The endocytic Rab5 effectors Ccz1-Mon1 complex and Rab7 promote autophagosome-lysosome fusion independent of Rab5, which facilitates a later step of autophagy: degradation of cargo within lysosomes.
Abstract
The small GTPase Rab5 promotes recruitment of the Ccz1-Mon1 guanosine exchange complex to endosomes to activate Rab7, which facilitates endosome maturation and fusion with lysosomes. How these factors function during autophagy is incompletely understood. Here we show that autophagosomes accumulate due to impaired fusion with lysosomes upon loss of the Ccz1-Mon1-Rab7 module in starved Drosophila fat cells. In contrast, autophagosomes generated in Rab5-null mutant cells normally fuse with lysosomes during the starvation response. Consistent with that, Rab5 is dispensable for the Ccz1-Mon1–dependent recruitment of Rab7 to PI3P-positive autophagosomes, which are generated by the action of the Atg14-containing Vps34 PI3 kinase complex. Finally, we find that Rab5 is required for proper lysosomal function. Thus the Ccz1-Mon1-Rab7 module is required for autophagosome-lysosome fusion, whereas Rab5 loss interferes with a later step of autophagy: the breakdown of autophagic cargo within lysosomes.
INTRODUCTION
Autophagy ensures the lysosomal degradation of self-material, including cytosol and organelles. During the main pathway, double-membrane autophagosomes serve as the transport vesicles (Mizushima et al., 2008). Endocytosis delivers plasma membrane, including transmembrane receptors, and exogenous substances taken up from the environment to lysosomes. Thus autophagy and endocytosis converge at the level of lysosomes, where degradation of cargo arriving from both routes takes place.
A critical event during these transport processes is vesicle maturation: how the newly formed vesicles acquire the molecular characteristics and protein complexes that establish their identity and determine the subsequent vesicle fusion events that often culminate in the lysosomal compartment. Several similarities between endosomes and autophagosomes are known. For example, both autophagosomes and endosomes are positive for phosphatidylinositol-3-phosphate (PI3P) due to localized vacuolar protein sorting 34 (Vps34) PI3 kinase activity, which we showed to be required for the generation of both types of vesicles in Drosophila larvae (Lindmo and Stenmark, 2006; Juhasz et al., 2008; Dooley et al., 2014). Autophagosomes can also fuse with endosomes to give rise to hybrid organelles termed amphisomes, which then fuse with lysosomes (Filimonenko et al., 2007; Rusten et al., 2007; Fader and Colombo, 2009).
Small GTPases of the Ras-related protein in brain (Rab) family are critical regulators of membrane trafficking in eukaryotic cells. An active, GTP-bound Rab protein binds to various effectors that usually regulate vesicle motility and fusion with the proper membrane compartment (Stenmark, 2009). In the endocytic pathway, Rab5 associates with early endosomes and activates a Vps34-containing phosphoinositide 3-kinase complex that generates PI3P on the surface of these vesicles. PI3P-binding domains such as the Fab-1, YGL023, Vps27, and EEA1 (FYVE) domain promote recruitment to early endosomes. Of importance, several proteins, including the vesicle tethers early endosomal antigen 1 (EEA1) and Rabenosin-5, have both FYVE and Rab5-binding domains, indicating that multiple interactions may play a role in the recruitment of effectors (Stenmark, 2009). Similarly, the Rab7 guanine nucleotide exchange factor (GEF) monensin sensitivity protein 1 (Mon1)–caffeine, calcium, and zinc 1 (Ccz1) complex binds to both the GTP-bound form of endosomal Rab5 and PI3P (Poteryaev et al., 2010; Cabrera et al., 2014; Cui et al., 2014). Rab7 is then activated by this complex and promotes fusion of late endosomes and lysosomes.
Others and we have shown that recruitment of the soluble N-methylamaleimide–sensitive factor attachment protein receptor (SNARE) Syntaxin 17 is a critical step in autophagosome maturation because these vesicles acquire fusion competence this way (Itakura et al., 2012; Takats et al., 2013). Interaction of Syntaxin 17 with the homotypic fusion and vacuole protein sorting (HOPS) tethering complex ensures efficient fusion between autophagosomes and lysosomes (Jiang et al., 2014; Takats et al., 2014). HOPS is believed to be a Rab7 effector, and Rab7 was indeed found to promote the formation of degradative autolysosomes in cultured cells (Gutierrez et al., 2004; McEwan et al., 2015), although it remains to be established whether this protein is already present on autophagosomes before the fusion with lysosomes. In theory, the binding of HOPS to lysosomal Rab7 and autophagosomal Syntaxin 17 (and other factors, such as phospholipids) may be sufficient for its tethering activity (Stroupe et al., 2006; Hickey et al., 2009; Jiang et al., 2014; Takats et al., 2014). In addition, autophagy-related gene 14 (Atg14), a Vps34 kinase complex subunit that is important for autophagosome formation, also functions as a tether and promotes autophagosome-lysosome fusion by directly binding to Syntaxin 17 (Diao et al., 2015).
In yeast, the fusion machinery differs somewhat from that of the animal cells because the SNAREs involved are not homologous (Dilcher et al., 2001; Ishihara et al., 2001; Ohashi and Munro, 2010). Still, autophagosome fusion with the vacuole (the equivalent of the lysosomal system in metazoan cells) requires HOPS, Ypt7/Rab7, and its GEF, the Mon1-Ccz1 complex (Rieder and Emr, 1997; Kim et al., 1999; Wang et al., 2002), and more recently, autophagosome-like structures were found to accumulate in yeast cells lacking the major Rab5 homologue Vps21 (Chen et al., 2014). Of interest, decreased Rab5 function attenuates the autophagic degradation of the pathogenic, mutant form of huntingtin in cultured human cells. This was attributed to impaired Vps34 lipid kinase activity and reduced formation of the Atg5-Atg12 conjugate, both of which are important for autophagosome formation (Ravikumar et al., 2008).
Thus the role of the Rab5-Ccz1-Mon1-Rab7 axis during autophagy is not clear. We set out to address this problem in the popular animal model Drosophila. Fruit flies offer certain advantages for such studies. First, there is only a single fly homologue of Rab5 (unlike in mammalian and yeast cells, which both have three different Rab5 proteins; Pereira-Leal and Seabra, 2001). Second, massive induction of autophagy is seen in the fat and liver tissue–like fat cells of starved larvae. Third, it is straightforward to carry out functional studies in mosaic animals, in which mutant cells are surrounded by control cells in the same tissue of the same animal, which reduces variability because one can compare the phenotype of neighboring control and loss-of-function cells (Mauvezin et al., 2014; Nagy et al., 2015). Using this system, we show that Ccz1, Mon1, and Rab7 are required for autophagosome-lysosome fusion in fat cells of starved animals independent of Rab5. Of interest, we find that Rab5 functions downstream of the Rab7 module by controlling a later step of autophagy: degradation of autophagic cargo within lysosomes.
RESULTS
Rab7 is important for autolysosome formation
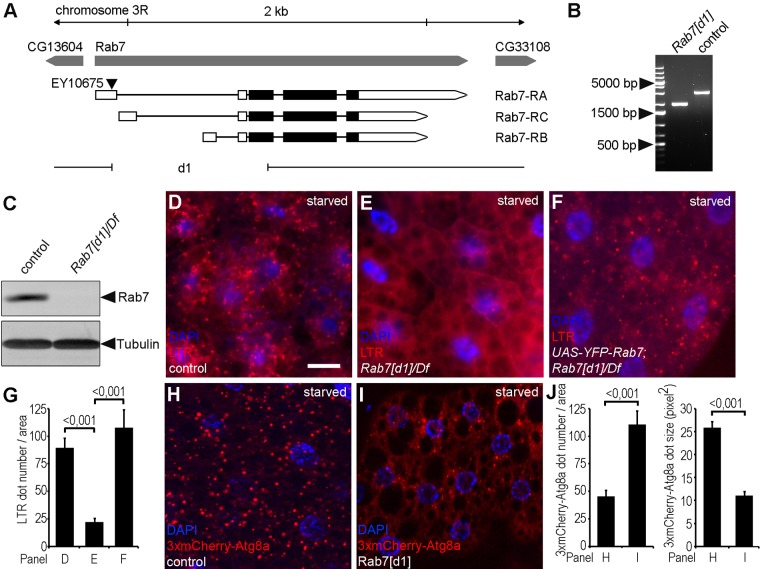
To characterize Rab7’s role in autophagy, we generated Rab7-null mutant flies by imprecise excision of the P-element EY10675. The resulting Rab7[d1] allele carries a 1025–base pair deletion, which removes most of the first protein-coding exon and the 5′ untranslated region (UTR) of all three predicted Rab7 isoforms (Figure 1, A and B). Animals homozygous or hemizygous for Rab7[d1] showed late pupal lethality, similar to another knockout allele published during the course of our work (Cherry et al., 2013). Western blots of third-instar larval lysates showed the absence of Rab7 protein in the mutant (Figure 1C), confirming that this allele represents a null mutant. Immunostaining developing eyes of control and Rab7[d1] mutant larvae revealed that down-regulation of endocytosed Notch and Boss receptors is impaired in Rab7 mutant tissue (Supplemental Figure S1, A, B, E, and F), consistent with the essential role of this protein for progression of late endosomes to endolysosomes.
FIGURE 1:
Rab7 is required for autophagy. (A) Genomic map of the Rab7 locus, showing the exon–intron structure, including 5′ and 3′ UTRs (open bars). The Rab7[d1]-null allele was generated by imprecise excision of the P-element EY10675. The extent of the deletion is indicated by the gap in the line. (B) PCR amplification from the Rab7 locus show the 1025–base pair deletion in Rab7[d1] mutants compared with controls. (C) Western blot shows the absence of Rab7 protein expression in samples prepared from larvae hemizygous for Rab7[d1]. Tubulin was used as loading control. (D–F) Many LTR-positive dots are present in fat cells of wild-type starved third-instar larvae (D). Rab7 mutants show almost complete lack of LTR dots (E). This phenotype can be rescued by transgenic expression of Rab7 (F). (G) Quantification of data shown in D–F; n = 10/genotype. (H, I) Large 3xmCherry-Atg8a–positive autolysosomes appear in starved control fat cells (H), whereas Rab7 mutant cells contain more but smaller autophagic structures (I). (J) Quantification of data shown in H and I; n = 10/genotype. Scale bar, 20 μm (D–F, H, I). Error bars denote SE in G and J, and the numbers above the clasps show p values in these and all subsequent charts.
