Abstract
Acute Intermittent Porphyria results from hydroxymethylbilane synthase (HMBS) mutations that markedly decrease HMBS enzymatic activity. This dominant disease is diagnosed when heterozygotes have life-threatening acute attacks, while most heterozygotes remain asymptomatic and undiagnosed. Although >400 HMBS mutations have been reported, the prevalence of pathogenic HMBS mutations in genomic/exomic databases, and the actual disease penetrance are unknown. Thus, we interrogated genomic/exomic databases, identified non-synonymous variants (NSVs) and consensus splice-site variants (CSSVs) in various demographic/racial groups, and determined the NSV’s pathogenicity by prediction algorithms and in vitro expression assays. Caucasians had the most: 58 NSVs and two CSSVs among ~92,000 alleles, a 0.00575 combined allele frequency. In silico algorithms predicted 14/58 NSVs as “likely-pathogenic”. In vitro expression identified 10/58 NSVs as likely-pathogenic (seven predicted in silico), which together with two CSSVs had a combined allele frequency of 0.00056. Notably, six presumably pathogenic mutations/NSVs in the Human Gene Mutation Database were benign. Compared to the recent prevalence estimate of symptomatic European heterozygotes (~0.000005), the prevalence of likely-pathogenic HMBS mutations among Caucasians was >100 times more frequent. Thus, the estimated penetrance of acute attacks was ~1% of heterozygotes with likely-pathogenic mutations, highlighting the importance of predisposing/protective genes and environmental modifiers that precipitate/prevent the attacks.
Keywords: allele frequency, allele prevalence, disease penetrance, in silico prediction, in vitro expression
Introduction
Current efforts in genomic medicine are focused on determining which non-synonymous variants identified by genomic/exomic sequencing are pathogenic or benign. Various approaches to evaluate the pathogenicity of these variants include an array of in silico prediction programs and for certain proteins functional expression assays. These methods allow the assessment of the numerous non-synonymous variants that are discovered for a given gene and provide the clinical relevance of these variants as well as information of the prevalence of disease causing mutations, some of which may have allele frequencies >0.001 [Xue et al., 2012; Amendola et al. 2015].
Here, we investigated the pathogenicity of all non-synonymous variants and consensus splice-site variants in genomic/exomic databases for autosomal dominant Acute Intermittent Porphyria (AIP; MIM# 176000), the most common acute hepatic porphyria [Puy et al., 2010; Anderson et al., 2014]. AIP is an inborn error of heme biosynthesis resulting from the reducedactivity of the heme biosynthetic enzyme, hydroxymethylbilane synthase (HMBS; MIM# 609806, EC 4.3.1.8; also known as porphobilinogen deaminase (PBGD)). Heterozygotes experience potentially life-threatening acute neurovisceral attacks precipitated by certain porphyrinogenic drugs (e.g., P450 inducers), dieting or fasting, and hormonal changes which induce the hepatic expression of the first and rate-limiting enzyme in the pathway, 5′-aminolevulinic acid synthase (ALAS1; MIM# 125290, EC 2.3.1.37) [Granick, 1963; Granick, 1966; Sassa et al., 1970]. Induction of the ALAS1 mRNA is regulated in the liver by a negative feedback repression mechanism that depends on the amount of free hepatic heme. Due to the increased ALAS1 enzymatic activity, the reducedHMBS activity becomes rate-limiting and the neurotoxic porphyrin precursors, 5′-aminolevulinic acid (ALA) and porphobilinogen (PBG), accumulate systemically and cause the acute neurovisceral attacks.
Clinically, the life-threatening acute attacks are characterized by excruciating abdominal pain, nausea, vomiting, hypertension, tachycardia, and central and peripheral nervous system manifestations, including motor/sensory neuropathy and psychiatric symptoms. If untreated by infusion of hemin to replenish the hepatic heme pool and down-regulate ALAS1, severe attacks can progress to advanced motor neuropathy, respiratory muscle paralysis, and bulbar palsy [Puy et al., 2010; Anderson et al., 2014]. Typically, the attacks occur after puberty and can be episodic or recurrent and ~90% occurs in women. Affected women may have monthly attacks due to hormonal changes in the luteal phase of their menstrual cycles [Hift et al., 2005; Innala et al., 2010]. It is estimated that about 10% of AIP heterozygotes have acute attacks [McColl et al., 1982]. In fact, most patients are diagnosed during or following an acute attack, while most AIP heterozygotes remain asymptomatic throughout their lives.
To date, over 400 presumably pathogenic HMBS mutations have been reported and catalogued in the Human Gene Mutation Database (HGMD) [Stenson et al., 2014]. Although there are no documented genotype/phenotype correlations, certain mutations are more frequent among patients with multiple or recurrent attacks (e.g. those encoding p.R149X, p.R173Q) [von und zu Fraunberg et al., 2005]. A recent estimate of the prevalence of AIP, based on newly diagnosed patients in Western European countries who sought medical attention for their attack, was ~0.000005 (or ~1 in 200,000) [Elder et al., 2013], with the notable exception of the Scandinavian countries where the disease prevalence is most frequent due to a founder effect for the mutation encoding p.W198X [Mustajoki et al., 1976; Lee et al., 1991; Floderus et al., 2002; Mykletun et al., 2014]. To our knowledge, the only other systematic study of AIP prevalence was the cross-sectional study of 3,350 French healthy blood donors, which identified two unrelated healthy individuals with decreased (~50%) erythrocytic HMBS activity [Nordmann et al., 1997], who were then confirmed to have known HMBS pathogenic mutations encoding p.D178N and p.L244fs for a frequency of ~0.0006 (1 in 1,675). Thus, these findings indicate a marked discrepancy in the estimated prevalence of HMBS heterozygotes and the occurrence of acute attacks in AIP patients, the penetrance, in the general Caucasian population.
To address this discrepancy, we interrogated genomic/exomic databases with a combined total of 45,955 Caucasians to identify non-synonymous and consensus splice site variants. We used in silico pathogenicity prediction programs and in vitro enzyme expression studies to assess if these HMBS variants were likely pathogenic or benign.
Materials and Methods
Database Variant Collection
The nomenclature of the HMBS variants reported is based on the cDNA sequence NM_000190.3 and in accordance with the standards of the Human Genome Variation Society (HGVS; http://www.HGVS.org/varnomen). Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in the reference sequence. Non-synonymous variants and consensus splice site HMBS mutations in various demographic and racial/ethnic groups including Caucasian or Western European populations were identified in public genomic/exomic databases using primarily the Diseases Variant Store (DIVAS) as the portal to other databases (Disease Variant Store, https://rvs.u.hpc.mssm.edu/divas/) [Cheng et al., 2016]. The DIVAS contains five major databases that include population ethnicity and allele frequencies: the 1000 Genomes Project [Genomes Project C et al., 2015], the NHLBI Exome Sequencing Project (ESP) (Exome Variant Server, http://evs.gs.washington.edu/EVS/), the Exome Aggregation Consortium (ExAC) (http://exac.broadinstitute.org), UK10K (http://www.uk10k.org/) and the Scripps Wellderly Cohort (https://genomics.scripps.edu/browser/) [Erikson et al., 2016]. Reported presumably pathogenic HMBS mutations causing AIP were obtained in the Human Gene Mutation Database (HGMD Professional 2016.1, https://portal.biobase-international.com/hgmd/pro/search_gene.php), which listed 403 mutations [Stenson et al., 2014].
