Abstract
Acetylcholinesterase (AChE; EC 3.1.1.7), an essential enzyme of cholinergic neurotransmission in vertebrates, is a primary target in acute nerve agent and organophosphate (OP) pesticide intoxication. Catalytically inactive OP–AChE conjugates formed between the active-center serine and phosphorus of OPs can, in principle, be reactivated by nucleophilic oxime antidotes. Antidote efficacy is limited by the structural diversity of OP–AChE conjugates resulting from differences in the structure of the conjugated OP, the different active-center volumes they occupy when conjugated to the active-center serine of AChE, and the distinct chemical characteristics of both OPs and oximes, documented in numerous X-ray structures of OP-conjugated AChEs. Efforts to improve oxime reactivation efficacy by AChE structure–based enhancement of oxime structure have yielded only limited success. We outline here potential limitations of available AChE X-ray structures that preclude an accurate prediction of oxime structures, which are necessary for association in the OP–AChE gorge and nucleophilic attack of the OP-conjugated phosphorus.
Keywords: acetylcholinesterase, oxime antidote, organophosphate, nucleophilic reactivation, X-ray structure, backbone flexibility
Introduction
Toxicity from exposure to organophosphosphate (OP) nerve agents and pesticides arises from covalent conjugation with the active site serine of acetylcholinesterase (AChE), resulting in life-threatening consequences.1 In higher organisms, AChE inhibition leads to compromised cholinergic neurotransmission. The only accepted therapeutic approach for recovering the acetylcholine hydrolyzing activity of inactive OP–AChE conjugates is by intramuscular or intravenous administration of nucleophilic pyridinium aldoxime reactivators, such as pralidoxime (2PAM), obidoxime, or HI-6.2,3 These initial antidotes were developed on the basis of directing a quaternary aldoxime nucleophile to the site of catalysis of quaternary acetylcholine hydrolysis, decades before AChE was purified and characterized and its structure was solved.4
Progress in development of enhanced, more efficient, and more universal oxime antidote reactivators has been slow and limited.3 One reason is due to the structural diversity of the OPs forming the conjugated enzyme. An effective oxime antidote has to approach the AChE-conjugated phosphorus of the OP to within a reactivation-productive distance, which must be less than a few Å. This approach appears differentially difficult to achieve for OPs of differing structures. The non-inhibited mammalian AChE gorge volume of ~ 300 Å3 is reduced upon covalent OP binding by ~ 156 Å3 (cyclosarin), ~ 121 Å3 (paraoxon), ~ 115 Å3 (sarin), ~ 105 Å3 (VX), and ~ 90 Å3 (dichlorvos (DDVP)).
A rational way to understand and overcome this structural barrier is to redesign oxime antidote structures by studying active-center space limitations revealed in X-ray structures of apo AChE, OP–AChE conjugates, and AChE complexes with reversible ligands, including oximes. Those X-ray structures, continuously increasing in number, provide a growing database of information critical for structure-based drug development. Nevertheless, despite more than 100 related X-ray structures deposited in the Protein Data Bank (PDB) since 1991, including 16 oxime-containing structures, only limited success in structure-based antidote design has been reported. In this short review, we reflect on possible limitations of available structures that could compromise the AChE structure–based oxime antidote drug design.
Experimental conditions for X-ray structure determinations
Although X-ray crystallography is a powerful tool in structure-based drug design, a reasonable question to ask is, Are the determined X-ray structures of AChE conjugated by OPs and in complex with oximes sufficiently similar to physiologically active structures to serve as an appropriate structural template for drug design? To date, all AChE structures deposited in the PDB were solved from X-ray diffraction on crystals at very low temperatures (100 K), 210 degrees below physiological temperature. Not only are molecular dynamics of proteins significantly reduced at low temperatures, but, importantly, the relationship of temperature to reduction in dynamics is not linear, and for many proteins this relationship shows at least one break point at ~ 200 K (−73 °C).5 Thus, the time and space average of electron density determined from X-ray diffraction for a protein at 100 K may be both quantitatively and qualitatively different from the physiologically functional state. The vast majority of AChE structures reveal significant, ligand-independent similarity of alpha carbon (Cα) protein backbone conformations that even extend to the vast majority of side chain orientations.6 The absence of X-ray–based evidence for noticeable conformational diversity also precludes explanation of how some reversible AChE complexes with large ligands such as huperzine A7,8 (PDB structure IDs 1VOT, 4EY5) or galantamine7,9 (PDB IDs 1QTI, 4EY6) are able to form, or how some large alkylating ligands with bulky leaving groups (used for in crystallo covalent inhibition) find their way into the catalytic gorge, and how their leaving groups (not detected in corresponding structures) diffuse out of the tight and tortuous, 20 Å–deep gorge leading to a functional AChE active center.10 In terms of structure-based oxime design, existing X-ray structures neither reveal the molecular motions of AChE that may be critical for allowing an oxime antidote to approach the OP-conjugated phosphorus within a reactivation-productive distance nor delineate the specific interactions between protein and oxime that stabilize a productive antidote orientation.
