Clathrin-mediated endocytosis sorts the bulk of membrane proteins and is a process that starts with adaptor-induced clathrin assembly. Real-time fluorescence analysis shows that adaptor sorting is determined not by the extent of clathrin recruited or the degree of clathrin clustered but instead by the rate of clathrin assembly.
Abstract
Clathrin-mediated endocytosis (CME) manages the sorting and uptake of the bulk of membrane proteins (or cargo) from the plasma membrane. CME is initiated by the formation of clathrin-coated pits (CCPs), in which adaptors nucleate clathrin assembly. Clathrin adaptors display diversity in both the type and number of evolutionarily conserved clathrin-binding boxes. How this diversity relates to the process of adaptor clustering as clathrin assembles around a growing pit remains unclear. Using real-time, fluorescence microscopy–based assays, we compare the formation kinetics and distribution of clathrin assemblies on membranes that display five unique clathrin adaptors. Correlations between equilibrium and kinetic parameters of clathrin assembly to the eventual adaptor distribution indicate that adaptor clustering is determined not by the amount of clathrin recruited or the degree of clathrin clustered but instead by the rate of clathrin assembly. Together our results emphasize the need to analyze kinetics of protein interactions to better understand mechanisms that regulate CME.
INTRODUCTION
Clathrin-mediated endocytosis (CME) is initiated by the formation of clathrin-coated pits (CCPs), sites where adaptor complexes nucleate clathrin assembly (Bonifacino and Traub, 2003; McMahon and Boucrot, 2011). Adaptors recognize sorting motifs on membrane proteins (cargoes), bind phosphoinositide lipids on the membrane, and recruit clathrin (Traub, 2009). Clathrin self-assembly in turn leads to the clustering of adaptors and their cognate cargoes. The heterotetrameric adaptor protein 2 (AP2) is the central player in this process, as it orchestrates myriad interactions with accessory proteins and clathrin, thus marking sites where CCPs are formed (Schmid and McMahon, 2007). Clathrin-associated sorting proteins (CLASPs) are an evolutionarily conserved set of adaptors that bind AP2 and expand the repertoire of cargoes internalized by CME (Traub, 2003). Thus AP2 recognizes YXXØ- and [DE]XXXL[LI]-based sorting motifs present on cargoes such as the transferrin receptor, mannose-6-phosphate receptor, lysosomal-associated membrane protein 1, and the CD4 receptor. CLASPs like disabled 2 (Dab2) and the low-density lipoprotein receptor adapter protein 1 or autosomal recessive hypercholesterolemia (ARH) recognize FXNPXY motifs present in members of the low-density-lipoprotein receptor superfamily. In addition, adaptor protein 180 (AP180) and its nonneuronal homologue, phosphatidylinositol-binding clathrin assembly protein, recognize soluble N-ethylmaleimide–sensitive factor attachment protein receptors, and epsins recognize ubiquitinylated proteins.
Cargo sorting in CME reflects the tendency for CLASPs to bind AP2 as the latter organizes growth of CCPs. AP2 is sufficient in assembling clathrin into coated buds (Kelly et al., 2014). The synergy between CLASPs and AP2 function in CME is unclear. Although they recognize distinct sets of cargoes, CLASPs confer nonredundant functions on AP2, such as facilitating the generation of membrane curvature (Ford et al., 2001, 2002; Miller et al., 2015; Skruzny et al., 2015) and reducing heterogeneity in CCP size (Zhang et al., 1998; Nonet et al., 1999; Bao et al., 2005; Meyerholz et al., 2005; Petralia et al., 2012; Miller et al., 2015). CLASPs contain evolutionarily conserved clathrin-binding sites/boxes whose contribution to the overall process of CCP formation is ill defined. Most cell types express a combination of CLASPs. Thus it is difficult to address their specific contribution to clathrin assembly. To address these issues, we use a reconstitution approach to analyze adaptor-mediated clathrin assembly.
RESULTS
Membrane curvature facilitates clathrin assembly
Adaptor-mediated clathrin recruitment on membranes has traditionally been assayed using electron microscopy (EM; Ford et al., 2002; Dannhauser and Ungewickell, 2012; Kelly et al., 2014), leaving dynamics of this process largely unclear. The analysis of dynamics using fluorescence microscopy on lipid vesicles, as recently reported (Saleem et al., 2015), is complicated because 1) as with EM studies, these reactions inherently suffer from clathrin assembly in solution into empty cages, since the excess unbound adaptors cannot be washed off before clathrin is introduced, 2) freely floating vesicles in solution prevent inflowing clathrin from triggering the assembly process, and 3) assembled clathrin on a spherical vesicle surface is subject to Brownian diffusion, which necessitates complex particle-tracking algorithms to analyze the dynamics of clathrin assembly.
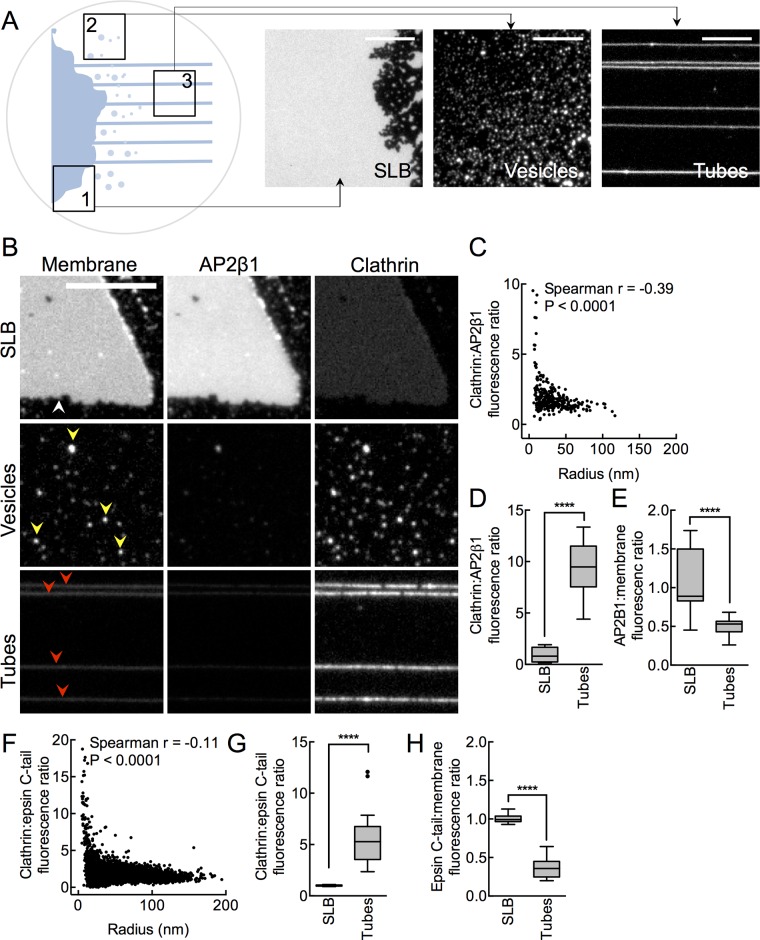
In our efforts at designing polymer-supported membrane tubes (Dar et al., 2015; Holkar et al., 2015), we discovered an assay platform that simultaneously displays a wide range of membrane shapes. Briefly, when a PEGylated glass coverslip containing a spot of dry lipid is assembled in a flow cell and allowed to hydrate under controlled buffer flow, we find three easily discernible membrane templates. The region where lipids are spotted shows up as a planar supported lipid bilayer (SLB; Figure 1A, region of interest 1 [ROI1], and Supplemental Figure S1A), adjacent to which are numerous lipid vesicles (Figure 1A, ROI2) and arrays of membrane tubes (Figure 1A, ROI3). Unlike free-standing vesicles, which are subject to thermal fluctuations, all three templates on the coverslip remain pinned to the surface and resist deformation even under the flow of buffer, affording a detailed analysis of how membrane shape affects clathrin assembly.
