Abstract
Homologous recombination is an important DNA repair pathway that plays key roles in maintaining genome stability. Escherichia coli RecA is an ATP-dependent DNA-binding protein that catalyzes the DNA strand exchange reactions in homologous recombination. RecA assembles into long helical filaments on single-stranded DNA, and these presynaptic complexes are responsible for locating and pairing with a homologous duplex DNA. Recent single molecule studies have provided new insights into RecA behavior, but the potential influence of ATP in the reactions remains poorly understood. Here we examine how ATP influences the ability of the RecA presynaptic complex to interact with homologous dsDNA. We demonstrate that over short time regimes, RecA presynaptic complexes sample heterologous dsDNA similarly in the presence of either ATP or ATPγS, suggesting that initial interactions do not depend on ATP hydrolysis. In addition, RecA stabilizes pairing intermediates in three-base steps, and stepping energetics is seemingly unaltered in the presence of ATP. However, the overall dissociation rate of these paired intermediates with ATP is ∼4-fold higher than with ATPγS. These experiments suggest that ATP plays an unanticipated role in promoting the turnover of captured duplex DNA intermediates as RecA attempts to align homologous sequences during the early stages of recombination.
Keywords: ATP, DNA recombination, DNA repair, homologous recombination, single-molecule biophysics
Introduction
Homologous recombination allows for the regulated exchange of genetic information between two different DNA molecules of identical or nearly identical sequence composition (1–4). Homologous recombination contributes to double-strand DNA break repair, the rescue of stalled or collapsed replication forks, programmed and aberrant chromosomal rearrangements, horizontal gene transfer, and meiosis (5–9). The importance of homologous recombination is underscored by its broad conservation across all kingdoms of life and the findings that defects in key recombination proteins can result in a loss of genome integrity and lead to gross chromosomal rearrangements that are a hallmark of cancer in eukaryotes.
Many of the key reactions in homologous recombination are catalyzed by the Rad51/RecA family of DNA recombinases (1–4). These recombinases are broadly conserved, and prominent family members include bacterial RecA, the archeal protein RadA, and the eukaryotic recombinases Rad51 and Dmc1. RecA is the archetypal recombinase originally identified in genetic screens for Escherichia coli mutants defective in recombination (10), and much of our current understanding of recombination mechanisms can be attributed to studies of bacterial RecA (4).
During recombination, RecA bound to the presynaptic ssDNA must first locate a homologous duplex DNA template. This process is referred to as the homology search, and it is conceptually similar to the target searches of other site-specific DNA-binding proteins (11–14). Once aligned, the presynaptic ssDNA can be paired with the complementary strand of a homologous dsDNA, resulting in the displacement of the non-complementary strand from the duplex to generate a D-loop intermediate (1–3). This intermediate can then be channeled through a number of related pathways, any of which can allow for the repair of the originally broken DNA molecule using information derived from the template (5, 15, 16).
Rad51/RecA family members are ATP-dependent DNA-binding proteins that form extended helical filaments on DNA (1–4). The bound DNA is extended by ∼50% relative to the contour length of B-form DNA, and crystal structures of RecA-ssDNA and RecA-dsDNA pre- and postsynaptic complexes reveal that the bound DNA is organized into near B-form base triplets separated by ∼8 Å between adjacent triplets (17). We have referred to this unique DNA architecture as RS-DNA (Rad51/RecA stretched-DNA) to help distinguish it from other forms of mechanically stretched DNA and as a reflection of its unique mechanistic implications.
RecA is a fairly robust ATPase, exhibiting a catalytic rate constant (kcat) of ∼30 min−1 when bound to ssDNA, and this value drops to ∼20 min−1 when homologous dsDNA is present (18). However, RecA is still capable of supporting the homology search and strand invasion in the absence of active ATP hydrolysis (19–21). This observation raises the question of why the nucleoprotein filament seems to hydrolyze so much ATP under normal reaction conditions (18). There are currently at least two mechanistic explanations for why RecA hydrolyzes ATP. First, ATP hydrolysis is intimately linked to end-dependent disassembly of the nucleoprotein filament (22); hence a primary role of ATP is to promote dissociation of RecA from DNA once strand invasion is completed. Second, ATP hydrolysis is required for RecA to bypass sequence heterology during strand invasion (21, 23), although the mechanism by which this takes place remains poorly understood. It remains unknown whether ATP hydrolysis by RecA may play other as yet unidentified roles during recombination.
To help better understand the mechanisms of genetic recombination, we have established a single-molecule DNA curtain assay for studying how presynaptic complexes interact with duplex DNA during the early stages of homologous recombination. Our work reveals the existence of two distinct time regimes for dsDNA interrogation (24). Duplex DNA sequences bearing fewer than 8 bp of microhomology are rapidly sampled and rejected within seconds. In contrast, dsDNA molecules bearing 8 bp of microhomology are more tightly bound, and the resulting intermediates exhibit single exponential kinetics with lifetimes corresponding to several minutes. Increasing the length of microhomology from 8 to 15 bp in single-nucleotide increments reveals that prominent changes of lifetimes in 3-bp increments. This finding suggests that strand exchange takes place in 3-bp steps, with each step exhibiting a characteristic energetic signature that is broadly conserved among the Rad51/RecA family members (25).
Our previous experiments with RecA were restricted to reactions using ATPγS (24, 25).2 Therefore an unresolved question relates to the potential role(s), if any, of ATP hydrolysis in the search for homology, strand invasion, or both. Here we examine RecA-ssDNA presynaptic complexes in the presence of ATP, allowing us to observe dsDNA sampling and capture under conditions where the presynaptic complex can hydrolyze ATP. Our results show that ATP hydrolysis has no apparent impact upon the short time regime sampling of duplex DNA molecules. However, we find that ATP hydrolysis specifically enhances turnover of early recombination intermediates bound by short tracts of microhomology 8–15 bp in length. These findings suggest an unanticipated role for ATP hydrolysis in the homology search.
Results
Visualizing RecA Presynaptic Filaments with ssDNA Curtains
We used ssDNA curtains and total internal refection fluorescence microscopy to visualize RecA presynaptic complexes (Fig. 1A). The ssDNA substrate was generated using M13 as a template for rolling circle replication and then anchored to a lipid bilayer within a microfluidic chamber through a biotin-streptavidin linkage and aligned along chromium barriers by application of hydrodynamic force, as previously described (24–26). The ssDNA unravels when incubated with RPA-eGFP, and the downstream ends of the RPA-ssDNA are anchored to exposed chromium pedestals through nonspecific adsorption. These assembly stages of the experiment rely upon the use of eukaryotic RPA because RPA does not extensively compact ssDNA, allowing preparation of double-tethered ssDNA curtains. In contrast, bacterial SSB causes extensive ssDNA compaction in our assays (27), and we have not yet established conditions under which we can readily observe double-tethered ssDNA molecules in the presence of SSB.
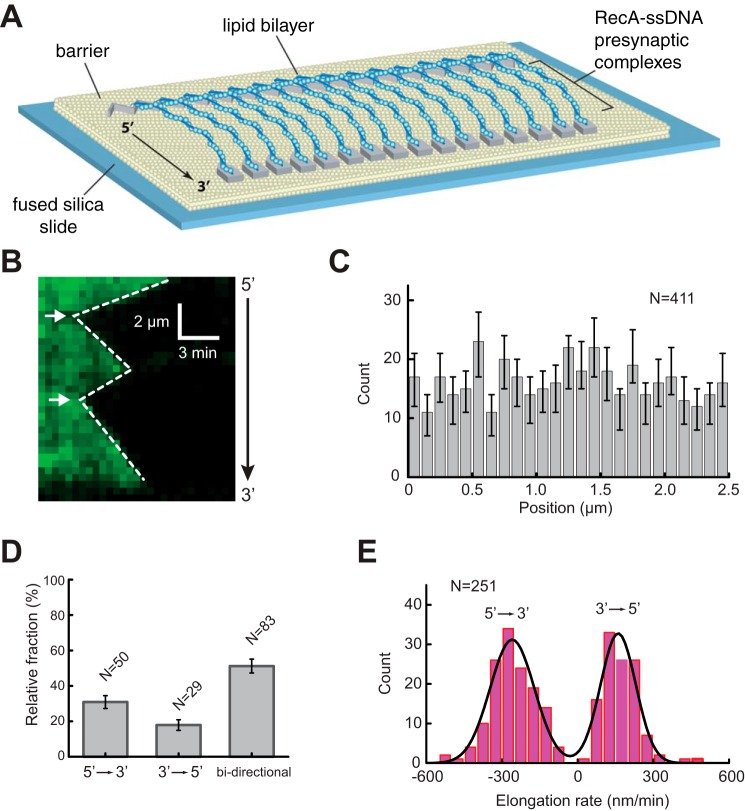
FIGURE 1.
RecA presynaptic complex assembly. A, schematic of double-tethered ssDNA curtain assay with RecA presynaptic complexes. B, representative kymograph showing the displacement of RPA-eGFP by unlabeled RecA. White arrowheads highlight the RecA filament nucleation sites for this particular ssDNA molecule. C, histogram showing the relative positions of observed nucleation events along one unit length of the M13 ssDNA substrate. The error bars represent 70% confidence intervals obtained through bootstrap analysis. D, fractions of filaments that exhibited 5′ → 3′, 3′ → 5′, and bi-directional elongation. The error bars represent standard deviation. E, observed RecA filament elongation rates for 5′ → 3′ and 3′ → 5′ assembly, as indicated. The black line corresponds to a double Gaussian fit to the data.