An added experimental difficulty in a crystallographic experiment is the need to use precipitants to promote the growth of protein crystals of a size suitable for X-ray diffraction. Some precipitants, such as polyethylene glycol (PEG), of selected chain lengths have been repeatedly observed (one of many examples is an AChE structure with PDB ID 3M3D) to associate both outside and inside the AChE active center gorge, serving to stabilize the protein conformation, but also interfere with binding of ligand.
Dimensions of the active-center gorge opening
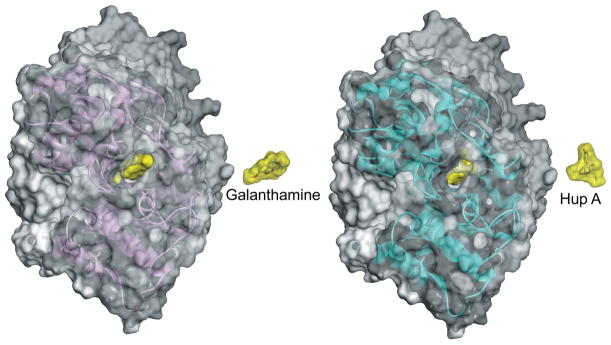
The geometry of the active-center gorge openings in all solved AChE structures appears narrow, with barely enough width to enable an acetylcholine molecule to approach the catalytic site. Variation of geometries among AChEs of different species is also evident. For example, mammalian (human and mouse) and fish (Torpedo californica) AChEs appear more similar, while insect (Drosophila melanogaster) and snake (Bungarus fasciatus) AChEs differ somewhat in the distribution of solvent-accessible volumes in the active center. Nevertheless, the overall protein fold and location of the buried catalytic site, generally ~20 Å deep in the active center, are largely conserved among AChEs, with an root mean square deviation (RMSD) of pairwise Cα backbone overlays not exceeding 1.5 Å.6 Molecules of the most structurally diverse reversible and covalent ligands of AChEs are larger in size and volume than ACh, yet they still efficiently bind inside the active center, associating with the enzyme at or near diffusion-limited rates11 despite the apparently constrained access to the active-center binding site seen in X-ray structures. Size constraints appear particularly critical for binding of two large ligands, huperzine A (volume ~ 234 Å3; diameter ~ 12 Å) and galantamine (volume ~ 269 Å3; diameter ~ 13 Å). Crystal structures of their reversible complexes with human (PDB IDs 4EY5, 4EY6) and fish (PDB IDs 1VOT, 1QTI) AChEs reveal available active-center openings (diameter ~ 5–10 Å), smaller than the size of bound ligands (Figure 1). None of the PDB-deposited AChE conformations, which exhibit limited structural variability, reveal how those ligands access their binding sites. Association rate constants for huperzine A11 are 4–5 orders of magnitude slower than diffusion limitation, indicating that very wide-open AChE conformations occur infrequently. In terms of structure-based oxime antidote design, conformations of native or OP-conjugated AChEs available in the PDB do not necessarily reflect the actual geometric configurations experienced by an oxime molecule accessing its target, the phosphorus of the conjugated OP. Consequently, more open, but yet unseen, AChE conformations must exist to both allow large reversible ligands to enter the AChE active center and to allow oxime antidotes to access the phosphorus in the additionally space-restricted gorge of OP-conjugated AChEs. The available geometries are most likely governed not only by concerted amino acid side chain rotations, but also by the flexibility of the AChE protein backbone.
Figure 1.
A view down the human AChE active-center gorge in X-ray structures of complexes with galantamine (left) and huperzine A (Hup A; right). Bound ligands are represented as yellow Connolly surfaces in the hAChE active-center gorge. Additional copies of ligands are shown next to hAChE molecules to better illustrate their relative sizes. Human AChE molecules are represented as red (galantamine complex; PDB ID 4EY6) and blue (huperzine A complex; PDB ID 4EY5) transparent Connolly surfaces with corresponding Cα backbone ribbons. Hydrogen atoms are included in all surface calculations with 1.4-Å probe size.