In Silico Prediction of Variant Pathogenicity
All 58 HMBS non-synonymous variants in Caucasian and Western European populations were assessed to determine if they were deleterious or benign using 16 algorithms evaluating each variant. These programs (in Supp. Table S1) included: CADD [Kircher et al., 2014], CONDEL [Gonzalez-Perez et al., 2011], I-Mutant [Capriotti et al., 2005], MAPP [Stone et al., 2005], MutationAssessor [Reva et al., 2011], MutPred [Li et al., 2009], nsSNPAnalyzer [Bao et al., 2005], PANTHER [Tang et al., 2016], PhD-SNP [Capriotti et al., 2006], PolyPhen-2 [Adzhubei et al., 2010], PON-P2 [Niroula et al., 2015], PoPMuSiC [Dehouck et al., 2011], PredictSNP [Bendl et al., 2014], PROVEAN [Choi et al., 2015], SIFT [Kumar et al., 2009], and SNAP2 [Hecht et al., 2015]. For CADD, deleterious was defined as a CADD Phred score >25 (The Phred quality score (Q) is logarithmically related to the error probability (E). Q=−10logE). For I-Mutant and PoPMuSiC, a variant was considered deleterious when it decreased the structural stability. For MutationAssessor, a Functional Impact (FI) score >2.0 was considered deleterious. For MutPred, a variant with probability of deleterious mutation >0.8 was considered deleterious. For PANTHER, Pdeleterious >0.8 was considered deleterious. For SNAP2, a score >10 and expected accuracy >70% were considered deleterious. For all other tools, the recommended default setting was used for calling deleterious/pathogenic lesions. For each variant, the “Consensus Deleterious Score” was the ratio of the number of programs predicting “deleterious” over the total number of programs with a predicted result, (i.e. predicting deleterious/total number of predictions). This approach of “Consensus Deleterious Score” from all 16 in silico prediction algorithms was used since it has been shown that combining multiple tools tends to provide a more reliable prediction of the “likely pathogenicity” [Polikar, 2006]. “Unknown” was not considered a prediction. Arbitrary cutoffs of <25% and >75% for deleterious were used to define each variant as “likely benign” and “likely pathogenic”, respectively, and as “ambiguous”, if the score fell between 25–75%.
In Vitro Expression Studies: Activity and Thermostability
HMBS cDNA constructs for each of the 58 HMBS non-synonymous variants were individually generated using the QuikChange Lightning Single-Site Mutagenesis kit (Agilent Genomics, Santa Clara, CA) to alter the HMBS wild-type (WT) cDNA in the pKK223 vector [Chen et al., 1994]. All constructs were re-sequenced to confirm their respective authenticities. Since HMBS is a cytosolic enzyme that does not undergo any post-translational modifications, WT and mutant constructs were each expressed in E. coli strain BL21(DE3)pLysS (Promega, Madison, WI), which had low endogenous HMBS background activity and produced large quantity of the recombinant human enzyme as used previously for mutational and crystalization analyses [Chen et al., 1994; Gill et al., 2009; Song et al., 2009]. The enzymatic activity of each mutant enzyme in the lysate was calculated as the percent of the expressed WT activity, which was expressed in the same experiment, as described previously [Chen et al., 1994]. All results are presented as the mean activities and standard deviations of at least three independent experiments. For the enzyme thermostability studies, the expressed recombinant enzyme in the lysate was assayed for HMBS activity after incubation at 65°C for 90 min at pH 8.0. The mean thermostability of each mutant HMBS enzyme was calculated as the percent of the initial expressed WT activity after heat treatment and was based on at least three independent experiments.
Results
Identification and Characterization of HMBS Variants in Genomic Databases
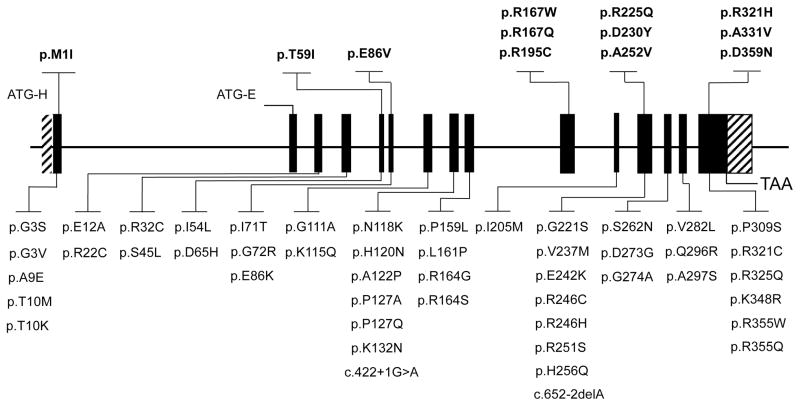
Genomic databases that included 69,530 individuals from various demographic and ethnic/racial groups, including 45,955 Caucasians, were interrogated for HMBS non-synonymous and consensus splice-site variants (Table 1). All 101 identified HMBS variants (98 non-synonymous and three consensus splice-site variants) are listed in Supplementary Materials (Supp. Table S2) by demographic/racial group. The consensus splice-site variants c.422+1G>A and c.652-2delAwere identified only in Caucasians, and c.613-1G>Awas present only in Africans (Table 1). Overall, a high total allele frequency of non-synonymous HMBS variants was found in each demographic/racial group (Table 1): 58 non-synonymous and two consensus splice-site variants with a total allele frequency of ~0.00575 (~1/174) for Caucasians (Figure 1); 16 non-synonymous variants with a total allele frequency of ~0.00271 (~1/369) for Latinos; 20 non-synonymous and one consensus splice-site variant with a total allele frequency of ~0.00634 (~1/157) for Africans; 20 non-synonymous variants with a total allele frequency of ~0.007 (~1/140) for South Asians; 10 non-synonymous variants with a total allele frequency of ~0.00176 (~1/568) for East Asians. Notably, only one variant (encoding p.R321H) occurred in all five groups, ranging from 0.00012 in East Asians to 0.00192 in Caucasians. The variant encoding p.R246C was present in four groups, variants encoding p.P127Q and p.R225Q were shared by three groups, and 16 others were present in two groups (see Supp. Table S2). Of the 20 non-synonymous variants shared among two or more groups, 9 (45%) were at CpG dinucleotides, hotspots for mutation [Cooper et al., 1988] (Supp. Table S2).
Table 1.
