Summary
Somatic mutation of the DNMT3A gene at the arginine R882 site is common in acute myeloid leukaemia (AML). The prognostic significance of DNMT3A R882 mutation clearance, using traditional diagnostic next generation sequencing (NGS) methods, during complete remission (CR) in AML patients is controversial. We examined the impact of clearing DNMT3A R882 mutations at diagnosis to the detectable threshold of <3% during CR on outcome in 56 adult AML patients. Mutational remission, defined as clearance of pre-treatment DNMT3A R882 and all other AML-associated mutations to a variant allele frequency <3%, occurred in 14 patients whereas persistent DNMT3A R882 mutations were observed in 42 patients. There were no significant differences in disease-free or overall survival between patients with and without DNMT3A R882 mutation clearance. Patients with persistent DNMT3A R882 who cleared all other AML mutations and did not acquire new mutations (n = 30), trended towards longer disease-free survival (1·6 vs. 0·6 years, P = 0·06) than patients with persistence of DNMT3A R882, in addition to other mutations or acquisition of new AML-associated mutations, such as those in TET2, JAK2, ASXL1 and TP53 (n = 12). These data demonstrate that DNMT3A R882 mutations, as assessed by traditional NGS methods, persist in the majority of AML patients in CR.
Keywords: acute myeloid leukaemia, DNA (cytosine-5)-methyltransferase 3 alpha, mutation clearance, survival, prognosis
Acute myeloid leukaemia (AML) is a clinically and biologically heterogeneous disease characterized by cytogenetic, molecular and epigenetic alterations. Cytogenetic risk stratification of AML is well established (Grimwade et al, 1998; Byrd et al, 2002; Mrόzek et al, 2004). Other prognostic factors including mutational aberrations that are identified with next generation sequencing (NGS) methods are utilized to predict outcome or direct therapy. One such aberration, mutation of DNA (cytosine-5)-methyltransferase 3 alpha (DNMT3A), is reported to occur in 23–36% of patients (Thol et al, 2011; Hou et al, 2012; Marcucci et al, 2012; Renneville et al, 2012; Ribeiro et al, 2012; Gaidzik et al, 2013; Gale et al, 2015). Nearly two-thirds of these DNMT3A mutations affect the R882 codon in exon 23 of the gene’s methyltransferase domain (Ley et al, 2010; Thol et al, 2011; Hou et al, 2012; Marcucci et al, 2012; Renneville et al, 2012; Ribeiro et al, 2012; Gaidzik et al, 2013; Gale et al, 2015). The DNMT3A R882 mutations exert a dominant-negative effect, which inhibits the residual de novo methyltransferase activity of the wild-type DNMT3A enzyme, leading to aberrant methylation patterns and genomic instability (Kim et al, 2013; Russler-Germain et al, 2014). In contrast, the mechanism of myeloid transformation for mutations not involving codon R882 of DNMT3A is not well defined. The DNMT3A gene itself is ubiquitously expressed in embryonic stem cells and known to be an important mediator of haematopoietic stem cell (HSC) differentiation (Challen et al, 2012; Mayle et al, 2015).
Acute myeloid leukaemia patients with DNMT3A mutations have distinct clinical features, including older age, association with cytogenetically normal AML (CN-AML), as well as concurrent presence of internal tandem duplications of the FLT3 gene (FLT3-ITDs) and NPM1 and IDH1/IDH2 mutations (Ley et al, 2010; Thol et al, 2011; Hou et al, 2012; Marcucci et al, 2012; Renneville et al, 2012; Ribeiro et al, 2012; Gaidzik et al, 2013; Gale et al, 2015). Additionally, most (Ley et al, 2010; Thol et al, 2011; Hou et al, 2012; Marcucci et al, 2012; Renneville et al, 2012; Ribeiro et al, 2012; Gale et al, 2015), but not all (Gaidzik et al, 2013), studies have shown DNMT3A mutations to be associated with inferior overall survival (OS) in AML patients. Recent reports describe DNMT3A mutations to be present in preleukaemic HSCs that are capable of surviving chemotherapy and persisting in remission samples (Corces-Zimmerman et al, 2014; Shlush et al, 2014). Mutations in DNMT3A, as well as in ASXL1, JAK2, TP53, TET2 and several other genes, have also been detected in healthy persons with evidence of clonal haematopoiesis that have a predisposition towards developing malignant haematological disorders including AML (Genovese et al, 2014; Jaiswal et al, 2014; Steensma et al, 2015). As such, mutation of DNMT3A is hypothesized to be a founding mutation, with serial acquisition of other mutations, such as FLT3-ITD, NPM1 (Krönke et al, 2013) and IDH1/IDH2 occurring at later time points and ultimately leading to clinical evidence of AML. These findings have raised important questions regarding the necessity of DNMT3A R882 mutational clearance for achievement of long-term remission following induction chemotherapy.
To date, only four studies have indirectly addressed these questions and report conflicting results regarding the frequency of DNMT3A mutation persistence during complete remission (CR) and the associated prognostic significance of these persistent mutations (Hou et al, 2012; Pløen et al, 2014; Debarri et al, 2015; Klco et al, 2015). Thus, the impact of eliminating DNMT3A R882 mutations following treatment is unclear. Herein, we examine the largest, to our knowledge, cohort of serially monitored DNMT3A R882 patients and demonstrate that persistence of DNMT3A R882 during remission at a detectable level (i.e. sensitivity of 3%) with traditional NGS does not adversely impact long-term outcome.
Methods
Patients and treatment
This study examined 57 de novo AML patients, median age 51 years (range, 28–72 years) with a dominant DNMT3A R882 mutation at time of diagnosis (variant allele frequency [VAF] ≥40%), who attained a first complete remission (CR1) and had at least one CR1 bone marrow (BM) sample available for analysis. Most CR samples were collected within 1 week of attainment of morphological CR, and the majority was collected on the same day that CR was documented. Nineteen patients had CR samples collected more than a month after CR was confirmed. Fifty-six out of the 57 patients could be successfully analysed for their mutational status in CR and were included in the statistical analyses. The sequencing data quality of one patient did not allow accurate variant calling, and the sample was excluded from all further statistical analyses (Patient 57; see Table SI for details). All patients received intensive cytarabine and daunorubicin-based induction treatment on Cancer and Leukemia Group B (CALGB) trials. Cancer and Leukemia Group B is now part of the Alliance for Clinical Trials in Oncology. Of the 56 patients included in the outcome analyses, those younger than 60 years were treated on CALGB/Alliance protocols 10503 (n = 17) (Blum et al, 2012), 19808 (n = 14) (Kolitz et al, 2010), 10603 (n = 9) (Stone et al, 2012), 9222 (n = 2) (Moore et al, 2005), and 9621 (n = 2) (Kolitz et al, 2004). Patients 60 years or older were treated on CALGB 10201 (n = 6) (Marcucci et al, 2007), 9720 (n = 5) (Baer et al, 2002) and 10502 (n = 1) (Attar et al, 2013). Specific details regarding these trials and dosing schedules are provided in the Supporting Information.
Three patients received allogeneic haematopoietic stem-cell transplantation (HSCT) in CR1 and had remission marrow samples obtained before the transplant. Seventeen patients underwent autologous HSCT in first CR and all but four patients had remission marrow samples collected before transplant.
Institutional review board approval of all protocols was obtained before any research was performed. All patients gave written informed consent for treatment and research use of their specimens, in accordance with the Declaration of Helsinki.
