Abstract
Whole genome analyses were performed to test the hypothesis that temporal cortical gene expression differs between epilepsy patients rendered seizure-free versus non-seizure-free following anterior temporal lobectomy with amygdalohippocampectomy (ATL/AH). Twenty four patients underwent ATL/AH to treat medically intractable seizures of temporal lobe origin (mean age 35.5 years, mean follow up 42.2 months), they were then dichotomized into seizure-free and non-seizure-free groups. Tissue RNA was isolated from the lateral temporal cortex and gene expression analysis was performed. Whole genome data were analyzed for prognostic value for seizure-free outcome following ATL/AH by logistic regression. Genes that could distinguish seizure outcome groups were identified based on providing an accuracy of >0.90 judging by area under the receiver operating characteristic curve, AUC, with a P value of the slope coefficient of <0.05. Four genes and seven RNA probes were with prognostic value for post-operative seizure-free outcome. Gene expression associated with seizure-free outcome included relative down-regulation of: zinc finger protein 852 (ZNF852); CUB domain containing protein 2 (CDCP2); proline-rich transmembrane protein 1 (PRRT1); hypothetical LOC440200 (FLJ41170); RNA probe 8047763; RNA probe 8126238; RNA probe 8113489; RNA probe 8092883; RNA probe 7935228; RNA probe 806293 and RNA probe 8104131. This study describes the predictive value of temporal cortical gene expression for seizure-free outcome after ATL/AH. Four genes and seven RNA probes were found to predict post-operative seizure-free outcome. Future prospective investigation of these genes and probes in human brain tissue and blood could establish new biomarkers predictive of seizure outcome following ATL/AH.
Keywords: Gene expression, temporal lobectomy, prognostic factor, amygdalohippocampectomy, neurosurgical genomics
Introduction
Epilepsy is one of the most common neurological disorders affecting 2 to 4 million people or approximately 1% of the population of the United States [1]. Treatment of epilepsy may include antiepileptic medications, diet modifications, vagus nerve stimulation, surgical disconnection of epileptic pathways, or resective surgery. Appropriate medications can control seizures in approximately 70% of cases [2,3]. The remaining 30% of patients, with refractory seizures, may consider surgical intervention for treatment of their epilepsy resulting in 52%–84% of patients with remission of seizures [4–7]. The most commonly performed operation for treatment of medically intractable seizures is amygdalohippocampectomy with or without resection of additional temporal lobe tissue, such as anterior temporal lobectomy [8]. Approximately 65% of patients with medically intractable temporal lobe epilepsy treated with temporal lobectomy and amygdalohippocampectomy (ATL/AH) are rendered seizure-free [8]. Human brain tissue obtained during en bloc ATL/AH surgery provides unique opportunities to study fundamental biological processes involved in the pathophysiology of medically intractable temporal lobe epilepsy [9].
Gene expression analyses have been performed to elucidate molecular mechanisms involved in the pathophysiology of epilepsy [10,11]. However, to date there are no studies that investigate the differences in gene expression between surgical patients that are rendered seizure-free and those who continue to have seizures. The purpose of this study is to compare the gene expression of brain tissue in patients who are rendered seizure-free with the gene expression of those who continue to have seizures following ATL/AH. We propose the concept of “neurosurgical genomics” in which gene expression is used as a biomarker with prognostic value predicting successful outcome following operative neurosurgical intervention.
There has been debate regarding the prognostic value of lateral temporal cortical resection, supplementing amygdalohippocampectomy, for treatment of intractable temporal lobe epilepsy [12]. Lateral temporal cortical pathophysiology influences temporal lobe epileptogenicity and seizure-free outcome following ATL/AH. Long-term lateral temporal cortical cerebral blood flow recording has determined that temporal lobe epileptogenicity, as measured by seizure interval (1/seizure frequency), correlates inversely with cerebral perfusion [13]. Furthermore, lateral temporal cortical hypoperfusion is prognostic for seizure-free outcome following ATL/AH [13]. Based on this evidence correlating lateral temporal cortical pathophysiology with temporal lobe epileptogenicity and post-operative seizure-free outcome, the current study was performed to investigate lateral temporal cortical RNA expression prognostic value for seizure-free outcome following ATL/AH. The detection of temporal cortical genomic prognostic parameters for seizure-free outcome following ATL/AH should improve understanding of the lateral cortical pathophysiology in temporal lobe epilepsy.