Conformational changes associated with ligand binding
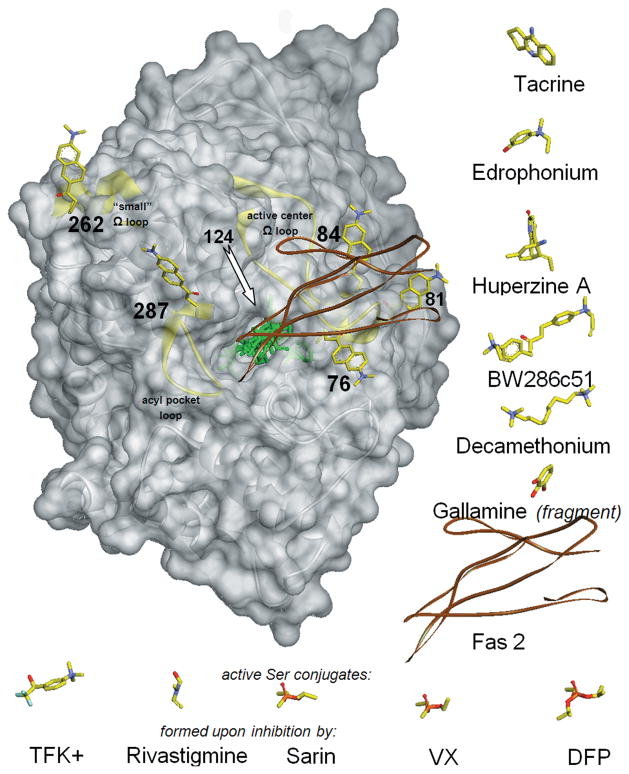
Conformational flexibility of the protein backbone in AChE appears essential for rapid substrate and other ligand association with AChE and seems to change locally, accommodating the formation of a reversible complex or covalent conjugate. By introducing the environmentally sensitive fluorophore acrylodan to specific sites in AChE, the regional flexibility of the AChE backbone can be spectroscopically probed in solution.12,13 This was achieved in mouse AChE, where acrylodan label was covalently attached to site-directed cysteine mutations at four structurally independent elements of the enzyme Cα backbone, in three surface loops, and in the interior of the active-center gorge (Fig. 2). Three positions were probed in the Ω loop that covers the active-center gorge: residue 76 at the rim of the gorge opening and residues 81 and 84 at the outer side of the loop, away from the gorge opening. One position (287) was probed in the outer side of the acyl pocket loop, close to Trp286 of the peripheral site, and another (262) in the small, flexible Ω loop, located on the surface, far from the catalytic or binding sites (Fig. 2); this loop is frequently disordered in X-ray structures.
Figure 2.
Structure of mouse AChE (gray transparent Connolly surface and ribbon) with covalently bound fluorescent acrylodan (represented by yellow sticks, docked to AChE manually) shown in each of six individually labeled positions (76, 81, 84,124, 262, and 287). In the centrally located active-center gorge, seven reversible ligands and five covalent conjugates are superimposed (represented by green sticks or brown ribbon for peptide Fas2) in positions resulting from overlays of corresponding ligand–AChE complexes or conjugates that include human, mouse, and Torpedo AChEs. To improve clarity, ligands are also shown next to the AChE molecule, in orientations identical or similar to the ones bound to AChE. Reversible ligands are shown to the right of AChE and covalent conjugates under AChE. Location of the acrylodan bound to position 124 is indicated by the arrow, but the attached acrylodan molecule is not shown, since it overlaps with most of the ligands bound in the center of the AChE gorge. Three structurally independent AChE surface loops (active center Ω loop, acyl pocket loop, and “small” ω loop) are highlighted as yellow ribbons.