FIGURE 1:
Membrane curvature facilitates clathrin assembly. (A) Schematic and corresponding fluorescence micrographs showing a supported lipid bilayer (SLB; ROI1), vesicles (ROI2), and tubes (ROI3) formed upon hydration of dried lipid spotted on a PEGylated coverslip and after flowing excess buffer. Scale bars,10 μm. (B) Representative images of AP2β1 and clathrin fluorescence on the SLB (white arrowhead), vesicles (yellow arrowheads), and tubes (red arrowheads). Scale bar, 10 μm. (C) Clathrin:AP2β1 fluorescence ratio as a function of vesicle size. Data represent values from 307 vesicles from five fields in one experiment. Spearman’s correlation coefficient r and p value. Box-and-whisker (Tukey) plots showing clathrin:AP2β1 (D) and AP2β1:membrane (E) fluorescence ratio on SLB and tubes. Data are normalized to the ratio seen on SLB and represent values from 15 SLB fields and 16 tubes in one experiment. ****p < 0.0001; Mann–Whitney test. (F) Clathrin:epsin C-tail fluorescence ratio as a function of vesicle size. Data represent values from 4879 vesicles from three fields from one experiment. Spearman’s r and p value. Box-and-whisker (Tukey) plots showing clathrin:epsin C-tail (G) and epsin C-tail:membrane (H) fluorescence ratio on SLB and tubes. Data are normalized to the ratio seen on SLB and represent values from 17 SLB fields and 13 tubes from one experiment. ****p < 0.0001; Mann–Whitney test.
We incorporated small amounts (5 mol%) of the chelator lipid (CL) DGS-NTA(Ni2+) and recruited a monomeric histidine (His)–tagged, monomeric enhanced green fluorescent protein (mEGFP)–AP2β1 (hinge plus appendage) construct (hereafter referred to as AP2β1) to the membrane. A similar construct was previously shown to mimic the open state of AP2 and, when recruited to lipid vesicles containing CL and mixed with clathrin, displayed clathrin-coated membrane buds under EM (Kelly et al., 2014). The excess unbound AP2β1 was washed off, and a low concentration (80 nM) of fluorescently labeled clathrin was introduced into the flow cell. EM studies showed that such low concentrations are sufficient to form clathrin lattices on adaptor-coated solid surfaces (Dannhauser et al., 2015). Reactions were then imaged after 10 min under a wide-field fluorescence microscope. Surprisingly, we find significant enrichment of clathrin on vesicles and tubes compared with the SLB (Figure 1B). Note that the adaptor fluorescence in micrographs shown in Figure 1B are equally scaled for contrast across the SLB, vesicles, and tubes and appear dim on vesicles and tubes because these are small, diffraction-limited objects whose fluorescence would be proportional to the net membrane area (Kunding et al., 2008). Control experiments in which the adaptor was omitted or was replaced with a His-tagged mEGFP showed no recruitment of clathrin to the membrane (unpublished data).
Using GFP fluorescence of the adaptor on SLB as an in situ calibration standard (see Materials and Methods and Supplemental Figure S1), we estimated dimensions of the vesicles and tubes. We found the tubes to be highly curved, with a mean radius of ∼26 nm, consistent with our previous estimates (Dar et al., 2015; Holkar et al., 2015), and vesicles had a wide range of sizes (Supplemental Figure S1E). The relative abundance of clathrin on tubes and vesicles suggests that high membrane curvature facilitates binding of clathrin to AP2β1. Indeed, plotting the clathrin:AP2β1 fluorescence ratio on each vesicle against its radius reveals a significant increase in clathrin abundance as the vesicle radius approaches ≤20 nm (Figure 1C). Consistently, the clathrin:AP2β1 fluorescence ratio was 10-fold higher on membrane tubes than on the SLB (Figure 1D). On the basis of data presented in Figure 1E, we find that adaptor density on tubes is approximately twofold lower than on SLBs, which is surprising because adaptors are recruited via a generic His tag–chelator lipid association. Of importance, however, despite the lower density of adaptors, clathrin appears enriched on highly curved membranes. In other words, when adaptor distribution is taken into account, the enrichment of clathrin on highly curved membranes is perhaps an underestimate. To determine whether this was a general property of clathrin, we used an alternate adaptor construct that contains the unstructured 144–575 residues of epsin1 (Kalthoff et al., 2002) appended to a His-tagged mEGFP. We find similar membrane curvature dependence on clathrin recruitment (Figure 1, F–H). Our results therefore indicate that clathrin–adaptor interactions are intrinsically sensitive to membrane curvature.