Methods
This study was performed in accordance with protocol and research consents approved by the University of Arizona Institutional Review Board. Informed consent was obtained from all individual participants included in the study. Brain tissue samples were obtained from subjects who underwent ATL/AH at the Banner University Medical Center (Tucson, AZ). The neurosurgical operative technique for anterior temporal lobectomy (ATL) involves en bloc resection of lateral temporal cortex without resection of underlying white matter. A standard technique was employed in which the left- and right-sided ATL extent of resection was 4.5 cm and at least 5.5 cm, measured from the anterior middle fossa posteriorly, respectively. Our neurosurgical operative protocol was designed in consultation with neuropathology to include en bloc resection of the hippocampus with only gross surgical pathology examination permitting storage of whole hippocampal specimens for future research.
All tissue samples were stored in RNAlater RNA stabilization Solution (Qiagen, Valencia, CA) at −80° C until RNA extraction was performed. RNA was extracted using the RNeasy lipid tissue mini kit (Qiagen, Valencia, CA) following manufacturer’s instructions. RNA was then stored at −80° C until RNA analysis was performed.
The isolated total RNA samples were used to produce labeled target, hybridized to Affymetrix GeneChip Human Gene 1.0 ST Array (Affymetrix, Santa Clara, CA), and read using the Agilent/Affymetrix 2500A (Affymetrix, Santa Clara, CA) scanner according to manufacturer’s protocols.
Statistical Analysis
Descriptive statistics were conducted using IBM SPSS statistic 22 (IBM corporation, Armonk, NY). Comparisons were made using χ2 analysis for gender and race. Student t-test was used to analyze the significance of age among the groups. The level of significance was established at p < 0.05.
Analysis of gene expression data was performed utilizing the bioinformatics and statistical tools provided by R programming (www.r-project.org). The analysis included assessment of data quality, positive and negative controls on the Affymetrix ST1.0 microarrays, and probe annotation. Whole genome data were analyzed for prognostic value for seizure-free outcome following ATL/AH by logistic regression using the likelihood ratio test. This analysis is a way to model gene expression data so that it is predictive of a yes/no response, in this case a seizure-free or non-seizure-free outcome. The logistic regression is useful for data that is not normally distributed, as in this case. Logistic regression criteria for significance were defined by a high quality regression model with an area under the receiver operator curve (AUC) > 0.9 and p values ≤ 0.05 for the slope coefficient of the logistic regression gene models. All genes on the Affymetrix microarrays were modeled using logistic regression. In addition, a leave one out cross validation (LOOCV) was performed for these probes. For each gene, individual samples were removed in turn and a logistic regression model was built. The model was then used to score the removed observation for validation. Conditional density plots that indicate the predictive value of increasing cutoff values for each gene for epilepsy group were also produced.
Results
Subject Characteristics
Twenty four patients underwent ATL/AH for medically intractable temporal lobe seizures. All patients had lateral temporal cortex removed for genetic analysis. The twenty four subjects were dichotomized into two groups, post-operatively seizure-free (N=17) or non-seizure-free (N=7) following ATL/AH. Clinical and demographic factors for the 24 subjects of this study are delineated in Table 1. In all 24 subjects, gross surgical pathology examination of the hippocampus was normal. Pre-operative MRI brain scans and, in one case CT head scan, delineated the temporal lobe anatomy in each study subject (Table 2). The antiepileptic drug regimens for all 24 subjects in this study are delineated in Table 2, along with the extent of the surgical resection. There were no differences in the pre-operative anticonvulsant medication administration between the seizure-free and non-seizure-free groups (data not shown).
Table 1.
Clinical and Demographic Data for 24 patients undergoing ATL/AH and Temporal Cortical RNA Expression Analysis
| Subject Number |
Seizure Frequency (/Month) |
Age (Years) |
Gender (M/F) |
Seizure Focus |
Duration (Years) |
Etiology | Outcome (SF/NSF) |
Follow- up (Months) |
|---|---|---|---|---|---|---|---|---|
| 1 | 60 | 19 | M | LMT* | 7 | TBI | NSF | 12 |
