Abstract
An HPLC method for the assay of an anticancer nucleoside, 4’-thio-2’-deoxycytidine (T–dCyd, NSC 764276), has been developed and validated. The stress testing of T-dCyd was carried out in accordance with ICH guidelines Q1A (R2) under acidic, alkaline, oxidative, thermolytic, and photolytic conditions. The separation of T-dCyd from its impurities and degradation products was achieved in 40 min on a Luna® Phenyl-Hexyl column (150 mm × 4.6 mm i.d., 3 µm) with a gradient elution using ammonium phosphate buffer (pH 3.85) and methanol as the mobile phase. The gradient starts from 2% and ends at 80% of methanol. Detection is by UV at 282 nm. LC-QTOF/MS was used to obtain mass data for characterization of impurities and degradation products. The proposed HPLC assay method was validated for specificity, linearity (concentration range 0.25–0.75 mg/mL, r ≥ 0.9998), accuracy (recovery 98.1–102.0%), precision (RSD ≤ 1.5%), and sensitivity (LOD 0.1 µg/mL). The developed method was suitable for the quality control and stability monitoring of the T-dCyd drug substance.
Keywords: 4’-thio-2’-deoxycytidine, T-dCyd, NSC 764276, forced degradation, HPLC validation, impurity and degradation product characterization
Graphical Abstract
1. Introduction
DNA methyltransferase I (DNMT1) is an enzyme that serves as a maintenance methyltransferase during cell replication and contributes to the hypermethylation of cytosines in 5'—C—phosphate—G—3' (CpG) islets in tumor suppressor genes, resulting in their silencing in a manner analogous to inactivating mutations [1–4]. Downgrading DNMT1 expression by genetic or pharmacological means decreases CpG methylation and, consequently, allows instead the expression of tumor suppressor genes, which diminishes proliferation and cellular transformation [5–7]. Reduction of DNMT1 levels has also been shown to suppress tumorigenesis in a manner independent of its DNA methyltransferase activity [8, 9]. DNMT1 protein stability is significantly increased in cancer cells, regardless of the mechanisms by which DNMT1 causes tumorigenesis [10].
Decitabine (aza-dCyd) and azacitidine (aza-Cyd) are known DNA-hypomethylating nucleosides that deplete DNMT1 and are currently approved for the treatment of myelodysplastic syndromes and other hematologic malignancies [10–13]. Two other nucleoside analogs, 5-F-2′-deoxycytidine (F-dCyd) and zebularine, are reported to deplete DNMT1 in cancer cells [14, 15]. In general, DNMT1 active site occupancy by the DNA-incorporated nucleotide analog is essential to cause DNMT1 trapping and ensuing degradation of the enzyme. The currently available agents have several unfavorable features, such as inefficient incorporation into DNA, DNA synthesis inhibition at high doses, chemical instability of the triazine ring, metabolic instability due to enzymatic deamination, and inhibition of other enzymes not related to DNA methylation [16]. These undesirable characteristics are responsible for decreased hypomethylation efficiency and preclude high-dose administration for extended periods. The continuing search for improved DNMT1-depleting agents has led to the discovery of the sulfur-containing deoxycytidine(dCyd) analog, 4′-thio-2′-deoxycytidine (T-dCyd)[17], which is currently being studied in a Phase I clinical trial [18]. In cell culture, T-dCyd is a substrate for dCyd kinases and the resultant triphosphate is efficiently incorporated into DNA without inhibition of DNA synthesis [19]. Furthermore, treatment with T-dCyd promotes re-expression of p15, a tumor suppressor gene, which is consequential to the depletion of DNMT-1. T-dCyd is cytotoxic to leukemia lines (CCRF-CEM and KG1a), with an IC50 of ca. 1 µM [16]. However, its activity towards solid tumor lines (NCI-H23, HCT-116, and IGROV-1) is negligible.
Unexpectedly, T-dCyd showed promising results in a lung adenocarcinoma xenograft model (NCI-H23). Intraperitoneal (i.p.) dosing at 5 mg/kg (0.56 MTD) in nu/nu mice on a daily dose five days per week for three weeks resulted in tumor stasis with acceptable toxicity. In contrast, the decitabine-treated control arm at MTD resulted in delayed tumor growth but not stasis, and an average of 10% weight loss was noted. Intratumoral levels of DNMT1 were reduced to undetectable levels in xenografts post administration of T-dCyd but were unaffected by decitabine treatment. Mass spectrometry analysis demonstrated incorporation of both T-dCyd and 4’-thiothymidine into cellular DNA [20].
In spite of a number of publications describing the synthesis, efficacy, and pharmacokinetics of T-dCyd, information on quantitative analysis and impurity characterization is lacking in the literature. Several HPLC methods of similar nucleoside analogs, including decitabine [21, 22], zebularine [23], fluorodeoxycytidine [24], 5-iodo-2-pyrimidinone-2’-deoxyribose [25], bromodeoxyuridine [26] and azacitidine [27], were reported in the literature. Most of these nucleosides have moderate to high polarities and were analyzed with C18 columns which were eluted with aqueous buffers containing low contents of methanol or acetonitrile as organic modifiers. Many of the reported methods were developed mostly for bioanalysis with LC/MS detection and not suitable for impurities and degradation products determination in the analysis of drug substances and products. The goal of this study was to develop and validate a stability-indicating HPLC method for T-dCyd in accordance with ICH guideline Q2 (R1) and to identify the impurities present in the drug substance as well as degradation products observed in stress testing. The study was carried out with HPLC and LC coupled with high-resolution mass spectrometry (HRMS).