Adaptor clustering upon clathrin assembly
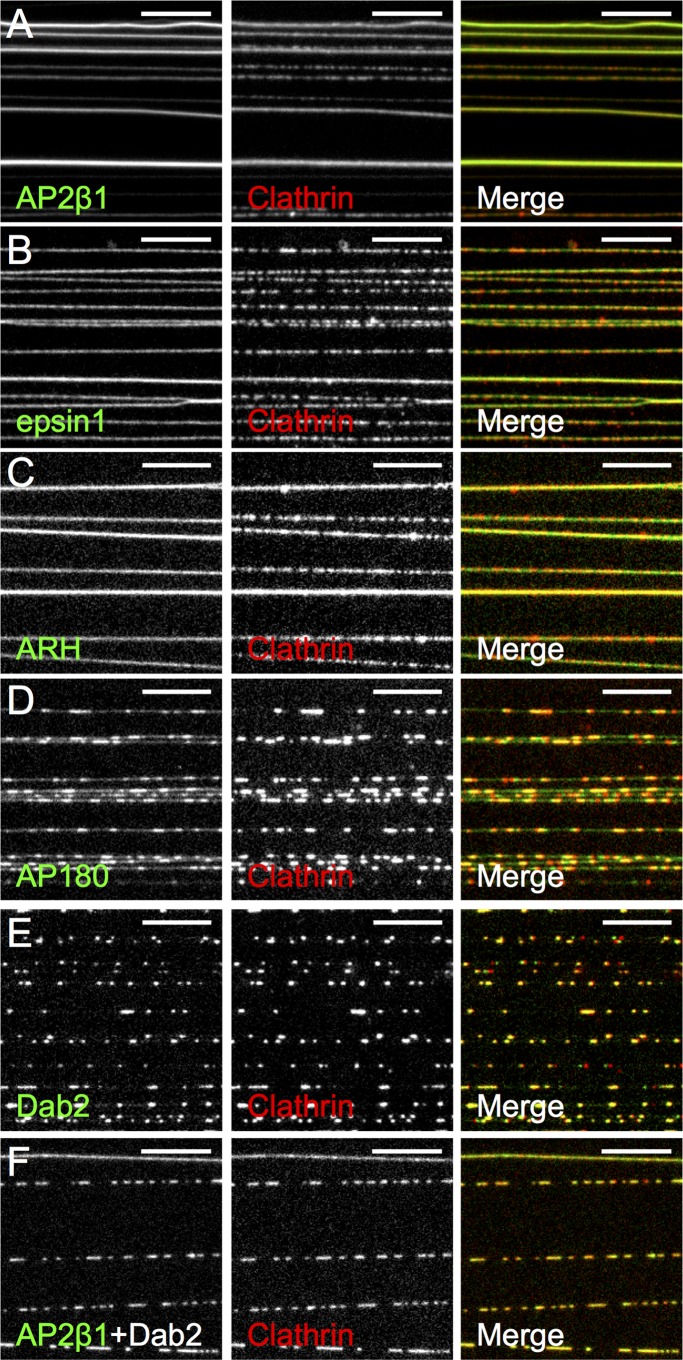
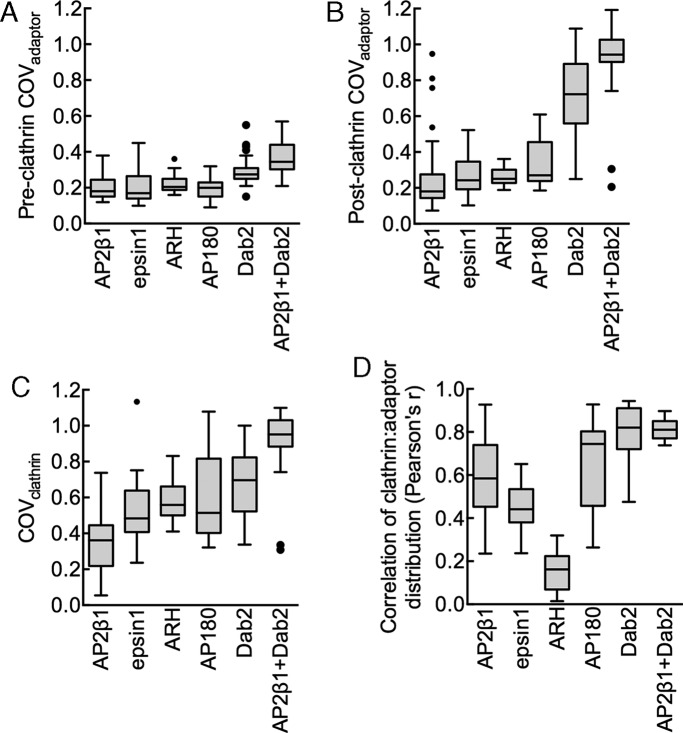
Despite the general notion that adaptors (and their cognate cargo) are clustered due to clathrin assembly, direct visualization of this process has not been demonstrated. We used membrane tubes as a starting template because the high radial curvature facilitates clathrin recruitment. At the same time, tubes are planar in the longitudinal axis and should allow diffusion-controlled clustering of adaptors in response to clathrin assembly. Membrane tubes are therefore the phenomenological equivalent of buds on a planar membrane. We separately recruited four different monomeric CLASPs (epsin1, ARH, AP180, and Dab2) and AP2β1 via an engineered N-terminal His tag to CL-containing tubes and analyzed their distribution in response to clathrin addition. This approach of recruiting proteins ensures similar membrane densities of adaptors and allows unambiguous assessment of their clathrin-assembling properties. However, this artificial mode of recruitment can compromise their intrinsic membrane curvature preferences (Holkar et al., 2015). Furthermore, clathrin-assembling properties seen here with the different adaptors might not fully mimic the cellular scenario in which the residence time of adaptors on the membrane is subject to regulation by multivalent interactions with cargo and phosphoinositides (Höning et al., 2005). The excess adaptor was washed off, and clathrin was flowed in, incubated for 10 min, and imaged. Remarkably, we found significant differences in the distribution of the adaptor and clathrin across these experiments. On AP2β1-coated tubes (Figure 2A), clathrin and adaptor distribution was relatively uniform, whereas on AP180- and Dab2-coated tubes (Figure 2, D and E), both clathrin and the adaptor appeared highly clustered. Reactions with epsin1- and ARH-coated tubes (Figure 2, B and C) showed an intermediate result in which clathrin appeared clustered, but the adaptor was relatively evenly distributed. These differences were quantified by estimating the coefficient of variation (COV) of fluorescence of adaptor before (Figure 3A) and after (Figure 3B) clathrin addition, as well as that of clathrin (Figure 3C) along the length of the tube. COV is defined as the ratio of the SD to the mean of fluorescence intensity and measures the extent of clustering of the fluorescent species. Before clathrin addition, all adaptors showed a uniform distribution, as seen by the relatively low COVadaptor (Figure 3A). However, after clathrin addition, whereas AP2β1 reactions showed a low value and AP180 and Dab2 showed a high value for COVadaptor and COVclathrin, both epsin1 and ARH showed a low COVadaptor and a high COVclathrin (Figure 3, B and C, and Supplemental Figure S2 for a pairwise comparison of significance). Consistently, a Pearson colocalization analysis (Figure 3D) revealed a relatively high degree of coincidence in clathrin and adaptor fluorescence for AP2β1, AP180, and Dab2 but a low degree for epsin1 and ARH. Together these results signify diversity in terms of how adaptors assemble clathrin on the membrane and their clustering in response to clathrin assembly.
FIGURE 2:
Adaptor clustering upon clathrin assembly. (A–F) Representative fluorescence micrographs showing distribution of the indicated adaptor (green) and clathrin (red) on tubes. Scale bars, 10 μm.
FIGURE 3:
Quantitation of adaptor clustering upon clathrin assembly. Box-and-whisker (Tukey) plots showing the COVadaptor before (A) and after (B) clathrin addition, COVclathrin (C), and Pearson’s r between adaptor and clathrin distribution (D). Data represent values from n ≥ 17 tubes from three experiments for each adaptor. All data sets differ significantly (p < 0.0001; nonparametric Kruskal–Wallis test; see Supplemental Figure S2 for a pairwise comparison of significance for each adaptor).
