Summary
Allografts of retinal pigment epithelial (RPE) cells have been considered for the treatment of ocular diseases. We recently started the transplantation of induced pluripotent stem cell (iPSC)-derived RPE cells for patients with age-related macular degeneration (autogenic grafts). However, there are at least two problems with this approach: (1) high cost, and (2) uselessness for acute patients. To resolve these issues, we established RPE cells from induced iPSCs in HLA homozygote donors. In vitro, human T cells directly recognized allogeneic iPSC-derived RPE cells that expressed HLA class I/II antigens. However, these T cells failed to respond to HLA-A, -B, and -DRB1-matched iPSC-derived RPE cells from HLA homozygous donors. Because of the lack of T cell response to iPSC-derived RPE cells from HLA homozygous donors, we can use these allogeneic iPSC-derived RPE cells in future clinical trials if the recipient and donor are HLA matched.
Highlights
-
•
We established human RPE cells from iPSCs in HLA homozygote donors
-
•
iPS-RPE cells uniquely expressed HLA class I and class II molecules
-
•
T lymphocytes responded to allogeneic iPS-RPE cells, but not iPSCs, in vitro
-
•
T lymphocytes failed to respond to allogeneic iPS-RPE cells from HLA homozygote donors
In this article, Takahashi and colleagues show that the allogeneic immune response to iPSC-derived retinal pigment epithelial (RPE) cells by T cells in vitro. However, HLA-restricted immune reaction (at least HLA-A, -B, and -DRB1-matched) does not occur when iPS-RPE cells established from HLA homozygous donors are used.
Introduction
Retinal pigment epithelial (RPE) cells play an important role in maintaining the immune privileged status of the eye. RPE cells have both proliferative and anti-proliferative effects on T cells, and these effects are regulated by cytokines (Streilein, 2003, Sugita, 2009). Interferon-γ (IFN-γ) inflammatory cytokines are upregulated in immunological processes such as transplant rejection (Huber and Irschick, 1988). IFN-γ induces the expression of major histocompatibility complex (MHC) class I and II (MHC-I, MHC-II) molecules on RPE cells (Enzmann et al., 1999, Sugita et al., 2009). T lymphocytes and inflammatory cytokines play the central effector role in cellular immune reactions including immune rejection. In addition to effective antigen recognition, the activation of these cells causes the secretion of inflammatory cytokines, i.e., IFN-γ. A complex network of helper CD4+ T cells (Th cells) is then initiated, and the lymphatic cell proliferation and immune reactions continue. This cascade may play a role in the rejection of allogeneic RPE transplants in the eye. Modulation of the transplanted cells leads to secretion of inflammatory cytokines that attract T cells and cause immune rejection. Therefore, the investigation of rejection mechanisms is important for the prevention of this process and prolonged graft survival.
RPE cell-associated allografts have been considered for the treatment of ocular diseases such as age-related macular degeneration (AMD). We successfully established human RPE cells from human iPSCs (Kamao et al., 2014, Sugita et al., 2015). In addition, we recently transplanted an iPSC-derived RPE (iPS-RPE) sheet into an AMD patient autograft. RPE cells including iPS-RPE cells have immunosuppressive properties; human RPE cells suppress T cell activation and can convert T cells to regulatory T cells (Horie et al., 2010, Imai et al., 2012, Sugita et al., 2015, Usui et al., 2008). However, several groups in human clinical trials found that RPE allografts did not survive because of immune rejection (Algvere, 1997, Algvere et al., 1999, Peyman et al., 1991, Weisz et al., 1999). Algvere et al. (1999) reported that immune rejection after RPE transplantation in humans includes loss of visual function over the transplant, development of an exudative response (e.g., serous retinal detachment), fluorescein leakage of the grafts, disruption of the grafts, depigmentation of the grafts, and encapsulation of the grafts. However, there have been no previous reports of how antigen and cell type affect the outcome of the retinal transplantation. In addition, as far as we know, no one has reported that RPE cells derived from embryonic stem cells (ESCs)/iPSCs are recognized by MHC-restricted immune cells, especially T cells.
Therefore, the purpose of the present study was to determine whether human RPE cells derived from iPSCs could be recognized by human leukocyte antigen (HLA)-restricted T cells. We used an in vitro model with human iPS-RPE cells from HLA-3 locus (A, B, DRB1) homozygote donors as target cells and allogeneic T cells as responder effector cells.
Results
Expression of HLA Class I and II on iPSC-Derived RPE Cells
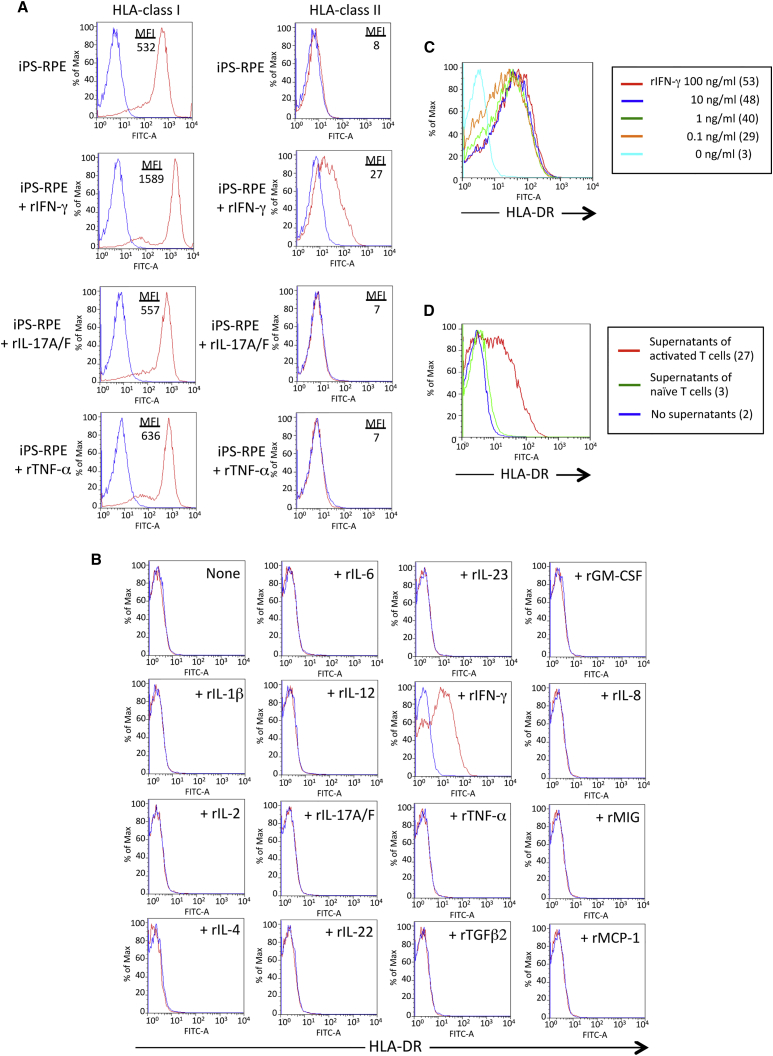
To confirm the expression of HLA molecules on human iPS-RPE cells, we prepared several iPS-RPE cell lines (Kamao et al., 2014, Sugita et al., 2015) and human control cells (ESC-derived RPE cells, ARPE-19 cell lines, fetal primary RPE cells, cornea endothelial cells, fibroblasts, and iPSCs). First, we examined the expression of HLA class I and II on iPS-RPE cells by flow cytometry. The iPS-RPE cells constitutively expressed HLA class I (A, B, C), but not class II (DR, DP, DQ, Figure 1A). IFN-γ-pretreated iPS-RPE cells expressed HLA class II, but interleukin-17A/F (IL-17A/F)-treated or tumor necrosis factor α (TNF-α)-treated cells did not. Conventional human RPE cell lines (ARPE-19) had similar results (data not shown). Other RPE cell lines also did not express class II under normal conditions, but class II expression was induced in the presence of IFN-γ (Figure S1). The expression pattern in control human RPE cells, such as ESC-derived RPE cells, ARPE-19 cells, and fetal RPE cells, and other control cells (cornea endothelial cells and fibroblasts) was similar. However, iPSCs did not express HLA class II molecules even when IFN-γ was added to the culture (Figure S1). During culture, the expression pattern of class I and II molecules in iPS-RPE cells was similar, but a slightly different (Figure S2). For example, iPS-RPE cells at the early stage (p1, day 14) expressed high levels of HLA class II when IFN-γ was added to the culture, but the expression was downregulated during the culture (p4, day 90). Importantly, the expression of HLA-DR on iPS-RPE cells was upregulated when the cells were pretreated with IFN-γ, but not with other recombinant proteins (IL-1β, IL-2, IL-4, IL-6, IL-12, IL-17A/F, IL-22, IL-23, TNF-α, transforming growth factor β2 [TGF-β2], granulocyte macrophage colony-stimulating factor [GM-CSF], IL-8, MIG, MCP-1 [Figure 1B], IL-10, IL-21, IL-27, TGF-β1, TNFRI, macrophage migration inhibitory factor, thrombospondin, and lipopolysaccharide; data not shown). The expression of HLA-DR in the presence of IFN-γ was upregulated in a cytokine dose-dependent manner (Figure 1C), and iPS-RPE cells exposed to supernatants from activated T cells, but not from naive T cells, clearly expressed HLA-DR (Figure 1D). Taken together, our experimental evidence indicates that HLA molecules are uniquely expressed on the surface of RPE cells, including iPS-RPE cells.