2. Materials and Methods
2.1. Chemicals and reagents
Three lots of T-dCyd (NSC 764276) drug substance and α-T-dCyd (NSC 776901) were provided by the National Cancer Institute (Bethesda, MD, USA). All the lots were prepared by Alchem Laboratories Corporation, 13305 Rachael Blvd, Alachua, FL 32615. Phosphoric acid and ammonium phosphate monobasic were purchased from Sigma-Aldrich (St. Louis, MO, USA). HPLC-grade methanol (MeOH), hydrogen peroxide (H2O2) 3% solution, and sodium hydroxide (NaOH) were purchased from Mallinckrodt (Paris, KY, USA). Water was purified through a Millipore Super-Q® Pure Water System (Waltham, MA, USA). The solution of hydrochloric acid (HCl) was prepared from DILUT-IT® Analytical Concentrate (J.T. Baker, Phillipsburg, NJ, USA).
2.2. HPLC
An Agilent 1100 HPLC system (Wilmington, DE, USA) equipped with a solvent degasser, quaternary pump, autosampler, and a diode array detector was used in the study. Agilent ChemStation® for LC 3D (Rev. A. 10.01) software was used for instrument operation control and data collection. The HPLC was performed on a Phenomenex, Luna® Phenyl-Hexyl column (150 mm × 4.6 mm i.d., 3 µm, Torrance, CA, USA). The column was held at 25 °C. The mobile phase was a combination of solvent A (10 mM ammonium phosphate monobasic in water adjusted pH to 3.85 with phosphoric acid) and solvent B (methanol). The following gradient program was used: 0–15 min, 2% solvent B and 98% solvent A; 15–20 min, linear gradient to 5% solvent B and 95% solvent A; 20–28 min, linear gradient to 80% solvent B and 20% solvent A; 28–33 min, hold at 80% solvent B and 20% solvent A; 33–34 min, step gradient to 2% solvent B and 98% solvent A; 34–40 min, re-equilibrate at 2% solvent B and 98% solvent A before the next injection. The injection volume was 10 µL. For all gradient segments, the elution flow rate was 1.0 mL/min, and the detection wavelength was set at 282 nm.
LC-MS was performed on an Agilent LC/MS system consisting of an Agilent 1200 binary LC pump, a temperature controlled autosampler, a photodiode array (PDA) UV detector, and a 6530 Accurate Mass Q-TOF mass spectrometer (Wilmington, DE, USA). The mass spectrometer was equipped with a JetStream® electrospray ionization (ESI) probe operating at atmospheric pressure. The ESI source parameter settings were: mass range m/z 100 – 1000, gas temperature 350 °C, gas flow 10 L/min, nebulizer 50 psi, sheath gas temperature 400 °C, sheath gas flow 12 L/min, capillary voltage (Vcap) 3500 V, nozzle voltage 500 V, fragmentor 200 V, skimmer1 65 V, and octopole r.f. (OCT 1 r.f. Vpp) 750 V. Tandem mass spectrometry was performed using ramped collision energy at slope 3 and offset 10. The LC conditions used in the LC-MS system were the same as those used in the Agilent 1100 LC-UV system except that the flow rate was reduced and the gradient time and injection volume were increased to gain ESI-MS sensitivity. The following gradient program was used: 0–30 min, 2% solvent B and 98% solvent A; 30–40 min, linear gradient to 5% solvent B and 95% solvent A; 40–56 min, linear gradient to 80% solvent B and 20% solvent A; 56–66 min, hold at 80% solvent B and 20% solvent A; 66–68 min, linear gradient to 2% solvent B and 98% solvent A; 68–80 min, re-equilibrate at 2% solvent B and 98% solvent A before the next injection. The injection volume was 10 µL. For all gradient segments, the elution flow rate was 0.5 mL/min.
2.3. Sample preparation
The system suitability standard, quality control standard and samples are prepared at a concentration of 0.5 mg/mL by adding 2-mL of water to dissolve accurately weighed amount of approximately 1-mg reference standard or the sample. The calibration standards were prepared at 0.25, 0.375, 0.5, 0.625 and 0.75 mg/mL by adding 2-mL of water to dissolve accurately weighed amounts of approximately 0.5, 0.75, 1, 1.25 and 1.5 mg of the reference standard. The forced degradation samples were prepared as shown in Supplementary Table 1. A stock solution was made by dissolving the T-dCyd solid in water at 1 mg/mL. The solution forced degradation samples were prepared by 1:1 mixing of the stock solution and the various reagents and treating with heat for a period of time (Supplementary Table 1). The solid forced degradation samples were prepared by heating the solid sample or exposing the sample to UV light (Supplementary Table1) and then dissolving the solid in water at 0.5 mg/mL.
