Abstract
The replication protein A (RPA) complex binds single-stranded DNA generated at stalled replication forks and recruits other DNA repair proteins to promote recovery of these forks. Here, we identify Ewing tumor-associated antigen 1 (ETAA1), which has been linked to susceptibility to pancreatic cancer, as a new repair protein that is recruited to stalled forks by RPA. We demonstrate that ETAA1 interacts with RPA through two regions, each of which resembles two previously identified RPA-binding domains, RPA70N-binding motif and RPA32C-binding motif, respectively. In response to replication stress, ETAA1 is recruited to stalled forks where it colocalizes with RPA, and this recruitment is diminished when RPA is depleted. Notably, inactivation of the ETAA1 gene increases the collapse level of the stalled replication forks and decreases the recovery efficiency of these forks. Moreover, epistasis analysis shows that ETAA1 stabilizes stalled replication forks in an ataxia telangiectasia and Rad3-related protein (ATR)-independent manner. Thus, our results reveal that ETAA1 is a novel RPA-interacting protein that promotes restart of stalled replication forks.
Keywords: DNA damage, DNA damage response, DNA recombination, DNA repair, DNA replication, ATR, ETAA1, Ewing tumor-associated antigen 1, RPA, stalled replication fork restart, ATR, replication stress
Introduction
The faithful replication of DNA is essential for the maintenance of genomic stability and the prevention of cancer-promoting mutations. Replication forks can be stalled by numerous obstacles on the DNA template, including unrepaired DNA damage, DNA-bound proteins, and secondary structures (1). Stalled replication forks are able to restart once the obstacles are removed or become broken (collapse) into DNA double strand breaks, which pose the most serious threat to genome integrity when fork protection fails (2, 3). However, how stalled replication forks are protected is not well understood.
The replication protein A (RPA)3 complex, which consists of RPA1 (RPA70), RPA2 (RPA32), and RPA3 (RPA14), plays crucial roles in a variety of DNA metabolic pathways, including DNA replication, recombination, repair, and DNA damage checkpoint (4–6). When replication forks stalled, single-stranded DNA (ssDNA) is generated and extended by minichromosome maintenance protein complex helicases (7, 8). The ssDNA is bound by RPA, which protects ssDNA from cleavage by nucleases and recruits repair proteins to initiate DNA damage responses. The RPA-ssDNA complex recruits and activates ATR/ATRIP thereby eliciting checkpoint signaling (9). In addition, RPA-ssDNA complex also recruits factors necessary for the stabilization and resumption of stalled replication forks, such as RAD51 (10, 11) and SMARCAL1 (12–15). Recently, several studies have also revealed a physical and functional interaction between RPA and the ubiquitin E3 ligases RFWD3 (16–18) and PRP19 (19), which ubiquitinate RPA and facilitate replication fork restart. Here, we identified a new RPA interaction protein, ETAA1, whose gene variation has been associated with susceptibility to pancreatic cancer (20). ETAA1 is recruited to stalled replication forks in response to replication stress, and the disruption of ETAA1 leads to fork collapse in an ATR-independent manner. These results suggest that ETAA1 is a new player involved in the stabilization of stalled replication forks.
Results
ETAA1 Is a Novel RPA-associated Protein
We transiently expressed FLAG-tagged RPA1 in HEK293 cells and immunoprecipitated the complexes with an anti-FLAG antibody (Fig. 1A). Mass spectrometry analysis revealed that, in addition to RPA2, RPA3, and the BLM complex, a novel protein, ETAA1, also immunoprecipitated with RPA1 (Fig. 1A). Immunoblotting confirmed this finding (Fig. 1B). To verify that ETAA1 indeed associates with the RPA complex, we performed a reciprocal immunoprecipitation using HEK293 cells expressing FLAG-tagged ETAA1; immunoblotting revealed that RPA was present in the ETAA1-associated complexes (Fig. 1C). To further confirm this interaction, we also performed immunoprecipitations with anti-RPA2 and anti-ETAA1 antibodies, and we found that endogenous ETAA1 strongly associated with the RPA complex in vivo (Fig. 1, D and E). Together, these data suggest that ETAA1 is a bona fide RPA-associated protein.