Figure 1.
Expression of HLA Class I and II on human iPSC-Derived RPE Cells
(A) iPS-RPE cells (836B1) were stained with anti-HLA class I antibody (A, B, C) or class II antibody (DR, DQ, DP). iPS-RPE cells in the presence of recombinant IFN-γ, IL-17A/F, or TNF-α were cultured for 48 hr (red). Numbers in the graphs indicate mean fluorescence intensity (MFI). Isotype control is shown in blue.
(B) iPS-RPE cells (454E2) were cultured with human recombinant proteins, such as IL-1β, IL-2, IL-4, IL-6, IL-12, IL-17A/F, IL-22, IL-23, TNF-α, TGFβ2, GM-CSF, IL-8/CXCL8, MIG/CXCL9, and MCP-1/CCL2, for 48 hr, and the cells were stained with anti-HLA-DR antibodies. We obtained similar results with anti-human HLA-DR, -DP, and -DQ antibodies.
(C) 454E2 iPS-RPE cells were cultured with recombinant IFN-γ in a dose-dependent manner (0, 0.1, 1, 10, 100 ng/mL), and the cells were stained with anti-HLA-DR antibody. Numbers in parentheses indicate MFI.
(D) iPS-RPE cells exposed to supernatants from activated T cells (agonistic anti-CD3 antibody-treated) or naive T cells were stained with anti-HLA-DR antibodies. Numbers in parentheses indicate MFI.
HLA Molecule Disparity between Lymphocytes and Allogeneic iPS-RPE Cells Induces Immune Responses
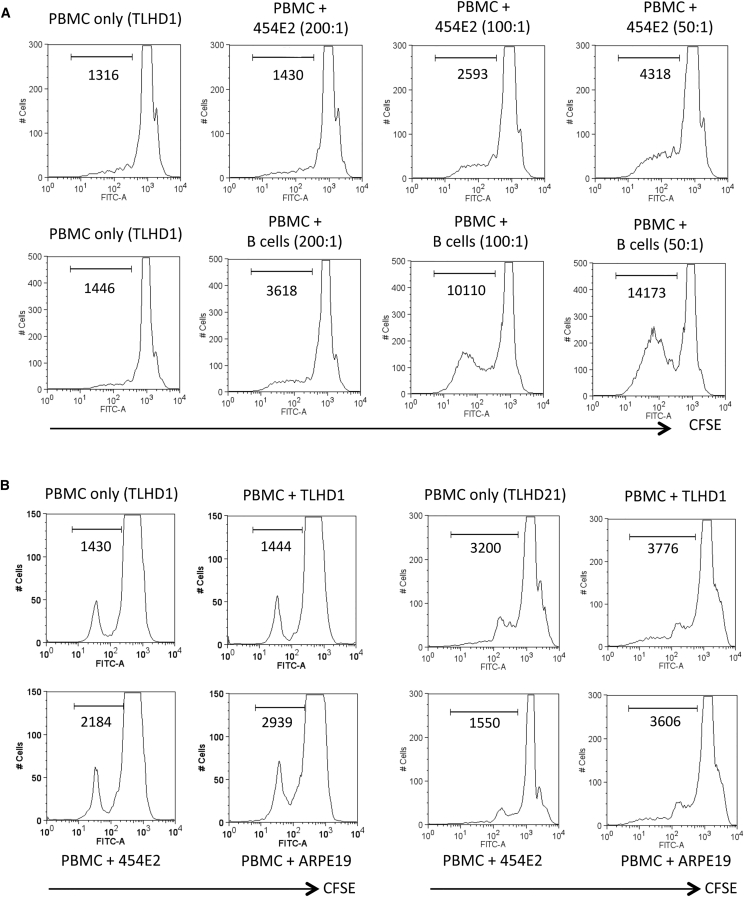
We confirmed that the HLA mismatch between peripheral blood mononuclear cells (PBMCs) and allogeneic iPS-RPE cells was sufficient to elicit allogeneic immune responses. We summarized the results of HLA-typing tests for HLA-A, -B, -C, -DRB1, -DQB1, and -DPB1, in T cells collected from healthy volunteers (n = 26, Table S1). MHC disparity between T cells and allogeneic iPS-RPE cells was previously shown to induce immune responses in animal models (Kamao et al., 2014). PBMCs from healthy donors were cultured with human iPS-RPE cells from the same donor (autogenic) or another donor (allogeneic). In a PBMC-RPE mixed lymphocyte reaction (MLR) assay, the PBMCs proliferated in vitro when co-cultured with allogeneic 454E2 iPS-RPE cells, as well as allogeneic B cells (positive control cells: Figure 2A). On the other hand, the PBMCs did not proliferate when co-cultured with allogeneic iPSCs (data not shown). PBMCs from a TLHD1 donor did not induce proliferation of TLHD1 iPS-RPE cells, whereas the PBMCs induced proliferation when co-cultured with allogeneic 454E2 iPS-RPE cells and ARPE-19 cells (Figure 2B). Allogeneic proliferation did not occur between TLHD21 or TLHD10 PBMCs and 454E2 RPE cells with completely matched HLA class I and II (HLA-A, -B, and -DRB1), whereas the PBMCs responded to other HLA-mismatched RPE cells (Figures 2B, S3A, and S3B). In addition, PBMCs from TLHD15, TLHD23, and TLHD24 donors proliferated when HLA homozygote iPS-RPE cells (454E2 and 453F2) and RPE cell lines were added to the cultures (Figure S3A).
Figure 2.
Lymphocyte Reactions of Allogeneic iPSC-Derived RPE Cells: PBMC-RPE MLR Assay
(A) CFSE-labeled PBMCs with iPS-RPE cells were analyzed by flow cytometry. iPS-RPE cells (target cells, 454E2 from HLA homozygote donors) or B cells (a positive control) were co-cultured with PBMCs (effector cells) from a TLHD1 donor for 120 hr (effector/target ratio = 200:1, 100:1, or 50:1).
(B) RPE cells (454E2 iPS-RPE, TLHD1 iPS-RPE, and ARPE-19 cells) were co-cultured with PBMCs from a TLHD1 donor (left panels) or a TLHD21 donor (right panels). The TLHD21 donor was completely matched for HLA-A, -B, and -DRB1 in 454E2 iPS-RPE cells, and the TLHD1 donor was mismatched. Numbers in the histogram indicate CFSE-positive cells.
iPS-RPE Cells from HLA Homozygote iPSCs Do Not Respond to HLA-Matched Allogeneic Immune Cells In Vitro
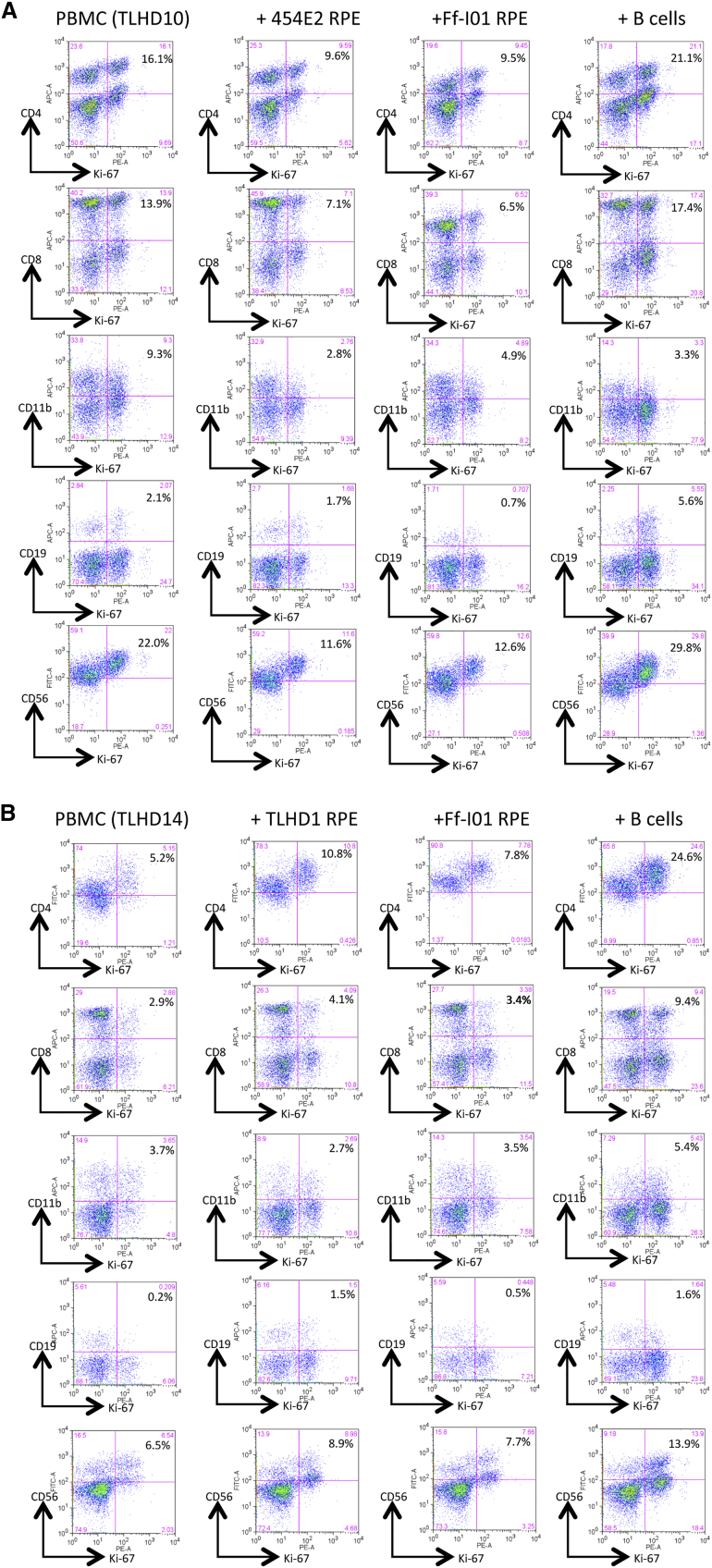
Next, we examined what kind of cell proliferates in cultures of PBMCs plus iPS-RPE cells. For the assay, we used anti-human Ki-67 antibody (a cell proliferation marker) in fluorescence-activated cell sorting (FACS) analysis. In the MLR assay with fresh PBMCs all cell types in PBMCs proliferated, except CD11b+ cells, when co-cultured with allogeneic B cells as a positive control compared with PBMCs only (Figure S4). The target B cells greatly expressed HLA class I, class II, CD40, CD80 (B7-1), and CD86 (B7-2) molecules (data not shown). By contrast, CD4+ cells (Th cells), CD8+ cells (cytotoxic T cells), CD11b+ cells (macrophages/monocytes), CD19+ cells (B cells), and CD56+ cells (natural killer [NK] cells) in TLHD1 PBMCs failed to proliferate when co-cultured with autogenic RPE cells (Figure S4).
