possibilities, which are not mutually exclusive, in the remainder of this work.
The effects of GTP and GDP on actin binding by Drp1
We examined the effect of guanine nucleotides on Drp1 binding to actin filaments. We first asked whether our purified Drp1 preparation contained bound nucleotide, using anion exchange chromatography. No signal corresponding to GTP or GDP is detected by this analysis (Figure 2A). The detection limit of the assay is ∼5% of the concentration of Drp1 analyzed (30 μM), meaning that our Drp1 preparation is essentially nucleotide-free.
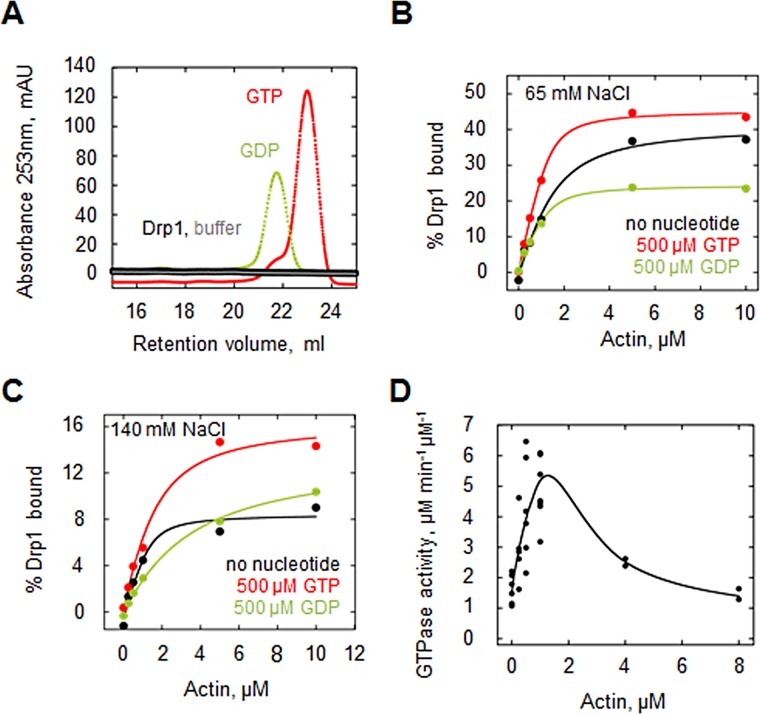
FIGURE 2:
Effects of GTP and GDP on Drp1/actin interaction. (A) Analysis of purified Drp1 for bound nucleotide. A fraction of purified Drp1 (30 μM) was boiled and centrifuged to remove the protein, and the supernatant was fractionated by anion exchange chromatography. No eluting peak corresponding to GTP or GDP was detected (analyzed at 30 and 15 μM, respectively). (B) Graph of Drp1 affinity for actin filaments (cosedimentation assay) in the absence of nucleotide (black curve) or in the presence of 500 μM GDP (green) or 500 μM GTP (red) at 65 mM NaCl. Assay used 1.3 μM Drp1 and 65 mM NaCl. (C) Drp1 affinity for actin filaments (cosedimentation assay) at high ionic strength (140 mM NaCl) in the absence of nucleotide (black curve) or in the presence of 500 μM GDP (green) or 500 μM GTP (red). Assay used 1.3 μM Drp1. (D) GTPase assays containing 1.3 μM Drp1 and 65 mM NaCl in the presence of actin filaments (prepolymerized for 1 h) for 5 min before GTP addition (250 μM). Activity expressed as micromoles phosphate released per minute per micromole Drp1. Seven experiments.
We measured Drp1/actin binding by cosedimentation assay in the presence of 500 μM GTP or GDP. This amount of GTP is sufficient to maintain GTP presence throughout the assay, with a calculated maximum of 160 μM GTP hydrolyzed. Both GTP and GDP cause apparent increases in Drp1 affinity for actin filaments, with Kd = 0.7, 0.2, and 0.2 μM for nucleotide-free, GTP, and GDP, respectively (Figure 2B). Of interest, however, GDP causes an approximately twofold decrease in the percentage of Drp1 bound to actin at saturating actin concentration, whereas GTP has negligible effect (41, 45, and 24% bound at saturation for nucleotide-free, GTP, and GDP, respectively; Figure 2B). At high ionic strength (140 mM NaCl), both GTP and GDP actually decrease Drp1 affinity for actin filaments with Kd = 0.2, 0.8, and 2.8 μM for nucleotide-free, GTP, and GDP, respectively (Figure 2C). Of interest, however, at this ionic strength, GTP causes an approximately twofold increase in the percentage of Drp1 bound to actin at saturating actin concentration (8, 16, and 13% bound for nucleotide-free, GTP, and GDP, respectively; Figure 2C).
Actin filaments have a biphasic effect on Drp1 GTP hydrolysis
In previous work, we showed that actin filaments stimulate Drp1 GTP hydrolysis (Ji et al., 2015). We expanded on this analysis and tested a wider range of actin filament concentrations (0–8 μM). Of interest, actin filaments exert a biphasic effect on Drp1 GTP hydrolysis: low actin filament concentrations stimulate GTP hydrolysis up to fourfold, but higher actin concentrations cause a return of hydrolysis rate to that of Drp1 alone (Figure 2D). One possibility is that Drp1 packs more tightly onto actin filaments at lower actin concentrations, facilitating GTP hydrolysis by bringing neighboring GTPase heads into close proximity (Chappie et al., 2010; Bui and Shaw, 2013).
Relationship between Drp1 oligomerization and actin binding
Purified Drp1 exists in several oligomeric states in solution (Frohlich et al., 2013; Macdonald et al., 2014). We find similar oligomeric heterogeneity for purified Drp1 by velocity analytical ultracentrifugation, with major species of ∼8, 10, and 15S when analyzed at 10 μM and 140 mM NaCl (Figure 3A). The 8S peak likely corresponds to Drp1 tetramers, based on past studies (Frohlich et al., 2013). The sedimentation profile changes appreciably at lower Drp1 concentration, with the 15S peak being negligible at 1.5 μM Drp1 and the 10S peak being substantially reduced at 0.75 μM Drp1 (Figure 3A). It is unclear whether the predominant peak at 0.75 μM (7.1S) represents Drp1 dimers (6S) or equilibrium between dimers and tetramers. However, these results show that there is clearly a redistribution to smaller oligomers at low Drp1 concentration, consistent with a dynamic equilibrium between the species. Of interest, there is no evidence of a monomer peak (4S) even at the lowest Drp1 concentration tested.
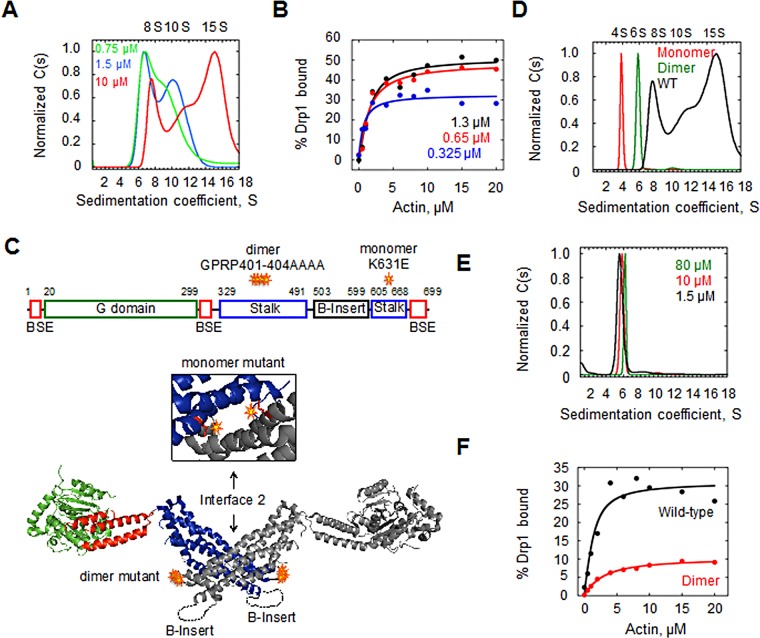
FIGURE 3:
Drp1 oligomeric state and actin binding. (A) Velocity analytical ultracentrifugation of WT Drp1 at concentrations 10, 1.5, and 0.75 μM (red, blue, and green, respectively) at 150 mM NaCl. (B) Binding of Drp1 to actin filaments (by cosedimentation assay) at 0.325, 0.67, and 1.3 μM Drp1 (blue, red, and black, respectively) and varying actin. Assay at 65 mM NaCl. (C) Positions of dimer and monomer mutants in Drp1. Top, bar diagram of Drp1 domain structure. Bottom, structural model from Protein Data Bank 4BEJ, showing the dimer mutations in the loop of the stalk domain, with inset showing an enlargement of the dimerization region (rotated 90° toward the viewer) and the position of the monomer mutations (red side chains). (D) Velocity analytical ultracentrifugation of 10 μM monomer mutant (red), dimer mutant (green), and WT Drp1 (black, same curve as in A). Assay, 150 mM NaCl. (E) Velocity analytical ultracentrifugation of dimer mutant at 1.5 (black), 10 (red, same curve as in D), and 80 μM. Assay at 150 mM NaCl. (F) Binding of 1.3 μM Drp1 WT (black) and dimer mutant (red) to actin filaments (by cosedimentation assay). Assay at 65 mM NaCl.
