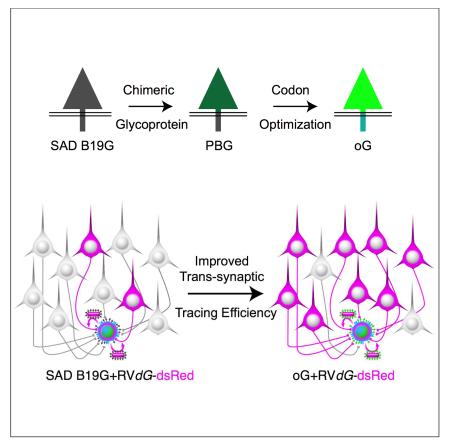
SUMMARY
Monosynaptic rabies virus tracing is a unique and powerful tool to identify neurons making direct presynaptic connections onto neurons of interest across the entire nervous system. Current methods utilize complementation of glycoprotein gene-deleted rabies of the SAD B19 strain with its glycoprotein, B19G, to mediate retrograde trans-synaptic spread across a single synaptic step. In most conditions this method labels only a fraction of input neurons and would thus benefit from improved efficiency of trans-synaptic spread. Here we report newly engineered glycoprotein variants to improve trans-synaptic efficiency. Among them, oG (optimized Glycoprotein) is a codon-optimized version of a chimeric glycoprotein consisting of the transmembrane/cytoplasmic domain of B19G and the extracellular domain of rabies Pasteur virus strain glycoprotein. We demonstrate that oG increases the tracing efficiency for long-distance input neurons up to 20-fold compared to B19G. oG-mediated rabies tracing will therefore allow identification and study of more complete monosynaptic input neural networks.
INTRODUCTION
Since its introduction in 2007, monosynaptic neural circuit tracing using glycoprotein (G)-deleted rabies virus (RVdG) has been a powerful and widely adopted tool for the study of neural circuit organization and function (Callaway and Luo, 2015; Wickersham et al., 2007b). Traditionally the intact rabies virus has been used as a retrograde trans-synaptic tracer (Callaway, 2008; Ugolini, 2008). However, since the intact rabies spreads across multiple synapses, it was challenging to identify clear input and output relationships between rabies labeled neurons. By using RVdG and pseudotyping it with EnvA, it is possible to target rabies virus infection to specific starter cell types of interest or even single neurons, and to trans-synaptically label only their direct presynaptic inputs throughout the mammalian brain (Wickersham et al., 2007b). With this approach, G, which is essential for the trans-synaptic spread of rabies virus, is deleted from the rabies genome and replaced by a transgene such as GFP. Consequently, RVdG infection is unable to spread except when G is provided in trans. Following trans-complementation, RVdG can utilize G to assemble functional rabies virus particles (G+RVdG) that can spread retrogradely across synapses and infect directly presynaptic neurons. The RVdG labels these presynaptic neurons by expressing transgenes of interest, such as GFP, but is unable to spread further due to the absence of G. Due to its ability to spread across only one synaptic step in the retrograde direction, RVdG has been used widely to delineate brain-wide monosynaptic connectivity to populations of neurons in both the central and peripheral nervous system (Callaway and Luo, 2015).
One important limitation of current monosynaptic rabies tracing is that only a fraction of presynaptic neurons are labeled (Callaway and Luo, 2015). Several lines of evidence suggest that G is a limiting factor (Callaway and Luo, 2015; Miyamichi et al., 2013); for example, increasing levels of G expression by either using a stronger promoter or using a cassette that expresses only G both result in improved tracing efficiency (Miyamichi et al., 2013; Watabe-Uchida et al., 2012). Therefore, improvements in labeling might be achieved by increasing the efficiency of packaging of G into rabies virus particles, improving the uptake of rabies particles by presynaptic neurons, or increasing levels of G expression in starter neurons. Improved efficiency of trans-synaptic spread using G-deleted rabies virus of the CVS-N2C strain is consistent with these possibilities (Reardon et al., 2016). Here we attempt to optimize all three of these factors. We designed additional rabies glycoproteins and tested their efficiency for transcomplementation and labeling of presynaptic neurons. We then codon optimized the best of these glycoproteins to maximize expression levels. This modified rabies glycoprotein, oG, significantly improves the efficiency of long distance trans-synaptic tracing with SAD B19 RVdG.
RESULTS
To improve the current monosynaptic rabies virus tracing system, we designed and engineered rabies glycoprotein variants, different from the original B19G. Various strains of rabies virus display different degrees of virulence and pathogenicity associated with synaptic spread (Schnell et al., 2010). Previous studies showed that retrograde infectivity and trans-synaptic spread could be improved in rabies glycoprotein pseudotyped lentivirus or HEP-Flury dG rabies system by using G from different strains of rabies (Kato et al., 2011b; Mori and Morimoto, 2014). Because G has a modular structure with distinct cytoplasmic and extracellular domains, we reasoned that efficiency of synaptic uptake would be most influenced by the extracellular domain, but packaging efficiency would be dependent on the cytoplasmic domain. To maintain optimized packaging efficiency, we therefore engineered chimeric glycoproteins that incorporated the B19G cytoplasmic domain (to match the SAD B19 RVdG) and the extracellular domain from G of other strains of rabies virus. NBG (N2C G and B19G chimeric protein) is a variant incorporating the extracellular domain from the challenge virus standard strain, CVS-N2C, whereas PBG (Pasteur strain G and B19G chimeric protein) is a variant incorporating the extracellular domain from the Pasteur virus strain of rabies (Kato et al., 2011a) (Figures 1A-B). If uptake efficiency by presynaptic terminals is derived from differences in amino acid sequences of extracellular domains between three strains of rabies (Figure 1A), trans-synaptic efficiency should be improved by incorporating extracellular domains from other strains having different pathogenicities. After initial assessment of trans-synaptic spreading efficiency, we decided to improve PBG further to produce oG (optimized Glycoprotein) via codon-optimization (Figures 1B and S1).