In the PBMC-RPE MLR assay with allogeneic HLA homozygote iPS-RPE cells (454E2 and Ff-I01), these immune cells in PBMCs (TLHD10) failed to proliferate when co-cultured with HLA-matched allogeneic iPS-RPE cells (HLA-A, -B, and -DRB1 matched: Figure 3A). However, these immune cells in PBMCs (TLHD14) induced the proliferation of HLA-mismatched iPS-RPE cells (TLHD1 and Ff-I01), i.e., all inflammatory cell types, especially CD4+ and CD8+ T cells, responded to TLHD1 iPS-RPE cells (all HLA mismatched) compared with PBMCs only (Figure 3B). Moreover, CD4+ and CD8+ T cells in a TLHD14 donor responded to Ff-I01 RPE lines that were HLA-A matched and HLA-B, -DRB1 mismatched. We had similar results with TLHD21 donor PBMCs plus HLA-mismatched TLHD1 RPE cells and HLA-matched Ff-I01 RPE cells; CD4 + and CD8+ T cells in TLHD21 donor PBMCs failed to respond to Ff-I01 RPE, but CD4+ and CD8+ T cells highly responded to TLHD1 RPE cells in vitro (Figure S5). These results imply that T lymphocytes may recognize MHC molecules on allogeneic iPS-RPE cells and then proliferate. However, an immune response cannot be induced in the PBMC-RPE MLR assay when the lymphocytes and RPE cells are HLA matched.
Figure 3.
Another PBMC-RPE MLR Assay with Allogeneic iPS-RPE Cells by Ki-67 Proliferation
To evaluate the PBMC-RPE MLR assay with allogeneic HLA homozygote iPS-RPE cells (454E2, 453F2, and Ff-I01) and B cells as positive control cells, we used Ki-67 proliferation by FACS analysis using antibodies against CD4+ cells (helper T cells), CD8+ cells (cytotoxic T cells), CD11b+ cells (macrophages/monocytes), CD19+ cells (B cells), and CD56+ (NK cells).
(A) TLHD10 PBMCs versus both 454E2 and Ff-I01 iPS-RPE cells = HLA-A, -B, -DRB1 matched.
(B) TLHD14 PBMCs versus TLHD1 iPS-RPE cells = HLA-A, -B, -DRB1 mismatched, and Ff-I01 iPS-RPE cells = HLA-A matched, and HLA-B and -DRB1 mismatched.
CD4+ T Cells Can Recognize HLA Molecules on Allogeneic iPS-RPE Cells, but Not MHC-Matched RPE Cells
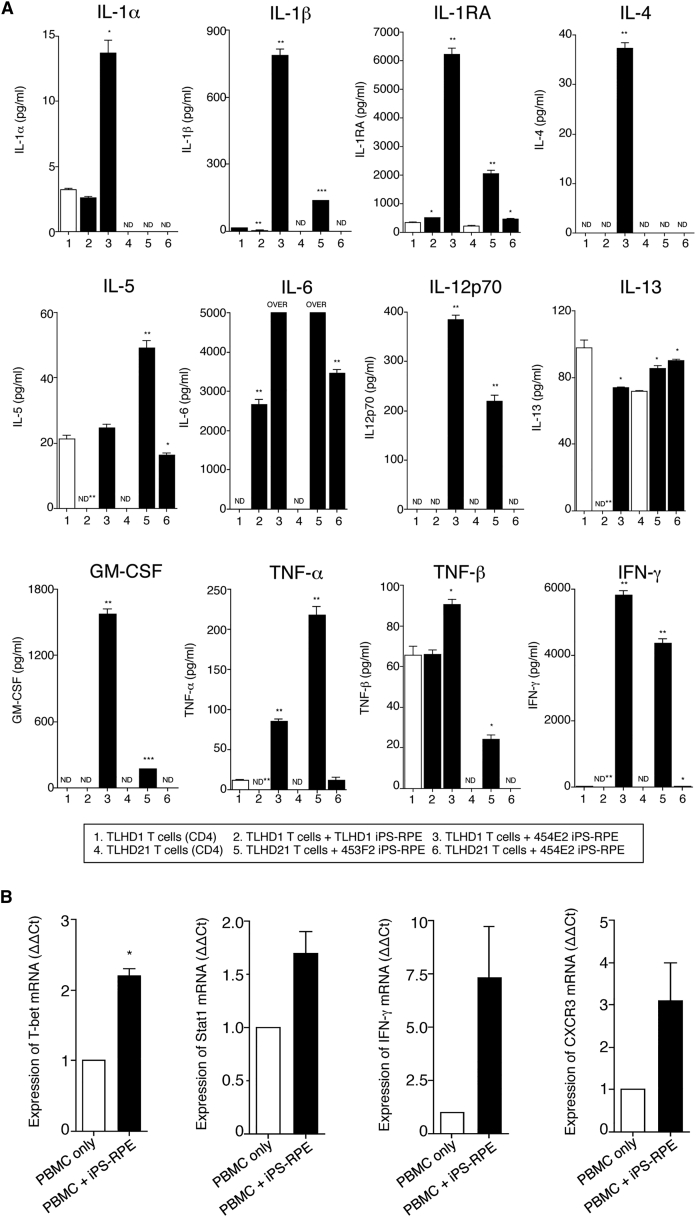
We next examined whether the RPE-direct recognition by T cells occurred in an HLA-restricted manner. We used purified CD4+ T cells co-cultured with iPS-RPE cells in the presence of recombinant IL-2 for a CD4-RPE cytokine assay. HLA homozygote 454E2 iPS-RPE cells and CD4+ T cells were co-cultured, and the supernatants were collected for 48 hr to measure the inflammatory cytokines.
We first examined which cytokines are released at high concentrations in the direct reactions between CD4+ T cells and allogeneic iPS-RPE cells. As revealed in Figure 4A, supernatants from the cultures of 454E2 iPS-RPE cells and CD4+ T cells (a TLHD1 MHC-mismatched donor) contained significant levels of IL-1α, IL-1β, IL-1RA, IL-4, IL-6, IL-12p70, GM-CSF, TNF-α, TNF-β, and IFN-γ, compared with that of T cells only or T cells exposed to autogenic TLHD1 RPE cells. When CD4+ T cells were prepared from an MHC-matched donor TLHD21 to 454E2 RPE cells, the T cells did not respond to the RPE cells. The T cells did not produce inflammatory cytokines, especially Th1-related cytokines (IL-1β, IL-12p70, GM-CSF, TNF-α, TNF-β, and IFN-γ), although the T cells are able to produce these cytokines in the presence of 453F2 RPE cells that are MHC-mismatched MHC homozygote cell lines (Figure 4A). These T cells in the cultures do not produce IL-7, IL-15, IL-31, or IFN-α (data not shown).
Figure 4.
