Female reproductive cells of most species lack centrosomes, but how spindles form in their absence is poorly understood. Study of oocytes in Caenorhabditis elegans uncovers new steps in this process and reveals mechanisms required for acentrosomal spindle bipolarity via studies of two proteins, KLP-18/kinesin-12 and MESP-1.
Abstract
Although centrosomes contribute to spindle formation in most cell types, oocytes of many species are acentrosomal and must organize spindles in their absence. Here we investigate this process in Caenorhabditis elegans, detailing how acentrosomal spindles form and revealing mechanisms required to establish bipolarity. Using high-resolution imaging, we find that in meiosis I, microtubules initially form a “cage-like” structure inside the disassembling nuclear envelope. This structure reorganizes so that minus ends are sorted to the periphery of the array, forming multiple nascent poles that then coalesce until bipolarity is achieved. In meiosis II, microtubules nucleate in the vicinity of chromosomes but then undergo similar sorting and pole formation events. We further show that KLP-18/kinesin-12 and MESP-1, previously shown to be required for spindle bipolarity, likely contribute to bipolarity by sorting microtubules. After their depletion, minus ends are not sorted outward at the early stages of spindle assembly and instead converge. These proteins colocalize on microtubules, are interdependent for localization, and can interact, suggesting that they work together. We propose that KLP-18/kinesin-12 and MESP-1 form a complex that functions to sort microtubules of mixed polarity into a configuration in which minus ends are away from the chromosomes, enabling formation of nascent poles.
INTRODUCTION
During mitosis and male meiosis, duplicated centrosomes nucleate microtubules and then separate to opposite sides of the cell, forming the poles of a spindle capable of aligning and segregating chromosomes (Walczak and Heald, 2008). However, in female reproductive cells (oocytes) of many species, centriole-containing centrosomes are degraded before the meiotic divisions (Dumont and Desai, 2012). We are interested in understanding the mechanisms by which microtubules are organized into a bipolar spindle in their absence.
In mouse oocytes, spindle assembly involves multiple acentriolar microtubule-organizing centers (MTOCs), which are believed to functionally replace centrosomes. These MTOCs are small asters of microtubules that contain pericentriolar material (PCM) components at their center, including pericentrin and γ-tubulin (Gueth-Hallonet et al., 1993; Palacios et al., 1993; Carabatsos et al., 2000), and they have been proposed to serve as major sites of microtubule nucleation. Live imaging studies have shown that >80 of these MTOCs form in the cytoplasm, coalesce to the outside of the nucleus, and then are incorporated into the meiotic spindle, leading to the model that self-organization of these structures drives acentrosomal spindle assembly in mammalian oocytes (Schuh and Ellenberg, 2007; Clift and Schuh, 2015). Similar asters have also been observed in Drosophila oocytes (Skold et al., 2005), raising the possibility that this feature of acentrosomal spindle assembly is conserved. However, several lines of evidence support the view that acentrosomal spindle assembly does not absolutely require participation from MTOCs. First, a number of studies in Drosophila have not reported the formation of MTOCs and instead demonstrate that microtubules are nucleated in the vicinity of chromosomes (Theurkauf and Hawley, 1992; Colombie et al., 2008). Moreover, MTOC asters have not been observed during acentrosomal spindle formation in either Xenopus egg extracts or human oocytes, strengthening the idea that alternative strategies exist. In Xenopus extracts, spindle assembly involves microtubule nucleation in the vicinity of chromatin followed by motor-driven reorganization of these microtubules into a bipolar structure (Heald et al., 1996; Walczak et al., 1998). However, because this is an in vitro system, it is possible that additional mechanisms may also contribute in vivo. In humans, microtubules also appear to nucleate in the vicinity of chromosomes (Holubcova et al., 2015), but the molecular mechanisms driving spindle assembly in these cells are poorly understood due to difficulties in obtaining and experimentally manipulating human oocytes.
Caenorhabditis elegans represents an ideal in vivo model in which to study acentrosomal spindle assembly since it is an experimentally tractable system that is amenable to live imaging of the meiotic divisions. To understand more about the molecular mechanisms driving spindle assembly in C. elegans oocytes, we previously performed an RNA interference (RNAi) screen and identified KLP-18 (kinesin-12) and the novel protein meiotic spindle 1 (MESP-1) as essential for acentrosomal spindle bipolarity. When either of these proteins was depleted, monopolar instead of bipolar spindles formed (Wignall and Villeneuve, 2009). However, when and how these proteins act to promote bipolarity during acentrosomal spindle assembly are not understood.
In this study, we gain insight into these questions by first defining how acentrosomal spindles form in C. elegans oocytes and then investigating the roles of KLP-18/kinesin-12 and MESP-1 in this process. We find that in contrast to a recently proposed model (Connolly et al., 2015; Severson et al., 2016), C. elegans oocyte spindles do not assemble via the nucleation and coalescence of MTOC asters, bolstering the idea that acentrosomal spindle formation does not universally require these structures. Instead, our findings are consistent with a model in which KLP-18 and MESP-1 sort microtubules of mixed polarity into a configuration in which their minus ends can be gathered into two oppositely oriented poles. We find that MESP-1 and KLP-18 are interdependent for localization and can interact, suggesting that they work together to perform this function. This work therefore sheds light on mechanisms by which acentrosomal spindles achieve bipolarity in C. elegans. Moreover, our findings establish C. elegans as a model for in vivo investigation of pathways of acentrosomal spindle assembly that do not involve MTOC asters, which may provide future insight into mechanisms used by human oocytes.
RESULTS AND DISCUSSION
Acentrosomal spindle assembly proceeds by microtubule nucleation followed by formation and coalescence of multiple poles
We set out to investigate the stages of acentrosomal spindle assembly in vivo using a strain expressing green fluorescent protein (GFP)–tubulin and GFP-histone to visualize microtubules and chromosomes, respectively (Figure 1A). Although meiotic spindle assembly was visualized in live worms in many previous studies (Yang et al., 2003, 2005; McNally et al., 2006; Connolly et al., 2014, 2015; Sumiyoshi et al., 2015), those experiments used conditions optimized to allow long-term filming, which limited the number of images that could be obtained at each time point without affecting viability, thereby limiting the number of z-stacks and the resolution of spindle structures. Therefore we took the complementary approach of acquiring high-resolution in vivo images representing each of the different stages of spindle assembly. We then ordered images into a spindle assembly pathway using three temporal markers: 1) the position of the oocyte within the gonad (since this position correlates with progression through meiosis), 2) the position of the chromosomes within the oocyte (since the chromosomes start in the center of the cell and move to the cortex as the spindle forms), and 3) the shape of the cell (since the morphology changes upon ovulation and again as the eggshell forms). We also quantified the types of microtubule structures we observed at specific locations in the gonad (Supplemental Figure S1) and compared our images to our (Figure 1B and Supplemental Movie S1) and published (Yang et al., 2003, 2005; McNally et al., 2006; Connolly et al., 2014, 2015; Sumiyoshi et al., 2015) lower-resolution time-lapse movies for additional confirmation. Our imaging has more precisely documented how microtubules form and reorganize during the process of acentrosomal spindle assembly in vivo.