We have previously reported experiments using E. coli RecA with the ssDNA curtains (24, 25). However, we could only observe stable RecA presynaptic complexes in the presence of the slowly hydrolyzed ATP analog ATPγS. It is possible that this limitation arises from our reliance upon RPA-eGFP during preparation of the ssDNA curtains. To overcome this problem, we sought to nucleate filament assembly in the presence of ATPγS and used ATP during the elongation phase of the reaction. This strategy was based upon the knowledge that nucleation is the rate-limiting step in RecA filament assembly and that ATPγS stimulates the rate of nucleation ∼5-fold relative to ATP (28, 29). In addition, free RecA was included in all reaction buffers to help ensure rapid replenishment of any RecA monomers that might dissociate from the filament during the observation windows. Under these conditions, presynaptic filament assembly is revealed as the loss of the RPA-eGFP signal as unlabeled RecA displaced the fluorescent proteins from the anchored ssDNA (Fig. 1B).
Characteristics of the ATP-mediated Assembly Reaction
Observation of the reactions revealed that RecA filament assembly was initiated by distinct nucleation events followed by rapid displacement of RPA-GFP as the RecA filaments covered the ssDNA (Fig. 1B). The location of the nucleation events appeared to be random (Fig. 1C), although we do not rule out the possible existence of preferred nucleation sites below our resolution limits. Interestingly, some of the filaments assembled in the 5′ → 3′ direction (∼30%, n = 50) and others in the 3′ → 5′ direction (∼18%, n = 29), but the largest fraction of the population exhibited bi-directional assembly (∼52%, n = 83) (Fig. 1D). Future work will be necessary to clarify these directional differences in elongation, but we speculate that the unidirectional filaments either have a defect at one end or may have encountered an obstacle on the ssDNA, such as small secondary structures or residual RPA that cannot be displaced. Analysis of the elongation rates revealed that filament elongation in the 5′ → 3′ direction was ∼70% faster than the rates observed for 3′ → 5′ elongation, yielding values of 259 ± 7.2 nm/s and 161 ± 6.1 nm/s, respectively at 37 °C (Fig. 1E). Together, these observations are consistent with previous single molecule imaging studies of RecA presynaptic complex assembly on SSB-coated ssDNA, which have also revealed bi-directional assembly that was faster in the 5′ → 3′ direction relative to the 3′ → 5′ (28–30). In addition, previous reports have also yielded pronounced phases of unidirectional assembly, which is also broadly consistent with our findings (28).
Although there are undoubtedly important differences between RecA presynaptic complex assembly on SSB-ssDNA (the physiological substrate) versus RPA-ssDNA (a non-physiological substrate), our findings suggest that the general characteristics of the assembly process are largely unaltered in these in vitro reaction conditions. From this, we infer that the presynaptic complexes prepared under these conditions can be used to probe the dsDNA binding properties of RecA in the presence of ATP.
RecA Turnover and Filament Stability
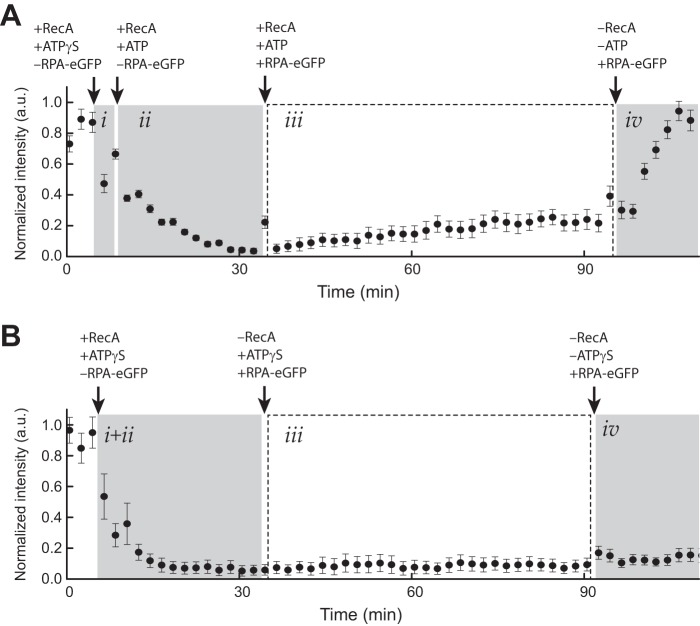
A crucial component of our dsDNA binding assay is the requirement that the presynaptic complexes remain intact over the time course of the observations, even if individual protein monomers exchange between free and bound states. To assess stability, RecA presynaptic complexes were assembled on ssDNA, as described above, and then observed over time in the presence of 1 mm ATP, 50 nm free RecA, and 0.2 nm free RPA-eGFP (Fig. 2A). Extensive dissociation of RecA from the ssDNA in the absence of replacement by free RecA in solution would be revealed as the appearance of RPA-eGFP on the ssDNA (24). These experiments revealed that the RecA remained intact for at least 60 min in the presence of 1 mm ATP (Fig. 2A). However, RecA rapidly dissociated from the ssDNA when ATP was replaced with buffer containing no nucleotide as evidenced by the recovery of the RPA-eGFP signal on the ssDNA (Fig. 2A), confirming that the continued presence of ATP was necessary to maintain presynaptic stability. In contrast, the RPA-eGFP signal was never recovered within our observation time windows when RecA filaments were assembled in the presence of ATPγS as the only available nucleotide cofactor (Fig. 2B).
FIGURE 2.
RecA presynaptic complex stability. A, graphical depiction of RPA-eGFP signal intensity (a.u.) averaged over 11 ssDNA molecules during assembly of the RecA presynaptic filament in the presence of ATP. Panel i corresponds to the brief nucleation phase conducted in the presence of 1 mm ATPγS and the absence of free RPA-eGFP. Panel ii shows nucleation was followed by filament elongation with 1 mm ATP, 50 nm RecA in the absence of free RPA-eGFP. In panel iii, the sample chamber was then flushed with buffer containing 1 mm ATP, 50 nm free RecA, and 0.2 nm free RPA-eGFP, which was used to monitor RecA dissociation from the ssDNA. Panel iv, finally, the sample chamber was flushed with buffer with no ATP and no free RecA causing the bound RecA to dissociate from the ssDNA, which is revealed as the reappearance of the RPA-eGFP signal. The error bars represent standard deviation measured from 11 individual ssDNA molecules. B, graphical depiction of RPA-eGFP signal intensity (a.u.) averaged over 11 ssDNA molecules during assembly of the RecA presynaptic filament in the presence of only ATPγS. Reactions were performed as in A with the exception that 1 mm ATPγS was substituted for ATP in all stages of the reaction.
Importantly, these assays reveal that the presynaptic complexes are stable in the presence of ATP, but they are not intended to address whether individual proteins within the filament are undergoing turnover between free and bound states. A requirement for free RecA (≥50 nm) to be present in solution in reactions with ATP strongly suggests that individual proteins within the filaments are in dynamic equilibrium between free and bound states, but future work would be necessary to fully evaluate this process. We conclude that the RecA presynaptic complexes prepared in the presence of ATP remain intact over time windows necessary to probe interactions with dsDNA substrates, even though the protein monomers within the filament may be subject to continual turnover.
ATP Hydrolysis Increases the Rate of dsDNA Turnover
We have previously used DNA curtain assays to show that RecA and related recombinases are capable of stably capturing small dsDNA fragments bearing internal tracts of microhomology that are complementary to sequences within presynaptic ssDNA (24, 25). We have also demonstrated that increases in binding energy occur in 3-bp increments, indicating that duplex DNA recognition during these early stages of D-loop formation occurs in 3-bp increments, consistent with the base triplet structural organization of RS-DNA (24, 25). We have previously hypothesized that base triplet stepping was driven by the structure of RS-DNA itself, which could imply that there might be little difference between observations made with ATP versus ATPγS. However, all of our prior experiments with RecA were limited to conditions using ATPγS (24, 25), potentially obscuring any possible role for ATP hydrolysis in these reactions.
