Abstract
Urinary extracellular vesicles (uEVs) are released from all regions of the kidney's nephron and from other cells that line the urinary tract. Extracellular vesicles retain proteomic and transcriptomic markers specific to their cell of origin and so represent a potential reservoir for kidney disease biomarker discovery. Exosomes, a subtype of uEVs, are distinguished from other vesicles by features related to their biogenesis within cells: mature multi‐vesicular bodies fuse with the cellular membrane to liberate exosomes into the extracellular space. uEVs represent a novel cell signalling mechanism because they can be shuttled to a recipient cell and, through a number of proposed mechanisms, affect the recipient cell's proteome and function. Here we review the current evidence for uEV signalling along the nephron, their role in health and disease of the kidney, and their potential for clinical translation as biomarkers and therapeutics.
Abbreviations
- AKI
acute kidney injury
- ESCRT
endosomal sorting complex required for transport
- EV
extracellular vesicle
- ILV
intraluminal vesicle
- MSC
mesenchymal stem cell
- MVB
multivesicular body
- PTM
post‐translational modification
- uEV
urinary extracellular vesicle
Introduction
The kidney is one of the most important regulators of the body's physiological state, manipulating filtration and reabsorption of solutes in order to maintain an optimal environment for health. It is vulnerable to a plethora of injury modalities. High oxygen demand and low tissue oxygen tensions in the renal parenchyma sensitize tubular cells to hypoxia and can lead to acute and chronic kidney injury, disease processes which are associated with substantial morbidity and mortality. Furthermore, tubular cells are vulnerable to the toxic effects of drugs. Increasing intra‐tubular drug concentrations as the filtrate moves along the nephron combined with reuptake mechanisms for solutes results in potentially toxic intracellular drug concentrations. As a result of the high morbidity associated with renal disease, and the limiting role of nephrotoxicity in translation of drug development to clinical practice, improving our understanding of the underlying molecular signalling would be of value to prevent toxicity and treat kidney injury (Lee et al. 2014).
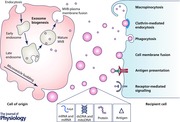
Urine, the excreted filtrate of the kidney, is unique in providing a non‐invasive snapshot of the kidney's function. This filtrate is composed of ions, inorganic and organic compounds (including proteins, hormones and metabolites) suspended in water. Investigation of the protein fraction of urine has demonstrated the presence of integral membrane proteins within small extracellular vesicles (EVs) (Wen et al. 1999). EVs are parcels of proteomic and transcriptomic information. Proteins, pre‐microRNA, mature microRNA, retrotransposon RNA transcripts, single‐stranded DNA (ssDNA), double‐stranded DNA (dsDNA) and mitochondrial DNA have all been identified within EVs (Abstract figure) (Guescini et al. 2010; Balaj et al. 2011; Thakur et al. 2014). Rigorous characterization of the protein and RNA content of the urinary EVs (uEVs) has been conducted, identifying molecules unique to all regions of the nephron and identifying products of genes associated with multiple disease processes (Pisitkun et al. 2004; Miranda et al. 2010). Several subtypes of vesicles have been identified in urine, including exosomes and microvesicles, the characteristics of which are described below (Pisitkun et al. 2004; Rood et al. 2010).
Composition and biogenesis of EVs
EVs encompass a vast heterogeneous and dynamic population of membrane bound vesicles; their content and membrane composition are not only dependent upon their cellular source but are also sensitive to cellular stress and environmental changes. Historically, naming of EVs was dependent upon their cell of origin, e.g. cardiosomes (cardiomyocyte origin) and prosatosomes (seminal fluid). Current nomenclature, however, largely distinguishes vesicles by their biogenesis. Criteria for EV classification have been proposed based on their origin, function or biogenesis; yet there is still no consensus about their nomenclature. Subpopulation categories range from three to six in number in most reviews and have been extensively discussed elsewhere (Thery et al. 2009; van der Pol et al. 2012). These categories are often difficult to use in practice, which results in confusing overlapping nomenclature. For the purposes of this review, we will consider three distinct subtypes with characteristics related to their origin, size and identifying markers.
Exosomes
Exosomes are derived from the endosomal pathway. Transmembrane proteins are trafficked to early endosomes by endocytosis. These early endosomes undergo sorting to late endosomes, which in turn become multivesicular bodies (MVBs). Formation of MVBs is directed by the recruitment of proteins and budding of intraluminal vesicles (ILVs), via the endosomal sorting complex required for transport (ESCRT). ILVs are released as exosomes upon fusion of MVBs with the plasma membrane (Fig. 1). Alternatively, MVBs can fuse with lysosomes, resulting in degradation of their contents.
Figure 1. EV biogenesis and role of ubiquitination .
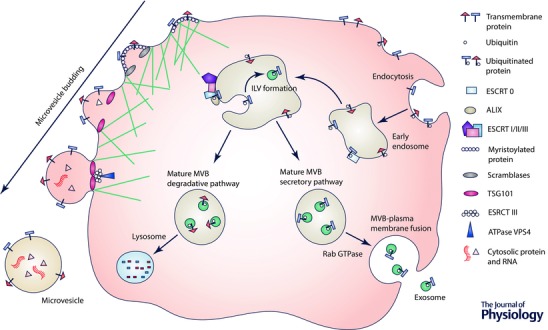
Exosome biogenesis is depicted on the right. Transmembrane proteins are internalized from the cell surface via endocytosis and their cytosolic domains ubiquinated. ESCRT 0 recognizes the ubiquitinated protein and segregates these proteins into microdomains. ESCRT I and II are subsequently recruited and initiate the reverse budding of intraluminal vesicles (ILVs). At this stage, a same amount of cytosol, and therefore cytosolic proteins and RNA, has access to the interior of the ILV. The ILV is cleaved from the bud, following recruitment of ESCRT III, by ESCRT II and ALIX. The mature multivesicular body (MVB) formed can either follow a degradation pathway, or proceed to fusion with the plasma membrane. The pathway for each MVB is likely determined by its contents. For example, ubiquitinated LMP2A has been shown to follow the secretory pathway, while MVBs containing ubiquitinated EGFR are degraded. The degradation pathway consists of fusion of the mature MVB with a lysosome. In the secretory pathway, MVBs fuse with the plasma membrane mediated by a Rab27A‐dependent pathway releasing the ILVs, now termed exosomes, into the extracellular environment. Note that not all proteins require ubiquitination to be targeted to ILVs. Other post‐translation modifications can result in recruitment to ILVs and have been reviewed in Moreno‐Gonzalo et al. (2014). Microvesicle assembly is illustrated on the left and modified from Cocucci & Meldolesi (2015). Transmembrane proteins cluster in membrane lipid microdomains during nucleation at the plasma membrane. Myristoylated protein contributes to membrane curvature; lipid distribution is randomized by calcium‐dependent scramblases and, concurrently, the cytoskeleton loosens. A member of the ESCRT I complex, TSG101, recruits ESCRT III to the plasma membrane, which promotes the assembly of a spiral, ultimately disassembled by ATPase VPS4. This process results in cleavage of the bud and release of a heterogeneous population of microvesicles.
Intriguingly, the relative abundance of cellular RNA and protein does not closely correlate with that of the exosome, suggesting a selective process of entry into vesicles (Valadi et al. 2007). This cargo sorting may allow the cell to generate exosomes of precisely defined biochemical composition. ESCRT, tetraspanins and lipid‐dependent mechanisms have all been implicated in the selective loading of proteins (Villarroya‐Beltri et al. 2014). A specific pattern of protein post‐translational modifications (PTMs) has been noted in exosomes and a potential role in exosome sorting has been suggested (Moreno‐Gonzalo et al. 2014). The most commonly described PTM, ubiquitination, involves the addition of ubiquitin to a target protein (Fig. 1). In MVBs, a ubiquitinated protein is sorted into ILVs by the ESCRT and the downstream fate of the MVB is dependent upon the type of ubiquitin modification on a specific substrate. Ubiquitin may be cleaved from its cargo protein during incorporation to the ILV but a recent in vivo study reported that 15% of proteins in uEVs are ubiquitinated, suggesting a significant proportion are sequestered into ILVs without deubiquitination (Agromayor & Martin‐Serrano, 2006; Huebner et al. 2016). Others PTMs that have been described include phosphorylation and glycosylation (Moreno‐Gonzalo et al. 2014).
