Abstract
Key points
MicroRNA (miRNA)‐based therapies are in development for numerous diseases, including heart disease. Currently, very limited basic information is available on the regulation of specific miRNAs in male and female hearts in settings of disease. The identification of sex‐specific miRNA signatures has implications for translation into the clinic and suggests the need for customised therapy.
In the present study, we found that a miRNA‐based treatment inhibiting miRNA‐34a (miR‐34a) was more effective in females in a setting of moderate dilated cardiomyopathy than in males. Furthermore, the treatment showed little benefit for either sex in a setting of more severe dilated cardiomyopathy associated with atrial fibrillation.
The results highlight the importance of understanding the effect of miRNA‐based therapies in cardiac disease settings in males and females.
Abstract
MicroRNA (miRNA)‐34a (miR‐34a) is elevated in the diseased heart in mice and humans. Previous studies have shown that inhibiting miR‐34a in male mice in settings of pathological cardiac hypertrophy or ischaemia protects the heart against progression to heart failure. Whether inhibition of miR‐34a protects the female heart is unknown. Furthermore, the therapeutic potential of silencing miR‐34a in settings of dilated cardiomyopathy (DCM) and atrial fibrillation (AF) has not been assessed previously. In the present study, we examined the effect of silencing miR‐34a in males and females in (1) a model of moderate DCM and (2) a model of severe DCM with AF. The cardiac disease models were administered with a locked nucleic acid‐modified oligonucleotide (LNA‐antimiR‐34a) at 6–7 weeks of age when the models display cardiac dysfunction and conduction abnormalities. Cardiac function and morphology were measured 6 weeks after treatment. In the present study, we show that inhibition of miR‐34a provides more protection in the DCM model in females than males. Disease prevention in LNA‐antimiR‐34a treated DCM female mice was characterized by attenuated heart enlargement and lung congestion, lower expression of cardiac stress genes (B‐type natriuretic peptide, collagen gene expression), less cardiac fibrosis and better cardiac function. There was no evidence of significant protection in the severe DCM and AF model in either sex. Sex‐ and treatment‐dependent regulation of miRNAs was also identified in the diseased heart, and may explain the differential response of males and females. These studies highlight the importance of examining the impact of miRNA‐based drugs in both sexes and under different disease conditions.
Keywords: atrial fibrillation, dilated cardiomyopathy, heart failure, microRNA, miR‐34a
Key points
MicroRNA (miRNA)‐based therapies are in development for numerous diseases, including heart disease. Currently, very limited basic information is available on the regulation of specific miRNAs in male and female hearts in settings of disease. The identification of sex‐specific miRNA signatures has implications for translation into the clinic and suggests the need for customised therapy.
In the present study, we found that a miRNA‐based treatment inhibiting miRNA‐34a (miR‐34a) was more effective in females in a setting of moderate dilated cardiomyopathy than in males. Furthermore, the treatment showed little benefit for either sex in a setting of more severe dilated cardiomyopathy associated with atrial fibrillation.
The results highlight the importance of understanding the effect of miRNA‐based therapies in cardiac disease settings in males and females.
Abbreviations
- AF
atrial fibrillation
- BNP
B‐type natriuretic peptide
- DCM
dilated cardiomyopathy
- dnPI3K
dominant negative phosphoinositide 3‐kinase
- FS
fractional shortening
- HR
heart rate
- LNA
locked nucleic acid
- LV
left ventricle/ventricular
- LVEDD
LV end‐diastolic dimension
- LVESD
LV end‐systolic dimension
- miRNA/miR
microRNA
- Mst1
mammalian sterile 20‐like kinase 1
- Ntg
non‐transgenic
- PI3K
phosphoinositide 3‐kinase
Introduction
Sex differences in the incidence, prevalence, symptoms, age at onset and severity of disease are now well documented in humans (Kim et al. 2010; Legato, 2016). This is particularly evident in cardiovascular disease. In both animal and human studies, it is apparent that there are sex differences in the degree of cardiac pathology in response to cardiac insults such as pressure overload (Bernardo et al. 2010). However, the molecular mechanisms underlying sex dimorphism are complex and are still not well understood. Data from clinical trials also demonstrate that males and females respond differently to cardiovascular drugs. For example, conventional pharmacological therapies including angiotensin‐converting enzyme inhibitors and β‐blockers are generally less effective in women than men, and show more side effects (Regitz‐Zagrosek, 2006). Furthermore, higher mortality has been reported in women treated with digoxin (Rathore et al. 2002). Despite these differences, biomedical research continues to use many more male subjects than females in both animal studies and human clinical trials (Editorial, 2010; Legato, 2016).
MicroRNAs (miRNAs) are a class of non‐coding RNAs that regulate gene expression by directing their target mRNAs for degradation or translational repression (Bernardo et al. 2012a). miRNAs are dysregulated in a number of disease settings, including cardiovascular disease (Hata, 2013). Targeting disease‐induced miRNAs is considered to be a promising therapeutic approach, with a number of drugs being tested in clinical trials and/or being developed (Janssen et al. 2013; Bernardo et al. 2015). Although sex differences are well recognized in human disease, relatively few studies have specifically examined miRNA sex differences in humans (Sharma & Eghbali, 2014). In the small number of human studies performed, sex‐based expression of miRNAs have been identified in disorders including metabolic syndrome, schizophrenia and cancer (Sharma & Eghbali, 2014). To date, the study of sex differences in miRNAs in humans with cardiovascular disease remains largely unexplored and represents a key knowledge gap (Sharma & Eghbali, 2014).
One miRNA that has received particular interest in the heart is miRNA‐34a (miR‐34a). The expression of miR‐34a is elevated in the mouse heart in a number of cardiac stress settings, including pathological hypertrophy, ischaemia as a result of myocardial infarction and ageing (Bernardo et al. 2012b; Boon et al. 2013; Huang et al. 2014; Bernardo et al. 2014a; Yang et al. 2015). Of clinical relevance, the expression of miR‐34a is also elevated in the human failing heart (Thum et al. 2007; Tabuchi et al. 2012). Therapeutic inhibition of miR‐34a has been shown to be beneficial by at least four independent laboratories in cardiac disease models (Bernardo et al. 2012b; Boon et al. 2013; Huang et al. 2014; Bernardo et al. 2012b; Yang et al. 2015). However, in each of these studies, only male mice were examined. To date, the therapeutic potential of silencing miR‐34a in females with cardiac pathology has not been explored. Furthermore, the role of miR‐34a in models of dilated cardiomyopathy (DCM) and atrial fibrillation (AF) had not previously been assessed. The present study aimed to examine whether inhibition of miR‐34a provides protection in mouse models with DCM and AF, in both males and females.
Methods
Ethical approval
Animal care and experimentation were conducted in accordance with the Australian code for the care and use of animals for scientific purposes (National Health and Medical Research Council, 2013) and approved by the Alfred Medical Research and Education Precinct Animal Ethics Committee. Age‐ and sex‐matched male and female heart disease mice of ∼13 weeks of age were killed by an i.p. injection of sodium pentobarbital (80 mg kg−1). Adequacy of anaesthesia was confirmed by the lack of pedal withdrawal reflex.
Experimental heart disease models
In the present study, we used two cardiac disease models: (1) a model of moderate dilated cardiomyopathy (referred to as DCM) and (2) a model of more severe DCM with atrial fibrillation (referred to as AF). These models are cardiac‐specific transgenic mouse models and were generated as described previously (Pretorius et al. 2009). In brief, mice develop DCM as a result of transgenic expression of mammalian sterile 20‐like kinase 1 (Mst1) (Yamamoto et al. 2003) and the AF model is generated by mating Mst1 mice with a model with reduced cardiac phosphoinositide 3‐kinase (PI3K) activity (as a result of the expression of dominant negative PI3K; dnPI3K) (Shioi et al. 2000). The Mst1 model is considered clinically relevant because the mice develop DCM as a consequence of increased apoptosis. Male hemizygous Mst1 mice and female hemizygous dnPI3K were bred to generate: (1) DCM mice (mice expressing Mst1 alone, Mst1+/−); (2) AF model (mice expressing both Mst1 and dnPI3K; i.e. Mst1+/−dnPI3K+/−); and (3) non‐transgenic littermate controls (Ntg; Mst1−/−dnPI3K−/−). All mice were littermates, on the same genetic background (C57BL/6‐FVB/N). Litters containing the range of models of both sexes were selected and mice were body weight‐matched before randomly being assigned to treatment. The animal groups comprised: Ntg LNA‐control (females n = 4, males n = 6); Ntg LNA‐antimiR‐34a (females n = 4, males n = 5); DCM LNA‐control (females n = 5, males n = 6); DCM LNA‐antimiR‐34a (females n = 8, males n = 7); AF LNA‐control (females n = 4, males n = 9); and AF LNA‐antimiR‐34a (females n = 5, males n = 5). No animals died before the end‐point of the study.
