Summary
Upon growth factor stimulation or in some EGFR mutant cancer cells, PKM2 translocates into the nucleus to induce glycolysis and cell growth. Here, we report that nuclear PKM2 binds directly to poly-ADP ribose and this PAR-binding capability is critical for its nuclear localization. Accordingly, PARP inhibition prevents nuclear retention of PKM2 and therefore suppresses cell proliferation and tumor growth. In addition, we found that PAR level correlates with nuclear localization of PKM2 in EGFR-mutant brain and lung cancers, suggesting that PAR-dependent nuclear localization of PKM2 likely contributes to tumor progression in EGFR-mutant glioblastoma and lung cancers. In addition, some EGFR inhibitor-resistant lung cancer cells are sensitive to PARP inhibitors. Taken together, our data indicate that suppression of PKM2 nuclear function by PARP inhibitors represents a treatment strategy for EGFR inhibitor-resistant cancers.
Introduction
Poly(ADP-robose) polymerase-1 (PARP1) or ADP-ribosyltransferase diphtheria toxin-like 1 (ARTD1) (Hottiger et al., 2010) is the most abundant and the best understood member of the 17 PARP family proteins. PARP1 binds to both single strand breaks (SSBs) (Fisher et al., 2007; Okano et al., 2003) and double-strand breaks (DSBs) (Ali et al., 2012) and participates in the recognition, excision and repair of DNA damage (Kim et al., 2005). The most extensively studied role of PARP1 is its involvement in base excision repair (BER) (Dantzer et al., 1999; de Murcia et al., 1997; Masson et al., 1998; Strom et al., 2011; Trucco et al., 1998). Moreover, suppression of PARP1/2 leads to synthetic lethality in BRCA1/2-defecient tumors, indicating that PARP1-dependent BER and BRCA-dependent homologous repair pathway have overlapping and redundant functions in DNA repair (Bryant et al., 2005; Farmer et al., 2005; Fong et al., 2009; Rouleau et al., 2010). Recent studies have also pointed to a broader utility of PARP inhibitors beyond hereditary BRCA-deficient cancers (Gelmon et al., 2011; Inbar-Rozensal et al., 2009; Telli, 2011).
Pyruvate kinase isoform M2 (PKM2) is a glycolysis enzyme that converts phosphoenolpyruvate (PEP) into pyruvate (Luo and Semenza, 2012). Up-regulation of PKM2 has been shown recently to be an important feature of tumorigenesis (Hacker et al., 1998; Mazurek et al., 2005). In tumor cells, PKM2 forms a dimmer that is catalytically inactive as a glycolysis enzyme (Mazurek et al., 2005), but provides advantage for tumor progression due to Warburg effect (Christofk et al., 2008). Moreover, recent studies have revealed that various stimuli and post-translational modifications including FGFR1-mediated Y105 phosphorylation (Hitosugi et al., 2009), ERK1/2-dependent S37 phosphorylation (Yang et al., 2012b), and P300-dependent K433 acetylation (Lv et al., 2013) trigger PKM2 translocation into nucleus. Once in the nucleus, PKM2 acts as co-activator for several transcription factors like HIF-1α (Luo et al., 2011) and β-catenin (Yang et al., 2011). In addition, nuclear PKM2 also functions as a protein kinase to phosphorylate STAT3 (Gao et al., 2012), Histone H3 (Yang et al., 2012a), and Bub3 (Jiang et al., 2014), all of which contribute to promoting tumor growth or proliferation.
In this report, we demonstrated that PARP1-dependent poly-ADP-ribose (PAR) is required for nuclear retention and nuclear function of PKM2. PKM2 translocates into nucleus and binds to PAR upon EGF stimulation. PARP inhibition, or the PKM2-C/A mutant, which abolishes the PKM2/PAR interaction, suppresses the nuclear function of PKM2. In addition, PARP inhibition also diminishes the nuclear PKM2-dependent glycolysis and tumor growth. Moreover, we showed that nuclear localization of PKM2 correlates with PAR expression in EGFR-mutant human Glioblastoma and lung cancer tissues. Interestingly, PARP inhibition leads to growth suppression of some EGFR-mutant lung cancer cells, which are resistant to EGFR inhibitor. These data together support an unexpected function of PARP inhibition in tumor suppression and indicate that nuclear PKM2 may serve as a promising biomarker for the further development of PARP inhibitor-based therapies.
Results
Nuclear PKM2 binds to PAR in vitro and in vivo
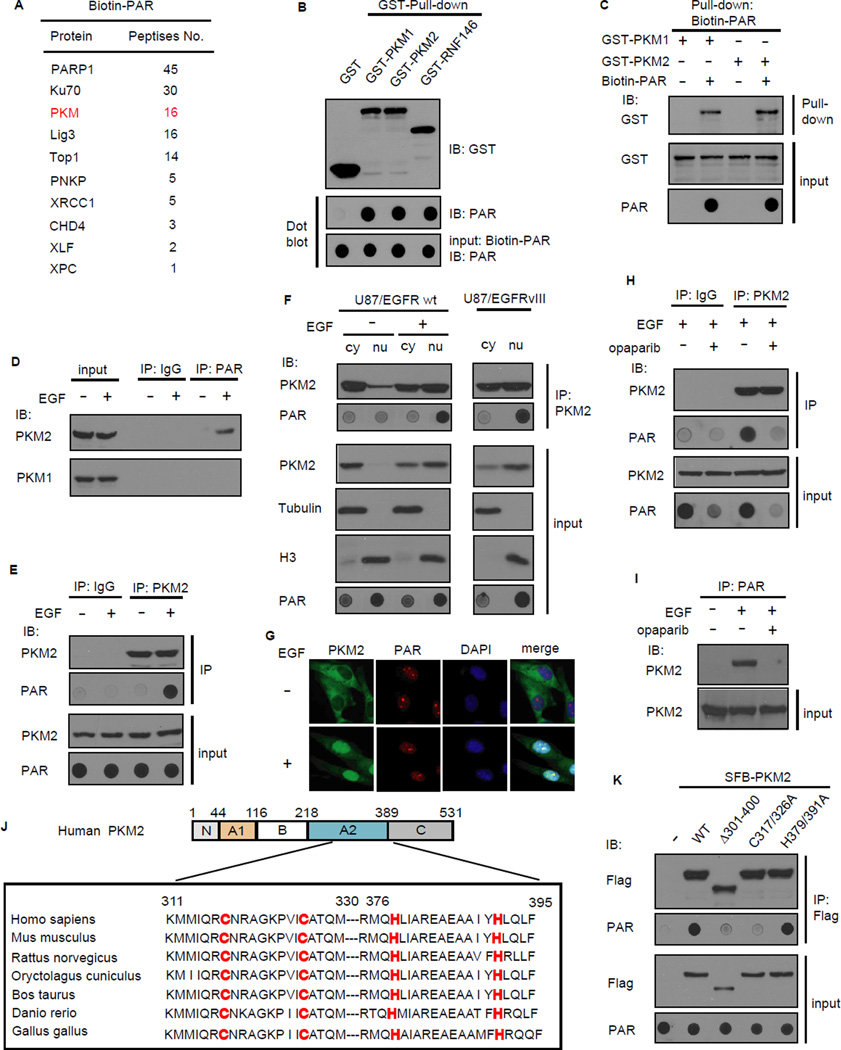
To understand how Poly-ADP-ribose (PAR) signaling participates in various cellular processes, we used a biotin-PAR pull-down coupled with mass spectrometry approach to identify new PAR-binding proteins. Mass spectrometry analysis revealed not only several known PAR-binding DNA repair proteins (e.g. Ku70, XRCC1, Lig3, TOP1, and PNKP), but also PKM as a major PAR-binding protein (Figure 1A). We performed in vitro PAR binding assays using dot blot. The result showed that just like the positive control RNF146, both PKM1 and PKM2 bind directly to PAR (Figure 1B). Reverse pull-down assays further confirmed the direct interaction between PAR and PKM1 or PKM2 (Figure 1C). To further demonstrate the direct binding between PKM2 and PAR, we performed the Biacore SPR assays and showed that the association between PAR and PKM2 is direct and specific (Figure S1A). To test the possibility that PKM2 itself may be PARylated by PARP1, in vitro PARP1 PARylation assay was performed and Histone-H3 was included as positive control for PARP1 substrate. We showed that PARP1 could ribosylate itself and Histone H3, but not PKM2 in vitro (Figure S1B), indicating that PKM2 is not a substrate of PARP1.
Figure 1. PKM2 binds to PAR in vitro and in vivo.
