Abstract
Bipolar disorder (BD), which is characterized by depression and mania, affects 1–2% of the world population. Current treatments are effective in only 40–60% of cases and cause severe side effects. Valproate (VPA) is one of the most widely used drugs for the treatment of BD, but the therapeutic mechanism of action of this drug is not understood. This knowledge gap has hampered the development of effective treatments. To identify candidate pathways affected by VPA, we performed a genome-wide expression analysis in yeast cells grown in the presence or absence of the drug. VPA caused up-regulation of FEN1 and SUR4, encoding fatty acid elongases that catalyze the synthesis of very long chain fatty acids (C24 to C26) required for ceramide synthesis. Interestingly, fen1Δ and sur4Δ mutants exhibited VPA sensitivity. In agreement with increased fatty acid elongase gene expression, VPA increased levels of phytoceramide, especially those containing C24–C26 fatty acids. Consistent with an increase in ceramide, VPA decreased the expression of amino acid transporters, increased the expression of ER chaperones, and activated the unfolded protein response element (UPRE), suggesting that VPA induces the UPR pathway. These effects were rescued by supplementation of inositol and similarly observed in inositol-starved ino1Δ cells. Starvation of ino1Δ cells increased expression of FEN1 and SUR4, increased ceramide levels, decreased expression of nutrient transporters, and induced the UPR. These findings suggest that VPA-mediated inositol depletion induces the UPR by increasing the de novo synthesis of ceramide.
Keywords: ceramide, fatty acid metabolism, lipid metabolism, sphingolipid, unfolded protein response (UPR), inositol
Introduction
Bipolar disorder (BD),2 one of the most severe forms of mood disorder, is characterized by recurrent episodes of depression and mania (1, 2). BD is ranked as the sixth leading cause of disability worldwide. It affects about 1–2% of the total world population (1–3) and leads to suicide in 15% of cases (4). Valproic acid (VPA), a branched short-chain fatty acid, is one of the most widely used drugs for the treatment of BD. However, it is effective in only 40–60% of cases and results in serious side effects, including hepatotoxicity and teratogenicity (5). Although many hypotheses have been postulated to explain its efficacy, the therapeutic mechanism of the drug is not understood, nor is the underlying cause of the disease (6, 7). This knowledge gap hampers the development of more effective drugs to treat BD.
The inositol depletion hypothesis has had a major impact on research in BD. Berridge (8) proposed that lithium, widely used to treat BD, inhibits inositol monophosphatase, causing inositol depletion and subsequently decreasing inositol 1,4,5-triphosphate-mediated signaling (8, 9). We have shown previously that VPA, similar to lithium, causes a decrease in intracellular inositol in yeast and mammalian cells (10–12). VPA indirectly inhibits myo-inositol-3-phosphate synthase (MIPS), the enzyme responsible for the rate-limiting step of de novo synthesis of inositol, suggesting that MIPS is post-translationally regulated (13). More recent findings show that yeast and human MIPS are phosphorylated (14, 15) and that phosphorylation of conserved sites affects enzymatic activity (14). These findings suggest that the mechanism by which VPA causes inositol depletion is conserved in yeast and mammals, supporting the yeast model for genetic and molecular studies of the mechanism of the drug.
In yeast, supplementation of inositol triggers a change in the expression of hundreds of inositol-regulated genes, including genes for lipid synthesis (16–18). Inositol-containing lipids, including inositol phosphoinositides, glycosylphosphatidylinositol, and sphingolipids, play crucial structural and functional roles in regulating membrane biogenesis, membrane trafficking, cytoskeletal organization, and gene expression (19–23). Hence, inositol depletion exerts profound effects on cellular function (24). Inositol depletion not only alters lipid biosynthesis (25) but also activates stress response pathways, including the protein kinase C and unfolded protein response (UPR) pathways (26–28). Cells grown in the absence of inositol exhibit induction of the UPR pathway (27, 29), which is reversed by inositol supplementation (16). These studies suggest that decreasing the intracellular levels of inositol induces the UPR pathway by a mechanism not yet characterized.
Interdependence of the UPR pathway and ceramide synthesis has been demonstrated in yeast and mammals. Induction of the UPR increases ceramide levels and viability in yeast (30). In mammals, activation of the UPR increases the expression of ceramide synthase CerS6, leading to increased synthesis of ceramides containing C16 fatty acids (30). Perturbation of de novo synthesis of sphingolipids activates the UPR in yeast (29, 31), and defective ceramide homeostasis leads to UPR failure (32). Ceramide also decreases the transcription of nutrient transporters, including amino acid transporter mCAT-1, glucose transporter GLUT-1 (33), glucose transporter HXT4, and uracil permease FUR4 (34). Therefore, the interrelationship between ceramide levels and the UPR pathway maintains cell homeostasis.
In the current study, we show for the first time that VPA-mediated inositol depletion induces the UPR pathway by increasing de novo synthesis of ceramide, especially C24–C26-containing phytoceramide. These findings have implications for the therapeutic mechanism of VPA.
Results
VPA Increases the Expression of Fatty Acid Elongases
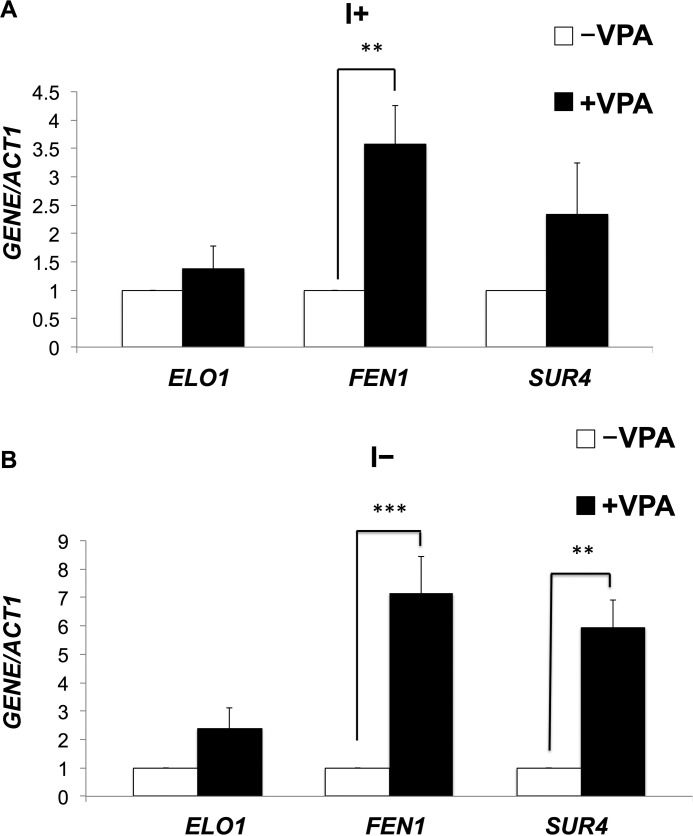
To determine candidate pathways that may be important for the therapeutic role of VPA, we performed a genome-wide microarray analysis of cells treated with 0.6 mm VPA for 5 h in the presence or absence of inositol, as described under “Experimental Procedures.” VPA treatment resulted in altered expression (>2-fold) of 324 genes in the presence of inositol and 413 genes in the absence of inositol (supplemental Table 1). Interestingly, fatty acid elongase genes FEN1 and SUR4 exhibited 2-fold increased expression in response to VPA (supplemental Table 1). qRT-PCR analysis of fatty acid elongase genes in WT cells treated with VPA validated these findings. As seen in Fig. 1, mRNA levels of FEN1 and SUR4 were increased 6- and 4-fold in the absence of inositol and to a lesser extent (3- and 2-fold) in the presence of inositol. Fen1 and Sur4 catalyze the synthesis of very long chain fatty acids, including C22 and C24 (Fen1) and C24 and C26 (Sur4) (35), which are used for the synthesis of ceramide. However, VPA did not alter the expression levels of ELO1, a fatty acid elongase that catalyzes the synthesis of C16 or C18 (Fig. 1). This indicates that VPA specifically increases the expression of fatty acid elongases catalyzing the synthesis of very long chain fatty acids (C24–C26). Mutants fen1Δ and sur4Δ exhibited sensitivity to VPA, as did the sphinganine C4-hydroxylase mutant, sur2Δ (Fig. 2). VPA sensitivity was partially rescued by inositol. The VPA-mediated increase in the expression of these genes and the VPA sensitivity of Fen1, Sur4, and Sur2 mutants suggested that VPA induces an increase in ceramide containing C24–C26 fatty acids and phytosphingosine (the product of Sur2).