3. Results and Discussion
3.1. HPLC method development and validation
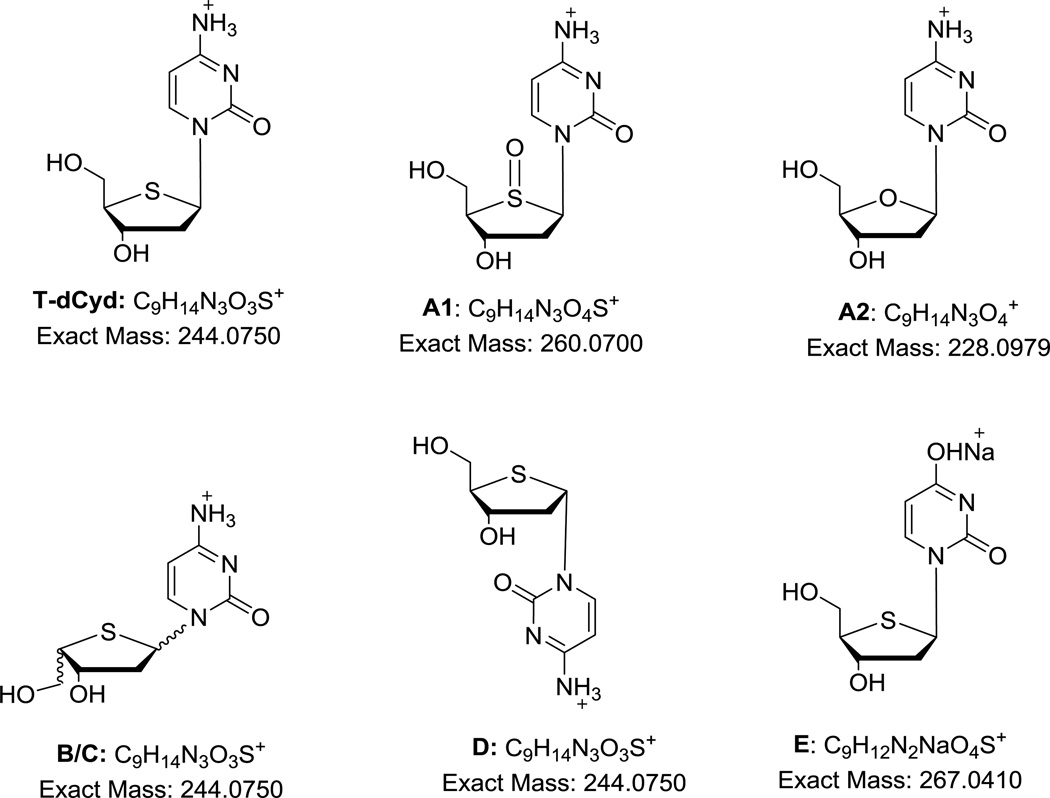
As the chemical structure of T-dCyd, or 4’-thio-2’-deoxycytidine indicates (Figure 1), it is a hydrophilic nucleoside with high polarity. Since reversed-phase liquid chromatography with a phosphate buffer mobile phase is commonly used for analysis of nucleosides, the effort to develop a method for T-dCyd was initiated with standard C18 columns. The following brands and configurations of C18 columns were readily available in our laboratory and were chosen for initial evaluation: Agilent Eclipse XDB C18 (5 µ, 150 × 4.6 mm i.d.), MAC MOD Ace C18 (3 µ, 150 × 4.6 mm i.d.) and Phenomenex Luna® C18 (2) (5 µ, 150 × 4.6 mm i.d.). These C18 columns have different properties (listed in the order of Eclipse, Ace and Luna): particle size (5, 3 and 5µm), pore size (80, 100 and 100Å), surface area (180, 300 and 400 m2/g) and carbon load (10, 15.5 and 17.5%). Thus, they offer variations in polarity, efficiency and separation performance, as presented in the literature [28]. The mobile phase was a combination of phosphate buffer and varying amounts of acetonitrile or methanol. Though the compound can be retained with a reasonable elution time of 4–5 min, the T–dCyd peak was quite broad on these C18 columns (plate number about 2500/column). The appearance of a broad major peak is an indication that the sensitivity of the method would be insufficient to detect impurities at 0.05% target level. Resolution of some of the impurities and degradation products was also marginal. To improve the peak shape and resolution, we examined the Phenomenex Luna® Phenyl-Hexyl column (3 µ, 150 × 4.6 mm) in which the phenyl ring was anticipated to provide substantial π-π interaction with the pyrimidine moiety of the nucleoside. As a result, the major peak was drastically sharpened (plate number about 12,000/column) and sensitivities doubled compared to those observed on the C18 columns. The peak shape was highly sensitive to the pH of the mobile phase: a mobile phase at pH < 3.7 would result in a tailing peak, but at pH > 4.1 a fronting peak was observed. The optimal pH was determined to be around pH 3.85. Using methanol as the organic modifier also provided better retention and peak shape than acetonitrile. The optimal mobile phase was a combination of 10 mM ammonium phosphate buffer at pH 3.85 and methanol. Forced degradation of the drug substance was performed, and the gradient program was optimized to provide the best separation of all impurities and degradation products.
Figure 1.
Structure of T-dCyd and proposed structures of its impurities and degradation products.