(A) PKM was identified as a PAR binding protein. Biotin-PAR (from Trevigen) was used as the bait for whole cell lysate pull-down and mass spectrometry analysis. The identified proteins and the number of peptides were listed. (B) PKM1/2 binds PAR in vitro. The recombinant GST, GST-PKM1/2 and GST-RNF146 (positive control) were incubated with Biotin-PAR, and protein-associated PAR was examined by dot blot using anti-PAR antibody. (C) The interaction between Biotin-PAR and GST-PKM1/2 was examined by the reciprocal pull-down. (D,E) EGF induced the PAR/PKM2 binding in vivo. U87/EGFR wt cells were serum starved for 12 hr and then treated without or with EGF (100ng/ml) for 6 h, Immunoprecipitation (IP) was carried out using anti-PAR antibody (D) or anti-PKM2 antibody (E) and then subjected to immunoblotting. (F) Nuclear PKM2 binds to PAR in vivo. Nuclear and cytoplasm fractions were prepared and IP/Western was conducted using indicated antibodies. (G) PKM2 and PAR localized in the nucleus after EGF treatment. U87/EGFR wt cells were serum starved and then treated without or with EGF (100 ng/ml). Cells were prepared for immunofluorescence staining. (H,I) The PAR/PKM2 interaction was abolished by PARP inhibitor. U87/EGFR wt cells were serum starved and treated without or with PARP inhibitor olaparib (0.5uM) and then treated with EGF. Immunoprecipitation (IP) was carried out using anti-PKM2 antibody (H) or anti-PAR antibody (I) and then subjected to immunoblotting. (J) Schematic diagram of the putative PAR-binding motif (C2H2 motif) in PKM2, and key residues of the motif were highly conserved among various species. (K) The C2H2 motif of PKM2 is critical for PKM2/PAR binding. U87/EGFR wt cells stably expressed the indicated constructs were serum starved and then treated with EGF and cell lysates were subjected to IP/dot blot analysis.
We next explored whether PKM2 or PKM1 would associate with PAR in vivo. As cellular PAR is dominantly localized in nucleus and PKM2 could translocate from cytoplasm to nucleus upon EGF stimulation (Yang et al., 2011), we speculated that the PAR-binding activity of PKM2 may promote its nuclear localization. We thus used U87 human glioblastoma (GBM) cells, which express wild-type EGFR with or without EGF treatment. Immunoprecipitation (IP) with anti-PAR antibody followed by Western blotting with anti-PKM1 and anti-PKM2 antibodies revealed that only the M2, but not M1, isoform of PKM binds to PAR in vivo after EGF treatment (Figure 1D). Reciprocal co-IP using anti-PKM2 antibody confirmed this observation (Figure 1E). In addition, cell fractionation co-IP and immunofluorescence staining experiments indicated only PKM2, but not PKM1, could translocate into the nucleus and bind to PAR upon EGF stimulation (Figure 1F, G and Figure S1C-F). We also used micro-irradiation to show that PKM2 is indeed localized to where PAR is induced, but only after EGF stimulation (Figure S1G). In U87/EGFRvIII cells, which express the EGFRvIII mutant and display constitutively active EGFR signaling, PKM2 partially localizes in the nucleus and the nuclear PKM2 binds to PAR even without EGF treatment (Figure 1F). Olaparib is a widely used PARP1/2 specific inhibitor that suppresses PAR synthesis (Menear et al., 2008). PKM2 could no longer bind to PAR following EGF treatment when PAR level was significantly reduced upon olaparib treatment (Figure 1H, I). Taken together, these data indicate that EGF stimulation triggers PKM2 translocation into nucleus and allows it to bind to PAR in the nucleus.
The above demonstrated in vitro and in vivo PAR-binding activity of PKM2 prompted us to search for PAR-binding motif in PKM2. There are several known PAR-binding motifs or domains, which include the PAR-binding motif (PBM) (Gagne et al., 2008), macrodomain (Pehrson and Fried, 1992), PAR-binding zinc finger (PBZ) (Ahel et al., 2008), WWE domain (Aravind, 2001), BRCT and FHA domains (Li et al., 2013; Li and Yu, 2013), and PAR-binding regulatory motif (PbR) (Min et al., 2013). We detected a putative C2H2 motif in PKM1/2 (Figure 1J and Figure S1D), which is distinct from the PBZ C2H2 and the PbR C2H2. This motif is highly conserved among various species (Figure 1J), indicating that it may serve as a PAR-binding motif, which is important for PKM2 function. To determine whether the C2H2 motif of PKM2 is indeed critical for its binding to PAR, we generated an internal deletion mutant of PKM2 (PKM2Δ301-400), which lacks the C2H2 motif, and two point mutants of PKM2 within the C2H2 motif (i.e. PKM2-C317/326A and PKM2-H379/391A). Co-IP and dot blot results showed that PKM2Δ301-400 and PKM2-C317/326A mutants failed to bind to PAR (Figure 1K), indicating that this putative C2H2 motif may be responsible for its interaction with PAR. These data together suggest that PKM2 is a PAR-binding protein.
PAR binding is required for PKM2 nucleus retention
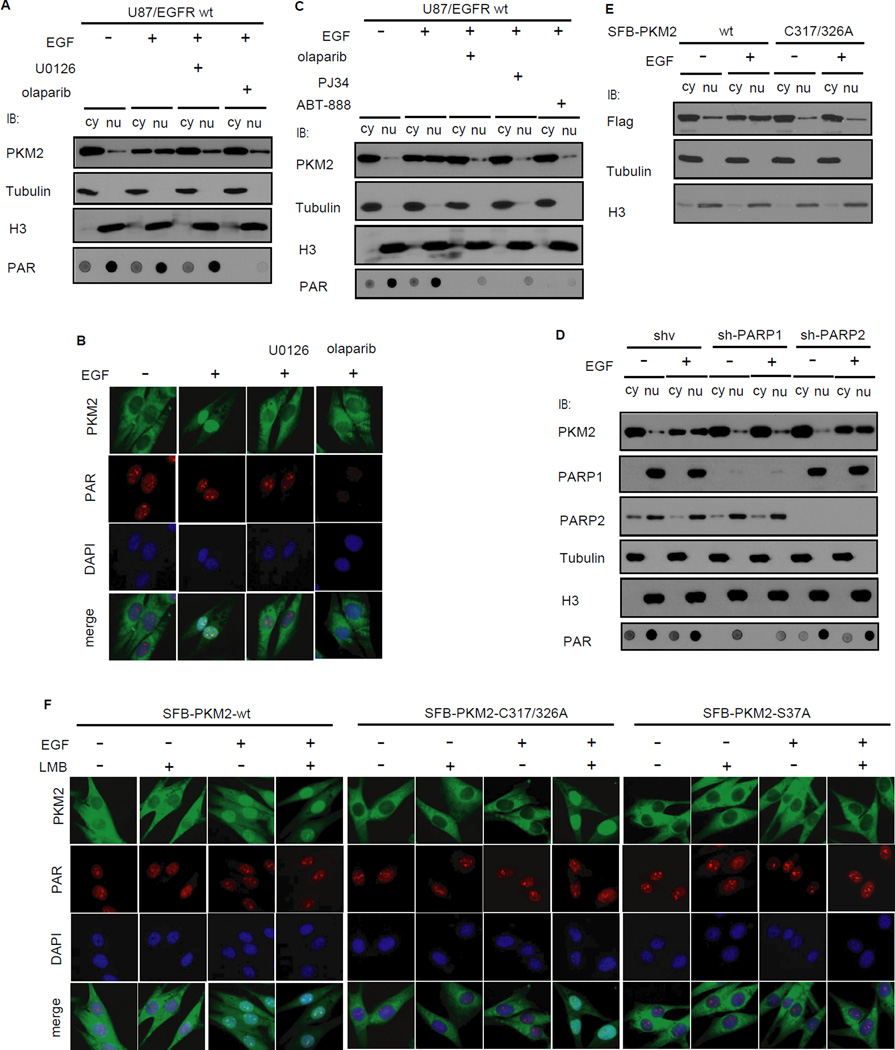
PKM2 normally functions as a glycolic enzyme in the cytoplasm. However, recent studies have demonstrated that PKM2 could translocate into nucleus and function as a protein kinase involved in gene transcription and tumorigenesis (Gao et al., 2012; Yang et al., 2012a). Since PKM2 binds to PAR in the nucleus, we asked whether PAR would regulate nuclear localization of PKM2. Cell fractionation and immunofluorescence staining experiments indicated that PARP inhibitor could abolish the EGF-induced nuclear translocation of PKM2 (Figure 2A, B). In these experiments, we used MEK/ERK inhibitor U0126 as a positive control, which is known to block PKM2 translocation (Yang et al., 2012b). Moreover, all three PARP inhibitors that we tested (i.e. olaparib, PJ34, ABT-888) could inhibit PKM2 nuclear translocation (Figure 2C). Olaparib inhibits both PARP1 and PARP2 (Menear et al., 2008). We stably knocked down PARP1 or PARP2 using small hairpin RNAs (shRNA) in cells and found that only knockdown of PARP1, but not that of PARP2, diminished EGF-induced nuclear localization of PKM2, as PARP1 but not PARP2 is predominantly responsible for PAR formation in the cell (Figure 2D). This may due to the fact that the abundance of PARP1 is much higher than that of PARP2 in vivo (Figure S2). Moreover, the PKM2-C317/326A mutant, which could not bind to PAR, also failed to translocate to nucleus following EGF treatment (Figure 2E). These data suggest that PAR-binding is critical for PKM2 nuclear localization.