LNA‐oligonucleotide synthesis and in vivo delivery
A 15‐mer antimiR‐34a was synthesized as a LNA:DNA mixmer with a complete phosphorothioate backbone (5′‐AgCtaAGacACTgCC‐3′; LNA indicated by uppercase letters, DNA indicated by lowercase letters; Roche Innovation Centre, Copenhagen, Denmark), as described previously (Bernardo et al. 2012b; Bernardo et al. 2014a). The LNA‐control sequence (5′‐TcAtaCTatAtGaCA‐3′) is a random sequence designed to have no perfect match binding sites in the transcriptome and has been validated in vivo as not differing from untreated or saline treated animals (Obad et al. 2011; Bernardo et al. 2012b; Bernardo et al. 2014a; Bernardo et al. 2014b; Bernardo et al. 2016). In the present study, there were no differences observed between mice administered LNA‐control or saline (data not shown).
At 6–7 weeks of age (after assessment of cardiac function by echocardiography and ECG), mice (Ntg, DCM model, AF model) were randomized to receive a loading dose (25 mg kg−1 s.c.) of LNA‐control or LNA‐antimiR‐34a (Monday) and two subsequent doses at 10 mg kg−1 (Wednesday and Friday), which were followed by treatment three times per week (10 mg kg−1 s.c.; Monday, Wednesday and Friday) for an additional 5 weeks (i.e. a total treatment period of 6 weeks) (Fig. 1 A). This was followed by a final assessment of cardiac function and ECG prior to tissue collection (measurement of body weight, heart weight, atria weight, lung weight and tibia length).
Figure 1. miR‐34a is elevated in the heart in a setting of DCM and AF, and silenced with LNA‐antimiR‐34a .
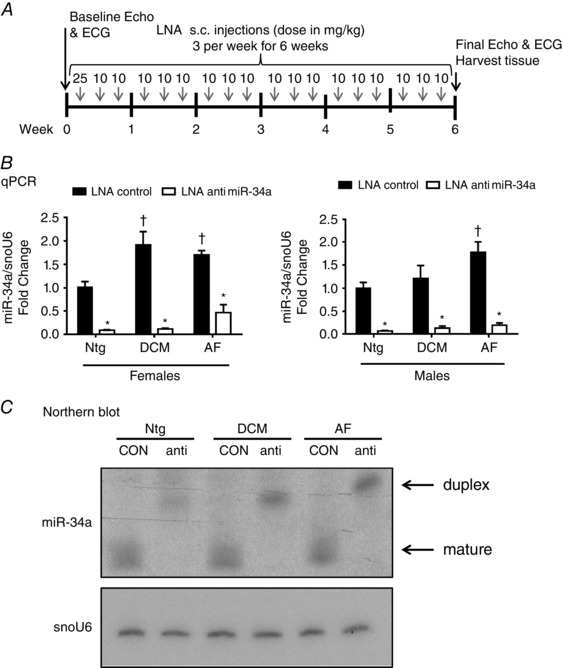
A, experimental time line. B, quantification of miR‐34a relative to snoU6 by quantitative PCR. Data are expressed as the mean ± SEM. Ntg: n = 4–5 per group, DCM: n = 4–6 per group, AF: n = 4–9 per group. * P < 0.05 vs. LNA‐control of the same group; † P < 0.05 vs. Ntg LNA‐control. One‐way ANOVA with Fisher's post hoc test. C, Northern blot of miR‐34a and snoU6 in hearts of Ntg, DCM and AF 6 weeks post administration with LNA‐control (con) or LNA‐antimiR‐34a (anti).
Assessment of left ventricular dimensions and function
Cardiac function was assessed non‐invasively before and after treatment by echocardiography (two‐dimensional M‐mode) in anaesthetized mice (1.8% isoflurane) using an iE33 ultrasound machine (Philips, Amsterdam, Netherlands) with a 15 MHz linear array transducer. LV wall thickness (LV posterior wall: LVPW), LV chamber dimensions (LV end‐diastolic dimension: LVEDD; LV end‐systolic dimension: LVESD), heart rate (HR) and fractional shortening (FS) ([(LVEDD – LVESD)/LVEDD] × 100%) were measured from the short‐axis view. All measurements were performed with an off‐line analysis system (ProSolv Cardiovascular Analyser 3.5; Fuji Film, Tokyo, Japan) from at least three beats that were averaged. Prior to treatment, cardiac function was comparable in the DCM groups of the same sex (e.g. male DCM LNA‐control vs. male DCM LNA‐antimiR‐34a) and AF groups (data not shown).
Cardiac conduction by ECG
Direct ECGs were recorded from anaesthetized mice (1.8% isoflurane) by placing a pair of 27 gauge needle electrodes s.c. (right arm and chest lead equivalent to V5). Signals were sampled for at least 5 min using the Powerlab system and BioAmp (ADInstruments, Sydney, Australia). ECG parameters (PR, RR, P and R‐amplitudes, HR) were measured using Chart 5 (ADInstruments) (ECG analysis module). Because the shape of P waves can be abnormal in the DCM and AF models (making detection by the ECG analysis module difficult), PR intervals were also confirmed manually.
Histological analysis
Ventricle samples were fixed in 4% paraformaldehyde, dehydrated and embedded in paraffin. Cardiac collagen deposition/interstitial fibrosis was assessed by a Masson's trichrome stain (Alfred Pathology, Melbourne, Australia). Images of the LV were obtained using a light microscope (Olympus, Tokyo, Japan) at 40× magnification. Collagen stained blue, which was measured and analysed using Image‐Pro Analyser, version 7.0 (Meyer Instruments, Houston, TX, USA). Percentage fibrosis was calculated by dividing the total area of collagen by the total area of the LV and multiplying by 100%. Photogaphs of whole hearts were taken for visualization of cardiac chambers.
RNA extraction and mRNA gene expression analyses
Total RNA was extracted from mouse ventricles using TRI‐reagent (Sigma Aldrich, St Louis, MI, USA). For mRNA Northern blotting, 20 μg of total RNA was electrophoresed in a 1.3% denaturing formaldehyde agarose gel in 1 × MOPS, and transferred onto Hybond N membranes (GE Healthcare, Waukesha, WI, USA). Membranes were blocked in hybridization solution (50% deionized formamide, 6X SSC, 5X Denhardt's solution, 0.5% SDS) containing 1 mg ml−1 denatured salmon sperm DNA at 42°C for 1 h prior to hybridization. Radiolabelled probes ([α‐32P]dCTP) were prepared using a Prime‐a‐Gene Labelling System (Promega, Madison, WI, USA). Membranes were hybridized with denatured radiolabelled probes at 42°C overnight (2 × 106 cpm ml−1 of hybridization solution). Membranes were rinsed twice at room temperature in 2X SSC, twice at 42°C in 2X SSC/1% SDS for 5 min and twice in 0.1X SSC for 30 min. Amersham Hyperfilm MP autoradiography films (GE Healthcare) were exposed to membranes with an intensifying screen at −80°C. Membranes were probed for B‐type natriuretic peptide (Nppb/Bnp) and collagen 3 (Col3a1), as well as glyceraldehyde‐3‐phosphate dehydrogenase (Gapdh).
miRNA expression
For Northern blotting, 15 μg of total RNA was electrophoresed in a 20% TBE acrylamide gel using high‐density TBE sample buffer and transferred to Amersham Hybond‐N™+ nylon membrane (GE Healthcare) by electrophoresis. The membrane was cross‐linked in an UV cross‐linker, pre‐hybridized (45°C for 30 min in ULTRAhyb®‐Oligo Hybridization Buffer; Life Technologies, Carlsbad, CA, USA) and probed with LNA‐modified oligonucleotides (Exiqon, Vedbæk, Denmark), complementary to mature miR‐34a and sno‐U6 (used as a loading control). Next, 10 pmol of each LNA oligonucleotide was end‐labelled with [γ‐32P]ATP (Perkin Elmer, Waltham, MA, USA) using T4 polynucleotide kinase (New England Biolabs, Ipswich, MA, USA) for 1 h at 37°C. Hybridization with labelled probes (5 min at 100°C prior to addition) was conducted at 45°C overnight in ULTRAhyb®‐Oligo Hybridization Buffer (Life Technologies). After hybridization, the membranes were washed for 30 min in NorthernMax® Low Stringency Wash Buffer (Life Technologies) at 45°C, a high stringency wash in 0.5X SSC for 30 min at 45°C (for LNA‐oligonucleotide miR‐34a) or 0.1X SSC for 30 min at 45°C (for LNA‐oligonucleotide sno‐U6). The membranes were exposed using autoradiography film.