FIGURE 1:
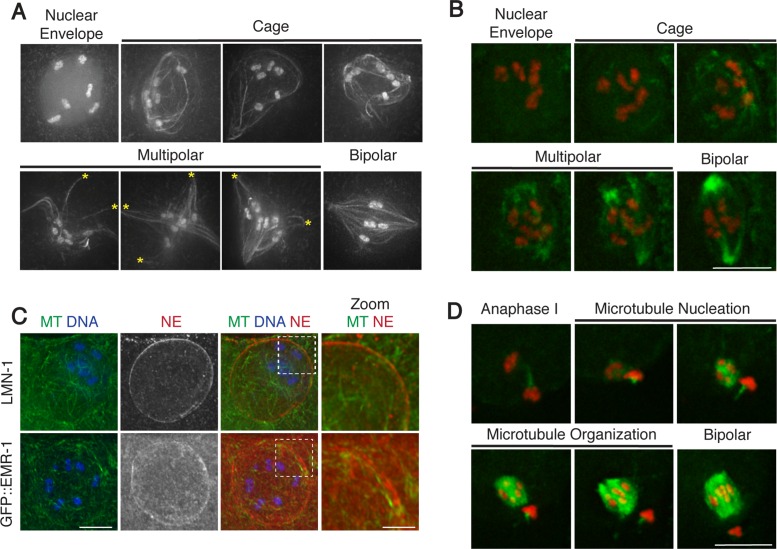
Acentrosomal spindle assembly in meiosis I proceeds through a cage and multipolar stage. (A) In vivo imaging of oocytes expressing GFP-tubulin and GFP-histone. After NEBD, a microtubule cage forms before microtubules are sorted into multiple nascent poles (asterisks) that coalesce until bipolarity is achieved. (B, D) Stills from movies capturing meiosis I (B) and meiosis II (D) spindle assembly in oocytes expressing GFP-tubulin and mCherry-histone. A microtubule cage does not form in meiosis II. (C) Fixed oocytes stained for tubulin (green), DNA (blue), and a nuclear envelope (NE) marker (red); top, LMN-1; bottom, GFP::EMR-1. Zooms show cage microtubule bundles adjacent to and within the nuclear envelope. Scale bars, 5 μm (full images), 1.25 μm (zoom).
First, we observed the early stages of spindle formation, beginning with nuclear envelope breakdown (NEBD). Oocytes progress through the C. elegans gonad in a production line manner, approaching the spermatheca, where sperm entry triggers NEBD and initiation of the meiotic divisions (McCarter et al., 1999). Oocytes with intact nuclear envelopes could be distinguished by hazy GFP-histone fluorescence confined within that area (Figure 1A). Whereas most oocytes adjacent to the spermatheca had intact nuclear envelopes (Supplemental Figure S1A), oocytes that had initiated NEBD were discernible due to dispersal of the hazy GFP signal and the presence of a microtubule array. This array was similar to the size and shape of the previously intact nuclear envelope, forming a “microtubule cage” comprising bundles of microtubules with the chromosomes contained inside (Figure 1, A and B, and Supplemental Movies S2, S4, and S5). Imaging of nuclear lamin (LMN-1) and the integral nuclear membrane protein emerin (EMR-1) revealed that a significant amount of nuclear envelope material was still present at this stage, suggesting that this structure may be analogous to the array that forms in mouse oocytes, where microtubules concentrate on the outside of the nuclear envelope before NEBD (Schuh and Ellenberg, 2007). However, we instead found that the majority of the prominent microtubule bundles forming the cage were on the inside of the LMN-1/EMR-1 signal (Figure 1C and Supplemental Movie S2), with microtubules both concentrated near the nuclear envelope remnants and also projecting inward toward the chromosomes. These results suggest that after nucleation, microtubules are constrained into the cage-like array by the presence of the disassembling envelope, which likely accounts for the “spherical” appearance of microtubules during spindle assembly previously documented in these cells (Sumiyoshi et al., 2015). Consistent with this idea, we did not observe formation of a microtubule cage during meiosis II (which is not preceded by NEBD, as the NE does not reform between the meiotic divisions), and microtubules instead appeared to nucleate in a smaller array in the vicinity of the chromosomes (Figure 1D and Supplemental Movie S3).
After cage formation in meiosis I oocytes, we found that microtubule ends began to appear focused at multiple sites on the periphery of a large array, forming nascent poles (Figure 1A, asterisks). Quantification of spindle morphologies observed at various locations in the gonad supports the view that this multipolar stage precedes the bipolar stage (Supplemental Figure S1A), suggesting that the nascent poles then coalesce until bipolarity is achieved. Lower-resolution time-lapse imaging also supported this ordering of events (Figure 1B and Supplemental Movie S1; Yang et al., 2003, 2005; McNally et al., 2006; Connolly et al., 2014, 2015; Sumiyoshi et al., 2015). After microtubule nucleation around the chromosomes in meiosis II, we observed a similar progression of events (Figure 1D and Supplemental Movie S3). Together our observations indicate that acentrosomal spindle assembly in C. elegans proceeds by microtubule nucleation followed by the formation and subsequent coalescence of multiple poles, as has been proposed previously (Connolly et al., 2015).
Microtubules of mixed polarity are nucleated and then sorted during acentrosomal spindle assembly
As previously mentioned, spindle assembly in mouse oocytes involves the nucleation and coalescence of many small MTOC asters containing PCM components at their centers (Skold et al., 2005; Schuh and Ellenberg, 2007; Clift and Schuh, 2015). In C. elegans oocytes, live imaging of the spindle pole protein ASPM-1, which is presumed to mark microtubule minus ends, revealed that this protein forms foci during spindle assembly, leading to a model in which acentrosomal spindle assembly in this system might also be driven by the nucleation and coalescence of MTOC-like structures (Connolly et al., 2015; Severson et al., 2016). However, previous work demonstrated that C. elegans oocyte spindles do not have the PCM components γ-tubulin, SPD-2, or SPD-5 at their poles (Bobinnec et al., 2000; Pelletier et al., 2004). Moreover, we did not detect MTOC-like asters in either our high-resolution live or fixed imaging (Figure 1 and Supplemental Movies S1 and S2). Thus we set out to examine what these previously observed ASPM-1 foci represent.
To address this question, we performed fixed imaging of ASPM-1 throughout the process of spindle assembly (Figure 2A, Supplemental Figure S2, and Supplemental Movies S4 and S5), achieving a higher level of resolution than the published live imaging. Consistent with our other images (Figure 1 and Supplemental Movie S2), we did not find evidence for MTOC-like microtubule asters, indicating that such structures do not form or are very transient and/or unstable. Instead, in early spindle assembly, we observed small foci of ASPM-1 dispersed throughout the cage structure that often appeared to localize at the tips of individual microtubule bundles (Figure 2A, Supplemental Figure S2, asterisks, and Supplemental Movies S4 and S5), suggesting that these foci mark minus ends. Many ASPM-1 foci were in the vicinity of the disassembling envelope, but some were also near the chromosomes, suggesting that these minus ends are distributed throughout the cage structure. Further, some foci appeared to colocalize with microtubule bundles but were not at the ends (Figure 2A, Supplemental Figure S2, arrows, and Supplemental Movies S4 and S5), suggesting that the bundles themselves likely are composed of multiple microtubules and that the ends are not all focused together at this stage.