FIGURE 1.
VPA up-regulates expression of fatty acid elongase genes. WT cells were precultured in I+ medium, harvested, washed twice with sterile water, and grown in I+ or I− medium until cells reached the mid-log phase (A550 = 0.5). Cells were pelleted and suspended in fresh I− medium with or without 0.6 mm VPA and incubated for 5 h. mRNA levels of fatty acid elongases were quantified by qRT-PCR in WT cells grown in the presence or absence of VPA. Values are reported as -fold change in expression in cells grown in VPA relative to cells grown without VPA, in the absence (A) or presence of inositol (B). Expression was normalized to the mRNA levels of the internal control ACT1. Data shown are mean ± S.D. (error bars) (n = 6) (**, p < 0.01; ***, p < 0.001).
FIGURE 2.
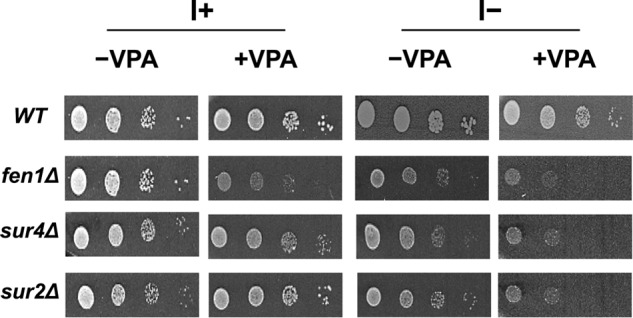
VPA sensitivity of fatty acid elongase and sphinganine C4-hydroxylase mutants. Cells were precultured in I+, counted using a hemocytometer, and washed with sterile water. 3-μl aliquots of a series of 10-fold dilutions were spotted onto I+ or I− plates in the presence or absence of 1 mm VPA and incubated for 3 days at 30 °C.
VPA Increases Ceramide Levels and Down-regulates Amino Acid Transporters
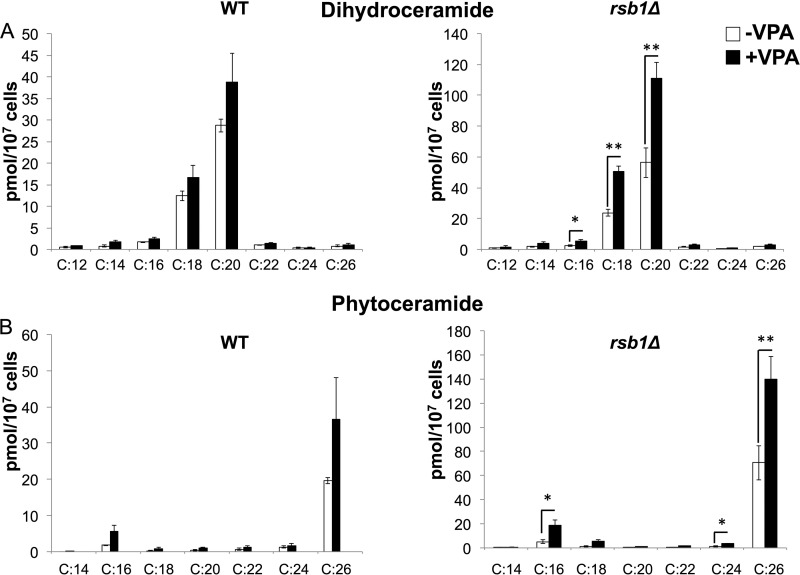
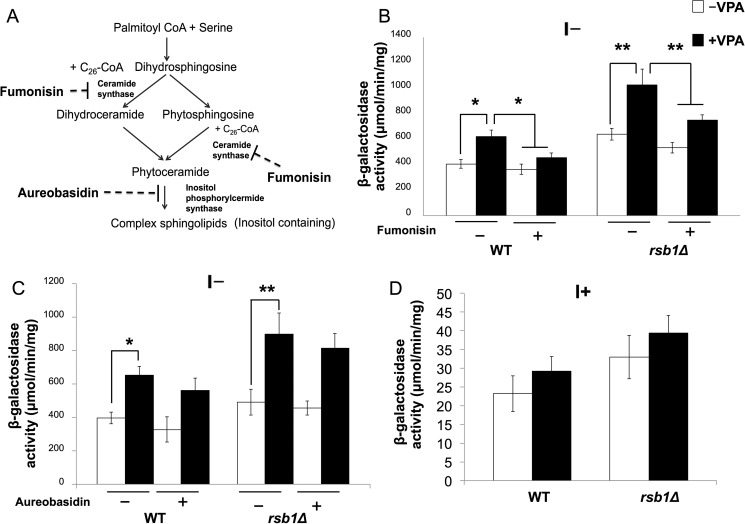
To determine whether VPA increases ceramide levels, dihydroceramide (DHC) and phytoceramide (PHC) ceramide species were analyzed by mass spectrometry in WT cells. VPA showed a trend of increase in ceramide levels, especially C26 phytoceramide, but this increase was not statistically significant (Fig. 3B). Interestingly, VPA treatment for 30 min up-regulated the expression of RSB1 (data not shown), which transports long chain bases (LCBs) (including dihydrosphingosine and phytosphingosine) across the plasma membrane from the inside to the outside of the cell (36). In WT cells, up-regulation of RSB1 increases the transport of LCBs, reducing the intracellular levels of these ceramide precursors. Therefore, we measured the effect of VPA on ceramide levels with varying fatty acids in rsb1Δ cells. VPA increased both DHC and PHC in rsb1Δ (Fig. 3). Specific ceramide species were increased, including DHC with C16, C18, and C20 and PHC with C16, C24, and C26 in rsb1Δ cells (Fig. 3). The highest increase was observed in C26 species of PHC. The likely explanation for this is that the increase in RSB1 in WT cells reduces the available LCB precursor for ceramide synthesis. However, when RSB1 is deleted, the increase in ceramide levels is more significant.