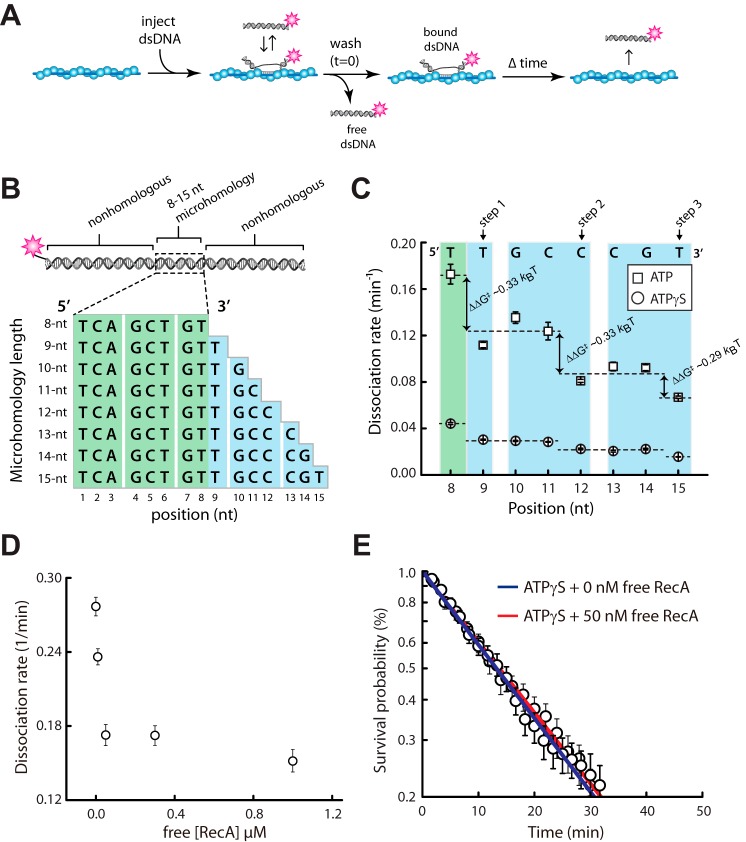
To assess duplex DNA binding, RecA presynaptic complexes were assembled in the presence of ATP, as described above, and then 70-bp dsDNA oligonucleotides labeled at one end with a single Atto565 fluorophore were injected into the sample chamber (Fig. 3A). The dsDNA oligonucleotides harbored a tract of microhomology ranging from 8- to 15-bp in length, which was complimentary to a single location on the presynaptic ssDNA (Fig. 3B) (24, 25). The dsDNA fragments were incubated with the presynaptic complexes for 10 min, unbound dsDNA was quickly flushed from the sample chamber, and the lifetimes of the bound dsDNA was determined by monitoring the reactions over time. Remarkably, these experiments revealed an ∼4-fold increase in the overall rate of dsDNA turnover in reactions conducted with ATP relative to those preformed in ATPγS (Fig. 3C). Despite the overall increase in dissociation rates, the pattern of binding was still consistent with a base triplet stepping mechanism, and each individual step was still characterized by a free energy change (ΔΔG‡) of ∼0.3 kBT (Fig. 3C), which was essentially indistinguishable from values obtained for RecA in the presence of ATPγS (25). Together, these observations suggest that the ability of RecA to hydrolyze ATP promotes the turnover of these metastable binding intermediates, without impacting the relative change in free energy that is observed from each triplet step.
FIGURE 3.
Influence of ATP on base triplet recognition by RecA. A, experimental schematic for measuring dsDNA binding by the RecA presynaptic complex. B, schematic representation of the 70-bp dsDNA substrates used in the binding assays. The dsDNA is labeled at one end with a single Atto565 fluorophore and contains a single internal tract of microhomology ranging from 8 to 15 nt in length, as indicated, which are targeted to only one site on the M13 presynaptic ssDNA. C, dissociation rate data for oligonucleotides bearing different lengths of microhomology in reactions with either 1 mm ATP or 1 mm ATPγS; note that the data for experiments with ATPγS have been previously published (25) and are reproduced here for direct comparison with the ATP data. D, dissociation rate data for the dsDNA substrate bearing a single 8-bp tract of microhomology in the presence of varying concentrations of free RecA. E, dissociation rate data for reactions with the dsDNA substrate bearing a single 8-bp tract of microhomology in the presence of 1 mm ATPγS with and without 50 nm free RecA.
Effect of Free RecA on dsDNA Dissociation
One important difference between the reactions with ATP versus ATPγS was the presence of 50 nm free RecA in the ATP-containing reactions (Fig. 3C). Therefore we next sought to establish the extent to which the presence of free RecA influenced dsDNA release from the presynaptic complex. First, we determined the dissociation rates for the dsDNA substrate bearing a single 8-bp tract of microhomology in the presence of ATP and varying concentrations of free RecA. These experiments revealed that dsDNA dissociation rates were more rapid at either 0 or 10 nm free RecA, which we attribute to dissociation of RecA from the ssDNA, and the dissociation rates plateau from 50 nm to 1 μm free RecA (Fig. 3D). We conclude that the addition of a large excess of free RecA has no appreciable impact on the release of bound dsDNA intermediates.
To rule out the possibility that the free RecA alone was responsible to differences in dissociation kinetics, we next tested the effect of free RecA in reactions with ATPγS. Comparison of dsDNA dissociation rates for reactions with ATPγS and either 0 nm free RecA or 50 nm free RecA revealed no significant effect of the free protein under these conditions (Fig. 3E). This finding argues against the possibility that the presence of free RecA affects the difference in dissociation rates for reactions with ATP versus ATPγS.
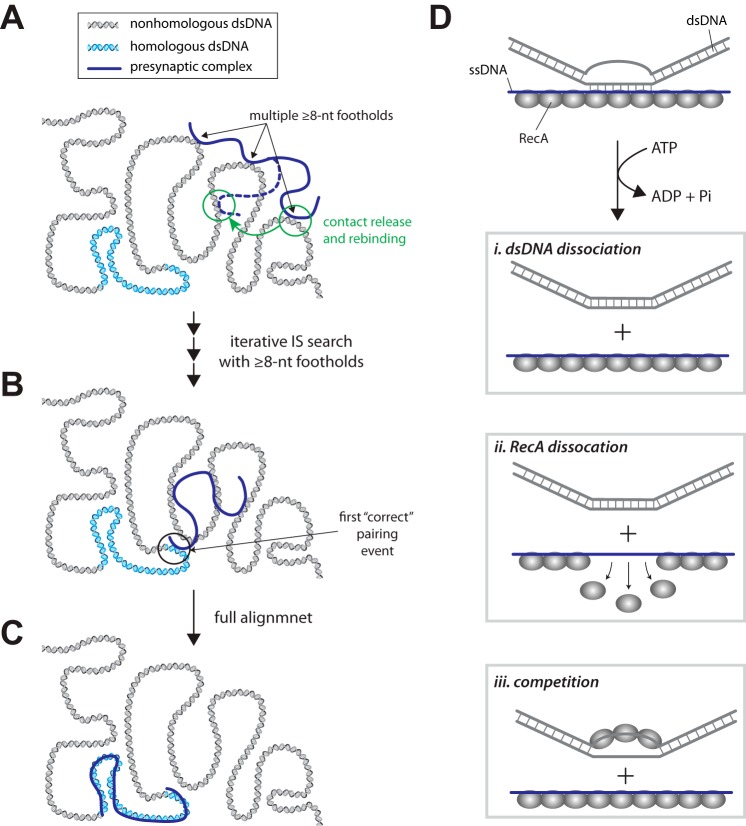
Increased dsDNA Turnover Is Not Due to Filament Dissociation
One possible explanation for the more rapid dsDNA turnover is that RecA itself was dissociating from the ssDNA and provoking concurrent release of the bound dsDNA molecules. To test this possibility we first measured the dissociation rate of RecA from the ssDNA when quickly chased with buffer containing no nucleotide. These experiments revealed an apparent dissociation rate constant of k = 0.38 ± 0.08 min−1 (Fig. 4A). We next incubated RecA presynaptic complexes with 70-bp duplex DNA oligonucleotides bearing either 8- or 15-bp of microhomology and then measured the lifetimes of the bound dsDNA after quickly flushing the sample chambers with buffer containing no nucleotide. As shown in Fig. 4 (B and C), both oligonucleotides quickly dissociated from the presynaptic complex with nearly identical kinetics when ATP was removed from the reactions, and the observed dissociation kinetics were essentially indistinguishable from the RecA dissociation rate from ssDNA (Fig. 4A). On the other hand, in the presence of ATP, the dsDNA dissociation was slower than in the absence of ATP. The dissociation rates of microhomologies are distinguishable (Figs. 3C and 4, B and C). When RecA dissociates internally from the presynaptic complex, the dissociation occurs stochastically at random places, and several RecA molecules might dissociate at one time. In our experiments, short tracks of microhomologies were tested, and at most five RecA molecules were involved. If the dsDNA dissociation is dominated by the random internal RecA dissociation, we would not observe the well defined stepwise behavior in Fig. 3C. Therefore we suggest that the dsDNA binding intermediates probed by our experiments require the continued presence of RecA for stability and that the rate constants measured for dsDNA dissociation from intact filaments reflect a process that is distinct from that which takes place when RecA itself dissociates from the ssDNA. These findings argue that the ∼4-fold increase in dsDNA dissociation rates observed in the presence of ATP is not the result of protein turnover but instead arises from some other ATP-dependent change in the nucleoprotein filament.
FIGURE 4.
Duplex DNA release upon removal of ATP. A, plot showing the normalized RPA-eGFP signal intensity when preassembled RecA presynaptic complexes are chased with buffer containing no nucleotide cofactor. The increased signal intensity reflects dissociation of unlabeled RecA from the ssDNA and its replacement by RPA-eGFP. B, survival probability plot for the 70-bp dsDNA harboring a single 8-bp microhomology insert when chased with buffer containing either 1 mm ATP or no nucleotide cofactor. C, survival probability plot for the 70-bp dsDNA harboring a single 15-bp microhomology insert when chased with buffer containing either 1 mm ATP or no nucleotide cofactor.