The sorting of RNA into exosomes is less well understood; specific small ubiquitin‐related modifier (SUMO) proteins are hypothesized to interact with cis‐acting elements within RNA, such as hnRNPA2B1, to selectively load vesicles (Villarroya‐Beltri et al. 2013). miRNA sorting to exosomes may be modulated by the dynamic transcriptomic changes seen in cell activation, differentially engaging miRNAs at P bodies or MVBs (Squadrito et al. 2014).
As a result of the above processes, exosomes represent a parcel of protein, DNA and various RNA species (Sheldon et al. 2010; Mittelbrunn et al. 2011). The majority of RNA species isolated from exosomes are small RNAs including miRNAs (Cheng et al. 2013). Exosomes are identified by their lipid bilayer membrane, 30–120 nm size and density of 1.15–1.19 g ml−1 in continuous sucrose gradient (Thery et al. 2006). Their often cited cup‐shaped morphology is likely artificial, attributed to collapse during drying (Raposo & Stoorvogel, 2013). An exosome's lipid bilayer outwardly displays the apical surface of the membrane from which the vesicle was formed, therefore displaying the same extracellular surface markers as the cell of origin (Fig. 1) (Thery et al. 2009). This membrane orientation has led to the postulation of inward budding, from the limiting membrane of endosomes, as the mechanism of exosome biogenesis. Exosomal protein markers mainly relate to intracellular vesicle trafficking and exosome biogenesis, i.e. ESCRT components, tetraspanins and flotillin. Although there are signature protein profiles for exosomes from defined tissue, there is no single unifying marker. Specific to the urinary exosome population, CD24 has been postulated as a suitable biomarker (Keller et al. 2007; Oosthuyzen et al. 2013).
Microvesicles
Microvesicles are shed directly from the plasma membrane via detachment of small cytoplasmic protrusions (Yanez‐Mo et al. 2015) in response to cell stress. This process is dependent on calcium influx, calpain and cytoskeleton reorganization (Fig. 1) (Cocucci et al. 2009). As a result of this outward budding, microvesicles contain a small volume of cytoplasm and are enriched with membrane markers from their cell of origin including proteins associated with membrane lipid rafts (Del Conde et al. 2005). Microvesicles are characteristically 50–1000 nm in size and specific markers proposed for their identification are ADP‐ribosylation factor 6 (ARF6; implicated in endocytosis of protein) and vesicle‐associated membrane protein 3 (VAMP3) (Muralidharan‐Chari et al. 2009).
Consideration can also be given to demonstrating the absence of non‐EV protein markers when screening exosome and microvesicle populations. For example transferrin receptors are enriched in the exosomes but are absent in the microvesicle population from the same tissue (Muralidharan‐Chari et al. 2010).
Apoptotic bodies
Apoptotic bodies are blebs containing cytoplasm and densely packed organelles. They are extensively liberated from the plasma membrane in the later stages of apoptosis. Although apoptotic bodies are generally considered to be larger in size than other vesicles (500–4000 nm), a smaller subpopulation of 50–500 nm has been proposed (Thery et al. 2009; Akers et al. 2013). The translocation of phosphatidylserine onto the outer cell membrane during apoptosis has been previously described and it is unsurprising that apoptotic bodies are enriched for this phospholipid. Binding of annexin V to phosphatidylserines therefore can be used as a protein marker for apoptotic bodies. Phosphatidylserines are also exposed by microvesicles and, to a lesser extent, exosomes. Increased binding sites for thrombospondin and C3b allow these proteins to be used as markers (van Engeland et al. 1998). Given the circumscribed pathophysiological role of apoptotic bodies and their efficient local clearance by phagocytosis in vivo, apoptotic bodies will not be discussed in further detail in this review.
Pisitkun et al. first described the presence of uEVs, identifying these as exosomes due to their small size and biogenesis, and describing the proteome of these vesicles (Pisitkun et al. 2004). Since this seminal work was published, the presence of different vesicle subtypes has also been described (Rood et al. 2010). Over recent years, a wealth of studies have gone on to describe both the proteome and the transcriptome of the uEV population (Gonzales et al. 2009; Miranda et al. 2010), culminating in the development of public access online databases (http://www.exocarta.org, https://hpcwebapps.cit.nih.gov/ESBL/Database/Exosome/).
Isolation of a pure vesicle subpopulation is notoriously difficult, in part because of the size overlap between the vesicle subpopulations, and other vesicles are often co‐purified along with the specific type of interest. Due to inconsistency in EV isolation protocols and often‐incomplete vesicle characterization, there is an inherent difficultly in differentiating between the subpopulations in published work. For clarity, in this review vesicle‐like structures that are not rigorously defined will be referred to as EVs or uEVs unless clearly stated.
uEV concentration and quantification
Technical standardization of sample processing is an area of controversy in EV research, although the influence of varying practice on downstream outputs remains unclear. Variability exists in storage, handling and characterization of uEVs between published works; fortunately guidance is now available through position papers of best practice and consensus statements on EV isolation and minimal experimentation requirements for EV definition (Witwer et al. 2013; Lotvall et al. 2014).
Storage and handling of uEVs
Fresh urine samples are preferable for isolation of uEVs, although for practicality frozen samples may need to be used with like‐for‐like being compared (i.e. fresh to fresh, frozen to frozen). EVs are relatively insensitive to freeze–thaw cycles and may even resist bursting in a hypotonic environment (Witwer et al. 2013). Storage of urine at −80°C is appropriate and uEVs have been concentrated from samples after 7 months in storage (Zhou et al. 2006; Oosthuyzen et al. 2013). Use of protease inhibitors to preserve samples has been described, but opinion remains inconsistent regarding their use (Zhou et al. 2006; Oosthuyzen et al. 2013). Calcium oxalate and amorphous calcium crystal precipitates can be present after thawing, macroscopically forming a cloudy sample. Vigorous vortexing can re‐dissolve these salts (Saetun et al. 2009).
Tamm‐Horsfall protein (THP; also known as uromodulin) is an abundant protein in urine and leads to uEV entrapment by polymerizing and co‐precipitating at low speed centrifugation, leading to a reduced yield of uEVs in the final ultracentrifugation pellet (Fernandez‐Llama et al. 2010; Kosanovic & Jankovic, 2014). Removal of THP is therefore recommended either with dithiothreitol (DTT), which disrupts the zona pellucida disulfide bonds of THP, or 3‐[(3‐cholamidopropyl)dimethylammonio]‐1‐propanesulfonate (CHAPS), a mild solubilizing detergent (Fernandez‐Llama et al. 2010; Musante et al. 2012; Witwer et al. 2013; Lotvall et al. 2014). Although the latter preserves protein conformation and enzymatic activity, it is significantly more time consuming to use.
Ultracentrifugation
Differential ultracentrifugation, with or without a size exclusion technique, remains the most accepted method of exosome isolation from biological fluids (Harding & Stahl, 1983; Johnstone et al. 1984). Following cell depletion of a biological fluid by slow speed centrifugation, 10,000–20,000 g is used to pellet larger EVs. Subsequently, smaller EVs, including exosomes, are pelleted from this supernatant using an ultracentrifuge (100,000–200,000 g). The limitations of this technique have previously been well described in other reviews; ultracentrifugation introduces variability, has a low throughput, and is heavily operator and rotor dependent (Cvjetkovic et al. 2014). Importantly, not all exosomes are recovered following ultracentrifugation; 40% remain in the supernatant although it is unknown if this subpopulation contains unique features of biological or clinical relevance (Musante et al. 2012). Size exclusion chromatography can be used in conjunction with ultracentrifugation to remove protein contaminants and is considered the gold standard for isolation of a highly purified population of morphologically intact exosomes (Rood et al. 2010, Boing et al. 2014).
Alternatively, filtration through a nano‐membrane, using slow centrifugation or gravity, can further aid size exclusion and theoretically improve purity of the vesicle subpopulation. Limitations of this technique include a lower EV recovery, vesicle fragmentation and contamination with proteins, with further protein retention on the membrane (Cheruvanky et al. 2007; Witwer et al. 2013). Addition of a sucrose gradient can further improve purity and isolate subpopulations, but this process is lengthy and associated with a very low EV yield (<1% of the initial crude pellet) (Hogan et al. 2014). Interestingly, sucrose gradient ultracentrifugation has been used to elucidate two different subtypes of exosome population within melanoma cell supernatant. The populations were distinct in their proteome, transcriptome and effect on recipient cell gene expression, raising the possibility of a heterogeneous exosome population capable of exerting different effects on downstream cell physiology (Willms et al. 2016).