Figure 2. PAR binding is required for PKM2 retention in nucleus.
(A,B) PARP inhibitor olaparib blocked EGF-induced nuclear localization of PKM2. U87/EGFR wt cells were serum starved for 12 hrs and then pretreated with U0126 (20 µM) or olaparib (0.5 µM) for 2 hrs before treating with EGF (100 ng/ml) for 6 hrs. Nuclear and cytoplasm fractions were prepared for immunoblotting (A) and immunofluorescence analyses were carried out using indicated antibodies (B). (C) Three different PARP inhibitors blocked PKM2 nuclear localization. U87/EGFR wt cells were serum starved for 12 hrs and then pretreated with olaparib (0.5 µM), PJ34 (2.5 µM) and ABT-888 (1 µM) for 2 hrs before treating with EGF (100 ng/ml) for 6 hrs. Nuclear and cytoplasm fractions were prepared and analyzed by immunoblotting (D) EGF-induced nuclear localization of PKM2 requires PARP1. PARP1 or PARP2 stable knockdown and control U87/EGFR wt cells were serum starved and treated without or with EGF. Cell fractions were subjected to immunoblotting analysis using indicated antibodies. (E) EGF fails to trigger the nuclear localization of PKM2-C/A mutant. U87/EGFR wt cells stably expressing wild-type PKM2 or the PKM2-C/A mutant were serum starved and treated without or with EGF. Cell fractions were prepared and analyzed by Western blotting. (F) PAR binding is required for PKM2 nuclear retention. U87/EGFR wt cells stably expressed S-Flag-SBP (SFB)-tagged PKM2 or PKM2-C317/326A were serum starved and then treated with EGF, LMB (2 ng/ml for 1 h) or combination with the two and immunofluorescence analyses were carried out using indicated antibodies.
PKM2 only associates with PAR in the nucleus, which ought to happen after the translocation of PKM2 from cytoplasm. We suspected that PAR-binding should affect the nuclear retention, but not nuclear translocation, of PKM2. To test this hypothesis, we generated the PKM2-S37A mutant, which was resistant to EGF-induced nuclear translocation (Yang et al., 2012b). This PKM2 mutant associated with PAR just like wild-type PKM2 (Figure S3A). While wild-type PKM2 displayed EGF-induced nuclear translocation in U87/EGFR wt cells, both the PKM2-S37A and the PKM2-C317/326A PAR-binding mutants failed to do so (Figure 2F). Moreover, LMB treatment, which abolishes the export of nuclear proteins, led to the nuclear localization of the PKM2-C317/326A mutant, but not the PKM2-S37A mutant after EGF treatment (Figure 2F). These observations indicate that while the S37 phosphorylation is important for PKM2 nuclear translocation as previously reported (Yang et al., 2012b), the PAR-binding activity of PKM2 is critical for PKM2 nuclear retention.
PAR regulates nuclear functions of PKM2
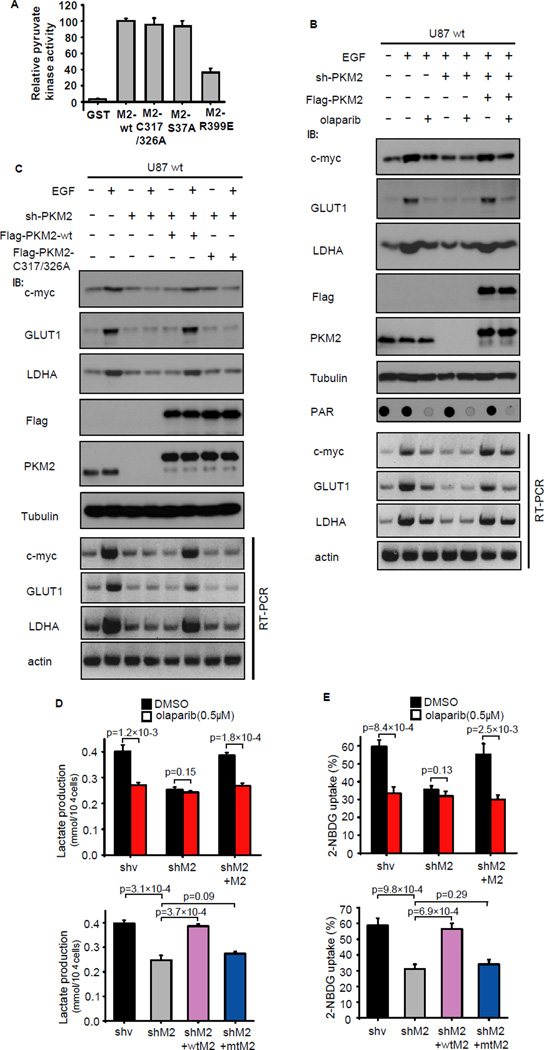
Cytoplasmic PKM2 normally function as a glycolysis enzyme, while nuclear PKM2 was reported as a coactivator for HIF-1α (Luo et al., 2011) and β-catenin (Yang et al., 2011). It was previously reported that the dimer and tetramer forms of PKM2 coexist in proliferation cells (Mazurek et al., 2005), the tetramer form of PKM2 was responsible of pyruvate kinase activity in the cytosol and the dimer form was responsible for PKM2 nuclear function (Gao et al., 2012). Since PAR binds to PKM2 in the nucleus, we suspected that PAR binding would be required for PKM2 functions in the nucleus, but not in the cytosol. We therefore generated bacteria expressed and purified wild type PKM2, PKM2-C317/326A, PKM2-S37A and PKM2-R399E proteins and performed in vitro pyruvate kinase assays. The PKM2-R399E mutant was reported to promote the tetramer to dimer transition and thus reduce the pyruvate kinase activity of PKM2 (Gao et al., 2012), and the PKM2-S37A mutant was thought to have no effect on PKM2 pyruvate kinase activity (Yang et al., 2012b). In agreement with these early reports, we showed that both the PAR-binding defective PKM2-C317/326A mutant and the PKM2-S37A mutant exhibited similar pyruvate kinase activity as wild-type PKM2, while the PKM2-R399E mutant displayed reduced pyruvate kinase activity (Figure 3A and S3B), indicating that PAR-binding is not required for PKM2 pyruvate kinase activity.
Figure 3. PAR regulates nuclear functions of PKM2.
(A) The activity of GST, GST-PKM2 and mutants of PKM2 towards PEP was measured using in vitro pyruvate kinase assays. (B) Effect of olaparib on glycolysis gene expression requires PKM2. U87/EGFR wt cells with stable PKM2 knockdown or reintroduction of PKM2 were serum starved and 26 treated without or with EGF and olaparib. Cells were collected and examined by Western blotting and RT-PCR as indicated. (C) EGF fails to trigger glycolysis gene up-regulation in PKM2- C317/326A cells. U87/EGFR wt cells with stable PKM2 knockdown followed by reintroduction of wild-type PKM2 or PKM2- C317/326A mutant were serum starved and treated without or with EGF. Cells were collected and examined by Western blotting and RT-PCR for the detection of c-Myc and glycolysis genes such as GLUT1 and LDHA. (D,E) Lactate production (D) and glucose uptake (E) were measured in the indicated U87/EGFRvIII cells, wtM2 is identical to wild-type PKM2 and mtM2 is identical to PKM2-C317/326A mutant. Data are means ±SD (n=3 independent experiments).
We further examined the effect of PAR on PKM2 nuclear functions in vivo. Since nuclear PKM2 is required for expression of several glycolysis genes c-Myc, GLUT and LDHA (Luo et al., 2011);(Yang et al., 2011; Yang et al., 2012b). Indeed, PKM2 depletion and rescue experiments demonstrated that olaparib suppressed expression of c-Myc, GLUT, LDHA (Figure 3B) and thus reduced lactate production and glucose uptake in PKM2-dependent manner (Figure 3D,E). Consistently, the PKM2-C317/326A mutant, which could not bind to PAR, failed to reverse the decrease of glycolysis induced by PKM2 knockdown (Figure 3C-E). These results demonstrated that PAR-binding is required for nuclear PKM2-induced glycolysis.