No Discernable Difference in the Turnover of Fully Homologous Substrates
The dsDNA substrates described above mimic early stages of strand exchange when the presynaptic complex has captured short tracts of microhomology embedded within an otherwise nonhomologous sequence. Using the same approach, we next sought to determine whether there was a measurable difference in the dissociation rates for perfectly matched 70-bp substrates in reactions with either ATP or ATPγS. These experiments demonstrate that the fully homologous 70-bp substrate remained tightly bound to the RecA presynaptic complex under both conditions (not shown), revealing dissociation rates of 0.0130 ± 0.0028 min and 0.0138 ± 0.0013 min−1 for reactions with ATP and ATPγS, respectively (not shown). These findings indicate that once bound, the fully paired 70-bp strand exchange intermediates are similarly stable, regardless of which nucleotide cofactor is used in the reactions.
Mismatch Discrimination Is Not Affected by ATP Hydrolysis
We have previously shown that RecA presynaptic complexes assembled in the presence of ATPγS can discriminate against terminal base triplets bearing a single mismatched nucleotide regardless of mismatch position or identity (25). In addition, we have shown that in the presence of ATPγS, RecA is capable of stepping over mismatched triplets located at internal positions within a tract of microhomology. Interestingly, previous bulk biochemical work has demonstrated that ATP hydrolysis is required to bypass heterologous insertions (21, 23), suggesting that the ability to hydrolyze ATP may alter interactions with imperfectly matched DNA sequences (18, 31). To explore this hypothesis further, we next sought to determine whether ATP influenced the ability of RecA to discriminate against single base mismatches at the level of an individual base triplet.
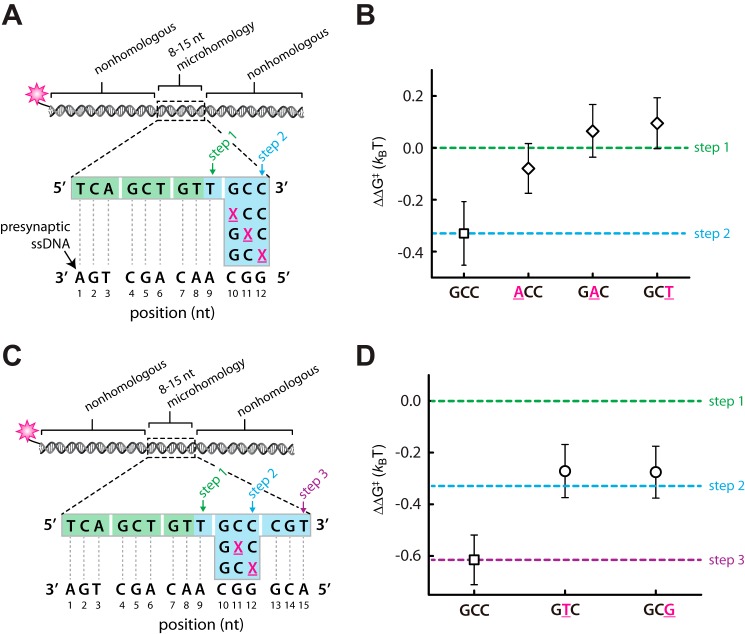
We first tested a series of oligonucleotides in which a single mismatch was marched across the 3′ terminal base triplet of an internal 12-bp tract of microhomology (Fig. 5A) (25). As expected, in the absence of a mismatch, this substrate yields a binding free energy consistent with having taken two base triplet steps (Fig. 5B). In contrast, a single base mismatch located at any position within the terminal triplet appeared to abolish any increase in binding free energy that could be attributed to this terminal base triplet. This observation is consistent with the conclusion that the mismatched terminal base triplet remains unpaired within the observed binding intermediates. We next asked whether RecA could step over an internal base triplet bearing a single mismatch. For these experiments, mismatches were introduced into a single base triplet with a 15-bp tract of microhomology (Fig. 5C). In the absence of a mismatch, this substrate yields a binding free energy consistent with having taken three base triplet steps; however, a single mismatch at either the 11th or 12th nucleotide position reduces the observed binding free energy by an amount consistent with loss of one triplet pairing interaction (Fig. 5D). It should be noted that we cannot test mutations at the 10th position with this set of oligonucleotides because mutations at this position target introduce new tracts of microhomology targeted to other regions of the presynaptic ssDNA, which would confound data analysis. These results are all similar to previous observations made for RecA presynaptic complexes prepared with only ATPγS (25). We conclude that ATP hydrolysis has no perceptible effect on mismatch discrimination by the RecA presynaptic complex at the level of individual base triplets, and although ATP promotes bypass of longer regions of heterology, it does not seem to impact discrimination against individual mispaired base triplets.
FIGURE 5.
Mismatch discrimination in the presence of ATP. A, schematic illustration of a dsDNA substrate harboring a 12-bp microhomology insert in which the 3′ most terminal base triplet has single nucleotide mismatches (indicated by an underlined magenta X) at each of the three possible positions. B, corresponding dissociation rate data expressed as the relative difference in binding free energy (ΔΔG‡) compared with a substrate harboring a 9-bp microhomology insert (step 1). C, schematic of a dsDNA substrate harboring a 15-bp microhomology insert in which the 3′ most penultimate base triplet has single nucleotide mismatches (indicated by an underlined magenta X) at one of two different positions. D, corresponding dissociation rate data expressed as the relative difference in binding free energy (ΔΔG‡) compared with a substrate harboring either a 9-bp (step 1), 12-bp (step 2), or 15-bp (step 3) microhomology insert.
Duplex DNA Sampling in the Presence of ATP
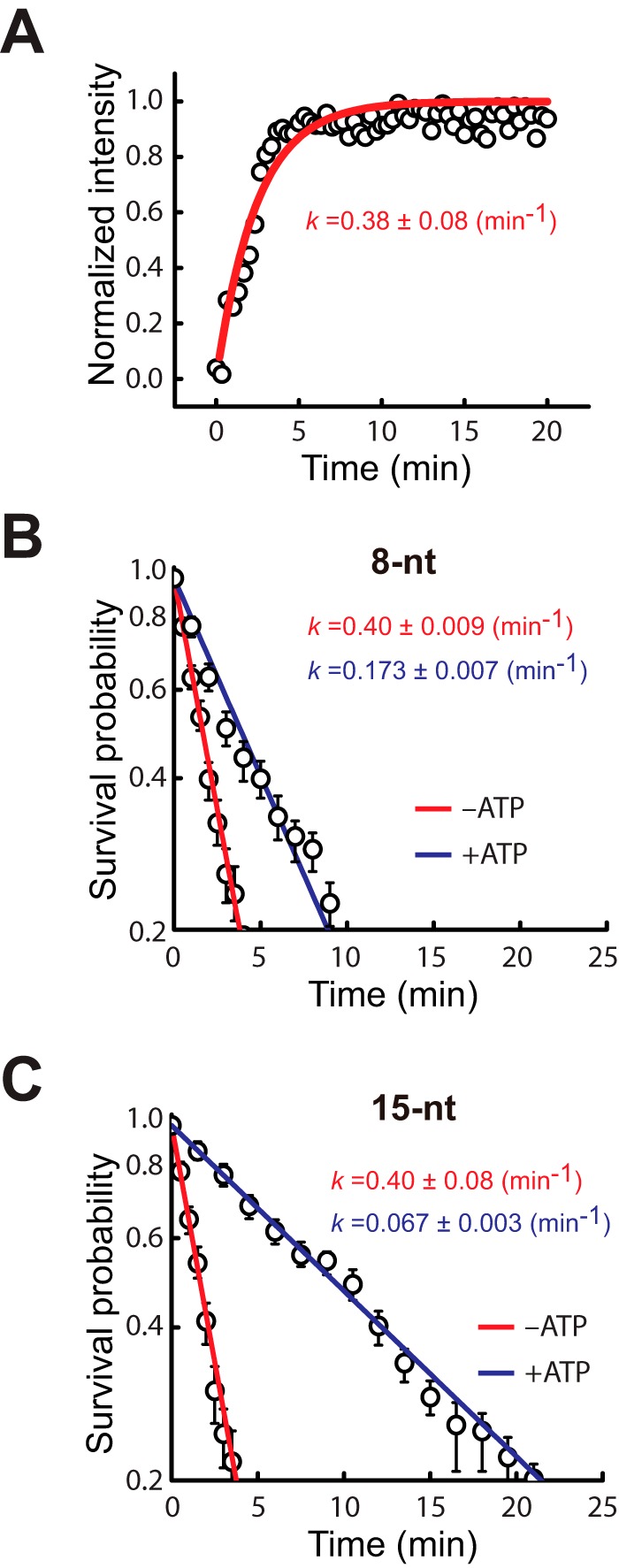
We have previously shown that heterologous dsDNA lacking tracts of microhomology ≥8 bp in length are sampled and rejected over very short time scales, and dsDNA bearing ≥8-bp tracts of microhomology are also sampled through a similar process when observed over short time scales (24). In both instances, the observed transient intermediates display power law distributed kinetics, indicating that the transient sampling events cannot be ascribed to a single conformational state but rather reflect the existence of a highly diverse ensemble of states with a correspondingly broad distribution of dissociation rates. The physical basis for this power law dependence can be rationalized by considering the vast number of potential intermediates even in our highly simplified system. If one assumes recognition involving 8-nt sequence motifs, then a 70-bp dsDNA can be misaligned with a total of 453,652 distinct sites on M13mp18, each of which can potentially give rise to energetically distinct states based on differences in sequence composition (24).