Cytogenetic and mutational analyses
Pre-treatment cytogenetic analyses of BM and/or blood samples were performed in the CALGB institutional cytogenetics laboratories and the results were confirmed by central karyotype review as previously reported (Mrόzek et al, 2008). Viable cryopreserved BM or blood cells of patients enrolled onto the CALGB 9665 tissue bank protocol were stored for future analyses prior to starting treatment. Mononuclear cells from BM or blood were enriched by Ficoll-Hypaque gradient and cryopreserved in liquid nitrogen until thawed at 37°C for analysis. DNA extractions were performed using the DNeasy Blood and Tissue Kit (QIAGEN, Hilden, Germany). The mutational status of 34 genes (see Supporting Information for list of genes), the details of which are provided in Table SI, was determined by targeted amplicon sequencing using the Miseq platform (Illumina, San Diego, CA). Briefly, DNA library preparations were performed according to the manufacturer’s instructions. Samples were pooled and run on the Miseq machine using the Illumina Miseq Reagent Kit v3. Sequenced reads were aligned to the hg19 genome build using the Illumina Isis Banded Smith-Waterman aligner. Single nucleotide variant and indel calling were performed using MuTect and Varscan2 (Broad Institute, Cambridge, MA), respectively (DePristo et al, 2011; Cibulskis et al, 2013), and these calls were sorted and aggregated with Mucor (Kroll et al, 2016). All called variants underwent visual inspection of the aligned reads using the Integrative Genomics Viewer (Broad Institute) (Robinson et al, 2011). In addition to the genes analysed with the targeted sequencing panel, testing for the presence or absence of biallelic CEBPA mutations was performed as previously reported (Marcucci et al, 2008). Thus, both methods combined enabled us to analyse the mutational status of 35 genes. The lower limit of detection for the DNMT3A R882 mutation in our NGS method of sequencing was determined by titration assays of a cell line (MOLM-13) containing this aberration. Depth of coverage at DNMT3A R882 in the standard curve ranged from 298 to 606 reads with a median of 396 reads. All standard curve samples below 3% VAF were not detectable by our methods (see Table SII). A VAF of ≥3% was therefore used as a cutoff for analyses related to detecting DNMT3A and other mutations. Variants were considered mutations if they were not reported in the 1000 Genome database (ftp://ftp. 1000genomes.ebi.ac.uk/vol1/ftp/), dbSNP137 or dbSNP142 (http://www.ncbi.nlm.nih.gov/SNP/).
Definition of clinical endpoints and statistical analysis
Definitions of clinical endpoints, i.e., CR, disease-free survival (DFS) and OS, are provided in the Supporting Information. The main objective of this study was to evaluate clinical outcomes in DNMT3A R882-mutated AML patients in whom DNMT3A R882 and other AML mutations either persisted at the time of CR achievement or, within 1 year, were no longer detectable above a VAF cut-off of 3%. Demographic and clinical features of patients in whom DNMT3A R882 mutations cleared below the VAF cut-off of 3% at CR (Group 1) were compared with those of patients whose DNMT3A R882 mutations persisted at a VAF ≥3% at CR (Group 2) using the Fisher’s exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively (Vittinghoff et al, 2005). Group 2 was further subdivided as follows: Group 2a comprised patients with persistence of DNMT3A R882 at a VAF cut-off of ≥3% but no other AML-associated mutations during CR, and Group 2b included patients who, in addition to persistent DNMT3A R882 at a VAF cut-off of ≥3%, had (i) persistence of one or more other AML-associated mutations, or (ii) acquired additional AML mutations that were not present at diagnosis. Demographic and clinical features of these two groups were also compared using the Fisher’s exact and Wilcoxon rank-sum tests for categorical and continuous variables, respectively (Vittinghoff et al, 2005). Estimated probabilities of DFS and OS were calculated using the Kaplan–Meier method (Kaplan & Meier, 1958), and the log-rank test evaluated differences between survival distributions. Cox proportional hazards model was used to calculate hazard ratios (HR) for DFS and OS (Vittinghoff et al, 2005). All analyses were performed by the Alliance for Clinical Trials in Oncology Statistics and Data Center on a database locked on 8 July 2015.
Results
Clinical and molecular characteristics of patients who cleared DNMT3A R882 mutations at CR and of those who did not
Among 56 AML patients with DNMT3A R882 at diagnosis, 14 cleared the DNMT3A R882 mutation, as well as all other AML mutations, below the VAF cut-off of 3% during CR (Group 1), whereas in 42 patients, DNMT3A R882 persisted at a VAF ≥3% (Group 2; see Table SI for details). This 3% cut-off was derived from our sensitivity studies described in the methods. The demographic and clinical features of patients assigned to Groups 1 and 2 are shown in Table I. Molecular data for individual patients are provided in Table SI. There were no differences between groups with respect to age, sex, race, platelet counts, white blood cell counts or percentages of presenting blood and BM blasts (Table I). Consistent with prior reports (Ley et al, 2010; Thol et al, 2011; Hou et al, 2012; Ribeiro et al, 2012; Gaidzik et al, 2013; Gale et al, 2015) the majority of patients, 86% in both groups, had CN-AML, whereas eight patients had an abnormal karyotype at diagnosis. With regard to the European LeukaemiaNet (ELN) genetic categories (Döhner et al, 2010; Mrόzek et al, 2012) all patients in Group 1 (n = 14) were classified in the Favourable (n = 7) or Intermediate-I (n = 7) ELN Genetic Group, whereas three (8%) of the 39 classifiable patients in Group 2 were included in the ELN Intermediate-II or Adverse Genetic Group. Ten (71%) patients in Group 1 underwent either allogeneic (n = 2) or autologous (n = 8) HSCT in first CR, compared with 18 (43%) patients in Group 2 who underwent HSCT in first CR; all except one patient in Group 2 received autologous HSCT. The median VAFs of DNMT3A R882 for Groups 1 and 2 at diagnosis were similar [45% (range, 40–53%) vs. 46% (range, 41–50%), P = 0·58; see Table SI for details). Among patients in Group 2, VAFs of DNMT3A R882 mutations were generally lower at time of CR than at diagnosis.
Table I.
Comparison of pre-treatment clinical characteristics of de novo AML patients who cleared DNMT3A R882 mutations at complete remission and of those who did not.