FIGURE 2:
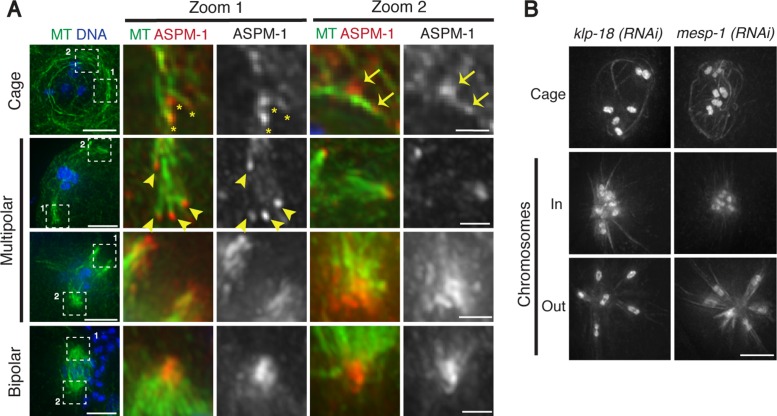
KLP-18 and MESP-1 sort microtubules during spindle assembly. (A) Fixed wild-type oocytes stained for tubulin (green), DNA (blue), and microtubule minus end marker ASPM-1 (red). All images are projections encompassing the entire spindle structure, except for the cage zoom images, which are a single z-plane to better show direct ASPM-1 and MT colocalization. ASPM-1 forms puncta at the ends of microtubule bundles at the cage and multipolar stages (asterisks and arrowheads, respectively) and sometimes within or along the side of a bundle (arrows), and then ASPM-1 marks larger stretches associated with spindle poles as spindle assembly proceeds. Minus ends are distributed throughout the microtubule cage and then are sorted away from the chromosomes as spindle assembly proceeds. (B) In vivo imaging of klp-18(RNAi) and mesp-1(RNAi) worms expressing GFP-tubulin and GFP-histone. Microtubules form a cage, but then minus ends collapse into a single aster. Scale bars, 5 μm (full images), 1.25 μm (zoom).
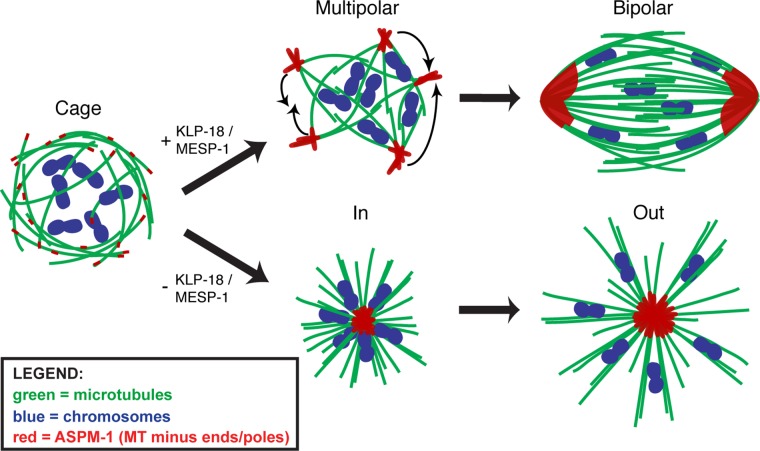
At later stages, ASPM-1 foci continued to decorate the ends of microtubules. In the multipolar stage, these foci were enriched on the periphery of the microtubule array, away from the chromosomes (Figure 2A and Supplemental Figure S2, arrowheads), and then began to form larger stretches connecting multiple microtubule bundles and forming the nascent poles (Figure 2A and Supplemental Figure S2). Taken together, our data suggest that MTOC asters (which are undetectable in either of our imaging conditions) do not play a major role in acentrosomal spindle assembly in C. elegans oocytes. Instead, we propose that spindle assembly proceeds through 1) formation of a disordered array of microtubules within the remnants of the nuclear envelope, 2) sorting of microtubule minus ends away from the chromosomes to the periphery of the array, 3) organization of these ends into nascent poles, and 4) progressive coalescence of these poles until bipolarity is achieved.
KLP-18/kinesin-12 and MESP-1 are required to sort microtubule minus ends away from the chromosomes
Next we sought to investigate how bipolarity is established, given this pathway of spindle assembly. To address this question, we initiated analysis of KLP-18 (kinesin-12 family) and MESP-1, two proteins that we previously found to be required for spindle bipolarity in an RNAi screen (Wignall and Villeneuve, 2009). Using high-resolution in vivo imaging, we found that the microtubule cage forms normally after depletion of either KLP-18 or MESP-1 (Figure 2B). However, after cage formation (at the stage at which multipolar spindles form in wild-type oocytes), microtubule ends converge into one central point, forming a microtubule aster with the chromosomes located close to the center; chromosomes then move outward, away from the pole (Figure 2B, Supplemental Figure S2B, and Supplemental Movie S6). ASPM-1 localizes to the center of these asters, confirming that they represent monopolar spindles (Wignall and Villeneuve, 2009; McNally and McNally, 2011; Connolly et al., 2014). Therefore KLP-18 and MESP-1 are not required for the earliest steps in the assembly pathway but are required to establish acentrosomal spindle bipolarity. Our observations are consistent with the idea that in the absence of these proteins, microtubule minus ends fail to be sorted away from the chromosomes after cage formation; consequently, factors that focus minus ends organize all of these ends into a single pole, bypassing the multipolar stage. These data therefore suggest that KLP-18 and MESP-1 are required to provide an outward force to sort microtubule minus ends away from the chromosomes, enabling bipolar spindle formation.
KLP-18/kinesin-12 and MESP-1 are interdependent for localization and can interact
Because both KLP-18 and MESP-1 are required for acentrosomal spindle bipolarity, we investigated the relationship between the two proteins. First, we assessed the localization of each during wild-type spindle assembly. KLP-18 was shown previously to localize to the poles of the bipolar spindle (Segbert et al., 2003). Using high-resolution microscopy, we confirmed this localization and also determined that KLP-18 and MESP-1 colocalize at all stages of acentrosomal spindle assembly (Figure 3A). Before NEBD and during early stages of spindle assembly, both proteins are broadly distributed in areas of high microtubule density (i.e., on microtubules adjacent to the nuclear envelope and also on the cage structure). As microtubules are further bundled and organized during spindle formation, KLP-18 and MESP-1 become enriched at the nascent poles of multipolar spindles and at the poles of the bipolar spindle, although they are also present at lower levels in the middle region of the spindle.