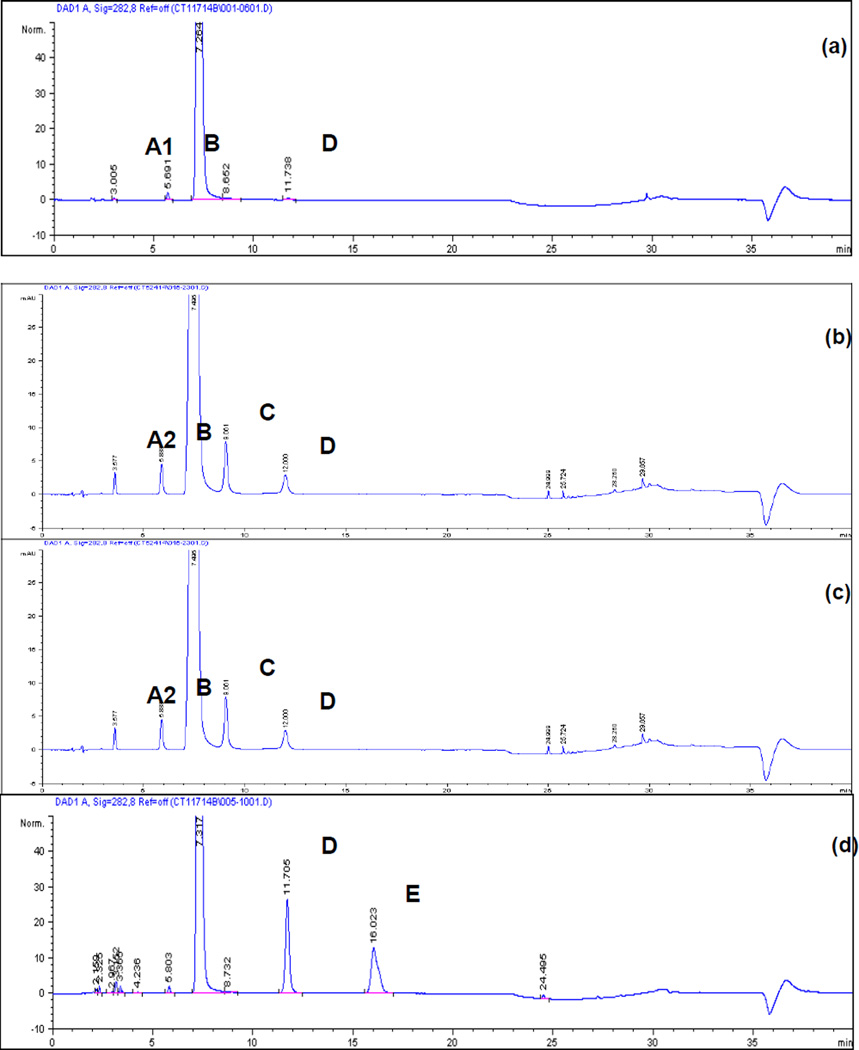
The final method provides a tailing factor of 1.0 for T-dCyd and a resolution greater than 3 for impurities and degradation products peaks from the T-dCyd peak. The optimized LC condition (provided in Section 2.2) was validated in accordance with the current ICH guideline Q2 (R1). A typical HPLC assay chromatogram is presented in Figure 2a. The method’s specificity was verified by a UV peak purity check (Agilent ChemStation® software). Resolution of the peaks preceding and following the major peak in all forced degradation samples was found to be greater than 3.0. Validation was conducted in three days by two analysts with two separate instruments to evaluate intra-day and intermediate precision as recommended by ICH guideline Q2 (R1). The system suitability and validation results are provided in Table 1. Linearity of the method was demonstrated by standard curve in the range of 0.25–0.75 mg/mL. The sample peak area (A, mAU) versus drug concentration (C, mg/mL) was analyzed by linear least square regression (A = Slope × C + Intercept). The linearity range was validated at 50 – 150 % of the target assay concentration, which satisfied the ICH Q2 (R1) requirement for both drug substance and drug product.
Figure 2.
Chromatograms of NSC 764276:
(a) 0.5 mg/mL S/D5 in water, expanded scale.
(b) 0.5 mg/mL S/D9 in water, expanded scale.
(c) 0.5 mg/mL S/D10 in water, expanded scale.
(d) 0.5 mg/mL S/D5 in 0.1 N NaOH, heated at 80°C for 4 hours, expanded scale.
Table 1.
System suitability and validation results.
| Day 1 Analyst 1 Equipment 1 |
Day 2 Analyst 1 Equipment 2 |
Day 3 Analyst 2 Equipment 1 |
||
|---|---|---|---|---|
| System Suitability | ||||
| Retention Time (min) | 7.4 | 7.7 | 8.0 | |
| Retention Time Variation (RSD) |
0.8% | 0.0% | 0.1% | |
| Peak Area Variation (RSD) | 0.3% | 0.1% | 0.6% | |
| USP Tailing Factor | 1.0 | 0.9 | 1.4 | |
| QC Recovery | 99.9–102.7% | 100.0–100.7% | 98.0–98.4% | |
| Resolution to Peak D | 15 | 13 | 14 | |
| Sensitivity (S/N) | 25 | 30 | 26 | |
| Linearity | ||||
| Slope | 25070 | 25076 | 25154 | |
| Intercept | 206.92 | −71.60 | −72.60 | |
| Correlation Coefficient (r) | 0.9998 | 0.9999 | 0.9999 | |
| Back Calculated Conc. Recovery |
99.3–100.8% | 99.3–100.5% | 99.3–100.6% | |
| Accuracy(%Recovery) | ||||
| 0.25mg/mL | 99.7, 99.6, 98.8 | 99.0, 100.1, 101.8 | 98.7, 100.0, 101.7 | |
| 0.5mg/mL | 98.1, 99.1, 100.8 |
100.2, 101.1, 100.2 | 100.7, 102.0, 101.1 |
|
| 0.75mg/mL | 99.3, 99.7, 99.6 | 99.0, 99.0, 98.3 | 99.5, 99.3, 98.9 | |
| Precision(%RSD) | ||||
| 0.25mg/mL | 0.5 | 1.4 | 1.5 | |
| 0.5mg/mL | 1.3 | 0.5 | 0.7 | |
| 0.75mg/mL | 0.2 | 0.4 | 0.3 | |
|
Intra-day precision (%RSD of 9 determinations each day) |
0.7 | 1.1 | 1.2 | |
|
Inter-day precision (%RSD of 27 determinations in 3 days) |
1.1 | |||
Accuracy and precision were established by evaluation of recoveries and RSD values obtained each day with three test solutions each at concentration of 0.25, 0.50, and 0.75 mg/mL corresponding to 50%, 100%, and 150% of the target assay concentration, respectively. Recovery was calculated by comparing the theoretical concentration calculated from each day's calibration curve and the nominal concentration. The accuracy results showed recoveries between 98.1% and 102.0%. Intra-day precision (repeatability) was validated to be no greater than 1.2% RSD from nine determinations on each day, and inter-day precision (intermediate precision) was validated with 1.1% RSD from 27 determinations in three days. The limit of detection and LOQ were shown to be 0.1 µg/mL and 0.25 µg/mL, respectively, using the criteria of signal to noise (S/N) > 3 for LOD and S/N > 10 for LOQ. The sample solution stability was tested and shown to be stable at 5 °C and at room temperature for 7 days with 100.5% and 100.8% recovery, respectively.