Figure 1. Rabies virus glycoprotein from B19 strain and its three engineered variants.
(A) Multiple protein sequence alignment for B19G, PBG, and NBG. ’*’ indicates a single fully conserved amino acid residue. ’:’ indicates conservation between amino acid groups of strongly similar properties, whereas ’.’ indicates conservation between amino acid groups of weakly similar properties (the Gonnet PAM 250 matrix score >0.5 and =<0.5, respectively; (Sievers et al., 2011)). (B) Schematic representation illustrating glycoprotein from B19 strain and three other chimeric variants: NBG, PBG and oG. Yellow highlighted sequences indicate stalk domain, red indicate transmembrane domain, and green indicate cytoplasmic domain of glycoproteins.
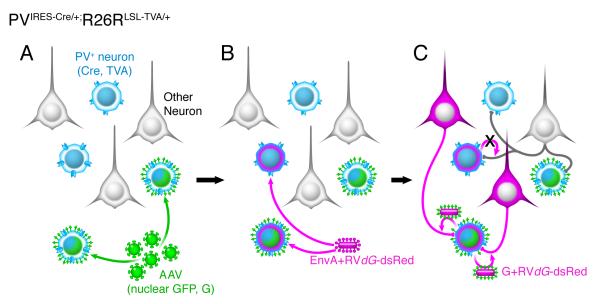
We made direct comparisons of in vivo trans-synaptic labeling efficiency using the three chimeric glycoproteins, NBG, PBG, oG, or the original B19G. We quantified how many long-distance presynaptic neurons per postsynaptic neuron in known circuits were traced using RVdG in combination with each glycoprotein variant. First, to utilize Cre recombinase technology to label and express genes in specific cell types, we developed AAV helper virus expressing glycoprotein and nuclear GFP (H2B-GFP) with the FLEX cassette in a Cre dependent manner (AAV-FLEX-H2B-GFP-2A-G). Co-expression of nuclear GFP and glycoprotein enables us to label and count the exact numbers of post-synaptic starter neurons (RVdG-infected cells that also express G in trans), unambiguously. We chose to examine retrograde labeling in two thalamic areas, the dorsal lateral geniculate nucleus (dLGN) and lateral posterior nucleus (LP), that provide long-distance inputs to Parvalbumin+ (PV+) interneurons in primary visual cortex (V1) of transgenic mouse, PVIRES-Cre/+;R26LSL-TVA/+. In PVIRES-Cre/+;R26LSL-TVA/+ mice, Cre recombinase and the avian viral receptor TVA are expressed only in PV-expressing neurons, including PV-expressing inhibitory neurons in V1 (Figure 2A). At postnatal day 38-78 (P38-78), we injected AAV-FLEX-H2B-GFP-2A-G into V1 of PVIRES-Cre/+;R26LSL-TVA/+ mice. Although different versions of AAV-FLEX-H2B-GFP-2A-G were used, each expressing a different glycoprotein variant, all injections were matched for viral titer (1.5×1012 GC/ml, except for oG which was 1.15×1012 GC/ml) and injection volume (100 nl). Only AAV-infected PV+ interneurons in V1 express nuclear GFP, glycoprotein, and TVA (blue/green neurons, Figure 2A). Two or three weeks later, we injected 200 nl of EnvA-pseudotyped dsRed expressing rabies virus (EnvA+RVdG-dsRed) into the same location (Figure 2B). EnvA-pseudotyped rabies can infect only neurons expressing its cognate receptor, TVA (PV+, TVA+ neurons) and primary infection is therefore restricted to PV+ neurons. PV+ neurons infected by both AAV helper and EnvA+RVdG-dsRed are designated as starter neurons and uniquely express both dsRed and nuclear GFP. In starter neurons, RVdG-dsRed can assemble functional virus particles due to trans-complementation with G and these particles can then spread monosynaptically and retrogradely (Figure 2C). Seven days after rabies virus injections, we harvested, sectioned, and stained the brains. dsRed+ input neurons and dsRed+ GFP+ starter neurons were analyzed in every section of thalamic regions and V1 for quantification, respectively.
Figure 2. Monosynaptic rabies virus tracing of inputs to PV interneurons.
Schematic illustration of the scheme for virus infection and spread of rabies virus from PV interneurons in primary visual cortex (V1) of PVIRES-Cre/+;R26LSL-TVA/+ mouse. (A) PVIRES-Cre/+;R26LSL-TVA/+ mice were injected in V1 with AAV-FLEX-H2B-GFP-2A-GWPRE. All PV interneurons express TVA (blue) and those that are starter cells also express nuclear GFP (H2B-GFP) and rabies glycoprotein (G). (B) After two or three weeks, EnvA+RVdG-dsRed was injected into the same site in V1. EnvA+RVdG-dsRed infects TVA-expressing PV interneurons including starter cells. (C) After one week, monosynaptic input neurons to PV starter neurons were labeled with dsRed+ due to retrograde trans-synaptic spread of rabies virus.