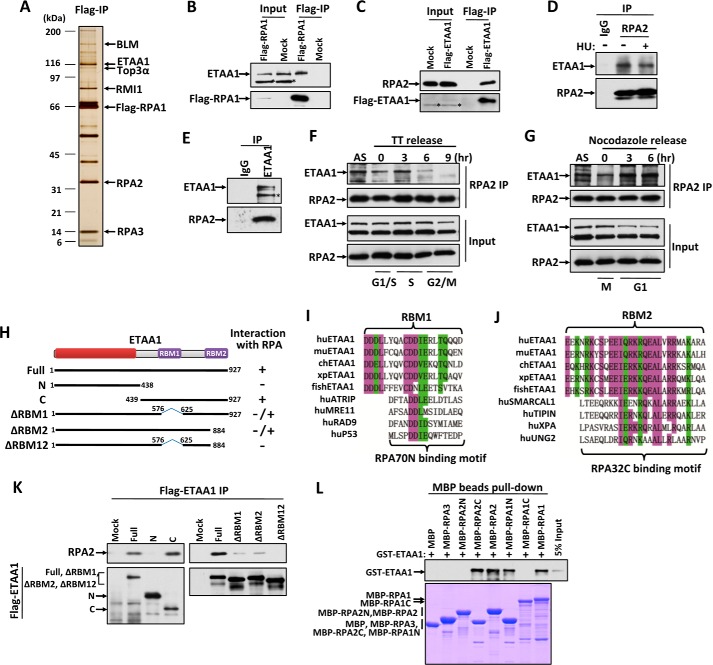
FIGURE 1.
ETAA1 associates with the RPA complex. A, silver-stained SDS-polyacrylamide gel showing the polypeptides that were immunopurified from extracts of HEK293 cells expressing FLAG-tagged RPA1 using the anti-FLAG antibody. The major polypeptides on the gel (arrows) were identified by mass spectrometry. B and C, immunoblot showing the immunoprecipitation (IP) of FLAG-tagged RPA1 (B) and ETAA1 (C). Asterisks indicate cross-reactive polypeptides. D and E, immunoblot showing the endogenous RPA2 (D) and ETAA1 (E) immunoprecipitation. HEK293 cells were treated with or without 4 mm HU for 3 h before harvest in the RPA2 immunoprecipitation. F and G, interaction of ETAA1 with RPA during the cell cycle. HEK293 cells were synchronized at the G1-S boundary by a double thymidine treatment (TT), released into fresh medium, and collected at the indicated times (F). Alternatively, cells were synchronized at prometaphase by a nocodazole block, released into fresh medium, and harvested at the indicated times (G). The cell cycle profile was analyzed by flow cytometry with the DNA content determined propidium iodide-staining (data not shown). Cell lysates and the anti-RPA immunoprecipitates (IP) were analyzed by Western blotting. AS, asynchronous cells. The asterisk indicates a cross-reactive polypeptide. H, schematic representation of the different ETAA1 deletion mutants (left) and their ability to coimmunoprecipitate with RPA from HEK293 extracts (right). I and J, sequence alignment of RBM1 and the RPA70N-binding motif (I) or RBM2 and the RPA32C-binding motif (J). K, immunoprecipitation and Western blotting to assess whether the various deletion mutants of ETAA1 described in H coimmunoprecipitated with RPA. L, direct binding between recombinant GST-tagged ETAA1 and MBP-tagged RPA. Upper panel, GST-ETAA1 was detected by immunoblotting with anti-GST antibodies. Lower panel, purified MBP-fused proteins was visualized by Coomassie staining.
To examine the interaction during the cell cycle, cells were synchronized at the G1-S boundary by a double thymidine treatment or at metaphase by a nocodazole treatment and then released. ETAA1 showed the strongest interaction with RPA at S phase and the lowest at G2/M phase (Fig. 1, F and G), suggesting that ETAA1 may function with RPA in DNA replication. Additionally, the binding of RPA2 to ETAA1 was not changed after cells were exposed to HU (Fig. 1D).
ETAA1 Contains Two RPA-binding Motifs
ETAA1 is expressed only in vertebrates. Sequence analyses showed that ETAA1 contains three conserved regions (Fig. 1H) as follows. The N-terminal conserved region contains no well known domains; the middle conserved region, RBM1, shows weak similarity to the RPA70N-binding motifs of ATRIP, MRE11, RAD9, and P53 (Fig. 1I) (21); and the C-terminal conserved region, RBM2, is similar to the RPA32C-terminal binding motifs of SMARCAL1, TIPIN, XPA, and UNG2 (Fig. 1J) (22, 23). To identify the region(s) of ETAA1 responsible for its interaction with RPA, we generated a series of ETAA1 truncation mutants (Fig. 1H). As shown in Fig. 1, H and K, the N-terminal region (residues 1–438, containing the first conserved region) of ETAA1 is dispensable for its interaction with RPA. The mutant lacking the RBM1 motif (ΔRBM1) or the RBM2 motif (ΔRBM2) dramatically decreased the ETAA1-RPA interaction, thus indicating that both RBM1 and RBM2 regions of ETAA1 are important for binding to RPA. Moreover, deletion of both RBM1 and RBM2 motifs (ΔRBM12) completely lost its interaction with RPA. These results are consistent with the sequence analyses that ETAA1 contains two RPA-binding motifs, RBM1 and RBM2.