| 2 | 4 | 59 | F | RLT* | 11 | TBI | SF | 33 |
| 3 | 4 | 30 | M | RT# | 8 | Unk | SF | 34 |
| 4 | 10 | 46 | F | RT# | 5 | Unk | SF | 23 |
| 5 | 4 | 16 | M | RT# | 10 | Unk | SF | 12 |
| 6 | 6 | 12 | F | RMT* | 6 | Unk | SF | 23 |
| 7 | 2 | 58 | F | LT# | 22 | Unk | SF | 20 |
| 8 | 2 | 14 | F | RT# | 8 | TBI | NSF | 78 |
| 9 | 12 | 48 | F | LRT* | 35 | Unk | NSF | 23 |
| 10 | 8 | 31 | F | RT# | 16 | Unk | NSF | 33 |
| 11 | 10 | 38 | M | RT# | 37 | Feb | SF | 30 |
| 12 | 2 | 44 | F | LT# | 23 | TBI | NSF | 83 |
| 13 | 1 | 50 | F | LT# | 12 | Unk | SF | 42 |
| 14 | 4 | 32 | M | RT# | 29 | Unk | NSF | 5 |
| 15 | 1 | 29 | F | RT# | 23 | JD | SF | 72 |
| 16 | 4 | 32 | M | RT# | 17 | Unk | NSF | 94 |
| 17 | 20 | 41 | F | RT# | 21 | Unk | SF | 53 |
| 18 | 1 | 41 | M | RT# | 34 | Unk | SF | 53 |
| 19 | 105 | 24 | M | LT# | 23 | I | SF | 23 |
| 20 | 3 | 37 | M | LT# | 6 | CAV | SF | 37 |
| 21 | 2 | 36 | M | RT# | 18 | Unk | SF | 79 |
| 22 | 4 | 29 | F | LT# | 28 | ABI | SF | 83 |
| 23 | 4 | 48 | F | RLT* | 31 | Unk | SF | 47 |
| 24 | 30 | 38 | M | RT# | 33 | Unk | SF | 39 |
Subject No. = Subject number in the study; Seizure Freq = Pre-operative seizure frequency (average number of seizures/month); Age (yrs.) = Age in years at time of anterior temporal lobectomy with amygdalohippocampectomy (ATL/AH); M/F = Male/Female; Duration = pre-operative duration of epilepsy diagnosis; Seizure Focus = Seizure Focus (L = left, R = right, MT = medial temporal, LT = lateral temporal, RT = regional temporal lobe; RT= right temporal lobe, LT = left temporal lobe) localized by * = ictal subdural EEG recording or # = ictal scalp-EEG recording; Etiology = etiology of epilepsy, TBI = traumatic brain injury, Feb = febrile illness, JD = juvenile diabetes, I = DPT immunization, CAV = cavernous malformation, Unk = unknown; ANI = anoxic brain injury (botulism), Outcome = Post-operative ATL/AH seizure outcome (SF = seizure-free, NSF = not seizure-free); Follow-up (mos.) = post-operative follow-up after ATL/AH in months at which time seizure outcome was determined.
Table 2.
Clinical Data for 24 patients undergoing ATL/AH and Temporal Cortical RNA Expression Analysis
| Subject Number | Seizure Focus | AED | MRI/CT | Extent Resection# (cm) |
|---|---|---|---|---|
| 1 | LMT* | P,V,S | MTS | 4.5 |
| 2 | RLT* | P,Z,L,LA,LAC,VI | TBI* | 5.5# |
| 3 | RT# | C,P,L,VI | MTS | 5.5# |
| 4 | RT# | C,O, Z, LA, VI | N | 5.5# |
| 5 | RT# | C, P, O, T, L, LA, LO |
N | 5.5# |
| 6 | RMT* | C, V, O, T, Z, L, Cl, LA, B, LO |
N | 5.5 # |
| 7 | LT# | P, L, LA, D, LO | MTS | 4.5 |
| 8 | RT# | O, L, LA | MTS | 5.5# |
| 9 | LRT* | P, G, Ph, VI, A | N | 4.5 |
| 10 | RT# | O, Z, L | MTS | 5.5# |
| 11 | RT# | L, LO, VI | HA | 5.5# |
| 12 | LT# | C, T, AC, Ci | MTS | 4.5 |
| 13 | LT# | P, O, T, L | MTS | 4.5 |
| 14 | RT# | P, V, T, LO | N | 5.5# |
| 15 | RT# | V, T, Z | MTS | 5.5# |
| 16 | RT# | C, P, V, O, T, Ph, L, TR, LY |
HA | 5.5# |
| 17 | RT# | O, Z, TR | MTS | 5.5# |
| 18 | RT# | C, P, O, L, TR | HA | 5.5# |
| 19 | LT# | LA, VI | MTS | 4.5 |
| 20 | LT# | P, L | CAV | 4.5 |
| 21 | RT# | C, P, Z, L | N | 5.5# |
| 22 | LT# | C, P, V, G, Z, LP | MTS | 4.5 |
| 23 | RLT* | C, L, LO | H | 5.5# |
| 24 | RT# | C | MTS | 5.5# |
Subject No. = Subject number in the study; Seizure Focus = Seizure Focus (first letter indicates lateraliaty: L = left, R = right, localization within temporal lobe indicated subsequently by: MT = medial temporal, LT = lateral temporal, RT = regional temporal lobe; RT= right temporal lobe, LT = left temporal lobe) localized by * = ictal subdural EEG recording or # = ictal scalp-EEG recording; AED = Anti-epileptic medication use pre-operatively (A = alprazolam, AC = acetazolamide, B = banzel, C = carbamaxepine, Ci = citalopram, Cl = clonazepam, D = diazepam, G = gabapentin, L = levetiracetam, LA = lamotrigine, LAC = lacosimide, LO = lorazepam, LP = loperamide, LY = lyrica, O = oxcarbazepine, P = phenytoin, Ph = phenobarbital, S = sabril, T = topiramate, TR = Trileptal, V = valproic acid, VI = vimpat, Z = zonisamide); MRI/CT = magnetic resonance imaging or computed tomography scan anatomic diagnosis pre-operatively, CT scan findings are indicated by “*” (MTS = medial temporal sclerosis, HA = hippocampal atrophy, CAV = cavernous malformation, N = normal temporal lobe anatomy, H = trigone heterotopia, TBI* = CT scan findings of traumatic brain injury), Extent Resection = extent of lateral temporal cortical resection in cm from anterior to posterior in the middle fossa, # = indicates a minimum of 5.5 cm anterior temporal lobectomy on the right side.