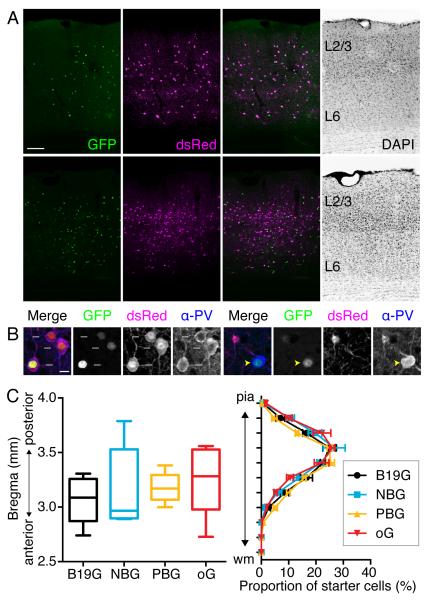
In all mouse brains included for analysis (Table S1), we observed that nuclear GFP and dsRed neurons were spread across the cortical layers within V1. Figure 3A shows examples of experiments in which B19G or oG were expressed in starter cells. We counted GFP+ dsRed+ neurons as PV+ starter neurons (Figure 3B, white arrows in left four panels). Neither GFP+ dsRed− nor GFP− dsRed+ neurons were counted as starter neurons, since lack of either GFP or dsRed indicates either the absence of G or the absence of rabies infection in those cells (Figure 3B, right four panels). We do not observe nuclear GFP expression in PV− cells, indicating that nuclear GFP and G expression are tightly controlled and only expressed in the PV-Cre+ neurons (Figure S2A). As a further control to test for possible “leak” expression of TVA in R26LSL-TVA/+ mouse neurons in the absence of Cre, we injected EnvA+RVdG-GFP or EnvA+RVdGH2B-mCherry into the visual cortex of PVIRES-Cre/+;R26LSL-TVA/+ or Tlx3-Cre/+; R26LSL-TVA/+ mice, respectively. Tlx-3 Cre mice express Cre only in cortical layer 5 pyramidal neurons (Gerfen et al., 2013). In both control experiments, fluorescent reporters expressed from the rabies genome were restricted to the Cre+ neurons, indicating that expression of TVA in the absence of Cre, if any, was insufficient to mediate infection with the EnvA+RVdG (Figure S2B-C). The locations of starter neurons are similar between all the experimental groups, since the coordinates of injection sites were the same in all brains; all GFP+ dsRed+ starter neurons are restricted to V1. The positions of starter neuron populations along the anterior-posterior axis of the brains are also similar (median 3.09 mm for B19G, 2.97 mm for NBG, 3.17 mm for PBG, and 3.28 mm for oG, Kruskal-Wallis test with Dunn's multiple comparisons test as a post hoc, p=0.5288, Figure 3C left panel). Along the dorso-ventral axis from pia to white matter, the distributions of starter neurons are also similar between the four groups (Kruskal-Wallis test with Dunn's multiple comparisons test as a post hoc, p=0.9992, Figure 3C right panel). In addition, we did not observe any effects of the ages of animals when AAV was injected, the timing between AAV and rabies virus injections, or the numbers of starter neurons on viral tracing efficiency (Pearson correlation analysis, Figure S3).
Figure 3. PV starter neuron distributions in mouse V1.
(A) Coronal sections of mouse V1 show starter neurons expressing both GFP from AAV-FLEX-H2B-GFP-2A-G and dsRed from EnvA+RVdG-dsRed. Top panels illustrate a mouse V1 section when B19G was used, whereas bottom panels illustrate use of oG. (B) Confocal images showing PV starter neurons (left panels, white arrows indicate GFP+ dsRed+ α-PV+ neurons) and PV+ non-starter neurons (right panels, yellow arrowheads indicate GFP+ α-PV+, but dsRed− neurons) (C) Starter cell distributions. (left panel) the median position of starter neuron populations expressed as center of gravity along the anterior-posterior axis for each of the four experimental groups labeled according the glycoprotein used for transcomplementation. Values (mm) indicate distances posterior to bregma. Boxes extend from the 25th to 75th percentile. Whiskers indicate ranges from smallest to largest values. There are no statistical differences between groups (Kruskal-Wallis test with Dunn’s multiple comparisons test, p=0.5288). (right panel) Distributions of starter neuron proportions plotted according to distance from pia to white matter (wm) (Kruskal-Wallis test with Dunn’s multiple comparisons test, p=0.9992). The values are reported as mean ± SEM. Scale bars = 100 μm (A), 10 μm (B).