To determine whether the interaction between RPA and ETAA1 is direct, we expressed and purified recombinant MBP-tagged RPA1, RPA2, RPA3, and GST-tagged ETAA1 from Escherichia coli. Pulldown experiments revealed that ETAA1 binds strongly with RPA1 and RPA2 but not with RPA3 (Fig. 1L), indicating that two independent ETAA1-binding sites exist on RPA1 and RPA2. Moreover, mapping analysis revealed that the RPA1 N-terminal region (residues 1–120) and RPA2 C-terminal region (residues 204–270) directly interacted with ETAA1 (Fig. 1L), thus agreeing that RBM1 and RBM2 of ETAA1 are RPA70N-terminal and RPA32C-terminal binding motifs, respectively.
ETAA1 Is Recruited to DNA Damage Sites by RPA
The RPA complex binds to ssDNA generated at stalled replication forks or DNA damage sites and forms an RPA-ssDNA platform, which facilitates the recruitment of many repair proteins. The interaction of ETAA1 and RPA suggests that ETAA1 may colocalize with RPA at ssDNA regions in vivo. As shown in Fig. 2A, FLAG-ETAA1 rarely formed foci in untreated cells. After HU treatment, ETAA1 was recruited to nuclear foci, where it colocalized with RPA, thus suggesting that ETAA1 localizes to stalled replication forks and may play a role in cellular responses to replication stress.
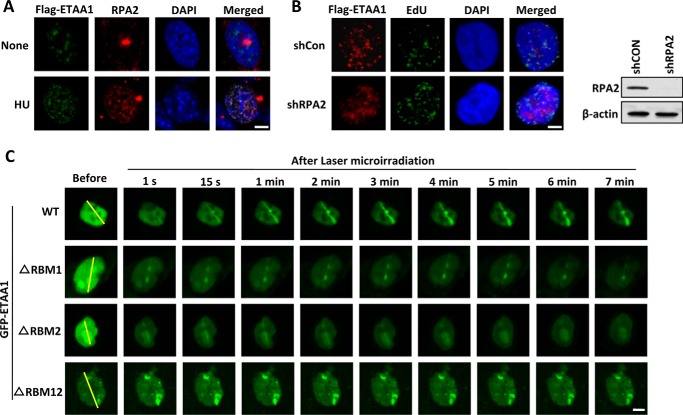
FIGURE 2.
ETAA1 colocalizes with RPA at stalled replication forks. A, ETAA1 localizes to stalled replication forks in response to replication stress. U2OS cells expressing FLAG-ETAA1 were mock-treated or treated with 5 mm HU for 6 h. Immunostaining was performed using anti-FLAG and anti-RPA2 antibodies. (Scale bar, 5 μm.) B, localization of ETAA1 to stalled replication forks requires RPA. U2OS cells expressing FLAG-ETAA1 were transfected with control or RPA2 shRNA. The cells were pulse-labeled with 20 μm EdU for 30 min and then treated with 5 mm HU for 6 h. Scale bar, 5 μm. Immunoblotting shows knockdown efficiency in the right panel. C, cells expressing wild-type or mutant GFP-ETAA1 were treated with laser microirradiation and monitored via live-cell imaging. The yellow lines indicate the positions for laser microirradiation. (Scale bar, 5 μm.)
We then determined whether the localization of ETAA1 at stalled replication forks was dependent on the RPA complex. As shown in Fig. 2B, ETAA1 exhibited dramatically decreased foci formation in RPA2-depleted cells but not in control cells after HU treatment, thereby indicating that ETAA1 is mainly recruited to stalled replication forks by the RPA complex.
We then examined the dynamic recruitment of GFP-ETAA1 at microirradiation-induced DNA damage sites in time-lapse experiments. GFP-ETAA1 accumulated at laser tracks very quickly, within 1 s, and the signal peaked at ∼2 min and then persisted for more than 1 h (Fig. 2C and data not shown). To determine whether this recruitment was also dependent on RPA, we tested ETAA1 mutants lacking the RBM1 or RBM2 motif. Deletion of RBM1 or RBM2 did not affect ETAA1 recruitment at the early stage (<1 s) but significantly reduced its recruitment to and (or) retention on DNA damage sites at the late stage (Fig. 2C). The deletion of both RBM1 and RBM2 completely abolished the localization of ETAA1 to DNA damage sites (Fig. 2C). These results suggest that the localization of ETAA1 at DNA damage sites is dependent on its interaction with RPA through the RBM1 and RBM2 motifs.
ETAA1 Is Required for Stalled Replication Fork Restart
The RPA complex plays a key role in replication and in stalled replication restart. Thus, we generated ETAA1-knock-out HCT116 cells using two different CRISPR target sites (Fig. 3A) and performed single DNA fiber analysis. Cells were pulse-labeled with CldU for 30 min and then incubated with HU (5 mm) and aphidicolin (5 μm) for 5 h to arrest replication forks. After a washout step, cells were incubated with IdU for 20 min. Restarted replication forks were visualized as tracks of CldU incorporation followed by tracks of IdU incorporation, whereas stalled or collapsed replication forks appeared as tracks of only CldU incorporation. We found that two ETAA1−/− clones both exhibited significantly decreased fork restart, as indicated by an approximate 2-fold reduction (Fig. 3B). These results suggest that ETAA1 promotes stalled fork restart upon replication stress.