CLASPs function in a complex with AP2 during CME. Is the clustering of isolated adaptors therefore relevant to the cellular scenario? To test this, we analyzed clathrin assembly–induced clustering of a complex of AP2β1 and Dab2 (since they independently showed opposite clustering tendencies in our assay). Consistent with previous results (Schmid et al., 2006), a GST-AP2β1 appendage construct was efficiently recruited to Dab2-coated membrane tubes and confirms the interaction between AP2β1 and Dab2 (Supplemental Figure S3). We recruited an equimolar mixture of AP2β1 and unlabeled Dab2 to membrane tubes, added clathrin, and monitored fluorescence distribution of the mEGFP-tagged AP2β1. Remarkably, on AP2β1 plus Dab2–coated tubes, both the adaptor and clathrin appeared clustered (Figure 2F), similar to what was seen with Dab2 alone (Figure 2E and Supplemental Figure S2) but different from AP2β1 alone (Figure 2A and Supplemental Figure S2). These results indicate that the clustering behavior of Dab2 prevails even when it is present as a complex with AP2β1. This is apparent from the high COVadaptor (Figure 3B) and COVclathrin (Figure 3C) values and a high colocalization index of AP2β1 plus Dab2 and clathrin fluorescence (Figure 3D). The clustering of CLASP-AP2 complexes therefore mirrors the behavior of the participant CLASP, which displays a high tendency to cluster in response to clathrin assembly.
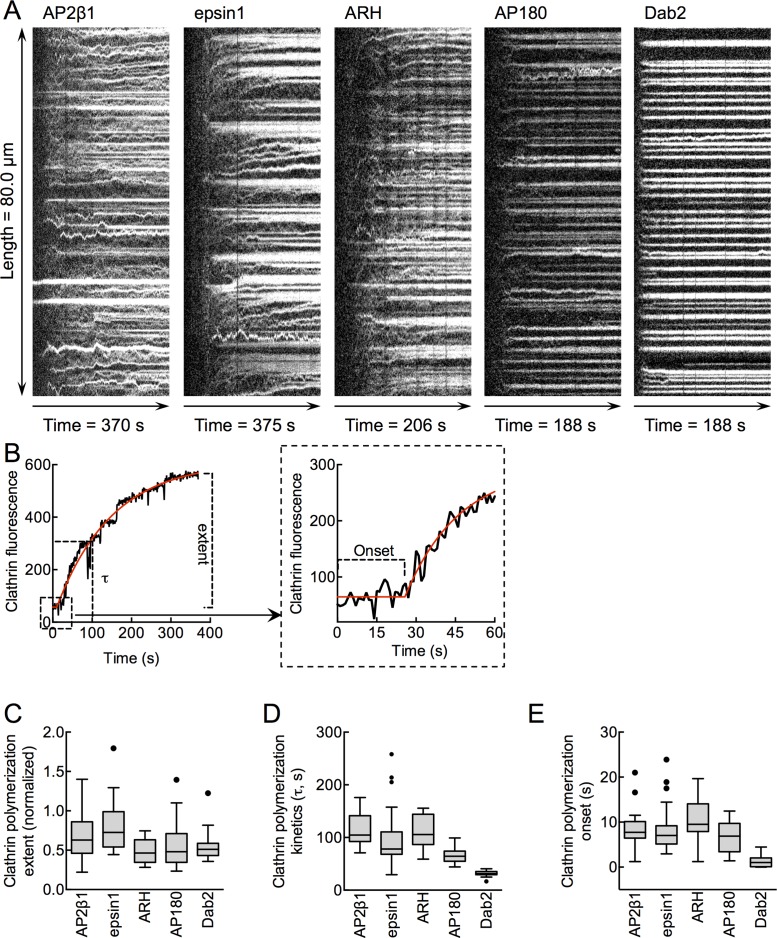
Kinetics of adaptor-mediated clathrin assembly
The foregoing experiments were carried out with similar membrane densities of adaptors and bulk clathrin concentrations. Therefore the determinants for efficient adaptor clustering must lie not just in the intrinsic self-assembling properties of clathrin but also in how adaptors engage with clathrin. To decipher these determinants, we acquired time-lapse images of reactions in which fluorescent clathrin is flowed over adaptor-coated tubes. Typical kymographs of clathrin recruitment along the tube length for each of the five adaptors are shown in Figure 4A. For all of the adaptors tested, clathrin fluorescence first appears as foci whose intensity increases until saturation is reached. Remarkably, however, we find adaptor-specific differences in how these foci behave with time. Clathrin foci formed with AP2β1 (Supplemental Video S1), epsin1, and ARH diffuse laterally on the tube and merge with time. In contrast, foci formed with AP180 and Dab2 maintain a discrete appearance and remain stationary throughout the course of the reaction. Data on mean clathrin intensity along the length of the tube versus time were extracted from kymographs and fitted to a plateau followed by a single-exponential rise function, which gave kinetic parameters such as the onset, rate (or t-half, τ), and extent of clathrin assembly (Figure 4B). Although the extents to which each adaptor recruited clathrin were largely similar (Figure 4C and Supplemental Figure S2), we find significant differences in the kinetic parameters of clathrin assembly. Dab2 displayed the shortest onset time and the fastest rate of clathrin assembly (Figure 4, D and E, and Supplemental Figure S2). The rates of clathrin assembly follow the order Dab2 > AP180 > ARH ≈ AP2β1 > epsin1.
FIGURE 4:
Kinetics of adaptor-mediated clathrin assembly. (A) Representative kymographs showing clathrin assembling on membrane tubes coated with the indicated adaptors. (B) Representative trace of the buildup of mean clathrin fluorescence on an AP2β1-coated membrane tube obtained from kymographs showing the various kinetic parameters analyzed. The red line represents the fit to a plateau followed by one-phase exponential-rise equation. Box-and-whisker (Tukey) plots showing the (C) extent, (D) rate (t-half, τ), and (E) onset of clathrin polymerization on adaptor-coated tubes. All data represent values from n ≥ 15 tubes from three experiments for each adaptor. Data sets differ significantly (p < 0.0001; nonparametric Kruskal–Wallis test; see Supplemental Figure S2 for pairwise comparison of significance for each adaptor).
Determinants of clathrin assembly–induced adaptor clustering
The extent of adaptor clustering at equilibrium (Figures 2 and 3) appears to be linked to the distribution and/or dynamics of clathrin assembly. Adaptors like Dab2 and AP180 that rapidly assemble clathrin and maintain discrete clathrin foci are efficiently clustered. AP2β1, epsin1, and ARH assemble clathrin relatively slowly and form clathrin foci that merge and get clustered less efficiently. Because clathrin has an intrinsic tendency to assemble when past a critical concentration, it is possible that in experiments with epsin1 and ARH, which bind clathrin with slow kinetics, we see a run-off reaction in which clathrin self-assembly takes over and leads to the formation of clathrin punctae.