FIGURE 3.
VPA increases levels of DHC and PHC. WT and rsb1Δ cells were grown as described in the legend to Fig. 1. Cells were pelleted, and DHC (A) and PHC (B) levels with varying fatty acids were quantified by mass spectrometry. Data shown are mean ± S.D. (error bars) (n = 6; *, p < 0.05; **, p < 0.01).
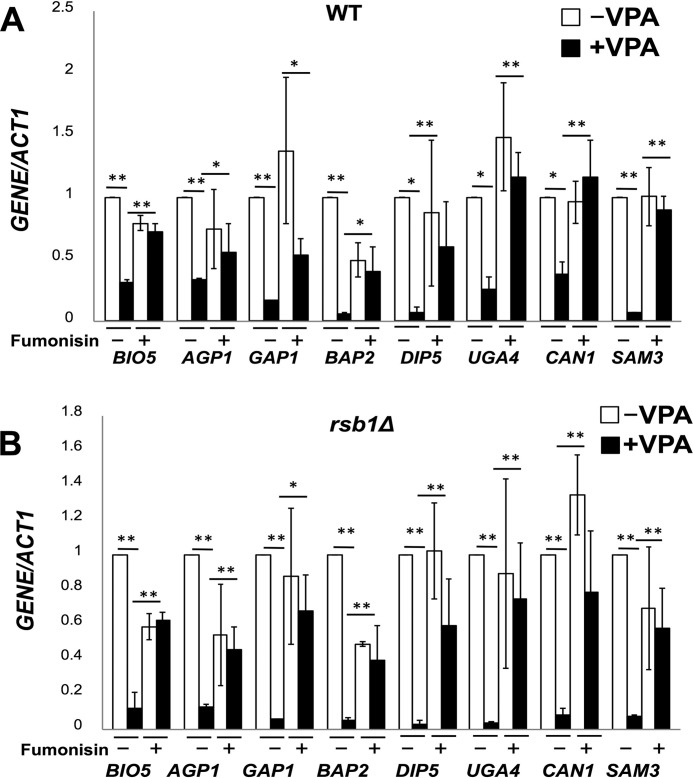
Previous studies have shown that increased levels of ceramide cause a decrease in the transcription of nutrient permeases, leading to reduced intake of nutrients and induction of stress (33, 34). Consistent with this, the microarray analysis revealed decreased expression of amino acid transporter genes BIO5, AGP1, GAP1, BAP2, DIP5, UGA4, CAN1, and SAM3 in response to VPA in the absence of inositol. The expression of these transporters was not altered in I+ medium (supplemental Table 1). Decreased expression of amino acid transporters was confirmed by qRT-pCR in both WT and rsb1Δ cells (Fig. 4). Down-regulation was greater in rsb1Δ than in WT cells, consistent with the increase in ceramide levels. To determine whether decreased expression of amino acid transporters resulted from increased ceramide, cells were treated with 100 μm fumonisin, a ceramide synthase inhibitor (37, 38). Fumonisin reversed the down-regulation of the amino acid transporters in both WT and rsb1Δ cells (Fig. 4). Taken together, these findings suggest that VPA increases levels of specific ceramide species, resulting in decreased expression of nutrient permeases.
FIGURE 4.
VPA down-regulates expression of nutrient transporters via ceramide. mRNA levels of nutrient transporters were quantified by qRT-PCR in WT (A) and rsb1Δ (B) cells grown in the presence or absence of VPA and fumonisin, as indicated. Values are reported as -fold change in expression in cells grown in VPA relative to cells grown without VPA. Expression was normalized to the mRNA levels of the internal control ACT1. Data shown are mean ± S.D. (error bars) (n = 6) (*, p < 0.05; **, p < 0.01).
VPA Induces the UPR by Increasing Ceramide Levels
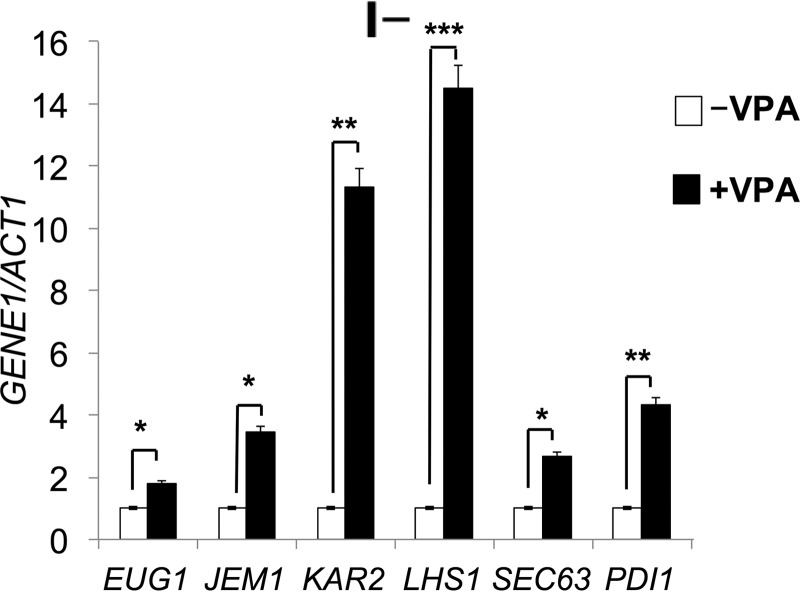
Decreased expression of nutrient transporters is expected to induce stress due to nutrient starvation (33). In agreement with this, VPA increased the expression of ER chaperone genes EUG1, JEM1, KAR2, LHS1, SEC63, and PDI1 in I− medium (supplemental Table 1 and Fig. 5), suggesting that the UPR pathway was induced. To test this, we analyzed the UPR response in WT and rsb1Δ cells expressing a UPRE-lacZ reporter plasmid. Increased lacZ activity was observed in response to VPA in I− medium (Fig. 6B). The increase was more pronounced in rsb1Δ than in WT cells. Fumonisin, a ceramide synthase inhibitor (Fig. 6, A and B) restored lacZ activity to normal levels. In contrast, aureobasidin, which inhibits the conversion of ceramide to complex sphingolipids (Fig. 6A), did not affect UPRE expression (Fig. 6C). Increased UPRE expression was not observed in I+ medium (Fig. 6D). These findings suggest that VPA induced the UPR pathway via increased de novo synthesis of ceramide and not by inhibiting the conversion of ceramide to complex sphingolipids.
FIGURE 5.
VPA increases expression of ER chaperones. WT cells were grown as described in the legend to Fig. 1. mRNA levels of chaperone genes were quantified by qRT-PCR in wild type cells grown in I− medium in the presence or absence of VPA. Values are reported as -fold change in expression in cells grown in VPA relative to cells grown without VPA. Expression was normalized to the mRNA levels of the internal control ACT1. Data shown are mean ± S.D. (error bars) (n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 6.