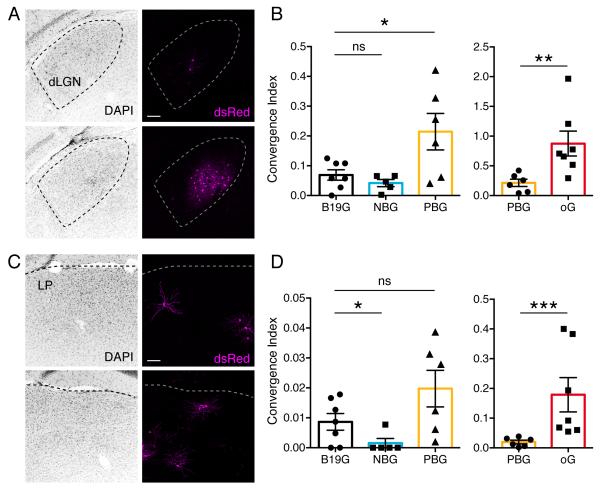
Next we compared trans-synaptic efficiency of rabies tracing using B19G, NBG and PBG. For most PVIRES-Cre/+;R26LSL-TVA/+ brains where we find GFP+ dsRed+ starter neurons in V1, we observe that both dLGN and LP are prominent thalamic structures where long-distance inputs are reliably labeled (Figures 4A and B). To assess the trans-synaptic spreading efficiency to dLGN and LP, we calculated the convergence index for each animal (Miyamichi et al., 2013). The convergence index is calculated as the number of dsRed+ input neurons divided by the number of GFP+ dsRed+ starter neurons. In dLGN, animals in which PBG was used for trans-complementation displayed significantly higher convergence indices compared to B19G (0.214 ± 0.06 vs. 0.069 ± 0.02 respectively, Mann-Whitney test, p<0.05), whereas the convergence index for NBG was not higher than that of B19G (0.042 ± 0.01 vs. 0.069 ± 0.02 respectively, Mann-Whitney test; Figure 4B). In LP, animals in which PBG was used for transcomplementation had a 2-fold higher convergence index than with B19G (0.020 ± 0.01 vs. 0.009 ± 0.01; Figure 4D) but this difference was not statistically significant (Mann-Whitney test, p=0.1113). The convergence index with NBG was significantly lower than for B19G (0.001± 0.00 vs. 0.009 ± 0.01; Mann-Whitney test, p<0.05; Figure 4D).
Figure 4. Efficiency of trans-synaptic spread following trans-complementation with different glycoproteins.
Coronal sections showing dsRed+ monosynaptic input neurons in dLGN (A) and LP (C) to PV interneurons in V1 when using B19G (top panels) and oG (bottom panels). (B and D) Convergence indices for long-distance presynaptic inputs in dLGN (B) and LP (D) when using B19G, NBG, PBG, or oG rabies glycoproteins. Values are reported as mean ± SEM. Statistics were calculated from Mann-Whitney test for non-parametric comparisons. Significant differences between pairs are indicated by the p value. *p < 0.05, **p < 0.01, ***p < 0.001. Abbreviations are: dLGN, dorsal lateral geniculate nucleus; LP, lateral posterior thalamic nucleus. Scale bars = 100 μm (A, C).
Based on these comparisons we conclude that PBG mediates trans-synaptic spread more efficiently than B19G, but NBG does not. We therefore aimed to further improve trans-synaptic spread by increasing PBG expression levels via codon optimization. We named the codon optimized PBG, oG (optimized glycoprotein). We expected that optimizing factors related to codon usage, including codon usage bias, GC content, and undesirable cis-acting elements for Mus musculus, would increase the expression level of glycoprotein by at least two-fold in our mouse system (Genscript). We conducted in vivo experiments using oG for trans-complementation and quantified the convergence indices for dLGN and LP thalamic inputs onto V1 PV+ neurons as described above. Notably, comparisons between PBG and oG showed that oG trans-complementation resulted in significantly higher convergence indices in both dLGN (0.214 ± 0.06 vs. 0.875 ± 0.21 for PBG and oG respectively, Mann-Whitney test, p < 0.01) and LP (0.020 ± 0.01 vs. 0.179 ± 0.06 for PBG and oG respectively, Mann-Whitney test, p < 0.001; Figures 4B and D, right panels).
When comparing convergence indices for oG versus the original glycoprotein, SAD B19G, the difference is even more striking, demonstrating an improvement of more than 12-fold for LGN labeling (0.875 ± 0.21 vs. 0.069 ± 0.02 for oG and SAD B19G) and nearly 20-fold for LP labeling (0.179 ± 0.06 vs. 0.009 ± 0.01 for oG and SAD B19G). In conclusion, our results demonstrate that oG is the most efficient of the glycoproteins tested and increases the efficiency of trans-synaptic spread by more than an order of magnitude compared to SAD B19G.
DISCUSSION
In this report, we investigated whether newly generated rabies glycoprotein variants can improve the current monosynaptic rabies tracing system by labeling a greater proportion of the neurons that are presynaptic to starter neurons. Our results demonstrated that a glycoprotein consisting of the extracellular domain of Pasteur strain glycoprotein and the transmembrane/cytoplasmic domain of SAD B19 glycoprotein (PBG), improves trans-complementation and trans-synaptic labeling by up to three-fold relative to the SAD B19 glycoprotein. Furthermore, oG, a codon-optimized version of PBG, resulted in an additional four- to nine-fold improvement for an overall improvement of up to 20-fold relative to the original SAD B19 glycoprotein. This reagent therefore provides a more powerful means to identify and study brain-wide presynaptic inputs to neurons of interest.