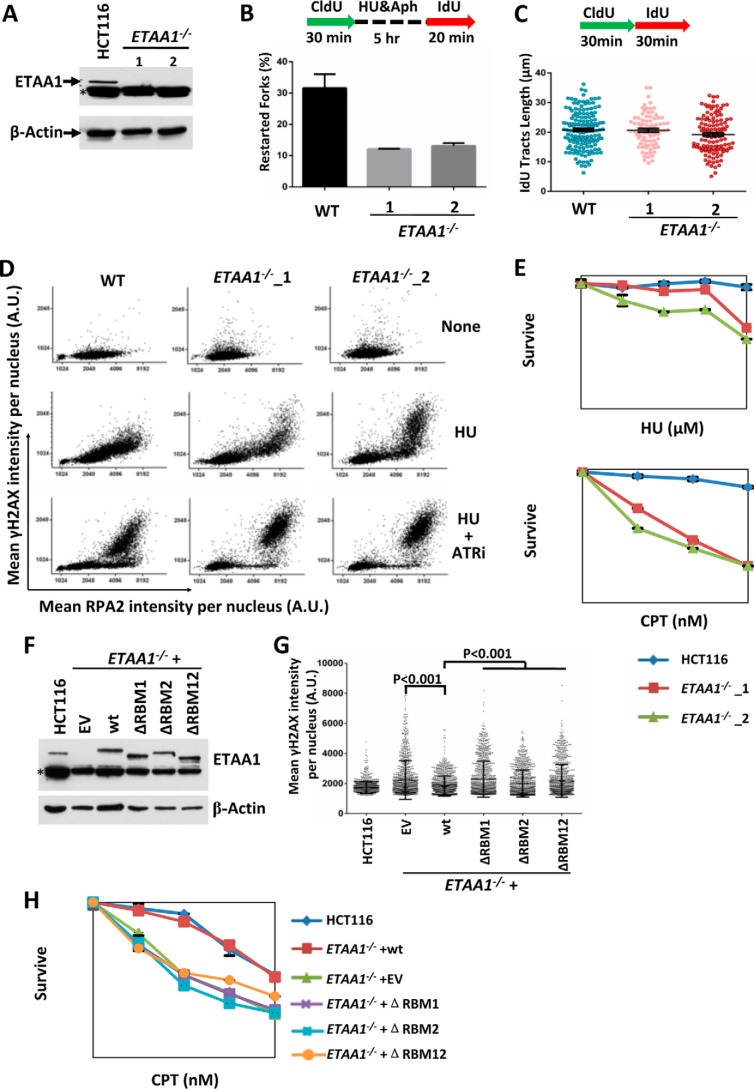
FIGURE 3.
ETAA1 stabilizes stalled replication forks. A, immunoblot shows that ETAA1 protein is absent in two ETAA−/− HCT116 cell clones. β-Actin is included as a control. The asterisk indicates a cross-reactive polypeptide. B, ETAA1 is required for stalled replication restart. Cells were pulse-labeled and treated as outlined in the top panel. DNA fibers were stained with antibodies recognizing IdU (red) and CldU (green). Restarted forks are indicated by green tracts followed by red tracts. Data represent the mean and S.D. C, ETAA1 is not required for normal replication. Cells were pulse-labeled as outlined in the top panel. The lengths of the IdU tracks were measured and are presented in the graph. D, QIBC of immunolabeled wild-type or ETAA1−/− HCT116 cells. Cells were treated with/without HU (2 mm) and the ATR inhibitor VE821 (10 μm) for 3 h, pre-extracted, and immunostained with the indicated antibodies. Mean nuclear intensities for RPA2 and γ-H2AX were determined for each of >5000 individual cells and are plotted in a scatter diagram. E, cell survival assay of the wild-type and ETAA1−/− HCT116 cells to HU and CPT. F–H, complementation experiments show that RPA-binding motifs of ETAA1 are required for its function in response to replication stress. The expression levels of ETAA1 in different cells were determined by immunoblotting (F). γH2AX intensity per nucleus was determined by QIBC (G). CPT sensitivity was measured by colony formation assay (H). EV, empty vector.
We also examined replication rate under normal conditions by measuring IdU track length (Fig. 3C). The ETAA1−/− cells showed similar track length as that of wild-type cells, indicating that ETAA1 is not required for normal replication.