Array quality control
RNA A260/A280 was 1.3 to 2.1 and yield was 3.7 to 30.2 µg from 100 mg of brain tissue. The density plot showed no outlying arrays. Positive and Negative control probes showed good separation in expression values. Background and non-background corrected values were similar. Overall these arrays had consistently high quality data. Positive and negative controls were left in the data to be analyzed to assess further the role of gene probes in distinguishing the seizure outcome groups.
Predictive Genes
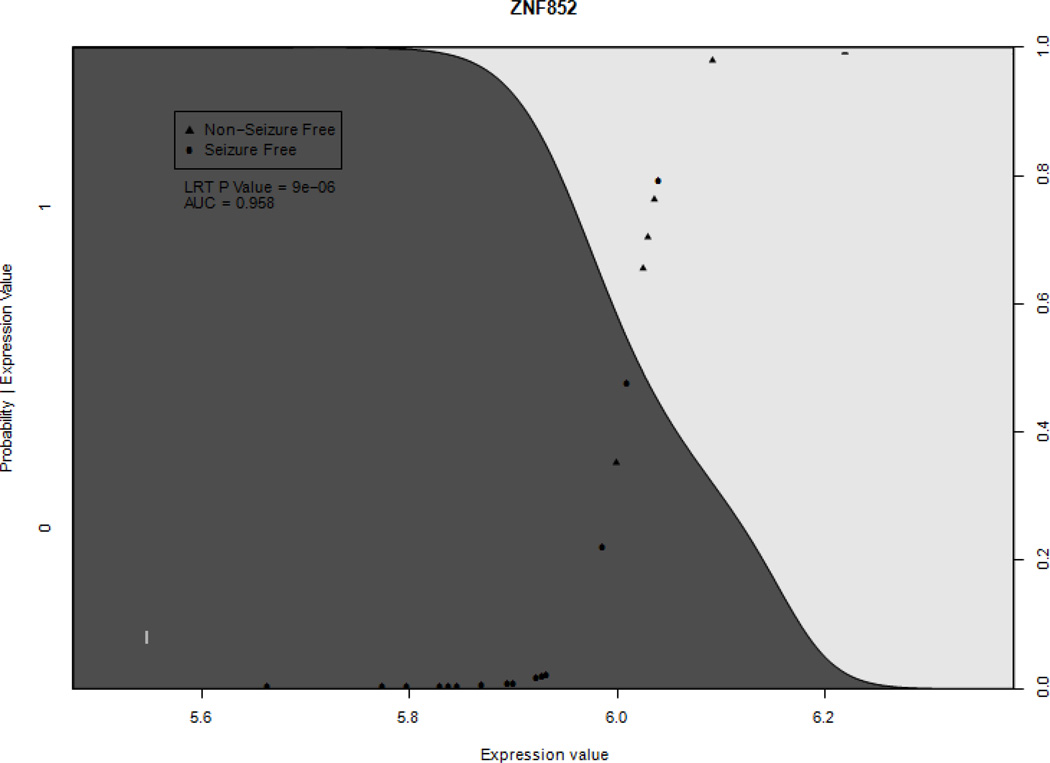
Logistic regression analysis identified four genes and seven RNA probes that met criteria for having significantly different expression levels between seizure-free and non-seizure-free groups (AUC>0.9 & P<0.05) (Figure 1). These four genes and seven RNA probes were identified as having predictive value for post-operative seizure-free outcome. Seizure-free subjects had significant relative down-regulation of the following genes: zinc finger protein 852 (ZNF852) (AUC 0.958, p value = 8.93 × 10−6, sensitivity = 0.857, specificity = 0.941); CUB domain containing protein 2 (CDCP2) (AUC 0.941, p value = 0.0002, sensitivity = 0.571, specificity = 0.941); proline-rich transmembrane protein 1 (PRRT1) (AUC 0.942, p value = 0.0077, sensitivity = 0.429, specificity = 0.941) and; hypothetical LOC440200 (FLJ41170) (AUC 0.908, p value = 0.000154, sensitivity = 0.857, specificity = 1.0) (Table 3). There was also a significant down-regulation of the following probes in association with seizure-free outcome: RNA probe 8047763; RNA probe 8126238; RNA probe 8113489; RNA probe 8092883; RNA probe 7935228; RNA probe 806293 and RNA probe 810413 (Table 4). An example of a conditional density plot for the genes and probes is presented in Figure 1. Finally, there were two negative controls, RNA probes 7895980 and 7893221, that showed significant differences between the seizure-free and non-seizure-free groups (AUC= 0.933, p=0.000135374 and AUC=0.915, p=0.000910822, respectively). The occurrence of these control probes among the results most likely results from the low expression values of the negative controls and the resulting variability in these values. Thus, a random assortment of low expression probes between groups could be expected to occur. Alternatively, the sequences of these probes could be detecting a gene that distinguishes the post-operative outcome groups. By contrast, the other genes found to distinguish the groups showed a larger change between groups, decreasing the chance of a random assortment. To examine more closely the role of individual samples in each post-operative outcome group in the gene models, a ‘leave one out’ analysis was performed. P values for the slope coefficients, determined by the likelihood ratio test, range between 6.6 × 10−7 and .030, showing that no individual sample is overly influencing the p values.