Robustness was evaluated with variations of column temperature, mobile phase pH and flow rate. Column efficiency (plate numbers/column), tailing factor and resolutions were evaluated and presented in Table 2. Varying temperature (± 2°C) and flow rate (± 0.1 mL/min) did not affect the method’s performance as gauged by plate number, tailing factor and resolution. However, as discussed in the method development section, pH had a significant effect on the method’s performance. When it deviated from pH 3.85, the plate number, tailing factor and resolution significantly deteriorated. Therefore, the precise pH of the mobile phase must be tightly controlled.
Table 2.
Robustness test results.
| Method | Temperature (°C) |
pH | Flow Rate (mL/min) |
RT (min) |
Plate Number Per Column |
USP Tailing |
Resolution to RRT 1.60 (Impurity D) |
|---|---|---|---|---|---|---|---|
| Standard | 25 | 3.85 | 1.0 | 8.11 | 14246 | 1.57 | 15.33 |
| Robustness 1 | 23 | 3.85 | 1.0 | 8.24 | 14092 | 1.52 | 14.86 |
| Robustness 2 | 27 | 3.85 | 1.0 | 7.70 | 15288 | 1.32 | 14.74 |
| Robustness 3 | 25 | 3.35 | 1.0 | 6.81 | 5310 | 2.28 | 11.25 |
| Robustness 4 | 25 | 4.35 | 1.0 | 8.76 | 11766 | 0.94 | 13.96 |
| Robustness 5 | 25 | 3.85 | 0.9 | 8.08 | 14748 | 1.32 | 14.73 |
| Robustness 6 | 25 | 3.85 | 1.1 | 8.05 | 15257 | 1.30 | 14.76 |
3.2. Characterization of impurities and degradation products by LC/MS
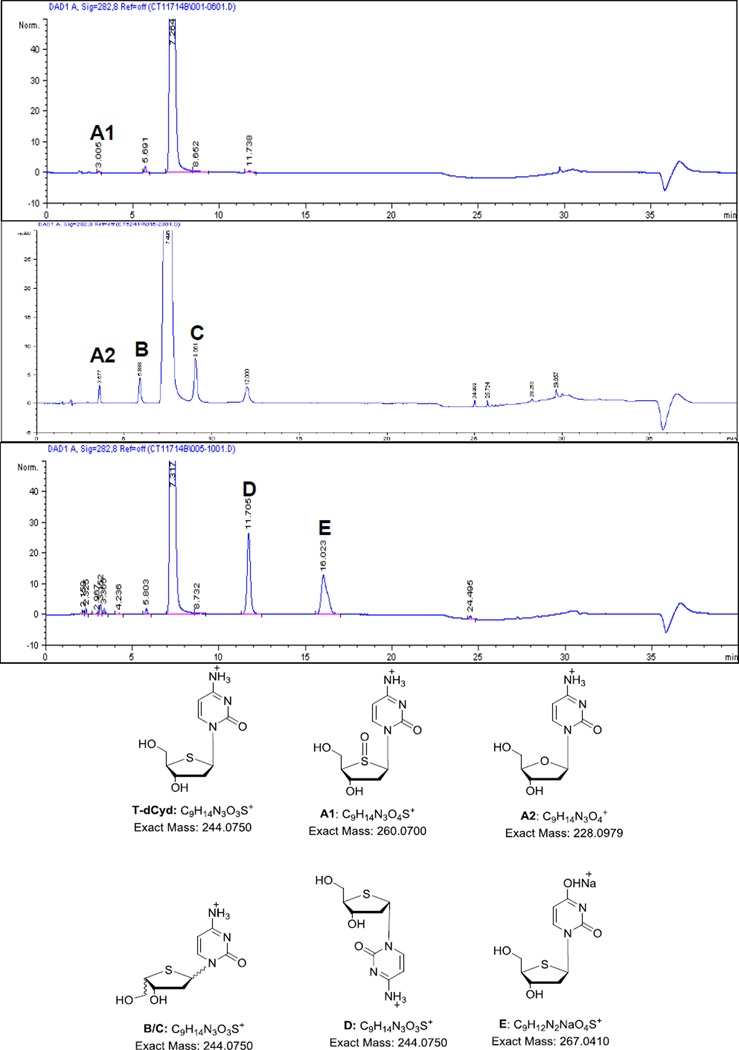
Figure 2a–c shows chromatograms of three drug substance lots –S/D5, –S/D9, and –S/D10. Representative retention times for all detected peaks are listed in Table 3, along with their mass and UV features. Two impurities, A1 and A2, eluted closely between 3–4 min. Sulfoxide A1, also an oxidative degradant of T-dCyd, was observed only in lot –S/D5. Impurity A2, the natural 2’-deoxycytidine, was present in lots –S/D9 and –S/D10. Impurities B, C, and D, which are diastereomers related to T-dCyd, are by-products generated in the manufacturing process. They were found at varying levels in the available lots of T-dCyd.