| Characteristic | Group 1* (n = 14) |
Group 2* (n = 42) |
P |
|---|---|---|---|
| Age, years | |||
| Median | 48 | 52 | 0·55 |
| Range | 28–65 | 28–72 | |
| Age group, n (%) | |||
| Younger (<60 years) | 11 (79) | 33 (79) | 1·00 |
| Older (≥60 years) | 3 (21) | 9 (21) | |
| Sex, n (%) | |||
| Male | 8 (57) | 25 (60) | 1·00 |
| Female | 6 (43) | 17 (40) | |
| Race, n (%) | |||
| White | 13 (93) | 39 (97) | 1·00 |
| Non-white | 1 (7) | 1 (3) | |
| Haemoglobin, g/l | |||
| Median | 83 | 94 | 0·06 |
| Range | 68–132 | 71–251 | |
| Platelet count, ×109/l | |||
| Median | 57 | 81 | 0·12 |
| Range | 13–266 | 16–347 | |
| WBC count, ×109/l | |||
| Median | 30–6 | 45–5 | 0·76 |
| Range | 5·3–131·7 | 2·0–248·0 | |
| Percentage of blood blasts | |||
| Median | 68 | 62 | 0·64 |
| Range | 6–97 | 0–96 | |
| Percentage of bone marrow blasts | |||
| Median | 69 | 75 | 0·86 |
| Range | 24–91 | 6–91 | |
| Extramedullary involvement, n (%) |
3 (25) | 16 (41) | 0·33 |
| Pretreatment cytogenetic findings, n (%) | |||
| Normal karyotype | 12 (86) | 36 (86) | 0·61 |
| Sole trisomy† | 1 (7) | 2 (5) | |
| Two numerical abnormalities‡ | 1 (7) | 0 (0) | |
| Sole deletion§ | 0 (0) | 3 (7) | |
| Complex karyotype with >5 abnormalities |
0 (0) | 1 (2) | |
| ELN Genetic Group¶ | |||
| Favourable | 7 (50) | 18 (46) | 1·00 |
| Intermediate-I | 7 (50) | 18 (46) | |
| Intermediate-II | 0 (0) | 2 (5) | |
| Adverse | 0 (0) | 1 (3) | |
| Transplantation in 1st CR, n (%) | |||
| Allogeneic | 2 (14) | 1 (2) | 0·06 |
| Autologous | 8 (57) | 17 (40) | |
| No transplantation | 4 (29) | 24 (57) | |
| Time from 1st CR to 1st CR sample (d) | |||
| Median | 0 | 0 | 0·17 |
| Range | 0–185 | 0–333 | |
| Time from 1st CR to 1st CR sample, n (%) | |||
| Same day | 10 (71) | 22 (52) | 0·10 |
| Within 1–7 d after CR date | 1 (7) | 3 (7) | |
| Within 7–30 d after CR date | 1 (7) | 0 (0) | |
| More than 30 d after CR date | 2 (14) | 17 (40) | |
| DNMT3A R882 VAF % at diagnosis | |||
| Median | 45 | 46 | 0·58 |
| Range | 40–50 | 41–50 | |
AML, acute myeloid leukaemia; CR, complete remission with remission sample obtained at any time point after morphological remission is achieved; ELN, European LeukaemiaNet; n, number; WBC, white blood cell; VAF, variant allele frequency.
Group 1 is defined as patients whose DNMT3A R882 mutation cleared below the VAF cut-off of 3% in their remission sample and who had no other AML mutation, and Group 2 is defined as patients with a DNMT3A R882 mutation with a VAF >3% with or without other AML mutations in their remission sample.
This category includes single patients with sole +4, +8, and +21, respectively.
The patient in this category had +Y and +8.
This category includes single patients with sole add(7)(q22), del(9) (q13q22) and del(20)(q12), respectively.
Disease outcomes of patients who cleared DNMT3A R882 mutation and all other studied AML mutations at CR and of those who did not
We next sought to determine if clearance of DNMT3A R882 and mutations in 34 other AML-associated genes included in our panel was associated with an improved outcome. With a median follow-up time for living patients of 5·5 years (range: 3·7–11·2 years), no significant differences in DFS (median, 0·6 vs. 1·1 years, P = 0·52) or OS (median, 1·3 vs. 3·1 years; P = 0·47) were observed between patients in Group 1 and Group 2, respectively (Table II, Fig 1A, B). Because a minority of patients had BM samples collected later than 1 month after achieving a CR, we performed a subset analysis of only those 37 patients for whom BM samples were collected within 1 month of documented CR to exclude a potential bias related to the time of sample collection. As shown in Table III, this analysis again did not demonstrate any difference in DFS or OS between 12 patients with and 25 without clearance of all mutations (Table III, Fig 1C, D).
Table II.
Outcomes of de novo AML patients who cleared DNMT3A R882 mutations at CR and of those who did not.
| End point |
DNMT3A CR Group 1* (n = 14) |
DNMT3A CR Group 2* (n = 42) |
P | HR (95% CI) |
|---|---|---|---|---|
| Complete remission, n (%) | 14 (100) | 42 (100) | – | – |
| Disease-free survival | ||||
| Median, years | 0·6 | 11 | 0·52 | 1·26 (0·61–2·60) |
| % disease-free at 3 years (95% CI) | 29 (9–52) | 33 (20–48) | ||
| % disease-free at 5 years (95% CI) | 29 (9–52) | 31 (18–45) | ||
| Overall survival | ||||
| Median, years | 1·3 | 31 | 0·47 | 1·33 (0·62–2·83) |
| % alive at 3 years (95% CI) | 36 (13–59) | 50 (34–64) | ||
| % alive at 5 years (95% CI) | 36 (13–59) | 40 (25–54) |
AML, acute myeloid leukaemia; CI, confidence interval; CR, complete remission; HR, hazard ratio; n, number.
Group 1 is defined as patients whose DNMT3A R882 mutation cleared below the VAF cut-off of 3% in their remission sample and who had no other AML mutation, and Group 2 is defined as patients with a DNMT3A R882 mutation with a VAF >3% with or without other AML mutations in their remission sample.
Fig 1.
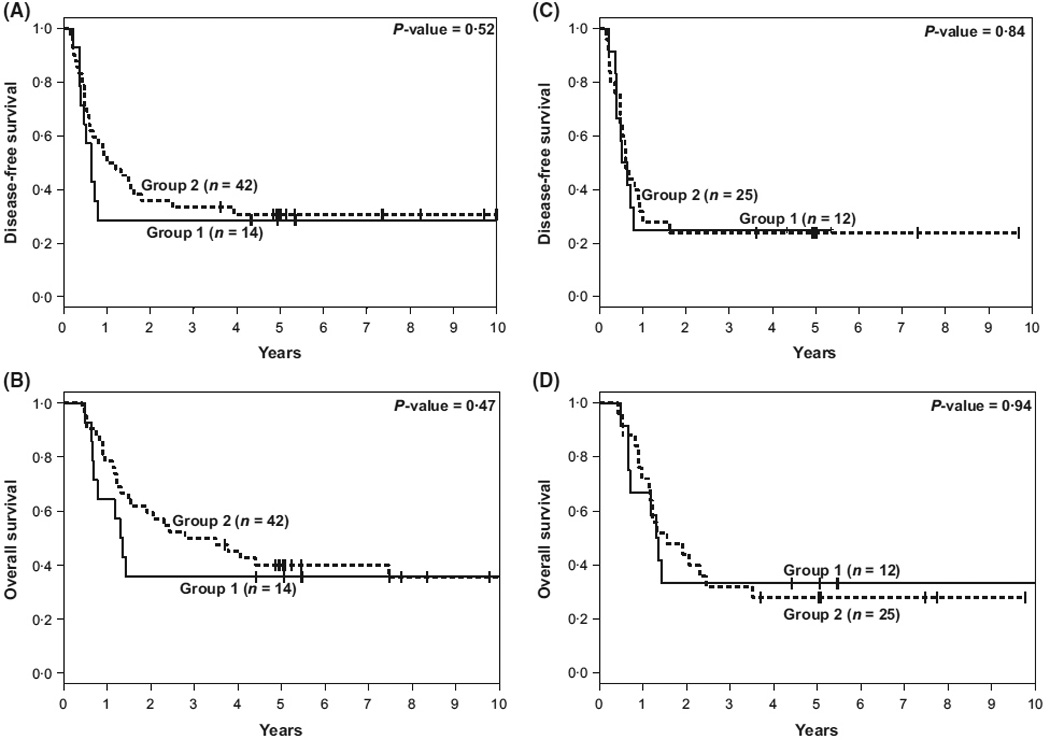
Kaplan–Meier survival plots of AML patients with pre-treatment DNMT3A R882 mutations who cleared DNMT3A R882 mutation at complete remission (CR) and those who did not. (A) Disease-free survival. (B) Overall survival. (C) Disease-free survival for patients with remission samples collected within 30 d of morphological CR date. (D) Overall survival for patients with remission samples collected within 30 d of morphological CR date. Group 1 includes patients whose DNMT3A R882 mutations cleared below the variant allele frequency (VAF) cut-off of 3% in their remission sample and who had no other AML mutation. Group 2 includes patients with a DNMT3A R882 mutation with a VAF ≥3% in their remission sample with or without other AML mutations.