Figure 1.
Example conditional probability plots of the ZNF852 gene. Significantly different gene expression was defined as the area under receiver operating characteristic curve > 0.9 and p value < 0.05. Positive (triangle) points represent patients where seizures persisted after AHL/ATL. Shaded curve represents probability of seizure free outcome dependent on expression level
Table 3.
Genes Down-Regulated in Seizure-Free Subjects
| Genes/Probes | AUC | P value | Sensitivity* | Specificity* |
|---|---|---|---|---|
| ZNF852 | 0.958 | 8.93 × 10−6 | 0.857 | 0.941176471 |
| CDCP2 | 0.941 | 0.0002 | 0.571428571 | 0.941176471 |
| PRRT1 | 0.9415 | 0.0077 | 0.428571429 | 0.941176471 |
| FLJ41170 | 0.908 | 0.000154 | 0.857142857 | 1 |
= Sensitivity and specificity of RNA expression for post-ATL/AH seizure-free outcome.
Table 4.
RNA Probes Down-Regulated in Seizure-Free Subjects
| Probe | Chromosome Locus |
AUC | P value | Sensitivity* | Specificity* |
|---|---|---|---|---|---|
| 8047763 | Chr 2 (q33.3) | 0.983193 | 1.37 × 10−06 | 0.857142857 | 1 |
| 8126238 | Chr 6 (p21.2) | 0.97479 | 1.87 × 10−05 | 0.714285714 | 0.941176471 |
| 8113489 | Chr 5 (q22.1) | 0.957983 | 7.26 × 10−06 | 0.714285714 | 0.882352941 |
| 8092883 | Chr 3 (q29) | 0.94958 | 6.15 × 10−05 | 0.857142857 | 0.882352941 |
| 7935228 | Chr 10 (q24.1) | 0.915966 | 0.000133 | 0.714285714 | 0.941176471 |
| 8062693 | Chr 20 (q13.11) | 0.907563 | 0.000189 | 0.571428571 | 0.941176471 |
| 8104131 | Chr 4 | 0.907563 | 0.000224 | 0.571428571 | 1 |
= Sensitivity and specificity of RNA expression for post-ATL/AH seizure-free outcome.
Discussion
Temporal lobectomy and amygdalohippocampectomy (ATL/AH) will render patients with medically intractable temporal lobe epilepsy seizure-free in approximately 65% of all cases [8]. The fact that approximately one-third of patients continue to have post-operative seizures emphasizes the need for improved prognostic value of selection criteria for ATL/AH candidates. This current work demonstrates the potential value of temporal cortical gene expression for predicting seizure-free outcome following ATL/AH. A total of four genes and seven RNA probes were identified that predict seizure-free outcome following ATL/AH.
Zinc Finger 852 (ZNF 852)
The ZNF852 gene is located on chromosome 3p21.31. Relatively lower expression levels of the gene ZNF 852 are predictive of seizure-free outcome following ATL/AH. Gene Ontology annotations related to this gene include nucleic acid binding [14]. Zinc finger proteins contain one or more short regions called zinc finger domains which have one of the most abundant DNA binding motifs in the eukaryotic genome recognizing any DNA sequence [14]. These regions include a specific pattern of amino acids and one or more zinc ions [14]. Specifically, the Cys2-His2 zinc finger domain consists of a chain of two cysteines and two histidines, which fold around a single zinc ion contacting between 3 to 18 base pairs of DNA [14]. ZNF852 Contains 12 C2H2-type zinc fingers [14].