For quantitative PCR analysis, 2 μg of total RNA was DNAse treated with a TURBO DNA‐free kit (Life Technologies) in accordance with the manufacturer's instructions. To detect the level of miR‐27a, miR‐34a, miR‐34b and miR‐34c, a quantitative RT‐PCR was performed using TaqMan® MicroRNA Assays (Life Technologies) using 50 ng of total RNA on an Applied Biosystems Quant Studio 6 and 7 real‐time PCR instrument (Life Technologies). Expression was normalized against snoU6 using the 2−ΔΔCt method of quantification.
miRNA‐Seq for miRNA expression analysis
Two samples from the following groups were used for miRNA‐Seq: male Ntg control (one saline treated mouse, one LNA‐control), female Ntg control (two LNA‐control), male DCM control (two LNA‐control), male DCM antimiR‐34a (two LNA‐antimiR‐34a), female DCM control (two LNA‐control), female DCM antimiR‐34a (two LNA‐antimiR‐34a), male AF control (two LNA‐control), male AF antimiR‐34a (two LNA‐antimiR‐34a), female AF control (two LNA‐control), female AF antimiR‐34a (two LNA‐antimiR‐34a).
Standard miRNA‐Seq libraries were prepared in accordance with the manufacturer's instructions (Illumina, Inc., San Diego, CA, USA). Samples (24 per lane) were multiplexed and sequenced on the Illumina Genome Analyser HiSeq 2000 (read length: 50 bp, Ramaciotti Centre for Genomics, University of NSW, Australia). MiRNA‐Seq reads were aligned to the mouse reference genome (mm10) using Partek Flow, version 3.0 (Partek Inc., Chesterfield, MO, USA) with default settings unless otherwise indicated. After removing adaptor sequences, the bases of unaligned reads were trimmed for a minimum Phred quality score of 20 (default settings used except minimum read length = 17). Fastq files were then aligned to the mm10 reference genome with Bowtie 1 (default parameters used except seed mismatch limit = 0, seed length = 22, alignments reported per read = 10) and Bowtie 2 (default parameters except interval between seeds S,1,1.25, ambiguous characters function L,O.15, default reporting mode = false, max alignments reported per read = 10; http://bowtie-bio.sourceforge.net/index.shtml) (Langmead et al. 2009). Prealignment and postalignment quality control was performed, and read counts were filtered for reads with minimum mapping quality of 20 (99% of reads met this criteria). The trimmed, aligned and filtered data were quantitated to known miRNAs using the miRBase mature miRNA database, version 21 (released 3 July 2014; http://www.mirbase.org). Reads are presented as log2 transcript counts from miRNA sequencing normalized to reads per kilobase of transcript per million mapped reads (Genomics Suite, version 6.6; Partek Inc.). For comparisons between the two Ntg groups, a t test was performed. For comparisons between the four DCM/AF groups, a one‐way ANOVA was performed. Unadjusted P‐values are presented in the Supporting information. Heat maps were generated using MeV, version 4.8.1 (TIGR MultiExperiment Viewer; http://www.tm4.org) with the specific parameters (t test based on Welch approximation, P‐value based on t‐distribution and hierarchical clustering using Pearson correlation) (Saeed et al. 2003).
Statistical analysis
Results are presented as the mean ± SEM. All statistics were performed using StatView (SAS Institute Inc., Cary, NC, USA) and Prism, version 6.02 (GraphPad Software Inc., San Diego, CA, USA), unless otherwise indicated. Differences between groups were identified using one‐way ANOVA followed by Fisher's least significant difference post hoc pairwise tests (when the ANOVA was significant). Unpaired t tests were used when comparing two groups for a single measure. For small animal groups (n = 4) in which the data were not normally distributed, differences between groups were identified using a Mann–Whitney non‐parametric t test. P < 0.05 was considered statistically significant. All relative units are expressed as the fold change with the relevant control group normalized to 1.
Results
miR‐34a levels are elevated in the DCM and AF cardiac disease models and inhibited by LNA‐antimiR‐34a
Tissue was harvested from mice 6 weeks after commencing treatment with LNA‐antimiR‐34a or LNA‐control (Fig. 1 A). miR‐34a expression levels were increased in the hearts of the female DCM and AF models compared to Ntg (Fig. 1 B; see LNA‐control groups). A similar observation was made in the male disease models, although miR‐34a was only significantly elevated in the hearts of the male AF model compared to Ntg (Fig. 1 B). Administration of LNA‐antimiR‐34a significantly reduced miR‐34a levels in both males and females (Fig. 1 B). Inhibition of miR‐34a was confirmed in a subset of samples by Northern blotting (Fig. 1 C). The presence of the shifted miR‐34a:LNA‐antimiR‐34a band (duplex) on the Northern blot indicates that LNA‐antimiR‐34a has sequestered miR‐34a in a stable heteroduplex, inhibiting its function, as reported previously (Bernardo et al. 2012b).
Inhibition of miR‐34a is more effective at attenuating DCM‐induced morphological changes in female mice than male mice
Prior to administration of LNA‐antimiR‐34a or LNA‐control, the DCM and AF models displayed cardiac dysfunction, atrial enlargement and ECG abnormalities (Fig. 2 A and B; arrhythmia also present in the AF model). At tissue dissection (6 weeks after LNA‐antimiR‐34a/control), the DCM and AF models were associated with dilatation of LV chambers and thinning of ventricular walls (Fig. 2 C). Female DCM mice displayed heart enlargement compared to Ntg controls (Fig. 3 A) and this was accompanied by atrial enlargement and increased lung weights (both markers of a failing heart) (Fig. 3 B and C and Table 1). Male DCM mice also had increased heart weights, atria weights and lung weights compared to Ntg controls (Fig. 3 and Table 2); however, the percentage increases in males were generally more modest than that observed in females (e.g. HW/TL increased ∼33% in females and 9% in males). LNA‐antimiR‐34a treatment prevented significant increases in normalized heart, atria and lung weights in the DCM female model (Fig. 3 and Table 1). Although LNA‐antimiR‐34a also prevented significant increases in normalized heart and lung weights in the DCM male model, it had no effect on atrial weight (Fig. 3 and Table 2). The AF model was also associated with increased atrial and lung weights (Fig. 3 B and C). Treatment with LNA‐antimiR‐34a was not associated with significant improvements in cardiac morphology or lung weights in female and male AF mice (Fig. 3 and Tables 1 and 2).
Figure 2. The DCM and AF models are associated with cardiac dysfunction, ECG abnormalities and dilatation of cardiac chambers .
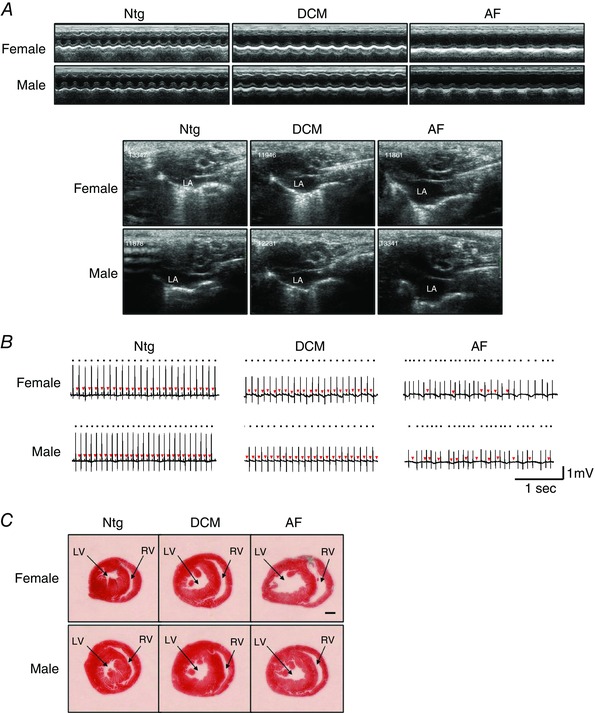
A, representative echocardiography images (upper panels, M‐mode; lower panels, long‐axis view highlighting differences in left atrial size, LA). B, ECG traces of Ntg, DCM and AF mice prior to treatment. Dots on the ECG traces are positioned above the R waves and highlight the irregular rhythm of the AF model. Red arrows highlight P‐waves. C, transverse sections of hearts highlighting dilated chambers in the female and male DCM and AF models. LV, left ventricle chamber; RV, right ventricle chamber. Scale bar = 1 mm.
Figure 3. Inhibition of miR‐34a improves cardiac morphology more in female DCM mice than male DCM mice .
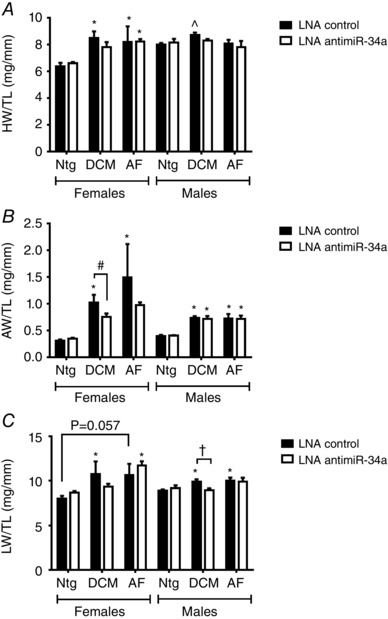
Graphs of (A) heart weight/tibia length (HW/TL), (B) atria weight/TL (AW/TL) and (C) lung weight/TL (LW/TL) in female and male Ntg, DCM and AF mice. Data are expressed as the mean ± SEM. Female Ntg: n = 4 per group, female DCM: n = 5–8 per group, female AF: n = 4–5 per group. Male Ntg: n = 5–6 per group, male DCM: n = 6–7 per group, male AF: n = 5–9 per group. * P < 0.05 vs. Ntg of same treatment group; † P < 0.05 (one‐way ANOVA with Fisher's post hoc test of Ntg, DCM and AF groups); ^P < 0.05 vs. Ntg of same treatment group (unpaired t test); # P < 0.05 (one‐way ANOVA with Fisher's post hoc test of Ntg and DCM groups only), P = 0.057 (Mann–Whitney non‐parametric t test).