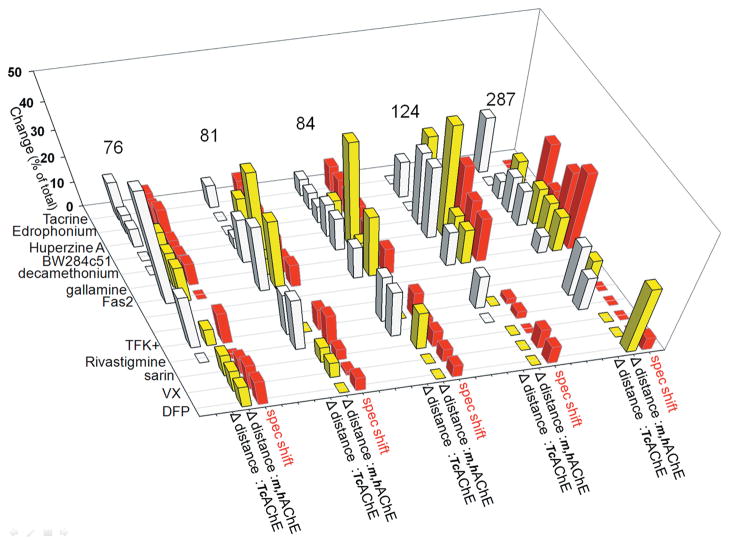
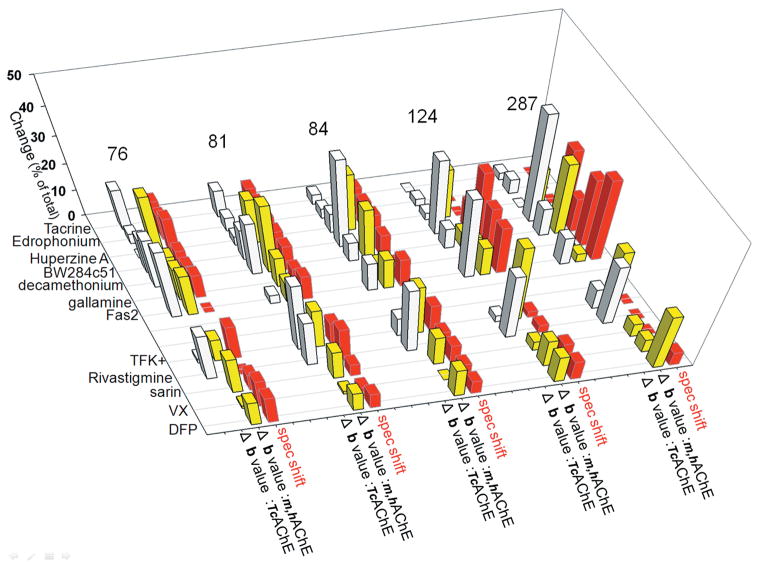
Except for position 262 in the distal and disordered small Ω surface loop, the Cα backbone conformations at all labeled positions were sensitive to either reversible or covalent ligand binding. That was detected by either red (+) or blue (−) shifts of the acrylodan fluorescence emission peak (Tables 1 and 2), respectively, indicative of more hydrophilic or hydrophobic acrylodan environments in the ligand-bound AChE. None of the bound ligands was large enough to sterically overlap with the acrylodan label, except when it was placed at position 124, since all remaining positions are significantly (> 10 Å) far away from the active center. The observed shifts in emission peaks of acrylodan fluorescence are therefore indicative of backbone movements associated with ligand binding. The movements were nicely illustrated in the large Ω loop covering the active-center gorge where the magnitude of spectral shifts was largest at position 84, coinciding with the loop tip, and smallest at position 76 closer to the loop base, indicating a concerted movement of residues on Ω the loop upon ligand binding. The exception was the peptide ligand Fas2 that binds in close enough proximity to five positions (76, 81, 84, 124, and 287) to influence acrylodan fluorescence by direct contact. This clear experimental evidence of ligand-associated movements in the AChE backbone in solution (at near physiological conditions) could not be readily predicted from comparison of the corresponding unbound and ligand-bound AChE X-ray structures. Based on fluorescence experiments, the Cα backbone of AChE exhibits well-defined and structurally detectable motions. It could be expected that distances between the backbone Cα atoms at five of the studied positions (76, 81, 84, 124, and 287) and the active serine (Ser203) Cα (taken as a point of reference) become different between unbound and ligand-bound X-ray structures, in agreement with acrylodan fluorescence–shift observations (Table 1; Fig. 3). Although, on average, both the smallest spectral shifts and the smallest distance differences were observed for sarin- and VX-generated covalent conjugates, and both the largest spectral shifts (next to Fas2) and the largest distance differences (next to Fas2) were observed for the bisquaternary reversible ligand decamethonium, more detailed correlations were limited. For example, the largest magnitudes of spectral shifts were observed at positions 84, then 81 > 124 > 287 > 76, while the largest magnitudes in distance differences were in position 76, then 81 > 84 > 287 = 124 for mammalian AChEs and at 81 then 84 = 287 > 76 > 124 for TcAChE. Also, the direction of the shift (indicating more hydrophilic or hydrophobic environment) and the distance to the active serine did not correspond. Similarly, comparison of structural parameters indicating thermal motions of atoms (i.e., B factor values) could be expected to detectably change upon ligand binding. Indeed, the data summarized in Table 2 and Figure 4 revealed, on average, the smallest B factor value change for VX- and sarin-generated covalent conjugates and the largest for decamethonium, consistent with the magnitudes of spectral shifts. However, the relative order of B factor change was not further preserved at discrete amino acid positions, as the B value change was observed for position 287 > 76 > 81 > 124 > 84 for mammalian AChEs and the order was 81 > 76 > 287 > 124 > 84 for TcAChE (compared to the spectral shift order of 84 then 81 > 124 > 287 > 76 in mAChE).
Table 1.