Table III.
Outcomes of de novo AML patients whose CR sample was analyzed molecularly within a month after CR date according to whether their DNMT3A R882 mutations were cleared at CR or not.
| End point | DNMT3A CR Group 1* (n = 12) |
DNMT3A CR Group 2* (n = 25) |
P | HR (95% CI) |
|---|---|---|---|---|
| Complete remission, n (%) | 12 (100) | 25 (100) | – | – |
| Disease-free survival | ||||
| Median, years | 0·6 | 0·6 | 0·84 | 1·08 (0·49–2·40) |
| % disease-free at 3 years (95% CI) | 25 (6–50) | 24 (10–42) | ||
| % disease-free at 5 years (95% CI) | 25 (6–50) | 24 (10–42) | ||
| Overall survival | ||||
| Median, years | 1·3 | 1·5 | 0·94 | 1·03 (0·45–2·37) |
| % alive at 3 years (95% CI) | 33 (10–59) | 32 (15–50) | ||
| % alive at 5 years (95% CI) | 33 (10–59) | 28 (12–46) |
AML, acute myeloid leukaemia; CI, confidence interval; CR, complete remission; HR, hazard ratio; n, number.
Group 1 is defined as patients whose DNMT3A R882 mutation cleared below the VAF cut-off of 3% in their remission sample and who had no other AML mutation, and Group 2 is defined as patients with a DNMT3A R882 mutation with a VAF >3% with or without other AML mutations in their remission sample.
Disease outcomes of patients with persistence of DNMT3A R882 only and of patients with persistent DNMT3A R882 and other AML mutations at CR
Some patients in Group 2 with persistent DNMT3A R882 mutations during CR had no other analysed AML mutations present, whereas others did. We hypothesized that the absence of established, AML-associated mutations, outside of DNMT3A R882 mutations, might influence outcome. We performed a subset analysis of Group 2 patients, comparing those who had no other mutations associated with AML at CR (Group 2a, n = 30) with those who carried at least one persistent or newly acquired AML-associated mutation in addition to DNMT3A R882 (Group 2b, n = 12). The pretreatment features of these two patient groups are shown in Table SIII. Patients in Group 2a were younger, more often had normal cytogenetics, and differed by ELN group as compared to Group 2b. The VAFs of DNMT3A R882 for Groups 2a and 2b at diagnosis were similar [median, 46% (range 41–50%), vs. median 46% (range 41–50%), P = 0·90]. As shown in Table IV and Fig 2, patients with persistent DNMT3A R882 who had no other AML mutations displayed a trend toward improved DFS compared to patients with additional mutations (median, 1·6 vs. 0·6 years, P = 0·06). Although OS was longer for Group 2a patients than for Group 2b patients, this difference did not meet statistical significance (median, 3·8 vs. 1·3 years, P = 0·14).
Table IV.
Outcomes of de novo AML patients who at CR cleared DNMT3A R882 and all other mutations, patients who retained only a DNMT3A R882 mutation and patients who retained a DNMT3A R882 mutation and had one or more other mutations.
| End point |
DNMT3A CR Group 1* (n = 14) |
DNMT3A CR Group 2a* (n = 30) |
DNMT3A CR Group 2b* (n = 12) |
P† |
P‡ Gl vs. G2a |
HR (95% CI) Gl vs. G2a |
P‡ Gl vs. G2b |
HR (95% CI) Gl vs. G2b |
P‡ G2a vs. G2b |
HR (95% CI) G2a vs. G2b |
|---|---|---|---|---|---|---|---|---|---|---|
| Complete remission, n (%) | 14 (100) | 27 (100) | 15 (100) | – | – | – | – | – | – | – |
| Disease-free survival | ||||||||||
| Median, years | 0·6 | 1·6 | 0·6 | 0·06 | 0·35 | 1·63 (0·63–4·21) | 0·99 | 0·68 (0·24–1·95) | 0·06 | 0·42 (0·17–1·05) |
| % disease-free at 3 years (95% CI) | 29 (9–52) | 40 (23–57) | 17 (3–41) | |||||||
| % disease-free at 5 years (95% CI) | 29 (9–52) | 40 (23–57) | 8 (0–31) | |||||||
| Overall survival | ||||||||||
| Median, years | 1·3 | 3·8 | 1·3 | 014 | 0·40 | 1·67 (0·61–4·53) | 0·99 | 0·78 (0·26–2·35) | 014 | 0·47 (0·18–1·23) |
| % alive at 3 years (95% CI) | 36 (13–59) | 57 (37–72) | 33 (10–59) | |||||||
| % alive at 5 years (95% CI) | 36 (13–59) | 46 (27–63) | 25 (6–50) |
AML, acute myeloid leukaemia; CI, confidence interval; CR, complete remission; HR, hazard ratio; n, number.
Group 1 (G1) is defined as patients whose DNMT3A R882 mutation cleared below the VAF cut-off of 3% in their remission sample and who had no other AML mutation. Group 2a (G2a) is defined as patients with a DNMT3A R882 mutation with a VAF >3% in their remission sample but who cleared all other AML mutations. Group 2b (G2b) is defined as patients with a DNMT3A R882 mutation with a VAF >3% in their remission sample and persistent or newly acquired other AML mutations.
Overall P-value pertaining to comparison of all three patients groups.
Pairwise comparisons are adjusted from multiple comparisons using the Bonferroni method (Westfall et al, 1999).
Fig 2.