Table 1.
Morphological data of Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a female mice after 6 weeks of treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Females | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 4 | 4 | 5 | 8 | 4 | 5 |
| Age (weeks) | 13.2 ± 0.4 | 14.0 ± 0.9 | 13.1 ± 0.6 | 12.4 ± 0.5 | 12.9 ± 0.5 | 12.4 ± 0.3 |
| Body weight (g) | 24.1 ± 0.8 | 24.9 ± 0.5 | 26.0 ± 1.0 | 25.5 ± 0.7 | 25.2 ± 1.1 | 26.8 ± 0.3 |
| Tibial length (mm) | 16.7 ± 0.1 | 16.7 ± 0.1 | 16.4 ± 0.3 | 16.6 ± 0.1 | 16.7 ± 0.2 | 16.8 ± 0.1 |
| HW (mg) | 106.7 ± 3.6 | 110.4 ± 1.6 | 138.5 ± 8.3* | 130.4 ± 7.0 | 136.8 ± 20.1* | 138.0 ± 3.2* |
| AW (mg) | 5.3 ± 0.4 | 5.9 ± 0.3 | 16.7 ± 2.5* | 12.6 ± 1.3 | 25.0 ± 10.6* | 16.4 ± 0.8 |
| LW (mg) | 133.7 ± 3.9 | 144.6 ± 3.2 | 176.6 ± 24.2* | 155.4 ± 6.8 | 177.8 ± 21.4* | 196.6 ± 7.9* |
| HW/TL (mg/mm) | 6.4 ± 0.2 | 6.6 ± 0.1 | 8.5 ± 0.5* | 7.8 ± 0.4 | 8.2 ± 1.2* | 8.2 ± 0.2* |
| AW/TL (mg/mm) | 0.31 ± 0.02 | 0.35 ± 0.02 | 1.02 ± 0.15* | 0.75 ± 0.07 | 1.49 ± 0.63* | 0.98 ± 0.05 |
| LW/TL (mg/mm) | 8.0 ± 0.3 | 8.7 ± 0.2 | 10.8 ± 1.4* | 9.3 ± 0.3 | 10.7 ± 1.2† | 11.7 ± 0.5* |
BW, body weight; TL, tibial length; HW, heart weight; AW, atrial weight; LW, lung weight. Data are shown as the mean ± SEM. * P < 0.05 vs. Ntg of the same treatment. One‐way ANOVA with Fisher's post hoc test. † P = 0.057 vs. Ntg of same treatment (Mann–Whitney non‐parametric t test).
Table 2.
Morphological data of Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a male mice after 6 weeks of treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Males | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 6 | 5 | 6 | 7 | 9 | 5 |
| Age (weeks) | 13.6 ± 0.6 | 13.6 ± 0.7 | 13.3 ± 0.5 | 13.6 ± 0.5 | 13.7 ± 0.3 | 13.3 ± 0.7 |
| Body weight (g) | 32.6 ± 1.0 | 31.7 ± 1.0 | 32.3 ± 0.5 | 31.7 ± 0.6 | 32.6 ± 0.6 | 33.4 ± 0.6 |
| Tibia length (mm) | 16.6 ± 0.1 | 16.8 ± 0.1 | 16.9 ± 0.1 | 16.8 ± 0.1 | 16.8 ± 0.2 | 17.0 ± 0.2 |
| HW (mg) | 132.9 ± 3.0 | 136.6 ± 4.8 | 146.8 ± 3.4 | 139.9 ± 2.0 | 135.6 ± 4.0 | 133.0 ± 8.9 |
| AW (mg) | 6.7 ± 0.4 | 6.9 ± 0.2 | 12.5 ± 0.7* | 12.1 ± 0.9* | 12.2 ± 1.4* | 12.3 ± 1.2* |
| LW (mg) | 147.6 ± 1.7 | 154.2 ± 5.2 | 166.5 ± 4.7* | 150.8 ± 3.2† | 168.0 ± 5.2* | 168.3 ± 8.8 |
| HW/TL (mg/mm) | 8.0 ± 0.1 | 8.1 ± 0.3 | 8.7 ± 0.2^ | 8.3 ± 0.1 | 8.1 ± 0.3 | 7.8 ± 0.4 |
| AW/TL (mg/mm) | 0.40 ± 0.02 | 0.41 ± 0.01 | 0.74 ± 0.04* | 0.72 ± 0.05* | 0.73 ± 0.09* | 0.72 ± 0.06* |
| LW/TL (mg/mm) | 8.9 ± 0.1 | 9.2 ± 0.3 | 9.9 ± 0.3* | 9.0 ± 0.2† | 10.0 ± 0.3* | 9.9 ± 0.4 |
BW, body weight; TL, tibia length; HW, heart weight; AW, atrial weight; LW, lung weight. Data are shown as the mean ± SEM. One‐way ANOVA with Fisher's post hoc test. * P < 0.05 vs. Ntg of the same treatment; ^P < 0.05 vs. Ntg of the same treatment (unpaired t test); † P < 0.05 vs. DCM LNA‐control.
Inhibition of miR‐34a was associated with better cardiac function in the DCM model but not the AF model
Hearts of the DCM and AF models display cardiac dilatation, thinning of the ventricular walls and a fall in cardiac function (assessed by calculating FS). The models are also associated with ECG abnormalities. On the majority of parameters, abnormalities in cardiac dimensions, function and ECG parameters are more severe in the AF model than the DCM model (Tables 3, 4, 5, 6, see LNA‐control groups for Ntg, DCM and AF). Administration of LNA‐antimiR‐34a for 6 weeks was associated with more favourable cardiac function and LV wall thickness in female DCM mice compared to female DCM mice administered LNA‐control (Fig. 4 and Table 3). Cardiac function also tended to be greater in LNA‐antimiR‐34a treated male DCM mice compared to LNA‐control male DCM mice, although this was not statistically significant. There was also no notable improvement in other echocardiography parameters (Table 4). LNA‐antimiR‐34a provided no benefit in the AF model of either sex (Fig. 4 and Tables 3 and 4). ECG parameters were not significantly changed by LNA‐antimiR‐34a in the disease models (Tables 5 and 6).
Table 3.
Echocardiography measurements in Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a female mice 6 weeks post‐treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Females | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 4 | 4 | 5 | 7 | 4 | 5 |
| HR (beats min–1) | 622 ± 22 | 628 ± 12 | 578 ± 14 | 524 ± 26 | 515 ± 44 | 448 ± 69* |
| LVPW (mm) | 0.80 ± 0.01 | 0.81 ± 0.00 | 0.65 ± 0.02* | 0.69 ± 0.01*† | 0.53 ± 0.02*‡ | 0.50 ± 0.01*‡ |
| LVEDD (mm) | 3.76 ± 0.02 | 3.76 ± 0.07 | 4.28 ± 0.17* | 4.29 ± 0.10* | 4.66 ± 0.29* | 4.54 ± 0.18* |
| LVESD (mm) | 2.01 ± 0.08 | 2.14 ± 0.03 | 3.20 ± 0.23* | 2.99 ± 0.09* | 3.89 ± 0.35*‡ | 3.84 ± 0.19*‡ |
| FS (%) | 46 ± 2 | 43 ± 1 | 26 ± 2* | 30 ± 1*† | 17 ± 2*§ | 16 ± 1*‡ |
HR, heart rate; LV, left ventricular; LVPW, LV posterior wall thickness; LVEDD, LV end‐diastolic dimension; LVESD, LV end‐systolic dimension; FS, fractional shortening. Data are shown as the mean ± SEM. * P < 0.05 vs. Ntg of same treatment; † P < 0.05 vs. LNA‐control of same model; ‡ P < 0.05 vs. DCM of same treatment. One‐way ANOVA with Fisher's post hoc test. § P = 0.063 vs. DCM of same treatment (Mann–Whitney non‐parametric t test).
Table 4.