To improve G-deleted rabies tracing efficiency, we first compared glycoproteins derived from different rabies strains before then using codon optimization to increase the expression levels of the best glycoprotein. Based on previous examples of successful improvements in various applications, particularly for lentiviruses pseudotyped with rabies glycoproteins (Hirano et al., 2013; Kato et al., 2011a; Kato et al., 2011b), we chose to investigate chimeric glycoproteins based on the CVS-N2C and Pasteur rabies viruses. In the case of Pasteur strain-derived G (PBG), we observed significant improvement of trans-synaptic efficiency. In contrast, we did not observe any statistically significant improvement when using CVS-N2C-derived G (NBG). There are several possible reasons for this difference. First, it is possible that CVS-N2C G does not mediate uptake at axon terminals as efficiently as the Pasteur virus G. But it is also possible that either the chimeric CVS-N2C G (NBG) does not interact efficiently with the SAD B19 viral core, despite incorporation of the SAD B19 transmembrane/cytoplasmic domain, or that the NBG chimeric protein compromises the native ability of CVS-N2C G to mediate infection of axon terminals. This last possibility is considered further (see below) in the context of a recent study utilizing the CVS-N2C strain of RVdG for trans-synaptic tracing (Reardon et al., 2016). It should be also noted that glycoprotein variants derived from other untested rabies virus strains might enhance the trans-synaptic efficiency even more effectively. Future studies should investigate other strains that have not been included in this study. It also remains to be understood by which mechanism PBG improves circuit tracing with the RVdG trans-complementation system: packaging efficiency, uptake efficiency or other factors. Detailed functional assessments of rabies biology at each step should be undertaken to assess these questions.
We aimed to further improve trans-synaptic labeling efficiency by using codon optimization for Mus musculus to produce oG and increase expression levels of the chimeric PBG rabies glycoprotein in starter cells. This resulted in a four- to nine-fold improvement in trans-synaptic labeling, adding further support to the hypothesis that glycoprotein expression should be maximized for efficient tracing (Miyamichi et al., 2013; Watabe-Uchida et al., 2012). In previous experiments this has been achieved through the use of stronger promoters in helper viruses (Miyamichi et al., 2013) or by expressing G independently from other genes (e.g. TVA and/or fluorescent proteins) that are typically used in RVdG tracing experiments; separate helper viruses are used for expressing G and other proteins rather than linking genes with 2A elements, since 2A elements appear to reduce expression levels (Wall et al., 2010; Watabe-Uchida et al., 2012). Here we incorporated both nuclear GFP and oG, linked by 2A elements, into the same helper virus in order to facilitate accurate and unambiguous quantification of starter cells. We therefore expect that expression levels and efficiency of trans-synaptic spread would be improved still further by using a helper virus in which oG is expressed independently. Future studies would benefit from a high quality, specific and sensitive antibody against rabies glycoprotein, obviating the need for expression of a linked fluorescent protein to indicate cells expressing G. It should also be noted that the AAV vector used for expression of oG had a 25% lower titer than for the other glycoproteins tested. While this difference is modest, there is probably not a linear relationship between viral titer and transduction efficiency (Nathanson et al., 2009). It is therefore possible that the improvement in gene expression levels and trans-synaptic labeling achieved by codon optimization (oG versus PBG) might be somewhat greater than indicated by the direct comparison of our results.
It has recently been reported that RVdG derived from the CVS-N2C strain of rabies virus has roughly 10 to 20-fold better trans-synaptic tracing efficiency than the original system using SAD B19 rabies virus (Reardon et al., 2016). This improvement is similar to the improvement that we have demonstrated here based on modifications to the glycoprotein used for trans-complementation of SAD B19 RVdG. Taken at face value this would suggest the two systems are similar in tracing efficiency and that the choice of system for use might be based on other factors such as convenience of virus production (easier for SAD B19), cytotoxicity and survival times (less and longer for CVS-N2C), or speed and levels of gene expression (faster and higher for SAD B19). However, it is difficult to predict relative trans-synaptic efficiency of these different systems (CVS-N2C versus SAD B19 with oG) that might be observed with direct comparisons. Differences in receptors that mediate rabies virus uptake from axon terminals of different cell types might result in different relative efficiencies depending on the circuits being investigated. Our experiments comparing different glycoproteins in the context of SAD B19 RVdG were conducted in different circuits (thalamocortical circuits in our study) than those comparing CVS-N2C to SAD B19 (corticostriatal or premotor-motor neuronal circuits). Future studies should not only make direct comparisons of the same circuits but should also explore the mechanisms responsible for any differences observed in order to facilitate further improvements in both systems.
In comparing these systems it is noteworthy that use of the chimeric glycoprotein NBG, derived from the extracellular domain of the CVS-N2C glycoprotein, did not improve trans-synaptic efficiency. While this might suggest that differences in efficiency between the original SAD B19 RVdG system and the CVS-N2C RVdG system are not due to improved uptake mediated by the N2C glycoprotein, this might not be the case. As noted above, it is possible that the chimeric NBG glycoprotein might fail to conserve the structure of the extracellular domain required for efficient uptake and would therefore perform worse than the native N2C glycoprotein when used to trans-complement CVS-N2C RVdG. Without such disruption (if present) it is theoretically possible that the native N2C glycoprotein might mediate viral uptake by axon terminals as efficiently as Pasteur strain glycoprotein (PV). This possibility might be tested directly by comparing tracing efficiency with the native N2C glycoprotein versus NBG glycoprotein in the context of the SAD B19 strain of RVdG. It would also be informative to generate a chimeric PV N2C glycoprotein for trans-complementation of CVS-N2C RVdG and test whether it can further improve trans-synaptic spread of that strain.