ETAA1 Stabilizes Stalled Replication Forks
Combined treatment with HU and an ATR inhibitor exhausts RPA and triggers fork breakage, and it is accompanied by hallmarks of ataxia telangiectasia-mutated (ATM) activity such as H2AX hyperphosphorylation (24). We examined fork stability during replication stress in ETAA1-defective cells by measuring H2AX hyperphosphorylation using a previously described quantitative image-based cytometry (QIBC) method (24). HU treatment induced H2AX hyperphosphorylation in ETAA1−/− cells but not in wild-type cells, a result similar to that induced in wild-type cells after treatment with both HU and an ATR inhibitor (Fig. 3D). These results suggest that ETAA1 has a similar function to that of ATR in preventing stalled fork collapse. Indeed, the loss of ETAA1 also caused hypersensitivity to HU and CPT (Fig. 3E), a topoisomerase I inhibitor that induces topoisomerase I-DNA adducts and blocks replication, thus further supporting a role for ETAA1 in fork protection upon replication stress. Although the expression of the wild-type ETAA1 completely rescued stabilization of stalled replication fork and CPT resistance in the ETAA1−/− cells, reconstitution with ETAA1 mutants lacking RBM1, RBM2, or both failed to do so (Fig. 3, F–H). This result suggests that the interaction of ETAA1 with RPA is important for its function in response to replication stress.
Moreover, the combination of the ATR inhibitor and HU treatment induced a higher γH2AX signal in ETAA1−/− cells than in wild-type cells, or in ETAA1−/− cells treated with HU alone (Fig. 3D), suggesting that ETAA1 has an ATR-independent role in protecting the stalled replication forks.
Discussion
The RPA complex is a key player in initiating DNA-damage checkpoint signaling, replication fork stabilization, and DNA repair. The binding of RPA to ssDNA not only protects ssDNA from degradation by nucleases but also forms a platform facilitating the recruitment of many binding partners for diverse functions. Here, we identified a novel RPA-binding protein, ETAA1, which is required for the stability of the stalled replication fork.
ETAA1 contains two RPA-binding motifs, RBM1 and RBM2. RBM1 shows weak similarity to the RPA70N-binding motif of ATRIP, MRE11, RAD9, and p53, whereas RBM2 is similar to the RPA32C-binding motifs of SMARCAL1, TIPIN, XPA, and UNG2. The presence of either RBM1 or RBM2 is sufficient for ETAA1 to bind RPA at ssDNA regions, although the signal is weaker, thus allowing one RPA complex to bind other proteins together with ETAA1. This implies that ETAA1 can be orchestrated with other RPA-binding proteins on the RPA-ssDNA platform, which coordinates different DNA damage responses.
RPA complexes protect stalled replication forks through at least three mechanisms, First, RPA directly binds and protects ssDNA generated at stalled replication forks from nucleolytic digestion. ETAA1 probably does not contribute to this step because ETAA1 did not affect the ssDNA binding activity of RPA, and ETAA1−/− cells even showed more ssDNA-bound RPA under replication stress than did wild-type cells (Fig. 3D). Second, RPA activates the ATR-dependent DNA replication checkpoint to stabilize stalled replication forks. However, our epistasis analysis using an ATR inhibitor in ETAA1-null cells showed that ETAA1 and ATR function in two parallel pathways that protect stalled replication forks. Third, RPA recruits repair proteins, which remodel and protect stalled replication forks. Whether ETAA1 facilitates remodeling of stalled replication forks, thereby promoting fork stabilization, remains to be determined.
Moreover, GFP-ETAA1 was recruited to laser-induced DNA damage sites in both G1 and G2/S phase cells (data not shown), thus suggesting that ETAA1 functions not only in stalled replication fork restart but also in other DNA repair pathways. Repair of the double strand breaks, which are commonly induced by laser treatment, also recruits RPA after end resection, which normally takes more than a few seconds. ETAA1 was recruited quickly to DNA damage sites (within 1 s), a result suggesting that ETAA1 may be recruited by RPA-independent mechanisms in the early stage after laser-induced DNA damage. The RPA-dependent and (or) independent functions of ETAA1 in other DNA damage repair processes require further study.
In summary, we identified ETAA1 as a novel RPA-binding protein, which protects stalled replication forks and maintains genomic stability. Our results provide insights into the function of ETAA1 in preventing pancreatic cancer.
Experimental Procedures
Cell Culture
HeLa, U2OS, and HEK293T cells were cultured in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum (FBS; Invitrogen). HCT116 cells were cultured in RPMI 1640 medium with 10% FBS (Invitrogen). HEK293 suspension cells were cultured in Freestyle medium (Invitrogen) supplemented with 1% FBS (Gibco) and 1% glutamine in an incubator with shaking at 130 rpm.
Coimmunoprecipitation
The immunoprecipitation of the complexes was performed as described previously (25).