Total HMBS Non-Synonymous & Consensus Splice-Site Variants in Genomic/Exomic Databases in Various Demographic/Racial Groups
| Caucasiansa (91,919 alleles) | Latinos (11,578 alleles) | Africans (10,406 alleles) | South Asians (16,512 alleles) | East Asians (8,654 alleles) | |
|---|---|---|---|---|---|
| A. Non-Synonymous Variants: | |||||
| Total Number of Variants | 58 | 16 | 20 | 20 | 10 |
| Listed in HGMD (N) | 12 | 3 | 6 | 4 | 1 |
| Allele Frequency < 0.0002(N,% of Total) | 55 (95%) | 13 (81%) | 14 (70%) | 13 (65%) | 7 (70%) |
| Allele Frequency Range | 0.00192 – 0.00001 | 0.00052 – 0.00009 | 0.0017 – 0.0001 | 0.00367 – 0.00006 | 0.000462 – 0.00012 |
| Total Non-Synonymous Allele Frequency | 0.00573 | 0.00271 | 0.00624 | 0.007 | 0.00176 |
| B. Consensus Splice-Site Variantsb: | |||||
| c.422+1G>A | 0.00001 | 0 | 0 | 0 | 0 |
| c.613-1G>T | 0 | 0 | 0.0001 | 0 | 0 |
| c.652-2delA | 0.00001 | 0 | 0 | 0 | 0 |
|
| |||||
| Total Variant Frequency: | 0.00575 (1 in 174) | 0.00271 (1 in 369) | 0.00634 (1 in 157) | 0.007 (1 in 140) | 0.00176 (1 in 568) |
Caucasian sequences include European and Finnish data
Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in NM_000190.3
Figure 1.
HMBS Non-Synonymous & Consensus Splice-Site Variants Identified in Caucasians in Genomic/Exomic Databases. Variants in the upper panel have been listed in HGMD as causing AIP. Variants in the lower panel are novel and identified only in the genomic/exomic databases.
To determine the predicted pathogenicity of the 58 non-synonymous HMBS variants in Caucasians, as a relevant group for an estimate of AIP prevalence, each non-synonymous variant was evaluated by in silico pathogenicity prediction programs and by in vitro expression assays. Most (95%) of the Caucasian variants had an allele frequency 0.0002 or less, while three variants encoding p.S45L, p.E86V, p.R321H, had allele frequencies of 0.0004, 0.001 and 0.0019, respectively.
Predicted Pathogenicity of the Caucasian HMBS Non-Synonymous Variants
The functional impact of each of the 58 non-synonymous variants on HMBS activity and stability was assessed by 16 different in silico pathogenicity prediction programs (Supp. Table S1) and the results were combined to generate a “Consensus Deleterious Score” or “CDS.” Based on the CDS, each variant was classified as likely “deleterious”, “benign” or, if most results were unknown or could not assess the variant, they were classified as “ambiguous”. Supplementary Table 3 (Supp. Table S3) summarizes the results of each prediction program for each variant. Of the 58 variants, 12 were previously listed in HGMD as causing AIP, and five were predicted as “deleterious” (variants encoding p.R167W, p.R167Q, p.R195C, p.R225Q and p.A331V), while seven were considered “ambiguous”(Table 2). Of the remaining 46 non-synonymous variants, 9 were predicted as “deleterious” (variants encoding p.R22C, p.D65H, p.I71T, p.G72R, p.A122P, p.L161P, p.R251S, p.V282L and p.R355W), 12 were “benign”, and the remaining 25 were designated as “ambiguous” and classified as unknown (Table 2).
Table 2.
HMBS Non-Synonymous & Consensus Splice-Site Variants in Caucasians in Genomic/Exomic Databases
| Genomic Position (GRCh37/hg19) | cDNAa/Amino Acidb Changes | Allele Frequency | In Silico Deleterious Scorec/Prediction | Consensus | Percent of Expressed WT Activity (Mean±SD) | Thermostability: % of Initial Expressed WT Activity after 65°C, 90min: (Mean±SD) | Predicted Pathogenicity |
|---|---|---|---|---|---|---|---|
| A. HMBS Non-Synonymous Variants Published in HGMD Reported Causing AIP: Arranged by In Vitro Expressed Activity | |||||||
| 118955746 | c.3G>A/p.M1I | 0.00001 | 0.38 | ambigious | 0±1% | - | Pathogenic |
| 118962124 | c.500G>A/p.R167Q | 0.0002 | 0.94 | deleterious | 1±1% | - | Pathogenic |
| 118962123 | c.499C>T/p.R167W | 0.0001 | 0.94 | deleterious | 3±1% | - | Pathogenic |
| 118962207 | c.583C>T/p.R195C | 0.0001 | 1 | deleterious | 3±1% | - | Pathogenic |
| 118963217 | c.755C>T/p.A252V | 0.00001 | 0.64 | ambigious | 61±12% | 10±2% | Pathogenic |
| 118963899 | c.992C>T/p.A331V | 0.00003 | 0.79 | deleterious | 62±5% | 12±2% | Pathogenic |
| 118959807 | c.176C>T/p.T59I | 0.00004 | 0.29 | ambigious | 77±3% | 51±16% | Benign |
| 118963150 | c.688G>T/p.D230Y | 0.00001 | 0.5 | ambigious | 88±4% | 50±16% | Benign |
| 118963982 | c.1075G>A/p.D359N | 0.00001 | 0.29 | ambigious | 86±14% | 42±13% | Benign |
| 118959973 | c.257A>T/p.E86V | 0.00113 | 0.43 | ambigious | 94±16% | 45±15% | Benign |
| 118963136 | c.674G>A/p.R225Q | 0.00026 | 0.93 | deleterious | 102±19% | 31±10% | Benign |
| 118963869 | c.962G>A/p.R321H | 0.00188 | 0.38 | ambigious | 122±24% | 31±10% | Benign |
| B. Non-Synonymous HMBS Variants Identified in Genomic/Exomic Databases: Arranged by In Vitro Expressed Activity | |||||||
| 118960959 | c.482T>C/p.L161P | 0.00001 | 1 | deleterious | 1±1% | - | Pathogenic |
| 118963215 | c.753G>C/p.R251S | 0.00001 | 1 | deleterious | 1±0% | - | Pathogenic |
| 118960719 | c.364G>C/p.A122P | 0.00001 | 0.93 | deleterious | 2±0% | - | Pathogenic |
| 118960709 | c.354C>G/p.N118K | 0.00001 | 0.57 | ambigious | 73±14% | 2±1% | Pathogenic |
| 118955751 | c.8G>T/p.G3V | 0.00001 | 0.54 | benign | 34±6% | 25±8% | Benign |
| 118960967 | c.490A>G/p.R164G | 0.00022 | 0.47 | ambigious | 61±4% | 27±9% | Benign |
| 118959928 | c.212T>C/p.I71T | 0.00003 | 1 | deleterious | 62±13% | 62±20% | Benign |
| 118959930 | c.214G>A/p.G72R | 0.00004 | 1 | deleterious | 69±1% | 32±10% | Benign |
| 118963123 | c.661G>A/p.G221S | 0.00001 | 0.67 | ambigious | 69±15% | 38±12% | Benign |
| 118959417 | c.160A>C/p.I54L | 0.00013 | 0.29 | ambigious | 71±25% | 52±17% | Benign |
| 118963517 | c.821G>C/p.G274A | 0.00001 | 0.27 | ambigious | 72±9% | 35±11% | Benign |