We tested the effect of varying Drp1 concentration on its ability to bind actin filaments. Partial binding occurs at three Drp1 concentrations tested (Figure 3B), with 0.325 μM Drp1 displaying the lowest actin-binding percentage at saturation (33%) and the 0.65 and 1.3 μM concentrations displaying 49 and 51% binding, respectively. The affinity for actin is in a similar range for the three Drp1 concentrations (Kdapp = 0.6, 1.1, and 0.9 μM for 0.325, 0.65, and 1.3 μM Drp1, respectively). Thus the degree of Drp1 binding is reduced at lower Drp1 concentration.
To measure directly the affinity of specific Drp1 oligomers for actin, we used Drp1 mutants previously shown to behave as monomer (K642E) and dimer (K401-404A) in solution (Frohlich et al., 2013; Figure 3C). In our Drp1-000 isoform, the analogous mutants maintain these properties, with the monomer mutant sedimenting as a single 4S species and the dimer mutant as a single 6S species (Figure 3D). The dimer mutant is resistant to higher-order oligomerization, as judged by two criteria. First, no higher S peaks occur at high Drp1 dimer concentration (80 μM; Figure 3E). Second, in the presence of GMPPCP, the dimer does not measurably sediment, whereas the wild-type (WT) protein is primarily in the pellet (Supplemental Figure S3). Of interest, the dimer mutant also does not display any measurable tendency to dissociate to monomers, since no 4S species appears even at 1.5 μM (Figure 3E).
We then tested the dimer mutant for actin filament binding by cosedimentation assay. The dimer mutant displays a lower affinity for actin filaments (Kdapp = 0.7 and 2.3 μM for WT and dimer mutant, respectively), as well as an approximately threefold lower percentage binding at saturation (31 vs. 10% binding for WT and dimer mutant; Figure 3F). Thus the inability to oligomerize affects both affinity and ability to saturate actin filaments. We cannot exclude the possibility that the mutated residues mediate direct contact with actin. The monomer mutant displays significantly lower affinity than both WT and dimer mutant Drp1 (unpublished data).
These results show that decreasing the oligomerization state of Drp1 causes a reduction in the maximal amount of Drp1 bound to actin. This effect is not simply due to reduced affinity, because the assays attain apparent saturated binding in all cases. An alternate explanation is that Drp1 exhibits noncanonical actin binding kinetics, which we examine in the next section.
Effect of unbound Drp1 on actin-bound Drp1
To examine the kinetics of the Drp1/actin interaction, we used total internal reflection fluorescence (TIRF) microscopy and green fluorescent protein (GFP)–tagged Drp1, similar to the system used previously (Ji et al., 2015). GFP-Drp1 binds filaments with similar affinity to untagged Drp1 but with lower percentage bound at saturation (Supplemental Figure S2G). To use defined concentrations of both Drp1 and actin, we modified our previous TIRF system by premixing GFP-Drp1 and tetramethylrhodamine isothiocyanate (TRITC)-phalloidin–stabilized filaments in solution for 1 h to reach equilibrium and then adding this mixture to the flow chamber. The inclusion of TRITC-phalloidin does not affect the Drp1/actin interaction significantly (Supplemental Figure S2F).
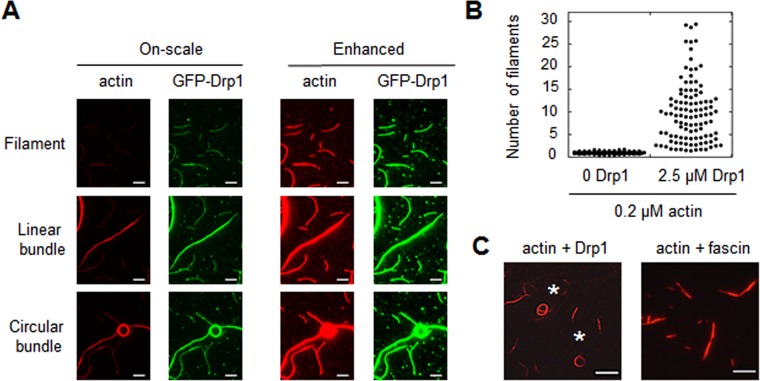
Previously we showed evidence that Drp1 can bundle actin filaments (Ji et al., 2015). Our modified TIRF system, in which binding reaches equilibrium in solution, reveals bundling more clearly than in the previous study and allows monitoring of single actin filaments and bundles in the same TIRF field (Figure 4A). Quantification of actin intensity reveals a wide distribution of bundle sizes, from single filaments to bundles containing ∼30 filaments (Figure 4B). Low-speed sedimentation assays, in which only actin bundles sediment, confirm the ability of Drp1 to bundle actin over a range of concentrations (Supplemental Figure S4, A and B). Curiously, a subset (26%; 386 bundles counted) of the bundles assumes a circular appearance (Figure 4A), with aspect ratios close to 1 (Supplemental Figure S4C) and diameters generally in the 4- to 6-μm range (Supplemental Figure S4D). These circular bundles are not present using another bundling protein, fascin (Figure 4C, 0 circular bundles from 375 total bundles).
FIGURE 4:
Actin bundling by Drp1. (A) TIRF microscopy images of 0.2 μM TRITC-phalloidin-stabilized actin filaments with 2.5 μM GFP-Drp1. On left, all images processed similarly, such that bundle intensity is within the linear range (“on-scale”). On right, images are processed with enhanced brightness, to reveal single filaments. Scale bar, 5 μm. (B) Graph quantifying number of filaments per bundle, based on TRITC-phalloidin fluorescence, for 0.2 μM actin in the absence or presence of 2.5 μM Drp1. 112 and 107 particles measured for actin alone and actin + Drp1, respectively. (C) Bundles assembled by Drp1 (2.5 μM) or fascin (2.5 μM) on 0.2 μM actin filaments (TRITC-phalloidin stabilized). Asterisks denote two circular bundles for Drp1. Scale bar, 10 μm.
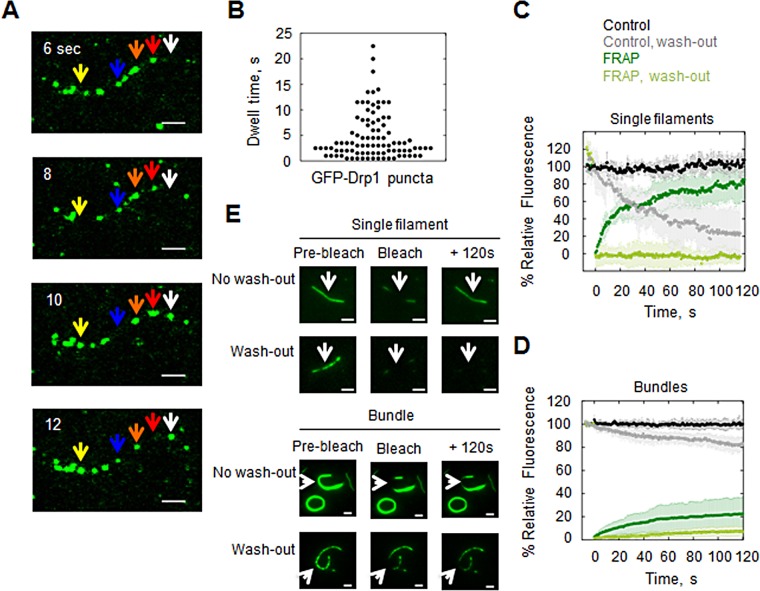
We examined Drp1 binding kinetics on both single actin filaments and large bundles (bundles >10 filaments). First, we observed GFP-Drp1 at steady state on single filaments when mixed at 1:99 ratio with unlabeled Drp1 to allow detection of individual GFP-Drp1 puncta. The overall Drp1:actin ratio at saturation is 20:1. These puncta are highly dynamic on the filament (Figure 5A), with a broad dwell-time distribution of 4.8 ± 4.6 s (Figure 5B and Supplemental Movie S1).
FIGURE 5:
Dynamics of GFP-Drp1 on actin filaments. (A) Time-lapse montage of steady-state GFP-Drp1 dynamics on single actin filament. Assay used 0.2 μM actin (TRITC-phalloidin stabilized) with 1 μM Drp1 (1% GFP-Drp1). Arrows denote examples of dynamic GFP-Drp1 puncta: yellow, binds between 8 and 10 s; blue, releases at 10 s and reappears at 12 s; orange, varies in size, or red, increases in size, and then disappears at 12 s; white, appears at 10 s. Scale bar, 2 μm. See Supplemental Movie S1. (B) The GFP-Drp1 dwell time on actin filaments. Mean dwell time is 4.8 ± 4.6 s (seven filaments, 92 punctae). (C) FRAP on single actin filaments saturated with GFP-Drp1 (2.5 μM GFP-Drp1, 0.2 μM actin) under two conditions: without washing out unbound GFP-Drp1 before photobleaching or with prior washout. (D) FRAP on actin bundles, following the same procedure as in C. Four to six filaments for each condition. (E) Representative images of each FRAP condition before photobleaching, immediately after bleaching, or at 120 s after photobleaching. Scale bar, 2 μm. See Supplemental Movies S2–S5.
Fluorescence recovery after photobleaching (FRAP) analysis of single actin filaments saturated with GFP-Drp1 shows that the majority GFP signal recovers (mobile fraction of 0.9 at 120 s; Figure 5, C and E, and Supplemental Movie S2). The recovery pattern does not fit a single exponential, suggesting that there are at least two populations of bound Drp1 (fast population, 43%, with t1/2 = 4.43 s; slow population, 47%, with t1/2 = 50.3 s). The recovery rate of GFP-Drp1 on actin bundles is much slower, with only partial recovery at 120 s (Figure 5, D and E, and Supplemental Movie S4).