To evaluate autophagy in our mutant, we stained fat cells of 4-h-starved, early L3-stage larvae with LysoTracker Red (LTR). LTR is an acidophilic dye commonly used for staining autolysosomes in this Drosophila tissue and detects many autolysosomes in control animals (Figure 1D). In contrast, Rab7 mutants showed a greatly reduced number of LTR-positive dots, which could be rescued by expressing YFP-Rab7 in the mutants (Figure 1, E–G). Because acidic autolysosomes were absent from Rab7 loss-of-function fat cells, we tested whether earlier steps of the autophagic pathway are also affected. We used our novel, 3xmCherry-tagged Atg8a reporter driven by its endogenous promoter to find that the number of autophagic structures is increased and their average size is decreased in fat cells of starved Rab7 mutants compared with control animals (Figure 1, H–J). These data suggested that Rab7 is required for autolysosome formation in fat cells.
The Ccz1-Mon1-Rab7 module regulates autophagosome-lysosome fusion
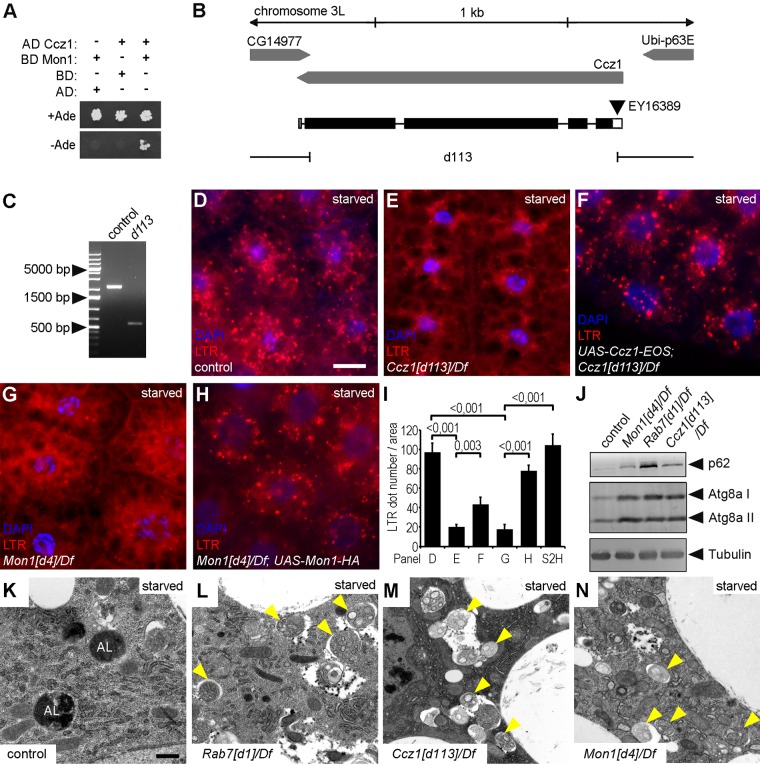
In yeast and mammalian cells, Rab7 is activated by the Ccz1-Mon1 heterodimer, which acts as a GEF (Nordmann et al., 2010; Gerondopoulos et al., 2012). To confirm the existence of this complex in Drosophila, we carried out yeast two-hybrid (Y2H) experiments, which showed a strong interaction between these two proteins (Figure 2A). A recent Drosophila study demonstrated that both Mon1 and Ccz1 are required for Rab7 recruitment to endosomes (Yousefian et al., 2013). Thus the Drosophila Ccz1-Mon1 complex functions similarly to its yeast and mammalian homologues.
FIGURE 2:
Rab7 module mutants are defective in autophagosome-lysosome fusion. (A) Y2H experiment shows the direct interaction of Drosophila Mon1 and Ccz1. (B) Genomic map of the Ccz1 locus. The Ccz1[d113] allele was generated by imprecise excision of the P-element EY16389. (C) PCR amplification of the Ccz1 locus shows the 1644–base pair deletion in Ccz1[d113] mutant animals relative to controls. (D–H) Many LTR-positive dots are seen in fat cells of starved control larvae (D). The formation of such acidic compartments is inhibited in Ccz1 (E) and Mon1 (G) mutants and can be restored by expression of wild-type Ccz1 and Mon1, respectively (F, H). (I) Quantification of LTR data shown in D–H and Supplemental Figure S2H; n = 10/genotype. (J) Western blots from starved larvae reveal accumulation of the selective autophagic cargo p62 and lipidated, autophagosome-associated Atg8a II in Ccz1, Mon1, and Rab7 mutants. Tubulin serves as loading control. (K–N) Electron micrographs of fat cells dissected from starved third-instar larvae. Control cells (K) contain large, electron-dense autolysosomes (AL), whereas Rab7 module mutants (L–N) show accumulation of autophagosomes (arrowheads) and lack of autolysosomes. Scale bar, 20 μm (in D, for D–H), 1 μm (in K, for K–N).
To evaluate the role of the Ccz1-Mon1 complex in autophagy, we first generated a Ccz1-null mutant line by imprecise excision of the P-element EY16389. The resulting Ccz1[d113] allele carries a 1644–base pair deletion that removes almost the entire coding region of this gene (Figure 2, B and C). We also obtained the recently published Mon1[d4]-null mutant Drosophila line (Yousefian et al., 2013) and investigated the effect of Ccz1 and Mon1 mutations on autophagy by LTR staining of fat cells from starved larvae. The lack of these genes strongly reduced the number of acidic autolysosomes (Figure 2, D, E, G, and I), and this phenotype could be rescued by expression of wild-type transgenes (Figure 2, F, H, and I), respectively. Mon1-null mutant fat cells of starved animals contained more but smaller 3xmCherry-Atg8a structures than did control cells (Supplemental Figure S2, A and B), similar to Rab7 mutants. In addition, Ccz1 and Mon1 mutant cells of the developing eye accumulated endocytosed but nondegraded Notch and Boss receptors, which is again similar to the situation in Rab7 mutants (Supplemental Figure S1, C, D, G, and H).
p62 (also known as Ref2P in flies) is a specific autophagic cargo commonly used to measure autophagic degradation (flux; Nezis et al., 2008; Pircs et al., 2012), along with the autophagosome-associated, lipidated form of Atg8a (Atg8a II). Western blots of lysates prepared from starved larvae showed that Rab7, Ccz1, and Mon1 mutants accumulate both p62 and Atg8a II (Figure 2J). A similar buildup of these proteins was seen in well-fed early-L3 and late-L3 wandering mutant larvae, respectively (Supplemental Figure S2C). These results indicate that the Ccz1/Mon1/Rab7 module is required for autophagic degradation during starvation-induced basal and developmental autophagy.
We next tested the colocalization of the autophagy reporter 3xmCherry-Atg8a with lysosomal-associated membrane protein 1 (Lamp1)–green fluorescent protein (GFP), a marker for late endosomes and lysosomes. As expected, large autolysosomes were positive for both 3xmCherry-Atg8a and Lamp1 in wild-type cells (Supplemental Figure S2D). In contrast, the Lamp1-GFP and 3xmCherry-Atg8a structures were much smaller in animals lacking Rab7 or Ccz1, and their colocalization was strongly reduced (Supplemental Figure S2, E and F). In line with this, ultrastructural analysis of fat cells from starved larvae revealed that Rab7, Ccz1, and Mon1 mutants accumulate autophagosomes and that autolysosomes are absent from these cells, unlike control cells (Figure 2, K–N). Thus Rab7 and its GEF, the Ccz1-Mon1 complex, are required for autophagosome-lysosome fusion.
Ccz1-Mon1 recruits Rab7 to autophagosomes
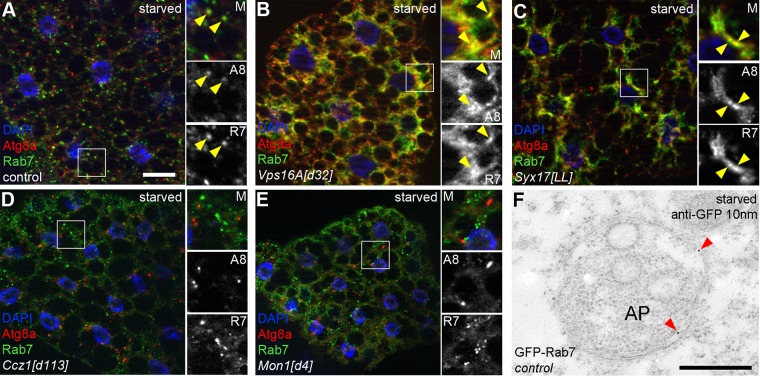
Others and we have shown that a Syx17-Snap29-Vamp7 SNARE complex mediates autophagosome-lysosome fusion (Itakura et al., 2012; Takats et al., 2013), which is promoted by the HOPS tethering complex, an interacting partner of Syx17 (Jiang et al., 2014; Takats et al., 2014). Rab7 is known to localize to the lysosome (Bucci et al., 2000). Because HOPS is considered to be an effector of Rab7, we reasoned that Rab7 must be present on autophagosomes. Indeed, 56% (112 of 200; n = 10) of dots positive for endogenous Atg8a colocalized with endogenous Rab7 in fat cells of starved control animals (Figure 3A). To confirm that these structures correspond to autophagosomes, we repeated this experiment using null mutants lacking Syx17 or an essential subunit of HOPS, Vps16A. These cells show large-scale accumulation of Atg8a-positive autophagosomes concentrated in the perinuclear region, as described earlier (Takats et al., 2013, 2014), and 84% (168 of 200; n = 10) and 84.5% (169 of 200; n = 10) of Atg8a-positive structures overlapped with the Rab7 signal in Vps16A and Syx17 mutants (Figure 3, B and C), respectively. In contrast, the mutation of either Ccz1 or Mon1 prevented the colocalization of Atg8a with Rab7: only 8% (16 of 200; n = 10) and 11% (22 of 200; n = 10) of the dots overlapped (Figure 3, D and E), respectively. We also carried out immunogold labeling to show that GFP-Rab7 is associated with the autophagosomal membrane (Figure 3F).