Other concentration techniques
A number of commercial platforms support rapid EV precipitation. These proprietary polymers pellet EVs, with the aid of a slow centrifugation step, and have been used for a wide variety of downstream applications. This approach is attractive due to the rapid aggregation, low user variability and relative simplicity (Schageman et al. 2013; Musante et al. 2014). However, caution must be exercised as these techniques also concentrate larger vesicles and protein aggregates, forming an impure pellet (Alvarez et al. 2012). Immunoaffinity precipitation has been used in a number of studies to concentrate purified subpopulations of EVs, utilizing magnetic beads coated with antibody against proteins of interest (Kalra et al. 2013; Wang & Sun, 2014). Similar peptide‐based isolation techniques have also been described (Ghosh et al. 2014). Although these techniques have the capability to achieve an adaptable, rapid platform for EV isolation, they do, inherently, introduce population bias due to targeting only vesicles that express a certain protein marker (Kowal et al. 2014).
Novel microfluidic devices utilize a variety of techniques; namely, immunoaffinity, sieving (through nanoporous membranes) and trapping of vesicle structures, to identify a highly purified EV population (Kanwar et al. 2014; Santana et al. 2014; Liga et al. 2015). Although in its infancy, this technique could be used in point‐of‐care rapid exosome isolation and has been comprehensively reviewed elsewhere (Liga et al. 2015). There remain a number of hurdles to employment of this technique, mainly low recovery of exosomes, sheer stress to structures, and requirement for prior sample preparation.
Normalization
Total EV number can be ascertained by nanotracking analysis (NTA) without or with fluorophores to known protein targets, with the additional benefit of using unprocessed urine samples (Oosthuyzen et al. 2013). This methodology could be used to sample two populations (the subpopulations of interest and a control) or even dichromatous populations (Hogan et al. 2014). Alternatively, manipulation of pore size of a stretchable nanopore membrane, utilized in resistive pulse sensing, can allow for measurement size distribution and concentration but this approach is limited by the inability to provide phenotypic information (Liga et al. 2015). The primary issues with NTA and nanopore technologies include lengthy processing and inter‐assay variability. Until recently conventional flow cytometry could only phenotype EVs down to ∼500 nm in size, restricting its use to the study of larger vesicle populations. The advent of newer instruments has raised the exciting possibility of discriminating between particles as small as 100 nm in diameter (Witwer et al. 2013). The impact of different refractive indices between biological, silica and polystyrene microparticles, and the resultant possibility of confounding results, remains debatable (Mullier et al. 2011, Witwer et al. 2013). This issue is further complicated in the study of uEVs, as calcium phosphate microprecipitates have been shown to overlap the EV population on the flow cytometry signal (Larson et al. 2013).
Given the limitations of describing a total uEV population, alternative normalization methods across samples are needed to allow valid analysis of proteomic and transcriptomic changes. This has previously been described ‘the holy grail’ of uEV study, with a number of well‐described limitations (Salih et al. 2014). Theoretically, two broad approaches exist. Firstly, normalization with a defined time period (time normalization) or secondly, by a housekeeping marker (protein normalization). Instinctively, the former approach would appear to be optimal, but obtaining a timed urine collection can be difficult in clinical practice. Therefore, urinary creatinine is commonly used as a surrogate with the assumption that uEV concentration is correlated to urine concentration. Protein normalization (normalizing to a target housekeeping protein) can also be used. Example proteins include CD9, CD63, CD81, TSG‐101 and ALIX (Street et al. 2011, Alvarez et al. 2012). This approach assumes the normalizing protein urinary excretion does not change in different disease states and there is no biological or clinical relevance to a change in the number of vesicles within the sample. Despite the different underlying principles, there is no consensus regarding the optimal approach for normalization.
Biomarker discovery in EVs
In both acute and chronic kidney disease, current biomarkers, such as creatinine, focus on the recognition of established disease rather than early detection of imminent renal dysfunction. These investigations provide little information about the underlying pathophysiology, and renal biopsy is considered the gold‐standard investigation. Renal biopsy is an invasive technique with a number of adverse events such as infection and haemorrhage. Also biopsy cannot be performed serially (daily) and is prone to sampling error. There is hope that harnessing the proteomic and transcriptomic changes of uEVs in varying disease states will present a non‐invasive alternative to biopsy. As discussed, uEVs are released into urine from all regions of the nephron and are readily identified by proteins specific to the cell of origin, potentially providing a non‐invasive snapshot of the nephron's physiological state (Pisitkun et al. 2004; Alvarez et al. 2012). uEVs represent a remarkably stable, easily accessible biomarker reservoir, which protects its cargo from the harsh extra‐vesicular environment. uEVs are promising biomarker candidates, with the potential to predict disease, define mechanisms and prognosticate.
Three fundamental biomarker development stages have previously been described: biomarker discovery, validation of the markers’ predictive value within the population and implementation of a clinically approved assay (Granger et al. 2004; Pisitkun et al. 2012). In relation to uEVs, biomarker discovery in well‐defined populations has identified a number of targets. Table 1 summarizes the candidate EV biomarkers to date, distinguishing biomarkers of interest by disease process. In models of kidney injury, the proteins in the exosomal fraction have been reported to change prior to elevation in the ‘free’ fraction in urine and, importantly, prior to traditional biochemical and histological diagnostic tests (Zhou et al. 2006; Alvarez et al. 2013). Lower EV recovery rates in patients with heavy proteinuria and the outlined difficulties in normalization have led investigators to pursue qualitative targets, i.e. the presence or absence of the target defines disease or health. Ultimately, quantitative measurements would be desirable, allowing the ability to track deterioration or improvement in the clinical condition. The validation phase of study and the challenges to further progress perhaps best reflect the current situation. Large population studies are required, outwith the population of interest, to identify both the positive and negative predictive value of the EV‐based biomarker. Several studies have investigated the selectivity of uEVs but few have interrogated the specificity. The practicality and cost of the necessary large‐scale clinical studies limits further progress, as sample processing and quantification are not currently translatable to hundreds or thousands of samples.
Table 1.
Examples of uEV biomarkers and proposed clinical usage
| Potential uEV biomarker | ||
|---|---|---|
| Disorder | Protein | miRNA/mRNA |
| Acute kidney injury | Fetuin ‐A*, activating transcription factor‐3*, Na+/H+ exchanger type 3* | CD2AP*† |
| Ischaemia–reperfusion injury | Aquaporin‐1*, activating transcription factor‐3* | |
| Glomerular injury | ||
| Diabetic nephropathy | Dipeptidyl peptidase IV†, podocalyxin*, Wilm's tumour‐1*, histone‐lysine N‐methyltransferase*, voltage‐dependent anion‐selective channel protein 1*, α1‐microglobulin/bikunin precursor* | miR‐145*, miR‐130*, miR‐155*, miR‐424* |
| Focal segmental glomerular sclerosis | Wilm's tumour 1†,‡, podocalyxin* | |
| Autoimmune glomerulonephritis | miR‐26a† | |
| Lupus nephritis | A disintegrin and metalloprotease 10* | miR‐29c*†, miR‐26a*, miR‐146a* |
| Glomerular disease (mixed population) | A disintegrin and metalloprotease 10* | |
| IgA nephropathy | α1‐antitrypsin*, aminopeptidase N*, vasorin precursor*, ceruloplasmin* | |
| Glomerular fibrosis | CD2AP*† | |
| Other disorders | ||
| Polycystic kidney disease | Polycyctin‐1*†, polycyctin‐2*†, polyductin*, transmembrane protein 2*† | |
| Primary aldosteronism | Phosphorlyated Na+−Cl− cotransporter*, prostasin* | |
| Obstructive nephropathy | Transforming growth factor β* | |
| Bartter syndrome | Na+, K+, Cl– cotransporter type 2* | |
| Gitelman syndrome | Phosphorlyated Na+−Cl− cotransporter* | |
| Renal fibrosis | miR‐29c*, miR‐200* | |
| Chronic kidney disease | Neutrophil gelatinase‐associated lipocalin* | |
| Transplant | Neutrophil gelatinase‐associated lipocalin† | mRNA Il‐18† |
| Cancer | ||
| Prostate cancer | Integrin β1*, integrin α3*, prostate specific antigen*,‡, prostate specific membrane antigen* | miR‐34a‡ |
| Bladder cancer | EGF‐like repeats and discoidin I‐like domains 3*, tumour‐associated calcium‐signal transducer 2*, mucin 4, epidermal growth factor receptor pathway substrate 8‐related protein 2* | LASS2*, GALNT1* |
| Renal cell carcinoma | Matrix metalloproteinase 9*, ceruloplasmin*, podocalyxin*, dickkopf‐related protein 4*, carbonic anhydrase IX*, aquaporin‐1*, extracellular matrix metalloproteinase inducer*, neprilysin*, dipeptidase 1*, syntenin‐1* | |
| Systemic illness | ||
| Acute myocardial infarction | miR‐1*, miR‐208* | |
| Parkinson's disease | Protein deglycase DJ‐1† | |
| Ovarian serous adenocarcinoma | miR‐30a‐59* | |
| Lupus erythematosus | miR‐146a* | |
| Autoimmune encephalomyelitis | miR‐155–5p*,‡ | |
| Type 2 diabetes | miR‐143* | |
| Non‐small cell lung cancer | Leucine‐rich α‐2‐glycoprotein* | |
*Diagnostic; †prognostic; ‡response to treatment.