In these FRAP experiments, excess GFP-Drp1 is in solution (2.5 μM). We also measured FRAP after washout of unbound GFP-Drp1. For both single filaments and actin bundles, no recovery occurs in the absence of unbound GFP-Drp1 (Figure 5, C–E, and Supplemental Movies S4 and S5). This result suggests that individual GFP-Drp1 units do not exchange laterally along the filament under these conditions.
We were surprised that GFP-Drp1 remained on actin filaments for a sufficiently long time after washout of free GFP-Drp1 to conduct FRAP experiments, since the FRAP rate in the presence of free GFP-Drp1 suggests that 90% of bound Drp1 should release from the filament within 120 s. We hypothesized that the off-rate of actin-bound GFP-Drp1 might be influenced by the GFP-Drp1 in solution. To test this hypothesis, we measured GFP-Drp1 dissociation rate from actin filaments after washing out unbound GFP-Drp1 under two conditions: washing out with buffer alone or buffer containing excess unlabeled Drp1. On washout with buffer alone, GFP-Drp1 signal from single filaments decays with t1/2 = 40 s (Figure 6, Table 1, and Supplemental Movie S6). In washout buffer containing unlabeled Drp1, the off-rate is fivefold faster, with a t1/2 = 8.1 s (Figure 6, Table 1, and Supplemental Movie S7). This result suggests that the association and dissociation reactions for Drp1 with actin filaments are not independent processes, in that Drp1 in solution accelerates the release of filament-bound Drp1. A similar property has been observed for fascin (Courson and Rock, 2010).
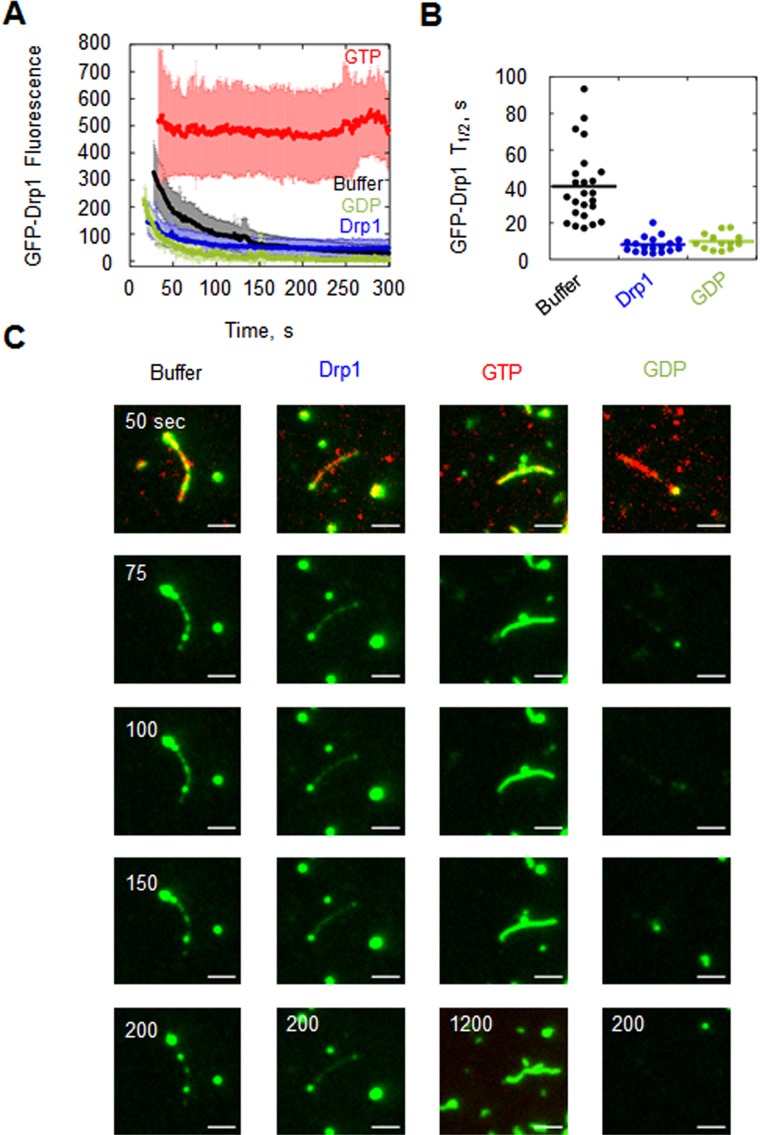
FIGURE 6:
Dissociation kinetics of GFP-Drp1 from actin is influenced by unbound Drp1 and by nucleotide. (A) GFP-Drp1 fluorescence decay from single actin filaments when free GFP-Drp1 is washed out with buffer alone (black curve), buffer containing 2.5 μM unlabeled Drp1 (blue), 500 μM GTP (red), or 500 μM GDP (green). Twenty-four, 19, 15, and 13 filaments were quantified, respectively. (B) GFP-Drp1 t1/2 values. (C) Representative images of GFP-Drp1 dissociation from single filaments. The 1200-s point is shown for GTP only, to show the persistence of binding. Scale bar, 2 μm. See Supplemental Movies S6–S9.
TABLE 1:
Dissociation parameters of GFP-Drp1 from actin filaments.
| Single filaments | Bundles | |||
|---|---|---|---|---|
| t½, s | Number of filaments | t½, s | Number of bundles | |
| Buffer | 40.0 ± 20.3 | 24 | 93.1 ± 88.0 | 13 |
| 2.5 μM Drp1 | 8.1 ± 4.3 | 18 | 84.1 ± 93.6 | 11 |
| 500 μM GTP | — | 15 | 93.2 ± 94.5 | 8 |
| 500 μM GDP | 9.9 ± 4.4 | 13 | 48.7 ± 20.6 | 9 |
See also Figure 6 (single filaments) and Supplemental Figure S5 (bundles).
GTP and GDP differentially influence Drp1 binding dynamics on actin filaments
We also measured off-rate upon washout in buffer containing 500 μM GTP or GDP. In washout buffer containing GTP, the off-rate is immeasurably slow, with >90% of the GFP-Drp1 remaining after 5 min (Figure 6A and Supplemental Movie S8) and significant fluorescence remaining after 20 min (Figure 6B). In washout buffer containing GDP, the off-rate is fourfold faster than buffer alone, with t1/2 = 9.8 s (Figure 6, Table 1, and Supplemental Movie S9). We also conducted a variation of this experiment, preloading filaments with GFP-Drp1 and GTP and then washing out free GFP-Drp1 with a variety of buffers (no nucleotide, unlabeled Drp1, GTP, unlabeled Drp1 plus GTP, GDP, or unlabeled Drp1 plus GDP). In this experiment, the off-rates are similar to those determined when filaments are preloaded in the absence of GTP, suggesting that the effect of GTP on off-rate is readily reversible (Supplemental Figure S6, A–C, and Table 2). The combination of GTP and unlabeled Drp1 in the washout buffer produces an intermediate dissociation rate of GFP-Drp1. The off-rates from actin bundles are slower those for than single filaments under all conditions. However, addition of GDP increases t1/2 1.9-fold (Supplemental Figure S5 and Table 1). The nucleotide results support differential roles of GTP and GDP in Drp1 interaction kinetics with actin. Of interest, washout with GTP induces “morphology” changes in otherwise stable bundles, in which the filaments in bundles appear to reorganize or leave the bundle completely (Supplemental Movie S10).
TABLE 2:
Dissociation parameters of GFP-Drp1 from actin filaments after charging with GTP.
| Single filaments | ||
|---|---|---|
| t½, s | Number of filaments | |
| Buffer | 26.9 ± 20.6 | 19 |
| 2.5 μM Drp1 | 4.1 ± 1.4 | 12 |
| 2.5 μM Drp1 + 500 μM GTP | 114.6 ± 43.5 | 8 |
| 500 μM GTP | — | 11 |
| 2.5 μM Drp1 + 500 μM GDP | 5.4 ± 2.0 | 9 |
| 500 μM GDP | 16.5 ± 2.5 | 5 |
See also Supplemental Figure S6, A–C.
We also used FRAP analysis to test whether GTP changes Drp1 binding dynamics when GTP is present with GFP-Drp1 in solution. GTP causes a slight reduction in recovery, reducing the mobile fraction to 66%, and a single recovery rate is observed with t1/2 = 25 s (Supplemental Figure S6, D and E). Similar to the off-rate results in Supplemental Figure S6, A–C, these results suggest that free Drp1 in solution mitigates the stabilizing effect of GTP on actin binding, increasing interaction dynamics.
We also tested the effect of ionic strength on GFP-Drp1 binding dynamics to actin using TIRF microscopy. GFP-Drp1 is not readily detectable on actin filaments at high ionic strength (150 mM KCl; Figure 7A). However, the presence of actin bundles suggests Drp1 binding below the detection limit of our system. In the presence of GTP, GFP-Drp1 is readily visible on actin bundles but still not visible on single filaments (Figure 7B and Supplemental Figure S7). Of interest, when buffer containing GTP and GFP-Drp1 is washed into a mix containing GFP-Drp1 and actin (without nucleotide), GFP-Drp1 rapidly accumulates on actin bundles (Figure 7C). FRAP analysis of GFP-Drp1–bound actin bundles at 150 mM KCl in the presence of GTP shows a biphasic recovery, including a fast component (20%) with t1/2 = 10 s and a slower component that does not reach a plateau during the assay time (Figure 7, D and E). Of interest, inclusion of GTP at 50 mM KCl renders GFP-Drp1 actin bundles more dynamic than without nucleotide (compare Figure 7D with Figure 5D).