Comparison of ligand-associated spectral shifts of acrylodan attached at one of five positions in mAChE with ligand-associated variability of distances between backbone Cα atoms at each position and the Cα of the active serine observed from X-ray structures.
| Ligand | Leu 76 (Gln 74) Ω Loop gorge rim |
Glu 81 (Ser 79) Ω Loop top outside |
Glu 84 (Glu 82) Ω Loop top outside |
Tyr 124 (Tyr 121) Gorge “choke point” |
His 287 (Asn 280) Acyl pocket loop, PAS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spectral shift (nm) |
Δ Distance (complex-apo) (Å) |
Spectral shift (nm) |
Δ Distance (complex-apo) (Å) |
Spectral shift (nm) |
Δ Distance (complex-apo) (Å) |
Spectral shift (nm) |
Δ Distance (complex-apo) (Å) |
Spectral shift (nm) |
Δ Distance (complex-apo) (Å) |
||||||
| mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | |
| Tacrine | 4 | — | −0.2 | 21 | — | −0.2 | 33 | — | −0.1 | −3 | — | 0.0 | 0 | — | −0.3 |
| Edrophonium | 4 | — | 0.1 | 21 | — | 0.0 | 33 | — | 0.1 | 0 | — | 0.1 | 0 | — | 0.0 |
| Huperzine A | 6 | −0.6 | −0.1 | 21 | −0.2 | 0.0 | 33 | −0.1 | 0.1 | 0 | −0.2 | 0.0 | 0 | −0.1 | −0.1 |
| BW284c51 | 3 | — | 0.0 | 21 | — | −0.1 | 35 | — | −0.2 | −13 | — | 0.2 | −14 | — | 0.2 |
| Decamethonium | 3 | −0.7 | 0.0 | 21 | −0.5 | −0.4 | 28 | −0.5 | −0.2 | −35 | −0.3 | 0.2 | −7 | −0.1 | 0.2 |
| Gallamine | −4 | −0.6 | 14 | −0.1 | 9 | −0.1 | −25 | −0.1 | −14 | −0.1 | |||||
| Fas2 | 0 | −0.6 | −0.6 | 21 | −0.4 | −0.6 | 35 | −0.3 | −0.2 | −23 | −0.1 | 0.1 | −17 | −0.1 | −0.1 |
| TFK+ | 6 | −0.3 | 0.3 | 21 | 0.0 | −0.4 | 35 | 0.0 | −0.3 | 3 | 0.0 | 0.1 | 0 | 0.1 | 0.3 |
| Rivastigmine | 0 | — | 0.0 | −30 | — | −0.5 | 20 | — | −0.3 | −4 | — | 0.0 | 0 | — | 0.2 |
| Sarin | 3 | −0.4 | — | 11 | −0.1 | — | 23 | −0.2 | — | 0 | 0.0 | — | 0 | 0.0 | — |
| VX | −5 | −0.4 | — | 4 | 0.1 | — | 19 | 0.0 | — | −11 | 0.0 | — | 0 | 0.0 | — |
| DFP | −5 | −0.4 | — | −14 | 0.0 | — | 18 | 0.0 | — | −10 | 0.0 | — | −3 | 0.2 | — |
Note: Comparison of ligand-associated spectral shifts of acrylodan12,13 covalently attached at one of five positions in mAChE (76,81,84,124, and 287; TcAChE numbering is shown in parentheses) with ligand-associated variability of distances between backbone Cα atoms at each position and the Cα of the active serine (Δdistance = (distance between Cαposition and CαSer203)complex − (distance between Cαposition and CαSer203)apo) observed from X-ray structures. Distance differences for mouse AChE (mAChE) and human AChE (hAChE) X-ray structures were similar and were averaged (for complexes with same ligand) and are shown in the same column (as m,hAChE). Data for Torpedo californica AChE (TcAChE) structures are shown in a separate column consistent with higher similarity in the Cα backbone between mammalian AChEs (Cα RMSD ~ 0.6) compared to TcAChE and mAChE (Cα RMSD ~ 0.8).6 A list of the structures used in analysis is given in Table S1.
Table 2.
Comparison of ligand-associated spectral shifts of acrylodan covalently attached at one of five positions in mAChE with ligand-associated variability of B factors observed from X-ray structures.