Function
Intercellular communication depends upon an EV's ability to influence a recipient cell, by receptor‐mediated interaction, endocytosis of the EV or fusion of the vesicle membrane to the plasma membrane. This latter signalling mechanism results in delivery of an EV's contents directly into the cytoplasm, including transcription factors, miRNA, mature RNA and infective particles (Ratajczak et al. 2006; Valadi et al. 2007; Cocucci et al. 2009; Camussi et al. 2010).
Physiology
Along the nephron, uEV‐mediated intercellular signalling has been postulated to explain why proximal tubule proteins are present in downstream nephron segments (van Balkom et al. 2011; Dear et al. 2013; Okada 2013), but this has not yet been conclusively demonstrated in vivo (van Balkom et al. 2011). uEVs are released into urine from all regions of the nephron and can be readily identified by transcriptomic and proteomic markers specific to the cell of origin (Alvarez et al. 2012; Pisitkun et al. 2004). Hypothetically, via uEV release and downstream reuptake, uEV contents could affect the function of a downstream recipient cell (Dimov et al. 2009). Notably, the exosomal fraction of aquaporin‐2 (AQP2) increases in response to desmopressin and transfer of uEVs from desmopressin‐treated cells to untreated cells results in an increase of functional AQP2 expression in the recipient cell (Street et al. 2011; Higashijima et al. 2013). Cortical collecting duct cells stimulated with vasopressin take up ECVs, in vitro and in vivo. This process can be manipulated to deliver miRNA to collecting duct cells resulting in downregulation of target transcripts (Oosthuyzen et al. 2016). This study demonstrated that uEV signalling is a physiologically regulated process, which can be manipulated to deliver miRNA. Interaction of uEVs with recipient cells may involve specific interaction with primary cilia, as reported with polycystic disease‐positive vesicles using transmission electron microscopy images (Woollard et al. 2007; Hogan et al. 2009). This observation is supported by data from a biliary model that demonstrates exosome signalling affects ERK signalling, miRNA expression and cell proliferation, with abolition of this signal following removal of cilia (Masyuk et al. 2010). This work, in conjunction with the observation of multiple protein products of genes known to be responsible for renal and systemic diseases in normal urine, raises interesting questions about the role of EV signalling in health and disease (Pisitkun et al. 2004).
Pathophysiology
uEV signalling has been implicated in the pathogenesis of acute kidney injury (AKI): exosomes from injured tubular cells transfer TGF‐β1 mRNA into fibroblasts, resulting in cell activation (Borges et al. 2013). In vitro, vesicles appear to be important in mediating vascular smooth muscle cell calcification, a potential mechanism for accelerated vascular calcification in end stage renal disease (Reynolds et al. 2004). Interestingly, the paracrine effect of liver stem cells has been shown to aid regeneration of renal tubular injury via release of EVs, highlighting the possible beneficial role of exosome signalling in systemic illness (Herrera Sanchez et al. 2014).
Mesenchymal stem cells (MSCs) accelerate recovery and repair tissue following AKI, an effect demonstrated using diverse injury modalities (Asanuma et al. 2010; Reinders et al. 2010; Togel & Westenfelder, 2012; Wise & Ricardo, 2012; Fleig &Humphreys, 2014). Whether this reflects direct cell engraftment and differentiation or is mediated through release of paracrine factors is unclear. However, the observation that the MSC supernatant conferred a beneficial effect, equal to that of MSCs themselves, has opened the field to EV research (Bi et al. 2007). Seminal work by Bruno et al. supported the claim that this positive result was largely due to EV transfer, likely to be related to RNA interference (Gatti et al. 2011; Biancone et al. 2012; Bruno et al. 2012; Cantaluppi et al. 2012; Zhou et al. 2013). Recent work suggests EV miRNA has a significant role in this MSC effect on kidney cell injury (Collino et al. 2015). Notably, miRNAs associated with EV endothelial progenitor cells reduced apoptosis and promoted cell regeneration in ischaemia–reperfusion injury (Bitzer et al. 2012; Cantaluppi et al. 2012; Chen et al. 2014). This possible role of EV signalling in the delivery of functional miRNA and pathogenesis of AKI highlights their potential as a therapeutic intervention. Abnormal levels of miRNA could be one of the mechanisms explaining dysregulated protein expression during kidney disease progression and interference with this process represents a potential therapeutic target (Ho & Kreidberg, 2012).
uEVs, in addition, may act as antibacterial immune effectors, mediating the host response to urinary tract infection by inhibiting growth of pathogenic and commensal Escherichia coli and inducing bacterial lysis (Hiemstra et al. 2014). This highlights the multimodality capacity of EVs in therapeutics, specifically their potential as novel antibiotics for urinary tract infections, a common illness affecting 150 million patients annually (Flores‐Mireles et al. 2015). Furthermore, renal brush border‐derived exosomes can induce calcium oxalate crystallization in nephrolithiasis and may have a role in renal stone disease, although this mechanism is yet to be demonstrated in vivo (Khan 2004).
Potential as therapeutics
The contribution of EV signalling in health and disease highlights their potential as attractive therapeutic targets and there are a number of on‐going phase I and II clinical trials harnessing EV‐based therapeutics. Although we remain in the early phase of such studies, theoretical clinical utility could be mediated by interfering with EV biogenesis or the manipulation of EVs as therapeutics vectors.
Vectors for drug delivery
EVs are candidate drug delivery systems; they are stable vehicles with a wide biodistribution. They can be selectively loaded and can deliver functional RNA into cells. The integrity of RNA isolated from vesicles is similar to that of tissue and far higher than RNA in whole urine, as the membrane protects the RNA cargo from RNase degradation (Miranda et al. 2010; Cheng et al. 2013). Interestingly, EVs have natural targeting capacity, presumably by receptor–ligand binding (Sun et al. 2010; Zhuang et al. 2011; Tian et al. 2014). Recent work, conducted by Hoshino et al. (2015) demonstrated that tissue‐specific uptake of EVs is mediated by distinct integrins via their interaction with the extracellular matrix of the target tissue. Manipulation of this mechanism, through therapeutic targeting of these integrins, reduced EV uptake and impeded metastatic spread of cancer. This ability to predict the metastatic course of cancer raises the exciting possibility of prediction and redirection of tumour progression. A similar mechanism could also be responsible for EV signalling along the kidney explaining the observation of proximal tubular specific proteins in distal segments. EV signal manipulation in vivo, to target exogenous vesicles to the tissue of interest through delivery of miRNA and siRNA, has already been demonstrated, ultimately affecting downstream gene expression (Alvarez‐Erviti et al. 2011; Bryniarski et al. 2013). Furthermore, bioengineered nanoparticles can serve as exosome mimics, recreating these functions and delivering targeted chemotherapeutics (Jang et al. 2013).