FIGURE 7:
Dynamics of GFP-Drp1/actin interaction at high ionic strength. (A, B) TIRF microscopy images of 0.2 μM TRITC-phalloidin–stabilized actin filaments with 2.5 μM GFP-Drp1 in buffer containing 150 mM KCl in the absence (A) or presence (B) of 500 μM GTP. Scale bar, 5 μm. (C) Representative TIRF microscopy images of GFP-Drp1 association to actin bundles in the presence of GTP at 150 mM KCl, where an initial condition of 0.2 μM TRITC-phalloidin–stabilized actin filaments and 2.5 μM GFP-Drp1 (no GTP) is followed by wash-in of buffer containing 500 μM GTP and 2.5 μM GFP-Drp1. Scale bar, 5 μm. (D) FRAP on actin bundles saturated with GFP-Drp1 (2.5 μM GFP-Drp1, 0.2 μM actin) in the presence of 500 μM GTP at 50 and 150 mM KCl. Six and three bundles, respectively. (E) Representative images of each FRAP condition before photobleaching, immediately after bleaching, or at 120 s after photobleaching. Scale bar, 2 μm.
Nonhydrolyzable GTP eliminates dynamics of Drp1 binding to actin
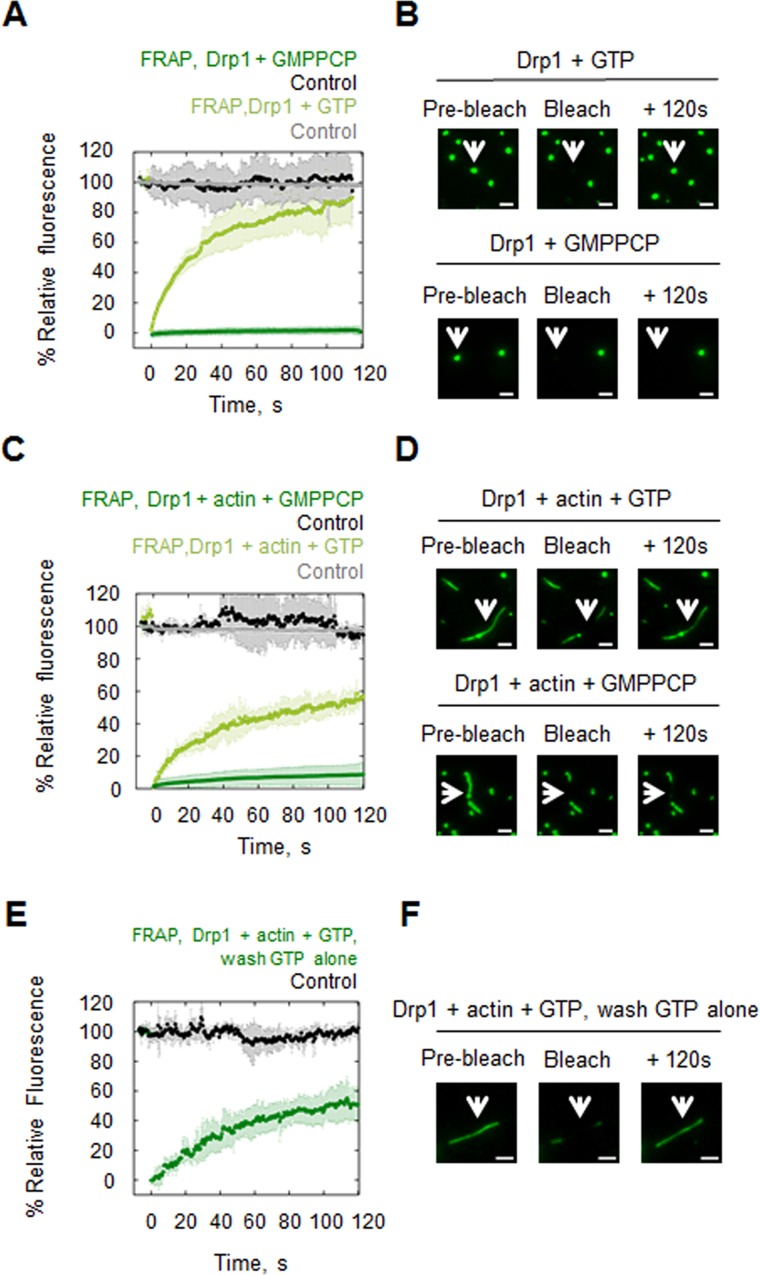
Our data clearly show that the presence of GTP significantly slows but does not eliminate the dynamics of the Drp1/actin interaction. We sought to test whether the residual kinetics was dependent upon GTP hydrolysis. To this end, we investigated the effect of the nonhydrolyzable analogue GMP-PCP on Drp1/actin interaction. Owing to the fact that GMP-PCP rapidly induces assembly of large Drp1 oligomers (Ingerman et al., 2005; Ji et al., 2015), we could not analyze actin binding by cosedimentation. Instead, we used TIRF and FRAP to examine recovery rates from both GFP-Drp1 oligomers induced by GTP or GMP-PCP, as well as from actin-bound GFP-Drp1 in the presence of these nucleotides. For Drp1 oligomers assembled in the presence of GTP without actin present, FRAP recovery follows a single exponential, with t1/2 = 19 s and mobile fraction of 83% (Figure 8, A and B), demonstrating that these are highly dynamic structures. In contrast, Drp1 oligomers assembled in the presence of GMP-PCP without actin present display no recovery after bleaching (Figure 8, A and B). Similarly, actin-bound GFP-Drp1 recovers rapidly after photobleaching when GTP is present (Figure 8, C and D). In contrast, there is no recovery of GFP-Drp1 on actin filaments in the presence of GMP-PCP (Figure 8, C and D). These experiments were conducted without washing out unbound GFP-Drp1. Of interest, after washout of unbound GFP-Drp1 with buffer containing GTP, recovery of actin-bound GFP-Drp1 occurs appreciably (t1/2 = 35 s and mobile fraction 57%; Figure 8, E and F), whereas no recovery is observed in the absence of nucleotide (Figure 5C). We postulate that recovery is due to GFP-Drp1 release from unbleached filaments or Drp1 oligomers and rebinding to bleached regions. Because this effect is not observed in the absence of nucleotide, GTP is likely to increase on-rate of the Drp1/actin interaction.
FIGURE 8:
Effects of GTP and GMP-PCP on Drp1 oligomer dynamics and actin binding dynamics. (A) FRAP on GFP-Drp1 puncta (2.5 μM GFP-Drp1, no actin present) in the presence of 500 μM GTP or 500 μM GMPPCP. Six Drp1 puncta for each condition. (B) Representative images of each FRAP condition before photobleaching, immediately after bleaching, or at a late time point after photobleaching. Scale bar, 2 μm. (C) FRAP on actin filaments saturated with GFP-Drp1 in the presence of GTP or GMPPCP (2.5 μM GFP-Drp1, 0.2 μM actin, 500 μM nucleotide). Four and 10 filaments, respectively. (D) Representative images of each FRAP condition before photobleaching, immediately after bleaching, or at a late time point after photobleaching. Scale bar, 2 μm. (E) FRAP on actin filaments initially saturated with GFP-Drp1 in the presence of GTP (2.5 μM GFP-Drp1, 0.2 μM actin, 500 μM GTP); then, free GFP-Drp1 was washed out with buffer containing GTP. Four filaments. (F) Representative images before photobleaching, immediately after bleaching, or at a late time point after photobleaching. Scale bar, 2 μm.
DISCUSSION
In this article, we show that the interaction between Drp1 and actin filaments is unusual in that it is strongly influenced by several factors, including additional Drp1 molecules in solution and guanine nucleotides. Drp1 also bundles actin filaments, and bundling predominates at physiological ionic strength. These factors generally affect the kinetics of the Drp1/actin interaction more strongly than they affect its equilibrium. This subtle feature may have profound cellular consequences.
One very general conclusion from this study is that the Drp1/actin interaction is complex, being a combination of four dynamic processes: 1) Drp1 oligomerization, 2) Drp1 GTP binding, hydrolysis, and release, 3) the actual Drp1/actin binding step, and 4) bundling of Drp1-bound actin filaments. Addition of any factors that influence any of these processes will influence the system as a whole. Drp1 oligomerization itself is a multistep process that involves at least two binding interfaces (Bui and Shaw, 2013; Frohlich et al., 2013). Complete understanding of the dynamics will require more knowledge about the structural changes that occur during oligomerization and actin binding and how these changes influence downstream interactions.
An exciting implication of this work is that actin filaments may serve as a dynamic reservoir for Drp1, which can be accessed for rapid assembly on mitochondria during mitochondrial fission. Our previous work shows that actin filaments accumulate before Drp1 during mitochondrial fission and that inhibition of actin polymerization blocks Drp1 oligomeric maturation at the fission site (Ji et al., 2015). Therefore actin filaments may serve to recruit Drp1 to the fission site by direct binding. If so, the Drp1 could transition from actin bound to mitochondria bound in order to exert its constrictive force.
In this light, the Drp1 receptors on mitochondria may have profound effects on Drp1/actin interactions. Overexpression of MiD51 causes aberrant mitochondrial actin accumulation (Palmer et al., 2011). Our previous work showed synergistic interplay between actin and Mff in stimulating Drp1 GTP hydrolysis (Ji et al., 2015). MiD49, MiD51, Mff, and Fis1, alone or in combination, might access the actin-bound Drp1 by influencing Drp1/actin interaction properties. Similar issues are likely to influence the interaction between Drp1 and cardiolipin, which also can have synergistic effects with Mff under certain conditions (Macdonald et al., 2016).