We next sought to determine whether the initial sampling of dsDNA by the RecA presynaptic complex was affected by ATP. To monitor initial dsDNA sampling, fluorescently tagged dsDNA substrates were injected into a sample chamber containing RecA presynaptic complexes that had been assembled in the presence of ATP (Fig. 6A), and videos were collected at a 60-millsecond frame rate with continuous laser illumination, as previously described (24, 25). These experiments revealed that all duplex DNA substrates bearing tracts of microhomology ranging from 8- to 15-bp in length exhibited power law sampling behavior when visualized over the short time scale regimes (not shown). Most importantly, side by side comparison of dsDNA substrates bearing <8-bp of microhomology reveals no difference in transient sampling kinetics for reactions conducted with either ATP or ATPγS (Fig. 6B). These results indicate that the presence of ATP, instead of ATPγS, has no perceptible impact on the short time regime sampling of heterologous dsDNA sequences, suggesting that kinetic discrimination against short tracts of microhomology (≤7 bp) during the homology search is not affected by ATP hydrolysis.
FIGURE 6.
Transient dsDNA sampling over short time regimes. A, schematic illustration of experiment for monitoring real time sampling of fluorescent dsDNA fragments by the RecA presynaptic complex. B, comparison of data obtained for a 70-bp dsDNA fragment harboring only a 7-bp microhomology insert for reactions conducted in the presence of either 1 mm ATP or 1 mm ATPγS, as indicated. Dashed red lines correspond to a power law fit to the resulting data.
Strand Exchange Kinetics with ATP versus ATPγS
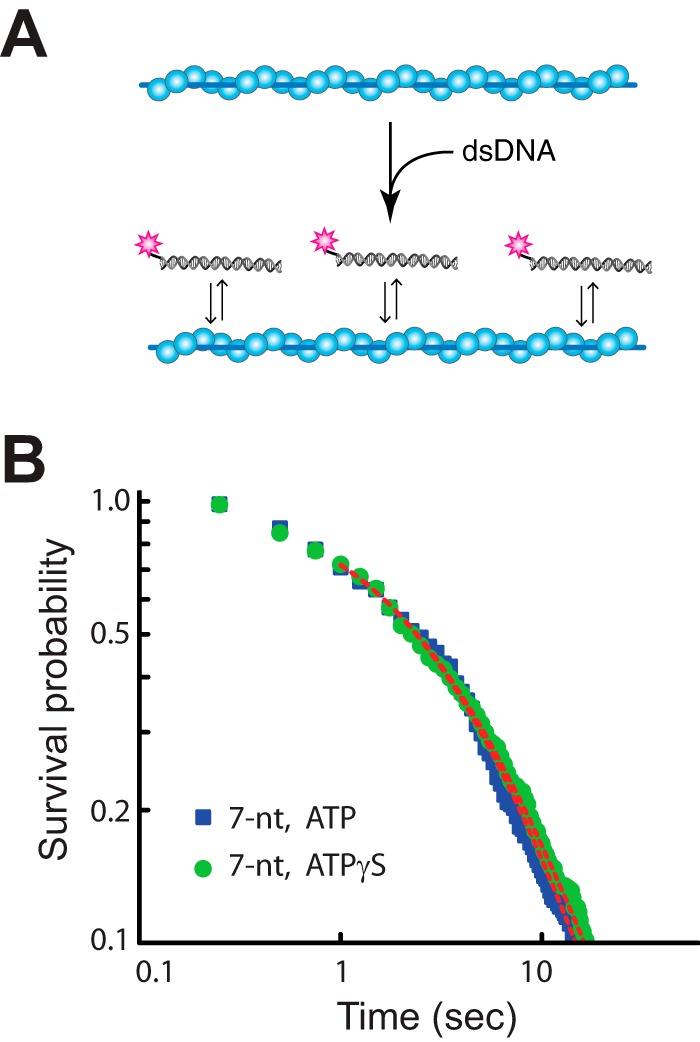
Our single molecule experiments reveal that the ability of RecA to hydrolyze ATP promotes the turnover of short (8–15 bp) strand exchange intermediates by a factor of ∼4 relative to reactions with ATPγS. These findings suggest the possibility that ATP hydrolysis might enhance the homology search by promoting turnover of misaligned intermediates trapped at tracts of microhomology. One prediction from this hypothesis is that the rate of product formation in bulk biochemical strand exchange assays should be faster with ATP compared with reactions with ATPγS. To test this prediction, we conducted strand exchange assays using circular φX174 ssDNA and linearized φX174 dsDNA. In additional to the single correct alignment register, the circular φX174 ssDNA can also be misaligned with the linearized φX174 dsDNA at over 1000 distinct tracts of microhomology ≥8 bp in length, each of which has the potential to slow down the homology search process as RecA sorts through these sequences (Fig. 7A). This graphical representation of the microhomology content of the φX174 substrates provides a good illustration of the challenge faced by RecA when trying to correctly align DNA sequences even in this simple in vitro assay. As predicted, the reactions performed with ATP yielded strand exchange products more rapidly than reactions performed with ATPγS, corresponding to a ∼3.4-fold difference in the apparent rate of product formation (Fig. 7, B and C). We conclude that RecA strand exchange products are formed more rapidly in reactions performed with ATP compared with reactions performed with ATPγS. These differences in product formation for reactions with ATP versus ATPγS are in agreement with previously published findings (23, 32), suggesting that the presence of ATP somehow promotes the overall efficiency of the strand exchange reaction. These findings, together with our single molecule results, suggest a model in which ATP hydrolysis by RecA enhances the turnover of short tracts of misaligned microhomology, which may in turn enhance the ability of the presynaptic complex to correctly align and pair with the dsDNA template.
FIGURE 7.
Strand exchange kinetics with ATP and ATPγS. A, microhomology analysis of the φX174 plasmid DNA substrates showing the number of potential sites (count) at which a given length of φX174 ssDNA microhomology, ranging from 5 to 12 bp, can be misaligned with the φX174 dsDNA. Numbers above the bar graph are the exact number of microhomology tracts of each given length. B, RecA strand exchange assays conducted with either ATP or ATPγS, as indicated. DNA products are labeled as jm (joint molecule), nc (nicked circle), and lds (linear double-stranded DNA). C, quantitation of total strand exchange products (jm + nc) for reactions conducted with either ATP or ATPγS. The error bars represent standard deviation based on three separate experiments. The rates of product formation are obtained by fitting the data with linear functions (black dashed lines).
Discussion
We have established single molecule DNA curtain methods for probing the properties of recombination intermediates, in particular, the presynaptic complex and its interactions with duplex DNA substrates. Here we examine how the RecA presynaptic complex interacts with dsDNA in the presence of ATP and compare these findings to reactions with ATPγS. Previous studies have demonstrated that ATP hydrolysis promotes the disassembly of RecA filaments after the completion of strand exchange, and ATP hydrolysis also enables the bypass of heterology during strand invasion (18–20, 22, 23). Our work suggests that ATP hydrolysis may also play an unanticipated role in the homology search process by promoting more rapid turnover of short tracts of microhomology from the presynaptic complex.
ATP Hydrolysis and the Homology Search
Our work demonstrates that the reactions that take place during the early stages of duplex DNA interrogation and capture by the RecA presynaptic complex are qualitatively similar in the presence of ATP or the slowly hydrolyzed analog ATPγS. Similarities between the reactions include (i) transient sampling over short time regimes through a mechanism that follows power law distributed kinetics, (ii) the need for 8 bp of microhomology for initial stable capture, and (iii) 3-bp stepping behavior consistent with the structure of RS-DNA. The only difference we find is that the presence of ATP promotes the turnover of duplex DNA fragments bound to the RecA presynaptic complex through 8–15-bp tracts of microhomology. Interestingly, ATP has no impact on initial rapid phase of the search that occurs over short time scales, nor does it appear to affect turnover of a fully homologous 70-bp substrate, indicating that its effects on duplex DNA turnover are confined to the metastable intermediates that form at captured tracts of microhomology.