Table 3.
Summary of chromatographic and spectral data for impurities and degradation products.
| Peak Label | RT (min) |
λmax, nm | Measured m/z |
Calculated m/z |
Error (ppm) |
Area % Lot S/D5 |
Area % Lot S/D9 |
Area % Lot S/D10 |
|---|---|---|---|---|---|---|---|---|
| T-dCyd | 7.5 | 275 | 244.0752 | 244.0750 | −0.82 | 99.71 | 98.27 | 99.02 |
| A1 | 3.0 | 205/275 | 260.0706 | 260.0700 | −2.31 | 0.01 | ND | ND |
| A2 | 3.6 | 270 | 228.0977 | 228.0979 | 0.88 | ND | 0.14 | 0.20 |
| B | 5.9 | 202/270 | 244.0753 | 244.0750 | −1.23 | 0.11 | 0.29 | 0.18 |
| C | 9.1 | 275 | 244.0750 | 244.0750 | 0.00 | 0.12 | 0.78 | 0.26 |
| D, α- T-dCyd | 12.0 | 275 | 244.0750 | 244.0750 | 0.00 | 0.06 | 0.37 | 0.12 |
| E | 16.0 | 205/265 | 267.0412 | 267.0410 | −0.75 | ND | ND | ND |
ND = not detected.
Supplementary Figure 1 presents chromatograms of T-dCyd in various forced degradation solutions. The base-treated sample (Supplementary Figure 1c) shows a degradation product E resulting from the hydrolysis of the amino group in the pyrimidine ring as well as an increased level of impurity D, which was shown to be a related diastereomer. The sulfoxide impurity A1, as expected, is also a major degradation product in hydrogen peroxide-treated samples (Supplementary Figure 1d). The hydrolytic degradant, E, was also observed in a heated aqueous sample solution, with or without acid (Supplementary Figures 1a and 1b).
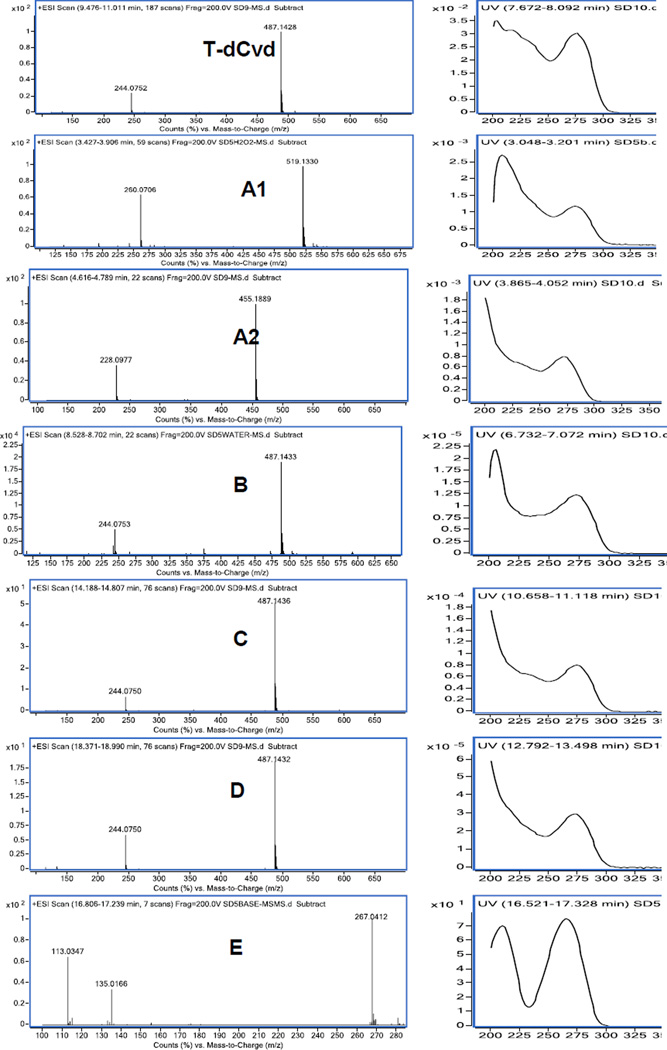
LC/MS was performed to characterize the major impurities and degradation products. Mass and UV spectra were obtained through LC with MS and PDA detections and are provided in Figure 3. Dimers and sodium adducts of many impurities are commonly observed in their mass spectra. Based on the high-resolution accurate mass data and UV data, the proposed chemical structures of the major impurities and degradation products are provided in Figure 1. The measured masses are within 3 ppm of the calculated masses of all proposed compounds (Table 3).
Figure 3.
Mass and UV spectra of T-dCyd and peaks A1, A2, B, C, D, and E.