Detection of Inflammatory Cytokines by CD4+ T Cells from HLA-Matched or -Mismatched Donors when Co-cultured with Homozygote iPS-RPE Cells
(A) Supernatants of CD4+ T cells (a TLHD1 donor) exposed to TLHD1 iPS-RPE cells (autogenic) or 454E2 iPS-RPE cells (allogeneic) were collected for a multiplex cytokine array assay (IL-1α, IL-1β, IL-1RA, IL-4, IL-5, IL-6, IL-12p70, IL-13, GM-CSF, TNF-α, TNF-β, and IFN-γ). In addition, data from TLHD21 CD4+ T cells plus 454E2 iPS-RPE cells (allogeneic reaction, but HLA-A, -B, -DRB1 matched) or 453F2 iPS-RPE cells (allogeneic reaction, HLA-A, -B, -DRB1 mismatched) are also presented. Data represent the mean ± SEM of three independent experiments. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005 compared with the positive control (CD4+ T cells alone, open bars). ND, not detected.
(B) Detection of mRNA for Th1-related genes in PBMCs in the presence of iPS-RPE cells by qRT-PCR. Total RNA was extracted from PBMCs exposed to allogeneic iPS-RPE cells (453F2 MHC homozygote lines). PBMCs without RPE cells were also prepared as a control. For PCR amplification, cDNA was amplified by using primers for T-bet (transcription factors T box), Stat1 (signal transducer and activator of transcription 1), CXCR3 (chemokine C-X-C motif receptor 3), IFN-γ, and β-actin. Results indicate the relative expression of the molecules (ΔΔCt for control PBMCs = 1). We obtained similar results with other iPS-RPE cell lines (836B1 and 454E2). Data represent the mean ± SEM of three independent experiments. ∗p < 0.05 compared with the control (PBMCs alone, open bars).
We next examined whether T cells exposed to allogeneic iPS-RPE cells can exhibit the Th1 phenotype. In qRT-PCR analysis, PBMCs in the presence of iPS-RPE cells expressed higher levels of mRNA for Th1-related genes such as T-bet, Stat1, IFN-γ, and CXCR3 compared with PBMCs alone without RPE cells (Figure 4B).
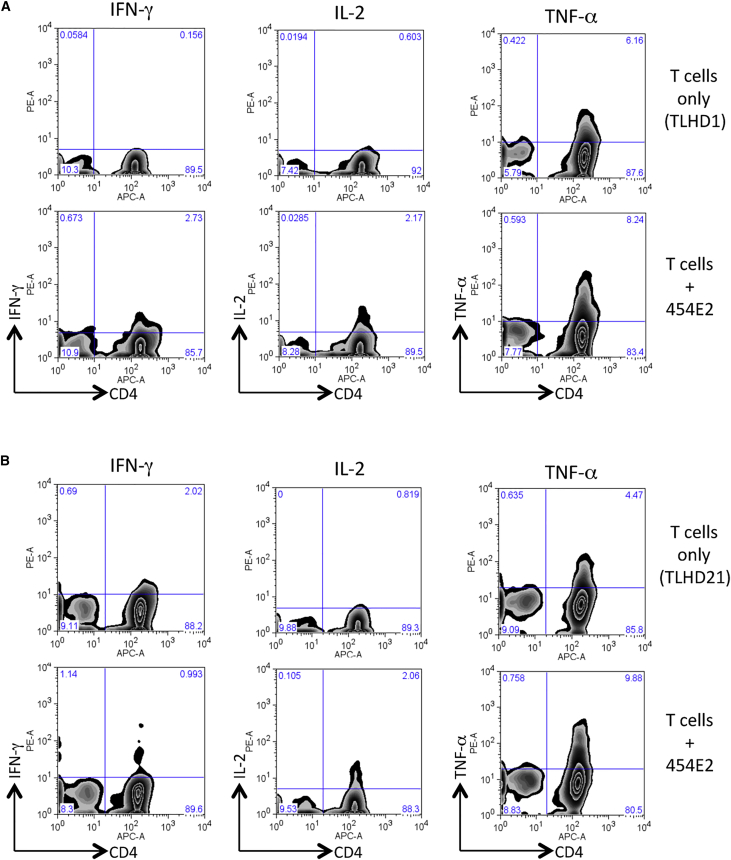
We then confirmed these results by performing FACS analysis with antibodies for Th1 cytokines, IFN-γ, IL-2, and TNF-α. Compared with control cultures (T cells without iPS-RPE cells), CD4+ T cells exposed to 454E2 iPS-RPE cells expressed IFN-γ, IL-2, and TNF-α (Figure 5A). However, TLHD21 T cells failed to express IFN-γ (but not IL-2 and TNF-α) when co-cultured with MHC-matched RPE cells (Figure 5B). Therefore, we chose IFN-γ evaluation for the following experiments.
Figure 5.
Expression of Th1-Associated Cytokines in Allogeneic iPS-RPE Cell-Exposed Helper T Cells
In FACS analysis, purified CD4+ T cells exposed to allogeneic HLA homozygous 454E2 iPS-RPE cells were stained with anti-human CD4/IFN-γ, IL-2, or TNF-α antibody for 48 hr.
(A) TLHD1 donor (versus 454E2 MHC-mismatched).
(B) TLHD21 donor (versus 454E2 MHC-matched).
CD4+ T Cells Cannot Recognize HLA-DRB1-Matched Allogeneic iPS-RPE Cells from HLA Homozygous Donors
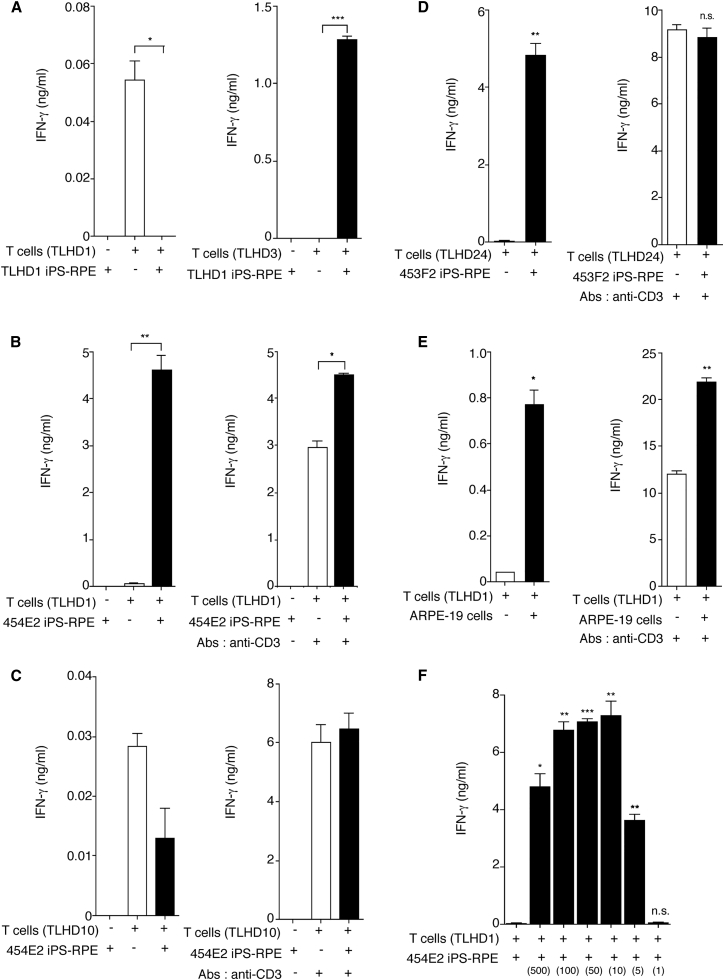
To examine the allogeneic recognition of T cells to HLA homozygous iPS-RPE cells, we next prepared CD4+ T cells from several healthy volunteers. First, we confirmed that CD4+ T cells recognize autogenic iPS-RPE cells. TLHD1 T cells did not produce IFN-γ when co-cultured with TLHD1 RPE cells, and the T cells produced significant levels of IFN-γ without RPE cells (control culture, Figure 6A). On the other hand, TLHD3 T cells recognized TLHD1 iPS-RPE cells (HLA mismatched), and the T cells produced significant amounts of IFN-γ. Unlike TLHD1 RPE cells, TLHD1 T cells greatly produced IFN-γ when co-cultured with HLA-mismatched 454E2 RPE cells (Figure 6B). Importantly, there was a statistically significant difference between T cells and RPE cells, but the extent of the difference was reduced when we added anti-CD3 antibody (first signal blocking) to the cultures (Figure 6B). On the other hand, there was no difference between T cells and RPE cells when TLHD10 T cells were co-cultured with 454E2 RPE cells (HLA-A, -B, -DRB1 matched) in the presence or absence of anti-CD3 antibody (Figure 6C). Moreover, we obtained similar results when using other HLA homozygote lines 453F2 (Figure 6D) and human RPE cell lines (Figure 6E). These results suggest that T cells directly recognize MHC molecules on RPE cells via the first signal.
Figure 6.
Production of IFN-γ in CD4+ T cells Exposed to Allogeneic iPS-RPE Cells
(A) Purified CD4+ T cells (TLHD1, left panel; TLHD3, right panel) were cultured with TLHD1 iPS-RPE cells for 48 hr, and the levels of IFN-γ in the supernatants were measured. The graph indicates data for IFN-γ production by CD4+ T cells exposed to iPS-RPE cells. Left bar, RPE cells only (RPE cells do not secrete IFN-γ); middle bar, T cells only; right bar, T cells plus RPE cells.