Echocardiography measurements in Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a male mice 6 weeks post‐treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Males | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 5 | 5 | 6 | 7 | 9 | 5 |
| HR (beats min–1) | 588 ± 16 | 609 ± 22 | 560 ± 21 | 562 ± 20 | 597 ± 19 | 567 ± 26 |
| LVPW (mm) | 0.86 ± 0.02 | 0.84 ± 0.02 | 0.69 ± 0.01* | 0.70 ± 0.02* | 0.53 ± 0.01*‡ | 0.53 ± 0.01*‡ |
| LVEDD (mm) | 4.04 ± 0.05 | 3.81 ± 0.16 | 4.45 ± 0.09* | 4.19 ± 0.10* | 4.63 ± 0.08* | 4.63 ± 0.2*‡ |
| LVESD (mm) | 2.29 ± 0.03 | 2.13 ± 0.13 | 3.17 ± 0.06* | 2.82 ± 0.08*† | 3.67 ± 0.11*‡ | 3.69 ± 0.27*‡ |
| FS (%) | 43 ± 1 | 44 ± 2 | 29 ± 1* | 33 ± 1*† | 21 ± 1*‡ | 21 ± 2*‡ |
HR, heart rate; LV, left ventricular; LVPW, LV posterior wall thickness; LVEDD, LV end‐diastolic dimension; LVESD, LV end‐systolic dimension; FS, fractional shortening. Data are shown as the mean ± SEM. One‐way ANOVA with Fisher's post hoc test. * P < 0.05 vs. Ntg of same treatment, † P = 0.06 vs. LNA‐control of same model, ‡ P < 0.05 vs. DCM of same treatment.
Table 5.
Electrocardiography measurements in Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a female mice 6 weeks post‐treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Females | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 3 | 3 | 4 | 5 | 4 | 4 |
| HR (beats min–1) | 581 ± 9 | 534 ± 3 | 520 ± 16 | 509 ± 17 | 533 ± 31 | 514 ± 23 |
| RR (ms) | 104 ± 2 | 112 ± 1 | 116 ± 4 | 120 ± 4 | 115 ± 8 | 118 ± 5 |
| PR (ms) | 34 ± 1 | 36 ± 1 | 49 ± 1* | 51 ± 3* | 53 ± 1* | 58 ± 4* |
| R amplitude (mV) | 1.64 ± 0.13 | 1.77 ± 0.08 | 1.17 ± 0.11* | 1.13 ± 0.12* | 0.96 ± 0.22* | 0.89 ± 0.12* |
Data are shown as the mean ± SEM. One‐way ANOVA with Fisher's post hoc test. * P < 0.05 vs. Ntg of same treatment; † P < 0.05 vs. LNA‐control of same model.
Table 6.
Electrocardiography measurements in Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a male mice 6 weeks post‐treatment
| Ntg | DCM | AF | ||||
|---|---|---|---|---|---|---|
| Males | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a | LNA‐control | LNA‐antimiR‐34a |
| Number of animals | 5 | 5 | 4 | 4 | 4 | 4 |
| HR (beats min–1) | 569 ± 7 | 574 ± 16 | 572 ± 2 | 538 ± 28 | 547 ± 2 | 529 ± 20 |
| RR (ms) | 106 ± 1 | 105 ± 3 | 105 ± 0 | 113 ± 6 | 110 ± 1 | 116 ± 5 |
| PR (ms) | 33 ± 1 | 34 ± 1 | 44 ± 2* | 43 ± 1* | 55 ± 2*‡ | 53 ± 6*‡ |
| R amplitude (mV) | 1.63 ± 0.08 | 1.70 ± 0.08 | 1.11 ± 0.05* | 1.22 ± 0.08* | 0.97 ± 0.07* | 0.91 ± 0.08*‡ |
Data are shown as the mean ± SEM. One‐way ANOVA with Fisher's post hoc test. * P < 0.05 vs. Ntg of same treatment; ‡ P < 0.05 vs. DCM of same treatment.
Figure 4. Administration of LNA‐antimiR‐34a is associated with better cardiac function in the DCM model, but not the AF model .
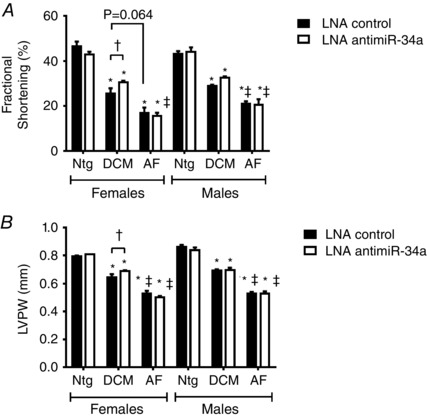
Quantification of (A) fractional shortening and (B) left ventricular posterior wall thickness (LVPW) 6 weeks post‐treatment. Data are the mean ± SEM. Ntg n = 4–5 per group, DCM n = 5–7 per group, AF n = 4–9 per group. * P < 0.05 vs. Ntg of same treatment; † P < 0.05 vs. LNA‐control of same model; ‡ P < 0.05 vs. DCM of same treatment (one‐way ANOVA with Fisher's post hoc test of Ntg, DCM and AF groups). P = 0.064 (Mann–Whitney non‐parametric t test).
LNA‐antimiR‐34a induced protection in the female DCM model is associated with an improved cardiac molecular profile
The DCM and AF models have previously been shown to be associated with increased expression of cardiac stress and collagen genes, which probably contribute to cardiac dysfunction and conduction abnormalities (Pretorius et al. 2009; Sapra et al. 2014). In the present study, BNP and collagen 3 gene expression levels were elevated in the hearts of both male and female DCM and more significantly in AF mice (Fig. 5 A and B, see LNA‐control groups). BNP was elevated less in LNA‐antimiR‐34a treated female DCM hearts than LNA‐control female DCM hearts (Fig. 5 A). Collagen 3 gene expression was lower in LNA‐antimiR‐34a treated female DCM mice than LNA‐control female DCM mice (Fig. 5 B). Consistent with this finding, LNA‐antimiR‐34a treated female DCM mice displayed less cardiac fibrosis than LNA‐control female DCM mice (Fig. 5 C). Inhibition of miR‐34a had no effect on gene expression in any other group (Fig. 5 A and B).
Figure 5. LNA‐antimiR‐34a treatment is associated with a more favourable cardiac molecular profile in female DCM mice but not male DCM mice .
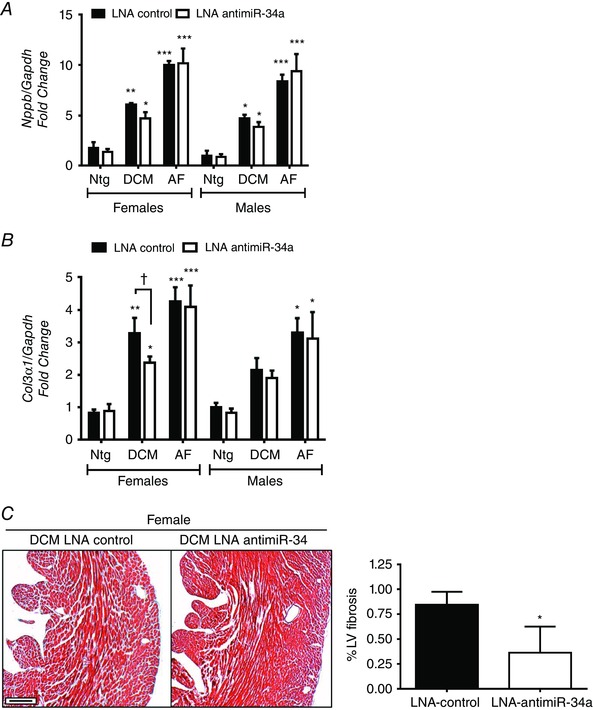
A, quantification of BNP (Nppb) and (B) collagen 3 (Col3a1) relative to Gapdh in hearts of female and male Ntg, DCM and AF LNA‐control and LNA‐antimiR‐34a treated mice. Data are the mean ± SEM. Ntg: n = 3–4 per group, DCM: n = 4–6 per group, AF: n = 4–5 per group. * P < 0.05, ** P < 0.005 and *** P < 0.001 vs. Ntg of same treatment group (one‐way ANOVA with Fisher's post hoc test of Ntg, DCM and AF groups). † P < 0.05 (one‐way ANOVA with Fisher's post hoc test of Ntg and DCM groups only). C, LV cross‐sections stained with Masson's trichrome and quantification of LV fibrosis in female DCM LNA control and LNA antimiR‐34a treated hearts. n = 4 per group. * P < 0.05 (unpaired t test). Scale bar = 200 μm.
Sex‐ and treatment‐dependent regulation of miRNAs in the diseased heart
To assess whether differential expression of other miRNAs might contribute to the phenotype of male and female DCM and AF models, we examined the expression of other miR‐34 family members (miR‐34b and miR‐34c) by quantitative PCR, as well as other miRNAs by miRNA‐Seq. Expression of miR‐34b and miR‐34c was previously shown to be elevated in other cardiac disease settings and to contribute to cardiac pathology (Bernardo et al. 2012b). Furthermore, although miR‐34 family members share some common targets, miR‐34b and miR‐34c are also predicted to target genes that are distinct from miR‐34a (see Supporting information, Fig. S1 and Tables S1 and S2). Cardiac expression levels of miR‐34b and miR‐34c were generally higher in the DCM and AF models of both sexes (Fig. 6). However, there were no distinct differences related to sex or the disease model to explain why LNA‐antimiR‐34a treatment provided more protection in the female DCM model, as well as no protection in the AF model.