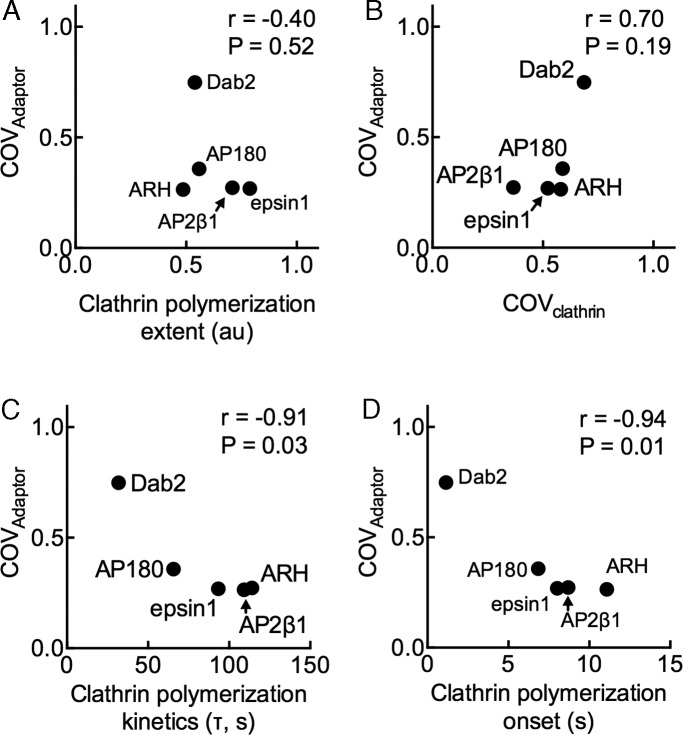
To arrive at a general and quantitative description of this relationship, we correlated the degree of adaptor clustering (COVadaptor) with the equilibrium and kinetic parameters of clathrin assembly for all five adaptors. As seen in Figure 5, COVadaptor correlates poorly (p = 0.52) with the extent of clathrin recruited. Surprisingly, COVadaptor correlates poorly (p = 0.19) with COVclathrin, indicating that the clustering of adaptor and that of clathrin are not necessarily linked events. This indicates that the process of clathrin self-assembly does not necessarily incorporate the adaptor that recruited clathrin in the first place, suggesting a possible run-off reaction in which clathrin self-assembles independently. However, COVadaptor correlates quite well with the rate (p = 0.03) and onset (p = 0.01) of clathrin assembly. A weak dependence of COVadaptor on COVclathrin but a strong dependence on onset and rate of clathrin assembly indicates that adaptor clustering is determined not by the extent to which clathrin clusters but instead on how rapidly adaptors engage with and assemble clathrin. Thus efficient clustering of adaptors follows from a kinetic fine-tuning of the early phases of clathrin self-assembly.
FIGURE 5:
Determinants of clathrin assembly–induced adaptor clustering. Correlation between the mean COVadaptor and (A) mean extent of clathrin polymerization, (B) mean COVclathrin, (C) mean rate of clathrin polymerization, and (D) mean onset of clathrin polymerization for each adaptor tested. Pearson’s r and p value.
DISCUSSION
Our results from quantitative fluorescence microscopy elucidate fundamental design principles for adaptor-mediated clathrin assembly. We find, using two different adaptors—one with a folded domain and the other unstructured—that adaptor-mediated clathrin assembly is intrinsically sensitive to the underlying membrane curvature. Thus, whereas clathrin recruitment to membranes is strictly dependent on adaptors, the high membrane curvature facilitates clathrin recruitment to the membrane. The curvature sensitivity for clathrin binding is consistent with a recent study describing the preferential polymerization of clathrin on adaptor-coated bead surfaces (Dannhauser et al., 2015). The origin of this curvature sensitivity probably lies in the intrinsic curvature of clathrin lattices, as suggested by cryo-EM reconstruction of the clathrin coat (Fotin et al., 2004) and theoretical modeling approaches (Nossal, 2001; Ferguson et al., 2006), or could arise from the intrinsic pucker in the clathrin triskelia. Our results are the first to systematically test the contribution of membrane curvature to the success of clathrin assembly using an experimental scheme that avoids membrane perturbation. Previous in vitro reconstitution schemes on adaptor-coated lipid vesicles and assayed with EM indicated robust (Ford et al., 2002; Dannhauser and Ungewickell, 2012; Kelly et al., 2014) to no deformation (Saleem et al., 2015) of the membrane in response to clathrin assembly. We believe that this inconsistency arises from variable sample processing methods used in EM, which is not the case in the present experiments. Taking the results together, it appears that an energetic input toward membrane bending, by membrane-active mechanisms (Ford et al., 2002; Miller et al., 2015), protein crowding (Busch et al., 2015), or simply reducing membrane tension (Saleem et al., 2015), could provide the necessary localized stimulus to recruit and assemble clathrin, which in turn could stabilize the bent membrane through a Brownian ratchet–like mechanism (Ungewickell and Hinrichsen, 2007). Of interest, recent proteomic analysis indicates that the bulk concentrations of clathrin and AP2 in synaptic boutons are in the micromolar range, (Wilhelm et al., 2014), far exceeding the nanomolar concentrations necessary for clathrin self-assembly (Dannhauser et al., 2015; present study). In this regard, our finding of a dramatic enhancement (∼10-fold) in clathrin abundance on curved membranes suggests that induction of membrane curvature could be an important factor in restricting clathrin assembly to sites where CCPs are formed.
We find tremendous diversity in how adaptors regulate clathrin assembly, both from a kinetic standpoint and in terms of the eventual distribution of these assemblies on the membrane. The evolutionary conservation of adaptors could therefore be explained not just by their ability to recognize specific cargoes, but also by their unique clathrin-assembling properties. Adaptors bind clathrin via consensus clathrin boxes/motifs (Lafer, 2002). The terminal domain of clathrin is a seven-blade β-propeller structure that defines seven grooves between the blades (ter Haar et al., 1998, 2000). Hydrophobic residues present in clathrin boxes fit within these groves. The adaptors tested here vary widely with respect to the type and numbers of clathrin boxes present in them. The clathrin-binding site on AP2β1 is a composite of a type I clathrin box (631LLNLD) present in the hinge region and residues in the appendage domain (Owen et al., 2000). On the other hand, epsin1 has two binding sites—a type I (480LVDLD) and a type II (257LMDLADV) clathrin box (Drake et al., 2000; Drake and Traub, 2001), ARH has a single type I (212LLDLE) clathrin box (He et al., 2002; Mishra et al., 2002b), and Dab2 has two binding sites—a type I (235LLVDLN) and a type II (363PWPYP) box (Mishra et al., 2002a). The construct of AP180 used here displays five noncanonical clathrin boxes containing DLL motifs (Morgan et al., 2000). The number of clathrin-binding sites present on adaptors cannot explain the diversity we observe. Despite there being two clathrin boxes on AP2β1, epsin1, ARH, and Dab2, these adaptors differ significantly in rates of clathrin assembly. In addition, AP180, with five DLL motifs, displays slower rates of clathrin assembly than Dab2, with two clathrin-binding sites. Because the accessibility, affinity, and number of clathrin-binding sites collectively contribute to the rate of clathrin assembly, our results indicate that predictions on how an adaptor fares in assembling clathrin based on any one of these parameters can be misleading.