(B) TLHD1 CD4+ T cells were cultured with MHC homozygote 454E2 iPS-RPE cells, and the levels of IFN-γ in the supernatants were measured. Anti-human CD3 antibody was used to block the first signal between RPE cells and T cells (right panel).
(C) TLHD10 CD4+ T cells were cultured with MHC homozygote 454E2 iPS-RPE cells (HLA-A, -B, -DRB1 matched).
Data are the mean ± SEM of three ELISA determinations. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005 compared with positive control (CD4+ T cells alone, open bar).
(D) Purified CD4+ T cells (TLHD24) were cultured with 453F2 MHC homozygote iPS-RPE cells, and the levels of IFN-γ in the supernatants were measured. Anti-human CD3 antibody was used to block the first signal between RPE cells and T cells (right panel).
(E) TLHD1 CD4+ T cells were cultured with ARPE-19 heterozygote RPE cell lines in the presence (right panel) or absence (left panel) of anti-CD3 antibodies.
(F) For determination of the RPE cell number, CD4+ T cells (TLHD1) were cultured with allogeneic 454E2 iPS-RPE cells in a cell number-dependent manner.
Data represent the means ± SEM of three or four independent experiments. ∗p < 0.05, ∗∗p < 0.005, ∗∗∗p < 0.0005 compared with the control (CD4+ T cells alone, open bar). n.s., not significant.
To confirm the ratio of T cells to RPE cells in the cultures, we prepared 5 × 105 T cells and from 1 × 103 (500:1) to 5 × 105 RPE cells (1:1). T cells significantly produced IFN-γ in an RPE cell number-dependent manner, whereas T cell activation was suppressed with 5 × 105 confluent RPE cells (Figure 6F), indicating that human RPE cells also have anti-inflammatory properties (Horie et al., 2010, Imai et al., 2012, Sugita et al., 2015, Usui et al., 2008).
To confirm these results, we prepared T cells from several HLA-mismatched donors. Representative results indicate that supernatants contained significant levels of IFN-γ in the T cell RPE cultures when the tested T cells were mismatched to HLA class II (DRB1) of 454E2 RPE cells (Figures S6A and S6B) or 453F2 RPE cells (Figures S6D and S6F). In contrast, the RPE recognition by T cells was reduced if the tested T cells (TLHD4 or TLHD6) matched the HLA-DRB1 on these RPE cells (Figures S6C and S6E). These results suggest that CD4+ helper T cells can recognize MHC molecules, probably HLA class II such as HLA-DRB1, on allogeneic iPS-RPE cells. We summarize the results of the T cell response to MHC homozygote 454E2 iPS-RPE cells in Table S2. In the tested donors, six types of T cells from TLHD6 (HLA-B∗52:01 and DRB1∗15:02 matched), TLHD10 (HLA-A∗24:02, B∗52:01, and DRB1∗15:02 matched), TLHD11 (HLA-A∗24:02 matched), TLHD18 (HLA-B∗52:01 and DRB1∗15:02 matched), TLHD19 (HLA-DRB1∗15:02 matched), and TLHD21 (HLA-A∗24:02, B52:01, and DRB1∗15:02 matched) did not respond to the RPE cells in vitro.
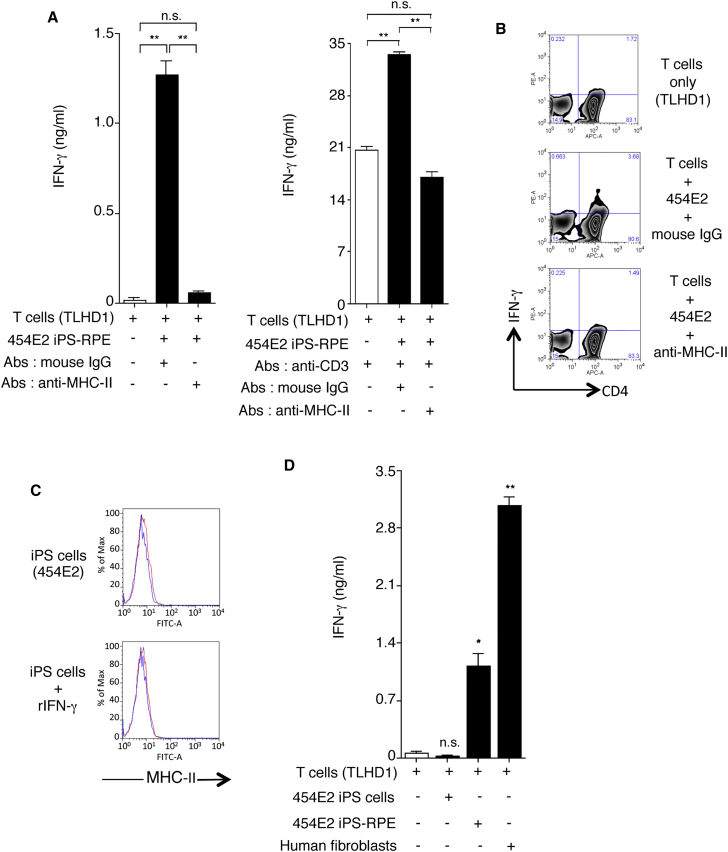
To confirm this finding, we examined whether CD4+ T cells can recognize allogeneic iPS-RPE cells in the presence of anti-HLA class II (MHC-II) blocking antibody. T cells significantly produced IFN-γ when co-cultured with 454E2 RPE cells, but the T cells failed to respond when anti-HLA class II antibodies were added to the cultures (Figure 7A). In addition, FACS analysis exhibited similar results (Figure 7B). These results indicate that the helper T cell response to allogeneic iPS-RPE cells is HLA class II restricted.
Figure 7.
Recognition of MHC-II Molecules on Allogeneic iPS-RPE Cells by CD4+ T Cells, but Not iPSCs
(A) The graph depicts data for IFN-γ ELISA by CD4+ T cells (TLHD1) exposed to 454E2 iPS-RPE cells. Anti-MHC-II (HLA-DR, -DP, -DQ) or isotype control antibody (mouse IgG) was used for the T cell RPE culture. Anti-human CD3 antibody was also used (right panel). Data represent the mean ± SEM of three independent experiments.
(B) FACS data for IFN-γ by CD4+ T cells (TLHD1) exposed to 454E2 iPS-RPE cells in the presence of anti-MHC-II blocking antibodies.
(C) 454E2 human iPSCs were stained with anti-MHC-II antibody. Cells were also cultured with recombinant IFN-γ for 48 hr. Red curve represents data for MHC-II molecules. Blue curve represents data for isotype controls.
(D) Purified CD4+ T cells (TLHD1) were cultured with 454E2 iPSCs, 454E2 iPS-RPE cells, and 454E2 fibroblasts for 48 hr, and the supernatants were measured for IFN-γ production by T cells.
Data represent the mean ± SEM of four independent experiments. ∗p < 0.05, ∗∗p < 0.005 compared with the positive control (open bars). n.s., not significant.
We next examined whether T cells are able to recognize iPSCs in vitro. First, we examined the expression of HLA class II (MHC-II) on human iPSCs, 454E2 homozygote lines. The iPSCs did not express HLA class II even when IFN-γ was added to the culture (Figure 7C). In an in vitro rejection assay using iPSCs, CD4+ T cells produced less IFN-γ even if co-cultured with allogeneic iPSCs (Figure 7C). In contrast, T cells produced large amounts of IFN-γ when co-cultured with allogeneic iPS-RPE cells or human skin fibroblasts (both from a 454E2 donor). Therefore, it is assumed that RPE cells are highly immunogenic compared with iPSCs.
We also examined whether IFN-γ-pretreated RPE cells have more immunogenicity because these iPS-RPE cells inducibly express HLA class II (see Figure 1). As expected, supernatants from T-RPE cultures contained high levels of IFN-γ when 454E2 or 453F2 RPE cells were pretreated with recombinant IFN-γ, compared with the results for untreated RPE cells (Figure S7A). We also verified that the T cell recognition is required for antigen-presenting cell (APC) immunity. Autogenic APCs were used in the CD4-RPE rejection assay. T cells produced significant amounts of IFN-γ in cultures of 454E2 RPE cells with or without APCs, but there was more effective production when APCs were added (Figure S7B). These results imply that T cell recognition by RPE cells might be required for APC activation.
Discussion
In the present study, we prepared human RPE cells derived from iPSCs as stimulators and T cells as responders to evaluate immune rejection in vitro. The ability of RPE cells to stimulate the bystander T cells was analyzed using MLR and IFN-γ production by T cells. In some experiments, APCs and anti-CD3 antibody (first signal blocking) were also used for the in vitro rejection assay. For the assay, we used T cell culture medium containing recombinant human IL-2. IL-2 is a growth factor for all subpopulations of T cells and acts as an antigen-nonspecific factor in the proliferation of these T cells. Th1 cells produce IL-2 as well as IFN-γ. The MLR assay is a useful tool with which to investigate the mechanisms of allogeneic responsiveness in vitro (allo-MLR) including the response to RPE cells (Kamao et al., 2014). In addition, we purified CD4+ T cells co-cultured with iPS-RPE cells in the presence of recombinant IL-2. To evaluate the recognition of T cells, we collected the supernatants to measure inflammatory cytokines such as IFN-γ for CD4+ T cells. We then analyzed whether the T cell recognition of RPE cells was HLA restricted. In this study, we focused on HLA-A, -B, and -DRB1 among the six HLA loci antigens, although we tested all six HLA genotypes, A, B, C, DRB1, DQB1, and DPB1, in PBMCs and RPE cells.