| Ligand | Leu 76 (Gln 74) Ω Loop gorge rim |
Glu 81 (Ser 79) Ω Loop top outside |
Glu 84 (Glu 82) Ω Loop top outside |
Tyr 124 (Tyr 121) Gorge “choke point” |
His 287 (Asn 280) Acyl pocket loop, PAS |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Spectral shift (nm) |
Δ B value (complex-apo) |
Spectral shift (nm) |
Δ B value (complex-apo) |
Spectral shift (nm) |
Δ B value (complex-apo) |
Spectr al shift (nm) |
Δ B value (complex-apo) |
Spectral shift (nm) |
Δ B value (complex-apo) |
||||||
| mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | mAChE | m,hAChE | TcAChE | |
| Tacrine | 4 | — | −8 | 21 | — | −6 | 33 | — | −1 | −3 | — | 0 | 0 | — | 1 |
| Edrophonium | 4 | — | 2 | 21 | — | −3 | 33 | — | −1 | 0 | — | 1 | 0 | — | 2 |
| Huperzine A | 6 | −20 | 0 | 21 | −13 | 2 | 33 | −11 | 2 | 0 | −10 | 1 | 0 | −13 | 0 |
| BW284c51 | 3 | 7 | 21 | 7 | 35 | 9 | −13 | 11 | −14 | 15 | |||||
| Decamethonium | 3 | −10 | −10 | 21 | −20 | −12 | 28 | −10 | −2 | −35 | 3 | 3 | −7 | −25 | −4 |
| Gallamine | −4 | 11 | 14 | 9 | 9 | 2 | −25 | −5 | −14 | −1 | |||||
| Fas2 | 0 | −14 | −14 | 21 | 6 | 2 | 35 | 6 | −3 | −23 | 6 | −12 | −17 | −3 | −4 |
| TFK+ | 6 | 8 | 1 | 21 | 12 | −16 | 35 | 10 | −2 | 3 | 16 | −1 | 0 | 21 | −3 |
| Rivastigmine | 0 | 10 | −30 | 11 | 20 | 7 | −4 | 10 | 0 | 9 | |||||
| Sarin | 3 | −13 | — | 11 | −9 | — | 23 | −6 | — | 0 | 2 | — | 0 | −5 | — |
| VX | −5 | −1 | — | 4 | 1 | — | 19 | 0 | — | −11 | 6 | — | 0 | 5 | — |
| DFP | −5 | −9 | — | −14 | −6 | — | 18 | −6 | — | −10 | −6 | — | −3 | 21 | — |
Note: Comparison of ligand-associated spectral shifts of acrylodan12,13 covalently attached at one of five positions in mAChE (76, 81, 84,124, and 287; TcAChE numbering is shown in parentheses) with ligand-associated variability of B factors (Δ B value = (B value: Cαposition)complex − (B value: Cαposition)apo) observed from X-ray structures. B value differences observed for mouse AChE (mAChE) and human AChE (hAChE) X-ray structures were similar (for complexes with the same ligand) and are shown in the same column (as m,hAChE), while data for Torpedo californica AChE (TcAChE) structures are shown in a separate column. A list of the structures used in the analysis is given in Table S1.
Figure 3.
Graphical comparison of ligand-associated spectral shifts of acrylodan with ligand-associated variability of distances (Δ distance) to the active serine observed from X-ray structures (cf. Fig. 2; Table 1). Absolute values of magnitude changes in both spectral shifts and Δ distance are represented as the percentage of total change observed at each of the acrylodan-labeling sites to facilitate their comparison. Adapted from data published in Refs. 12 and 13.
Figure 4.
Graphical comparison of ligand-associated spectral shifts of acrylodan with ligand-associated variability of X-ray B factor values (Δ B value) to the active-center serine observed from X-ray structures (cf. Fig. 2, Table 2). Absolute values of the magnitude changes in both spectral shifts and Δ B value are represented as the percentage of total change observed at each of the acrylodan-labeling sites to facilitate their comparison. Adapted from data published in Refs. 12 and 13.
Similar conclusions were obtained in different and more complex experiments examining the decay of fluorescence anisotropy of the fluorophore-substituted side chain (quantitative comparisons are not shown here) where enhanced segmental motion is evident, when the fluorophore is in a more hydrophilic environment.14,15
Some similarity, thus, exists in the general AChE backbone flexibilities detected in solution and deduced from the analysis of X-ray structures. Predominant differences are found in more detailed, pointwise comparisons. Accordingly, X-ray structures obtained at low temperatures, when examined alone, may not be optimal templates for structure-based design of AChE ligands that are able to approach the AChE active center serine to serve as effective substrates, covalent inhibitors, or nucleophilic oxime reactivator antidotes. Interaction of those ligands may be limited by AChE backbone motions determined by energy landscapes dictated by physiological conditions in solution, but are even more limited under the energetically and dynamically restricted conditions necessary for resolving low-temperature X-ray structures.