The elucidation of the zinc finger protein structure and the mechanism by which zinc finger proteins target a particular DNA sequence transformed the early field of genetic engineering, allowing specific DNA sequences of choice to be modified [15,16]. The first proof of principle of this technique in humans was applied to X-linked severe combined immune deficiency (SCID) in which fusion of zinc finger proteins to a nuclease domain caused a double stranded break creating a specific DNA sequence alteration stimulating homologous recombination between the chromosome and an extrachromosomal DNA donor [17]. Genome correction with engineered zinc finger nucleases has also been achieved in Down syndrome, alpha-1 antitrypsin deficiency and chronic granulomatous disease [14]. The current finding that this DNA-binding transcription factor’s RNA expression has prognostic value for seizure-free outcome following ATL/AH represents the first known function for ZNF852.
Cystathione Beta-synthase domain-containing protein 2 (CDCP2)
The CDCP2 gene is located on chromosome 1p32.3. A relatively lower expression level of CDCP2 is predictive of seizure-free outcome following ATL/AH. CDCP2 is a protein-coding gene and the cystathione Beta-synthase (CBS) domain is a highly evolutionarily-conserved protein domain identified in all three kingdoms of life [18]. CBS sequence pairs form a discrete structural domain, termed a Bateman domain, which binds adenosyl compounds and regulates IMP dehydrogenase, CBS, chloride channels and AMP-activated protein kinase [19]. CUB domain proteins mediate extracellular protein-protein interactions and exist as tandem repeats in cytosolic and membrane proteins and, as binding sites for adenosine derivatives, may serve as sensors of cellular energy status [20,18]. CUB proteins have diverse functions ranging from metabolic enzymes and transcription regulators to ion channels (including chloride) and transporters and are involved in a variety of major biological processes including complement activation, developmental patterning, tissue repair, axon guidance and angiogenesis, cell signaling, fertilization, haemostasis, inflammation, neurotransmission, receptor-mediated endocytosis and tumor suppression [20,18]. Point mutations in the CBS domain impair specific protein functions and are responsible for many human hereditary diseases including homocystinuria, Wolff-Parkinson-White syndrome, retinitis pigmentosa, congenital myotonia, idiopathic generalized epilepsy, hypercalciuric nephrolithiasis and classic Bartter syndrome [18,19]. There has been conflicting evidence regarding involvement of CDCP2 in Parkinson Disease. Maraganore et al. identified the CDCP2 gene, designated LOC200008, within the PARK10 locus for late-onset Parkinson disease (PD) susceptibility on chromosome 1 [20]. Maraganore et al. stated that the CDCP2 protein had inferred oxidoreductase activity and was potentially involved in cholesterol biosynthesis and electron transport and mapped the CDCP2 gene to chromosome 1p32 [20]. By examining CDCP2 SNPs in an expanded PD family dataset (293 multiplex and 467 singleton families) and a discordant sib-pair dataset, Li et al. found no significant association of CDCP2 with PD [21]. They noted that the results confirmed previous negative findings for CDCP2 as a candidate PARK10 gene [21]. CDCP2 may influence seizure outcomes in numerous ways. It may be the effect that this gene has on ion channels, or the effect the gene has on inflammation. At this time it is too early to speculate on the pathophysiological influences of this gene on seizure outcomes following surgery. The finding that CDCP2 RNA expression has prognostic value for seizure-free outcome following ATL/AH represents the first known function in partial epilepsy for CDCP2.
Proline-rich Transmembrane Protein 1 (PRRT1)
PRRT1 is located on chromosome 6p21.32. Relatively lower expression levels of PRRT1 are predictive of seizure-free outcomes following ATL/AH. PRRT1 encodes a transmembrane protein whose function enables cells to respond and interact with the environment [22]. Specifically, parotid gland PRRT1 expression is decreased by a macronutrient diet [22]. In humans, parotid gland proteins appear to exert a protective role against dietary phenols with an inhibitory effect on astringency perception [23]. The finding that PRRT1 RNA expression has prognostic value for seizure-free outcome following ATL/AH represents the first known function in epilepsy for PRRT1.
Hypothetical LOC440200 (FLJ41170)
FLJ41170 is located on chromosome 14q32.31. Relatively lower gene expression levels of FLJ41170 are predictive of seizure free outcome following ATL/AH. While no functional characterization has been performed, Hypothetical LOC440200 (FLJ41170) is a novel gene which is highly selectively expressed in fetal cartilage [24]. The finding that FLJ41170 RNA expression has prognostic value for seizure-free outcome following ATL/AH represents the first known function for FLJ41170.