A question that arises from these results is, why is Drp1 not bound to all actin filaments? Our quantification of cytosolic Drp1 (∼0.5 μM) and actin (∼200 μM) would suggest that a substantial portion of Drp1 should be bound to filaments throughout the cell. However, we observe no evidence for Drp1 enrichment at lamellipodia, filopodia, or stress fibers in our cellular studies. Other factors must influence Drp1/actin binding in cells. There is a growing appreciation for mechanisms that confine specific actin-binding proteins on specific actin-based structures, including competition or cooperativity between actin-binding proteins, local chemical environments that favor specific actin-binding proteins, differential actin-binding protein localization, and differential effects of actin nucleation factors (Clayton et al., 2010; Skau and Kovar, 2010; Michelot and Drubin, 2011; Rotty and Bear, 2014). Additional factors, such as myosin II, might influence Drp1 affinity for actin filaments. We showed that myosin IIA and B play a role in mitochondrial fission (Korobova et al., 2014) and that myosin IIA suppression inhibits Drp1 accumulation on mitochondria without affecting actin accumulation (Ji et al., 2015). Myosin II could orient actin filaments in an optimal manner for Drp1 binding in addition to our previously proposed role in promoting preconstriction (Hatch et al., 2014). Other factors that may influence Drp1/actin binding are posttranslational modifications such as phosphorylation (Otera et al., 2013; Wilson et al., 2013).
A curious feature of the Drp1/actin interaction is the partial Drp1 binding that occurs even at saturating actin concentrations. We postulate that two properties contribute to this phenomenon: the oligomeric equilibrium of Drp1, and the effects of free Drp1 on the off-rate of bound Drp1. Drp1 in solution is in equilibrium among at least three oligomeric states. If larger Drp1 oligomers have higher actin avidity, depletion of oligomers from solution will shift the remaining solution Drp1 to smaller oligomers, stalling binding. However, increasing actin concentration should still increase Drp1 binding in this case, albeit at lower affinity. The further contribution of accelerated off-rate from the remaining Drp1 in solution could at this point contribute to the plateau.
The fundamental oligomeric unit of Drp1 in solution is still in question. Our analytical ultracentrifugation analysis suggests that the smallest unit of Drp1 is dimeric, based on the absence of a monomer peak when wild-type Drp1 or dimer mutant Drp1 is analyzed at low concentration (0.75 and 1.5 μM, respectively). Our estimate for cytoplasmic Drp1 concentration is 0.5 μM, based on quantitative Western blotting of U2OS extracts. Therefore it is possible that a monomer pool of Drp1 exists at this concentration (below the detection limit of our analytical ultracentrifuge). In addition, it is formally possible that other factors might serve to dissociate dimeric Drp1 in the cytoplasm. We tested one possible factor, guanine nucleotide, and found that neither GDP nor GTP alters the sedimentation profile of Drp1 at 1.5 μM (unpublished data). Thus our conclusion at this point is that the minimal Drp1 unit is the dimer.
Although actin has a stimulatory effect on Drp1 GTPase activity (Ji et al., 2015), Drp1 has little effect on actin polymerization under most conditions. We find no effect of Drp1 on actin polymerization kinetics (Ji et al., 2015) or the equilibrium of actin polymerization/depolymerization (this work), suggesting that Drp1 does not significantly alter nucleation, elongation, or depolymerization of actin alone. GTP and GDP do not induce Drp1-mediated changes in actin polymerization, although nonhydrolyzable GTP analogues cause Drp1 to have an inhibitory effect on polymerization (unpublished data). The major structural effect of Drp1 on actin is its ability to assemble actin bundles of up to 30 actin filaments. The cellular significance of bundling is unclear, but the predominance of bundle-bound Drp1 at physiological ionic strength suggests that this might be the predominant form of Drp1-bound actin. We wonder whether the biphasic effect of actin filaments on Drp1 GTP hydrolysis rate might be related to the degree of bundling. Similar multiphasic effects of actin on nucleotide hydrolysis have been observed for myosin I (Albanesi et al., 1985).
Other dynamin family proteins have been shown to interact with actin filaments, both biochemically and in cells (Lee and De Camilli, 2002; Orth et al., 2002; Schafer et al., 2002; Mooren et al., 2009; Gu et al., 2010; Grassart et al., 2014; Palmer et al., 2015). Dynamin2 has been shown to have effects on actin polymerization and bundling in the presence of cortactin and Arp2/3 complex (Schafer et al., 2002; Mooren et al., 2009), and actin polymerization precedes dynamin2 recruitment during endocytosis (Grassart et al., 2014). Dynamin1 binds actin filaments directly and can bundle filaments (Gu et al., 2010). Actin filaments stimulate dynamin1 oligomerization and GTPase activity, and oligomerized dynamin1 in turn affects actin polymerization by inhibiting the ability of gelsolin to cap barbed ends (Gu et al., 2010). Dynamins also decorate actin comet tails assembled by Listeria or other mechanisms (Lee and De Camilli, 2002; Orth et al., 2002). The yeast dynamin Vps1 binds and bundles filaments, and the Vps1/actin interaction is important in the scission step of yeast endocytosis (Palmer et al., 2015). Taken together, these results suggest that actin interaction is a common and important property of dynamin GTPases. Although much remains to be learned about the mechanistic implications of this interaction for membrane dynamics, our work shows that the interaction is highly dynamic and that the dynamics is acutely sensitive to additional binding factors.
MATERIALS AND METHODS
DNA constructs
Human Drp1 isoform 000 (NP_055681.2) follows the nomenclature described in Strack et al. (2013) and corresponds to the “short” isoform (Macdonald et al., 2016) or isoform 3 (Koirala et al., 2013). hDrp1-000 was cloned with an N-terminal Strep-tag followed by an HRV3C protease site into the pET16b vector (Novagen, EMD Millipore, Billerica, MA). For GFP-Drp1, GFP was inserted between the HRV3C and Drp1 sequences in this vector. The GFP contains the A206K mutation, which significantly reduces GFP dimerization (Zacharias et al., 2002). Drp1 point mutations were made using QuikChange mutagenesis (Stratagene, Santa Clara, CA). We created monomer (K631E) and dimer (GPRP401-404AAAA) Drp1 mutants. The Drp1 K631E monomer mutation is the analogous residue to the human Drp1 isoform 2 K642E monomer mutant (Frohlich et al., 2013).
Protein purification
Drp1 was expressed in One Shot BL21 Star (DE3) Escherichia coli (C6010-03; Life Technologies, Carlsbad, CA) by isopropyl-β-d-thiogalactoside induction at 20°C for 16 h. Cell pellets were resuspended in lysis buffer (100 mM Tris-Cl, pH 8.0, 500 mM NaCl, 1 mM dithiothreitol [DTT], 1 mM EDTA, 2 μg/ml leupeptin, 10 μg/ml aprotinin, 2 μg/ml pepstatin A, 2 mM benzamidine, 1 μg/ml calpain inhibitor I [ALLN], and 1 μg/ml calpeptin) and lysed using a high-pressure homogenizer (M-110L Microfluidizer Processor; Microfluidics, Newton, MA). The lysate was clarified by centrifugation at 40,000 rpm (type 45 Ti rotor; Beckman, Brea, CA) for 1 h at 4°C. Avidin (20 μg/ml; PI-21128; ThermoFisher Scientific, Waltham, MA) was added, and then the supernatant was loaded onto Strep-Tactin Superflow resin (2-1206-025; IBA, Göttingen, Germany) by gravity flow. The column was washed with 20-column volumes of lysis buffer without protease inhibitors. To elute Drp1, 0.01 mg/ml HRV3C protease in lysis buffer without protease inhibitors was added for 16 h at 4°C. The Strep-Tactin Superflow eluate was further purified by size exclusion chromatography on Superdex200 (GE Biosciences, Piscataway, NJ), spin concentrated, frozen in liquid nitrogen, and stored at −80°C.
Rabbit skeletal muscle actin was extracted from acetone powder as previously described (Spudich and Watt, 1971), and further purified by size exclusion chromatography on Superdex 75 (GE Biosciences). Actin was stored at 4°C in G-buffer (2 mM Tris, pH 8.0, 0.5 mM DTT, 0.2 mM ATP, 0.1 mM CaCl2, and 0.01% NaN3).