| 118963663 | c.844G>C/p.V282L | 0.00003 | 0.86 | deleterious | 73±2% | 20±6% | Benign |
| 118960469 | c.343A>C/p.K115Q | 0.00008 | 0.29 | ambigious | 76±10% | 40±13% | Benign |
| 118960458 | c.332G>C/p.G111A | 0.00002 | 0.43 | ambigious | 76±3% | 32±10% | Benign |
| 118963708 | c.889G>T/p.A297S | 0.00001 | 0.2 | benign | 76±8% | 49±16% | Benign |
| 118963868 | c.961C>T/p.R321C | 0.00001 | 0.27 | ambigious | 77±12% | 33±11% | Benign |
| 118963514 | c.818A>G/p.D273G | 0.00003 | 0.21 | benign | 78±15% | 38±12% | Benign |
| 118955769 | c.26C>A/p.A9E | 0.00008 | 0.2 | benign | 79±0% | 53±17% | Benign |
| 118963832 | c.925C>T/p.P309S | 0.00007 | 0.13 | benign | 79±6% | 59±19% | Benign |
| 118955772 | c.29C>T/p.T10M | 0.00002 | 0.33 | benign | 79±12% | 45±14% | Benign |
| 118963970 | c.1063C>T/p.R355W | 0.00006 | 0.87 | deleterious | 80±8% | 26±8% | Benign |
| 118958995 | c.64C>T/p.R22C | 0.00003 | 1 | deleterious | 80±13% | 38±12% | Benign |
| 118960969 | c.492G>T/p.R164S | 0.00003 | 0.27 | ambigious | 81±18% | 30±10% | Benign |
| 118960953 | c.476C>T/p.P159L | 0.00001 | 0.73 | deleterious | 81±20% | 32±10% | Benign |
| 118962837 | c.615C>G/p.I205M | 0.00003 | 0.43 | ambigious | 83±9% | 24±8% | Benign |
| 118963186 | c.724G>A/p.E242K | 0.00001 | 0.36 | ambigious | 83±13% | 22±7% | Benign |
| 118960734 | c.379C>G/p.P127A | 0.00001 | 0.2 | benign | 84±19% | 37±12% | Benign |
| 118955772 | c.29C>A/p.T10K | 0.00003 | 0.2 | benign | 86±25% | 60±19% | Benign |
| 118960735 | c.380C>A/p.P127Q | 0.0002 | 0.27 | ambigious | 87±2% | 47±15% | Benign |
| 118960751 | c.396G>C/p.K132N | 0.00001 | 0.29 | ambigious | 87±13% | 43±14% | Benign |
| 118959391 | c.134 C>T/p.S45L | 0.00043 | 0.19 | benign | 88±17% | 63±20% | Benign |
| 118963881 | c.974G>A/p.R325Q | 0.00003 | 0.25 | ambigious | 88±4% | 22±7% | Benign |
| 118963706 | c.887A>G/p.Q296R | 0.00001 | 0.21 | benign | 92±6% | 50±16% | Benign |
| 118959824 | c.193G>C/p.D65H | 0.00001 | 0.77 | deleterious | 93±25% | 38±12% | Benign |
| 118963230 | c.768C>A/p.H256Q | 0.00001 | 0.27 | ambigious | 93±6% | 40±13% | Benign |
| 118960713 | c.358C>A/p.H120N | 0.00001 | 0.33 | ambigious | 95±2% | 45±15% | Benign |
| 118963481 | c.785G>A/p.S262N | 0.00001 | 0.6 | ambigious | 96±28% | 73±23% | Benign |
| 118959351 | c.94 C>T/p.R32C | 0.00003 | 0.73 | ambigious | 98±18% | 43±14% | Benign |
| 118963971 | c.1064G>A/p.R355Q | 0.00001 | 0.6 | ambigious | 98±10% | 31±10% | Benign |
| 118963950 | c.1043A>G/p.K348R | 0.00004 | 0.13 | benign | 100±8% | 35±11% | Benign |
| 118959972 | c.256G>A/p.E86K | 0.00001 | 0.43 | ambigious | 104±27% | 40±13% | Benign |
| 118958966 | c.35A>C/p.E12A | 0.00003 | 0.2 | benign | 106±23% | 53±17% | Benign |
| 118963199 | c.737G>A/p.R246H | 0.00006 | 0.5 | ambigious | 122±24% | 29±9% | Benign |
| 118963171 | c.709G>A/p.V237M | 0.00001 | 0.36 | ambigious | 128±6% | 45±15% | Benign |
| 118963198 | c.736C>T/p.R246C | 0.00001 | 0.43 | ambigious | 130±1% | 53±17% | Benign |
| 118955750 | c.7G>A/p.G3S | 0.00003 | 0.21 | benign | 211±36% | 150±48% | Benign |
| C. Consensus Splicing Variants Identified in Genomic/Exomic Databases: | |||||||
| 118960778 | c.422+1G>A | 0.00001 | - | - | - | - | Pathogenic |
| 118963111 | c.652-2delA | 0.00001 | - | - | - | - | Pathogenic |
Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in NM_000190.3;
amino acid numbering uses +1 as the first amino acid of the NP_000181.2
Consensus Deleterious score = (number of programs predicting deleterious/total number of programs with a predicted result), see Materials and Methods for details
In Vitro Activity and Thermostability of HMBS Variant Enzymes
All 58 non-synonymous variants were expressed in vitro and their enzymatic activities and thermostabilities were determined (Table 2). Of the 12 variants listed in HGMD as pathogenic, only four (encoding p.M1I, p.R167W, p.R167Q and p.R195C) had enzymatic activities of ≤3% of the expressed WT activity. Notably, the eight variants encoding p.T59I, p.E86V, p.R225Q, p.D230Y, p.A252V, p.R321H, p.A331V and p.D359N listed in HGMD all had ~60–100% of the expressed WT activity. Of the remaining 46 variants in genomic/exomic databases, three encoding p.A122P, p.L161P and p.R251S had expressed enzymatic activities of ≤2% of expressed WT activity (but have not been reported in AIP patients to date) and 41 had normal or near normal enzymatic activities (62–130% of expressed WT activity), while the variants encoding p.G3V and p.G3S had ~35% and ~200% of expressed WT activity, respectively (Table 2).
The 51 variants with residual HMBS activities >10% of expressed WT activity were subjected to heat inactivation at 65°C, pH 8.0 for 90 min. While 48 variants were relatively thermostable compared to the expressed WT enzyme (Table 2), three encoding p.N118K, p.A252V and p.A331V had markedly reduced enzyme activities after heat inactivation (2%, 10% and 12% of initial expressed WT activity, respectively), indicating that their respective amino acid substitutions destabilized the HMBS mutant protein, and suggesting that they may be pathogenic. Of note, variants encoding p.A252V and p.A331V were found in AIP patients and listed in HGMD as pathogenic.
Thus, the likely pathogenic non-synonymous variants with <3% of expressed WT activity included four variants listed in HGMD, and three novel non-synonymous variants from the genomic/exomic databases with a combined allele frequency of ~0.000477. The addition of the two consensus splice-site variants increased the likely pathogenic HMBS allele frequency to ~0.000504. Finally, when the three variants with markedly decreased thermostabilities were included, the total likely pathogenic allele frequency, or prevalence of autosomal dominant AIP, was ~0.00056 or 1 in ~1,782 (Table 3).