FIGURE 3:
The Mon1-Ccz1 complex is necessary for Rab7 recruitment to autophagic structures. (A–E) Endogenous Atg8a and Rab7 immunolabeling of fat cells in starved larvae. Insets, boxed areas enlarged (M, merged image; A8, Atg8a channel; R7, Rab7 channel; arrowheads mark colocalization). Colocalizing dots are seen in wild-type larvae (A). The Atg8a and Rab7 signals largely overlap in the perinuclear regions of Vps16A (B) and Syx17 (C) mutant fat cells. Loss of Ccz1 (D) or Mon1 (E) prevents the colocalization of Rab7 and Atg8a. (F) Immuno–electron microscopy shows that GFP-Rab7 is associated with the surface of an autophagosome (AP). Immunogold particles are highlighted by arrowheads. Scale bar, 20 μm (in A, for A–E), 0.5 μm (F).
These data suggested that Rab7 is recruited to autophagosomes in a Ccz1-Mon1 (GEF)–dependent manner, independent of HOPS or Syx17. Of importance, a constitutively active, GTP-locked mutant form of Rab7 was detected on endogenous Atg8a-positive autophagosomes even in the absence of Mon1 (Supplemental Figure S2G), and Rab7-GTP expression rescued starvation-induced punctate LTR/autolysosome staining in Mon1 mutant fat cells (Supplemental Figure S2H and Figure 2I). This is in line with yeast genetic data, which showed that the vacuole fragmentation phenotype of yeast Ccz1 mutants can be suppressed by point mutations in Ypt7/Rab7 (Kucharczyk et al., 2001).
Rab5 is dispensable for autophagosome-lysosome fusion
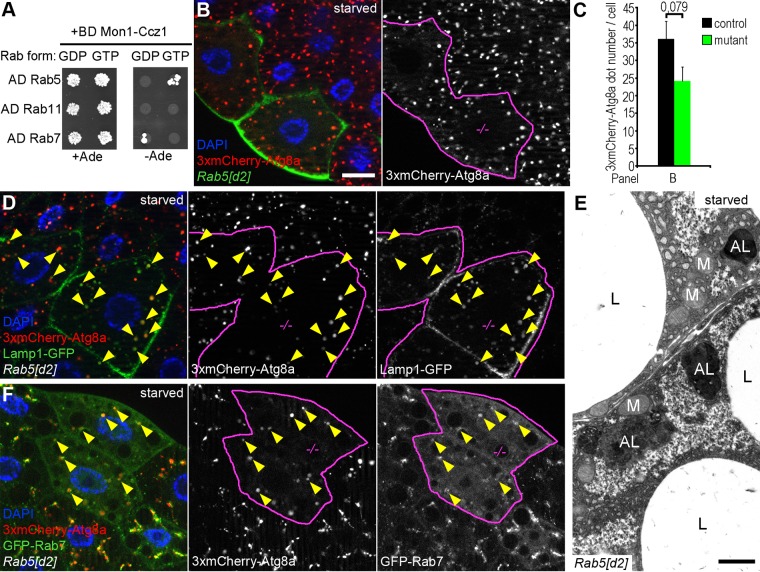
Rab5 promotes the recruitment of the Ccz1-Mon1 complex to endosomal membranes. Because Ccz1 and Mon1 are both required for the autophagosomal localization of Rab7 and the subsequent fusion to lysosomes, we asked whether Rab5 plays a role in this process. We first confirmed that the Drosophila Ccz1-Mon1 complex preferentially binds to GTP-locked Rab5 and GDP-locked Rab7 in Y2H experiments (Figure 4A). Next we generated mosaic animals expressing the 3xmCherry-tagged Atg8a reporter in all cells and containing clones of cells homozygous for Rab5[d2], a widely used null mutant allele (Wucherpfennig et al., 2003). Surprisingly, Rab5 mutant cells (marked by the expression of GFP) contained large 3xmCherry-Atg8a–positive dots in similar numbers and size as surrounding control cells did after a 4-h starvation (Figure 4, B and C). Because these autophagic structures colocalized with lysosomal Lamp1-GFP (Figure 4D), this suggested that autophagosomes normally fuse with lysosomes to generate autolysosomes in the absence of Rab5. We further confirmed these findings by ultrastructural analysis of Rab5 mutant fat cell clones, which also revealed the presence of large, electron-dense autolysosomes and no accumulation of autophagosomes (Figure 4E and Supplemental Figure S3A). In line with this, Rab5 was dispensable for the recruitment of GFP-Rab7 to autophagic structures labeled by 3xmCherry-Atg8a (Figure 4F). It is worth noting that the cytoplasmic signal of GFP-Rab7 is slightly elevated in starved Rab5-null mutant fat cell clones (Figure 4F and Supplemental Figure S3B). We also tested the colocalization of Rab7 with the late endosome and lysosome marker Lamp, using our new reporter: endogenous promoter–driven, full-length Drosophila dLamp-3xmCherry. In fat cells of well-fed animals that show minimal autophagic activity, loss of Rab5 caused a striking decrease in the number of GFP-Rab7 and dLamp-3xmCherry vesicles relative to surrounding wild-type cells, which contain abundant Lamp1- and Rab7-positive dots that often overlap (Supplemental Figure S3C). However, in starved animals, many colocalizing GFP-Rab7 and dLamp-3xmCherry vesicles formed in Rab5 mutant cells (Supplemental Figure S3D), potentially due to the induction of autophagy.
FIGURE 4:
Rab5 is dispensable for autophagosome-lysosome fusion. (A) Y2H assays show that the Mon1-Ccz1 complex interacts with GTP-bound Rab5 and GDP-bound Rab7. Rab11 was used as negative control. (B) GFP-positive fat cell clones homozygous for the Rab5[d2]-null mutation show a similar pattern of 3xmCherry-Atg8a granules as seen in GFP-negative control cells. (C) Quantification of data shown in B; n = 10/genotype. (D) 3xmCherry-Atg8a and Lamp1 colocalize in intracellular structures (arrowheads) in Rab5 mutant clones, and the accumulation of Lamp1-GFP in vesicles and at the plasma membrane is obvious in Rab5[d2] cells. (E) Large, dense autolysosomes (ALs) are visible in an electron micrograph from a Rab5 mutant fat cell of a starved larva. L, lipid droplet; M, mitochondrion. (F) 3xmCherry-Atg8a colocalizes with GFP-Rab7 (arrowheads) in starved Rab5 mutant clone cells, and the diffuse cytoplasmic level of Rab7 is increased in mutant cells. Scale bar, 20 μm (B, D, F). Clone cells in grayscale images of D and F are outlined in magenta. Note that Rab5 mutant cells were recognized based on their larger size and nucleus and different reporter expression pattern in D and F. See also Figure 7B and Supplemental Figure S3B for further data. Scale bar, 1 μm (E).
We noticed that Rab5 mutant fat cells are larger than neighboring control cells, which is in line with a report showing that the loss of Rab5 function confers a selective growth advantage in cells of the developing wing (Lu and Bilder, 2005). We carried out epistasis analysis to find out how the different phenotypes behave in Rab5 and Mon1 double-mutant cells. First, we performed LTR staining of fat cells from starved larvae containing Rab5 or Mon1 single-mutant and Rab5/Mon1 double-mutant clones. We found that cells homozygous for Rab5[d2] contain large LTR-positive autolysosomes with similar size and number as in surrounding control cells (Supplemental Figure S4, A and D). In contrast, Mon1[d4] mutant clones had fewer and fainter acidic, LTR-positive structures (Supplemental Figure S4, B and D). Double-mutant fat cells lacking both Rab5 and Mon1 appeared similar to Mon1 single-mutant cells regarding the lack of LTR dots (Supplemental Figure S4, C and D). Ultrastructural analysis revealed the accumulation of autophagosomes and lack of autolysosomes in double-mutant cells (Supplemental Figure S4E), as seen in Mon1 single mutants (Figure 2N). In addition, both Rab5 single- and Rab5/Mon1 double-mutant cells are larger than neighboring control cells, unlike Mon1 single-mutant clone cells, which have a normal size (Supplemental Figure S4, A–C).
Taken together, our results indicate that the autophagosome fusion defect of Ccz1/Mon1/Rab7 module mutants is independent of Rab5, whereas the large–cell size phenotype caused by Rab5 deletion is epistatic to the mutation of Mon1.
Rab5 is dispensable for the generation of PI3P-positive autophagosomes
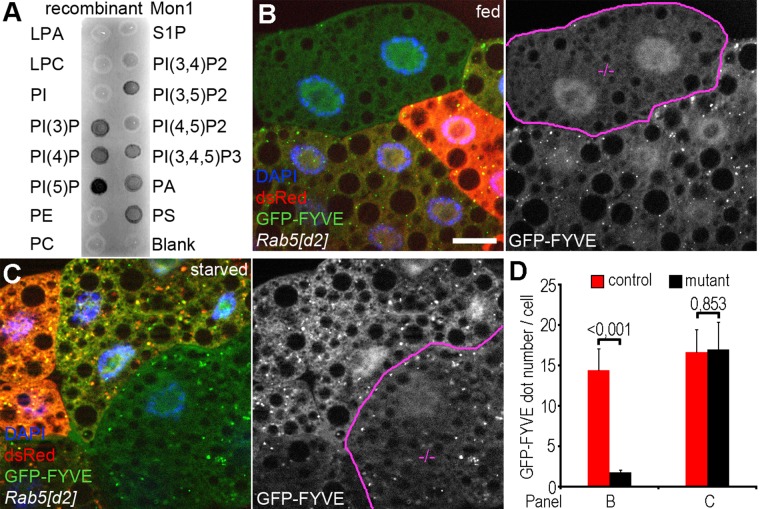
The Ccz1-Mon1 complex strongly binds to membrane lipids, including PI3P (Poteryaev et al., 2010; Cabrera et al., 2014), which is enriched in early endosomal and autophagosomal membranes. To test whether Drosophila Mon1 also has an affinity for PI3P, we purified recombinant Mon1 and performed a protein–lipid overlay assay. Indeed, Mon1 bound to PI3P with high affinity, among other phospholipids (Figure 5A). These data raised the possibility that the Ccz1-Mon1 complex is recruited to autophagosomes through binding to PI3P.