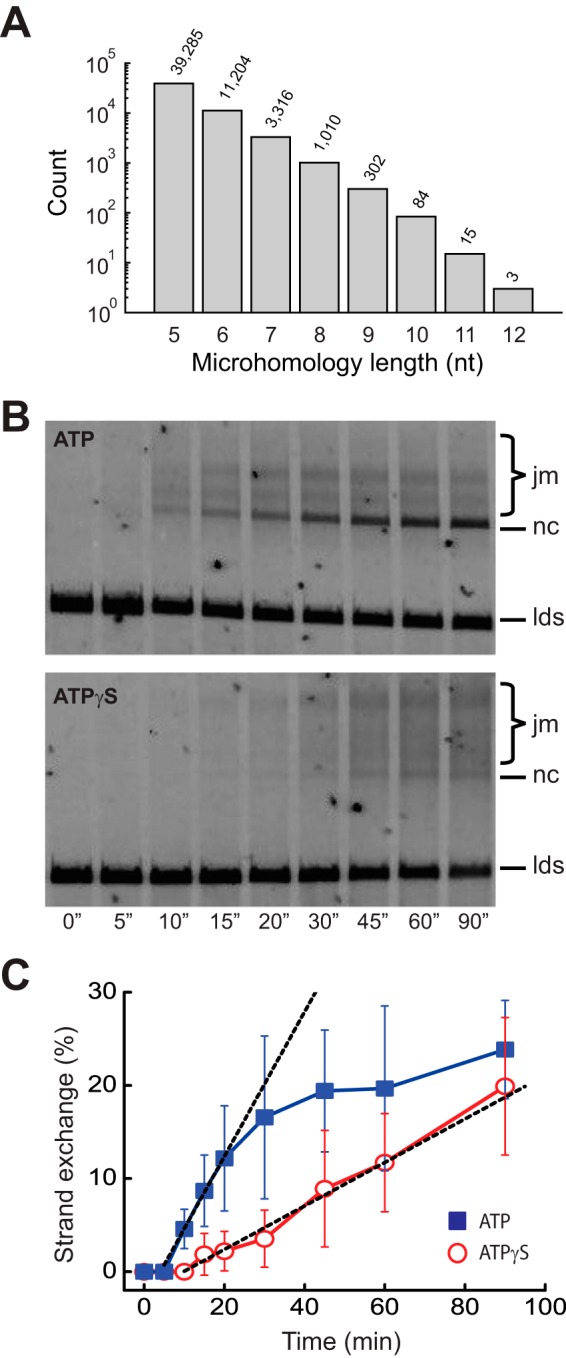
These findings have specific implications for how ATP hydrolysis might contribute to DNA sequence alignment by RecA during the homology search (Fig. 8). Several models have been proposed for the homology search by RecA (33–35). On the other hand, our overall model for the homology search is based upon the premise that the RecA presynaptic complex will rapidly engage duplex DNA and undergo an overall search process characterized by intersegmental transfer, which takes advantage of the polyvalent nature of the RecA presynaptic complex (Fig. 8A) (36). We propose that as this search takes place, the presynaptic complex establishes multiple ≥8-bp footholds mediated by Watson-Crick pairing interactions at short tracts of microhomology within the dsDNA substrate. Iterative binding and release events involving ≥8-bp footholds allow the presynaptic complex to sample multiple regions of the duplex DNA by intersegmental transfer (Fig. 8, A and B). Our data suggest that the turnover of these “microhomology footholds” is the step during the homology search that is enhanced by ATP hydrolysis. The first correct pairing event between the presynaptic ssDNA and the dsDNA would then allow the presynaptic complex to promote more extensive strand invasion, which is in turn anticipated to quickly disrupt any remaining interactions involving microhomology footholds, allowing for complete pairing (Fig. 8, B and C).
FIGURE 8.
A role for ATP hydrolysis during the homology search. A, initial interactions between the RecA presynaptic complex and the dsDNA involve multiple ≥8-bp footholds while minimizing interactions with shorter tracts of microhomology. These ≥8-bp footholds are released more rapidly in the presence of ATP compared with reactions with ATPγS allowing for a more efficient search process. B, iterative binding and release events allow the presynaptic complex to search the dsDNA by intersegmental transfer (IS), as previously demonstrated. C, full alignment can take place quickly once the first correct pairing event has been established. Additional details are presented in the text. D, models for the role of ATP hydrolysis in the release of a single dsDNA foothold. Details are presented under “Discussion.”
Models for dsDNA Release
Our findings raise the important question of exactly how ATP hydrolysis might promote more rapid turnover of the bound dsDNA footholds. Here we consider three possible models (Fig. 8D). One model that we favor is that ATP-dependent conformational changes within the presynaptic complex promote release of the complementary dsDNA strand (Fig. 8D, panel i). Interestingly, RecA and related recombinases for filaments have different pitches; the active ATP-bound form of the filament has a longer helical pitch, whereas the inactive ADP-bound form of the filament has a shorter helical pitch (37–40). ATP hydrolysis may cause local changes in the RecA filament pitch as it shifts between ATP- and ADP-bound states, which in turn may result in more rapid dissociation of the bound dsDNA. Alternatively, ATP hydrolysis may drive local RecA monomer dissociation, which may cause concomitant dsDNA release (Fig. 8D, panel ii), or free RecA compete for binding to the dsDNA that has already been captured by the presynaptic complex, which may somehow drive release of the bound dsDNA (Fig. 8D, panel iii). Notably, these two later models are both predicted to exhibit a strong dependence upon free RecA, and the observation that up to 1 μm free RecA does not alter the dsDNA rate would seemingly argue against both alternative models. Similarly, the dsDNA dissociation rates observed in the presence of ATPγS were similar in the presence and absence of free RecA, which would argue against any model invoking enhanced dsDNA dissociation because of competing binding interactions. Future work will be essential to more fully understand how ATP hydrolysis is coupled to dsDNA turnover.
Base Triplet Stepping during Recombination
We have previously shown that RecA presynaptic complexes assembled with ATPγS can stably capture duplex DNA fragments bearing as few as 8 bp of microhomology (24), which is also consistent with previous reports (35, 41), and that subsequent changes in binding free energy take place with a 3-bp periodicity that is thought to reflect the base triplet architecture of RS-DNA (25). These general attributes are conserved between E. coli RecA, Saccharomyces cerevisiae Rad51, human Rad51, S. cerevisiae Dmc1, and human Dmc1 (24, 25). Here we have extended these findings to show that strand exchange intermediates are also stabilized in 3-bp steps for RecA presynaptic complexes assembled with ATP. Even though the overall rate of dsDNA turnover is ∼4 times faster in reactions with ATP compared with ATPγS, our findings suggest that stepping energetics remain unaltered by ATP hydrolysis, yielding a characteristic free energy change of ∼0.3 kBT per base triplet step. Similarly, we have shown that this characteristic free energy change remains unaltered for S. cerevisiae Rad51 in reactions with either ATP or the nonhydrolyzable analog AMP-PNP (25). Taken together, these findings suggest that during strand exchange the relative change in free energy change associated with each base triplet step remains unaffected by ATP hydrolysis.
Interestingly, previous theoretical studies have suggested that the net change in binding free energy for each correctly paired triplet should be on the order of thermal energy (42–44) and that the fidelity of base triplet pairing during DNA strand exchange would benefit from relatively low changes in binding free energy for each triplet step (42–44). These studies have also suggested that the structural deformation necessary to extend B-DNA to match the much longer extension of RS-DNA may enhance the ability of the presynaptic complex to reject nonhomologous sequences during the homology search (44–46). This enhancement arises because the energetic penalty associated with dsDNA extension reduces the net binding free energies of successive sets of base triplets, such that the energetic penalty for stretching the incoming dsDNA is optimized against the gain in binding free energy for each correctly paired triplet (44). In contrast, mispaired triplets are not sufficiently stable to overcome the energetic penalty associated with dsDNA extension, allowing the presynaptic complex to quickly discriminate against nonhomologous triplets. Consistent with these theoretical predictions, our experimental results reveal only small changes in binding free energy with each successive base triplet, and we also show that these changes in free energy are the same for reactions conducted with either ATP or ATPγS.
Potential Implications for Eukaryotic Recombinases
E. coli RecA can more quickly release short tracts of microhomology in the presence of ATP, suggesting that ATP hydrolysis enhances turnover of these search intermediates. However, we have also reported that there is no substantial difference in dsDNA turnover for S. cerevisiae Rad51 in reactions with either ATP or AMP-PNP, arguing against a role for ATP hydrolysis in dsDNA turnover for this eukaryotic recombinase (25). Bacterial RecA tends to have much higher ATP hydrolysis rates than any of the eukaryotic recombinases (1), and it is possible that this higher level of ATP hydrolysis is necessary to promote duplex DNA release. However, the eukaryotic recombinases interact with a number of different protein factors, including robust ATP-dependent motor proteins such as Rad54 and Rdh54, both of which are thought to contribute to the homology search (13, 47). It is possible that that instead of relying only upon the relatively weak ATPase activity of Rad51, the turnover of short tracts of microhomology bound to the Rad51 presynaptic filaments may instead be enhanced through the action of Rad54, Rdh54, or perhaps some additional protein cofactor. It will be a continuing challenge to marry in vivo and in vitro results to help more fully understand the biological implications of these biochemical and physical studies.
Experimental Procedures
Microscopy and Flow Cell Preparation
All experiments were conducted with a custom-built prism-type total internal reflection fluorescence microscope (Nikon) equipped with a 488-nm laser (Coherent Sapphire, 200 milliwatt) and a 561-nm laser (Coherent Sapphire, 200 milliwatt). The 488-nm laser was used for RPA-eGFP imaging and was set to 20 milliwatt at the prism face. The 561-nm laser was used for detecting Atto565-labeled dsDNA and was set to 30 milliwatt at the prism face. Chromium barriers were deposited onto the surface of a fused silica slide glass by electron beam lithography, which were then assembled into a sample chamber using doubled-sided tape and a borosilicate cover slide, as described (48, 49). Bilayers were prepared with 91.5% 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 0.5% 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine-N-(biotinyl) (biotinylated DPPE), and 8% mPEG 2000 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). The biotinylated ssDNA molecules were injected into the sample chamber and attached to the bilayer through a biotin-streptavidin linkage. Buffer was delivered to the sample chambers using a syringe pump (Kd Scientific), and sample delivery was controlled by two SCIVEX HPLC valves equipped with either a 50-μl loop or a 500-μl loop. The anchored ssDNA molecules were then aligned at the barriers by application of flow in buffer containing 40 mm Tris-HCl (pH 7.5), 2 mm MgCl2, 1 mm DTT, 50 mm NaCl, 0.2 mg/ml BSA, and 0.2 nm RPA-eGFP for a total period of 15 min (1 ml/min flow rate). At the 3-min time point, 500 μl of 7 m urea was injected into the flow cell to remove any remaining φ29 DNA polymerase (see below), followed by continued rinse with the buffer containing 40 mm Tris-HCl (pH 7.5), 2 mm MgCl2, 1 mm DTT, 50 mm NaCl, 0.2 mg/ml BSA, and 0.2 nm RPA-eGFP.