Table 3.
Summary of Known and Predicted Pathogenic HMBS Variants in Caucasians in Genomic/Exomic Databases
| cDNA Changesa | Amino Acid Changesb | CpG dinucleotide | Reported in HGMD | HMBS Activity | Allele Frequency |
|---|---|---|---|---|---|
| c.3G>A | p.M1I | + | 0±1% | 0.000014 | |
| c.499C>T | p.R167W | + | + | 1±1% | 0.0001 |
| c.500G>A | p.R167Q | + | + | 3±1% | 0.0002 |
| c.583C>T | p.R195C | + | + | 3±1% | 0.0001 |
| c.755C>T | p.A252V | + | 10±2% c | 0.000014 | |
| c.992C>T | p.A331V | + | 12±2% c | 0.000027 | |
| c.364G>C | p.A122P | 1±1% | 0.000014 | ||
| c.482T>C | p.L161P | 1±0% | 0.000014 | ||
| c.753G>C | p.R251S | 2±0% | 0.000014 | ||
| c.354C>G | p.N118K | 2±1% c | 0.000014 | ||
| c.422+1G>A | - | 0.000014 | |||
| c.652-2delA | - | 0.000014 | |||
|
| |||||
| Total Variant Allele Frequency = | 0.00056 | ||||
Nucleotide numbering uses +1 as the A of the ATG translation initiation codon in
amino acid numbering uses +1 as the first amino acid of the NP_000181.2A
Variants encoding p.N118K, p.A252V and p.A331V are thermolabile and activities reported are percent of initial WT activity after 65°C at 90 min, pH 8.0
Discussion
The acute hepatic and erythropoietic porphyrias are among a unique group of monogenic disorders (e.g. G6PD deficiency, malignant hyperthermia, dihydropyrimidine dehydrogenase deficiency) that require additional genetic and/or environmental triggering factors (e.g., drugs, metabolites, etc.) for their clinical expression, which has led to their being referred to as pharmacogenetic and/or ecogenetic disorders [Meyer, 2004; van Kuilenburg, 2004; Stowell, 2008; Puy et al., 2010; Anderson et al., 2014]. For AIP and the other acute hepatic porphyrias, the potentially life-threatening acute attacks are precipitated by porphyrinogenic drugs (typically P450 inducers), dieting/fasting or hormonal changes. Because of these triggering factors, most probands are diagnosed only during or after an acute attack, while the disease remains clinically asymptomatic and undiagnosed in most heterozygotes. Thus, the actual incidence, prevalence, and penetrance of autosomal dominant AIP remain unknown.
To date, the estimated prevalence of AIP in Western European countries, based on the frequency of newly diagnosed symptomatic patients was at ~0.000005 or 1 in ~200,000 individuals [Elder et al., 2013]. Estimates of the penetrance of AIP range from ~10% in Western Europe to 30–50% in Scandinavia [Andersson et al., 2000, Schuurmans et al., 2001; Bylesjö et al., 2009; Mykletun et al., 2014]. But since the prevalence of all individuals carrying a pathogenic mutation for AIP has not been determined, the penetrance of the disease remains unknown. Therefore, we determined the prevalence of pathogenic HMBS mutations by interrogating the non-synonymous and consensus splice-site variants in genomic/exomic databases that included ~46,000 Caucasians and assessed which were likely pathogenic. A total of 58 HMBS non-synonymous variants were identified with a combined allele frequency of ~0.00575 (1 in ~174 individuals). In vitro expression of each variant identified only seven with expressed activities that were <3% of expressed WT activity. In addition, there were two pathogenic consensus splice-site variants and three thermolabile non-synonymous variants, yielding a combined allele frequency, or prevalence of autosomal dominant AIP, of ~0.00056 or 1 in ~1,782. Interestingly, the estimated prevalence of healthy French blood donors (3,350 individuals) with reducedHMBS activity was similar, 1 in 1675 [Nordmann et al., 1997].
Over 400 reported HMBS mutations causing AIP patients have been listed in HGMD, including 162 (~40.2%) non-synonymous (missense) mutations [Stenson et al., 2014]. Of particular note, 12 of the 162 non-synonymous variants listed in HGMD were present in genomic/exomic databases. However, only six of these had <3% of WT expressed activity or reduced thermostability. Review of the original reports for the other six variants identified 13 reportedly symptomatic AIP patients who had variants encoding p.T59I [Schneider-Yin et al., 2008], p.E86V [Floderus et al., 2002], p.R225Q [Floderus et al., 2002; von Brasch et al., 2004], or p.R321H [Schuurmans et al., 2001; von Brasch et al., 2004; Anyaegbu et al., 2012; Cerbino et al., 2015]. However, the pathogenicity of most of these patients was not verified by elevated urinary ALA or PBG levels with the exception of (1) three patients with p.R321H who also had a second and pathogenic HMBS mutation [von Brasch et al., 2004; Cerbino et al., 2015]; and (2) one patient with p.R321H who had significantly increased urinary ALA or PBG concentrations, suggesting the presence of an unidentified second and pathogenic mutation [Anyaegbu et al., 2012] (Supp. Table S4).
Major sources of DNA sequence variations are replication errors and deamination at CpG dinucleotides, the latter being hotspots for mutation [Cooper et al., 1988]. Of the 58 HMBS non-synonymous genomic/exomic variants in Caucasians, 18 (~31%) occurred at CpG dinucleotides, including the most common benign variants p.R321H (allele frequency = 0.0019). Of the HMBS 1,086 base-pair coding sequence, there are 31 CpG dinucleotides. Interestingly, among the pathogenic variants causing biochemically confirmed AIP, the most common non-synonymous variants in unrelated probands occurred at CpG dinucleotides that encoded p.R167, p.R173, p.R225 and p.R325, representing ~27% of all identified HMBS mutations causing AIP (unpublished data, Desnick, RJ and Doheny, D, Mount Sinai Porphyria Diagnostic Laboratory).
A current focus of human genomics is to identify which non-synonymous variants in each disease-causing gene are pathogenic or benign. Many genomicists use in silico predictive tools to evaluate pathogenicity, as Whately and Badminton previously did to identify four putative pathogenic non-synonymous HMBS variants in the 1,092 individuals of all ethnic/demographic groups from the 1000 Genomes Project [Whatley and Badminton, 2013]. Although the in silico approach has proven useful to predict if a missense variant is damaging, several studies have demonstrated their limitations when used as stand-alone tools [e.g., Dorfman et al., 2010; van der Velde et al., 2015]. Specifically, a performance plateau of ~80% in their success rates [Riera et al. 2014] has been noted, a value below that required for clinical use [Richards et al., 2015]. In addition, in silico predictability may be gene/disease-specific [e.g., Leong et al., 2015; Adebali et al., 2016]. To circumvent these limitations, various studies suggest that prediction performance can be improved by combining the predictions of a number of tools [e.g., Chan et al., 2007; Konig et al., 2016]. Here sixteen predictive tools were employed to evaluate the HMBS non-synonymous variants. The “consensus deleterious score” predicted 14 variants to be deleterious, and 12 to be benign, but was unable to classify 32 variants, which were designated as “ambiguous” or “unknown.” Of the 14 variants classified as deleterious, only 6 (43%) had <3% expressed enzyme activities, while three (considered ambiguous) had markedly decreased thermostability in vitro. Based on these results, the functional in vitro expression and thermostability studies were more predictive of variant pathogenicity than the combined in silico analyses and should be used when appropriate to validate all new non-synonymous variants in patients with or without biochemical evidence of AIP.