MBP Pulldown Assay
MBP-tagged RPA proteins were expressed in E. coli. Cells were harvested and resuspended in lysis buffer (20 mm Tris-HCl, pH 7.0, 300 mm NaCl, 1% Triton X-100, and 1 μg/ml each of leupeptin, aprotinin, and pepstatin). After sonicating, the extract was centrifuged at 18,000 rpm for 40 min. The supernatant was collected and incubated with amylose resins for 2 h at 4 °C. After washing the beads with washing buffer (20 mm Tris-HCl, 500 mm NaCl, 0.5% Nonidet P-40, 1 mm DTT), the protein-bound beads were incubated with the extract of E. coli cells expressing GST-ETAA1 for 2 h at 4 °C. After washing with binding buffer (20 mm Tris-HCl, pH 7.0, 150 mm NaCl, 0.5% Nonidet P-40, 1 mm DTT), the proteins were eluted with sample buffer (63 mm Tris-HCl, pH 6.8, 10% glycerol, 2% SDS, 0.0025% bromphenol blue) and analyzed with SDS-PAGE.
Laser Microirradiation
U2OS cells expressing GFP-ETAA1 were cultured at 37 °C in CO2-independent medium (Invitrogen) containing 10% FBS in a temperature-controlled container in glass-bottom dishes (MatTek). Laser microirradiation was carried out with the MicroPoint Laser Illumination and Ablation System coupled to a Nikon eclipse Ti microscope with a plan fluor 60 × 0.5–1.25 oil iris immersion objective. Time-lapse images were acquired with ANDOR IQ3 software through an ANDOR IXON camera.
Generation of ETAA1 Knock-out Cells
ETAA1-deficient HCT116 cells were generated using CRISPR. Briefly, two guide sequences, AGGAAACACCAAGATATCTG and GCTACAAAAAAGCCAATCAA, targeting two different sites of the human ETAA1 gene were inserted into the pX330 vector (26). The guide sequence containing pX330 plasmids were transfected into HCT116 cells. Single colonies were picked after 8–10 days of incubation. The genomic fragments of the ETAA1 gene were amplified by PCR using the following primers: GAGCTAGATGTGATTCAAGAGC and CTGTCCGCTACATTTCTGAG. The products were digested with EcoRV and BslI, respectively. Colonies containing the expected PCR fragments were then sequenced and examined by Western blotting.
DNA Fiber Assay
The restart efficiency of stalled replication forks was determined by using DNA fiber assays as described previously (27). Cells were first labeled with CldU (100 μm) for 30 min and then treated with HU (5 mm) and aphidicolin (5 μm) for 5 h. After being washed with PBS, cells were recovered in fresh medium with IdU (20 μm) for 20 min. Cells were then trypsinized and resuspended in PBS to a concentration of 2.5 × 105 cells/ml. Then the cells were diluted 1:4 with unlabeled cells at the same concentration, and 2.5 μl of cells was mixed with 7.5 μl of lysis buffer (200 mm Tris-HCl, pH 7.5, 50 mm EDTA, and 0.5% SDS) on a clean glass slide. After the edges were dried for 3–5 min, the slides were tilted at 15° to horizontal, allowing the DNA to slowly flow down along the slide. The slides were then air-dried, fixed in 3:1 methanol/acetic acid, and refrigerated overnight. The slides were treated with 2.5 m HCl for 1 h, neutralized in 0.1 m Na3B4O7, pH 8.5, and rinsed three times in PBST (PBS buffer with 0.1% Tween 20). The slides were then blocked in blocking buffer (PBST buffer containing 1% BSA) for 20 min and incubated with rat anti-BrdU antibody (Abcam BU1/75, 1:200) in blocking buffer at 37 °C for 1 h. After three washes, with PBST, slides were incubated with Alexa Fluor 488-conjugated anti-rat (Molecular Probes, 1:200 dilution) for 45 min. After additional washes, the slides were incubated with mouse anti-BrdU (BD Biosciences, B44, 1:40) for 1 h and then washed once with high-salt PBST (0.5 m NaCl) and three times with PBST. Then the slides were incubated with Alexa Fluor 549-conjugated anti-mouse (Molecular Probes, 1:200 dilution) for 45 min. After three washes with PBST, the slides were mounted in SlowFade Gold antifade reagent (Invitrogen). The slides were imaged on a Zeiss Axiovert microscope with a 100× objective.