Finally, Reardon et al. also compared the health of cells infected with CVS-N2C RVdG versus SAD B19 RVdG and reported that when potentially toxic gene products (ChR2 or GCaMP) are expressed, the CVS-N2C RVdG-infected cells are healthier. It is important to note that this difference is not likely to be related to tracing efficiency. They demonstrate toxic effects at early time points post-infection. But SAD B19 RVdG is not toxic at these time points when non-toxic gene products such as GFP are expressed (Wickersham et al., 2007a). The differences in toxicity may instead be attributed to differences in gene expression between the two viral strains; since CVS-N2C RVdG requires more time to express transgenes at high levels, the toxic effects of transgenes are not apparent as quickly (Reardon et al., 2016). It is also possible that differences in toxicity were related to the 50 to 100-fold higher viral titers used for SAD B19 RVdG. Previous studies have demonstrated that increased levels of glycoprotein expression from rabies virus do not increase pathogenicity (Wirblich and Schnell, 2011) and we have not observed any signs of increased toxicity when using oG.
It would also be useful to investigate whether oG can be used to improve other procedures and applications that are based on SAD B19 RVdG. If oG enhances the efficiency of rabies uptake by presynaptic neurons, oG+RVdG might function as a more efficient retrograde viral tracer than B19G+RVdG. It would therefore be interesting to examine not only whether oG+RVdG might label more input neurons, but also whether it might spread to neurons that were not labeled when using B19G+RVdG. oG should also be a valuable reagent for recovering new RVdG variants from plasmid DNA (Osakada and Callaway, 2013; Osakada et al., 2011). To rescue the RVdG from plasmid DNA, current systems use cell lines stably expressing B19G and T7 RNA polymerase, such as the B7GG cell line (Osakada et al., 2011). If oG can improve packaging efficiency of SAD B19 in producer cells, it should also enhance virus production, resulting in higher titers of RVdG. This approach might be particularly useful in cases when the recovery of rabies variants has been difficult (Osakada et al., 2011) or when rabies with high titer is required, for example, for single cell network tracing (Marshel et al., 2010; Wertz et al., 2015). In summary, with its simplicity and versatility, oG should provide strong advantages for the study of neural circuits and is expected to have a broad range of applications.
EXPERIMENTAL PROCEDURES
Transgenic mice
All experimental procedures were approved by the Salk Institute Animal Care and Use Committee. PVIRES-Cre/+, Tlx3-Cre PL56, and R26LSL-TVA/+ mouse lines have been described previously (Gerfen et al., 2013; Hippenmeyer et al., 2005; Seidler et al., 2008). Briefly, PVIRES-Cre/+ and Tlx3-Cre PL56 mouse lines express Cre only in PV+ neurons or cortical layer 5 pyramids respectively, whereas R26LSL-TVA/+ is a responder line expressing TVA in a Cre dependent manner. Homozygous PVIRES-Cre/IRES-Cre and R26LSL-TVA/LSL-TVA mice were bred to produce PVIRES-Cre/+;R26LSL-TVA/+ mice.
Chimeric glycoprotein design
NBG was cloned to consist of the extracellular domain of CVS-N2C rabies G and transmembrane/cytoplasmic domain of SAD B19G, using pCAGGS-FuG-B (Kato et al., 2011a) and pHCMV-B19G (Sena-Esteves et al., 2004) respectively. PBG was cloned to consist of the extracellular domain of Pasteur strain rabies G and transmembrane/cytoplasmic domain of SAD B19G, using pCAGGS-FuG-B2 (Kato et al., 2011a) and pHCMV-B19G (Sena-Esteves et al., 2004) respectively. oG was codon-optimized from PBG for Mus musculus (Genscript).
Virus preparation
Four different helper viruses were produced through the Salk Viral Vector core: AAV8.EF1α.FLEX.H2B-GFP-F2A-G.WPRE.hGH (AAV-FLEX-H2B-GFP-2A-G) expressing each rabies glycoprotein in a Cre-dependent manner. Four kinds of AAV-FLEX-H2B-GFP-2A-G express different glycoprotein variants: B19G, NBG, PBG, or oG each. Titers of AAVs expressing B19G, NBG, or PBG were adjusted to 1.5×1012 GC/ml, whereas the titer of AAV expressing oG was 1.15×1012 GC/ml. EnvA-pseudotyped G-deleted rabies virus, EnvA+RVdG-dsRed with the titer of 3.6×107 infectious units (IU/ml), EnvA+RVdG-GFP (4.3×108 IU/ml), and EnvA+RVdG-H2B-mCherry (1.6×108 IU/ml) were produced following the published protocol (Osakada and Callaway, 2013). The plasmid AAV-FLEX-H2B-GFP-2A-oG, as well as additional plasmids for expressing oG, have been depsosited at Addgene and AAVs can be purchased from the Salk Viral Vector Core.
Animal surgery for virus injection
PVIRES-Cre/+;R26LSL-TVA/+ mice received AAV helper injections at P38-78. Mice were anaesthetized with 100 mg/kg of ketamine and 10 mg/kg of xylazine cocktail via intraperitoneal injections and mounted in a stereotax (David Kopf Instruments Model 940 series, Tujunga, CA) for surgery and stereotaxic injections. Virus was injected into the center of V1, using the following coordinates: 3.4 mm rostral, 2.6 mm lateral relative to bregma and 0.5-0.7 mm ventral from the pia. We injected 100 nl of AAVs using air pressure by picospritzer (General Valve Corp, Fairfield, NJ). To prevent virus backflow, the pipette was left in the brain for 5-10 minutes after completion of injection. Two or three weeks after AAV helper injection, 200 nl of five-fold diluted EnvA+RVdG-dsRed (7.2×106 IU/ml) were injected into the same site in V1 using picospritzer-mediated air pressure. For control experiments, PVIRES-Cre/+;R26LSL-TVA/+ or Tlx3-Cre/+;R26LSL-TVA/+ mice received 200nl of EnvA+RVdG-GFP or EnvA+RVdG-H2B-mCherry injections at P70, respectively, and no helper virus was injected. Mice were housed for seven days to allow for trans-synaptic rabies spread and fluorescent protein expression.