In conclusion, our studies using genomic/exomic sequencing data from 45,955 Caucasians identified 10 non-synonymous and two consensus splice-site pathogenic variants for a combined prevalence of ~0.00056. Since the estimated prevalence of acute attacks is ~0.000005, and the estimated frequency of clinical pathogenic variants is ~0.00056, the penetrance of AIP is a surprising low 1% of all AIP heterozygotes. As AIP is a monogenic disorder, this extremely low penetrance suggests a critical role for modifying factors (environmental and/or genetic) in predisposing heterozygotes to acute attacks.
Supplementary Material
Acknowledgments
The authors thank Ms. Kiera Jaffin and Ms. Nirip Singh for excellent secretarial assistance.
Financial Support
This research was supported in part by Postdoctoral Training Grant (5T32 GM082773) to B.C., a Career Development Award (K23 DK095946) to M.B. and a Cooperative Grant (U54 DK083909) to R.J.D for the Porphyrias Consortium of the Rare Disease Clinical Research Network, all from the National Institutes of Health (NIH).
List of Abbreviations
- AIP
Acute Intermittent Porphyria
- HMBS
Hydroxymethylbilane synthase
- ALAS1
5′-aminolevulinate synthase
- ALA
5′-aminolevulinic acid
- PBG
porphobilinogen
- HGMD
Human Gene Mutation Database
- ESP
Exome Sequencing Project
- ExAC
Exome Aggregation Consortium
- DIVAS
Disease Variant Store
Footnotes
Conflict of Interest
MB and RJD serve as consultants to Alnylam Pharmaceuticals and Recordati Rare Diseases. All other authors report no conflict of interests.
Author Contributions
R.J.D. conceived and supervised the project. B.C. and C.S. designed and performed all in vitro experiments. B.C., J.H., W.Q., and R.S. interrogated the genomic/exomic databases and/or performed the bioinformatic and in silico pathogenic prediction analyses. M.Y., M.B., D.D., I.P., and R.C. contributed to the epidemiological and bioinformatic studies and B.C. and R.J.D. wrote the manuscript with input from all authors.
References
- Adebali O, Reznik AO, Ory DS, Zhulin B. Establishing the precise evolutionary history of a gene improves prediction of disease-causing missense mutations. Genet Med. 2016 doi: 10.1038/gim.2015.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amendola LM, Dorschner MO, Robertson PD, Salama JS, Hart R, Shirts BH, Murray ML, Tokita MJ, Gallego CJ, Kim DS, Bennett JT, Crosslin DR, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–15. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, Sassa S, Bishop DF, Desnick RJ. Disorders of Heme Biosynthesis: X-Linked Sideroblastic Anemia and the Porphyrias. In: Valle D, Beaudet AL, Vogelstein B, Kinzler KW, Antonarakis SE, Ballabio A, Gibson K, Mitchell G, editors. 8. New York, NY: McGraw-Hill; New York: McGraw-Hill; 2014. http://ommbid.mhmedical.com/content.aspx?bookid=971&Sectionid=62638866. 2001. [Google Scholar]
- Andersson C, Floderus Y, Wikberg A, Lithner F. The W198X and R173W mutations in the porphobilinogen deaminase gene in acute intermittent porphyria have higher clinical penetrance than R167W. A population-based study. Scand J Clin Lab Invest. 2000;60:643–8. doi: 10.1080/003655100300054891. [DOI] [PubMed] [Google Scholar]
- Anyaegbu E, Goodman M, Ahn SY, Thangarajh M, Wong M, Shinawi M. Acute intermittent porphyria: a diagnostic challenge. J Child Neurol. 2012;27:917–21. doi: 10.1177/0883073811427603. [DOI] [PubMed] [Google Scholar]
- Bao L, Zhou M, Cui Y. nsSNPAnalyzer: identifying disease-associated nonsynonymous single nucleotide polymorphisms. Nucleic Acids Res. 2005;33:W480–482. doi: 10.1093/nar/gki372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendl J, Stourac J, Salanda O, Pavelka A, Wieben ED, Zendulka J, Brezovsky J, Damborsky J. PredictSNP: robust and accurate consensus classifier for prediction of disease-related mutations. PLoS Comput Biol. 2014;10:e1003440. doi: 10.1371/journal.pcbi.1003440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkovsky HL, Maddukuri VC, Yazici C, Anderson KE, Bissell DM, Bloomer JR, Phillips JD, Naik H, Peter I, Baillargeon G, Bossi K, Gandolfo L, et al. Acute porphyrias in the USA: features of 108 subjects from porphyrias consortium. Am J Med. 2014;127:1233–1241. doi: 10.1016/j.amjmed.2014.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bylesjö I, Wikberg A, Andersson C. Clinical aspects of acute intermittent porphyria in northern Sweden: a population-based study. Scand J Clin Lab Invest. 2009;69:612–8. doi: 10.1080/00365510902935979. [DOI] [PubMed] [Google Scholar]
- Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capriotti E, Calabrese R, Casadio R. Predicting the insurgence of human genetic diseases associated to single point protein mutations with support vector machines and evolutionary information. Bioinformatics. 2006;22:2729–2734. doi: 10.1093/bioinformatics/btl423. [DOI] [PubMed] [Google Scholar]
- Cerbino GN, Gerez EN, Varela LS, Melito VA, Parera VE, Batle A, Rossetti MV. Acute intermittent porphyria in Argentina: an update. Biomed Res Int. 2015 doi: 10.1155/2015/946387. 946387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PA, Duraisamy S, Miller PJ, Newell JA, McBride C, Bond JP, Raevaara T, Ollila S, Nyström M, Grimm AJ, Christodoulou J, Oetting WS, et al. Interpreting missense variants: comparing computational methods in human disease genes CDKN2A, MLH1, MSH2, MECP2, and tyrosinase (TYR) Hum Mutat. 2007;28:683–93. doi: 10.1002/humu.20492. [DOI] [PubMed] [Google Scholar]
- Chen CH, Astrin KH, Lee G, Anderson KE, Desnick RJ. Acute intermittent porphyria: identification and expression of exonic mutations in the hydroxymethylbilane synthase gene. An initiation codon missense mutation in the housekeeping transcript causes “variant acute intermittent porphyria” with normal expression of the erythroid-specific enzyme. J Clin Invest. 1994;94:1927–1937. doi: 10.1172/JCI117543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng WY, Hakenberg J, Li SD, Chen R. DIVAS: a centralized genetic variant repository representing 150 000 individuals from multiple disease cohorts. Bioinformatics. 2016;32:151–153. doi: 10.1093/bioinformatics/btv511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. 2015;31:2745–2747. doi: 10.1093/bioinformatics/btv195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Youssoufian H. The CpG dinucleotide and human genetic disease. Hum Genet. 1988;78:151–15543. doi: 10.1007/BF00278187. [DOI] [PubMed] [Google Scholar]