Figure 6. Expression of miRNA‐34b and miRNA‐34c are elevated in the DCM and AF models .
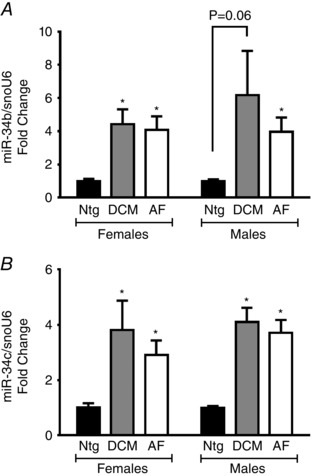
Quantification of (A) miR‐34b and (B) miR‐34c relative to snoU6 by quantitative PCR in LNA‐control hearts. Data are expressed as the mean ± SEM. The Ntg group for females and males has been normalized to 1.0. Ntg: n = 4 per group, DCM: n = 3–5 per group, AF: n = 4–5 per group. * P ≤ 0.05 vs. Ntg.
By miRNA‐Seq, the expression levels of miRNAs in hearts from normal adult mice which have been characterized as cardiac specific/selective were comparable in both sexes (Ntg; Fig. 7 A, e.g. miR‐133a‐3p, miR‐208a‐3p). Interestingly, there was distinct regulation of less well characterized miRNAs in male and female Ntg hearts (Fig. 7 B; see also Supporting information, Table S3). Next, we interrogated miRNAs in the DCM and AF models with and without LNA‐antimiR‐34a treatment. Presentation of differentially expressed miRNAs in heatmaps highlighted the distinct regulation of some miRNAs between sexes in the disease models, as well as with LNA‐antimiR‐34a treatment (Fig. 7 C and D; see also Supporting information, Tables S4 and S5). Expression of 3 miRNAs in Fig. 7 has been validated by quantitative PCR (see Supporting information, Fig. S2).
Figure 7. Sex‐ and treatment‐dependent regulation of miRNAs in the diseased heart .
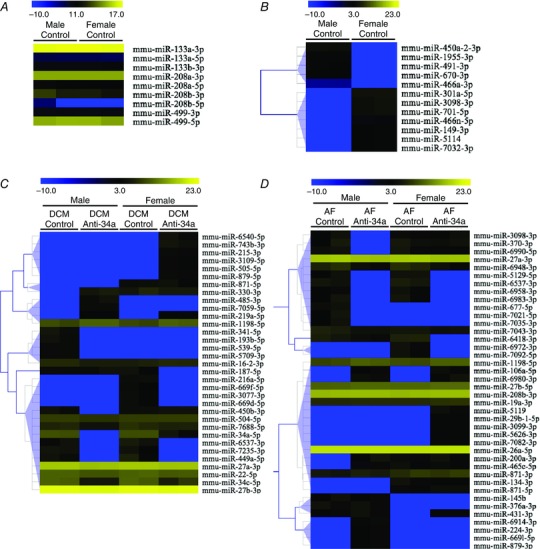
Expression of (A) cardiac specific/selective miRNAs and (B) differentially expressed miRNAs in hearts from male and female Ntg mice (control). No differentially expressed miRNAs identified in (A). Differentially expressed miRNAs in hearts from male and female DCM mice (C) and AF mice (D) with LNA‐control or LNA‐antimiR‐34a. Each row represents a miRNA and each column represents an individual heart sample. Yellow represents upregulation compared to male Ntg control and blue represents downregulation. Sex differences in the expression of miR‐34a‐5p in the DCM model (Fig. 7 C) are difficult to visualize in this heatmap because a scale was selected to encompass the wide variation in expression of numerous miRNAs within the DCM model. An additional heat map with a scale optimized for miR‐34a‐5p and a comparison of Ntg with DCM is provided in the Supporting information (Fig. S3). This visualization is consistent with the sex difference in miR‐34a in the DCM model presented in Figure 1 B.
Discussion
Despite numerous studies examining the role of miRNAs in settings of cardiac disease, and the development of miRNA‐based therapies, very little is known about whether these disease‐induced miRNAs are regulated similarly in both sexes (Sharma & Eghbali, 2014). The present study is the first to examine the effect of silencing miR‐34a in the female heart. To our knowledge, it also represents the only study to have specifically compared the effect of inhibiting any miRNA in the adult heart in both sexes. The results highlight the need to characterize the expression of miRNAs in both males and females, as well as in different cardiac disease settings. This information will be critical for determining which patient groups wil respond favourably to therapeutic interventions.
There are three major new findings arising from the present study. First, the inhibition of miR‐34a provided more protection in the female DCM model than the male DCM model. Second, silencing miR‐34a provided no significant benefit in the more severe model of DCM associated with AF in either males or females. Third, we identified miRNAs that were differentially regulated in the diseased heart of males and females, and some of these were differentially affected by LNA‐antimiR‐34a treatment. Collectively, these findings have important implications for the development of miRNA‐based therapies for heart disease.
In the present study, we assessed the potential of attenuating expression of miR‐34a in a model of DCM with moderate cardiac pathology and dysfunction, and a more severe model of DCM also associated with cardiac conduction abnormalities including AF (referred to as the AF model). LNA‐antimiR‐34a treatment was able to provide some benefit in the DCM model in both males and females but, interestingly, the therapeutic intervention was more effective in females than males. This could partly be a result of miR‐34a playing a more dominant role in the DCM‐induced phenotype in females than males. Consistent with this hypothesis, the fold increase in miR‐34a expression levels in the female DCM heart compared to Ntg was higher than those found in the male DCM heart.
The AF model is a more severe model than the DCM model, characterized by more dilatation of cardiac chambers, higher expression of cardiac stress genes and more severe cardiac conduction abnormalities (Pretorius et al. 2009). Even though miR‐34a was elevated in the hearts of the AF model in both sexes, the silencing of miR‐34a provided no protection against cardiac pathology and dysfunction. The therapeutic benefit of inhibiting miR‐34a in the moderate DCM model but not the more severe DCM model with AF is consistent with our previous study in which LNA‐antimiR‐34a provided protection in a setting of moderate pressure overload but not severe pressure overload (Bernardo et al. 2014a). In a more severe setting, additional miRNAs are probably dysregulated. Thus, it may be necessary to target a group of miRNAs concurrently rather than a single miRNA (Bernardo et al. 2012b; Bernardo et al. 2014a).
To further explore why LNA‐antimiR‐34a administration may have been more effective in the female DCM model than the male DCM model, and why the treatment was ineffective in the AF model, we undertook additional experiments. First, we assessed the expression of other miR‐34 family members that have also been associated with cardiac pathology. In mammals, the miR‐34 family consists of three homologous transcripts: miR‐34a, miR‐34b and miR‐34c. In the mouse, miR‐34a is located on chromosome 4, whereas miR‐34b/c are located on chromosome 9. In humans, the miR‐34a gene maps to chromosome 1p36.22, whereas the genes coding for both miR‐34b and miR‐34c map to chromosome 11q23.1 (Agostini & Knight, 2014). Expression levels of miR‐34b and miR‐34c were elevated in the hearts of the DCM and AF models, as reported previously in other cardiac stress models (pressure overload and myocardial infarction; higher expression in more severe cardiac stress settings) (Bernardo et al. 2012b; Bernardo et al. 2014a). However, there were no notable differences in the expression of miR‐34b and miR‐34c between sexes in the DCM model that might contribute to the better outcome in LNA‐antimiR‐34a treated female DCM mice. Furthermore, even though the AF model is a more severe model than the DCM model, miR‐34b and miR‐34c were expressed at comparable levels in the two models. Thus, heightened expression of miR‐34b and miR‐34c probably does not account for the differences observed. Next, we performed miRNA‐Seq to examine the global expression of all miRNAs in the diseased models with and without LNA‐antimiR‐34a. To our knowledge, a direct comparison of miRNAs regulated in the female and male adult heart in cardiac disease settings, together with a therapeutic intervention, has not been reported previously. Evangelista et al. (2013) profiled 609 miRNAs in hearts from 15‐week‐old adult male and female C57BL/6J mice under basal conditions using a miRNA microarray (Affymetrix, Santa Clara, CA, USA). Eight miRNAs were identified to be differentially regulated in the heart between the sexes; four miRNAs had higher relative expression in males than females (miR‐144, miR‐34b‐3p, miR‐205, miR‐222) and 4 miRNAs had higher relative expression in females (miR‐1, miR‐106b, miR‐720, miR‐29b) (Evangelista et al. 2013). In another study, the expression of miR‐1 was found to be expressed at lower levels in female but not male adult rat ventricular myocytes following stimulation with a pathological stimulus, phenylephrine (no significant differences were observed in unstimulated control myocytes) (Stauffer et al. 2011). A more recent study examined tissue and sex associated miRNAs in zebrafish under basal settings (Vaz et al. 2015). Sex‐associated miRNAs were identified in the brain and liver; hearts from male and female zebrafish were not examined specifically (Vaz et al. 2015). Queiros et al. (2013) examined the expression of a group of miRNAs (miR‐21, ‐24, ‐27a, ‐27b, ‐106a, ‐106b) in the hearts of male and female mice with pathological hypertrophy as a result of pressure overload induced by aortic constriction. Each of these miRNAs was more highly expressed in the hypertrophied male heart than the female heart. In humans, previous studies have identified miRNAs that are dysregulated in the heart or circulation in cardiac disease settings (Thum et al. 2007; Devaux et al. 2013; Matsumoto et al. 2013). However, these studies have not specifically examined sex differences in miRNA expression, or were heavily biased towards male subjects. In the present study, miRNA‐Seq analyses demonstrated that a number of miRNAs were differentially expressed in the male and female DCM and AF models, and LNA‐antimiR‐34a differentially regulated some of these miRNAs. The underlying mechanisms responsible for this finding will require further study. However, it is noteworthy that sex steroid hormones and X‐linked genes can influence the regulation of miRNAs (Sharma & Eghbali, 2014) and also that oestradiol can regulate miR‐34a (Li et al. 2014). LNA‐antimiR‐34a is reported to directly target miR‐34a specifically (Bernardo et al. 2012b). However, as a result of miR‐34a regulating the expression of numerous target genes, including transcription factors (which can regulate other miRNAs), and also as a result of feedback loops, other miRNAs can be differentially regulated as a ‘secondary’ response (Matkovich et al. 2013; Ooi et al. 2016). Altogether, this highlights the differential regulation and complexity of miRNA–drug interactions in the male and female heart.