Histology and image analysis
Brains were harvested after transcardial perfusion using phosphate-buffered saline (PBS) followed by 4% paraformaldehyde (PFA). Brains were dissected out from skulls and post-fixed with 2% PFA and 15% sucrose in PBS at 4°C for 16-20 hours, then immersed in 30% sucrose in PBS at 4°C before sectioning. 50 μm coronal brain sections were prepared using a freezing microtome and stored in PBS with 0.01% sodium azide at 4°C. To label PV+ neurons, free-floating sections were incubated at 4°C for 16-48 hours with mouse anti-PV (1:1000, Sigma-Aldrich P3088) primary antibodies in PBS/0.5% normal donkey serum/0.1% Triton-X 100, followed by the appropriate secondary antibodies conjugated with Alexa 647 (Molecular Probes) at room temperature for 2-3 hours. Sections were counterstained with 10 μM DAPI in PBS for 30 min to visualize cell nuclei. Immunostained tissue sections were mounted on slides with polyvinyl alcohol mounting medium containing DABCO and allowed to air-dry overnight. Confocal imaging was performed with Zeiss LSM780 confocal microscope. Confocal images were processed and analyzed in NIH ImageJ software (FIJI).
For rabies tracing analysis, every 50 μm coronal section from bregma −1.34 mm to −5.20 mm was collected. Individual tissue sections were scanned with a 10X objective using Olympus VS120 slide scanner or were directly examined with Olympus BX51 microscope. Cell counts and registrations were acquired using OlyVIA 2.6 software provided by Olympus. The section was then registered again using The Mouse Brain in Stereotaxic Coordinates atlas (Paxinos et al., 2001) fit to the scanned image using the first registration as a guide to adjust for minor tissue variation or deformation. Borders based on nuclear counterstain density were then added to denote layer boundaries. Lastly, dsRed and GFP channels were turned on and cell counts were recorded for each brain region.
For starter cell location analysis along the anterior-posterior axis, center of gravity analysis was performed by weighting starter neuron count for each coronal sections of V1 at the bregma stereotaxic coordinates. Center of gravity = Σ (number of starter neurons × bregma) / Σ number of starter neurons. For distribution analysis of starter neurons along the dorso-ventral axis, we used custom-made MATLAB code to calculate the proportion of starter neurons per bin from the pia to the white matter after dividing them into 11 bins.
Statistical analysis
For analysis of starter neuron distribution, Kruskal-Wallis test with Dunn's multiple comparisons test as a post hoc were used with all values reported as the median. For rabies traced input analysis, one-tailed Mann-Whitney test was used for comparisons with all values reported as the mean ± SEM.
Supplementary Material
Highlights.
Newly engineered glycoproteins improve monosynaptic rabies tracing
Previous methods labeled a small fraction of presynaptic neurons
oG, optimized glycoprotein, increases tracing efficiency up to 20-fold
ACKNOWLEDGEMENTS
We thank F. Osakada for discussion and help at the initial phase of project, K. Fischer for Matlab programming help and reading the manuscript, J. Neichin, S. Nopar for cell counting and histology help, Z. Zhang for advice on statistical analysis. We also thank the Salk viral vector and biophotonics core staff members. This work was supported by the National Institutes of Health grants EY022577 and MH063912, the Gatsby Charitable Foundation, and a Howard Hughes Medical Institute Collaborative Innovation Award (E.M.C). E.J.K. is a Biogen-IDEC Fellow of the Life Science Research Foundation and a recipient of 2012 NARSAD Young Investigator Award from Brain & Behavior Research Foundation.
Footnotes
CONFLICT OF INTEREST
The authors have no conflicts of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
AUTHOR CONTRIBUTIONS
All authors had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. Study concept and design: E.J.K. Acquisition of data: E.J.K., M.W.J., T.I. Analysis and interpretation of data: E.J.K. Drafting of the manuscript: E.J.K., E.M.C. Statistical analysis: E.J.K. Obtained funding: E.J.K., E.M.C. Study supervision: E.J.K., E.M.C.