VPA up-regulates UPRE expression. A, schematic of sphingolipid synthesis. Serine is condensed with palmitoyl-CoA to form 3-ketodihydrosphingosine, which is reduced to dihydrosphigosine. Dihydrosphingosine is hydroxylated on C4 to produce phytosphingosine. Ceramide synthase catalyzes the linkage of very long chain fatty acids produced by fatty acid elongases to dihydrosphingosine and phytosphingosine to form ceramide (dihydroceramide or phytoceramide). Ceramide is used as a precursor for the synthesis of inositol containing complex sphingolipids. Inositol phosphorylceramide synthase catalyzes the first step of complex sphingolipid synthesis. Fumonisin is a ceramide synthase inhibitor, and aureobasidin inhibits inositol phosphorylcermide synthase. B, WT and rsb1Δ cells expressing a UPRE-lacZ plasmid were precultured in Ura-I+ medium and then grown in Ura-I− medium with or without 100 μm fumonisin or 0.5 μg/μl aureobasidin (B and C) or Ura-I+ medium (D). 0.6 mm VPA was added when indicated. LacZ activity is quantified as indicated. Data shown are mean ± S.D. (error bars) (n = 6) (*, p < 0.05; **, p < 0.01).
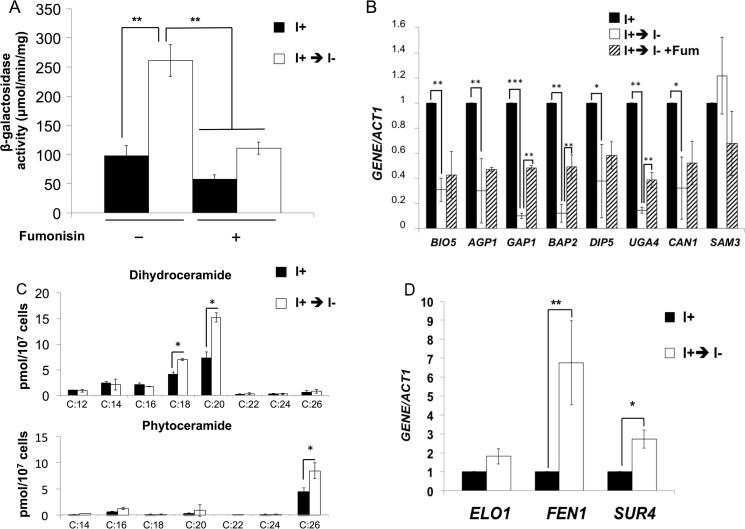
Consistent with previous reports showing that inositol starvation induces the UPR pathway (27, 29), UPRE expression was increased in inositol-starved ino1Δ mutant cells (Fig. 7A). Treatment with fumonisin decreased UPRE expression to levels observed in control cells, indicating that inositol starvation of ino1Δ cells induces the UPR pathway by increasing ceramide levels.
FIGURE 7.
VPA induces the UPR by inositol depletion. ino1Δ cells expressing the UPRE-lacZ reporter plasmid were precultured and grown in Ura-I+ medium until the cells reached mid-log phase (A550 = 0.5). Cells were washed and transferred to Ura-I− medium with or without 100 μm fumonisin for 3 h. A, LacZ activity. Data shown are mean ± S.D. (error bars) (n = 6) (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, DHC and PHC levels with varying fatty acids were quantified by mass spectrometry. Data shown are mean ± S.D. (n = 6) (*, p < 0.05; **, p < 0.01; ***, p < 0.001). mRNA levels of nutrient transporters (C) and fatty acid elongases (D) were quantified by qRT-PCR, as indicated. Values are reported as -fold change in expression in cells grown in I− medium relative to cells grown in I+ medium. Expression was normalized to the mRNA levels of the internal control ACT1. Data shown are mean ± S.D. (n = 6). *, p < 0.05; **, p < 0.01; ***, p < 0.001.
Similar to VPA treatment, ino1Δ cells starved for inositol for 3 h also exhibited decreased expression of amino acid transporter genes BIO5, AGP1, GAP1, BAP2, DIP5, UGA4, CAN1, and SAM3 (Fig. 7B). This decrease was ceramide-dependent because it was blocked by fumonisin. Decreased expression of amino acid transporter genes BAP2, GAP1, and UGA4 was partially rescued by fumonisin (Fig. 7B), suggesting that inositol depletion decreased the expression of amino acid transporter genes, at least in part, by increasing ceramide levels.
To directly test the hypothesis that inositol depletion decreased the expression of amino acid transporters and induced the UPR by increasing the intracellular ceramide levels, ino1Δ cells were starved for 3 h, and ceramide levels were quantified (Fig. 7C). Inositol starvation increased levels of some DHC and PHC species (Fig. 7C). Most notable were increases in DHC with C18 and C20 and PHC with C26 (Fig. 7C). The highest increase was observed in C26 species of PHC, similar to the response to VPA (Fig. 3). Expression of fatty acid elongases FEN1 and SUR4 was also increased during inositol starvation (Fig. 7D).
Taken together, our findings indicate that inositol depletion mediated by VPA or by starvation of ino1Δ cells led to increased expression of fatty acid elongases (elongating C24–C26), resulting in increasing ceramide levels with C24–C26 fatty acids. Increased ceramide synthesis led to down-regulation of transporters and induction of the UPR pathway. These findings are the first to show that VPA-mediated inositol depletion induces the UPR by increasing ceramide levels.
Discussion
In this study, we show that VPA induces the UPR by increasing ceramide levels via inositol depletion. Our specific findings indicated that VPA 1) increased expression of fatty acid elongases synthesizing C24 and C26 fatty acids; 2) increased ceramide levels, particularly PHC with C24–C26 fatty acids; 3) decreased the expression of amino acid transporters; and 4) induced the UPR pathway. These outcomes were abrogated by inhibition of de novo ceramide synthesis or by supplementation of inositol.
Ceramide levels can be increased via two routes, increased de novo synthesis by ceramide synthase or inhibition of inositol phosphorylceramide synthase, which converts ceramide to complex sphingolipids. Our findings suggest that VPA induces the UPR pathway by increasing de novo synthesis of ceramide (Fig. 6B) as a result of increased expression of fatty acid elongases, because induction did not occur in the presence of the ceramide synthase inhibitor fumonisin (37). In contrast, aureobasidin, which inhibits inositol phosphorylceramide synthase, did not affect VPA-mediated induction of the UPR (Fig. 6C).