It is important to acknowledge that no animal model will perfectly mimic human disease and that differences in species need to be considered (Riley et al. 2012). For example, the high heart rate and small atrial size of mice compared to humans are factors that contribute to the absence of permanent AF in mice. The disease animal models used in the present study (DCM and AF) were selected because they display some characteristics that are similar to human cardiac disease (e.g. left ventricular dilatation, fibrosis, lung congestion, molecular changes observed in human diseased hearts) (Yamamoto et al. 2003; Pretorius et al. 2009; Sapra et al. 2014). Furthermore, the genetic alterations in the hearts of the mouse models (i.e. increased Mst1 and decreased PI3K) are also features that have been observed in human DCM and AF (Pretorius et al. 2009; Maejima et al. 2013).
The success of miRNA therapeutics in preclinical models of disease has led to the development and translation of miRNA therapies into the clinic for hepatitis C virus (Janssen et al. 2013) and liver cancer (Bader, 2012). miRNA‐based therapies have also shown potential in preclinical models of cardiac pathology but are yet to enter the clinic for cardiovascular disease. One of the biggest steps for the translation of a miRNA‐based therapy for heart failure is identifying strategies for specific and efficient delivery to the heart (Ooi et al. 2014; Bernardo et al. 2015). Nonetheless, with improvements and developments in delivery tools (including the use of nanoparticles, liposomes and antibodies), miRNA‐based therapies remain a promising approach for the treatment of heart failure (Bernardo et al. 2015).
In conclusion, the present study contributes to the growing body of literature showing that inhibition of miR‐34a represents a promising therapeutic approach in a number of cardiac disease settings (Bernardo et al. 2012b; Boon et al. 2013; Huang et al. 2014; Bernardo et al. 2014a; Yang et al. 2015). However, the lack of benefit in the more severe DCM model with AF is also consistent with previous work showing that silencing miR‐34a is less effective in settings of severe cardiac pathology (Bernardo et al. 2012b; Bernardo et al. 2014a). The present study also uncovered differential responses of the male and female heart to LNA‐antimiR‐34a treatment in a setting of DCM. Our subsequent identification of sex‐, disease‐ and treatment‐regulated miRNAs has important and broad implications for the development of miRNA‐based therapies for cardiovascular disease, and potentially other diseases.
Additional information
Competing interests
SO is an employee of Roche Innovation Centre Copenhagen A/S, a clinical‐stage biopharmaceutical company that develops RNA‐targeted therapeutics.
Author contributions
Experiments were performed in the Cardiac Hypertrophy Research Laboratory at Baker IDI Heart and Diabetes Institute. JRM and BCB contributed to the experimental design and co‐ordinated the project. RCYL contributed to the experimental design and co‐ordinated the miRNA‐Seq experiment. BCB, JYYO, AM, YKT, SS, NLP, HK, RCYL and JRM contributed to the acquisition, analysis or interpretation of data. SO contributed to the experimental design and provided the LNA‐antimiRs. JS provided the Mst1 Tg model and expertise regarding this model. JRM and BCB wrote the paper. All authors approved the manuscript final version and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work were appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
This study was funded by National Health and Medical Research Council Project Grant 586603 (to JRM and RCYL), and was also supported in part by the Victorian Government's Operational Infrastructure Support Program. JRM is a National Health and Medical Research Council Senior Research Fellow (586604 & 1078985). JRM and RCYL were also supported by an Australian Research Council Future Fellowship (FT0001657) and a University of New South Wales Vice Chancellor Research Fellowship, respectively.
Supporting information
Figure S1: Predicted targets of the miR‐34 family members.
Figure S2: Validation of miR‐seq data by quantitative PCR.
Figure S3: Heat map of mmu‐miR‐34a‐5p.
Table S1: Predicted mRNA targets to miR‐34a but not miR‐34b and miR‐34c.
Table S2: Predicted mRNA targets common to both miR‐34b and miR‐34c.
Table S3: List of cardiac specific/selective miRNAs and differentially expressed miRNAs in hearts from male and female Ntg mice from miR‐seq data.
Table S4: List of differentially expressed miRNAs in hearts from male and female DCM mice with LNA‐control or LNA‐antimiR‐34a from miR‐seq data.
Table S5: List of differentially expressed miRNAs in hearts from male and female AF mice with LNA‐control or LNA‐antimiR‐34a from miR‐seq data.
Acknowledgements
We thank Geeta Sapra for ECG assistance and Nicole Jennings for assistance with echocardiography.
This is an Editor's Choice article from the 15 October 2016 issue.
Linked articles This article is highlighted by a Perspective by Tian ' Zucker. To read this Perspective, visit http://dx.doi.org/10.1113/JP272892.
Contributor Information
Bianca C. Bernardo, Email: bianca.bernardo@bakeridi.edu.au
Julie R. McMullen, Email: julie.mcmullen@bakeridi.edu.au
References
- Agostini M & Knight RA (2014). miR‐34: from bench to bedside. Oncotarget 5, 872–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader AG (2012). miR‐34 – a microRNA replacement therapy is headed to the clinic. Front Genet 3, 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo BC, Charchar FJ, Lin RC & McMullen JR (2012. a). A microRNA guide for clinicians and basic scientists: background and experimental techniques. Heart Lung Circ 21, 131–142. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Gao XM, Tham YK, Kiriazis H, Winbanks CE, Ooi JY, Boey EJ, Obad S, Kauppinen S, Gregorevic P, Du XJ, Lin RC & McMullen JR (2014. a). Silencing of miR‐34a attenuates cardiac dysfunction in a setting of moderate, but not severe, hypertrophic cardiomyopathy. PLoS ONE 9, e90337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo BC, Gao XM, Winbanks CE, Boey EJ, Tham YK, Kiriazis H, Gregorevic P, Obad S, Kauppinen S, Du XJ, Lin RC & McMullen JR (2012. b). Therapeutic inhibition of the miR‐34 family attenuates pathological cardiac remodeling and improves heart function. Proc Natl Acad Sci USA 109, 17615–17620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo BC, Nguyen SS, Gao XM, Tham YK, Ooi JY, Patterson NL, Kiriazis H, Su Y, Thomas CJ, Lin RC, Du XJ & McMullen JR (2016). Inhibition of miR‐154 protects against cardiac dysfunction and fibrosis in a mouse model of pressure overload. Sci Rep 6, 22442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo BC, Nguyen SS, Winbanks CE, Gao XM, Boey EJ, Tham YK, Kiriazis H, Ooi JY, Porrello ER, Igoor S, Thomas CJ, Gregorevic P, Lin RC, Du XJ & McMullen JR (2014. b). Therapeutic silencing of miR‐652 restores heart function and attenuates adverse remodeling in a setting of established pathological hypertrophy. FASEB J 28, 5097–5110. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Ooi JY, Lin RC & McMullen JR (2015). miRNA therapeutics: a new class of drugs with potential therapeutic applications in the heart. Future Med Chem 7, 1771–1792. [DOI] [PubMed] [Google Scholar]
- Bernardo BC, Weeks KL, Pretorius L & McMullen JR (2010). Molecular distinction between physiological and pathological cardiac hypertrophy: experimental findings and therapeutic strategies. Pharmacol Ther 128, 191–227. [DOI] [PubMed] [Google Scholar]
- Boon RA, Iekushi K, Lechner S, Seeger T, Fischer A, Heydt S, Kaluza D, Treguer K, Carmona G, Bonauer A, Horrevoets AJ, Didier N, Girmatsion Z, Biliczki P, Ehrlich JR, Katus HA, Muller OJ, Potente M, Zeiher AM, Hermeking H & Dimmeler S (2013). MicroRNA‐34a regulates cardiac ageing and function. Nature 495, 107–110. [DOI] [PubMed] [Google Scholar]