Retinal allografts have been considered for the treatment of ocular disease, and there is extensive evidence that allografts can survive in the retina without immunosuppression (Algvere et al., 1999). However, we previously showed that allogeneic transplants of iPS-RPE cells in animal models could elicit immune responses in the retina (Kamao et al., 2014), and several groups found that RPE allografts did not survive in human clinical trials (Algvere, 1997, Algvere et al., 1999, Peyman et al., 1991, Weisz et al., 1999). There have been no previous reports of how antigen (including MHC antigen) and cell type affect the outcome of retinal transplantation. In humans, allogeneic cell transplants almost always include a short course of immunosuppression, commonly cyclosporine A or tacrolimus, to minimize the chances of graft rejection through a T cell-mediated adaptive immune response (Clipstone and Crabtree, 1992). Eventually we might be able to control immune rejection by using T cell-specific medication. In addition, in immunohistochemistry with retinal sections, many T cells had invaded the retina after transplantation of iPS-RPE cells/sheets (allografts) in animal models, whereas control retinal sections examined by immunohistochemistry had no T cells in the retina (our unpublished data). Therefore, the T cell-mediated immune response plays a critical role in the pathogenesis of immune rejection after allogeneic RPE transplantation. As revealed in the present study, T cells exposed to iPS-RPE cells exhibited a Th1-type response (IFN-γ+ IL-2+ IL-1β+ T-bet+ STAT1+ CXCR3+). In fact, RPE cells, especially IFN-γ-treated RPE cells, can produce Th1-related chemokines (IFN-γ-related chemokines) such as CXCL9, CXCL10, and CXCL11 (Juel et al., 2012).
We have confirmed that established iPS-RPE cells constitutively express MHC-I but not MHC-II. However, the cytokines produced acutely in response to immune rejection may upregulate RPE MHC-II expression, as seen in vitro. This upregulation may be sufficient to initiate a recognition response in vivo, and it seems that infiltrating T cells would be strongly activated by the presentation of allospecific MHC. Since MHC-II was inducibly expressed by IFN-γ-treated RPE cells, but not by other cytokine-treated cells, it is assumed that the unique MHC-II expression on RPE cells might be IFN-γ specific, as shown in this study. In addition, RPE cells have been reported to act as APCs in the retina (Percopo et al., 1990). In previous reports (Drukker et al., 2002, Suarez-Alvarez et al., 2010), human iPSCs/ESCs have low MHC-I expression, undetectable MHC-II expression, and no expression of CD80 (B7-1)/CD86 (B7-2) co-stimulatory molecules, suggesting that the human iPSCs/ESCs have a limited capacity for antigen processing and presentation. However, RPE cells, including differentiated cells from human iPSCs, expressed MHC-I, MHC-II, and several co-stimulatory molecules (B7-H1, B7-H3, and B7-H5; data not shown), and the expression of these antigens in RPE cells determines the outcome of alloantigen-specific T cell responses in vitro.
Compared with the rejection of other graft types, corneal allograft rejection is delayed and less frequent, in part because the normal cornea lacks MHC-II-expressing APCs (Hamrah et al., 2002). Human RPE cells have a similar phenotype as corneal cells. As shown in the present study, iPS-RPE cells do not express MHC-II, and they constitutively express MHC-I. During the stress of immune rejection, MHC-II molecules are expressed by RPE cells, making these MHC-II-positive retinal cells potential targets for CD4+ effector T cells, especially Th1 cells. After the activation of helper T cells, CD8+ cytotoxic T cells (CTLs) can be activated by inflammatory cytokines such as IL-2 and IFN-γ, which are produced by Th1 cells. Eventually, CTLs attack and kill the target cells that express MHC-I. To avoid immune rejection without strong immunosuppression for successful transplantation, we should use HLA homozygous RPE cell lines to match the recipient. As revealed in the present study, HLA disparity between T cells and allogeneic iPS-RPE cells can induce immune responses, i.e., MLRs (allogeneic) and T cell activation (production of inflammatory cytokines). However, the HLA-restricted immune reaction did not occur when iPS-RPE cells from HLA homozygous donors were used. Although a more detailed analysis is needed with a large number of target cells (RPE cells) and effector cells (T cells or PBMCs), we note that effector T cells can recognize MHC molecules on allogeneic iPS-RPE cells, and the immune reaction by the T cells can be prevented after HLA blood tests.
In conclusion, T lymphocytes cannot recognize HLA molecules on allogeneic iPS-RPE cells established from HLA homozygote donors, as well as autogenic iPS-RPE cells in vitro. We plan to transplant human iPS-RPE cells using HLA homozygote allografts in our next clinical trial, and are now examining the HLA phenotype in patients with ocular retinal diseases such as AMD.
Experimental Procedures
Establishment of Human iPSCs
After informed consent was obtained, iPSCs (454E2, 453F2, and TLHD1) were established from skin fibroblasts or dental pulp cells of a patient with retinitis pigmentosa or a healthy donor by using an episomal vector of several genes, OCT3/4, SOX2, KLF4, L-MYC, LIN28, and p53 small hairpin RNA (Sugita et al., 2015). iPSCs were established from dermal fibroblasts of a patient with retinitis pigmentosa (101G26) or a healthy donor (836B1) by using an episomal vector of six genes, OCT3/4, SOX2, KLF4, L-MYC, LIN28, and GLIS1 (Sugita et al., 2015). Ff-I01 iPSCs (Okita et al., 2013) were also established from healthy donor PBMCs by pCE-hSK, pCE-hUL, pCE-hOCT3/4, pCE-mp53DD, and pCXB-EBNA1. 101G26, 836B1, and TLHD1 iPSCs were established from HLA heterozygous donors, and 454E2, 453F2, and Ff-I01 iPSCs were established from HLA homozygous donors. The research followed the tenets of the Declaration of Helsinki, and the study was approved by the Institutional Ethics Committees of the Center for Developmental Biology, RIKEN.
Preparation of iPSC-Derived RPE Cells
To differentiate into RPE cells, we cultured human iPSCs on gelatin-coated dishes using Glasgow’s minimal essential medium (GMEM) supplemented as previously described (Kamao et al., 2014, Sugita et al., 2015). Signal inhibitors Y-27632 (10 μM, Wako), SB431542 (5 μM, Sigma), and CKI-7 (3 μM, Sigma) were added to the GMEM (Kamao et al., 2014, Sugita et al., 2015). After the appearance of pigment epithelium-like colonies, the medium was switched to DMEM/F12 medium with B27 supplement (Invitrogen). iPS-RPE cells expressed specific makers for primary RPE cells such as RPE65, bestrophin, MiTF, ZO-1, TGF-β1, -β2, and -β3, pigment epithelium-derived factor, and vascular endothelial growth factor (Kamao et al., 2014, Sugita et al., 2015).
HLA Typing
HLA typing (A, B, C, DRB1, DQB1, and DPB1) of human iPS-RPE cells, control cells, or blood of healthy donors was performed with PCR reverse sequence-specific oligonucleotide probes using LABType SSO (One Lambda) or WAKFlow (Wakunaga Pharmaceutical) (Okita et al., 2011). The results of HLA-allele typing of blood from healthy donors are shown in Table S1. HLA-allele type in human iPS-RPE cells and control cells was as follows. (1) 836B1 iPS-RPE cells as HLA-A∗02:01/-; HLA-B∗27:03/50:01; HLA-C∗01:02/04:01; HLA-DRB1∗01:01/13:03; HLA-DQB1∗03:01/05:01; HLA-DPB1∗01:01/23:01. (2) 101G26 iPS-RPE cells as HLA-A∗11:01/24:02; HLA-B∗40:01/54:01; HLA-C∗01:02/07:02; HLA-DRB1∗04:05/08:03; HLA-DQB1∗06:01/-; HLA-DPB1∗02:01/05:01. (3) 454E2 iPS-RPE cells as HLA-A∗24:02/-; HLA-B∗52:01/-; HLA-C∗12:02/-; HLA-DRB1∗15:02/-; HLA-DQB1∗06:01/-; HLA-DPB1∗05:01/09:01. (4) 453F2 iPS-RPE cells as HLA-A∗11:01/-; HLA-B∗15:01/-; HLA-C∗04:01/-; HLA-DRB1∗04:06/-; HLA-DQB1∗03:02/-; HLA-DPB1∗02:01/04:02. (5) Ff-I01 iPS-RPE cells as HLA-A∗24:02/-; HLA-B∗52:01/-; HLA-C∗12:02/-; HLA-DRB1∗15:02/-; HLA-DQB1∗06:01/-; HLA-DPB1∗09:01/-. (6) TLHD1 iPS-RPE cells as HLA-A∗11:01/-; HLA-B∗15:01/67:01; HLA-C∗04:01/07:02; HLA-DRB1∗09:01/16:02; HLA-DQB1∗03:03/05:02; HLA-DPB1∗02:02/05:01. (7) RPE cell lines (ARPE19) as HLA-A∗02:01/03:01; HLA-B∗07:02/14:02; HLA-C∗07:02/08:02; HLA-DRB1∗13:02/15:01; HLA-DQB1∗06:02/06:09; HLA-DPB1∗02:01/04:01. (8) Human fibroblast cells as HLA-A∗24:02/26:02; HLA-B∗40:02/52:01; HLA-C∗03:04/12:02; HLA-DRB1∗09:01/15:02; HLA-DQB1∗04:01/-; HLA-DPB1∗02:01/05:01.