FIGURE 5:
Autophagosomal PI3P generation is independent of Rab5. (A) Mon1 binds to several phospholipids, with a preference for phosphatidyl-inositol monophosphates, including PI3P. (B) GFP-FYVE labels PI3P-positive endosomes in fat cells of well-fed larvae, which are almost completely absent from dsRed-negative Rab5 mutant clone cells. (C) GFP-FYVE dots are observed in large numbers during starvation in both Rab5[d2] homozygous mutant (dsRed–) and control (dsRed+) cells. (D) Quantification of data shown in B and C; n = 10/genotype. Clone cells in grayscale images of B and C are outlined in magenta. Scale bar, 20 μm (in B, for B, C). LPA, lysophosphatidic acid; LPC, lysophosphocholine; PA, phosphatidic acid; PC, phosphatidylcholine; PE, phosphatidylethanolamine; PI, phosphatidylinositol; PS, phosphatidylserine; S1P, sphingosine-1-phosphate.
We showed previously that the Vps34 lipid kinase complex is required for the generation of PI3P-positive endosomes and autophagosomes in Drosophila fat cells (Juhasz et al., 2008). Vps34 is activated by Rab5 on endosomes (Stenmark, 2009). In the present study, we found that autophagosome-lysosome fusion takes place in the absence of Rab5. Thus autophagosomal PI3P may be generated during autophagy induction in Rab5 mutant fat cells. We already showed that the PI3P reporter GFP-FYVE labels mostly endosomes in well-fed larval fat cells, whereas starvation results in the generation of autophagosomes that are also positive for GFP-FYVE (Juhasz et al., 2008). We quantified the autophagosomal localization of GFP-FYVE using our 3xmCherry-Atg8a reporter in fat cells of nutrient-replete and -starved animals. PI3P-positive structures very rarely colocalized with 3xmCherry-Atg8a in well-fed cells (9.5%; 19 of 200; n = 10), whereas during starvation, 89.5% (179 of 200; n = 10) of 3xmCherry-Atg8a vesicles were positive for PI3P (Supplemental Figure S5). This is likely explained by the metabolic and storage functions of the fat body in Drosophila, as starvation-induced inactivation of TOR kinase in fat cells not only triggers autophagy but also down-regulates endocytosis (Hennig et al., 2006).
In line with the importance of Rab5 for endocytic maturation, the loss of Rab5 completely suppressed PI3P generation in well-fed cells, based on the lack of GFP-FYVE puncta formation (Figure 5, B and D). However, GFP-FYVE–positive structures appeared in Rab5 mutant cells in similar numbers as in control cells during starvation (Figure 5, C and D). This is in line with the rest of our data and indicates that PI3P is generated on autophagosomes independently of Rab5.
Atg14, but not ultraviolet radiation resistance–associated gene, supports PI3P-positive autophagosome formation in starved fat cells
Two multisubunit Vps34 PI3 kinase complexes exist, which share three core subunits: the catalytic subunit, Vps34; the regulatory subunit, Vps15; and Atg6 (Beclin1). The mutually exclusive subunits that define the two distinct complexes are ultraviolet radiation resistance–associated gene (UVRAG) and Atg14, respectively (Itakura et al., 2008; Matsunaga et al., 2009). It is clear that Atg14 functions during autophagosome biogenesis in yeast (Obara et al., 2006), but both proteins were suggested to regulate autophagy in mammalian cells (Liang et al., 2006; Itakura et al., 2008).
We recently reported that UVRAG is dispensable for autophagosome formation and fusion with lysosomes in Drosophila fat cells (Takats et al., 2014). To clarify the role of the Atg14 and UVRAG complexes during PI3P formation, we targeted the Atg14 locus using Cas9 mutagenesis. A 790–base pair deletion was identified that begins 17 base pairs upstream of the start ATG and deletes nearly the first half of the protein-coding sequence (Supplemental Figure S6, A and B). Flies homozygous for this Atg14[d13] allele accumulated high amounts of the specific autophagic cargo p62, and mutant fat cell clones completely lacked LTR-positive autolysosomes (Supplemental Figure S6, C and D), similar to previously described Atg mutants. Of importance, the expression of an endogenous promoter–driven, C-terminally hemagglutinin (HA)-tagged Atg14 transgene on the Atg14 mutant background suppresses the buildup of p62 (Supplemental Figure S6C), indicating that the autophagy defect is solely due to loss of Atg14 in these animals.
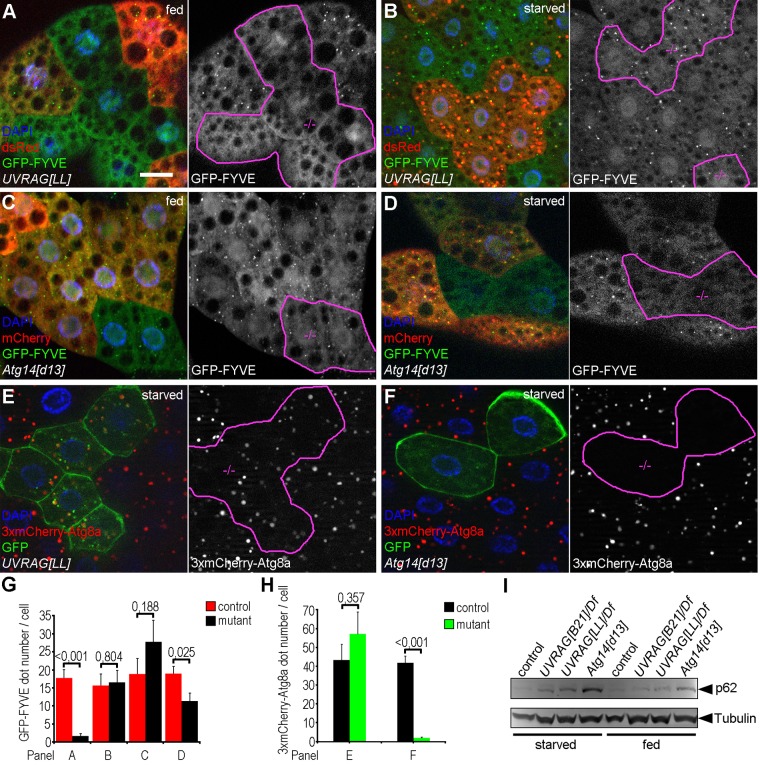
We used our new Atg14[d13] and the recently described UVRAG-null mutant (Takats et al., 2014) alleles to test how these affect Vsp34 kinase activity and autophagy. UVRAG[LL] mutant fat cells showed a complete lack of PI3P under well-fed conditions (Figure 6, A and G), whereas starvation readily induced GFP-FYVE dots both in control and UVRAG loss-of-function cells (Figure 6, B and G). Cells lacking Atg14 showed an opposite phenotype: the number of GFP-FYVE structures was not statistically significantly different from that in neighboring control cells in the fed state (Figure 6, C and G), whereas upon starvation, the generation of PI3P-positive vesicles was strongly decreased in Atg14[d13] mutant clones (Figure 6, D and G). We also tested whether the formation of 3xmCherry-Atg8a–positive structures is affected in UVRAG and Atg14 mutants. In cells lacking UVRAG, the number of 3xmCherry-Atg8a dots was comparable to that in control cells (Figure 6, E and H), in line with our recent report. In contrast, autophagic structures were absent from Atg14 mutant fat cell clones (Figure 6, F and H). We also tested the autophagosomal localization of Mon1 in wild-type and mutant backgrounds to more directly support these findings. As expected, we detected the presence of Mon1-HA on endogenous Atg8a-positive autophagosomes in both control and UVRAG mutant fat cells (Supplemental Figure S6, E and F). In contrast, Atg14 mutant cells lacked endogenous Atg8a-positive autophagosomes, whereas Mon1-HA dots were still observed (Supplemental Figure S6G).
FIGURE 6:
Different mechanisms of endosomal and autophagosomal PI3P formation. (A, B) GFP-FYVE–marked, PI3P-positive endosomes are absent from UVRAG mutant fat cell clones (dsRed– cells in A) of well-fed L3 larvae. However, starvation induces the generation of GFP-FYVE dots in UVRAG loss-of-function cells (dsRed–) in a similar number as in dsRed+ controls (B). (C, D) The number of GFP-FYVE–positive endosomes is similar in well-fed Atg14 mutant clones (dsRed–) and dsRed+ control cells (C). In contrast, Atg14 mutant cells contain fewer GFP-FYVE dots than control cells under starvation conditions (D). (E) Starvation-induced formation of 3xmCherry-Atg8a–labeled autophagic structures is similar in UVRAG-null mutant clones (GFP+) and control cells (GFP–). (F) On the contrary, complete inhibition of punctate 3xmCherry-Atg8a is observed in GFP+ Atg14 mutant cells compared with neighboring GFP– controls. (G, H) Quantification of data in A–D (G) and E–F (H); n = 10/genotype. (I) Western blots reveal that UVRAG mutants show mild, whereas Atg14 mutants show strong, accumulation of the autophagic cargo p62 in both well-fed and starved larvae. Tubulin serves as a loading control. Clone cells in grayscale images of A–F are outlined in magenta. Scale bar, 20 μm (in A, for A–F).