RecA Presynaptic Complexes
S. cerevisiae RPA-eGFP and E. coli RecA were prepared as previously described (25, 26). ssDNA substrates were prepared using circular M13 ssDNA as a template for rolling circle replication using φ29 DNA polymerase and a biotinylated oligonucleotide primer, as described (24, 26). DNA curtains were visualized by total internal reflection fluorescence microscopy as described (24). Free RPA was removed by flushing the sample chamber with reaction buffer containing 25 mm Tris acetate (pH 7.5), 4 mm magnesium acetate, 10 mm sodium acetate, 1 mm DTT, and 0.2 mg/ml BSA for 5 min at a flow rate of 1 ml/min. Filament nucleation was initiated by injecting 1 μm RecA in reactions buffer supplemented with 1 mm ATPγS. Buffer flow was then terminated, and the reactions were incubated for a period of 30 s at 37 °C. Unbound RecA and free ATPγS were removed by flushing the sample chamber with reaction buffer for a period of 2 min at a flow rate of 1 ml/min. Filament elongation was initiated by injecting 2 μm RecA in reaction buffer supplemented with 1 mm ATP, and an ATP regenerating system comprised of 20 mm creatine and 0.04 mg/ml creatine kinase. These reactions were incubated for an additional 20 min at 37 °C while observing the RecA-mediated displacement of RPA-eGFP from the ssDNA. Once the RecA presynaptic complexes were assembled, the sample chambers were flushed with 50 nm RecA in reaction buffer supplemented with 1 mm ATP plus the ATP regenerating system.
RecA Presynaptic Complex Stability
The stability of the RecA presynaptic complexes was assessed by flushing the sample chamber with reaction buffer supplemented with 0.2 nm RPA-eGFP and 50 nm RecA in reaction buffer supplemented with 1 mm ATP, plus the ATP regenerating system at 37 °C. Dissociation of unlabeled RecA from the ssDNA would be revealed as the appearance of fluorescent RPA-eGFP on the ssDNA molecules. We observed no significant dissociation of RecA from the ssDNA over a time period of 1 h when ATP, the ATP regenerating system, and ≥50 nm free RecA were maintained in the reaction buffer. In contrast, RecA was rapidly displaced from the ssDNA by RPA-eGFP if ATP was removed from the reactions.
Duplex DNA Binding Measurements
All duplex DNA binding measurements were performed at 30 °C to allow for direct comparison with our previously published experimental data (24, 25). For this work, presynaptic complexes were prepared with ATP at 37 °C, as described above, and the sample chambers were then cooled to 30 °C. Duplex DNA oligonucleotides (2 nm) labeled at one end with a single Atto565 dye were then injected into the sample chamber in reaction buffer containing 50 nm RecA, 1 mm ATP plus the ATP regenerating system, and the reactions were then incubated for 10 min at 30 °C. The sample chambers were then quickly flushed with reaction buffer containing 50 nm RecA, 1 mm ATP, plus the ATP regenerating system, to remove free dsDNA collecting images (100-ms exposures) at varying time intervals, as previously described (24, 25). The resulting data were rendered into kymographs each reflecting the dsDNA binding properties of a single presynaptic complex, and the survival probabilities of the bound dsDNA were determined from the kymographs as previously described (24, 25).
Nucleation Positions and Elongation Rates
Nucleation positions were defined by analysis of the fluorescence intensity profiles of kymographs generated from individual ssDNA molecules onto which had been assembled RecA presynaptic complexes. The nucleation positions were defined as minima in the RPA-eGFP fluorescence signal that took place during the first five frames of the assembly reactions, and all nucleation positions were further confirmed by visual inspection of the corresponding assembly kymographs. Each presynaptic complex contains approximately five repeats of M13 ssDNA sequence (24); therefore each molecule was subdivided into five equivalent increments for position analysis. Histograms for the resulting data were built with a 1-μm bin size, and error bars were obtained by bootstrap analysis with one sigma confidence interval. RecA filament elongation was observed as displacement of the fluorescent RPA-eGFP emanating from the initial nucleation sites. The RecA elongation rates were obtained from kymographs by manually fitting a line to the border between the dark RecA filaments and the adjacent regions of fluorescent RPA-eGFP. The slopes of the resulting lines correspond to the filament elongation rates, which are expressed as positive values for 3′ → 5′ elongation and negative values for 5′ → 3′ elongation.
Real Time DNA Binding Measurements
Real time dsDNA binding assays were performed using 70-bp dsDNA substrates labeled with a single Atto565 fluorophore, as previously described (24). In brief, the RecA presynaptic complexes were prepared as described above. The 70-bp dsDNA oligonucleotides (2 nm) were then injected into the sample chamber, buffer flow was terminated, and data collection was initiated at 16.7 frames/s (60-ms exposures) with continuous laser illumination. Kymographs were generated from the resulting video data, and the binding lifetimes of the dsDNA oligonucleotides were obtained from analysis of these kymographs and plotted as survival probability versus time, as described (24).
Strand Exchange Assays
Strand exchange assays were conducted with φX174 RFI DNA (NEB) that was linearized by digestion with PsiI (NEB) and purified by gel extraction. Strand exchange reactions were carried out in buffer containing 25 mm Tris acetate (pH 7.5), 4 mm magnesium acetate, 1 mm DTT, and either 2 mm ATP or 2 mm ATPγS, as indicated. Reactions with ATP also included 20 mm creatine phosphate and 0.04 mg/ml creatine kinase. RecA (11 μm) was preincubated with φX174 circular ssDNA (20 μm nucleotides) at 37 °C for 3 min. SSB (1.4 μm) was then added, and the reactions were incubated for an additional 3 min at 37 °C. The strand exchange reactions were then initiated by the addition of linearized φX174 (7 μm nucleotides). Aliquots were then removed at the indicated time intervals, and the reactions were terminated by mixing with 2% SDS, 0.5 mg/ml proteinase K, and 20 mm EDTA. Samples were incubated for an additional 20 min at 37 °C for deproteinization and then placed on ice. Samples were then resolved by electrophoresis on 1% agarose gels and stained with SYBRSafe (Invitrogen), and the resulting products quantified by ImageJ. The rates of product formation were obtained by fitting the initial linear region of the time course data with a linear function.
Author Contributions
J. Y. L. performed all of the experimental work and data analysis. Z. Q. assisted with the experimental design and data analysis. E. C. G. guided the project, interpreted data, and wrote the manuscript with input from all co-authors.
Acknowledgments
We thank Daniel Duzdevich, Frank Ma, Justin Steinfeld, Kyle Kaniecki, and Luisina de Tullio for comments on this manuscript.
This work was supported by National Institutes of Health Grant R356M118026 (to E. C. G.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ATPγS
- adenosine 5′-O-(thiotriphosphate)
- eGFP
- enhanced GFP
- RPA
- replication protein A
- SSB
- single-stranded DNA-binding protein.