FIGURE 3:
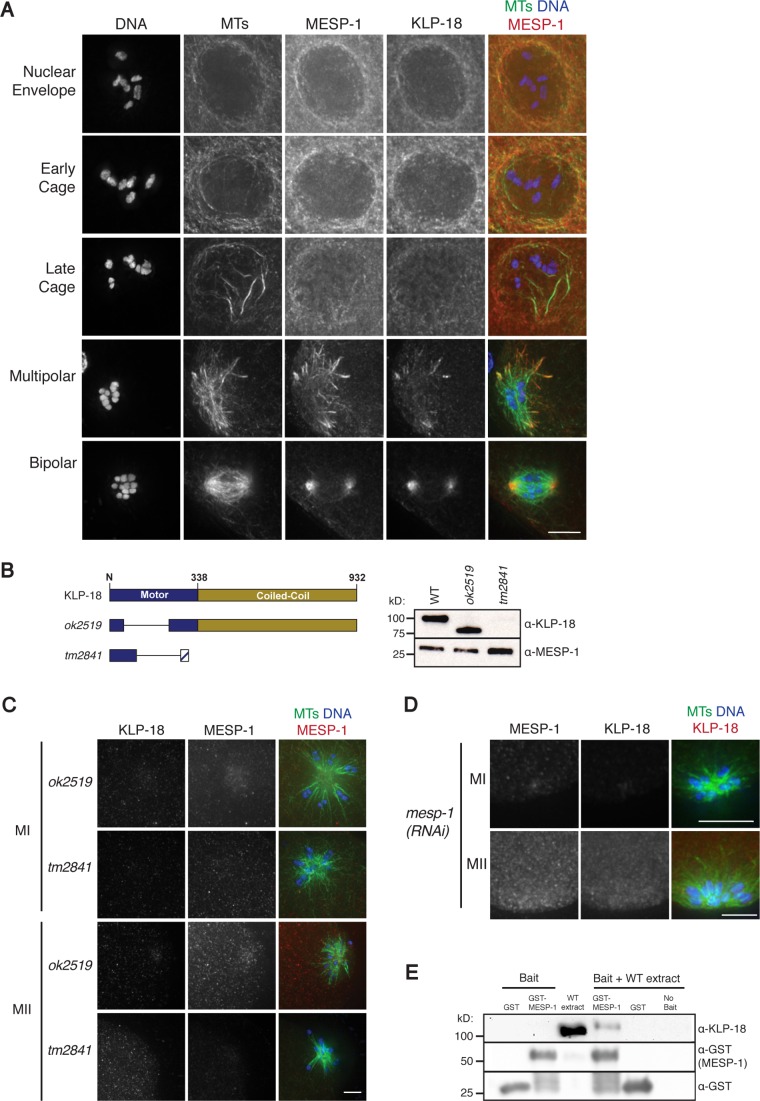
KLP-18 and MESP-1 colocalize and are interdependent for localization. (A) Images of fixed wild-type oocytes stained for DNA (blue), tubulin (green), MESP-1 (red in merge), and KLP-18 (not shown in merge). KLP-18 and MESP-1 colocalize along microtubules during the early cage and become enriched at poles during multipolar and bipolar stages. Pearson’s correlation coefficient for MESP-1/KLP-18 colocalization at cage, multipolar, and bipolar stages was 0.74, 0.78, and 0.92 respectively. (B) Schematic of full-length KLP-18 (top) and analyzed mutants (bottom). klp-18(ok2519) contains an in-frame 510–base pair deletion that removes 170 residues from the motor domain, and klp-18(tm2841) contains a 162–base pair deletion that results in a predicted early stop. Western blotting with a KLP-18 antibody shows that ok2519 results in a truncated protein with ∼25 kDa removed, but KLP-18 is not detected in tm2841, indicating that either it is not expressed or lacks the C-terminal domain. (C) ok2519 and tm2841 oocytes in meiosis I (MI) and meiosis II (MII) stained for DNA (blue), tubulin (green), KLP-18 (not shown in merge), and MESP-1 (red in merge). MESP-1 does not localize to the spindle in either mutant. (D) mesp-1(RNAi) oocytes stained for DNA (blue), tubulin (green), MESP-1 (not shown in merge), and KLP-18 (red in merge). KLP-18 does not localize after MESP-1 depletion. (E) GST pull down demonstrating interaction between KLP-18 and MESP-1. GST-MESP-1 or GST-alone was incubated with wild-type (WT) worm extract and then retrieved. Bait proteins are shown before incubation on the left, and the eluate after incubation is shown on the right. A KLP-18 antibody was used to visualize KLP-18, and a GST antibody was used to visualize both GST and GST-MESP-1. KLP-18 is present in eluate from the GST–MESP-1 pull down but not from GST-alone. Scale bars, 5 μm.
Given their colocalization, we next sought to investigate whether MESP-1 and KLP-18 are dependent on one another for spindle targeting. For this analysis, we took advantage of two klp-18 mutants, ok2519 and tm2841. ok2519 is an in-frame deletion that results in the production of a truncated protein missing part of the motor domain (Figure 3B), and tm2841 is a deletion that results in a predicted early stop. Because our antibody recognizes only the C-terminus of KLP-18 (amino acids 508–932; Segbert et al., 2003), we cannot determine whether a truncated form of KLP-18 is made or targeted to the spindle in this second mutant. As expected from the klp-18(RNAi) phenotype, both klp-18(ok2519) and klp-18(tm2841) homozygotes have monopolar oocyte spindles and 100% embryonic lethality. Moreover, in ok2519, we did not observe KLP-18 localization to microtubules (Figure 3C). Therefore the deleted portion of the motor domain is required for proper spindle localization. Of importance, we found that MESP-1 did not target properly to spindle microtubules in either mutant (Figure 3C). This represents defective localization and not decreased protein abundance, since MESP-1 is present at similar levels in both klp-18 mutants (Figure 3B). In the converse experiment, KLP-18 failed to target to microtubules after mesp-1 RNAi (Figure 3D). Therefore KLP-18 and MESP-1 are interdependent for localization.
Given this finding, we predicted that KLP-18 and MESP-1 might associate in vivo. Therefore, we used pull-down experiments to determine whether they are present in the same protein complex. Specifically, we incubated recombinant glutathione S-transferase (GST)–MESP-1 with wild-type worm extract, retrieved GST–MESP-1 with glutathione Sepharose beads, and eluted associated proteins. Endogenous KLP-18 was detected in the eluate by Western blot (Figure 3E), indicating that MESP-1 and KLP-18 can form a complex and suggesting that they work together in vivo.
MESP-1 is a rapidly evolving protein that may be performing the kinesin-12 targeting role of TPX2
Intriguingly, our findings regarding MESP-1 are reminiscent of a well-studied protein in vertebrates, TPX2, which performs many important functions during mitosis and meiosis (Gruss and Vernos, 2004; Neumayer et al., 2014). Significant to this study, TPX2 is required for kinesin-12 targeting to the spindle (Wittmann et al., 1998, 2000; Heidebrecht et al., 2003; Tanenbaum et al., 2009; Vanneste et al., 2009). A putative C. elegans TPX2 homologue, TPXL-1, was previously shown to perform some of the known functions of TPX2 (Ozlu et al., 2005). However, limited sequence homology and some functional differences between the two proteins has raised questions about whether TPXL-1 is a true TPX2 orthologue (Karsenti, 2005). Of interest, we found that TPXL-1 depletion by RNAi did not cause defects in oocyte spindle morphology (as shown in Ozlu et al., 2005) and also did not affect KLP-18 targeting to the spindle (Figure 4A) under conditions in which we recapitulated the published mitotic phenotype (Figure 4B). Therefore C. elegans TPXL-1 does not appear to facilitate kinesin-12 targeting, and we propose that instead MESP-1 has taken on this role. Our findings support the idea that TPX2 has multiple functional counterparts in C. elegans, with TPXL-1 and MESP-1 providing different essential functions of this important protein.