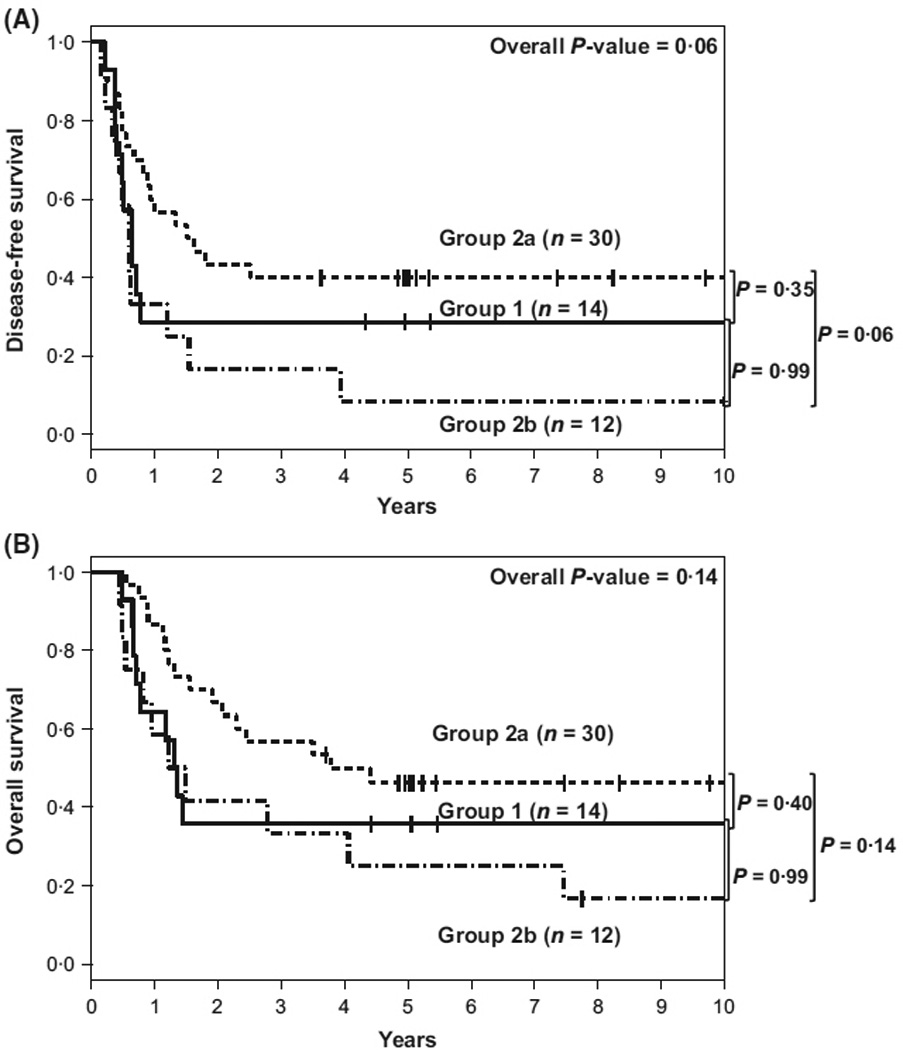
Kaplan–Meier survival plots of AML patients with pretreatment DNMT3A R882 mutations by three DNMT3A complete remission groups (A) Disease-free survival. (B) Overall survival. Group 1 includes patients whose DNMT3A R882 mutations cleared below the variant allele frequency (VAF) cut-off of 3% in their remission sample and who had no other AML mutation. Group 2a includes patients with a DNMT3A R882 mutation with a VAF ≥3% in their remission sample but with clearance of all other AML mutations. Group 2b includes patients with a DNMT3A R882 mutation with a VAF ≥3% and ≥1 other AML mutation in their remission sample.
We next compared the outcome of patients in Group 2a, who had persistence of only DNMT3A R882, with outcome of patients who lacked DNMT3A R882 mutations in remission (Group 1). No pretreatment feature separated these groups. The outcomes for patients in either group did not differ significantly with respect to either DFS (median, 1·6 vs. 0·6 years, P = 0·35) or OS (median, 3v8 vs. 1·3 years, P = 0·40).
Frequent identification of new mutations in remission samples in Group 2B
Six of the 12 patients in Group 2b acquired new AML-associated mutations at a VAF >3% (Pt ID 5, 11, 18, 21, 24, 29), in addition to persistent DNMT3A R882, during CR as shown in Table SI. Among these six patients, new mutations involved only ASXL1 (n = 2), only TET2 (n = 1), only JAK2 (n = 1), both TET2 and JAK2 (n = 1) and only TP53 (n = 1), all of which have been previously associated, not only with AML, but also with clonal haematopoiesis in healthy individuals without haematological disease (Xie et al, 2014). Of the six patients with new AML mutations, five ultimately relapsed.
Discussion
Herein, we report, to our knowledge, the largest series of sequentially analysed AML patients with DNMT3A R882 mutations who attained a CR and had remission samples examined for persistent DNMT3A R882 and other concurrent AML mutations with a traditional NGS sequencing assay typically utilized in medical practice. We demonstrate that clearance of other AML-associated mutations is common among patients with extended DFS, but DNMT3A R882 typically persists in the majority of patients. In contrast to previous publications suggesting that persistence of DNMT3A R882 mutations predicts for poor outcome (Hou et al, 2012; Klco et al, 2015), we found no significant differences in DFS or OS between patients who had cleared DNMT3A R882 mutations at the 3% detection during first CR and those who did not. A subset of our patients had marrow samples taken later than 30 d into treatment and we considered the possibility that this could explain, at least in part, the discordant findings. However, even after limiting our analysis to the 37 patients for whom BM samples were available within a month of their marrow CR date, our results were similar (Table III, Fig 1C, D). In a subset analysis of patients with persistent DNMT3A R882 mutations following induction chemotherapy (Group 2), we also demonstrated that patients who do not have other AML-associated mutations during CR (Group 2a) show a trend toward longer DFS compared with patients with new or persistent mutations in addition to DNMT3A R882 (Group 2b) (Table IV, Fig 2A). While it is possible that a more sensitive assay for detecting DNMT3A R882 mutations might identify a smaller subset whose disease has improved outcome, such assays are not widely utilized in general AML practice. Most importantly, our study demonstrates that persistence of DNMT3A R882A at levels of 3% or greater, as typically detected by NGS assays widely used in practice, bear no prognostic significance.
Our findings support those of two other studies in which persistent DNMT3A R882 mutations at the time of remission did not affect clinical course (Pløen et al, 2014; Debarri et al, 2015). Cell sorting experiments performed on non-leukaemic HSCs in the study by Pløen et al (2014) demonstrated the presence of mutant DNMT3A in T-cells and B-cells, at allele frequencies between 4% and 31%. Further, the DNMT3A allele burden in these cells was found to increase over time, thereby supporting the idea that acquisition of DNMT3A mutations occurs first in ancestral preleukaemic HSCs, as recently reported by two separate groups, where a growth advantage appears to exist (Corces-Zimmerman et al, 2014; Shlush et al, 2014).
Our study and that of Pløen et al (2014) have different conclusions relative to the importance of persistent DNMT3A R882 mutations at the time of remission than those of another recently published study Klco et al (2015). That study, in a manner similar to ours, used deep digital sequencing of paired samples obtained at diagnosis and remission in 50 AML patients and showed that mutation clearance in remission samples was associated with significantly improved event-free survival (EFS) (median, 17·9 vs. 6·0 months, hazard ratio [HR], 3·67) and OS (median, 42·2 vs. 10·5 months, HR, 2·86), compared with patients with persistent mutations in ≥5% bone marrow cells at CR. Within this group, 12 of 13 patients whose samples collected at remission had persistent DNMT3A mutations (type not specified) at a VAF >2·5% relapsed. Although the authors reported no association between EFS and mutation status of DNMT3A, OS was improved for patients, particularly those with intermediate-risk cytogenetics, who had cleared the DNMT3A mutation. Reasons for the discordant results between studies are uncertain but may reflect the fact that 10 of 23 (43%) patients with DNMT3A mutations analysed by Klco et al (2015) harboured non-R882 DNMT3A mutations, whereas our study and that of Pløen et al (2014) exclusively evaluated DNMT3A R882 mutations. Similar to the aforementioned study, we observed resolution of other common AML mutations in all but two of our patients with long DFS; these two patients had a JAK2 mutation (V617F) or two distinct mutations of the TET2 gene persisting at a high VAF.