Inhibiting EV biogenesis and uptake
The circulating concentration of exosomes has been correlated to cancer progression and overall survival, which suggests that reducing exosome numbers may be a potential therapeutic approach. Proof‐of‐concept using amiloride (an antihypertensive agent) to attenuate endocytic vesicle recycling increased the effect of chemotherapy agents in a murine model, speculatively as a result of reduced EV numbers in the circulation (Chalmin et al. 2010). Although precise regulation of exosome release remains unclear, a number of possible therapeutic targets have been identified. Rab27b interference inhibits exosome release and can reduce tumour progression (Ostrowski et al. 2010; Bobrie et al. 2012; Peinado et al. 2012). Other therapeutic targets of interest include P53 and GTPases, implicated in the cytoskeleton‐dependent mechanism underpinning exosome exocytosis (Savina et al. 2005; Hsu et al. 2010; Zhuang et al. 2011). Inhibition of EV uptake into cells is also possible by blocking surface phosphatidylserine; however, due to lack of specificity of this mechanism it is unlikely to translate into a therapeutic intervention. Regardless of the therapeutic strategy employed, there are a large number of limitations to targeting exosome biogenesis and uptake. In particular, currently elucidated mechanisms are not tissue specific and affect a number of complex, core functions in diseased and healthy tissue. For a future drug, this may manifest in a large number of off‐target effects, greatly affecting this approach as a therapeutic strategy.
Future focus
There is great potential for uEVs as both disease biomarkers and vectors for targeted therapeutic delivery, yet we remain some way from realizing this potential and improving patient outcomes. uEVs are abundant in urine, but the contribution of EVs to normal renal physiology and their ability to modulate pathophysiological processes has yet to be proven. We still are not clear how EVs enter the urine from circulation, what the mechanisms are for targeting and how these can be effectively manipulated. The role of EVs as clinical biomarkers is perhaps closer to clinical utility. Biomarker utility needs to be confirmed in larger studies and several key challenges remain such as the development of high throughput platforms for rapid EV quantification and a consensus on normalization across samples. These remain the same hurdles we identified previously in our 2013 review, but developments have been made through improvements to microfluidic devices and nanopore arrays (Dear et al. 2013). Significant challenges remain that need to be overcome if investigation and manipulation of EVs is to be translated into point‐of‐care diagnostics and therapeutic interventions.
Additional information
Competing interests
None declared.
Author contributions
All authors have approved the final version of the manuscript and agree to be accountable for all aspects of the work. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Funding
E.E.M. is funded by the MRC Scottish Clinical Pharmacology and Pathology Programme MRC 65368/1.
Biographies
James Dear has a PhD in Pharmacology from UCL and BM BCh from Oxford University. He is now Reader in Clinical Pharmacology at the University of Edinburgh. Emma Morrison received her MB from Glasgow University in 2008 and currently is working towards her PhD as an MRC funded clinical research fellow at the University of Edinburgh.
Matt Bailey has a PhD in renal physiology from the University of London. He trained at UCL, CNRS in Paris and Yale University. He is a Reader at the University of Edinburgh.

References
- Agromayor M & Martin‐Serrano J (2006). Interaction of AMSH with ESCRT‐III and deubiquitination of endosomal cargo. J Biol Chem 281, 23083–23091. [DOI] [PubMed] [Google Scholar]
- Akers JC, Gonda D, Kim R, Carter BS & Chen CC (2013). Biogenesis of extracellular vesicles (EV): exosomes, microvesicles, retrovirus‐like vesicles, and apoptotic bodies. J Neurooncol 113, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez ML, Khosroheidari M, Kanchi Ravi R & DiStefano JK (2012). Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int 82, 1024–1032. [DOI] [PubMed] [Google Scholar]
- Alvarez S, Suazo C, Boltansky A, Ursu M, Carvajal D, Innocenti G, Vukusich A, Hurtado M, Villanueva S, Carreno JE, Rogelio A & Irarrazabal CE (2013). Urinary exosomes as a source of kidney dysfunction biomarker in renal transplantation. Transplant Proc 45, 3719–3723. [DOI] [PubMed] [Google Scholar]
- Alvarez‐Erviti L, Seow Y, Yin H, Betts C, Lakhal S & Wood MJ (2011). Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol 29, 341–345. [DOI] [PubMed] [Google Scholar]
- Asanuma H, Meldrum DR & Meldrum KK (2010). Therapeutic applications of mesenchymal stem cells to repair kidney injury. J Urol 184, 26–33. [DOI] [PubMed] [Google Scholar]
- Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO & Skog J (2011). Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun 2, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi B, Schmitt R, Israilova M, Nishio H & Cantley LG (2007). Stromal cells protect against acute tubular injury via an endocrine effect. J Am Soc Nephrol 18, 2486–2496. [DOI] [PubMed] [Google Scholar]
- Biancone L, Bruno S, Deregibus MC, Tetta C & Camussi G (2012). Therapeutic potential of mesenchymal stem cell‐derived microvesicles. Nephrol Dial Transplant 27, 3037–3042. [DOI] [PubMed] [Google Scholar]
- Bitzer M, Ben‐Dov IZ & Thum T (2012). Microparticles and microRNAs of endothelial progenitor cells ameliorate acute kidney injury. Kidney Int 82, 375–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bobrie A, Krumeich S, Reyal F, Recchi C, Moita LF, Seabra MC, Ostrowski M & Thery C (2012). Rab27a supports exosome‐dependent and ‐independent mechanisms that modify the tumor microenvironment and can promote tumor progression. Cancer Res 72, 4920–4930. [DOI] [PubMed] [Google Scholar]
- Boing AN, van der Pol E, Grootemaat AE, Coumans FA, Sturk A & Nieuwland R (2014). Single‐step isolation of extracellular vesicles by size‐exclusion chromatography. J Extracell Vesicles 3, doi: 10.3402/jev.v3.23430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges FT, Melo SA, Ozdemir BC, Kato N, Revuelta I, Miller CA, Gattone VH 2nd, LeBleu VS & Kalluri R (2013). TGF‐β1‐containing exosomes from injured epithelial cells activate fibroblasts to initiate tissue regenerative responses and fibrosis. J Am Soc Nephrol 24, 385–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruno S, Grange C, Collino F, Deregibus MC, Cantaluppi V, Biancone L, Tetta C & Camussi G (2012). Microvesicles derived from mesenchymal stem cells enhance survival in a lethal model of acute kidney injury. PLoS One 7, e33115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryniarski K, Ptak W, Jayakumar A, Pullmann K, Caplan MJ, Chairoungdua A, Lu J, Adams BD, Sikora E, Nazimek K, Marquez S, Kleinstein SH, Sangwung P, Iwakiri Y, Delgato E, Redegeld F, Blokhuis BR, Wojcikowski J, Daniel AW, Groot Kormelink T & Askenase PW (2013). Antigen‐specific, antibody‐coated, exosome‐like nanovesicles deliver suppressor T‐cell microRNA‐150 to effector T cells to inhibit contact sensitivity. J Allergy Clin Immunol 132, 170–181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camussi G, Deregibus MC, Bruno S, Cantaluppi V & Biancone L (2010). Exosomes/microvesicles as a mechanism of cell‐to‐cell communication. Kidney Int 78, 838–848. [DOI] [PubMed] [Google Scholar]
- Cantaluppi V, Gatti S, Medica D, Figliolini F, Bruno S, Deregibus MC, Sordi A, Biancone L, Tetta C & Camussi G (2012). Microvesicles derived from endothelial progenitor cells protect the kidney from ischemia‐reperfusion injury by microRNA‐dependent reprogramming of resident renal cells. Kidney Int 82, 412–427. [DOI] [PubMed] [Google Scholar]
- Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy‐Martin JP, Boireau W, Rouleau A, Simon B, Lanneau D, De Thonel A, Multhoff G, Hamman A, Martin F, Chauffert B, Solary E, Zitvogel L, Garrido C, Ryffel B, Borg C, Apetoh L, Rebe C & Ghiringhelli F (2010). Membrane‐associated Hsp72 from tumor‐derived exosomes mediates STAT3‐dependent immunosuppressive function of mouse and human myeloid‐derived suppressor cells. J Clin Invest 120, 457–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Lai PF, Lan YF, Cheng CF, Zhong WB, Lin YF, Chen TW & Lin H (2014). Exosomal ATF3 RNA attenuates pro‐inflammatory gene MCP‐1 transcription in renal ischemia‐reperfusion. J Cell Physiol 229, 1202–1211. [DOI] [PubMed] [Google Scholar]
- Cheng L, Sun X, Scicluna BJ, Coleman BM & Hill AF (2013). Characterization and deep sequencing analysis of exosomal and non‐exosomal miRNA in human urine. Kidney Int 86, 433–444. [DOI] [PubMed] [Google Scholar]
- Cheruvanky A, Zhou H, Pisitkun T, Kopp JB, Knepper MA, Yuen PS & Star RA (2007). Rapid isolation of urinary exosomal biomarkers using a nanomembrane ultrafiltration concentrator. Am J Physiol Renal Physiol 292, F1657–F1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocucci E & Meldolesi J (2015). Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol 25, 364–372. [DOI] [PubMed] [Google Scholar]
- Cocucci E, Racchetti G & Meldolesi J (2009). Shedding microvesicles: artefacts no more. Trends Cell Biol 19, 43–51. [DOI] [PubMed] [Google Scholar]
- Collino F, Bruno S, Incarnato D, Dettori D, Neri F, Provero P, Pomatto M, Oliviero S, Tetta C, Quesenberry PJ & Camussi G (2015). AKI recovery induced by mesenchymal stromal cell‐derived extracellular vesicles carrying microRNAs. J Am Soc Nephrol 26, 2349–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvjetkovic A, Lotvall J & Lasser C (2014). The influence of rotor type and centrifugation time on the yield and purity of extracellular vesicles. J Extracell Vesicles 3, doi: 10.3402/jev.v3.23111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear JW, Street JM & Bailey MA (2013). Urinary exosomes: a reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 13, 1572–1580. [DOI] [PubMed] [Google Scholar]
- Del Conde I, Shrimpton CN, Thiagarajan P & Lopez JA (2005). Tissue‐factor‐bearing microvesicles arise from lipid rafts and fuse with activated platelets to initiate coagulation. Blood 106, 1604–1611. [DOI] [PubMed] [Google Scholar]
- Dimov I, Jankovic Velickovic L & Stefanovic V (2009). Urinary exosomes. ScientificWorldJournal 9, 1107–1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez‐Llama P, Khositseth S, Gonzales PA, Star RA, Pisitkun T & Knepper MA (2010). Tamm‐Horsfall protein and urinary exosome isolation. Kidney Int 77, 736–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleig SV & Humphreys BD (2014). Rationale of mesenchymal stem cell therapy in kidney injury. Nephron Clin Pract 127, 75–80. [DOI] [PubMed] [Google Scholar]
- Flores‐Mireles AL, Walker JN, Caparon M & Hultgren SJ (2015). Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13, 269–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatti S, Bruno S, Deregibus MC, Sordi A, Cantaluppi V, Tetta C & Camussi G (2011). Microvesicles derived from human adult mesenchymal stem cells protect against ischaemia‐reperfusion‐induced acute and chronic kidney injury. Nephrol Dial Transplant 26, 1474–1483. [DOI] [PubMed] [Google Scholar]
- Ghosh A, Davey M, Chute IC, Griffiths SG, Lewis S, Chacko S, Barnett D, Crapoulet N, Fournier S, Joy A, Caissie MC, Ferguson AD, Daigle M, Meli MV, Lewis SM & Ouellette RJ (2014). Rapid isolation of extracellular vesicles from cell culture and biological fluids using a synthetic peptide with specific affinity for heat shock proteins. PLoS One 9, e110443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS & Knepper MA (2009). Large‐scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20, 363–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granger CB, Van Eyk JE, Mockrin SC, Anderson NL; National Heart, Lung, And Blood Institute Clinical Proteomics Working Group (2004). National Heart, Lung, And Blood Institute Clinical Proteomics Working Group report. Circulation 109, 1697–1703. [DOI] [PubMed] [Google Scholar]
- Guescini M, Genedani S, Stocchi V & Agnati LF (2010). Astrocytes and glioblastoma cells release exosomes carrying mtDNA. J Neural Transm 117, 1–4. [DOI] [PubMed] [Google Scholar]
- Harding C & Stahl P (1983). Transferrin recycling in reticulocytes: pH and iron are important determinants of ligand binding and processing. Biochem Biophys Res Commun 113, 650–658. [DOI] [PubMed] [Google Scholar]
- Herrera Sanchez MB, Bruno S, Grange C, Tapparo M, Cantaluppi V, Tetta C & Camussi G (2014). Human liver stem cells and derived extracellular vesicles improve recovery in a murine model of acute kidney injury. Stem Cell Res Ther 5, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiemstra TF, Charles PD, Gracia T, Hester SS, Gatto L, Al‐Lamki R, Floto RA, Su Y, Skepper JN, Lilley KS & Karet Frankl FE (2014). Human urinary exosomes as innate immune effectors. J Am Soc Nephrol 25, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashijima Y, Sonoda H, Takahashi S, Kondo H, Shigemura K & Ikeda M (2013). Excretion of urinary exosomal AQP2 in rats is regulated by vasopressin and urinary pH. Am J Physiol Renal Physiol 305, F1412–F1421. [DOI] [PubMed] [Google Scholar]
- Ho J & Kreidberg JA (2012). The long and short of microRNAs in the kidney. J Am Soc Nephrol 23, 400–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan MC, Bakeberg JL, Gainullin VG, Irazabal MV, Harmon AJ, Lieske JC, Charlesworth MC, Johnson KL, Madden BJ, Zenka RM, McCormick DJ, Sundsbak JL, Heyer CM, Torres VE, Harris PC & Ward CJ (2014). Identification of biomarkers for PKD1 using urinary exosomes. J Am Soc Nephrol 26, 1661–1670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan MC, Johnson KL, Zenka RM, Charlesworth MC, Madden BJ, Mahoney DW, Oberg AL, Huang BQ, Leontovich AA, Nesbitt LL, Bakeberg JL, McCormick DJ, Bergen HR & Ward CJ (2014). Subfractionation, characterization, and in‐depth proteomic analysis of glomerular membrane vesicles in human urine. Kidney Int 85, 1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan MC, Manganelli L, Woollard JR, Masyuk AI, Masyuk TV, Tammachote R, Huang BQ, Leontovich AA, Beito TG, Madden BJ, Charlesworth MC, Torres VE, LaRusso NF, Harris PC & Ward CJ (2009). Characterization of PKD protein‐positive exosome‐like vesicles. J Am Soc Nephro l 20, 278–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino A, Costa‐Silva B, Shen TL, Rodrigues G, Hashimoto A, Tesic Mark M, Molina H, Kohsaka S, Di Giannatale A, Ceder S, Singh S, Williams C, Soplop N, Uryu K, Pharmer L, King T, Bojmar L, Davies AE, Ararso Y, Zhang T, Zhang H, Hernandez J, Weiss JM, Dumont‐Cole VD, Kramer K, Wexler LH, Narendran A, Schwartz GK, Healey JH, Sandstrom P, Jorgen Labori K, Kure EH, Grandgenett PM, Hollingsworth MA, de Sousa M, Kaur S, Jain M, Mallya K, Batra SK, Jarnagin WR, Brady MS, Fodstad O, Muller V, Pantel K, Minn AJ, Bissell MJ, Garcia BA, Kang Y, Rajasekhar VK, Ghajar CM, Matei I, Peinado H, Bromberg J & Lyden D (2015). Tumour exosome integrins determine organotropic metastasis. Nature 527, 329–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu C, Morohashi Y, Yoshimura S, Manrique‐Hoyos N, Sung J, Lauterbach MA, Bakhti M, Gronborg M, Mobius W, Rhee J, Barr FA & Simons M (2010). Regulation of exosome secretion by Rab35 and its GTPase‐activating proteins TBC1D10A‐C. J Cell Biol 189, 223–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huebner AR, Cheng L, Somparn P, Knepper MA, Fenton RA & Pisitkun T (2016). Deubiquitylation of protein cargo is not an essential step in exosome formation. Mol Cell Proteomics 15, 1556–1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jang SC, Kim OY, Yoon CM, Choi DS, Roh TY, Park J, Nilsson J, Lotvall J, Kim YK & Gho YS (2013). Bioinspired exosome‐mimetic nanovesicles for targeted delivery of chemotherapeutics to malignant tumors. ACS Nano 7, 7698–7710. [DOI] [PubMed] [Google Scholar]