MS/MS was performed to further characterize impurities and degradation products. The available MS/MS spectra are given in Supplementary Figure 2. MS/MS fragment interpretations are given in Supplementary Figures 3. A common fragment of m/z 112.0505 was observed in T-dCyd and all impurities and degradation products except for E. This fragment represents the cytosine aglycon and indicated the pyrimidine base was intact in all detected compounds except for E. The corresponding fragment for compound E was 113.0346 (or sodium adduct of m/z 135.0165), confirming the conversion of the amino group to a hydroxyl group. Another common fragment m/z 133.0318, observed in T-dCyd and compounds B, C, and D, was derived from their sugar moieties. Compounds B, C, and D have both fragments (m/z 112.0505 and 133.0318) identical to the two fragments of T-dCyd, confirming that they were stereoisomers. From the fragments of m/z 150.0345/131.0161 in A1, and m/z 117.0546 in A2, it was reasonable to conclude that compounds A1 and A2 differ from T-dCyd in the structure of the sugar moieties.
Identification of impurity D
Peak D (RRT 1.60) is an impurity observed in all three lots of drug substance. Accurate mass and MS/MS fragments suggest that it is an isomer of T-dCyd. Its identity was confirmed as the α-isomer of 4’-thio-2’-deoxycytidine (α-T-dCyd) by spiking with an authentic sample (Supplementary Figures 4 and 5).
3.3. Drug substance purity and stability study
The validated method was used in the GMP material release analysis of three lots of T-dCyd drug substance. The purity results (reported by peak area percentage) are provided in Table 3. The validated method was also used for the ICH stability study of the T-dCyd drug substance. The stability results, including assay, purity, and water content, are given in Supplementary Table 2. The stability results indicate that the compound is chemically stable under recommended storage conditions.
Conclusion
An HPLC assay method useful in the quality control of the drug substance and product has been developed and validated for T-dCyd. The HPLC method separates T-dCyd from its impurities and forced degradation products. Identities of the impurities and degradation products have been elucidated by mass spectral data. One major impurity was identified as the α-isomer of 4’-thio-2’-deoxycytidine (α– T–dCyd) by spiking with an authentic sample. The assay method has been validated to be specific, linear (r ≥ 0.9998), accurate (recovery 98.1–102.0%), precise (RSD ≤ 1.5%), and sensitive (LOD 0.1 µg/mL).
Supplementary Material
Highlights.
An HPLC method for a potent antitumor nucleoside T-dCyd was developed
The HPLC method was validated to be stability indicating per ICH guidelines
A total of six related compounds were characterized through LC/MS
One related compound was identified by spiking of the authentic compound.
Acknowledgments
This work was funded in whole or in part by Federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261201200028C. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001;61:3225–3229. [PubMed] [Google Scholar]
- 2.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baylin SB, Ohm JE. Epigenetic gene silencing in cancer— a mechanism for early oncogenic pathway addiction? Nat. Rev. Cancer. 2006;6:107–116. doi: 10.1038/nrc1799. [DOI] [PubMed] [Google Scholar]
- 4.Ooi SK, O’Donnell AH, Bestor TH. Mammalian cytosine methylation at a glance. J. Cell Sci. 2009;122:2787–2791. doi: 10.1242/jcs.015123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robert MF, Morin S, Beaulieu N, Gauthier F, Chute IC, Barsalou A, MacLeod AR. DNMT1 is required to maintain CpG methylation and aberrant gene silencing in human cancer cells. Nat. Genet. 2003;33:61–65. doi: 10.1038/ng1068. [DOI] [PubMed] [Google Scholar]
- 6.Laird PW, Jackson-Grusby L, Fazeli A, Dickinson SL, Jung WE, Li E, Weinberg RA, Jaenisch R. Suppression of intestinal neoplasia by DNA hypomethylation. Cell. 1995;81:197–205. doi: 10.1016/0092-8674(95)90329-1. [DOI] [PubMed] [Google Scholar]
- 7.Belinsky SA, Klinge DM, Stidley CA, Issa JP, Herman JG, March TH, Baylin SB. Inhibition of DNA methylation and histone deacetylation prevents murine lung cancer. Cancer Res. 2003;63:7089–7093. [PubMed] [Google Scholar]
- 8.Milutinovic S, Knox JD, Szyf M. DNA methyltransferase inhibition induces the transcription of the tumor suppressor p21(WAF1/CIP1) J. Biol. Chem. 2000;275:6353–6359. doi: 10.1074/jbc.275.9.6353. [DOI] [PubMed] [Google Scholar]
- 9.Milutinovic S, Brown SE, Zhuang Q, Szyf M. DNA methyltransferase 1 knock down induces gene expression by a mechanism independent of DNA methylation and histone deacetylation. J. Biol. Chem. 2004;279:27915–27927. doi: 10.1074/jbc.M312823200. [DOI] [PubMed] [Google Scholar]