PAR binding promotes nuclear PKM2-induced cell proliferation and tumor growth
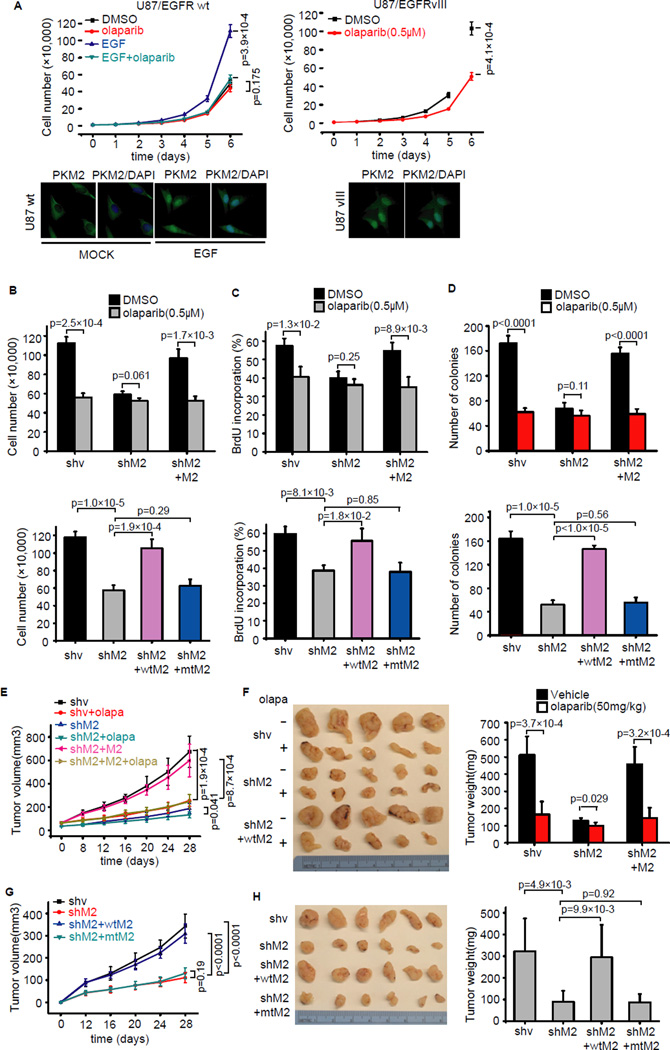
Nuclear PKM2 was reported to be involved in cell proliferation and Warburg effect (Gao et al., 2012; Yang et al., 2012b), which led us to determine whether PAR would affect cell proliferation. To this end, we first checked the effect of PARP inhibitor on the proliferation of U87/EGFR wt and U87/EGFRvIII cells. In U87/EGFR wt cells, PARP inhibitor olaparib only suppressed cell proliferation when these cells were stimulated with EGF, whereas in U87/EGFRvIII cells olaparib reduced cell proliferation even in the absence of EGF since EGF signaling is constitutively activated in these cells (Figure 4A). Correspondingly, we noticed that PKM2 only displayed nuclear localization in U87/EGFR wt cells upon EGF stimulation, while U87/EGFRvIII cells showed constitutive nuclear localization of PKM2 (Figure 4A), implying that the effect of PARP inhibitor on cell proliferation may depend on PKM2 nuclear localization. To test this possibility, we established PKM2 knockdown U87/EGFRvIII cells and reintroduced wild-type PKM2 or PKM2-C317/326A mutant in these knockdown cells. We found that olaparib did not significantly reduce cell proliferation in PKM2 knockdown cells, while reintroduction of shRNA-resistant PKM2 restored olaparib-induced growth inhibition (Figure 4B). Moreover, similar to olaparib treatment, PKM2 knockdown suppressed proliferation of U87/EGFRvIII cells, whereas re-expression of wild-type PKM2, but not the PKM2-C317/326A mutant, rescued this proliferation defect (Figure 4B). These results were confirmed by BrdU incorporation (Figure 4C) and colony formation assays (Figure 4D).
Figure 4. PAR binding promotes PKM2-induced cell proliferation and tumor growth.
(A) PARP inhibitor olaparib suppresses cell proliferation in U87 cells. U87/EGFR wt cells and U87/EGFRvIII cells were treated with EGF or olaparib, and cell proliferation was determined. Data are means ±SD (n=3 independent experiments), and immunofluorescence analyses were carried out using PKM2 antibody. (B) The indicated PKM2 stable knockdown cells and cells with reintroduction of wild-type PKM2 or PKM2-C317/326A mutant in U87/EGFRvIII were treated without or with olaparib and cell proliferation were measured at day 6 (n=3). (C) The BrdU staining of indicated U87/EGFRvIII cells were preformed and the percentages of positive cells are summarized. Data are means ±SD (n=3 independent experiments). (D) The indicated U87/EGFRvIII cells were tested in colony formation assay. Viable colonies after 12 days were counted and analyzed (n=4). (E,F) Tumor volume (E) and weight (F) of mice with subcutaneous injection of 5 × 106 indicated U87/EGFRvIII cells with vehicle or olaparib treatment were presented (n=5 mice, mean±SD). (G,H) Tumor volume (G) and weight (H) of mice with subcutaneous injection of 5 × 106 shv, shM2, shM2+wtM2 and shM2+mtM2 U87/EGFRvIII cells were presented (n=6 mice, mean±SD), wtM2 is identical to 27 wild-type PKM2 and mtM2 is identical to PKM2-C317/326A mutant.
In addition, we tested the effect of PARP inhibition on PKM2-dependent tumor growth using xenograft models. We showed that PARP inhibitor olaparib-treated tumors displayed smaller tumor volume and weight compared to vehicle-treated tumors (Figure 4E,F). However, this inhibitor effect of olaparib on tumor growth was dampened when we used PKM2 knockdown U87/EGFRvIII cells (Figure 4E,F). Moreover, mice injected with PKM2 knockdown U87/EGFRvIII cells showed significantly reduced tumor growth, while this reduction was rescued by reintroduction of wild-type PKM2, but not the PKM2-C317/326A mutant (Figure 4G,H). Taken together, these results demonstrated that PAR-binding is required for nuclear PKM2-induced cell proliferation and tumor growth.
PARP inhibitor suppresses growth of EGFR-mutant lung cancer cells by targeting nuclear PKM2
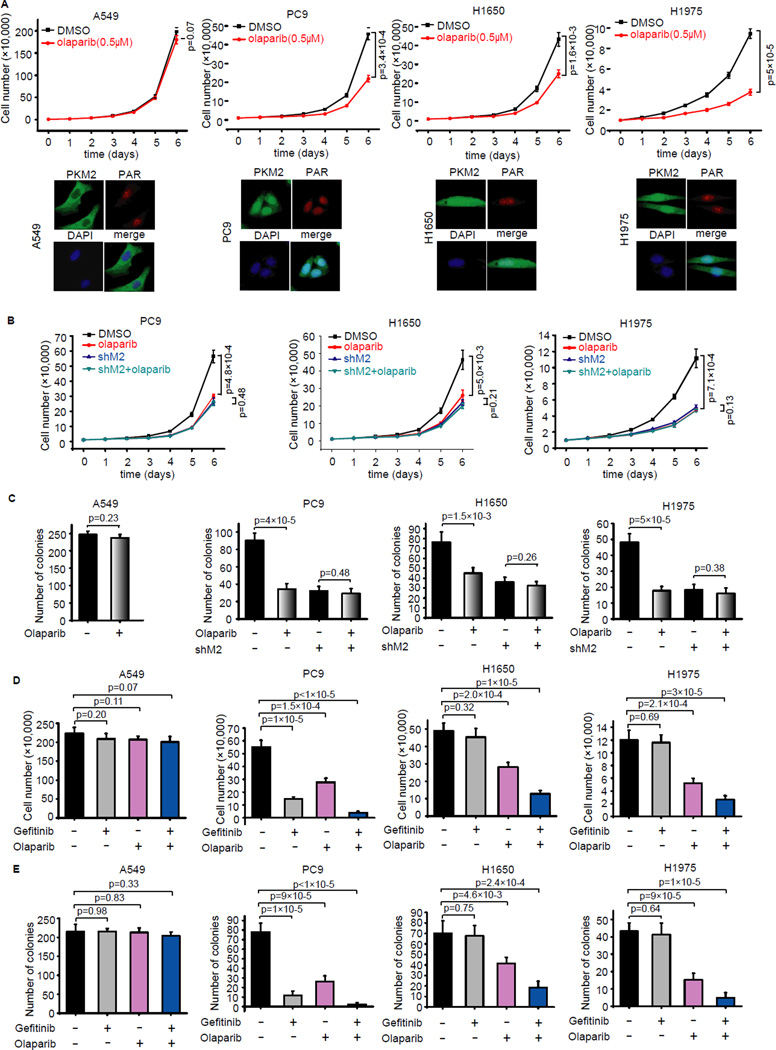
Somatic mutations of EGFR play a critical role in tumor progression and therapeutic response in human lung cancers (Paez et al., 2004). We showed above that PARP inhibitor suppressed nuclear PKM2-induced cell proliferation upon EGF stimulation or activation. Thus, we decided to test whether any of the EGFR-mutant lung cancer cells would be sensitive to PARP inhibition. We used 3 lung cancer cell lines with wild-type EGFR (i.e. A549, H358 and H460) and 10 EGFR-mutant lung cancer cell lines (i.e. H1650, HCC827, HCC2279, HCC2935, HCC4006, PC9, H820, H3255, HCC4011 and H1975) (Figure S4A). In EGFR wild-type A549, H358 and H460 cells, and EGFR-mutant HCC827, HCC2279, H820, H3255 and HCC40111 cells, PKM2 mainly localized in the cytoplasm and these cells were resistant to PARP inhibitor olaparib (Figure 5A, Figure S4B, Table S1), whereas in EGFR-mutant H1650, H1975, PC9, HCC2935 and HCC4006 cells, PKM2 localized in the nucleus and these cells were sensitive to olaparib treatment (Figure 5A, Figure S4C, Table S1). We further established H1650, H1975 and PC9 derivative cell lines with stable PKM2 knockdown and showed that depletion of PKM2 converted these two cell lines from olaparib-sensitive to olaparib resistant (Figure 5B, S5A). These results were confirmed by colony formation assays (Figure 5C), indicating that nuclear PKM2 may be a marker for cancer cells that are sensitive to PARP inhibition.