- Johnstone RM, Adam M & Pan BT (1984). The fate of the transferrin receptor during maturation of sheep reticulocytes in vitro. Can J Biochem Cell Biol 62, 1246–1254. [DOI] [PubMed] [Google Scholar]
- Kalra H, Adda CG, Liem M, Ang CS, Mechler A, Simpson RJ, Hulett MD & Mathivanan S (2013). Comparative proteomics evaluation of plasma exosome isolation techniques and assessment of the stability of exosomes in normal human blood plasma. Proteomics 13, 3354–3364. [DOI] [PubMed] [Google Scholar]
- Kanwar SS, Dunlay CJ, Simeone DM & Nagrath S (2014). Microfluidic device (ExoChip) for on‐chip isolation, quantification and characterization of circulating exosomes. Lab Chip 14, 1891–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller S, Rupp C, Stoeck A, Runz S, Fogel M, Lugert S, Hager HD, Abdel‐Bakky MS, Gutwein P & Altevogt P (2007). CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int 72, 1095–1102. [DOI] [PubMed] [Google Scholar]
- Khan SR (2004). Role of renal epithelial cells in the initiation of calcium oxalate stones. Nephron Exp Nephrol 98, e55–e60. [DOI] [PubMed] [Google Scholar]
- Kosanovic M & Jankovic M (2014). Isolation of urinary extracellular vesicles from Tamm‐Horsfall protein‐depleted urine and their application in the development of a lectin‐exosome‐binding assay. Biotechniques 57, 143–149. [DOI] [PubMed] [Google Scholar]
- Kowal J, Tkach M & Thery C (2014). Biogenesis and secretion of exosomes. Curr Opin Cell Biol 29, 116–125. [DOI] [PubMed] [Google Scholar]
- Larson MC, Luthi MR, Hogg N & Hillery CA (2013). Calcium‐phosphate microprecipitates mimic microparticles when examined with flow cytometry. Cytometry A 83, 242–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CG, Kim JG, Kim HJ, Kwon HK, Cho IJ, Choi DW, Lee WH, Kim WD, Hwang SJ, Choi S & Kim SG (2014). Discovery of an integrative network of microRNAs and transcriptomics changes for acute kidney injury. Kidney Int 86, 943–953. [DOI] [PubMed] [Google Scholar]
- Liga A, Vliegenthart AD, Oosthuyzen W, Dear JW & Kersaudy‐Kerhoas M (2015). Exosome isolation: a microfluidic road‐map. Lab Chip 15, 2388–2394. [DOI] [PubMed] [Google Scholar]
- Lotvall J, Hill AF, Hochberg F, Buzas EI, Di Vizio D, Gardiner C, Gho YS, Kurochkin IV, Mathivanan S, Quesenberry P, Sahoo S, Tahara H, Wauben MH, Witwer KW & Thery C (2014). Minimal experimental requirements for definition of extracellular vesicles and their functions: a position statement from the International Society for Extracellular Vesicles. J Extracell Vesicles 3, 26913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masyuk AI, Huang BQ, Ward CJ, Gradilone SA, Banales JM, Masyuk TV, Radtke B, Splinter PL & LaRusso NF (2010). Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am J Physiol Gastrointest Liver Physiol 299, G990–G999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda KC, Bond DT, McKee M, Skog J, Paunescu TG, Da Silva N, Brown D & Russo LM (2010). Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int 78, 191–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittelbrunn M, Gutierrez‐Vazquez C, Villarroya‐Beltri C, Gonzalez S, Sanchez‐Cabo F, Gonzalez MA, Bernad A & Sanchez‐Madrid F (2011). Unidirectional transfer of microRNA‐loaded exosomes from T cells to antigen‐presenting cells. Nat Commun 2, 282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno‐Gonzalo O, Villarroya‐Beltri C & Sanchez‐Madrid F (2014). Post‐translational modifications of exosomal proteins. Front Immunol 5, 383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullier F, Bailly N, Chatelain C, Dogne JM & Chatelain B (2011). More on: calibration for the measurement of microparticles: needs, interests, and limitations of calibrated polystyrene beads for flow cytometry‐based quantification of biological microparticles. J Thromb Haemost 9, 1679–1681. [DOI] [PubMed] [Google Scholar]
- Muralidharan‐Chari V, Clancy J, Plou C, Romao M, Chavrier P, Raposo G & D'Souza‐Schorey C (2009). ARF6‐regulated shedding of tumor cell‐derived plasma membrane microvesicles. Curr Biol 19, 1875–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muralidharan‐Chari V, Clancy JW, Sedgwick A & D'Souza‐Schorey C (2010). Microvesicles: mediators of extracellular communication during cancer progression. J Cell Sci 123, 1603–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Saraswat M, Duriez E, Byrne B, Ravida A, Domon B & Holthofer H (2012). Biochemical and physical characterisation of urinary nanovesicles following CHAPS treatment. PLoS One 7, e37279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musante L, Tataruch DE & Holthofer H (2014). Use and isolation of urinary exosomes as biomarkers for diabetic nephropathy. Front Endocrinol 5, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H (2013). A new look at tubulointerstitial communication with exosomes. J Am Soc Nephrol 24, 330–332. [DOI] [PubMed] [Google Scholar]
- Oosthuyzen W, Scullion KM, Ivy JR, Morrison EE, Hunter RW, Starkey Lewis PJ, O'Duibhir E, Street JM, Caporali A, Gregory CD, Forbes SJ, Webb DJ, Bailey MA & Dear JW (2016). Vasopressin regulates extracellular vesicle uptake by kidney collecting duct cells. J Am Soc Nephrol DOI:10.1681/ASN.2015050568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oosthuyzen W, Sime NE, Ivy JR, Turtle EJ, Street JM, Pound J, Bath LE, Webb DJ, Gregory CD, Bailey MA & Dear JW (2013). Quantification of human urinary exosomes by nanoparticle tracking analysis. J Physiol 591, 5833–5842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski M, Carmo NB, Krumeich S, Fanget I, Raposo G, Savina A, Moita CF, Schauer K, Hume AN, Freitas RP, Goud B, Benaroch P, Hacohen N, Fukuda M, Desnos C, Seabra MC, Darchen F, Amigorena S, Moita LF & Thery C (2010). Rab27a and Rab27b control different steps of the exosome secretion pathway. Nat Cell Biol 12, 19–30. [DOI] [PubMed] [Google Scholar]
- Peinado H, Aleckovic M, Lavotshkin S, Matei I, Costa‐Silva B, Moreno‐Bueno G, Hergueta‐Redondo M, Williams C, Garcia‐Santos G, Ghajar C, Nitadori‐Hoshino A, Hoffman C, Badal K, Garcia BA, Callahan MK, Yuan J, Martins VR, Skog J, Kaplan RN, Brady MS, Wolchok JD, Chapman PB, Kang Y, Bromberg J & Lyden D (2012). Melanoma exosomes educate bone marrow progenitor cells toward a pro‐metastatic phenotype through MET. Nat Med 18, 883–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T, Gandolfo MT, Das S, Knepper MA & Bagnasco SM (2012). Application of systems biology principles to protein biomarker discovery: urinary exosomal proteome in renal transplantation. Proteomics Clin Appl 6, 268–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisitkun T, Shen RF & Knepper MA (2004). Identification and proteomic profiling of exosomes in human urine. Proc Natl Acad Sci USA 101, 13368–13373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raposo G & Stoorvogel W (2013). Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 200, 373–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P & Ratajczak MZ (2006). Embryonic stem cell‐derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia 20, 847–856. [DOI] [PubMed] [Google Scholar]
- Reinders ME, Fibbe WE & Rabelink TJ (2010). Multipotent mesenchymal stromal cell therapy in renal disease and kidney transplantation. Nephrol Dial Transplant 25, 17–24. [DOI] [PubMed] [Google Scholar]
- Reynolds JL, Joannides AJ, Skepper JN, McNair R, Schurgers LJ, Proudfoot D, Jahnen‐Dechent W, Weissberg PL & Shanahan CM (2004). Human vascular smooth muscle cells undergo vesicle‐mediated calcification in response to changes in extracellular calcium and phosphate concentrations: a potential mechanism for accelerated vascular calcification in ESRD. J Am Soc Nephrol 15, 2857–2867. [DOI] [PubMed] [Google Scholar]
- Rood IM, Deegens JK, Merchant ML, Tamboer WP, Wilkey DW, Wetzels JF & Klein JB (2010). Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int 78, 810–816. [DOI] [PubMed] [Google Scholar]