Finally, we tested the level of p62 in fed and starved larvae in two independent UVRAG-null mutant lines, as well as in Atg14[d13] homozygotes. We found large-scale accumulation of p62 in Atg14 mutants under both conditions (Figure 6I), the results together indicating that the Atg14 complex is required for PI3P-positive autophagosome formation. UVRAG loss-of-function lines showed a moderate increase of p62 compared with control animals (Figure 6I). These findings are consistent with a recent study in which we showed that although the loss of UVRAG does not inhibit autophagosome formation or fusion, it impairs the lysosomal degradation of autophagic cargo (Takats et al., 2014), which is likely responsible for the increase in the amount of p62. Considering that UVRAG- but not Atg14-defective cells showed very similar phenotypes regarding GFP-FYVE distribution and 3xmCherry-Atg8a puncta formation to the ones observed in Rab5 mutants, we propose that Rab5 regulates endosomal traffic through an UVRAG-containing Vps34 complex in Drosophila fat cells. Moreover, Rab5 is dispensable for the activity of the Atg14-containing Vps34 complex, which promotes autophagosome formation.
Rab5 facilitates autophagic cargo degradation by regulating lysosomal function
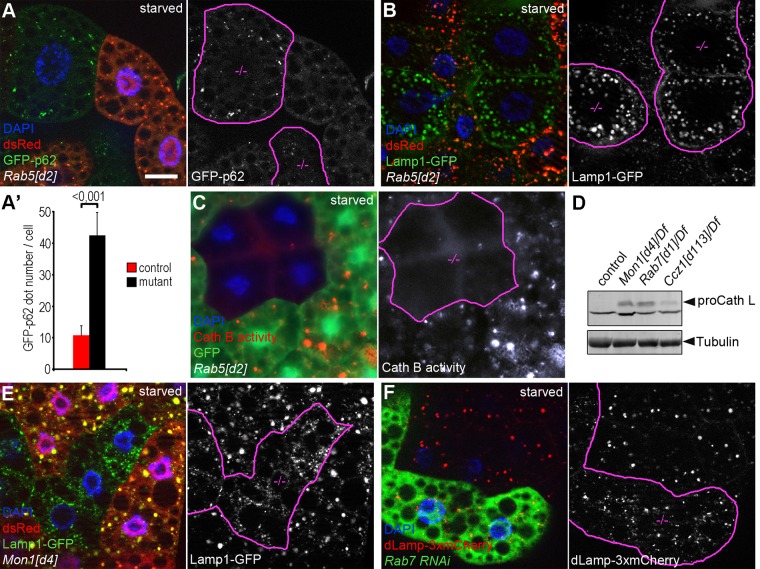
Considering our recent findings that UVRAG regulates endocytosis and lysosomal function (Takats et al., 2014) and that Rab5 appears to act on endosomal membrane trafficking at least in part through the Vps34-UVRAG complex, we hypothesized that Rab5 may also have a role in maintaining proper lysosomal function. To evaluate autophagic degradation (flux) in Rab5 mutants, we generated a novel, tubulin promoter–driven GFP-p62 reporter. Because the tubulin promoter is insensitive for autophagy induction signals, unlike that of endogenous p62, it ensures a constant and relatively low expression level in fat cells, so this reporter may be even better for measuring autophagic flux than the endogenous protein. An elevated level of GFP-p62 was seen in Rab5[d2] homozygous cell clones compared with neighboring control cells (Figure 7, A and A′).
FIGURE 7:
Rab5 is required for proper autolysosomal degradation. (A) The selective autophagic cargo p62 accumulates in Rab5 mutant (dsRed–) clones compared with dsRed+ control cells. Note that GFP-p62 is expressed in all cells by the constitutive tubulin promoter, which excludes transcriptional changes. (A′) Quantification of data shown in A; n = 10/genotype. (B) In Rab5 mutant clone cells (dsRed–), the late endosomal and lysosomal marker Lamp1-GFP accumulates both intracellularly and at the plasma membrane compared with surrounding dsRed+ control cells. (C) Strongly reduced cathepsin B activity is observed in GFP– Rab5 mutant clone cells compared with GFP+ control cells. (D) Western blot reveals the obvious accumulation of pro–cathepsin L in lysates of Rab7, Ccz1, and Mon1 mutant larvae compared with controls. (E) In contrast to dsRed+ control cells, dsRed– Mon1 mutant clones show highly fragmented Lamp1-GFP positive lysosomes. (F) Similarly, the dLamp-3xmCherry compartment is dispersed in Rab7 RNAi clones (GFP+) compared with surrounding GFP– control cells. Clone cells in grayscale images of A–C, E, and F are outlined in magenta. Scale bar, 20 μm (in A, for A–C, E, F).
To test whether this is due to impaired lysosomal digestion, we measured the level of Lamp1-GFP, a reporter that consists of GFP linked to only one transmembrane domain followed by the cytoplasmic tail of human Lamp1 protein. As a result, GFP is constantly degraded within lysosomes, and thus its levels inversely correlate with lysosome function (Pulipparacharuvil et al., 2005). Rab5 mutant clones showed a large-scale accumulation of Lamp1-GFP on intracellular structures and in the plasma membrane relative to surrounding control cells (Figure 7B). Of interest, vesicles positive for dLamp-3xmCherry are fainter in Rab5-null mutant cells than in control cells (Supplemental Figure S3, C and D). This is likely because in this case, full-length Drosophila dLamp protein is tagged with 3xmCherry on its C-terminus facing the cytoplasm, so this reporter is better suited for following trafficking to, but not turnover in, lysosomes. These data taken together indicate defective trafficking and turnover of lysosomal membrane proteins in Rab5 mutant cells. Finally, we examined the activity of the lysosomal hydrolase cathepsin B by incubating fat cells dissected from starved larvae in a medium containing its substrate Magic Red. In control cells, Magic Red penetrates into cells and stains degrading lysosomes that contain active cathepsin B. Rab5 mutant fat cells showed highly reduced cathepsin B activity (Figure 7C), further supporting our model that Rab5 promotes the degradation of autophagic cargo by facilitating the targeting of lysosomal proteins.
Of note, the Rab7 module also appears to promote lysosomal function because the lysosomal processing of pro–cathepsin L was clearly perturbed in Mon1, Rab7, and Ccz1 mutant animals (Figure 7D). Mon1 and Rab7 loss-of-function fat cells of starved larvae showed the accumulation of small Lamp1-GFP or dLamp-3xmCherry puncta compared with surrounding control cells (Figure 7, E and F, and Supplemental Figure S7, A and B), which likely represent primary lysosomes that are unable to fuse with autophagosomes. Of interest, this phenotype is clearly different from that seen in Rab5 mutant cells, which showed large-scale accumulation of Lamp1-GFP in large autolysosomes and at the plasma membrane (Figure 7B). These observations again support that Rab5 and Rab7 modules play different roles during autophagy, as well as in lysosomal membrane protein trafficking and turnover.
DISCUSSION
In this study, we show that the Rab7 module and Rab5 control different steps of autophagy. Rab7 mediates autophagosome-lysosome fusion together with its GEF, the Ccz1-Mon1 complex. This is likely achieved by the recruitment of Rab7 to autophagosomes in a Ccz1-Mon1–dependent manner. Although Drosophila Mon1 binds to the active, GTP-locked form of Rab5 as in other organisms (Poteryaev et al., 2010; Cabrera et al., 2014), Rab5 is dispensable for the fusion of autophagosomes with lysosomes and for Rab7 localization to autophagosomes and autolysosomes. The question is then: what is the signal that recruits Ccz1-Mon1 and Rab7 to autophagic structures?
Mon1 and Ccz1 bind to phospholipids, including PI3P, in yeast (Cabrera et al., 2014), and we find that Drosophila Mon1 has similar features. This raises the possibility that the Ccz1-Mon1 complex is recruited to the PI3P-positive surface of autophagosomes through this interaction. Vps34-dependent PI3P generation is required for autophagosome formation and endosome maturation. Vps34 is activated by Rab5 (Stenmark, 2009). Of interest, our data suggest that loss of Rab5 inhibits PI3P generation only on endosomes but not on autophagosomes. Loss of UVRAG but not Atg14 inhibits PI3P generation on endosomes, whereas loss of Atg14 leads to complete inhibition of PI3P-positive autophagosome biogenesis. Thus UVRAG is dispensable for Vps34 activity during autophagosome formation, and its loss causes a defect in autolysosomal degradation (Takats et al., 2014). Similarly, Rab5 mutant cells showed accumulation of autophagic cargo due to impaired lysosomal degradation.
Recently the Rab5-related Vps21 small GTPase was suggested to control the fusion of autophagosome with the vacuole (lysosome) in yeast cells (Chen et al., 2014). In this study, clusters of autophagic structures were found to accumulate near the vacuole. However, these vesicles were positive for both the autophagy marker GFP-Atg8 and the vacuolar marker FM4-64, suggesting that some sort of fusion must have occurred in this case, too.
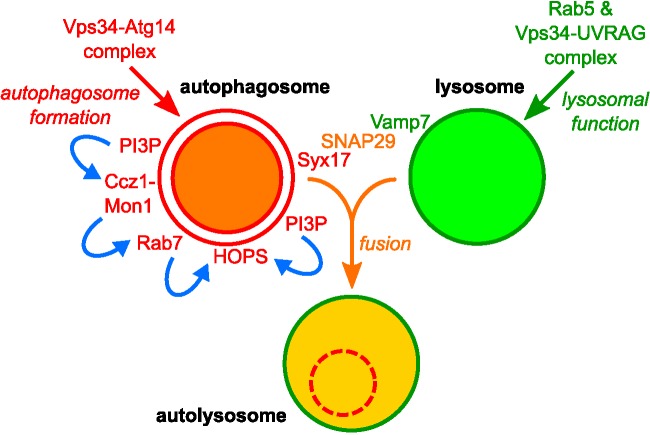
On the basis of our results, we propose the following model of autolysosome formation in fat cells of starved Drosophila larvae (Figure 8). PI3P-positive autophagosomes are generated through the action of an Atg14-containing Vps34 PI3 kinase complex. PI3P attracts Ccz1-Mon1, which promotes Rab7 recruitment to autophagosomes. Both PI3P and Rab7 bind to the HOPS tethering complex, and thus these factors promote the tethering of autophagosomes with late endosomes and lysosomes. The membrane fusion is then executed by the Syx17-Snap29-Vamp7 SNARE complex. Autophagic cargo is broken down in autolysosomes, and their full degradative capacity requires the function of Rab5 and the UVRAG-containing Vps34 complex for the proper delivery of lysosomal proteins, likely including both acidic hydrolases and membrane proteins. This is in line with the finding that simultaneous knockdown of all three Rab5 homologues leads to a collapse of the endolysosomal system in mouse liver cells (Zeigerer et al., 2012).