Drp1 and actin quantification in U2OS cells
Quantitative Western blotting was used to determine endogenous Drp1 concentration in U2OS cells. Cells growing logarithmically were trypsinized, washed multiple times in phosphate-buffered saline (PBS), and then resuspended in PBS, and aliquots were taken for cell counts (hemocytometer) and protein quantification (Bradford, 500-0006; Bio-Rad, Hercules, CA). Cellular protein was calculated at 0.397 ng/cell, similar to values obtained for other adherent cell types growing in monolayer (Galacci and Higuchi, 1966; Nicholson-Dykstra and Higgs, 2008). Protein was extracted by addition of 9 volumes of 4% SDS and 10 mM DTT followed by immediate boiling. N-ethylmaleimide (Sigma-Aldrich, St. Louis, MO) was added to 30 mM from a 300 mM stock. Extract was mixed 1:1 with 2xDB (250 mM Tris-HCl, pH 6.8, 2 mM EDTA, 20% glycerol, 0.8% SDS, 0.02% bromophenol blue, 1 M NaCl, 4 M urea) and further diluted with 1xDB to obtain the desired concentration. Quantitative Western blotting was performed by running 5 μg of U2OS cell extract on gel with varying amounts (0.1–6.4 ng) of purified Drp1 spiked into extract. A separate set of samples contained a range of U2OS cell extract concentrations (4–10 μg). Proteins were separated by 7.5% SDS–PAGE and transferred to a polyvinylidene difluoride membrane (EMD Millipore, Billerica, MA). The membrane was blocked with TBS-T (20 mM Tris-HCl, pH 7.6, 136 mM NaCl, and 0.1% Tween-20) containing 3% bovine serum albumin (BSA; Research Organics, Cleveland, OH) for 30 min and then incubated with primary antibody to detect Drp1 (D6C7; Cell Signaling, Danvers, MA) at 4°C overnight. After being washed with TBS-T, the membrane was incubated with horseradish peroxidase (HRP)–conjugated secondary antibody (1706515; Bio-Rad) overnight at 4°C. Signals were detected by chemiluminescence (Pierce, ThermoFisher Scientific, Rockford, IL). Antibody signal was quantified using ImageJ (National Institutes of Health, Bethesda, MD). We included all three U2OS Drp1 bands observed on the Western blot in our quantification. The initial sample set containing 5 μg of U2OS extract with various amounts of purified Drp1 (0.1–6.4 ng) was used to generate a standard curve by subtracting the average band intensity of the 5-μg U2OS extract sample. The standard curve was then used to determine the amount of Drp1 present in each U2OS extract sample (4–10 μg). The amount of cellular Drp1 was calculated using the mean cellular volume and nuclear volume of Swiss 3T3 cells (Nicholson-Dykstra and Higgs, 2008), which are of similar size to U2OS cells. The SD represents the error from Western blotting (Drp1) or Coomassie staining (actin). We did not calculate the error from our cytoplasmic volume estimate (which does not reflect excluded volume from organelles other than the nucleus).
U2OS cytosolic actin concentration was measured by Coomassie staining of varying microgram amounts of U2OS extracts separated by SDS–PAGE. The position of actin was determined from purified actin run in parallel. The actin signal intensity was linear with protein load for both the U2OS extract and the purified actin in the concentration ranges used here.
Actin and Drp1 preparation for biochemical assays
Actin filaments were assembled from monomers (20–40 μM) for 1 h at 23°C by addition of a 10× stock of polymerization buffer (500 mM NaCl, 10 mM MgCl2, 10 mM ethylene glycol tetraacetic acid [EGTA], 100 mM imidazole, pH 7.0) to a 1× final concentration. To maintain ionic strength across all samples, an actin blank was prepared in parallel using G-buffer in place of actin monomers and used to dilute actin filaments as needed for each sample. Drp1 was diluted to 10 μM in 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.0, and then centrifuged at 100,000 rpm for 20 min at 4°C in a TLA-120 rotor (Beckman). The supernatant was stored on ice, and its protein concentration was determined by Bradford assay (500-0006; Bio-Rad).
High-speed cosedimentation assay
Drp1 (varying concentrations, but typically 1.3 μM) was incubated with varying concentrations of actin filaments (0.25–10 μM) for varying times (1 h unless otherwise stated) at 23°C in a 200-μl volume. The final ionic strength was adjusted to 75 mM using 5 M NaCl. Unless otherwise stated, NaCl was added to 65 mM. The calculated ionic contribution of the other buffer components (imidazole, MgCl2, EGTA, ATP, NaN3) is the equivalent of ∼10 mM monovalent ion. After incubation, samples were centrifuged at 80,000 rpm for 20 min at 4°C in a TLA-100.1 rotor (Beckman). The supernatant was carefully removed. Pellets were washed briefly and gently with 100 μl of 50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.4, and then resuspended in 100 μl of SDS–PAGE sample buffer and resolved by SDS–PAGE. Gels were stained with Colloidal Blue Staining SDS–PAGE (LC6025; Invitrogen, Carlsbad, CA), and band intensity was analyzed using ImageJ software, deriving Kdapp and maximal percentage bound from single-exponential fits. Shown in Supplemental Figure S2A are the results from six independent assays under identical conditions (1.3 μM Drp1 in 65 mM NaCl), with Kdapp varying from 0.25 to 0.87 μM (mean 0.59 ± 0.20 μM) and maximum percentage Drp1 bound in a range of 31.1 to 51.2% (mean 42.3 ± 6.7%). The fluctuating values rendered problematic a truly accurate comparison of affinity and percentage bound between conditions due to the number of replicates and amount of protein that would be required. As a more attainable solution, all comparisons of conditions are conducted against standard control (1.3 μM Drp1 in 65 mM NaCl) carried out on the same day.
In the supernatant rebinding assay (Figure 1C), Drp1 (1.3 μM) was incubated with 5 μM actin filaments for 1 h at 23°C in a 200-μl volume. The final ionic strength was adjusted to 75 mM using NaCl. After incubation, samples were centrifuged at 80,000 rpm for 20 min at 4°C in a TLA-100.1 rotor (Beckman). The supernatant was carefully removed and mixed with actin filaments (5 μM) for 1 h at 23°C in a 100-μl volume and then recentrifuged. The supernatant was carefully removed. Pellets from both centrifugations were washed briefly and gently with 100 μl of 50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.4, and then resuspended in 50 μl of SDS–PAGE sample buffer and resolved by SDS–PAGE.
Drp1 oligomerization assay/high-speed sedimentation assay
Drp1 (0.5–2 μM) was incubated with 500 μM GMPPCP (Sigma-Aldrich) for 1 h at 23°C in a 200-μl volume. The final ionic strength was adjusted to 75 mM using NaCl. After incubation, samples were centrifuged at 80,000 rpm for 20 min at 4°C in a TLA-100.1 rotor (Beckman). The supernatant was carefully removed, and 100 μl was mixed with SDS–PAGE sample buffer. Pellets were washed briefly and gently with 100 μl of 50 mM NaCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, pH 7.4, and then resuspended in 100 μl of SDS–PAGE sample buffer and resolved by SDS–PAGE. Gels were stained with Colloidal Blue Staining SDS–PAGE, and band intensity was analyzed using ImageJ software.
Low-speed cosedimentation (bundling) assay
Samples were prepared by mixing varying concentrations of Drp1 (0–1.5 μM) with actin filaments (0–4 μM) in buffer as described for high-speed pelleting (65 mM NaCl final) in 1.5-ml screw-cap microcentrifuge tubes. Pipette tips were cut to minimize actin filament shearing. Samples were incubated for 1 h at 23°C in a 300-μl volume and then centrifuged at 13,000 rpm and 4°C in a microcentrifuge. The samples were returned to ice, and the upper 225 μl of supernatant was carefully removed. The pellets were washed twice by carefully adding 300 μl of 150 mM NaCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole, recentrifuging, and again carefully removing the top 300 μl of supernatant. The pellets were mixed with 75 μl of 2× SDS–PAGE sample buffer, resolved by SDS–PAGE, stained by colloidal Coomassie, and analyzed using ImageJ.
Velocity analytical ultracentrifugation
Drp1 was diluted to the desired concentration in 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), pH 7.4, 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 1 mM DTT. Sample was allowed to equilibrate to 20°C under vacuum for 1 h and then centrifuged in an An60 rotor and TLA centrifuge (Beckman) at 35,000 rpm and 20°C for 12 h or until complete pelleting occurred. Drp1 was monitored at 280 nm for 10 μM, 293 nm for 20, 40, and 80 μM, and 225 nm for 0.75 and 1.5 μM. Results were analyzed using SedFit (National Institutes of Health open access software) after calculation of buffer viscosity and density using Sednterp (open access software from University of New Hampshire, Durham, NH).
GTPase assay
To remove ATP, actin monomers in G-buffer were incubated with AG1-X2 100–200 mesh anion exchange resin (Dowex; 1401241; Bio-Rad) rotating at 4°C for 5 min, followed by low-speed centrifugation. Actin filaments (20 μM) were polymerized in 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM HEPES, pH 7.4, for 1 h at 23°C. Drp1 was diluted in 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM HEPES, pH 7.4, and then centrifuged to remove aggregates as described.
Drp1 (1.3 μM) was mixed with the indicated concentration of actin filaments, and the final ionic strength was adjusted to the equivalent of 65 mM KCl using 4 M KCl stock. Samples were incubated at 37°C for 5 min. At this point, GTP was added to a final concentration of 250 μM to start reactions at 37°C. Reactions were quenched at various time points by mixing 20 μl of sample with 5 μl of 125 mM EDTA in a clear, flat-bottomed, 96-well plate (Greiner, Monroe, NC). Six time points were acquired for all conditions. Inorganic phosphate was determined by addition of 150 μl of malachite green solution (1 mM Malachite green [2290105–100 g; Sigma-Aldrich] and 10 mM ammonium molybdate [A7302–100g Sigma-Aldrich] in 1 N HCl) to 25-μl quenched reactions. Absorbance at 650 nm was measured with a 96-well fluorescence plate reader (Infinite M1000; Tecan, Mannedorf Switzerland). GTP hydrolysis rates were determined by plotting phosphate concentration as a function of time.
Total internal reflection microscopy imaging
Actin filaments (5 μM) were assembled from monomers for 1 h as described and then stabilized with TRITC-phalloidin (5.5 μM; P1951; Sigma-Aldrich) for 10 min. GFP-Drp1 was diluted to 10 μM in 50 mM KCl, 1 mM MgCl2, 1 mM EGTA, and 10 mM imidazole and then centrifuged at 100,000 rpm for 20 min at 4°C in a TLA-120 rotor (Beckman). The supernatant was stored on ice, and its protein concentration was determined by Bradford assay (500–0006; Bio-Rad). Phalloidin-stabilized actin filaments (200 nM) were incubated in the presence or absence of saturating amounts of GFP-Drp1 (2.5 μM) for 1 h at 23°C.