Previous results from cells indicate that CLASPs function in association with AP2. Our results and their interpretation are not to be confused with the notion that CLASPs can manage clathrin assembly by themselves in cells, since they would presumably always be associated with AP2. However, we believe that the analyses of AP2:CLASP complexes would take us no closer to understanding determinants of adaptor clustering without separately understanding the contribution of each adaptor to clathrin assembly. For this reason, and to increase the robustness of correlation between clathrin assembly and adaptor clustering, we carried out an independent analysis of AP2 and CLASPs. From such an analysis (specifically comparing our results on AP2β1, Dab2, and AP2β1:Dab2), we infer that the clathrin-assembling properties of CLASPs would dominate over those of AP2 if they were better at assembling clathrin than AP2.
A large literature points to the role of AP180 and its nonneuronal homologue, CALM, in reducing heterogeneity in CCP sizes in cells (Zhang et al., 1998; Nonet et al., 1999; Bao et al., 2005; Meyerholz et al., 2005; Petralia et al., 2012; Miller et al., 2015). Our results indicate that the mechanistic basis for such an influence on CCP structure lies in the unique dynamic attributes of AP180-induced and Dab2-induced, clathrin assemblies. Despite previous work on the role of Dab2 in affecting CCP size from overexpression studies (Chetrit et al., 2009), our results are the first to reveal similarities between Dab2 and AP180 in how they organize clathrin lattices. AP180- and Dab2-induced assemblies display greatly reduced tendencies to merge on the plane of the membrane. Such behavior is reminiscent of the differences seen in dynamics of clathrin on coated buds and plaques in cells (Saffarian et al., 2009). The dome-shaped architecture of clathrin assemblies on buds reduces their tendency to merge, whereas the relatively planar assemblies on plaques allow merging, possible because lattice edges are more accessible in this configuration. Thus it is possible that the AP180- and Dab2-induced clathrin assemblies remain as discrete foci because of a dome-shaped structure, whereas those formed with AP2β1, epsin1, and ARH merge because of a relatively planar structure. Despite our best efforts using negative-stain EM and stimulated emission depletion microscopy, we have not been successful in determining the structure of these lattices.
The typical lifetime of a clathrin-coated pit is ∼30–60 s (Loerke et al., 2009; Doyon et al., 2011; Taylor et al., 2011; Cocucci et al., 2012), during which time, adaptors and their cognate cargoes are sorted into a growing pit. The typical time scales of adaptor-mediated clathrin assembly in our assays (∼30–100 s) fall within the estimated lifetime of coated pits. The sorting of adaptors into coated pits therefore must be strongly influenced by the kinetics of adaptor and clathrin interaction. Our results (Figure 5, C and D) indicate that an important determinant for efficient clustering of adaptors is the rapidity with which they assemble clathrin. The onset and rate are indicators of the strength of interaction of the adaptor for clathrin triskelia. Our results therefore indicate that adaptors that bind clathrin strongly would sort more efficiently into clathrin-coated pits. Current models proposed to explain mechanisms of adaptor sorting during CME have evolved from the discovery of motifs that govern protein–protein interactions. On the basis of such models, one would have anticipated the adaptors tested here to have clustered equally well since all of them contain clathrin boxes, which is clearly not the case. Our results therefore emphasize the need to go beyond equilibrium models and invoke kinetic parameters of adaptor–clathrin interactions in order to understand mechanisms that regulate CME.
MATERIALS AND METHODS
Expression, purification, and fluorescence labeling of proteins
Rat epsin1 (O88339: Isoform 1), mouse Dab2 (P98078: Isoform p96), human ARH (Q5SW96), and mouse AP180 (Q61548: Isoform short) were cloned as N-terminal hexahistidine (6xHis) and C-terminal StrepII-tag fusions. Residues 584–951 of the human AP2β1 (P63010: Isoform 2) and residues 144–575 of rat epsin1 were cloned as N-terminal 6xHis-mEGFP and C-terminal StrepII-tag fusions. All constructs were confirmed by sequencing. Proteins were expressed in BL21(DE3) at 18°C in autoinduction medium (Formedium, Norfolk, United Kingdom) and purified by tandem affinity as described previously (Holkar et al., 2015). Clathrin was extracted from clathrin-coated vesicles isolated from goat brains and purified as described previously (Holkar et al., 2015). Clathrin and purified adaptors (except AP2β1 and epsin C-tail constructs, which contained mEGFP) were labeled with cysteine-reactive fluorescent probes as described previously (Holkar et al., 2015).
Membrane template preparation
The composition of the lipid mix was 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1,2-dioleoyl-sn-glycero-3-phospho-l-serine (DOPS), 1,2-dioleoyl-sn-glycero-3-[(N-(5-amino-1-carboxypentyl)iminodiacetic acid)succinyl] (DGS-NTA[Ni2+]), and 1.1’-dioctadecyl-3,3,3’,3’-tetramethylindocarbocyanine (DiD; 79.5:15:5:0.5 mol%). Appropriate volumes of lipid stocks (Avanti Polar Lipids, Alabaster, AL) were aliquoted into a glass vial, diluted to a final concentration of 1 mM in chloroform, and stored at −80°C. Before use, the vial was brought to room temperature. A small aliquot (1–5 nmol of total lipid) was spread on a PEGylated glass coverslip (Dar et al., 2015) and assembled in a flow cell that was filled with filtered and degassed assay buffer. The flow cell was left undisturbed for 10 min at room temperature before flowing assay buffer to remove excess membrane reservoir.
Clathrin assembly assays
Membrane templates were first equilibrated in filtered and degassed assay buffer, after which, 200 μl of assay buffer containing 1% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) was flowed in and incubated for 10 min. Subsequently, 200 μl of 200 nM each adaptor (diluted in assay buffer) was introduced and incubated with templates for 10 min. Excess adaptor was rinsed with 2 × 200 μl of assay buffer and 1 × 200 μl of assay buffer containing an oxygen scavenger cocktail of 0.2 mg/ml glucose oxidase (G-2133; Sigma-Aldrich), 0.035 mg/ml catalase (C-40; Sigma-Aldrich), 4.5 mg/ml glucose, and 1 mM dithiothreitol. The clathrin assembly reaction was monitored in real time by flowing in 200 μl of 80 nM Texas red–labeled clathrin and freshly diluted in assay buffer containing the oxygen scavenger cocktail. All reactions were carried out at 25°C.
Fluorescence microscopy
Fluorescence imaging was carried out on an Olympus IX71 inverted microscope through a 100×/numerical aperture 1.4 oil-immersion objective equipped with an Evolve 512 electron-multiplying charge-coupled device camera (Photometrics, Tucson, AZ) as described by Holkar et al. (2015).
Image analysis
Fluorescence micrographs and time-lapse sequences were analyzed using Fiji (Schindelin et al., 2012). Nonlinear regression and statistical analyses were carried out using GraphPad Prism. The dead time of the flow cell was estimated by calculating the onset of fluorescent clathrin into the microscope field. Frames before the calculated onset were removed from time-lapse sequences. Background-corrected kymographs were generated from lines placed across the entire length of the membrane tube. The mean pixel intensity versus time data in kymographs were exported and fitted to a plateau followd by one-phase exponential rise function.