Figure 5. PARP inhibitor suppresses growth of EGFR-mutant lung cancer cells.
(A) A549, PC9, H1650 and H1975 cells were treated without or with olaparib (0.5 µM), and cell proliferation was determined. Data are means ±SD (n=3 independent experiments). Immunofluorescence analyses were carried out using PKM2 and PAR antibodies. (B) PC9, H1650 and H1975 with PKM2 stable knockdown or control cells were treated without or with olaparib and examined to determine the rate of cell proliferation. Data are means ±SD (n=3 independent experiments). (C) The indicated cells were treated without or with olaparib and tested in colony formation assay. Viable colonies after 12 days were counted and analyzed (n=4). (D) The A549, PC-9, H1650 and H1975 cells were treated with gefitinib or olaparib and cell proliferation were measured at day 6 (n=4). (E) The indicated cells were treated with gefitinib or olaparib and tested in colony formation assay. Viable colonies after 12 days were counted and analyzed (n=4).
H1975 is a NSCLC cell line that was reported to contain L858R/T790M EGFR mutations and H1650 had a deletion on Exon19 of EGFR and they were highly resistant to EGFR inhibitor gefitinib (Pao et al., 2005). We used EGFR inhibitor gefitinib and PARP inhibitor olaparib and found that as expected A549 cells were resistant to single or combination treatment of gefitinib/olaparib (Figure 5D, E, Figure S5B). While PC9, HCC2935 and HCC4006 cells were very sensitive to gefitinib and combination of these two inhibitors showed at least additive effect (Figure 5D,E, Figure S4C, Table S1). Interestingly, H1650 and H1975 cells were resistant to gefitinib, but sensitive to olaparib, and the combination of these two inhibitors led to significant growth inhibition (Figure 5D,E, Figure S5B, Table S1). These data suggest that by targeting nuclear functions of PKM2, PARP inhibitors may be used to treat EGFR-mutant lung cancers, especially some of the EGFR inhibitor-resistant cancers, which provides a way to overcome therapeutic resistance for EGFR mutant cancers.
Nuclear localization of PKM2 is associated with PAR expression in EGFR-mutant tumors
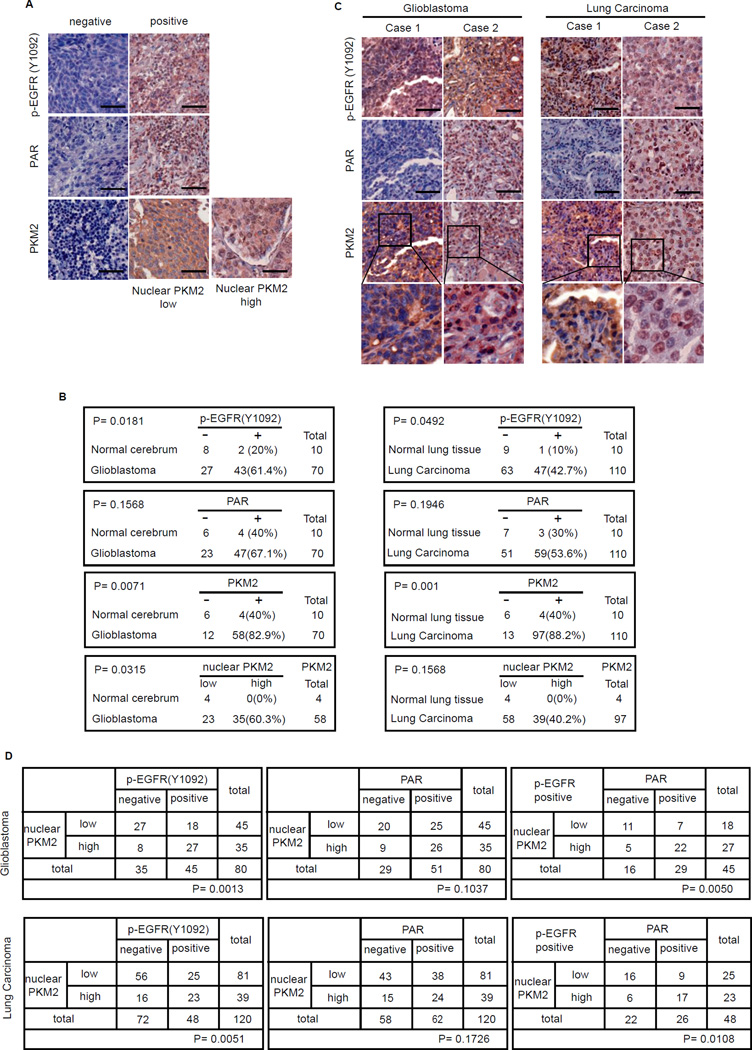
Our results so far have indicated that inhibition of PAR reduces growth of EGFR-mutant cancers by suppressing PKM2 nuclear localization and PKM2 nuclear functions. To determine the clinical relevance of PKM2 regulation by PAR in human cancers, we first validated that the anti-PKM2, PAR and p-EGFR antibodies are suitable for immunohistochemistry (IHC) staining (Figure S6A). We then determined the expression of p-EGFR, PKM2, and PAR in patient-derived Glioblastoma and lung carcinoma tissue samples (Figure 6A). We found that p-EGFR and PKM2 expressions were up-regulated in Glioblastoma (20% positive p-EGFR staining in normal cerebrum vs 61.4% positive p-EGFR staining in Glioblastoma, and 40% positive PKM2 staining in normal cerebrum vs 82.9% positive PKM2 staining in Glioblastoma respectively) and lung cancers (10% positive p-EGFR staining in normal lung tissue vs 42.7% positive p-EGFR staining in lung carcinoma, and 40% positive PKM2 staining in normal lung tissue vs 88.2% positive PKM2 staining in lung carcinoma), while there was no significant change in PAR expression in normal tissue versus in tumor tissue in both Glioblastoma and lung carcinoma TMA samples (Figure 6B). Moreover, in all PKM2 positive samples, nuclear PKM2 expression was up-regulated (none in normal cerebrum vs 60.3% in Glioblastoma, and none in normal lung tissue vs 40.2% in lung cancer) (Figure 6B).
Figure 6. Nuclear localization of PKM2 is correlated with PAR level in EGFR-mutant cancers.
(A) Immunohistochemical (IHC) staining of PKM2, PAR and p-EGFR in Glioblastoma and lung carcinoma specimens was conducted on the US Biomax tissue microarrays. Brown staining indicates positive immunoreactivity. Scale bars, 50 µM. (B) PAR, p-EGFR and PKM2 expression status in Glioblastoma and lung carcinoma tissue samples. (C) IHC staining of p-EGFR, PAR and PKM2 of representative human Glioblastoma and lung cancer samples. Scale bars, 50 µM. (D) Correlation between expression status of nuclear PKM2/p-EGFR, nuclear PKM2/PAR, p values were determined by chi-square analysis.
Interestingly, PKM2 nuclear localization correlated with EGFR activation (Figure 6D), which is consistent with the previous study (Yang et al., 2012a), but PAR levels shown no correlation with EGFR activation (Figure S6B). Notably, nuclear expression of PKM2 correlated with PAR only in p-EGFR positive tumor samples (Figure 6C,D, Figure S6B), indicating that PAR might be required for nuclear localization of PKM2 when EGF signal was activated. These results supported a role for PAR-dependent nuclear function of PKM2 in Glioblastoma and lung Carcinoma, and partially explained why only about 40–60% of EGFR-mutant tumors showed nuclear PKM2.