FIGURE 4:
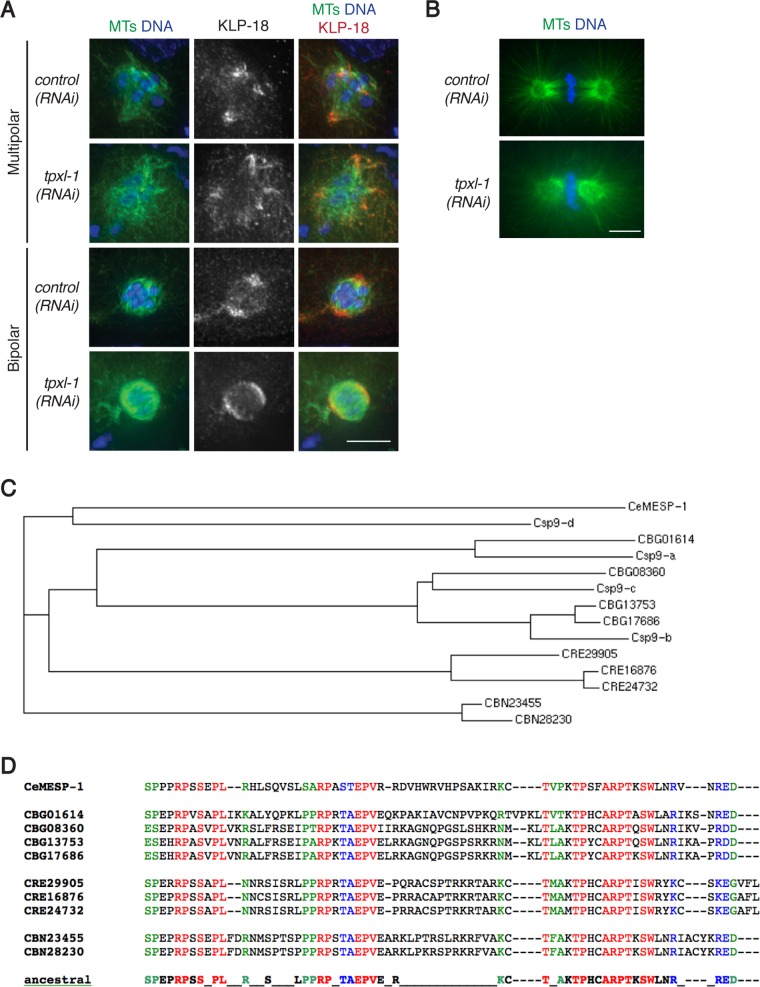
MESP-1 is a rapidly evolving protein that may perform the kinesin-12–targeting role of TPX2. (A, B) Control and tpxl-1(RNAi) worms were fixed and stained for DNA (blue), tubulin (green), and KLP-18 (red). KLP-18 targeting is unaffected by tpxl-1(RNAi) (A), whereas the mitotic spindle in the one-cell-stage embryo is shorter than in wild type (B), confirming the previously described TPXL-1–depletion phenotype (Ozlu et al., 2005). Scale bars, 5 μm. (C) Divergence tree of MESP-1 paralogues from five Caenorhabditis species (C. elegans, C. species 9, C. briggsae, C. remanei, and C. brenneri) based on a ClustalW alignment (generated using the TimeLogic DeCypher server at Stanford University) of the regions corresponding to the C-terminal 65 amino acids from C. elegans MESP-1. The tree structure indicates that paralogues arose both via gene duplication events occurring specifically in the C. briggsae, C. remanei, and C. brenneri lineages and via earlier gene duplication events that occurred before divergence of C. briggsae and C. species 9 (aka C. nigoni). (D) ClustalW alignment of the C-terminal domains of the MESP-1 paralogues from C. elegans, C. briggsae, C. remanei, and C. brenneri. Red indicates identity among all paralogues, blue indicates conservative substitutions, and green indicates semiconservative substitutions.
Although the kinesin-12–targeting function of MESP-1 suggests that it may serve as a functional counterpart of TPX2, sequence analysis of MESP-1 did not identify homology with TPX2 or any previously studied proteins. However, TPX2 is not well conserved in invertebrates. For example, Drosophila Mei-38, identified as a putative TPX2 homologue based on only a few short conserved regions, lacks several key domains present in vertebrate TPX2, suggesting that TPX2 function may commonly be compartmentalized to multiple proteins and that these functional counterparts may not share strong sequence homology (Goshima, 2011).
Independent of the question of homology to known proteins, our MESP-1 sequence analysis revealed two notable features. First, MESP-1 is a member of a rapidly evolving protein family. Homologues are detected only within the genus Caenorhabditis, and the number of paralogues varies among species (Figure 4C). Diversification of this family appears to be an ongoing process, as gene duplications occurring both before and after speciation events have been detected. Second, the MESP-1 protein sequence is enriched for prolines, with proline residues accounting for 12% of its 193 amino acid residues. Because conservation among all family members is concentrated predominantly in the C-terminal 70–80 amino acids, we used a ClustalW alignment of this portion of the homologues to deduce an ancestral protein motif (Figure 4D); 9 of the 31 amino acid residues in this motif are prolines, suggesting that this proline-rich character may be important for MESP-1 function.
New insights into C. elegans acentrosomal spindle assembly
In summary, we propose that in C. elegans oocytes, acentrosomal spindle assembly proceeds by the initial formation of a microtubule array of mixed polarity, followed by sorting of microtubule minus ends toward the periphery of this array and then gathering of minus ends into nascent poles. These poles then progressively coalesce until bipolarity is achieved (Figure 5). Although the coalescence of multiple spindle poles was previously observed (Connolly et al., 2015), our work sheds light on the earliest events of spindle formation, as our studies do not favor the current idea that the nucleation and then coalescence of small MTOC asters drive the initial stages of spindle assembly (Connolly et al., 2015; Severson et al., 2016), as occurs in mouse and possibly fly oocytes (Skold et al., 2005; Schuh and Ellenberg, 2007; Clift and Schuh, 2015). In contrast, our model aligns well with previous studies of acentrosomal spindle formation in Xenopus egg extracts, where microtubules are nucleated and then sorted by motors and organized into poles, as we proposed (Heald et al., 1996, 1997; Walczak et al., 1998). Of note, whereas microtubules seem to be nucleated primarily in the vicinity of the chromatin in the extract system (Heald et al., 1996) and in C. elegans oocytes during meiosis II (Figure 1D), we observe a high concentration of microtubule bundles underlying the disassembling nuclear envelope in meiosis I, forming a cage-like structure (Figure 1, A and B). Although this finding does not rule out the possibility that chromosomes also contribute to spindle assembly in these oocytes, it raises the intriguing possibility that there may be additional regions where microtubules can be nucleated and/or stabilized.
FIGURE 5:
Model for MTOC-independent acentrosomal spindle assembly. Acentrosomal spindle assembly in C. elegans depicting microtubules (green), chromosomes (blue), and ASPM-1 (representing microtubule minus ends; red). After NEBD, microtubules of mixed polarity are organized adjacent to the disassembling nuclear envelope into a cage structure surrounding the chromosomes. The microtubule bundles are then sorted by KLP-18/MESP-1 such that the minus ends are oriented at the periphery of the array and bundled into nascent poles, forming a multipolar spindle (top). The nascent poles then coalesce to achieve bipolarity. In the absence of KLP-18/MESP-1, outward sorting of microtubule bundles is lost, and the minus ends collapse into a single aster and form a monopolar spindle (bottom).
We also find that KLP-18/kinesin-12 and MESP-1 appear to collaborate to establish spindle bipolarity in this system. These proteins localize to microtubules early during spindle assembly, are interdependent for localization, and inhibition of either causes monopolar spindle formation. Our data are consistent with a model in which KLP-18 and MESP-1 are involved in the initial sorting of microtubules such that the minus ends are pushed to the outside of the microtubule array. Without this contribution, factors responsible for organizing minus ends dominate, and all of the minus ends collapse into a single pole (Figure 5).