- Dehouck Y, Kwasigroch JM, Gilis D, Rooman M. PoPMuSiC 2.1: a web server for the estimation of protein stability changes upon mutation and sequence optimality. BMC Bioinformatics. 2011;12:151. doi: 10.1186/1471-2105-12-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delfau MH, Picat C, de Rooij FW, Hamer K, Bogard M, Wilson JH, Deybach JC, Nordmann Y, Gradchamp B. Two different point G to A mutations in exon 10 of the porphobilinogen deaminase gene are responsible for acute intermittent porphyria. J Clin Invest. 1990;86:1511–1516. doi: 10.1172/JCI114869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pierro E, Besana V, Moriondo V, Brancaleoni V, Cappellini MD. Hum Genet. 2006;119:364. doi: 10.1007/s00439-006-0139-8. [DOI] [PubMed] [Google Scholar]
- Dorfman R, Nalpathamkalam T, Taylor C, Gonska T, Keenan K, Yuan XW, Corey M, Tsui LC, Zielenski J, Durie P. Do common in silico tools predict the clinical consequences of amino-acid substitutions in the CFTR gene? Clin Genet. 2010;77:464–73. doi: 10.1111/j.1399-0004.2009.01351.x. [DOI] [PubMed] [Google Scholar]
- Elder G, Harper P, Badminton M, Sandberg S, Deybach JC. The incidence of inherited porphyrias in Europe. J Inherit Metab Dis. 2013;36:849–857. doi: 10.1007/s10545-012-9544-4. [DOI] [PubMed] [Google Scholar]
- Erikson GA, Bodian DL, Rueda M, Molparia B, Scott ER, Scott-Van Zeeland AA, Topol SE, Wineinger NE, Niederhuber JE, Topol EJ, Torkamani A. Whole-Genome Sequencing of a Healthy Aging Cohort. Cell. 2016;165:1–10. doi: 10.1016/j.cell.2016.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floderus Y, Shoolingin-Jordan PM, Harper P. Acute intermittent porphyria in Sweden. Molecular, functional and clinical consequences of some new mutations found in the porphobilinogen deaminase gene. Clin Genet. 2002;62:288–297. doi: 10.1034/j.1399-0004.2002.620406.x. [DOI] [PubMed] [Google Scholar]
- Genomes Project C, Auton A, Brooks LD, Dubin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill R, Kolstoe SE, Mohammed F, Al D-Bass A, Mosely JE, Sarwar M, Cooper JB, Wood SP, Shoolingin-Jordan PM. Structure of human porphobilinogen deaminase at 2.8Å: the molecular basis of acute intermittent porphyria. Biochem J. 2009;420:17–25. doi: 10.1042/BJ20082077. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Perez A, Lopez-Bigas N. Improving the assessment of the outcome of nonsynonymous SNVs with a consensus deleteriousness score, Condel. Am J Hum Genet. 2011;88:440–449. doi: 10.1016/j.ajhg.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granick S. Induction of the synthesis of delta-aminolevulinic acid synthetase in liver parenchyma cells in culture by chemical that induce acute porphyria. J Biol Chem. 1963;238:2247–2249. [PubMed] [Google Scholar]
- Granick S. The induction in vitro of the synthesis of delta-aminolevulinic acid synthetase in chemical porphyria: a response to certain drugs, sex hormones, and foreign chemicals. J Biol Chem. 1966;241:1359–1375. [PubMed] [Google Scholar]
- Hecht M, Bromberg Y, Rost B. Better prediction of functional effects for sequence variants. BMC Genomics. 2015;16(Suppl 8):S1. doi: 10.1186/1471-2164-16-S8-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hift RJ, Meissner PN. An analysis of 112 acute porphyric attacks in Cape Town, South Africa: Evidence that acute intermittent porphyria and variegate porphyria differ in susceptibility and severity. Medicine (Baltimore) 2005;84:48–60. doi: 10.1097/01.md.0000152454.56435.f3. [DOI] [PubMed] [Google Scholar]
- Innala E, Backstrom T, Bixo M, Anderson C. Evaluation of gonadotropin-releasing hormone agonist treatment for prevention of menstrual-related attacks in acute porphyria. Acta Obstet Gynecol Scand. 2010;89:95–100. doi: 10.3109/00016340903390729. [DOI] [PubMed] [Google Scholar]
- Kauppinen R, Mustajoki S, Pihlaja H, Peltonen L, Mustajoki P. Acute Intermittent porphyria in Finland: 19 mutations in the porphobilinogen deaminase gene. Hum Mol Genet. 1995;4:215–222. doi: 10.1093/hmg/4.2.215. [DOI] [PubMed] [Google Scholar]
- Kircher M, Witten DM, Jain P, O’Roak BJ, Cooper GM, Shendure J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat Genet. 2014;46:310–315. doi: 10.1038/ng.2892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König E, Rainer J, Domingues FS. Computational assessment of feature combinations for pathogenic variant prediction. Mol Genet Genomic Med. 2016;14:431–46. doi: 10.1002/mgg3.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- Lee J, Anvret M. Identification of the most common mutation within the porphobilinogen deaminase gene in Swedish patients with acute intermittent porphyria. Proc Natl Acad Sci USA. 1991;88:10912–5. doi: 10.1073/pnas.88.23.10912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leong IU, Stuckey A, Lai D, Skinner JR, Love DR. Assessment of the predictive accuracy of five in silico prediction tools, alone or in combination, and two metaservers to classify long QT syndrome gene mutations. BMC Med Genet. 2015;16:34. doi: 10.1186/s12881-015-0176-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Krishnan VG, Mort ME, Xin F, Kamati KK, Cooper DN, Mooney SD, Radivojac P. Automated inference of molecular mechanisms of disease from amino acid substitutions. Bioinformatics. 2009;25:2744–2750. doi: 10.1093/bioinformatics/btp528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn DH, Smyth SJ, Elder GH, Hutchesson AC, Rattenbury JM. Homozygous acute intermittent porphyria: compound heterozygosity for adjacent base transitions in the same codon of the porphobilinogen deaminase gene. Hum Genet. 1992;89:97–98. doi: 10.1007/BF00207051. [DOI] [PubMed] [Google Scholar]
- McColl KE, Moore MR, Thompson GG, Goldberg A. Screening for latent acute intermittent porphyria: the value of measuring both leucocyte delta-aminolaevulinic acid synthase and erythrocyte uroporphyrinogen-1-synthase activities. J Med Genet. 1982;19:271–276. doi: 10.1136/jmg.19.4.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer UA. Pharmacogenetics – five decades of therapeutic lessons from geentic diversity. Nat Rev Genet. 2004;5:669–76. doi: 10.1038/nrg1428. [DOI] [PubMed] [Google Scholar]