Discussion
In this study, we identified nuclear PKM2 as a target of PARP inhibition. Our results demonstrated that PKM2 contains a conserved PAR-binding C2H2 motif, which is similar to PBZ (Ahel et al., 2008) and PbR (Min et al., 2013) C2H2 motif. Upon growth stimuli, cytoplasmic PKM2 translocates into nucleus and binds to PAR through this C2H2 motif. We showed that the PKM2/PAR interaction is required for PKM2 nuclear retention and functions. Moreover, PAR level positively correlates with PKM2 nuclear expression in EGFR-mutant patient samples, and cancers contain nuclear PKM2 show significant sensitivity to PARP inhibition. These data not only revealed a mechanism of PAR-dependent regulation of nuclear PKM2, but also highlighted the potential application of PARP inhibitors in cancer therapy beyond BRCA mutant cancers.
Many PAR-binding domains and PAR-binding proteins have been identified recently and shown to act as important mediators involved in various signaling pathways and cellular processes (Li and Chen, 2014). The PAR-binding motif (PBM) (Gagne et al., 2008)-containing protein p53 and p21 are involved in cell cycle control and proliferation (Fahrer et al., 2007; Pleschke et al., 2000), while the binding of PAR to the PBM of Ku70 and XRCC1 is believed to regulate cellular response to DNA damage (Pleschke et al., 2000). In addition, the macrodomain-containing proteins macroH2A1 and macroH2A2 function in transcription regulation (Changolkar et al., 2007; Costanzi and Pehrson, 2001). The PBZ motifs of APLF and CHFR interact with PAR to control DNA damage response and mitotic checkpoint respectively (Ahel et al., 2008). The WWE domain of RNF146 also recognizes PAR and participates in RNF146- and PAR-mediated degradation of Axin (Huang et al., 2009), 3BP2 (Levaot et al., 2011), and PTEN (Li et al., 2015a). Here, we identified a putative PAR-binding C2H2 motif in PKM2 and demonstrated that PAR-binding is critical for the nuclear retention and nuclear functions of PKM2. This study further expands our understanding of PAR signaling in diverse cellular processes.
The M2 isoform of pyruvate kinase (PKM2) contributes to tumorigenesis and Warburg effect (Christofk et al., 2008; Lim et al., 2012). Cancers often evolve complex regulation of PKM2 to meet their needs for energy and cellular building blocks. For examples, HIF-1 and c-Myc regulate transcription and splicing of PKM2 (David et al., 2010; Luo et al., 2011), while v-Src and FGFR1 (fibroblast growth factor receptor type1) control the tetramer-to-dimer transition of PKM2 via phosphorylating PKM2 (Hitosugi et al., 2009; Presek et al., 1988). As a matter of fact, various post-translational modifications are known to occur on and regulate PKM2. These modifications on PKM2 include its K305 acetylation (Lv et al., 2011), K433 acetylation (Lv et al., 2013), C358 oxidation (Anastasiou et al., 2011), P403/408 hydroxylation (Luo et al., 2011), and S37 phosphorylation (Yang et al., 2012b). In our study, we demonstrated a mechanism for PKM2 regulation via PAR signaling, which is critical for PKM2 nuclear retention and nuclear function.
While the glycolytic function of PKM2 in cytosol is well studied, the nuclear functions of PKM2 as coactivator for HIF-1α (Luo et al., 2011) and β-catenin (Yang et al., 2011), and as nuclear protein kinase (Gao et al., 2012; Jiang et al., 2014; Yang et al., 2012a) were widely reported in the past few years. Although PKM2 protein kinase activity was questioned by a recent study (Hosios et al., 2015), a more recent publication demonstrated that yeast PKM2 can phosphorylate histone H3 both in vivo and in vitro (Li et al., 2015b). Thus, it is possible that while the protein kinase activity of PKM2 may be low, due to the high abundance of PKM2 in tumor cells, its protein kinase activity could still contribute to the nuclear functions of PKM2 by regulating glycolysis gene expression, Warberg effect and tumor growth.
Nuclear PKM2 is activated following the stimulation of human epidermal growth factor receptor (EGFR), while EGFR belongs to a family of receptor tyrosine kinases (TKs) including EGFR, ERBB2 (HER2), ERBB3 (HER3), and ERBB4 (HER4) (Avraham and Yarden, 2011). When binding to ligands such as EGF or transforming growth factor, EGFR is activated and promotes downstream events including cell proliferation and survival (Hynes and Lane, 2005). EGFR is frequently over-expressed or mutated in Glioma and lung cancers (Oxnard et al., 2011; Taylor et al., 2012), but only few high-grade glioma show strong clinical response to EGFR inhibitor like gefitinib (Taylor et al., 2012). This is not due to the failure of this drug to cross blood-brain barrier (Hegi et al., 2011), but due to the expression of extracellular-domain EGFR mutant, EGFRvIII (Gan et al., 2009). On the other hand, lung cancers also frequently express mutant EGFR. These mutants of EGFR, commonly within EGFR kinase domain, endow these lung cancers hypersensitive to EGFR inhibitors. However, treatment with EGFR TKIs in lung cancer is not curative. These tumors often acquire secondary mutations within EGFR kinase domain (normally the T790M mutation), which causes therapeutic resistance (Oxnard et al., 2011). Therefore, new generations of EGFR inhibitors or combinatory therapies are being developed and tested to improve treatment for lung cancer. The particularly interesting observation presented in our study is that PARP inhibitor treatment suppresses EGFR-mutant tumor growth, including the EGFRvIII mutant in Glioblastoma (Figure 4) and EGFR-T790M mutant in lung cancer cells (Figure 5). Moreover, combination of PARP inhibitor olaparib with EGFR inhibitor Gefitinib significantly overcomes the acquired resistance in some lung cancer cells (Figure 5D,E). Our findings suggest that PARP inhibitors may be used not only for the treatment of BRCA-deficient breast cancer and ovarian cancer (Scott et al., 2015), but also for other cancers that carry EGFR mutations and/or nuclear PKM2. Steering besides BRCA1/2, nuclear PKM2 as a secondary biomarker for using in clinical treatment.
Experimental Procedures
The information about antibody, constructs, cell culture, immunoprecipitation and immunofluorescence, GST pull-down and mass spectrometry analysis, pyruvate kinase assay, glucose uptake assay, lactate production assay, BrdU incorporation, laser microirradiation, surface Plasmon resonance, and in vitro PARsylation assay in this study is described in the Supplemental Experimental Procedures.
Dot blot assay
GST fusion proteins (10 pmol) were conjugated to the glutathione beads and incubated with Biotin-PAR (50 pmol, Trevigen) for 2 hrs at 4°C, The beads were washed by NETN buffer and proteins from beads were eluted by glutathione and spotted onto a nitrocellulose membrane. The membrane was blocked by TBST buffer with 5% milk and was examined by anti-PAR antibody.
Cell proliferation and Colony formation assays
For proliferation assay, equal numbers of cells were plated in 6-cm dished. From the next day, cells were collected and counted every day.
For colony formation assay, cells were seeded at 1000 cells (for H1975 cells, seed 5000 cells per dish) into 6-cm dishes. 12 hours after seeding, cells were treated with olaparib (0.5 uM) or gefitinib (50 nM). Medium was replaced every 2 days and cells were incubated for 12 days. Resulting colonies were fixed and stained with Coomassie blue. Numbers of colonies were counted with a GelDoc with Quantity One software (BioRad).
Human tissue IHC analysis
Human Glioblastoma multiforme (GBM) tissue arrays and Human lung cancer tissue array (BC041115a) were purchased from US Biomax. GL805a and GL806c all contain 35 cases of Glioblastoma and 5 cases of normal cerebrum. BC041115a contains 110 cases of lung carcinoma and 10 cases of normal lung tissue. Samples were deparaffinized and rehydrated, and antigens were retrieved by using 0.01 M sodium-citrate buffer (pH 6.0) in a microwave oven. To block endogenous peroxidase activity, the sections were treated with 1% hydrogen peroxide in methanol for 30 min. After 1h pre-incubation in 10% normal goat serum to prevent nonspecific staining, the samples were incubated with antibodies to PAR (4335-MC-100, Trevigen, 1:50 dilution), PKM2 (4053S, Cell Signaling Technology, 1:100 dilution) or EGFR-pY1172 (SAB400177, Sigma-Aldrich, 1:50 dilution) at 4°C overnight. The sections were then incubated with a biotinylated secondary antibody (Vector Laboratories, PK-6101, 1:200) for 3 hrs and then incubated with avidin-biotin peroxidase complex solution (1:100) for 30 min at RT. Color was developed with the 3-amino-9-ethylcarbazole (AEC) solution. Sections were counterstained with Mayer's haematoxylin. All immunostained slides were scanned on the Automated Cellular Image System III (ACIS III, Dako, Denmark) for quantification by digital image analysis. The correlation between proteins was determined by the chi-square analysis.