Our model fits well with work in vertebrate cell lines that has implicated kinesin-12–family motors in spindle assembly. Although kinesin-5–family motors are the dominant motors promoting bipolarity in those cells, when kinesin-5 is inhibited/depleted, bipolar spindles form using a kinesin-12–mediated mechanism (Tanenbaum et al., 2009; Vanneste et al., 2009, 2011; Florian and Mayer, 2011; Raaijmakers et al., 2012; Sturgill and Ohi, 2013; Sturgill et al., 2016) that is believed to act by promoting the formation of parallel microtubule bundles (Drechsler and McAinsh, 2016). Because it has been demonstrated that kinesin-5 is not an essential motor in C. elegans (Bishop et al., 2005), KLP-18/kinesin-12 instead appears to be the dominant motor promoting spindle bipolarity in worm oocytes. In this context, MESP-1 may contribute to this process by taking on the kinesin-12–targeting role performed by TPX2 in other organisms, providing KLP-18 with the opportunity to sort microtubules. Future work determining the mechanisms by which KLP-18 and MESP-1 act to organize microtubules and generate bipolarity will shed light on this important but poorly understood specialized cell division, as well as on overall kinesin-12 function during cell division.
MATERIALS AND METHODS
Worm strains
In this work, “wild type” refers to N2 (Bristol) worms. For RNAi experiments and high-resolution in vivo imaging, strain EU1067 (unc-119(ed3) ruIs32[unc-119(+) pie-1promoter::GFP::H2B] III; ruIs57[pie-1promoter::GFP::tubulin + unc-119(+)]) expressing GFP::tubulin and GFP::histone (gift of Bruce Bowerman, University of Oregon; Wignall and Villeneuve, 2009) was used. VC1915 (klp-18(ok2519)IV/nT1[qIs51]), XA3504 (unc-119(ed3)III; qaEx3504[pie-1 promoter::GFP::emr-1 + unc-119(+)]), and EU2876 (or1935[GFP::aspm-1] I; itIs37[pie-1promoter::mCherry::H2B::pie-1 3′UTR + unc-119(+)]IV) were obtained from the C. elegans Genetics Center. klp-18(tm2841) was obtained from the National Bioresource Project of Japan and balanced to create strain SMW9 (klp-18(tm2841)IV/nT1[qIs51]). SMW13 was created by crossing SMW9 into EU1067 to generate klp-18(tm2841)IV/nT1[qIs51] expressing GFP::histone and GFP::tubulin. For each klp-18 mutant strain, homozygotes grew to adulthood but displayed 100% embryonic lethality. For movies of wild-type spindle formation, strain OD57 expressing GFP::tubulin and mCherry::histone (gift of Jon Audhya, University of Wisconsin; McNally et al., 2006) was used.
Immunofluorescence
Immunofluorescence was performed as described previously (Oegema et al., 2001; Monen et al., 2005), with a fixation time of 40 min in methanol at −20°C. The following primary antibodies were used: Alexa Fluor 488–conjugated mouse anti–α-tubulin (1:500, DM1α; Sigma-Aldrich, St. Louis, MO), Alexa Fluor 596-conjugated goat anti-GFP (used for GFP::EMR-1, 1:250, ab6660; Abcam, Cambridge, MA), monoclonal mouse anti-GFP (used for GFP::ASPM-1, 1:200, A11120; Invitrogen, Waltham, MA), rabbit anti-KLP-18 and rat anti-KLP-18 (1:10,000 and 1:200, respectively, gifts of Olaf Bossinger, RWTH Aachen University; Segbert et al., 2003), rabbit anti–ASPM-1 (1:5000; gift of Arshad Desai, Ludwig Institute for Cancer Research; Wignall and Villeneuve, 2009), rat anti-LMN-1 (1:500; gift of Katherine Wilson, Johns Hopkins University; Gruenbaum et al., 2002), and rabbit anti–MESP-1 (1:3000). Anti–MESP-1 antibody was raised through genomic antibody technology (Strategic Diagnostics, Newark, DE) against amino acids 60–159 and then affinity purified. Secondary antibodies used were Alexa Fluor 555–conjugated goat anti-rabbit and Alexa Fluor 647–conjugated goat anti-rat (both at 1:500; Invitrogen).
Image acquisition and processing
All imaging was performed at the Northwestern Biological Imaging Facility supported by the Northwestern University Office for Research. High-resolution imaging was performed as described previously (Wignall and Villeneuve, 2009; Muscat et al., 2015) using a DeltaVision Core microscope with a 100× objective (numerical aperture [NA], 1.40). Image stacks were obtained at 0.2 μm z-steps for all live and fixed high-resolution images and at 2.0 μm with a 60× objective (NA 1.40) and 1.6× optical zoom for lower-resolution movies. All image acquisition, processing, and analysis was performed with softWoRx software (GE Heathcare Life Sciences, Pittsburgh, PA). All images in this study were deconvolved and displayed as full maximum-intensity projections of data stacks encompassing the entire spindle structure unless otherwise indicated.
Two-color live imaging was performed using a spinning-disk confocal microscope with a 63× HC PL APO 1.40 NA objective lens. A spinning-disk confocal unit (CSU-X1; Yokogawa Electric Corporation, Sugar Land, TX) attached to an inverted microscope (Leica DMI6000 SD; Leica, Wetzlar, Germany) and a Spectral Applied Imaging laser merge ILE3030 and a back-thinned electron-multiplying charge-coupled device camera (Evolve 521 Delta; Photometrics, Tucson, AZ) were used for image acquisition. The microscope and attached devices were controlled using MetaMorph Image Series Environment software (Molecular Devices, Sunnyvale, CA). Twelve z-stacks at 1 μm increments were taken every 20–30 s at room temperature. Image deconvolution was done using AutoQuant X3 (Media Cybernetics, Rockville, MD). Images are shown as maximum-intensity projections of the entire data stack. Live, intact worms were mounted on 5% agarose, M9 pads in 50% live imaging solution (modified S-basal [50 mM KH2PO4, 10 mM K-citrate, 0.1 M NaCl, 0.025 mg/ml cholesterol, 3 mM MgSO4, 3 mM CaCl2, 20 mM serotonin-HCl, 0.1% tricaine, 0.01% levamisole]), and 50% 0.1 μm polystyrene Microspheres (Polysciences, Warrington, PA) and covered with a coverslip.
Spindle assembly stages were quantified by scoring live worms mounted in anesthetic (0.2% tricaine, 0.02% levamisole in M9) with a Leica DM5500B fluorescence microscope.
RNAi
Individual RNAi clones picked from an RNAi feeding library (Fraser et al., 2000; Kamath et al., 2003) were used to inoculate Luria broth (LB) plus ampicillin (100 μg/ml) and grown overnight at 37°C. These cultures were used to seed nematode growth medium (NGM)/ampicillin (100 μg/ml)/1 mM isopropyl-β-d-thiogalactoside (IPTG) plates, and the plates were then left overnight at room temperature to induce RNA expression. Synchronized L1 worms (EU1067) were plated on induced plates and grown at 15°C for 5 d until they became gravid adults. Control plates were seeded with bacteria containing empty vector L4440 (designated as control(RNAi) throughout the article). For tpxl-1(RNAi) (which has multiple available clones in the library), multiple clones were tried (and yielded identical results), but clone Y39G10A_246.k was used for the experiments displayed.