References
- 1. Bianco P. R., Tracy R. B., and Kowalczykowski S. C. (1998) DNA strand exchange proteins: a biochemical and physical comparison. Front. Biosci. 3, D570–D603 [DOI] [PubMed] [Google Scholar]
- 2. Morrical S. W. (2015) DNA-pairing and annealing processes in homologous recombination and homology-directed repair. Cold Spring Harb. Perspect. Biol. 7, a016444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Kowalczykowski S. C. (2015) An overview of the molecular mechanisms of recombinational DNA repair. Cold Spring Harb. Perspect. Biol. 7, a016410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Prentiss M., Prévost C., and Danilowicz C. (2015) Structure/function relationships in RecA protein-mediated homology recognition and strand exchange. Crit. Rev. Biochem. Mol. Biol. 50, 453–476 [DOI] [PubMed] [Google Scholar]
- 5. San Filippo J., Sung P., and Klein H. (2008) Mechanism of eukaryotic homologous recombination. Annu. Rev. Biochem. 77, 229–257 [DOI] [PubMed] [Google Scholar]
- 6. Cromie G. A., Connelly J. C., and Leach D. R. (2001) Recombination at double-strand breaks and DNA ends: conserved mechanisms from phage to humans. Mol. Cell 8, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 7. Hunter N. (2015) Meiotic recombination: the essence of heredity. Cold Spring Harb. Perspect. Biol. 7, a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cox M. M. (2001) Historical overview: searching for replication help in all of the rec places. Proc. Natl. Acad. Sci. U.S.A. 98, 8173–8180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox M. M., Goodman M. F., Kreuzer K. N., Sherratt D. J., Sandler S. J., and Marians K. J. (2000) The importance of repairing stalled replication forks. Nature 404, 37–41 [DOI] [PubMed] [Google Scholar]
- 10. Clark A. J., and Margulies A. D. (1965) Isolation and Characterization of Recombination-deficient Mutants of Escherichia coli K12. Proc. Natl. Acad. Sci. U.S.A. 53, 451–459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Weiner A., Zauberman N., and Minsky A. (2009) Recombinational DNA repair in a cellular context: a search for the homology search. Nat. Rev. Microbiol. 7, 748–755 [DOI] [PubMed] [Google Scholar]
- 12. Barzel A., and Kupiec M. (2008) Finding a match: how do homologous sequences get together for recombination? Nat. Rev. Genet. 9, 27–37 [DOI] [PubMed] [Google Scholar]
- 13. Renkawitz J., Lademann C. A., and Jentsch S. (2014) Mechanisms and principles of homology search during recombination. Nat. Rev. Mol. Cell Biol. 15, 369–383 [DOI] [PubMed] [Google Scholar]
- 14. von Hippel P. H., and Berg O. G. (1989) Facilitated target location in biological systems. J. Biol. Chem. 264, 675–678 [PubMed] [Google Scholar]
- 15. Pâques F., and Haber J. E. (1999) Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 63, 349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Symington L. S., Rothstein R., and Lisby M. (2014) Mechanisms and regulation of mitotic recombination in Saccharomyces cerevisiae. Genetics 198, 795–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chen Z., Yang H., and Pavletich N. P. (2008) Mechanism of homologous recombination from the RecA-ssDNA/dsDNA structures. Nature 453, 489–494 [DOI] [PubMed] [Google Scholar]
- 18. Cox M. M. (1994) Why does RecA protein hydrolyse ATP? Trends Biochem. Sci. 19, 217–222 [DOI] [PubMed] [Google Scholar]
- 19. Menetski J. P., Bear D. G., and Kowalczykowski S. C. (1990) Stable DNA heteroduplex formation catalyzed by the Escherichia coli RecA protein in the absence of ATP hydrolysis. Proc. Natl. Acad. Sci. U.S.A. 87, 21–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rehrauer W. M., and Kowalczykowski S. C. (1993) Alteration of the nucleoside triphosphate (NTP) catalytic domain within Escherichia coli recA protein attenuates NTP hydrolysis but not joint molecule formation. J. Biol. Chem. 268, 1292–1297 [PubMed] [Google Scholar]
- 21. Rosselli W., and Stasiak A. (1991) The ATPase activity of RecA is needed to push the DNA strand exchange through heterologous regions. EMBO J. 10, 4391–4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Menetski J. P., and Kowalczykowski S. C. (1985) Interaction of recA protein with single-stranded DNA: quantitative aspects of binding affinity modulation by nucleotide cofactors. J. Mol. Biol. 181, 281–295 [DOI] [PubMed] [Google Scholar]
- 23. Kim J. I., Cox M. M., and Inman R. B. (1992) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange. I. Bypassing a short heterologous insert in one DNA substrate. J. Biol. Chem. 267, 16438–16443 [PubMed] [Google Scholar]
- 24. Qi Z., Redding S., Lee J. Y., Gibb B., Kwon Y., Niu H., Gaines W. A., Sung P., and Greene E. C. (2015) DNA sequence alignment by microhomology sampling during homologous recombination. Cell 160, 856–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lee J. Y., Terakawa T., Qi Z., Steinfeld J. B., Redding S., Kwon Y., Gaines W. A., Zhao W., Sung P., and Greene E. C. (2015) Base triplet stepping by the Rad51/RecA family of recombinases. Science 349, 977–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Gibb B., Silverstein T. D., Finkelstein I. J., and Greene E. C. (2012) Single-stranded DNA curtains for real-time single-molecule visualization of protein-nucleic acid interactions. Anal. Chem. 84, 7607–7612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gibb B., Ye L. F., Gergoudis S. C., Kwon Y., Niu H., Sung P., and Greene E. C. (2014) Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLoS One 9, e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bell J. C., Plank J. L., Dombrowski C. C., and Kowalczykowski S. C. (2012) Direct imaging of RecA nucleation and growth on single molecules of SSB-coated ssDNA. Nature 491, 274–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Galletto R., Amitani I., Baskin R. J., and Kowalczykowski S. C. (2006) Direct observation of individual RecA filaments assembling on single DNA molecules. Nature 443, 875–878 [DOI] [PubMed] [Google Scholar]
- 30. Joo C., McKinney S. A., Nakamura M., Rasnik I., Myong S., and Ha T. (2006) Real-time observation of RecA filament dynamics with single monomer resolution. Cell 126, 515–527 [DOI] [PubMed] [Google Scholar]
- 31. Cox M. M. (2007) Motoring along with the bacterial RecA protein. Nat. Rev. Mol. Cell Biol. 8, 127–138 [DOI] [PubMed] [Google Scholar]
- 32. Kim J. I., Cox M. M., and Inman R. B. (1992) On the role of ATP hydrolysis in RecA protein-mediated DNA strand exchange: II. Four-strand exchanges. J. Biol. Chem. 267, 16444–16449 [PubMed] [Google Scholar]
- 33. Gonda D. K., and Radding C. M. (1983) By searching processively RecA protein pairs DNA molecules that share a limited stretch of homology. Cell 34, 647–654 [DOI] [PubMed] [Google Scholar]
- 34. Gonda D. K., and Radding C. M. (1986) The mechanism of the search for homology promoted by recA protein. Facilitated diffusion within nucleoprotein networks. J. Biol. Chem. 261, 13087–13096 [PubMed] [Google Scholar]
- 35. Ragunathan K., Liu C., and Ha T. (2012) RecA filament sliding on DNA facilitates homology search. Elife 1, e00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Forget A. L., and Kowalczykowski S. C. (2012) Single-molecule imaging of DNA pairing by RecA reveals a three-dimensional homology search. Nature 482, 423–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Conway A. B., Lynch T. W., Zhang Y., Fortin G. S., Fung C. W., Symington L. S., and Rice P. A. (2004) Crystal structure of a Rad51 filament. Nat. Struct. Mol. Biol. 11, 791–796 [DOI] [PubMed] [Google Scholar]
- 38. Yu X., Jacobs S. A., West S. C., Ogawa T., and Egelman E. H. (2001) Domain structure and dynamics in the helical filaments formed by RecA and Rad51 on DNA. Proc. Natl. Acad. Sci. U.S.A. 98, 8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Sheridan S. D., Yu X., Roth R., Heuser J. E., Sehorn M. G., Sung P., Egelman E. H., and Bishop D. K. (2008) A comparative analysis of Dmc1 and Rad51 nucleoprotein filaments. Nucleic Acids Res. 36, 4057–4066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. VanLoock M. S., Yu X., Yang S., Lai A. L., Low C., Campbell M. J., and Egelman E. H. (2003) ATP-mediated conformational changes in the RecA filament. Structure 11, 187–196 [DOI] [PubMed] [Google Scholar]
- 41. Hsieh P., Camerini-Otero C. S., and Camerini-Otero R. D. (1992) The synapsis event in the homologous pairing of DNAs: RecA recognizes and pairs less than one helical repeat of DNA. Proc. Natl. Acad. Sci. U.S.A. 89, 6492–6496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kates-Harbeck J., Tilloy A., and Prentiss M. (2013) Simplified biased random walk model for RecA-protein-mediated homology recognition offers rapid and accurate self-assembly of long linear arrays of binding sites. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 88, 012702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jiang L., and Prentiss M. (2014) RecA-mediated sequence homology recognition as an example of how searching speed in self-assembly systems can be optimized by balancing entropic and enthalpic barriers. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 90, 022704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Savir Y., and Tlusty T. (2010) RecA-mediated homology search as a nearly optimal signal detection system. Mol. Cell 40, 388–396 [DOI] [PubMed] [Google Scholar]
- 45. Danilowicz C., Feinstein E., Conover A., Coljee V. W., Vlassakis J., Chan Y. L., Bishop D. K., and Prentiss M. (2012) RecA homology search is promoted by mechanical stress along the scanned duplex DNA. Nucleic Acids Res. 40, 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Danilowicz C., Peacock-Villada A., Vlassakis J., Facon A., Feinstein E., Kleckner N., and Prentiss M. (2014) The differential extension in dsDNA bound to Rad51 filaments may play important roles in homology recognition and strand exchange. Nucleic Acids Res. 42, 526–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Renkawitz J., Lademann C. A., Kalocsay M., and Jentsch S. (2013) Monitoring homology search during DNA double-strand break repair in vivo. Mol. Cell 50, 261–272 [DOI] [PubMed] [Google Scholar]
- 48. Greene E. C., Wind S., Fazio T., Gorman J., and Visnapuu M. L. (2010) DNA curtains for high-throughput single-molecule optical imaging. Methods Enzymol. 472, 293–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Qi Z., and Greene E. C. (2016) Visualizing recombination intermediates with single-stranded DNA curtains. Methods 105, 62–74 [DOI] [PMC free article] [PubMed] [Google Scholar]