- Mgone CS, Lanyon WG, Moore MR, Louie GV, Connor JM. Detection of a high mutation frequency in exon 12 of the porphobilinogen deaminase gene in patients with acute intermittent porphyria. Hum Genet. 1993;92:619–622. doi: 10.1007/BF00420949. [DOI] [PubMed] [Google Scholar]
- Mustajoki P, Koskelo P. Hereditary hepatic porphyrias in Finland. Acta Med Scand. 1976;200:171–178. doi: 10.1111/j.0954-6820.1976.tb08216.x. [DOI] [PubMed] [Google Scholar]
- Mykletun M, Aarsand AK, Stole E, Villanger JH, Tollanes MC, Baravelli C, Sandberg S. Porphyrias in Norway. Tidsskr Nor Laegeforen. 2014;134:831–836. doi: 10.4045/tidsskr.13.0649. [DOI] [PubMed] [Google Scholar]
- Niroula A, Urolagin S, Vihinen M. PON-P2: prediction method for fast and reliable identification of harmful variants. PLoS One. 2015;10:e0117380. doi: 10.1371/journal.pone.0117380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann Y, Puy H, Da Silva V, Simonin S, Robreau AM, Bonaiti C, Phung LN, Deybach JC. Acute intermittent porphyria: prevalence of mutations in the porphobilinogen deaminase gene in blood donors in France. J Intern Med. 1997;242:213–217. doi: 10.1046/j.1365-2796.1997.00189.x. [DOI] [PubMed] [Google Scholar]
- Polikar R. Ensemble based systems in decision making. IEEE Circuits Syst Mag. 2006;6:21–45. [Google Scholar]
- Puy H, Gouya L, Deybach JC. Porphyrias. Lancet. 2010;375:924–937. doi: 10.1016/S0140-6736(09)61925-5. [DOI] [PubMed] [Google Scholar]
- Reva B, Antipin Y, Sander C. Predicting the functional impact of protein mutations: application to cancer genomics. Nucleic Acids Res. 2011;39:e118. doi: 10.1093/nar/gkr407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riera C, Lois S, de la Cruz X. Prediction of pathological mutations in proteins: the challenge of integrating sequence conservation and structure stability principles. Wiley Interdiscip Rev Comput Mol Sci. 2014;4:249–268. [Google Scholar]
- Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–423. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassa S, Granick S. Induction of -aminolevulinic acid synthetase in chick embryo liver cells in cluture. Proc Natl Acad Sci U S A. 1970;67:517–522. doi: 10.1073/pnas.67.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider-Yin X, Ulbrichova D, Mamet R, Martasek P, Marohnic CC, Goren A, Minder El, Schoenfeld N. Characterization of two missense variants in the hydroxymethylbilane synthase gene in the Israeli population, which differ in their associations with acute intermittent porphyria. Mol Genet Metab. 2008;94:343–346. doi: 10.1016/j.ymgme.2008.03.001. [DOI] [PubMed] [Google Scholar]
- Schuurmans MM, Schneider-Yin X, Rufenacht UB, Schnyder C, Minder CE, Puy H, Deybach JC, Minder El. Influence of age and gender on the clinical expression of acute intermittent porphyria based on molecular study of porphobilinogen deaminase gene among Swiss patients. Mol Med. 2001;7:535–542. [PMC free article] [PubMed] [Google Scholar]
- Song G, Li Y, Cheng C, Zhao Y, Gao A, Zhang R, Joachimiak A, Shaw N, Liu ZJ. Structural insight into acute intermittent porphyria. FASEB J. 2009;23:396–40. doi: 10.1096/fj.08-115469. [DOI] [PubMed] [Google Scholar]
- Stenson PD, Mort M, Ball EV, Shaw K, Phillips A, Cooper DN. The Human Gene Mutation Database: building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum Genet. 2014;133:1–9. doi: 10.1007/s00439-013-1358-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone EA, Sidow A. Physicochemical constraint violation by missense substitutions mediates impairment of protein function and disease severity. Genome Res. 2005;15:978–986. doi: 10.1101/gr.3804205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stowell KM. Malignant hyperthermia: a pharmacogenetic disorder. Pharmacogenomics. 2008;9:1657–72. doi: 10.2217/14622416.9.11.1657. [DOI] [PubMed] [Google Scholar]
- Tang H, Thomas PD. PANTHER-PSEP: predicting disease-causing genetic variants using position-specific evolutionary preservation. Bioinformatics. 2016 doi: 10.1093/bioinformatics/btw222. [DOI] [PubMed] [Google Scholar]
- von Brasch L, Zang C, Haverkamp T, Schlechte H, Heckers H, Petrides PE. Molecular analysis of acute intermittent porphyria: mutation screening in 20 patients in Germany reveals 11 novel mutations. Blood Cells Mol Dis. 2004;32:309–14. doi: 10.1016/j.bcmd.2003.12.003. [DOI] [PubMed] [Google Scholar]
- van der Velde KJ, Kuiper J, Thompson BA, Plazzer JP, van Valkenhoef G, de Haan M, Jongbloed JD, Wijmenga C, de Koning TJ, Abbott KM, Sinke R, Spurdle AB, et al. Evaluation of CADD Scores in Curated Mismatch Repair Gene Variants Yields a Model for Clinical Validation and Prioritization. Hum Mutat. 2015;36:712–9. doi: 10.1002/humu.22798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Kuilenburg AB. Dihydropyrimidine dehydrogenase and the efficacy and toxicity of 5-fluorouracil. Eur J Cancer. 2004;40:939–50. doi: 10.1016/j.ejca.2003.12.004. [DOI] [PubMed] [Google Scholar]
- von und zu Fraunberg M, Pischik E, Udd L, Kauppinen R. Clinical and biochemical characteristics and genotype-phenotype correlation in 143 Finnish and Russian patients with acute intermittent porphyria. Medicine (Baltimore) 2005;84:35–47. doi: 10.1097/01.md.0000152455.38510.af. [DOI] [PubMed] [Google Scholar]
- Whatley SD, Mason NG, Woolf JR, Newcombe RG, Elder GH, Badminton MN. Diagnostic strategies for autosomal dominant acute porphyrias: retrospective analysis of 467 unrelated patients referred for mutational analysis of the HMBS, CPOX, or PPOX gene. Clin Chem. 2009;55:1406–1414. doi: 10.1373/clinchem.2008.122564. [DOI] [PubMed] [Google Scholar]
- Whatley S, Badminton M. Prevalence of Potentially Pathogenic Mutations in the HMBS Gene in Subjects Enrolled in the 1000 Genomes Project. Clin Chem Lab Med. 2013:eA1–eA64. [Google Scholar]
- Xue Y, Chen Y, Ayub Q, Huang N, Ball EV, Mort M, Phillips AD, Shaw K, Stenson PD, Cooper DN, Tyler-Smith C. Deleterious- and Disease-Allele Prevalence in Healthy Individuals: insights from Current Predictions, Mutation Databases, and Population-Scale Resequencing. Am J Hum Genet. 2012;91:1022–1032. doi: 10.1016/j.ajhg.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.