Our findings are consistent with studies showing that inositol starvation induces the UPR. UPR target genes were found to be significantly up-regulated in cells grown in the absence of inositol (16). In addition, inositol supplementation alters the expression of UPR pathway genes (39). Although initial studies showed that accumulation of unfolded proteins in the ER induces the UPR pathway (40, 41), it has since been determined that Ire1p, the transmembrane kinase that senses ER stress (27, 42), induces the UPR pathway in response to changes in membrane lipid composition (29). In this light, inositol starvation may trigger the UPR as a result of lipid-related membrane changes in the ER, including perturbation of sphingolipid metabolism. The addition of inositol induces changes in the synthesis and levels of numerous lipids, including sphingolipids (28, 43). Recent studies show that perturbation of sphingolipid metabolism may affect ER membrane homeostasis. Mutant cells lacking ORM1 and ORM2, negative regulators of sphingolipid metabolism (31, 44), exhibit constitutive induction of the UPR response (31). In addition, orm1Δ orm2Δ cells contain elevated levels of sphingolipids (31, 44) and are hypersensitive to stress induced by inositol starvation (31). Similarly, UPR expression is constitutive in isc1Δ mutant cells, which contain elevated sphingolipids due to a block in sphingolipid turnover (45, 46). Conversely, inhibiting sphingolipid synthesis with myriocin treatment suppresses activation of the UPR induced by inositol starvation (29). Elevated sphingolipids in the ER may lead to membrane aberrancy that activates the UPR pathway independent of accumulation of unfolded protein. The role of specific sphingolipids in activation of the UPR is not known. Our study suggests that accumulation of ceramide, specifically C24–C26-containing PHC, induces the UPR upon inositol depletion.
We have previously shown that VPA decreases intracellular inositol levels (10–13, 47). VPA induction of the UPR occurred only in I− conditions, suggesting that the response to VPA was mediated by inositol depletion. Consistent with this, inositol starvation of ino1Δ cells caused similar effects, including increased expression of fatty acid elongases, increased intracellular levels of ceramides, especially PHC with C24 and C26, down-regulation of nutrient transporters, and induction of the UPR. Based on these data, we propose the following model (Fig. 8). VPA decreases intracellular inositol levels, causing an increase in C24 and C26 PHC species as a result of up-regulation of expression of fatty acid elongases (FEN1 and SUR4). This results in increased ceramide synthesis, which causes down-regulation of expression of nutrient transporters, inducing stress and the UPR.
FIGURE 8.
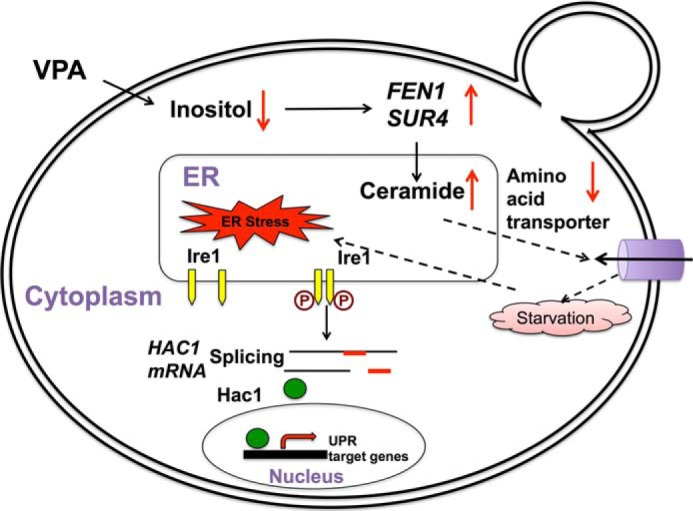
Model; VPA induces the UPR pathway by increasing intracellular ceramide levels. In the proposed model, VPA-mediated inositol depletion leads to increased expression of fatty acid elongases, specifically FEN1 and SUR4, which catalyze the synthesis of C24 and C26 fatty acids and ceramides, especially PHC containing C24–C26 fatty acids. Increased ceramide levels decrease expression of nutrient transporters, stressing the cell due to lack of nutrients and inducing the UPR pathway.
Several studies suggest that the UPR pathway may be involved in the pathophysiology of BD. Lymphoblasts from bipolar patients show an aberrant ER stress response (48, 49). Compared with cells from healthy controls, lymphoblasts from bipolar patients had lower expression of XBP1 (mammalian homologue of yeast HAC1), the spliced form of which binds the UPRE element and induces the UPR pathway in response to ER stress inducers (49). A polymorphism (−116C → G) in the XBP1 promoter that is associated with lower gene transcription was observed in BD lymphoblastoid cells (50). These findings suggest that the UPR pathway may be perturbed in bipolar patients, and activation of this pathway may be important for the therapeutic action of VPA. In this light, the UPR pathway may be a possible new target for BD therapy.
Experimental Procedures
Yeast Strains, Growth Media, and Conditions
Strains used in this study are summarized in Table 1. Cells were maintained on YPD medium (2% glucose, 1% yeast extract, 2% bactopeptone). Deletion mutants were maintained on medium supplemented with G418 (200 μg/ml). Synthetic minimal medium without inositol (I−) contained all of the essential components of Difco yeast nitrogen base (minus inositol); 2% glucose; 0.2% ammonium sulfate; vitamins; the four amino acids histidine (20 mg/liter), methionine (20 mg/liter), leucine (60 mg/liter), and lysine (20 mg/liter); and the nucleobase uracil (40 mg/liter). Where indicated, inositol (I) was added at a concentration of 75 μm. For selection of plasmids, uracil was omitted. Liquid and solid media were supplemented with 0.6 and 1 mm VPA, respectively, when indicated. Fumonisin B1 (Sigma) and aureobasidin A (Clontech) were used at a concentration of 100 μm and 0.5 μg/μl, respectively. For solid media, 2% agar was added. Absorbance was measured at 550 nm to monitor growth in liquid cultures. All incubations were at 30 °C.
TABLE 1.
Yeast strains and plasmids used in this study
| Strain/Plasmid | Genotype/Description | Source/Reference |
|---|---|---|
| Wild type | MATa, his 3Δ1, leu 2Δ0, met 15Δ0, ura3Δ0 | Invitrogen |
| fen1Δ | MATa, his 3Δ1, leu 2Δ0, met 15Δ0, ura 3Δ0, fen1Δ::KanMX4 | Invitrogen |
| sur4Δ | MATa, his 3Δ1, leu 2Δ0, met 15Δ0, ura 3Δ0, sur4Δ::KanMX4 | Invitrogen |
| rsb1Δ | MATa, his 3Δ1, leu 2Δ0, met 15Δ0, ura 3Δ0, rsb1Δ::KanMX4 | Invitrogen |
| ino1Δ | MATa, his 3Δ1, leu 2Δ0, met 15Δ0, ura 3Δ0, ino1Δ::KanMX4 | Invitrogen |
| RSB1-HA | pRS316-RSB1Δ335–382–3 × HA | Johnson et al. (55) |
| UPRE-LacZ | pJC104, containing UPRE-CYC-lacZ | Chang et al. (53) |
VPA Treatment
WT cells were precultured in synthetic minimal medium with inositol (I+), harvested, washed twice with sterile water, and grown in I+ until the cells reached the mid-log phase (A550 = 0.5). Cells were pelleted, washed twice with sterile water and inoculated in I+ or I− to a final A550 of 0.05 and cultured until the cells reached the mid-log phase (A550 = 0.5). Cells were then pelleted and suspended in fresh I− or I+ medium with or without 0.6 mm VPA and incubated for 5 h.
ino1Δ Starvation
ino1Δ cells were precultured in I+, harvested, washed twice with sterile water, and grown in I+ until the cells reached mid-log phase (A550 = 0.5). Cells were pelleted, washed twice with sterile water, and transferred to fresh I− (inositol starvation) or I+ (control) for 3 h.