Protein expression and purification
GST–MESP-1 and GST were cloned into pGEX 6P-1 (GE) and expressed in BL21 Escherichia coli cells. Cultures were grown at 37ºC to an OD of 0.6–0.8 and then induced at 28ºC for 4 h with 0.1 mM IPTG. After harvesting, cells were lysed at room temperature for 15 min with B-PER Protein Extraction Reagent (ThermoFisher Scientific, Waltham, MA) including 100 μg/ml lysozyme, 20 μl DNase I (ThermoFisher), and EDTA-free protease inhibitor cocktail tablets (Roche, Indianapolis, IN). The lysate was then cleared by centrifugation at 40,000 rpm for 35 min. Lysate was applied to glutathione Sepharose resin (GE) and then washed with wash buffer (phosphate-buffered saline, pH 7.4, 250 mM NaCl, 0.1% Tween 20, 2 mM benzamidine-HCl, 1 mM dithiothreitol [DTT]). Bound protein was then eluted with elution buffer (50 mM Tris, pH 8.1, 75 mM KCl, 10 mM reduced glutathione). Protein was dialyzed into BRB80 (80 mM 1,4-piperazinediethanesulfonic acid [PIPES], pH 6.8, 1 mM ethylene glycol tetraacetic acid [EGTA], 1 mM MgCl2) overnight and stored at −80ºC. For all recombinant protein purifications, purity was confirmed with SDS–PAGE and concentration determined by Bradford assay (Bio-Rad, Hercules, CA).
Large-scale worm growth and protein extraction
Large-scale worm growth was performed similarly to previous work (Zanin et al., 2011). Briefly, wild-type (N2) young adult worms were picked onto NGM plates seeded with OP50, and cultures were allowed to starve. Between 10 and 12 of these plates were then washed into 500 ml of S-Complete (50 mM KH2PO4, 10 mM K-citrate, 0.1 M NaCl, 0.025 mg/ml cholesterol, 3 mM MgSO4, 3 mM CaCl2, 0.05 mM disodium EDTA, 0.025 mM FeSO4*7H2O, 0.01 mM MnCl2*4H2O, 0.01 mM ZnSO4*7H2O, 0.001 mM CuSO4*5H2O) seeded with OP50-1, and the culture was shaken at 16–22°C (depending on desired growth rate) until the worms were gravid adults. The culture was then bleached, and the resulting embryos were hatched in 500 ml of S-Complete shaken overnight at 20ºC. OP50-1 was added to the arrested L1 larvae, and the culture was again shaken at 16–22°C until the worms were gravid adults. The culture was harvested and washed with M9 and lysis buffer (50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid [HEPES], pH 7.4, 1 mM EGTA, 1 mM MgCl2, 100 mM KCl, 10% glycerol). Worm pellets were snap frozen by dropping them into liquid nitrogen and stored at –80ºC. Protein was extracted from frozen worm pellets as previously described (Sonneville et al., 2012). Pellets were thawed on ice and washed in lysis buffer with protease inhibitors (EDTA-free cocktail; Roche). A 0.5 ml volume of Zirconia beads (Biospec, Bartlesville, OK) was added to resuspended worms in 750 μl of lysis buffer. The samples were vortexed six times (1 min vortexing with 20 s rest) at 4ºC, and the efficiency of lysis was confirmed using a dissecting microscope. Lysate was then separated from beads, incubated on ice for 30 min, and cleared by centrifugation at 4ºC (25,000 rpm for 10 min and 50,000 rpm for 20 min). Protein concentration was measured using Bradford assay (Bio-Rad), and worm lysate was kept on ice until use.
GST pull-down assay
GST-tagged bait protein was incubated with worm extract for 30 min at 4ºC, and then glutathione Sepharose (equilibrated with wash buffer: 50 mM HEPES, pH 7.4, 1 mM EGTA, 1 mM MgCl2, 300 mM KCl, 0.05% NP-40, 10% glycerol) was added, and the mixture was incubated at 4ºC overnight. Beads were pelleted at 16,000 × g for 1 min and then resuspended in wash buffer and incubated for 10 min while being rotated. Protease inhibitors (EDTA free; Roche) and 0.5 mM DTT were added to the wash buffer. After two washes, bound protein was eluted by mixing for 10 min with 100 μl of elution buffer (50 mM Tris-HCl, pH 8.0, 75 mM KCl, 20 mM reduced glutathione) and removed from beads after centrifugation. Eluate was added to 100 μl 2× Laemmli Sample Buffer (Bio-Rad), boiled at 95ºC for 10 min, and used for SDS–PAGE analysis. For Western analysis, we used the antibodies anti–KLP-18 (1:5000) and anti-GST (1:2000; gift of Jason Brickner, Northwestern University).
Supplementary Material
Acknowledgments
We thank members of the Wignall and Villeneuve labs for support and discussions, Amanda Davis for critical reading of the manuscript, Bohan Xing for help with worm growth, and Jon Audhya, Olaf Bossinger, Bruce Bowerman, Jason Brickner, Arshad Desai, Sarah Rice, and Katherine Wilson for reagents. In addition, some strains were provided by the National Bioresource Project of Japan and by the Caenorhabditis Genetics Center, which is funded by the National Institutes of Health Office of Research Infrastructure Programs (P40 OD010440). This work was supported by the Chicago Biomedical Consortium with support from the Searle Funds at the Chicago Community Trust (to S.M.W.), Grant 253551 from the American Cancer Society, Illinois Division (to S.M.W.), American Cancer Society Grant ACS-IRG 93-037-18 (to S.M.W.), National Institutes of Health/National Institute of General Medical Sciences Training Grant T32GM008382 (to I.D.W.), National Institutes of Health/National Cancer Institute Training Grant T32CA009560 (to T.J.M.), and National Institutes of Health Grant RO1 GM53804 (to A.M.V.). A.M.V. is an American Cancer Society Research Professor.
Abbreviations used:
- GFP
green fluorescent protein
- GST
glutathione S-transferase
- KLP
kinesin-like protein
- MESP
meiotic spindle
- MTOC
microtubule-organizing center
- NE
nuclear envelope
- NEBD
nuclear envelope breakdown
- PCM
pericentriolar material
- RNAi
RNA interference.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-05-0291) on August 24, 2016.