- Devaux Y, Vausort M, McCann GP, Kelly D, Collignon O, Ng LL, Wagner DR & Squire IB (2013). A panel of 4 microRNAs facilitates the prediction of left ventricular contractility after acute myocardial infarction. PLoS ONE 8, e70644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Editorial (2010). Putting gender on the agenda. Nature 465, 665. [DOI] [PubMed] [Google Scholar]
- Evangelista AM, Deschamps AM, Liu D, Raghavachari N & Murphy E (2013). miR‐222 contributes to sex‐dimorphic cardiac eNOS expression via ets‐1. Physiol Genomics 45, 493–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A (2013). Functions of microRNAs in cardiovascular biology and disease. Annu Rev Physiol 75, 69–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Qi Y, Du JQ & Zhang DF (2014). MicroRNA‐34a regulates cardiac fibrosis after myocardial infarction by targeting Smad4. Expert Opin Ther Targets 18, 1355–1365. [DOI] [PubMed] [Google Scholar]
- Janssen HL, Reesink HW, Lawitz EJ, Zeuzem S, Rodriguez‐Torres M, Patel K, van der Meer AJ, Patick AK, Chen A, Zhou Y, Persson R, King BD, Kauppinen S, Levin AA & Hodges MR (2013). Treatment of HCV infection by targeting microRNA. N Engl J Med 368, 1685–1694. [DOI] [PubMed] [Google Scholar]
- Kim AM, Tingen CM & Woodruff TK (2010). Sex bias in trials and treatment must end. Nature 465, 688–689. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M & Salzberg SL (2009). Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legato MJ (2016). Gender‐specific medicine in the genomic era. Clin Sci (Lond) 130, 1–7. [DOI] [PubMed] [Google Scholar]
- Li XJ, Ren ZJ & Tang JH (2014). MicroRNA‐34a: a potential therapeutic target in human cancer. Cell Death Dis 5, e1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maejima Y, Kyoi S, Zhai P, Liu T, Li H, Ivessa A, Sciarretta S, Del Re DP, Zablocki DK, Hsu CP, Lim DS, Isobe M & Sadoshima J (2013). Mst1 inhibits autophagy by promoting the interaction between Beclin1 and Bcl‐2. Nat Med 19, 1478–1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matkovich SJ, Hu Y & Dorn GW, 2nd (2013). Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res 113, 62–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto S, Sakata Y, Suna S, Nakatani D, Usami M, Hara M, Kitamura T, Hamasaki T, Nanto S, Kawahara Y & Komuro I (2013). Circulating p53‐responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ Res 113, 322–326. [DOI] [PubMed] [Google Scholar]
- National Health and Medical Research Council (2013). Australian code for the care and use of animals for scientific purposes, 8th ed. Canberra: National Health and Medical Research Council. [Google Scholar]
- Obad S, dos Santos CO, Petri A, Heidenblad M, Broom O, Ruse C, Fu C, Lindow M, Stenvang J, Straarup EM, Hansen HF, Koch T, Pappin D, Hannon GJ & Kauppinen S (2011). Silencing of microRNA families by seed‐targeting tiny LNAs. Nat Genet 43, 371–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi JY, Bernardo BC & McMullen JR (2014). The therapeutic potential of miRNAs regulated in settings of physiological cardiac hypertrophy. Future Med Chem 6, 205–222. [DOI] [PubMed] [Google Scholar]
- Ooi JY, Bernardo BC, Singla S, Patterson NL, Lin RC & McMullen JR (2016). Identification of miR‐34 regulatory networks in settings of disease and antimiR‐therapy: Implications for treating cardiac pathology and other diseases. RNA Biol, April 28.0. E‐pub ahead of print. PMID: 27124358, DOI: 10.1080/15476286.2016.1181251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pretorius L, Du XJ, Woodcock EA, Kiriazis H, Lin RC, Marasco S, Medcalf RL, Ming Z, Head GA, Tan JW, Cemerlang N, Sadoshima J, Shioi T, Izumo S, Lukoshkova EV, Dart AM, Jennings GL & McMullen JR (2009). Reduced phosphoinositide 3‐kinase (p110alpha) activation increases the susceptibility to atrial fibrillation. Am J Pathol 175, 998–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queiros AM, Eschen C, Fliegner D, Kararigas G, Dworatzek E, Westphal C, Sanchez Ruderisch H & Regitz‐Zagrosek V (2013). Sex‐ and estrogen‐dependent regulation of a miRNA network in the healthy and hypertrophied heart. Int J Cardiol 169, 331–338. [DOI] [PubMed] [Google Scholar]
- Rathore SS, Wang Y & Krumholz HM (2002). Sex‐based differences in the effect of digoxin for the treatment of heart failure. N Engl J Med 347, 1403–1411. [DOI] [PubMed] [Google Scholar]
- Regitz‐Zagrosek V (2006). Therapeutic implications of the gender‐specific aspects of cardiovascular disease. Nat Rev Drug Discov 5, 425–438. [DOI] [PubMed] [Google Scholar]
- Riley G, Syeda F, Kirchhof P & Fabritz L (2012). An introduction to murine models of atrial fibrillation. Front Physiol 3, 296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M, Sturn A, Snuffin M, Rezantsev A, Popov D, Ryltsov A, Kostukovich E, Borisovsky I, Liu Z, Vinsavich A, Trush V & Quackenbush J (2003). TM4: a free, open‐source system for microarray data management and analysis. Biotechniques 34, 374–378. [DOI] [PubMed] [Google Scholar]
- Sapra G, Tham YK, Cemerlang N, Matsumoto A, Kiriazis H, Bernardo BC, Henstridge DC, Ooi JY, Pretorius L, Boey EJ, Lim L, Sadoshima J, Meikle PJ, Mellet NA, Woodcock EA, Marasco S, Ueyama T, Du XJ, Febbraio MA & McMullen JR (2014). The small‐molecule BGP‐15 protects against heart failure and atrial fibrillation in mice. Nat Commun 5, 5705. [DOI] [PubMed] [Google Scholar]
- Sharma S & Eghbali M (2014). Influence of sex differences on microRNA gene regulation in disease. Biol Sex Differ 5, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shioi T, Kang PM, Douglas PS, Hampe J, Yballe CM, Lawitts J, Cantley LC & Izumo S (2000). The conserved phosphoinositide 3‐kinase pathway determines heart size in mice. EMBO J 19, 2537–2548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauffer BL, Sobus RD & Sucharov CC (2011). Sex differences in cardiomyocyte connexin43 expression. J Cardiovasc Pharmacol 58, 32–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi T, Satoh M, Itoh T & Nakamura M (2012). MicroRNA‐34a regulates the longevity‐associated protein SIRT1 in coronary artery disease: effect of statins on SIRT1 and microRNA‐34a expression. Clin Sci 123, 161–171. [DOI] [PubMed] [Google Scholar]
- Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G & Bauersachs J (2007). MicroRNAs in the human heart: a clue to fetal gene reprogramming in heart failure. Circulation 116, 258–267. [DOI] [PubMed] [Google Scholar]
- Vaz C, Wee CW, Lee GP, Ingham PW, Tanavde V & Mathavan S (2015). Deep sequencing of small RNA facilitates tissue and sex associated microRNA discovery in zebrafish. BMC Genomics 16, 950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto S, Yang G, Zablocki D, Liu J, Hong C, Kim SJ, Soler S, Odashima M, Thaisz J, Yehia G, Molina CA, Yatani A, Vatner DE, Vatner SF & Sadoshima J (2003). Activation of Mst1 causes dilated cardiomyopathy by stimulating apoptosis without compensatory ventricular myocyte hypertrophy. J Clin Invest 111, 1463–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Cheng HW, Qiu Y, Dupee D, Noonan M, Lin YD, Fisch S, Unno K, Sereti KI & Liao R (2015). MicroRNA‐34a plays a key role in cardiac repair and regeneration following myocardial infarction. Circ Res 117, 450–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1: Predicted targets of the miR‐34 family members.
Figure S2: Validation of miR‐seq data by quantitative PCR.
Figure S3: Heat map of mmu‐miR‐34a‐5p.
Table S1: Predicted mRNA targets to miR‐34a but not miR‐34b and miR‐34c.
Table S2: Predicted mRNA targets common to both miR‐34b and miR‐34c.
Table S3: List of cardiac specific/selective miRNAs and differentially expressed miRNAs in hearts from male and female Ntg mice from miR‐seq data.
Table S4: List of differentially expressed miRNAs in hearts from male and female DCM mice with LNA‐control or LNA‐antimiR‐34a from miR‐seq data.
Table S5: List of differentially expressed miRNAs in hearts from male and female AF mice with LNA‐control or LNA‐antimiR‐34a from miR‐seq data.