Further, we did in fact see mutations in TET2, ASXL1, TP53 or JAK2 emerge during remission in six patients, all six of whom also harboured DNMT3A R882 mutations. In such a case, the non-DNMT3A R882 clone would be more likely to contribute to relapse. Analysis of relapse samples (which were not available in our series) would be required to determine this. Interrogation of this potential change with new primary ancestral clonal cells, and clonal architecture contributing to relapse should be considered as part of prospective AML trials in the future.
Limitations of our study include the retrospective nature of the analysis and the limited sample size, although ours is the largest patient cohort with DNMT3A R882 mutations followed serially reported to date. Moreover, a minority of our patients were analysed after CR1 was documented, although even when this group was excluded from the analysis, the results were similar.
In conclusion, we demonstrate that persistence of DNMT3A R882 mutations during CR does not adversely impact long-term DFS and OS in AML patients. Future studies examining other possible mechanisms that are modulated by persistent DNMT3A R882 expression have the potential to provide insights regarding its significance.
Supplementary Material
Acknowledgments
The authors are grateful to the patients who consented to participate in these clinical trials and the families who supported them; to Donna Bucci and the CALGB/Alliance Leukemia Tissue Bank at The Ohio State University Comprehensive Cancer Center, Columbus, OH, for sample processing and storage services and Lisa J. Sterling and Chris Finks for data management. This research was supported by National Institutes of Health (NIH) grants R35 CA197734, 5P30 CA016058, U10 CA180861, CA101140, CA140158, CA196171, CA180821, CA180882, and CA077658, the Coleman Leukemia Research Foundation (A.-K.E.), the Pelotonia Fellowship Program (A.-K.E.), the D Warren Brown Foundation, the NCCN Foundation (J.S.B.) and by an allocation of computing resources from The Ohio Supercomputer Center.
Footnotes
Authorship contributions
B.B., A.-K.E., K.M., J.C.B. and C.D.B. contributed to the design and analysis of the study and the writing of the manuscript; A.-K.E. and S.O. performed laboratory-based research; D.N., J.K., and J.B. performed statistical analysis; J.C.B., W.B., R.L., J.K., R.S. and C.D.B. were involved directly or indirectly in the care of patients and/or sample procurement. All authors read and agreed on the final version of the manuscript.
Disclosure of conflicts of interest
The authors declare no conflicts of interest.
Additional Supporting Information may be found in the online version of this article:
Data S1. Supporting methods.
Table SI. Genes mutated at diagnosis and remission in individual patients.
Table SII. Standard curve detection sensitivity study of DNMT3A R882 mutated versus DNMT3A wild-type cell lines.
Table SIII. Comparison of pretreatment clinical characteristics of de novo AML patients who at CR cleared DNMT3A R882 and all other mutations with patients who retained only a DNMT3A R882 mutation and patients who retained a DNMT3A R882 mutation and had one or more other mutations.
References
- Attar EC, Johnson JL, Amrein PC, Lozanski G, Wadleigh M, DeAngelo DJ, Kolitz JE, Powell BL, Voorhees P, Wang ES, Blum W, Stone RM, Marcucci G, Bloomfield CD, Moser B, Larson RA. Bortezomib added to daunorubicin and cytarabine during induction therapy and to intermediate-dose cytarabine for consolidation in patients with previously untreated acute myeloid leukemia age 60 to 75 years: CALGB (Alliance) study 10502. Journal of Clinical Oncology. 2013;31:923–929. doi: 10.1200/JCO.2012.45.2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baer MR, George SL, Dodge RK, O’Loughlin KL, Minderman H, Caligiuri MA, Anastasi J, Powell BL, Kolitz JE, Schiffer CA, Bloomfield CD, Larson RA. Phase 3 study of the multidrug resistance modulator PSC-833 in previously untreated patients 60 years of age and older with acute myeloid leukemia: Cancer and Leukemia Group B Study 9720. Blood. 2002;100:1224–1232. [PubMed] [Google Scholar]
- Blum W, Sanford B, Klisovic RB, DeAngelo DA, Uy G, Powell BL, Stock W, Baer MR, Kolitz JE, Wetzler M, Hoke E, Bloomfield CD, Geyer S, Marcucci G, Stone RM, Larson RA. Maintenance therapy with decitabine in younger adults with acute myeloid leukemia (AML) in first remission: a phase II Cancer and Leukemia Group B study (CALGB 10503, Alliance) [abstract] Blood. 2012:120. doi: 10.1038/leu.2016.252. Abstract 44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrd JC, Mrόzek K, Dodge RK, Carroll AJ, Edwards CG, Arthur DC, Pettenati MJ, Patil SR, Rao KW, Watson MS, Koduru PR, Moore JO, Stone RM, Mayer RJ, Feldman EJ, Davey FR, Schiffer CA, Larson RA, Bloomfield CD. Pretreatment cytogenetic abnormalities are predictive of induction success, cumulative incidence of relapse, and overall survival in adult patients with de novo acute myeloid leukemia: results from Cancer and Leukemia Group B (CALGB 8461) Blood. 2002;100:4325–4336. doi: 10.1182/blood-2002-03-0772. [DOI] [PubMed] [Google Scholar]
- Challen GA, Sun D, Jeong M, Luo M, Jelinek J, Berg JS, Bock C, Vasanthakumar A, Gu H, Xi Y, Liang S, Lu Y, Darlington GJ, Meissner A, Issa JP, Godley LA, Li W, Goodell MA. Dnmt3a is essential for hematopoietic stem cell differentiation. Nature Genetics. 2012;44:23–31. doi: 10.1038/ng.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nature Biotechnology. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces-Zimmerman MR, Hong WJ, Weissman IL, Medeiros BC, Majeti R. Preleu-kemic mutations in human acute myeloid leukemia affect epigenetic regulators and persist in remission. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:2548–2553. doi: 10.1073/pnas.1324297111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debarri H, Lebon D, Roumier C, Cheok M, Marceau-Renaut A, Nibourel O, Geffroy S, Helevaut N, Rousselot P, Gruson B, Gardin C, Chretien ML, Sebda S, Figeac M, Berthon C, Quesnel B, Boissel N, Castaigne S, Dombret H, Renneville A, Preudhomme C. IDH1/2 but not DNMT3A mutations are suitable targets for minimal residual disease monitoring in acute myeloid leukemia patients: a study by the Acute Leukemia French Association. Oncotarget. 2015;6:42345–42353. doi: 10.18632/oncotarget.5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Döhner H, Estey EH, Amadori S, Appel-baum FR, Büchner T, Burnett AK, Dom-bret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tall-man MS, Löwenberg B, Bloomfield CD. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- Gaidzik VI, Schlenk RF, Paschka P, Stölzle A, Späth D, Kuendgen A, von Lilienfeld-Toal M, Brugger W, Derigs HG, Kremers S, Greil R, Raghavachar A, Ringhoffer M, Salih HR, Wattad M, Kirchen HG, Runde V, Heil G, Petzer AL, Girschikofsky M, Heuser M, Kayser S, Goehring G, Teleanu MV, Schlegelberger B, Ganser A, Krauter J, Bullinger L, Döhner H, Döhner K. Clinical impact of DNMT3A mutations in younger adult patients with acute myeloid leukemia: results of the AML Study Group (AMLSG) Blood. 2013;121:4769–4777. doi: 10.1182/blood-2012-10-461624. [DOI] [PubMed] [Google Scholar]