Relative under-expression of seven RNA probes, heretofore lacking any known function as non-protein transcribing RNA, from temporal cortical tissue were found to have prognostic value for seizure outcome following anterior temporal lobectomy with amygdalohippocampectomy. These RNA probes are listed in Table 4.
Study Limitations
Limitations of this study include a small sample size limiting the statistical evaluation to univariate analysis. While the sample is large enough for logistic regression, it is not large enough to investigate combinations of these genes and probes in a multivariate predictive model. Therefore, a larger study will be required to develop a multivariate model with prognostic value for post-operative seizure outcome. While temporal cortical tissue was acquired prospectively, the study also has inherit limits associated with the analysis of a selected series of patients from a single institution in a retrospective manner. Finally, future studies may benefit from gene expression analysis from additional regions of the resected temporal lobe, including the amygdala and hippocampus.
Conclusion
This study describes the predictive value of temporal cortical gene expression for seizure outcome after ATL/AH. Four genes and seven RNA probes of unknown genetic function were found to be associated with seizure-free outcome after ATL/AH. All of the genes and RNA probes are novel as prognostic factors for post-operative seizure-free outcome. Future prospective investigation of these and other genes and RNA probes in human brain tissue and blood could establish new biomarkers predictive of seizure outcome following ATL/AH. Ultimately, preoperative analysis of neurosurgical genomics could lead to improved prognostication and patient selection for respective treatments.
Acknowledgments
The authors would like to acknowledge the following funding used to make this manuscript possible.
Funding: Weinand M, Co-PI, RO1MH065151 (Yuri Persidsky, Temple University) (HCS 04-42), National Institutes of Health subcontract from Temple University to University of Arizona. Support for purchase of laboratory materials.
Gallek M, Robert Wood Johnson Nurse Faculty Scholar Grant # 70318. Salary support.
Footnotes
Disclosure of Potential Conflicts of Interest:
Conflict of Interest: Drs. Gallek, Skoch and Weinand are co-inventors of U.S. Provisional Patent 61/828,596, “Neurosurgical Genomics”.
Ethical approval: All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. IRB# 1401194084, The University of Arizona.
References
- 1.Schachter SC. Seizure disorders. Med Clin North Am. 2009;93(2):343–351. doi: 10.1016/j.mcna.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 2.Kwan P, Brodie MJ. Early identification of refractory epilepsy. N Engl J Med. 2000;342(5):314–319. doi: 10.1056/NEJM200002033420503. [DOI] [PubMed] [Google Scholar]
- 3.Brodie MJ. Diagnosing and predicting refractory epilepsy. Acta Neurol Scand Suppl. 2005;181:36–39. doi: 10.1111/j.1600-0404.2005.00507.x. [DOI] [PubMed] [Google Scholar]
- 4.Cohen-Gadol AA, Wilhelmi BG, Collignon F, White JB, Britton JW, Cambier DM, Christianson TJ, Marsh WR, Meyer FB, Cascino GD. Long-term outcome of epilepsy surgery among 399 patients with nonlesional seizure foci including mesial temporal lobe sclerosis. J Neurosurg. 2006;104(4):513–524. doi: 10.3171/jns.2006.104.4.513. [DOI] [PubMed] [Google Scholar]
- 5.Jeong SW, Lee SK, Kim KK, Kim H, Kim JY, Chung CK. Prognostic factors in anterior temporal lobe resections for mesial temporal lobe epilepsy: multivariate analysis. Epilepsia. 1999;40(12):1735–1739. doi: 10.1111/j.1528-1157.1999.tb01591.x. [DOI] [PubMed] [Google Scholar]
- 6.Jutila L, Immonen A, Mervaala E, Partanen J, Partanen K, Puranen M, Kalviainen R, Alafuzoff I, Hurskainen H, Vapalahti M, Ylinen A. Long term outcome of temporal lobe epilepsy surgery: analyses of 140 consecutive patients. J Neurol Neurosurg Psychiatry. 2002;73(5):486–494. doi: 10.1136/jnnp.73.5.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McIntosh AM, Kalnins RM, Mitchell LA, Fabinyi GC, Briellmann RS, Berkovic SF. Temporal lobectomy: long-term seizure outcome, late recurrence and risks for seizure recurrence. Brain. 2004;127(Pt 9):2018–2030. doi: 10.1093/brain/awh221. [DOI] [PubMed] [Google Scholar]