FIGURE 8:
Proposed model of autophagosome maturation. PI3P is a key membrane lipid regulating both endocytosis and autophagy, the two main lysosomal degradation pathways. Rab5, together with its effector, the Vps34-UVRAG complex, promotes endosomal maturation and proper degradative capacity of lysosomes through the generation of PI3P on endosomal membranes. In parallel, the Vps34-Atg14 complex produces PI3P during autophagosome formation in a Rab5-independent manner. PI3P-positive autophagosomes mature to gain fusion competence by acquiring Rab7 via the action of the PI3P-binding Ccz1-Mon1 GEF complex. Both Rab7 and PI3P are likely important for recruiting the HOPS tethering complex to promote autophagosome-lysosome fusion, together with the Syx17-Snap29-Vamp7 SNARE complex.
Others and we have already demonstrated that autophagosome-lysosome fusion is mediated by the HOPS tethering complex and the SNAREs Syx17, Snap29, and Vamp7/8 (Itakura et al., 2012; Takats et al., 2013, 2014; Jiang et al., 2014). It is not yet clear how these fusion factors are recruited to the autophagosomal membrane. HOPS is known as a Rab7 effector (Stenmark, 2009), and according to our findings, Rab7 is present on autophagosomes. We propose that autophagosomal PI3P recruits the Ccz1-Mon1-Rab7 module to facilitate the loading of HOPS and subsequent tethering of vesicles.
Vps34 is considered as a bona fide Rab5 effector (Stenmark, 2009). Surprisingly, we found that whereas Rab5 mediates only the generation of PI3P on endosomes mainly through the action of a UVRAG-containing Vps34 complex, it is dispensable for PI3P-positive autophagosome biogenesis, which depends on the Atg14-containing Vps34 complex. Thus the current concept that Vps34 is a Rab5 effector must be revisited: it is true for endocytosis but not applicable for autophagy in fat cells of starved Drosophila larvae.
A previous study showed that Rab5 promotes autophagy-mediated huntingtin clearance in cultured human cells and Drosophila eyes (Ravikumar et al., 2008). Simultaneous small interfering RNA knockdown of all three Rab5 genes (Rab5a, Rab5b, Rab5c) reduced the level of Atg5-Atg12 conjugate and autophagosome formation. Although we did not see perturbations of autophagosome biogenesis and fusion in Rab5 mutant fat cells, these discrepancies may be due to the different models used. In our experiments, starvation induces a massive wave of autophagy in larval Drosophila fat cells that entirely relies on the activity of the Rab5-independent Atg14-Vps34 PI3 kinase complex. It is possible that during the basal, nonstarved conditions studied by Ravikumar et al. (2008), Rab5 can contribute to autophagosome formation. In fact, UVRAG has also been suggested to control autophagosome formation in cultured cells, which is compatible with this model (Liang et al., 2006).
In summary, Rab7 is recruited to autophagosomes by the Ccz1-Mon1 complex to promote autophagosome-lysosome fusion. We show that autophagosome formation and fusion is independent of Rab5 and the UVRAG-containing Vps34 PI3 kinase complex but requires the action of the Atg14-Vps34 complex. Rab5, similar to UVRAG, is necessary for proper lysosomal function by promoting the trafficking of lysosomal proteins.
MATERIALS AND METHODS
Fly work
Flies were kept on standard yeast/cornmeal medium. For starvation experiments, well-fed mid L3-stage larvae (aged 80–88 h after egg laying) were floated in a 20% sucrose solution for 4 h. The Rab7[d1] and Ccz1[d113] null alleles were generated by imprecise excision of the transposable elements Rab7[EY10675] and Ccz1[EY16389], respectively (both obtained from the Bloomington Drosophila Stock Center [BDSC], Bloomington, IN). The Atg14[d13] allele was generated by clustered regularly interspaced short palindromic repeats/Cas9 mutagenesis using a double gRNA approach as described previously (Kondo and Ueda, 2013). In all three cases, null mutant lines were identified by PCR screening and sequencing of candidates. The deficiencies Df(3R)Exel6196, Df(3L)Exel6098, and Df(2L)ED7853 were obtained from BDSC and Df(2L)ED784 from Drosophila Genomics and Genetic Resources (Kyoto, Japan). UAS-YFP-Rab7, UAS-YFP-Rab7[Q67L], UAS-GFP-Rab7, hs-Gal4, da-Gal4, and the Rab5-null mutant FRT40A Rab5[d2] lines came from BDSC. The RNA interference line UAS-Rab7[GD11800] was purchased from the Vienna Drosophila Resource Center (VDRC; Vienna, Austria). Vps16A[d32], Syx17[LL06330] and UVRAG[LL03097], UVRAG[B21], and UAS-GFP-FYVE lines were described earlier (Wucherpfennig et al., 2003; Juhasz et al., 2008; Lee et al., 2011; Takats et al., 2013, 2014). The FRT40A Mon1[d4], UAS-Mon1-HA, and UAS-Ccz1-EosFP stocks were kindly provided by Thomas Klein (Heinrich Heine University, Düsseldorf, Germany). We generated Gal4-expressing fat cell clones using hs-Flp[22]; dLamp-3xmCherry, UAS-GFP; Act>CD2>Gal4, UAS-Dcr2 and positively marked mutant clones using the lines hs-Flp; FRT40A tub-QS; et49-QF, QUAS-mCD8-GFP[5B] and hs-Flp[22]; QUAS-mCD8-GFP[5J]; ET49-QF, FRT82B tub-QS[21]/TM6 (all QF, QS, and QUAS transgenes came from BDSC). For the analysis of negatively marked mutant clones, we used the genotypes hs-Flp[22]; Fb-Gal4, FRT40A UAS-dsRed and hs-Flp[22]; Fb-Gal4, UAS LAMP1-GFP, FRT40A UAS-dsRed and hs-Flp[22]; Fb-Gal4, FRT40A UAS-GFP. For two experiments with Rab5[d2] mutant clones, we used clonal systems without a mutant cell marker: hs-Flp[22]; Fb-Gal4, UAS-LAMP1-GFP, FRT40A and hs-Flp[22]; Fb-Gal4, FRT40A; UAS-GFP-Rab7. Because fat cells homozygous mutant for Rab5[d2] are larger than control cells and have a bigger nucleus, these cells can be easily identified without using an additional marker. Detailed genotype information is shown in Supplemental Table S1.
Molecular cloning and generation of transgenic animals
For generating N-terminally 3xmCherry tagged Atg8a under the control of the endogenous Atg8a promoter and containing all introns and 3′ UTR, mCherry coding sequence was amplified from UAS-mCherry-GGG vector (kindly provided by Thomas Neufeld, University of Minnesota, Minneapolis, MN) by using primers AGAGGTACCAGAAGGGTGGCGGAAGTGGCATGGTGAGCAAGGGCGAGGA and TCCGGTACCCGGATCCACCTCCCTTGTACAGCTCGTCCATGCCG. The resulting PCR fragment was cut with Acc65I and cloned into a BsrGI site located at the end of mCherry coding sequence in the UAS-mCherry-GGG vector. By repeating another round of cloning on the resulting vector, a tandem 3xmCherry vector was made. Then a 1700–base pair fragment upstream of the translation start ATG sequence was amplified from the genomic Atg8a locus using primers GACTGAATTCGATTGCAATGAAGAGGTAATTGGC and GAGCAGCATGCGAATGTGATTGAT. This extended Atg8a promoter region was cloned into our 3xmCherry vector as an SphI-EcoRI fragment, replacing the UAS sequences and minimal Hsp70 promoter. Finally, the genomic region of Atg8a was PCR amplified using the primers CTCGAGGTACCAAAACTGCGAGGCCAACGAAC and TATAGCGGCCGCGGAGGTGGCATGAAGTTCCAATACAAGGAGGAGCA and cloned as a NotI-Acc65I fragment into our new vector to generate pAtg8apromoter-3xmCherry-Atg8a. To clone the genomic promoter–driven, C-terminally 3xmCherry-tagged dLamp reporter, the Drosophila Lamp locus was PCR amplified, including 403 base pairs upstream of the translation start ATG codon. using primers TATATCAATTGCATGCTGCAGTTTCCACTGTGTTATAAACCCTGTGTG and catctgaattcGAAGCTCATGTAACCGCGGGAG. This PCR product was cut into two fragments by SphI-EcoRI digestion (this gene contains an SphI digestion site 854 base pairs downstream of the translation start codon). The 3′ fragment was ligated into an SphI-EcoRI–digested UAS-3xmCherry-GGG vector, replacing UAS sequences and the Hsp70 promoter. Then a 5′ SphI fragment was inserted into the partial 3′ Lamp-3xmCherry construct SphI to yield pdLamppromoter-dLamp-3xmCherry. The tubulin promoter–driven, N-terminally GFP-tagged p62 construct was made by cloning a 518–base pair promoter region upstream of the translation start codon of alphaTub84B gene using primers GTATGCGGCCGCATGCAAGGGAGAGGGGAAGTTATGGAGTT and TATAGCGGCCGCTGCCGAATTCATTGAGTTTTTATTGGAAGTGTTTCACACG as an SphI-EcoRI fragment into pUAST, replacing UAS sequences and the Hsp70 promoter. GFP-p62 coding sequences were cut out from pUAS-GFP-p62 (Chang and Neufeld, 2009) and inserted downstream of the tubulin promoter using NotI and XbaI. To generate genomic promoter–driven, C-terminally 3xHA-tagged Atg14 rescue transgene, we first replaced the EcoRI-Acc65I mCherry coding fragment in our pGen3xmCherry vector (Takats et al., 2014) with that of 3xHA annealed from synthetic oligos. Next we amplified the Atg14 locus, including the promoter region 629 base pairs upstream of the translation start codon, using GGCGCGCCGCATGCGGCCGCCCATGCCCTATGCCAAGAC and TCTAGAGCTAGCGGTACCTTTGATCCAGCGCAGCACCGA. This fragment was cloned into pGen3xHA upstream of the 3xHA sequence after NotI-Acc65I digestion. To generate a double gRNA construct to target the Atg14 locus, two pairs of oligos (CTTCGCTTGCAGCTTTCGTCGGAGC, AAACGCTCCGACGAAAGCTGCAAGC and CTTCGAGCGCCTCCACAAACGGCG, AAACCGCCGTTTGTGGAGGCGCTC) were annealed and cloned into the pBFv-U6.2B vector as described (Kondo and Ueda, 2013). Stable transgenic fly lines were established by microinjection of Drosophila embryos for all constructs (BestGene, Chino Hills, CA).