- Gale RE, Lamb K, Allen C, El-Sharkawi D, Stowe C, Jenkinson S, Tinsley S, Dickson G, Burnett AK, Hills RK, Linch DC. Simpson’s paradox and the impact of different DNMT3A mutations on outcome in younger adults with acute myeloid leukemia. Journal of Clinical Oncology. 2015;33:2072–2083. doi: 10.1200/JCO.2014.59.2022. [DOI] [PubMed] [Google Scholar]
- Genovese G, Kahler AK, Handsaker RE, Lind-berg J, Rose SA, Bakhoum SF, Chambert K, Mick E, Neale BM, Fromer M, Purcell SM, Svantesson O, Landen M, Hoglund M, Lehmann S, Gabriel SB, Moran JL, Lander ES, Sullivan PF, Sklar P, Gronberg H, Hultman CM, McCarroll SA. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. New England Journal of Medicine. 2014;371:2477–2487. doi: 10.1056/NEJMoa1409405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimwade D, Walker H, Oliver F, Wheatley K, Harrison C, Harrison G, Rees J, Hann I, Stevens R, Burnett A, Goldstone A. The importance of diagnostic cytogenetics on outcome in AML: analysis of 1,612 patients entered into the MRC AML 10 trial. Blood. 1998;92:2322–2333. [PubMed] [Google Scholar]
- Hou HA, Kuo YY, Liu CY, Chou WC, Lee MC, Chen CY, Lin LI, Tseng MH, Huang CF, Chiang YC, Lee FY, Liu MC, Liu CW, Tang JL, Yao M, Huang SY, Ko BS, Hsu SC, Wu SJ, Tsay W, Chen YC, Tien HF. DNMT3A mutations in acute myeloid leukemia: stability during disease evolution and clinical implications. Blood. 2012;119:559–568. doi: 10.1182/blood-2011-07-369934. [DOI] [PubMed] [Google Scholar]
- Jaiswal S, Fontanillas P, Flannick J, Manning A, Grauman PV, Mar BG, Lindsley RC, Mermel CH, Burtt N, Chavez A, Higgins JM, Moltchanov V, Kuo FC, Kluk MJ, Henderson B, Kinnunen L, Koistinen HA, Ladenvall C, Getz G, Correa A, Banahan BF, Gabriel S, Kathiresan S, Stringham HM, McCarthy MI, Boehnke M, Tuomilehto J, Haiman C, Groop L, Atzmon G, Wilson JG, Neuberg D, Altshuler D, Ebert BL. Age-related clonal hematopoiesis associated with adverse outcomes. New England Journal of Medicine. 2014;371:2488–2498. doi: 10.1056/NEJMoa1408617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of American Statistical Association. 1958;53:457–481. [Google Scholar]
- Kim SJ, Zhao H, Hardikar S, Singh AK, Goodell MA, Chen T. A DNMT3A mutation common in AML exhibits dominantnegative effects in murine ES cells. Blood. 2013;122:4086–4089. doi: 10.1182/blood-2013-02-483487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klco JM, Miller CA, Griffith M, Petti A, Spencer DH, Ketkar-Kulkarni S, Wartman LD, Christopher M, Lamprecht TL, Helton NM, Duncavage EJ, Payton JE, Baty J, Heath SE, Griffith OL, Shen D, Hundal J, Chang GS, Fulton R, O’Laughlin M, Fron-ick C, Magrini V, Demeter RT, Larson DE, Kulkarni S, Ozenberger BA, Welch JS, Walter MJ, Graubert TA, Westervelt P, Radich JP, Link DC, Mardis ER, DiPersio JF, Wilson RK, Ley TJ. Association between mutation clearance after induction therapy and outcomes in acute myeloid leukemia. JAMA. 2015;314:811–822. doi: 10.1001/jama.2015.9643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolitz JE, George SL, Dodge RK, Hurd DD, Powell BL, Allen SL, Velez-Garcia E, Moore JO, Shea TC, Hoke E, Caligiuri MA, Vardiman JW, Bloomfield CD, Larson RA. Dose escalation studies of cytarabine, daunorubicin, and etoposide with and without multidrug resistance modulation with PSC-833 in untreated adults with acute myeloid leukemia younger than 60 years: final induction results of Cancer and Leukemia Group B Study 9621. Journal of Clinical Oncology. 2004;22:4290–4301. doi: 10.1200/JCO.2004.11.106. [DOI] [PubMed] [Google Scholar]
- Kolitz JE, George SL, Marcucci G, Vij R, Powell BL, Allen SL, DeAngelo DJ, Shea TC, Stock W, Baer MR, Hars V, Maharry K, Hoke E, Vardiman JW, Bloomfield CD, Larson RA, for the Cancer Leukemia Group B. P-glycoprotein inhibition using valspodar (PSC-833) does not improve outcomes for patients younger than age 60 years with newly diagnosed acute myeloid leukemia: Cancer and Leukemia Group B study 19808. Blood. 2010;116:1413–1421. doi: 10.1182/blood-2009-07-229492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroll KW, Eisfeld AK, Lozanski G, Bloomfield CD, Byrd JC, Blachly JS. MuCor: mutation aggregation and correlation. Bioinformatics. 2016;32:1557–1558. doi: 10.1093/bioinformatics/btw028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krönke J, Bullinger L, Teleanu V, Tschürtz F, Gaidzik VI, Kühn MW, Rücker FG, Holz-mann K, Paschka P, Kapp-Schwörer S, Späth D, Kindler T, Schittenhelm M, Krau-ter J, Ganser A, Göhring G, Schlegelberger B, Schlenk RF, Döhner H, Döhner K. Clonal evolution in relapsed NPM1-mutated acute myeloid leukemia. Blood. 2013;122:100–108. doi: 10.1182/blood-2013-01-479188. [DOI] [PubMed] [Google Scholar]
- Ley TJ, Ding L, Walter MJ, McLellan MD, Lamprecht T, Larson DE, Kandoth C, Pay-ton JE, Baty J, Welch J, Harris CC, Lichti CF, Townsend RR, Fulton RS, Dooling DJ, Koboldt DC, Schmidt H, Zhang Q, Osborne JR, Lin L, O’Laughlin M, McMichael JF, Delehaunty KD, McGrath SD, Fulton LA, Magrini VJ, Vickery TL, Hun-dal J, Cook LL, Conyers JJ, Swift GW, Reed JP, Alldredge PA, Wylie T, Walker J, Kalicki J, Watson MA, Heath S, Shannon WD, Varghese N, Nagarajan R, Westervelt P, Tomasson MH, Link DC, Graubert TA, DiPersio JF, Mardis ER, Wilson RK. DNMT3A mutations in acute myeloid leukemia. New England Journal of Medicine. 2010;363:2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Moser B, Blum W, Stock W, Wetzler M, Kolitz JE, Thakuri M, Carter T, Stuart RK, Larson RA. A phase III randomized trial of intensive induction and consolidation chemotherapy ± oblimersen, a pro-apopatientotic Bcl-2 antisense oligonucleotide in untreated acute myeloid leukemia patients >60 years old [abstract] Journal of Clinical Oncology. 2007;25(suppl):360s. Abstract 7012. [Google Scholar]