In vivo xenograft study
All animal experiments were performed in accordance with a protocol approved by the Institutional Animal Care and Use Committee (IACUC) of MD Anderson Cancer Center. Our experiments indicated that 5 or 6 mice per group could identify the expected effect of PKM2 or olaparib on tumor size and weight with 100% power. Animals were randomly assigned to different groups. Tumor cells (5 × 106 in 100 µl of medium) were injected into six- to eight-week-old female BALB/c nude mice. Tumor size were measured as indicated using a caliper and tumor volume was calculated using the formula: 0.5×L×W2. Mice were euthanized when they met the institutional euthanasia criteria for tumor size and overall health condition. The tumors were removed, photographed and weighted.
For olaparib-treated tumor study, we injected (5 × 106 in 100 µl of medium) tumor cells into xixto eight-week-old female BALB/c nude mice. After about 10 days, the tumor volume of sh-vector group reached a calculated average volume of 65 mm3, measured the tumor volume of all 3 groups (shv, shM2 and shM2+M2) and the animals in each group were randomized into 2 sub-groupps (5 mice/sub-group): (1) vehicle control. (2) olaparib. Mice were treated for 4 weeks, and olaparib was dosed (50mg/kg i.p. injection) once daily for 5 days/week. Tumor volume was measured at indicated days after treatment. Mice were euthanized and tumors were removed, photographed and weighted 4 weeks after treatment.
Statistical analysis
Each experiment was repeated there times or more, unless otherwise noticed. No samples were from the excluded analysis, and samples were randomly assigned to different groups. Data were analyzed by the One-way ANOVA test and Pearson chi-square analysis. We used an F-test to compare variances, and the variances were not significantly different. A p value <0.05 was considered as statistically significant.
Supplementary Material
Highlights.
Nuclear PKM2 binds to poly-ADP-ribose
PAR-binding is required for PKM2 nuclear retention
PARP inhibition suppresses growth of EGFR-mutant cancers
PAR level correlates with nuclear localization of PKM2 in EGFR-mutant cancers
Acknowledgments
We thank all our colleagues in Dr. Chen’s laboratory for insightful discussions and technical assistance. We thank Dr. zhimin Lu (MD Anderson Cancer Center) for reagents and suggestions for this work. We also thank Dr. Mien-Chie Hung (MD Anderson Cancer Center) for providing reagents. This work was supported in part by MD Anderson Start-up fund to JC. JC is also a recipient of an Era of Hope Scholar Research Award (W81XWH-09-1-0409). This work was also supported in part by MD Anderson’s Cancer Center Support Grant (CA016672).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
N.L and J.C designed the experiments. N.L performed all the experiments with assistance from F.L, J.W, M.K and M.K.T, D.J.T; H.L and S.L provided reagents and helpful suggestions and discussions on the experiment. J.C supervised the study. N.L and J.C wrote the manuscript. All authors commented on the manuscript.
The authors declare no conflict of interest.
References
- Ahel I, Ahel D, Matsusaka T, Clark AJ, Pines J, Boulton SJ, West SC. Poly(ADP-ribose)-binding zinc finger motifs in DNA repair/checkpoint proteins. Nature. 2008;451:81–85. doi: 10.1038/nature06420. [DOI] [PubMed] [Google Scholar]
- Ali AA, Timinszky G, Arribas-Bosacoma R, Kozlowski M, Hassa PO, Hassler M, Ladurner AG, Pearl LH, Oliver AW. The zinc-finger domains of PARP1 cooperate to recognize DNA strand breaks. Nat Struct Mol Biol. 2012;19:685–692. doi: 10.1038/nsmb.2335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang JK, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, et al. Inhibition of pyruvate kinase M2 by reactive oxygen species contributes to cellular antioxidant responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Lyon CE, Fox AH, Leung AK, Lam YW, Steen H, Mann M, Lamond AI. Directed proteomic analysis of the human nucleolus. Curr Biol. 2002;12:1–11. doi: 10.1016/s0960-9822(01)00650-9. [DOI] [PubMed] [Google Scholar]
- Aravind L. The WWE domain: a common interaction module in protein ubiquitination and ADP ribosylation. Trends Biochem Sci. 2001;26:273–275. doi: 10.1016/s0968-0004(01)01787-x. [DOI] [PubMed] [Google Scholar]
- Avraham R, Yarden Y. Feedback regulation of EGFR signalling: decision making by early and delayed loops. Nat Rev Mol Cell Biol. 2011;12:104–117. doi: 10.1038/nrm3048. [DOI] [PubMed] [Google Scholar]
- Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913–917. doi: 10.1038/nature03443. [DOI] [PubMed] [Google Scholar]
- Changolkar LN, Costanzi C, Leu NA, Chen D, McLaughlin KJ, Pehrson JR. Developmental changes in histone macroH2A1-mediated gene regulation. Mol Cell Biol. 2007;27:2758–2764. doi: 10.1128/MCB.02334-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- Costanzi C, Pehrson JR. MACROH2A2, a new member of the MARCOH2A core histone family. J Biol Chem. 2001;276:21776–21784. doi: 10.1074/jbc.M010919200. [DOI] [PubMed] [Google Scholar]
- Dantzer F, Schreiber V, Niedergang C, Trucco C, Flatter E, De La Rubia G, Oliver J, Rolli V, Menissier-de Murcia J, de Murcia G. Involvement of poly(ADP-ribose) polymerase in base excision repair. Biochimie. 1999;81:69–75. doi: 10.1016/s0300-9084(99)80040-6. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M, et al. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303–7307. doi: 10.1073/pnas.94.14.7303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahrer J, Kranaster R, Altmeyer M, Marx A, Burkle A. Quantitative analysis of the binding affinity of poly(ADP-ribose) to specific binding proteins as a function of chain length. Nucleic Acids Res. 2007;35:e143. doi: 10.1093/nar/gkm944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- Fisher AE, Hochegger H, Takeda S, Caldecott KW. Poly(ADP-ribose) polymerase 1 accelerates single-strand break repair in concert with poly(ADP-ribose) glycohydrolase. Mol Cell Biol. 2007;27:5597–5605. doi: 10.1128/MCB.02248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ, et al. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123–134. doi: 10.1056/NEJMoa0900212. [DOI] [PubMed] [Google Scholar]
- Gagne JP, Isabelle M, Lo KS, Bourassa S, Hendzel MJ, Dawson VL, Dawson TM, Poirier GG. Proteome-wide identification of poly(ADP-ribose) binding proteins and poly(ADP-ribose)-associated protein complexes. Nucleic Acids Res. 2008;36:6959–6976. doi: 10.1093/nar/gkn771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan HK, Kaye AH, Luwor RB. The EGFRvIII variant in glioblastoma multiforme. J Clin Neurosci. 2009;16:748–754. doi: 10.1016/j.jocn.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. 2012;45:598–609. doi: 10.1016/j.molcel.2012.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelmon KA, Tischkowitz M, Mackay H, Swenerton K, Robidoux A, Tonkin K, Hirte H, Huntsman D, Clemons M, Gilks B, et al. Olaparib in patients with recurrent high-grade serous or poorly differentiated ovarian carcinoma or triple-negative breast cancer: a phase 2, multicentre, open-label, non-randomised study. Lancet Oncol. 2011;12:852–861. doi: 10.1016/S1470-2045(11)70214-5. [DOI] [PubMed] [Google Scholar]
- Hacker HJ, Steinberg P, Bannasch P. Pyruvate kinase isoenzyme shift from L-type to M2-type is a late event in hepatocarcinogenesis induced in rats by a choline-deficient/DL-ethionine-supplemented diet. Carcinogenesis. 1998;19:99–107. doi: 10.1093/carcin/19.1.99. [DOI] [PubMed] [Google Scholar]
- Hegi ME, Diserens AC, Bady P, Kamoshima Y, Kouwenhoven MC, Delorenzi M, Lambiv WL, Hamou MF, Matter MS, Koch A, et al. Pathway analysis of glioblastoma tissue after preoperative treatment with the EGFR tyrosine kinase inhibitor gefitinib--a phase II trial. Mol Cancer Ther. 2011;10:1102–1112. doi: 10.1158/1535-7163.MCT-11-0048. [DOI] [PubMed] [Google Scholar]