Histology, imaging, and statistics
For LTR staining, we dissected and incubated the fat body from 4-h-starved early L3-stage larvae for 5 min in 100 nM LTR (Invitrogen, Budapest, Hungary) diluted in phosphate-buffered saline (PBS). Then we transferred specimens to mounting solution (0.2 μg/ml 4′,6-diamidino-2-phenylindole in a 1:1 mixture of PBS and glycerin). Magic Red staining was performed as described (Mauvezin et al., 2015). For immunostaining of developing eyes, late-L3-stage wandering larvae were inverted and fixed for 30 min in 3.7% paraformaldehyde in PBS at room temperature. Specimens were washed for 3 × 20 min in PBTX-DOC buffer (0.1% Triton X-100 and 0.5% sodium deoxycholate in PBS) and incubated overnight in 10% goat serum–containing PBTX-DOC at 4°C. Specimens were transferred to primary antibody solution diluted in 5% goat serum–containing PBTX-DOC for 90 min at room temperature. After 3 × 20 min washes in PBTX-DOC, specimens were incubated in secondary antibody solution diluted in 5% goat serum–containing PBTX-DOC for 90 min at room temperature. Finally, specimens were washed once in PBTX-DOC and twice in PBS and mounted as described. The protocol for immunostaining of 4-h-starved early-L3 larval fat bodies was described earlier (Takats et al., 2013). We used the primary antibodies rabbit anti-Boss (1:1000; Sevrioukov et al., 1999), mouse anti-Notch (1:50; C458.2H-c; Developmental Studies Hybridoma Bank [DSHB], Iowa City, Iowa), rat anti-Atg8a (1:300; Takats et al., 2013), and rabbit anti-Rab7 (1:500; Tanaka and Nakamura, 2008) and the secondary antibodies Alexa Fluor 488–, 568–, and 647–conjugated anti-rabbit, anti-rat, and anti-mouse (all 1:1500; Invitrogen). Pictures were taken using a Zeiss Axio Imager M2 microscope equipped with an Apotome2 grid confocal unit and AxioCam MRm. We quantified fluorescence structures from original, unmodified pictures using ImageJ (National Institutes of Health, Bethesda, MD). The threshold was set manually by the same person working in a darkroom. The quantified data were evaluated by performing the appropriate statistical tests as described previously (Takats et al., 2013, 2014).
Biochemistry
For Y2H assays, Mon1 and Ccz1 cDNAs were amplified using the primer pairs ATGGAAGTAGAGCAGACGTCAGTCAG and TTAGAATGTGGCATGGTTTCGTATAAA, and ATGGCTAAATTATTGCAACGCGTA and TTATTTGTCAAAAAATACATCATCTGTGAGC, respectively. Rab5, Rab7, and Rab11 constitutive active and dominant negative versions were amplified using genomic DNA extracted from transgenic fly stocks. The primer pairs ATGGCAACCACTCCACGCAGCG and TCACTTGCAGCAGTTGTTCGTCG were used for Rab5, ATGTCCGGACGTAAGAAATCC and TTAGCACTGACAGTTGTCAGGA for Rab7, and ATGGGTGCAAGAGAAGACGAGTA and TCACTGACAGCACTGTTTGCG for Rab11. The fragments were cloned into pGADT7 AD (Gal4 DNA–activation domain) and pGBKT7 BD (Gal4 DNA–binding domain) vectors (Clontech/Central European Biosystems, Budapest, Hungary) and then transformed into the yeast strain PJ69-4A using the Frozen-EZ Yeast Transformation II kit (Zymo Research/Biocenter, Szeged, Hungary). The transformants were selected by growth in minimal medium (Trp−, Leu−), and to assay activation of the reporter gene and hence interaction, transformants were selected by growth on Trp−, Leu−, Ade− plates. Empty vectors were used as negative controls. At least three colonies were checked for interaction for each transformation.
For the protein–lipid overlay assay, recombinant Drosophila Mon1 protein was cloned into the pET15b vector and expressed in Escherichia coli Rosetta cells (Millipore/Biocenter, Szeged, Hungary) and purified as described (Takats et al., 2013). The manufacturer’s instructions were followed for the PIP Strips Membrane (Invitrogene, Budapest, Hungary) experiment.
Western blotting was carried out as described before (Pircs et al., 2012; Takats et al., 2013). Primary antibodies were rabbit anti-Rab7 (1:5000; Tanaka and Nakamura, 2008), rabbit anti-p62 (1:5000; Pircs et al., 2012), rabbit anti-Atg8a (1:5000; Takats et al., 2013), rabbit anti–cathepsin L (1:500; ab58991; Abcam, Cambridge, MA), and mouse anti-tubulin (1:1000; AA4.3; DSHB). Secondary antibodies were alkaline phosphatase–conjugated anti-rabbit and anti-mouse (both 1:5000; Sigma-Aldrich, Budapest Hungary).
Electron microscopy
Dissected fat bodies were processed for ultrastructural analysis as before (Takats et al., 2013). For correlative light and electron microscopy, fat bodies were adhered to a poly-l-lysine–coated glass slide in a drop of PBS. Images were taken by a fluorescence microscope in different magnifications to facilitate subsequent recognition of the fat body region containing the clone cells. Then fat bodies were fixed and embedded on the slide. Embedded clones were located in semithick sections stained with toluidine blue, followed by ultrasectioning. Images were taken by a JEOL JEM-1011 transmission electron microscope equipped with an Olympus Morada camera and iTEM software.
Supplementary Material
Acknowledgments
We thank Sarolta Pálfia for technical assistance and colleagues and stock centers listed in the Materials and Methods section for providing reagents. This work was supported by the Wellcome Trust (087518/Z/08/A to G.J.), the Hungarian Scientific Research Fund (PD112632 to K.H.), and the Hungarian Academy of Sciences (Lendulet LP2014-2 to G.J.).
Abbreviations used:
- Atg
autophagy-related gene
- Ccz1
caffeine, calcium, and zinc 1
- FYVE
Fab-1, YGL023, Vps27, and EEA1
- GEF
guanine nucleotide exchange factor
- HOPS
homotypic fusion and vacuole protein sorting
- Lamp
lysosomal-associated membrane protein
- LTR
LysoTracker Red
- Mon1
monensin sensitivity protein 1
- PI3P
phosphatidylinositol-3-phosphate
- Rab
Ras-related protein in brain
- SNARE
soluble N-methylamaleimide–sensitive factor attachment protein receptor
- UVRAG
ultraviolet radiation resistance–associated gene
- Vps
vacuolar protein sorting
- Y2H
yeast two-hybrid.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-03-0205) on August 24, 2016.