- 10.Agoston AT, Argani P, Yegnasubramanian S, De Marzo AM, Ansari-Lari MA, Hicks JL, Davidson NE, Nelson WG. Increased protein stability causes DNA methyltransferase 1 dysregulation in breast cancer. J. Biol. Chem. 2005;280:18302–18310. doi: 10.1074/jbc.M501675200. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Agoston AT, Atadja P, Nelson WG, Davidson NE. Inhibition of histone deacetylases promotes ubiquitin-dependent proteasomal degradation of DNA methyltransferase 1 in human breast cancer cells. Mol. Cancer Res. 2008;6:873–883. doi: 10.1158/1541-7786.MCR-07-0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghoshal K, Datta J, Majumder S, Bai S, Kutay H, Motiwala T, Jacob ST. 5-Azadeoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol. Cell Biol. 2005;25:4727–4741. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 13.Patel K, Dickson J, Din S, Macleod K, Jodrell D, Ramsahoye B. Targeting of 5-aza-2′-deoxycytidine residues by chromatin-associated DNMT1 induces proteasomal degradation of the free enzyme. Nucleic Acids Res. 2010;38:4313–4324. doi: 10.1093/nar/gkq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beumer JH, Parise RA, Newman EM, Doroshow JH, Synold TW, Lenz HJ, Egorin MJ. Concentrations of the DNA methyltransferase inhibitor 5-fluoro-2′-deoxycytidine (FdCyd) and its cytotoxic metabolites in plasma of patients treated with FdCyd and tetrahydrouridine. Cancer Chemother. Pharmacol. 2008;62:363–368. doi: 10.1007/s00280-007-0603-8. [DOI] [PubMed] [Google Scholar]
- 15.Marquez VE, Barchi JJ, Jr, Kelley JA, Rao KV, Agbaria R, Ben-Kasus T, Cheng JC, Yoo CB, Jones PA. Zebularine: a unique molecule for an epigenetically based strategy in cancer chemotherapy. The magic of its chemistry and biology. Nucleosides, Nucleotides Nucleic Acids. 2005;24:305–318. doi: 10.1081/ncn-200059765. [DOI] [PubMed] [Google Scholar]
- 16.Thottassery JV, Sambandam V, Allan PW, Maddry JA, Maxuitenko YY, Tiwari K, Hollingshead M, Parker WB. Novel DNA methyltransferase-1 (DNMT1) depleting anticancer nucleosides, 4′-thio-2′-deoxycytidine and 5-aza-4′-thio-2′- deoxycytidine. Cancer Chemother. Pharmacol. 2014;74:291–302. doi: 10.1007/s00280-014-2503-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Secrist JA, III, Tiwari KN, Riordan JM, Montgomery JA. Synthesis and biological activity of 2’-deoxy-4’-thio pyrimidine nucleosides. J. Med. Chem. 1991;34:2361–2366. doi: 10.1021/jm00112a007. [DOI] [PubMed] [Google Scholar]
- 18.National Cancer Institute. Phase I trial of 4’-thio-2’-deoxycytidine (TdCyd) in patients with advanced solid tumors. ClinicalTrial.gov, Identifier: NCT02423057.
- 19.Parker WB, Shaddix SC, Rose LM, Waud WR, Shewach DS, Tiwari KN, Secrist JA., III Metabolism of 4′-thio-beta-D-arabinofuranosylcytosine in CEM cells. Biochem. Pharmacol. 2000;60:1925–1932. doi: 10.1016/s0006-2952(00)00520-7. [DOI] [PubMed] [Google Scholar]
- 20.Kinders R, Hollingshead M, Thottassery J, Parker W, Pfister T, Anderson L, Tomaszewski J, Collins J, Doroshow J. Pre-clinical development of 4'-thio-2'-deoxycytidine (T-dCyd) as a DNA-demethylating agent for use in treating solid tissue tumors. Proceedings of the 105th Annual Meeting of the American Association for Cancer Research; 2014 Apr 5–9; San Diego, CA. [Google Scholar]
- 21.Hepsiba G, Teja D, Kumar K, Reddy Y, Panigrahi U, Reddy t. Stability indicating RPHPLC method development and validation of decitabine drug in formulation. Int. J. Pharm. Tech. Res. 2011;3:237–243. [Google Scholar]
- 22.Das H. Analytical method development and validation of decitabine in API and bulk dosage forms by using rapid and sensitive RP-HPLC. Int. J. Med. Nanotech. 2014;2:54–63. [Google Scholar]
- 23.Beumer J, Joseph E, Egorin M, Covey J, Eiseman J. Quantitative determination of zebularine (NSC 309132), a DNA methyltransferase inhibitor, and three metabolites in murine plasma by high-performance liquid chromatography coupled with on-line radioactivity detection. J Chromatogr B Analyt Technol Biomed Life Sci. 2006;831:147–55. doi: 10.1016/j.jchromb.2005.11.048. [DOI] [PubMed] [Google Scholar]
- 24.Beumer J, Eiseman J, Parise R, Joseph E, Holleran J, Covey J, Egorin M. Pharmacokinetisc, metabolism, and oral bioavailability of the DNA methyltransferase inhibitor 5-fluoro-2'-deoxycytidine in mice. Clin. Cancer Res. 2006;12:7483–7491. doi: 10.1158/1078-0432.CCR-06-1250. [DOI] [PubMed] [Google Scholar]
- 25.Kummar S, Anderson L, Hill K, Majerova E, Allen D, Horneffer Y, Ivy S, Rubistein L, Harris P, Doroshow J, Collins J. First-in-human phase 0 trial of oral 5-iodo-2-pyrimidinone-2’-deoxyribose in patients with advanced malignancies. Clin. Cancer Res. 2013;19:1852–1857. doi: 10.1158/1078-0432.CCR-12-3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheung A, He J, Ha Y. New hydrolytic products detected in aqueous solution of bromodeoxyuridine. J. Chromatogr. A. 1998;797:283–293. [Google Scholar]
- 27.Marineni B, Reddy T. Development and validation of stability-indicating RP-HPLC assay method for azacitidine and its bulk drug. Int. J. Pharmacy and Pharmaceutical Sci. 2014;6:240–244. [Google Scholar]
- 28.Mac Moc Analytic Inc. Comparison Guide to C18 Reversed Phase HPLC Columns. 2008 Jun; [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.