Preparation of T Cells and Antigen-Presenting Cells
T cells were established from autogenic or allogeneic T cells from PBMCs. CD4+ T cells were prepared separately by using separation beads (MACS cell isolation kit, Miltenyi Biotec). These cells were more than 94% CD4-positive. These T cells were co-cultured with RPMI-1640 medium containing 10% fetal bovine serum (BioWhittaker), human recombinant IL-2 (Becton Dickinson), 10 mM HEPES (Sigma), 0.1 mM nonessential amino acids (Sigma), 1 mM sodium pyruvate (Sigma), penicillin-streptomycin (Gibco), and 1 × 10−5 M 2-mercaptoethanol (Sigma). APCs from PBMCs of healthy donors were also prepared. The X-irradiated APCs (20 Gy) were cultured with T cells plus iPS-RPE cells in some experiments.
Mixed Lymphocyte Reactions with iPS-RPE Cells
After informed consent was obtained, PBMCs were isolated from healthy donors, and allogeneic immune responses were assessed for proliferation by carboxyfluorescein succinimidyl ester (CFSE; Cayman Chemical) incorporation by the PBMCs. CFSE-labeled PBMCs were cultured with iPS-RPE cells (454E2, 453F2, Ff-I01, and TLHD1) and ARPE-19 cell lines. As a positive control, Epstein-Barr virus-transformed B cells from a healthy donor that are HLA class I+, HLA class II+, CD40+, B7-1 (CD80)+, and B7-2 (CD86)+ were also prepared. The culture medium used was RPMI-1640. Before the assay, the target RPE cells or B cells were irradiated (20 Gy). After 96–120 hr, CFSE-labeled PBMCs were washed and analyzed by flow cytometry. The PBMC-RPE MLR assay in monkey cells was also performed using similar methods (Kamao et al., 2014).
In the Ki-67 proliferation assay by FACS analysis, the following antibodies were used: anti-human CD4 (BioLegend, catalog #317416), anti-human CD8 (eBioscience, #17-0088), anti-human CD11b (Miltenyi Biotec, #130-091-241), anti-human CD19 (BD PharMingen, #561742), anti-human CD56 (BioLegend, #304604), and anti-human Ki-67 (BioLegend, #350504). The following isotype control antibodies were used: mouse immunoglobulin 2a (IgG2a), κ isotype control fluorescein isothiocyanate (FITC) (BioLegend, #400208), mouse IgG1, κ isotype control APC, and mouse IgG1 (BioLegend, #400122), κ isotype control phycoerythrin (PE) (BioLegend, #400112). The harvested PBMCs and PBMCs co-cultured with human iPS-RPE cells (or B cells) were stained with these antibodies at 4°C for 30 min. For intracellular staining by Ki-67, staining was performed after cell fixation and permeabilization (BioLegend). All samples were analyzed on a FACSCanto flow cytometer (BD). Data were analyzed by using FlowJo software (version 9.3.1).
In Vitro CD4-RPE Rejection Assay
Effector T cells from PBMCs were collected from healthy volunteers (n = 26, Table S1). Purified CD4+ T cells (5–8 × 105 cells/well in 96-well plates) from the PBMCs of healthy donors were co-cultured with target iPS-RPE cells (5–8 × 103 cells/well: effector/target ratio = 100:1) for 48 hr. T cell activation was assessed for IFN-γ production (R&D Systems) by the CD4+ T cells. The target iPS-RPE cells were prepared from three HLA homozygote cell lines, 454E2, 453F2, and Ff-I01, and three HLA heterozygote cell lines, TLHD1, 836B1, and 101G26. As controls, human iPSCs (454E2), human fibroblasts (454E2), and human RPE cell lines (ARPE-19) were also prepared for the assay. In company experiments, blocking antibodies for MHC-II (HLA-DR, DQ, DP: 10 μg/mL; BD PharMingen, #555557) or isotype control (mouse IgG2a: 10 μg/mL; BD PharMingen, #555571) were used in some cultures of T cells plus iPS-RPE cells. Anti-human CD3 antibody (1 μg/mL; Ancell, #144-020) was also used in the CD4-RPE rejection assay.
Flow Cytometry
Expression of MHC-I (HLA-A, -B, -C) and MHC-II (HLA-DR, -DQ, -DP) on several human iPS-RPE cells and control human cells (ES-RPE cells, ARPE-19 cell lines, primary fetal RPE cells, cornea endothelial cells, fibroblasts, and 836B1 or 454E2 iPSCs) was examined by FACS analysis. Before staining, these cells were incubated with a human Fc block (Miltenyi Biotec) at 4°C for 15 min. The cells were stained with anti-HLA class I antibody (FITC anti-human HLA-A, -B, -C; Sigma-Aldrich, #F 5662), anti-HLA class II antibody (FITC anti-human HLA-DR, -DP, -DQ; DakoCytomation, #F0817 or BD PharMingen, #555558), anti-HLA-DR antibody (FITC; eBioscience, #11-9956), or isotype controls (mouse IgG2a, κ isotype control, FITC; BD PharMingen, #555573 or mouse IgG2b, κ isotype control, FITC; eBioscience, #11-4732) at 4°C for 30 min. iPS-RPE cells or control cells co-cultured with recombinant IFN-γ (100 ng/mL) for 48 hr were also prepared. RPE cells were also treated with other recombinant proteins and supernatants of T cells (agonistic anti-human CD3 antibody-treated activated T cells or naive untreated T cells). We used recombinant human proteins such as IL-1β (50 ng/mL; R&D Systems), IL-2 (100 U/ml; BD Biosciences), IL-6 (50 ng/mL; BD PharMingen), IL-8/CXCL8 (50 ng/mL; R&D Systems), IL-10 (50 ng/mL; R&D Systems), IL-12 (50 ng/mL; R&D Systems), IL-17A/F (50 ng/mL; R&D Systems), IL-21 (100 ng/mL; R&D Systems), IL-22 (50 ng/mL; eBioscience), IL-23 (50 ng/mL; R&D Systems), IL-27 (50 ng/mL; R&D Systems), IFN-γ (0.1–100 ng/mL; R&D Systems), TNF-α (100 ng/mL; R&D Systems), TNFR1 (50 ng/mL; R&D Systems), TGF-β1 (50 ng/mL; R&D Systems), TGF-β2 (50 ng/mL; R&D Systems), GM-CSF (50 ng/mL; R&D Systems), MIG/CXCL9 (50 ng/mL; BD PharMingen), MCP-1/CCL2 (50 ng/mL; R&D Systems), MIP-3α/CCL20 (50 ng/mL; R&D Systems), macrophage migration inhibitory factor (50 ng/mL: R&D Systems), thrombospondin-1 (50 ng/mL: R&D Systems), and lipopolysaccharide (1,000 ng/mL: Sigma-Aldrich).
The expression of Th1 cytokines, such as IFN-γ, IL-2, and TNF-α, in CD4+ T cells exposed to allogeneic iPS-RPE cells was assessed by intracellular staining (BD Cytofix/Cytoperm kits, BD PharMingen) followed by flow cytometry. Before staining for these cytokines, cells were pre-stimulated with T cell-stimulation materials (CytoStim, Miltenyi Biotec) for 1–2 hr, and cells were incubated for an additional 5 hr in the presence of a protein transport inhibitor (BD GolgiStop, BD). After Fc block staining, the T cells were stained with anti-human IFN-γ (PE-labeled; R&D Systems, #25723), anti-human IL-2 (PE-labeled; BD PharMingen, #560902), and anti-human TNF-α (PE-labeled; R&D Systems, #IC210P) at 4°C or room temperature for 30 min. T cells were also stained with anti-human CD4 antibody (APC-labeled; eBioscience, #17-0049) or isotype control antibody (mouse IgG2b, isotype control, PE; R&D Systems, #IC0041P) at 4°C for 30 min. All samples were analyzed on a FACSCanto flow cytometer.