Microarray Analysis
Total RNA was isolated by hot phenol extraction (51) and purified using an RNeasy kit from Qiagen. Quality of RNA was determined using Agilent 2100 Bioanalyzer. RNA was labeled using the Agilent Low Input Quick-Amp labeling kit (Agilent Technologies). Cy3-labeled cRNA was then hybridized to the 8 × 15K Agilent Yeast V2 Arrays (design ID 016322). Slides were scanned on an Agilent G2505B microarray scanner, and the resulting image files were processed with Agilent Feature Extraction software (version 9.5.1). All procedures were carried out according to the manufacturer's protocols. Subsequent analysis was performed using GeneSpring (version 10.0) software. Microarray analysis was carried out at the Research Technology Support Facility at Michigan State University.
qRT-PCR Analysis
Total RNA was extracted using the hot phenol method (51) and purified using an RNeasy mini plus kit (Qiagen, Valencia, CA). cDNA was synthesized using the first strand cDNA synthesis kit from Roche Applied Science as described in the manufacturer's manuals. qRT-PCRs were done in a 20-μl volume reaction using Brilliant III Ultra-Faster SYBR Green qPCR master mix (Agilent Technologies, Santa Clara, CA). Each reaction was done in triplicate. The primers used for the qRT-PCRs are listed in Table 2. RNA levels were normalized to ACT1 levels (internal control). Relative values of mRNA transcripts are shown as -fold change relative to the indicated controls. Primers were validated as suggested in the Methods and Applications Guide (Agilent Technologies). All primers used in this study had primer efficiency between 85 and 105%. Optimal primer concentrations were determined, and primer specificity of a single product was monitored by a melt curve following the amplification reaction. PCRs were initiated at 95 °C for 10 min for denaturation followed by 40 cycles consisting of 30 s at 95 °C and 60 s at 55 °C.
TABLE 2.
Real-time PCR primers used in this study
| Gene | Primer | Sequence (5′–3′) |
|---|---|---|
| ACT1 | Forward | ACGTTCCAGCCTTCTACGTTTCCA |
| ACT1 | Reverse | ACGTGAGTAACACCATCACCGGAA |
| FEN1 | Forward | TGGGTTCAACAACTGCCACCTTTG |
| FEN1 | Reverse | TCATTAACCTTTGCGGCAACACCG |
| SUR4 | Forward | TGTTATGGTACTCAGGCTGCTGCT |
| SUR4 | Reverse | AGTAGAAGAACCGGATGCAACGGA |
| RSB1 | Forward | TTGCCCTCTCCAATGGCGTATTCT |
| RSB1 | Reverse | ACATGATTGCCGGTTGTTGTGGAC |
| ELO1 | Forward | AGAAAGCCTCTAGGTTTCGCCCAA |
| ELO1 | Reverse | AAAGGCTGCTTCCCAACGGTAAAC |
| BIO5 | Forward | GCATCCGGACTACGAGTTAAAG |
| BIO5 | Reverse | GGGCAACGGAGTTGAATAAATG |
| AGP1 | Forward | GAACGATCTTACGTCGGCTATC |
| AGP1 | Reverse | GACCTGTATTAGCGCCTATGTT |
| GAP1 | Forward | GTGACACTCCAGGTGCTAAA |
| GAP1 | Reverse | GCAGCAAGACCAACCAATTC |
| BAP2 | Forward | GAGGATGGCGTTGAGTCTATC |
| BAP2 | Reverse | GTCCCAATACCTGTCCCTAAAG |
| DIP5 | Forward | TCATTTCTTGGGCTGGTTACA |
| DIP5 | Reverse | GGTCCTTCATTCTTCCCTCTTC |
| UGA4 | Forward | TGGTGGTCCAGCAACATTAG |
| UGA4 | Reverse | AGCGGTAGGAATGGAACTTG |
| CAN1 | Forward | GAACGCTGAAGTGAAGAGAGAG |
| CAN1 | Reverse | GTTGGTCAGAGGTGTGGATAAA |
| SAM3 | Forward | GATGTATCTGCCTCTCCCTTTG |
| SAM3 | Reverse | CACAACCGCAACAAGGATAAC |
| EUG1 | Forward | TGGTCAAGTCTATCGCGGTGTCAA |
| EUG1 | Reverse | CATTCAAGCCTGTCAAGCCTCTGT |
| JEM1 | Forward | TGGGACAAGGTGCATCAGAAGGAT |
| JEM1 | Reverse | GCGTTATGCGTAGCAGCTCAGAAA |
| KAR2 | Forward | AAAGATGGGAAGCCCGCTGTAGAA |
| KAR2 | Reverse | ACAGCATGGGTAACCTTAGTGCCT |
| LHS1 | Forward | GCGCGGAAGTGCTTATCCAAACAA |
| LHS1 | Reverse | ACGCAACTCCTGACGAGCACTTAT |
| SEC63 | Forward | AGCAAAGGGCCTAACACCTGATGA |
| SEC63 | Reverse | TGGGCCATCTGGATGACCGTATTT |
| PDI1 | Forward | TGCCATCCACGACATGACTGAAGA |
| PDI1 | Reverse | ACTCCAACACGATCTTGTCGCTCA |
Ceramide Measurement
Cells were grown and treated with VPA as described above for 5 h, pelleted, and stored at −80 °C. Extraction of lipids from yeast pellets and lipid quantification by LC/MS/MS was performed as described previously (52).
β-Galactosidase Assays
Cells expressing the UPRE-LacZ reporter plasmid provided by Dr. Susan Henry (53) were precultured in Ura-I+ and grown in Ura-I+ to an A550 of 0.5, washed, and transferred to Ura-I− medium with or without VPA for 5 h at 30 °C. Cells were harvested, and β-galactosidase was assayed as described (54).
Author Contributions
S. J. and M. L. G. designed the research and wrote the manuscript; S. J. carried out all of the experiments; S. R., S. C., R. S., and A. C. performed ceramide measurements; S. J., A. C., R. S., and M. L. G. carried out the data analysis. All authors reviewed the results and approved the final version of the manuscript.
Supplementary Material
Acknowledgments
We thank Dr. Susan Henry for the UPRE-lacZ reporter plasmid, Dr. Jeff Landgraf and the Research Technology Support Facility at Michigan State University for assistance in carrying out the microarray analysis, and Dr. Yusuf Hannun for valuable insights.
This work was supported by National Institutes of Health Grant DK081367 (to M. L. G.), the Swiss National Science Foundation (to R. S.), and the Graduate School of Wayne State University (to S. J.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.

This article contains supplemental Table 1.
- BD
- bipolar disorder
- VPA
- valproic acid
- ER
- endoplasmic reticulum
- UPR
- unfolded protein response
- PHC
- phytoceramide
- DHC
- dihydroceramide
- LCB
- long chain base
- MIPS
- myo-inositol-3-phosphate synthase
- qRT-PCR
- quantitative RT-PCR.