- Hitosugi T, Kang S, Vander Heiden MG, Chung TW, Elf S, Lythgoe K, Dong S, Lonial S, Wang X, Chen GZ, et al. Tyrosine phosphorylation inhibits PKM2 to promote the Warburg effect and tumor growth. Sci Signal. 2009;2:ra73. doi: 10.1126/scisignal.2000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosios AM, Fiske BP, Gui DY, Vander Heiden MG. Lack of Evidence for PKM2 Protein Kinase Activity. Mol Cell. 2015;59:850–857. doi: 10.1016/j.molcel.2015.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hottiger MO, Hassa PO, Luscher B, Schuler H, Koch-Nolte F. Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci. 2010;35:208–219. doi: 10.1016/j.tibs.2009.12.003. [DOI] [PubMed] [Google Scholar]
- Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, et al. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of targeted inhibitors. Nat Rev Cancer. 2005;5:341–354. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- Inbar-Rozensal D, Castiel A, Visochek L, Castel D, Dantzer F, Izraeli S, Cohen-Armon M. A selective eradication of human nonhereditary breast cancer cells by phenanthridine-derived polyADP-ribose polymerase inhibitors. Breast Cancer Res. 2009;11:R78. doi: 10.1186/bcr2445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Li X, Yang W, Hawke DH, Zheng Y, Xia Y, Aldape K, Wei C, Guo F, Chen Y, Lu Z. PKM2 regulates chromosome segregation and mitosis progression of tumor cells. Mol Cell. 2014;53:75–87. doi: 10.1016/j.molcel.2013.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: 'PAR-laying' NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Levaot N, Voytyuk O, Dimitriou I, Sircoulomb F, Chandrakumar A, Deckert M, Krzyzanowski PM, Scotter A, Gu S, Janmohamed S, et al. Loss of Tankyrase-mediated destruction of 3BP2 is the underlying pathogenic mechanism of cherubism. Cell. 2011;147:1324–1339. doi: 10.1016/j.cell.2011.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Lu LY, Yang CY, Wang S, Yu X. The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 2013;27:1752–1768. doi: 10.1101/gad.226357.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu X. Function of BRCA1 in the DNA damage response is mediated by ADP-ribosylation. Cancer Cell. 2013;23:693–704. doi: 10.1016/j.ccr.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Chen J. ADP-ribosylation: activation, recognition, and removal. Mol Cells. 2014;37:9–16. doi: 10.14348/molcells.2014.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang Y, Han X, Liang K, Wang J, Feng L, Wang W, Songyang Z, Lin C, Yang L, et al. Poly-ADP ribosylation of PTEN by tankyrases promotes PTEN degradation and tumor growth. Genes Dev. 2015a;29:157–170. doi: 10.1101/gad.251785.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Swanson SK, Gogol M, Florens L, Washburn MP, Workman JL, Suganuma T. Serine and SAM Responsive Complex SESAME Regulates Histone Modification Crosstalk by Sensing Cellular Metabolism. Mol Cell. 2015b;60:408–421. doi: 10.1016/j.molcel.2015.09.024. [DOI] [PubMed] [Google Scholar]
- Lim JY, Yoon SO, Seol SY, Hong SW, Kim JW, Choi SH, Cho JY. Overexpression of the M2 isoform of pyruvate kinase is an adverse prognostic factor for signet ring cell gastric cancer. World J Gastroenterol. 2012;18:4037–4043. doi: 10.3748/wjg.v18.i30.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, Cole RN, Pandey A, Semenza GL. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. 2011;145:732–744. doi: 10.1016/j.cell.2011.03.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. 2012;23:560–566. doi: 10.1016/j.tem.2012.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Li D, Zhao D, Lin R, Chu Y, Zhang H, Zha Z, Liu Y, Li Z, Xu Y, et al. Acetylation targets the M2 isoform of pyruvate kinase for degradation through chaperone-mediated autophagy and promotes tumor growth. Mol Cell. 2011;42:719–730. doi: 10.1016/j.molcel.2011.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, Jiang Y, Zhou X, Li TT, Guan KL, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. 2013;52:340–352. doi: 10.1016/j.molcel.2013.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson M, Niedergang C, Schreiber V, Muller S, Menissier-de Murcia J, de Murcia G. XRCC1 is specifically associated with poly(ADP-ribose) polymerase and negatively regulates its activity following DNA damage. Mol Cell Biol. 1998;18:3563–3571. doi: 10.1128/mcb.18.6.3563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- Menear KA, Adcock C, Boulter R, Cockcroft XL, Copsey L, Cranston A, Dillon KJ, Drzewiecki J, Garman S, Gomez S, et al. 4-[3-(4-cyclopropanecarbonylpiperazine-1-carbonyl)-4-fluorobenzyl]-2H-phthalazin- 1-one: a novel bioavailable inhibitor of poly(ADP-ribose) polymerase-1. J Med Chem. 2008;51:6581–6591. doi: 10.1021/jm8001263. [DOI] [PubMed] [Google Scholar]
- Min W, Bruhn C, Grigaravicius P, Zhou ZW, Li F, Kruger A, Siddeek B, Greulich KO, Popp O, Meisezahl C, et al. Poly(ADP-ribose) binding to Chk1 at stalled replication forks is required for S-phase checkpoint activation. Nat Commun. 2013;4:2993. doi: 10.1038/ncomms3993. [DOI] [PubMed] [Google Scholar]
- Okano S, Lan L, Caldecott KW, Mori T, Yasui A. Spatial and temporal cellular responses to single-strand breaks in human cells. Mol Cell Biol. 2003;23:3974–3981. doi: 10.1128/MCB.23.11.3974-3981.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oxnard GR, Arcila ME, Chmielecki J, Ladanyi M, Miller VA, Pao W. New strategies in overcoming acquired resistance to epidermal growth factor receptor tyrosine kinase inhibitors in lung cancer. Clin Cancer Res. 2011;17:5530–5537. doi: 10.1158/1078-0432.CCR-10-2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez JG, Janne PA, Lee JC, Tracy S, Greulich H, Gabriel S, Herman P, Kaye FJ, Lindeman N, Boggon TJ, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pehrson JR, Fried VA. MacroH2A, a core histone containing a large nonhistone region. Science. 1992;257:1398–1400. doi: 10.1126/science.1529340. [DOI] [PubMed] [Google Scholar]
- Pleschke JM, Kleczkowska HE, Strohm M, Althaus FR. Poly(ADP-ribose) binds to specific domains in DNA damage checkpoint proteins. J Biol Chem. 2000;275:40974–40980. doi: 10.1074/jbc.M006520200. [DOI] [PubMed] [Google Scholar]
- Presek P, Reinacher M, Eigenbrodt E. Pyruvate kinase type M2 is phosphorylated at tyrosine residues in cells transformed by Rous sarcoma virus. FEBS Lett. 1988;242:194–198. doi: 10.1016/0014-5793(88)81014-7. [DOI] [PubMed] [Google Scholar]
- Rouleau M, Patel A, Hendzel MJ, Kaufmann SH, Poirier GG. PARP inhibition: PARP1 and beyond. Nat Rev Cancer. 2010;10:293–301. doi: 10.1038/nrc2812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott CL, Swisher EM, Kaufmann SH. Poly (ADP-Ribose) Polymerase Inhibitors: Recent Advances and Future Development. J Clin Oncol. 2015 doi: 10.1200/JCO.2014.58.8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strom CE, Johansson F, Uhlen M, Szigyarto CA, Erixon K, Helleday T. Poly (ADP-ribose) polymerase (PARP) is not involved in base excision repair but PARP inhibition traps a single-strand intermediate. Nucleic Acids Res. 2011;39:3166–3175. doi: 10.1093/nar/gkq1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor TE, Furnari FB, Cavenee WK. Targeting EGFR for treatment of glioblastoma: molecular basis to overcome resistance. Curr Cancer Drug Targets. 2012;12:197–209. doi: 10.2174/156800912799277557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Telli ML. PARP inhibitors in cancer: moving beyond BRCA. Lancet Oncol. 2011;12:827–828. doi: 10.1016/S1470-2045(11)70236-4. [DOI] [PubMed] [Google Scholar]
- Trucco C, Oliver FJ, de Murcia G, Menissier-de Murcia J. DNA repair defect in poly(ADP-ribose) polymerase-deficient cell lines. Nucleic Acids Res. 1998;26:2644–2649. doi: 10.1093/nar/26.11.2644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, Aldape K, Hunter T, Alfred Yung WK, Lu Z. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. 2012a;150:685–696. doi: 10.1016/j.cell.2012.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, Gao X, Aldape K, Lu Z. Nuclear PKM2 regulates beta-catenin transactivation upon EGFR activation. Nature. 2011;480:118–122. doi: 10.1038/nature10598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, Lyssiotis CA, Aldape K, Cantley LC, Lu Z. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. 2012b;14:1295–1304. doi: 10.1038/ncb2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.