REFERENCES
- Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobinnec Y, Fukuda M, Nishida E. Identification and characterization of Caenorhabditis elegans gamma-tubulin in dividing cells and differentiated tissues. J Cell Sci. 2000;113:3747–3759. doi: 10.1242/jcs.113.21.3747. [DOI] [PubMed] [Google Scholar]
- Carabatsos MJ, Combelles CM, Messinger SM, Albertini DF. Sorting and reorganization of centrosomes during oocyte maturation in the mouse. Microsc Res Tech. 2000;49:435–444. doi: 10.1002/(SICI)1097-0029(20000601)49:5<435::AID-JEMT5>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Clift D, Schuh M. A three-step MTOC fragmentation mechanism facilitates bipolar spindle assembly in mouse oocytes. Nat Commun. 2015;6:7217. doi: 10.1038/ncomms8217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombie N, Cullen CF, Brittle AL, Jang JK, Earnshaw WC, Carmena M, McKim K, Ohkura H. Dual roles of Incenp crucial to the assembly of the acentrosomal metaphase spindle in female meiosis. Development. 2008;135:3239–3246. doi: 10.1242/dev.022624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AA, Osterberg V, Christensen S, Price M, Lu C, Chicas-Cruz K, Lockery S, Mains PE, Bowerman B. Caenorhabditis elegans oocyte meiotic spindle pole assembly requires microtubule severing and the calponin homology domain protein ASPM-1. Mol Biol Cell. 2014;25:1298–1311. doi: 10.1091/mbc.E13-11-0687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly AA, Sugioka K, Chuang CH, Lowry JB, Bowerman B. KLP-7 acts through the Ndc80 complex to limit pole number in C. elegans oocyte meiotic spindle assembly. J Cell Biol. 2015;210:917–932. doi: 10.1083/jcb.201412010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsler H, McAinsh AD. Kinesin-12 motors cooperate to suppress microtubule catastrophes and drive the formation of parallel microtubule bundles. Proc Natl Acad Sci USA. 2016;113:E1635–E1644. doi: 10.1073/pnas.1516370113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont J, Desai A. Acentrosomal spindle assembly and chromosome segregation during oocyte meiosis. Trends Cell Biol. 2012;22:241–249. doi: 10.1016/j.tcb.2012.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florian S, Mayer TU. Modulated microtubule dynamics enable Hklp2/Kif15 to assemble bipolar spindles. Cell Cycle. 2011;10:3533–3544. doi: 10.4161/cc.10.20.17817. [DOI] [PubMed] [Google Scholar]
- Fraser AG, Kamath RS, Zipperlen P, Martinez-Campos M, Sohrmann M, Ahringer J. Functional genomic analysis of C. elegans chromosome I by systematic RNA interference. Nature. 2000;408:325–330. doi: 10.1038/35042517. [DOI] [PubMed] [Google Scholar]
- Goshima G. Identification of a TPX2-like microtubule-associated protein in Drosophila. PLoS One. 2011;6:e28120. doi: 10.1371/journal.pone.0028120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Lee KK, Liu J, Cohen M, Wilson KL. The expression, lamin-dependent localization and RNAi depletion phenotype for emerin in C. elegans. J Cell Sci. 2002;115:923–929. doi: 10.1242/jcs.115.5.923. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, Vernos I. The mechanism of spindle assembly: functions of Ran and its target TPX2. J Cell Biol. 2004;166:949–955. doi: 10.1083/jcb.200312112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, Maro B. gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105:157–166. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- Heald R, Tournebize R, Habermann A, Karsenti E, Hyman A. Spindle assembly in Xenopus egg extracts: respective roles of centrosomes and microtubule self-organization. J Cell Biol. 1997;138:615–628. doi: 10.1083/jcb.138.3.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidebrecht HJ, Adam-Klages S, Szczepanowski M, Pollmann M, Buck F, Endl E, Kruse ML, Rudolph P, Parwaresch R. repp86: a human protein associated in the progression of mitosis. Mol Cancer Res. 2003;1:271–279. [PubMed] [Google Scholar]
- Holubcova Z, Blayney M, Elder K, Schuh M. Human oocytes. Error-prone chromosome-mediated spindle assembly favors chromosome segregation defects in human oocytes. Science. 2015;348:1143–1147. doi: 10.1126/science.aaa9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- Karsenti E. TPX or not TPX. Mol Cell. 2005;19:431–432. doi: 10.1016/j.molcel.2005.08.002. [DOI] [PubMed] [Google Scholar]
- McCarter J, Bartlett B, Dang T, Schedl T. On the control of oocyte meiotic maturation and ovulation in Caenorhabditis elegans. Dev Biol. 1999;205:111–128. doi: 10.1006/dbio.1998.9109. [DOI] [PubMed] [Google Scholar]
- McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally KP, McNally FJ. The spindle assembly function of Caenorhabditis elegans katanin does not require microtubule-severing activity. Mol Biol Cell. 2011;22:1550–1560. doi: 10.1091/mbc.E10-12-0951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- Muscat CC, Torre-Santiago KM, Tran MV, Powers JA, Wignall SM. Kinetochore-independent chromosome segregation driven by lateral microtubule bundles. eLife. 2015;4:e06462. doi: 10.7554/eLife.06462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumayer G, Belzil C, Gruss OJ, Nguyen MD. TPX2: of spindle assembly, DNA damage response, and cancer. Cell Mol Life Sci. 2014;71:3027–3047. doi: 10.1007/s00018-014-1582-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozlu N, Srayko M, Kinoshita K, Habermann B, O’Toole TE, Muller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- Palacios MJ, Joshi HC, Simerly C, Schatten G. Gamma-tubulin reorganization during mouse fertilization and early development. J Cell Sci. 1993;104:383–389. doi: 10.1242/jcs.104.2.383. [DOI] [PubMed] [Google Scholar]
- Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- Raaijmakers JA, van Heesbeen RG, Meaders JL, Geers EF, Fernandez-Garcia B, Medema RH, Tanenbaum ME. Nuclear envelope-associated dynein drives prophase centrosome separation and enables Eg5-independent bipolar spindle formation. EMBO J. 2012;31:4179–4190. doi: 10.1038/emboj.2012.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans. Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severson AF, von Dassow G, Bowerman B. Oocyte meiotic spindle assembly and function. Curr Top Dev Biol. 2016;116:65–98. doi: 10.1016/bs.ctdb.2015.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skold HN, Komma DJ, Endow SA. Assembly pathway of the anastral Drosophila oocyte meiosis I spindle. J Cell Sci. 2005;118:1745–1755. doi: 10.1242/jcs.02304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonneville R, Querenet M, Craig A, Gartner A, Blow JJ. The dynamics of replication licensing in live Caenorhabditis elegans embryos. J Cell Biol. 2012;196:233–246. doi: 10.1083/jcb.201110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Norris SR, Guo Y, Ohi R. Kinesin-5 inhibitor resistance is driven by kinesin-12. J Cell Biol. 2016;213:213–227. doi: 10.1083/jcb.201507036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturgill EG, Ohi R. Kinesin-12 differentially affects spindle assembly depending on its microtubule substrate. Curr Biol. 2013;23:1280–1290. doi: 10.1016/j.cub.2013.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumiyoshi E, Fukata Y, Namai S, Sugimoto A. Caenorhabditis elegans Aurora A kinase is required for the formation of spindle microtubules in female meiosis. Mol Biol Cell. 2015;26:4187–4196. doi: 10.1091/mbc.E15-05-0258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum ME, Macurek L, Janssen A, Geers EF, Alvarez-Fernandez M, Medema RH. Kif15 cooperates with eg5 to promote bipolar spindle assembly. Curr Biol. 2009;19:1703–1711. doi: 10.1016/j.cub.2009.08.027. [DOI] [PubMed] [Google Scholar]
- Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste D, Ferreira V, Vernos I. Chromokinesins: localization-dependent functions and regulation during cell division. Biochem Soc Trans. 2011;39:1154–1160. doi: 10.1042/BST0391154. [DOI] [PubMed] [Google Scholar]
- Vanneste D, Takagi M, Imamoto N, Vernos I. The role of Hklp2 in the stabilization and maintenance of spindle bipolarity. Curr Biol. 2009;19:1712–1717. doi: 10.1016/j.cub.2009.09.019. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Heald R. Mechanisms of mitotic spindle assembly and function. Int Rev Cytol. 2008;265:111–158. doi: 10.1016/S0074-7696(07)65003-7. [DOI] [PubMed] [Google Scholar]
- Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/s0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Boleti H, Antony C, Karsenti E, Vernos I. Localization of the kinesin-like protein Xklp2 to spindle poles requires a leucine zipper, a microtubule-associated protein, and dynein. J Cell Biol. 1998;143:673–685. doi: 10.1083/jcb.143.3.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–457. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C elegans. Dev Biol. 2003;260:245–259. doi: 10.1016/s0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- Zanin E, Dumont J, Gassmann R, Cheeseman I, Maddox P, Bahmanyar S, Carvalho A, Niessen S, Yates JR, 3rd, Oegema K, et al. Affinity purification of protein complexes in C. elegans. Methods Cell Biol. 2011;106:289–322. doi: 10.1016/B978-0-12-544172-8.00011-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.