- 8.Weinand ME, Wyler AR, Richey ET, Phillips BB, Somes GW. Long-term ictal monitoring with subdural strip electrodes: prognostic factors for selecting temporal lobectomy candidates. J Neurosurg. 1992;77(1):20–28. doi: 10.3171/jns.1992.77.1.0020. [DOI] [PubMed] [Google Scholar]
- 9.Fiala M, Avagyan H, Merino JJ, Bernas M, Valdivia J, Espinosa-Jeffrey A, Witte M, Weinand ME. Chemotactic and mitogenic stimuli of neuronal apoptosis in patients with medically intractable temporal lobe epilepsy. Pathophysiology. 2012 doi: 10.1016/j.pathophys.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Okamoto OK, Janjoppi L, Bonone FM, Pansani AP, da Silva AV, Scorza FA, Cavalheiro EA. Whole transcriptome analysis of the hippocampus: toward a molecular portrait of epileptogenesis. BMC Genomics. 2010;11:230. doi: 10.1186/1471-2164-11-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xi ZQ, Xiao F, Yuan J, Wang XF, Wang L, Quan FY, Liu GW. Gene expression analysis on anterior temporal neocortex of patients with intractable epilepsy. Synapse. 2009;63(11):1017–1028. doi: 10.1002/syn.20681. [Erratum appears in Synapse. 2010 Apr;64(4):339] [DOI] [PubMed] [Google Scholar]
- 12.Ramey WL, Martirosyan NL, Lieu CM, Hasham HA, Lemole GM, Jr, Weinand ME. Current management and surgical outcomes of medically intractable epilepsy. Clin Neurol Neurosurg. 2013;115(12):2411–2418. doi: 10.1016/j.clineuro.2013.09.035. doi: http://dx.doi.org/10.1016/j.clineuro.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 13.Weinand ME, Carter LP, el-Saadany WF, Sioutos PJ, Labiner DM, Oommen KJ. Cerebral blood flow and temporal lobe epileptogenicity. J Neurosurg. 1997;86(2):226–232. doi: 10.3171/jns.1997.86.2.0226. [DOI] [PubMed] [Google Scholar]
- 14.Ain QU, Chung JY, Kim YH. Current and future delivery systems for engineered nucleases: ZFN, TALEN and RGEN. Journal of controlled release : official journal of the Controlled Release Society. 2014 doi: 10.1016/j.jconrel.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 15.Gersbach CA. Genome engineering: the next genomic revolution. Nature methods. 2014;11(10):1009–1011. doi: 10.1038/nmeth.3113. [DOI] [PubMed] [Google Scholar]
- 16.Jabalameli HR, Zahednasab H, Karimi-Moghaddam A, Jabalameli MR. Zinc finger nuclease technology: advances and obstacles in modelling and treating genetic disorders. Gene. 2015;558(1):1–5. doi: 10.1016/j.gene.2014.12.044. [DOI] [PubMed] [Google Scholar]
- 17.Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435(7042):646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 18.Jeong BC, Yoo KS, Jung KW, Shin JS, Song HK. Purification, crystallization and preliminary X-ray diffraction analysis of a cystathionine beta-synthase domain-containing protein, CDCP2, from Arabidopsis thaliana. Acta crystallographica Section F, Structural biology and crystallization communications. 2008;64(Pt 9):825–827. doi: 10.1107/S1744309108025128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kemp BE. Bateman domains and adenosine derivatives form a binding contract. J Clin Invest. 2004;113(2):182–184. doi: 10.1172/JCI20846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maraganore DM, de Andrade M, Lesnick TG, Strain KJ, Farrer MJ, Rocca WA, Pant PV, Frazer KA, Cox DR, Ballinger DG. High-resolution whole-genome association study of Parkinson disease. Am J Hum Genet. 2005;77(5):685–693. doi: 10.1086/496902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li YJ, Deng J, Mayhew GM, Grimsley JW, Huo X, Vance JM. Investigation of the PARK10 gene in Parkinson disease. Annals of human genetics. 2007;71(Pt 5):639–647. doi: 10.1111/j.1469-1809.2007.00353.x. [DOI] [PubMed] [Google Scholar]
- 22.Simon J, DiCarlo LM, Kruger C, Johnson WD, Kappen C, Richards BK. Gene expression in salivary glands: effects of diet and mouse chromosome 17 locus regulating macronutrient intake. Physiological reports. 2015;3(2) doi: 10.14814/phy2.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dinnella C, Recchia A, Vincenzi S, Tuorila H, Monteleone E. Temporary modification of salivary protein profile and individual responses to repeated phenolic astringent stimuli. Chem Senses. 2010;35(1):75–85. doi: 10.1093/chemse/bjp084. [DOI] [PubMed] [Google Scholar]
- 24.Day A, Dong J, Funari VA, Harry B, Strom SP, Cohn DH, Nelson SF. Disease gene characterization through large-scale co-expression analysis. PLoS ONE. 2009;4(12):e8491. doi: 10.1371/journal.pone.0008491. [DOI] [PMC free article] [PubMed] [Google Scholar]