REFERENCES
- Bucci C, Thomsen P, Nicoziani P, McCarthy J, van Deurs B. Rab7: a key to lysosome biogenesis. Mol Biol Cell. 2000;11:467–480. doi: 10.1091/mbc.11.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabrera M, Nordmann M, Perz A, Schmedt D, Gerondopoulos A, Barr F, Piehler J, Engelbrecht-Vandre S, Ungermann C. The Mon1-Ccz1 GEF activates the Rab7 GTPase Ypt7 via a longin-fold-Rab interface and association with PI3P-positive membranes. J Cell Sci. 2014;127:1043–1051. doi: 10.1242/jcs.140921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YY, Neufeld TP. An Atg1/Atg13 complex with multiple roles in TOR-mediated autophagy regulation. Mol Biol Cell. 2009;20:2004–2014. doi: 10.1091/mbc.E08-12-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhou F, Zou S, Yu S, Li S, Li D, Song J, Li H, He Z, Hu B, et al. A Vps21 endocytic module regulates autophagy. Mol Biol Cell. 2014;25:3166–3177. doi: 10.1091/mbc.E14-04-0917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherry S, Jin EJ, Ozel MN, Lu Z, Agi E, Wang D, Jung WH, Epstein D, Meinertzhagen IA, Chan CC, et al. Charcot-Marie-Tooth 2B mutations in rab7 cause dosage-dependent neurodegeneration due to partial loss of function. Elife. 2013;2:e01064. doi: 10.7554/eLife.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L. Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell. 2014;26:2080–2097. doi: 10.1105/tpc.114.123141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, et al. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilcher M, Kohler B, von Mollard GF. Genetic interactions with the yeast Q-SNARE VTI1 reveal novel functions for the R-SNARE YKT6. J Biol Chem. 2001;276:34537–34544. doi: 10.1074/jbc.M101551200. [DOI] [PubMed] [Google Scholar]
- Dooley HC, Razi M, Polson HE, Girardin SE, Wilson MI, Tooze SA. WIPI2 links LC3 conjugation with PI3P, autophagosome formation, and pathogen clearance by recruiting Atg12–5-16L1. Mol Cell. 2014;55:238–252. doi: 10.1016/j.molcel.2014.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fader CM, Colombo MI. Autophagy and multivesicular bodies: two closely related partners. Cell Death Differ. 2009;16:70–78. doi: 10.1038/cdd.2008.168. [DOI] [PubMed] [Google Scholar]
- Filimonenko M, Stuffers S, Raiborg C, Yamamoto A, Malerod L, Fisher EM, Isaacs A, Brech A, Stenmark H, Simonsen A. Functional multivesicular bodies are required for autophagic clearance of protein aggregates associated with neurodegenerative disease. J Cell Biol. 2007;179:485–500. doi: 10.1083/jcb.200702115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerondopoulos A, Langemeyer L, Liang JR, Linford A, Barr FA. BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 2012;22:2135–2139. doi: 10.1016/j.cub.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117:2687–2697. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hennig KM, Colombani J, Neufeld TP. TOR coordinates bulk and targeted endocytosis in the Drosophila melanogaster fat body to regulate cell growth. J Cell Biol. 2006;173:963–974. doi: 10.1083/jcb.200511140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey CM, Stroupe C, Wickner W. The major role of the Rab Ypt7p in vacuole fusion is supporting HOPS membrane association. J Biol Chem. 2009;284:16118–16125. doi: 10.1074/jbc.M109.000737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara N, Hamasaki M, Yokota S, Suzuki K, Kamada Y, Kihara A, Yoshimori T, Noda T, Ohsumi Y. Autophagosome requires specific early Sec proteins for its formation and NSF/SNARE for vacuolar fusion. Mol Biol Cell. 2001;12:3690–3702. doi: 10.1091/mbc.12.11.3690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi C, Inoue K, Mizushima N. Beclin 1 forms two distinct phosphatidylinositol 3-kinase complexes with mammalian Atg14 and UVRAG. Mol Biol Cell. 2008;19:5360–5372. doi: 10.1091/mbc.E08-01-0080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juhasz G, Hill JH, Yan Y, Sass M, Baehrecke EH, Backer JM, Neufeld TP. The class III PI(3)K Vps34 promotes autophagy and endocytosis but not TOR signaling in Drosophila. J Cell Biol. 2008;181:655–666. doi: 10.1083/jcb.200712051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Dalton VM, Eggerton KP, Scott SV, Klionsky DJ. Apg7p/Cvt2p is required for the cytoplasm-to-vacuole targeting, macroautophagy, and peroxisome degradation pathways. Mol Biol Cell. 1999;10:1337–1351. doi: 10.1091/mbc.10.5.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo S, Ueda R. Highly improved gene targeting by germline-specific Cas9 expression in Drosophila. Genetics. 2013;195:715–721. doi: 10.1534/genetics.113.156737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharczyk R, Kierzek AM, Slonimski PP, Rytka J. The Ccz1 protein interacts with Ypt7 GTPase during fusion of multiple transport intermediates with the vacuole in S. cerevisiae. J Cell Sci. 2001;114:3137–3145. doi: 10.1242/jcs.114.17.3137. [DOI] [PubMed] [Google Scholar]
- Lee G, Liang C, Park G, Jang C, Jung JU, Chung J. UVRAG is required for organ rotation by regulating Notch endocytosis in Drosophila. Dev Biol. 2011;356:588–597. doi: 10.1016/j.ydbio.2011.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang C, Feng P, Ku B, Dotan I, Canaani D, Oh BH, Jung JU. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nat Cell Biol. 2006;8:688–699. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- Lindmo K, Stenmark H. Regulation of membrane traffic by phosphoinositide 3-kinases. J Cell Sci. 2006;119:605–614. doi: 10.1242/jcs.02855. [DOI] [PubMed] [Google Scholar]
- Lu H, Bilder D. Endocytic control of epithelial polarity and proliferation in Drosophila. Nat Cell Biol. 2005;7:1232–1239. doi: 10.1038/ncb1324. [DOI] [PubMed] [Google Scholar]
- Matsunaga K, Saitoh T, Tabata K, Omori H, Satoh T, Kurotori N, Maejima I, Shirahama-Noda K, Ichimura T, Isobe T, et al. Two Beclin 1-binding proteins, Atg14L and Rubicon, reciprocally regulate autophagy at different stages. Nat Cell Biol. 2009;11:385–396. doi: 10.1038/ncb1846. [DOI] [PubMed] [Google Scholar]
- Mauvezin C, Ayala C, Braden CR, Kim J, Neufeld TP. Assays to monitor autophagy in Drosophila. Methods. 2014;68:134–139. doi: 10.1016/j.ymeth.2014.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauvezin C, Nagy P, Juhasz G, Neufeld TP. Autophagosome-lysosome fusion is independent of V-ATPase-mediated acidification. Nat Commun. 2015;6:7007. doi: 10.1038/ncomms8007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan DG, Popovic D, Gubas A, Terawaki S, Suzuki H, Stadel D, Coxon FP, Miranda de Stegmann D, Bhogaraju S, Maddi K, et al. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol Cell. 2015;57:39–54. doi: 10.1016/j.molcel.2014.11.006. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy P, Varga A, Kovacs AL, Takats S, Juhasz G. How and why to study autophagy in Drosophila: it’s more than just a garbage chute. Methods. 2015;75:151–161. doi: 10.1016/j.ymeth.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nezis IP, Simonsen A, Sagona AP, Finley K, Gaumer S, Contamine D, Rusten TE, Stenmark H, Brech A. Ref(2)P, the Drosophila melanogaster homologue of mammalian p62, is required for the formation of protein aggregates in adult brain. J Cell Biol. 2008;180:1065–1071. doi: 10.1083/jcb.200711108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Brocker C, Ostrowicz C, Engelbrecht-Vandre S, Ungermann C. The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol. 2010;20:1654–1659. doi: 10.1016/j.cub.2010.08.002. [DOI] [PubMed] [Google Scholar]
- Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes–Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–1539. doi: 10.1091/mbc.E05-09-0841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. doi: 10.1091/mbc.E10-05-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. Evolution of the Rab family of small GTP-binding proteins. J Mol Biol. 2001;313:889–901. doi: 10.1006/jmbi.2001.5072. [DOI] [PubMed] [Google Scholar]
- Pircs K, Nagy P, Varga A, Venkei Z, Erdi B, Hegedus K, Juhasz G. Advantages and limitations of different p62-based assays for estimating autophagic activity in Drosophila. PLoS One. 2012;7:e44214. doi: 10.1371/journal.pone.0044214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A. Identification of the switch in early-to-late endosome transition. Cell. 2010;141:497–508. doi: 10.1016/j.cell.2010.03.011. [DOI] [PubMed] [Google Scholar]
- Pulipparacharuvil S, Akbar MA, Ray S, Sevrioukov EA, Haberman AS, Rohrer J, Kramer H. Drosophila Vps16A is required for trafficking to lysosomes and biogenesis of pigment granules. J Cell Sci. 2005;118:3663–3673. doi: 10.1242/jcs.02502. [DOI] [PubMed] [Google Scholar]
- Ravikumar B, Imarisio S, Sarkar S, O’Kane CJ, Rubinsztein DC. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of Huntington disease. J Cell Sci. 2008;121:1649–1660. doi: 10.1242/jcs.025726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SE, Emr SD. A novel RING finger protein complex essential for a late step in protein transport to the yeast vacuole. Mol Biol Cell. 1997;8:2307–2327. doi: 10.1091/mbc.8.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusten TE, Vaccari T, Lindmo K, Rodahl LM, Nezis IP, Sem-Jacobsen C, Wendler F, Vincent JP, Brech A, Bilder D, et al. ESCRTs and Fab1 regulate distinct steps of autophagy. Curr Biol. 2007;17:1817–1825. doi: 10.1016/j.cub.2007.09.032. [DOI] [PubMed] [Google Scholar]
- Sevrioukov EA, He JP, Moghrabi N, Sunio A, Kramer H. A role for the deep orange and carnation eye color genes in lysosomal delivery in Drosophila. Mol Cell. 1999;4:479–486. doi: 10.1016/s1097-2765(00)80199-9. [DOI] [PubMed] [Google Scholar]
- Stenmark H. Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10:513–525. doi: 10.1038/nrm2728. [DOI] [PubMed] [Google Scholar]
- Stroupe C, Collins KM, Fratti RA, Wickner W. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p. EMBO J. 2006;25:1579–1589. doi: 10.1038/sj.emboj.7601051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Nagy P, Varga A, Pircs K, Karpati M, Varga K, Kovacs AL, Hegedus K, Juhasz G. Autophagosomal Syntaxin17-dependent lysosomal degradation maintains neuronal function in Drosophila. J Cell Biol. 2013;201:531–539. doi: 10.1083/jcb.201211160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takats S, Pircs K, Nagy P, Varga A, Karpati M, Hegedus K, Kramer H, Kovacs AL, Sass M, Juhasz G. Interaction of the HOPS complex with Syntaxin 17 mediates autophagosome clearance in Drosophila. Mol Biol Cell. 2014;25:1338–1354. doi: 10.1091/mbc.E13-08-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Nakamura A. The endocytic pathway acts downstream of Oskar in Drosophila germ plasm assembly. Development. 2008;135:1107–1117. doi: 10.1242/dev.017293. [DOI] [PubMed] [Google Scholar]
- Wang CW, Stromhaug PE, Shima J, Klionsky DJ. The Ccz1-Mon1 protein complex is required for the late step of multiple vacuole delivery pathways. J Biol Chem. 2002;277:47917–47927. doi: 10.1074/jbc.M208191200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wucherpfennig T, Wilsch-Brauninger M, Gonzalez-Gaitan M. Role of Drosophila Rab5 during endosomal trafficking at the synapse and evoked neurotransmitter release. J Cell Biol. 2003;161:609–624. doi: 10.1083/jcb.200211087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefian J, Troost T, Grawe F, Sasamura T, Fortini M, Klein T. Dmon1 controls recruitment of Rab7 to maturing endosomes in Drosophila. J Cell Sci. 2013;126:1583–1594. doi: 10.1242/jcs.114934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigerer A, Gilleron J, Bogorad RL, Marsico G, Nonaka H, Seifert S, Epstein-Barash H, Kuchimanchi S, Peng CG, Ruda VM, et al. Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature. 2012;485:465–470. doi: 10.1038/nature11133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.