Immunostaining and Immunoblotting
U2OS or HCT16 cells were cultured on polylysine-coated coverslips 24 h before the experiments. After washing with PBS, the cells were pre-extracted with 0.5% Triton X-100 in CSK buffer (20 mm HEPES, pH 7.0, 100 mm NaCl, 300 mm sucrose, and 3 mm MgCl2). The cells were then washed three times with PBS and fixed with 3% paraformaldehyde for 10 min at room temperature. Before staining, the cells were permeabilized for 10 min with PBS, 0.5% Triton X-100, and washed three times. For EdU staining, the cells were incubated with Click-iT reaction buffer (PBS with 50 μm Alexa Fluor® 488 azide, 10 mm sodium ascorbate, and 2 mm CuSO4) for 30 min at room temperature. After washing, the cells were blocked with 5% BSA (Sigma) in PBS for 15 min. The primary antibodies were diluted in PBS containing 1% BSA and incubated with the cells for 90 min. After washing, secondary antibodies diluted in PBS containing 1% BSA were added to the cells for 30 min. The cells were washed three times and mounted with ProLong Gold antifade reagent with DAPI (Invitrogen). Images were acquired with an LSM710 confocal microscope (Zeiss) using a 100×/1.4 NA objector. For immunoblotting, primary antibodies were incubated for 1.5 h at room temperature in PBST containing 5% powder milk. Secondary peroxidase-coupled antibodies (Jackson ImmunoResearch) were incubated at room temperature for 45 min. ECL-based chemiluminescence was detected by using film. Primary antibodies were used at the following dilutions: RPA2/RPA32 (Bethyl, A300-244A, WB, 1:2000; IF, 1:500), ETAA1 (Abcam, ab192402, WB, 1:1000), H2AX-pS139 (Millipore, 05-636, IF, 1:5000), FLAG (MBL, M185–3L, WB, 1:2500; IF, 1:250).
QIBC
QIBC was performed as described previously (24). Briefly, images were acquired in an unbiased fashion with an Image Xpress Micro XL microscope (Molecular Devices) with a 10×/0.3 NA objective and a scientific CMOS camera. For every sample, 30–50 images were acquired, containing a total of 5000 to 10,000 cells per condition. After acquisition, the images were processed for automated analysis with the MetaXpress High Content Image Acquisition and Analysis software.
Cell Survival Assay
Cell survival curves for HCT116 cells treated with HU and CPT were generated as described previously (28). An appropriate number of cells were plated in 6-well plates and cultured for 24 h, and then the indicated dose of HU or CPT was added to the medium. After an additional 9–14 days of incubation, the colonies were stained with methylene blue and counted.
Author Contributions
S. F., Y. Z., Y. X., S. N., W. H., and M. H. conducted the experiments. G. G., J. J., R. G., and D. X. supervised the project. S. F., Y. X., S. N., W. H., G. G., J. J., R. G., and D. X. analyzed the data. S. F., R. G., and D. X. wrote the manuscript with input from the other authors.
Acknowledgments
We thank Weidong Wang and Lee Zou for their advice and revisions to the manuscript, Jun Huang for providing MBP-tagged RPA2- and RPA3-expressing plasmids, and Xingzhi Xu for providing the laser microirradiation system. We thank the mass spectrometry facility at the National Center for Protein Sciences, Peking University, for assistance with the identification of the proteins, and the Core Facility of Life Sciences, Peking University, for assistance with the imaging.
This work was supported in part by the National Basic Research Program of China Grant 2013CB911002 and the National Natural Science Foundation of China Grants 31271435 and 31370836. The authors declare that they have no conflicts of interest with the contents of this article.
- RPA
- replication protein A
- HU
- hydroxyurea
- MBP
- maltose-binding protein
- ATR
- ataxia telangiectasia and Rad3-related protein
- ssDNA
- single-stranded DNA
- IdU
- 5-iodo-2′-deoxyuridine
- QIBC
- quantitative image-based cytometry
- WB
- Western blot
- IF
- immunofluorescence
- CldU
- 5-chloro-2′-deoxyuridine
- CPT
- camptothecin.