REFERENCES
- Callaway EM. Transneuronal circuit tracing with neurotropic viruses. Current Opinion in Neurobiology. 2008;18:617–623. doi: 10.1016/j.conb.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, Luo L. Monosynaptic Circuit Tracing with Glycoprotein-Deleted Rabies Viruses. J Neurosci. 2015;35:8979–8985. doi: 10.1523/JNEUROSCI.0409-15.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerfen CR, Paletzki R, Heintz N. GENSAT BAC Cre-Recombinase Driver Lines to Study the Functional Organization of Cerebral Cortical and Basal Ganglia Circuits. Neuron. 2013;80:1368–1383. doi: 10.1016/j.neuron.2013.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippenmeyer S, Vrieseling E, Sigrist M, Portmann T, Laengle C, Ladle DR, Arber S. A developmental switch in the response of DRG neurons to ETS transcription factor signaling. PLoS Biol. 2005;3:e159. doi: 10.1371/journal.pbio.0030159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano M, Kato S, Kobayashi K, Okada T, Yaginuma H, Kobayashi K. Highly efficient retrograde gene transfer into motor neurons by a lentiviral vector pseudotyped with fusion glycoprotein. PLoS One. 2013;8:e75896. doi: 10.1371/journal.pone.0075896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Kuramochi M, Kobayashi K, Fukabori R, Okada K, Uchigashima M, Watanabe M, Tsutsui Y, Kobayashi K. Selective Neural Pathway Targeting Reveals Key Roles of Thalamostriatal Projection in the Control of Visual Discrimination. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2011a;31:17169–17179. doi: 10.1523/JNEUROSCI.4005-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S, Kuramochi M, Takasumi K, Kobayashi K, Inoue K.-i., Takahara D, Hitoshi S, Ikenaka K, Shimada T, Takada M, et al. Neuron-Specific Gene Transfer Through Retrograde Transport of Lentiviral Vector Pseudotyped with a Novel Type of Fusion Envelope Glycoprotein. Human Gene Therapy. 2011b;22:1511–1523. doi: 10.1089/hum.2011.111. [DOI] [PubMed] [Google Scholar]
- Marshel JH, Mori T, Nielsen KJ, Callaway EM. Targeting Single Neuronal Networks for Gene Expression and Cell Labeling In Vivo. Neuron. 2010;67:562–574. doi: 10.1016/j.neuron.2010.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamichi K, Shlomai-Fuchs Y, Shu M, Weissbourd BC, Luo L, Mizrahi A. Dissecting local circuits: parvalbumin interneurons underlie broad feedback control of olfactory bulb output. Neuron. 2013;80:1232–1245. doi: 10.1016/j.neuron.2013.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori T, Morimoto K. Rabies virus glycoprotein variants display different patterns in rabies monosynaptic tracing. Frontiers in Neuroanatomy. 2014;7 doi: 10.3389/fnana.2013.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nathanson JL, Yanagawa Y, Obata K, Callaway EM. Preferential labeling of inhibitory and excitatory cortical neurons by endogenous tropism of adeno-associated virus and lentivirus vectors. NSC. 2009;161:441–450. doi: 10.1016/j.neuroscience.2009.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Callaway EM. Design and generation of recombinant rabies virus vectors. Nature protocols. 2013;8:1583–1601. doi: 10.1038/nprot.2013.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakada F, Mori T, Cetin AH, Marshel JH, Virgen B, Callaway EM. New Rabies Virus Variants for Monitoring and Manipulating Activity and Gene Expression in Defined Neural Circuits. Neuron. 2011;71:617–631. doi: 10.1016/j.neuron.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ, Franklin KBJ. The mouse brain in stereotaxic coordinates. 2nd edn Academic Press; San Diego: 2001. [Google Scholar]
- Reardon TR, Murray AJ, Turi GF, Wirblich C, Croce KR, Schnell MJ, Jessell TM, Losonczy A. Rabies Virus CVS-N2c(DeltaG) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron. 2016;89:711–724. doi: 10.1016/j.neuron.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnell MJ, McGettigan JP, Wirblich C, Papaneri A. The cell biology of rabies virus: using stealth to reach the brain. Nat Rev Microbiol. 2010;8:51–61. doi: 10.1038/nrmicro2260. [DOI] [PubMed] [Google Scholar]
- Seidler B, Schmidt A, Mayr U, Nakhai H, Schmid RM, Schneider G, Saur D. A Cre-loxP-based mouse model for conditional somatic gene expression and knockdown in vivo by using avian retroviral vectors. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:10137–10142. doi: 10.1073/pnas.0800487105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sena-Esteves M, Tebbets JC, Steffens S, Crombleholme T, Flake AW. Optimized large-scale production of high titer lentivirus vector pseudotypes. J Virol Methods. 2004;122:131–139. doi: 10.1016/j.jviromet.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugolini G. Use of rabies virus as a transneuronal tracer of neuronal connections: implications for the understanding of rabies pathogenesis. Dev Biol (Basel) 2008;131:493–506. [PubMed] [Google Scholar]
- Wall NR, Wickersham IR, Cetin A, De La Parra M, Callaway EM. Monosynaptic circuit tracing in vivo through Cre-dependent targeting and complementation of modified rabies virus. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:21848–21853. doi: 10.1073/pnas.1011756107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, Uchida N. Whole-Brain Mapping of Direct Inputs to Midbrain Dopamine Neurons. Neuron. 2012;74:858–873. doi: 10.1016/j.neuron.2012.03.017. [DOI] [PubMed] [Google Scholar]
- Wertz A, Trenholm S, Yonehara K, Hillier D, Raics Z, Leinweber M, Szalay G, Ghanem A, Keller G, Rozsa B, et al. PRESYNAPTIC NETWORKS. Single-cell-initiated monosynaptic tracing reveals layer-specific cortical network modules. Science. 2015;349:70–74. doi: 10.1126/science.aab1687. [DOI] [PubMed] [Google Scholar]
- Wickersham IR, Finke S, Conzelmann KK, Callaway EM. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat Methods. 2007a;4:47–49. doi: 10.1038/NMETH999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzelmann K-K, Young JAT, Callaway EM. Monosynaptic Restriction of Transsynaptic Tracing from Single, Genetically Targeted Neurons. Neuron. 2007b;53:639–647. doi: 10.1016/j.neuron.2007.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirblich C, Schnell MJ. Rabies virus (RV) glycoprotein expression levels are not critical for pathogenicity of RV. J Virol. 2011;85:697–704. doi: 10.1128/JVI.01309-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.