Multiplex Cytokine Array
Supernatants of CD4+ T cells (from TLHD1 or TLHD21 donors) exposed to TLHD1, 454E2, or 453F2 iPS-RPE cells were prepared for multiplex cytokine array assay (Procarta Immunoassay Kit, Filgen). The following 16 factors were measured: IL-1α, IL-1β, IL-1RA, IL-4, IL-5, IL-6, IL-7, IL-12p70, IL-13, IL-15, IL-31, GM-CSF, TNF-α, TNF-β, IFN-α, and IFN-γ. A significant concentration for each cytokine is >10.0 pg/mL, and the undetectable level is <1.0 pg/mL. The assay was performed twice, and the supernatants were measured in triplicate each time that the assay was performed.
qRT-PCR
mRNA expression for Th1-related genes such as T-bet, Stat1, IFN-γ, and CXCR3 in PBMCs in the presence of iPS-RPE cells was evaluated by qRT-PCR. Total RNA was isolated from PBMCs with and without exposure to iPS-RPE cells (453F2 or 836B1). After cDNA synthesis, the expression of Th1-related genes and β-actin in triplicate samples was analyzed by qRT-PCR. The primers were as follows: tgtggtccaagtttaatcagca (left) and gacaggaatgggaacatcc (right) for T-bet; tgagttgatttctgtgtctgaagtt (left) and acacctcgtcaaactcctcag (right) for Stat1; ggcattttgaagaattggaaag (left) and tttggatgctctggtcatctt (right) for IFN-γ; and ccatggtccttgaggtgag (left) and tccatagtcataggaagagctgaa (right) for CXCR3. The probes (Universal Probe Library) were #9 for T-bet, #32 for Stat1, #21 for IFN-γ, and #79 for CXCR3. The results indicate the relative expression of the molecules (ΔΔCt for control cells = 1).
Statistical Evaluation
At least three independent experiments were performed for all in vitro assays. All statistical analyses were performed with the Student's t test (paired or unpaired, as appropriate). Values were considered statistically significant if p was less than 0.05.
Author Contributions
S.S. was the principal investigator who designed and performed experiments, and wrote the manuscript. Y.I. and K.M. cultured and prepared RPE cells and collected blood samples. T.K., T.F., and S.S. performed human HLA analysis. M.T. designed and conceptualized the study, and drafted and edited the manuscript.
Acknowledgments
We appreciate the expert technical assistance of N. Hayashi, K. Iseki, T. Hashiguchi, W. Ohashi, M. Kawahara, C. Yamada, H. Kitajima, and C. Morinaga (Laboratory for Retinal Regeneration, Center for Developmental Biology, RIKEN, Kobe) and Drs. M. Sawada and T. Maeda (Healios KK, Kobe). We appreciate Dr. S. Okamoto (Department of Physiology, Keio University School of Medicine, Tokyo) for the establishment of iPSCs and RPE cells. This work was supported by a Scientific Research Grant (B, 25293357) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, by the Charitable Trust Fund for Ophthalmic Research in Commemoration of Santen Pharmaceutical's Founder, and by a grant from the Project for Realization of Regenerative Medicine from MEXT.
Published: September 15, 2016
Footnotes
Supplemental Information includes seven figures and two tables and can be found with this article online at http://dx.doi.org/10.1016/j.stemcr.2016.08.011.
Supplemental Information
References
- Algvere P.V. Clinical possibilities in retinal pigment epithelial transplantations. Acta Ophthalmol. Scand. 1997;75:1. doi: 10.1111/j.1600-0420.1997.tb00239.x. [DOI] [PubMed] [Google Scholar]
- Algvere P.V., Gouras P., Dafgard Kopp E. Long-term outcome of RPE allografts in non-immunosuppressed patients with AMD. Eur. J. Ophthalmol. 1999;9:217–230. doi: 10.1177/112067219900900310. [DOI] [PubMed] [Google Scholar]
- Clipstone N.A., Crabtree G.R. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Drukker M., Katz G., Urbach A., Schuldiner M., Markel G., Itskovitz-Eldor J., Reubinoff B., Mandelboim O., Benvenisty N. Characterization of the expression of MHC proteins in human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2002;99:9864–9869. doi: 10.1073/pnas.142298299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enzmann V., Kaufmann A., Hollborn M., Wiedemann P., Gemsa D., Kohen L. Effective chemokines and cytokines in the rejection of human retinal pigment epithelium (RPE) cell grafts. Transpl. Immunol. 1999;7:9–14. doi: 10.1016/s0966-3274(99)80013-6. [DOI] [PubMed] [Google Scholar]
- Hamrah P., Zhang Q., Liu Y., Dana M.R. Novel characterization of MHC class II-negative population of resident corneal Langerhans cell-type dendritic cells. Invest. Ophthalmol. Vis. Sci. 2002;43:639–646. [PubMed] [Google Scholar]
- Horie S., Sugita S., Futagami Y., Yamada Y., Mochizuki M. Human retinal pigment epithelium-induced CD4+CD25+ regulatory T cells suppress activation of intraocular effector T cells. Clin. Immunol. 2010;136:83–95. doi: 10.1016/j.clim.2010.03.001. [DOI] [PubMed] [Google Scholar]
- Huber C., Irschick E. Cytokines in the regulation of allograft rejection. Bibl. Cardiol. 1988:103–110. [PubMed] [Google Scholar]
- Imai A., Sugita S., Kawazoe Y., Horie S., Yamada Y., Keino H., Maruyama K., Mochizuki M. Immunosuppressive properties of regulatory T cells generated by incubation of peripheral blood mononuclear cells with supernatants of human RPE cells. Invest. Ophthalmol. Vis. Sci. 2012;53:7299–7309. doi: 10.1167/iovs.12-10182. [DOI] [PubMed] [Google Scholar]
- Juel H.B., Faber C., Udsen M.S., Folkersen L., Nissen M.H. Chemokine expression in retinal pigment epithelial ARPE-19 cells in response to coculture with activated T cells. Invest. Ophthalmol. Vis. Sci. 2012;53:8472–8480. doi: 10.1167/iovs.12-9963. [DOI] [PubMed] [Google Scholar]
- Kamao H., Mandai M., Okamoto S., Sakai N., Suga A., Sugita S., Kiryu J., Takahashi M. Characterization of human induced pluripotent stem cell-derived retinal pigment epithelium cell sheets aiming for clinical application. Stem Cell Rep. 2014;2:205–218. doi: 10.1016/j.stemcr.2013.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita K., Matsumura Y., Sato Y., Okada A., Morizane A., Okamoto S., Hong H., Nakagawa M., Tanabe K., Tezuka K. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- Okita K., Yamakawa T., Matsumura Y., Sato Y., Amano N., Watanabe A., Goshima N., Yamanaka S. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- Percopo C.M., Hooks J.J., Shinohara T., Caspi R., Detrick B. Cytokine-mediated activation of a neuronal retinal resident cell provokes antigen presentation. J. Immunol. 1990;145:4101–4107. [PubMed] [Google Scholar]
- Peyman G.A., Blinder K.J., Paris C.L., Alturki W., Nelson N.C., Jr., Desai U. A technique for retinal pigment epithelium transplantation for age-related macular degeneration secondary to extensive subfoveal scarring. Ophthalmic Surg. 1991;22:102–108. [PubMed] [Google Scholar]
- Streilein J.W. Ocular immune privilege: therapeutic opportunities from an experiment of nature. Nat. Rev. Immunol. 2003;3:879–889. doi: 10.1038/nri1224. [DOI] [PubMed] [Google Scholar]
- Suarez-Alvarez B., Rodriguez R.M., Calvanese V., Blanco-Gelaz M.A., Suhr S.T., Ortega F., Otero J., Cibelli J.B., Moore H., Fraga M.F. Epigenetic mechanisms regulate MHC and antigen processing molecules in human embryonic and induced pluripotent stem cells. PLoS One. 2010;5:e10192. doi: 10.1371/journal.pone.0010192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita S. Role of ocular pigment epithelial cells in immune privilege. Arch. Immunol. Ther. Exp. (Warsz) 2009;57:263–268. doi: 10.1007/s00005-009-0030-0. [DOI] [PubMed] [Google Scholar]
- Sugita S., Usui Y., Horie S., Futagami Y., Aburatani H., Okazaki T., Honjo T., Takeuchi M., Mochizuki M. T-cell suppression by programmed cell death 1 ligand 1 on retinal pigment epithelium during inflammatory conditions. Invest. Ophthalmol. Vis. Sci. 2009;50:2862–2870. doi: 10.1167/iovs.08-2846. [DOI] [PubMed] [Google Scholar]
- Sugita S., Kamao H., Iwasaki Y., Okamoto S., Hashiguchi T., Iseki K., Hayashi N., Mandai M., Takahashi M. Inhibition of T-cell activation by retinal pigment epithelial cells derived from induced pluripotent stem cells. Invest. Ophthalmol. Vis. Sci. 2015;56:1051–1062. doi: 10.1167/iovs.14-15619. [DOI] [PubMed] [Google Scholar]
- Usui Y., Okunuki Y., Hattori T., Kezuka T., Keino H., Ebihara N., Sugita S., Usui M., Goto H., Takeuchi M. Functional expression of B7H1 on retinal pigment epithelial cells. Exp. Eye Res. 2008;86:52–59. doi: 10.1016/j.exer.2007.09.007. [DOI] [PubMed] [Google Scholar]
- Weisz J.M., Humayun M.S., De Juan E., Jr., Del Cerro M., Sunness J.S., Dagnelie G., Soylu M., Rizzo L., Nussenblatt R.B. Allogenic fetal retinal pigment epithelial cell transplant in a patient with geographic atrophy. Retina. 1999;19:540–545. doi: 10.1097/00006982-199911000-00011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.