- Marcucci G, Maharry K, Radmacher MD, Mrozek K, Vukosavljevic T, Paschka P, Whitman SP, Langer C, Baldus CD, Liu CG, Ruppert AS, Powell BL, Carroll AJ, Caligiuri MA, Kolitz JE, Larson RA, Bloomfield CD. Prognostic significance of, and gene and microRNA expression signatures associated with, CEBPA mutations in cytogenetically normal acute myeloid leukemia with high-risk molecular features: a Cancer and Leukemia Group B Study. Journal of Clinical Oncology. 2008;26:5078–5087. doi: 10.1200/JCO.2008.17.5554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcucci G, Metzeler KH, Schwind S, Becker H, Maharry K, Mrozek K, Radmacher MD, Kohlschmidt J, Nicolet D, Whitman SP, Wu YZ, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Moore JO, Caligiuri MA, Larson RA, Bloomfield CD. Age-related prognostic impact of different types of DNMT3A mutations in adults with primary cytogenetically normal acute myeloid leukemia. Journal of Clinical Oncology. 2012;30:742–750. doi: 10.1200/JCO.2011.39.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayle A, Yang L, Rodriguez B, Zhou T, Chang E, Curry CV, Challen GA, Li W, Wheeler D, Rebel VI, Goodell MA. Dnmt3a loss predisposes murine hematopoietic stem cells to malignant transformation. Blood. 2015;125:629–638. doi: 10.1182/blood-2014-08-594648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore JO, George SL, Dodge RK, Amrein PC, Powell BL, Kolitz JE, Baer MR, Davey FR, Bloomfield CD, Larson RA, Schiffer CA. Sequential multiagent chemotherapy is not superior to high-dose cytarabine alone as postremission intensification therapy for acute myeloid leukemia in adults under 60 years of age: Cancer and Leukemia Group B Study 9222. Blood. 2005;105:3420–3427. doi: 10.1182/blood-2004-08-2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrόzek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Reviews. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- Mrόzek K, Carroll AJ, Maharry K, Rao KW, Patil SR, Pettenati MJ, Watson MS, Arthur DC, Tantravahi R, Heerema NA, Koduru PRK, Block AW, Qumsiyeh MB, Edwards CG, Sterling LJ, Holland KB, Bloomfield CD. Central review of cytogenetics is necessary for cooperative group correlative and clinical studies of adult acute leukemia: the Cancer and Leukemia Group B experience. International Journal of Oncology. 2008;33:239–244. [PMC free article] [PubMed] [Google Scholar]
- Mrόzek K, Marcucci G, Nicolet D, Maharry KS, Becker H, Whitman SP, Metzeler KH, Schwind S, Wu YZ, Kohlschmidt J, Pettenati MJ, Heerema NA, Block AW, Patil SR, Baer MR, Kolitz JE, Moore JO, Carroll AJ, Stone RM, Larson RA, Bloom-field CD. Prognostic significance of the European LeukemiaNet standardized system for reporting cytogenetic and molecular alterations in adults with acute myeloid leukemia. Journal of Clinical Oncology. 2012;30:4515–4523. doi: 10.1200/JCO.2012.43.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pløen GG, Nederby L, Guldberg P, Hansen M, Ebbesen LH, Jensen UB, Hokland P, Aggerholm A. Persistence of DNMT3A mutations at long-term remission in adult patients with AML. British Journal of Haematology. 2014;167:478–486. doi: 10.1111/bjh.13062. [DOI] [PubMed] [Google Scholar]
- Renneville A, Boissel N, Nibourel O, Berthon C, Helevaut N, Gardin C, Cayuela JM, Hayette S, Reman O, Contentin N, Bordessoule D, Pautas C, Botton S, Revel T, Terre C, Fenaux P, Thomas X, Castaigne S, Dombret H, Preudhomme C. Prognostic significance of DNA methyltransferase 3A mutations in cytogenetically normal acute myeloid leukemia: a study by the Acute Leukemia French Association. Leukemia. 2012;26:1247–1254. doi: 10.1038/leu.2011.382. [DOI] [PubMed] [Google Scholar]
- Ribeiro AFT, Pratcorona M, Erpelinck-Verschueren C, Rockova V, Sanders M, Abbas S, Figueroa ME, Zeilemaker A, Melnick A, Löwenberg B, Valk PJM, Delwel R. Mutant DNMT3A: a marker of poor prognosis in acute myeloid leukemia. Blood. 2012;119:5824–5831. doi: 10.1182/blood-2011-07-367961. [DOI] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nature Biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russler-Germain DA, Spencer DH, Young MA, Lamprecht TL, Miller CA, Fulton R, Meyer MR, Erdmann-Gilmore P, Townsend RR, Wilson RK, Ley TJ. The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers. Cancer Cell. 2014;25:442–454. doi: 10.1016/j.ccr.2014.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlush LI, Zandi S, Mitchell A, Chen WC, Brandwein JM, Gupta V, Kennedy JA, Schimmer AD, Schuh AC, Yee KW, McLeod JL, Doedens M, Medeiros JJF, Marke R, Kim HJ, Lee K, McPherson JD, Hudson TJ HALTPan-Leukemia Gene Panel Consortium. Brown AMK, Yousif F, Trinh QM, Stein LD, Minden MD, Wang JCY, Dick JE. Identification of preleukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506:328–333. doi: 10.1038/nature13038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steensma DP, Bejar R, Jaiswal S, Lindsley RC, Sekeres MA, Hasserjian RP, Ebert BL. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126:9–16. doi: 10.1182/blood-2015-03-631747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RM, Fischer T, Paquette R, Schiller G, Schiffer CA, Ehninger G, Cortes J, Kantarjian HM, DeAngelo DJ, Huntsman-Labed A, Dutreix C, del Corral A, Giles F. Phase IB study of the FLT3 kinase inhibitor midostaurin with chemotherapy in younger newly diagnosed adult patients with acute mye-loid leukemia. Leukemia. 2012;26:2061–2068. doi: 10.1038/leu.2012.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thol F, Damm F, Lüdeking A, Winschel C, Wagner K, Morgan M, Yun H, Göhring G, Schlegelberger B, Hoelzer D, Lübbert M, Kanz L, Fiedler W, Kirchner H, Heil G, Krauter J, Ganser A, Heuser M. Incidence and prognostic influence of DNMT3A mutations in acute myeloid leukemia. Journal of Clinical Oncology. 2011;29:2889–2896. doi: 10.1200/JCO.2011.35.4894. [DOI] [PubMed] [Google Scholar]
- Vittinghoff E, Glidden DV, Shiboski SC, McCulloch CE. Regression Methods in Biostatistics: Linear, Logistic, Survival and Repeated Measures Models. New York, NY: Springer; 2005. [Google Scholar]
- Westfall PH, Tobias RD, Rom D, Wolfinger RD, Hochberg Y. Multiple Comparisons and Multiple Tests Using the SAS System. Cary, NC: SAS Institute; 1999. [Google Scholar]
- Xie M, Lu C, Wang J, McLellan MD, Johnson KJ, Wendl MC, McMichael JF, Schmidt HK, Yellapantula V, Miller CA, Ozenberger BA, Welch JS, Link DC, Walter MJ, Mardis ER, Dipersio JF, Chen F, Wilson RK, Ley TJ, Ding L. Age-related mutations associated with clonal hematopoietic expansion and malignancies. Nature Medicine. 2014;20:1472–1478. doi: 10.1038/nm.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