- Saetun P, Semangoen T & Thongboonkerd V (2009). Characterizations of urinary sediments precipitated after freezing and their effects on urinary protein and chemical analyses. Am J Physiol Renal Physiol 296, F1346–1354. [DOI] [PubMed] [Google Scholar]
- Salih M, Zietse R & Hoorn EJ (2014). Urinary extracellular vesicles and the kidney: biomarkers and beyond. Am J Physiol Renal Physiol 306, F1251–1259. [DOI] [PubMed] [Google Scholar]
- Santana SM, Antonyak MA, Cerione RA & Kirby BJ (2014). Microfluidic isolation of cancer‐cell‐derived microvesicles from hetergeneous extracellular shed vesicle populations. Biomed Microdevices 16, 869–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savina A, Fader CM, Damiani MT & Colombo MI (2005). Rab11 promotes docking and fusion of multivesicular bodies in a calcium‐dependent manner. Traffic 6, 131–143. [DOI] [PubMed] [Google Scholar]
- Schageman J, Zeringer E, Li M, Barta T, Lea K, Gu J, Magdaleno S, Setterquist R & Vlassov AV (2013). The complete exosome workflow solution: from isolation to characterization of RNA cargo. Biomed Res Int 2013, 253957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheldon H, Heikamp E, Turley H, Dragovic R, Thomas P, Oon CE, Leek R, Edelmann M, Kessler B, Sainson RC, Sargent I, Li JL & Harris AL (2010). New mechanism for Notch signaling to endothelium at a distance by Delta‐like 4 incorporation into exosomes. Blood 116, 2385–2394. [DOI] [PubMed] [Google Scholar]
- Squadrito ML, Baer C, Burdet F, Maderna C, Gilfillan GD, Lyle R, Ibberson M & De Palma M (2014). Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep 8, 1432–1446. [DOI] [PubMed] [Google Scholar]
- Street JM, Birkhoff W, Menzies RI, Webb DJ, Bailey MA & Dear JW (2011). Exosomal transmission of functional aquaporin 2 in kidney cortical collecting duct cells. J Physiol 589, 6119–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D, Zhuang X, Xiang X, Liu Y, Zhang S, Liu C, Barnes S, Grizzle W, Miller D & Zhang HG (2010). A novel nanoparticle drug delivery system: the anti‐inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol Ther 18, 1606–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur BK, Zhang H, Becker A, Matei I, Huang Y, Costa‐Silva B, Zheng Y, Hoshino A, Brazier H, Xiang J, Williams C, Rodriguez‐Barrueco R, Silva JM, Zhang W, Hearn S, Elemento O, Paknejad N, Manova‐Todorova K, Welte K, Bromberg J, Peinado H & Lyden D (2014). Double‐stranded DNA in exosomes: a novel biomarker in cancer detection. Cell Res 24, 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thery C, Amigorena S, Raposo G & Clayton A (2006). Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr Protoc Cell Biol Chapter 3, Unit 3.22; doi: 10.1002/0471143030.cb0322s30. [DOI] [PubMed] [Google Scholar]
- Thery C, Ostrowski M & Segura E (2009). Membrane vesicles as conveyors of immune responses. Nat Rev Immunol 9, 581–593. [DOI] [PubMed] [Google Scholar]
- Tian Y, Li S, Song J, Ji T, Zhu M, Anderson GJ, Wei J & Nie G (2014). A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 35, 2383–2390. [DOI] [PubMed] [Google Scholar]
- Togel FE & Westenfelder C (2012). Kidney protection and regeneration following acute injury: progress through stem cell therapy. Am J Kidney Dis 60, 1012–1022. [DOI] [PubMed] [Google Scholar]
- Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ & Lotvall JO (2007). Exosome‐mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9, 654–659. [DOI] [PubMed] [Google Scholar]
- van Balkom BW, Pisitkun T, Verhaar MC & Knepper MA (2011). Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int 80, 1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Pol E, Boing AN, Harrison P, Sturk A & Nieuwland R (2012). Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol Rev 64, 676–705. [DOI] [PubMed] [Google Scholar]
- van Engeland M, Nieland LJ, Ramaekers FC, Schutte B & Reutelingsperger CP (1998). Annexin V‐affinity assay: a review on an apoptosis detection system based on phosphatidylserine exposure. Cytometry 31, 1–9. [DOI] [PubMed] [Google Scholar]
- Villarroya‐Beltri C, Baixauli F, Gutierrez‐Vazquez C, Sanchez‐Madrid F and Mittelbrunn M (2014). Sorting it out: regulation of exosome loading. Semin Cancer Biol 28, 3–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villarroya‐Beltri C, Gutierrez‐Vazquez C, Sanchez‐Cabo F, Perez‐Hernandez D, Vazquez J, Martin‐Cofreces N, Martinez‐Herrera DJ, Pascual‐Montano A, Mittelbrunn M & Sanchez‐Madrid F (2013). Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat Commun 4, 2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D & Sun W (2014). Urinary extracellular microvesicles: isolation methods and prospects for urinary proteome. Proteomics 14, 1922–1932. [DOI] [PubMed] [Google Scholar]
- Wen H, Frokiaer J, Kwon TH & Nielsen S (1999). Urinary excretion of aquaporin‐2 in rat is mediated by a vasopressin‐dependent apical pathway. J Am Soc Nephrol 10, 1416–1429. [DOI] [PubMed] [Google Scholar]
- Willms E, Johansson HJ, Mager I, Lee Y, Blomberg KE, Sadik M, Alaarg A, Smith CI, Lehtio J, El Andaloussi S, Wood MJ & Vader P (2016). Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci Rep 6, 22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise AF & Ricardo SD (2012). Mesenchymal stem cells in kidney inflammation and repair. Nephrology 17, 1–10. [DOI] [PubMed] [Google Scholar]
- Witwer KW, Buzas EI, Bemis LT, Bora A, Lasser C, Lotvall J, Nolte‐'t Hoen EN, Piper MG, Sivaraman S, Skog J, Thery C, Wauben MH & Hochberg F (2013). Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J Extracell Vesicles 2; doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woollard JR, Punyashtiti R, Richardson S, Masyuk TV, Whelan S, Huang BQ, Lager DJ, vanDeursen J, Torres VE, Gattone VH, LaRusso NF, Harris PC & Ward CJ (2007). A mouse model of autosomal recessive polycystic kidney disease with biliary duct and proximal tubule dilatation. Kidney Int 72, 328–336. [DOI] [PubMed] [Google Scholar]
- Yanez‐Mo M, Siljander PR, Andreu Z, Zavec AB, Borras FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colas E, Cordeiro‐da Silva A, Fais S, Falcon‐Perez JM, Ghobrial IM, Giebel B, Gimona M, Graner M, Gursel I, Gursel M, Heegaard NH, Hendrix A, Kierulf P, Kokubun K, Kosanovic M, Kralj‐Iglic V, Kramer‐Albers EM, Laitinen S, Lasser C, Lener T, Ligeti E, Line A, Lipps G, Llorente A, Lotvall J, Mancek‐Keber M, Marcilla A, Mittelbrunn M, Nazarenko I, Nolte‐'t Hoen EN, Nyman TA, O'Driscoll L, Olivan M, Oliveira C, Pallinger E, Del Portillo HA, Reventos J, Rigau M, Rohde E, Sammar M, Sanchez‐Madrid F, Santarem N, Schallmoser K, Ostenfeld MS, Stoorvogel W, Stukelj R, Van der Grein SG, Vasconcelos MH, Wauben MH & De Wever O (2015). Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles 4, 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Pisitkun T, Aponte A, Yuen PS, Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA & Star RA (2006). Exosomal Fetuin‐A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int 70, 1847–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JW, Gross P, Knepper MA & Star RA (2006). Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int 69, 1471–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Xu H, Xu W, Wang B, Wu H, Tao Y, Zhang B, Wang M, Mao F, Yan Y, Gao S, Gu H, Zhu W & Qian H (2013). Exosomes released by human umbilical cord mesenchymal stem cells protect against cisplatin‐induced renal oxidative stress and apoptosis in vivo and in vitro. Stem Cell Res Ther 4, 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuang X, Xiang X, Grizzle W, Sun D, Zhang S, Axtell RC, Ju S, Mu J, Zhang L, Steinman L, Miller D & Zhang HG (2011). Treatment of brain inflammatory diseases by delivering exosome encapsulated anti‐inflammatory drugs from the nasal region to the brain. Mol Ther 19, 1769–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]