Glass flow chambers for TIRF were assembled using VWR microcover glasses (22 × 22 and 18 × 18 mm, No 1.5) with double-stick Scotch tape to hold 10-μl volume. Before chamber assembly, the coverglasses were washed with acetone (50 min), ethanol (10 min), and water (1 min), and incubated in a solution containing a 1:2 ratio of 30% H2O2:concentrated H2SO4 for 1 h. Coverglasses were then rinsed with water, 0.1 M KOH, and water again. Inert gas was used to completely dry cover glasses before silanization. Glasses were silanized overnight in a solution of 0.0025% dichlorodimethyl silane (85126; Sigma-Aldrich) in chloroform and then washed with methanol and dried with inert gas. Glasses were stored in a clean, sealed container. Immediately before starting an assay, chambers were incubated with 1% Pluronic F127 (P2443; Sigma-Aldrich) in BRB80 buffer (80 mM 1,4-piperazinediethanesulfonic acid/KOH, pH 6.9, 1 mM EGTA, 1 mM MgCl2) for 1 min and then equilibrated with TIRF buffer.
Before imaging, samples were mixed 1:1 with 2× TIRF buffer (50 mM KCl, 1 mM MgCl2, 1 mM EGTA, 10 mM HEPES, pH 7.4, 100 mM DTT, 0.2 mM ATP, 15 mM glucose, 0.5% methyl cellulose, 0.01 mg/ml catalase [C3515; Sigma-Aldrich], 0.05 mg/ml glucose oxidase [G6125; Sigma-Aldrich], 0.1% BSA) and added to glass flow chambers. Filaments were visualized using a Nikon Eclipse Ti inverted microscope using the 488- and 561-nm lasers and driven by Nikon Elements software. Simultaneous dual-color images were acquired with TIRF objective (60×, 1.49 numerical aperture) and two Andor electron-multiplying charge-coupled device cameras with an Andor TuCam adapter using Perfect Focus.
GFP-Drp1 off-rate experiments
To measure GFP-Drp1 off-rate from actin filaments, TIRF buffer alone or containing Drp1, GDP, or GTP was manually added to the flow cell in a 10 μl volume while the current flow cell solution was removed with Whatman paper. Images were acquired before, during, and after buffer wash. The “dead time” of the assay (time spent out of focus during buffer addition) is 25.8 ± 10.5 s. The ImageJ plug-in Time Series Analyzer V3 was used to measure fluorescence changes over time. Surprisingly, we found the filament-bound GFP-Drp1 fluorescence does not decrease significantly after washing with GTP (Figure 6), with >90% of signal remaining 5 min after washout. Therefore we used the average GFP-Drp1 fluorescence after washout with GTP to normalize the initial fluorescence intensity of our other conditions and determine t1/2.
Fluorescence recovery after photobleaching
As described earlier, phalloidin-stabilized actin filaments (200 nM) were incubated in the presence of saturating amounts of GFP-Drp1 (2.5 μM) for 1 h at 23°C to allow binding to reach steady-state equilibrium. Immediately before imaging, samples were diluted with 2× TIRF buffer to a final concentration of 1× TIRF buffer, introduced into the flow chamber, and imaged for 2 min. The 405-nm laser was pulsed for 1 s at 40% power to single-point bleach a region of a GFP-Drp1–bound actin filament or bundle. Fluorescence recovery was measured for 2 min after photobleaching. The ImageJ plug-in Time Series Analyzer V3 was used to measure fluorescence over time in the bleached filament/bundle region as well as a control “unbleached” filament. Background-subtracted fluorescence was normalized to 100% before bleaching.
Actin bundle analysis
To determine the number of actin filaments in a bundle, phalloidin-TRITC (220 nM)–stabilized actin filaments (200 nM) were incubated with or without unlabeled Drp1 (2.5 μM) for 1 h at 23°C and then imaged by TIRF microscopy, using the same exposure times and laser intensities for both conditions, such that the TRITC and GFP levels were always in the linear range. Background-subtracted fluorescence intensity was quantified from a 108-pixel2 rectangle including the filament using ImageJ. The sample incubated without Drp1 was used to calibrate the average intensity of single actin filaments. ImageJ was used to measure x- and y-diameters of “circular” bundles and subsequently to determine the aspect ratio.
Drp1 nucleotide state
To determine the nucleotide state of purified Drp1, samples were prepared of 150 μM GTP, 75 μM GDP, and 160 μM Drp1. Samples were diluted into 20 mM HEPES, pH 7.5, 150 mM KCl, 2 mM MgCl2, 1 mM DTT, and 0.5 mM EDTA and then boiled for 10 min and centrifuged for 10 min at 13,000 rpm in a microcentrifuge to denature and remove protein. The supernatant was diluted 1:5 with 10 mM Tris, pH 8.0, and 1 mM MgCl2. We loaded 400 μl of sample onto a 1-ml SourceQ15 column (GE Biosciences). A linear gradient from 0 to 150 mM was used to elute nucleotides. Absorbance at 253 nm was used to detect nucleotide.
Supplementary Material
Acknowledgments
We thank Ann Lavanway for help with TIRF microscopy, Laura Lackner and Janet Shaw for advice on Drp1 expression and purification, Dorothy Schafer for discussions on dynamin, Roberto Dominguez for fascin, Mike Ostap for the information on multiphasic myosin I activity, Dean Madden, Jon Kull, and Jason McClellen for brainstorming sessions on Drp1's unusual actin-binding properties, River Rose for her accumulated resources, and Lori Schoenfeld for molecular biology help. This work was supported by National Institutes of Health Grants GM069818 and GM106000 to H.N.H. and NS056244 and NS087908 to S.S. and a National Science Foundation Predoctoral Fellowship to A.L.H.
Abbreviations used:
- Drp1
dynamin-related protein-1
- Fis1
fission factor 1
- FRAP
fluorescence recovery after photobleaching
- GFP
green fluorescent protein
- GMPPCP
β-γ-methyleneguanosine 5’-triphosphate sodium salt
- Mff
mitochondrial fission factor
- MiD49
mitochondrial dynamics protein of 49 kDa
- MiD51
mitochondrial dynamics protein of 51 kDa
- TIRF
total internal reflection fluorescence
- TRITC
tetramethyl rhodamine isothiocyanate
- WT
wild type.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0193) on August 24, 2016.
REFERENCES
- Albanesi JP, Coue M, Fujisaki H, Korn ED. Effect of actin filament length and filament number concentration on the actin-activated ATPase activity of Acanthamoeba myosin I. J Biol Chem. 1985;260:13276–13280. [PubMed] [Google Scholar]
- Bui HT, Shaw JM. Dynamin assembly strategies and adaptor proteins in mitochondrial fission. Curr Biol. 2013;23:R891–R899. doi: 10.1016/j.cub.2013.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustillo-Zabalbeitia I, Montessuit S, Raemy E, Basanez G, Terrones O, Martinou JC. Specific interaction with cardiolipin triggers functional activation of dynamin-related protein 1. PLoS One. 2014;9:e102738. doi: 10.1371/journal.pone.0102738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlier MF, Pantaloni D. Binding of phosphate to F-ADP-actin and role of F-ADP-Pi-actin in ATP-actin polymerization. J Biol Chem. 1988;263:817–825. [PubMed] [Google Scholar]
- Chang CR, Blackstone C. Dynamic regulation of mitochondrial fission through modification of the dynamin-related protein Drp1. Ann NY Acad Sci. 2010;1201:34–39. doi: 10.1111/j.1749-6632.2010.05629.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chappie JS, Acharya S, Leonard M, Schmid SL, Dyda F. G domain dimerization controls dynamin's assembly-stimulated GTPase activity. Nature. 2010;465:435–440. doi: 10.1038/nature09032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Chan DC. Mitochondrial dynamics–fusion, fission, movement, and mitophagy–in neurodegenerative diseases. Hum Mol Genet. 2009;18:R169–176. doi: 10.1093/hmg/ddp326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JE, Sammons MR, Stark BC, Hodges AR, Lord M. Differential regulation of unconventional fission yeast myosins via the actin track. Curr Biol. 2010;20:1423–1431. doi: 10.1016/j.cub.2010.07.026. [DOI] [PubMed] [Google Scholar]
- Courson DS, Rock RS. Actin cross-link assembly and disassembly mechanics for alpha-Actinin and fascin. J Biol Chem. 2010;285:26350–26357. doi: 10.1074/jbc.M110.123117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vos KJ, Allan VJ, Grierson AJ, Sheetz MP. Mitochondrial function and actin regulate dynamin-related protein 1-dependent mitochondrial fission. Curr Biol. 2005;15:678–683. doi: 10.1016/j.cub.2005.02.064. [DOI] [PubMed] [Google Scholar]
- DuBoff B, Gotz J, Feany MB. Tau promotes neurodegeneration via DRP1 mislocalization in vivo. Neuron. 2012;75:618–632. doi: 10.1016/j.neuron.2012.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman JR, Nunnari J. Mitochondrial form and function. Nature. 2014;505:335–343. doi: 10.1038/nature12985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich C, Grabiger S, Schwefel D, Faelber K, Rosenbaum E, Mears J, Rocks O, Daumke O. Structural insights into oligomerization and mitochondrial remodelling of dynamin 1-like protein. EMBO J. 2013;32:1280–1292. doi: 10.1038/emboj.2013.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galacci RR, Higuchi K. Differences in protein contents among fibroblast cell lines grown in monolayer cultures. 1966. Available at www.dtic.mil/dtic/tr/fulltext/u2/481027.pdf (accessed 24 March 2016)