References
- 1. Petermann E., and Helleday T. (2010) Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell Biol. 11, 683–687 [DOI] [PubMed] [Google Scholar]
- 2. Tercero J. A., Longhese M. P., and Diffley J. F. (2003) A central role for DNA replication forks in checkpoint activation and response. Mol. Cell 11, 1323–1336 [DOI] [PubMed] [Google Scholar]
- 3. Lambert S., and Carr A. M. (2005) Checkpoint responses to replication fork barriers. Biochimie 87, 591–602 [DOI] [PubMed] [Google Scholar]
- 4. Wold M. S. (1997) Replication protein A: a heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu. Rev. Biochem. 66, 61–92 [DOI] [PubMed] [Google Scholar]
- 5. Iftode C., Daniely Y., and Borowiec J. A. (1999) Replication protein A (RPA): the eukaryotic SSB. Crit. Rev. Biochem. Mol. Biol. 34, 141–180 [DOI] [PubMed] [Google Scholar]
- 6. Fanning E., Klimovich V., and Nager A. R. (2006) A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 34, 4126–4137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sogo J. M., Lopes M., and Foiani M. (2002) Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 [DOI] [PubMed] [Google Scholar]
- 8. Byun T. S., Pacek M., Yee M. C., Walter J. C., and Cimprich K. A. (2005) Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 19, 1040–1052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zou L., and Elledge S. J. (2003) Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science 300, 1542–1548 [DOI] [PubMed] [Google Scholar]
- 10. Jackson S. P. (2002) Sensing and repairing DNA double-strand breaks. Carcinogenesis 23, 687–696 [DOI] [PubMed] [Google Scholar]
- 11. Stauffer M. E., and Chazin W. J. (2004) Physical interaction between replication protein A and Rad51 promotes exchange on single-stranded DNA. J. Biol. Chem. 279, 25638–25645 [DOI] [PubMed] [Google Scholar]
- 12. Bansbach C. E., Bétous R., Lovejoy C. A., Glick G. G., and Cortez D. (2009) The annealing helicase SMARCAL1 maintains genome integrity at stalled replication forks. Genes Dev. 23, 2405–2414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ciccia A., Bredemeyer A. L., Sowa M. E., Terret M. E., Jallepalli P. V., Harper J. W., and Elledge S. J. (2009) The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 23, 2415–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Driscoll R., and Cimprich K. A. (2009) HARPing on about the DNA damage response during replication. Genes Dev. 23, 2359–2365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan J., Ghosal G., and Chen J. (2009) The annealing helicase HARP protects stalled replication forks. Genes Dev. 23, 2394–2399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gong Z., and Chen J. (2011) E3 ligase RFWD3 participates in replication checkpoint control. J. Biol. Chem. 286, 22308–22313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liu S., Chu J., Yucer N., Leng M., Wang S. Y., Chen B. P., Hittelman W. N., and Wang Y. (2011) RING finger and WD repeat domain 3 (RFWD3) associates with replication protein A (RPA) and facilitates RPA-mediated DNA damage response. J. Biol. Chem. 286, 22314–22322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Elia A. E., Wang D. C., Willis N. A., Boardman A. P., Hajdu I., Adeyemi R. O., Lowry E., Gygi S. P., Scully R., and Elledge S. J. (2015) RFWD3-dependent ubiquitination of RPA regulates repair at stalled replication forks. Mol. Cell 60, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maréchal A., Li J. M., Ji X. Y., Wu C. S., Yazinski S. A., Nguyen H. D., Liu S., Jiménez A. E., Jin J., and Zou L. (2014) PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 53, 235–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Childs E. J., Mocci E., Campa D., Bracci P. M., Gallinger S., Goggins M., Li D., Neale R. E., Olson S. H., Scelo G., Amundadottir L. T., Bamlet W. R., Bijlsma M. F., Blackford A., Borges M., et al. (2015) Common variation at 2p13.3, 3q29, 7p13, and 17q25.1 associated with susceptibility to pancreatic cancer. Nat. Genet. 47, 911–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Xu X., Vaithiyalingam S., Glick G. G., Mordes D. A., Chazin W. J., and Cortez D. (2008) The basic cleft of RPA70N binds multiple checkpoint proteins, including RAD9, to regulate ATR signaling. Mol. Cell. Biol. 28, 7345–7353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feldkamp M. D., Mason A. C., Eichman B. F., and Chazin W. J. (2014) Structural analysis of replication protein A recruitment of the DNA damage response protein SMARCAL1. Biochemistry 53, 3052–3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xie S., Lu Y., Jakoncic J., Sun H., Xia J., and Qian C. (2014) Structure of RPA32 bound to the N terminus of SMARCAL1 redefines the binding interface between RPA32 and its interacting proteins. FEBS J. 281, 3382–3396 [DOI] [PubMed] [Google Scholar]
- 24. Toledo L. I., Altmeyer M., Rask M. B., Lukas C., Larsen D. H., Povlsen L. K., Bekker-Jensen S., Mailand N., Bartek J., and Lukas J. (2013) ATR prohibits replication catastrophe by preventing global exhaustion of RPA. Cell 155, 1088–1103 [DOI] [PubMed] [Google Scholar]
- 25. Xing M., Yang M., Huo W., Feng F., Wei L., Jiang W., Ning S., Yan Z., Li W., Wang Q., Hou M., Dong C., Guo R., Gao G., Ji J., Zha S., Lan L., Liang H., and Xu D. (2015) Interactome analysis identifies a new paralogue of XRCC4 in non-homologous end joining DNA repair pathway. Nat. Commun. 6, 6233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Cong L., Ran F. A., Cox D., Lin S., Barretto R., Habib N., Hsu P. D., Wu X., Jiang W., Marraffini L. A., and Zhang F. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Davies S. L., North P. S., and Hickson I. D. (2007) Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 14, 677–679 [DOI] [PubMed] [Google Scholar]
- 28. Katsube T., Mori M., Tsuji H., Shiomi T., Shiomi N., and Onoda M. (2011) Differences in sensitivity to DNA-damaging agents between XRCC4- and Artemis-deficient human cells. J. Radiat. Res. 52, 415–424 [DOI] [PubMed] [Google Scholar]