Estimation of tube and vesicle dimensions
The SLB formed during SMrT template preparation was used as an in situ calibration standard to estimate tube and vesicle dimensions and is based on the premise that fluorescence of diffraction-limited membrane-bound objects is proportional to the net membrane area (Kunding et al., 2008). We first validated that the source indeed represents an SLB. Typical fluorescence micrographs of a background-corrected SLB image doped with 0.5 mol% DiD (Supplemental Figure S1A) shows regions of varying fluorescence, marked as regions of interest (ROIs) 1–3. A histogram of fluorescence intensities in the entire field shows three peaks in fluorescence, associated with ROIs 1–3 (Supplemental Figure S1B), that scale linearly (Supplemental Figure S1C). These results validate the assumption that ROIs 1–3 represent a stack of one, two, and three SLBs, respectively. Because the vesicles formed in our preparations could be multilamellar, we considered fluorescence of the externally added His-tagged AP2β1/epsin C-tail for estimating tube and vesicle dimensions, since these would only bind the outer lamellae. For this, fluorescence micrographs of SLBs coated with AP2β1/epsin1 C-tail were corrected for background. The integrated fluorescence intensity in different ROIs placed on regions showing a single stack of SLB was then plotted against its area to give a slope K. The integrated fluorescence intensities of vesicles, collected using an appropriate threshold, were converted to area by dividing by K. The radius (r) of the vesicle was then calculated from r = sqrt(area/π). The integrated fluorescence intensities of tubes, collected from ROIs of a defined length (l), were converted to area by dividing by K. The radius of the tube was then calculated from r = area/(2πl). Tube dimensions were corrected for the approximately twofold enrichment of His-tagged AP2β1/epsin1 C-tail fluorescence seen on SLBs (Figure 1, E and H).
Supplementary Material
Acknowledgments
We thank members of the Pucadyil laboratory for comments on the manuscript. We also thank Linton Traub, Bernard Kelly, and David Owen for DNA constructs, Srishti Dar for help with GST-AP2β ear binding experiments, and Devika Andhare for essential controls in the clathrin assembly assays. T.J.P. is an Intermediate Fellow of the Wellcome Trust-DBT India Alliance and thanks the Alliance and IISER Pune for funds. S.H. thanks the Council of Scientific and Industrial Research, India, for a senior research fellowship.
Abbreviations used:
- AP180
adaptor protein 180
- AP2
adaptor protein 2
- ARH
autosomal recessive hypercholesterolemia
- CALM
phosphatidylinositol-binding clathrin assembly protein
- CCP
clathrin-coated pits
- CD4
cluster of differentiation 4
- CLASP
clathrin-associated sorting proteins
- CME
clathrin-mediated endocytosis
- COV
coefficient of variation
- Dab2
disabled-2
- EM
electron microscopy
- ROI
region of interest
- SLB
supported lipid bilayer.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E16-06-0399) on August 24, 2016.
REFERENCES
- Bao H, Daniels RW, MacLeod GT, Charlton MP, Atwood HL, Zhang B. AP180 maintains the distribution of synaptic and vesicle proteins in the nerve terminal and indirectly regulates the efficacy of Ca2+-triggered exocytosis. J Neurophysiol. 2005;94:1888–1903. doi: 10.1152/jn.00080.2005. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Traub LM. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu Rev Biochem. 2003;72:395–447. doi: 10.1146/annurev.biochem.72.121801.161800. [DOI] [PubMed] [Google Scholar]
- Busch DJ, Houser JR, Hayden CC, Sherman MB, Lafer EM, Stachowiak JC. Intrinsically disordered proteins drive membrane curvature. Nat Commun. 2015;6:7875. doi: 10.1038/ncomms8875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chetrit D, Ziv N, Ehrlich M. Dab2 regulates clathrin assembly and cell spreading. Biochem J. 2009;418:701–715. doi: 10.1042/BJ20081288. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Aguet F, Boulant S, Kirchhausen T. The first five seconds in the life of a clathrin-coated pit. Cell. 2012;150:495–507. doi: 10.1016/j.cell.2012.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannhauser PN, Ungewickell EJ. Reconstitution of clathrin-coated bud and vesicle formation with minimal components. Nat Cell Biol. 2012;14:634–639. doi: 10.1038/ncb2478. [DOI] [PubMed] [Google Scholar]
- Dar S, Kamerkar SC, Pucadyil TJ. A high-throughput platform for real-time analysis of membrane fission reactions reveals dynamin function. Nat Cell Biol. 2015;17:1588–1596. doi: 10.1038/ncb3254. [DOI] [PubMed] [Google Scholar]
- Doyon JB, Zeitler B, Cheng J, Cheng AT, Cherone JM, Santiago Y, Lee AH, Vo TD, Doyon Y, Miller JC, et al. Rapid and efficient clathrin-mediated endocytosis revealed in genome-edited mammalian cells. Nat Cell Biol. 2011;13:331–337. doi: 10.1038/ncb2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake MT, Downs MA, Traub LM. Epsin binds to clathrin by associating directly with the clathrin-terminal domain. Evidence for cooperative binding through two discrete sites. J Biol Chem. 2000;275:6479–6489. doi: 10.1074/jbc.275.9.6479. [DOI] [PubMed] [Google Scholar]
- Drake MT, Traub LM. Interaction of two structurally distinct sequence types with the clathrin terminal domain beta-propeller. J Biol Chem. 2001;276:28700–28709. doi: 10.1074/jbc.M104226200. [DOI] [PubMed] [Google Scholar]
- Ferguson ML, Prasad K, Sackett DL, Boukari H, Lafer EM, Nossal R. Conformation of a clathrin triskelion in solution. Biochemistry. 2006;45:5916–5922. doi: 10.1021/bi052568w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ford MGJ, Mills IG, Peter BJ, Vallis Y, Praefcke GJK, Evans PR, McMahon HT. Curvature of clathrin-coated pits driven by epsin. Nature. 2002;419:361–366. doi: 10.1038/nature01020. [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns (4, 5) P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Fotin A, Cheng Y, Sliz P, Grigorieff N, Harrison SC, Kirchhausen T, Walz T. Molecular model for a complete clathrin lattice from electron cryomicroscopy. Nature. 2004;432:573–579. doi: 10.1038/nature03079. [DOI] [PubMed] [Google Scholar]
- He G, Gupta S, Yi M, Michaely P, Hobbs HH, Cohen JC. ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J Biol Chem. 2002;277:44044–44049. doi: 10.1074/jbc.M208539200. [DOI] [PubMed] [Google Scholar]