References
- 1. Belmaker R. H. (2004) Bipolar disorder. N. Engl. J. Med. 351, 476–486 [DOI] [PubMed] [Google Scholar]
- 2. Goodwin G. M. (2007) Quetiapine more effective than placebo for depression in bipolar I and II disorder. Evid. Based Ment. Health 10, 82. [DOI] [PubMed] [Google Scholar]
- 3. Cheng R. S., Lin C., Fok M. L., and Leung C. M. (2005) Shoplifting in the mentally ill: the role of bipolar affective disorder and mental retardation. Med. Sci. Law 45, 317–320 [DOI] [PubMed] [Google Scholar]
- 4. Bostwick J. M., and Pankratz V. S. (2000) Affective disorders and suicide risk: a reexamination. Am. J. Psychiatry 157, 1925–1932 [DOI] [PubMed] [Google Scholar]
- 5. Henry C. A., Zamvil L. S., Lam C., Rosenquist K. J., and Ghaemi S. N. (2003) Long-term outcome with divalproex in children and adolescents with bipolar disorder. J. Child Adolesc. Psychopharmacol. 13, 523–529 [DOI] [PubMed] [Google Scholar]
- 6. Gould T. D., Quiroz J. A., Singh J., Zarate C. A., and Manji H. K. (2004) Emerging experimental therapeutics for bipolar disorder: insights from the molecular and cellular actions of current mood stabilizers. Mol. Psychiatry 9, 734–755 [DOI] [PubMed] [Google Scholar]
- 7. Williams R. S., Cheng L., Mudge A. W., and Harwood A. J. (2002) A common mechanism of action for three mood-stabilizing drugs. Nature 417, 292–295 [DOI] [PubMed] [Google Scholar]
- 8. Berridge M. J. (1989) Inositol 1,4,5-trisphosphate-induced calcium mobilization is localized in Xenopus oocytes. Proc. R. Soc. Lond. B Biol. Sci. 238, 235–243 [DOI] [PubMed] [Google Scholar]
- 9. Hallcher L. M., and Sherman W. R. (1980) The effects of lithium ion and other agents on the activity of myo-inositol-1-phosphatase from bovine brain. J. Biol. Chem. 255, 10896–10901 [PubMed] [Google Scholar]
- 10. Shaltiel G., Shamir A., Shapiro J., Ding D., Dalton E., Bialer M., Harwood A. J., Belmaker R. H., Greenberg M. L., and Agam G. (2004) Valproate decreases inositol biosynthesis. Biol. Psychiatry 56, 868–874 [DOI] [PubMed] [Google Scholar]
- 11. Vaden D. L., Ding D., Peterson B., and Greenberg M. L. (2001) Lithium and valproate decrease inositol mass and increase expression of the yeast INO1 and INO2 genes for inositol biosynthesis. J. Biol. Chem. 276, 15466–15471 [DOI] [PubMed] [Google Scholar]
- 12. Ye C., and Greenberg M. L. (2015) Inositol synthesis regulates the activation of GSK-3α in neuronal cells. J. Neurochem. 133, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ju S., Shaltiel G., Shamir A., Agam G., and Greenberg M. L. (2004) Human 1-d-myo-inositol-3-phosphate synthase is functional in yeast. J. Biol. Chem. 279, 21759–21765 [DOI] [PubMed] [Google Scholar]
- 14. Deranieh R. M., He Q., Caruso J. A., and Greenberg M. L. (2013) Phosphorylation regulates myo-inositol-3-phosphate synthase: a novel regulatory mechanism of inositol biosynthesis. J. Biol. Chem. 288, 26822–26833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Parthasarathy R. N., Lakshmanan J., Thangavel M., Seelan R. S., Stagner J. I., Janckila A. J., Vadnal R. E., Casanova M. F., and Parthasarathy L. K. (2013) Rat brain myo-inositol 3-phosphate synthase is a phosphoprotein. Mol. Cell Biochem. 378, 83–89 [DOI] [PubMed] [Google Scholar]
- 16. Jesch S. A., Zhao X., Wells M. T., and Henry S. A. (2005) Genome-wide analysis reveals inositol, not choline, as the major effector of Ino2p-Ino4p and unfolded protein response target gene expression in yeast. J. Biol. Chem. 280, 9106–9118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Santiago T. C., and Mamoun C. B. (2003) Genome expression analysis in yeast reveals novel transcriptional regulation by inositol and choline and new regulatory functions for Opi1p, Ino2p, and Ino4p. J. Biol. Chem. 278, 38723–38730 [DOI] [PubMed] [Google Scholar]
- 18. Henry S. A., Gaspar M. L., and Jesch S. A. (2014) The response to inositol: regulation of glycerolipid metabolism and stress response signaling in yeast. Chem. Phys. Lipids 180, 23–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol. Rev. 93, 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fagone P., and Jackowski S. (2009) Membrane phospholipid synthesis and endoplasmic reticulum function. J. Lipid Res. 50, S311–S316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knödler A., Konrad G., and Mayinger P. (2008) Expression of yeast lipid phosphatase Sac1p is regulated by phosphatidylinositol-4-phosphate. BMC Mol. Biol. 9, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mayinger P. (2009) Regulation of Golgi function via phosphoinositide lipids. Semin. Cell Dev. Biol. 20, 793–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Villa-García M. J., Choi M. S., Hinz F. I., Gaspar M. L., Jesch S. A., and Henry S. A. (2011) Genome-wide screen for inositol auxotrophy in Saccharomyces cerevisiae implicates lipid metabolism in stress response signaling. Mol. Genet. Genomics 285, 125–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deranieh R. M., and Greenberg M. L. (2009) Cellular consequences of inositol depletion. Biochem. Soc. Trans. 37, 1099–1103 [DOI] [PubMed] [Google Scholar]
- 25. Gaspar M. L., Aregullin M. A., Jesch S. A., and Henry S. A. (2006) Inositol induces a profound alteration in the pattern and rate of synthesis and turnover of membrane lipids in Saccharomyces cerevisiae. J. Biol. Chem. 281, 22773–22785 [DOI] [PubMed] [Google Scholar]
- 26. Chang H. J., Jones E. W., and Henry S. A. (2002) Role of the unfolded protein response pathway in regulation of INO1 and in the sec14 bypass mechanism in Saccharomyces cerevisiae. Genetics 162, 29–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cox J. S., Chapman R. E., and Walter P. (1997) The unfolded protein response coordinates the production of endoplasmic reticulum protein and endoplasmic reticulum membrane. Mol. Biol. Cell 8, 1805–1814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jesch S. A., Gaspar M. L., Stefan C. J., Aregullin M. A., and Henry S. A. (2010) Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J. Biol. Chem. 285, 41947–41960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Promlek T., Ishiwata-Kimata Y., Shido M., Sakuramoto M., Kohno K., and Kimata Y. (2011) Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 22, 3520–3532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Epstein S., Kirkpatrick C. L., Castillon G. A., Muñiz M., Riezman I., David F. P., Wollheim C. B., and Riezman H. (2012) Activation of the unfolded