Nonproductive oxime orientations in X-ray structures of OP–AChE conjugates
Catalytically inactive, OP-conjugated AChE formed upon exposure to nerve agent and pesticide OPs can be reactivated by small molecule nucleophilic oxime antidotes. In vitro reactivation efficacy of oxime antidotes depend on at least two factors: (1) the nucleophilicity of the attacking oximate group and (2) the geometry of the oximate oxygen approach to phosphorus of the conjugated OP, which ideally should conform to the SN2 in-line attack. Oximate nucleophilicity depends not only on the intrinsic electronic properties of an oxime molecule, but also on interaction with proton-withdrawing influences of AChE residues in the vicinity of the catalytic site, assuming that the oxime binds the active site in the protonated form. The angle of attack, on the other hand, depends on the geometry of the oxime molecule and the geometry of the AChE backbone and side chains surrounding the phosphorus. Structural information on oxime interaction with OP–AChE conjugates is thus critical for design of efficient reactivators from both electronic and steric points of view. The current 16 AChE structures in complex with oximes, five of which were of non-aged OP–AChE conjugates, do not appear to be very informative in that regard. Table 3 lists distances of oximate oxygens from the conjugated phosphorus that range from 4.9 Å to 7.6 Å, a distance too large to determine the reactive oxime conformation and orientation. On the other hand, the distance of the oximate to the most likely proton-withdrawing group in the vicinity, the catalytic triad His, varies from 2.8 Å to 7.9 Å. The lower distance limit may be sufficient for the formation of a hydrogen bond instrumental in influencing oxime-to-oximate dissociation, but it does not show proximity of the oximate to the phosphorus. Thus, none of the low-temperature structures illustrate an in-line attack for reactivation-reactive conformation of the oximate. The existing X-ray structures may well be influenced by restricted and reduced dynamics of the backbone and side chain in AChE protein at the low temperature.
Table 3.
Reactivation potential of crystallographic orientations of OP-AChE or AChE bound oximes.
| X-ray structure | P ···· O-N=C-R (Å) | His N ···· O-N=C-R (Å) |
|---|---|---|
| 2WHP : (mAChE+Sarin) + HI6 | ~ 4.9 | ~ 5.7 |
| 2WU3 : (mAChE+Fenamiphos) + HI6 | ~ 7.6 | ~ 7.9 |
| 2WU4 : (mAChE+Fenamiphos) + ortho7 | 5.7 5.7 | 2.8 2.8 |
| 2JEZ : (mAChE+Tabun) + HL07 | 5.6 5.1 | 6.1 4.5 |
| 2JF0 : (mAChE+Tabun) + ortho7 | 6.7 6.5 | 3.4 2.8 |
| 2VQ6 : TcAChE + 2PAM | (~ 3) | 6.4 |
| 5BWB : TcAChE + 2BIM-7 | (~ 10) | 10.6 |
| 5BWC: TcAChE + ortho7 | (~ 4) | 5.2 |
| 2WHR : mAChE + KO27 | (~ 5) | 5.1 5.1 |
| 2JEY : mAChE + HL07 | (~ 1) | 3.8 4.0 |
| 2GYU : mAChE + HI6 | (~ 9) | 9.6 9.6 |
| 2GYV : mAChE + ortho7 | (~ 6) | 5.7 5.6 |
| 2GYW : mAChE + obidoxime | (~ 5) | 4.3 4.5 |
| 2WHQ : mAChE (aged) + HI6 | 8.4 8.5 | 6.1 6.4 |
| 2WG1 : TcAChE (aged) + 2PAM | 7.5 | 4.9 |
Note: Reactivation potential of crystallographic orientations of OP-AChE or AChE bound oximes evaluated by distances between oximate oxygens and phosphorus atoms and between oximate oxygens and catalytic triad histidines. Obtained for a series of oximes (listed in bold italics) in interaction with mouse AChE (mAChE) and Torpedo californica AChE (TcAChE). Two parallel values for some of the structures reflect slight heterogeneity in catalytic centers of two monomers in crystallographic dimers. The approximate values given for the first two structures refer to distances between modeled oxime groups, inserted in place of missing atoms in the X-ray structure. Numbers in parentheses refer to approximate distance to a hypothetic P atom modeled atop Ser:O:gamma.
Conclusions
Structure-based design of functional AChE ligands is currently based on a database of more than 100 PDB-deposited X-ray structures obtained at low temperatures, which are all surprisingly similar in backbone conformation despite considerable structural diversity of bound ligands. While highly valuable for design of high-affinity reversible inhibitors where the precise point of binding is not critical, these structures are less useful as structural templates for design of substrates, covalent inhibitors, or nucleophilic oxime reactivators where the precise point of interaction between AChE and ligand is critical. It seems that efficacy of those ligands under physiological conditions (i.e., physiological temperatures in physiological media) relies on dynamics of AChE backbone motions that cannot be observed in the existing structures or precisely extrapolated from low-temperature X-ray data to physiological conditions. Advanced biophysical approaches are therefore necessary to reveal AChE structures in solution at close to physiological temperatures. High-resolution data from X-ray data collection at room temperature coupled with neutron diffraction experiments to resolve proton positions in the AChE active center are essential for understanding the mechanism of catalysis and role of specific AChE residues in nucleophilic oxime reactivation of OP–AChE conjugates. A combination of small-angle scattering of X-rays and neutrons in solution with room temperature X-ray diffraction should lead to constructing dynamic structural AChE templates for improved design of novel reactivator antidotes.