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grassart A, Cheng AT, Hong SH, Zhang F, Zenzer N, Feng Y, Briner DM, Davis GD, Malkov D, Drubin DG. Actin and dynamin2 dynamics and interplay during clathrin-mediated endocytosis. J Cell Biol. 2014;205:721–735. doi: 10.1083/jcb.201403041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu C, Yaddanapudi S, Weins A, Osborn T, Reiser J, Pollak M, Hartwig J, Sever S. Direct dynamin-actin interactions regulate the actin cytoskeleton. EMBO J. 2010;29:3593–3606. doi: 10.1038/emboj.2010.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatch AL, Gurel PS, Higgs HN. Novel roles for actin in mitochondrial fission. J Cell Sci. 2014;127:4549–4560. doi: 10.1242/jcs.153791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingerman E, Perkins EM, Marino M, Mears JA, McCaffery JM, Hinshaw JE, Nunnari J. Dnm1 forms spirals that are structurally tailored to fit mitochondria. J Cell Biol. 2005;170:1021–1027. doi: 10.1083/jcb.200506078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji WK, Hatch AL, Merrill RA, Strack S, Higgs HN. Actin filaments target the oligomeric maturation of the dynamin GTPase Drp1 to mitochondrial fission sites. Elife. 2015;4:e11553. doi: 10.7554/eLife.11553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashatus DF, Lim KH, Brady DC, Pershing NL, Cox AD, Counter CM. RALA and RALBP1 regulate mitochondrial fission at mitosis. Nat Cell Biol. 2011;13:1108–1115. doi: 10.1038/ncb2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koirala S, Guo Q, Kalia R, Bui HT, Eckert DM, Frost A, Shaw JM. Interchangeable adaptors regulate mitochondrial dynamin assembly for membrane scission. Proc Natl Acad Sci USA. 2013;110:E1342–1351. doi: 10.1073/pnas.1300855110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Gauvin TJ, Higgs HN. A role for myosin II in mammalian mitochondrial fission. Curr Biol. 2014;24:409–414. doi: 10.1016/j.cub.2013.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korobova F, Ramabhadran V, Higgs HN. An actin-dependent step in mitochondrial fission mediated by the ER-associated formin INF2. Science. 2013;339:464–467. doi: 10.1126/science.1228360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lackner LL, Horner JS, Nunnari J. Mechanistic analysis of a dynamin effector. Science. 2009;325:874–877. doi: 10.1126/science.1176921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, De Camilli P. Dynamin at actin tails. Proc Natl Acad Sci USA. 2002;99:161–166. doi: 10.1073/pnas.012607799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xu S, Roelofs BA, Boyman L, Lederer WJ, Sesaki H, Karbowski M. Transient assembly of F-actin on the outer mitochondrial membrane contributes to mitochondrial fission. J Cell Biol. 2015;208:109–123. doi: 10.1083/jcb.201404050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loson OC, Song Z, Chen H, Chan DC. Fis1, Mff, MiD49, and MiD51 mediate Drp1 recruitment in mitochondrial fission. Mol Biol Cell. 2013;24:659–667. doi: 10.1091/mbc.E12-10-0721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PJ, Francy CA, Stepanyants N, Lehman L, Baglio A, Mears JA, Qi X, Ramachandran R. Distinct splice variants of dynamin-related protein 1 differentially utilize mitochondrial fission factor as an effector of cooperative GTPase activity. J Biol Chem. 2016;291:493–507. doi: 10.1074/jbc.M115.680181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macdonald PJ, Stepanyants N, Mehrotra N, Mears JA, Qi X, Sesaki H, Ramachandran R. A dimeric equilibrium intermediate nucleates Drp1 reassembly on mitochondrial membranes for fission. Mol Biol Cell. 2014;25:1905–1915. doi: 10.1091/mbc.E14-02-0728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manor U, Bartholomew S, Golani G, Christenson E, Kozlov M, Higgs H, Spudich J, Lippincott-Schwartz J. A mitochondria-anchored isoform of the actin-nucleating Spire protein regulates mitochondrial division. Elife. 2015;4 doi: 10.7554/eLife.08828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelot A, Drubin DG. Building distinct actin filament networks in a common cytoplasm. Curr Biol. 2011;21:R560–R569. doi: 10.1016/j.cub.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mooren OL, Kotova TI, Moore AJ, Schafer DA. Dynamin2 GTPase and cortactin remodel actin filaments. J Biol Chem. 2009;284:23995–24005. doi: 10.1074/jbc.M109.024398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicholson-Dykstra SM, Higgs HN. Arp2 depletion inhibits sheet-like protrusions but not linear protrusions of fibroblasts and lymphocytes. Cell Motil Cytoskeleton. 2008;65:904–922. doi: 10.1002/cm.20312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Suomalainen A. Mitochondria: in sickness and in health. Cell. 2012;148:1145–1159. doi: 10.1016/j.cell.2012.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orth JD, Krueger EW, Cao H, McNiven MA. The large GTPase dynamin regulates actin comet formation and movement in living cells. Proc Natl Acad Sci USA. 2002;99:167–172. doi: 10.1073/pnas.012607899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osellame LD, Singh AP, Stroud DA, Palmer CS, Stojanovski D, Ramachandran R, Ryan MT. Cooperative and independent roles of Drp1 adaptors Mff and MiD49/51 in mitochondrial fission. J Cell Sci. 2016;129:2170–2181. doi: 10.1242/jcs.185165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Ishihara N, Mihara K. New insights into the function and regulation of mitochondrial fission. Biochim Biophys Acta. 2013;1833:1256–1268. doi: 10.1016/j.bbamcr.2013.02.002. [DOI] [PubMed] [Google Scholar]
- Otera H, Miyata N, Kuge O, Mihara K. Drp1-dependent mitochondrial fission via MiD49/51 is essential for apoptotic cristae remodeling. J Cell Biol. 2016;212:531–544. doi: 10.1083/jcb.201508099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otera H, Wang C, Cleland MM, Setoguchi K, Yokota S, Youle RJ, Mihara K. Mff is an essential factor for mitochondrial recruitment of Drp1 during mitochondrial fission in mammalian cells. J Cell Biol. 2010;191:1141–1158. doi: 10.1083/jcb.201007152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Elgass KD, Parton RG, Osellame LD, Stojanovski D, Ryan MT. Adaptor proteins MiD49 and MiD51 can act independently of Mff and Fis1 in Drp1 recruitment and are specific for mitochondrial fission. J Biol Chem. 2013;288:27584–27593. doi: 10.1074/jbc.M113.479873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer CS, Osellame LD, Laine D, Koutsopoulos OS, Frazier AE, Ryan MT. MiD49 and MiD51, new components of the mitochondrial fission machinery. EMBO Rep. 2011;12:565–573. doi: 10.1038/embor.2011.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer SE, Smaczynska-de R, II, Marklew CJ, Allwood EG, Mishra R, Johnson S, Goldberg MW, Ayscough KR. A dynamin-actin interaction is required for vesicle scission during endocytosis in yeast. Curr Biol. 2015;25:868–878. doi: 10.1016/j.cub.2015.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Rotty JD, Bear JE. Competition and collaboration between different actin assembly pathways allows for homeostatic control of the actin cytoskeleton. Bioarchitecture. 2014;5:27–34. doi: 10.1080/19490992.2015.1090670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schafer DA, Weed SA, Binns D, Karginov AV, Parsons JT, Cooper JA. Dynamin2 and cortactin regulate actin assembly and filament organization. Curr Biol. 2002;12:1852–1857. doi: 10.1016/s0960-9822(02)01228-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz TL. Mitochondrial trafficking in neurons. Cold Spring Harb Perspect Biol. 2013;5:a011304. doi: 10.1101/cshperspect.a011304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Q, Yamano K, Head BP, Kawajiri S, Cheung JT, Wang C, Cho JH, Hattori N, Youle RJ, van der Bliek AM. Mutations in Fis1 disrupt orderly disposal of defective mitochondria. Mol Biol Cell. 2014;25:145–159. doi: 10.1091/mbc.E13-09-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skau CT, Kovar DR. Fimbrin and tropomyosin competition regulates endocytosis and cytokinesis kinetics in fission yeast. Curr Biol. 2010;20:1415–1422. doi: 10.1016/j.cub.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- Stepanyants N, Macdonald PJ, Francy CA, Mears JA, Qi X, Ramachandran R. Cardiolipin's propensity for phase transition and its reorganization by dynamin-related protein 1 form a basis for mitochondrial membrane fission. Mol Biol Cell. 2015;26:3104–3116. doi: 10.1091/mbc.E15-06-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strack S, Wilson TJ, Cribbs JT. Cyclin-dependent kinases regulate splice-specific targeting of dynamin-related protein 1 to microtubules. J Cell Biol. 2013;201:1037–1051. doi: 10.1083/jcb.201210045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguchi N, Ishihara N, Jofuku A, Oka T, Mihara K. Mitotic phosphorylation of dynamin-related GTPase Drp1 participates in mitochondrial fission. J Biol Chem. 2007;282:11521–11529. doi: 10.1074/jbc.M607279200. [DOI] [PubMed] [Google Scholar]
- Wilson TJ, Slupe AM, Strack S. Cell signaling and mitochondrial dynamics: Implications for neuronal function and neurodegenerative disease. Neurobiol Dis. 2013;51:13–26. doi: 10.1016/j.nbd.2012.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youle RJ, van der Bliek AM. Mitochondrial fission, fusion, and stress. Science. 2012;337:1062–1065. doi: 10.1126/science.1219855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias DA, Violin JD, Newton AC, Tsien RY. Partitioning of lipid-modified monomeric GFPs into membrane microdomains of live cells. Science. 2002;296:913–916. doi: 10.1126/science.1068539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.