protein response pathway causes ceramide accumulation in yeast and INS-1E insulinoma cells. J. Lipid Res. 53, 412–420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Han S., Lone M. A., Schneiter R., and Chang A. (2010) Orm1 and Orm2 are conserved endoplasmic reticulum membrane proteins regulating lipid homeostasis and protein quality control. Proc. Natl. Acad. Sci. U.S.A. 107, 5851–5856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mousley C. J., Tyeryar K., Ile K. E., Schaaf G., Brost R. L., Boone C., Guan X., Wenk M. R., and Bankaitis V. A. (2008) Trans-Golgi network and endosome dynamics connect ceramide homeostasis with regulation of the unfolded protein response and TOR signaling in yeast. Mol. Biol. Cell 19, 4785–4803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Guenther G. G., Peralta E. R., Rosales K. R., Wong S. Y., Siskind L. J., and Edinger A. L. (2008) Ceramide starves cells to death by downregulating nutrient transporter proteins. Proc. Natl. Acad. Sci. U.S.A. 105, 17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Payet L. A., Pineau L., Snyder E. C., Colas J., Moussa A., Vannier B., Bigay J., Clarhaut J., Becq F., Berjeaud J. M., Vandebrouck C., and Ferreira T. (2013) Saturated fatty acids alter the late secretory pathway by modulating membrane properties. Traffic 14, 1228–1241 [DOI] [PubMed] [Google Scholar]
- 35. Oh C. S., Toke D. A., Mandala S., and Martin C. E. (1997) ELO2 and ELO3, homologues of the Saccharomyces cerevisiae ELO1 gene, function in fatty acid elongation and are required for sphingolipid formation. J. Biol. Chem. 272, 17376–17384 [DOI] [PubMed] [Google Scholar]
- 36. Kihara A., and Igarashi Y. (2002) Identification and characterization of a Saccharomyces cerevisiae gene, RSB1, involved in sphingoid long-chain base release. J. Biol. Chem. 277, 30048–30054 [DOI] [PubMed] [Google Scholar]
- 37. He Q., Suzuki H., Sharma N., and Sharma R. P. (2006) Ceramide synthase inhibition by fumonisin B1 treatment activates sphingolipid-metabolizing systems in mouse liver. Toxicol. Sci. 94, 388–397 [DOI] [PubMed] [Google Scholar]
- 38. Wu W. I., McDonough V. M., Nickels J. T. Jr, Ko J., Fischl A. S., Vales T. R., Merrill A. H. Jr., and Carman G. M. (1995) Regulation of lipid biosynthesis in Saccharomyces cerevisiae by fumonisin B1. J. Biol. Chem. 270, 13171–13178 [DOI] [PubMed] [Google Scholar]
- 39. Jesch S. A., Liu P., Zhao X., Wells M. T., and Henry S. A. (2006) Multiple endoplasmic reticulum-to-nucleus signaling pathways coordinate phospholipid metabolism with gene expression by distinct mechanisms. J. Biol. Chem. 281, 24070–24083 [DOI] [PubMed] [Google Scholar]
- 40. Chapman R., Sidrauski C., and Walter P. (1998) Intracellular signaling from the endoplasmic reticulum to the nucleus. Annu. Rev. Cell Dev. Biol. 14, 459–485 [DOI] [PubMed] [Google Scholar]
- 41. Pahl H. L. (1999) Signal transduction from the endoplasmic reticulum to the cell nucleus. Physiol. Rev. 79, 683–701 [DOI] [PubMed] [Google Scholar]
- 42. Mori K., Ogawa N., Kawahara T., Yanagi H., and Yura T. (2000) mRNA splicing-mediated C-terminal replacement of transcription factor Hac1p is required for efficient activation of the unfolded protein response. Proc. Natl. Acad. Sci. U.S.A. 97, 4660–4665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Alvarez-Vasquez F., Sims K. J., Cowart L. A., Okamoto Y., Voit E. O., and Hannun Y. A. (2005) Simulation and validation of modelled sphingolipid metabolism in Saccharomyces cerevisiae. Nature 433, 425–430 [DOI] [PubMed] [Google Scholar]
- 44. Breslow D. K., Collins S. R., Bodenmiller B., Aebersold R., Simons K., Shevchenko A., Ejsing C. S., and Weissman J. S. (2010) Orm family proteins mediate sphingolipid homeostasis. Nature 463, 1048–1053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Gururaj C., Federman R. S., and Chang A. (2013) Orm proteins integrate multiple signals to maintain sphingolipid homeostasis. J. Biol. Chem. 288, 20453–20463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sawai H., Okamoto Y., Luberto C., Mao C., Bielawska A., Domae N., and Hannun Y. A. (2000) Identification of ISC1 (YER019w) as inositol phosphosphingolipid phospholipase C in Saccharomyces cerevisiae. J. Biol. Chem. 275, 39793–39798 [DOI] [PubMed] [Google Scholar]
- 47. Ju S., and Greenberg M. L. (2003) Valproate disrupts regulation of inositol responsive genes and alters regulation of phospholipid biosynthesis. Mol. Microbiol. 49, 1595–1603 [DOI] [PubMed] [Google Scholar]
- 48. Hayashi A., Kasahara T., Kametani M., Toyota T., Yoshikawa T., and Kato T. (2009) Aberrant endoplasmic reticulum stress response in lymphoblastoid cells from patients with bipolar disorder. Int. J. Neuropsychopharmacol. 12, 33–43 [DOI] [PubMed] [Google Scholar]
- 49. So J., Warsh J. J., and Li P. P. (2007) Impaired endoplasmic reticulum stress response in B-lymphoblasts from patients with bipolar-I disorder. Biol. Psychiatry 62, 141–147 [DOI] [PubMed] [Google Scholar]
- 50. Kakiuchi C., Iwamoto K., Ishiwata M., Bundo M., Kasahara T., Kusumi I., Tsujita T., Okazaki Y., Nanko S., Kunugi H., Sasaki T., and Kato T. (2003) Impaired feedback regulation of XBP1 as a genetic risk factor for bipolar disorder. Nat. Genet. 35, 171–175 [DOI] [PubMed] [Google Scholar]
- 51. Köhrer K., and Domdey H. (1991) Preparation of high molecular weight RNA. Methods Enzymol. 194, 398–405 [DOI] [PubMed] [Google Scholar]
- 52. Brice S. E., Alford C. W., and Cowart L. A. (2009) Modulation of sphingolipid metabolism by the phosphatidylinositol-4-phosphate phosphatase Sac1p through regulation of phosphatidylinositol in Saccharomyces cerevisiae. J. Biol. Chem. 284, 7588–7596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Chang H. J., Jesch S. A., Gaspar M. L., and Henry S. A. (2004) Role of the unfolded protein response pathway in secretory stress and regulation of INO1 expression in Saccharomyces cerevisiae. Genetics 168, 1899–1913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fu Y., and Xiao W. (2006) Study of transcriptional regulation using a reporter gene assay. Methods Mol. Biol. 313, 257–264 [DOI] [PubMed] [Google Scholar]
- 55. Johnson S. S., Hanson P. K., Manoharlal R., Brice S. E., Cowart L. A., and Moye-Rowley W. S. (2010) Regulation of yeast nutrient permease endocytosis by ATP-binding cassette transporters and a seven-transmembrane protein, RSB1. J. Biol. Chem. 285, 35792–35802 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.