Supplementary Material
Footnotes
Conflicts of interest
The authors declare no conflicts of interest.
References
- 1.Taylor P. In: Goodman & Gilman’s The Pharmacological Basis of Therapeutics. 13. Brunton LL, Chabner B, Knollman B, editors. McGraw-Hill Inc; New York: 2011. pp. 239–254. [Google Scholar]
- 2.Jokanović M. Pyridinium oximes in the treatment of poisoning with organophosphorus compounds. In: Gupta R, editor. Handbook of Toxicology of Chemical Warfare Agents. 2. Academic Press; London: 2015. pp. 1057–1070. [Google Scholar]
- 3.Thiermann H, Worek F, Kehe K. Limitations and challenges in treatment of acute chemical warfare agent poisoning. Chem-Biol Interact. 2013;206:435–443. doi: 10.1016/j.cbi.2013.09.015. [DOI] [PubMed] [Google Scholar]
- 4.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 5.Ringe D, Petsko GA. The ‘glass transition’ in protein dynamics: what it is, why it occurs, and how to exploit it. Biophys Chem. 2003;105:667–680. doi: 10.1016/s0301-4622(03)00096-6. [DOI] [PubMed] [Google Scholar]
- 6.Radić Z, Taylor P. Structure and Function of Cholinesterases. In: Gupta R, editor. Toxicology of Organophosphate and Carbamate Compounds. Elsevier; Amsterdam: 2006. pp. 161–186. [Google Scholar]
- 7.Cheung J, Rudolph M, Burshteyn F, Cassidy M, Gary E, Love J, Height J, Franklin M. Structures of human acetylcholinesterase in complex with pharmacologically important ligands. J Med Chem. 2012;55:10282–10286. doi: 10.1021/jm300871x. [DOI] [PubMed] [Google Scholar]
- 8.Raves ML, Harel M, Silman I, Sussman JL. Structure of acetylcholinesterase complexed with the nootropic alkaloid, (−)-huperzine A. Nat Struct Biol. 1997;4:57–63. doi: 10.1038/nsb0197-57. [DOI] [PubMed] [Google Scholar]
- 9.Bartolucci C, Perola E, Pilger C, Fels G, Lamba D. Three-dimensional structure of a complex of galantamine (Nivalin) with acetylcholinesterase from Torpedo californica: implications for the design of new anti-Alzheimer drugs. Proteins. 2001;42:182–191. doi: 10.1002/1097-0134(20010201)42:2<182::aid-prot50>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- 10.Bartolucci C, Perola E, Cellai L, Brufani M, Lamba D. “Back door” opening implied by the crystal structure of a carbamoylated acetylcholinesterase. Biochemistry. 1999;38:5714–5719. doi: 10.1021/bi982723p. [DOI] [PubMed] [Google Scholar]
- 11.Radić Z, Taylor P. Interaction kinetics of reversible inhibitors and substrates with acetylcholinesterase and its fasciculin 2 complex. J Biol Chem. 2001;276:4622–4633. doi: 10.1074/jbc.M006855200. [DOI] [PubMed] [Google Scholar]
- 12.Shi J, Boyd E, Radić Z, Taylor P. Reversibly bound and covalently attached ligands induce conformational changes in the omega loop, Cys69-Cys96, of mouse acetylcholinesterase. J Biol Chem. 2001;276:42196–42204. doi: 10.1074/jbc.M106896200. [DOI] [PubMed] [Google Scholar]
- 13.Shi J, Radić Z, Taylor P. Inhibitors of different structure induce distinguishing conformations in the omega loop, Cys69-Cys96, of mouse acetylcholinesterase. J Biol Chem. 2002;277:43301–43308. doi: 10.1074/jbc.M204391200. [DOI] [PubMed] [Google Scholar]
- 14.Shi J, Tai K, McCammon JA, Taylor P, Johnson DA. Nanosecond dynamics of the mouse acetylcholinesterase omega loop. J Biol Chem. 2003;278:30906–30911. doi: 10.1074/jbc.M303730200. [DOI] [PubMed] [Google Scholar]
- 15.Boyd AE, Dunlop CS, Wong L, Radic Z, Taylor P, Johnson DA. Nanosecond dynamics of acetylcholinesterase near the active center gorge. J Biol Chem. 279:20612–20618. doi: 10.1074/jbc.M401482200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.