- Holkar SS, Kamerkar SC, Pucadyil TJ. Spatial control of epsin-induced clathrin assembly by membrane curvature. J Biol Chem. 2015;290:14267–14276. doi: 10.1074/jbc.M115.653394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höning S, Ricotta D, Krauss M, Späte K, Spolaore B, Motley A, Robinson M, Robinson C, Haucke V, Owen DJ. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol Cell. 2005;18:519–531. doi: 10.1016/j.molcel.2005.04.019. [DOI] [PubMed] [Google Scholar]
- Kalthoff C, Alves J, Urbanke C, Knorr R, Ungewickell EJ. Unusual structural organization of the endocytic proteins AP180 and epsin 1. J Biol Chem. 2002;277:8209–8216. doi: 10.1074/jbc.M111587200. [DOI] [PubMed] [Google Scholar]
- Kelly BT, Graham SC, Liska N, Dannhauser PN, Honing S, Ungewickell EJ, Owen DJ. AP2 controls clathrin polymerization with a membrane-activated switch. Science. 2014;345:459–463. doi: 10.1126/science.1254836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunding AH, Mortensen MW, Christensen SM, Stamou D. A fluorescence-based technique to construct size distributions from single-object measurements: application to the extrusion of lipid vesicles. Biophys J. 2008;95:1176–1188. doi: 10.1529/biophysj.108.128819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafer EM. Clathrin–protein interactions. Traffic. 2002;3:513–520. doi: 10.1034/j.1600-0854.2002.30801.x. [DOI] [PubMed] [Google Scholar]
- Loerke D, Mettlen M, Yarar D, Jaqaman K, Jaqaman H, Danuser G, Schmid SL. Cargo and dynamin regulate clathrin-coated pit maturation. PLoS Biol. 2009;7:e57. doi: 10.1371/journal.pbio.1000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Boucrot E. Molecular mechanism and physiological functions of clathrin-mediated endocytosis. Nat Rev Mol Cell Biol. 2011;12:517–533. doi: 10.1038/nrm3151. [DOI] [PubMed] [Google Scholar]
- Meyerholz A, Hinrichsen L, Groos S, Esk P-C, Brandes G, Ungewickell EJ. Effect of clathrin assembly lymphoid myeloid leukemia protein depletion on clathrin coat formation. Traffic. 2005;6:1225–1234. doi: 10.1111/j.1600-0854.2005.00355.x. [DOI] [PubMed] [Google Scholar]
- Miller SE, Mathiasen S, Bright NA, Pierre F, Kelly BT, Kladt N, Schauss A, Merrifield CJ, Stamou D, Höning S, et al. CALM regulates clathrin-coated vesicle size and maturation by directly sensing and driving membrane curvature. Dev Cell. 2015;33:163–175. doi: 10.1016/j.devcel.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Keyel PA, Hawryluk MJ, Agostinelli NR, Watkins SC, Traub LM. Disabled-2 exhibits the properties of a cargo-selective endocytic clathrin adaptor. EMBO J. 2002a;21:4915–4926. doi: 10.1093/emboj/cdf487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra SK, Watkins SC, Traub LM. The autosomal recessive hypercholesterolemia (ARH) protein interfaces directly with the clathrin-coat machinery. Proc Natl Acad Sci USA. 2002b;99:16099–16104. doi: 10.1073/pnas.252630799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan JR, Prasad K, Hao W, Augustine GJ, Lafer EM. A conserved clathrin assembly motif essential for synaptic vesicle endocytosis. J Neurosci. 2000;20:8667–8676. doi: 10.1523/JNEUROSCI.20-23-08667.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonet ML, Holgado AM, Brewer F, Serpe CJ, Norbeck BA, Holleran J, Wei L, Hartwieg E, Jorgensen EM, Alfonso A. UNC-11, a Caenorhabditis elegans AP180 homologue, regulates the size and protein composition of synaptic vesicles. Mol Biol Cell. 1999;10:2343–2360. doi: 10.1091/mbc.10.7.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nossal R. Energetics of clathrin basket assembly. Traffic. 2001;2:138–147. doi: 10.1034/j.1600-0854.2001.020208.x. [DOI] [PubMed] [Google Scholar]
- Owen DJ, Vallis Y, Pearse BM, McMahon HT, Evans PR. The structure and function of the beta 2-adaptin appendage domain. EMBO J. 2000;19:4216–4227. doi: 10.1093/emboj/19.16.4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petralia RS, Wang Y-X, Indig FE, Bushlin I, Wu F, Mattson MP, Yao PJ. Reduction of AP180 and CALM produces defects in synaptic vesicle size and density. Neuromol Med. 2012;15:49–60. doi: 10.1007/s12017-012-8194-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saffarian S, Cocucci E, Kirchhausen T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 2009;7:e1000191. doi: 10.1371/journal.pbio.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Morlot S, Hohendahl A, Manzi J, Lenz M, Roux A. A balance between membrane elasticity and polymerization energy sets the shape of spherical clathrin coats. Nat Commun. 2015;6:6249. doi: 10.1038/ncomms7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, Ford MGJ, Burtey A, Praefcke GJK, Peak-Chew S-Y, Mills IG, Benmerah A, McMahon HT. Role of the AP2 β-appendage hub in recruiting partners for clathrin-coated vesicle assembly. PLoS Biol. 2006;4:e262. doi: 10.1371/journal.pbio.0040262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid EM, McMahon HT. Integrating molecular and network biology to decode endocytosis. Nature. 2007;448:883–888. doi: 10.1038/nature06031. [DOI] [PubMed] [Google Scholar]
- Skruzny M, Desfosses A, Prinz S, Dodonova SO, Gieras A, Uetrecht C, Jakobi AJ, Abella M, Hagen WJ, Schulz J, et al. An organized co-assembly of clathrin adaptors is essential for endocytosis. Dev Cell. 2015;33:150–162. doi: 10.1016/j.devcel.2015.02.023. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc Natl Acad Sci USA. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ter Haar E, Musacchio A, Harrison SC, Kirchhausen T. Atomic structure of clathrin: a beta propeller terminal domain joins an alpha zigzag linker. Cell. 1998;95:563–573. doi: 10.1016/s0092-8674(00)81623-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. Sorting it out: AP-2 and alternate clathrin adaptors in endocytic cargo selection. J Cell Biol. 2003;163:203–208. doi: 10.1083/jcb.200309175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traub LM. Tickets to ride: selecting cargo for clathrin-regulated internalization. Nat Rev Mol Cell Biol. 2009;10:583–596. doi: 10.1038/nrm2751. [DOI] [PubMed] [Google Scholar]
- Ungewickell EJ, Hinrichsen L. Endocytosis: clathrin-mediated membrane budding. Curr Opin Cell Biol. 2007;19:417–425. doi: 10.1016/j.ceb.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Wilhelm BG, Mandad S, Truckenbrodt S, Kröhnert K, Schäfer C, Rammner B, Koo SJ, Claßen GA, Krauss M, Haucke V, et al. Composition of isolated synaptic boutons reveals the amounts of vesicle trafficking proteins. Science. 2014;344:1023–1028. doi: 10.1126/science.1252884. [DOI] [PubMed] [Google Scholar]
- Zhang B, Koh YH, Beckstead RB, Budnik V, Ganetzky B, Bellen HJ. Synaptic vesicle size and number are regulated by a clathrin adaptor protein required for endocytosis. Neuron. 1998;21:1465–1475. doi: 10.1016/s0896-6273(00)80664-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.